Abstract
In insects, antimicrobial humoral immunity is governed by two distinct gene cascades, IMD pathway mainly targeting Gram-negative bacteria and Toll pathway preferentially targeting Gram-positive bacteria, which are widely conserved among diverse metazoans. However, recent genomic studies uncovered that IMD pathway is exceptionally absent in some hemipteran lineages like aphids and assassin bugs. How the apparently incomplete immune pathways have evolved with functionality is of interest. Here we report the discovery that, in the hemipteran stinkbug Plautia stali, both IMD and Toll pathways are present but their functional differentiation is blurred. Injection of Gram-negative bacteria and Gram-positive bacteria upregulated effector genes of both pathways. Notably, RNAi experiments unveiled significant functional permeation and crosstalk between IMD and Toll pathways: RNAi of IMD pathway genes suppressed upregulation of effector molecules of both pathways, where the suppression was more remarkable for IMD effectors; and RNAi of Toll pathway genes reduced upregulation of effector molecules of both pathways, where the suppression was more conspicuous for Toll effectors. These results suggest the possibility that, in hemipterans and other arthropods, IMD and Toll pathways are intertwined to target wider and overlapping arrays of microbes, which might have predisposed and facilitated the evolution of incomplete immune pathways.
Keywords: insect immunity, innate immunity, antimicrobial peptide, IMD pathway, Plautia stali
1. Introduction
The innate immune system is highly conserved among metazoans, which enables prompt defense against microbial intruders [1,2]. In insects, the innate immunity is often classified into three categories: physical, cellular and humoral [3]. The humoral immunity rapidly and transiently activates immune genes upon microbial infections and elicits the release of an array of antimicrobial peptides (AMPs) to haemolymph [4]. Humoral immunity is triggered by sensing of microbial infections via interactions of peptidoglycan recognition proteins (PGRP) and Gram-negative binding proteins (GNBP) with microbial surface molecules [2]. In general, each of the recognition proteins induces subsequent reactions of one of the two signalling pathways, namely immune deficiency (IMD) pathway, which is preferentially activated by Gram-negative bacteria, and Toll pathway, which is preferentially activated by Gram-positive bacteria and fungi. In the fruit fly Drosophila melanogaster, most of the AMPs are preferentially activated by either IMD pathway or Toll pathway, but exceptionally, some AMPs (such as Drosomycin) are activated by both IMD and Toll pathways [5,6]. Despite the presence of some minor crosstalk, the two immune pathways are generally recognized as independent.
Genome-wide analyses have shown that IMD pathway and Toll pathway are both highly conserved in holometabolous insects including the fruit flies Drosophila spp. [7–9], the mosquito Anopheles gambiae [10], the silkmoth Bombyx mori [11], the honeybee Apis mellifera [12] and the flour beetle Tribolium castaneum [13]. These immune pathways correspond to the mammalian signalling pathways of interleukin-1 receptor and tumour necrosis factor receptor [14]. Hence, it is presumed that primitive forms of IMD pathway and Toll pathway must have arisen in the common ancestor of arthropods and vertebrates.
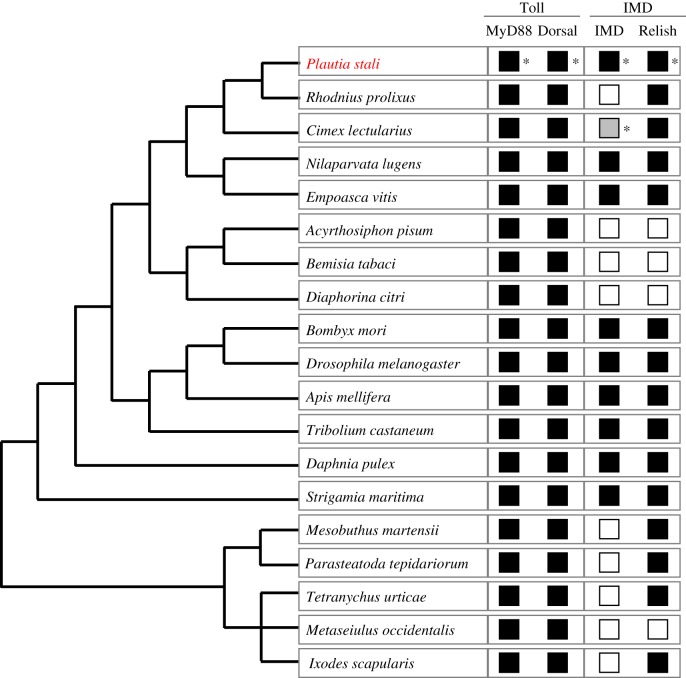
However, in some insects belonging to the Hemiptera, recent studies uncovered the absence or incompleteness of IMD pathway. The pea aphid Acyrthosiphon pisum lacks all IMD pathway genes including PGRP-encoding gene, Imd and Relish [14] and the assassin bug Rhodnius prolixus lacks key components of IMD pathway-like Imd but retains Relish [15]. Lack of Imd was also reported from the bedbug Cimex lectularius and other assassin bugs Triatoma pallidipennis, T. dimidiata and T. infestans [16,17]. On the other hand, the brown planthopper Nilaparvata lugens possesses a conserved IMD pathway [18]. The conserved IMD pathway was also found in non-insect arthropods like the water flea Daphnia pulex [19] and the coastal centipede Strigamia maritima [20], but the Chinese scorpion Mesobuthus martensii and the deer tick Ixodes scapularis do not possess Imd [21,22] and the western orchard predatory mite Metaseiulus occidentalis lacks almost all IMD pathway components including Imd and Relish [20]. Hence, phylogenetic mapping suggests repeated and independent losses of IMD pathway components in the evolutionary course of arthropods (figure 1), but it has been poorly understood how the erosion of IMD pathway has proceeded.
Figure 1.
A schematic phylogenetic tree of arthropods on which presence/absence of humoral immunity-related genes is mapped. Black and white squares indicate the presence and absence of humoral immunity-related genes, respectively. There is a grey square that indicates uncertainty (see text for details). The phylogeny was drawn on the basis of Misof et al. [23], Kolokotronis et al. [24], Sharma et al. [25] and Palmer & Jiggins [20]. Presence/absence of the genes were based on Mesquita et al. [15] for R. prolixus, Benoit et al. [16] and Zumaya-Estrada et al. [17] for C. lectularius, Bao et al. [18] for N. lugens, Shao et al. [26] for E. vitis, Gerardo et al. [14] for A. pisum, Zhang et al. [27] for B. tabaci, Arp et al. [28] for D. citri, Sackton et al. [9] for D. melanogaster, Tanaka et al. [11] for B. mori, Evans et al. [12] for A. mellifera, Zou et al. [13] for T. castaneum, McTaggart et al. [19] for D. pulex, Palmer & Jiggins [20] for S. maritima, P. tepidariorum and M. occidentalis, Cao et al. [21] for M. martensii, Grbíc et al. [29] for T. urticae and Smith & Pal [22] for I. scapularis. Squares with asterisks indicate the results of this study (see Results and discussion). (Online version in colour.)
