Significance
Plant seedlings encounter increasing levels of UV-B irradiation during their emergence from soil as they undergo a light-induced developmental process termed photomorphogenesis. However, less is known about the molecular mechanisms underlying plant responses to this invisible part of the light spectrum. Here, we reveal that the UV-B light signal is interpreted coordinately by two sets of E3 ligases and a transcription factor in Arabidopsis. The UV-B–inducible E3 CUL4-DDB1–RUP1/RUP2 complex targets the master transcription factor HY5 for proteolysis. COP1 directly interacts with and degrades RUP1/RUP2 to stabilize HY5, ultimately promoting photomorphogenesis under UV-B light. These findings uncover the mechanism of plant responses to UV-B irradiation, and are critical to develop methods to improve plant growth and light energy utilization.
Keywords: light signaling, UV-B, E3 ligase
Abstract
Photomorphogenesis is a pivotal developmental strategy used by plants to respond to environmental light levels. During emergence from the soil and the establishment of photomorphogenesis, seedlings encounter increasing levels of UV-B irradiation and develop adaptive responses accordingly. However, the molecular mechanisms that orchestrate UV-B signaling cascades remain elusive. Here, we provide biochemical and genetic evidence that the prolonged signaling circuits of UV-B–induced photomorphogenesis involve two sets of E3 ligases and a transcription factor in Arabidopsis thaliana. The UV-B–inducible protein RUP1/RUP2 associates with the CUL4-DDB1 scaffold to form an E3 ligase, which represses photomorphogenesis by mediating the degradation of HY5, the hub transcription factor in the light signaling pathway. Conversely, COP1 directly targets RUP1/RUP2 for ubiquitination and degradation, leading to balanced RUP1/RUP2 accumulation, alleviation of the COP1–HY5 interaction, and stabilization of HY5 protein. Therefore, our study reveals that these two E3-substrate modules, CUL4-DDB1-RUP1/RUP2-HY5 and COP1-RUP1/RUP2, constitute the repression and derepression machinery by which plants respond to prolonged UV-B irradiation in photomorphogenic development.
Plants are inevitably exposed to UV-B irradiation, which has been increasing due to ozone depletion over the past few decades. Exposure to UV-B light (280–315 nm) brings about different physiological responses in plants, depending on its wavelength, fluence rate, and duration. As young seedlings emerge from the soil, they perceive low levels of UV-B irradiation, which causes them to develop photomorphogenic responses and undergo UV-B acclimation (1).
UV RESISTANCE LOCUS 8 (UVR8), which was initially identified in Arabidopsis thaliana, shares sequence similarity with Regulator of Chromosome Condensation 1 (RCC1) (2). UVR8 is a highly conserved UV-B photoreceptor among plant species (3, 4). It forms a symmetrical homodimer in the absence of UV-B light. Upon UV-B irradiation, the tryptophan residues on the surface of dimeric UVR8 collectively serve as an intrinsic chromophore to sense UV-B light. This light-sensing step intermediately disrupts the homodimeric interface and results in UVR8 monomerization (5, 6). Monomerized UVR8 subsequently interacts with CONSTITUTIVELY PHOTOMORPHOGENIC 1 (COP1) (7), and accumulates in the nucleus with the aid of COP1 (8, 9). Although critically repressing photomorphogenesis under light conditions without UV-B, COP1 is a pivotal positive regulator of UV-B–induced photomorphogenesis (7). The nuclear UVR8-COP1 module governs photomorphogenic UV-B signal transduction (10) by triggering the dissociation of the COP1–SUPPRESSOR OF PHYA (SPA) core complex from the CULLIN 4-DAMAGED DNA BINDING PROTEIN 1 (CUL4-DDB1) E3 apparatus, ultimately stabilizing the central photomorphogenesis-promoting transcription factor ELONGATED HYPOCOTYL 5 (HY5) (11, 12). In addition, the nuclear UVR8 interacts with the transcription factors WRKY DNA-BINDING PROTEIN 36 (WRKY36), BRI1-EMS-SUPPRESSOR 1 (BES1), and BES1-INTERACTING MYC-LIK1 (BIM1) to regulate UV-B responsive gene expression (13, 14). On the other hand, two homologous proteins, REPRESSOR OF UV-B PHOTOMORPHOGENESIS 1 (RUP1) and RUP2, function redundantly to negatively regulate UV-B–induced photomorphogenesis by facilitating UVR8 redimerization and disturbing the UVR8–COP1 interaction (15–18).
Although the initiation of UV-B light signaling has been extensively studied, the mechanism underlying the coordinated regulation of this process downstream of the photoreceptor remains poorly understood, particularly at the level of protein stability regulation. In this study, we identified the UV-B–inducible E3 ligase CUL4-DDB1-RUP1/RUP2 and found that it represses photomorphogenesis by mediating the degradation of HY5. Conversely, COP1 directly targets RUP1/RUP2 for ubiquitination and degradation, leading to balanced accumulation of RUP1/RUP2, alleviation of the COP1–HY5 interaction, and stabilization of HY5. By adjusting HY5 and RUP1/RUP2 protein levels, these two E3-substrate modules, CUL4-DDB1-RUP1/RUP2-HY5 and COP1-RUP1/RUP2, coordinate the antagonistic regulatory machinery that controls plant photomorphogenesis under UV-B light.
Results
RUP1 and RUP2 Directly Interact with HY5.
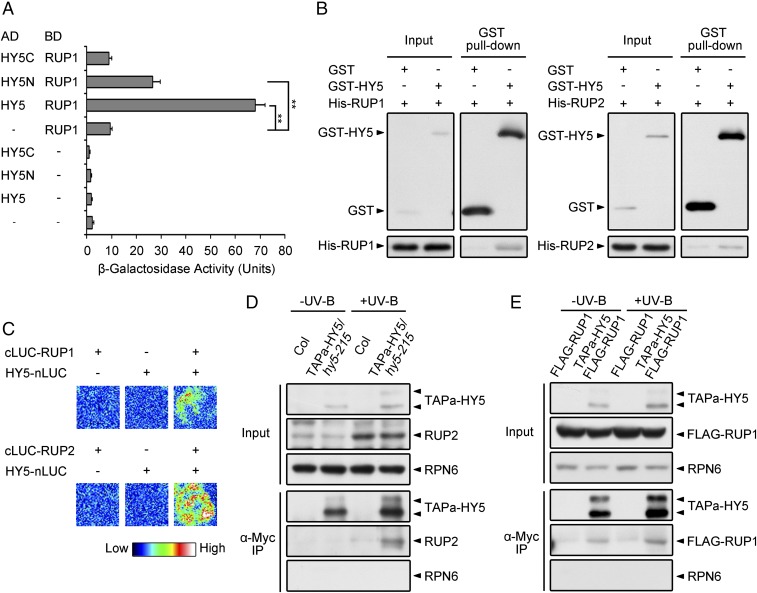
To study the mechanism repressing UV-B–induced photomorphogenesis, we hypothesized that central repressive machinery might directly regulate the transcription factor HY5, a signaling hub required for genome-wide transcriptional reprogramming in response to UV-B light (19). We therefore performed yeast two-hybrid assays to examine the interaction of HY5 with UV-B–inducible proteins and found that RUP1 directly interacts with HY5 (Fig. 1A). Using truncation constructs of HY5, the primary RUP1-interacting domain of HY5 was mapped to its N-terminal region (Fig. 1A and SI Appendix, Fig. S1A), which is known to mediate the COP1–HY5 interaction (20). We then confirmed the HY5–RUP1 interaction using in vitro pull-down assays and found that recombinant GST-tagged HY5 (GST-HY5) was able to pull down His-tagged RUP1 (His-RUP1) and His-RUP2 (Fig. 1B). In addition, we performed firefly luciferase complementation imaging (LCI) in Nicotiana benthamiana leaves to test the HY5–RUP1/RUP2 interaction in planta. The full-length sequences of HY5 and RUP1/RUP2, respectively, were fused to split N-terminal luciferase (nLUC) and C-terminal luciferase (cLUC). The luciferase activity was reconstituted when HY5-nLUC and cLUC-RUP1/RUP2 were coexpressed in N. benthamiana leaves (Fig. 1C). To further investigate how UV-B light regulates the HY5–RUP1/RUP2 interaction in vivo, we performed coimmunoprecipitation (co-IP) assays using transgenic Arabidopsis plants overexpressing HY5 fused with an alternative tandem affinity purification tag (TAPa-HY5) consisting of two copies of the protein A IgG binding domain, an 8-aa sequence corresponding to the 3C protease cleavage site, a six-histidine stretch, and nine repeats of the myc epitope (21). The UV-B irradiation strongly increased the protein level of endogenous RUP2, and RUP2 was obviously coimmunoprecipitated by TAPa-HY5 under UV-B light (Fig. 1D). The overexpressed FLAG-RUP1 fusion protein can be coimmunoprecipitated by TAPa-HY5 independent of UV-B light (Fig. 1E). These results suggest that RUP1 and RUP2 may be involved in the regulation of HY5 under UV-B light based on their in vivo association.
