Significance
A properly functioning primary cilium is prerequisite for both normal development and aging of all ciliated organisms, including humans. In vertebrates, the signaling of Hedgehog family morphogens depends entirely on primary cilium. Recently, we reported that fibroblast growth factors (FGF) signaling interacts with that of Hedgehog, and that this is a consequence of FGF regulating length of the cilium and speed of processes that happen therein. In this report, we provide a molecular mechanism of such interaction, identifying intestinal cell kinase as a mediator of the FGF-induced changes in the ciliary morphology and function. This expands our understanding how FGF signaling regulates intracellular processes, and how aberrant FGF signaling contributes to diseases, such as achondroplasia and cancer.
Keywords: fibroblast growth factor, FGFR, intestinal cell kinase, ICK, cilia length
Abstract
Vertebrate primary cilium is a Hedgehog signaling center but the extent of its involvement in other signaling systems is less well understood. This report delineates a mechanism by which fibroblast growth factor (FGF) controls primary cilia. Employing proteomic approaches to characterize proteins associated with the FGF-receptor, FGFR3, we identified the serine/threonine kinase intestinal cell kinase (ICK) as an FGFR interactor. ICK is involved in ciliogenesis and participates in control of ciliary length. FGF signaling partially abolished ICK’s kinase activity, through FGFR-mediated ICK phosphorylation at conserved residue Tyr15, which interfered with optimal ATP binding. Activation of the FGF signaling pathway affected both primary cilia length and function in a manner consistent with cilia effects caused by inhibition of ICK activity. Moreover, knockdown and knockout of ICK rescued the FGF-mediated effect on cilia. We provide conclusive evidence that FGF signaling controls cilia via interaction with ICK.
In vertebrates, the signaling of Hedgehog (Hh) morphogens depends entirely on primary cilium. The cilia provide a structural and functional compartment that integrates a series of intricate molecular mechanisms allowing cells to process Hh-target transcriptional regulators and alter gene-expression programs in response to Hh (1). Giving the rapidly growing importance of primary cilia in the regulation of physiologic and pathologic cellular functions (2), many other signaling systems are anticipated to work through the cilia.
Several recent lines of evidence demonstrate that fibroblast growth factors (FGF) regulate primary cilia. Inactivation of the FGF-receptor Fgfr1 or its FGF ligands lead to shorter cilia in zebrafish and Xenopus (3). In mammals, FGF signaling regulates the length of primary cilia in skin fibroblasts, lung, kidney, and liver cells, human embryonic stem cells and human induced pluripotent stem cells, embryonal fibroblasts, and mesenchymal cells (4). In addition, the human skeletal dysplasias caused by activating FGFR3 mutations, such as achondroplasia, manifest by abnormal cilia (4, 5). Evidence strongly suggests that FGF signaling integrates cilia into the canonical FGF signaling pathway. However, the mechanism through which FGFs regulate primary cilia is not known.
Several serine/threonine kinases control ciliogenesis or other specific functions of primary cilia. These “ciliary kinases” include TTBK2 and GSK3β, involved in initiation of ciliogenesis and assembly of the ciliary membrane (6, 7), NEK2, which regulates cilia disassembly (8), and CK1 and GRK2, which are important for Smoothened (SMO) translocation into the cilia (9). The MAP-kinase superfamily kinase intestinal cell kinase (ICK) is another well-known regulator of primary cilia, conserved in this function from single-cell organisms to mammals. Deletion of ICK or its homologs increases the cilia length in green algae, protists, and nematodes in vivo (10–12). In cultured mammalian cells, down-regulations of ICK kinase activity lead to extended and abnormal cilia, demonstrating that ICK is an essential regulator of the length of primary cilia (13–16).
As the activity of kinases is frequently modulated by transphosphorylation by unrelated kinases, the ciliary kinases represent potential sites of interaction of primary cilia with other signaling systems. In this study, we describe one such mechanism. We unravel how FGF signaling regulates primary cilia length, leading to direct downstream consequences. Using proteomics to characterize the FGFR3 interactome in cells, we identified ICK as an FGFR interactor (17). Here, we demonstrate that FGFRs phosphorylate ICK and partially suppress ICK kinase activity and thus employ ICK to regulate the length and function of primary cilia in cells.
Results and Discussion
FGFR1, -3, and -4, but Not FGFR2, Interacts with ICK.
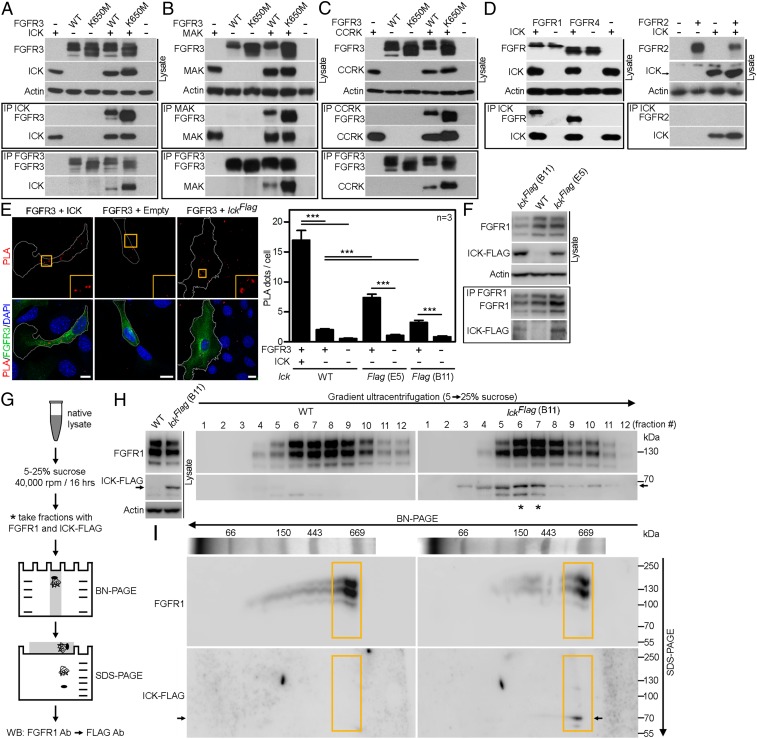
Tandem mass-spectrometry (MS) was used to identify novel FGFR3 interactors among proteins coimmunoprecipitated (co-IP) with FGFR3 from cells, or among phosphotyrosine proteins isolated from cells with activated FGFR3 signaling. In a total of 26 experiments carried out in 293T cells overexpressing FGFR3, ICK and its homolog male germ cell-associated kinase (MAK) were found in 10 (38%) and 12 (46%) of experiments, respectively (17). Additionally, the ICK-activating kinase, CCRK (18), was identified in 10 (38%) experiments. The ICK association with FGFR3 was confirmed by co-IPs of wild-type FGFR3 and ICK expressed in 293T cells (Fig. 1A). The active FGFR3 mutant K650M that associates with thanatophoric dysplasia (19) also coimmunoprecipitates with ICK. FGFR3 coimmunoprecipitates with MAK and CCRK (Fig. 1 B and C). ICK coimmunoprecipitates with FGFR1 and FGFR4; no association with FGFR2 was found (Fig. 1D).
Fig. 1.
FGFRs interact with ICK, MAK, and CCRK. (A) IP of FLAG-tagged ICK with V5-tagged wild-type (WT) FGFR3 or activating FGFR3 mutant K650M in 293T cells, or (B and C) FLAG-tagged MAK or CCRK with V5-tagged wild-type FGFR3 or FGFR3-K650M in 293T cells. Actin serves as a loading control. (D) IP of ICK with FGFR1, FGFR2, and FGFR4 demonstrating the ICK association with FGFR1 and FGFR4 but not FGFR2. (E) Wild-type NIH 3T3 cells were transfected with FLAG-tagged ICK together with V5-tagged FGFR3; IckFlag NIH 3T3 cells were transfected only with V5-tagged FGFR3. The antibodies against protein tags were used in the PLA (red); FGFR3 antibody was used to counterstain the transfected cells (green). As a negative control, cells were transfected with FGFR3 and an empty vector (WT), or by GFP (WT and IckFlag). Numbers of PLA dots per cell were calculated and plotted (Student’s t test, ***P < 0.001). (Scale bars, 10 µm.) Two clones of IckFlag NIH 3T3 cells, B11, and E5, were analyzed. (F) IP of endogenous FLAG-tagged ICK with endogenous FGFR1 in IckFlag NIH 3T3 cells; actin serves as a loading control. (G–I) Endogenous ICK forms a complex with endogenous FGFR1 in NIH 3T3 cells. (G) Scheme of the procedure, comprising ultracentrifugation, BN-PAGE, SDS/PAGE, and Western blot. (H) Cofractionalization of FGFR1 and ICK-FLAG in IckFlag(B11) NIH 3T3 cells (*). WT NIH 3T3 cells were used as a control. (I) Native complexes (fractions #6 and/or #7) were separated using BN-PAGE, followed by second dimension SDS/PAGE. Orange box shows separation of the ∼669-kDa complex containing FGFR1 and ICK-FLAG (arrow).
