Summary
Introduction
Anatomic characteristics of kidneys derived from ultrasound images are potential biomarkers of children with congenital abnormalities of the kidney and urinary tract (CAKUT), but current methods are limited by the lack of automated processes that accurately classify diseased and normal kidneys.
Objective
We sought to evaluate the diagnostic performance of deep transfer learning techniques to classify kidneys of normal children and those with CAKUT.
Study design
A transfer learning method was developed to extract features of kidneys from ultrasound images obtained during routine clinical care of 50 children with CAKUT and 50 controls. To classify diseased and normal kidneys, support vector machine classifiers were built on the extracted features using 1) transfer learning imaging features from a pre-trained deep learning model, 2) conventional imaging features, and 3) their combination. These classifiers were compared and their diagnosis performance was measured using area under the receiver operating characteristic curve (AUC), accuracy, specificity, and sensitivity.
Results
The AUC for classifiers built upon the combination features were 0.92, 0.88, and 0.92 for discriminating left, right, and bilateral abnormal kidney scans from controls with classification rates of 84%, 81%, 87%, specificity of 84%, 74%, 88%, and sensitivity of 85%, 88%, 86%, respectively. These classifiers performed better than classifiers built on either the transfer learning features or the conventional features alone (p<0.001).
Discussion
The present study validated transfer learning techniques for imaging feature extraction of ultrasound images to build classifiers for distinguishing children with CAKUT from controls. The experiments have demonstrated that the classifiers built on the transfer learning features and conventional image features could distinguish abnormal kidney images from controls with AUCs greater than 0.88, indicating that classification of ultrasound kidney scans has a great potential to aid kidney disease diagnosis. A limitation of the present study is the moderate number of patients that contributed data to the transfer learning approach.
Conclusions
The combination of transfer learning and conventional imaging features yielded the best classification performance for distinguishing children with CAKUT from controls based on ultrasound images of kidneys.
Keywords: Transfer learning, congenital abnormalities of the kidney and urinary tract, convolutional neural networks, ultrasound images, kidney
Introduction
Congenital abnormalities of the kidney and urinary tract (CAKUT), including posterior urethral valves (PUV) and kidney dysplasia, account for 50–60% of chronic kidney disease (CKD) in children and are the most common cause of end-stage renal disease (ESRD) in this age group [1, 2]. The widespread use of ultrasound imaging facilitates early detection of CAKUT, but current methods are limited by the lack of automated processes that accurately classify diseased and normal kidneys.
Recent studies have demonstrated that features computed from imaging data have promising performance for predicting risk of ESRD in boys with posterior urethral valves and decline in kidney function among adults with autosomal dominant polycystic kidney disease [3, 4]. Therefore, imaging features computed from ultrasound scans might facilitate automated detection of CAKUT. However, existing studies typically adopt imaging features that were empirically defined and therefore may not fully harness the predictive power of all data contained in ultrasound images.
In recent years, deep learning techniques have demonstrated promising performance in learning imaging features for a variety of pattern recognition tasks [5]. In these studies, convolutional neural networks (CNNs) have been applied to learn informative imaging features by directly optimizing the pattern recognition performance. Therefore, it is expected that incorporating the deep learning techniques into ultrasound imaging data analysis would improve ultrasound image classification and subsequently allow automated CAKUT diagnosis in children based on images obtained routinely for clinical care.
With the ultimate objective of developing anatomic biomarkers of CKD progression for children with CAKUT, we developed a pattern recognition method for distinguishing children with CAKUT from controls based on their ultrasound kidney images. Building on successful deep learning and transfer learning techniques [6, 7], this transfer learning method for extracting imaging features from 2D ultrasound kidney images uses a pre-trained model of convolutional neural networks (CNNs), namely imagenet-caffe-alex. In this study, we determined the capacity of these features to correctly classify abnormal and normal kidneys [6]. We hypothesized that integration of deep learning based imaging features and conventional imaging features better distinguishes CAKUT patients from controls based on their ultrasound kidney images than conventional and deep learning based imaging features alone.
Materials and Methods
Setting and Participants
Ultrasound kidney images were obtained from 50 controls and 50 patients with CAKUT evaluated at The Children’s Hospital of Philadelphia. Participants were randomly sampled from patients enrolled in the Registry of Urologic Disease, a comprehensive patient registry that includes 90% of patients seen in the Urology clinic since 2000. Controls were children with unilateral mild hydronephrosis (Society of Fetal Urology grade I-II). Both kidneys were included as control data to increase the generalizability of the feature extraction algorithm by ensuring that the full spectrum of “normal” kidneys were considered. We considered these kidneys normal because mild hydronephrosis is generally a benign finding that stabilizes or resolves spontaneously in 98% of patients and does not affect the echogenicity, growth, or function of the affected or contralateral normal kidney [8, 9].
