Abstract
Melatonin, the hormone of darkness, is a peculiar molecule found in most living organisms. Emerging as a potent broad-spectrum antioxidant, melatonin was repurposed into extra roles such as the modulation of circadian and seasonal rhythmicity, affecting numerous aspects of physiology and behaviour, including sleep entrainment and locomotor activity. Interestingly, the pineal gland—the melatonin synthesising organ in vertebrates—was suggested to be absent or rudimentary in some mammalian lineages, including Cetacea. In Cetacea, pineal regression is paralleled by their unique bio-rhythmicity, as illustrated by the unihemispheric sleeping behaviour and long-term vigilance. Here, we examined the genes responsible for melatonin synthesis (Aanat and Asmt) and signalling (Mtnr1a and Mtnr1b) in 12 toothed and baleen whale genomes. Based on an ample genomic comparison, we deduce that melatonin-related gene modules are eroded in Cetacea.
Keywords: gene loss, Cetacea, melatonin, circadian rhythms
1. Introduction
Circadian rhythmicity is critical for broad organism homeostasis. In mammals, a complex network of brain anatomical structures, such as the suprachiasmatic nuclei (SCN), the master clock, and the pineal gland, as well as a set of specific molecular pathways, optimise physiological timekeeping [1,2]. First described in the 1950s, melatonin has since been suggested to act as a multi-purpose hormone, with important roles in SCN excitability and sleep modulation [3,4,5,6,7]. In vertebrates, endogenous melatonin levels exhibit daily rhythmicity and a secretory peak at night. Melatonin is predominantly synthesised in the pineal gland, with the canonical synthesis pathway initiating from a precursor molecule, the amino acid tryptophan [3]. The two final steps of the enzymatic cascade are sequentially controlled by the circadian regulated aralkylamine N-acetyltransferase (AANAT) and by N-acetylserotonin methyltransferase (ASMT) enzymes, respectively, converting the intermediate serotonin into melatonin [8,9]. Subsequent hormone signalling in target cells occurs via two high affinity G-protein couple receptors, (MTNR1A) and (MTNR1B) [10]. While MTNR1A is mostly expressed in the SCN and pars tuberalis of the pituitary gland, MTNR1B is predominantly found in the retina, with lower levels detected in the hippocampus and whole brain [10]. Thus, by acting through MTNR1A and MTNR1B receptors, melatonin participates in the light-dependent modulation of SCN activity and sleep/wake cycles [2,11]. Interestingly, vertebrate AANAT functional evolution, towards the conversion of serotonin into the substrate of ASMT, N-acetyl serotonin, was suggested to parallel the emergence of the vertebrate pineal gland and melatonin rhythms [12]. In agreement, AANAT is also expressed in the retina, reflecting an ancient role in photoreception [13,14]. Despite the clear presence of a pineal gland in most mammalian species, in some lineages such as mole rats, sirenians and cetaceans, pineal organs were suggested to be absent or vestigial [15,16,17,18]. Consistently, in the naked mole rat (Heterocephalus glaber), a subterranean rodent inhabiting a dark ecosystem, an aberrant melatonin system has been described with both melatonin cognate receptors found with inactivating mutations in the coding region [15,19]. Thus, pineal atrophy, such as that described in mole rats, is apparently correlated with genomic gene loss signatures [15,20]. In this context, Cetacea provide a valuable model to track the molecular signature of a macroevolutionary transition, notably regarding the adaptation to a distinct photic and thermal environment [21,22,23]. In fact, the existence of a functional pineal gland in these aquatic mammals is still contentious, even among individuals of the same species: with conflicting reports from various whale species (e.g., Megaptera novaeangliae (humpback whale)) or the commonly studied Tursiops truncatus (bottlenose dolphin) [16]. To further address the parallelism between pineal regression and melatonin synthesis and signalling, we extracted genomic information and addressed the coding status of Aanat, Asmt and the melatonin receptors Mtnr1a and Mtnr1b in Cetacea and in Hippopotamus amphibius (common hippopotamus), the closest extant lineage of Cetacea. Further comparative analysis using Trichechus manatus latirostris (Florida manatee), also lacking a pineal gland, provided additional insight into the convergent adaptation to fully aquatic lifestyles [18].
2. Materials and Methods
2.1. Synteny Maps
To perform synteny analyses, the human and Bos Taurus (domestic cow) loci of Aanat, Mtnr1a, Mtnr1b and Asmt genes were used as reference. Importantly, in B. taurus two Aanat genes were found, and thus we considered as main reference the gene with NM_177509.2 accession number. After collecting the genomic regions of the reference species, several Cetacea genome assemblies and annotations were inspected and scrutinised using the NCBI browser (Homo sapiens—GCF_000001405.38, B. taurus—GCF_002263795.1, Orcinus orca (orca whale)—GCF_000331955.1, Balaenoptera acutorostrata (common minke whale)—GCF_000493695.1, Delphinapterus leucas (beluga whale)—GCF_002288925.1, T. truncatus—GCF_001922835.1, Lagenorhynchus obliquidens (Pacific white-sided dolphin)—GCF_003676395.1, Physeter catodon (sperm whale)—GCF_002837175.1, Lipotes vexillifer (Yangtze river dolphin)—GCF_000442215.1, Neophocaena asiaeorientalis (Indo-Pacific finless porpoise)—GCF_003031525.1). To perform the synteny maps the following procedure was used: (1) five flanking genes from each side of the target gene were collected, (2) only genes neighbouring genes classified as coding were considered (Gene Type: Protein Coding), (3) if the target gene was not found, the genomic region between the direct neighbouring genes (first gene at left and right side) was inspected. The inspection was done in two ways: firstly, via blast searches to determine the total or partial absence of the gene and after, manually to ensure the continuity or disruption of the genomic region (with presence or absence of N’s).
2.2. Sequence Retrieval
Genomic sequence retrieval for gene annotation was performed using two strategies: (1) In species with fully annotated genomes (O. orca, T. truncatus, D. leucas, L. vexillifer, N. asiaeorientalis, P. catodon, B. acutorostrata and T. manatus latirostris), the genomic locus of the target gene was collected directly from NCBI. The genomic sequence was inspected, from the 5′ neighbouring gene to the 3′neighbouring gene of the target gene. For example, in the case of Aanat the genomic sequence was collected ranging from UBE20 to RHBDF. (2) For species whose genomes were not annotated, the corresponding genomic sequence of the target gene was retrieved, from full genome assemblies, via blastn searches using human and B. taurus sequences as query.