Fig. 1.
HY5 interacts with RUP1 and RUP2. (A) HY5 interacts with RUP1 in yeast two-hybrid assays. β-Galactosidase activity was quantified using o-nitrophenyl-β-d-galactopyranoside as a substrate (mean ± SD, n = 3). The asterisks indicate a significant difference by Student’s t test (**P < 0.01). (B) HY5 interacts with RUP1 and RUP2 in vitro. Purified GST or GST-HY5 was incubated with His-RUP1 or His-RUP2 before being pulled down by Glutathione Sepharose 4B. His-RUP1 and His-RUP2 were detected by anti-RUP1 and anti-RUP2 antibodies, respectively. (C) HY5 interacts with RUP1 and RUP2 in N. benthamiana, as assayed by firefly LCI. The color bar shows the range of luminescence intensity. The minus symbols (−) indicate empty vectors of nLUC or cLUC. (D) TAPa-HY5 associates with RUP2 in Arabidopsis. Total proteins were extracted from 4-d-old seedlings grown under −UV-B and +UV-B light conditions for co-IP with Anti–c-Myc Affinity Gel. Proteins were analyzed by immunoblotting with anti–c-Myc, anti-RUP2, and anti-RPN6 antibodies. RPN6 was used as a loading and negative control. (E) TAPa-HY5 associates with FLAG-RUP1 in Arabidopsis. Total proteins were extracted from 4-d-old seedlings grown under −UV-B and +UV-B light conditions for co-IP with Anti–c-Myc Affinity Gel. Proteins were analyzed by immunoblotting with anti–c-Myc, anti-FLAG, and anti-RPN6 antibodies. RPN6 was used as a loading and negative control.
RUP1 and RUP2 Associate with the E3 Ligase CUL4-DDB1.
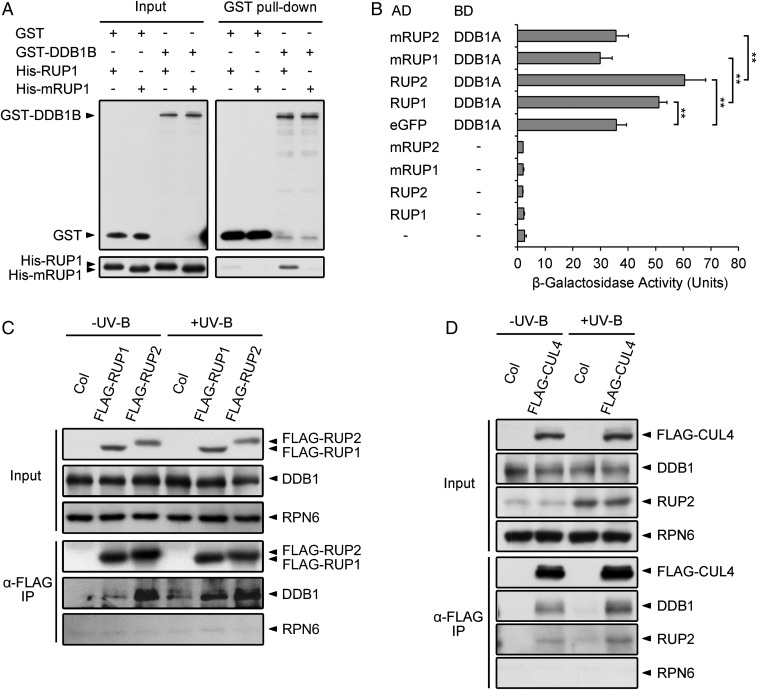
RUP1 and RUP2 are two homologous proteins containing WD40-repeat domains. We closely analyzed their protein sequences and found that each of them contains three typical WDXR motifs (SI Appendix, Fig. S1B). The WDXR motif is a determinant for DDB1-BINDING WD40 (DWD) proteins in both plants and animals, allowing them to act as substrate receptors for CUL4-DDB1–based E3 ligases (22, 23). To determine whether RUP1 and RUP2 are bona fide DWD proteins, we performed in vitro pull-down and yeast two-hybrid assays to examine the interaction between DDB1 and RUP1/RUP2. Recombinant His-RUP1 and His-RUP2 were pulled down by GST-DDB1B in vitro, but the physical binding was impaired by mutations in the WDXR motifs of RUP1 and RUP2 (Fig. 2A and SI Appendix, Fig. S2A). In addition, although DDB1A showed some β-galactosidase activity when used as bait, as previously reported (23, 24), significantly increased activity was observed when RUP1 or RUP2 was introduced as prey. The activation of β-galactosidase, however, was again abolished by mutations in the WDXR motifs of RUP1 and RUP2 (Fig. 2B). Collectively, these results demonstrate that RUP1 and RUP2 directly interact with DDB1 via their WDXR motifs.
Fig. 2.
RUP1 and RUP2 interact with DDB1 via their DWD motifs. (A) RUP1 interacts with DDB1B in vitro. Purified GST or GST-DDB1B was incubated with His-RUP1 or His-mRUP1 before being pulled down by Glutathione Sepharose 4B. His-RUP1 and His-mRUP1 were detected by anti-RUP1 antibodies. (B) RUP1 and RUP2 interact with DDB1A in yeast two-hybrid assays. β-Galactosidase activity was quantified using o-nitrophenyl-β-d-galactopyranoside as a substrate (mean ± SD, n = 3). The asterisks indicate significant differences by Student’s t test (**P < 0.01). (C) FLAG-RUP1 and FLAG-RUP2 associate with DDB1 in Arabidopsis. Total proteins were extracted from 4-d-old seedlings grown under −UV-B and +UV-B light conditions for co-IP with ANTI-FLAG Magnetic Beads. Proteins were analyzed by immunoblotting with anti-FLAG, anti-DDB1, and anti-RPN6 antibodies. RPN6 was used as a loading and negative control. (D) FLAG-CUL4 associates with DDB1 and RUP2 in Arabidopsis. Total proteins were extracted from 4-d-old seedlings grown under −UV-B and +UV-B light conditions for co-IP with ANTI-FLAG Magnetic Beads. Proteins were analyzed by immunoblotting with anti-FLAG, anti-DDB1, anti-RUP2, and anti-RPN6 antibodies. RPN6 was used as a loading and negative control.
Next, we tested the in vivo association of RUP1/RUP2 with CUL4-DDB1 using transgenic Arabidopsis plants. The seedlings overexpressing FLAG-RUP1 or FLAG-RUP2 were not only hyposensitive to photomorphogenic UV-B light, as revealed by examining UV-B–induced hypocotyl growth and gene expression, but also showed longer hypocotyls than Columbia (Col) under various light conditions (SI Appendix, Fig. S3). Our co-IP assays found that the endogenous DDB1 was coimmunoprecipitated by the overexpressed FLAG-RUP1 and FLAG-RUP2 (Fig. 2C). Similarly, DDB1 and RUP2 can be coimmunoprecipitated by the overexpressed FLAG-CUL4 (Fig. 2D). Together, these results suggest that the CUL4-DDB1–RUP1/RUP2 complex potentially functions as an E3 ligase that is associated with the repression of UV-B–induced photomorphogenesis.
The E3 Ligase CUL4-DDB1–RUP1/RUP2 Complex Mediates HY5 Degradation Under UV-B Light.
