Abstract
Intraspecific genetic structure in widely distributed marine species often mirrors the boundaries between temperature-defined bioregions. This suggests that the same thermal gradients that maintain distinct species assemblages also drive the evolution of new biodiversity. Ecological speciation scenarios are often invoked to explain such patterns, but the fact that adaptation is usually only identified when phylogenetic splits are already evident makes it impossible to rule out the alternative scenario of allopatric speciation with subsequent adaptation. We integrated large-scale genomic and environmental datasets along one of the world's best-defined marine thermal gradients (the South African coastline) to test the hypothesis that incipient ecological speciation is a result of divergence linked to the thermal environment. We identified temperature-associated gene regions in a coastal fish species that is spatially homogeneous throughout several temperature-defined biogeographic regions based on selectively neutral markers. Based on these gene regions, the species is divided into geographically distinct regional populations. Importantly, the ranges of these populations are delimited by the same ecological boundaries that define distinct infraspecific genetic lineages in co-distributed marine species, and biogeographic disjunctions in species assemblages. Our results indicate that temperature-mediated selection represents an early stage of marine ecological speciation in coastal regions that lack physical dispersal barriers.
Keywords: adaptation, dispersal barrier, ecological speciation, marine biogeography, seascape genomics, thermal selection
1. Introduction
Molecular phylogenies of marine species present along continuous coastlines have revealed that spatial disjunctions between distinct evolutionary lineages are often associated with the boundaries between different marine biogeographic regions [1,2], but such genetic patterns tend to be present in only a fraction of species [1,3–6] (figure 1). This discrepancy is often attributed to life history: actively dispersing species, and those with extended planktonic dispersal phases, cross the boundaries between bioregions more frequently than species with short propagule duration, making them less likely to diverge in spatial isolation [5,13]. However, support for this paradigm is not consistent, as numerous studies from North America [6,14], South Africa [1], and Australasia [4] have failed to identify a clear link between genetic structure and dispersal potential.
Figure 1.
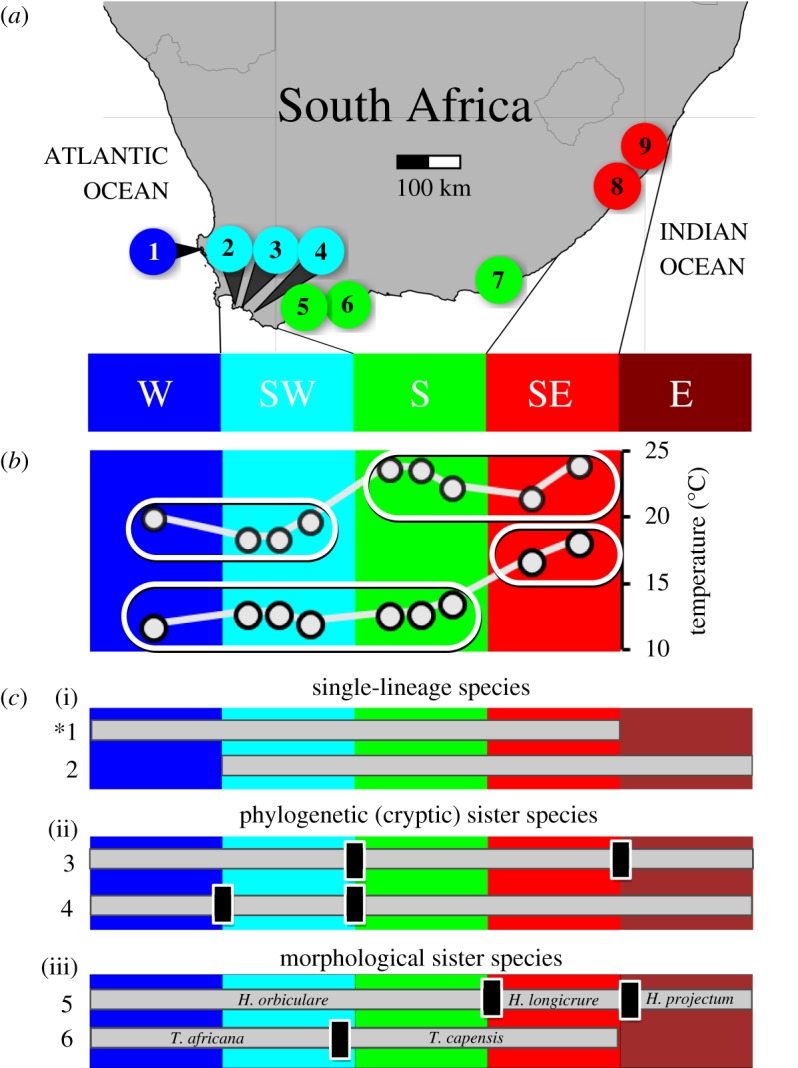
Sampling sites, marine bioregions, and examples of genetic breaks in South African coastal animals; (a) a map indicating the location of sampling sites within southern Africa's temperature-defined marine bioregions; (b) maximum and minimum sea surface temperatures (SSTs) at each sampling site; these temperatures divide bioregions into two groups each (indicated by ellipses), and by themselves only partially explain the region's biogeography; (c) examples of distribution ranges (grey horizontal bars) and location of genetic breaks (black vertical bars) in coastal South African animals, arranged hierarchically. (i) Species that occur as a single phylogenetic lineage in multiple bioregions: 1, Psammogobius knysnaensis (the study species, marked with an asterisk) [7] and 2, Scutellastra longicosta [8]. (ii) Species with phylogenetically distinct sister lineages that are not distinguishable morphologically (cryptic species): 3, Callichirus kraussi [9] and 4, Palaemon peringueyi [10]. (iii) Morphologically distinguishable sister species: 5, Hymenosoma spp. [11] and 6, Tricolia spp. [12]. W, cool-temperate west coast; SW, transition zone on the southwest coast; S, warm-temperate south coast; SE, transition zone on the southeast coast; E, subtropical east coast.
