Significance
Measles virus is a highly contagious virus, with recent resurgence posing serious problems for human health. Replication of paramyxoviral RNA genomes involves assembly into helical nucleocapsids that protect the genome from the host immune system. Genome encapsidation is essential to the viral cycle and represents a powerful and largely unexplored target for viral treatment. To understand genome protection, we determined the near-atomic resolution structures of nucleocapsid particles assembled on specific RNA sequences using cryoelectron microscopy. The 3′ end of the RNA genome is found to be accessible to the viral polymerase, possibly enabling efficient RNA processing. We identify an amino acid that stabilizes the RNA within the binding cleft that is shown to be essential for encapsidation.
Keywords: cryoelectron microscopy, measles virus, nucleocapsids, assembly, NMR
Abstract
Assembly of paramyxoviral nucleocapsids on the RNA genome is an essential step in the viral cycle. The structural basis of this process has remained obscure due to the inability to control encapsidation. We used a recently developed approach to assemble measles virus nucleocapsid-like particles on specific sequences of RNA hexamers (poly-Adenine and viral genomic 5′) in vitro, and determined their cryoelectron microscopy maps to 3.3-Å resolution. The structures unambiguously determine 5′ and 3′ binding sites and thereby the binding-register of viral genomic RNA within nucleocapsids. This observation reveals that the 3′ end of the genome is largely exposed in fully assembled measles nucleocapsids. In particular, the final three nucleotides of the genome are rendered accessible to the RNA-dependent RNA polymerase complex, possibly enabling efficient RNA processing. The structures also reveal local and global conformational changes in the nucleoprotein upon assembly, in particular involving helix α6 and helix α13 that form edges of the RNA binding groove. Disorder is observed in the bound RNA, localized at one of the two backbone conformational switch sites. The high-resolution structure allowed us to identify putative nucleobase interaction sites in the RNA-binding groove, whose impact on assembly kinetics was measured using real-time NMR. Mutation of one of these sites, R195, whose sidechain stabilizes both backbone and base of a bound nucleic acid, is thereby shown to be essential for nucleocapsid-like particle assembly.
Measles virus (MeV) is a highly contagious human pathogen that, despite extensive vaccination, still kills nearly 100,000 people per year worldwide. MeV is a member of the Paramyxoviridae family that also includes a number of zoonoses, such as Nipah and Hendra viruses. Replication and transcription of paramyxoviral genomes by the viral polymerase (L) requires an RNA template that is encapsidated by thousands of nucleoproteins (N) in the ribonucleoprotein, forming long helical nucleocapsids (NCs) (1, 2). Genome encapsidation is thought to protect the viral RNA from recognition and degradation by the innate immune system. The paramyxoviral replication machinery also includes the tetrameric phosphoprotein (P) (3–5), a cofactor of L that also maintains N in a monomeric state (6, 7).
Three-dimensional structures of MeV NC-like particles (NCLPs) have been determined using electron microscopy (EM) (8–10) and more recently to near-atomic resolution using cryo-EM (11), while mimics of negative-strand RNA virus NCLP complexes have been crystallized in the form of closed rings (12–15). In all cases, the particles were purified in the assembled form, following spontaneous assembly of N on cellular RNA when overexpressed in bacterial or eukaryotic expression systems. The RNA in these complexes is therefore effectively random in sequence, and structural information could only be interpreted in terms of conformationally averaged positions of generic bases. Nevertheless the 4.3-Å resolution cryo-EM structure of MeV (11), revealed N- and C-terminal subdomains (NTDARM and CTDARM) (see SI Appendix, Fig. S1 for a schematic representation of N and its subdomains) that interact with adjacent N molecules in the NCLP, a mode of stabilization observed in crystal structures of parainfluenza virus 5 (PIV5) (15), pneumovirus respiratory syncytial virus (14), Rhabdoviridae vesicular stomatitis virus (VSV) (13), rabies (12), and Ebola virus (16, 17). The position of the RNA with respect to MeV N within the NCLP was found to follow the “rule of six” nucleotides per monomer (18, 19), consisting of three bases pointing into the RNA binding groove and three bases pointing away from the groove (11, 15). However, further insight into the precise binding mechanism was hampered by the lack of specific information concerning nucleotide–protein interaction, due both to the random nature of the encapsidated RNA, and the absence of higher-resolution NCLP structures.
