Abstract
Non-invasive imaging has the potential to play a crucial role in the characterization and translation of experimental animal models to investigate human brain development and disorders, especially when employed to study animal models that more accurately represent features of human neuroanatomy. The purpose of this study was to build and make available MRI and DTI templates and analysis tools for the ferret brain as the ferret is a well-suited species for pre-clinical MRI studies with folded cortical surface, relatively high white matter volume and body dimensions that allow imaging with pre-clinical MRI scanners. Four ferret brain templates were built in this study – in-vivo MRI and DTI and ex-vivo MRI and DTI – using brain images across many ferrets and region of interest (ROI) masks corresponding to established ferret neuroanatomy were generated by semi-automatic and manual segmentation. The templates and ROI masks were used to create a web-based ferret brain viewing software for browsing the MRI and DTI volumes with annotations based on the ROI masks. A second objective of this study was to provide a careful description of the imaging methods used for acquisition, processing, registration and template building and to demonstrate several voxelwise analysis methods including Jacobian analysis of morphometry differences between the female and male brain and bias-free identification of DTI abnormalities in an injured ferret brain. The templates, tools and methodological optimization presented in this study are intended to advance non-invasive imaging approaches for human-similar animal species that will enable the use of pre-clinical MRI studies for understanding and treating brain disorders.
Keywords: Ferret, Neuroanatomy, Template, Atlas, DTI, Voxelwise
Introduction
Non-invasive brain imaging and especially MRI has become increasingly employed to characterize brain development and disorders in animal models with the goal to identify common markers and target features of experimental models that overlap with imaging findings in humans. Furthermore, improvements to the quality of pre-clinical image acquisition and the evolution of sophisticated image processing, modeling and analysis tools are promising for providing outcome measures in basic brain research and in the development of therapies. An additional avenue to improve the effectiveness of pre-clinical studies is the extension of experimental models to species with brains that more closely resemble the human brain and may offer a more relevant system for understanding features of the normal and dis ordered brain. This is especially evident for approaches that are sensitive to tissue features that are absent in rodent models such as a folded cortical geometry or complex white matter systems. The combination of appropriate animal models with outcome measures that also apply to human subjects and patients has the potential to identify and target the features most relevant for understanding and treating human disorders.
The ferret, of the mustelid genus, has been identified as a potentially informative model system in neuroscience research as the ferret brain cortex is folded, or gyrencephalic, and contains a relatively greater white mater volume than rodent species (Fox and Marini, 2014). The primary brain research application for the ferret has been for the study of cortical development as the ferret is altricial and born before gyrification of the cortex allowing for postnatal investigation of the mechanisms that underlie cortical folding (Empie et al., 2015; Jackson et al., 1989; Noctor et al., 1997; Poluch and Juliano, 2015). A number of studies of acquired neurologic disorders including cortical dysplasia (Poluch and Juliano, 2015; Poluch et al., 2008; Abbah et al., 2014), epilepsy (Youngblood et al., 2015) and traumatic brain injury (Hutchinson et al., 2016; Schwerin et al., 2014) have taken advantage of the distinct neuroanatomical features of the ferret brain. Several basic neuroscience research approaches have also successfully employed the ferret brain to make important advances such as understanding cortical neurophysiology by slice recordings (Shu et al., 2003; Sanchez-Vives and McCormick, 2000) and mapping of cortical neural activity by in-vivo optical imaging (Schwartz and Bonhoeffer, 2001). Efforts toward mapping the ferret genome (Peng et al., 2014) and the development genetic tools in the ferret (Kou et al., 2015) demonstrate the potential for generating meaningful experimental models that are highly relevant to human neurologic disorders.
MRI methods are well suited to the study of the ferret brain not only for their non-invasive characterization of the whole brain and in vivo applications, but also for the rich foundation of quantitative tools that have been developed to study the anatomy, morphology and microstructure of the human brain. These may be readily adapted for use in the ferret, which is one of the only gyrencephalic animals with body dimensions that allow imaging in specialized pre-clinical scanners. Several MRI studies of the ferret brain have been performed to characterize the anatomical changes during developmental gyrification by conventional MRI approaches (Neal et al., 2007) as well as more advanced quantitative morphological analysis (Knutsen et al., 2010; Knutsen et al., 2013) and diffusion tensor imaging (DTI) (Barnette et al., 2009; Kroenke et al., 2009; Jespersen et al., 2012). These studies have provided an important basis to study the effects of developmental injury or disorder and several initial studies of enucleation (Bock et al., 2010; Bock et al., 2012) and chronic hypoxia (Tao et al., 2012) demonstrate the utility of quantitative MRI for characterizing the effects of disruptions in normal brain development. MRI studies in the adult ferret may also be beneficial for disorders that are influenced by brain anatomy and white matter content such as traumatic brain injury, for which several T2 and DTI abnormalities have been identified acutely following injury (Hutchinson et al., 2016).
Increasingly, advanced MRI analysis approaches have made use of voxelwise techniques that depend on the accurate warping of multiple individual brains into a common template space and subsequent statistical analysis at each voxel as is common for fMRI and morphometry studies. Advances in registration algorithms for structural MRI (Avants et al., 2008) have improved reliable warping of neuroanatomical regions across brains into a common space and provided template building methods to generate average brain volumes that preserve anatomical boundaries and edges. Recently, the ability of correcting EPI distortions in diffusion weighted images and tensor-based approaches for registration of DTI data (Irfanoglu et al., 2016; Zhang et al., 2006) have made possible template generation techniques for DTI that are morphologically faithful and high quality. This in turn has allowed improved voxelwise analysis and morphometric studies to be performed in DTI data taking advantage of the intrinsic anatomical landmarks that DTI is able to detect. Migration of advanced registration and voxelwise analysis methods into pre-clinical MRI studies provides a useful methodological basis for quantitative analysis methods that is high-throughput, unbiased and systematic. In light of these advantages, brain templates have been generated for several species including rodents (Aggarwal et al., 2009; Dorr et al., 2008; Papp et al., 2014; Johnson et al., 2010), nonhuman primates (Black et al., 2001; Black et al., 2001; Frey et al., 2011; McLaren et al., 2009; Calabrese et al., 2015; Hikishima et al., 2011), sheep (Nitzsche et al., 2015) and zebrafish (Ullmann et al., 2010) among others. These normative templates have been used for the consistent registration of MRI and DTI data to a common space, which allows anatomical localization of structures, quantitative analysis of morphological features and voxel-wise statistical analysis of quantitative metrics. As the quality of pre-clinical MRI data improves and the ability of increasingly sophisticated registration algorithms to faithfully coregister image data is realized, voxelwise MRI tools will provide a new perspective to understand neuroanatomical, physiological and structural features of brain tissue in animal model studies.
The objectives of this study were to generate and make available high quality population based templates for in-vivo and ex-vivo structural and diffusion tensor MRI of the ferret brain and to optimize MRI processing pipelines, registration tools and voxelwise analysis for ferret brain images. Diffeomorphic scalar and tensor based registration techniques were implemented to warp multiple structural and DTI images of each modality to a common space resulting in four templates: in-vivo MRI, in-vivo DTI, ex-vivo MRI and ex-vivo DTI. Based on these and with reference to known ferret neuroanatomy (Fox and Marini, 2014), the brain volume was segmented into 48 regions of interest (ROIs). The resulting template and ROI masks were then incorporated into a web-based image viewing software for visualization and annotated with neuroanatomical information using the ROI masks. In addition to providing templates, ROI tools and a visualization software, this study applied these tools to optimize and demonstrate several template based analysis approaches. First, the variability of DTI and morphometry metrics across normal ferret brains were evaluated using ROI analysis in template space. Next, as an example of potential applications of the use of the template for single subject analysis, voxelwise morphometric analysis was used to show differences between a single female ferret brain and the male ferret template. Finally, tensor-based registration and warping of an injured ferret brain to the DTI template was used for voxelwise detection of post-traumatic DTI abnormalities. The primary goal of this work is to provide a methodological framework for building a set of high quality in-vivo and ex-vivo MRI templates the ferret brain. We hope that the use of these templates in conjunction with the ROI and voxelwise analysis techniques we propose will facilitate research that will advance our understanding of brain development and brain disorders for which the ferret is an ideal model.
Methods
MRI and DTI acquisition
All animals were housed and treated in accordance with national guidelines (i.e. the NIH guide) and adhering to an animal study protocol that was approved by the Uniformed Services University of Health Sciences institutional animal care and use committee. It should be noted that a subset of the raw data included in this study (n=10 invivo MRI scans) were previously used in a separate study of brain injury to provide baseline and normative MRI and DTI values (Hutchinson et al., 2016). Adult male ferrets (Mustela putorius furo) were used in this study with an age range of 5–10 months and weight range of 1.3–2.4 kg.
In-vivo MRI and DTI acquisition
Ferrets underwent in-vivo MRI scanning to obtain structural images (n=26) and DTI data sets (n=12). During each MRI session the ferret was anesthetized with inhaled isoflurane (5.0% induction, 1–3% maintenance) and warmed by a circulating water heating pad. Anesthesia level and water pad temperature were adjusted according to physiological monitoring of temperature and respiration rate.
In-vivo ferret imaging was conducted using a horizontal bore Bruker 7T MRI system with either a 6 cm (Doty, Columbia, SC) or 8.6 cm (Bruker, Billerica, MA) quadrature volume coil for transmit and receive and ParaVision software to acquire the structural T2 weighted MRI (versions 5.1, n=12 and 6.0, n=14) and DTI (Paravision version 6.0 only) as described below:
T2 weighted MRI.
A multi-echo Rapid Acquisition with Relaxation Enhancement (RARE) pulse sequence was used to acquire 2D coronal structural MRI scans with two similar acquisition schemes for different software versions. For Paravision 5.1 the following parameters were used: TE=20, 60, 100 and 140 ms, TR=8 s, RARE factor=4, nex=2, 1 repetition, FOV=60mmx70mm, matrix=120×140, slices=40, slice thickness=0.5mm without FOV saturation. Following an upgrade to ParaVision 6.0 the following parameters were used for structural imaging: TE=12, 36, 60, 84, 108 and 132 ms, TR=10 s, RARE factor=4, FOV=48mmx48mm, matrix=96×96, slice thickness=0.5mm, number of slices =42, nex=2, 1 repetition and saturation bands were placed outside of the FOV to suppress non-brain tissue. Both schemes resulted in isotropic resolution of 0.5mm and similar tissue contrast for the lowest TE value MRI volume and these volumes were used for T2 template generation, however for generation of the multi-echo template set only volumes acquired with ParaVision 6.0 were used.
