Abstract
A number of studies have reported that aberrant expression of microRNAs (miRNAs) closely correlates with the bone metastasis of prostate cancer (PCa). However, clinical significance and functional roles of both strands of a single miRNA in bone metastasis of PCa remain undefined. Here, we reported that miR-582-3p and miR-582-5p expression were simultaneously reduced in bone metastatic PCa tissues compared with non-bone metastatic PCa tissues. Downexpression of miR-582-3p and miR-582-5p strongly and positively correlated with advanced clinicopathological characteristics and shorter bone metastasis-free survival in PCa patients. Upregulating miR-582-3p and miR-582-5p inhibited invasion and migration abilities of PCa cells in vitro, as well as repressed bone metastasis in vivo. Our results further revealed that miR-582-3p and miR-582-5p attenuated bone metastasis of PCa via inhibiting transforming growth factor β (TGF-β) signaling by simultaneously targeting several components of TGF-β signaling, including SMAD2, SMAD4, TGF-β receptor I (TGFBRI), and TGFBRII. Moreover, deletion contributes to miR-582-3p and miR-582-5p downexpression in PCa tissues. Finally, clinical negative correlations of miR-582-3p and miR-582-5p with SMAD2, SMAD4, TGFBRI, and TGFBRII were demonstrated in PCa tissues. Thus, our findings explore a novel tumor-suppressive miRNA with its both strands implicated in bone metastasis of PCa, suggesting its potential therapeutic value in treatment of PCa bone metastasis.
Introduction
Prostate cancer (PCa) is the most frequently diagnosed malignancy and the second leading cause of cancer-associated death in men worldwide.1 Most PCa patients do not die of the primary tumor, but rather due to the metastatic progression of the tumor to distant organs, predominantly to the bone.2 Although local tumors can be well controlled through surgery, hormone therapy, and radiation, metastatic bone tumor is still incurable and contributes to the poor survival time of PCa patients.3 Therefore, early detection and treatment before the tumors colonize the bone are of utmost importance for prognostic and therapeutic purposes in PCa.
It is well recognized that transforming growth factor β (TGF-β) signaling has a complex and sometimes paradoxical role in cancer: in early stages, it inhibits tumor cell proliferation and growth as a tumor suppressor, whereas TGF-β signaling promotes cancer cell invasion and metastasis to several distant organs in later stages, particularly metastasis to bone.4 In bone metastasis of cancer, multiple downstream bone metastasis-associated target genes of the TGF-β signaling pathway are significantly upregulated.5, 6, 7 Importantly, TGF-β signaling has also been demonstrated to play an important role in the bone metastasis of PCa.8, 9, 10 Our previous study has shown that upregulation of Protein Interacting with PRKCA 1 (PICK1), an important negative regulator of TGF-β signaling, suppressed invasion and migration in vitro, as well as bone metastasis in PCa cells in vivo, through repression of TGF-β signaling.11 These studies indicate that activation of TGF-β signaling is crucial for bone metastasis of cancers.
From a molecular perspective, activation of TGF-β signaling starts with TGF-β binding to the TGF-β receptor complex, including the TGF-β receptor types I and II, which further phosphorylates SMAD2 and SMAD3 and promotes their nuclear translocation, where they form a complex with SMAD4 and cooperatively regulate transcription of TGF-β target genes.12, 13 Evidence indicates that disruption of any step may influence the cascade transduction of TGF-β signaling, leading to inactivation of TGF-β signaling. Silencing of the TGF-β receptor I (TGFBRI) attenuated the response of cells to ectopic TGF-β treatment, which further inhibited cell-migratory capacity and recovered the epithelial phenotype.14, 15 Furthermore, Kang et al.5 have demonstrated that Smad4 was indispensable for the induction of IL-11, a gene implicated in bone metastasis of breast cancer in genetic depletion experiments of the mouse model system. However, how these components of TGF-β signaling are simultaneously disrupted in cancers, which commonly repress activity of TGF-β signaling, remains undefined.
By simultaneously exhibiting potent repressive activity on target genes by binding to the 3′ UTR of mRNA, microRNAs (miRNAs) can potentially affect multiple steps of cancer progression and metastasis.16, 17, 18, 19, 20 Here, we found that miR-582-3p and miR-582-5p expression were elevated in PCa tissues compared with the adjacent normal tissues. Strikingly, their expression levels were simultaneously decreased in bone metastatic PCa tissues compared with non-bone metastatic PCa tissues. Downexpression of miR-582-3p and miR-582-5p was correlated with serum prostate-specific antigen (PSA) levels, Gleason grade, and bone metastasis status, as well as predicting shorter bone metastasis-free survival in PCa patients. Upregulating miR-582-3p and miR-582-5p inhibited the invasion, migration abilities, and bone metastasis in PCa cells in vitro and in vivo. Our results further revealed that miR-582-3p and miR-582-5p inhibited bone metastasis of PCa via simultaneously targeting several components of TGF-β signaling, including SMAD2, SMAD4, TGFBRI, and TGFBRII, leading to inactivation of TGF-β signaling. The analysis of clinical correlation reveals that miR-582-3p and miR-582-5p inversely correlated with SMAD2, SMAD4, TGFBRI, and TGFBRII expression in human PCa tissues. Taken together, our results indicate that miR-582-3p and miR-582-5p simultaneously play tumor-suppressive roles in bone metastasis of PCa via inhibiting TGF-β signaling.
Results
Both miR-582-3p and miR-582-5p Expression Are Reduced in Bone-Metastatic PCa Tissues
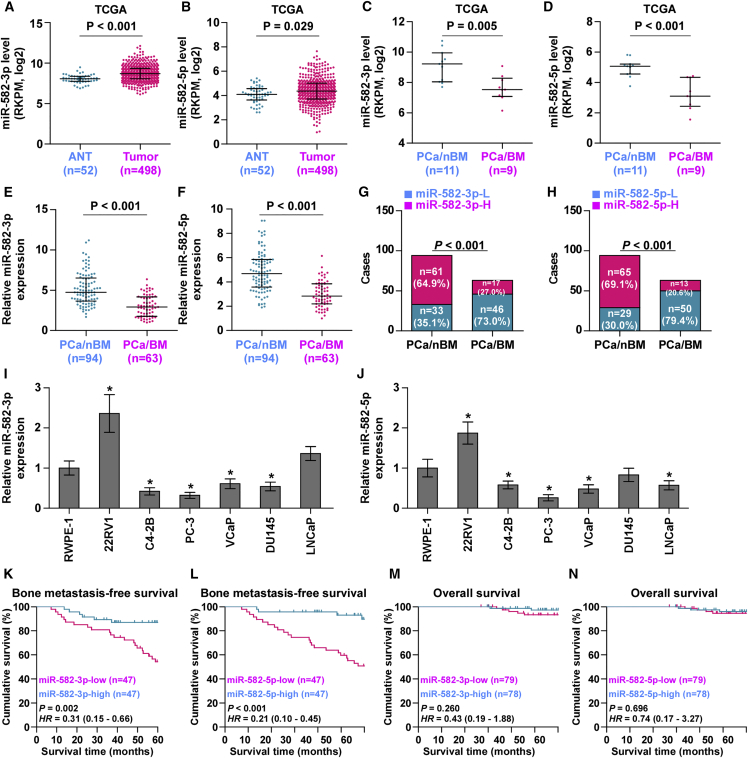
To evaluate the expression levels of miR-582-3p and miR-582-5p in PCa, we first analyzed the miRNA sequencing dataset of PCa from The Cancer Genome Atlas (TCGA) and found that miR-582-3p and miR-582-5p were upregulated in PCa tissues compared with those in the adjacent normal tissues (ANTs) (Figures 1A and 1B; Figures S1A and S1B). Paradoxically, both miR-582-3p and miR-582-5p expression were dramatically downregulated in bone metastatic PCa tissues compared with that in non-bone metastatic PCa tissues (Figures 1C and 1D). Strikingly, miR-582-3p and miR-582-5p expression in bone metastatic PCa tissues were even lower than that in ANTs (Figures S1C and S1D). We further examined the expression levels of miR-582-3p and miR-582-5p in our 157 individual PCa tissues and found that the miR-582-3p and miR-582-5p expression levels in bone metastatic PCa tissues were robustly reduced compared with those in non-bone metastatic PCa tissues and ANTs (Figures 1E and 1F). The percentage of low expression of miR-582-3p and miR-582-5p was remarkably higher in bone metastatic PCa tissues than in non-bone metastatic PCa tissues (Figures 1G and 1H). Interestingly, miR-582-3p and miR-582-5p expression in six paired non-bone metastatic PCa tissues were elevated compared with those in the matched ANTs (Figures S1E and S1F); conversely, miR-582-3p and miR-582-5p expression in four paired bone metastatic PCa tissues were differentially reduced compared with those in the matched ANTs (Figures S1E and S1F). Consistently, miR-582-3p and miR-582-5p expression were dramatically elevated in primary PCa cell 22RV1 compared with those in normal prostate epithelial cells RWPE-1, but were differentially decreased in bone metastatic PCa cell lines (PC-3, C4-2B, and VCaP) (Figures 1I and 1J). Statistical analysis of PCa tissue samples revealed that miR-582-3p and miR-582-5p expression inversely correlated with serum PSA levels, Gleason grade, and bone metastasis status in PCa (Tables S1 and S2). Kaplan-Meier survival analysis indicated that PCa patients with low miR-582-3p or miR-582-5p expression presented shorter bone metastasis-free survival compared with those with high miR-582-3p or miR-582-5p expression (Figures 1K and 1L), but had no significant effect on overall survival (Figures 1M and 1N). Collectively, these results indicate that low expression of miR-582-3p and miR-582-5p is simultaneously implicated in the bone metastasis of PCa.