Because 293T cells do not form cilia, we asked if ICK interacts with FGFR3 in ciliated NIH 3T3 cells. Expressed V5-tagged FGFR3 and FLAG-tagged ICK interacted in intact NIH 3T3 cells by proximity ligation assay (PLA) (Fig. 1E). Next, we used CRISPR/Cas9 to insert FLAG epitope into the Ick locus in NIH 3T3 cells, to generate cells expressing C-terminally 3xFLAG-tagged endogenous ICK (IckFlag cells). PLA showed interaction of endogenous ICK with expressed FGFR3 in two independent IckFlag clones (Fig. 1E). Importantly, IP of endogenous FGFR1 from IckFlag cells demonstrated that endogenous ICK interacts with endogenous FGFR1 (Fig. 1F). In native lysates of IckFlag cells separated at 5–25% sucrose gradients, a cofractionation of FGFR1 with ICK was observed (Fig. 1 G and H). Fractions rich in both FGFR1 and ICK were resolved by blue-native (BN)-PAGE to separate protein complexes, which were then analyzed by second-dimension SDS/PAGE to obtain their individual components. Immunoblotting revealed an ∼669-kDa protein complex containing FGFR1 and ICK (Fig. 1I).
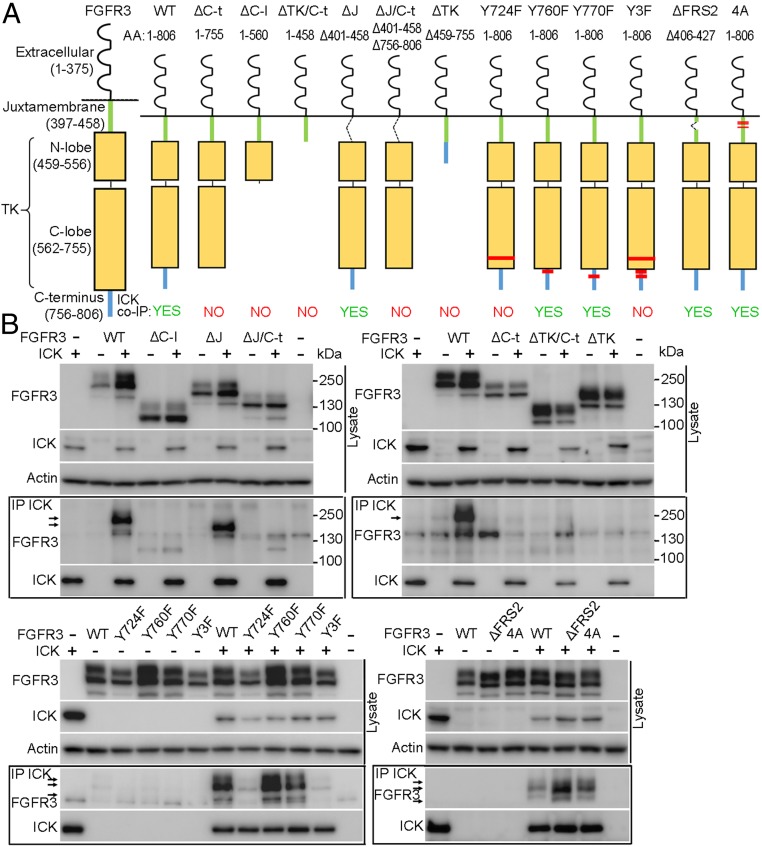
To further characterize the FGFR–ICK interaction, we generated a series of FGFR3 constructs with truncations in their intracellular domain (Fig. 2A). Mutated variants of FGFR3 were also prepared, by targeting Y724 in the tyrosine kinase (TK) domain, and Y760 and Y770 both located in the C-terminal tail. The Y724/Y760/Y770 mediate interaction with signaling intermediates SH2-βB, p85 PI3K, PLCγ, and GRB14 (20–22). The binding site for the FRS2 adapter was also targeted, either by replacing P418, L419, R425, and V427 with alanines, or by removing the entire region implicated in the interaction (amino acids 406–427) (23). FGFR3 variants were coexpressed with ICK in 293T cells, and analyzed by co-IP. All FGFR3 variants with deleted C terminus did not interact with ICK (Fig. 2B). Similarly, FGFR3-Y724F and -3YF (containing triple substitution Y724F/Y760F/Y770F) did not interact, despite having intact C termini. Deletion of FRS2 binding site had no effect on FGFR3 interaction with ICK, similar to Y760F or Y770F substitutions, which coimmunoprecipitated with ICK normally.
Fig. 2.
FGFR3 interacts with ICK via C terminus and Y724. (A) Truncated or mutated FGFR3 variants prepared for this study. Positions of point-mutated residues are indicated in red. (B) IP of FGFR3 with FLAG-tagged ICK in 293T cells. Actin serves as loading control. FGFR3 variants lacking C terminus or carrying the Y724F mutation did not co-IP with ICK. Arrows indicate FGFR3.
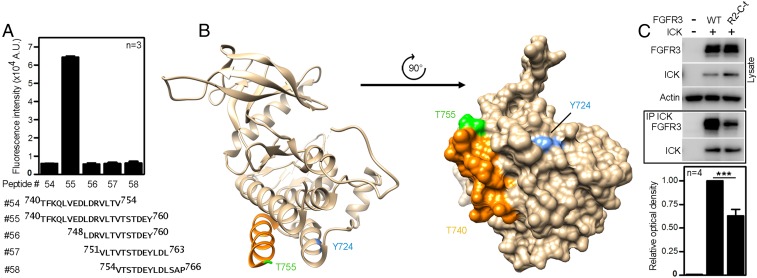
Fig. 2 shows that both the C terminus of FGFR3 and Y724 are required for ICK binding. To characterize in detail the FGFR3 epitopes involved in interaction with ICK, a peptide microarray technology was used. Short peptides (7–22 aa) covering the intracellular part of FGFR3 were synthetized and immobilized on a glass slide, incubated with recombinant ICK, and the peptide–ICK interaction was analyzed as detailed in Materials and Methods. Strong ICK binding was obtained for the FGFR3 peptide 740TFKQLVEDLDRVLTVTSTDEY760, located at the boundary between the TK domain and the C terminus (SI Appendix, Fig. S1A). Interestingly, shorter versions of this peptide, specifically 748LDRVLTVTSTDEY760 and 740TFKQLVEDLDRVLTV754, did not show any ICK binding (Fig. 3A and SI Appendix, Fig. S1B). Crystal structure of the FGFR3 TK domain (PDB ID code 4K33) shows that 740TFKQLVEDLDRVLTV754 forms an α-helix followed by an intrinsically disordered 755TSTDEY760 sequence (Fig. 3B). It is thus possible that ICK binds only to a correctly assembled secondary structure in FGFR3, and not to the peptides lacking either the helical or the unstructured parts of the 740TFKQLVEDLDRVLTVTSTDEY760 motif. This is supported by the IP data, where the C-lobe of the TK domain alone or the C terminus alone did not interact with ICK (Fig. 2B) (FGFR3-ΔC-t and FGFR3-ΔTK). Next, we asked whether differences in the sequence of FGFR3 motif involved in ICK interaction could account for the lack of FGFR2 association with ICK, observed in co-IP experiments (Fig. 1D). We replaced the 751VLTVTSTDEY760 in FGFR3 with homologous FGFR2 sequence 760ILTLTTNEEY769, and determined the interaction of chimeric FGFR3 (FGFR3-R2-C-t) with ICK using co-IP. Fig. 3C demonstrates that FGFR3-R2-C-t capacity to co-IP with ICK diminished by ∼40%, compared with the wild-type FGFR3.
Fig. 3.
The 751VLTVTSTDEY760 motif in FGFR3 is required for the interaction with ICK. (A) Averaged fluorescence intensities from three replicates of the peptide microarray involving peptides from FGFR3 C-terminal region. (B) Ribbon and surface representations of the crystal structure of the TK domain of FGFR3 (PDB ID code 4K33). Residue Y724 and C-terminal region implicated in ICK binding by co-IP experiments and peptide microarray analysis are highlighted in blue and orange/green, respectively. Orange, α-helix; green, the residue T755 (unstructured). Note that the absence of structural information for residues 756STDEY760 is suggestive of structural disorder. (C) The putative ICK interacting motif on FGFR3 (751VLTVTSTDEY760) was replaced by the analogous sequence from FGFR2 (760ILTLTTNEEY769), creating the FGFR3-R2-C-t chimera. The co-IP of FGFR3-R2-C-t with ICK compared with wild-type FGFR3 (Student’s t test; ***P < 0.001).
Our data indicate that the Y724 and C terminus of the FGFR3 are both essential for ICK binding; FGFR3-Y724F has an intact C terminus but does not bind ICK. Similarly, the FGFR3 constructs with a deleted C terminus did not bind ICK, despite having the Y724 intact (Fig. 2B). Thus, the ICK binding to either site is rather weak and cooperativity between these two sites is required in the context of the 3D FGFR3 structure (Fig. 3B). Alternatively, the Y724 mediates ICK binding indirectly, acting as an allosteric element controlling accessibility of the C terminus for ICK.
FGF Signaling Triggers Cytoplasmic Accumulation of ICK.
