Abstract
Ancestral environmental conditions can impact descendant phenotypes through a variety of epigenetic mechanisms. Previous studies on transgenerational effects in Drosophila melanogaster suggest that parental nutrition may affect the body size, developmental duration and egg size of the next generation. However, it is unknown whether these effects on phenotype remain stable across generations, or if specific generations have general responses to ancestral diet. In the current study, we examined the effect on multiple life-history phenotypes of changing diet quality across three generations. Our analysis revealed unforeseen patterns in how phenotypes respond to dietary restriction. Our generalized linear model showed that when considering only two generations, offspring phenotypes were primarily affected by their own diet, and to a lesser extent by the diet of their parents or the interaction between the two generations. Surprisingly, however, when considering three generations, offspring phenotypes were primarily impacted by their grandparents' diet and their own diet. Interactions among different generations’ diets affected development time, egg volume and pupal mass more than ovariole number or wing length. Furthermore, pairwise comparisons of diet groups from the same generation revealed commonalities in strong responses to rich versus poor diet: ovariole number, pupal mass and wing length responded more strongly to poor diet than to rich diet, while development time responded strongly to both rich and poor diets. To improve investigations into the mechanisms and consequences of transgenerational, epigenetic inheritance, future studies should closely examine how phenotypes change across a higher number of generations, and consider responses to broader variability in diet treatments.
Keywords: transgenerational epigenetic, inheritance, phenotype, nutrition, starvation, Drosophila
1. Introduction
For many decades, the consequences of ancestral experiences on the performance and survival of descendants in plants and animals have been a dynamic area of research. Biologists have come to realize that the non-genetic inheritance of environment-dependent effects may represent a significant source of variation for many organismal traits. Recent studies and reviews on humans and mice have highlighted the importance of these phenomena in mediating disease phenotypes, or phenotypes deviating from a defined norm, such as diabetes [1], autism [2] and cancer [3]. While a substantial portion of effort is directed at piecing together mechanisms of epigenetic gene regulation, biologists have also re-considered the ecological and evolutionary implications of transgenerational effects, by considering the relationship between epigenetic variation and fitness in natural populations [4] and how the timing at which a transgenerational effect occurs may determine whether an epigenetic effect is functional versus an impairment [5]. For example, diversity in the location of methylation marks among populations of bat species may allow them to rapidly buffer changes in crowdedness, meteorological conditions (e.g. temperature), noise and light disturbances [6]. In a second example, early-life grooming of rat pups is associated with changes in methylation of hypothalamic-pituitary-adrenal axis related genes, which are associated with low corticosterone levels and lowered anxiety [7]. Female offspring with these modifications groom pups at the same time they receive maternal care, which perpetuates transgenerational transmission of the behaviour [8].
Researchers from various biological disciplines have been compelled to understand the broad and mechanistic significance of ‘epigenetic’ phenomena, and consequently, the literature has been peppered with confusing definitions and co-opted terminology [9]. Definitions typically favour a particular organizational level of study, ranging from a strict focus on underlying molecular mechanisms (e.g. DNA methylation, histone acetylation) to the outcome on phenotype (e.g. developmental plasticity; [10]). For the purposes of this study, we use the phrase ‘transgenerational epigenetic inheritance’ to mean any time a form of gene regulation that is not coded by the genomic DNA sequence itself (e.g. factors bound to DNA or freely floating) is inherited by one or more descendant generations, with the inheritance mechanism occurring at some time between germ cell formation and birth of the descendant generation. Different described mechanisms of transgenerational epigenetic inheritance may actually work in concert, so we can imagine a maternal supply of mRNA (influenced or not by the maternal environment), RNA feedback loops and chromatin modifications as ultimate sources of epigenetic variation that influence phenotypes.
A small number of previous studies have, to our knowledge, investigated the patterns of non-genetic inheritance of dietary effects on various traits in Drosophila melanogaster (electronic supplementary material, table S1). Here, we briefly review their findings, which suggest some patterns of transgenerational inheritance over one generation, but also leave a number of questions unanswered. Reduction of specific dietary nutrients such as sugar [11,12], yeast protein [11,13] or fat [14], or a dilution of the standard diet [15,16], consistently have effects on egg size, duration of L1 (first instar) -adult development, metabolic pools (concentration of a specific macronutrient in the haemolymph) and body mass in descendant generations. However, the generality of these patterns is questionable owing to differences in experimental design between studies, including wide variability in diet recipes, whether diets differ between parents, and how different diets are between generations. In general, females fed a dilution of standard diet lay larger eggs, from which emerge slowly developing larvae that reach a small body size [15,16]. When specific nutrient content is altered, however, these life-history traits are differentially affected. For instance, F1 females with mothers that ate high protein/low sugar versus low protein/high-sugar were heavier as adults, with more protein, glycogen and triglycerides in their haemolymph, and laid more eggs, but this pattern changes among genotypes [11]. When sugar content was unchanged in a high- versus low-fat diet, F1 females with mothers that ate high-fat food versus low-fat food stored fat more rapidly, stored less triacyl glycerides (TAG), had higher concentration of circulating sugars, increased expression of fat lipolysis and gluconeogenesis genes, and decreased expression of fatty-acid synthesis, sugar transport and glycolysis genes. These changes in circulating sugar and TAG persisted to the second generation [12]. In the only other study we are aware of in which two generations of potential epigenetic inheritance were considered, the effect of high fat in the grandparental generation on the metabolic pools (macronutrient concentrations in the haemolymph), pupal mass and egg size of the next two generations, as well as ancestry-independent effects of nutrition, were largely dependent upon genotype and sex [14]. In addition, some evidence suggests that offspring ovariole number is influenced by diet restriction in the previous generation: mothers that were deprived of all nutrients birthed daughters that developed more ovarioles than unstarved mothers [17]. These studies provided strong support for parental diet influencing some offspring life-history traits. However, all but two of these studies [12,14] considered potential effects across only one generation, and none considered the relative strength of a specific generation's diet versus ancestral diets on different traits.