Figure 1.
miR-582-3p and miR-582-5p Are Downregulated in Bone Metastatic PCa Tissues and Cells
(A and B) miR-582-3p (A) and miR-582-5p (B) expression levels were increased in PCa tissues compared with that in the adjacent normal tissues (ANTs) as assessed by analyzing the PCa miRNA sequencing dataset from TCGA (PCa, n = 498; ANT, n = 52). (C and D) miR-582-3p (C) and miR-582-5p (D) expression levels were reduced in bone metastatic PCa tissues (PCa/BM) compared with that in non-bone metastatic PCa tissues (PCa/nBM) as assessed by analyzing the PCa miRNA sequencing dataset from TCGA (PCa/nBM, n = 11; PCa/BM, n = 9). (E and F) Real-time PCR analysis of (E) miR-582-3p and (F) miR-582-5p expression in 94 non-bone metastatic and 63 bone metastatic PCa samples. Transcript levels were normalized to U6 expression. (G and H) Percentages and number of samples showed high or low (G) miR-582-3p and (H) miR-582-5p expression in bone metastatic and non-bone metastatic PCa tissues in our PCa tissues. (I and J) Real-time PCR analysis of (I) miR-582-3p and (J) miR-582-5p expression levels in normal prostate epithelial cell (RWPE-1), primary PCa cell 22RV1, bone metastatic PCa cell lines (PC-3, C4-2B, and VCaP), brain metastatic cell line DU145, and lymph node metastatic cell line LNCaP. Transcript levels were normalized to U6 expression. *p < 0.05. (K and L) Kaplan-Meier analysis of bone metastasis-free survival curves of the PCa patients stratified by (K) miR-582-3p and (L) miR-582-5p expression. (M and N) Kaplan-Meier analysis of overall survival curves of the PCa patients stratified by (M) miR-582-3p and (N) miR-582-5p expression.
Overexpression of pri-miR-582 Represses Bone Metastasis of PCa Cells In Vivo
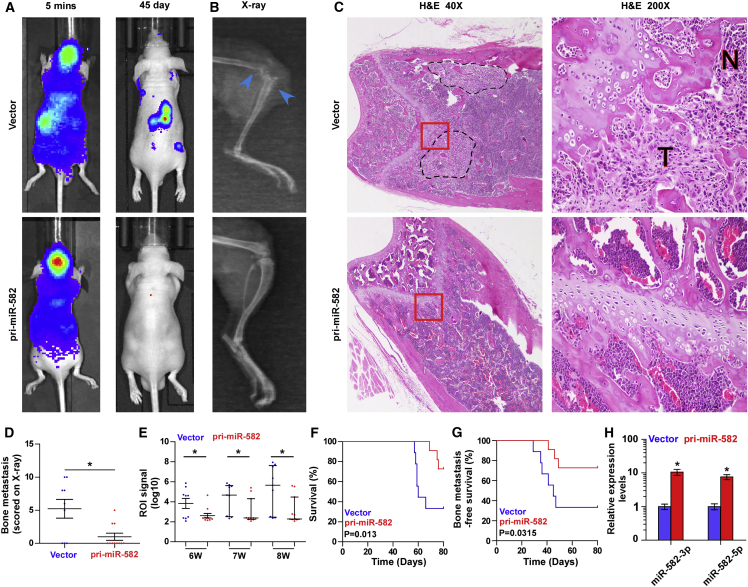
To address whether miR-582-3p and miR-582-5p might play a key role in bone metastasis of PCa, a pri-miR-582 construct containing both sequences of miR-582-3p and miR-582-5p was stably transduced into PCa cells, where miR-582-3p and miR-582-5p expression were dramatically upregulated in PCa cells compared with the vector PCa cells (Figures S2A–S2C). We further examined the effect of miR-582-3p and miR-582-5p on the bone metastasis of PCa using a mouse model of left cardiac ventricle inoculation in vivo, where the luciferase-labeled vector or pri-miR-582-overexpressing PC-3 cells were inoculated, respectively, into the left cardiac ventricle of male nude mice to monitor the progression of bone metastasis by bioluminescence imaging (BLI) and X-ray. As shown in Figures 2A and 2B, upregulating miR-582-3p and miR-582-5p effectively repressed bone metastasis ability compared with the vector group by X-ray and BLI. H&E staining of the tumor sections from the indicated tibias of injected mice revealed that upregulating miR-582-3p and miR-582-5p reduced the tumor burden in bone (Figure 2C). Moreover, cells transduced with pri-miR-582 exhibited fewer bone metastatic sites and smaller osteolytic area of metastatic tumors, as well as longer survival and bone metastasis-free survival compared with the control group (Figures 2D–2G). The miR-582-3p and miR-582-5p expression levels were dramatically upregulated in the tumor tissues of the mice inoculated with PC-3 cells stably transfected with pri-miR-582 compared with those in the vector at the end of the experiments (Figure 2H). Furthermore, a gene set enrichment analysis (GSEA) of miR-582-3p and miR-582-5p expression levels based on miRNA expression data of PCa from TCGA was performed, and the results showed that low expression of miR-582-3p and miR-582-5p positively correlated with metastatic propensity (Figures S2D–S2I). Invasion and migration assays showed that upregulating miR-582-3p and miR-582-5p inhibited invasion and migration abilities of PCa cells (Figure S2J). Taken together, these findings demonstrate that upregulating miR-582-3p and miR-582-5p inhibits the bone metastasis of PCa in vivo.
Figure 2.
Upregulation of miR-582-3p and miR-582-5p Represses Bone Metastasis of PC-3 Cells In Vivo
(A) Representative BLIs signal of bone metastasis of a mouse from the vector or pri-miR-582 groups of mice at 5 min and 45 days, respectively. (B) Representative radiographic images of bone metastases in the indicated mice (blue arrows indicate osteolytic lesions). (C) Representative H&E-stained sections of tibias from the indicated mouse. (D) The sum of bone metastasis scores for each mouse in tumor-bearing mice inoculated with vector (n = 9) or pri-miR-582 (n = 11) cells. (E) Quantification of the ROI signaling in the vector and pri-miR-582 groups at 6, 7, and 8 weeks, respectively. (F and G) Kaplan-Meier analysis of mouse survival (F) and bone metastasis-free survival (G) in the vector and pri-miR-582 groups. (H) Real-time PCR analysis of miR-582-3p and miR-582-5p expression in the tumor tissues of the mice inoculated with PC-3 cells stably transfected with pri-miR-582 compared with those in the vector at the end of the experiments. Transcript levels were normalized by U6 expression. Error bars represent the mean ± SD of three independent experiments. *p < 0.05.
Ectopic Expression of miR-582-3p or miR-582-5p Represses Migration and Invasion Abilities in PCa Cells
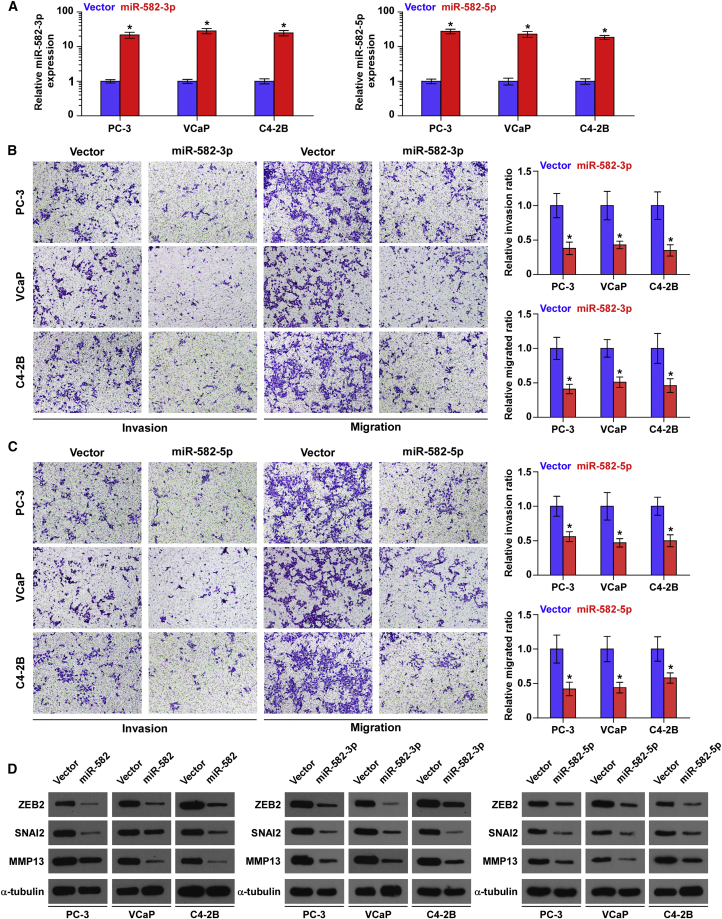
We further evaluated the effects of miR-582-3p or miR-582-5p on invasion and migration abilities in PCa cells via transfecting miR-582-3p or miR-582-5p mimics, respectively (Figure 3A). As shown in Figure 3B, upregulating miR-582-3p attenuated the invasion and migration abilities of PCa cells. Consistently, upregulating miR-582-5p reduced the invasion and migration abilities of PCa cells (Figure 3C). Furthermore, upregulating miR-582-3p or miR-582-5p, or both, differentially inhibited the expression of several TGF-β signaling downstream genes that regulate epithelial-mesenchymal-transition (EMT) or metastasis and/or invasion in PCa cells, such as ZEB2, SNAI2, and MMP13 (Figure 3D). These results indicate that the presence of either miR-582-3p or miR-582-5p is sufficient to repress tumor invasiveness in PCa cells.