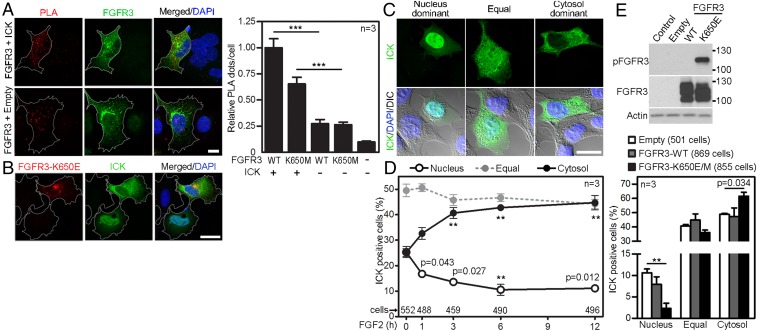
The association of wild-type FGFR3 and FGFR3-K650M with ICK was confirmed by PLA, carried out with 293T cells expressing V5-tagged FGFR3 and FLAG-tagged ICK (Fig. 4A). Immunocytochemistry revealed an overlap of FGFR3 signal with ICK in the cytoplasm, which contrasted with predominant nuclear ICK localization in cells not expressing FGFR3 (Fig. 4B), suggesting that FGFRs could cause ICK’s retention in the cytoplasm. We tested this hypothesis by determining the ICK localization in 293T cells, where the signaling of endogenous FGFR1–4 (24) was activated by addition of FGFR ligand FGF2. Progressive cytoplasmic accumulation of ICK was found in cells treated with FGF2 (Fig. 4 C and D). Expression of active FGFR3-K650E or -K650M also retained ICK in the cytoplasm (Fig. 4E). Thus, the activation of FGF signaling alters subcellular localization of ICK, causing its cytoplasmic accumulation.
Fig. 4.
FGF signaling alters ICK’s subcellular distribution. (A) 293T cells were transfected with FLAG-tagged ICK together with V5-tagged wild-type FGFR3 or its active mutant K650M. The antibodies against protein tags were used in the PLA (red); FGFR3 antibody was used to counterstain the transfected cells (green). As a negative control, cells were transfected with FGFR3 and an empty vector. Numbers of PLA dots per cell were calculated and plotted (Student’s t test, ***P < 0.001). (Scale bar, 10 µm.) (B) Increased cytosolic localization of transfected ICK in a 293T cell cotransfected with FGFR3-K650E, determined by ICK and FGFR3 immunocytochemistry (Scale bar, 20 µm.). (C and D) Altered ICK subcellular distribution in 293T cells expressing FLAG-tagged ICK, treated with FGF2; ICK was visualized by FLAG immunocytochemistry. (C) Typical localization patterns of ICK (DIC, differential interference contrast). (Scale bar, 20 µm.) (D) Percentages of cells in each category of ICK localization (Student’s t test, **P < 0.01). (E) 293T cells were transfected with FLAG-tagged wild-type FGFR3, active FGFR3-K650E, or K650M, or empty vector, and immunoblotted for phosphorylated (p) FGFR3. ICK and FGFR3 were visualized by immunocytochemistry, and ICK subcellular localization was determined.
FGFRs Phosphorylate ICK and Inhibit ICK Kinase Activity.
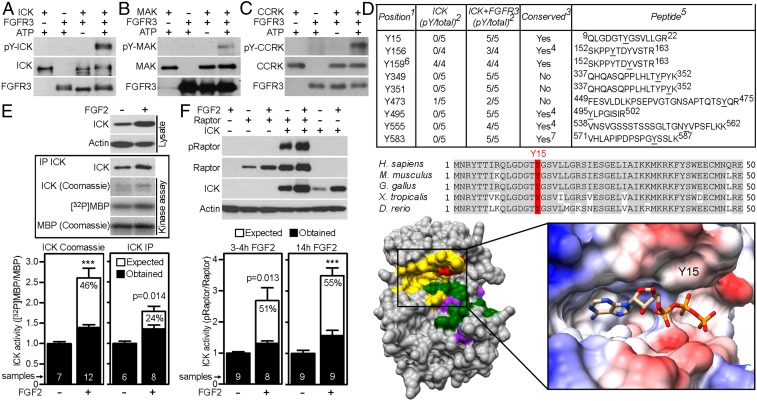
ICK shuttles between the cytoplasm and nucleus (25) and this is affected by kinase activity, as demonstrated by cytoplasmic accumulation of the partially inactive ICK mutant R272Q (26). Because FGF signaling altered ICK subcellular localization (Fig. 4), we asked whether it affected ICK activity through phosphorylation. Kinase assays utilizing recombinant FGFR3 and ICK revealed that FGFR3 phosphorylated ICK at tyrosine residues (Fig. 5A, lane 4). Kinase assays carried out with recombinant MAK and CCRK yielded similar results (Fig. 5 B and C, lane 4). Phosphotyrosine mapping identified several ICK tyrosines phosphorylated by FGFR3, among which Y15, Y156, Y495, and Y555 are conserved in ICK and MAK in human, mouse, chick, Xenopus, and zebrafish (Fig. 5D). Y495 and Y555 localized to an unstructured regulatory region of ICK (amino acids 320–632 for human ICK), making it impossible to predict the effect of their phosphorylation on ICK function. In contrast, the Y15 lies within the highly structured ICK kinase domain. In silico modeling revealed that phosphorylation at Y15 positions a negatively charged phosphate moiety in immediate proximity to the pocket used for binding of an ATP phosphate group, suggesting that phosphorylation at Y15 down-regulates ICK kinase activity via interference with optimal ATP binding (Fig. 5D). We tested this prediction by determining the kinase activity of ICK, immunopurified from 293T cells in which endogenous FGFR was activated by treatment with FGF2. FGF2 induced accumulation of expressed ICK, but its kinase activity diminished by ∼30% at the same time, as determined in a kinase assay utilizing myelin basic protein (MBP) as a substrate and P32-ATP to visualize ICK phosphorylation (Fig. 5E). Based on FGF treatment producing accumulated cytoplasmic ICK, it predicts that the degree of ICK accumulation should induce MBP phosphorylation at commensurate levels. However, the results show little induction of MBP phosphorylation after FGF2 treatment, suggesting partial inhibition of ICK activity by FGF (Fig. 5E). Because the MBP kinase assay is a cell-free experiment, we tested whether the FGF signaling inhibited ICK activity in cells. In 293T cells coexpressing ICK and mammalian target of rapamycin (mTOR) complex 1 protein Raptor, the levels of ICK kinase activity were determined by detecting previously established ICK-mediated Raptor phosphorylation at T908 (27). Treatment with FGF2 caused accumulation of ICK, but the pRaptor(T908) levels increased only weakly, corresponding to 51–55% inhibition of relative ICK activity (Fig. 5F). These experiments demonstrate that interaction with FGFRs stabilizes cytoplasmic ICK while partially downregulating its kinase activity.
Fig. 5.
FGFRs phosphorylate and partially inactivate ICK. (A–C) Tyrosine phosphorylation of ICK, MAK, or CCRK in cell-free kinase assay with recombinant FGFR3 and recombinant ICK, MAK or CCRK, detected by Western blot with pY antibody. (D) FGFR3-mediated tyrosine phosphorylation of ICK in cell-free kinase assay analyzed by MS. Footnotes: (1) Tyrosine position relative to human ICK sequence (NP_055735.1); (2) number of experiments varies due to limited sequence coverage in some MS samples; (3) conservation in Homo sapiens ICK/MAK, Mus musculus Ick/Mak, Gallus gallus ICK/MAK, Xenopus laevis ick/mak, Danio rerio mak, Drosophila melanogaster DmeI_CG42366; (4) conservation in H. sapiens, M. musculus, G. gallus, X. laevis, D. rerio but not in D. melanogaster; (5) representative peptide sequence found in MS, pY underlined; (6) ICK activating dual phosphorylation motif T157-D-Y159; (7) conservation only in ICK, not in MAK. (Middle) Sequence alignment of N-terminal region of ICK, conserved residues in gray; Y15 in red. (Bottom Left) Three-dimensional homology model of ICK kinase domain showing ATP binding site, activation loop, and substrate binding site in yellow, green, and purple, respectively, Y15 in red. (Bottom Right) Electrostatic representation of ATP binding site with bound ATP. Electronegative and electropositive sites are in blue and red, respectively. (E) The kinase activity of ICK immunoprecipitated from FGF2-treated 293T cells, measured using MBP as a substrate, in the presence of [32P]-ATP. ICK activity is presented as a relative [32P]MBP/MBP ratio (“Obtained”), and compared with the expected values based on ICK amounts entering the kinase assay quantified using Coomassie stained gel (ICK Coomassie) or ICK immunoblot (ICK IP) (Student’s t test, ***P < 0.001). The percentages express the extent of inhibition of the ICK kinase activity in FGF2-treated cells. (F) ICK kinase activity in 293T cells treated with FGF2, determined as a degree of ICK-mediated Thr908 phosphorylation of expressed Raptor. The ICK activity is presented as a relative pRaptor/Raptor ratio (“Obtained”) and compared with the expected values based on ICK levels. The percentages express the extent of inhibition of the ICK kinase activity in FGF2-treated cells.
FGF Signaling Regulates the Cilia Length via ICK.