In this study, we demonstrate that a hemipteran insect, the brown-winged green stinkbug Plautia stali, retains both IMD pathway and Toll pathway but their functional differentiation has been blurred, which provides insight into the evolutionary process of incomplete immune pathways in hemipteran insects and other arthropods.
2. Material and methods
(a). Insects
Adult insects of P. stali, collected at the forest edge in the National Institute of Advanced Industrial Science and Technology, Tsukuba, Japan, were used to establish an inbred strain. A mass-reared colony of the strain was used as the source of the experimental insects. The insects were reared in plastic containers (150 mm in diameter, 60 mm high) supplied with raw peanuts, dry soya beans and water supplemented with 0.05% ascorbic acid at 25°C ± 1°C under a long-day regime of 16 L : 8 D dark following the previous study [30].
(b). RNA-seq analysis
Total RNAs were extracted from fat body, midgut or whole body of adult females approximately 5 days after ecdysis using RNeasy Mini Kit (Qiagen, Hilden, Germany). To efficiently obtain humoral immunity-related genes, RNAs were also extracted from fat body of adult insects injected with Gram-negative Escherichia coli or Gram-positive Micrococcus luteus. The cDNAs were sequenced by Illumina HiSeq 2500 with paired-end 101 bp (Macrogen Japan Corp., Kyoto, Japan), and the generated raw reads (Accession nos. DRR118501–DRR118507; electronic supplementary material, table S1) were cleaned by trimming low-quality regions and adapter sequences using Trimmomatic version 0.36 with minimum length setting as 50 bp [31]. Reference contigs were constructed by de novo assembly of all the cleaned paired-end reads using Trinity v. 2.4.0 [32]. After automatic assembly, we manually checked and corrected the target gene sequences using the Integrative Genomics Viewer [33]. For annotation of the reference contigs, each contig sequence was compared with NCBI-nr protein database by blastx (e-value less than <1 × 10−3), and a summary of the top-hit result was added to each contig. In addition, CDS region of each contig was predicted by TransDecoder v. r20140704 [32], and conserved domains in each predicted amino acid sequence were searched by HMMER v. 3.1b2 [34] using Pfam protein families database [35]. For searching orthologous genes related to Toll and IMD pathways, blastp search using Drosophila Toll and IMD genes as query sequences were performed against a database of the predicted amino acid sequences of P. stali. For transcript quantification of the reference contigs for each fat body sample, align_and_estimate_abundance.pl script bundled with Trinity v. r20140707 was used for the cleaned reads of each fat body sample with –est_method RSEM, –aln_method bowtie2 and –trinity_mode options, and the calculated expression levels of the reference contigs were normalized to fragments per kilobase per million mapped fragments (FPKM) values with Trimmed mean of M values (TMM) normalization among all the fat body samples by abundance_estimate_to_matrix.pl script bundled with Trinity v. r20140707. The normalized FRKM values were used for comparing gene (contig) expression level between control and bacteria-treated fat body samples.
(c). Septic shock experiments
Adult female P. stali approximately 5 days after ecdysis were injected by glass capillary tubes (size: 100 µl; Drummond, Alabama, USA). The females (approximately 7.0 mm in thorax width and 0.13 mg in body weight) were injected with ca 5 µl of 0.9% saline (Otsuka Pharmaceutical, Tokyo, Japan) or 108 µl−1 heat-killed bacteria (E. coli or M. luteus), into the ventral septum between thoracic and abdominal segments. As controls, non-treated females and mock-injected (wounded by glass capillary tubes only) females were used. The fat body was excised 1, 4, 8 or 24 h after injection and total RNA was extracted (see below; electronic supplementary material, figure S1a).
(d). Quantitative RT-PCR
Total RNAs extracted from abdominal fat bodies using TRIzol (Life Technologies Japan Ltd, Tokyo, Japan) were reverse transcribed into cDNA using High-Capacity cDNA Reverse Transcription Kit (Life Technologies Japan Ltd, Tokyo, Japan). Quantitative RT-PCR of humoral immunity-related genes (PGRP-encoding genes, GNBPs, Imd, Relish, myeloid differentiation protein-88 (MyD88), Dorsal, Defensin, Hemiptericin and Lysozyme) was conducted with primer pairs designed on the basis of RNA-seq data (electronic supplementary material, table S2) using LightCycler 480 and LightCycler 480 SYBR Green Master (Roche Diagnostics, Tokyo, Japan).
(e). RNAi experiments
To knockdown the mRNA levels of humoral immunity-related genes, adult females approximately 5 days after ecdysis were injected with roughly 5 µl of double-stranded RNA (dsRNA) solution (100 ng µl−1) into the ventral septum between thoracic and abdominal segments. dsRNA was synthesized from a PCR product using primers listed in electronic supplementary material, table S3, and MEGAscript RNAi Kit (Thermo Fisher Inc., Kanagawa, Japan). Three days after dsRNA injection, several individuals were subjected to RNA extraction to examine the effects of RNAi (electronic supplementary material, figure S1b). Other individuals were injected with heat-killed E. coli or M. luteus 3 days after dsRNA injection, and then, subjected to RNA extraction on the following day (electronic supplementary material, figure S1c). The extracted RNAs were subjected to quantitative RT-PCR (as above) to infer the expression levels of AMPs.
(f). Phylogenetic analysis
The amino acid sequences inferred from PGRP, GNBP and lysozyme genes were aligned by ClustalW [36] and subjected to phylogenetic analyses by the maximum-likelihood method using MEGA7 program [37,38].
3. Results
(a). Genes of Plautia stali upregulated by septic shock
Compared with non-treated control, 650 RNA-seq contigs showed 10-fold or higher expression upon injection of heat-killed E. coli (a representative of Gram-negative bacteria), whereas 554 contigs showed 10-fold or higher expression upon injection of heat-killed M. luteus (a representative of Gram-positive bacteria). Notably, 274 contigs were commonly identified between them (i.e. upregulated by both E. coli and M. luteus) (electronic supplementary material, table S4).
(b). Immune pathway genes of Plautia stali
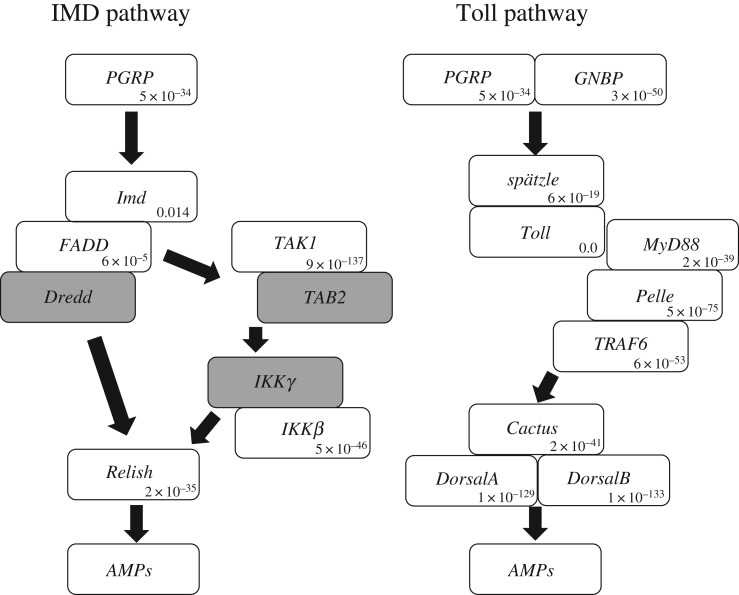
Blastp searches on transcriptomes of P. stali (both septic shocked and non-treated ones) using immunity-related genes of D. melanogaster as queries suggested that P. stali possesses nearly complete IMD pathway and Toll pathway (figure 2; electronic supplementary material, figure S2).