Figure 3.
Upregulation of miR-582-3p or miR-582-5p Represses Invasion and Migration Abilities of PCa Cells In Vitro
(A) Real-time PCR analysis of miR-582-3p or miR-582-5p expression in PCa cells transduced with miR-582-3p or miR-582-5p mimics, respectively, compared with the vector controls. Transcript levels were normalized by U6 expression. Error bars represent the mean ± SD of three independent experiments. (B) Overexpression of miR-582-3p inhibited invasion and migration abilities in PCa cells. Error bars represent the mean ± SD of three independent experiments. (C) Overexpression of miR-582-5p inhibited invasion and migration abilities in PCa cells. Error bars represent the mean ± SD of three independent experiments. (D) Western blotting of ZEB2, SNAI2, and MMP13 expression in the indicated PCa cells. α-Tubulin served as the loading control. *p < 0.05.
miR-582-3p and miR-582-5p Inhibit TGF-β Signaling Activity
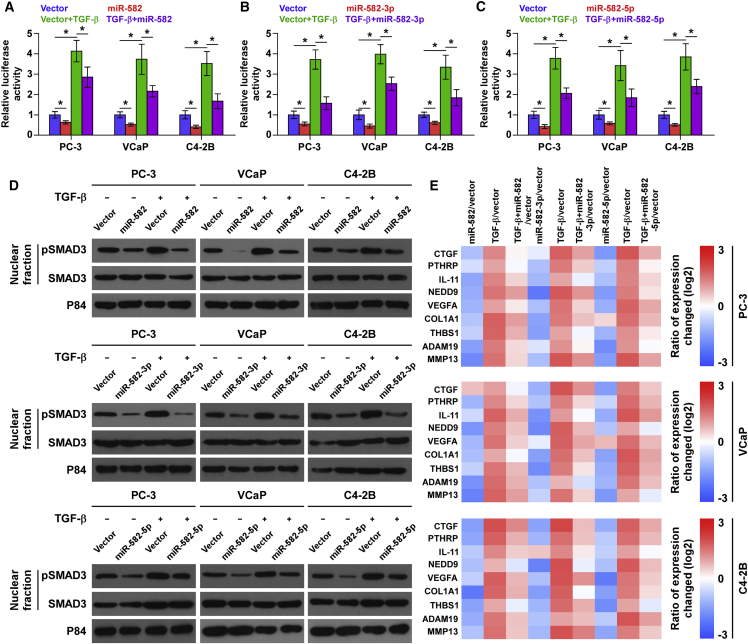
To determine the underlying mechanism of the inhibitory role of miR-582-3p and miR-582-5p in the bone metastasis PCa, GSEA analysis of miR-582-3p and miR-582-5p expression against the oncogenic signatures collection of the Molecular Signatures Database (MSigDB) was performed, and the result showed that low expression of miR-582-3p and miR-582-5p significantly and positively correlated with TGF-β signaling (Figures S3A–S3F). These results suggest that miR-582-3p and miR-582-5p may negatively regulate the activity of TGF-β signaling that has been reported to play a crucial role in bone metastasis in a variety of cancers.8, 11 Therefore, we further investigated the effects of miR-582-3p or miR-582-5p, or both, on the activity of TGF-β signaling in PCa cells. As shown in Figures 4A–4C, we found that upregulating miR-582-3p or miR-582-5p, or both, suppressed the transcriptional activity of the TGF-β/Smad-responsive luciferase reporter plasmid CAGA12, which consists of 12 tandem copies of the Smad/DNA binding motif CAGAC, in the presence or absence of ectopic TGF-β. Cellular fractionation and western blotting analysis revealed that upregulating miR-582-3p or miR-582-5p, or both, reduced pSMAD3 nuclear translocation in PCa cells in the presence or absence of ectopic TGF-β (Figure 4D). Moreover, real-time PCR analysis showed that upregulating miR-582-3p or miR-582-5p, or both, repressed multiple downstream bone metastasis-related genes of the TGF-β pathway in both the absence and presence of ectopic TGF-β (Figure 4E). The miR-582-3p or miR-582-5p expression level was not affected by TGF-β treatment (Figures S3G and S3H). Thus, these results demonstrate that miR-582-3p and miR-582-5p inhibit TGF-β signaling activity in PCa cells.
Figure 4.
miR-582-3p and miR-582-5p Repress TGF-β Signaling Activity in PCa Cells
(A–C) Upregulation of miR-582-3p (A), miR-582-5p (B), or both (C) attenuated transcriptional activity based on a TGF-β/Smad-responsive luciferase reporter in the absence or presence of TGF-β (5 ng/mL). *p < 0.05. (D) Western blot analysis showed that upregulation of miR-582-3p, miR-582-5p, or both decreased nuclear translocation of pSMAD3 in PCa cells in the absence or presence of TGF-β (5 ng/mL). The nuclear protein p84 was used as a nuclear protein marker. (E) Upregulation of miR-582-3p, miR-582-5p, or both reduced downstream bone metastasis-related genes of the TGF-β pathway, including CTGF, PTHRP, IL-11, NEDD9, MMP13, ADAM19, THBS1, COL1A1, and VEGFA, in the absence or presence of TGF-β (5 ng/mL). Transcript levels were normalized to GAPDH expression. Error bars represent the mean ± SD of three independent experiments.
miR-582-3p and miR-582-5p Target Several Components of TGF-β Signaling
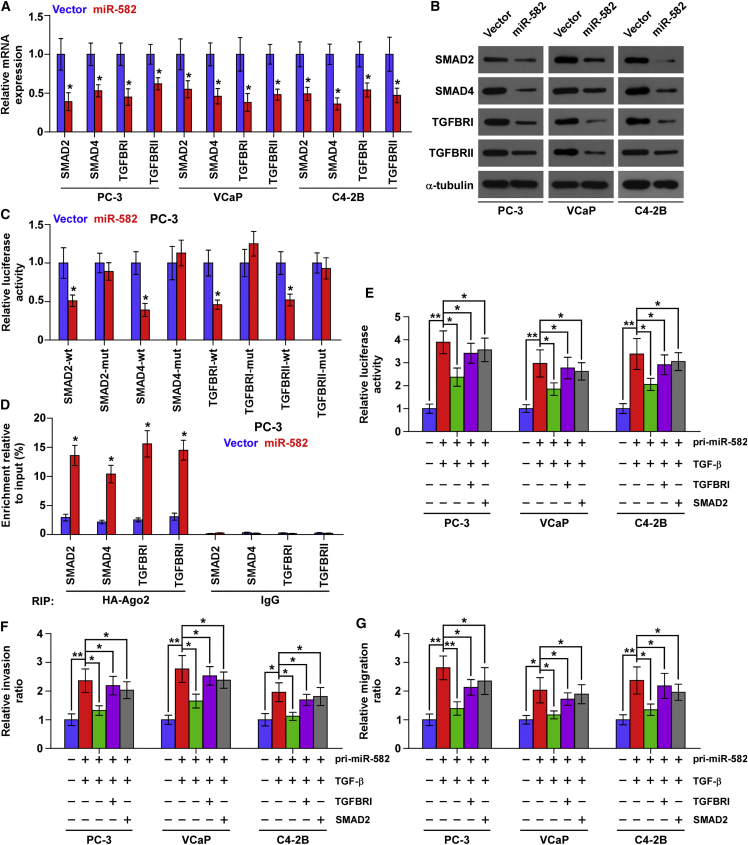
Through analyzing the publicly available algorithms, including TargetScan, miRWalk, and miRanda, we found that several components of TGF-β signaling, including SMAD2, SMAD4, and TGFBRI, may be potential targets of miR-582-3p, and SMAD2, SMAD4, TGFBRI, and TGFBRII for miR-582-5p (Figures S4A–S4C). Real-time PCR and western blotting analysis revealed that simultaneously upregulating miR-582-3p and miR-582-5p reduced the expression levels of SMAD2, SMAD4, TGFBRI, and TGFBRII (Figures 5A and 5B). However, upregulating miR-582-3p reduced the expression levels of SMAD2, SMAD4, and TGFBRI, but not of TGFBRII, in PCa cells (Figures S5A and S5B); upregulating miR-582-5p reduced the expression levels of SMAD2, TGFBRI, and TGFBRII, but not of SMAD4, in PCa cells (Figures S5C and S5D). Luciferase assay revealed that upregulating miR-582-3p repressed the reporter activity in the 3′ UTRs of SMAD2, SMAD4, and TGFBRI, but not in TGFBRII; Figures S6A–S6C); upregulating miR-582-5p repressed the reporter activity in the 3′ UTRs of SMAD2, TGFBRI, and TGFBRII, but not in SMAD4 (Figures S6D–S6F); upregulating both miR-582-3p and miR-582-5p simultaneously reduced the reporter activity in the 3′ UTRs of SMAD2, SMAD4, TGFBRI, and TGFBRII (Figure 5C; Figures S6G and S6H). However, upregulating miR-582-3p or miR-582-5p, or both, had no significant effect on the mutant 3′ UTR of SMAD2, SMAD4, TGFBRI, and TGFBRII within the miR-582-3p and miR-582-5p-binding seed regions in PCa cells (Figure 5C; Figures S6A–S6H). miRNA immunoprecipitation (RIP) assay showed a direct association of miR-582-3p with SMAD2, SMAD4, and TGFBRI transcripts (Figures S7A–S7C), miR-582-5p with SMAD2, TGFBRI, and TGFBRII transcripts (Figures S7D–S7F), and both with SMAD2, SMAD4, TGFBRI, and TGFBRII (Figure 5D; Figures S7G and S7H). Therefore, our results demonstrate that miR-582-3p and miR-582-5p directly target SMAD2, SMAD4, TGFBRI, and TGFBRII in PCa cells.