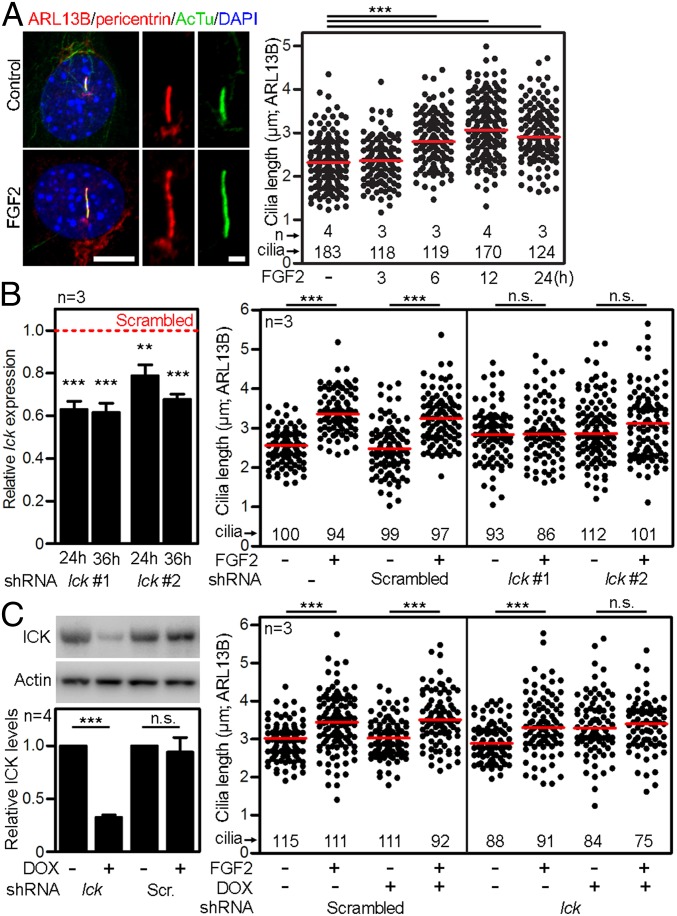
Because both ICK and FGF regulate cilia length (4, 15), we asked whether FGF signaling regulates cilia via ICK. NIH 3T3 cells were serum-starved for 12 h to produce primary cilia, treated with FGF2 for up to 24 h, and cilia were visualized by immunostainings for the axoneme (acetylated tubulin), ciliary membrane (ARL13B), and centrioles (pericentrin). FGF2 triggered progressive increase in cilia length peaking at 12 h (3.13 ± 0.05 µm vs. 2.33 ± 0.04 µm in controls) (Fig. 6A). Transfection of two independent Ick short-hairpin (sh)RNAs resulted in ∼20–40% knockdown of Ick expression in NIH 3T3 cells, with corresponding (11–18%) extension of primary cilia length, compared with nontransfected controls or cells transfected with scrambled shRNA (Fig. 6B). Importantly, the cilia in Ick shRNA cells were resistant to FGF2-mediated elongation, in contrast to scramble shRNA or control cells, which responded to FGF2 with cilia elongation (31–33%). Next, we down-regulated ICK in IckFlag(B11) NIH 3T3 cells by stable transfection of doxycycline (DOX)-inducible lentiviral shRNA construct. DOX caused ∼67% down-regulation of the ICK protein. DOX-induced cells were resistant to FGF2-mediated cilia elongation, in contrast to controls, which responded to FGF2 with cilia elongation (Fig. 6C). Finally, we tested whether chemical inhibition of ICK kinase activity affects cilia length. According to the DrugKiNET database (www.drugkinet.ca), flavopiridol, AT7519, and lestaurtinib act as chemical ICK inhibitors, with KD values of 0.69 nM, 8.3 nM, and 39 nM, respectively (28). NIH 3T3 cells treated with flavopiridol, AT7519 or lestaurtinib showed concentration-dependent deregulation of cilia length. Importantly, FGF2 failed to elongate these cilia (SI Appendix, Fig. S2).
Fig. 6.
FGF regulates the length of primary cilia via ICK. (A) Primary cilia length extension in NIH 3T3 cells treated with FGF2. Cilia were visualized by ARL13B, acetylated tubulin (AcTu) and pericentrin immunostaining, measured in 3D and plotted. Black dots, individual cilia; red bars, medians. [Scale bars: 5 µm (cells) and 1 µm (cilia).] (B) Rescue of FGF2-mediated cilia extension with two independent Ick shRNAs (Ick #1 and Ick #2). Ick transcript levels were monitored by qPCR at 24 h (beginning of serum starvation) and 36 h (FGF2 treatment) after transfection, and normalized to Gapdh expression. The columns show Ick expression levels relative to the scrambled control (red dashed line). Cilia length was measured 48 h after transfection and graphed. (C) Rescue of FGF2-mediated cilia extension in IckFlag(B11) NIH 3T3 cells stably transfected with DOX-inducible shRNA-expressing construct targeting ICK expression. ICK protein levels were monitored by immunoblot after 4 d with DOX (beginning of FGF2 treatment), normalized to actin, and plotted. Cells expressing scrambled (Scr.) shRNA upon DOX were used as a control. Student’s t test, **P < 0.01, ***P < 0.001; n.s., not significant.
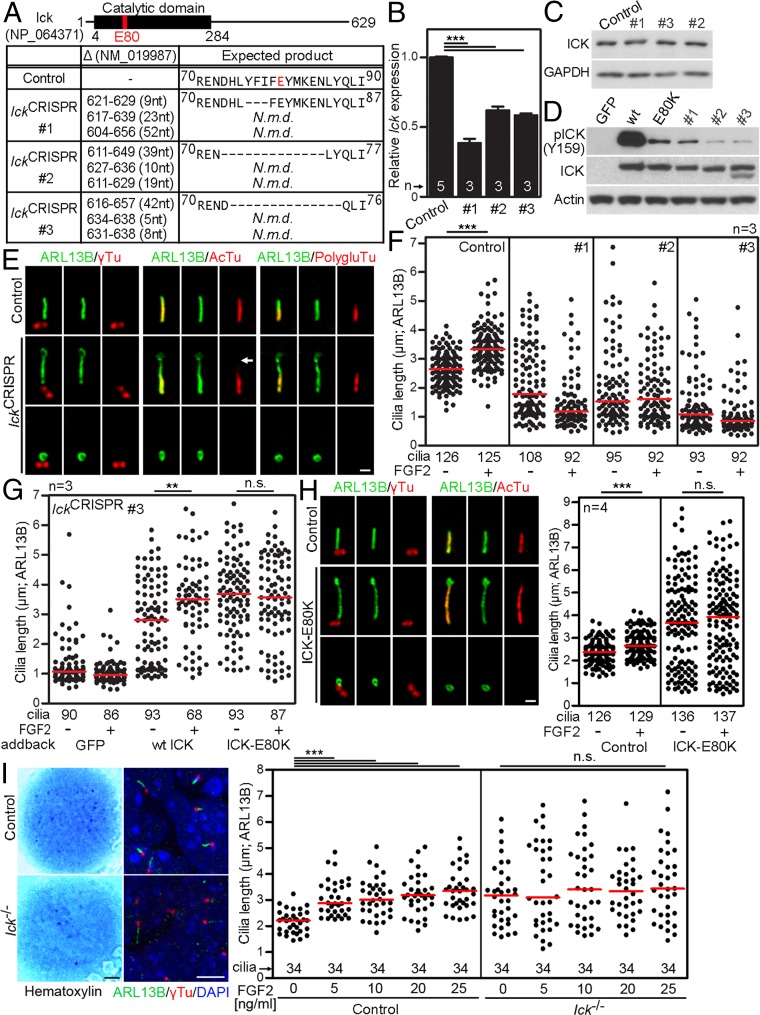
Next, the Ick locus in NIH 3T3 cells was inactivated by CRISPR/Cas9. A sequence corresponding to Glu80 in ICK was targeted, because it localizes to the ATP binding pocket and its substitution with Lys abolishes ICK kinase activity (16). We failed to generate IckE80K cells, but nevertheless produced clones with disrupted Ick. Interestingly, none of the 302 clones obtained in three rounds of CRISPR targeting possessed complete inactivation of all three Ick loci in triploid NIH 3T3 cells (29), suggesting that some level of ICK protein is essential for NIH 3T3 growth. Three clones were selected for further analyses, in which the two Ick alleles were inactivated, and the remaining one allele contained in frame deletions ranging from 3 to 14 residues surrounding the Glu80 (Fig. 7A). Ick mRNA was down-regulated to about 40–60% among the selected clones, but the protein levels remained unchanged (Fig. 7 B and C), again suggesting that presence of the ICK protein is necessary for NIH 3T3 survival. The ICK deletions generated by CRISPR were introduced into the wild-type ICK via site-directed mutagenesis, and the resulting ICK variants were evaluated for kinase activity in 293T cells. We observed a dramatic reduction in ICK activating autophosphorylation at the Y159 (25) in all three CRISPR variants, compared with wild-type ICK, demonstrating that IckCRISPR cells express normal levels of kinase-dead ICK (Fig. 7D).
Fig. 7.
FGF increases cilia length via ICK. (A) CRISPR/Cas9 targeting of Ick in triploid NIH 3T3 cells. IckCRISPR #1–3, three clones with all three alleles targeted; the positions of deletions are indicated (N.m.d., nonsense mediated decay). (B) Ick expression was analyzed by qPCR, and normalized to Gapdh. (C) ICK protein levels were analyzed by Western blot. (D) 293T cells were transfected with FLAG-tagged wild-type ICK, kinase-dead ICK-E80K or ICK variants harboring in-frame deletions as shown in A, and Western blot for ICK activating phosphorylation (p) at Y159 was used to determine the kinase activity. (E) Cilia in IckCRISPR cells were visualized by ARL13B, acetylated tubulin (AcTu), polyglutamylated tubulin (PolygluTu) or γ-tubulin (γTu) immunostaining. Both long and extremely short cilia signals are shown for IckCRISPR cells. (Scale bar, 1 µm.) Missing AcTu staining is indicated (arrow). (F) IckCRISPR cells were treated with FGF2 for 12 h and the cilia length was measured; black dots, individual cilia; red bars, medians. (G) IckCRISPR #3 cells were transfected with wt ICK or ICK-E80K, treated with FGF2, and the cilia length was measured. GFP transfection was used as a control. FGF2 extended primary cilia length in wild-type ICK add-back cells but not in ICK-E80K cells. (H) Human control and ICK-E80K fibroblasts were serum starved and immunostained to visualize cilia. (Scale bar, 1 µm.) Cells were treated with FGF2 and the cilia length was measured. (I) Micromasses produced from limb buds of either Ick+/+ (control) or Ick−/− E12 mouse littermates were treated with FGF2 for 24 h and stained with hematoxylin or ARL13B/γ-tubulin. [Scale bars: 1 mm (Left) and 5 µm (Right).] The cilia lengths were measured and graphed. Student’s t test, **P < 0.01, ***P < 0.001; n.s., nonsignificant.