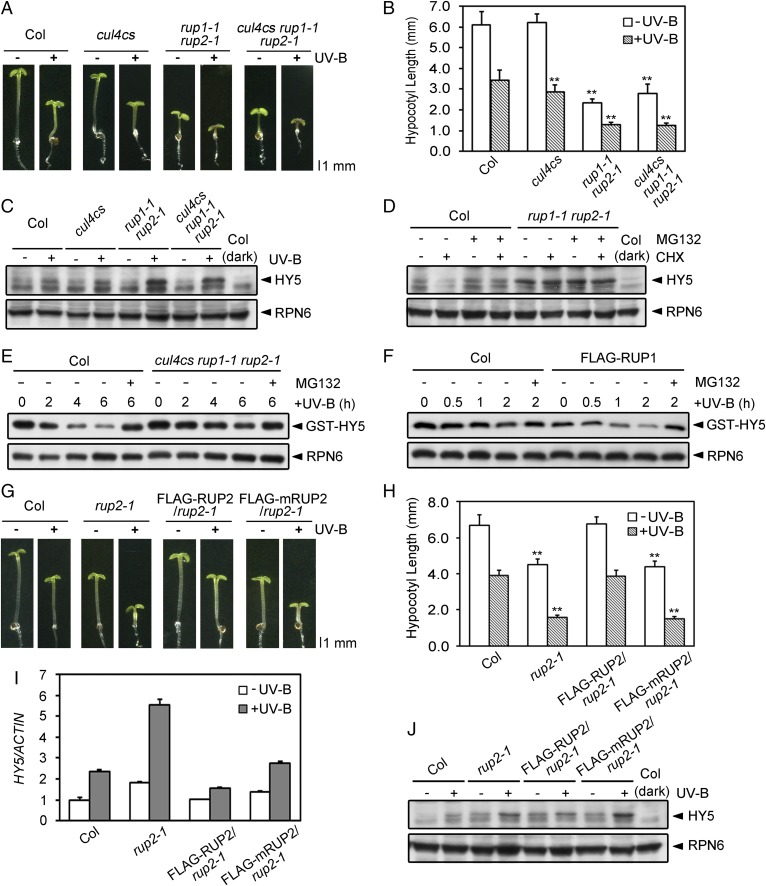
To explore the biological significance of the association between RUP1/RUP2 and CUL4, we analyzed the genetic interaction between CUL4 and RUP1/RUP2 by crossing cul4cs (a cul4 cosuppression allele) with rup1-1 rup2-1. Compared with their wild-type counterparts, cul4cs rup1-1 rup2-1 seedlings phenocopied rup1-1 rup2-1 (Fig. 3A), showing shorter hypocotyls under both −UV-B and +UV-B conditions (Fig. 3B), as well as enhanced UV-B–induced anthocyanin accumulation and gene expression (SI Appendix, Fig. S4). Additionally, cul4cs rup1-1 rup2-1 seedlings resembled rup1-1 rup2-1 in that they accumulated much more HY5 protein than the wild type under UV-B light (Fig. 3C). We also examined the effect of RUP2 overexpression on HY5 protein, which is constitutively expressed under the control of the 35S promoter. Compared with TAPa-HY5, TAPa-HY5 FLAG-RUP2 harbored a decreased level of TAPa-HY5 only under UV-B light (SI Appendix, Fig. S5A). Collectively, these results demonstrate that RUP1/RUP2 represses UV-B–induced photomorphogenesis downstream of CUL4 and inhibits HY5 protein accumulation at a posttranscriptional level.
Fig. 3.
CUL4-DDB1-RUP1/RUP2 E3 ligase mediates UV-B–induced degradation of HY5. (A) Phenotypes of 4-d-old Col, cul4cs, rup1-1 rup2-1, and cul4cs rup1-1 rup2-1 seedlings grown under −UV-B and +UV-B light conditions. (B) Hypocotyl length of the seedlings shown in A (mean ± SD, n ≥ 30). The asterisks indicate significant differences by Student’s t test (**P < 0.01) compared with Col under each light condition. (C) HY5 protein levels in 4-d-old Col, cul4cs, rup1-1 rup2-1, and cul4cs rup1-1 rup2-1 seedlings grown under −UV-B and +UV-B light conditions. Proteins were analyzed by immunoblotting with anti-HY5 and anti-RPN6 antibodies. RPN6 was used as a loading control. (D) Effect of RUP1 and RUP2 on HY5 stability in Arabidopsis under UV-B light. Immunoblot analysis of HY5 proteins in 4-d-old Col and rup1-1 rup2-1 seedlings grown under +UV-B light and treated with 500 μM CHX and/or 50 μM MG132 for 3 h. HY5 was detected with anti-HY5 antibodies. RPN6 was used as a loading and negative control. (E) Effect of CUL4, RUP1, and RUP2 on HY5 stability in vitro, as analyzed using cell-free degradation assays. Purified GST-HY5 was incubated with total proteins extracted from 4-d-old UV-B light-grown Col and cul4cs rup1-1 rup2-1 seedlings for 6 h. The degradation mixture was treated with or without 50 μM MG132. GST-HY5 was detected with anti-GST antibody. RPN6 was used as a loading and negative control. (F) Effect of FLAG-RUP1 on HY5 stability in vitro, as analyzed by cell-free degradation assays. Purified GST-HY5 was incubated with total proteins extracted from 4-d-old +UV-B light-grown Col and FLAG-RUP1 seedlings for 2 h. The degradation mixture was treated with or without 50 μM MG132. GST-HY5 was detected with anti-GST antibody. RPN6 was used as a loading and negative control. (G) Phenotypes of 4-d-old Col, rup2-1, FLAG-RUP2/rup2-1, and FLAG-mRUP2/rup2-1 seedlings grown under −UV-B and +UV-B light conditions. (H) Hypocotyl length of the seedlings shown in G (mean ± SD, n ≥ 30). The asterisks indicate significant differences by Student’s t test (**P < 0.01) compared with Col under each light condition. The mRNA levels (I) and protein levels (J) of HY5 in the seedlings shown in G are illustrated (mean ± SD, n = 3).
Next, we investigated how RUP1/RUP2 regulates HY5 protein levels. In the wild type, HY5 protein levels decreased after cycloheximide (CHX; a protein synthesis inhibitor) treatment and increased after MG132 (a proteasome inhibitor) treatment, whereas HY5 was maintained at higher levels in rup1-1 rup2-1 under continuous +UV-B conditions instead of −UV-B conditions (Fig. 3D and SI Appendix, Fig. S5B). The result suggests that RUP1/RUP2 mediates HY5 degradation through the ubiquitin/proteasome pathway specifically under UV-B light. We further monitored the degradation of recombinant HY5 using cell-free degradation assays in which GST-HY5 was incubated with protein extracts from +UV-B–grown wild-type, mutant, or overexpression plants. Consistent with the previous report (12), GST-HY5 was relatively stable in the first 2 h of reaction (Fig. 3 E and F and SI Appendix, Fig. S5C). Its degradation was later detected as the reaction was prolonged to 4–6 h, and it was abolished by MG132, suppressed in cul4cs rup1-1 rup2-1 (Fig. 3E), and slightly enhanced in FLAG-RUP1 (Fig. 3F) and FLAG-RUP2 (SI Appendix, Fig. S5C).
We then generated transgenic plants expressing FLAG-RUP2 with the wild-type or mutated WDXR motif at comparable protein levels to further analyze the role of RUP1/RUP2 in HY5 stability (SI Appendix, Fig. S5D). Unlike FLAG-RUP2/rup2-1, which complemented the rup2-1 mutant phenotype and exhibited normal UV-B–induced photomorphogenesis, FLAG-mRUP2/rup2-1 failed to rescue rup2-1 (Fig. 3 G and H). HY5 protein levels, instead of HY5 mRNA levels, were much higher in FLAG-mRUP2/rup2-1 than those in Col and FLAG-RUP2/rup2-1 under UV-B light (Fig. 3 I and J), demonstrating the action of WDXR motifs of RUP1 and RUP2 in HY5 degradation. However, the WDXR mutation exerted little influence on UVR8 conformation examined as previously described (3). UVR8 conformation did not show obvious differences in Col, rup2-1, FLAG-RUP2/rup2-1, and FLAG-mRUP2/rup2-1 under either UV-B–removed or continuous −UV-B/+UV-B conditions (SI Appendix, Fig. S5 E and F). Together with the observation of reduced photomorphogenic responses to various light conditions without UV-B in FLAG-RUP1/RUP2 (SI Appendix, Fig. S3), these data demonstrate that HY5 is subjected to UV-B–induced proteolysis mediated by the E3 ligase CUL4-DDB1-RUP1/RUP2, with RUP1/RUP2 functioning independent of UVR8 conformation regulation.
COP1 Directly Interacts with RUP1 and RUP2 and Mediates Their Degradation Under UV-B Light.