Figure 5.
miR-582-3p and miR-582-5p Target Several Components of TGF-β Signaling
(A) Real-time PCR analysis of SMAD2, SMAD4, TGFBRI, and TGFBRII expression in the pri-miR-582-transfecting or vector PCa cells. Transcript levels were normalized by GAPDH expression. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (B) Western blotting of SMAD2, SMAD4, TGFBRI, and TGFBRII expression in the pri-miR-582-transfecting or vector PCa cells. α-Tubulin served as the loading control. (C) Luciferase assay of the cells transfected with pmirGLO-3′ UTR reporter of SMAD2, SMAD4, TGFBRI, and TGFBRII in the pri-miR-582-transfecting or vector PC-3 cells. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (D) Microribunocleoprotein (miRNP) IP assay showing the association between miR-582-3p and miR-582-5p and SMAD2, SMAD4, TGFBRI, and TGFBRII transcripts in the pri-miR-582-transfecting or vector PC-3 cells. Pulldown of IgG antibody served as the negative control. Error bars represent the mean ± SD of three independent experiments. *p < 0.05. (E–G) Upregulating TGFBRI or SMAD2 partially rescued the (E) TGF-β signaling activity, (F) invasion, and (G) migration abilities in the pri-miR-582-transfecting PCa cells in the presence of TGF-β (5 ng/mL). *p < 0.05; **p < 0.01.
TGF-β Signaling Activity Restores Invasion and Migration Abilities in miR-582-3p and miR-582-5p-Overexpressing PCa Cells
Next, we further investigated whether TGF-β signaling activity enhances invasion and migration abilities repressed by miR-582-3p and miR-582-5p overexpression via upregulating TGFBRI or SMAD2 in PCa cells. Our results showed that upregulating TGFBRI or SMAD2 did not restore TGF-β signaling activity, invasion, and migration abilities in miR-582-transfected PCa cells in the absence of TGF-β (Figures S8A–S8C). This finding may be explained by that after binding of TGF-β to the TGF-β receptor complex, phosphorylation of SMAD2 and SMAD3 is a critical step for their nuclear translocation, where they form a complex with SMAD4 and cooperatively regulate transcription of TGF-β target genes. Therefore, it is conceivable that without TGF-β stimulation, only upregulating the components of TGF-β signaling, such as TGFBRI or SMAD2, does not affect the phosphorylation levels of SMAD2 and SMAD3, and thereby does not affect the TGF-β signaling activity. So, we further evaluated the effects of TGFBRI or SMAD2 overexpression on TGF-β signaling activity, invasion, and migration abilities in miR-582-transfected PCa cells in the presence of ectopic TGF-β. As shown in Figures 5E–5G, upregulating TGFBRI or SMAD2 enhanced the TGF-β signaling activity, invasion, and migration abilities in miR-582-transfected PCa cells in the presence of ectopic TGF-β. Thus, these results reveal that miR-582-3p and miR-582-5p repress invasion and migration abilities via inhibiting TGF-β signaling in PCa cells.
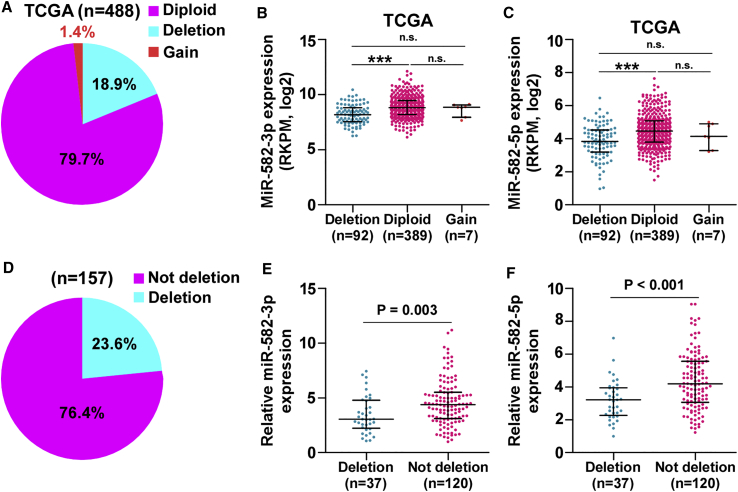
Deletion Contributes to miR-582-3p and miR-582-5p Downexpression in PCa Tissues
To further explore the underlying mechanism of miR-582-3p and miR-582-5p downexpression in PCa tissues, we analyzed the miRNAs PCa dataset from TCGA and found that deletion occurred in 18.9% of PCa tissues (Figure 6A). The expression levels of miR-582-3p and miR-582-5p in PCa tissues with deletion were dramatically reduced compared with those without deletion (Figures 6B and 6C). We further measured the deletion levels in our own PCa tissues and found that deletion was found in 37/157 PCa tissues (23.6%) (Figure 6D). Furthermore, the expression levels of miR-582-3p and miR-582-5p in PCa tissues with the deletion were robustly downregulated (Figures 6E and 6F). Thus, these results indicate that deletion is involved in miR-582-3p and miR-582-5p downexpression in a portion of PCa tissues.
Figure 6.
Deletion Is Implicated in Downexpression of miR-582-3p and miR-582-5p in PCa Tissues
(A) Deletion, diploid, and gains levels of MIR582 in the PCa dataset from TCGA (deletion: n = 92; diploid: n = 389; gains = 7). (B and C) The average expression levels of (B) miR-582-3p and (C) miR-582-5p in PCa tissues with deletion was lower than those without deletion in the PCa dataset from TCGA. Each bar represents the median values ± quartile values. ***p < 0.001. (D) Deletion levels of MIR582 in our PCa samples (deletion: n = 37; not deletion: n = 120). (E and F) The average expression levels of (E) miR-582-3p and (F) miR-582-5p in PCa tissues with deletion were lower than those without deletion in our PCa samples. Each bar represents the median values ± quartile values.
Clinical Correlation of miR-582-3p and miR-582-5p with TGFBRI, TGFBRII, SMAD2, and SMAD4 in Human PCa Tissues
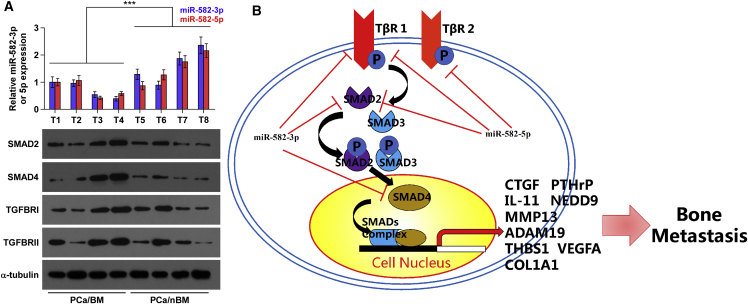
Finally, the clinical significance of miR-582-3p and miR-582-5p with TGFBRI, TGFBRII, SMAD2, and SMAD4 was examined in clinical PCa tissues. As shown in Figure 7A, miR-582-3p and miR-582-5p expression in bone metastatic PCa tissues (T1–4) were reduced compared with that in non-bone metastatic PCa tissues (T5–8). Conversely, protein expression of TGFBRI, TGFBRII, SMAD2, and SMAD4 were elevated in bone metastatic PCa tissues (Figure 7A). Pearson analysis revealed that miR-582-3p and miR-582-5p expression inversely correlated with SMAD2, SMAD4, TGFBRI, and TGFBRII (Figures S9A–S9H). Interestingly, the negative correlations of miR-582-3p with TGFBRII (r = −0.828, p < 0.05; Figure S9D) and miR-582-5p with SMAD4 (r = −0.711, p < 0.05; Figure S9F) were observed in PCa tissues. This finding did not mean that miR-582-3p directly inhibited TGFBRII expression and miR-582-5p directly inhibited SMAD4 expression in PCa tissues. It may be due to downregulation of TGFBRII repressed by miR-582-5p and downregulation of SMAD4 repressed by miR-582-3p in PCa tissue samples. Taken together, our results indicate that expression levels of miR-582-3p and miR-582-5p are negatively associated with TGFBRI, TGFBRII, SMAD2, and SMAD4 in clinical PCa tissues.