The N-terminal 40 amino acids of the disordered domain of P are known to interact with N, forming a heterodimeric chaperone (N0P) that inhibits NC assembly. Molecular constructs of the minimal N0P complex have been engineered, comprising peptides from P and the folded domains of N (NCORE, with the NTDs and CTDs of N deleted), allowing stabilization and crystallization of RNA-free N0P (20–23). The N terminus of P binds to NCORE in a helix-kink-helix conformation. Comparison with the cryo-EM structure (11) of MeV NCLP revealed that the NTDARM and CTDARM binding sites in the NCLPs overlap almost perfectly with the binding sites of the two α-helices of the P protein in the N0P complex. The relative orientation of the N- and C-terminal lobes of NCORE differs by ∼20° between N0P and NCLPs, with the assembled state adopting a more “closed” and the chaperoned form a more “open” conformation. Notably, however, deletion of the NTDARM and CTDARM domains of N in these N0P constructs prevents their assembly into NCLPs.
We recently overcame this lack of functionality by designing a MeV N0P construct comprising NTDARM and CTDARM (with or without NTAIL) (SI Appendix, Fig. S1), fused to the first 50 residues of P via a cleavable linker (24). This construct can be purified as a nucleotide-free heterodimer that self-assembles into highly regular, micrometer-length NCLPs in vitro upon addition of RNA as short as six nucleotides in length. NMR, fluorescence anisotropy, and negative-stain EM were used to follow the kinetics of assembly. Remarkably, NCLP assembly was found to depend on RNA sequence, with polyA or 5′ viral genomic RNA hexanucleotides inducing assembly into long linear rods, and polyU hexanucleotides failing to measurably induce assembly (24). This observation may be related to a similar sequence-dependence of reencapsidation of RNA in VSV capsids (25).
Here, we exploit this tool to determine cryo-EM structures of MeV NCLPs polymerized on polyA and viral genomic 5′ (ACCAGA) sequences. These structures, bound on specific RNA sequences, allow us to unambiguously identify the initial binding position of the RNA strand in the RNA-binding groove and to determine the register of RNA within the NCLP, which in turn reveals enhanced RNA accessibility at the 3′ end. This insight allows us to propose a model for localization of the polymerase complex during replication and transcription via N–P interactions. The higher resolution available from these structures also identifies local conformational disorder in the bound RNA at an important conformational switch site and reveals local and global conformational changes in N that accompany NCLP formation. In particular helix α6, which forms the lower edge of the RNA binding pocket, exhibits large-scale degrees-of-freedom that may play a role in accommodating different RNA sequences. The structures also reveal more detail about base-specific protein:RNA interactions, and we use NMR and negative-staining EM to determine the impact of these interaction sites on NCLP assembly, identifying a single residue (R195) whose mutation effectively abrogates this essential process.
Results
High-Resolution Structures of NCLP6A and NCLP5′.
NCLPs were formed from P1–50N1–405 (including NTDARM and CTDARM but not NTAIL) and 5′ viral genomic RNA (HO-ACCAGA-OH) or a PolyA (HO-AAAAAA-OH) sequence. Structures of NCLP6A and NCLP5′ were determined at 3.3-Å resolution (Fig. 1 and SI Appendix, Fig. S2) as described in the Materials and Methods. Although the conformations of NCLPs assembled in vitro are very similar to the previously determined structure from NCLPs assembled in insect cells and bound to random RNA sequences (11) (1.18-Å RMSD over backbone residues 1–371), deviations are nevertheless observed throughout the chain (SI Appendix, Fig. S3). Differences are clustered in four regions, two of which may be expected to be disordered: the loop region of the N-terminal arm and the variable domain at the extreme end of the N-terminal lobe. However, the remaining differences are clustered in two helices (α6 and α13) that both border the RNA binding pocket. These differences may reflect the better structural definition concerning protein–RNA interactions available from NCLPs assembled in vitro on identical strands of RNA, compared with the structures that are assembled in cellulo on random RNA.
Fig. 1.
Cryo-EM structure of NCLP6A and NCLP5′. (A and B) Representative cryo-EM micrographs of NCLP6A (A) and NCLP5′ (B), with a representative class average shown as Inset. (Scale bar: 20 nm.) (C and D) Isosurface representation of the cryo-EM maps of NCLP6A (C) and NCLP5′ (D) colored as in the previously published work (11): NTDARM in blue navy, NTD in blue, CTD in salmon, CTDARM in yellow, RNA in green. A front view and a cutaway view are shown. (E and F) Zoom of three consecutive protomers of NCLP6A (E) and NCLP5′ (F) viewed from the helix exterior. Each RNA molecule is colored in different shades of green and the gaps between the RNA molecules are highlighted by red arrows. (G and H) Zoom of protein–RNA interaction in NCLP6A (G) and NCLP5′ (H), with asterisks to highlight gaps between the RNA molecules. Some clearly visible side chains described in the text are indicated.