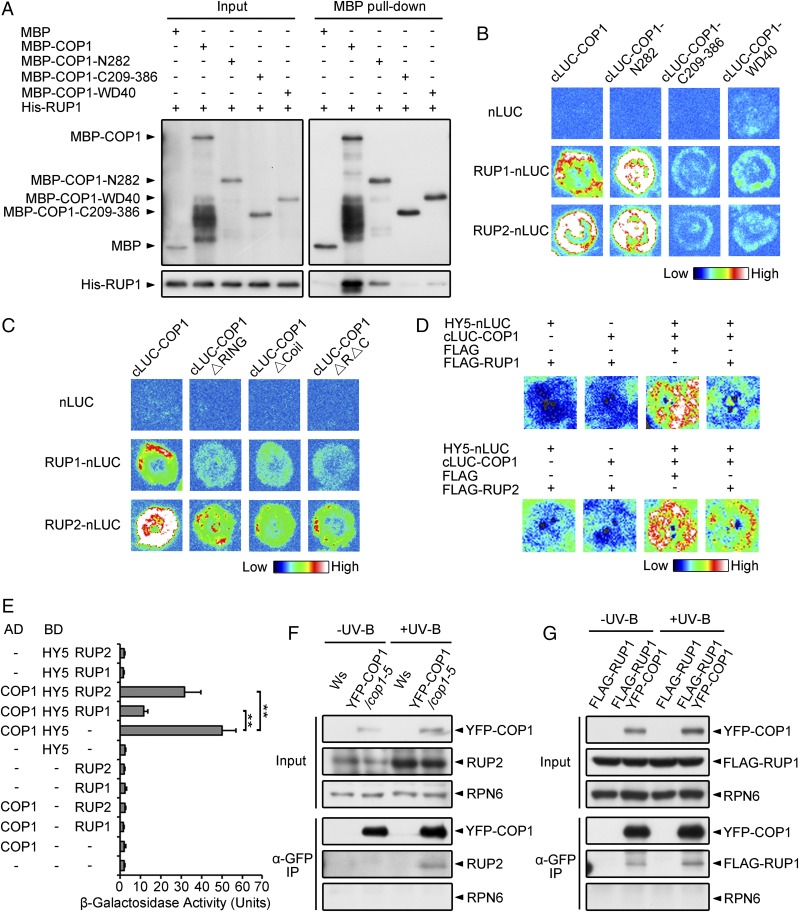
RUP1 and RUP2 negatively regulate UV-B–induced photomorphogenesis (15). However, how these proteins are regulated during this process is largely unknown. Given that COP1 and RUP1/RUP2 are both WD40 proteins with protein–protein interaction domains, we examined whether they interact with each other using various systems. In in vitro pull-down assays, His-RUP1 and His-RUP2, respectively, were precipitated by maltose binding protein-tagged COP1 (MBP-COP1) (Fig. 4A and SI Appendix, Fig. S2B). In LCI assays, luciferase activity was reconstituted when RUP1/RUP2-nLUC and cLUC-COP1 were coexpressed (Fig. 4B). Using truncation constructs of COP1, the primary RUP1-interacting region of COP1 was mapped to its N-terminal 282-aa region (COP1-N282), which includes the RING domain and the coiled-coil domain (Fig. 4 A and B). Deletion of either domain within COP1-N282 led to almost abolished COP1 binding to RUP1/RUP2 (Fig. 4C). In addition, a mild interaction was detected between RUP1 and the WD40 repeats of COP1, which mediates COP1–HY5 interaction (Fig. 4 A and B). Together with the observation that RUP1 directly interacts with the COP1-interacting region in HY5 (Fig. 1A), it is probable that RUP1 may disturb COP1–HY5 interaction. We then verified this hypothesis in LCI and yeast two-hybrid assays and found that the COP1–HY5 interaction was indeed impaired by RUP1/RUP2 (Fig. 4 D and E, and SI Appendix, Fig. S5G). Moreover, our co-IP experiments showed the in vivo association of yellow fluorescent protein-fused COP1 (YFP-COP1) with endogenous RUP2 specifically under UV-B light (Fig. 4F) and constitutively overexpressed FLAG-RUP1 independent of UV-B light (Fig. 4G). These in vitro and in vivo results demonstrate that COP1 directly interacts with RUP1/RUP2, implying that COP1 may act as an E3 ligase for RUP1/RUP2.
Fig. 4.
COP1 Interacts with RUP1 and RUP2. (A) RUP1 interacts with full-length and truncated COP1 in vitro. Purified MBP, MBP-COP1, MBP-COP1-N282, MBP-COP1-C209-386, or MBP-COP1-WD40 was incubated with His-RUP1 before being pulled down by Amylose Resin. His-RUP1 was detected by anti-RUP1 antibodies. (B) RUP1 and RUP2 interact with full-length and truncated COP1 in N. benthamiana, as assayed by firefly LCI. The color bar shows the range of luminescence intensity. (C) COP1 interacts with RUP1 and RUP2 primarily through the RING domain and Coil domain in N. benthamiana, as assayed by firefly LCI. The color bar shows the range of luminescence intensity. (D) RUP1/RUP2 impairs the COP1–HY5 interaction in N. benthamiana, as assayed by firefly LCI. The color bar below shows the range of luminescence intensity in each image. The minus symbols (−) indicate empty vectors of nLUC, cLUC, or FLAG. (E) RUP1/RUP2 impairs the COP1–HY5 interaction in yeast. β-Galactosidase activity was quantified using o-nitrophenyl-β-d-galactopyranoside as a substrate (mean ± SD, n = 3). The asterisks indicate significant differences by Student’s t test (**P < 0.01). (F) YFP-COP1 associates with RUP2 in Arabidopsis. Total proteins were extracted from 4-d-old seedlings grown under −UV-B and +UV-B light conditions for co-IP with Dynabeads Protein G and anti-GFP antibodies. Proteins were analyzed by immunoblotting with anti-GFP, anti-RUP2, and anti-RPN6 antibodies. RPN6 was used as a loading and negative control. (G) YFP-COP1 associates with FLAG-RUP1 in Arabidopsis. Total proteins were extracted from 4-d-old seedlings grown under −UV-B and +UV-B light conditions for co-IP with Dynabeads Protein G and anti-GFP antibodies. Proteins were analyzed by immunoblotting with anti-GFP, anti-FLAG, and anti-RPN6 antibodies. RPN6 was used as a loading and negative control.
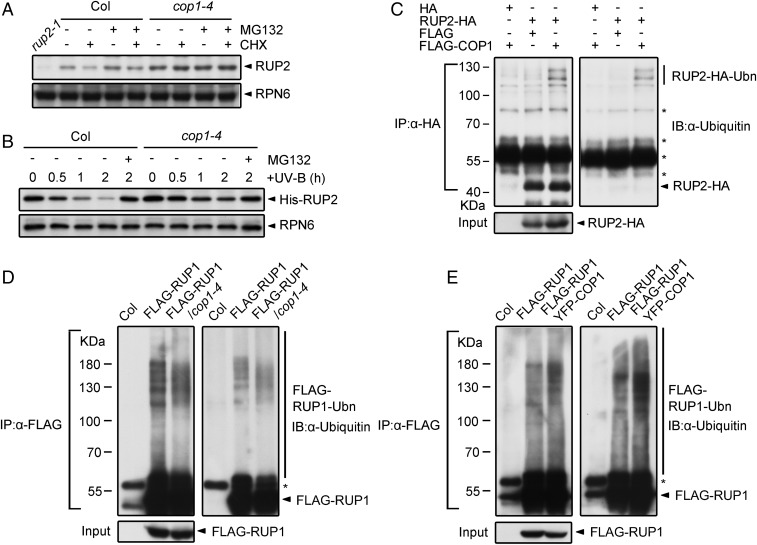
To study whether COP1 regulates RUP1/RUP2, we measured RUP2 mRNA and protein levels in Col and cop1-4 seedlings. Under −UV-B conditions, the cop1-4 mutation led to an increase in RUP2 protein level probably due to an increased RUP2 mRNA level. With UV-B treatment, cop1-4 resulted in strong accumulation of RUP2 proteins, without altering RUP2 mRNA levels (SI Appendix, Fig. S6 A and B), suggesting that COP1 plays a negative role in the posttranscriptional regulation of RUP2 predominantly under UV-B light. We then found that the protein levels of endogenous RUP2 decreased after CHX treatment and increased after MG132 treatment in Col grown under +UV-B conditions, but not −UV-B conditions (Fig. 5A and SI Appendix, Fig. S6C). Cell-free degradation assays using recombinant RUP2 further verified that RUP2 degradation occurred predominantly under +UV-B rather than −UV-B conditions (Fig. 5B). Such degradation was not detected in cop1-4 and uvr8-6 (SI Appendix, Fig. S6 D and E), suggesting the UV-B–induced RUP2 degradation is mediated by COP1 and dependent on UVR8.
Fig. 5.