An alternative explanation for this paradox is offered by ecological divergence that preceded the allopatric distribution patterns evident on the basis of selectively neutral genetic markers. This is primarily supported by ‘phylogenetic shifts approaches', in which phylogenetic splits coincide with ecological divergence [15]. The evidence for ecological speciation is particularly strong when phylogenetic splits are not associated with physical dispersal barriers that can completely isolate sister lineages [15], when contact zones are located in regions where environmental conditions are intermediate [1,16], and when each lineage displays reduced fitness in the habitat of its sister lineage [17,18]. Phylogenetic splits that are shared by multiple species across the same boundary may differ considerably in age [1,8], and by extension, this supports the hypothesis that species in which phylogenetic divergence is not yet evident have undergone ecological differentiation very recently.
The phylogenetic evidence for ecological drivers of speciation is nonetheless circumstantial, because divergence events mostly occurred during the Pleistocene or earlier [8,19], and it is difficult to extrapolate from contemporary conditions when species’ historical distribution patterns are unknown and past oceanographic conditions not well understood. Because of such uncertainties, it is controversial to ascertain whether adaptation to divergent environments that reduced levels of gene flow because of the maladaptation of migrants was the primary driver of divergence, or whether it occurred after a phylogenetic split that may very well have evolved during an extended period of physical isolation.
More compelling evidence for ecological speciation in the sea would come from scenarios in which there is support for genetic differentiation that coincides with biogeography, but in which phylogenetic divergence indicative of speciation has not yet occurred [20]. The fact that phylogeographic breaks tend to be present in only a fraction of the species whose ranges span the boundaries between ecologically distinct marine regions [1,3,4] suggests that the condition of recent divergence may be met by those species that display no genetic divergence on the basis of the selectively neutral datasets typically employed in phylogeographic studies [21].
The South African coastline is characterized by ecologically distinct marine bioregions (figure 1) that are arranged along a thermal gradient [1]. This provides a unique opportunity for studying the importance of incipient environmentally driven parapatric speciation in the sea, as biogeography (and, by extension, ecological speciation) is believed to be primarily a function of species' thermal tolerance ranges [22]. Numerous species complexes exist along this coastline that comprise cryptic species whose ranges are limited by the boundaries between bioregions [1], and which exhibit distinct temperature preferences [17,18]. This suggests that thermal adaptation contributes towards limiting gene flow between biogeographic regions by reducing migrant fitness and by subjecting migrants to competitive exclusion [1]. However, in some species, a single evolutionary lineage is found across multiple bioregions (figure 1). The latter are suitable candidates for determining whether diversifying selection driven by the environment, and corresponding reductions in gene flow, may have preceded phylogenetic splits.
We tested this hypothesis by generating genome-wide data from one of these phylogenetically homogeneous species [7], the Knysna sandgoby, Psammogobius knysnaensis (figure 1). This numerically dominant estuarine fish, which ranges from the west coast to the southeast coast and which disperses by means of planktonic larvae (electronic supplementary material), was selected because the purity of extracted DNA was much higher than that of other candidate species. We expect population divergence that mirrors coastal biogeography to be evident based only on temperature-associated genes. This would support the idea that in coastal regions lacking physical dispersal barriers, thermal selection plays a defining role in the early stages of parapatric ecological speciation.
2. Methods
(a). Sampling procedure
Tissue samples from a total of 312 individuals of P. knysnaensis were collected from at least three different locations in the mouth areas of nine estuaries throughout the species’ range over a period of 2 years (electronic supplementary material, table S1), using a pushnet. Upon capture, a fin clip was obtained from one of the pectoral fins using sterilized fingernail scissors, and immediately preserved in 100% ethanol. The fish were subsequently released.
(b). Generation and processing of genomic data
Double digest restriction site-associated DNA (ddRAD) libraries were constructed for a subset of 109 individuals and 12 replicates with particularly high-quality DNA, following the protocol described in Sandoval-Castillo et al. [23]. Libraries were pooled in groups of 48 or 93 samples per lane and sequenced on an Illumina HiSeq 2000 (100 bp paired-end reads) platform at the McGill University and Genome Québec Innovation Centre. Raw sequences were processed as described in the electronic supplementary material.