Diffusion weighted MRI.
A 2D EPI pulse sequence was used to acquire diffusion weighted image (DWI) volumes with TE/TR=40/5000ms, segments=1 and nex=1. Image slices were acquired in the coronal plane with FOV = 3.6mmx4.8mm and matrix = 48×64 to yield 0.75mmx0.75mm in-plane resolution with 48 slices of 0.5mm slice thickness. To reduce the matrix size, saturation bands were placed around the extracranial tissue (i.e. muscle and skin of the head). The diffusion experimental design included two repetitions of 3 unweighted (b=0) images, 30 DWIs with b=700 s/mm2 and 30 b=1000 s/mm2. A second repetition of these 126 DWIs was collected with the opposite phase encoding direction, which required a minor adjustment to the standard Bruker DtiEpi pulse sequence. DWIs with opposite phase encoding direction were acquired for use with the diffeomorphic registration for Blip-Up Blip-Down Diffusion Imaging (DRBUDDI) algorithm which corrected for geometric distortions and combined the opposite phase repetitions (Irfanoglu et al., 2015).
Ex-vivo MRI and DTI acquisition
Brain specimens from normal adult male ferrets (n=8) were prepared for ex-vivo MRI scanning following cardiac perfusion of the ferrets. On the day of perfusion each animal was deeply anesthetized by isoflurane inhalation (5% in oxygen) and an i.p. overdose of Euthasol (50 mg/kg). Upon cessation of reflexes, ferrets were transcardially perfused with 1 L of ice-cold phosphate buffered saline (PBS) (pH 7.4) followed by 1 L of 4% paraformaldehyde solution in PBS (Santa Cruz Biotechnology) containing 47.6 mg of heparin (Sigma-Aldrich). The brains were harvested, post-fixed in 4% paraformaldehyde for 8–10 days, and then transferred to a storage solution containing 0.03% sodium azide in PBS. Following at least one week of rehydration in this solution, specimens were immersed in Fluorinert (FC-3283, 3 M, St. Paul, MN) in a 25 mm glass NMR tube for imaging.
Ferret brain specimens were imaged using a Bruker 7 T vertical wide-bore magnet equipped with micro2.5 and 25 mm rf coil micro-imaging system, Avance III spectrometer running Paravision 5.1 software and three GREAT60 gradient amplifiers. For T2 imaging, a 3D multi-slice multi-echo (MSME) pulse sequence was used with TE/TR=10–100/3000 ms, nex=1, reps=1 with the following spatial parameters: FOV=26 mm×40 mm×20 mm, matrix=104×160×80 for isotropic voxel dimensions of 250 μm. Total scan time for this acquisition was 6 hours 57 min.
A total of 88 diffusion weighted image (DWI) volumes were acquired with the same spatial geometry and dimensions as the T2 MRI. A 3D EPI pulse sequence – modified slightly from the standard DtiEpi pulse sequence to exclude gradient crushers. The imaging parameters were: TE/TR=36/700 ms, nex=1, 8 segments and two repetitions for each DWI with opposite phase encode directions. The DWI sampling scheme was b(in s/mm2)/# gradient directions = 100/6, 200/6, 500/6, 1000/6, 1700/32 and 3800/32. The acquisition time for a single image was 7.5 min and the total acquisition time for all DWIs and repetitions was 22 h.
MRI and DTI processing and template generation
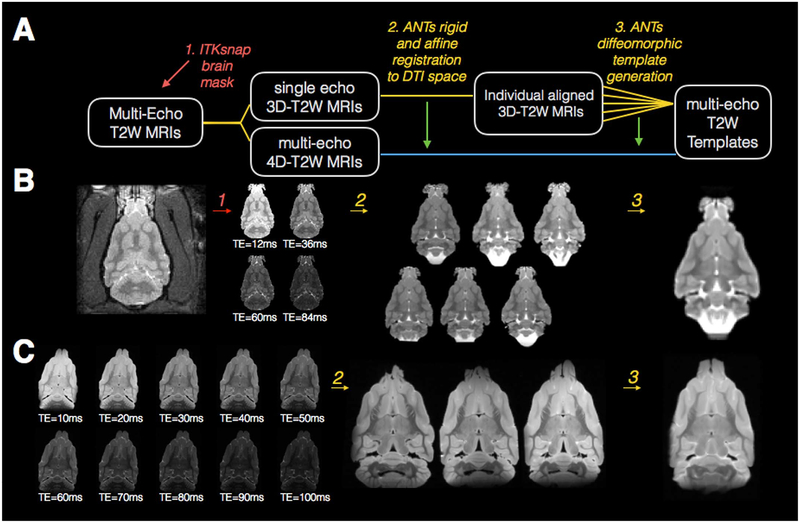
Methods for image processing and template generation are provided below and organized according to the numbering of Figs. 2 and 3. In addition to the below methods for constructing each type of template, it is notable that the order of template generation and the registration targets were chosen so that all templates were ultimately generated in a comparable space with the exception of non-linear differences between DTI and T2 and resolution differences between in vivo and ex-vivo templates.
Fig. 2.
Processing pipeline and template generation for T2 images of the in-vivo and ex-vivo ferret brain. The diagram drawing (A) explains each step of processing that is performed for in-vivo and ex-vivo T2 template generation where rounded boxes describe the T2 image(s) at each step, numbered italicized text indicates the software and processing implemented for each step and the same-colored lines connect the input and output images for each step. Red and green arrows in (A) indicate the indirect use of data: the use of an ROI from brain segmentation to mask the 3D and 4D T2W images (red) and the application of the transforms derived from the 3D template generation to the 4D images (green). Representative in-vivo (B) and ex-vivo (C) images are shown for each processing step described in (A) including different contrast images labeled with corresponding TE values.
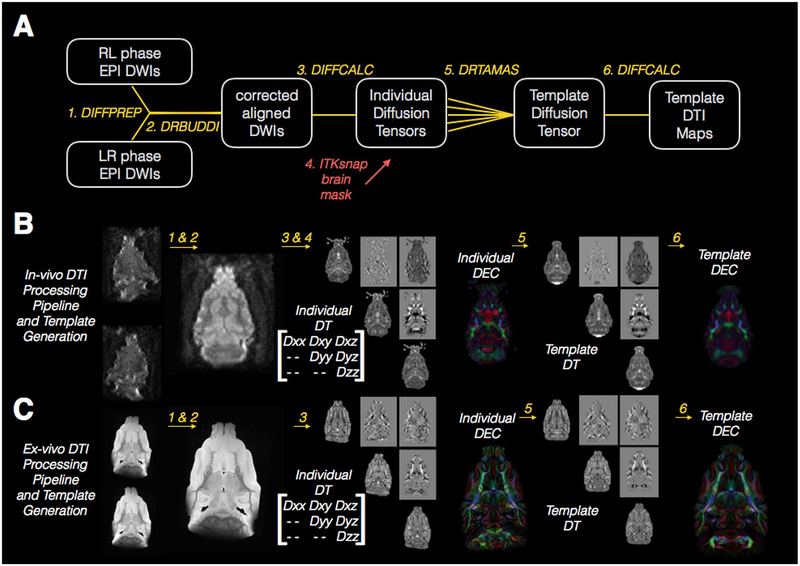
Fig. 3.
Processing pipeline and template generation for DTI images of the in-vivo and ex-vivo ferret brain. The diagram drawing (A) explains each step of processing that is performed for in-vivo and ex-vivo DTI template generation where rounded boxes describe the DWI images and diffusion tensor images at each step, numbered italicized text indicates the software and processing implemented for each step and the same-colored lines connect the input and output images for each step. The red arrow in (A) indicates the indirect use of an ROI from brain segmentation to mask the diffusion tensor volumes prior to template generation. Representative in-vivo (B) and ex-vivo (C) images are shown for each processing step described in (A) with the elements of the diffusion tensor (DT) labeled to correspond to the images from each of these in the individual and template DT volumes.
Multiple contrast T2weighted MRI template generation
Masking of the brain from surrounding tissue. This step was only performed for in-vivo images to extract the brain from other parts of the image (e.g. muscle, skin, eyes). Raw multi-echo T2 images were imported using Matlab tools (R2016a, Natick, MA) and a single 3D volume was extracted for each brain. Brain segmentation masks were generated using the ITK-snap semi-automatic active contour segmentation algorithm (Yushkevich et al., 2006). In particular, the image was loaded in ITKsnap and the snake tool was launched. A threshold was selected for the image and seed regions were placed within the brain. The snake algorithm was initiated and once the brain region was filled, the process was halted resulting in semi-automatic generation of brain masks.
Rigid and Affine Alignment of the structural images. All registration and template building involved with the structural template generation was performed using the ANTs software package (Avants et al., 2008). In this step, the masked 3D T2W volumes were rigidly aligned with the preliminary template1 and then they underwent 12-parameter affine registration to the same template.
Diffeomorphic warping and template generation. The “buildtemplateparallel.sh” script that is included with the ANTs software package was used with the cross correlation similarity metric, symmetric normalization transformation model and N4 Bias Field Correction.
DTI processing and template generation
Both ex-vivo and in-vivo DWI processing was performed using the TORTOISE pipeline (Pierpaoli et al., 2010).
Motion and Eddy-current corrections. Following importation with the DIFFPREP module of TORTOISE, DWIs were rigidly aligned to correct for artifacts. The DWIs were also rigidly registered to the realigned T2W structural image. Realignment of the structural image for this purpose was accomplished in mipav software either using midsagittal plane realignment (for in-vivo processing) or using landmark registration (for ex-vivo processing).
Geometric distortion correction using DRBUDDI. The DRBUDDI algorithm (Irfanoglu et al., 2015) was used to correct for geometric distortions using the structural image as a target. The transformations from these two initial steps were combined and applied once to the original DWIs to minimize interpolation. Orientation information was preserved by reorienting the b-matrix according to the preprocessing transformations and the ex-vivo data dimensions remained 250 μm isotropic while the in-vivo spatial dimensions were resampled to 400 μm isotropic.
Individual tensor calculation. Corrected DWIs were fit to the nonlinear DTI model (Basser et al., 1994) and for in-vivo DTI the resulting trace map was used to generate a brain mask (see next step) and the masked DWIs were re-fit. The masked DT volumes were used for template generation as described in the next section.
Masking of the brain from surrounding tissue. For the ex-vivo DWIs, masking by simple thresholding in the TORTOISE software prior to DTI fit was sufficient, but for in-vivo DWIs, brain masking was performed using the same procedure described for T2 images above (step 1) applied to the trace map as explained in the previous step.