Figure 2.
Genes cascades of IMD and Toll pathways. White and grey boxes show presence and presumable absence of the genes in P. stali, respectively. The number in each white box is blastp e-value of the orthologue in P. stali using the amino acid sequence of D. melanogaster as a query. For PGRP and GNBP family members, those with lowest e-value are shown (For PGRP, PsPGRP-L1b and DmPGRP-LE exhibit lowest e-value (5 × 10−34); for GNBP, PsGNBP1 and DmGNBP1 exhibit lowest e-value (3 × 10−50)).
(i). Pattern recognition molecules
Three P. stali orthologues of a peptidoglycan recognition protein (i.e. PsPGRP-L1a, PsPGRP-L1b and PsPGRP-L2) and an orthologue of a lysin motif protein (PsLysM), a member of peptidoglycan recognition proteins known to bind to peptidoglycans in plants [39], were found. The three PGRPs in P. stali were named as PsPGRP-L1a, PsPGRP-L1b and PsPGRP-L2, respectively, because all of them were categorized as long type and the two PGRPs (PsPGRP-L1a and PsPGRP-L1b) were considered as isoforms. Predicted amino acid sequences of PsPGRP-L1a, PsPGRP-L1b and PsPGRP-L2 exhibited transmembrane domains and were presumably without amidase activities for scavenging digested peptidoglycan (electronic supplementary material, figures S3–S5). Two orthologues of GNBPs (PsGNBP1 and PsGNBP2) were also found (electronic supplementary material, figure S6).
(ii). Signalling pathway components
Orthologues of all major IMD pathway and Toll pathway components were found in the RNA-seq data (figure 2). However, we failed to detect several IMD component genes such as death-related ced-3/Nedd2-like caspase (Dredd), TAK1-binding protein (TAB2) and IEC-intrinsic IκB kinase (IKKγ), although full genomic sequencing of P. stali is needed to verify the absence of these genes. PsImd and Fas-associated protein with death domain (PsFADD) exhibited relatively low homology values to DmImd and DmFADD, respectively (e-values by blastp were 0.014 and 6 × 10−5, respectively; note that PsImd appeared to be functional in IMD pathway; see below).
(iii). Immune-responsive effectors
As immune-responsive effector genes, two orthologues of defensin (PsDefensin1 and PsDefensin2), one orthologue of hemiptericin (PsHemiptericin) and six orthologues of lysozyme (PsLysozyme b-1, PsLysozyme b-2, PsLysozyme c-1, PsLysozyme c-2, PsLysozyme c-3 and PsLysozyme i-1) were identified in the RNA-seq data (electronic supplementary material, figure S7).
(c). Expression of immune pathway genes upon septic shock
(i). Pattern recognition molecules
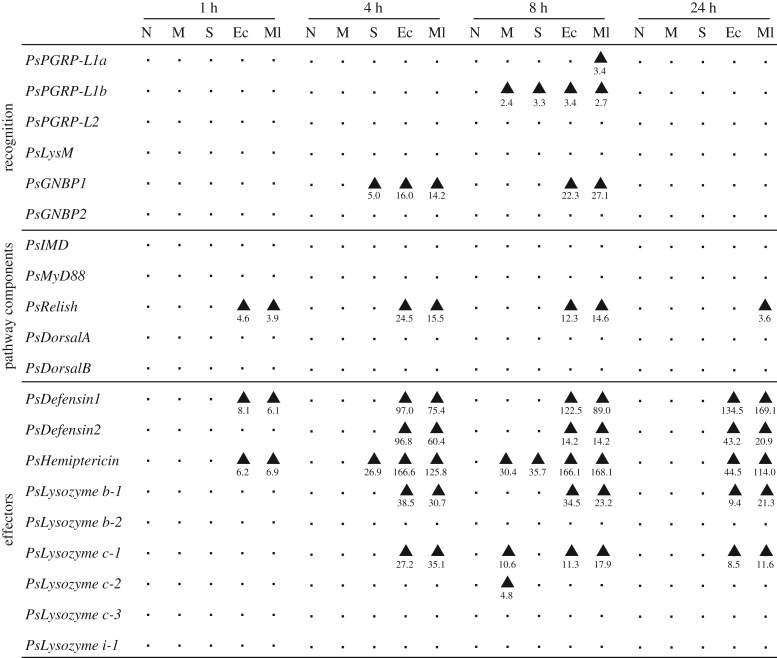
PsPGRP-L1a was significantly upregulated by M. luteus 8 h after injection but was not by E. coli (×3.4) (figure 3; electronic supplementary material, figure S8). PsPGRP-L1b was significantly upregulated not only by E. coli (×3.4) and M. luteus (×2.7) but also by mock (×2.4) and saline (×3.3) 8 h after injection (figure 3; electronic supplementary material, figure S8). By contrast, PsPGRP-L2 and PsLysM were not affected by the septic shock treatments (figure 3; electronic supplementary material, figure S8). PsGNBP1 was upregulated by both E. coli (×16.0–22.3) and M. luteus (×14.2–27.1) 4 and 8 h after injection, whereas PsGNBP2 was not affected by the septic shock treatments (figure 3; electronic supplementary material, figure S8).
Figure 3.
Effects of septic shock treatments on the expression levels of immune pathway genes in P. stali. Adult females were subjected to the following treatments: N, no treatment; M, mock injection; S, saline injection; Ec, E. coli injection; Ml, M. luteus injection. The abdominal fat body was excised 1, 4, 8 and 24 h after injection (n = 5–8) and quantitative RT-PCR was performed for the samples. Significantly upregulated cohorts in comparison to non-treated cohorts were shown by arrowheads (Steel-Dwass test, p < 0.05). Numbers below the arrowheads indicate the fold-change value for the upregulation compared with non-treated cohorts. Detailed data are shown in electronic supplementary material, figures S7–S10.
(ii). Signalling pathway components
Among five pathway component genes, only PsRelish (an IMD pathway component) was upregulated by both E. coli (×4.6–24.5) and M. luteus (×3.6–15.5) 1, 4, 8 and 24 h after injection, while the others (i.e. PsImd, PsMyD88, PsDorsalA and PsDorsalB) were not affected by the septic shock treatments (figure 3; electronic supplementary material, figure S9).
(iii). Immune-responsive effectors
PsDefensin1 and PsHemiptericin were upregulated by both E. coli (×8.1–134.5 and ×6.2–166.6) and M. luteus (×6.1–169.1 and ×6.9–168.1) 1 h after injection and on, while Defensin2 was upregulated by E. coli (×14.2–96.8) and M. luteus (×14.2–60.4) 4 h after injection and on (figure 3; electronic supplementary material, figure S10). Among six lysozyme genes, Lysozyme b-1 and Lysozyme c-1 were upregulated by both E. coli (×9.4–38.5 and ×8.5–27.2) and M. luteus (×21.3–30.7 and ×11.6–35.1) 4 h after injection and on, while the remaining four lysozyme genes were not upregulated by the septic shock treatments (figure 3; electronic supplementary material, figure S11).