In this study, we were interested in the following questions: how does the effect of parental nutrition change between generations?; are responses of phenotype to diet similar in direction and magnitude?; how do two generations of ancestral nutrition affect pupal mass, development duration, ovariole number, egg size and wing length? We tested the following specific hypotheses about the effects of F0 nutrition on F1 phenotypes, as predicted by previous studies on D. melanogaster: when F0 females experience dietary restriction versus yeast supplementation, (1a) their descendant generations will lay eggs of increased size, (1b) from which will emerge more slowly developing larvae, (1c) with lower pupal mass. Lower pupal mass may indicate (1d) lower ovariole number and (1e) shorter wings, but unknown trait linkages that produce trade-offs between traits may lead to alternative changes in certain phenotypes.
Our results show support for some of these predictions. In addition, we unexpectedly found that the likelihood of a transgenerational effect varies between generations, and that these effects are apparent after a specific direction of dietary shift (rich to poor or poor to rich).
2. Material and methods
(a). Husbandry
All flies (D. melanogaster) used in the experiment originate from an Oregon R-C stock (Bloomington Drosophila Stock Center (BDSC) no. 5) that was maintained in the laboratory at room temperature (approx. 23°C) for approximately 5 years before the study was conducted. Flies were maintained at 25°C and 60% relative humidity (rh) for the duration of the experiment. Stock larvae were reared on ‘standard’ diet: 8500 ml of water, 79 g agar (0.9% w/v), 275 g torula yeast (3.2% w/v), 520 g cornmeal (6.1% w/v), 1100 g dextrose (12.9% w/v) and 23.8 g methyl p-hydroxybenzoate (an antifungal agent) dissolved in 91.8 ml of 95% ethanol. Experimental larvae of each generation were fed ‘rich’, standard or ‘poor’ diet. The rich diet consisted of solid standard food supplemented with approximately 30 µl of a torula yeast slurry of density 2.86 µg µl−1 (1 mg of yeast dissolved in 350 µl water), that was pipetted atop the food. Poor diet was made with freshly cooked standard food diluted with boiled 3% agar in a 1 : 3 ratio (25% concentration of standard diet) without yeast supplementation.
(b). Experimental design
Three groups of 25 females and 14 males were placed in egg collection cages with apple juice plates (90 g agar, 100 g sugar, 1 l apple juice, 3 l water and 6 g Nipagin dissolved in 60 ml ethanol). Each cage consisted of a ventilated 60 mm Petri dish bottom three-quarters full of apple juice medium, and a dab of yeast paste. First instar larvae were collected over 3 days, replacing the yeast-supplemented apple juice cage each day. Forty larvae at a time were placed in one vial of a set of 8–10 replicate vials for each dietary treatment. All adults, eggs and larvae were maintained at 25°C and 60% rh throughout the experiment.
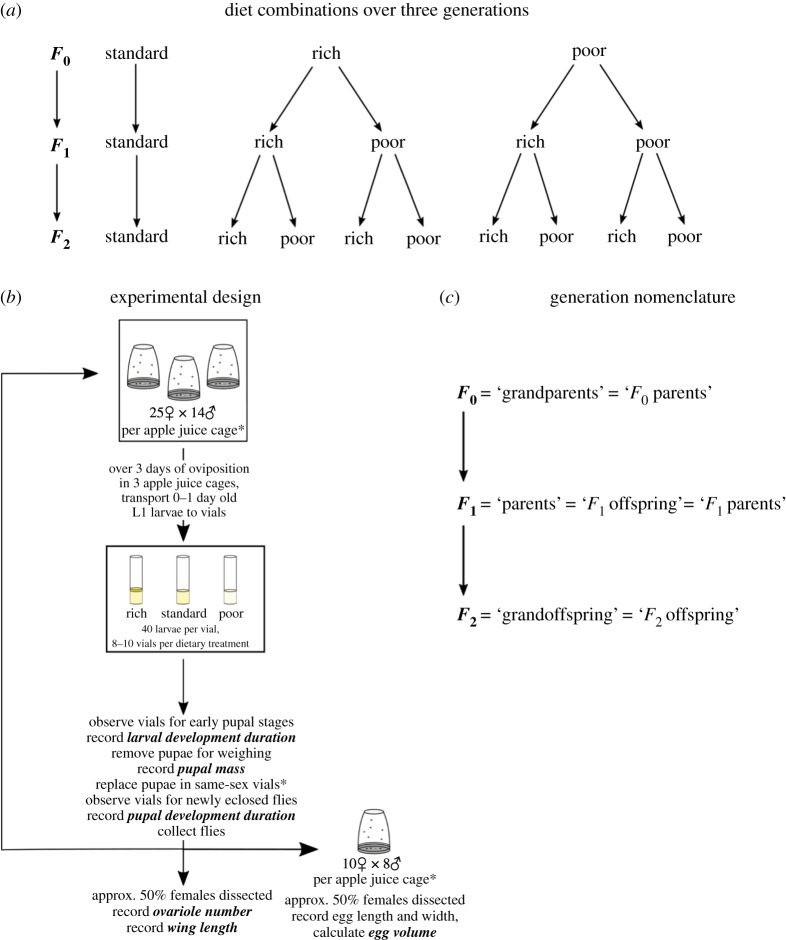
We examined a total of three generations, recording five life-history phenotypes (described in ‘Phenotype pipeline’ below) for each generation, using one or more of three different diets at each generation (described in ‘Husbandry’ above; figure 1). The design was aimed at examining the different effects of rich and poor diets in each of the three generations. The effects of ‘standard’ diet were not specifically examined; rather, the standard diet was included only for normalization. Throughout the remainder of the article, we refer to the first of these three generations with the label ‘F0’, in some cases referring to them as ‘F0 parents’ (of F1 offspring), and in other cases, referring to them as ‘grandparents’ (of grandoffspring). We refer to the second generation with the ‘F1’ label, considering them either as ‘F1 offspring’ (of F0 parents) or as ‘parents’ (of F2 offspring), and the third generation with the ‘F2’ label or ‘grandoffspring’ (figure 1c).
Figure 1.
Experimental design and generation nomenclature for the present study. (a) Three generations of flies were observed, subjected to various combinations of diets across each generation. (b) Schematic of the experimental set-up for a given generation. See Methods for details. (c) Nomenclature used herein to refer to each generation of the study.
(c). Phenotype pipeline
The metamorphosis from the first instar to adult was recorded every 12 h. We binned the entire developmental period into three phenotypes, larval development (L1-LP), pupal development (LP-adult) and L1-adult development. Pupae were sexed, and individual female pupal mass recorded, then all pupae were placed into fresh vials and allowed to develop to eclosion.