To scrutinise the status of the Aanat, Mtnr1a, Mtnr1b and Asmt gene sequences in H. amphibius we built an in-house blast database with the genome assembly (GCA_002995585.1), directly retrieved from the NCBI database. Next, interrogated the in-house database using the human and B. taurus, to retrieve full or partial gene sequences for H. amphibius (blast-n: -word_size 10, -outfmt 6, -num_threads 50) [24]. The output results were manually scrutinised and, using the qstart, qend, sstart send and bit score options of outfmt6 format of blast software, we identified the scaffolds containing each gene. All scaffolds or sets of scaffolds, with evidence of containing the target genes, were gathered and submitted to AUGUSTUS webserver gene prediction [25]. Finally, the annotation files with the CDS and protein sequence of Aanat, Mtnr1a and Mtnr1b genes were generated (Table S1 and Supplementary Gene Annotations). For the Asmt gene, only exons 2, 5, 7, 8 and 9 were recovered from the genome assembly. Additional searches in sequencing read archives (SRAs) yielded exon 1 (Figure S21).
2.3. Gene Annotation and Mutational Validation
For gene annotation, the collected genomic sequences were uploaded into Geneious7.1.9. (https://www.geneious.com/). The annotation was performed using as reference the corresponding B. taurus gene sequences, with the exception of Asmt gene, for which the human gene sequence was used as reference due to the presence of multiple expressed isoforms. For each reference sequence, all exons were individualised and mapped to the corresponding genomic region in Cetacea and T. manatus latirostris, using the built-in map to the reference tool. The resulting aligned regions were individually inspected to identify ORF abolishing mutations (frameshift, exon deletion, non-canonical splice site and premature stop codons), and concatenated to obtain the predicted sequence. Next, at least one conserved ORF-abolishing mutation was further validated by the identification of the same mutation in different genomic projects or individual samples of the same species. For this, the predicted exonic sequence for each species containing the selected ORF disrupting mutation was used as query in blastn searches in the corresponding available SRAs and Trace Archive in NCBI (only available for T. truncatus). Blast hits were collected and uploaded to Geneious and mapped to the corresponding exon. Reads poorly aligned or with identity scores below 95% were removed and the mutational status was confirmed when possible, in at least 2 distinct sequencing projects or individuals from the same species. Validation was performed for all species with the exception of L. vexillifer for which only one genomic resource resulting from a single genome sequencing project is presently available in NCBI.
2.4. Phylogenetic Analysis
To infer the orthology of H. amphibius sequences we performed a phylogenetic analysis using sequences from species belonging to three taxonomic orders: Perissodactyla, Artiodactyla and Primates. Primate sequences were used to root the phylogenetic trees. Briefly, to collect the sequences, blastn searches were done (defaults) in NCBI using as reference the annotated human MTNR1A (NM_005958.4), MTNR1B (NM_005959.3) and AANAT (NM_001166579.1) nucleotide sequences [24]. H. amphibius sequences were obtained in the previous analyses (see sequence retrieval section). After sequence collection, an initial alignment was performed and partial and low-quality sequences were removed with Jalview v2.10.5 [26]. A total of 23 Aanat, 17 Mtnr1a and 18 Mtnr1b orthologues were included for further analysis (accession numbers of all datasets can be retrieved from Tables S2 to S4). The final orthologous gene sets were aligned independently using MAFFT (setting the parameter—auto mode method selection) [27,28], with the method L-INS-I being automatically selected for all datasets. Using the previous alignments as input, the maximum likelihood phylogenetic analyses were done using the PhyML v3.1 software (http://www.atgc-montpellier.fr/phyml/) [29].To determine the best evolutionary model for each set of sequences, the smart model selection (SMS) option was used [30]: selecting GTR + G + I model for the Aanat dataset, and GTR + G model for the Mtnr1a and Mtnr1b datasets. In addition, to calculate the branch support for each dataset, the aBayes method was selected. Finally, the resulting trees were submitted to the Dendroscope software [31], and the primate clades were used to root the trees.
3. Results
3.1. Aanat Exhibits Conserved Disruptive Mutations in Cetacea
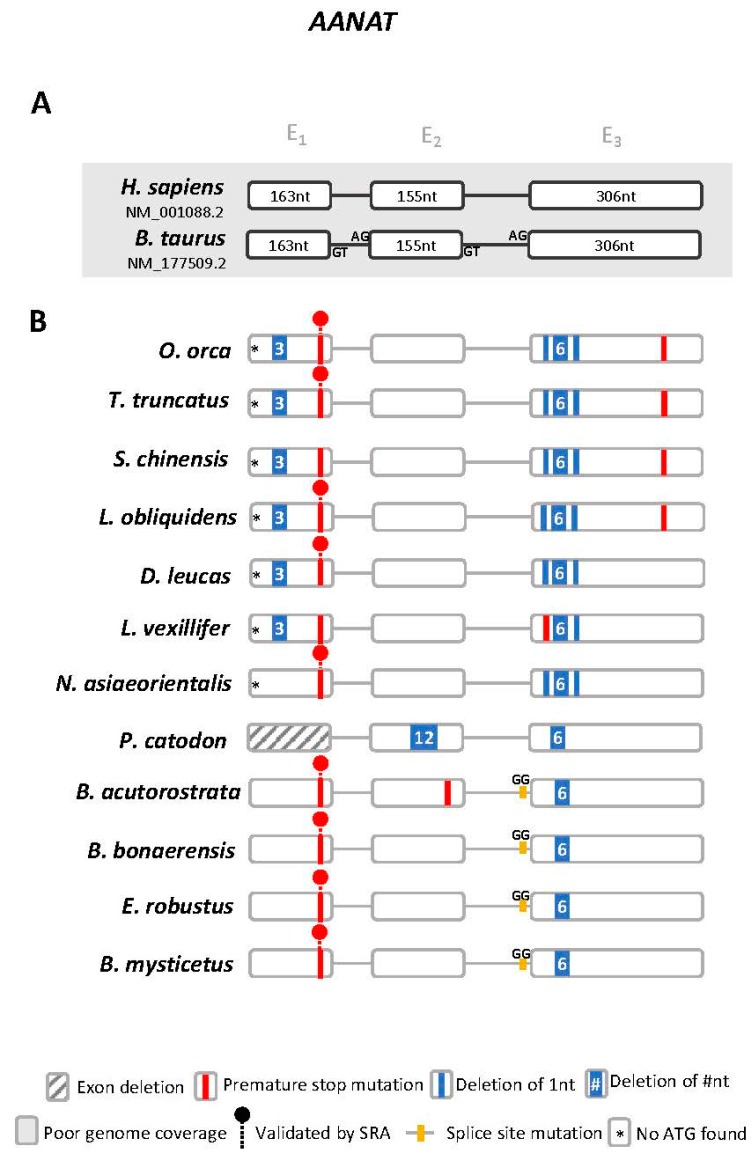
Using the available genomic data we examined the genomic regions of melatonin synthesis and signalling modules in 12 Cetacea species (O. orca, T. truncatus, L. obliquidens, S. chinensis (Indo-Pacific humpbacked dolphin), D. leucas, L. vexillifer, N. asiaeorientalis, P. catodon, B. acutorostrata, Balaenoptera bonaerensis (Antarctic minke whale), Eschrichtius robustus (grey whale), Balaena mysticetus (bowhead whale)) [32,33,34,35,36,37,38,39,40]. We started by analysing the Aanat gene. The Aanat locus-of-origin was first identified using the human (NM_001088.2) and the B. taurus (NM_177509.2) gene sequences (Figure S1). In the case of T. truncatus, the Aanat gene was not annotated as a coding open reading frame (ORF), yet the gene locus and corresponding neighbouring gene families were found to be conserved (Figure S1). Next, we performed a comparative analysis of the exonic sequences, using the cow orthologue as reference, to identify potential mutations [41,42,43,44]. In summary, we were able to deduce the presence of multiple and shared disruptive mutations (Figure 1 and Figure S2). These included a stop codon in exon 1 present in all analysed Cetacea with the exception of P. catodon (exon 1 was not found in the current assembly), an absent starting codon and frameshift deletions in exon 3 in Odontoceti (toothed whales), and a disruptive intron 2/exon 3 boundary mutation in Mysticeti (baleen whales). In P. catodon a unique deletion of 12 nucleotides in exon 2 was observed, which, combined with an additional 6 nucleotide deletion in exon 3, strongly suggests an inactivated ORF. In O. orca and T. truncatus, a premature stop codon truncates exon 3. Other species-specific mutations were identified. To further confirm our findings, we selected the most critical and evolutionary conserved ORF-disruptive mutation for validation using SRAs. As such, we confirmed the presence of the premature stop codon in exon 1 across 9 Cetacea species (Figure S3). Regarding the Odontoceti O. orca, T. truncatus, D. leucas, N. asiaeorientalis and the Mysticeti B. acutorostrata and E. robustus, we were able to validate the conserved premature stop using two independent sequenced samples per species.