For abnormal kidneys, we used ultrasound images from 50 children with CAKUT (e.g. PUV, kidney dysplasia), who have varying degrees of increased cortical echogenicity, decreased corticomedullary differentiation, and hydronephrosis (please see kidney ultrasound images in online supplement). Of the 50 patients with CAKUT, 35 had bilateral renal anomalies, 4 patients had abnormal left kidneys, and 11 patients had abnormal right kidneys.
2D kidney ultrasound images of these patients were obtained using a Philips IU22 Ultrasound system at a frequency of 55HZ using an abdominal transducer. All images were obtained for routine clinical care. The first ultrasound after birth was used and all identifying information was removed. The work described has been carried out in accordance with the Declaration of Helsinki. The study has been reviewed and approved by the Institutional Review Board at the Children’s Hospital of Philadelphia.
Outcome
The clinical diagnosis of CAKUT was the outcome and gold standard against which the computer-aided diagnosis of CAKUT was compared. These clinical diagnoses were recorded prospectively in the Registry of Urology Disease and were independently verified by a pediatric urologist, who was not involved in the application of the feature extraction algorithms to the images.
Computer aided diagnosis of CAKUT in children based on ultrasound imaging data
The computer aided diagnosis of CAKUT in children based on ultrasound imaging data is formulated as an image classification problem, i.e., classifying ultrasound images into CAKUT cases and controls. Our method consists of 3 components, namely: 1) kidney segmentation, 2) feature extraction, and 3) support vector machine (SVM) based classification, as illustrated in Figure 1.
Figure 1.
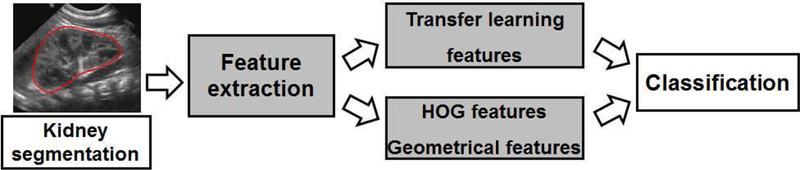
Flowchart of classification of ultrasound kidney images for computer aided diagnosis of kidney disorders.
Kidney Segmentation
We adopted a graph-cuts method to segment kidneys in ultrasound images [10]. Particularly, the graph-cuts method segments kidneys based on both image intensity information and texture features extracted using Gabor filters, which are orientation-sensitive image filters generally used for characterizing image texture in a similar way as the human visual system [11, 12]. This image segmentation method has achieved promising performance for segmenting kidneys in ultrasound images [10]. To make ultrasound kidney images of different subjects directly comparable, we normalized ultrasound kidney images of different subjects as detailed in the supplementary material.
Feature Extraction
From the normalized images, we then extracted transfer learning features based on a deep CNN model and conventional imaging features. Particularly, the CNN model is a deep learning method to learn image filters for exacting image features by optimizing their discriminative performance and have been successfully applied to a variety of image analysis problems [5], while conventional imaging features are computed using image filters manually designed, including histogram of oriented gradients (HOG) [13] features and geometrical features. Details about the feature extraction are presented in the supplementary document.
Classification of kidney images based on imaging features
The diagnosis of CAKUT was modeled as a pattern classification problem based on ultrasound imaging features. Particularly, SVM classifiers [14] were built on the transfer learning imaging features and conventional image features to distinguish US images of children with CAKUT from those of controls. Details about the SVM based classification are presented in the supplementary material. In order to investigate if the transfer learning based features and the conventional image features could provide complementary information for distinguishing abnormal kidney images from controls, we compared SVM classifiers built on different sets of features, including the transfer learning features (referred to as CNN features), the conventional imaging features, and their combination. Particularly, there were 7 settings of features, including CNN features, HOG features, Geometrical features, HOG+Geometrical features, CNN+Geometrical features, CNN+HOG features, CNN+HOG+Geometrical features. We built SVM classification models based on all the available imaging data for the bilateral kidney images, left kidney images, and right kidney images separately.