Upon eclosion, females were kept separated from males in standard food vials supplemented with yeast to stimulate egg production. Three to 6 days later, approximately 50% of these females were dissected in 1x phosphate buffered saline (PBS) and their ovaries harvested, which were fixed in 4% paraformaldehyde in 1x PBS, then stored in methanol at −20°C until full ovariole number counts could be made. Heads and thoraxes were stored in tubes of 70% ethanol for later measurements. After one to three months of storage, ovarioles were gradually rehydrated in a mixture of 1x PBS with 0.02% Triton-X with DAPI at a 1 : 500 dilution of a 10 mg ml−1 stock solution, then teased apart using minuten pins. Ovariole number for a given treatment group was calculated as the average ovariole number per ovary over all ovaries scored for a given dietary treat.
Facing the ventral side of a dissected thorax, we dissected the right wing, and flattened it in a drop of ethanol on a labelled section of microscope slide. Photos of wings were taken using an eyepiece camera (DinoXcope 7023M) placed in the eyepiece of a Zeiss Stemi DV4 stereo microscope at 25× magnification. Wing photos were viewed in DinoXcope software v. 1.16 for Mac OS X. Wing length was measured as the distance between the humeral-costal break and the end of vein L3 (fig. 1 of [18]).
Egg volume was estimated by inserting the width and length of an egg into an equation for estimated egg volume: (1/6)πW2L [19]. These values were averaged for each treatment group. From each treatment group, approximately 10 adult females and eight adult males, both aged for 4 days following eclosion in single-sex vials, were placed in caged, apple-juice plates with a smear of yeast paste to mate and lay eggs. Approximately 20 eggs were collected per diet treatment per generation (treatment groups). Photos of eggs were taken using an eyepiece camera (DinoXcope 7023M) placed in the eyepiece of a Zeiss Stemi DV4 stereo microscope at 25× magnification.
Sample sizes for all phenotypes measured at each generation and diet treatment are included in the electronic supplementary material, table S4. Raw data for all phenotypes scored are available at https://extavourlab.github.io/TransgenerationalEffectOfNutrition/.
(d). Statistical analysis
All statistical analyses were performed using R v. 3.4.3 [20]. For each generation and phenotype, we constructed a general additive model for location scale and shape (‘gamlss’ package; [21]) to analyse the interactive effect of a specific generational group's experimental diet, and its ancestors' experimental diet, on different phenotypic responses. We used this model because it allowed us to test each of our response distributions against a larger, general family of model distributions. Moreover, under this model, data transformation is unnecessary and one is not restricted to subsets of the exponential family of model distributions. Phenotypes of individuals of the F2 generation were potentially affected by three generations of diet (F0 diet × F1 diet × F2 diet), those of the F1 generation were potentially affected by two generations of diet (F0 diet × F1 diet), while those of the F0 generation were potentially affected by one generation of diet (F0 diet). We used the R package ‘fitdistrplus’ [22] for fitting model distributions against the observed distribution of a specific phenotype and generation. These distributions are listed in the electronic supplementary material, table S2. We selected the one with the lowest Akaike information criterion (AIC) value [23] for use in our GAMLSS model. This was verified by comparing the AIC values of GAMLSS models (with ‘gamlss’ package) using the distributions with the three lowest AIC values. We also checked whether vial identity should be included as a random effect. In most instances, leaving out vial identity produced a lower AIC value, so we left it out.
We also created an interactive heatmap tool that allows users to visualize the significance of pairwise comparisons of trait values of interest between each of the treatment groups. This heatmap can be used to explore patterns of phenotype differences among any desired set of treatment groups. For example, users can visualize the effect of two generations of poor versus rich diet on grandoffspring ovariole number, of an increase or decrease in food quality on larval development duration, or of any other combination of ancestral diets on the trait of interest. The interactive heatmap of pairwise comparisons, as well as the raw data, are freely available at https://extavourlab.github.io/TransgenerationalEffectOfNutrition/.
For each fly of each generation, we normalized each phenotype response by dividing it by the response of the control (standard food) from the same generation (SS for F1, SSS for F2). We used this ratio as the response variable for our GAMLSS analyses. We then took the base 10 logarithm of this ratio for use in our graphs (figures 2–4). Visualizing the normalized responses relative to each other allowed us to see whether there were general differences in treatment effects. The distribution of values for each normalized phenotype was normal or close to normal using Cullen and Frey plots, so we performed linear regressions. We then assessed for statistical significance using Tukey's test for multiple comparisons using the ‘multcomp’ package [24].
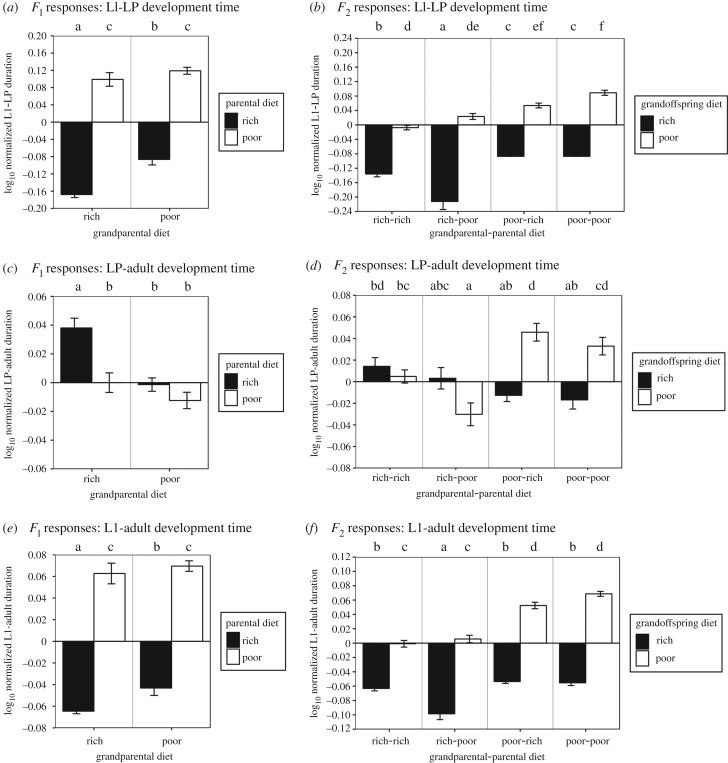
Figure 2.