DRTAMAS template generation. The masked diffusion tensor volumes for each brain were used to generate in-vivo and ex-vivo DTI templates using DRTAMAS tools (Irfanoglu et al., 2016). Briefly, this procedure first rigidly aligns all DT volumes with the preliminary template,1 then performs affine registration followed by diffeomorphic tensor-based registration using a symmetric normalization transformation model, combined metric similarity of trace and the DT images and the constraint that the average of all displacement fields across the brains used for each template must be zero.
Template DTI map generation. The template diffusion tensor map generated in the previous map was imported into DIFFCALC software and the eigenvalues computed. From these, DTI scalar maps were generated including fractional anisotropy (FA) and Trace (TR) as well as the directionally encoded color (DEC) map, which is a visual representation of the orientation of the primary eigenvector (Pajevic and Pierpaoli, 1999).
Brain segmentation for neuroanatomical ROI masks
A single volume containing 48 labeled masks was generated to define anatomically relevant divisions of the ferret brain according to the book chapter by Kroenke, Mills, Olavarria and Neil (Fox and Marini, 2014). Masks were created using the ex-vivo DTI templates and a combination of semi-automatic and manual segmentation tools. The atropos tissue segmentation algorithm of the ANTs software package (Avants et al., 2011) was used to classify voxels in the masked template brain based on FA and TR values into 2 compartments that were categorized as Gray Matter (GM) or White Matter (WM). This initial segmentation was manually corrected in ITKsnap software. Next, GM and WM brain regions were manually delineated with reference to the Figures from Neal and Kroenke using ITKsnap manual segmentation tools to preserve the tissue boundaries determined by automatic segmentation while allowing manual determination of boundaries within GM or WM regions.
Web-based template viewer
The templates and ROI masks were used to create a web-based software that allows the user to browse template maps and navigate through the images in an orthographic view. A 3D viewer is included to visualize the cursor location within the 3D brain volume. An information window is included in the software that reports neuroanatomical information about the cursor location including the coarse brain region category (e.g. white matter or cortex) and neuroanatomical region (e.g. corpus callosum or anterior ectosylvian gyrus) as well as quantitative information such as DTI metrics. The layout and basic functions of this viewer may be explored publically at the web address: www.mriatlases.nih.gov.
Template-based analysis approaches
Template-based ROI analysis of DTI metric variance in the ex-vivo ferret brain
The eight individual brains used to generate the ex-vivo DTI atlas were warped into the common template space using the transformations generated during template construction. Mean values for FA and Trace in each brain were extracted and graphed for each region of the ROI mask set using the R statistical package (version 3.3.1, www.r-project.org/) and the ANTsR library (version 0.3.3, http://stnava.github.io/ANTsR/).
Voxelwise morphometric analysis of sexual dimorphism
Based on existing studies reporting neuroanatomical differences between female and male ferrets (Sawada et al., 2013; Sawada et al., 2015) a demonstration was performed in which a single in-vivo T2W brain volume for a female ferret was warped to the in-vivo structural template of male brains. The single brain volume was first masked to include only brain voxels as described previously. Next, the “antsRegistration” command line tool was used to perform rigid, then affine, then diffeomorphic registration of the brain volume to the template using same or similar parameters as those used for template generation. The transformation information from either the difeomorphic registration alone or the combined transform of the affine and diffeomorphic registration were used to generate maps for the log of the determinant of the Jacobian (henceforth “LogJ”) of the deformation field using the ANTs command “ANTSJacobian” with the uselog option of 1. The values in a LogJ map are the log of the ratiometric scaling that an individual voxel from the template space must undergo to match the native image such that negative values indicate a smaller local volume in the registered image and positive values indicate larger local volume in the registered image compared to the template. Visualization of LogJ information was performed using ITKsnap software to overlay the LogJ maps on the template image and quantitative thresholds were used with color-coding for positive values with warm colors and negative values with cool colors as indicated by color bars.
Detection of abnormal DTI values following injury using voxelwise statistics
Ex-vivo DTI images from an injured ferret brain were acquired with the same DTI methods described earlier in this section. This brain was obtained from a ferret one week following controlled cortical impact (CCI) (Lighthall, 1988), which has recently been adapted for the ferret (Schwerin et al., 2014). Briefly, the ferret was anesthetized and monitored under aseptic conditions and a cranial window was made in the skull over the intended injury site, then a stereotaxic impactor device (Leica Biosystems, Buffalo Grove, IL) was used to induce focal CCI by a 3 mm diameter cylindrical bit with velocity of 5 m/s, penetration depth of 4 mm and dwell time of 100 ms. One week following CCI, the ferret underwent cardiac perfusion and the brain extracted and prepared for ex-vivo imaging as described above.
Diffeomorphic tensor-based registration (Irfanoglu et al., 2016) was used to register the single injured brain to the ex-vivo DTI template using an exclusion mask to remove the effects of the lesion on the registration. Next, Z-score maps were generated using the ANTsR package to compare the DTI value of the injured brain with the mean and standard deviation of the template population using the equation shown in Fig. 9. The Z-score map was visualized in ITKsnap as an overlay on the template FA and trace maps with color scale and window levels to show differences in the injured brain from normative values.
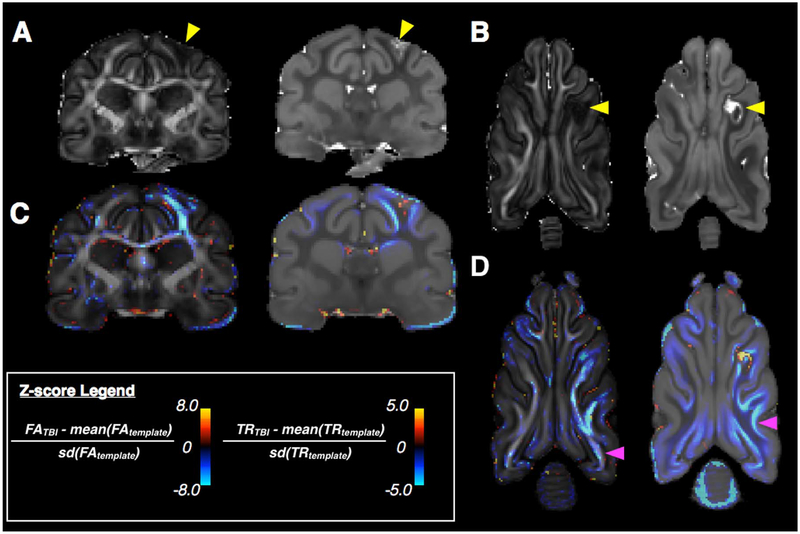
Fig. 9.
Demonstration for voxelwise Z-score analysis to identify DTI abnormalities following traumatic brain injury (TBI). Slices from the FA and Trace maps of an injured ex-vivo ferret brain are shown (A and B) and abnormalities near the site of injury (arrowhead) are readily identifiable including decreased FA and abnormal Trace. Voxelwise Z-score maps comparing DTI values from single injured brain with normative template values for FA and Trace (C and D) are shown for similar slice levels as an overlay on the template with negative values (cool) indicating a reduction of the DTI metric in the TBI injured brain and positive values (warm) indicating an increase. In addition to the abnormalities near the injury site (yellow arrowhead), several more subtle differences (magenta arrowheads) are found by this method including extensive FA reduction in the body of the white matter and regions of decreased Trace that extend beyond the gray matter white matter boundaries into the gray matter.
Ancillary methods for visualization of cross-species brain differences
In order to provide an illustration of brain data across mice, ferrets and humans, DTI brain volumes for each species were obtained including an ex-vivo mouse brain volume imaged with similar acquisition parameters as for the ferret brains described above but resolution of 100 μm3 upsampled to 50 μm3 and DWI sampling scheme optimized for tractography with 12 reference images and 114 DWI volumes with b=4800 s/mm2, ferret data from a single brain was acquired with the same acquisition parameters as described above, but with DWI sampling scheme of 105 directions with b=10,000 s/mm2 and a single subject pre-processed diffusion MRI human data set was obtained from the Human Connectome Project database (Van Essen et al., 2013). For DWI data of all species, figures showing cortical surface geometry were created by generation of a whole brain mask using the active contour tool in ITKsnap (Yushkevich et al., 2006) and image capture of the 3D visualization of the mask volume from the ITKsnap 3D viewer. Tract weighted images (Calamante et al., 2012) were generated for each set in a similar manner using MRtrix3 tools (http://www.mrtrix.org/) and pipelines including constrained spherical deconvolution (Tournier et al., 2004), streamlines whole brain tractography (Tournier et al., 2012) and tract density imaging (Calamante et al., 2010). To demonstrate several features of the ferret brain, orientation distribution functions were visualized and screen captured using MRview and a U-fiber tractogram was generated from DTI data using DTK and TrackVis software with appropriate seed and exclusion masks to illustrate the structure.
Results
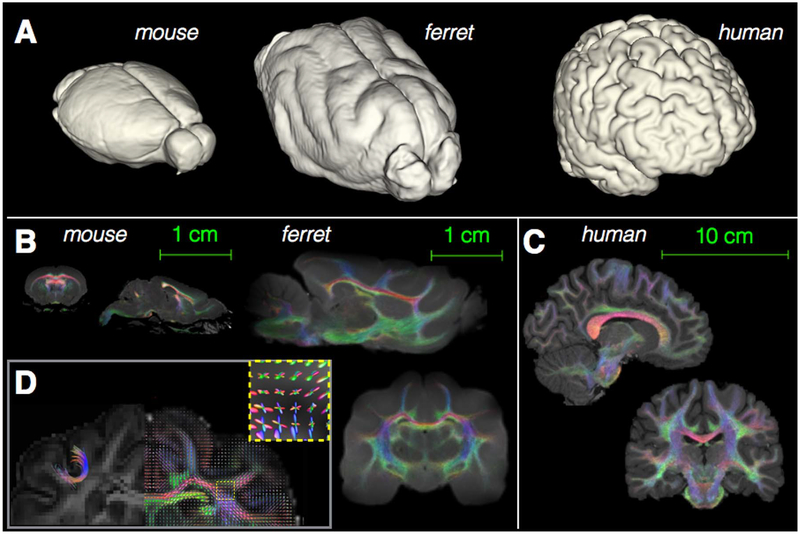
Broad neuroanatomical differences across species are illustrated in Fig. 1 to provide a sense of scale and comparative anatomy for the ferret brain in the context of the more commonly investigated brain anatomy of the mouse and human. The intermediate nature of the ferret brain between mouse and human cortical anatomy can be observed from 3D surface rendering across species, which shows the ferret brain and human brain are gyrencephalic or folded, while the mouse brain is lissencephalic or smooth (Fig. 1a). However, the ferret brain anatomy is simplified compared with the human having far fewer sulci and little frontal or associative cortical regions (note: a more detailed description of ferret neuroanatomy alongside volume renderings may be found in the book chapter by Kroenke et al., 2014 (Fox and Marini, 2014)). Tract-weighted images (Calamante et al., 2012) (Figs. 1b and c) show white matter content and complexity across species. While the ferret brain white matter is considerably more complex than the mouse with U-fibers and regions of multiple fiber tract crossings (Fig. 1d) that are similar to human white matter anatomy, it is also a simplified system compared to the human with fewer associative pathways (Fox and Marini, 2014; Neal et al., 2007).