Figure 1.
Aanat gene annotation. (A) Schematic representation of the gene structure of human and Bos taurus Aanat gene, each box represents an exon and lines represent intronic regions (not to scale). (B) Schematic representation of the corresponding Aanat genes identified in Cetacea and location of the identified mutations. Non-canonical splice sites are indicated above the corresponding annotation. SRA: sequencing read archives.
3.2. Cetacea Asmt Reveals Conserved Frameshift Mutation in Exon 1
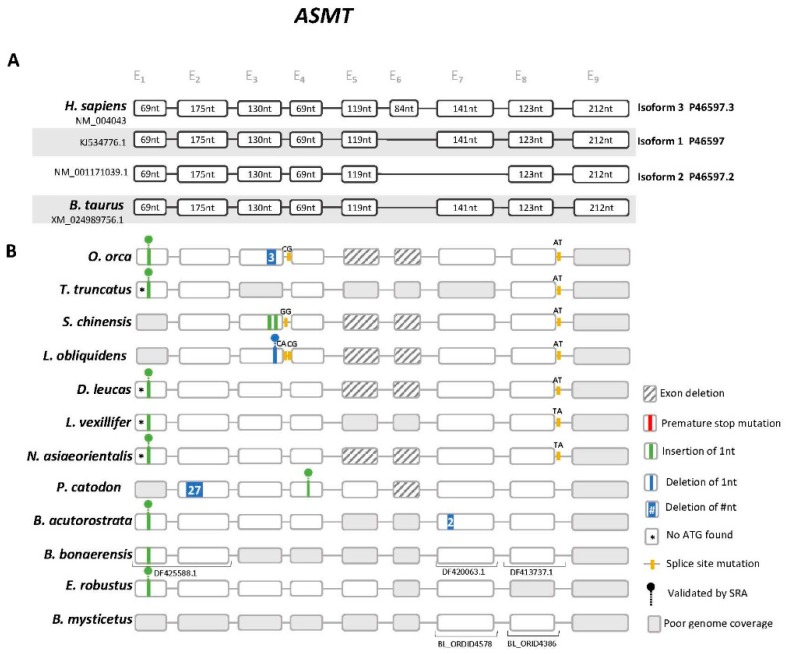
Next, we examined the Asmt gene (Figure S4). In human, 3 distinct isoforms of Asmt resulting from the alternative splicing of exon 6 and 7 were identified [45]. Yet, only isoform 1 (P46597) has been reported to display ASMT enzymatic activity (Figure S5) [45]. Thus, both human and B. taurus genes were used as reference. Gene annotation revealed the presence of a conserved 1 nucleotide insertion in exon 1, validated by independent SRAs in O. orca, T. truncatus, D. leucas, N. asiaeorientalis, B. acutorostrata and E. robustus (Figure 2, Figures S6 and S7). In L. obliquidens, S. chinensis, P. catodon and B. mysticetus exon 1 could not be retrieved using the current assemblies. Complete erosion of exon 5 was also detected in O. orca, L. obliquidens, S. chinensis, N. asiaeorientalis and D. leucas, for which full genomic sequences ranging from exon 4 to exon 7 were available; for P. catodon and E. robustus exon 5 was identified with no deleterious mutations, while for the remaining species the presence of sequencing gaps within this region impaired further assessment. The deletion of exon 5 is expected to abolish enzyme activity given that this exon contains a highly conserved glycine (Gly187), participating in the active site of ASMT [45] (Figures S5 and S8). Also, aside from the conserved non-canonical splice site observed in exon 8 of Odonoceti, other non-conserved ORF disrupting mutations were found (Figure 2): notably, a 1 nucleotide insertion in exon 4 of P. catodon, validated by independent SRAs (Figure S9), and a 1 nucleotide deletion in exon 3 of L. obliquidens, also validated by SRA (Figure S10). Finally, exon 9 was not found in the present genome assemblies of all analysed species (Figure 2).
Figure 2.
Asmt gene annotation. (A) Schematic representation of the gene structure and distinct isoforms of human and B. taurus Asmt gene (not to scale), each box represents an exon and lines represent intronic regions. (B) Schematic representation of the corresponding Asmt genes in Cetacea and location of the identified mutations. Non-canonical splice sites are indicated above the corresponding annotation.
3.3. Signalling Gene Modules are Absent or Mutated in Cetacea
We next examined Mtnr1a and Mtnr1b genes, which are the receptors responsible for transducing melatonin signalling. Here, we did not detect Mtnr1a-like sequences in Odontoceti genomes. Comparative synteny analysis revealed that Fat1 and F11, which are adjacent to Mtnr1a in human and cow, are also side-by-side in Odontoceti (Figure 3).
Figure 3.
Comparative synteny maps of Mtnr1a genomic locus.