(c). Identification of loci under thermal selection and neutral loci
We assessed the contribution of coastal sea surface temperature (SST) to the overall pattern of genetic differentiation using the R package gINLAnd [24]. This software uses a spatial generalized linear mixed model to quantify the correlation between genotypes and environmental variables, while controlling for the effects of spatial population structure and population history. As the application of satellite-based SST data is often problematic when studying coastal biogeography, because it includes data from offshore regions [25], we used southern African temperature data based on in situ measurements, as described in the electronic supplementary material. To ascertain that the study species does not yet exhibit genetic divergence based on putatively neutral data, which could indicate that geographical isolation preceded thermal adaptation, we also created a reduced dataset comprising selectively neutral data. In addition to excluding loci identified as being under thermal selection, we excluded additional outlier loci from FST-based genome scans, using BayeScan v. 2.1 [26], as temperature may only be one of a number of drivers of selection.
(d). Functional annotation
To identify the possible functions of genes under thermal selection, we blasted the flanking sequences of temperature-associated loci against the National Center for Biotechnology Information (NCBI) non-redundant nucleotide database. The resulting reads were then annotated against the UniProtKB/Swiss-Prot database [27]. We then performed a gene ontology term analysis in topGO 2.24.0 [28]. Genes whose function indicates an influence of thermal selection were identified by searching the relevant literature.
(e). Population genetic structure
Genetic structure was investigated using both clustering and phylogenetic approaches. The clustering approaches were used to analyse both loci under selection and neutral loci, and included discriminant analysis of principal components (DAPC), which was performed with the R package adegenet v. 2.1.0 [29], and inference of population structure using fastStructure 1.0 [30]. DAPC was used to explore various combinations of maximum or minimum temperature as the environmental variable (with covariance factors that included geographical distance, biogeographic boundaries, and a combination of the two), while fastStructure was used to confirm the DAPC results for the most informative dataset identified in gINLAnd based on minimum SST (with geographical distance as the covariance factor). Phylogenetic analyses were performed in BEAST v. 2.4.7 [31] using (i) the most informative dataset of loci under thermal selection identified in gINLAnd using minimum SST (see Results) and (ii) previously published mtDNA COI data [7]. In both cases, maximum clade credibility (MCC) trees were reconstructed using a discrete phylogeographic analysis [32].
3. Results
A total of 8532 single nucleotide polymorphisms (SNPs) were generated for 109 individuals of P. knysnaensis, of which up to 239 were identified as being under thermal selection (electronic supplementary material, table S2). SNPs from ddRADseq originate from all genomic regions, and some may be located on protein-coding genes that are strongly affected by temperature. While such associations may not necessarily imply a causal relationship, identifying their function may contribute towards an improved understanding of possible drivers of genetic divergence between temperature-defined bioregions. Although no fully annotated transcriptome for the family Gobiidae is presently available, nine of the loci (identified using either maximum or minimum temperature, with geographical distance as the covariance factor) could be annotated as genes involved in mitigating thermal stress (electronic supplementary material, table S3). Three of these (14-3-3 gene, tyrosine protein kinase, and tubulin beta chain) are of particularly interest because they were involved in heat stress responses in a species of goby [33], or cold stress adaptation/acclimation in other teleosts [34,35]. In all but two cases, loci that were identified using minimum temperature data were also identified using maximum temperatures (data available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.ns790j4 [36]). This suggests that even though most experimental studies investigated responses to heat stress, genetically fixed differences of these genes between temperature-defined marine bioregions may reflect general adaptations to different thermal environments, and thus play a role in determining thermal tolerance ranges.
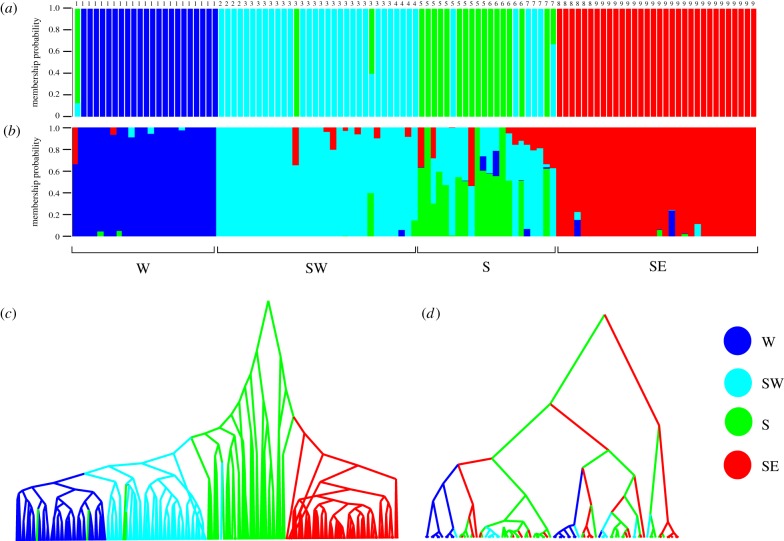
Clustering methods found high statistical support for three to four clusters (K) when analysing the temperature-associated loci, while a single cluster (K = 1) was found for the neutral loci (electronic supplementary material, figures S2 and S3), confirming that thermal selection preceded geographical divergence. Even though temperature alone only accounts for two marine bioregions (figure 1), and despite the fact that geographical distances and/or the boundaries between bioregions were controlled for when identifying temperature-associated loci, an affiliation of genetic clusters with up to four bioregions was found for the temperature-associated loci (figure 2a; electronic supplementary material, figure S4). The distinctness of subtropical (SE) individuals from the temperate (W, SW, and S) sites was evident in all analyses, and most analyses using minimum temperature as the environmental variable also identified the W coast as a distinct cluster. There was even evidence for distinct SW and S coast clusters, although these were comparatively poorly differentiated. This result was robust, and was also recovered using fastStructure (figure 2b).