The conformation of N is highly conserved between NCLP6A and NCLP5′ (0.45-Å backbone RMSD) (SI Appendix, Fig. S3). The orientation of the CTDARM that precedes the intrinsically disordered NTAIL domain is better defined in NCLP6A than in NCLP5′, with detectable Coulomb potential up to I396 in the former, but only until S377 in the latter (for the sake of simplicity, in the rest of the article we will refer to Coulomb potential as “density”). The conformation of the RNA backbone is also closely reproduced (0.47 Å over the backbone of the six nucleotides) (Fig. 2A).
Fig. 2.
Cryo-EM structure of RNA and NCLP. (A) Conformation of RNA in NCLP5′ and NCLP6A showing clear nucleobase stacking interactions between triplets comprising (6-1-2) and (3-4-5) that point toward solvent and protein, respectively. The gap between nucleotides 6 and 1 (asterisk) is evident in both structures. Weaker density is also seen at the C5′-O5′ atoms comprising the switch between the triplets (6-1-2) and (3-4-5) (orange arrows) but not between (3-4-5) and (6-1-2) (blue dashed arrows). (B) Representation of three N protomers within the NCLPs. Ni is represented in ribbon format (NCLP5′ and NCLP6A are superimposed and shown in orange and blue, respectively). The neighboring N molecules are shown in accessible surface format. The five phosphate groups of the RNA molecule bound to protomer Ni are shown as red spheres, while the position of the equivalent phosphate group at position one in the NCLPs assembled in vivo is shown in yellow (11) (PDB ID code 4UFT, absent in the NCLPs assembled in vitro). The orange arrow indicates the three accessible nucleotides in the final protomer of the assembled NCLP that are not buried in the cleft formed by a neighboring protomer. (C) Depiction of the N-terminal (blue) and C-terminal (pink) domains of Ni shown relative to the position of the bound RNA (orange), illustrating the overhang at the 3′ end. Residues 377–396 are not shown for clarity. The orange arrow again indicates the three accessible nucleotides.
RNA–Protein Binding Register.
The RNA molecules (polyA, HO-A1-A2-A3-A4-A5-A6-OH and 5′ genomic, HO-A1-C2-C3-A4-G5-A6-OH) are numbered from 5′ (position 1) to 3′ (position 6). The arrangement of both polyA and 5′ genomic RNA within the NCLPs shows a clear lack of density between two bases, situated at six nucleic acid intervals (Fig. 1 E and F arrows, Fig. 1 G and H, asterisks, and Fig. 2A, asterisks). Examination of the cryo-EM map at different thresholds, and comparison with the density measured form the protein, demonstrates that the NCLPs formed in vitro are fully occupied by the RNA (SI Appendix, Fig. S4), and that the RNA is the best defined region of the structure. The base at the 5′ end of the RNA hexamer (A1) binds at one end of the RNA binding cleft. This base faces toward the solvent, and stacks with base A2/C2. This is followed by a switch in the backbone conformation (Fig. 2A, orange arrows) leading to the following three bases facing into the cleft, and another switch (Fig. 2A, blue arrows) resulting in the base at the 3′ end (A6), again facing the exterior and stacking with base A1. The current model unambiguously determines the binding register, and confirms that this is retained throughout the assembled NCLP. The RNA molecule used here comprises one OH rather than a phosphate group at one end of the hexamer, leading to an unoccupied phosphate-binding site at position 1 (shown as yellow spheres in Fig. 2B), compared with the RNA upon which the structure is assembled in cellular expression systems. The replacement of the phosphate group with the OH results in a slight increase in distance between O3′ from position 6 in one hexamer to the O5′ of position 1 in the following hexamer (2.5 Å to 3.5 Å), mainly due to reorientation of the OH group, although stacking between the base of position 6 in one hexamer with the base in position 1 of the following hexamer is maintained with optimal geometry. Note that NCLP assembly is also successful when adding the (Phos-ACCAGA-OH) to N (SI Appendix, Fig. S5), although no structure of these NCLPs has been determined.