Evaluation of diagnostic performance
We tested the diagnostic performance of the above SVM classification algorithms using the clinical classification as the gold standard. We carried out classification experiments for bilateral (35 CAKUT vs. 50 controls) kidney images, left kidney images (39 CAKUT vs. 50 controls), and right kidney images (46 CAKUT vs. 50 controls) separately. The feature extraction algorithms were applied to images without knowledge of the clinical diagnosis.
Classification results were validated using 10-fold cross-validation. Briefly, we first randomly partitioned the whole dataset into 10 subsets, and then selected one subset for testing and used the remaining subsets for training. We repeated the process 100 times to avoid the possible bias during dataset partitioning for cross-validation. In the cross-validation experiments, the algorithms were trained to build classifiers based on training data. Only after the algorithms classified the images as CAKUT or normal were the clinical diagnoses assigned to the testing images. The diagnostic performance was measured by correct classification rate, specificity, sensitivity, and area under the receiver operating characteristic (ROC) curve (AUC). Classification models built on different features were compared using Wilcoxon signed-rank test based on their classification performance measures determined by the cross-validation procedure.
RESULTS
Patient characteristics
The demographic and clinical data are summarized in Table 1. There were no differences in age, sex, or race between patients with CAKUT and controls. The CAKUT subjects had heterogenous abnormalities, as summarized in Table 2.
Table 1.
Demographic information.
| Controls(n=50) | CAKUT (n=50) | p-value | |
|---|---|---|---|
| Sex; n (%) | |||
| Male | 34 (68) | 34 (68) | 1 |
| Female | 16 (32) | 16 (32) | |
| Age (days, mean±std) | 73.2±69.0 | 111.0±262.1 | 0.327 |
| Race; n (%) | |||
| Asian | 3 (6) | 1 (2) | |
| Black or African American | 7 (14) | 14 (28) | 0.260 |
| Native Hawaiian or other pacific | 0 (0) | 1 (2) | |
| islander | 31 (62) | 22 (44) | |
| White | 9 (18) | 12 (24) | |
| Unknown | |||
| Ethnicity | 0 (0) | 6 (12) | |
| Hispanic or Latino | 35 (70) | 44 (88) | <0.001 |
| Not Hispanic or Latino | 15 (30) | 0 (0) | |
| Not Recorded | |||
Age: Two-sample t-test
Sex, Race, Ethnicity: Chi-square test
Table 2.
Characteristics of CAKUT subjects (n=50).
| Characteristics | number |
|---|---|
| Posterior urethral valves | 24 |
| Multicystic dysplastic kidney | 6 |
| Renal hypodysplasia | 11 |
| Ureterocele | 3 |
| Vesicoureteral reflux | 2 |
| Ectopic ureter | 3 |
| Autosomal recessive polycystic kidney disease | 1 |
Classification performance of the transfer learning features and conventional image features
Figure 2 shows ROC curves of one run of 10-fold cross-validation for the classification of the left, right, and bilateral kidney scans using different imaging features. As summarized in Table 3, the transfer learning features had better classification performance than the conventional features (Wilcoxon signed-rank test, p<0.001), the combination of the HOG features and the geometrical features had better classification performance than the HOG or geometrical features alone (p<0.001), and the integration of the transfer learning and conventional image features had the best performance (p<0.001).
Figure 2.
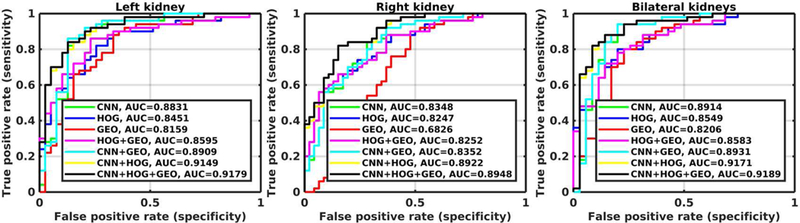
ROC curves of one run of 10-fold cross-validation for classification models built on different imaging features, including CNN features, HOG features, geometrical (GEO) features, HOG + GEO features, CNN + GEO features, CNN + HOG features, and CNN + HOG + GEO features.
Table 3.