Severe abnormalities were found in the cilia of IckCRISPR cells, which we termed a “cilia disaster” phenotype. These cilia were highly variable in length (0.4–6.9 μm; CV 65.14%; n = 296), compared with relatively narrow range in control cells (1.2–4.4 μm; 22.05%; n = 409) (SI Appendix, Fig. S3). The IckCRISPR cilia displayed abnormal morphology, often manifested as rudiments negative for acetylated and polyglutamylated tubulin (Fig. 7E, Bottom). Long and twisted cilia in some IckCRISPR cells also appeared less stable than wild-type cilia, as suggested by lesser axoneme staining for acetylated tubulin (30) (Fig. 7E, arrow). IckCRISPR cells showed profound disruption of Hh signaling, manifested as failure to process the GLI3 transcriptional regulator in response to Hh agonist SAG (SMO agonist) (31), and to induce expression of Hh target genes Gli1 and Ptch1 (SI Appendix, Fig. S4 A–C). Mislocalization of GLI3 and the components of the cilia transport BBS8 and intraflagellar transport (IFT)172 was found in the IckCRISPR cilia tips (SI Appendix, Fig. S4D). Finally, the Hh coreceptor SMO localized into the cilia in SAG-naïve IckCRISPR cells, in contrast to wild-type controls, which localized SMO to cilia only when treated with SAG (SI Appendix, Fig. S4E), suggesting increased permeability in IckCRISPR cilia.
FGF2 did not elongate cilia in any of the three IckCRISPR clones; however, the informative value of this data may be affected by the profound ciliary dysregulation (Fig. 7F). Addition of wild-type ICK into the IckCRISPR background partially reversed the cilia disaster phenotype, resulting in formation of many cilia of normal length, which responded to FGF2 with usual elongation (Fig. 7G). Addition of kinase-dead ICK-E80K also rescued the cilia disaster; however, these cilia were longer than those in wild-type ICK add-back cells and, importantly, were resistant to FGF2-mediated elongation (Fig. 7G).
We next evaluated the effect of FGF2 on cilia length in fibroblasts established from an individual with lethal short rib polydactyly syndrome due to an inactivating E80K mutation in ICK (16). Compared with control fibroblasts, ICK-E80K cells exhibited greater range in their cilia length distribution, with many long and twisted cilia (Fig. 7H). Treatment with FGF2 caused statistically significant cilia elongation in control fibroblasts but not in ICK-E80K cells.
Finally, the limb bud micromass cultures were used to evaluate the FGF2 effect on cilia. FGF2 caused primary cilia extension in micromasses established from embryonic day 12 (E12) mouse limb buds isolated from wild-type or Ick+/− mice (Fig. 7I) (4). Importantly, this effect depended on ICK, as cells derived from Ick−/− mouse limb buds (13) were insensitive to FGF2-mediated cilia elongation. Taken together, the ICK deletion or down-regulation of ICK kinase activity rescued the FGF-mediated elongation of primary cilia. These findings establish that FGF signaling regulates cilia length via ICK.
Because ICK inactivation inhibits Hh signaling, we analyzed the Hh activity in several models to ICK inhibition. The Sonic hedgehog (Shh)-LIGHT2 NIH 3T3 cells stably express the GLI-driven Firefly luciferase reporter capable of monitoring the Hh pathway activity (32). We inhibited endogenous Ick activity by treatment with FGF2 in Shh-LIGHT2 cells, and analyzed the transactivation of the Hh reporter. At the maximal induction obtained with 100 nM SAG, FGF2 inhibited the luciferase activity by 37% (SI Appendix, Fig. S5A). Similarly, inhibition of Ick activity in NIH 3T3 cells using flavopiridol, AT7519, or lestaurtinib abolished the SAG-mediated GLI1 up-regulation, as did FGF2 (SI Appendix, Fig. S5B). Finally, micromasses derived from murine Ick−/− limb buds failed to efficiently up-regulate Gli1 and Ptch1 expression and ciliary GLI2 localization upon SAG, similarly to the FGF2-treated control micromasses (SI Appendix, Fig. S5 C and D).
Regulation of Primary Cilia by FGF–ICK Pathway.
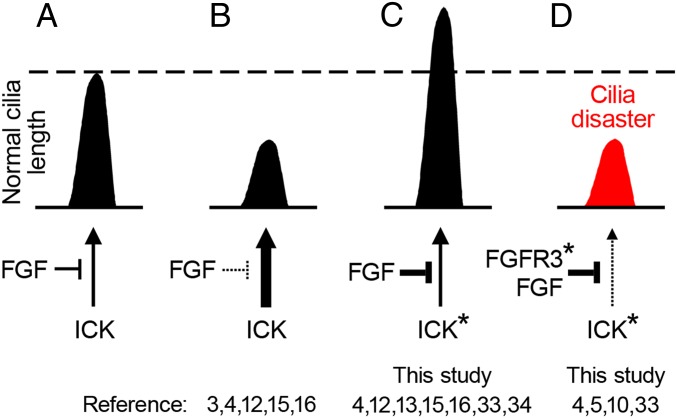
The regulation of primary cilia length and function by FGF signaling has recently emerged as a new paradigm in cell biology (3–5). However, the molecular mechanism by which FGFs regulate cilia remains unclear. In this article, we uncover that FGFRs interact with and phosphorylate an important and conserved ciliary kinase, ICK, leading to partial inhibition of its kinase activity and altered subcellular localization (Figs. 1–5). Modulations of FGF signaling regulate primary cilia length, IFT velocity, and Hh signaling consistent with effects caused by its inhibition of ICK activity (3, 4, 15, 16, 33). Moreover, ICK removal or down-regulation abolished the FGF-mediated effect on ciliary length (Figs. 6 and 7), demonstrating that FGF signaling regulates primary cilia via ICK. In the following section, we describe known cilia phenotypes regulated by FGF signaling, and explain these phenotypes on the basis of the FGF–ICK interaction (Fig. 8).
Fig. 8.
Model describing regulation of primary cilia length by the FGF–ICK pathway. (A) Under basal conditions, endogenous FGF signaling restricts ICK activity to the level required for optimal length of the cilia. (B) Experimental up-regulation of ICK activity, by increased expression of active ICK, shortens the primary cilia (12, 15, 16). Similar effect is achieved in cells with abolished FGF signaling, which are unable to inhibit ICK, leading to up-regulation of ICK kinase activity and ciliary shortening (3, 4). (C) Down-regulation of ICK activity, either by transient increase in activity of FGF signaling, or by small chemicals, expression of kinase-inactive ICK mutants (*), Ick knockout or knockdown, extends primary cilia (4, 12, 13, 15, 16, 33). (D) Under certain conditions, inactivation of ICK results in profound dysregulation of primary cilia (cilia disaster), manifested by low cilia stability and extreme length variability (10, 33). A similar phenotype is achieved by strong activation of FGF signaling via disease-associated FGFR3 mutations (*) (4, 5), suggesting that the strength of the FGF stimulus regulates optimal ciliary length through ICK.
First, the ICK is a sensitive regulator of primary cilia length. Deletion of ICK homologs LF4, LmxMPK9, and DYF-5 increased the length of cilia in green algae (Chlamydomonas reinhardtii), protists (Leishmania mexicana), and worms (Caenorhabditis elegans) (10–12). Similarly, Ick−/− mice or humans carrying partially or completely inactivating ICK mutations R272Q and E80K showed elongated primary cilia (13, 16, 34). Expression of these variants and other inactive ICK mutants in cultured cells, or partial down-regulation of Ick expression via RNA interference also elongated primary cilia (13–16, 35). Because FGFRs phosphorylate and inactivate ICK (Fig. 5), a down-regulation of FGF signaling should relieve this inhibition, resulting in elevated ICK activity and shorter cilia (Fig. 8B). Indeed, mice treated with chemical inhibitors of FGFR activity showed shorter cilia in the biliary ducts, kidney, and lung (4). This is corresponding to the evidence obtained in zebrafish and Xenopus, where inactivation of Fgf signaling led to shorter cilia in multiple tissues (3). Similar to ICK inactivation, stimulation of FGF signaling elongated primary cilia in NIH 3T3 cells, primary mouse embryonic and human fibroblasts, epithelial IMCD3 cells, and mouse limb bud mesenchymal cells (4), but failed to do so in Ick−/− background or in cells endogenously expressing inactive ICK (Fig. 7). Similarly, cells with diminished Ick expression or activity were insensitive to the FGF-mediated cilia elongation (Fig. 6), demonstrating that FGF signaling elongates primary cilia via inhibition of ICK kinase activity (Fig. 8C).
While inhibition of ICK activity extended primary cilia, experimental up-regulation of ICK activity produced shorter cilia (11–13, 16, 33). Interestingly, ICK inactivation also led to cilia shortening in at least two instances, in the neural tube and embryonic fibroblasts isolated from an Ick−/− mice (33), and in the NIH 3T3 IckCRISPR cells reported here (Fig. 7 A–G and SI Appendix, Fig. S3) (the cilia disaster phenotype). In our previous work, we showed that FGF signaling can also shorten primary cilia (4, 5), as observed in cells expressing constitutively active FGFR3-K650E or -K650M mutants. Interestingly, these FGFR3 mutants interacted with ICK (Fig. 1). Thus, the FGF-mediated ICK inactivation may lead to profound dysregulation of ciliogenesis manifested as cilia shortening or extension, depending on the strength and duration of the FGF stimulus (Fig. 8D).