Fig. 1.
3D rendering of the cortical surface and WM DTI representations of the ferret brain alongside mouse and human. Visualization of the cortical surface by 3D rendering of segmented brain images (A) shows size and gross morphological differences across the three species, especially the lissencephalic cortex of the mouse and gyrencephalic cortex of the ferret and human brains (images are scaled differently). Comparative anatomical differences of white matter content and complexity are illustrated by the Tract Weighted Imaging maps in the mouse and ferret (B) using the same spatial scale (scale bar 1 cm) and for the human brain (C), which has been scaled differently (scale bar 10 cm). An inset (D) is provided to show details of the ferret brain white matter that are similar to human features, but absent in the rodent brain including U-fibers and regions of multi-fiber crossings in the cortex.
Key observations on MRI and DTI processing and template generation pipelines
Image processing and template generation pipelines used in this work are shown alongside image slices representing the MRI data at each step in the process in Figs. 2 and 3 for structural MRI and DTI respectively. While this work was primarily methodological in nature and described in greater detail in the methods section, several key observations about the templates and image processing steps are reported below.
Notable observations about the generation of multi-contrast anatomical MRI templates using the pipeline presented in Fig. 2 include the importance of consistent segmentation of brain tissue from surrounding muscle or skin tissue for which active contour segmentation (Yushkevich et al., 2006) was able to reliably extract ferret brain tissue for the in-vivo images in this study. Also the combination of multiple registration transformations (i.e. rigid, affine and diffeomorphic) into a single applied transformation minimized interpolation effects. The resulting structural MRI templates demonstrated tissue boundaries between gray matter, white matter and cerebrospinal fluid (CSF) with high contrast, which is beneficial for structural MRI registration algorithms.
The pipeline for DTI processing and template generation is shown in Fig. 3 and several observations about EPI acquisition, DTI processing and template building are given that are relevant for the best use of template tools. First, the image quality of raw EPI images was very different between in-vivo images (e.g. Fig. 3B) and ex-vivo images (Fig. 3C). Ex-vivo 3D EPI was relatively free of imaging distortions and artifacts with high signal to noise ratio (SNR) of 49.8 for b=1700 in a single raw DWI made possible by the use of a segmented 3D EPI sequence, small RF coil and long imaging times. In contrast, the in-vivo 2D EPI images had lower SNR of 2.9 for b=1000 in a single raw DWI and were subject to considerable geometric distortions and more vulnerable to motion and eddy current artifacts, which required more extensive pre-processing corrections (e.g. corrections for motion, eddy currents and geometric distortions were essential). In particular, the in-vivo images suffered from considerable geometric distortions due to the single segment acquisition and thin slice profile, which are described in the methods section. In this case, DRBUDDI corrections were essential and remarkably effective for correction of geometric distortions. As described for the structural images, brain segmentation was also an important part of DTI processing for in-vivo data and active contour segmentation tool of the trace map prior to template building was essential for reliable registration. For both in-vivo and ex vivo diffusion tensor registration using the parameters described in the methods section, DRTAMAS template generation resulted in DTI templates that preserved many relevant anatomical features without substantial spatial smoothing.
In-vivo and ex-vivo ferret brain MRI and DTI templates
Four templates were generated in this work and are shown in Figs. 4–6. These templates are also available for download or online viewing at the website: www.mriatlases.nih.gov/ferret. In addition to the templates, a set of 48 region of interest (ROI) masks were generated (Fig. 7) to provide delineation and labeling according to anatomical regions including the gyri and sulci of the cortex, major white matter tracts and subcortical regions described elsewhere (Fox and Marini, 2014). The templates and mask overlays were combined for use with the web-based viewing software to allow navigation through the templates accompanied by anatomical annotation.
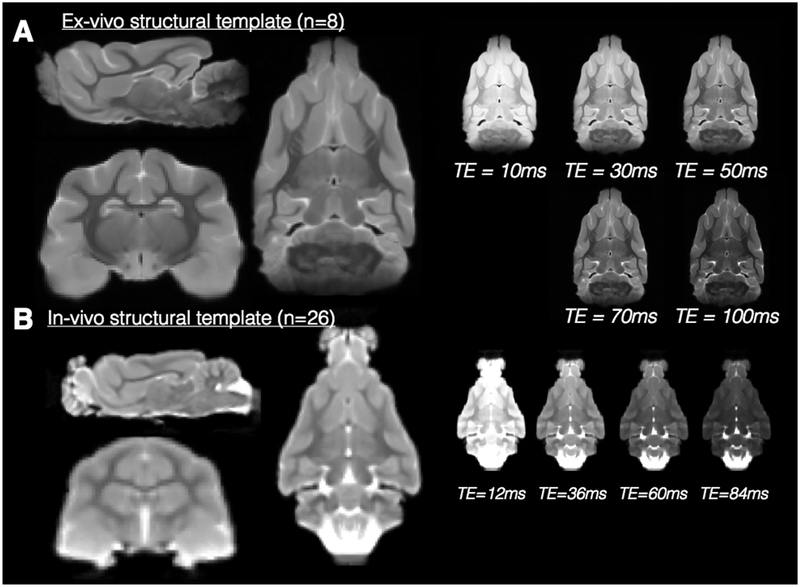
Fig. 4.
Multiple contrast structural templates in for in-vivo and ex-vivo ferret brain MRI. Orthographic views of the in-vivo (A) and ex-vivo (B) structural templates generated from the number of individual brain volumes given. The templates demonstrate the resolution of neuroanatomical structures and the right of the orthographic views, a single slice from the multi-echo volumes are shown to demonstrate the range of tissue contrasts labeled according to TE value.
Fig. 6.
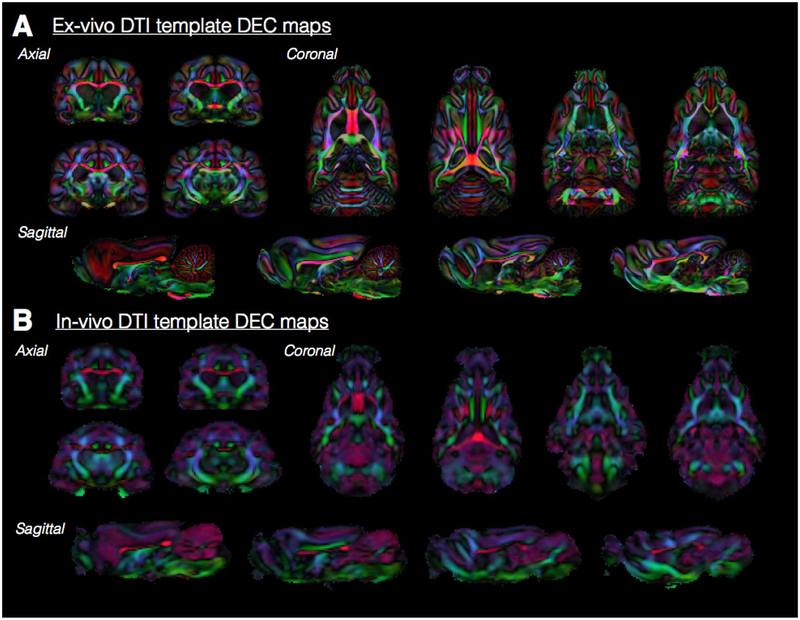
Ex-vivo and in-vivo DEC maps throughout the ferret brain. Representative slices from DEC maps for each of the orthogonal orientations are shown to demonstrate the anatomical resolution especially of white matter fiber tracts of the ex-vivo (A) and in-vivo (B) 3D DTI templates. The color coding for primary eigenvector orientations is the same as for Fig. 5.
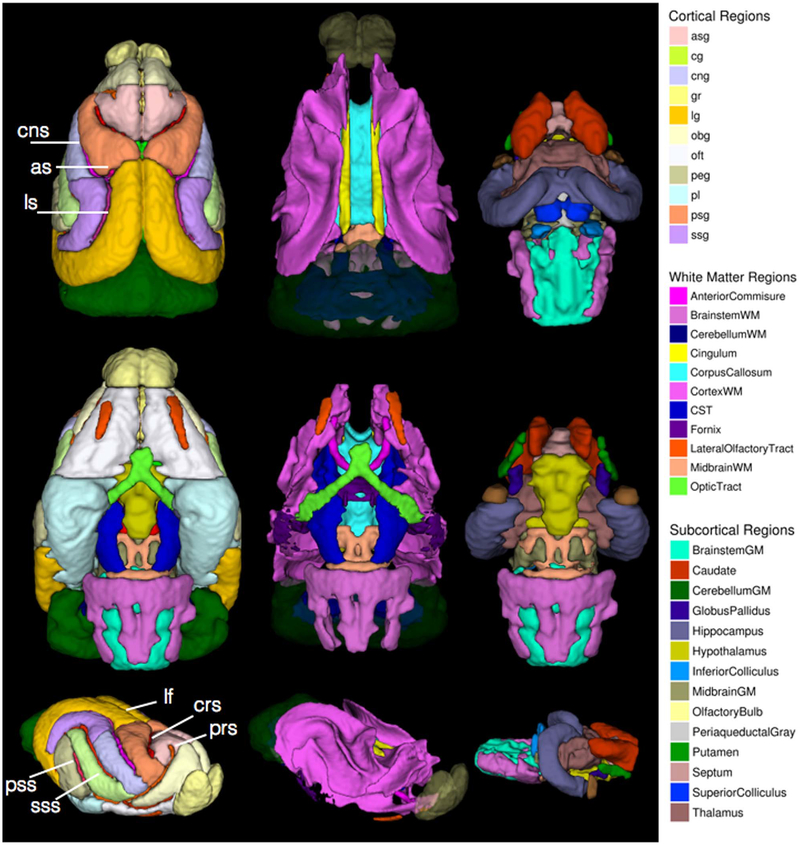
Fig. 7.