Classification performance estimated from 100 runs of 10-fold cross-validation (Mean±std).
| CNN | HOG | Geometrical | HOG + Geometrical |
CNN + Geometrical |
CNN + HOG |
CNN + HOG + Geometrical |
||
|---|---|---|---|---|---|---|---|---|
| Accuracy | ||||||||
| Left | 0.85±1.8e-2 | ^#*0.77±2.3e-2 | ^*0.78±1.5e-2 | ^*0.79±2.7e-2 | 0.87±1.5e-2 | *0.84±1.4e-2 | 0.84±1.6e-2 | |
| Right | *0.75±2.0e-2 | *0.74±2.5e-2 | ^#*0.67±1.7e-2 | *0.74±2.4e-2 | *0.74±2.1e-2 | 0.81±1.8e-2 | 0.81±1.8e-2 | |
| Bilateral | *0.85±1.2e-2 | ^#*0.78±2.4e-2 | ^#*0.76±1.8e-2 | ^*0.79±2.3e-2 | *0.85±1.4e-2 | *0.86±2.1e-2 | 0.87±2.1e-2 | |
| AUC | ||||||||
| Left | *0.88±0.8e-2 | ^#*0.84±1.5e-2 | ^#*0.80±0.9e-2 | ^*0.86±1.5e-2 | *0.88±0.9e-2 | *0.92±0.8e-2 | 0.92±0.7e-2 | |
| Right | *0.83±1.3e-2 | *0.83±1.6e-2 | ^#*0.67±1.7e-2 | *0.83±1.6e-2 | *0.83±1.2e-2 | 0.88±1.0e-2 | 0.88±1.1e-2 | |
| Bilateral | *0.89±0.8e-2 | ^#*0.87±1.6e-2 | ^#*0.82±1.3e-2 | ^*0.87±1.5e-2 | *0.89±0.8e-2 | *0.92±0.7e-2 | 0.92±0.7e-2 | |
| Specificity | ||||||||
| Left | *0.80±2.6e-2 | ^*0.70±3.9e-2 | ^#*0.63±2.9e-2 | ^*0.71±4.3e-2 | *0.82±2.6e-2 | *0.83±1.9e-2 | 0.84±2.2e-2 | |
| Right | *0.66±2.6e-2 | ^*0.63±4.3e-2 | ^#*0.45±2.3e-2 | ^*0.62±4.2e-2 | *0.65±2.7e-2 | 0.74±2.6e-2 | 0.74±2.6e-2 | |
| Bilateral | *0.83±1.4e-2 | ^#*0.78±3.3e-2 | ^#*0.62±3.2e-2 | ^*0.79±3.3e-2 | *0.82±1.8e-2 | 0.88±2.6e-2 | 0.88±2.6e-2 | |
| Sensitivity | ||||||||
| Left | 0.89±2.2e-2 | ^#*0.82±2.6e-2 | 0.90±1.4e-2 | ^0.84±2.7e-2 | 0.91±1.4e-2 | *0.84±1.7e-2 | 0.85±1.8e-2 | |
| Right | *0.83±2.8e-2 | *0.84±2.5e-2 | 0.88±2.1e-2 | *0.85±2.4e-2 | *0.83±3.1e-2 | 0.88±2.0e-2 | 0.88±2.1e-2 | |
| Bilateral | 0.86±1.9e-2 | ^#*0.78±2.9e-2 | 0.86±1.8e-2 | ^*0.79±2.9e-2 | 0.87±1.9e-2 | *0.85±2.6e-2 | 0.86±2.6e-2 | |
CNN + HOG + Geometrical better than others; Wilcoxon signed-rank test, p<0.001
CNN better than HOG, Geometrical, and HOG + Geometrical; Wilcoxon signed-rank test, p<0.001
HOG + Geometrical better than HOG or Geometrical alone; Wilcoxon signed-rank test, p<0.001
DISCUSSION
In this study, we found that the performance of machine learning algorithms in correctly classifying kidneys of children with CAKUT and normal controls was high. In particular, the combination of transfer learning and conventional image features performed best, with sensitivity, specificity, discrimination, and accuracy all exceeding 87% for bilateral kidney images. The relatively small standard deviations of classification performance measures in different runs of the cross-validation experiments demonstrated that the classification performance was relatively stable in terms of changes in training samples. These results suggest that the classifiers built upon the combination of transfer learning and conventional imaging features could robustly distinguish children with CAKUT from controls based on their ultrasound kidney image, despite the heterogeneity of diagnoses and kidney characteristics of individuals with CAKUT. In addition, transfer learning classifiers were able to successfully discriminate individuals with CAKUT from individuals with mild hydronephrosis, which would increase the difficulty of classification. These results have importance for the ultimate development of models incorporating anatomic information obtained during routine clinical care that predict important outcomes such as CKD progression.