Rich food shortens larval development and lengthens pupal development, regardless of grandparental or grandparental-parental diet. Log-normalized response of developmental duration (y-axis) of parental (a) and grandoffspring (b) generations to larval diet, during the larval (a,b), pupal (c,d) and entire developmental period (e,f). Log-normalization was calculated as the base 10 logarithm of the ratio of parental or grandparental experimental response/control group response (two generations of standard diet for parental, three generations of standard diet for grandparental). Diet treatments are colour-coded: rich (black), standard (grey) and poor (white). Letters represent how different responses are from each other (using Tukey HSD). Non-significant responses share letters. Error bars represent standard error.
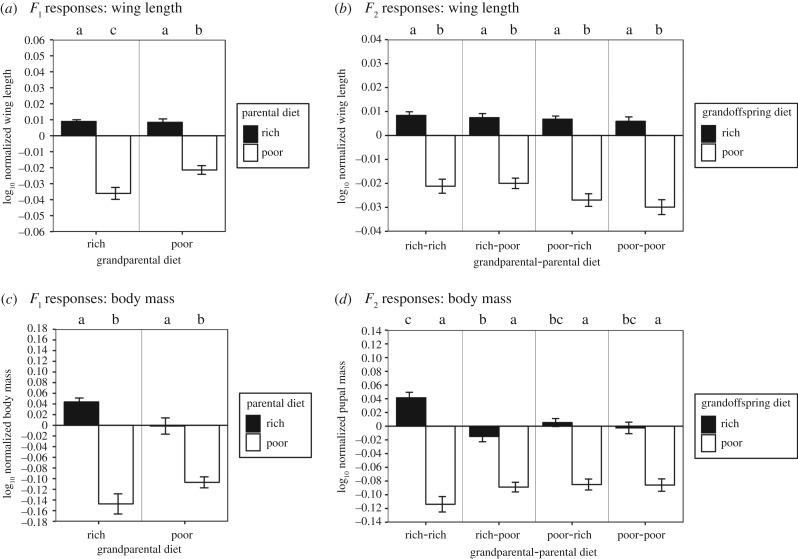
Figure 3.
Poor diet is more influential than rich diet in changing wing length across two generations; poor-fed flies from otherwise strictly rich-fed lineages have the lowest pupal mass at each generation. Response of wing length of parental (a) and grandoffspring (b) generations to larval diet. Response of pupal mass of parental (c) and grandoffspring (d) generations to larval diet. Log-normalization was calculated as in figure 2. Colour-coding, letters of significance and error bars were set as in figure 2.
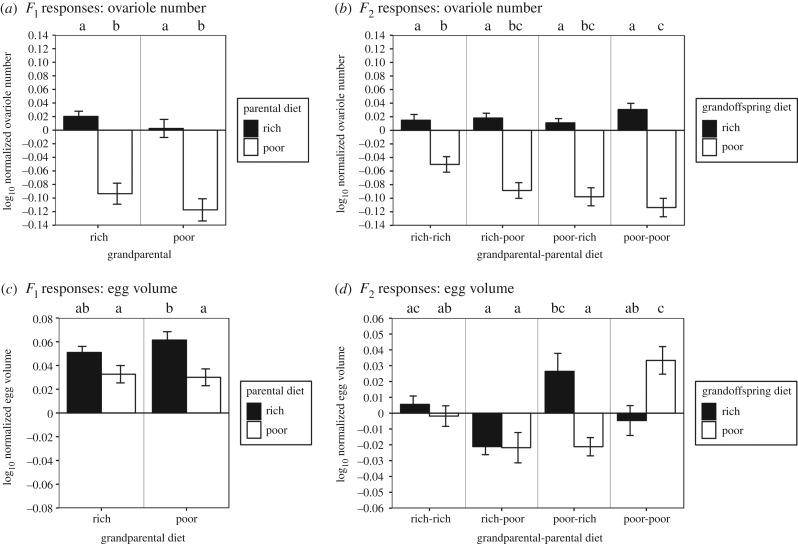
Figure 4.
Ovariole number is lowered by ancestral poor diets; switching diets among generations causes disordered effects of diet on egg volume. Response of ovariole number of parental (a) and grandoffspring (b) generations to larval diet. Response of egg volume of parental (c) and grandoffspring (d) generations to larval diet. Log-normalization was calculated as in figure 2. Colour-coding, letters of significance and error bars were set as in figure 2.
3. Results
For the three generations examined in this study, for consistency throughout the manuscript, we use the naming convention schematized in figure 1. We refer to the first of these three generations as the F0 generation and call them ‘grandparents’ (even when discussing them in relation to their offspring, the F1 generation). We refer to the second generation as F1, calling them ‘parents’ (of F2 offspring). Finally, we refer to the third generation as F2 and call them ‘grandoffspring’ (even when discussing them in relation to their parents, the F1 generation).
Results of all GAMLSS analyses are reported in table 1 and the electronic supplementary material, table S1. We interpreted the regression coefficients from each predictor (F0 diet and F1 diet for the F1 generation; F0 diet, F1 diet and F2 diet for the F2 generation) as the predicted magnitude of change in the value of the measured phenotype. Relative to each other, these coefficients tell us about the relative strength of the effect of the predictor (diet) on a given phenotype. Below, we discuss ways in which our observations supported or deviated from our initial predictions about the effect of diet quality on several different life-history phenotypes across two generations. Results of pairwise comparisons are reported in the electronic supplementary material, table S3.
Table 1.