A notable lack of density is observed in the bound RNA of both NCLP6A and NCLP5′, concerning the C5′ and O5′ atoms of A/C3 around the backbone α/β dihedral angles (Fig. 2A, orange arrows). The absence of density for these atoms appears to reflect local conformational flexibility associated with the RNA backbone flip between trinucleotides pointing out of (6-1-2), then into (3-4-5), the RNA-binding cleft. Density at the C5′ and O5′ atoms of A6 (Fig. 2A, dashed blue arrows) is seen in both cases, suggesting the additional degrees of conformational freedom are significantly enhanced in the (6-1-2):(3-4-5) flip compared with (3-4-5):(6-1-2). Conformational freedom at the flip sites is also reflected in variability in backbone RNA dihedral angles over the 13 RNA conformers present in the PIV5 N–RNA ring structure, although in this case both switch sites exhibit the same disorder (SI Appendix, Fig. S6) (15). The PIV5 N–RNA rings spontaneously assemble on RNA strands of random length encountered within the cellular expression system, suggesting that the disorder observed in MeV NCLPs is not a consequence of the length of the RNA used to assemble NCLPs in vitro.
Conformational Changes Between N0P and NCLP.
Comparison of the structure of NCORE in the NCLPs with the recently determined N0P heterodimers from MeV and NiV (20, 21) reveals a significant reorientation of the N- and C-terminal lobes. Overall, the NCORE region (31–371), comprising both lobes, has a backbone RMSD of 3.8 Å compared with N0P, reflecting the previously observed change in orientation of the two domains of N upon NCLP assembly (11, 15, 20). Fig. 2C illustrates the strongly asymmetric aspect of N protomers in assembled NCLPs, leading to considerable overhang of the RNA hexamer at the 3′ end relative to the protomer to which it is bound (positions 4, 5, and 6) (arrows in Fig. 2 B and C). Because we now know the correct binding register, and invoking the rule of six (18, 19) that stipulates that the genome contains a multiple of six nucleotides, this observation reveals that, at the 3′ end of the entire NCLP, these three nucleotides are very likely to be accessible to the viral polymerase in the assembled viral NCs (because no N + 1 protomer will be present to form the upper part of the RNA binding cleft).
The N-terminal domain (31–261) of NCLP6A has an RMSD of 1.95 Å with respect to MeV N0P (21), with changes in the orientation of helix 186–203 (α6) constituting the lower lip of the RNA binding cleft (Fig. 3A). This results in W196 occupying an unusual solvent-exposed position in N0P that is stabilized in the crystal lattice by occupation of a hydrophobic pocket on the surface of the neighboring protein (SI Appendix, Fig. S7). The corresponding W194 of N0P from the Nipah virus (NiV) (20) superimposes closely with our NCLP structures, suggesting that the helix rotation in MeV N0P results from crystal packing, but nevertheless reflects the high level of conformational freedom accessible to the entire helix within correctly folded N. This is supported by further observations, in particular that α6 is tilted in the NCLP structure relative to NiV and MeV N0P (Fig. 3A), and is more structured and longer by between one and two turns at the N terminus. The first turn, D186-A189, resembles a 3-10 helix, resulting in a slight kink (Fig. 3B), and a reorganization of the binding groove allowing optimal positioning of sidechains (R194, R195, Q202) to interact with the RNA backbone. Numerous amino acids both within, or in loops adjacent to α6, are completely absent or exhibit elevated B-factors in N0P and ring crystal structures (15, 20, 21, 26) of different viruses. The position of the bound RNA in the ring crystal structure of PIV5 and in MeV NCLPs indeed follows the curvature of the α6 helix in the associated N protomers (SI Appendix, Fig. S8). Taken together, these observations suggest that α6 exhibits high intrinsic flexibility, possibly facilitating accommodation of different RNA sequences.
Fig. 3.
Differences between NCLP5′ structures and N°P complexes. (A) N-terminal domains of N from PIV5, within the N0P complex (23), and within the crystallized ring structure of N:RNA (15), NiV N0P complex (20), and MeV N0P (21) (PDB ID codes 5WKN, 4XJN, 4CO6, and 5E4V, respectively) and NCLP5′ (present study). The α6-helix is shown in red (MeV numbering 186–203). (B) Density of the α6-helix. Orientation is ∼90°(x axis) compared with the same structures in A. (C) Comparison of the CTDs of MeV N0P (21) (green) and NCLP5′ (pink, present study). The induced helical turn between residues S346 and L350 is indicated. Further conformational rearrangements associated with domain reorientation upon NCLP formation are seen at the lower limb of the domain.