Figure 2.
Population genetic structure inferred for temperature-associated loci, and reconstruction of phylogenetic relationships between individuals of Psammogobius knysnaensis from four South African marine bioregions; (a) DAPC compoplot for minimum SST data using geography as the covariate (results for alternative combinations of temperature data and covariates are shown in electronic supplementary material, figure S4); (b) corresponding consensus fastStructure barplot for four genetic clusters (K) (for comparison, barplots for K = 2–5 are shown in electronic supplementary material, figure S5); (c) MCC tree of 218 sequences from phased SNP data (same as in a and b), with location state reconstructions of ancestral nodes; and d corresponding phylogenetic tree based on mtDNA COI data [7]. In c, clear regional structure is evident, but there are possible migrants. The south coast cluster was identified as being the oldest. By contrast, there are no clear regional clades in d, and there was no evidence for any cluster being the oldest. Site numbers and abbreviations correspond to those in figure 1 and electronic supplementary material, table S1, and trees are not drawn to scale.
Congruent with the clustering methods, an MCC tree (figure 2c) of temperature-associated loci recovered both the western and southeastern group as mostly distinct but poorly differentiated clusters nested within a tree whose oldest nodes were inferred to have existed on the south coast. Some branches are nested within clades that mostly have location states from other regions, which may reflect migration between adjacent marine bioregions, and further supports the idea that thermal selection drove ecological speciation in the absence of geographical isolation. For comparison, an MCC tree reconstructed from mtDNA COI sequences [7] shows no clear regional structure (figure 2d).
4. Discussion
Speciation is a continuous process comprising a number of evolutionary stages that range from adaptive differentiation to complete reproductive isolation between populations [37]. Identifying the primary drivers of speciation is challenging because a considerable amount of time has often already passed by the time incipient speciation becomes evident. This makes it difficult to distinguish ecologically driven divergence from allopatric speciation with subsequent adaptation [38,39].
Marine biogeography is often considered to be a function of species' thermal tolerance ranges [22,40–43]. The fact that South Africa's coastal biogeography is mirrored by intraspecific spatial genetic structure suggests that species present in more than one province should comprise multiple evolutionary lineages that represent cryptic species [1]. The goby P. knysnaensis is one of a number of coastal southern African species that occur in multiple marine bioregions, but which displays no regional divergence on the basis of selectively neutral markers [7] of the type that are primarily used in phylogeographic research [21]. Here, we reject the previous finding of genetic homogeneity and show that this species is in fact represented by multiple regional groups delimited by temperature-defined bioregions. The fact that this is only evident for temperature-associated loci, and not for putatively neutral loci, confirms that divergence must have taken place in the absence of an interruption of gene flow due to physical dispersal barriers. Under these conditions, a pattern of isolation-by-adaptation [44] can be expected to eventually evolve, as migrants dispersing into adjacent bioregions will have fewer surviving offspring and reduced survival rates compared to residents. West-to-east thermal differentiation was evident particularly for the minimum temperatures, where the two easternmost sites had much warmer water than the other sites (figure 1). It is not clear why minimum temperature would have more explanatory power than maximum temperature, but its importance to southern Africa's marine biogeography has also been reported in a study on seaweed β-diversity [45]. Experiments on crustacean larvae from the southeast coast indicate that development is considerably delayed at lower temperatures typical of upwelling events in the temperate bioregions [17,18]. It is possible that, in addition to metabolic constraints at both high and low temperatures [46], recruitment success of the larvae of warm-adapted species is reduced at low temperatures because of the increased likelihood of predation, advection away from suitable habitat, and inability to capture prey [17,18]. There was no indication that marine bioregions could be identified based on high or low temperatures alone, and the identification of loci under thermal selection and subsequent detection of up to four genetic clusters cannot be explained as being an artefact of the temperature variables used in the gINLAnd analyses.
Adaptations to the thermal environment are complex and ubiquitous in nature. Temperature affects many different biological pathways, with strong effects on the integrity of proteins and cellular structures and on the rates of physiological processes, particularly in ectotherms [47]. The thermal environment can promote partial reproductive isolation between populations, which might drive them along the speciation continuum [48]. This is particularly true for organisms that (i) have distinct populations with parapatric distributions along the thermal gradient, (ii) do not maintain a stable internal temperature (poikilotherms), and (iii) are found across stable thermal gradients (e.g. aquatic environments), which are regions where exogenous divergent selection is not expected to weaken due to marked temperature fluctuations [48]. Our study system meets all these conditions and represents an example of parapatric ecological divergence with genomic hallmarks of incipient evolutionary divergence driven by the thermal environment.