Summary of transgenerational effects of nutrition on life-history traits. (The first column indicates the generation/diet effect, and the remaining columns indicate the phenotypes. p-values from the GAMLSS have been summarized in this table. Statistically significant (p-value < 0.05) effects are indicated in italics.)
| generation/diet effect | L1-LP development (days) | LP-adult development (days) | L1-adult development (days) | wing length (mm) | pupal mass (mg) | ovariole number | egg volume (mm3) |
|---|---|---|---|---|---|---|---|
| F0 diet | <0.0001 | 0.0548 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0724 |
| F1 diet | <0.0001 | 0.1437 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0088 |
| F0 diet | 0.2452 | 0.1766 | 0.4700 | <0.0001 | 0.1381 | 0.4550 | 0.7916 |
| F1 diet × F0 diet | 0.0072 | 0.0263 | 0.2430 | 0.0104 | 0.0108 | 0.9670 | 0.2971F0 |
| F2 diet | <0.0001 | 0.0009 | <0.0001 | <0.0001 | <0.0001 | <0.0001 | 0.0028 |
| F0 diet | <0.0001 | <0.0001 | <0.0001 | 0.0004 | 0.7707 | 0.0999 | <0.0001 |
| F1 diet | 0.0001 | 0.0891 | 0.0220 | 0.6826 | 0.9954 | 0.4501 | <0.0001 |
| F2 diet × F0 diet | 0.8534 | 0.0026 | 0.0444 | 0.0945 | 0.5814 | 0.0746 | 0.0364 |
| F2 diet × F1 diet | 0.0123 | 0.2701 | 0.1041 | 0.6701 | 0.6653 | 0.1321 | <0.0001 |
| F0 diet × F1 diet | 0.5780 | 0.7654 | 0.3402 | 0.1207 | 0.2799 | 0.3018 | <0.0001 |
| F2 diet × F0 diet × F1 diet | 0.1463 | 0.669132 | 0.109 | 0.4106 | 0.00241 | 0.9439 | 0.0048 |
(a). Predictions supported for egg volume and development time
Our first prediction was that poor-fed ancestors would lay large eggs (prediction 1a), from which would emerge slowly developing (1b), low-weight (1c) flies with low ovariole number (1d) and small wings (1e). Indeed, we observed that when grandparents were poor-fed versus rich-fed, parents laid eggs of marginally significantly increased size (p = 0.0548; table 1 and figure 4c; electronic supplementary material, table S3), but only if these parents ate rich food. Also consistent with our prediction, flies emerging from these larger eggs (poor-fed grandparents, rich-fed parents) had longer larval development (F2 × F1 diet, p = 0.0123; table 1 and figure 2b; electronic supplementary material, table S3). However, their pupal mass (F2 × F1 diet, p = 0.6653; table 1), wing length (F2 × F1 diet, p = 0.6701; table 1) and ovariole number (F2 × F1 diet, p = 0.1321; table 1) were not significantly different from grandoffspring after two generations of rich food (figures 3b and 4b,d; electronic supplementary material, table S3). Consistent with predictions, poor-fed grandoffspring whose grandparents were also poor-fed laid larger eggs (figure 4d; electronic supplementary material, table S3), but we did not measure traits for the F3 generation.
(b). Select phenotype responses are sensitive to ancestral shifts in diet
Having found that our results largely confirmed our initial predictions regarding the general transgenerational effects of poor or rich diets, we then asked whether life-history phenotypes responded to specific combinations of ancestral poor or rich food, relative to conditions where all generations ate standard food. We found that specific hierarchical patterns of diet and ancestry that led to transgenerational effects were not always consistent for rich- or poor-fed parents or grandoffspring, or across generations for the same phenotype. Instead, we found that for some but not all phenotypes, a shift from rich to poor diet or vice versa in ancestors led to significant changes in the phenotypes of offspring. Rich-fed grandparents produced parents that had slower larval and overall development, and faster pupal development than parents from poor-fed grandparents, but only when those parents ate rich food (figure 2a,c,e; electronic supplementary material, table S3). This difference was absent among poor-fed parents of rich-fed grandparents, but they had longer wings than if they were produced by poor-fed grandparents (figure 3a; electronic supplementary material, table S3). By contrast, grandparental diet had no effect on parental ovariole number (F1 p = 0.455, F1 × F0 diet, p = 0.967; table 1 and figure 4a; electronic supplementary material, table S3).
We also found that rich diet in grandparents (F0) could often improve these life-history phenotypes for grandoffspring (F2). These phenotypic effects were particularly notable when the parents (F1) of those grandoffspring were poor-fed. Poor-fed grandoffspring had larger pupal mass (figure 3d; electronic supplementary material, table S3), lower ovariole number (figure 4b; electronic supplementary material, table S3) and longer overall development (figure 2f; electronic supplementary material, table S3) if their F0 grandparents were poor-fed versus rich-fed. Rich-fed grandoffspring had longer development duration (figure 2f; electronic supplementary material, table S3), and decreased pupal mass if their grandparents were poor-fed versus rich-fed (figure 3d; electronic supplementary material, table S3). Two generations of rich-fed ancestors yielded grandoffspring that developed more quickly to pupation than grandoffspring descended from two generations of poor-fed ancestors (figure 3b; electronic supplementary material, table S3). If the grandoffspring were poor-fed, their overall development was quicker (figure 3b; electronic supplementary material, table S3), they had more ovarioles (figure 4b; electronic supplementary material, table S3) and laid smaller eggs (figure 4d; electronic supplementary material, table S3).
4. Discussion
In summary, our study uniquely reveals clear differences in how the phenotypes of each generation are affected by different predictors, and correlative, transgenerational effects of nutrition on sets of phenotypes. Our observations were consistent with our predictions for transgenerational effects on egg volume in the F2 generation, and on development time in F1 and F2 generations, but were not consistent for the remaining traits. However, a more thorough analysis revealed key additional insights: namely, that F1 phenotypes were primarily affected by their own diet, while F2 phenotypes were primarily affected by grandparents' diet and their own diet; pupal mass and ovariole number were less likely to be affected by transgenerational effects; larval and pupal development times respond in opposing ways; and egg volume is sensitive to many different diet interactions. F1 phenotypes were primarily affected by their own diet, and to a lesser extent, the diet of their parents or the interaction between the diets of the two generations. However, F2 phenotypes were primarily impacted by their grandparents’ diet and their own diet. F1 diet individually had little effect on larval and overall development, and no effect on other phenotypes. Interactions among the three diet/generation groups had stronger effects on development time and egg volume than on pupal mass, ovariole number or wing length. When we examined differences among diet groups from the same generation using pairwise comparisons, we found that rich grandparental diet generally decreased the development time, increased the wing length and decreased the egg size of grandoffspring. Two generations of rich diet yielded grandoffspring that, when poor-fed, developed more quickly, had more ovarioles, and smaller eggs. Thus, the complex diet interactions among generations and phenotypes demonstrate a sensitivity and dynamism in transgenerational effects that makes it difficult to extract general principles or trends.