(d). RNAi-mediated knockdown of immune pathway genes and expression levels of effector genes
In P. stali, RNAi generally worked efficiently and stably by injection of dsRNA. For example, injection of dsPsRelish into adult insects suppressed the expression levels of PsRelish to approximately one-tenth in comparison with the dsEGFP-injected control insects during at least 14 days after injection (electronic supplementary material, figure S12). All the other immune component genes we examined, namely PsImd, PsMyD88, PsDorsal, PsPGRP-L1a, PsPGRP-L1b, PsPGRP-L2, PsLysM, PsGNBP1 and PsGNBP2, were also suppressed efficiently by RNAi (electronic supplementary material, figures S13–S16).
(i). Which immune pathway controls AMPs and lysozymes?
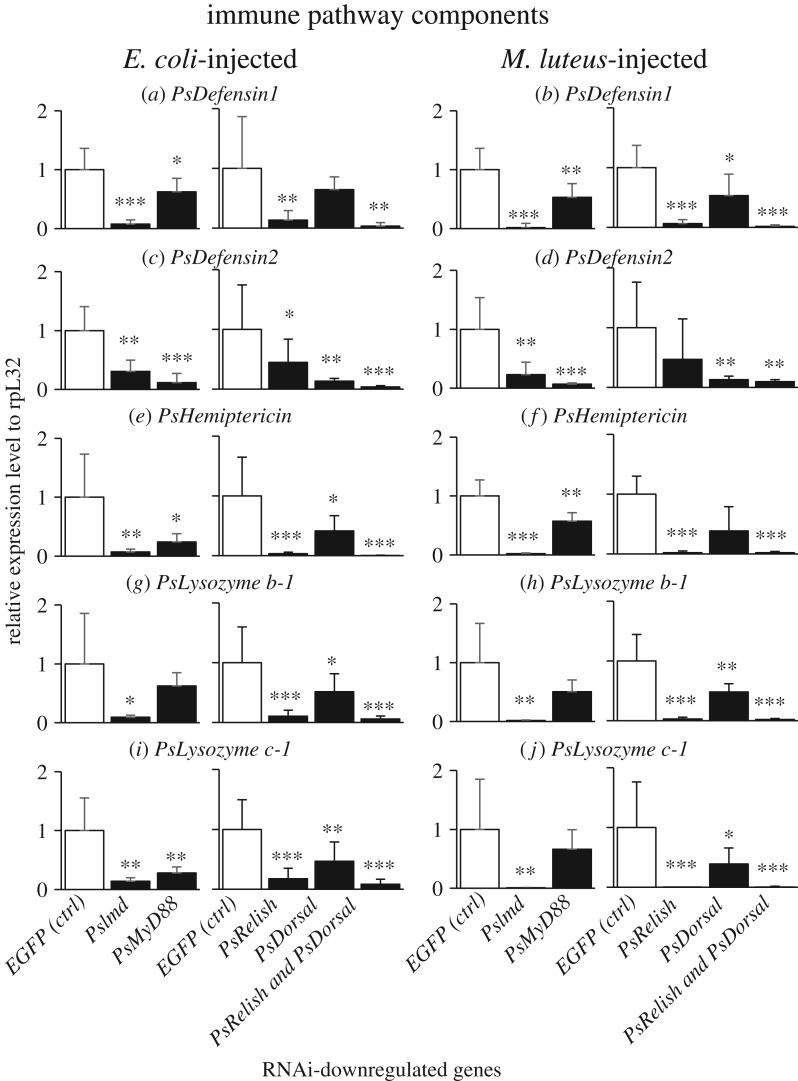
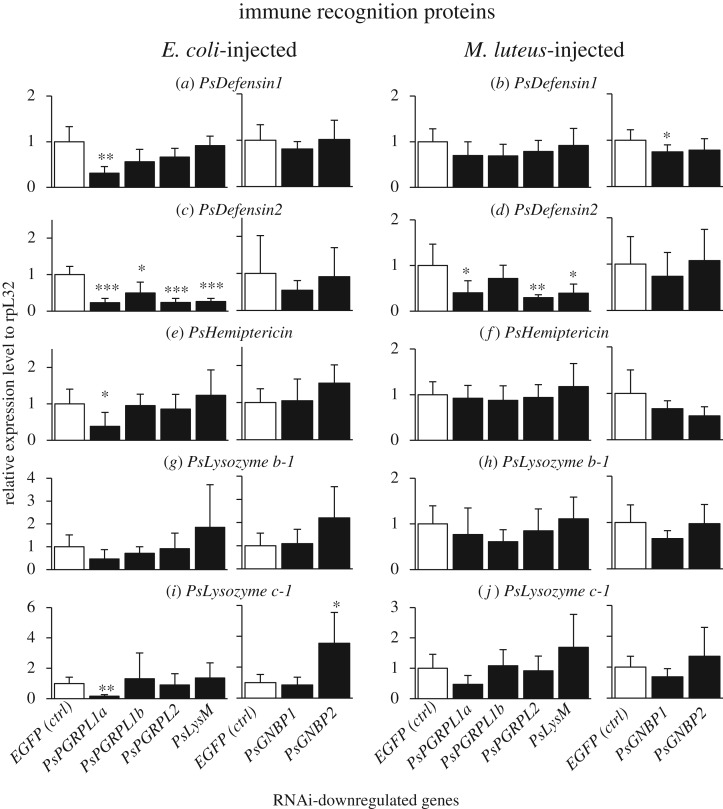
Blocking of IMD pathway by RNAi of PsImd or PsRelish resulted in suppressed upregulation of PsDefensin1, PsHemiptericin, PsLysozyme b-1 and PsLysozyme c-1 (figure 4a,b,e–j), indicating that these immune effector molecules are mainly under the control of IMD pathway. By contrast, blocking of Toll pathway by RNAi of PsMyD88 or PsDorsal caused suppressed upregulation of PsDefensin2 (figure 4c,d), indicating that this effector is mainly under the control of Toll pathway. Here it should be noted that the RNAi blocking of IMD pathway also resulted in, though less remarkable, suppressed upregulation of PsDefensin2 (figure 4c,d). Likewise, the RNAi blocking of Toll pathway caused, though less remarkable, suppressed upregulation of PsDefensin1, PsHemiptericin, PsLysozyme b-1 and PsLysozyme c-1 (figure 4a,b,e–j). Simultaneous blocking of IMD pathway and Toll pathway by RNAi of PsRelish and PsDorsal was most effective in all effector genes (figure 4a–j).
Figure 4.
Effects of RNAi knockdown of immune pathway component genes on septic shock-induced upregulation of immune-responsive effector genes in P. stali. The expression levels of the immune effector genes were standardized to the expression levels of ribosomal protein L32 (rpL32). Adult females were injected with heat-killed E. coli or M. luteus 3 days after dsRNA injection and subjected to RNA extraction from fat bodies on the following day. Significant downregulation compared with EGFP injection is shown by asterisks (t-test: *p < 0.05; **p < 0.01; ***p < 0.001). The error bar indicates the standard deviation (s.d.).