Machine learning techniques have been adopted in studies of ultrasound images. Prior studies have used decision support systems to classify ultrasound images of normal patients and patients with kidney disease based on second order grey level co-occurrence matrix statistical features [15]. In addition, neural networks in conjunction with principal component analysis have been used to classify ultrasound kidney images [16], and SVM classifiers have been built on texture features extracted from regions of interest of ultrasound images to classify kidney images [17, 18]. These studies have demonstrated that pattern classifiers built on imaging features could obtain promising performance for classifying ultrasound imaging data.
In this study, we extend the findings from these prior studies by exploring the deep learning method for feature extraction on ultrasound imaging data because it can discover the representations needed for computer aided diagnosis from raw data automatically by replacing manual feature engineering. Since a large dataset is typically needed to build a generalizable deep learning based classification model, for applications with small datasets the deep learning tools are often adopted in a transform learning setting (i.e., applying deep learning models trained based on a large dataset for one problem to a different but related problem with relatively small training dataset) [19]. The transfer learning strategy by applying trained CNNs for feature extraction to small datasets has achieved promising performance in pattern recognition studies of medical image data [7, 20]. Furthermore, it has been demonstrated that classifiers built on combination of transfer learning features and hand-crafted features typically achieve better pattern recognition performance than those built on transfer learning features and hand-crafted features alone [7, 21].
The present study validated the transfer learning techniques for imaging feature extraction to build classifiers for distinguishing children with CAKUT from controls based on ultrasound images, aiming to discover anatomic biomarkers of CKD progression for children with CAKUT. The experiments have demonstrated that the classifiers built on the transfer learning features and conventional image features could distinguish abnormal kidney images from controls with AUCs greater than 0.88, indicating that classification of ultrasound kidney scans has a great potential to aid kidney disease diagnosis. Since the imaging feature extraction and SVM classification can be finished within seconds, this progress suggests real-time of CKD progression using machine learning of ultrasound images may be possible. The pattern classification performance might be further improved by building pattern recognition models based on a larger dataset and using advanced deep learning techniques [22], and fully automated deep learning based image segmentation tools may further improve the kidney segmentation efficiency [23, 24].
A limitation of the present study is the moderate number of patients that contributed data to the transfer learning approach. Based on the available dataset, we carried out pattern classification studies for distinguishing left, right, and bilateral abnormal kidneys from controls. Although the classification performance varied across these settings, the results demonstrated that classification models built on the transfer learning features had overall better classification performance than those built on conventional imaging features and the integration of the transfer learning based features and the conventional imaging features had the overall best classification performance. We will further validate our method based on larger datasets.
Conclusions
This pattern classification study of ultrasound kidney data has demonstrated that transfer learning for feature extraction based on deep CNN models could improve pattern classification models that are built on conventional imaging features, including texture features and geometric features. Our results suggest that promising classification performance could be achieved by the classification model built on a combination of the transfer learning features and the conventional imaging features. The promising classification performance also indicates that the imaging features might be informative for predicting CKD progression.
Supplementary Material
Acknowledgements
Blind information
This work was supported by in part by China Postdoctoral Science Foundation [grant number 2015M581203]; the International Postdoctoral Exchange Fellowship Program [grant number 20160032]; National Institutes of Health grants [grant numbers DK114786]; the National Center for Advancing Translational Sciences of the National Institutes of Health under Award Number UL1TR001878; and the Institute for Translational Medicine and Therapeutics’ (ITMAT) Transdisciplinary Program in Translational Medicine and Therapeutics.
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final citable form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Disclosures
The authors declare that they have no relevant financial interests.