COP1 degrades RUP1 and RUP2 under UV-B light. (A) Effect of COP1 on RUP2 stability in Arabidopsis under UV-B light. Immunoblot analysis of RUP2 proteins in 4-d-old Col and cop1-4 seedlings grown under +UV-B light and treated with 500 μM CHX and/or 50 μM MG132 for 3 h. RUP2 was detected with anti-RUP2 antibodies. RPN6 was used as a loading and negative control. (B) Effect of COP1 on RUP2 stability in vitro under UV-B light, as analyzed by cell-free degradation assays. Purified His-RUP2 was incubated with total proteins extracted from 4-d-old Col and cop1-4 seedlings grown under +UV-B light for 2 h. The degradation mixture was treated with or without 50 μM MG132. His-RUP2 was detected with anti-RUP2 antibodies. RPN6 was used as a loading and negative control. (C) Effect of FLAG-COP1 on the ubiquitination of RUP2-HA in HEK293T cells. Total proteins were extracted from HEK293T cells that were transfected with FLAG/FLAG-COP1 and HA/RUP2-HA for co-IP with Dynabeads Protein G and anti-HA antibodies. Proteins were analyzed by immunoblotting with anti-HA and anti-Ubiquitin antibodies. Ubn, ubiquitin chain. The asterisks indicate nonspecific bands. (D and E) Effect of COP1 on the ubiquitination of FLAG-RUP1 in vivo. Total proteins were extracted from 4-d-old Col, FLAG-RUP1, FLAG-RUP1/cop1-4 (D), or FLAG-RUP1 YFP-COP1 (E) seedlings grown under +UV-B light and treated with 50 μM MG132 for 24 h before co-IP with ANTI-FLAG Magnetic Beads. Proteins were analyzed by immunoblotting with anti-FLAG and anti-Ubiquitin antibodies. The asterisks indicate nonspecific bands.
Furthermore, we utilized human and plant cells to analyze RUP1/RUP2 ubiquitination regulated by COP1. In human embryonic kidney (HEK) 293T cells expressing RUP2-HA and/or FLAG-COP1, RUP2-HA enriched by IP showed obvious ubiquitination when coexpressed with FLAG-COP1 (Fig. 5C). In Arabidopsis seedlings, the cop1-4 mutation alleviated the ubiquitination of FLAG-RUP1 (Fig. 5D), whereas the constitutive expression of YFP-COP1 led to enhanced ubiquitination of FLAG-RUP1 (Fig. 5E). Genetically, crossing the progenies of cop1-4 with rup1-1 rup2-1 or FLAG-RUP1/RUP2 phenocopied cop1-4, as they failed to exhibit UV-B–induced hypocotyl growth (SI Appendix, Fig. S6 F and G) and anthocyanin accumulation (SI Appendix, Fig. S6H). This finding is consistent with a previous report that the action of RUP1/RUP2 in photomorphogenesis is dependent on COP1 (15). Taken together, these results demonstrate that COP1 directly interacts with RUP1/RUP2 and mediates the ubiquitination and degradation of RUP1 and RUP2 under UV-B light.
Discussion
During their emergence from the soil to establish photomorphogenesis, plant seedlings encounter increasing levels of UV-B irradiation and develop adaptive responses accordingly. In the past decade, the perception of UV-B light and the initiation of UV-B signaling by UVR8 have been extensively studied. Two recent studies have illustrated that UVR8 directly interacts with transcription factors WRKY36 and BES1/BIM1 to regulate UV-B–responsive transcription (13, 14). However, the mechanisms by which the UV-B signaling cascades are orchestrated downstream of the UV-B photoreceptor are still largely unknown. In this study, we identify RUP1/RUP2 as a substrate receptor of the E3 ligase CUL4-DDB1 for HY5 destabilization (Figs. 1–3). Disruption of the RUP1/RUP2–DDB1 interaction led to increased HY5 accumulation and enhanced UV-B–induced photomorphogenesis (Fig. 3 G and H). This finding reveals that the E3 ligase CUL4-DDB1-RUP1/RUP2 is a crucial molecular brake of UV-B signaling by modulating HY5 protein levels. On the other hand, COP1 directly targets RUP1/RUP2 for degradation, leading to balanced accumulation of RUP1/RUP2, alleviation of the COP1–HY5 interaction, and stabilization of HY5 (Figs. 4 and 5). As HY5 and RUP1/RUP2 are the central promoter and repressor, respectively, of UV-B–induced photomorphogenesis, these two sets of E3 ligases constitute antagonistic regulatory circuits of prolonged UV-B light signaling, allowing the abundance of HY5 and RUP1/RUP2 and their downstream events to be tightly controlled at appropriate levels for photomorphogenesis (Fig. 6).
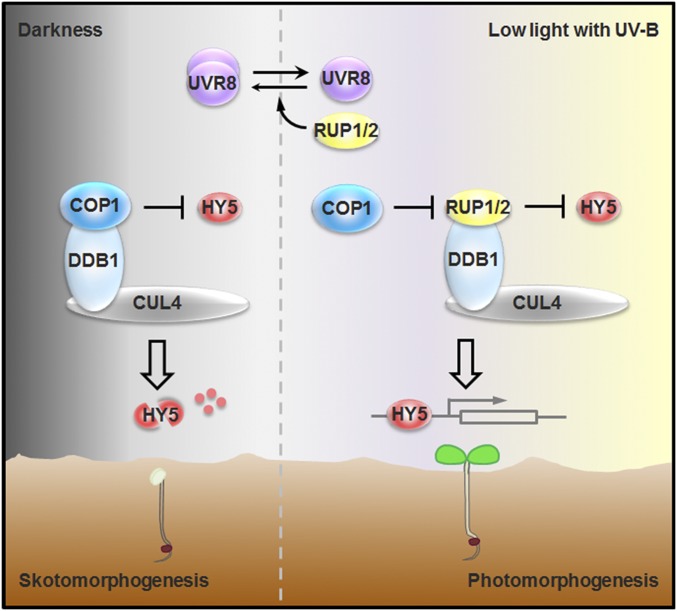
Fig. 6.
Working model for the coordinated regulation of photomorphogenesis by COP1, HY5, RUP1/RUP2, and CUL4-DDB1. During their emergence from the soil to establish photomorphogenesis, seedlings encounter increasing levels of UV-B light. In darkness, COP1, together with CUL4-DDB1, represses photomorphogenesis by mediating HY5 degradation. In a prolonged response to low light with UV-B, the E3 ligase responsible for HY5 degradation switches from CUL4-DDB1-COP1 to CUL4-DDB1-RUP1/RUP2. Meanwhile, COP1 directly targets RUP1/RUP2 for degradation, leading to balanced accumulation of RUP1/RUP2, alleviation of the COP1–HY5 interaction, and stabilization of HY5. Once UV-B light is removed, RUP1 and RUP2 function redundantly to mediate UVR8 redimerization to halt UVR8 signaling. As a result, RUP1/RUP2, CUL4-DDB1, COP1, and HY5 constitute the repression and derepression machinery by which plants develop photomorphogenesis.
Several studies have demonstrated that RUP1 and RUP2 play key roles in the negative feedback regulation of UV-B–induced photomorphogenesis via facilitating UVR8 redimerization and disturbing the UVR8–COP1 interaction (15, 16, 18). RUP2 has lately been revealed to repress UVR8-mediated flowering under short-day conditions (25). Here, we uncover the action of RUP1/RUP2 in the modulation of protein stability, further substantiating the multifunctional nature of RUP1/RUP2 in UV-B–dependent plant development. As RUP1 and RUP2 function redundantly to mediate UVR8 redimerization for photoreceptor inactivation, UVR8 undergoes normal redimerization as long as one of these two proteins is functional (16). Without affecting the conformational change of UVR8, the WDXR mutation of RUP2 in rup2-1 results in increased HY5 stability (Fig. 3 and SI Appendix, Fig. S5). Not merely under UV-B light, the seedlings overexpressing FLAG-RUP1/RUP2 exhibit hyposensitivity of photomorphogenesis under various light conditions (SI Appendix, Fig. S3). As hy5 mutant seedlings show reduced photomorphogenesis under all light conditions (26, 27), these results substantiate the role of RUP1/RUP2 in the degradation of HY5 and establish the differentiated contributions of RUP1/RUP2 in UVR8 inactivation and HY5 destabilization. Moreover, HY5 and RUP1/RUP2 are UV-B–inducible genes, and their proteins accumulate within the first few hours of photomorphogenic UV-B irradiation (7, 15). Here, we demonstrate that in a prolonged response to UV-B irradiation, HY5 and RUP1/RUP2 are subjected to degradation after they accumulate to a relatively high level. Once UV-B light is removed, RUP1 and RUP2 mediate UVR8 redimerization to halt UVR8 signaling. Therefore, RUP1 and RUP2 are multifunctional for multistage regulation of UV-B light signaling (Fig. 6).