(ii). Which recognition protein triggers the immune pathways?
RNAi of PsPGRP-L1a resulted in suppressed upregulation of PsDefensin1, PsHemiptericin and PsLysozyme c-1 when challenged by Gram-negative E. coli (figure 5a,e,i). For the effectors that are mainly under the control of IMD pathway (figure 4), the suppression was not observed when challenged by Gram-positive M. luteus (figure 5b,f,j).
Figure 5.
Effects of RNAi knockdown of immune recognition genes on the septic shock-induced upregulation of immune-responsive effector genes. The expression levels of the immune effector genes were standardized to the expression levels of ribosomal protein L32 (rpL32). Adult females were injected with heat-killed E. coli or M. luteus 3 days after dsRNA injection and subjected to RNA extraction from fat bodies on the following day. Significant downregulation compared with EGFP injection is shown by asterisks (t-test: *p < 0.05; **p < 0.01; ***p < 0.001). The error bar indicates the standard deviation (s.d.).
Meanwhile, the suppression patterns observed with the effector PsDefensin2, which is mainly under the control of Toll pathway (figure 4), were observed by RNAi of PsPGRP-L1a, PsPGRP-L2 and PsLysM (and marginally PsPGRP-L1b), irrespective of bacterial challenges by Gram-negative E. coli or Gram-positive M. luteus (figure 5c,d).
4. Discussion
(a). Immune pathway genes of Plautia stali
Our RNA-seq analysis showed that P. stali possesses a repertoire of well-conserved genes of IMD pathway and Toll pathway, but some components of IMD pathway (Dredd, TAB2 and IKKγ) had not been found. The lack of Dredd, TAB2 and IKKγ was also reported in the bedbug C. lectularius and the assassin bug R. prolixus, in which Imd was also assumed to be absent [15,17]. Using PsImd as a query, however, we found a homologous sequence in C. lectularius (uncharacterized protein, XP_014246002; e-values were 4e-5 by blastp and 0.084 by tblastx) but not in R. prolixus (lowest e-values were 0.15 to mapmodulin-like protein [AY340275] by blastp and 0.19 to Krüppel gene [JN092576] by tblastx). It is likely that the barely homologous sequence in C. lectularius may function as Imd. We speculate that the Imd sequence is so divergent that it is extremely difficult to find by homology search in some hemipteran species, mites and ticks (figure 1) [20].
(b). Induction of immune pathway genes in response to Gram-negative and Gram-positive bacteria
Almost all immune pathway genes upregulated by septic shock treatments in P. stali, including PsPGRP-L1b, PsGNBP1, PsRelish, PsDefensin1, PsDefensin2, PsHemiptericin, PsLysozyme b-1 and PsLysozyme c-1, responded to Gram-negative E. coli and Gram-positive M. luteus indiscriminately, although E. coli and M. luteus represent different cell wall components on their surface, namely E. coli with DAP-type peptidoglycan and M. luteus with Lys-type peptidoglycan. These upregulation patterns are distinct from the well-known patterns established for D. melanogaster: most of the AMPs are preferentially activated by either IMD pathway or Toll pathway (but with lesser degree for Drosomycin [6]), whereas IMD pathway and Toll pathway are preferentially activated by Gram-negative bacteria and Gram-positive bacteria, respectively [40–42]. In T. castaneum [43] and A. mellifera [44], it has been reported that some AMPs ambiguously respond to various bacteria while other AMPs respond to specific bacteria in a canonical manner. Our results in P. stali is peculiar in that, in all the affected genes, the degree of response was similar between the insects challenged by Gram-negative bacteria and those challenged by Gram-positive bacteria.
Notably, upregulation of these responded effectors was maintained for at least 24 h, which was in contrast to most AMPs in D. melanogaster that exhibit transient monomodal upregulation within 12 h after bacterial challenge [40,41,45]. The long-lasting expression of AMPs might be relevant to the presumable lack of amidase activities in the PGRPs of P. stali. Because the amino acid sequences of three PGRP orthologues in P. stali assumed to have no amidase activity, three PGRP orthologues were considered presumably to play some roles in recognizing invading bacteria. PGRPs with amidase activity (amidase PGRPs), such as PGRP-LB and PGRP-SC in D. melanogaster, play important roles in downregulation of IMD pathway by degrading bacterial peptidoglycans [46,47].
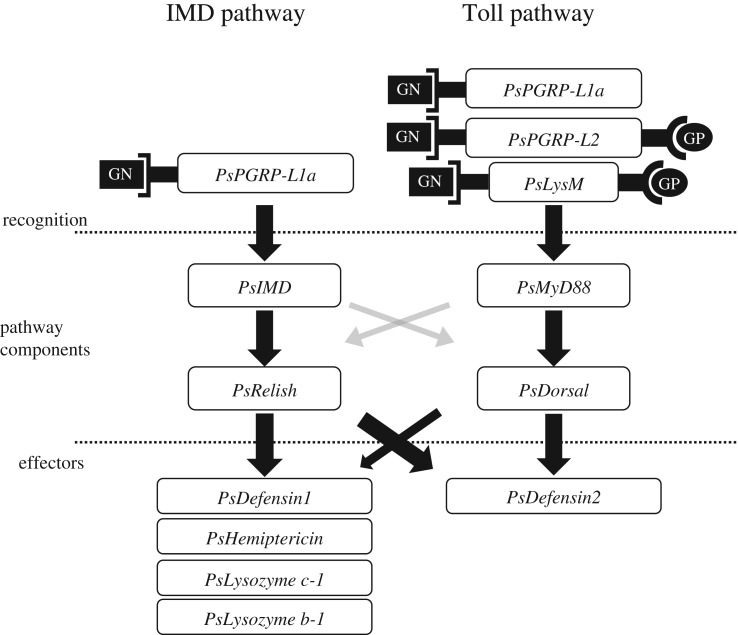
(c). Functional crosstalk across IMD and Toll pathways in Plautia stali
The knockdown of IMD pathway components reduced the upregulation of effectors of both pathways (but more remarkable for those of IMD pathway), and the knockdown of Toll pathway components also reduced the upregulation of effectors of both pathways (but more remarkable for those of Toll pathways). These results suggest that IMD pathway and Toll pathway are not distinct but intertwined in P. stali. Figure 6 shows a hypothetical model of innate immune pathways in P. stali. The crosstalk between IMD pathway and Toll pathway is mainly mediated by PsRelish and PsDorsal, although the possibility of crosstalk mediated by upstream genes cannot be excluded. The conspicuous crosstalk between IMD pathway and Toll pathway in P. stali counter the well-established notion in D. melanogaster that IMD pathway and Toll pathway are preferentially activated by and acting on Gram-negative bacteria and Gram-positive bacteria, respectively [40,41]. Meanwhile, several studies reported slight interactive effects between the immune pathways in D. melanogaster, which are mediated by NF-κB family transcription factors that bind to promoters of immune effector genes [48,49]. The NF-κB-mediated interactive effects were also found in the honeybee A. mellifera [44]. Because Toll pathway component MyD88 and Imd share a death domain, MyD88 might activate IMD pathway in D. melanogaster [50]. It was also reported that, in the flour beetle T. castaneum, IMD pathway and Toll pathway are activated and used more promiscuously than in D. melanogaster [43].