Unlike previous spatial demographic inferences from coastal southern Africa, which typically reflect the influence of past climatic changes [8,49], the spatial genetic patterns identified here can be explained by present-day environmental conditions. On the east coast, northward dispersal in the near shore area is facilitated by wind-driven circulation [50], but this is unlikely to occur beyond site 9 (the northern distribution limit of P. knysnaensis) [49,51] because under contemporary conditions, the southward-flowing Agulhas Current flows very close to the coast and causes the parallel southward flow of near shore circulation [52]. In the western portion of the species' range, gene flow between the south and west coast is primarily facilitated by the westward drift of surface water [53]. The limited evidence for gene flow in both cases would be difficult to explain if one exclusively invoked physical isolation, given the high dispersal potential of the species’ larvae coupled with the region's strong ocean circulation. It suggests that migrants from a particular bioregion are maladapted to the environmental conditions in adjacent bioregions. For example, the distinctness of the west coast population from those on the southwest and south coast may reflect the influence of cold-water upwelling in the west [54].
We hypothesize that thermal selection, perhaps in combination with factors such as oceanography and primary productivity that covary with temperature to influence local adaptation [48], acts primarily on the sensitive larvae. Under this scenario, ecologically diverging populations are limited in their ability to exchange genes and, as such, reproductive isolation is expected to ensue [48]. There are no known conspicuous phenotypes that differ between the presumably locally adapted P. knysnaensis populations, but this is unsurprising because thermal adaptation often initially creates cryptic changes at the level of cell membranes or thermal stability of enzymes [55]. Studies that combine information from population genomics and controlled laboratory experiments using temperature-defined populations along an evolutionary continuum of speciation are expected to improve the identification of phenotypes enriched for selection signals of thermal adaptation.
5. Conclusion
Allopatric speciation in the marine environment is often invoked along continuous but ecologically subdivided coastlines, despite evidence that the physical dispersal barriers to whom this is attributed are insufficient to completely isolate regional populations [56,57]. Our study contributes to the growing evidence that in adjacent, temperature-defined marine provinces, divergence of loci linked to the thermal environment can precede significant spatial divergence of selectively neutral markers [58]. This strongly favours a scenario of parapatric ecological divergence over one in which allopatric divergence is followed by thermal adaptation. In the context of larger biogeographic patterns, where range boundaries in the sea often coincide with the boundaries between temperature-defined bioregions [59], this evidence suggests that temperature-driven diversifying selection may be an important early-stage factor in the evolution of marine biodiversity.
Supplementary Material
Acknowledgements
The authors are grateful to Robert Schlegel and Albertus Smit for providing the SST data, and to the Centre for High Performance Computing (CHPC), particularly Dane Kennedy, for supercomputer resources and bioinformatics support.
Ethics
Ethical clearance for this sampling approach was granted by the University of Johannesburg Faculty of Science Ethics Committee (Ethics reference no.: 2015-11-01/Teske). The samples in the West Coast National Park (Langebaan Lagoon) were collected under research permit no. CRC/2015/033-2015/V1.
Data accessibility
All data are available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.ns790j4 [36].
Competing interests
We declare we have no competing interests.
Funding
This study was funded by the PADI Foundation (grant no. 10981 to P.R.T.), the National Research Foundation (CSUR grant no. 87702 to P.R.T.), the University of Johannesburg (URC/FRC grant to P.R.T.), and the Australian Research Council (FT130101068 and DP110101275 to L.B.B.). M.T., T.R.G., and A.E.-K. acknowledge the University of Johannesburg for Global Excellence and Stature (GES) fellowships.