We found that transgenerational effects were distinct across generations for different phenotypes. For example, we found that poor-fed grandoffspring had higher ovariole numbers when their grandparents were rich-fed versus poor-fed. This does not agree with a previous report that food deprivation in previous generations (at least one) increases ovariole number in the next generation [17]. A likely explanation for the discrepancy between our results and those of Wayne et al. [17] is the use of different genotypes. This explanation is consistent with the observations of Matzkin et al. [11] and Dew-Budd et al. [14], who both found that the magnitude and direction of effect of ancestral diet varied by sex and genotype. Another possibility is the difference in how we imposed our manipulations. Wayne et al. [17] imposed starvation specifically in the adult generation after larvae had developed on standard food, while we imposed starvation during larval development. This difference in experimental design would be expected to yield different effects on ovariole number, because larval nutrition has been shown to be a major contributor to ovariole number in multiple previous studies [25–28]. Wayne et al. [17] also imposed starvation selection on both their ‘selected’ and ‘control’ lines (as opposed to imposing selection on just one line), which may explain why they found effects of starvation on ovariole number in both lines. Lastly, food deprivation in their study consisted of food absence for a specific duration as adults, whereas in our study food deprivation represented a consistent dilution of diet over the entire L1-adult experimental period. Whether food is absent for a short period or in diluted quantities over a long period is likely to interact with the time-specific development of different phenotypes. If development duration is affected in any way by either manipulation, then this may also have cascading effects on other phenotypes.
An under-addressed issue in transgenerational effect studies is how we should interpret dietary manipulations and the effects they have on the organism. Different sugars and yeasts have different effects on life-history phenotypes. Flies fed high levels of sucrose suffer reduced fecundity [29], while flies fed sucrose but not fructose have reduced lifespan and reduced fecundity [30], both under normal conditions [30,31] and with a starvation period [31]. In Drosophila, it has been shown that lifespan and fecundity of experimental animals in response to dietary restriction varies depending upon what type of active yeast is used, as well as the amount of carbohydrate relative to protein. This suggests that flies may actually be responding to the restriction of a specific nutrient as opposed to the amount of food [29]. Likewise, it is possible that the generation-by-diet-dependent changes in egg volume that we observed across generations reflect, in some cases, the rescue of the potential volume (under non-manipulated conditions) from the effects of sub-optimal food types that increase or decrease egg size. To further complicate the issue, there is evidence suggesting that a factor other than amino acids or sugar is present in (at least one species of) yeast, and that this factor is a major nutrient cue for the insulin and TOR signalling pathways that are responsible for nutrient-dependent growth and development [32].
One important model for measuring the impact of dietary components on an organism is the geometric framework of nutrition. Nutritional geometry is used to test how combinations of nutrients influence phenotype outcomes, as opposed to considering a specific nutrient in isolation. Principle findings to date suggest that each animal has an intake target (IT), which is the amount and balance of protein and carbohydrates that an animal needs to consume within a specific period to achieve maximal fitness [33]. Bondurianksy et al. [34] found that maternal protein influenced offspring pupal mass and head length (a secondary sexual trait) in both sons and daughters, and paternal carbohydrate influenced offspring pupal mass and head length differently between sons and daughters. An alternative hypothesis that could account for these observations, however, is that the IT shifted in offspring as they aged, which influenced foraging preferences and feeding behaviour [35]. In our feeding experiments, it is formally possible that quantified phenotypes were altered not by poor diet per se, but rather by a change in feeding rate in response to ‘missing’ or suboptimal nutrients in the modified diets.
We have been cautious in our language regarding the interpretation of effects on phenotype. When a non-human animal has increased glucose and TAG in their haemolymph, larger body size and/or lower survival, there is a tendency in the literature to explain these results within the context of metabolic disorders in humans (obesity and diabetes) [36–40]. We would argue that these are medical terms with negative health and socio-economic connotations that could reflect societal biases in how we perceive changes in human physiology, and that making evolutionary arguments about relative fitness gains or costs based on such physiological data may not be appropriate.
For example, an increase in adipose tissue or decrease in lifespan as a result of an increase in sugar or fat content in parental or grandparental diets may, in the end, increase the likelihood of reproducing, regardless of or within specific environmental contexts. The existence of similar regulatory machinery across organisms (e.g. insulin and insulin-like signalling pathways) does not imply that this machinery achieves the same goals for reproductive fitness across different organisms. Kuo et al. [41] found that chico and Akt mutants had different cuticular hydrocarbon (CHC) profiles and were less attractive to males than control females. Schultzhaus et al. [42] found that males were less attracted to high-fat-fed females, which had different CHC profiles, than they were to low fat-fed females, in both light and dark conditions. These females also had altered CHC profiles. High fat-fed females are also more fecund [43]. However, Lin et al. [44] found that females which ate a high yeast diet were heavier, more fecund, more immobile, had shorter lifespans, and had different CHC profiles, but were also more attractive to males than low yeast-fed females, or than high yeast-fed mutant females (hypomorphic for insulin peptides) or females experiencing oenocyte-specific gene disruption of insulin signalling. These studies illustrate that ‘attractiveness’ of females, as a measure of reproductive fitness, is represented by CHC profile; these profiles are strongly influenced by high yeast or high-fat diet, as well as the disruption of insulin signalling. High yeast and high fat both make female flies heavier, more immobile, and more fecund, but the former are more ‘attractive’ in this context, meaning likely to be mated. We acknowledge that transgenerational effects mediated by nutrition may be important with regards to metabolic disorders such as diabetes or obesity in humans. However, we see our results as applying to D. melanogaster and other organisms with more similar behaviour and physiology, and do not believe it would be wise to speculate beyond these life histories to suggest that our results imply anything predictive or proscriptive about human obesity, fitness, or sugar metabolism.