Figure 6.
Schematic overview of IMD and Toll pathways in P. stali. Although the two conserved pathways are present, considerable crosstalk occurs across the two pathways. The left and right arms on the recognition proteins depict receptors for Gram-negative (GN) bacteria and Gram-positive (GP) bacteria, respectively.
(d). Which recognition protein triggers the immune pathways?
RNAi of PsPGRP-L1a resulted in suppressed upregulation of effectors that are mainly under the control of IMD pathway when challenged by Gram-negative E. coli but not by the challenge of Gram-positive M. luteus. Meanwhile, the suppression patterns observed with the effector that is mainly under the control of Toll pathway was observed by RNAi of PsPGRP-L1a, PsPGRP-L2 and PsLysM (and marginally PsPGRP-L1b). These results indicate that PsPGRP-L1a recognizes Gram-negative E. coli, thereby upregulating the downstream effector molecules PsDefensin1, PsHemiptericin and PsLysozyme c-1. PsPGRP-L2 and PsLysM (and possibly PsPGRP-L1b) are located at the upstream of the pathway(s) leading to upregulation of PsDefensin2 and might be capable of recognizing not only Gram-positive M. luteus but also Gram-negative E. coli. On the other hand, the suppressive effect of PsPGRP-L1a RNAi on upregulation of PsDefensin2 when challenged by Gram-positive M. luteus (p = 0.032) is confusing and difficult to interpret, deserving detailed analyses in future studies. Taken together, these results suggest that the specificity of the recognition proteins to Gram-positive and Gram-negative bacteria may be also blurred in P. stali.
This blurred recognition of Gram-negative and Gram-positive bacteria might cause intertwining across IMD pathway and Toll pathway. While IMD pathway is mainly induced by Gram-negative bacteria via PsPGRP-L1a, Toll pathway is stimulated not only by Gram-positive bacteria but also by Gram-negative bacteria via PsPGRP-L2 and PsLysM (and possibly PsPGRP-L1b).
(e). PsLysM as a presumable immune recognition protein
Our observation that RNAi of PsLysM suppressed the septic shock-induced upregulation of PsDefensin2 (figure 5c,d) suggests that PsLysM may function as an immune recognition protein in P. stali. PsLysM contains a lysin motif (LysM), which is a ubiquitous protein module identified in many prokaryotes and eukaryotes [40]. In some plants, LysM-containing genes recognize bacteria, especially symbiotic ones [51]. To our knowledge, this study is the first to show an immunity-related role of a LysM-containing gene in insects. It is notable that LysM-containing genes have been identified in other hemipteran insects such as Riptortus pedestris [52] and Diaphorina citri [28], but the function has not been examined.
(f). Evolutionary insight into the loss of IMD pathway genes in some arthropods
In previous studies on the genomics of the pea aphid A. pisum, the following aspects were argued in relation to the loss of IMD pathway genes [14,53]. First, the aphid feeds on plant phloem sap that is substantially sterile [54] and thus may entail the relatively low risk of encountering microbial pathogens. Second, the aphid often harbours facultative bacterial symbionts (including Regiella, Hamiltonella, Rickettsia, Rickettsiella and Spiroplasma) that exhibit defensive activities against microbial pathogens and insect parasitoids [55–57], which may at least partially compensate for the host's innate immune functions. Notably, P. stali have obligate symbionts but its defensive function had not been explored [30,58,59]. Finally, upon injury, the aphid was reported to increase terminal reproductive investment, presumably at the expense of humoral immunity [60], which may to some extent attenuate dependence on its own innate immune system. In this study, we propose a hypothesis that the functional redundancy between IMD pathway and Toll pathway may have predisposed and facilitated the loss of some or entire IMD pathway genes. It should be noted that this hypothesis is, although more generally applicable, not mutually exclusive with the above-mentioned hypotheses and might be strengthened by functional analysis of IMD pathway and Toll pathway in other hemipteran insects and non-insect arthropods that have incomplete IMD pathway. The functional crosstalk across IMD pathway and Toll pathway may be more common than generally recognized, and it might be the evolutionarily intermediate step towards the loss of IMD-related genes.
Supplementary Material
Supplementary Material
Acknowledgements
We thank Masae Takashima for technical assistance.
Data accessibility
The supporting datasets are available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.t50q216 [61].
Authors' contributions
Y.N. performed all experiments, analysed the data and wrote the draft manuscript. Y.N. and H.T. conceived of and designed the experiments. K.Y., A.J. and R.F. analysed RNA-seq data. D.K., R.F and T.F. revised the manuscript. All authors discussed the results and contributed to writing the article.
Competing interests
We have no competing interests.
Funding
This study was supported by the JSPS KAKENHI grant nos. JP16K21613 to Y.N. and JP25221107 and JP17H06388 to T.F.
References
- 1.Hultmark D. 2003. Drosophila immunity: paths and patterns. Curr. Opin Immunol. 15, 12–19. ( 10.1016/S0952-7915(02)00005-5) [DOI] [PubMed] [Google Scholar]
- 2.Lemaitre B, Hoffmann J. 2007. The host defense of Drosophila melanogaster. Annu. Rev. Immunol. 25, 697–743. ( 10.1146/annurev.immunol.25.022106.141615) [DOI] [PubMed] [Google Scholar]
- 3.Gillespie JP, Kanost MR, Trenczek T. 1997. Biological mediators of insect immunity. Annu. Rev. Entomol. 42, 611–643. ( 10.1146/annurev.ento.42.1.611) [DOI] [PubMed] [Google Scholar]
- 4.Engström Y. 1999. Induction and regulation of antimicrobial peptides in Drosophila. Dev. Comp. Immunol. 23, 345–358. ( 10.1016/S0145-305X(99)00016-6) [DOI] [PubMed] [Google Scholar]
- 5.Kounatidis I, Ligoxygakis P. 2012. Drosophila as a model system to unravel the layers of innate immunity to infection. Open Biol. 2, 120075 ( 10.1098/rsob.120075) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Valanne S, et al. 2010. Genome-wide RNA interference in Drosophila cells identifies G protein-coupled receptor kinase 2 as a conserved regulator of NF-κB signaling. J. Immunol. 184, 6188–6198. ( 10.4049/jimmunol.1000261) [DOI] [PubMed] [Google Scholar]