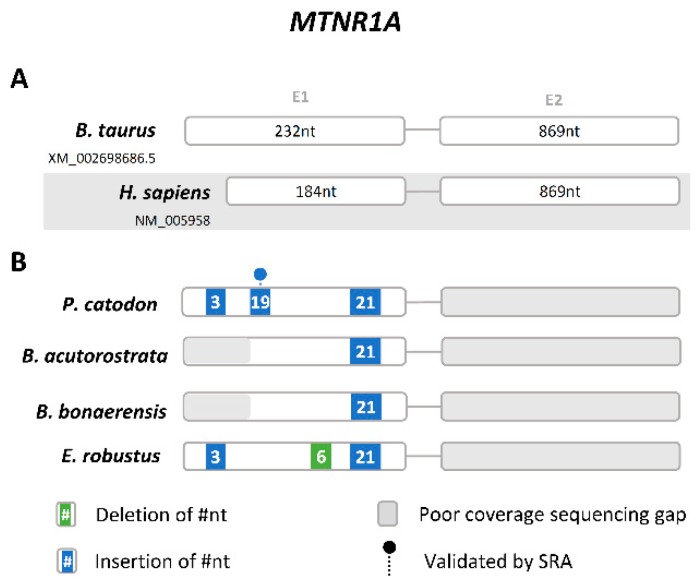
To further validate this observation, we carefully examined the genomic sequences flanked by Fat1 and F11 genes. While in some species we observed the occurrence of sequencing gaps, impairing a robust conclusion regarding Mtnr1a absence, in L. obliquidens, D. leucas and N. asiaeorientalis the genomic sequences between Fat1 and F11 genes are complete, and yield no remnants of Mtnr1a. In Mysticeti, we were able to deduce the presence of relics of exon 1, yet presenting potentially disruptive mutations (Figure 4). For instance, in P. catodon, a 19-nucleotide deletion in exon 1 was found and validated by SRAs (Figures S11 and S12). Regarding exon 2, the current assemblies are incomplete and did not allow a decisive conclusion. Yet, overall, the data strongly suggests that Mtnr1a is most likely lost or inactivated across Cetacea.
Figure 4.
Gene annotation of Mtnr1a relics in Mysticeti. (A) Schematic representation of the gene structure of human and B. taurus Mtnr1a gene (not to scale), each box represents an exon and lines represent intronic region. (B) Schematic representation of the corresponding Mtnr1a genes identified in Cetacea and location of the identified insertions and deletions.
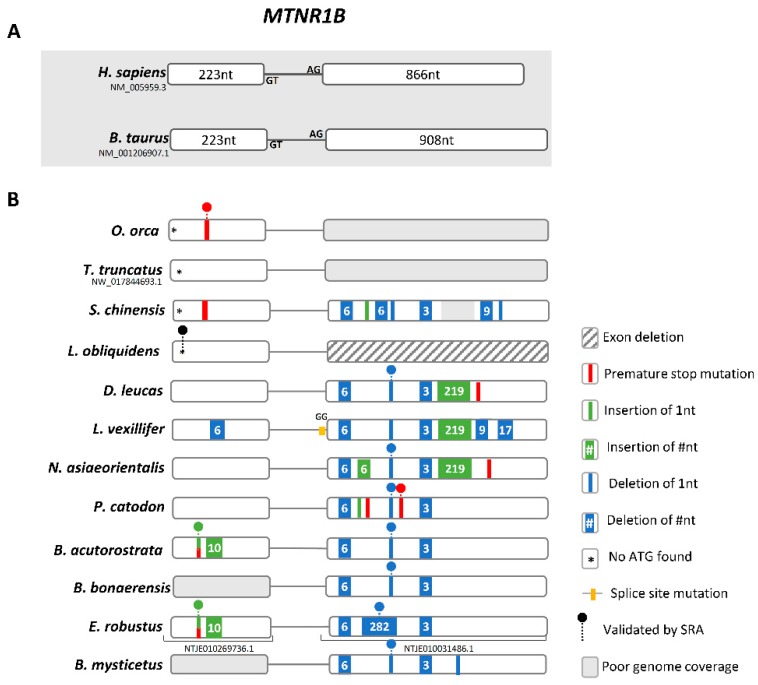
In the case of Mtnr1b, we retrieved Mtnr1b-like sequences in all analysed genomes (Figure 5 and Figure S13). Yet, for L. obliquidens, only the first exon was identified, while no sequencing gaps were detected between the retrieved exon and the neighbouring Slc36a4 gene (Figure S13); exon 2 also remained undetected in O. orca and T. truncatus, yet the presence of sequencing gaps, between the Mtnr1b-like sequence and Slc36a4, impaired a final conclusion regarding these species. Further examination of retrieved exon 1 and exon 2 sequences indicated a spectrum of mutations with predicted disruptive effects (Figure 5). In exon 1, a conserved stop codon is present in O. orca and S. chinensis, validated by SRA in the former (Figures S14 and S15); a 1 nucleotide insertion in exon 1, generating a premature stop codon, was also found and validated for B. acutorostrata and E. robustus (Figures S14 and S16); and the absence of start codon was detected in O. orca, T. truncatus, L. obliquidens and S. chinensis, and further validated by SRA in L. obliquidens (Figures S14 and S17). In exon 2, a trans-species deletion of 1 nucleotide was found in all species with an intact exon, and confirmed by SRA in all species with the exception of S. chinensis, shifting the coding frame from this section of the gene (Figures S18 and S19). A 282-nucleotide deletion was also identified in E. robustus (Figure S20).
Figure 5.
Mtnr1b gene annotation. (A) Schematic representation of the gene structure of human and B. taurus Mtnr1b gene (not to scale), each box represents an exon and lines represent intronic region. (B) Schematic representation of the corresponding Mtnr1b genes identified in Cetacea and location of the identified mutations. Non-canonical splice sites are indicated above the corresponding annotation.
3.4. Hippopotamuses Have Coding Melatonin Synthesis and Signalling Orthologues
To further refine our analysis, we next investigated the recently released genome of the H. amphibius [39], the closest extant lineage of Cetacea. We were able to deduce the complete ORFs for Aanat, Mtnr1a and Mtnr1b gene orthologues (gene annotations in Supplementary Information). In contrast, in the case of Asmt we were able to retrieve partial sequences (exons 2, 5, 7, 8 and 9). Furthermore, searches in SRAs allowed the recovery of exon 1 (Figure S21). Regarding Aanat, two Aanat-like genes were retrieved for H. amphibius as well as B. taurus and Capra aegagrus hircus (domestic goat), but these are lineage-specific duplications (Figure S22). Phylogenetic analysis of Aanat, Mtnr1a and Mtnr1b gene sequences revealed that H. amphibius orthologues clustered within Artiodactyls (even-toed ungulates), as expected (Figures S22–S24). These findings indicate that the inactivation of melatonin-related pathways most likely took place in the common ancestor of Cetacea, after the divergence of Hippopotamidae and Cetacea.
3.5. Convergent Disruption of Melatonin Metabolism and Signalling Modules in the Fully Aquatic Manatee
We next examined an additional exclusive aquatic non-cetacean mammal, T. manatus latirostris and identified ORF-abolishing mutations in all melatonin synthesis and signalling module genes (Figure S25). Regarding Aanat, these included an absent start codon and a canonical splice site mutation in exon 3, whereas in Asmt, a premature stop codon was identified in exon 5. In Mtnr1a, a single nucleotide deletion was identified at the exon 2 and a set of frameshift mutations including a premature stop codon were identified in the exon 2 of Mtnr1b. The identified mutations were further validated by SRAs (Figures S26–S29).