Figure 7.
Clinical Relevance of miR-582-3p and miR-582-5p with SMAD2, SMAD4, TGFBRI, and TGFBRII in Human PCa Tissues
(A) Analysis of miR-582-3p and miR-582-5p expression with SMAD2, SMAD4, TGFBRI, and TGFBRII in four bone metastatic PCa tissues (T1–4) and four non-bone metastatic PCa tissues (T5–8). U6 was used as the control for miRNA loading. miR-582-3p or miR-582-5p expression levels were normalized to that of miR-582-3p or miR-582-5p expression of T1. Each bar represents the mean ± SD of three independent experiments. Loading controls were α-tubulin and p84 for the cytoplasmic and nuclear fractions. ***p < 0.001. (B) Hypothetical model illustrating that miR-582-3p and miR-582-5p repress bone metastasis of PCa via inhibiting the TGF-β signaling pathway by directly targeting SMAD2, SMAD4, TGFBRI, and TGFBRII.
Discussion
Evidence has shown that TGF-β signaling functions as an oncogenic or tumor-suppressive pathway depending on the developmental stage of tumor: in early stages, TGF-β signaling inhibits cell growth and is a tumor suppressor, whereas in later stages, TGF-β signaling promotes invasion and metastasis, in particular, metastasis to bone.4, 21 Furthermore, TGF-β signaling has also played a paradoxical role in PCa. In the early stage of PCa, TGF-β signaling functions as a tumor-suppressive signaling to inhibit PCa cell proliferation and growth via inducing growth barrier.22 However, in a later stage of PCa, TGF-β signaling induces a pro-bone metastasis program in PCa cells.8, 23, 24 This interesting finding well explains the discrepancy of expression pattern of miR-582-3p and miR-582-5p in primary PCa tissues and cells compared with ANTs and normal prostate epithelial cells, and in bone metastatic PCa tissues and cells compared with primary PCa tissues and cells shown in this study.
Activity of TGF-β signaling has been demonstrated to be pivotal in bone metastasis of PCa.8, 11 Accumulating evidence has shown that miRNAs can affect TGF-β signaling activity via negative regulation of the components of TGF-β signaling. Inhibition of the TGFBRI or TGFBRII by different miRNAs induced mesenchymal epithelial transition, and it represses cell-migratory capacity and stemness in cancer cells.25, 26, 27, 28 Wang et al.29 reported that ectopic expression of miR-323-3p inhibited invasion and metastasis of pancreatic ductal adenocarcinoma cells via directly targeting SMAD2 and SMAD3, both key components of TGF-β signaling. In addition, the upregulation of miRNA-146a suppressed proliferation and migration of glioblastoma cells via reducing SMAD4 expression, leading to inactivation of TGF-β signaling.30 However, how these components of TGF-β signaling are simultaneously disrupted by miRNA in cancers, which commonly represses activity of TGF-β signaling, remains undefined. In this study, we found that miR-582-3p targeted SMAD2, SMAD4, and TGFBRI, and miR-582-5p directly inhibited SMAD2, TGFBRI, and TGFBRII. Importantly, ectopic expression of miR-582-3p or miR-582-5p, or both, dramatically repressed TGF-β signaling activity in PCa cells, indicating that miR-582-3p and miR-582-5p inhibit the TGF-β signaling pathway via simultaneously targeting multiple components of TGF-β signaling. Therefore, our results unveil a novel mechanism for constitutive activation of TGF-β signaling in bone metastasis of PCa.
As one of the originally identified miRNAs, aberrant expression of miR-582-3p and miR-582-5p has been reported to be implicated in several diseases, including drug addiction,31 hyperlipidemia and hepatic insulin resistance,32 cerebral amyloid angiopathy,33 amyotrophic lateral sclerosis,34 tuberculosis,35 and ischemia and/or reperfusion injury.36 In cancers, numerous studies have shown that miR-582-3p and miR-582-5p function as oncogenic or tumor-suppressive miRNA dependent on tumor types: miR-582-3p was upregulated in colon cancer,37 non-small cell lung carcinoma,38 and non-functioning pituitary adenomas,39 but reduced in Hodgkin’s lymphoma;40 however, miR-582-5p expression was elevated in colorectal cancer,41 castration-resistant PCa,42 oral cancer,43 and non-functioning pituitary adenomas,39 but decreased in hepatocellular carcinoma44 and bladder cancer.45 However, the clinical significance and functional roles of miR-582-3p and miR-582-5p in bone metastasis of PCa remain not reported. In this study, our results revealed that miR-582-3p and miR-582-5p were upregulated in PCa tissues compared with ANTs. Strikingly, miR-582-3p and miR-582-5p expression levels were simultaneously reduced in bone metastatic PCa tissues, even lower than ANTs. Furthermore, low expression of miR-582-3p and miR-582-5p strongly and positively correlated with advanced clinicopathological characteristics and poor bone metastasis-free survival. Gain-of-function assays demonstrated that upregulation of miR-582-3p and miR-582-5p inhibited invasion and bone metastasis via simultaneously targeting multiple components of TGF-β signaling in PCa cells. Thus, our results demonstrate that miR-582-3p and miR-582-5p both play tumor-suppressive roles in bone metastasis of PCa. Interestingly, several lines of evidence have reported that miR-582-3p and miR-582-5p simultaneously act as oncogene or tumor suppressor in different types of cancer. In colorectal cancer and non-functioning pituitary adenomas, miR-582-3p and miR-582-5p expression were simultaneously upregulated and function as oncogenic miRNA.37, 39, 41 Conversely, miR-582-3p and miR-582-5p expression have been reported to be strongly decreased in bladder cancer tissues, in which they played tumor-suppressive roles.45 These studies in combination with ours suggest that miR-582-3p and miR-582-5p play similar roles in the same tumor type. This may be true because no studies in the literature have yet reported the opposite roles of miR-582-3p and miR-582-5p in the same tumor type.
miRNA-based preclinical and clinical trials have grown over the past several years and presented favorable prospects in cancer.46, 47 For example, Cai et al.48 reported that miR-128-3p sponge and antagomir strikingly diminished primary tumor growth and spontaneous lung metastasis of the subcutaneously inoculated or intravenously injected A549; furthermore, Liu et al.’s49 study has shown that upregulation of miR-141 via lentiviral infection dramatically reduced primary tumors and lung metastasis. These studies indicate that a strategy targeting miRNAs is an effective therapeutic avenue in the treatment of cancer. In this study, our results demonstrated that upregulation of miR-582-3p and miR-582-5p effectively inhibited bone metastasis of PCa cells in a mouse model of bone metastasis in vivo. Therefore, our results provide evidence for the potential therapeutic efficacy of a signal miRNA with its both strands in bone metastasis of PCa.
Interestingly, a study by Qin et al.50 has reported that expression levels of miR-582 in the serum of patients yielded an excellent AUC (the areas under the receiver operating characteristic [ROC] curve) in discriminating deep vein thrombosis from the healthy controls. This finding suggested that serum miR-582 holds great potential applicable value as a minimally invasive diagnostic marker. However, there is no definitive evidence as yet to investigate the role of serum miR-582-3p or miR-582-5p level in the early diagnosis of cancer, which will require careful controlled studies in large numbers of cancer patients. As well, it is clinically significant to determine whether serum levels of miR-582-3p or miR-582-5p, or both, together could serve as a diagnostic marker in bone metastasis of PCa.
In summary, our results demonstrate that miR-582-3p and miR-582-5p inhibit bone metastasis of PCa via simultaneously repressing multiple components of TGF-β signaling, resulting in the inactivation of TGF-β signaling. Furthermore, our findings first provide evidence of a novel regulatory pathway of both strands of tumor-suppressive miR-582 in bone metastasis of PCa. Therefore, in-depth understanding of the molecular mechanisms of miR-582 in bone metastasis of PCa will provide novel insights to facilitate the development of anti-bone metastasis therapeutic strategies against PCa.
Materials and Methods
Cell Culture
The human PCa cell lines 22RV1, PC-3, VCaP, DU145, and LNCaP and normal prostate epithelial cells RWPE-1 were obtained from the Shanghai Chinese Academy of Sciences cell bank (China). RWPE-1 cells were grown in defined keratinocyte-serum-free medium (SFM) (1×) (Invitrogen). PC-3, LNCaP, and 22Rv1 cells were cultured in RPMI-1640 medium (Life Technologies, Carlsbad, CA, USA) supplemented with penicillin G (100 U/mL), streptomycin (100 mg/mL), and 10% fetal bovine serum (FBS; Life Technologies). DU145 and VCaP cells were grown in DMEM (Invitrogen) supplemented with 10% FBS. The C4-2B cell line was purchased from the MD Anderson Cancer Center and maintained in T-medium (Invitrogen) supplemented with 10% FBS.51 All cell lines were grown under a humidified atmosphere of 5% CO2 at 37°C.