Ick−/− mouse models described by both Chaya et al. (33) and Moon et al. (13) displayed preaxial polydactyly and dwarfism, a similar phenotype to human short-rib polydactyly syndrome associated with an inactivating ICK mutation (16), or to mice and humans with endocrine-cerebro-osteodysplasia syndrome coupled with mildly inactivating ICK mutations (26, 35, 36). These phenotypes suggest impaired Hh signaling, as polydactyly stems from defective Hh function in early limb patterning (37), while the shortened appendicular skeletons likely result from disrupted Hh regulation of chondrocyte proliferation in the growth plate cartilage (38). Indeed, poor and mislocalized expression of Hh target genes Gli1, Ptch1, and HoxD13 was found in the growing skeleton of the Ick−/− mice (13, 16). However, the two Ick−/− mouse models had differing ciliary length consequences. Ick−/− mice reported by Chaya et al. (33) had shortened primary cilia, yet the Moon et al. (13) mouse model showed extended primary cilia, demonstrating that both extension and shortening of primary cilia due to ICK insufficiency adversely affects Hh signaling. Similar effects were induced by experimental activation of FGF signaling, when both extension of primary cilia caused by transient FGFR activation, or shortening of cilia induced by constitutive FGF signaling, lead to similar attenuation of cell response to Hh signal (4).
In this study, we demonstrate that FGFR interacts with ICK to regulate cilia length. Several questions remain to be addressed to fully understand the process. First, it is unclear where in cell the FGFR–ICK interaction takes place. Because ICK, FGFR1, and FGFR3 are known to localize to the cilia (5, 13–16, 33, 35, 39, 40), it is possible that they interact in the cilium or in its basal body. Alternatively, the FGFRs may interact with ICK outside of the cilia. We and others (16, 25) show that the majority of the ICK protein shuttles between nucleus and cytosol, and activation of FGF signaling causes ICK retention in the cytoplasm. This suggests that FGFRs may alter intracellular distribution of ICK, thus impeding ICK cilia function by its sequestration away from cilia, in addition to partial inactivation. Second, despite numerous reports describing an essential role of ICK in maintenance of proper cilia form and function, the molecular mechanism of this phenotype is poorly known. ICK phosphorylates Kif3a (13), but how this phosphorylation affects Kif3a function or IFT velocity is not known. Further understanding of the FGF–ICK regulation of cilia will provide insights into the pathology of Mendelian inherited disorders and cancers caused by defective FGF signaling.
Materials and Methods
Cell Culture, Vectors, Transfection, RNAi, CRISPR/Cas9, Lentiviruses, and qPCR.
The 293T and NIH 3T3 cells were obtained from ATCC. Shh-LIGHT2 cells were a kind gift from P. Beachy, Stanford University School of Medicine, Stanford, CA (32). Control and ICK-E80K fibroblasts were established by an International Skeletal Dysplasia Registry, and obtained from the Registry under an approved University of California, Los Angeles human subjects protocol involving informed consent. Cells were propagated in DMEM media, supplemented with 10% FBS and antibiotics (Invitrogen). FGF2 was from R&D Systems, SAG from Millipore, leastaurtinib and flavopiridol from Tocris, AT7519 from Selleckchem. Cells were transfected using the FuGENE6 (Promega), Lipofectamine 2000 (Invitrogen), or by electroporation using the Neon Transfection System (Invitrogen). The following vectors were used: FGFR1–4 (41, 42); pmaxGFP (Lonza); Raptor (43); ICK, MAK, and CCRK (Origene). The ICK mutation E80K was described previously (16); the ICK mutants were generated by site-directed mutagenesis (Agilent Technologies). Truncated and mutated FGFR3 variants were generated by PCR mutagenesis. SureSilencing shRNA plasmid (KM37622G, clones #1 and #2; Qiagen) and scrambled control NEG4-G were used for RNAi; the GFP expression was used to identify shRNA-expressing cells. CRISPR/Cas9 was used as described previously (44). A pair of SpCas9n (D10A) nickases targeting 5′-ACTCGAAGATAAAGTAAAGATGG-3′, 5′-CTTTATCTTCGAGTACATGAAGG-3′ sites in the fourth exon of Ick were used, and cotransfected with pmaxGFP. GFP+ cells were picked to grow single-cell clones. Targeting was confirmed by PCR sequencing. To generate IckFlag cells, NIH 3T3 cells were transfected with a pair of nickases targeting 5′-AGTACCCATCCCGGCGGTGA-3′ and 5′-GCGGTGACTGTCCCGGCCA-3′ sites at the end of the last exon together with synthetic DNA (gBlock, IDT) containing 3xFlag sequence surrounded with 451- and 491-bp long homology arms at left and right. Single-cell colonies which were screened by Western blot for the presence of Flag tag at Ick. Targeting was verified by PCR sequencing. A lentiviral vector containing DOX-inducible U6 promoter and TetRep-P2A-Puro-P2A-mCherry (45) was modified to express shRNA by introducing oligonucleotides (shIck forward: ccggcacaaccacgaggcggtgtaactcgagttacaccgcctcgtggttgtgtttttg, reverse: attcaaaaacacaaccacgaggcggtgtaactcgagttacaccgcctcgtggttgtg; Scrambled forward: ccgggcgtaccaaccggaactgagactcgagtctcagttccggttggtacgtttttg; reverse: aattcaaaaagcgtaccaaccggaactgagactcgagtctcagttccggttggtacgc). Lentiviral particles were generated as described previously (46) using pMD2.G (Addgene #12259) and psPAX2 (Addgene #12260) (gift from Didier Trono, École Polytechnique Fédérale de Lausanne, Lausanne, Switzerland). IckFlag NIH 3T3 cells were transduced using generated lentiviral particles and mCherry+ cells were sorted using BD FACSAria (BD Biosciences). Cells were treated with 1 µg/mL DOX (Sigma-Aldrich) to induce the shRNA expression for 4 d. Total RNA was isolated using RNeasy Mini Kit (Qiagen) and reverse-transcribed by First Strand cDNA Synthesis Kit (Roche). For qPCR, the following QuantiTect Primers were used (Qiagen): Mm_Ick_1_SG (QT01037729), Mm_Gapdh_3_SG (QT01658692), Mm_Ptch1_1_SG (QT00149135), and Mm_Gli1_1_SG (QT00173537). For micromasses, the following primers were used in qPCR: Gli1 (5′-CTTGTGGTGGAGTCATTGGA-3′, 5′-GAGGTTGGGATGAAGAAGCA-3′), Ptch1 (5′-AATTCTCGACTCACTCGTCCA-3′, 5′-CTCCTCATATTTGGGGCCTT-3′), Gapdh (5′-CCTGTTGCTGTAGCCGTATT-3′, 5′-AACAGCAACTCCCACTCTTC-3′).
Gradient Ultracentrifugation, BN-PAGE, Western Blot, IP, Kinase Assays, and Peptide Microarrays.
Native cell lysates (50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 0.5% Igepal CA-630, 1 mM EDTA pH 8, 0.25% sodium deoxycholate, 1 mM Na2VO4, proteinase inhibitors) were cleared, loaded on 5–25% sucrose gradient (1 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 3 mM MgCl2, proteinase inhibitors), and centrifuged at 40,000 rpm/4 °C/16 h using SW 40 Ti swinging bucket rotor (Beckman Coulter). Gradient fractions were either precipitated with 10% TCA or used for BN-PAGE. For BN-PAGE, fractions were mixed with solubilization buffer (50 mM NaCl, 50 mM imidazole/HCl, 1 mM EDTA, pH 7) and concentrated using Spin-X UF 500 (cut-off 30 kDa; Corning). Before sample loading, the wells of the 4–15% native tricine-imidazole gel were washed with cathode buffer (50 mM tricine, 7.5 mM imidazole, 0.02% Coomassie blue G250, pH ∼ 7), and the gel run at constant 15 mA/4 °C. The 29- to 669-kDa native protein ladder was from Sigma. Individual lanes were cut from the gel, placed over the SDS gel, and protein complexes were separated using second dimension SDS/PAGE, and analyzed by Western blot. For Western blot, lysates were resolved by SDS/PAGE, transferred onto a PVDF membrane and visualized by chemiluminiscence (Thermo). The following antibodies were used: GAPDH (5174), GLI1 (2643), FGFR1 (9740), actin (3700; Cell Signaling); HA (sc-805), FGFR3 (sc-123), pFGFR3Y724 (sc-33041; Santa Cruz Biotechnology); FLAG (F1804), GST (G1160; Sigma); V5 (R960-25; Invitrogen), 4G10 (05-321; Millipore), pICKY159 (ab138435; Abcam), GLI3 (AF3690; RnD Systems), ICK (43), and pRaptorT908 (27). For IP, cells were extracted in buffer containing 50 mM Tris⋅HCl pH 7.4, 150 mM NaCl, 0.5% Nonidet P-40, 0.1% sodium deoxycholate, 2 mM EDTA pH 8.0, 0.5 mM DTT, proteinase inhibitors; immunocomplexes were collected on protein A/G agarose (Santa Cruz). Kinase assays were performed using recombinant FGFR3 together with ICK, MAK, or CCRK (SignalChem), in kinase buffer (60 mM Hepes pH 7.5, 3 mM MgCl2, 3 mM MnCl2, 3 µM Na3VO4, 1.2 mM DTT) in the presence of 10 µM ATP. Tyrosine phosphorylation was determined by Western blot with 4G10 antibody (Millipore). [32P]-ATP kinase assays were carried out with IP ICK and 4 μg of recombinant MBP (Sigma) as a substrate, in a kinase buffer (50 mM Hepes pH 7.5, 10 mM MnCl2, 10 mM MgCl2, 8 mM β-glycerophosphate, 1 mM DTT, 0.1 mM Na3VO4, 0.1 mM PMSF), in the presence of 1 μCi [32P]-ATP (Izotop). Samples were resolved by SDS/PAGE and visualized by autoradiography. Band intensities were quantified in ImageJ. Peptide libraries corresponding to FGFR3 intracellular domain were synthetized and immobilized on a glass slide via hydrophilic linker (Pepstar microarray; JPT Peptide Technologies). The library contained overlapping peptides (13-aa long with 10-aa overlap) covering disordered parts of FGFR3 in the juxtamembrane and C-terminal regions, and nonoverlapping peptides of varying length that emulated elements of the secondary structure exposed on the surface of FGFR3 TK domain (FGFR3: 4K33) (SI Appendix, Fig. S1B). Microarrays were incubated with recombinant ICK (I01-10G; SignalChem), labeled by DyLigh 650 (ThermoFisher Scientific). Fluorescence intensities were obtained using laser scanner Axon Genepix Scanner 4300 SL50, and quantified using GenePix (Molecular Devices). The peptides with fluorescence intensity at least 10-fold above nonspecific were considered as potential binding sites. Microarray preparation, data acquisition, and analysis were described previously (47).