- 7.Tauszig S, Jouanguy E, Hoffmann JA, Imler J-L. 2000. Toll-related receptors and the control of antimicrobial peptide expression in Drosophila. Proc. Natl Acad. Sci. USA 97, 10520–10525. ( 10.1073/pnas.180130797) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Irving P, Troxler L, Heuer TS, Belvin M, Kopczynski C, Reichhart J-M, Hoffmann JA, Hetru C. 2001. A genome-wide analysis of immune responses in Drosophila. Proc. Natl Acad. Sci. USA 98, 15 119–15 124. ( 10.1073/pnas.261573998) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Sackton TB, Lazzaro BP, Schlenke TA, Evans JD, Hultmark D, Clark AG. 2007. Dynamic evolution of the innate immune system in Drosophila. Nat. Genet. 39, 1461–1468. ( 10.1038/ng.2007.60) [DOI] [PubMed] [Google Scholar]
- 10.Waterhouse RM, et al. 2007. Evolutionary dynamics of immune-related genes and pathways in disease-vector mosquitoes. Science 316, 1738–1743. ( 10.1126/science.1139862) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Tanaka H, et al. 2008. A genome-wide analysis of genes and gene families involved in innate immunity of Bombyx mori. Insect. Biochem. Mol. Biol. 38, 1087–1110. ( 10.1016/j.ibmb.2008.09.001) [DOI] [PubMed] [Google Scholar]
- 12.Evans JD, et al. 2006. Immune pathways and defence mechanisms in honey bees Apis mellifera. Insect. Mol. Biol. 15, 645–656. ( 10.1111/j.1365-2583.2006.00682.x) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Zou Z, Evans JD, Lu Z, Zhao P, Williams M, Sumathipala N, Hetru C, Hultmark D, Jiang H. 2007. Comparative genomic analysis of the Tribolium immune system. Genome Biol. 8, R177 ( 10.1186/gb-2007-8-8-r177) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Gerardo NM, et al. 2010. Immunity and other defenses in pea aphids, Acyrthosiphon pisum. Genome Biol. 11, R21 ( 10.1186/gb-2010-11-2-r21) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Mesquita RD, et al. 2015. Genome of Rhodnius prolixus, an insect vector of Chagas disease, reveals unique adaptations to hematophagy and parasite infection. Proc. Natl Acad. Sci. USA 112, 14 936–14 941. ( 10.1073/pnas.1506226112) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Benoit JB, et al. 2016. Unique features of a global human ectoparasite identified through sequencing of the bed bug genome. Nat. Commun. 7, 10165 ( 10.1038/ncomms10165) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zumaya-Estrada FA, Martínez-Barnetche J, Lavore A, Rivera-Pomar R, Rodríguez MH. 2018. Comparative genomics analysis of triatomines reveals common first line and inducible immunity-related genes and the absence of Imd canonical components among hemimetabolous arthropods. Parasit. Vect. 11, 48 ( 10.1186/s13071-017-2561-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao Y-Y, Qu L-Y, Zhao D, Chen L-B, Jin H-Y, Xu L-M, Cheng J-A, Zhang C-X. 2013. The genome- and transcriptome-wide analysis of innate immunity in the brown planthopper, Nilaparvata lugens. BMC Genomics 14, 160 ( 10.1186/1471-2164-14-160) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.McTaggart SJ, Conlon C, Colbourne JK, Blaxter ML, Little TJ. 2009. The components of the Daphnia pulex immune system as revealed by complete genome sequencing. BMC Genomics 10, 175 ( 10.1186/1471-2164-10-175) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Palmer WJ, Jiggins FM. 2015. Comparative genomics reveals the origins and diversity of arthropod immune systems. Mol. Biol. Evol. 32, 2111–2129. ( 10.1093/molbev/msv093) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cao Z, et al. 2013. The genome of Mesobuthus martensii reveals a unique adaptation model of arthropods. Nat. Commun. 4, 2602 ( 10.1038/ncomms3602) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Smith AA, Pal U. 2014. Immunity-related genes in Ixodes scapularis—perspectives from genome information. Front. Cell. Infect. Microbiol. 4, 116 ( 10.3389/fcimb.2014.00116) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Misof B, et al. 2014. Phylogenomics resolves the timing and pattern of insect evolution. Science 346, 763–767. ( 10.1126/science.1257570) [DOI] [PubMed] [Google Scholar]
- 24.Kolokotronis S-O, et al. 2016. The mitogenome of the bed bug Cimex lectularius (Hemiptera: Cimicidae). Mitochondrial DNA B 1, 425–427. ( 10.1080/23802359.2016.1180268) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Sharma PP, Kaluziak ST, Pérez-Porro AR, González VL, Hormiga G, Wheeler WC, Giribet G. 2014. Phylogenomic interrogation of Arachnida reveals systemic conflicts in phylogenetic signal. Mol. Biol. Evol. 31, 2963–2984. ( 10.1093/molbev/msu235) [DOI] [PubMed] [Google Scholar]
- 26.Shao E, et al. 2017. Identification of transcripts involved in digestion, detoxification and immune response from transcriptome of Empoasca vitis (Hemiptera: Cicadellidae) nymphs. Genomics 109, 58–66. ( 10.1016/j.ygeno.2016.11.006) [DOI] [PubMed] [Google Scholar]
- 27.Zhang C-R, Zhang S, Xia J, Li F-F, Xia W-Q, Liu S-S, Wang X-W. 2014. The Immune strategy and stress response of the mediterranean species of the Bemisia tabaci complex to an orally delivered bacterial pathogen. PLoS ONE 9, e94477 ( 10.1371/journal.pone.0094477) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Arp AP, Hunter WB, Pelz-Stelinski KS. 2016. Annotation of the Asian citrus psyllid genome reveals a reduced innate immune system. Front. Physiol. 7, 570 ( 10.3389/fphys.2016.00570) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Grbić M, et al. 2011. The genome of Tetranychus urticae reveals herbivorous pest adaptations. Nature 479, 487–492. ( 10.1038/nature10640) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nishide Y, Onodera NT, Tanahashi M, Moriyama M, Fukatsu T, Koga R. 2017. Aseptic rearing procedure for the stinkbug Plautia stali (Hemiptera: Pentatomidae) by sterilizing food-derived bacterial contaminants. Appl. Entomol. Zool. 52, 407–415. ( 10.1007/s13355-017-0495-y) [DOI] [Google Scholar]
- 31.Bolger AM, Lohse M, Usadel B. 2014. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics 30, 2114–2120. ( 10.1093/bioinformatics/btu170) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Haas BJ, et al. 2013. De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512. ( 10.1038/nprot.2013.084) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Thorvaldsdóttir H, Robinson JT, Mesirov JP. 2013. Integrative genomics viewer (IGV): high-performance genomics data visualization and exploration. Brief. Bioinform. 14, 178–192. ( 10.1093/bib/bbs017) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Eddy SR. 2011. Accelerated profile HMM searches. PLoS Comput. Biol. 7, e1002195 ( 10.1371/journal.pcbi.1002195) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Finn RD, et al. 2016. The Pfam protein families database: towards a more sustainable future. Nucleic Acids Res. 44, D279–D285. ( 10.1093/nar/gkv1344) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Thompson JD, Higgins DG, Gibson TJ. 1994. CLUSTAL W: improving the sensitivity of progressive multiple sequence alignment through sequence weighting, position-specific gap penalties and weight matrix choice. Nucleic Acids Res. 22, 4673–4680. ( 10.1093/nar/22.22.4673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Jones DT, Taylor WR, Thornton JM. 1992. The rapid generation of mutation data matrices from protein sequences. Bioinformatics 8, 275–282. ( 10.1093/bioinformatics/8.3.275) [DOI] [PubMed] [Google Scholar]