Plasmid, Small Interfering RNA, and Transfection
The human pri-miR-582 gene was PCR-amplified from genomic DNA and cloned into a pMSCV-puro retroviral vector (Clontech, Japan). The (CAGAC) 12/pGL3 TGF-β/Smad-responsive luciferase reporter plasmid and control plasmids (Clontech, Japan) were used to quantitatively assess the transcriptional activity of TGF-β signaling components. The 3′ UTRs of SMAD2, SMAD4, TGFBRI, and TGFBRII were PCR-amplified from genomic DNA and cloned into pmirGLO vectors (Promega, USA), and the list of primers used in cloning reactions is presented in Table S3. miR-582-3p mimics, miR-582-5p mimic, the plasmids for the TGFBRI and SMAD2 overexpression, and the corresponding vector control were synthesized and purified by RiboBio. Transfection of miRNA and plasmids was performed using Lipofectamine 3000 (Life Technologies, USA) according to the manufacturer’s instructions.
RNA Extraction, Reverse Transcription, and Real-Time PCR
Total RNA from tissues or cells was extracted using the RNA Isolation Kit (QIAGEN, USA) according to the manufacturer’s instructions. mRNA and miRNA were reverse transcribed from total mRNA using the RevertAid First Strand cDNA Synthesis Kit (Thermo Fisher, USA) according to the manufacturer’s protocol. cDNA was amplified and quantified on the CFX96 system (Bio-Rad, USA) using iQ SYBR Green (Bio-Rad, USA). The primers are provided in Table S4. Real-time PCR was performed according to a standard method, as described previously.52 Primers for U6, miR-582-3p, and miR-582-5p were synthesized and purified by RiboBio (Guangzhou, China). U6 or glyceraldehyde-3-phosphate dehydrogenase (GAPDH) was used as the endogenous controls. Relative fold expressions were calculated with the comparative threshold cycle (2−ΔΔCt) method.
Patients and Tumor Tissues
A total of 157 archived PCa tissues, including 94 non-bone metastatic PCa tissues and 63 bone metastatic PCa tissues, were obtained during surgery or needle biopsy at the Second Affiliated Hospital of Guangzhou Medical University (Guangdong, China) between January 2008 and October 2012. Patients were diagnosed based on clinical and pathological evidence, and the specimens were immediately snap-frozen and stored in liquid nitrogen tanks. For the use of these clinical materials for research purposes, prior patient consents and approval from the Institutional Research Ethics Committee were obtained. The clinicopathological features of the patients are summarized in Table S5. The median of miR-582-3p or miR-582-5p expression in PCa tissues was used to stratify high and low expression of miR-582-3p or miR-582-5p, respectively.
Western Blotting
Nuclear and/or cytoplasmic fractionation was separated using the Cell Fractionation Kit (Cell Signaling Technology, USA) according to the manufacturer’s instructions, and the whole-cell lysates were extracted with radio-immunoprecipitation assay (RIPA) Buffer (Cell Signaling Technology). Western blotting was performed according to a standard method, as described previously.53 Antibodies against SMAD2, SMAD4, TGFBRI, TGFBRII, pSMAD3, and SMAD3 were purchased from Cell Signaling Technology, and p84 from Invitrogen. The membranes were stripped and reprobed with an anti-α-tubulin antibody (Sigma-Aldrich, USA) as the loading control.
Luciferase Assay
Cells (4 × 104) were seeded in triplicate in 24-well plates and cultured for 24 h, and luciferase assay was performed as previously described.54 Cells were transfected with 250 ng (CAGAC) of 12/pGL3 reporter luciferase plasmid, or pmirGLO-SMAD2-3′ UTR, pmirGLO-SMAD4-3′ UTR, pmirGLO-TGFBRI-3′ UTR, or pmirGLO-TGFBRII-3′ UTR luciferase plasmid, plus 5 ng of pRL-TK the Renilla plasmid (Promega) using Lipofectamine 3000 (Invitrogen) according to the manufacturer’s recommendations. Luciferase and Renilla signals were measured 36 h after transfection using a Dual Luciferase Reporter Assay Kit (Promega) according to the manufacturer’s protocol.
miRNA Immunoprecipitation
Cells were co-transfected with human influenza hemagglutinin (HA)-Ago2, followed by HA-Ago2 immunoprecipitation using anti-HA-antibody. Real-time PCR analysis of the immunoprecipitation (IP) material was performed to test the association of the mRNA of SMAD2, SMAD4, TGFBRI, and TGFBRII with the RNA-induced silencing complex (RISC). The specific processes were performed as previously described.55
Invasion and Migration Assays
The invasion and migration assays were performed using a Transwell chamber consisting of 8-mm membrane filter inserts (Corning) with or without coated Matrigel (BD Biosciences), respectively, as described previously.56 In brief, the cells were trypsinized and suspended in serum-free medium. Then, 1.5 × 105 cells were added to the upper chamber, and the lower chamber was filled with the culture medium supplemented with 10% FBS. After incubation for 24–48 h, cells passed through the coated membrane to the lower surface, where cells were fixed with 4% paraformaldehyde and stained with hematoxylin. The cell count was performed under a microscope (×100).
Animal Study
All mouse experiments were approved by The Institutional Animal Care and Use Committee of Sun Yat-sen University, and the approval no. was L102012016110D. For the bone metastasis study, BALB/c-nu mice (5–6 weeks old, 18–20 g) were anaesthetized and inoculated into the left cardiac ventricle with 1 × 105 PC-3 cells in 100 μL of PBS. Bone metastases were monitored by bioluminescent imaging (BLI) as previously described.57 Osteolytic lesions were identified on radiographs as radiolucent lesions in the bone. The area of the osteolytic lesions was measured using the MetaMorph image analysis system and software (Universal Imaging Corporation), and the total extent of bone destruction per animal was expressed in square millimeters. Each bone metastasis was scored based on the following criteria: 0, no metastasis; 1, bone lesion covering <1/4 of the bone width; 2, bone lesion involving 1/4–1/2 of the bone width; 3, bone lesion across 1/2–3/4 of the bone width; and 4, bone lesion >3/4 of the bone width. The bone metastasis score for each mouse was the sum of the scores of all bone lesions from four limbs. For survival studies, mice were monitored daily for signs of discomfort, and were either euthanized all at one time or individually when presenting signs of distress, such as a 10% loss of body weight, paralysis, or head tilting.
Statistical Analysis
All values are presented as the mean ± SD. Significant differences were determined using the GraphPad 5.0 software (USA). Student’s t test was used to determine statistical differences between two groups. The chi-square test was used to analyze the relationship between miR-582-3p or miR-582-5p expression and clinicopathological characteristics. Survival curves were plotted using the Kaplan-Meier method and compared by log rank test. p < 0.05 was considered significant. All experiments were repeated three times.
Author Contributions
J.P. and X.Z. developed ideas and drafted the manuscript. Shuai Huang, C.Z., and Y.T. conducted the experiments and contributed to the analysis of data. Q.W., X.P., and X.C. contributed to the analysis of data. C.Y. and D.R. conducted the experiments. Y.H. and Z.L. contributed to the analysis of data and revised the manuscript. Sheng Huang edited the manuscript. All authors contributed to revising the manuscript and approved the final version for publication.
Conflicts of Interest
The authors declare no competing interests.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (31430030, 81872172, 81503281, 81702671, 81502219, and 81660362), the Guangdong Natural Science Foundation (2016A030313199), and a Talent Grant of Sun Yat-sen University (17ykpy30).
Footnotes
Supplemental Information includes nine figures and five tables and can be found with this article online at https://doi.org/10.1016/j.omtn.2019.01.004.
Contributor Information
Shuai Huang, Email: huang-shuai@hotmail.com.
Xuenong Zou, Email: zxnong@hotmail.com.
Jincheng Pan, Email: panjinchengos@163.com.