References
- 1.Teske PR, Von der Heyden S, McQuaid CD, Barker NP. 2011. A review of marine phylogeography in southern Africa. South Afr. J. Sci. 107, 1–11. ( 10.4102/sajs.v107i5/6.514) [DOI] [Google Scholar]
- 2.Ayre DJ, Minchinton TE, Perrin C. 2009. Does life history predict past and current connectivity for rocky intertidal invertebrates across a marine biogeographic barrier? Mol. Ecol. 18, 1887–1903. ( 10.1111/j.1365-294X.2009.04127.x) [DOI] [PubMed] [Google Scholar]
- 3.von der Heyden S. 2009. Why do we need to integrate population genetics into South African marine protected area planning? Afr. J. Mar. Sci. 31, 263–269. ( 10.2989/AJMS.2009.31.2.14.886) [DOI] [Google Scholar]
- 4.Dawson MN. 2012. Parallel phylogeographic structure in ecologically similar sympatric sister taxa. Mol. Ecol. 21, 987–1004. ( 10.1111/j.1365-294X.2011.05417.x) [DOI] [PubMed] [Google Scholar]
- 5.Pelc RA, Warner RR, Gaines SD. 2009. Geographical patterns of genetic structure in marine species with contrasting life histories. J. Biogeogr. 36, 1881–1890. ( 10.1111/j.1365-2699.2009.02138.x) [DOI] [Google Scholar]
- 6.Kelly RP, Palumbi SR. 2010. Genetic structure among 50 species of the northeastern Pacific rocky intertidal community. PLoS ONE 5, e8594 ( 10.1371/journal.pone.0008594) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Drost E, Golla TR, Heyden S, Teske PR. 2016. No divergent evolution, despite restricted connectivity, between Atlantic and Indian Ocean goby populations. Mar. Biodivers. 46, 465–471. ( 10.1007/s12526-015-0389-6) [DOI] [Google Scholar]
- 8.Mmonwa KL, Teske PR, McQuaid CD, Barker NP. 2015. Historical demography of southern African patellid limpets: congruence of population expansions, but not phylogeography. Afr. J. Mar. Sci. 37, 11–20. ( 10.2989/1814232X.2015.1009165) [DOI] [Google Scholar]
- 9.Teske PR, Winker H, McQuaid CD, Barker NP. 2009. A tropical/subtropical biogeographic disjunction in southeastern Africa separates two evolutionarily significant units of an estuarine prawn. Mar. Biol. 156, 1265–1275. ( 10.1007/s00227-009-1168-3) [DOI] [Google Scholar]
- 10.Teske PR, Froneman PW, Barker NP, McQuaid CD. 2007. Phylogeographic structure of the caridean shrimp Palaemon peringueyi in South Africa: further evidence for intraspecific genetic units associated with marine biogeographic provinces. Afr. J. Mar. Sci. 29, 253–258. ( 10.2989/AJMS.2007.29.2.9.192) [DOI] [Google Scholar]
- 11.Teske PR, et al. 2009. Tri-locus sequence data reject a ‘Gondwanan origin hypothesis' for the African/South Pacific crab genus Hymenosoma. Mol. Phylogenet. Evol. 53, 23–33. ( 10.1016/j.ympev.2009.05.031) [DOI] [PubMed] [Google Scholar]
- 12.Nangammbi TC, Herbert DG, Teske PR. 2016. Molecular insights into species recognition within southern Africa's endemic Tricolia radiation (Vetigastropoda: Phasianellidae). J. Molluscan Stud. 82, 97–103. ( 10.1093/mollus/eyv037) [DOI] [Google Scholar]
- 13.Haye PA, Segovia NI, Muñoz-Herrera NC, Gálvez FE, Martínez A, Meynard A, Pardo-Gandarillas MC, Poulin E, Faugeron S. 2014. Phylogeographic structure in benthic marine invertebrates of the southeast Pacific coast of Chile with differing dispersal potential. PLoS ONE 9, 1–15. ( 10.1371/journal.pone.0088613) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Weersing K, Toonen RJ. 2009. Population genetics, larval dispersal, and connectivity in marine systems. Mar. Ecol. Prog. Ser. 393, 1–12. ( 10.3354/meps08287) [DOI] [Google Scholar]
- 15.Nosil P. 2012. Ecological speciation. Oxford, UK: Oxford University Press. [Google Scholar]
- 16.Rocha LA, Robertson DR, Roman J, Bowen BW. 2005. Ecological speciation in tropical reef fishes. Proc. R. Soc. B 272, 573–579. ( 10.1098/2004.3005) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Teske PR, Papadopoulos I, Newman BK, Dworschak PC, McQuaid CD, Barker NP. 2008. Oceanic dispersal barriers, adaptation and larval retention: an interdisciplinary assessment of potential factors maintaining a phylogeographic break between sister lineages of an African prawn. BMC Evol. Biol. 8, 341 ( 10.1186/1471-2148-8-341) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Papadopoulos I, Teske PR. 2014. Larval development reflects biogeography in two formerly synonymised southern African coastal crabs. Afr. J. Aquat. Sci. 39, 347–350. ( 10.2989/16085914.2014.938600) [DOI] [Google Scholar]
- 19.Waltari E, Hickerson MJ. 2013. Late Pleistocene species distribution modelling of North Atlantic intertidal invertebrates. J. Biogeogr. 40, 249–260. ( 10.1111/j.1365-2699.2012.02782.x) [DOI] [Google Scholar]
- 20.Beheregaray LB, Sunnucks P. 2001. Fine-scale genetic structure, estuarine colonization and incipient speciation in the marine silverside fish Odontesthes argentinensis. Mol. Ecol. 10, 2849–2866. ( 10.1046/j.1365-294X.2001.t01-1-01406.x) [DOI] [PubMed] [Google Scholar]