References
- [1].Dodson JL, Jerry-Fluker JV, Ng DK, Moxey-Mims M, Schwartz GJ, Dharnidharka VR, et al. Urological disorders in chronic kidney disease in children cohort: clinical characteristics and estimation of glomerular filtration rate. The Journal of urology. 2011;186:1460–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Neild GH. What do we know about chronic renal failure in young adults? I. Primary renal disease. Pediatric nephrology 2009;24:1913–9. [DOI] [PubMed] [Google Scholar]
- [3].Kline TL, Korfiatis P, Edwards ME, Bae KT, Yu A, Chapman AB, et al. Image texture features predict renal function decline in patients with autosomal dominant polycystic kidney disease. Kidney International 2017;92:1206–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Pulido JE, Furth SL, Zderic SA, Canning DA, Tasian GE. Renal parenchymal area and risk of ESRD in boys with posterior urethral valves. Clinical journal of the American Society of Nephrology : CJASN 2014;9:499–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].LeCun Y, Bengio Y, Hinton G. Deep learning. Nature 2015;521:436–44. [DOI] [PubMed] [Google Scholar]
- [6].Krizhevsky A, Sutskever I, Hinton GE. ImageNet classification with deep convolutional neural networks. Advances in Neural Information Processing Systems 25 (NIPS 2012). 2012:1097–105. [Google Scholar]
- [7].Cheng PM, Malhi HS. Transfer learning with convolutional neural networks for classification of abdominal ultrasound images. Journal of Digital Imaging 2017;30:234–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Gokaslan F, Yalcinkaya F, Fitoz S, Ozcakar ZB. Evaluation and outcome of antenatal hydronephrosis: a prospective study. Ren Fail 2012;34:718–21. [DOI] [PubMed] [Google Scholar]
- [9].Longpre M, Nguan A, Macneily AE, Afshar K. Prediction of the outcome of antenatally diagnosed hydronephrosis: a multivariable analysis. J Pediatr Urol 2012;8:135–9. [DOI] [PubMed] [Google Scholar]
- [10].Zheng Q, Warner S, Tasian G, Fan Y. A dynamic graph-cuts method with integrated multiple feature maps for segmenting kidneys in ultrasound images. Academic Radiology 2018;25:1136–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Marcelja S Mathematical-Description of the Responses of Simple Cortical-Cells. J Opt Soc Am 1980;70:1297–300. [DOI] [PubMed] [Google Scholar]
- [12].Daugman JG. Uncertainty Relation for Resolution in Space, Spatial-Frequency, and Orientation Optimized by Two-Dimensional Visual Cortical Filters. J Opt Soc Am A 1985;2:1160–9. [DOI] [PubMed] [Google Scholar]
- [13].Dalal N, Triggs B. Histograms of Oriented Gradients for Human Detection. IEEE Conference on Computer Vision and Pattern Recognition (CVPR) 2005;II:886–93. [Google Scholar]
- [14].Fan RE, Chang KW, Hsieh CJ, Wang XR, Lin CJ. LIBLINEAR: A library for large linear classification. Journal of Machine Learning Research 2008;9:1871–4. [Google Scholar]
- [15].Sharma K, Virmani J. A dicision support system for classification of normal and medical renal disease using ultrasound images: A decision support system for medical renal diseases. International Journal of Ambient Computing and Intelligence 2017;8:52–69. [Google Scholar]
- [16].Abou-Chadi FEZ, Mekky N. Classification of ultrasound kidney images using PCA and neural networks. International Journal of Advanced Computer Science and Applications 2015;6:53–7. [Google Scholar]
- [17].Bama S, Selvathi D. Discrete tchebichef moment based machine learning method for the classification of disorder with ultrasound kidney images. Biomedical Research 2016:s223–s9.
- [18].Subramanya MB, Kumar V, Mukherjee S, Saini M. SVM-based CAC system for B-mode kidney ultrasound images. Journal of Digital Imaging 2015;28:448–58. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Pratt LY. Discriminability-based transfer between neural networks. NIPS Conference: Advances in Neural Information Processing Systems 1993:204–11.
- [20].Weiss K, Khoshgoftaar TM, Wang DD. A survey of transfer learning. Journal of Big Data 2016;3:9:1–40. [Google Scholar]
- [21].Ciompi F, de Hoop B, van Riel SJ, Chung K, Scholten ET, Oudkerk M, et al. Automatic classification of pulmonary peri-fissural nodules in computed tomography using an ensemble of 2D views and a convolutional neural network out of the box. Medical Image Analysis 2015;26:195–202. [DOI] [PubMed] [Google Scholar]
- [22].Li H, Habes M, Fan Y. Deep Ordinal Ranking for Multi-Category Diagnosis of Alzheimer’s Disease using Hippocampal MRI data. arXiv:170901599 2017.
- [23].Men K, Boimel P, Janopaul-Naylor J, Zhong H, Huang M, Geng H, et al. Cascaded atrous convolution and spatial pyramid pooling for more accurate tumor target segmentation for rectal cancer radiotherapy. Physics in medicine and biology 2018;63:185016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Zhao X, Wu Y, Song G, Li Z, Zhang Y, Fan Y. A deep learning model integrating FCNNs and CRFs for brain tumor segmentation. Medical Image Analysis 2018;43:98–111. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