4. Discussion
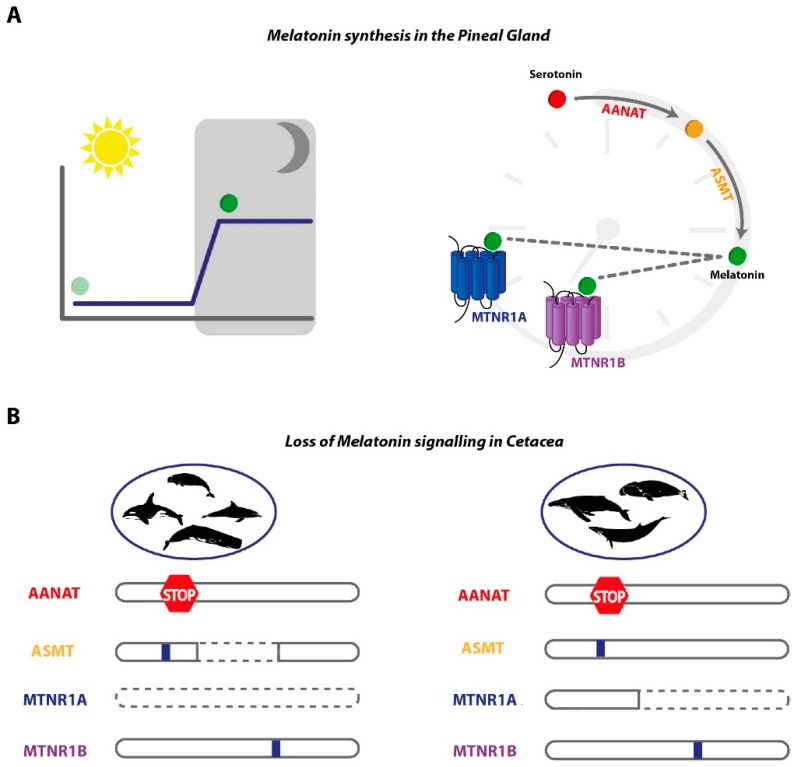
Taken together, our results strongly suggest that melatonin production and signalling is impaired in Cetacea (Figure 6). Despite the lower number of disrupting mutations found in Asmt, we still find indications of pseudogenisation in the analysed species with the exception of B. mysticetus, due to poor genome coverage. Furthermore, we were able to infer that ancestral inactivating mutations (shared between toothed and toothless whales), occurred in Mtnr1b and Aanat ORFs. In the absence of compensatory mechanisms, the observed pattern of gene loss, or coelimination of functionally related genes could represent a case of regressive evolution [46]. Convergent disruption of this set of genes was also observed in T. manatus latirostris. These evidence support the anatomical observations that the pineal gland is absent or vestigial and that classical melatonin-based circadian rhythmicity is lacking in these fully aquatic species [16,17].
Figure 6.
Melatonin synthesis and signalling modules are eroded in Cetacea. (A) Melatonin synthesis exhibits daily rhythmicity with a production peak at night (left); AANAT and ASMT sequentially convert serotonin into melatonin, which serves as ligand to two high affinity G-protein couple receptors, MTNR1A and MTNR1B (right). (B) Analysed genes from toothed whales (Odontoceti, left) and baleen whales (Mysticeti, right), premature stops, alternative splicing, indels (blue box), and complete gene erosion (dashed line), are depicted.
Yet, a paradoxical observation emerging from our findings relates to the reported presence of circulating melatonin in the bottlenose dolphin [16]. Without a functional AANAT or specific receptors, it seems unlikely that the reported melatonin levels in the bottlenose dolphin are endogenously and rhythmically produced by the canonical cascade and/or act on MTNR-dependent pathways. Thus, melatonin levels could be explained by alternative metabolic pathways: i.e., an increased activity of other enzyme cascades, including AANAT-independent pathways. In effect, AANAT-independent melatonin synthesis has been previously reported, notably in peripheral organs [47,48]. For instance, in mouse skin samples, serotonin acetylation into N-acetylserotonin was shown to be mediated by the more promiscuous arylamine N-acetyltransferase (NAT), in the absence of AANAT [47,48]. Moreover, an alternative cascade, with reversed enzymatic steps, has also been proposed. In this scenario, serotonin is initially methylated, yielding 5-methyltryptamide, which can serve as substrate for AANAT or NAT to be further N-acetylated into melatonin [47]. Given the circadian regulation of AANAT activity, and the lower stability of 5-methyltryptamide, when compared to N-acetylserotonin, the classical pathway is suggested to prevail regarding pineal melatonin build-up [13,14,47]. An ubiquitous ASMT-like gene has also been identified in vertebrates, with an unreported function so far [49,50]. The remarkable conservation of the putative catalytic domain for SAM binding supports the methyltransferase activity of this ASMT-like enzyme [50]. On the other hand, melatonin could be acquired from food sources [4]. In fact, no circadian melatonin fluctuations were indisputably acknowledged for bottlenose dolphin [16].
Regardless of the putative maintenance of endogenous, pineal-independent, melatonin sources, our results strongly support the inactivation of both MTNR1A and MTNR1B receptors in Cetacea, and in the Sirenia T. manatus latirostris. What would be the trade-off of losing melatonin synthesis and signalling modules? Melatonin is commonly known for participating in the maintenance of circadian rhythms. Yet, in animals with impaired melatonin synthesis, sleep/wake cycles are not fully disrupted [51]. Nonetheless, melatonin does seem to have synchronising and phase-locking effects, allowing an optimised adjustment to light/dark cycles [51]. In fact, deletion of both Mtnr1a and Mtnr1a in mice suggested a cooperative role in the regulation of sleep/wake cycles, notably through the increase in wakefulness [11]. In addition, Aanat knock-out mice were also shown to exhibit an increased diurnal activity [51]. Thus, the erosion of the melatonin synthesis and signalling modules could have accompanied cetacean idiosyncratic sleeping and vigilant behaviours [52,53]. This loss might also reflect other specific life-history traits. For instance, fully aquatic mammals dwell in light-limited, and wavelength-selective, environments with specific thermal constraints [23]. Both light intensity and spectral properties were suggested to modulate melatonin production [22,54]. In addition, melatonin-induced sleep entrainment promotes body temperature shifts [21]; thus, the offsetting of melatonin production and signalling in aquatic mammals could facilitate thermoregulatory mechanisms.
In the absence of melatonin receptors, circulating melatonin could act on MTNR-independent physiological processes, such as inflammatory and immune responses suggested to be modulated by melatonin-dependent gene-expression [4]. Also, one cannot disregard the ancient antioxidant capacities of melatonin [4]. In fact, cetaceans have evolved strategies to offset hypoxia-induced oxidative stress, mirrored by the mutational tinkering observed in mink whale’s genes participating in antioxidant systems (e.g., glutathione metabolism [40]). Overall, our findings provide a unique genomic signature suggesting a behavioural adaptation entailed by a cascade of gene loss events in Cetacea [55].