Atlas ROI masks in template space. 3D rendered images are shown for superior (top row), inferior (middle row) and oblique (bottom row) views of different sets of brain regions including cortical regions (left column), white matter regions (middle column) and subcortical regions (right column). Each region is color-coded according to the legend and with reference to the abbreviations in Table 1. Sulci of the cortex are labeled by white text annotation and lines. For the white matter masks, the olfactory bulbs and cerebellum are shown with transparency for anatomical reference.
Structural templates
Ex-vivo structural template (Fig. 4a).
Diffeomorphic registration of eight ex-vivo MRI ferret brain volumes was used to create a template with a range of contrasts from different TE values (10–100 ms) with isotropic voxel resolution of 250 μm. Many anatomical regions can be delineated in this template as blurring of the images was minimized by the quality of the registration across samples. The set of ten templates covers a range of tissue contrasts according to T2 weighting.
In-vivo structural template (Fig. 4b).
Diffeomorphic registration of 26 in-vivo MRI ferret brain volumes was used to create an anatomical template as well as 4 template volumes with different contrast having TE=12–84 ms. The isotropic resolution of the template is 500 μm and the diffeomorphic algorithm resulted in minimal blurring of structural features. Major anatomical regions are identifiable, although finer structures are unable to be resolved at this resolution.
In addition to anatomical resolution, several notable differences between the in-vivo and ex-vivo templates are evident that are consequential for their use as registration targets. First, the ventricular space is bright in the in-vivo template, especially for long TE images, but dark for the ex-vivo template due to filling of the space with Fluorinert. Because edge detection and signal intensity are primary drivers of most registration algorithms, this difference is quite consequential and appropriate brain masking should be used prior to registration of other structural volumes especially for the ex-vivo template.
DTI templates
Ex-vivo DTI template (Fig. 5a).
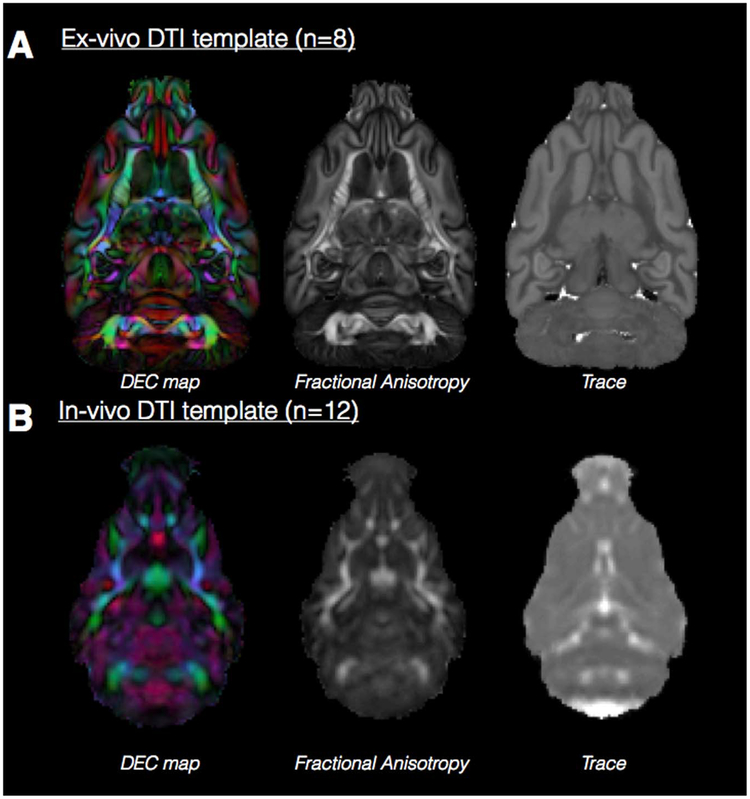
Fig. 5.
Ex-vivo and in-vivo template DTI maps of the ferret brain. A single slice at the same level is shown for Trace, FA and DEC maps derived from in-vivo (A) and ex-vivo (B) DTI templates generated from the number of individual DTI volumes indicated. The color coding for DEC maps, which report the orientation of the primary eigenvector is: red=left/right, green=anterior/posterior and blue=dorsal/ventral and maps are weighted by the linear anisotropy. The DTI maps demonstrate the relative resolution, smoothing and contrast features of each template.
The ex-vivo DTI template was generated from 8 ferret brain DTI volumes and provides high resolution (250 mm isotropic) with sharp contrast between anatomical regions and different tissue types. Slices at different orthogonal levels (Fig. 6a) show that fine-scale features of most anatomical regions are discernible such as small white matter tracts as well as layers of the hippocampus and cerebellum.
In-vivo DTI template (Fig. 5b).
The in-vivo DTI template was generated from 12 ferret brain DTI volumes with 400 mm isotropic resolution resulting in more blurred boundaries between anatomical structures and tissue types. Major white matter pathways may be identified in this template (Fig. 6b), however smaller tracts and anatomical information within gray matter structures is not apparent. Nevertheless, the isotropic dimensions allow for continuous orthogonal representation of major anatomical regions, which provides a suitable target for registration of in-vivo DTI data.
There are multiple differences between the in-vivo and ex-vivo templates beyond anatomical resolution that indicate they should only be used with data that is similar (i.e. it is not recommended to register in-vivo DTI data with the ex-vivo template or vice-versa). In particular, the contrast of the Trace map shows sharp boundaries for the ex-vivo template, but is homogeneous for gray and white matter for the in-vivo template.
Population features of MRI and DTI metrics across normal ferret brains
Descriptive statistics of anisotropy and diffusivity across regions of ex-vivo normal ferret brains used in this study were made using the template masks to extract quantitative values from the ex-vivo DTI maps for each of 8 brains warped into the common space of the ex-vivo DTI template. The mean and standard deviation for each region are reported in Table 1 for cortical regions and Table 2 for white matter regions and subcortical structures (Fig. 7). In general, the mean values and variance appear to be most similar across cortical regions and to differ according to ROI in the white matter and subcortical structures. For example, the Trace of the hypothalamus is considerably lower than for the hippocampus, while the FA values of these structures are more similar. Another comparative example is the greater metric variance across samples of Trace and FA in the smaller white matter structures (e.g. the optic and olfactory tracts) compared with larger regions (e.g. the corpus callosum and cortical white matter).
Table 1.
Cortical brain region names and DTI values. For the template ROI masks shown in Fig. 7 (left column), the full names and abbreviations are given for each regions as well as the mean and standard deviation of the FA and Trace values across all brains used to generate the ex-vivo template.
| Cortical Regions | Fissures and Sulci | ||||||
|---|---|---|---|---|---|---|---|
| FA ± std. | TR ± std. | FA ± std. | TR ± std. | ||||
| Anterior ectosylvian gyrus (aeg) | 0.21 ± 0.07 | 702 ± 135 | orbital gyrus (obg) | 0.22 ± 0.08 | 655 ± 154 | Ansinate sulcus (as) | Presylvian sulcus (prs) |
| Anterior sigmoid gyrus (asg) | 0.24 ± 0.07 | 679 ± 141 | olfactory tubercle (oft) | 0.21 ± 0.08 | 643 ± 138 | Coronal sulcus (cns) | Pseudosylvian sulcus (pss) |
| cingulate gyrus (cg) | 0.23 ± 0.1 | 706 ± 145 | Posterior ectosylvian gryus (peg) | 0.21 ± 0.08 | 666 ± 134 | Cruciate sulcus (crs) | Rhinal fissure (rf) |
| Coronal Gyrus (cng) | 0.18 ± 0.06 | 731 ± 119 | Piriform lobe (pl) | 0.23 ± 0.07 | 644 ± 148 | longitudinal fissure (lf) | Splenial sulcus (ss) |
| Gyrus rectus (gr) | 0.21 ± 0.1 | 640 ± 137 | Posterior sigmoid gyrus (psg) | 0.17 ± 0.08 | 711 ± 138 | lateral sulcus (ls) | Suprasylvian sulcus (sss) |
| Lateral gyrus (lg) | 0.2 ± 0.08 | 708 ± 148 | Suprasylvian gyrus (ssg) | 0.2 ± 0.07 | 707 ± 137 | ||
Table 2.
White matter and subcortical region names and DTI values. For the template ROI masks shown in Fig. 7 (middle and right columns), the name of each region is givan as well as the FA and Trace values across all brains used to generate the ex-vivo template.