Immunocytochemistry, PLA, and Cilia Length Measurements.
Cells were fixed in paraformaldehyde and incubated with the following antibodies: V5 (R960-25), acetylated α-tubulin (32-2700; Invitrogen), FLAG (F1804; Sigma), ARL13B (17711-1-AP; Proteintech), γ-tubulin (ab11316), pericentrin (ab4448; Abcam), polyglutamylated tubulin (AG-20B-0020-C100; AdipoGen), SMO (sc-166685), BBS8 (sc-271009), IFT172 (sc-398393; Santa Cruz), and GLI3 (AF3690; RnD Systems). Duolink PLA (Sigma) was used for PLA, with V5 (sc-83849; Santa Cruz) and FLAG (F1804; Sigma) antibodies; FGFR3 (sc-123; Santa Cruz) was used to counterstain the transfected cells. AlexaFluor488/594 secondary antibodies were from Invitrogen. PLA analysis was done in Fiji (fiji.sc/) using maximum projections of z-stacks. Cilia length in 3D were determined as described previously (16).
MS, Modeling, Animal Experiments, Immunohistochemistry, and Statistics.
Kinase reactions containing FGFR3 and ICK were subjected to reduction, alkylation, and in-solution digestion by trypsin. Samples were analyzed using nanoscale liquid chromatography connected to the tandem mass spectrometer (RSLCnano connected to Orbitrap Elite; Thermo Fisher). High-resolution HCD or ETD MS/MS spectra were acquired in the Orbitrap analyzer. The analysis of the MS RAW data files was carried out using the Proteome Discoverer software (v.1.4; Thermo) with a Mascot search engine (v.2.4.1; Matrix Science). Quantitative information assessment and phosphopeptide signal validation was done in Skyline. The 3D ICK model was obtained via template-based homology modeling using the PHYRE software (48). The ICK-specific functional elements, predicted using the National Center for Biotechnology Information Conserved Domain Database (49), were mapped onto a 3D model of the ICK using the CHIMERA software (50). Homology modeling (PDB ID codes 3C4W and 1JNK) was employed to dock ATP into the ICK ATP binding site. Ick−/− mice were described previously (16). Ick+/− mice were maintained in a C57/BL6N background and used as controls. For micromasses, the primary mesenchymal cells were harvested from the limb buds of E12 mouse embryos, digested with dispase II, and spotted in 10-μL aliquots at 2 × 107 cells/mL. Cells were allowed to adhere before differentiating media (60% F12/40% DMEM, 10% FBS, 50 μg/mL ascorbic acid, 10 mM β-glycerol phosphate, 1% l-glutamine) was added. Micromasses were grown for 1 d in media supplemented with FGF2 (Sigma), fixed with methanol and paraformaldehyde, and immunolabeled by ARL13B and γ-tubulin (T6557; Sigma) antibodies. For GLI2 detection, micromasses were serum-starved for 12 h, treated with FGF2 and SAG for additional 12 h, and immunostained using GLI2 and ARL13B antibodies (13). Animal experiments were approved by the Institutional Animal Care and Use Committee of Dongguk University, Korea (IACUC-2016-016-1). All experiments were performed at least in triplicate unless stated otherwise. The n values express the number of independent biological experiments. Data are presented as mean ± SEM. Two-tailed Student’s t test was used for statistical analysis of data. Brightness and contrast were adjusted in microphotographs, homogenously throughout each panel.
Supplementary Material
Acknowledgments
We thank Mikael Altun and Johan Boström for providing the U6 vector. This work was supported by the Ministry of Education, Youth and Sports of the Czech Republic (Grant KONTAKT II LH15231, National Program of Sustainability II Projects LQ1605 and LQ1601); the Agency for Healthcare Research of the Czech Republic (Grants 15-33232A, 15-34405A, and NV18-08-00567); and the Czech Science Foundation (Grants GA17-09525S, 14-31540S, and GJ16-24004Y). D.K. is supported by NIH Grants R01AR062651, R01AR066124, and R01DE019567, and by NIH/National Center for Advancing Translational Science University of California, Los Angeles Clinical and Translational Science Institute Grant UL1TR000124. Z.F. is supported by NIH Grant NIH-GM127690. H.W.K. is supported by Korea Mouse Phenotyping Project NRF2014M3A9D5A01073969 of the Ministry of Science, Information, and Communication Technology and Future Planning through the National Research Foundation. Czech Infrastructure for Integrative Structural Biology Research Infrastructure Project LM2015043 funded by the Ministery of Education, Youth and Sports of the Czech Republic is gratefully acknowledged for the financial support of the measurements at the CEITEC Proteomics Core Facility. M.K.B. was supported by the funds from Masaryk University (Grant MUNI/E/0563/2018). M.K.B. and B.F. are supported by a Junior Researcher award from the Faculty of Medicine, Masaryk University.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1800338116/-/DCSupplemental.
References
- 1.Bangs F, Anderson KV. Primary cilia and mammalian hedgehog signaling. Cold Spring Harb Perspect Biol. 2017;9:a028175. doi: 10.1101/cshperspect.a028175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Reiter JF, Leroux MR. Genes and molecular pathways underpinning ciliopathies. Nat Rev Mol Cell Biol. 2017;18:533–547. doi: 10.1038/nrm.2017.60. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Neugebauer JM, Amack JD, Peterson AG, Bisgrove BW, Yost HJ. FGF signalling during embryo development regulates cilia length in diverse epithelia. Nature. 2009;458:651–654. doi: 10.1038/nature07753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kunova Bosakova M, et al. Regulation of ciliary function by fibroblast growth factor signaling identifies FGFR3-related disorders achondroplasia and thanatophoric dysplasia as ciliopathies. Hum Mol Genet. 2018;27:1093–1105. doi: 10.1093/hmg/ddy031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Martin L, et al. Constitutively-active FGFR3 disrupts primary cilium length and IFT20 trafficking in various chondrocyte models of achondroplasia. Hum Mol Genet. 2018;27:1–13. doi: 10.1093/hmg/ddx374. [DOI] [PubMed] [Google Scholar]
- 6.Čajánek L, Nigg EA. Cep164 triggers ciliogenesis by recruiting Tau tubulin kinase 2 to the mother centriole. Proc Natl Acad Sci USA. 2014;111:E2841–E2850. doi: 10.1073/pnas.1401777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Zhang B, et al. GSK3β-Dzip1-Rab8 cascade regulates ciliogenesis after mitosis. PLoS Biol. 2015;13:e1002129. doi: 10.1371/journal.pbio.1002129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Kim S, Lee K, Choi J-H, Ringstad N, Dynlacht BD. Nek2 activation of Kif24 ensures cilium disassembly during the cell cycle. Nat Commun. 2015;6:8087. doi: 10.1038/ncomms9087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Chen Y, et al. Sonic hedgehog dependent phosphorylation by CK1α and GRK2 is required for ciliary accumulation and activation of smoothened. PLoS Biol. 2011;9:e1001083. doi: 10.1371/journal.pbio.1001083. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Berman SA, Wilson NF, Haas NA, Lefebvre PA. A novel MAP kinase regulates flagellar length in Chlamydomonas. Curr Biol. 2003;13:1145–1149. doi: 10.1016/s0960-9822(03)00415-9. [DOI] [PubMed] [Google Scholar]
- 11.Bengs F, Scholz A, Kuhn D, Wiese M. LmxMPK9, a mitogen-activated protein kinase homologue affects flagellar length in Leishmania mexicana. Mol Microbiol. 2005;55:1606–1615. doi: 10.1111/j.1365-2958.2005.04498.x. [DOI] [PubMed] [Google Scholar]
- 12.Burghoorn J, et al. Mutation of the MAP kinase DYF-5 affects docking and undocking of kinesin-2 motors and reduces their speed in the cilia of Caenorhabditis elegans. Proc Natl Acad Sci USA. 2007;104:7157–7162. doi: 10.1073/pnas.0606974104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Moon H, et al. Intestinal cell kinase, a protein associated with endocrine-cerebro-osteodysplasia syndrome, is a key regulator of cilia length and Hedgehog signaling. Proc Natl Acad Sci USA. 2014;111:8541–8546. doi: 10.1073/pnas.1323161111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Yang Y, Roine N, Mäkelä TP. CCRK depletion inhibits glioblastoma cell proliferation in a cilium-dependent manner. EMBO Rep. 2013;14:741–747. doi: 10.1038/embor.2013.80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Broekhuis JR, Verhey KJ, Jansen G. Regulation of cilium length and intraflagellar transport by the RCK-kinases ICK and MOK in renal epithelial cells. PLoS One. 2014;9:e108470. doi: 10.1371/journal.pone.0108470. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Paige Taylor S, et al. University of Washington Center for Mendelian Genomics An inactivating mutation in intestinal cell kinase, ICK, impairs hedgehog signalling and causes short rib-polydactyly syndrome. Hum Mol Genet. 2016;25:3998–4011. doi: 10.1093/hmg/ddw240. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Balek L, et al. Proteomic analyses of signalling complexes associated with receptor tyrosine kinase identify novel members of fibroblast growth factor receptor 3 interactome. Cell Signal. 2018;42:144–154. doi: 10.1016/j.cellsig.2017.10.003. [DOI] [PubMed] [Google Scholar]