The C-terminal (261–371) domain of NCLP6A exhibits a 1.74-Å backbone RMSD compared with N0P (21). Conformational changes are observed in the loop-α13 helix motif (L316-F324) that constitutes a hinge in the interface between the N and C lobes, and residues E344-R354, which constitutes the upper rim of the binding cleft. In particular a 3-10 helical turn between S346 and L350 appears to be induced upon binding the RNA (Fig. 3C).
Identification of Protein–RNA Interactions.
A number of contacts between protein and RNA are observed that are closely reproduced in both structures (Figs. 1 G and H and 4, and SI Appendix, Table S1). The 5′ nucleotide RNA backbone interacts with T183. Further interactions with the RNA phosphate backbone are found for K180, R194, R195, Y260, A267, and R354. These interactions have previously been observed in NCLPs assembled in vivo and in crystalline rings of PIV5. Remarkably, the three nucleotides (3-4-5) that point into the binding groove do not pack tightly with protein residues on the surface of the groove, but actually allow the formation of a large cavity between the bases and two adjacent proteins in the NC (Fig. 4 A–C). For this reason, interactions with RNA base moieties are confined to the lower edge of the binding groove, where they are clearly seen for R195, with detectable density of the side-chain showing that both phosphate backbone and A/G5 side-chain interact with the two NH2 groups (2.7 Å between A5 N7 and R195 Nη1 atoms in NCLP6A) (Fig. 4D). The sidechain of Q202 is also well-defined, and shows similar geometry with respect to N3 of A6 in NCLP6A as that found in PIV5, where a protein–base interaction was also evoked (15, 23, 27). E263 showed negligible density of the sidechains beyond Cβ, but three of 27 backbone-dependent sidechain rotameric states (28) for Glu place the hydrogen bonding moiety within 3.3 Å of the N6 of the RNA base in position 4. N351 also shows limited density beyond Cβ; in this case 3 of 18 backbone-dependent sidechain rotameric states (28) for Asn place the sidechain in a favorable position to form cation–π interactions that could stabilize the RNA base in position 2. On this basis, these four amino acids were targeted for mutagenesis studies and NCLP assembly assays. We note that the higher-resolution structure suggests that the Y260 ring does not stack with the 3-4-5 A-form helical strand, as was previously suggested, but rather adopts an edge-to-face orientation via energetically more favorable π–π interactions (29), as also seen in PIV5 (15).
Fig. 4.
N–RNA interaction sites within the binding groove in NCLP6A. (A–C) RNA-binding forms a cavity with amino acids from adjacent protomers. Adjacent protomers are shown in green and blue (ribbon) for the polyA NCLP. Bound RNA is shown in stick representation. The three bases pointing into the cavity are colored in red, while 1, 2, and 6 are colored in orange. (A) Cavities were calculated using a probe with radius 3 Å. Only one cavity (gray) is found. (B) Cavity surface is colored as a function of the amino acids in closest proximity (green and blue for adjacent protomers) (C) shows the view in B rotated by 90°. (D) Key interactions between a single polyA RNA molecule and a single nucleoprotein within the NCLP. RNA is shown in orange, and sites that were selected for mutation are shown in red (Q202, R195, E263, and N351). Sidechains of interacting protein residues are shown as sticks. Structures of protein and RNA in NCLP assembled on 5′ viral RNA are very similar to the conformation shown here (NCLP6A). The Inset shows the bipartite interaction of R195 with both backbone and sidechain of the nucleotide in position 5 (A in polyA).
Assembly Kinetics of NCLP5′ Binding Cleft Mutations.
Having identified these four potential protein–base interactions (R195, Q202, E263, and N351), the associated residues were mutated individually to alanine. NMR spectroscopy was used to quantitatively investigate their structural integrity as well as their assembly kinetics. Although only the flexible regions of the protein are observed by NMR, the similarity of the spectra (SI Appendix, Fig. S9), reporting on residues from NTDARM and CTDARM for the mutated and wild-type N0P, allows us to conclude that N is correctly folded in all cases and that the chaperoned heterodimer maintained. Size-exclusion profiles also confirm the integrity of these constructs.