Acknowledgments
We acknowledge the various genome consortiums for sequencing and assembling the genomes.
Supplementary Materials
The following are available online at http://www.mdpi.com/2073-4425/10/2/121/s1, Figure S1: Comparative synteny maps of Aanat genomic locus; Figure S2: Multiple alignment of the predicted exon 1 of Aanat in Cetacea; Figure S3: SRA validation for conserved stop mutation in exon 1 of Aanat in Cetacea; Figure S4: Comparative synteny maps of Asmt genomic locus; Figure S5: Schematic representation of the Human ASMT isoform 1; Figure S6: Multiple alignment of the predicted exon 1 of Asmt in Cetacea; Figure S7: SRA validation for conserved frameshift mutation in exon 1 of Asmt in Cetacea; Figure S8: Alignment of exon 5 of Asmt in multiple mammals; Figure S9: SRA validation for frameshift mutation in exon 4 of P. catodon Asmt; Figure S10: SRA validation for frameshift mutation in exon 3 of L. obliquidens Asmt; Figure S11: Multiple alignment of the predicted Mtnr1a relic sequences (exon 1) in Cetacea; Figure S12: SRA validation for 19 nucleotide deletion in exon 1 of P. catodon Mtnr1a; Figure S13: Comparative synteny maps of Mtnr1b genomic locus; Figure S14: Multiple alignment of the predicted exon 1 of Mtnr1b in Cetacea; Figure S15: SRA validation for stop mutation in exon 1 of Mntr1b from O. Orca; Figure S16: SRA validation for nucleotide insertion generating a premature stop mutation in exon 1 of the Mntr1b gene from B. acutorostrata and E. robustus; Figure S17: SRA validation for loss of start codon in exon 1 of the Mntr1b gene from L. obliquidens; Figure S18: Partial multiple alignment of the predicted exon 2 of Mtnr1b in Cetacea; Figure S19: SRA validation for conserved nucleotide deletion in exon 2 of Mtnr1b in Cetacea; Figure S20: SRA validation for nucleotide deletion in exon 2 of Mtnr1b from E. robustus; Figure S21: H. amphibius exon 1 and 5 assembly; Figure S22: Phylogenetic analysis of Perissodactyla, Artiodactyla and Primate Aanat genes; Figure S23: Phylogenetic analysis of Perissodactyla, Artiodactyla and Primate Mtnr1a genes; Figure S24: Phylogenetic analysis of Perissodactyla, Artiodactyla and Primate Mtnr1b genes; Figure S25: T. manatus latirostris gene annotations; Figure S26: SRA validation for loss of start codon in exon 1 of the Aanat gene from T. manatus latirostris; Figure S27: SRA validation for premature stop codon in exon 5 of the Asmt gene from T. manatus latirostris; Figure S28: SRA validation for single nucleotide insertion in exon 2 of the Mtnr1a gene from T. manatus latirostris; Figure S29: SRA validation for frameshift mutation in exon 2 of the Mtnr1b gene from T. manatus latirostris; Table S1: Blast-n output of Bos Taurus and Homo sapiens sequences against the H. amphibius genome; Table S2: Accession numbers of the Aanat orthologues sequences used in phylogenetic analyses; Table S3: Accession numbers of the Mtnr1a orthologues sequences used in phylogenetic analyses; Table S4: Accession numbers of the Mtnr1b orthologues sequences used in phylogenetic analyses; Gene annotations: In silico gene annotations of Aanat, Mtnr1a and Mtnr1b in H. amphibius.
Author Contributions
Conceptualisation, M.L.-M., R.R. and L.F.C.C.; Methodology, M.L.-M., R.R., L.Q.A., A.M.M. and L.F.C.C.; Software, L.Q.A. and A.M.M.; Validation, M.L.M., R.R. and L.F.C.C.; Formal Analysis, L.Q.A. and A.M.M.; Investigation, M.L.M., L.Q.A., N.S., A.M.M.; Resources, L.F.C.C.; Data Curation, M.L.M., L.Q.A., A.M.M.; Writing—Original Draft Preparation, M.L.-M., R.R., L.Q.A., N.S., A.M.M., L.F.C.C.; Writing—Review & Editing, R.R. and L.F.C.C.; Visualisation, M.L.-M., R.R., A.M.M., L.Q.A.; Supervision, R.R. and L.F.C.C.; Project Administration, L.F.C.C.; Funding Acquisition, L.F.C.C.
Funding
This work was supported by Project No. 031342 co-financed by COMPETE 2020, Portugal 2020 and the European Union through the ERDF, and by FCT through national funds.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Pevet P., Challet E. Melatonin: Both master clock output and internal time-giver in the circadian clocks network. J. Physiol.-Paris. 2011;105:170–182. doi: 10.1016/j.jphysparis.2011.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Moore R.Y. Suprachiasmatic nucleus in sleep-wake regulation. Sleep Med. 2007;8(Suppl. 3):27–33. doi: 10.1016/j.sleep.2007.10.003. [DOI] [PubMed] [Google Scholar]
- 3.Pévet P. Melatonin. Dialogues Clin. Neurosci. 2002;4:57–72. doi: 10.31887/DCNS.2002.4.1/ppevet. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Tan D.X., Hardeland R., Manchester L.C., Paredes S.D., Korkmaz A., Sainz R.M., Mayo J.C., Fuentes-Broto L., Reiter R.J. The changing biological roles of melatonin during evolution: From an antioxidant to signals of darkness, sexual selection and fitness. Biol. Rev. Camb. Philos. Soc. 2010;85:607–623. doi: 10.1111/j.1469-185X.2009.00118.x. [DOI] [PubMed] [Google Scholar]
- 5.Arendt J. Melatonin. Clin. Endocrinol. 1988;29:205–229. doi: 10.1111/j.1365-2265.1988.tb00263.x. [DOI] [PubMed] [Google Scholar]
- 6.Tosches M.A., Bucher D., Vopalensky P., Arendt D. Melatonin signaling controls circadian swimming behavior in marine zooplankton. Cell. 2014;159:46–57. doi: 10.1016/j.cell.2014.07.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Lerner A.B., Case J.D., Takahashi Y., Lee T.H., Mori W. Isolation of melatonin, the Pineal Gland factor that lightens melanocytes1. J. Am. Chem. Soc. 1958;80:2587. doi: 10.1021/ja01543a060. [DOI] [Google Scholar]
- 8.Klein D., Coon S., Roseboom P., Weller J.L., Bernard M., Gastel J.A., Zatz M., Iuvone P., Rodriguez I., Bégay V., et al. The melatonin rhythm-generating enzyme: Molecular regulation of serotonin N-acetyltransferase in the pineal gland. Recent Prog. Horm. Res. 1997;52:307–357. [PubMed] [Google Scholar]