COP1 is a master repressor of photomorphogenesis in the absence of UV-B light by associating with CUL4-DDB1 to destabilize HY5 in the dark (28, 29). By contrast, COP1 is a pivotal positive regulator of photomorphogenesis and is required for HY5 stability under UV-B light (7, 12). However, the molecular basis for the opposite roles of COP1 in photomorphogenesis is unknown. It has been demonstrated that UV-B light induces COP1 to physically and functionally dissociate with the E3 ligase scaffold CUL4-DDB1. Further, the reduced accumulation and stability of HY5 in the cop1-4 mutant under UV-B light implies the existence of an alternative E3 ligase of HY5 (7, 11, 12). In this study, we disclose that under UV-B light, the E3 ligase responsible for HY5 degradation switches from CUL4-DDB1-COP1-SPAs to CUL4-DDB1-RUP1/RUP2. However, the abundance and activity of HY5 are guaranteed in multiple ways. It has been proposed that upon UV-B irradiation, the monomerized UVR8 binds to COP1’s WD40 domain to disturb COP1-mediated HY5 degradation (11). Here, RUP1 and RUP2 are experimentally characterized as COP1-interacting proteins, and they occupy COP1’s WD40 domain, disrupting the COP1–HY5 interaction. The COP1-RUP1/RUP2 module becomes a second security measure that ensures HY5 accumulation after the initiation of UV-B light signaling. Therefore, the switchable roles of COP1 in plant photomorphogenesis strictly rely on its differential regulation of HY5 stability. Taken together, our findings provide insight that variable E3-substrate modules consisting of COP1, HY5, RUP1/RUP2, and CUL4-DDB1 constitute the repression and derepression machinery by which plants develop photomorphogenesis.
Methods
Plant Materials and Growth Conditions.
The wild-type A. thaliana lines used in this study are in the Col and Wassilewskija backgrounds. The following mutants and transgenic lines used in this study were described previously: cul4cs (30), rup1-1 and rup2-1 (15), cop1-4 (31), uvr8-6 (11), TAPa-HY5/hy5-215 (21), FLAG-CUL4 (30), and YFP-COP1/cop1-5 (12). Transgenic FLAG-RUP1, FLAG-RUP2/rup2-1, FLAG-mRUP2/rup2-1, and FLAG-RUP2 plants were generated by the floral dip method (32) using Agrobacterium strain GV3101. The following plant materials were generated by crossing: rup1-1 rup2-1, cul4cs rup1-1 rup2-1, rup1-1 rup2-1 cop1-4, TAPa-HY5, TAPa-HY5 FLAG-RUP1, TAPa-HY5 FLAG-RUP2, FLAG-RUP1 YFP-COP1, FLAG-RUP1/cop1-4, and FLAG-RUP2/cop1-4.
The seeds were surface-sterilized and sown on solid 1% Murashige and Skoog medium supplemented with 1% sucrose for molecular and biochemical assays or with 0.3% sucrose for phenotypic analysis, followed by cold treatment at 4 °C for 4 d before light treatment. For UV-B–induced photomorphogenesis, seedlings were grown at 22 °C under continuous low-white light (3 μmol⋅m−2⋅s−1, measured by an HR-350 Light Meter; Hipoint) supplemented with UV-B light from Philips TL20W/01RS narrowband UV-B tubes (1.5 μmol⋅m−2⋅s−1, measured by a UV-297 UV-B Light Meter; HANDY) under a 350-nm cutoff (half-maximal transmission at 350 nm) ZUL0350 filter (−UV-B light; Asahi spectra) or a 300-nm cutoff (half-maximal transmission at 300 nm) ZUL0300 filter (+UV-B light; Asahi spectra).
Plasmid Construction and Site-Directed Mutagenesis.
For plant transformation, to obtain transgenic plants expressing FLAG-RUP1 or FLAG-RUP2, the KpnI/XhoI fragment containing the full-length RUP1 coding sequence or the SalI/Sacl fragment containing the full-length RUP2 coding sequence was cloned into the pF3PZPY122 binary vector (33). To obtain transgenic plants expressing FLAG-mRUP2, the SalI/Sacl fragment containing the mRUP2 coding sequence from pB42AD-mRUP2 was introduced into the pF3PZPY122 vector. The XhoI/SpeI fragment containing the coding sequence of FLAG-mRUP2 from pF3PZPY122-mRUP2 was then cloned into the pJim19 (bar) binary vector.
For the yeast two-hybrid assays, the MfeI/XhoI fragment containing the full-length RUP1 or RUP2 coding sequence was cloned into the pB42AD or pLexA vector (Clontech). Site-directed mutagenesis by PCR was used to generate pB42AD-mRUP1 and pB42AD-mRUP2. The EcoRI/XhoI fragments containing the HY5N and HY5C coding sequences from pLexA-HY5N and pLexA-HY5C (20), respectively, were cloned into the pB42AD vector. The pLexA-DDB1A (30), pB42AD-eGFP (23), and pB42AD-HY5 (34) constructs were used as described previously.
For the in vitro pull-down assays, the pGEX-4T-1-DDB1B (29) and pGEX-4T-1-HY5 constructs (20) were used as described previously. The SalI/PstI fragment containing the full-length COP1 coding sequence and the EcoRI/PstI fragments encoding truncated fragments of COP1 (COP1-N282, COP1-C209-386, and COP1-WD40) were cloned into the pMAL-c2X vector (New England BioLabs). The MfeI/XhoI fragments containing the full-length RUP1 and RUP2 coding sequences were cloned into pET-28a (Novagen). Site-directed mutagenesis by PCR was used to generate pET-28a-mRUP1 and pET-28a-mRUP2.
For the firefly LCI, the KpnI/SalI fragment containing the full-length RUP1 coding sequence and the BglII/SalI fragment containing the full-length RUP2 coding sequence were cloned into the pCAMBIA1300-nLuc vector (35). The KpnI/SalI fragment containing the full-length RUP1 or COP1 coding sequence and the BglII/SalI fragment containing the full-length RUP2 coding sequence were cloned into pCAMBIA1300-cLuc (35). The fragment containing COP1-N282, COP1-C209-386, COP1-WD40, COP1ΔRING, COP1ΔCoil, or COP1ΔRΔC was cloned into pCAMBIA1300-cLuc by ligation-independent cloning. pCAMBIA1300-HY5-nLuc (34) was used as described previously.
For the yeast three-hybrid assays, the KpnI/EcoRI fragment containing the full-length RUP1 coding sequence was cloned into the modified pGADT7 vector (Clontech) with the AD fragment removed [i.e., pGADT7(−AD)]. The fragment containing the full-length RUP2 coding sequence was cloned into pGADT7(−AD) by ligation-independent cloning. The constructs of pLexA-HY5 and pB42AD-COP1 (20) were used as described previously.
For ubiquitination assays in HEK293T cells, the fragment containing the full-length COP1 coding sequence was cloned into pBobi-FLAG by ligation-independent cloning. The fragment containing the full-length RUP2 coding sequence was cloned into pcDNA3.1-HA by ligation-independent cloning.
All primers are listed in SI Appendix, Table S1, and all constructs were confirmed by sequencing.
Hypocotyl Length and Anthocyanin Measurements.
Hypocotyl length was measured as previously described (36). For each line grown under −UV-B light or +UV-B light for 4 d, hypocotyl length was analyzed in three biological replicates. In each replicate, at least 30 Arabidopsis seedlings were measured. Hypocotyl length was quantified using ImageJ software (https://imagej.nih.gov/ij/).
Anthocyanin was extracted and quantified as previously described (37). Briefly, Arabidopsis seedlings were harvested, placed in extraction solution (18% 1-propanol and 1% HCl), and incubated in the dark at 4 °C for at least 10 h. After a brief centrifugation to pellet the tissue debris, the supernatant was removed and diluted with extraction solution. The anthocyanin content was presented as A535 − 2(A650) g−1 fresh weight and was analyzed in three biological replicates.
qRT-PCR.
Total RNA was extracted from 4-d-old Arabidopsis seedlings using an Eastep Total RNA Extraction Kit (Promega). Reverse transcription was performed using a GoScript Reverse Transcription System (Promega). qRT-PCR analysis was performed using iTaq Universal SYBR Green Supermix (Bio-Rad) on a CFX Connect Real-Time PCR System (Bio-Rad). Plant materials were collected from three biological replicates, and three technical replicates were assayed per experiment. The primers used for the qRT-PCR assays are listed in SI Appendix, Table S1.
Yeast Two-Hybrid Assays.
The respective combinations of AD- and BD-fused constructs were cotransformed into yeast strain EGY48 containing the reporter plasmid p8op-LacZ according to the instructions provided with the Matchmaker LexA Two-Hybrid System (Clontech). Transformants were selected on minimal synthetic defined agar base plates supplemented with dropout (DO) supplement -His/-Trp/-Ura. β-Galactosidase activity was analyzed using o-nitrophenyl-β-d-galactopyranoside as a substrate [β-galactosidase units = 1,000 × OD420/(t × V × OD600), where t is elapsed time (in minutes) of incubation, V = 0.1 mL × concentration factor, and OD600 = A600 of 1 mL of culture]. Three biological replicates were analyzed for each interaction pair, and three technical replicates were assayed per experiment.