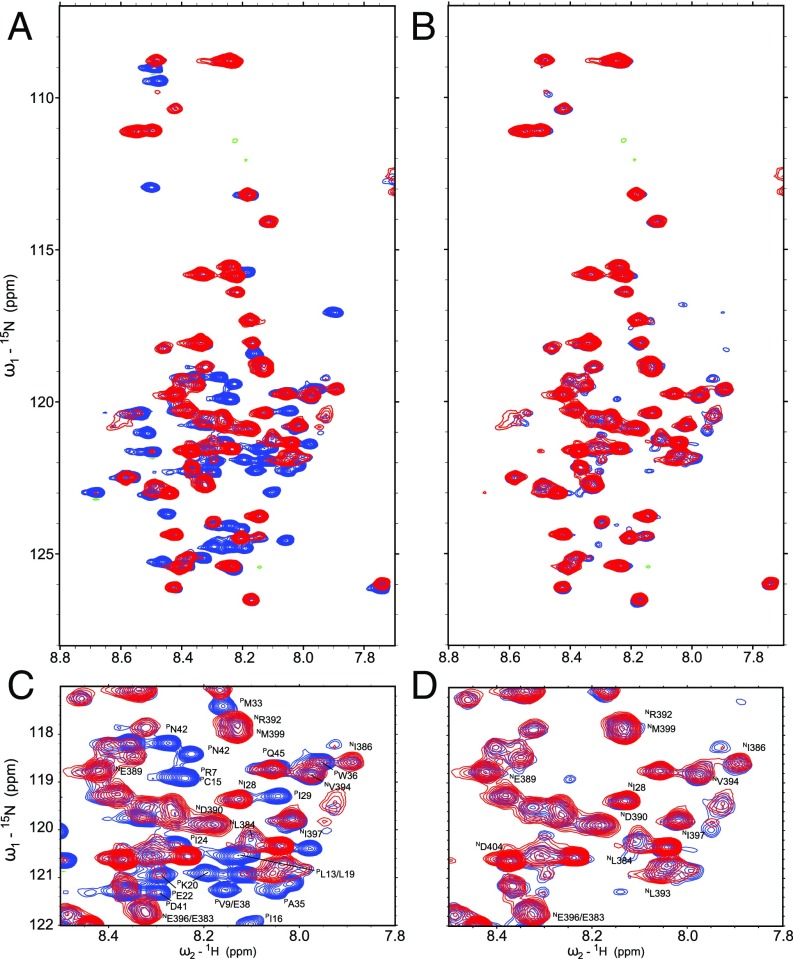
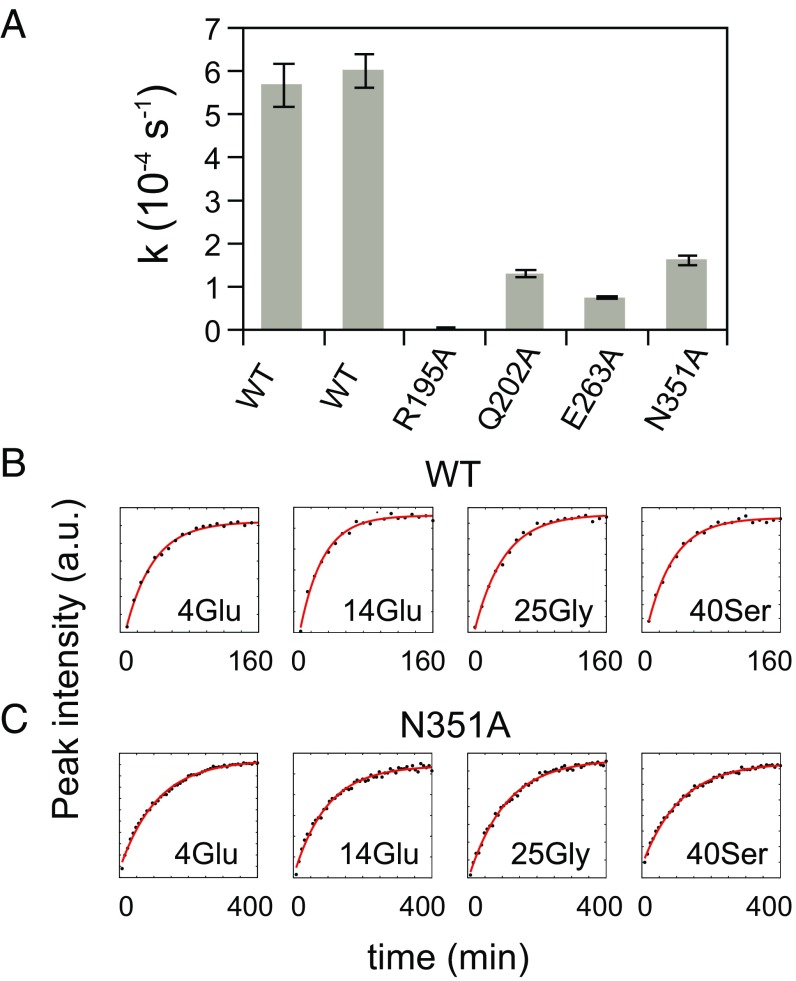
The kinetics of NCLP assembly were followed by observing amide resonances corresponding to NTDARM and CTDARM and P using heteronuclear NMR of 15N-labeled N0P. As shown recently (24), the former lose intensity as NCLPs are formed and the flexible arms become structured, while the latter gain intensity as the P peptide dissociates from N (Fig. 5). Rates of assembly of NCLPs can be determined by fitting the changes in resonance intensity of P as a function of time (Fig. 6 and SI Appendix, Fig. S10). The rate of assembly is found to be significantly different between the wild-type protein and all four mutants (Fig. 6). These experiments are quantitatively reproducible as illustrated in the example of wild-type N. Notably, the R195A mutant does not measurably assemble (Fig. 5), with negligible spectral changes compared with assembly of wild-type N. Negative-stain EM images show that NCLPs assemble poorly after 24-h incubation for R195A, Q202A, E263A mutants, while N351A exhibits NCLPs that resemble wild-type NCLPs (SI Appendix, Fig. S11).
Fig. 5.
Assembly of NCs from N0P. (A) Overlay of 1H-15N SOFAST HMQC experiments of P50N405 wild-type before (red) and 6 h after addition of HO-ACCAGA-OH (blue) showing the release of the P50 peptide giving rise to additional NMR signals. (B) Overlay of 1H-15N SOFAST HMQC experiments of N0P wild-type in the absence of RNA (red) and of N0P R195A 6 h after addition of HO-ACCAGA-OH (blue) demonstrating the absence of NCLP assembly. Note that the folded core domain of N is not visible in the spectrum. The mutated and the wild-type P1–50N1–405 spectra in the absence of RNA are therefore the same. (C and D) Zoom-ins into the respective spectra. The assignment of representative peaks corresponding to N and P residues are indicated in both zooms.
Fig. 6.
Kinetics of NC assembly. (A) Peak intensities from the 1H-15N SOFAST HMQC experiments during NCLP assembly were measured and plotted as a function of time. A set of peaks corresponding to P residues, which appear during assembly, were fitted with a monoexponential increase and then averaged. The average rate constants and corresponding SDs are plotted for each N0P mutant. The wild-type (wt) was repeated to assess reliability of the approach. Fits of representative kinetic traces (black: data points; red: exponential fit) of the wild-type and N351A assembly are shown in B and C, respectively.
Discussion