| White Matter | Subcortical Gray Matter | ||||
|---|---|---|---|---|---|
| FA ± std. | TR ± std. | FA ± std. | TR ± std. | ||
| Anterior Commisure | 0.57 ± 0.17 | 565 ± 95 | Amygdala | 0.19 ± 0.06 | 734 ± 125 |
| Brainstem White Matter | 0.56 ± 0.17 | 658 ± 157 | Caudate | 0.18 ± 0.06 | 737 ± 107 |
| Cerebellum White Matter | 0.5 ± 0.15 | 640 ± 115 | Globus Pallidus | 0.23 ± 0.06 | 693 ± 86 |
| Cingulum | 0.49 ± 0.12 | 564 ± 95 | Hippocampus | 0.21 ± 0.08 | 777 ± 130 |
| Corpus Callosum | 0.63 ± 0.16 | 540 ± 88 | Hypothalamus | 0.18 ± 0.07 | 556 ± 116 |
| CortexWM | 0.48 ± 0.13 | 584 ± 103 | Inferior Colliculus | 0.25 ± 0.06 | 678 ± 95 |
| Corticospinal Tract (CST) | 0.56 ± 0.16 | 599 ± 115 | Olfactory Bulb | 0.24 ± 0.06 | 599 ± 74 |
| Fornix | 0.61 ± 0.17 | 582 ± 147 | Periaqueductal Gray | 0.23 ± 0.09 | 561 ± 117 |
| Lateral Olfactory Tract | 0.49 ± 0.21 | 545 ± 236 | Putamen | 0.22 ± 0.06 | 735 ± 87 |
| Midbrain White Matter | 0.38 ± 0.08 | 619 ± 115 | Septum | 0.28 ± 0.08 | 615 ± 117 |
| Optic Nerve/Tract | 0.52 ± 0.24 | 580 ± 237 | Superior Colliculus | 0.18 ± 0.08 | 607 ± 120 |
| Thalamus | 0.24 ± 0.1 | 682 ± 103 | |||
Jacobian analysis of sexual dimorphism
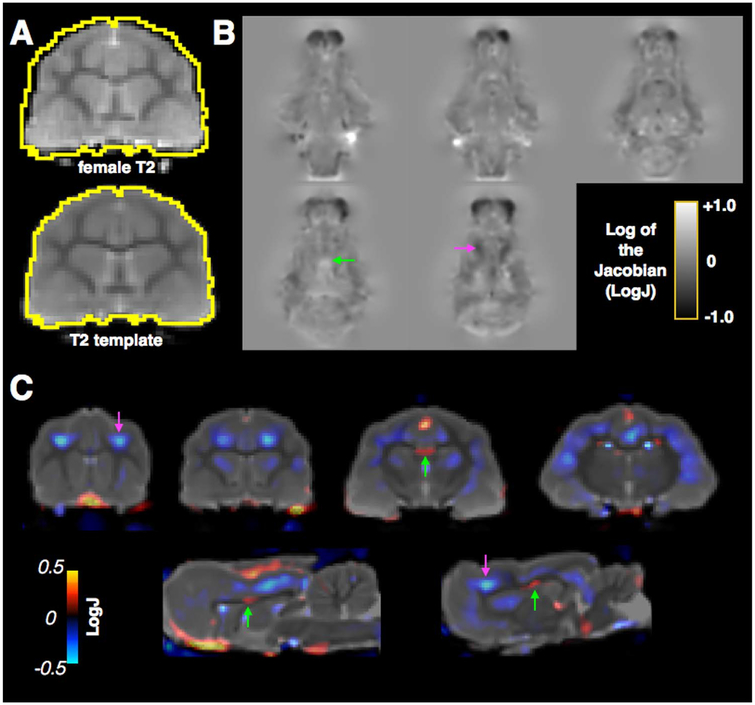
The ferret is known to be sexually dimorphic and brain differences between males and females have been identified using MRI by previous studies (Sawada et al., 2013; Sawada et al., 2015). To evaluate the feasibility of using templates from the current study with a LogJ voxelwise morphometric analysis for addressing this type of anatomical question, the in-vivo T2W brain volume of a female ferret was warped to the ferret male in-vivo template (Fig. 8). The global scaling differences between the female brain and template are shown in Fig. 8a and from the affine transformation a scaling value of 0.95 was determined relating the female brain to the template. The local morphometric differences after accounting for global scaling were shown using LogJ analysis of the diffeomorphic warping fields, which are shown for multiple coronal levels in the brain indicating several darker regions for which the female brain has a smaller relative volume and bright regions with a greater relative volume. Another representation of this data is shown as a colorized overlay of the LogJ map on the template image (Fig. 8c). From this image it is clear that the female brain exhibits smaller relative volume, indicated by cooler colors, in frontal cortical regions (green arrow) and greater relative volume, indicated by warm colors in midline regions including the white matter (magenta arrow).
Fig. 8.
Demonstration of registration and LogJ analysis to identify sex differences in the ferret brain. A single female brain (A, top) was registered to the adult male template (A, bottom) to demonstrate voxelwise analysis morphologic differences. LogJ maps are shown at several levels (B) indicating local volume increases (positive values, green arrow) or decreases (negative values, magenta arrow) in the female brain compared with the template. An overlay map of LogJ is shown with respect to the T2 template (C) for visualization of brain regions with local expansion or contraction required to warp the female brain to the template where negative values (blue, magenta arrow) correspond to regions that are smaller in the female brain and positive values (red, green arrow) correspond to regions that are larger.
Voxelwise analysis to identify DTI abnormalities after TBI
To identify regions of abnormal DTI values in a quantitative and operator independent (bias-free) way for a single TBI injured ferret brain, a single ex-vivo DT dataset of the injured brain was registered to the ex-vivo template and Z-score maps were generated for FA and Trace. Z-score values – which report for the registered brain the number of standard deviations away from the template value at each voxel – are shown as an overlay map in Fig. 9 alongside native space DTI maps for the individual injured brain. In regions of abnormality that are strongly evident by eye, the Z-score map reveals distinct subregions of high Trace as well as reduced Trace and regions of reduced FA.
Z-score maps identify additional abnormalities that are not as obvious by qualitative inspection of the native space DTI maps. In particular, FA is reduced in the body of the white matter over nearly the entire hemisphere ipsilateral to the injury site that is described quantitatively by the Z-score maps. A spatial pattern of decreased Trace localized to the tissue boundary between cortical gray and white matter is also evident in the Z-score maps and could be overlooked by visual inspection alone for the single brain DTI maps.
Discussion
A collection of ferret brain MRI and DTI templates and anatomical masks have been generated and made available both as downloadable image volumes and as an interactive web-based viewer (www.mriatlases.nih.gov). In addition to developing a robust and accurate approach for the creation of these tools, another goal of this work was to optimize and describe their use for performing ROI and voxelwise analyses to address neuroanatomical or neurobiological questions. Overall, this study was intended to provide a description and evaluation across the full process of image acquisition, image processing, template generation and application of template-based tools to advance and enable use of these approaches for pre-clinical research in experimental models.
Template-based analysis tools to investigate neuroanatomy and neurobiology of the ferret brain
The primary impetus for generation of ferret brain templates and anatomical masks in this study is for their use in template-based ROI analysis and voxelwise analysis of in-vivo and ex-vivo MRI and DTI modalities. The in-vivo and ex-vivo MRI and DTI templates, neuroanatomical masks and web-browser are intended to extend the analysis toolbox available for studying the ferret brain. The potential for these tools to be useful has been demonstrated in a handful of existing quantitative MRI studies including straightforward segmentation for volume comparisons, techniques to measure cortex gyrification, curvature and morphometry during development (Neal et al., 2007; Knutsen et al., 2013; Barnette et al., 2009) and disorder (Bock et al., 2012) and sophisticated voxelwise cortical surface representation methods (Knutsen et al., 2010). In the present study, several template ROI and voxelwise analysis techniques were selected to demonstrate approaches to address common questions including statistical analysis of DTI metrics in the whole brain across brain samples, detection of morphological differences between brains and quantitative detection of DTI abnormalities in an experimental model of brain injury.
Perhaps the most straightforward use of the template tools provided in this work is to extract quantitative values from a set of MRI maps using common ROI masks. This was accomplished to describe normative FA and Trace values and their variance across brain regions and could be reasonably extended by other studies to examine quantitative MRI values in a consistent manner for experimental models. While template-based tools and atlas masks are relatively recent for pre-clinical MRI, they are commonly used tools of human MRI (Mazziotta et al., 2001) that are widely integrated with image analysis software packages and provide automated anatomical localization brain coordinates, seed regions for connectivity analysis procedures and a means for other types of automatic regional analysis.
Whole-brain, voxelwise analysis of differences in brain structure morphology was accomplished in this study by evaluation of the deformation fields with LogJ analysis for the comparison of an individual female brain to the adult male template. Given that LogJ analysis is part of a well established set of voxelwise volumetric and morphometric tools (Ashburner and Friston, 2003; Ashburner et al., 1998) that are particularly relevant for identification of local shape and volume differences, it is expected to be sensitive to differences arising from experimental treatment or occurring naturally between different groups. LogJ analysis in this study confirmed differences found previously (Sawada et al., 2013; Sawada et al., 2015) between male and female brains by showing relatively lower brain volume in cortical regions and extended these findings to provide greater detail by finer spatial localization and by consideration of all brain voxels. In addition, this study identified regions of increased local volume in the female brain, such as the corpus callosum. This is not meant to provide new biological insight, but to demonstrate the potential use of this voxelwise tool.
Another demonstration of voxelwise analysis in this study was the detection of abnormalities in a single injured brain by registration of the ex-vivo DTI volume to the corresponding template and voxelwise calculation of the Z-score to compare single brain DTI values with the mean and standard deviation of the normal brains used to generate the DTI template. In addition to characterizing abnormal DTI values that are evident qualitatively (i.e. near the injury site), this approach also revealed more subtle abnormalities that may not have been apparent by qualitative visualization alone (i.e. extensive decreased FA in the ipsilateral WM and decreased Trace in the deep cortical GM). Voxelwise analysis has been a long-standing challenge for DTI methods due to fundamental differences in linear and non-linear registration methods, erroneous warping of scalar DTI maps and the effects of smoothing (Jones and Cercignani, 2010). Currently, the most widely used voxelwise analysis tool is tract-based spatial statistics (TBSS, (Smith et al., 2006)), which relies on a search for the highest FA value for each individual by searching perpendicularly to a white matter “skeleton” for the population. With these strategies, TBSS aims to avoid potential misregistration among individuals, but the skeletonization requirements limit the analysis to particular types of white matter pathology and preclude others by selecting only the highest FA values. Recent advances in DTI-specific registration tools have improved significantly our ability of coregistering corresponding anatomical regions among subjects (Irfanoglu et al., 2016) making the need for skeletonization less compelling. In the whole brain analysis employed in this study after DRTAMAS registration of DTI data, plausible results were demonstrated by Z-score analysis to identify TBI-related DTI abnormalities. Notably, this approach may also be extended beyond single brain registration to enable group analysis using voxelwise statistics.
Imaging methods for effective acquisition, pre-processing and registration
During the course of this study, each step of the imaging pipeline – from data acquisition, to image processing to registration algorithms – was optimized for enabling the types of analysis methods described above. Several factors at each step in this process can greatly influence the quality, reliability and reproducibility of the resulting analysis and ultimately the ability to make inferences using these tools in neurobiological study. Given the importance of these factors, several key observations including isotropic voxel dimensions, pre-processing correction and registration algorithms for template building are discussed below.
Whole-brain isotropic image acquisition.
The most effective use of template-based and voxelwise analysis requires that the input volumes have similar resolution in all planes and cover the entire volume. In human MRI and DTI, whole brain coverage and isotropic voxel dimensions are routinely acquired, but pre-clinical DWIs are most often collected using high in-plane resolution and thick slices such that the voxel dimensions are highly anisotropic. This approach can lead not only to partial volume effects, but also to incorrect fitting of the diffusion tensor in anisotropic voxels compared with isotropic voxels having the same volume resulting for example in anisotropy values that are dependent upon the plane of acquisition (Pierpaoli, Chapter 18, (Jones, 2010)). Furthermore, anisotropic voxels result in decreased anatomical information in the slice direction making template generation and the registration of brain volumes more challenging or even impossible. In this study, whole brain coverage and high resolution isotropic voxel dimensions were relatively straightforward to acquire for ex-vivo DWIs by employing a 3D EPI pulse sequence which has improved SNR over a 2D sequence and lower acquisition time than a spin echo pulse sequence. For in-vivo DTI, attaining isotropic voxel dimensions and whole brain coverage was considerably more challenging due to the shorter imaging times and the use of 2DEPI, which is limited for small slice thicknesses by the gradient profile. Also, the constraint of using single segmentation to avoid the effects of motion resulted in increased geometric distortions and longer TE with decreased SNR compared with segmented EPI. This was addressed by reducing the matrix size with the use of saturation bands in non-brain tissue. While the in-vivo DTI images acquired in this work were adequate for template generation and this type of acquisition is suitable for voxelwise studies, higher resolution is desireable and improvements to the quality of in-vivo DWI acquisition should be a goal of future work in the field.