- 9.Simonneaux V., Ribelayga C. Generation of the melatonin endocrine message in mammals: A review of the complex regulation of melatonin synthesis by norepinephrine, peptides, and other pineal transmitters. Pharmacol. Rev. 2003;55:325. doi: 10.1124/pr.55.2.2. [DOI] [PubMed] [Google Scholar]
- 10.Reppart S.M., Weaver D.R., Godson C. Melatonin receptors step into the light: Cloning and classification of subtypes. Trends Pharmacol. Sci. 1996;17:100–102. doi: 10.1016/0165-6147(96)10005-5. [DOI] [PubMed] [Google Scholar]
- 11.Comai S., Ochoa-Sanchez R., Gobbi G. Sleep–wake characterization of double MT1/MT2 receptor knockout mice and comparison with MT1 and MT2 receptor knockout mice. Behav. Brain Res. 2013;243:231–238. doi: 10.1016/j.bbr.2013.01.008. [DOI] [PubMed] [Google Scholar]
- 12.Falcón J., Coon S.L., Besseau L., Cazaméa-Catalan D., Fuentès M., Magnanou E., Paulin C.-H., Boeuf G., Sauzet S., Jørgensen E.H., et al. Drastic neofunctionalization associated with evolution of the timezyme AANAT 500 Mya. Proc. Natl. Acad. Sci. USA. 2014;111:314–319. doi: 10.1073/pnas.1312634110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Klein D.C. Evolution of the vertebrate pineal gland: The aanat hypothesis. Chronobiol. Int. 2006;23:5–20. doi: 10.1080/07420520500545839. [DOI] [PubMed] [Google Scholar]
- 14.Klein D.C. Arylalkylamine N-acetyltransferase: “The Timezyme”. J. Biol. Chem. 2007;282:4233–4237. doi: 10.1074/jbc.R600036200. [DOI] [PubMed] [Google Scholar]
- 15.Kim E.B., Fang X., Fushan A.A., Huang Z., Lobanov A.V., Han L., Marino S.M., Sun X., Turanov A.A., Yang P., et al. Genome sequencing reveals insights into physiology and longevity of the naked mole rat. Nature. 2011;479:223. doi: 10.1038/nature10533. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Panin M., Gabai G., Ballarin C., Peruffo A., Cozzi B. Evidence of melatonin secretion in cetaceans: Plasma concentration and extrapineal HIOMT-like presence in the bottlenose dolphin Tursiops truncatus. Gen. Comp. Endocrinol. 2012;177:238–245. doi: 10.1016/j.ygcen.2012.04.012. [DOI] [PubMed] [Google Scholar]
- 17.Ralph C.L. The pineal gland and geographical distribution of animals. Int. J. Biometeorol. 1975;19:289–303. doi: 10.1007/BF01451040. [DOI] [PubMed] [Google Scholar]
- 18.Ralph C.L., Young S., Gettinger R., O’Shea T.J. Does the manatee have a pineal body? Acta Zool. 1985;66:55–60. doi: 10.1111/j.1463-6395.1985.tb00647.x. [DOI] [Google Scholar]
- 19.Fang X., Seim I., Huang Z., Gerashchenko M.V., Xiong Z., Turanov A.A., Zhu Y., Lobanov A.V., Fan D., Yim S.H., et al. Adaptations to a subterranean environment and longevity revealed by the analysis of mole rat genomes. Cell Rep. 2014;8:1354–1364. doi: 10.1016/j.celrep.2014.07.030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Quay W.B. Pineal atrophy and other neuroendocrine and circumventricular features of the naked mole-rat, heterocephalusglaber (Rüppell), a fossorial, equatorial rodent. J. Neural Transm. 1981;52:107–115. doi: 10.1007/BF01253102. [DOI] [PubMed] [Google Scholar]
- 21.Cajochen C., Pache M., Flammer J., Wirz-Justice A. Thermoregulatory effects of melatonin in relation to sleepiness AU - Kräuchi, Kurt. Chronobiol. Int. 2006;23:475–484. doi: 10.1080/07420520500545854. [DOI] [PubMed] [Google Scholar]
- 22.Dauchy R.T., Wren-Dail M.A., Hoffman A.E., Hanifin J.P., Warfield B., Brainard G.C., Hill S.M., Belancio V.P., Dauchy E.M., Blask D.E. Effects of daytime exposure to light from blue-enriched light-emitting diodes on the nighttime melatonin amplitude and circadian regulation of rodent metabolism and physiology. Comp. Med. 2016;66:373–383. [PMC free article] [PubMed] [Google Scholar]
- 23.Fasick J.I., Robinson P.R. Adaptations of cetacean retinal pigments to aquatic environments. Front. Ecol. Evol. 2016;4 doi: 10.3389/fevo.2016.00070. [DOI] [Google Scholar]
- 24.Camacho C., Coulouris G., Avagyan V., Ma N., Papadopoulos J., Bealer K., Madden T.L. BLAST+: Architecture and applications. BMC Bioinform. 2009;10:421. doi: 10.1186/1471-2105-10-421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stanke M., Morgenstern B. AUGUSTUS: A web server for gene prediction in eukaryotes that allows user-defined constraints. Nucleic Acids Res. 2005;33:W465–W467. doi: 10.1093/nar/gki458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Waterhouse A.M., Procter J.B., Martin D.M.A., Clamp M., Barton G.J. Jalview Version 2—A multiple sequence alignment editor and analysis workbench. Bioinformatics. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Katoh K., Rozewicki J., Yamada K.D. MAFFT online service: Multiple sequence alignment, interactive sequence choice and visualization. Brief. Bioinform. 2017 doi: 10.1093/bib/bbx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Katoh K., Standley D.M. MAFFT Multiple Sequence Alignment Software Version 7: Improvements in performance and usability. Mol. Biol. Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Guindon S., Dufayard J.-F., Lefort V., Anisimova M., Hordijk W., Gascuel O. New algorithms and methods to estimate maximum-likelihood phylogenies: Assessing the performance of PhyML 3.0. Syst. Biol. 2010;59:307–321. doi: 10.1093/sysbio/syq010. [DOI] [PubMed] [Google Scholar]
- 30.Lefort V., Longueville J.-E., Gascuel O. SMS: Smart model selection in PhyML. Mol. Biol. Evol. 2017;34:2422–2424. doi: 10.1093/molbev/msx149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Huson D.H., Scornavacca C. Dendroscope 3: An interactive tool for rooted phylogenetic trees and networks. Syst. Biol. 2012;61:1061–1067. doi: 10.1093/sysbio/sys062. [DOI] [PubMed] [Google Scholar]