- 38.Kumar S, Stecher G, Tamura K. 2016. MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. ( 10.1093/molbev/msw054) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Buist G, Steen A, Kok J, Kuipers OP. 2008. LysM, a widely distributed protein motif for binding to (peptido)glycans. Mol. Microbiol. 68, 838–847. ( 10.1111/j.1365-2958.2008.06211.x) [DOI] [PubMed] [Google Scholar]
- 40.Lemaitre B, Reichhart J-M, Hoffmann JA. 1997. Drosophila host defense: differential induction of antimicrobial peptide genes after infection by various classes of microorganisms. Proc. Natl Acad. Sci. USA 94, 14 614–14 619. ( 10.1073/pnas.94.26.14614) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Rutschmann S, Jung AC, Hetru C, Reichhart J-M, Hoffmann JA, Ferrandon D. 2000. The Rel Protein DIF mediates the antifungal but not the antibacterial host defense in Drosophila. Immunity 12, 569–580. ( 10.1016/S1074-7613(00)80208-3) [DOI] [PubMed] [Google Scholar]
- 42.Hoffmann JA. 2003. The immune response of Drosophila. Nature 426, 33–38. ( 10.1038/nature02021) [DOI] [PubMed] [Google Scholar]
- 43.Yokoi K, Koyama H, Minakuchi C, Tanaka T, Miura K. 2012. Antimicrobial peptide gene induction, involvement of Toll and IMD pathways and defense against bacteria in the red flour beetle, Tribolium castaneum. Results Immunol. 2, 72–82. ( 10.1016/j.rinim.2012.03.002) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Lourenço AP, Guidugli-Lazzarini KR, Freitas FCP, Bitondi MMG, Simões ZLP. 2013. Bacterial infection activates the immune system response and dysregulates microRNA expression in honey bees. Insect. Biochem. Mol. Biol. 43, 474–482. ( 10.1016/j.ibmb.2013.03.001) [DOI] [PubMed] [Google Scholar]
- 45.Boutros M, Agaisse H, Perrimon N. 2002. Sequential activation of signaling pathways during innate immune responses in Drosophila. Dev. Cell 3, 711–722. ( 10.1016/S1534-5807(02)00325-8) [DOI] [PubMed] [Google Scholar]
- 46.Bischoff V, Vignal C, Duvic B, Boneca IG, Hoffmann JA, Royet J. 2006. Downregulation of the Drosophila immune response by peptidoglycan-recognition proteins SC1 and SC2. PLoS Pathog. 2, e14 ( 10.1371/journal.ppat.0020014) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Zaidman-Rémy A, et al. 2006. The Drosophila amidase PGRP-LB modulates the immune response to bacterial infection. Immunity 24, 463–473. ( 10.1016/j.immuni.2006.02.012) [DOI] [PubMed] [Google Scholar]
- 48.Han ZS, Ip YT. 1999. Interaction and specificity of Rel-related proteins in regulating Drosophila immunity gene expression. J. Biol. Chem. 274, 21 355–21 361. ( 10.1074/jbc.274.30.21355) [DOI] [PubMed] [Google Scholar]
- 49.Tanji T, Hu X, Weber ANR, Ip YT. 2007. Toll and IMD pathways synergistically activate an innate immune response in Drosophila melanogaster. Mol. Cell. Biol. 27, 4578–4588. ( 10.1128/MCB.01814-06) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Horng T, Medzhitov R. 2001. Drosophila MyD88 is an adapter in the Toll signaling pathway. Proc. Natl Acad. Sci. USA 98, 12 654–12 658. ( 10.1073/pnas.231471798) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Limpens E, Franken C, Smit P, Willemse J, Bisseling T, Geurts R. 2003. LysM Domain receptor kinases regulating rhizobial nod factor-induced infection. Science 302, 630–633. ( 10.1126/science.1090074) [DOI] [PubMed] [Google Scholar]
- 52.Futahashi R, Tanaka K, Tanahashi M, Nikoh N, Kikuchi Y, Lee BL, Fukatsu T. 2013. Gene expression in gut symbiotic organ of stinkbug affected by extracellular bacterial symbiont. PLoS ONE 8, e64557 ( 10.1371/journal.pone.0064557) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Consortium TIAG. 2010. Genome sequence of the pea aphid Acyrthosiphon pisum. PLoS Biol. 8, e1000313 ( 10.1371/journal.pbio.1000313) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Douglas AE. 2006. Phloem-sap feeding by animals: problems and solutions. J. Exp. Bot. 57, 747–754. ( 10.1093/jxb/erj067) [DOI] [PubMed] [Google Scholar]
- 55.Łukasik P, van Asch M, Guo H, Ferrari J, Godfray HCJ. 2013. Unrelated facultative endosymbionts protect aphids against a fungal pathogen. Ecol. Lett. 16, 214–218. ( 10.1111/ele.12031) [DOI] [PubMed] [Google Scholar]
- 56.Scarborough CL, Ferrari J, Godfray HCJ. 2005. Aphid protected from pathogen by endosymbiont. Science 310, 1781–1781 ( 10.1126/science.1120180) [DOI] [PubMed] [Google Scholar]
- 57.Oliver KM, Moran NA, Hunter MS. 2005. Variation in resistance to parasitism in aphids is due to symbionts not host genotype. Proc. Natl Acad. Sci. USA 102, 12 795–12 800. ( 10.1073/pnas.0506131102) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Hosokawa T, Ishii Y, Nikoh N, Fujie M, Satoh N, Fukatsu T. 2016. Obligate bacterial mutualists evolving from environmental bacteria in natural insect populations. Nat. Microbiol. 1, 15011 ( 10.1038/nmicrobiol.2015.11) [DOI] [PubMed] [Google Scholar]
- 59.Abe Y, Mishiro K, Takanashi M. 1995. Symbiont of brown-winged green bug, Plautia stali Scott. Jpn. J. Appl. Entomol. Zool. 39, 109–115. ( 10.1303/jjaez.39.109) [DOI] [Google Scholar]
- 60.Altincicek B, Gross J, Vilcinskas A. 2008. Wounding-mediated gene expression and accelerated viviparous reproduction of the pea aphid Acyrthosiphon pisum. Insect. Mol. Biol. 17, 711–716. ( 10.1111/j.1365-2583.2008.00835.x) [DOI] [PubMed] [Google Scholar]
- 61.Nishide Y, Kageyama D, Yokoi K, Jouraku A, Tanaka H, Futahashi R, Fukatsu T. 2019. Data from: Functional crosstalk across IMD and Toll pathways: insight into the evolution of incomplete immune cascades Dryad Digital Repository. ( 10.5061/dryad.t50q216) [DOI] [PMC free article] [PubMed]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Data Citations
- Nishide Y, Kageyama D, Yokoi K, Jouraku A, Tanaka H, Futahashi R, Fukatsu T. 2019. Data from: Functional crosstalk across IMD and Toll pathways: insight into the evolution of incomplete immune cascades Dryad Digital Repository. ( 10.5061/dryad.t50q216) [DOI] [PMC free article] [PubMed]
Supplementary Materials
Data Availability Statement
The supporting datasets are available in the Dryad Digital Repository: https://doi.org/10.5061/dryad.t50q216 [61].