Pre-processing corrections.
Prior to template generation, individual volumes are pre-processed to provide a set of input images that are best suited for registration with one-another and avoid misregistration artifacts. For anatomical image registration consistent segmentation of the brain from surrounding tissue is the most crucial step to ensure proper registration. While brain segmentation or “skull-stripping” tools have been well established for human MRI, their use is not robust for animal studies and the segmentation of the ferret brain is non-trivial, but in this study application of active contour segmentation (Yushkevich et al., 2006) was effective.
DTI data requires a greater amount of pre-processing than structural MRI due to the vulnerability of the EPI pulse sequence to artifacts and distortions (Andersson and Skare, Chapter 17, (Jones, 2010)) and also because the many DWIs collected must be properly aligned with one another for accurate modeling (Pierpaoli Chapter 18, (Jones, 2010)). In this study, the most consequential processing step was correction of geometric distortions by the recently developed “blip-up blip-down” approach of DRBUDDI (Irfanoglu et al., 2015). In particular, this was crucial for correcting severe distortions of the in-vivo DTI data thereby enabling the use of single-segmentation and thin slices in the acquisition protocol. It is notable that the use of a corrective post-processing technique not only improved the quality of the DWI data but also made possible the use of an acquisition scheme with less sensitivity to motion artifacts and dimensions to allow isotropic voxels.
Anatomical MRI and Diffusion tensor -based registration for template building.
Faithful anatomic registration of many brains into a common space has been a longstanding goal and challenge for medical image analysis and considerable improvements have been made in this field as non-linear warping techniques have evolved. In this study, diffeomorphic approaches were selected for template building and individual brain registration based on the past success of these methods for structural image registration (Klein et al., 2009) and their utility in combination with tensor-based registration algorithms (Irfanoglu et al., 2016). The templates that were generated by these methods demonstrated sharp boundaries between tissue types and clear delineation of the brain edges despite averaging across multiple brain volumes. Two aspects of the template building procedures that contributed greatly to the quality of the final template images were the symmetric normalization transformation model which is capable of accommodating large deformations between images (Avants et al., 2008) and the use of iterative averaging algorithms (Avants and Gee, 2004; Avants et al., 2010; Joshi et al., 2004; Wu et al., 2011) which are able to combine many images without introducing anatomical bias by arbitrary selection of template anatomy that can lead to blurring of anatomical boundaries in the resulting template. The DTI registration and template generation algorithms used in this study additionally benefitted from the recently developed DRTAMAS approach to combine scalar and tensor information to improve both global registration and local fiber alignment for white matter tracts (Zhang et al., 2006). This resulted in DTI templates with excellent localization of both gray and white matter anatomical features across all brains, which could enable new observations about ferret brain anatomy in the normal or disordered brain.
Conclusion
This work has provided a set of ferret brain MRI and DTI templates and anatomical ROI masks along with a careful description of the methodological aspects of acquisition, registration, template building and voxelwise analysis. As the availability and quality of small animal imaging systems improves, the ability to harness these tools to describe whole-brain and quantitative experimental outcomes will benefit the study of normal and disordered brain anatomy and microstructure using animal models. In addition to making these templates and ROI masks publically available for download, a web-based viewer was created to provide broad access to a 3D annotated representation of the ferret brain so that other researchers across many disciplines may learn more about ferret brain anatomy or use the annotations as a resource for locating brain structures of particular interest. The long term goal for this work is to maintain and improve these templates though our own efforts and by inviting the larger MRI and ferret neurobiology communities to interact via the website to help generate more detailed “crowd-sourced” annotation and new template volumes to address pertinent areas of ferret brain research including development and disorders.
Acknowledgements
The authors thank the Congressionally Directed Medical Research Programs (CDMRP) for funding this work (award numbers W81XWH-13-2-0019 and W81XWH-13-2-0018) and The Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. (HJF) for administration. We also thank the Center for Neuroscience and Regenerative Medicine (CNRM) for core facility support enabling this work, especially the translational imaging facility, Asamoah Bosomtwi and Alexandru Korotcov for in-vivo MRI support.
Funding source
Congressionally Directed Medical Research Programs. W81XWH-13-2-0019 | Recipient: Carlo Pierpaoli, M.D., Ph.D. and W81XWH-13-2-0018 | Recipient: Sharon Juliano, Ph.D.
Footnotes
The authors declare no relevant conflicts of interest.
A preliminary template was used for initial rigid registration of all structural and DT images. The choice of orientation for this preliminary template and consequently for the templates presented in this paper was guided by a photographic cross-section of the brain in skull from the ferret brainstem atlas of Hawthorn (1985). In a sagittal midline view, the corpus callosum and the dorsal cortical boundary are parallel to one another and parallel to the horizontal axis of the sagittal plane. Orientation in the axial and coronal planes was based on maintaining left-right symmetry. The preliminary templates were sampled to 500 and 250 μm isotropic resolution for use with the in-vivo and ex-vivo images respectively.
References
- Abbah J, Braga MF, Juliano SL, 2014. Targeted disruption of layer 4 during development increases GABAA receptor neurotransmission in the neocortex. J. Neurophysiol 111, 323–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aggarwal M, Zhang J, Miller MI, Sidman RL, Mori S, 2009. Magnetic resonance imaging and micro-computed tomography combined atlas of developing and adult mouse brains for stereotaxic surgery. Neuroscience 162, 1339–1350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ashburner J, Friston K, 2003., Morphometry, Morphometry
- Ashburner J, Hutton C, Frackowiak R, Johnsrude I, Price C, Friston K, 1998. Identifying global anatomical differences: deformation-based morphometry. Hum. Brain Mapp 6, 348–357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants B, Gee JC, 2004. Geodesic estimation for large deformation anatomical shape averaging and interpolation. NeuroImage 23 (Suppl 1), 50. [DOI] [PubMed] [Google Scholar]
- Avants BB, Epstein CL, Grossman M, Gee JC, 2008. Symmetric diffeomorphic image registration with cross-correlation: evaluating automated labeling of elderly and neurodegenerative brain. Med. Image Anal 12, 26–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Yushkevich P, Pluta J, Minkoff D, Korczykowski M, Detre J, Gee JC, 2010. The optimal template effect in hippocampus studies of diseased populations. NeuroImage 49, 2457–2466. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Avants BB, Tustison NJ, Wu J, Cook PA, Gee JC, 2011. An open source multivariate framework for n-tissue segmentation with evaluation on public data. Neuroinformatics 9, 381–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barnette AR, Neil JJ, Kroenke CD, Griffith JL, Epstein AA, Bayly PV, Knutsen AK, Inder TE, 2009. Characterization of brain development in the ferret via MRI. Pediatr. Res 66, 80–84. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Basser PJ, Mattiello J, LeBihan D, 1994. MR diffusion tensor spectroscopy and imaging. Biophys. J 66, 259–267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Black KJ, Snyder AZ, Koller JM, Gado MH, Perlmutter JS, 2001. Template images for nonhuman primate neuroimaging: 1. Baboon. NeuroImage 14, 736–743. [DOI] [PubMed] [Google Scholar]
- Black KJ, Koller JM, Snyder AZ, Perlmutter JS, 2001. Template images for nonhuman primate neuroimaging: 2. Macaque. NeuroImage 14, 744–748. [DOI] [PubMed] [Google Scholar]
- Bock AS, Olavarria JF, Leigland LA, Taber EN, Jespersen SN, Kroenke CD, 2010. Diffusion tensor imaging detects early cerebral cortex abnormalities in neuronal architecture induced by bilateral neonatal enucleation: an experimental model in the ferret. Front. Syst. Neurosci 4, 149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bock AS, Kroenke CD, Taber EN, Olavarria JF, 2012. Retinal input influences the size and corticocortical connectivity of visual cortex during postnatal development in the ferret. J. Comp. Neurol 520, 914–932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calabrese E, Badea A, Coe CL, Lubach GR, Shi Y, Styner MA, Johnson GA, 2015. A diffusion tensor MRI atlas of the postmortem rhesus macaque brain. NeuroImage 117, 408–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calamante F, Tournier J-DD, Jackson GD, Connelly A, 2010. Track-density imaging (TDI): super-resolution white matter imaging using whole-brain track-density mapping. NeuroImage 53, 1233–1243. [DOI] [PubMed] [Google Scholar]
- Calamante F, Tournier J-DD, Smith RE, Connelly A, 2012. A generalised framework for super-resolution track-weighted imaging. NeuroImage 59, 2494–2503. [DOI] [PubMed] [Google Scholar]
- Dorr AE, Lerch JP, Spring S, Kabani N, Henkelman RM, 2008. High resolution three-dimensional brain atlas using an average magnetic resonance image of 40 adult C57Bl/6J mice. NeuroImage 42, 60–69. [DOI] [PubMed] [Google Scholar]
- Empie K, Rangarajan V, Juul SE, 2015. Is the ferret a suitable species for studying perinatal brain injury? Int. J. Dev. Neurosci.: Off. J. Int. Soc. Dev. Neurosci 45, 2–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fox JG, Marini RP, 2014. Biology and diseases of the ferret. Biol. Dis. Ferret [Google Scholar]