In Vitro Pull-Down Assays.
The recombinant proteins were expressed in Escherichia coli strain Transetta (DE3). GST, GST-DDB1B, and GST-HY5 were purified with Glutathione Sepharose 4B (GE Healthcare). MBP, MBP-COP1, MBP-COP1-N282, MBP-COP1-C209-386, and MBP-COP1-WD40 were purified with Amylose Resin (New England BioLabs). His-RUP1/RUP2 and His-mRUP1/RUP2 were purified using Ni-NTA Agarose (Qiagen).
For the GST pull-down assays, 1 μg of GST, GST-DDB1B, or GST-HY5 was mixed with 1 μg of His-RUP1/RUP2 or His-mRUP1/RUP2 and 15 μL of Glutathione Sepharose 4B. For the MBP pull-down assays, 1 μg of MBP, MBP-COP1, MBP-COP1-N282, MBP-COP1-C209-386, or MBP-COP1-WD40 was mixed with 1 μg of His-RUP1 or His-RUP2 and 15 μL of Amylose Resin. The mixture was incubated in binding buffer [50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, and 0.1% Tween 20] at 4 °C for 3 h. The pellets were washed three times, eluted in 2× SDS loading buffer, and boiled at 95 °C for 10 min before immunoblotting. For each in vitro pull-down assay, three independent repetitions of experiments were performed, and one representative result was presented.
Firefly LCI.
Firefly LCI was performed in N. benthamiana leaves as described previously (35). Briefly, Agrobacterium strain GV2260 cells transformed with the nLUC- or cLUC-fused construct were infiltrated into N. benthamiana leaves. After infiltration, the N. benthamiana plants were grown for 3 d and injected with luciferin, followed by imagining using a Tanon 5200S Luminescent Imaging Workstation. Three biological replicates were analyzed for each interaction pair, and three technical replicates were assayed per experiment.
Co-IP Assays.
Total proteins (3–5 mg) were extracted from 4-d-old Arabidopsis seedlings in protein extraction buffer containing 50 mM Tris⋅HCl (pH 7.5), 150 mM NaCl, 1 mM EDTA, 10% glycerol, 0.1% Tween 20, 1 mM NaF, 2 mM Na3VO4, 50 mM β-glycerophosphate, 1 mM phenylmethylsulfonyl fluoride (PMSF), and 1× Complete Protease Inhibitor Mixture (Roche). The extracts were incubated with 35 μL of ANTI-FLAG Magnetic Beads (Sigma–Aldrich) for α-FLAG IP, 30 μL of Anti–c-Myc Affinity Gel (Sigma–Aldrich) for α-Myc IP, or 8 μL of anti-GFP antibodies (Thermo Fisher Scientific) coupled with 25 μL of Dynabeads Protein G (Thermo Fisher Scientific) for α-GFP IP at 4 °C for 3 h under the same light conditions (−UV-B or +UV-B) used for seedling growth. The pellets were washed three times and eluted with 2× SDS protein loading buffer or acid eluting buffer containing 100 mM glycine (pH 2.5) and 100 mM NaCl. The acid elutes were immediately neutralized with 2 M Tris⋅HCl (pH 9.0) and 100 mM NaCl and concentrated with StrataClean Resin (Agilent Technologies). All eluted products were boiled at 95 °C for 10 min before immunoblotting. For each in vivo co-IP assay, three independent repetitions of experiments were performed, and one representative result was presented.
Yeast Three-Hybrid Assays.
Yeast three-hybrid assays were performed using the Matchmaker LexA Two-Hybrid System following the manufacturer’s instructions, with some modifications. The respective combinations of AD- and BD-fused constructs, together with another construct expressing a third protein, were cotransformed into yeast strain EGY48 containing the reporter plasmid p8op-LacZ according to the manufacturer’s instructions. Transformants were selected on minimal synthetic defined agar base plates supplemented with DO supplement -His/-Leu/-Trp/-Ura. The positive transformants were transferred to synthetic defined agar base/Gal/Raf plates supplemented with DO supplement -His/-Leu/-Trp/-Ura and X-β-galactosidase for blue color development [β-galactosidase units = 1,000 × OD420/(t × V × OD600), t is elapsed time (in minutes) of incubation, V = 0.1 mL × concentration factor, and OD600 = A600 of 1 mL of culture]. Three biological replicates were analyzed for each interaction pair, and three technical replicates were assayed per experiment.
Cell-Free Protein Degradation Assays.
Cell-free protein degradation assays were performed as previously described (28). Total proteins were extracted from 4-d-old Arabidopsis seedlings grown under −UV-B or +UV-B light in degradation buffer containing 25 mM Tris⋅HCl (pH 7.5), 10 mM NaCl, 10 mM MgCl2, 4 mM PMSF, 5 mM DTT, and 10 mM ATP. Total protein (500 μg) was incubated with 300 ng of recombinant protein at 22 °C under the same light conditions (−UV-B or +UV-B) used for seedling growth, and the aliquots were harvested at different time points before boiling and immunoblotting. The proteasome inhibitor MG132 (Merck) was selectively added as indicated. For each cell-free protein degradation assay, three independent repetitions of experiments were performed, and one representative result was presented.
Ubiquitination Assays in HEK293T Cells.
HEK293T cells were transiently transfected with 10 μg of the respective combinations of constructs for 24 h. Ubiquitinated RUP2-HA was detected by IP with 2 μL of anti-HA (Sigma–Aldrich) coupled with 10 μL of Dynabeads Protein G (Thermo Fisher Scientific) at 4 °C for 5 h. The pellets were washed three times and eluted with 2× SDS protein loading buffer. All of the eluted products were boiled at 95 °C for 10 min before immunoblotting. Three independent repetitions of experiments were performed, and one representative result was presented.
UVR8 Dimer/Monomer Assays.
UVR8 dimer/monomer assays were performed as previously described (3). Total proteins were extracted from Arabidopsis seedlings in protein extraction buffer and were separated by SDS/PAGE without boiling of the samples. The protein gel was then transferred to a thin layer of SDS/PAGE running buffer to avoid drying, and was irradiated with UV-B light before the proteins were electrophoretically transferred onto a PVDF membrane. Anti-UVR8 antibodies (12) were used for immunoblotting. For each UVR8 dimer/monomer assay, three independent repetitions of experiments were performed, and one representative result was presented.
Antibodies Used for Immunoblotting.
The following primary antibodies used in this study are commercially available or were described previously: anti-GST (Sino Biological, Inc.), anti–c-Myc (Thermo Fisher Scientific), anti-FLAG (Sigma–Aldrich), anti-MBP (TransGen Biotech), anti-GFP (Thermo Fisher Scientific), anti-Ubiquitin (Cell Signaling Technology), anti-DDB1 (38), anti-RUP2 (17), anti-UVR8 (12), and anti-HA (Sigma–Aldrich). The anti-RUP1, anti-RPN6, and anti-HY5 antibodies were generated in this study by raising rabbit polyclonal antibodies against His-RUP1, His-RPN6, and His-HY5 recombinant proteins, respectively.
Quantification and Statistical Analysis.
Hypocotyl lengths were measured using ImageJ software. The data are shown as mean ± SD, and n indicates the number of the seedlings used for hypocotyl length measurements. In the yeast two-hybrid assays, n indicates the number of biological replicates. Statistical analysis was performed using Student’s t test, with a P value less than 0.05 considered significant (*P < 0.05; **P < 0.01).
Supplementary Material
Acknowledgments
We thank Dr. Jigang Li and Dr. Yi Tao for their valuable suggestions. This work was supported by grants from the National Key R&D Program of China (Grants 2017YFA0506100 and 2016YFA0502900), the National Natural Science Foundation of China (Grants 31771347 and 31471154), the National Natural Science Foundation of Fujian (Grant 2018J06009), the Fundamental Research Funds for the Central Universities (Grants 20720150098 and 20720170068), and the Recruitment Program of Global Youth Experts of China.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816268116/-/DCSupplemental.