- 21.Beheregaray LB. 2008. Twenty years of phylogeography: the state of the field and the challenges for the Southern Hemisphere. Mol. Ecol. 17, 3754–3774. ( 10.1111/j.1365-294X.2008.03857.x) [DOI] [PubMed] [Google Scholar]
- 22.Pörtner HO, Peck L, Somero G. 2007. Thermal limits and adaptation in marine Antarctic ectotherms: an integrative view. Phil. Trans. R. Soc. B 362, 2233–2258. ( 10.1098/rstb.2006.1947) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sandoval-Castillo J, Robinson NA, Hart AM, Strain LWS, Beheregaray LB. 2018. Seascape genomics reveals adaptive divergence in a connected and commercially important mollusc, the greenlip abalone (Haliotis laevigata), along a longitudinal environmental gradient. Mol. Ecol. 27, 1603–1620. ( 10.1111/mec.14526) [DOI] [PubMed] [Google Scholar]
- 24.Guillot G, Vitalis R, le Rouzic A, Gautier M. 2014. Detecting correlation between allele frequencies and environmental variables as a signature of selection. A fast computational approach for genome-wide studies. Spat. Stat. 8, 145–155. ( 10.1016/j.spasta.2013.08.001) [DOI] [Google Scholar]
- 25.Smit AJ, Roberts M, Anderson RJ, Dufois F, Dudley SFJ, Bornman TG, Olbers J, Bolton JJ. 2013. A coastal seawater temperature dataset for biogeographical studies: large biases between in situ and remotely-sensed data sets around the coast of South Africa. PLoS ONE 8, e81944 ( 10.1371/journal.pone.0081944) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Foll M, Gaggiotti O. 2008. A genome-scan method to identify selected loci appropriate for both dominant and codominant markers: a Bayesian perspective. Genetics 180, 977–993. ( 10.1534/genetics.108.092221) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boutet E, Lieberherr D, Tognolli M, Schneider M, Bairoch A. 2007. UniProtKB/Swiss-Prot. Methods Mol. Biol. Clifton NJ 406, 89–112. ( 10.1007/978-1-59745-535-0_4) [DOI] [PubMed] [Google Scholar]
- 28.Alexa A, Rahnenfuhrer J. 2016. topGO: enrichment analysis for gene ontology. R package version 2.24.0. [Google Scholar]
- 29.Jombart T, Ahmed I. 2011. adegenet 1.3-1: New tools for the analysis of genome-wide SNP data. Bioinformatics 27, 3070–3071. ( 10.1093/bioinformatics/btr521) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Raj A, Stephens M, Pritchard JK. 2014. FastSTRUCTURE: variational inference of population structure in large SNP data sets. Genetics 197, 573–589. ( 10.1534/genetics.114.164350) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Bouckaert R, Heled J, Kühnert D, Vaughan T, Wu C-H, Xie D, Suchard MA, Rambaut A, Drummond AJ. 2014. BEAST 2: a software platform for Bayesian evolutionary analysis. PLoS Comput. Biol. 10, e1003537 ( 10.1371/journal.pcbi.1003537) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lemey P, Rambaut A, Drummond AJ, Suchard MA. 2009. Bayesian phylogeography finds its roots. PLoS Comput. Biol. 5, e1000520 ( 10.1371/journal.pcbi.1000520) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Buckley BA, Gracey AY, Somero GN. 2006. The cellular response to heat stress in the goby Gillichthys mirabilis: a cDNA microarray and protein-level analysis. J. Exp. Biol. 209, 2660–2677. ( 10.1242/jeb.02292) [DOI] [PubMed] [Google Scholar]
- 34.Detrich HW, Parker SK. 1993. Divergent neural β tubulin from the Antarctic fish Notothenia coriiceps neglecta: potential sequence contributions to cold adaptation of microtubule assembly. Cytoskeleton 24, 156–166. ( 10.1002/cm.970240303) [DOI] [PubMed] [Google Scholar]
- 35.Castilho PC, Buckley BA, Somero G, Block BA. 2009. Heterologous hybridization to a complementary DNA microarray reveals the effect of thermal acclimation in the endothermic bluefin tuna (Thunnus orientalis). Mol. Ecol. 18, 2092–2102. ( 10.1111/j.1365-294X.2009.04174.x) [DOI] [PubMed] [Google Scholar]
- 36.Teske P, Sandoval-Castillo J, Golla T, Emami-Khoyi A, Tine M, von der Heyden S, Beheregaray L. 2019. Data from: Thermal selection as a driver of marine ecological speciation Dryad Digital Repository. ( 10.5061/dryad.ns790j4) [DOI] [PMC free article] [PubMed]
- 37.Wu CI. 2001. The genic view of the process of speciation. J. Evol. Biol. 14, 851–865. ( 10.1046/j.1420-9101.2001.00335.x) [DOI] [Google Scholar]
- 38.Nosil P. 2008. Speciation with gene flow could be common. Mol. Ecol. 17, 2103–2106. ( 10.1111/j.1365-294X.2008.03715.x) [DOI] [PubMed] [Google Scholar]
- 39.Bowen BW, Rocha LA, Toonen RJ, Karl SA. 2013. The origins of tropical marine biodiversity. Trends Ecol. Evol. 28, 359–366. ( 10.1016/j.tree.2013.01.018) [DOI] [PubMed] [Google Scholar]