Despite considerable progress in the determination of 3D structures of paramyxo- and related viral NCLPs using EM (8, 11, 16, 17, 30), or X-ray crystallography (12–15), all existing structures have been based on average nucleobase conformations, due to the random sequence of the bound RNA. The recent demonstration that NCLPs can be assembled in vitro by adding short RNA sequences to MeV N0P constructs provided the opportunity to study the structure of the N–RNA complex in much higher detail, because the RNA sequence is both known and specific. We have determined two structures, assembled on different RNA hexamers, polyA and the 5′ viral genomic RNA sequence, both of which result in long, highly ordered NCLPs.
This approach resulted in 3D structures determined to the highest resolution yet achieved using cryo-EM of paramyxoviral NCLPs. Although the structures are similar to the conformation determined from NCLPs that were purified from insect cells, significant differences are found, in particular in two helices that form the edges of the RNA binding pocket. It is not clear whether these differences derive from the elimination of averaging over different RNA sequences present in the previous structure, or the inherently higher resolution available from the present study. Both NCLP6A and NCLP5′ conformations reveal hexameric RNA binding to a single protomer. No significant conformational variations were observed in the RNA binding cleft of the two assembled complexes. The rule of six (18, 19) is respected, with the first two nucleotides at the 5′ terminus binding the RNA pocket, with their bases accessible to the solvent, followed by a flip in the RNA backbone conformation, allowing bases 3–5 to point toward the protein surface, and a second backbone flip, allowing base 6 to again bind toward the solvent and stack with bases 1 and 2 of the following hexamer. These two triplets of bases (A6-A1-A2/C2 and A3/C3-A4-A5/G5) stack in A-form conformation, as in previously determined structures. Comparison of the density measured from protein and RNA suggests that the RNA binding pocket is always occupied, indicating that the RNA is an essential component of assembly of each protomer and does not act as a simple seed for NCLP assembly. Weaker density is observed at the C5′-O5′ sites of nucleobase 3 (Fig. 2A), where the flip between the trinucleotides pointing out of (6-1-2) and into (3-4-5) of the RNA groove appears to allow for, or require, additional conformational freedom both compared with all other nucleotides, but also compared with the inverse flip, from (3-4-5) to (6-1-2). The presence of increased disorder in both RNA sequences suggests that the additional flexibility may be required for assembly or stability of NCLPs.