Supplemental Information
References
- 1.Ferlay J., Soerjomataram I., Dikshit R., Eser S., Mathers C., Rebelo M., Parkin D.M., Forman D., Bray F. Cancer incidence and mortality worldwide: sources, methods and major patterns in GLOBOCAN 2012. Int. J. Cancer. 2015;136:E359–E386. doi: 10.1002/ijc.29210. [DOI] [PubMed] [Google Scholar]
- 2.Heidenreich A., Bastian P.J., Bellmunt J., Bolla M., Joniau S., van der Kwast T., Mason M., Matveev V., Wiegel T., Zattoni F., Mottet N., European Association of Urology EAU guidelines on prostate cancer. Part II: treatment of advanced, relapsing, and castration-resistant prostate cancer. Eur. Urol. 2014;65:467–479. doi: 10.1016/j.eururo.2013.11.002. [DOI] [PubMed] [Google Scholar]
- 3.Bubendorf L., Schöpfer A., Wagner U., Sauter G., Moch H., Willi N., Gasser T.C., Mihatsch M.J. Metastatic patterns of prostate cancer: an autopsy study of 1,589 patients. Hum. Pathol. 2000;31:578–583. doi: 10.1053/hp.2000.6698. [DOI] [PubMed] [Google Scholar]
- 4.Pickup M., Novitskiy S., Moses H.L. The roles of TGFβ in the tumour microenvironment. Nat. Rev. Cancer. 2013;13:788–799. doi: 10.1038/nrc3603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Kang Y., He W., Tulley S., Gupta G.P., Serganova I., Chen C.R., Manova-Todorova K., Blasberg R., Gerald W.L., Massagué J. Breast cancer bone metastasis mediated by the Smad tumor suppressor pathway. Proc. Natl. Acad. Sci. USA. 2005;102:13909–13914. doi: 10.1073/pnas.0506517102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Sethi N., Dai X., Winter C.G., Kang Y. Tumor-derived JAGGED1 promotes osteolytic bone metastasis of breast cancer by engaging notch signaling in bone cells. Cancer Cell. 2011;19:192–205. doi: 10.1016/j.ccr.2010.12.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kang Y., Siegel P.M., Shu W., Drobnjak M., Kakonen S.M., Cordón-Cardo C., Guise T.A., Massagué J. A multigenic program mediating breast cancer metastasis to bone. Cancer Cell. 2003;3:537–549. doi: 10.1016/s1535-6108(03)00132-6. [DOI] [PubMed] [Google Scholar]
- 8.Fournier P.G., Juárez P., Jiang G., Clines G.A., Niewolna M., Kim H.S., Walton H.W., Peng X.H., Liu Y., Mohammad K.S. The TGF-β signaling regulator PMEPA1 suppresses prostate cancer metastases to bone. Cancer Cell. 2015;27:809–821. doi: 10.1016/j.ccell.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hu Z., Gupta J., Zhang Z., Gerseny H., Berg A., Chen Y.J., Zhang Z., Du H., Brendler C.B., Xiao X. Systemic delivery of oncolytic adenoviruses targeting transforming growth factor-β inhibits established bone metastasis in a prostate cancer mouse model. Hum. Gene Ther. 2012;23:871–882. doi: 10.1089/hum.2012.040. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wan X., Li Z.G., Yingling J.M., Yang J., Starbuck M.W., Ravoori M.K., Kundra V., Vazquez E., Navone N.M. Effect of transforming growth factor beta (TGF-β) receptor I kinase inhibitor on prostate cancer bone growth. Bone. 2012;50:695–703. doi: 10.1016/j.bone.2011.11.022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Dai Y., Ren D., Yang Q., Cui Y., Guo W., Lai Y., Du H., Lin C., Li J., Song L., Peng X. The TGF-β signalling negative regulator PICK1 represses prostate cancer metastasis to bone. Br. J. Cancer. 2017;117:685–694. doi: 10.1038/bjc.2017.212. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Shi Y., Massagué J. Mechanisms of TGF-beta signaling from cell membrane to the nucleus. Cell. 2003;113:685–700. doi: 10.1016/s0092-8674(03)00432-x. [DOI] [PubMed] [Google Scholar]
- 13.Huse M., Muir T.W., Xu L., Chen Y.G., Kuriyan J., Massagué J. The TGF beta receptor activation process: an inhibitor- to substrate-binding switch. Mol. Cell. 2001;8:671–682. doi: 10.1016/s1097-2765(01)00332-x. [DOI] [PubMed] [Google Scholar]
- 14.Bertran E., Crosas-Molist E., Sancho P., Caja L., Lopez-Luque J., Navarro E., Egea G., Lastra R., Serrano T., Ramos E., Fabregat I. Overactivation of the TGF-β pathway confers a mesenchymal-like phenotype and CXCR4-dependent migratory properties to liver tumor cells. Hepatology. 2013;58:2032–2044. doi: 10.1002/hep.26597. [DOI] [PubMed] [Google Scholar]
- 15.Ishimoto T., Miyake K., Nandi T., Yashiro M., Onishi N., Huang K.K., Lin S.J., Kalpana R., Tay S.T., Suzuki Y. Activation of transforming growth factor beta 1 signaling in gastric cancer-associated fibroblasts increases their motility, via expression of rhomboid 5 homolog 2, and ability to induce invasiveness of gastric cancer cells. Gastroenterology. 2017;153:191–204.e16. doi: 10.1053/j.gastro.2017.03.046. [DOI] [PubMed] [Google Scholar]
- 16.Ren D., Yang Q., Dai Y., Guo W., Du H., Song L., Peng X. Oncogenic miR-210-3p promotes prostate cancer cell EMT and bone metastasis via NF-κB signaling pathway. Mol. Cancer. 2017;16:117. doi: 10.1186/s12943-017-0688-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Guo W., Ren D., Chen X., Tu X., Huang S., Wang M., Song L., Zou X., Peng X. HEF1 promotes epithelial mesenchymal transition and bone invasion in prostate cancer under the regulation of microRNA-145. J. Cell. Biochem. 2013;114:1606–1615. doi: 10.1002/jcb.24502. [DOI] [PubMed] [Google Scholar]
- 18.Zhang X., Liu J., Zang D., Wu S., Liu A., Zhu J., Wu G., Li J., Jiang L. Upregulation of miR-572 transcriptionally suppresses SOCS1 and p21 and contributes to human ovarian cancer progression. Oncotarget. 2015;6:15180–15193. doi: 10.18632/oncotarget.3737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ren D., Lin B., Zhang X., Peng Y., Ye Z., Ma Y., Liang Y., Cao L., Li X., Li R. Maintenance of cancer stemness by miR-196b-5p contributes to chemoresistance of colorectal cancer cells via activating STAT3 signaling pathway. Oncotarget. 2017;8:49807–49823. doi: 10.18632/oncotarget.17971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ren D., Wang M., Guo W., Huang S., Wang Z., Zhao X., Du H., Song L., Peng X. Double-negative feedback loop between ZEB2 and miR-145 regulates epithelial-mesenchymal transition and stem cell properties in prostate cancer cells. Cell Tissue Res. 2014;358:763–778. doi: 10.1007/s00441-014-2001-y. [DOI] [PubMed] [Google Scholar]
- 21.Siegel P.M., Massagué J. Cytostatic and apoptotic actions of TGF-beta in homeostasis and cancer. Nat. Rev. Cancer. 2003;3:807–821. doi: 10.1038/nrc1208. [DOI] [PubMed] [Google Scholar]
- 22.Qin J., Wu S.P., Creighton C.J., Dai F., Xie X., Cheng C.M., Frolov A., Ayala G., Lin X., Feng X.H. COUP-TFII inhibits TGF-β-induced growth barrier to promote prostate tumorigenesis. Nature. 2013;493:236–240. doi: 10.1038/nature11674. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Waning D.L., Mohammad K.S., Reiken S., Xie W., Andersson D.C., John S., Chiechi A., Wright L.E., Umanskaya A., Niewolna M. Excess TGF-β mediates muscle weakness associated with bone metastases in mice. Nat. Med. 2015;21:1262–1271. doi: 10.1038/nm.3961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.TGFβ induces a pro-bone metastasis program in prostate cancer. Cancer Discov. 2015;5:OF23. doi: 10.1158/2159-8290.CD-RW2015-098. [DOI] [PubMed] [Google Scholar]
- 25.Yang H., Fang F., Chang R., Yang L. MicroRNA-140-5p suppresses tumor growth and metastasis by targeting transforming growth factor β receptor 1 and fibroblast growth factor 9 in hepatocellular carcinoma. Hepatology. 2013;58:205–217. doi: 10.1002/hep.26315. [DOI] [PubMed] [Google Scholar]
- 26.Hu S., Chen H., Zhang Y., Wang C., Liu K., Wang H., Luo J. MicroRNA-520c inhibits glioma cell migration and invasion by the suppression of transforming growth factor-β receptor type 2. Oncol. Rep. 2017;37:1691–1697. doi: 10.3892/or.2017.5421. [DOI] [PubMed] [Google Scholar]
- 27.Dickman C.T., Lawson J., Jabalee J., MacLellan S.A., LePard N.E., Bennewith K.L., Garnis C. Selective extracellular vesicle exclusion of miR-142-3p by oral cancer cells promotes both internal and extracellular malignant phenotypes. Oncotarget. 2017;8:15252–15266. doi: 10.18632/oncotarget.14862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Braun J., Hoang-Vu C., Dralle H., Hüttelmaier S. Downregulation of microRNAs directs the EMT and invasive potential of anaplastic thyroid carcinomas. Oncogene. 2010;29:4237–4244. doi: 10.1038/onc.2010.169. [DOI] [PubMed] [Google Scholar]