In conclusion, based on the results of our study and previous studies, we suggest three major potential improvements into future investigations into the transgenerational effects of nutrition on Drosophila life-history traits. First, owing to the variation in transgenerational effects after only one generation, measurements should be conducted for two or more generations. Second, while genotype- and sex-specific effects indicate two important sources of phenotypic variation, general principles are difficult to draw from these effects. A helpful experiment would be to run multiple trials of a well-defined dietary regime on both sexes of multiple genotypes (for example, using the Drosophila Genetic Reference Panel or a similar collection of population variants [45]), in order to discover genotypes with reproducible quantitative phenotypes. These ‘standard’ genotypes would then allow different types of experiments focused at lower organizational levels to be compared across studies. Finally, using a geometric framework of nutrition may give us a robust quantification of the combination of protein, fats and carbohydrates per genotype that are associated with specific quantitative phenotypes, thereby informing us on how nutritional ITs, and potentially also correspondent foraging behaviour, may play a role in mediating the effects of parental nutrition on offspring phenotypes.
Supplementary Material
Supplementary Material
Acknowledgements
We thank the Extavour Laboratory members for their creative and effective suggestions on experimental design and data analysis.
Data accessibility
All data generated in this study are available at https://extavourlab.github.io/TransgenerationalEffectOfNutrition.
Competing interests
We declare we have no competing interests.
Funding
This work was supported by NIH grant R01HD073499 to C.G.E. and funds from Harvard University. J.B.D. was partially supported by NIH Supplement 3R01HD073499-03S1.
References
- 1.Wei Y, Yang C, Wei Y, Zhao Z, Hou Y, Schatten H, Sun Q. 2014. Paternally induced transgenerational inheritance of susceptibility to diabetes in mammals. Proc. Natl Acad. Sci. USA 111, 1873–1878. ( 10.1073/pnas.1321195111) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Loke YJ, Hannan AJ, Craig JM. 2015. The role of epigenetic change in autism spectrum disorders. Front. Neurol. 6, 1–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Feinberg AP, Ohlsson R, Henikoff S. 2006. The epigenetic progenitor origin of human cancer. Nat. Rev. Genet. 7, 21–33. ( 10.1038/nrg1748) [DOI] [PubMed] [Google Scholar]
- 4.Kilvitis HJ, Alvarez M, Foust CM, Schrey AW, Robertson M, Richards CL. 2014. Ecological epigenetics. In Ecological genomics: ecology and the evolution of genes and genomes (eds Landry CR, Aubin-Horth N), pp. 191–203. Dordrecht, The Netherlands: Springer. [Google Scholar]
- 5.Kuzawa CW, Thayer ZM. 2011. Timescales of human adaptation: the role of epigenetic processes. Epigenomics 3, 221–234. ( 10.2217/epi.11.11) [DOI] [PubMed] [Google Scholar]
- 6.Liu S, Sun K, Jiang T, Feng J. 2015. Natural epigenetic variation in bats and its role in evolution. J. Exp. Biol. 218, 100–106. ( 10.1242/jeb.107243) [DOI] [PubMed] [Google Scholar]
- 7.Zhang T-Y, Meaney MJ. 2010. Epigenetics and the environmental regulation of the genome and its function. Annu. Rev. Psychol. 61, 439–466. ( 10.1146/annurev.psych.60.110707.163625) [DOI] [PubMed] [Google Scholar]
- 8.Champagne FA. 2008. Epigenetic mechanisms and the transgenerational effects of maternal care. Front. Neuroendocrinol. 29, 386–397. ( 10.1016/j.yfrne.2008.03.003) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Haig D. 2004. The (dual) origin of epigenetics. Cold Spring Harbor Symp. Quant. Biol. 69, 67–70. ( 10.1101/sqb.2004.69.67) [DOI] [PubMed] [Google Scholar]
- 10.Ho DH, Burggren WW. 2010. Epigenetics and transgenerational transfer: a physiological perspective. J. Exp. Biol. 213, 3–16. ( 10.1242/jeb.019752) [DOI] [PubMed] [Google Scholar]
- 11.Matzkin LM, Johnson S, Paight C, Markow TA. 2013. Preadult parental diet affects offspring development and metabolism in Drosophila melanogaster. PLoS ONE 8, e59530 ( 10.1371/journal.pone.0059530) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Buescher JL, Musselman LP, Wilson CA, Lang T, Keleher M, Baranski TJ, Duncan JG. 2013. Evidence for transgenerational metabolic programming in Drosophila. Dis. Model. Mech. 6, 1123–1132. ( 10.1242/dmm.011924) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Valtonen TM, Kangassalo K, Pölkki M, Rantala MJ. 2012. Transgenerational effects of parental larval diet on offspring development time, adult body size, and pathogen resistance in Drosophila melanogaster. PLoS ONE 7, e31611 ( 10.1371/journal.pone.0031611) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Dew-Budd K, Jarnigan J, Reed LK. 2016. Genetic and sex-specific transgenerational effects of a high fat diet in Drosophila melanogaster. PLoS ONE 11, e0160857 ( 10.1371/journal.pone.0160857) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Prasad NG, Shakarad M, Rajamani M, Joshi A. 2003. Interaction between the effects of maternal and larval levels of nutrition on pre-adult survival in Drosophila melanogaster. Evol. Ecol. Res. 5, 903–911. [Google Scholar]
- 16.Vijendravarma RK, Narasimha S, Kawecki TJ. 2010. Effects of parental larval diet on egg size and offspring traits in Drosophila. Biol. Lett. 6, 238–241. ( 10.1098/rsbl.2009.0754) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wayne ML, Soundararajan U, Harshman LG. 2006. Environmental stress and reproduction in Drosophila melanogaster: starvation resistance, ovariole numbers and early age egg production. BMC Evol. Biol. 6, 57 ( 10.1186/1471-2148-6-57) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Gilchrist AS, Partridge L. 1999. A comparison of the genetic basis of wing size divergence in three parallel body size clines of Drosophila melanogaster. Genetics 153, 1775–1787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Preston FW. 1974. The volume of an egg. The Auk 91, 132–138. [Google Scholar]