- 18.Fu Z, et al. Identification of yin-yang regulators and a phosphorylation consensus for male germ cell-associated kinase (MAK)-related kinase. Mol Cell Biol. 2006;26:8639–8654. doi: 10.1128/MCB.00816-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Naski MC, Wang Q, Xu J, Ornitz DM. Graded activation of fibroblast growth factor receptor 3 by mutations causing achondroplasia and thanatophoric dysplasia. Nat Genet. 1996;13:233–237. doi: 10.1038/ng0696-233. [DOI] [PubMed] [Google Scholar]
- 20.Kong M, Wang CS, Donoghue DJ. Interaction of fibroblast growth factor receptor 3 and the adapter protein SH2-B. A role in STAT5 activation. J Biol Chem. 2002;277:15962–15970. doi: 10.1074/jbc.M102777200. [DOI] [PubMed] [Google Scholar]
- 21.Salazar L, et al. A novel interaction between fibroblast growth factor receptor 3 and the p85 subunit of phosphoinositide 3-kinase: Activation-dependent regulation of ERK by p85 in multiple myeloma cells. Hum Mol Genet. 2009;18:1951–1961. doi: 10.1093/hmg/ddp116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Brewer JR, Molotkov A, Mazot P, Hoch RV, Soriano P. Fgfr1 regulates development through the combinatorial use of signaling proteins. Genes Dev. 2015;29:1863–1874. doi: 10.1101/gad.264994.115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ong SH, et al. FRS2 proteins recruit intracellular signaling pathways by binding to diverse targets on fibroblast growth factor and nerve growth factor receptors. Mol Cell Biol. 2000;20:979–989. doi: 10.1128/mcb.20.3.979-989.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gudernova I, et al. Multikinase activity of fibroblast growth factor receptor (FGFR) inhibitors SU5402, PD173074, AZD1480, AZD4547 and BGJ398 compromises the use of small chemicals targeting FGFR catalytic activity for therapy of short-stature syndromes. Hum Mol Genet. 2016;25:9–23. doi: 10.1093/hmg/ddv441. [DOI] [PubMed] [Google Scholar]
- 25.Fu Z, et al. Activation of a nuclear Cdc2-related kinase within a mitogen-activated protein kinase-like TDY motif by autophosphorylation and cyclin-dependent protein kinase-activating kinase. Mol Cell Biol. 2005;25:6047–6064. doi: 10.1128/MCB.25.14.6047-6064.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lahiry P, et al. A multiplex human syndrome implicates a key role for intestinal cell kinase in development of central nervous, skeletal, and endocrine systems. Am J Hum Genet. 2009;84:134–147. doi: 10.1016/j.ajhg.2008.12.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wu D, et al. Intestinal cell kinase (ICK) promotes activation of mTOR complex 1 (mTORC1) through phosphorylation of Raptor Thr-908. J Biol Chem. 2012;287:12510–12519. doi: 10.1074/jbc.M111.302117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Davis MI, et al. Comprehensive analysis of kinase inhibitor selectivity. Nat Biotechnol. 2011;29:1046–1051. doi: 10.1038/nbt.1990. [DOI] [PubMed] [Google Scholar]
- 29.Todaro GJ, Green H. Quantitative studies of the growth of mouse embryo cells in culture and their development into established lines. J Cell Biol. 1963;17:299–313. doi: 10.1083/jcb.17.2.299. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Janke C, Bulinski JC. Post-translational regulation of the microtubule cytoskeleton: Mechanisms and functions. Nat Rev Mol Cell Biol. 2011;12:773–786. doi: 10.1038/nrm3227. [DOI] [PubMed] [Google Scholar]
- 31.Chen JK, Taipale J, Young KE, Maiti T, Beachy PA. Small molecule modulation of Smoothened activity. Proc Natl Acad Sci USA. 2002;99:14071–14076. doi: 10.1073/pnas.182542899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Taipale J, et al. Effects of oncogenic mutations in Smoothened and patched can be reversed by cyclopamine. Nature. 2000;406:1005–1009. doi: 10.1038/35023008. [DOI] [PubMed] [Google Scholar]
- 33.Chaya T, Omori Y, Kuwahara R, Furukawa T. ICK is essential for cell type-specific ciliogenesis and the regulation of ciliary transport. EMBO J. 2014;33:1227–1242. doi: 10.1002/embj.201488175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Okamoto S, et al. Ick ciliary kinase is essential for planar cell polarity formation in inner ear hair cells and hearing function. J Neurosci. 2017;37:2073–2085. doi: 10.1523/JNEUROSCI.3067-16.2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Oud MM, et al. A novel ICK mutation causes ciliary disruption and lethal endocrine-cerebro-osteodysplasia syndrome. Cilia. 2016;5:8. doi: 10.1186/s13630-016-0029-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ding M, et al. A murine model for human ECO syndrome reveals a critical role of intestinal cell kinase in skeletal development. Calcif Tissue Int. 2018;102:348–357. doi: 10.1007/s00223-017-0355-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.te Welscher P, et al. Progression of vertebrate limb development through SHH-mediated counteraction of GLI3. Science. 2002;298:827–830. doi: 10.1126/science.1075620. [DOI] [PubMed] [Google Scholar]
- 38.Vortkamp A, et al. Regulation of rate of cartilage differentiation by Indian hedgehog and PTH-related protein. Science. 1996;273:613–622. doi: 10.1126/science.273.5275.613. [DOI] [PubMed] [Google Scholar]
- 39.Honda A, et al. FGFR1-mediated protocadherin-15 loading mediates cargo specificity during intraflagellar transport in inner ear hair-cell kinocilia. Proc Natl Acad Sci USA. 2018;115:8388–8393. doi: 10.1073/pnas.1719861115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ding VMY, et al. Tyrosine phosphorylation profiling in FGF-2 stimulated human embryonic stem cells. PLoS One. 2011;6:e17538. doi: 10.1371/journal.pone.0017538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Krejci P, et al. Bisindolylmaleimide I suppresses fibroblast growth factor-mediated activation of Erk MAP kinase in chondrocytes by preventing Shp2 association with the Frs2 and Gab1 adaptor proteins. J Biol Chem. 2007;282:2929–2936. doi: 10.1074/jbc.M606144200. [DOI] [PubMed] [Google Scholar]
- 42.Krejci P, et al. Receptor tyrosine kinases activate canonical WNT/β-catenin signaling via MAP kinase/LRP6 pathway and direct β-catenin phosphorylation. PLoS One. 2012;7:e35826. doi: 10.1371/journal.pone.0035826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Fu Z, Kim J, Vidrich A, Sturgill TW, Cohn SM. Intestinal cell kinase, a MAP kinase-related kinase, regulates proliferation and G1 cell cycle progression of intestinal epithelial cells. Am J Physiol Gastrointest Liver Physiol. 2009;297:G632–G640. doi: 10.1152/ajpgi.00066.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ran FA, et al. Genome engineering using the CRISPR-Cas9 system. Nat Protoc. 2013;8:2281–2308. doi: 10.1038/nprot.2013.143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Eshtad S, et al. hMYH and hMTH1 cooperate for survival in mismatch repair defective T-cell acute lymphoblastic leukemia. Oncogenesis. 2016;5:e275. doi: 10.1038/oncsis.2016.72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Barta T, Peskova L, Hampl A. miRNAsong: A web-based tool for generation and testing of miRNA sponge constructs in silico. Sci Rep. 2016;6:36625. doi: 10.1038/srep36625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Harnoš J, et al. Analysis of binding interfaces of the human scaffold protein AXIN1 by peptide microarrays. J Biol Chem. 2018;293:16337–16347. doi: 10.1074/jbc.RA118.005127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kelley LA, Mezulis S, Yates CM, Wass MN, Sternberg MJE. The Phyre2 web portal for protein modeling, prediction and analysis. Nat Protoc. 2015;10:845–858. doi: 10.1038/nprot.2015.053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Marchler-Bauer A, et al. CDD: A conserved domain database for the functional annotation of proteins. Nucleic Acids Res. 2011;39:D225–D229. doi: 10.1093/nar/gkq1189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.