- 29.Wang C., Liu P., Wu H., Cui P., Li Y., Liu Y., Liu Z., Gou S. MicroRNA-323-3p inhibits cell invasion and metastasis in pancreatic ductal adenocarcinoma via direct suppression of SMAD2 and SMAD3. Oncotarget. 2016;7:14912–14924. doi: 10.18632/oncotarget.7482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Lv S., Sun B., Dai C., Shi R., Zhou X., Lv W., Zhong X., Wang R., Ma W. Retraction. Mol. Neurobiol. 2017;54:2379. doi: 10.1007/s12035-017-0452-3. [DOI] [PubMed] [Google Scholar]
- 31.Cabana-Domínguez J., Roncero C., Pineda-Cirera L., Palma-Álvarez R.F., Ros-Cucurull E., Grau-López L., Esojo A., Casas M., Arenas C., Ramos-Quiroga J.A. Association of the PLCB1 gene with drug dependence. Sci. Rep. 2017;7:10110. doi: 10.1038/s41598-017-10207-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sud N., Zhang H., Pan K., Cheng X., Cui J., Su Q. Aberrant expression of microRNA induced by high-fructose diet: implications in the pathogenesis of hyperlipidemia and hepatic insulin resistance. J. Nutr. Biochem. 2017;43:125–131. doi: 10.1016/j.jnutbio.2017.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nicolas G., Wallon D., Goupil C., Richard A.C., Pottier C., Dorval V., Sarov-Rivière M., Riant F., Hervé D., Amouyel P. Mutation in the 3′untranslated region of APP as a genetic determinant of cerebral amyloid angiopathy. Eur. J. Hum. Genet. 2016;24:92–98. doi: 10.1038/ejhg.2015.61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Campos-Melo D., Droppelmann C.A., He Z., Volkening K., Strong M.J. Altered microRNA expression profile in Amyotrophic Lateral Sclerosis: a role in the regulation of NFL mRNA levels. Mol. Brain. 2013;6:26. doi: 10.1186/1756-6606-6-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Liu Y., Jiang J., Wang X., Zhai F., Cheng X. miR-582-5p is upregulated in patients with active tuberculosis and inhibits apoptosis of monocytes by targeting FOXO1. PLoS ONE. 2013;8:e78381. doi: 10.1371/journal.pone.0078381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zhang C., Huang J., An W. Hepatic stimulator substance resists hepatic ischemia/reperfusion injury by regulating Drp1 translocation and activation. Hepatology. 2017;66:1989–2001. doi: 10.1002/hep.29326. [DOI] [PubMed] [Google Scholar]
- 37.Bobowicz M., Skrzypski M., Czapiewski P., Marczyk M., Maciejewska A., Jankowski M., Szulgo-Paczkowska A., Zegarski W., Pawłowski R., Polańska J. Prognostic value of 5-microRNA based signature in T2-T3N0 colon cancer. Clin. Exp. Metastasis. 2016;33:765–773. doi: 10.1007/s10585-016-9810-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Fang L., Cai J., Chen B., Wu S., Li R., Xu X., Yang Y., Guan H., Zhu X., Zhang L. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/β-catenin signalling. Nat. Commun. 2015;6:8640. doi: 10.1038/ncomms9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Butz H., Likó I., Czirják S., Igaz P., Korbonits M., Rácz K., Patócs A. MicroRNA profile indicates downregulation of the TGFβ pathway in sporadic non-functioning pituitary adenomas. Pituitary. 2011;14:112–124. doi: 10.1007/s11102-010-0268-x. [DOI] [PubMed] [Google Scholar]
- 40.Paydas S., Acikalin A., Ergin M., Celik H., Yavuz B., Tanriverdi K. Micro-RNA (miRNA) profile in Hodgkin lymphoma: association between clinical and pathological variables. Med. Oncol. 2016;33:34. doi: 10.1007/s12032-016-0749-5. [DOI] [PubMed] [Google Scholar]
- 41.Shu Z., Chen L., Ding D. miR-582-5P induces colorectal cancer cell proliferation by targeting adenomatous polyposis coli. World J. Surg. Oncol. 2016;14:239. doi: 10.1186/s12957-016-0984-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Maeno A., Terada N., Uegaki M., Goto T., Okada Y., Kobayashi T., Kamba T., Ogawa O., Inoue T. Up-regulation of miR-582-5p regulates cellular proliferation of prostate cancer cells under androgen-deprived conditions. Prostate. 2014;74:1604–1612. doi: 10.1002/pros.22877. [DOI] [PubMed] [Google Scholar]
- 43.Lu Y.C., Chen Y.J., Wang H.M., Tsai C.Y., Chen W.H., Huang Y.C., Fan K.H., Tsai C.N., Huang S.F., Kang C.J. Oncogenic function and early detection potential of miRNA-10b in oral cancer as identified by microRNA profiling. Cancer Prev. Res. (Phila.) 2012;5:665–674. doi: 10.1158/1940-6207.CAPR-11-0358. [DOI] [PubMed] [Google Scholar]
- 44.Zhang Y., Huang W., Ran Y., Xiong Y., Zhong Z., Fan X., Wang Z., Ye Q. miR-582-5p inhibits proliferation of hepatocellular carcinoma by targeting CDK1 and AKT3. Tumour Biol. 2015;36:8309–8316. doi: 10.1007/s13277-015-3582-0. [DOI] [PubMed] [Google Scholar]
- 45.Uchino K., Takeshita F., Takahashi R.U., Kosaka N., Fujiwara K., Naruoka H., Sonoke S., Yano J., Sasaki H., Nozawa S. Therapeutic effects of microRNA-582-5p and -3p on the inhibition of bladder cancer progression. Mol. Ther. 2013;21:610–619. doi: 10.1038/mt.2012.269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Davidson B.L., McCray P.B., Jr. Current prospects for RNA interference-based therapies. Nat. Rev. Genet. 2011;12:329–340. doi: 10.1038/nrg2968. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Takeshita F., Ochiya T. Therapeutic potential of RNA interference against cancer. Cancer Sci. 2006;97:689–696. doi: 10.1111/j.1349-7006.2006.00234.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Cai J., Fang L., Huang Y., Li R., Xu X., Hu Z., Zhang L., Yang Y., Zhu X., Zhang H. Simultaneous overactivation of Wnt/β-catenin and TGFβ signalling by miR-128-3p confers chemoresistance-associated metastasis in NSCLC. Nat. Commun. 2017;8:15870. doi: 10.1038/ncomms15870. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Liu C., Liu R., Zhang D., Deng Q., Liu B., Chao H.P., Rycaj K., Takata Y., Lin K., Lu Y. MicroRNA-141 suppresses prostate cancer stem cells and metastasis by targeting a cohort of pro-metastasis genes. Nat. Commun. 2017;8:14270. doi: 10.1038/ncomms14270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Qin J., Liang H., Shi D., Dai J., Xu Z., Chen D., Chen X., Jiang Q. A panel of microRNAs as a new biomarkers for the detection of deep vein thrombosis. J. Thromb. Thrombolysis. 2015;39:215–221. doi: 10.1007/s11239-014-1131-0. [DOI] [PubMed] [Google Scholar]
- 51.Wu T.T., Sikes R.A., Cui Q., Thalmann G.N., Kao C., Murphy C.F., Yang H., Zhau H.E., Balian G., Chung L.W. Establishing human prostate cancer cell xenografts in bone: induction of osteoblastic reaction by prostate-specific antigen-producing tumors in athymic and SCID/bg mice using LNCaP and lineage-derived metastatic sublines. Int. J. Cancer. 1998;77:887–894. doi: 10.1002/(sici)1097-0215(19980911)77:6<887::aid-ijc15>3.0.co;2-z. [DOI] [PubMed] [Google Scholar]
- 52.Wang M., Ren D., Guo W., Huang S., Wang Z., Li Q., Du H., Song L., Peng X. N-cadherin promotes epithelial-mesenchymal transition and cancer stem cell-like traits via ErbB signaling in prostate cancer cells. Int. J. Oncol. 2016;48:595–606. doi: 10.3892/ijo.2015.3270. [DOI] [PubMed] [Google Scholar]
- 53.Zhang X., Ren D., Guo L., Wang L., Wu S., Lin C., Ye L., Zhu J., Li J., Song L. Thymosin beta 10 is a key regulator of tumorigenesis and metastasis and a novel serum marker in breast cancer. Breast Cancer Res. 2017;19:15. doi: 10.1186/s13058-016-0785-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Zhang X., Zhang L., Lin B., Chai X., Li R., Liao Y., Deng X., Liu Q., Yang W., Cai Y. Phospholipid Phosphatase 4 promotes proliferation and tumorigenesis, and activates Ca2+-permeable Cationic Channel in lung carcinoma cells. Mol. Cancer. 2017;16:147. doi: 10.1186/s12943-017-0717-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li X., Liu F., Lin B., Luo H., Liu M., Wu J., Li C., Li R., Zhang X., Zhou K., Ren D. miR-150 inhibits proliferation and tumorigenicity via retarding G1/S phase transition in nasopharyngeal carcinoma. Int. J. Oncol. 2017;50:1097–1108. doi: 10.3892/ijo.2017.3909. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ren D., Wang M., Guo W., Zhao X., Tu X., Huang S., Zou X., Peng X. Wild-type p53 suppresses the epithelial-mesenchymal transition and stemness in PC-3 prostate cancer cells by modulating miR-145. Int. J. Oncol. 2013;42:1473–1481. doi: 10.3892/ijo.2013.1825. [DOI] [PubMed] [Google Scholar]
- 57.Ren D., Dai Y., Yang Q., Zhang X., Guo W., Ye L., Huang S., Chen X., Lai Y., Du H. Wnt5a induces and maintains prostate cancer cells dormancy in bone. J. Exp. Med. 2018 doi: 10.1084/jem.20180661. jem.20180661. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.