- 32.Foote A.D., Liu Y., Thomas G.W.C., Vinař T., Alföldi J., Deng J., Dugan S., van Elk C.E., Hunter M.E., Joshi V., et al. Convergent evolution of the genomes of marine mammals. Nat. Genet. 2015;47:272–275. doi: 10.1038/ng.3198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Zimin A.V., Delcher A.L., Florea L., Kelley D.R., Schatz M.C., Puiu D., Hanrahan F., Pertea G., Van Tassell C.P., Sonstegard T.S., et al. A whole-genome assembly of the domestic cow, Bos taurus. Genome Biol. 2009;10:R42. doi: 10.1186/gb-2009-10-4-r42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Jones S.J.M., Taylor G.A., Chan S., Warren R.L., Hammond S.A., Bilobram S., Mordecai G., Suttle C.A., Miller K.M., Schulze A., et al. The genome of the beluga whale (Delphinapterus leucas) Genes. 2017;8:378. doi: 10.3390/genes8120378. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zhou X., Sun F., Xu S., Fan G., Zhu K., Liu X., Chen Y., Shi C., Yang Y., Huang Z., et al. Baiji genomes reveal low genetic variability and new insights into secondary aquatic adaptations. Nat. Commun. 2013;4:2708. doi: 10.1038/ncomms3708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.DeWoody J.A., Fernandez N.B., Bruniche-Olsen A., Antonides J.D., Doyle J.M., San Miguel P., Westerman R., Vertyankin V.V., Godard-Codding C.A.J., Bickham J.W. Characterization of the gray whale Eschrichtius robustus genome and a genotyping array based on single-nucleotide polymorphisms in candidate genes. Biol. Bull. 2017;232:186–197. doi: 10.1086/693483. [DOI] [PubMed] [Google Scholar]
- 37.Park J.Y., An Y.-R., Kanda N., An C.-M., An H.S., Kang J.-H., Kim E.M., An D.-H., Jung H., Joung M., et al. Cetaceans evolution: Insights from the genome sequences of common Minke whales. BMC Genom. 2015;16:13. doi: 10.1186/s12864-015-1213-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ming Y., Jian J., Yu F., Yu X., Wang J., Liu W. Molecular footprints of inshore aquatic adaptation in Indo-Pacific humpback dolphin (Sousa chinensis) Genomics. 2018 doi: 10.1016/j.ygeno.2018.07.015. [DOI] [PubMed] [Google Scholar]
- 39.Arnason U., Lammers F., Kumar V., Nilsson M.A., Janke A. Whole-genome sequencing of the blue whale and other rorquals finds signatures for introgressive gene flow. Sci. Adv. 2018;4 doi: 10.1126/sciadv.aap9873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Yim H.-S., Cho Y.S., Guang X., Kang S.G., Jeong J.-Y., Cha S.-S., Oh H.-M., Lee J.-H., Yang E.C., Kwon K.K., et al. Minke whale genome and aquatic adaptation in cetaceans. Nat. Genet. 2014;46:88–92. doi: 10.1038/ng.2835. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Jebb D., Hiller M. Recurrent loss of HMGCS2 shows that ketogenesis is not essential for the evolution of large mammalian brains. Elife. 2018;7 doi: 10.7554/eLife.38906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lopes-Marques M., Machado A.M., Barbosa S., Fonseca M.M., Ruivo R., Castro L.F.C. Cetacea are natural knockouts for IL20. Immunogenetics. 2018 doi: 10.1007/s00251-018-1071-5. [DOI] [PubMed] [Google Scholar]
- 43.Sharma V., Hecker N., Roscito J.G., Foerster L., Langer B.E., Hiller M. A genomics approach reveals insights into the importance of gene losses for mammalian adaptations. Nat. Commun. 2018;9:1215. doi: 10.1038/s41467-018-03667-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Springer M.S., Gatesy J. Evolution of the MC5R gene in placental mammals with evidence for its inactivation in multiple lineages that lack sebaceous glands. Mol. Phylogenet. Evol. 2018;120:364–374. doi: 10.1016/j.ympev.2017.12.010. [DOI] [PubMed] [Google Scholar]
- 45.Botros H.G., Legrand P., Pagan C., Bondet V., Weber P., Ben-Abdallah M., Lemière N., Huguet G., Bellalou J., Maronde E., et al. Crystal structure and functional mapping of human ASMT, the last enzyme of the melatonin synthesis pathway. J. Pineal Res. 2012;54:46–57. doi: 10.1111/j.1600-079X.2012.01020.x. [DOI] [PubMed] [Google Scholar]
- 46.Marti-Solans J., Belyaeva O.V., Torres-Aguila N.P., Kedishvili N.Y., Albalat R., Canestro C. Coelimination and survival in gene network evolution: Dismantling the RA-signaling in a chordate. Mol. Biol. Evol. 2016;33:2401–2416. doi: 10.1093/molbev/msw118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Tan D.-X., Hardeland R., Back K., Manchester L.C., Alatorre-Jimenez M.A., Reiter R.J. On the significance of an alternate pathway of melatonin synthesis via 5-methoxytryptamine: Comparisons across species. J. Pineal Res. 2016;61:27–40. doi: 10.1111/jpi.12336. [DOI] [PubMed] [Google Scholar]
- 48.Slominski A., Pisarchik A., Semak I., Sweatman T., Wortsman J. Characterization of the serotoninergic system in the C57BL/6 mouse skin. Eur. J. Biochem. 2003;270:3335–3344. doi: 10.1046/j.1432-1033.2003.03708.x. [DOI] [PubMed] [Google Scholar]
- 49.Ried K., Rao E., Schiebel K., Rappold G.A. Gene duplications as a recurrent theme in the evolution of the human pseudoautosomal region 1: isolation of the gene ASMTL. Hum. Mol. Genet. 1998;7:1771–1778. doi: 10.1093/hmg/7.11.1771. [DOI] [PubMed] [Google Scholar]
- 50.Zhang K., Ruan Z., Li J., Bian C., You X., Coon L.S., Shi Q. A comparative genomic and transcriptomic survey provides novel insights into N-acetylserotonin methyltransferase (ASMT) in Fish. Molecules. 2017;22:1653. doi: 10.3390/molecules22101653. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Pfeffer M., Korf H.-W., Wicht H. Synchronizing effects of melatonin on diurnal and circadian rhythms. Gen. Comp. Endocrinol. 2017 doi: 10.1016/j.ygcen.2017.05.013. [DOI] [PubMed] [Google Scholar]
- 52.Lyamin O.I., Manger P.R., Ridgway S.H., Mukhametov L.M., Siegel J.M. Cetacean sleep: An unusual form of mammalian sleep. Neurosci. Biobehav. Rev. 2008;32:1451–1484. doi: 10.1016/j.neubiorev.2008.05.023. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Sekiguchi Y., Arai K., Kohshima S. Sleep in continuously active dolphins. Nature. 2006;441:E9. doi: 10.1038/nature04898. [DOI] [PubMed] [Google Scholar]
- 54.Reiter R.J., Tan D.X., Galano A. Melatonin: Exceeding expectations. Physiology. 2014;29:325–333. doi: 10.1152/physiol.00011.2014. [DOI] [PubMed] [Google Scholar]
- 55.Albalat R., Canestro C. Evolution by gene loss. Nat. Rev. Genet. 2016;17:379–391. doi: 10.1038/nrg.2016.39. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.