References
- 1.Jenkins GI. Signal transduction in responses to UV-B radiation. Annu Rev Plant Biol. 2009;60:407–431. doi: 10.1146/annurev.arplant.59.032607.092953. [DOI] [PubMed] [Google Scholar]
- 2.Kliebenstein DJ, Lim JE, Landry LG, Last RL. Arabidopsis UVR8 regulates ultraviolet-B signal transduction and tolerance and contains sequence similarity to human regulator of chromatin condensation 1. Plant Physiol. 2002;130:234–243. doi: 10.1104/pp.005041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rizzini L, et al. Perception of UV-B by the Arabidopsis UVR8 protein. Science. 2011;332:103–106. doi: 10.1126/science.1200660. [DOI] [PubMed] [Google Scholar]
- 4.Fernández MB, Tossi V, Lamattina L, Cassia R. A comprehensive phylogeny reveals functional conservation of the UV-B photoreceptor UVR8 from green algae to higher plants. Front Plant Sci. 2016;7:1698. doi: 10.3389/fpls.2016.01698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Christie JM, et al. Plant UVR8 photoreceptor senses UV-B by tryptophan-mediated disruption of cross-dimer salt bridges. Science. 2012;335:1492–1496. doi: 10.1126/science.1218091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wu D, et al. Structural basis of ultraviolet-B perception by UVR8. Nature. 2012;484:214–219. doi: 10.1038/nature10931. [DOI] [PubMed] [Google Scholar]
- 7.Oravecz A, et al. CONSTITUTIVELY PHOTOMORPHOGENIC1 is required for the UV-B response in Arabidopsis. Plant Cell. 2006;18:1975–1990. doi: 10.1105/tpc.105.040097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Yin R, Skvortsova MY, Loubéry S, Ulm R. COP1 is required for UV-B-induced nuclear accumulation of the UVR8 photoreceptor. Proc Natl Acad Sci USA. 2016;113:E4415–E4422. doi: 10.1073/pnas.1607074113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Qian C, et al. Dual-source nuclear monomers of UV-B light receptor direct photomorphogenesis in Arabidopsis. Mol Plant. 2016;9:1671–1674. doi: 10.1016/j.molp.2016.10.005. [DOI] [PubMed] [Google Scholar]
- 10.Huang X, Yang P, Ouyang X, Chen L, Deng XW. Photoactivated UVR8-COP1 module determines photomorphogenic UV-B signaling output in Arabidopsis. PLoS Genet. 2014;10:e1004218. doi: 10.1371/journal.pgen.1004218. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Favory JJ, et al. Interaction of COP1 and UVR8 regulates UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. EMBO J. 2009;28:591–601. doi: 10.1038/emboj.2009.4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Huang X, et al. Conversion from CUL4-based COP1-SPA E3 apparatus to UVR8-COP1-SPA complexes underlies a distinct biochemical function of COP1 under UV-B. Proc Natl Acad Sci USA. 2013;110:16669–16674. doi: 10.1073/pnas.1316622110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Yang Y, et al. UVR8 interacts with WRKY36 to regulate HY5 transcription and hypocotyl elongation in Arabidopsis. Nat Plants. 2018;4:98–107. doi: 10.1038/s41477-017-0099-0. [DOI] [PubMed] [Google Scholar]
- 14.Liang T, et al. UVR8 interacts with BES1 and BIM1 to regulate transcription and photomorphogenesis in Arabidopsis. Dev Cell. 2018;44:512–523 e5. doi: 10.1016/j.devcel.2017.12.028. [DOI] [PubMed] [Google Scholar]
- 15.Gruber H, et al. Negative feedback regulation of UV-B-induced photomorphogenesis and stress acclimation in Arabidopsis. Proc Natl Acad Sci USA. 2010;107:20132–20137. doi: 10.1073/pnas.0914532107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Heijde M, Ulm R. Reversion of the Arabidopsis UV-B photoreceptor UVR8 to the homodimeric ground state. Proc Natl Acad Sci USA. 2013;110:1113–1118. doi: 10.1073/pnas.1214237110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ouyang X, et al. Coordinated photomorphogenic UV-B signaling network captured by mathematical modeling. Proc Natl Acad Sci USA. 2014;111:11539–11544. doi: 10.1073/pnas.1412050111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yin R, Arongaus AB, Binkert M, Ulm R. Two distinct domains of the UVR8 photoreceptor interact with COP1 to initiate UV-B signaling in Arabidopsis. Plant Cell. 2015;27:202–213. doi: 10.1105/tpc.114.133868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ulm R, et al. Genome-wide analysis of gene expression reveals function of the bZIP transcription factor HY5 in the UV-B response of Arabidopsis. Proc Natl Acad Sci USA. 2004;101:1397–1402. doi: 10.1073/pnas.0308044100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Ang LH, et al. Molecular interaction between COP1 and HY5 defines a regulatory switch for light control of Arabidopsis development. Mol Cell. 1998;1:213–222. doi: 10.1016/s1097-2765(00)80022-2. [DOI] [PubMed] [Google Scholar]
- 21.Rubio V, et al. An alternative tandem affinity purification strategy applied to Arabidopsis protein complex isolation. Plant J. 2005;41:767–778. doi: 10.1111/j.1365-313X.2004.02328.x. [DOI] [PubMed] [Google Scholar]
- 22.Higa LA, et al. CUL4-DDB1 ubiquitin ligase interacts with multiple WD40-repeat proteins and regulates histone methylation. Nat Cell Biol. 2006;8:1277–1283. doi: 10.1038/ncb1490. [DOI] [PubMed] [Google Scholar]
- 23.Lee JH, et al. Characterization of Arabidopsis and rice DWD proteins and their roles as substrate receptors for CUL4-RING E3 ubiquitin ligases. Plant Cell. 2008;20:152–167. doi: 10.1105/tpc.107.055418. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lau OS, et al. Interaction of Arabidopsis DET1 with CCA1 and LHY in mediating transcriptional repression in the plant circadian clock. Mol Cell. 2011;43:703–712. doi: 10.1016/j.molcel.2011.07.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Arongaus AB, et al. Arabidopsis RUP2 represses UVR8-mediated flowering in noninductive photoperiods. Genes Dev. 2018;32:1332–1343. doi: 10.1101/gad.318592.118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ang LH, Deng XW. Regulatory hierarchy of photomorphogenic loci: Allele-specific and light-dependent interaction between the HY5 and COP1 loci. Plant Cell. 1994;6:613–628. doi: 10.1105/tpc.6.5.613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Koornneef M, Rolff E, Spruit CJP. Genetic control of light-inhibited hypocotyl elongation in Arabidopsis thaliana (L.) Heynh. Z Pflanzenphysiol. 1980;100:147–160. [Google Scholar]
- 28.Osterlund MT, Hardtke CS, Wei N, Deng XW. Targeted destabilization of HY5 during light-regulated development of Arabidopsis. Nature. 2000;405:462–466. doi: 10.1038/35013076. [DOI] [PubMed] [Google Scholar]
- 29.Chen H, et al. Arabidopsis CULLIN4-damaged DNA binding protein 1 interacts with CONSTITUTIVELY PHOTOMORPHOGENIC1-SUPPRESSOR of PHYA complexes to regulate photomorphogenesis and flowering time. Plant Cell. 2010;22:108–123. doi: 10.1105/tpc.109.065490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen H, et al. Arabidopsis CULLIN4 forms an E3 ubiquitin ligase with RBX1 and the CDD complex in mediating light control of development. Plant Cell. 2006;18:1991–2004. doi: 10.1105/tpc.106.043224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.McNellis TW, von Arnim AG, Deng XW. Overexpression of Arabidopsis COP1 results in partial suppression of light-mediated development: Evidence for a light-inactivable repressor of photomorphogenesis. Plant Cell. 1994;6:1391–1400. doi: 10.1105/tpc.6.10.1391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Weigel D, et al. Activation tagging in Arabidopsis. Plant Physiol. 2000;122:1003–1013. doi: 10.1104/pp.122.4.1003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Feng S, et al. The COP9 signalosome interacts physically with SCF COI1 and modulates jasmonate responses. Plant Cell. 2003;15:1083–1094. doi: 10.1105/tpc.010207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li J, et al. Arabidopsis transcription factor ELONGATED HYPOCOTYL5 plays a role in the feedback regulation of phytochrome A signaling. Plant Cell. 2010;22:3634–3649. doi: 10.1105/tpc.110.075788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chen H, et al. Firefly luciferase complementation imaging assay for protein-protein interactions in plants. Plant Physiol. 2008;146:368–376. doi: 10.1104/pp.107.111740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Huang X, et al. Arabidopsis FHY3 and HY5 positively mediate induction of COP1 transcription in response to photomorphogenic UV-B light. Plant Cell. 2012;24:4590–4606. doi: 10.1105/tpc.112.103994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Noh B, Spalding EP. Anion channels and the stimulation of anthocyanin accumulation by blue light in Arabidopsis seedlings. Plant Physiol. 1998;116:503–509. doi: 10.1104/pp.116.2.503. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Zhang Y, et al. Arabidopsis DDB1-CUL4 ASSOCIATED FACTOR1 forms a nuclear E3 ubiquitin ligase with DDB1 and CUL4 that is involved in multiple plant developmental processes. Plant Cell. 2008;20:1437–1455. doi: 10.1105/tpc.108.058891. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.