- 40.Belanger CL, Jablonski D, Roy K, Berke SK, Krug AZ, Valentine JW. 2012. Global environmental predictors of benthic marine biogeographic structure. Proc. Natl Acad. Sci. USA 109, 14 046–14 051. ( 10.1073/pnas.1212381109) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Bowen BW, Gaither MR, DiBattista JD, Iacchei M, Andrews KR, Grant WS, Toonen RJ, Briggs JC. 2016. Comparative phylogeography of the ocean planet. Proc. Natl Acad. Sci. USA 113, 7962–7969. ( 10.1073/pnas.1602404113) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Tittensor DP, Mora C, Jetz W, Lotze HK, Ricard D, Berghe EV, Worm B. 2010. Global patterns and predictors of marine biodiversity across taxa. Nature 466, 1098–1101. ( 10.1038/nature09329) [DOI] [PubMed] [Google Scholar]
- 43.Stuart-Smith RD, Edgar GJ, Bates AE. 2017. Thermal limits to the geographic distributions of shallow-water marine species. Nat. Ecol. Evol. 1, 1846–1852. ( 10.1038/s41559-017-0353-x) [DOI] [PubMed] [Google Scholar]
- 44.Nosil P, Funk DJ, Ortiz-Barrientos D. 2009. Divergent selection and heterogeneous genomic divergence. Mol. Ecol. 18, 375–402. ( 10.1111/j.1365-294X.2008.03946.x) [DOI] [PubMed] [Google Scholar]
- 45.Smit AJ, Bolton JJ, Anderson RJ. 2017. Seaweeds in two oceans: beta-diversity. Front. Mar. Sci. 4, 404 ( 10.3389/fmars.2017.00404) [DOI] [Google Scholar]
- 46.Pörtner HO. 2001. Climate change and temperature-dependent biogeography: oxygen limitation of thermal tolerance in animals. Naturwissenschaften 88, 137–146. ( 10.1007/s001140100216) [DOI] [PubMed] [Google Scholar]
- 47.Porcelli D, Butlin RK, Gaston KJ, Joly D, Snook RR. 2015. The environmental genomics of metazoan thermal adaptation. Heredity 114, 502–514. ( 10.1038/hdy.2014.119) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Keller I, Seehausen O. 2012. Thermal adaptation and ecological speciation. Mol. Ecol. 21, 782–799. ( 10.1111/j.1365-294X.2011.05397.x) [DOI] [PubMed] [Google Scholar]
- 49.Teske PR, Papadopoulos I, Mmonwa KL, Matumba TG, McQuaid CD, Barker NP, Beheregaray LB. 2011. Climate-driven genetic divergence of limpets with different life histories across a southeast African marine biogeographic disjunction: different processes, same outcome. Mol. Ecol. 20, 5025–5041. ( 10.1111/j.1365-294X.2011.05307.x) [DOI] [PubMed] [Google Scholar]
- 50.Assis J, Zupan M, Nicastro KR, Zardi GI, McQuaid CD, Serrão EA. 2015. Oceanographic conditions limit the spread of a marine invader along southern African shores. PLoS One 10, e0128124 ( 10.1371/journal.pone.0128124) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Teske P, Bader S, Rao Golla T. 2015. Passive dispersal against an ocean current. Mar. Ecol. Prog. Ser. 539, 153–163. ( 10.3354/meps11516) [DOI] [Google Scholar]
- 52.Schumann EH, Cohen AL, Jury MR. 1995. Coastal sea surface temperature variability along the south coast of South Africa and the relationship to regional and global climate. J. Mar. Res. 53, 231–248. ( 10.1357/0022240953213205) [DOI] [Google Scholar]
- 53.Shannon LV, Chapman P. 1983. Suggested mechanism for the chronic pollution by oil of beaches east of Cape Agulhas, South Africa. Afr. J. Mar. Sci. 1, 231–244. ( 10.2989/025776183784447520) [DOI] [Google Scholar]
- 54.Teske PR, Zardi GI, McQuaid CD, Nicastro KR. 2013. Two sides of the same coin: extinctions and originations across the Atlantic/Indian Ocean boundary as consequences of the same climate oscillation. Front. Biogeogr. 5, 48–59. ( 10.21425/F55115591) [DOI] [Google Scholar]
- 55.Angilletta MJ. 2009. Thermal adaptation: a theoretical and empirical synthesis. Oxford, UK: Oxford University Press. [Google Scholar]
- 56.Apte S, Gardner JPA. 2002. Population genetic subdivision in the New Zealand greenshell mussel (Perna canaliculus) inferred from single-strand conformation polymorphism analysis of mitochondrial DNA. Mol. Ecol. 11, 1617–1628. ( 10.1046/j.1365-294X.2002.01554.x) [DOI] [PubMed] [Google Scholar]
- 57.Ayers KL, Waters JM. 2005. Marine biogeographic disjunction in central New Zealand. Mar. Biol. 147, 1045–1052. ( 10.1007/s00227-005-1632-7) [DOI] [Google Scholar]
- 58.Gleason LU, Burton RS. 2016. Genomic evidence for ecological divergence against a background of population homogeneity in the marine snail Chlorostoma funebralis. Mol. Ecol. 25, 3557–3573. ( 10.1111/mec.13703) [DOI] [PubMed] [Google Scholar]
- 59.Briggs JC. 1974. Marine zoogeography. New York, NY: McGraw-Hill. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Teske P, Sandoval-Castillo J, Golla T, Emami-Khoyi A, Tine M, von der Heyden S, Beheregaray L. 2019. Data from: Thermal selection as a driver of marine ecological speciation Dryad Digital Repository. ( 10.5061/dryad.ns790j4) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
All data are available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.ns790j4 [36].