- Frey S, Pandya DN, Chakravarty MM, Bailey L, Petrides M, Collins DL, 2011. An MRI based average macaque monkey stereotaxic atlas and space (MNI monkey space). NeuroImage 55, 1435–1442. [DOI] [PubMed] [Google Scholar]
- Hikishima K, Quallo MM, Komaki Y, Yamada M, Kawai K, Momoshima S, Okano HJ, Sasaki E, Tamaoki N, Lemon RN, Iriki A, Okano H, 2011. Population-averaged standard template brain atlas for the common marmoset (Callithrix jacchus). NeuroImage 54, 2741–2749. [DOI] [PubMed] [Google Scholar]
- Hutchinson EB, Schwerin SC, Radomski KL, Irganglu MO, Juliano SL, Pierpaoli CM, 2016. Quantitative MRI and DTI abnormalities during the acute period following CCI in the ferret. Shock 46, 167–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Modi P, Nayak A, Hutchinson EB, Sarlls J, Pierpaoli C, 2015. DRBUDDI (Diffeomorphic Registration for Blip-Up blip-Down Diffusion Imaging) method for correcting echo planar imaging distortions. NeuroImage 106, 284–299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Irfanoglu MO, Nayak A, Jenkins J, Hutchinson EB, Sadeghi N, Thomas CP, Pierpaoli C, 2016. DR-TAMAS: diffeomorphic Registration for Tensor Accurate Alignment of Anatomical Structures. NeuroImage 132, 439–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jackson CA, Peduzzi JD, Hickey TL, 1989. Visual cortex development in the ferret. I. Genesis and migration of visual cortical neurons. J. Neurosci.: Off. J. Soc. Neurosci 9, 1242–1253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jespersen SN, Leigland LA, Cornea A, Kroenke CD, 2012. Determination of axonal and dendritic orientation distributions within the developing cerebral cortex by diffusion tensor imaging. IEEE Trans. Med. imaging 31, 16–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson GA, Badea A, Brandenburg J, Cofer G, Fubara B, Liu S, Nissanov J, 2010. Waxholm space: an image-based reference for coordinating mouse brain research. NeuroImage 53, 365–372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones DK, Cercignani M, 2010. Twenty-five pitfalls in the analysis of diffusion MRI data. NMR Biomed. [DOI] [PubMed] [Google Scholar]
- Jones DK, 2010., Diffusion mri, Diffusion mri
- Joshi S, Davis B, Jomier M, Gerig G, 2004. Unbiased diffeomorphic atlas construction for computational anatomy. NeuroImage 23. [DOI] [PubMed] [Google Scholar]
- Klein A, Andersson J, Ardekani BA, Ashburner J, Avants B, Chiang M-CC, Christensen GE, Collins DL, Gee J, Hellier P, Song JH, Jenkinson M, Lepage C, Rueckert D, Thompson P, Vercauteren T, Woods RP, Mann JJ, Parsey RV, 2009. Evaluation of 14 nonlinear deformation algorithms applied to human brain MRI registration. NeuroImage 46, 786–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen AK, Chang YV, Grimm CM, Phan L, Taber LA, Bayly PV, 2010. A new method to measure cortical growth in the developing brain. J. Biomech. Eng 132, 101004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Knutsen AK, Kroenke CD, Chang YV, Taber LA, Bayly PV, 2013. Spatial and temporal variations of cortical growth during gyrogenesis in the developing ferret brain. Cereb. Cortex (New Y., N. Y.: 1991) 23, 488–498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kou Z, Wu Q, Kou X, Yin C, Wang H, Zuo Z, Zhuo Y, Chen A, Gao S, Wang X, 2015. CRISPR/Cas9-mediated genome engineering of the ferret. Cell Res 25, 1372–1375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroenke CD, Taber EN, Leigland LA, Knutsen AK, Bayly PV, 2009. Regional patterns of cerebral cortical differentiation determined by diffusion tensor MRI. Cereb. Cortex (New Y., N. Y.: 1991) 19, 2916–2929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lighthall JW, 1988. Controlled cortical impact: a new experimental brain injury model. J. neurotrauma 5, 1–15. [DOI] [PubMed] [Google Scholar]
- Mazziotta J, Toga A, Evans A, Fox P, Lancaster J, Zilles K, Woods R, Paus T, Simpson G, Pike B, Holmes C, Collins L, Thompson P, MacDonald D, Iacoboni M, Schormann T, Amunts K, Palomero-Gallagher N, Geyer S, Parsons L, Narr K, Kabani N, Le Goualher G, Boomsma D, Cannon T, Kawashima R, Mazoyer B, 2001. A probabilistic atlas and reference system for the human brain: international Consortium for Brain Mapping (ICBM). Philos. Trans. R. Soc. Lond. Ser. B, Biol. Sci 356, 1293–1322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McLaren DG, Kosmatka KJ, Oakes TR, Kroenke CD, Kohama SG, Matochik JA, Ingram DK, Johnson SC, 2009. A population-average MRI-based atlas collection of the rhesus macaque. NeuroImage 45, 52–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neal J, Takahashi M, Silva M, Tiao G, Walsh CA, Sheen VL, 2007. Insights into the gyrification of developing ferret brain by magnetic resonance imaging. J. Anat 210, 66–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nitzsche B, Frey S, Collins LD, Seeger J, Lobsien D, Dreyer A, Kirsten H, Stoffel MH, Fonov VS, Boltze J, 2015. A stereotaxic, population-averaged T1w ovine brain atlas including cerebral morphology and tissue volumes. Front. Neuroanat 9, 69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noctor SC, Scholnicoff NJ, Juliano SL, 1997. Histogenesis of ferret somatosensory cortex. J. Comp. Neurol 387, 179–193. [PubMed] [Google Scholar]
- Pajevic S, Pierpaoli C, 1999. Color schemes to represent the orientation of anisotropic tissues from diffusion tensor data: application to white matter fiber tract mapping in the human brain. Magn. Reson. Med 42, 526–540. [PubMed] [Google Scholar]
- Papp EA, Leergaard TB, Calabrese E, Johnson GA, Bjaalie JG, 2014. Waxholm Space atlas of the Sprague Dawley rat brain. NeuroImage 97, 374–386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng X, Alföldi J, Gori K, Eisfeld AJ, Tyler SR, Tisoncik-Go J, Brawand D, Law GL, Skunca N, Hatta M, Gasper DJ, Kelly SM, Chang J, Thomas MJ, Johnson J, Berlin AM, Lara M, Russell P, Swofford R, Turner-Maier J, Young S, Hourlier T, Aken B, Searle S, Sun X, Yi Y, Suresh M, Tumpey TM, Siepel A, Wisely SM, Dessimoz C, Kawaoka Y, Birren BW, Lindblad-Toh K, Di Palma F, Engelhardt JF, Palermo RE, Katze MG, 2014. The draft genome sequence of the ferret (Mustela putorius furo) facilitates study of human respiratory disease. Nat. Biotechnol 32, 1250–1255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pierpaoli C, Barnett A, Basser P, Chang L, Koay C, Pajevic S, Rohde G, Sarlls J, Wu M, 2010. TORTOISE: An Integrated Software Package for Processing of Diffusion MRI Data. ISMRM [Google Scholar]
- Poluch S, Juliano SL, 2015. Fine-tuning of neurogenesis is essential for the evolutionary expansion of the cerebral cortex. Cereb. Cortex (New Y., N. Y.: 1991) 25, 346–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poluch S, Jablonska B, Juliano SL, 2008. Alteration of interneuron migration in a ferret model of cortical dysplasia. Cereb. Cortex (New Y., N. Y.: 1991) 18, 78–92. [DOI] [PubMed] [Google Scholar]
- Sanchez-Vives MV, McCormick DA, 2000. Cellular and network mechanisms of rhythmic recurrent activity in neocortex. Nat. Neurosci 3, 1027–1034. [DOI] [PubMed] [Google Scholar]
- Sawada K, Horiuchi-Hirose M, Saito S, Aoki I, 2013. MRI-based morphometric characterizations of sexual dimorphism of the cerebrum of ferrets (Mustela putorius). NeuroImage 83, 294–306. [DOI] [PubMed] [Google Scholar]
- Sawada K, Horiuchi-Hirose M, Saito S, Aoki I, 2015. Sexual dimorphism of sulcal morphology of the ferret cerebrum revealed by MRI-based sulcal surface morphometry. Front. Neuroanat 9, 55. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schwartz TH, Bonhoeffer T, 2001. In vivo optical mapping of epileptic foci and surround inhibition in ferret cerebral cortex. Nat. Med 7, 1063–1067. [DOI] [PubMed] [Google Scholar]
- Schwerin S, Hutchinson E, Ngalula K, Pierpaoli C, Juliano S, 2014. Development of a ferret model of traumatic brain injury; preliminary imaging, behavioral and immunohistochemical findings. Soc. Neurosci. [Google Scholar]
- Shu Y, Hasenstaub A, McCormick DA, 2003. Turning on and off recurrent balanced cortical activity. Nature 423, 288–293. [DOI] [PubMed] [Google Scholar]
- Smith SM, Jenkinson M, Johansen-Berg H, Rueckert D, 2006. Tract-based spatial statistics: voxelwise analysis of multi-subject diffusion data. Neuroimage [DOI] [PubMed] [Google Scholar]
- Tao JD, Barnette AR, Griffith JL, Neil JJ, Inder TE, 2012. Histopathologic correlation with diffusion tensor imaging after chronic hypoxia in the immature ferret. Pediatr. Res 71, 192–198. [DOI] [PubMed] [Google Scholar]
- Tournier JD, Calamante F, Gadian DG, Connelly A, 2004. Direct estimation of the fiber orientation density function from diffusion-weighted MRI data using spherical deconvolution. NeuroImage 23, 1176–1185. [DOI] [PubMed] [Google Scholar]
- Tournier J, Calamante F, Connelly A, 2012. MRtrix: diffusion tractography in crossing fiber regions. Int. J. Imaging Syst. Technol 22, 53–66. [Google Scholar]
- Ullmann JF, Cowin G, Kurniawan ND, Collin SP, 2010. A three-dimensional digital atlas of the zebrafish brain. NeuroImage 51, 76–82. [DOI] [PubMed] [Google Scholar]
- Van Essen DC, Smith SM, Barch DM, Behrens TE, Yacoub E, Ugurbil K, Consortium WUM, 2013. The WU-Minn Human Connectome Project: an overview. NeuroImage 80, 62–79. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu G, Jia H, Wang Q, Shen D, 2011. SharpMean: groupwise registration guided by sharp mean image and tree-based registration. NeuroImage 56, 1968–1981. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Youngblood MW, Chen WC, Mishra AM, Enamandram S, Sanganahalli BG, Motelow JE, Bai HX, Frohlich F, Gribizis A, Lighten A, Hyder F, Blumenfeld H, 2015. Rhythmic 3–4 Hz discharge is insufficient to produce cortical BOLD fMRI decreases in generalized seizures. NeuroImage 109, 368–377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yushkevich PA, Piven J, Hazlett HC, Smith RG, Ho S, 2006. User-guided 3D active contour segmentation of anatomical structures: significantly improved efficiency and reliability. Neuroimage [DOI] [PubMed] [Google Scholar]
- Zhang H, Yushkevich PA, Alexander DC, Gee JC, 2006. Deformable registration of diffusion tensor MR images with explicit orientation optimization. Med. image Anal 10, 764–785. [DOI] [PubMed] [Google Scholar]