- 20.R Development Core Team. 2015. R: a language and environment for statistical computing. Vienna, Austria: R Foundation for Statistical Computing. [Google Scholar]
- 21.Rigby RA, Sasinopoulos DM. 2005. Generalized additive models for location, scale and shape (with discussion). J. R. Stat. Soc. C Appl. Stat. 54, 507–554. ( 10.1111/j.1467-9876.2005.00510.x) [DOI] [Google Scholar]
- 22.Delignette-Muller ML, Dutang C. 2015. fitdistrplus: an R package for fitting distributions. Journal of Statistical Software 64, 1–34. [Google Scholar]
- 23.Akaike H. 1973. Information theory and an extension of the maximum likelihood principle. In 2nd International Symposium on information theory (eds Petrov BN, Csaki F), pp. 267–281. Budapest, Hungary: Akademia Kiado. [Google Scholar]
- 24.Hothorn T, Bretz F, Westfall P. 2008. Simultaneous inference in general parametric models. Biom. J. 50, 346–363. ( 10.1002/bimj.200810425) [DOI] [PubMed] [Google Scholar]
- 25.Green DA II, Extavour CG. 2014. Insulin signaling underlies both plasticity and divergence of a reproductive trait in Drosophila. Proc. R. Soc. B 281, 20132673 ( 10.1098/rspb.2013.2673) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hodin J, Riddiford LM. 2000. Different mechanisms underlie phenotypic plasticity and interspecific variation for a reproductive character in Drosophilids (Insecta: Diptera). Evolution 5, 1638–1653. ( 10.1111/j.0014-3820.2000.tb00708.x) [DOI] [PubMed] [Google Scholar]
- 27.Mendes CC, Mirth CK. 2016. Stage-specific plasticity in ovary size is regulated by insulin/insulin-like growth factor and ecdysone signaling in Drosophila. Genetics 202, 703–719. ( 10.1534/genetics.115.179960) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sarikaya DP, Belay AA, Ahuja A, Green DA II, Dorta A, Extavour CG. 2012. The roles of cell size and cell number in determining ovariole number in Drosophila. Dev. Biol. 363, 279–289. ( 10.1016/j.ydbio.2011.12.017) [DOI] [PubMed] [Google Scholar]
- 29.Bass TM, Grandison RC, Wong R, Martinez P, Partridge L, Piper MDW. 2007. Optimization of dietary restriction protocols in Drosophila. J. Gerontol. A Biol. Sci. Med. Sci. 62, 1071–1081. ( 10.1093/gerona/62.10.1071) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lushchak OV, Gospodaryov DV, Rovenko BM, Yurkevych IS, Perkhulyn NV, Lushchak VI. 2014. Specific dietary carbohydrates differentially influence the life span and fecundity of Drosophila melanogaster. J. Gerontol. A. Biol. Sci. Med. Sci. 69, 3–12. ( 10.1093/gerona/glt077) [DOI] [PubMed] [Google Scholar]
- 31.Hassett CC. 1948. The utilization of sugars and other substances by Drosophila. Biol. Bull. 95, 114–123. ( 10.2307/1538158) [DOI] [PubMed] [Google Scholar]
- 32.Nagarajan S, Grewal SS. 2014. An investigation of nutrient-dependent mRNA translation in Drosophila larvae. Biol. Open 3, 1020–1031. ( 10.1242/bio.20149407) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Raubenheimer D, Simpson SJ, Mayntz D. 2009. Nutrition, ecology and nutritional ecology: toward an integrated framework. Funct. Ecol. 23, 4–16. ( 10.1111/j.1365-2435.2009.01522.x) [DOI] [Google Scholar]
- 34.Bonduriansky R, Runagall-McNaull A, Crean AJ. 2016. The nutritional geometry of parental effects: maternal and paternal macronutrient consumption and offspring phenotype in a neriid fly. Funct. Ecol. 30, 1675–1686. ( 10.1111/1365-2435.12643) [DOI] [Google Scholar]
- 35.Paoli PP, Donley D, Stabler D, Saseendranath A, Nicolson SW, Simpson SJ, Wright GA. 2014. Nutritional balance of essential amino acids and carbohydrates of the adult worker honeybee depends on age. Amino Acids 45, 1449–1458. ( 10.1007/s00726-014-1706-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Graham P, Pick L. 2017. Drosophila as a model for diabetes and diseases of insulin resistance. Curr. Top. Dev. Biol. 121, 397–419. ( 10.1016/bs.ctdb.2016.07.011) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hardy CM, Burke MK, Everett LJ, Han MV, Lantz KM, Gibbs AG. 2018. Genome-wide analysis of starvation-selected Drosophila melanogaster: a genetic model of obesity. Mol. Biol. Evol. 35, 50–65. ( 10.1093/molbev/msx254) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Musselman LP, Kuhnlein RP. 2018. Drosophila as a model to study obesity and metabolic disease. J. Exp. Biol. 221(Suppl. 1), pii: jeb163881 ( 10.1242/jeb.163881) [DOI] [PubMed] [Google Scholar]
- 39.Riddle MR, et al. 2018. Insulin resistance in cavefish as an adaptation to a nutrient-limited environment. Nature 555, 647–651. ( 10.1038/nature26136) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Teleman AA, Ratzenbock I, Oldham S. 2012. Drosophila: a model for understanding obesity and diabetic complications. Exp. Clin. Endocrinol. Diabetes 120, 184–185. ( 10.1055/s-0032-1304566) [DOI] [PubMed] [Google Scholar]
- 41.Kuo TH, Fedina TY, Hansen I, Dreisewerd K, Dierick HA, Yew JY, Pletcher SD. 2012. Insulin signaling mediates sexual attractiveness in Drosophila. PLoS Genet. 8, e1002684 ( 10.1371/journal.pgen.1002684) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schultzhaus JN, Bennett CJ, Iftikhar H, Yew JY, Mallett J, Carney GE. 2018. High fat diet alters Drosophila melanogaster sexual behavior and traits: decreased attractiveness and changes in pheromone profiles. Sci. Rep. 8, 5387 ( 10.1038/s41598-018-23662-2) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Schultzhaus JN, Nixon JJ, Duran JA, Carney GE. 2017. Diet alters Drosophila melanogaster mate preference and attractiveness. Anim. Behav. 123, 317–327. [Google Scholar]
- 44.Lin WS, Yeh SR, Fan SZ, Chen LY, Yen JH, Fu TF, Wu MS, Wang PY. 2018. Insulin signaling in female Drosophila links diet and sexual attractiveness. FASEB J. 32, 3870–3877. ( 10.1096/fj.201800067R) [DOI] [PubMed] [Google Scholar]
- 45.Mackay TF, et al. 2012. The Drosophila melanogaster genetic reference panel. Nature 482, 173–178. ( 10.1038/nature10811) [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are available at https://extavourlab.github.io/TransgenerationalEffectOfNutrition.