It is known that single-strand polyA RNA has a strong propensity to adopt an A-form helical structure in solution (31). Our results demonstrate that the binding event of each hexamer–protomer interaction involves two large-scale conformational switches (2-3) and (5-6) to adopt the final binding pose. The thermodynamic and kinetic details of this molecular recognition trajectory remain to be elucidated.
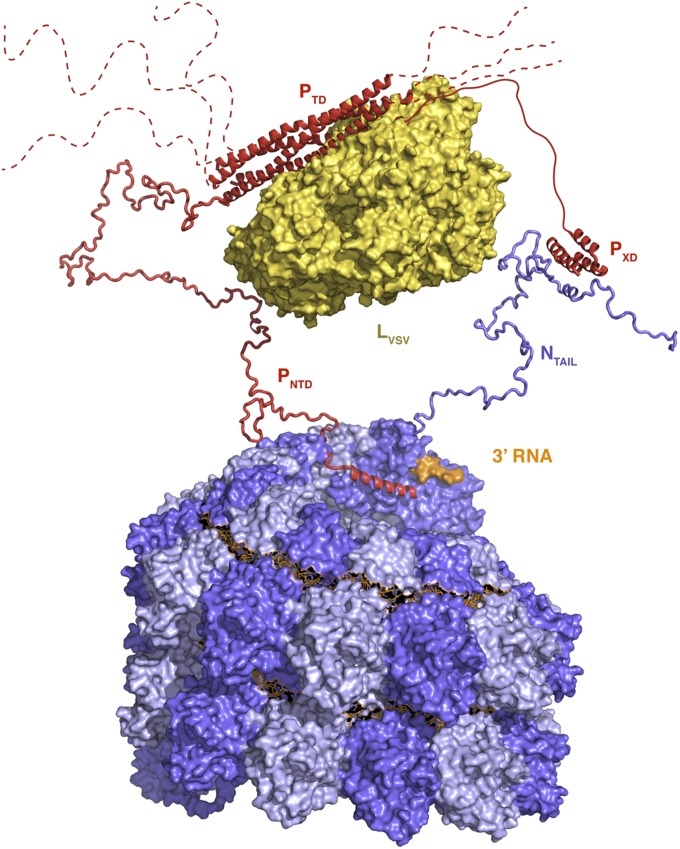
Due to the chemical composition of the added RNA (five PO4 groups for one hexamer), one phosphate binding site at the 5′ extremity of the RNA-binding pocket always remains unoccupied in NCLPs assembled in vitro (it is occupied in the equivalent site in NCLPs assembled on random RNA in insect cells). This demonstrates that the register is always maintained, likely reflecting the physiological binding mode in the initial stages of NCLP formation. The observed register would also explain the biochemical observation that positions 1 and 6 were the most accessible to chemical modification (32). Crucially, the particular shape of N when bound to RNA reveals a considerable overhang at the 3′ end with respect to the C-terminal, upper lobe of NCORE (Fig. 2 B and C). This overhang is compensated by the following (N + 1) protomer that continues the binding groove, except at the 3′ end of the NCLP, where no (N + 1) protomer is present. This observation, in combination with the known register of binding and the fact that the genome contains a multiple of six nucleotides (18, 19), reveals that the final three nucleotides at the 3′ terminus of the genome remain accessible. This characteristic may be important during transcription and replication, allowing the viral polymerase complex to access the RNA. The positioning of the viral polymerase (protein L) relative to the 3′ end of the RNA may be facilitated by the binding of P (a cofactor of L) either to NTAIL via the C-terminal XD domain of P, via the N-terminal peptide of P binding the terminal N protomer in the NC, or via the recently identified “HELL” motif 160 amino acids downstream of the N terminus of P (33). N-terminal binding of P would be further facilitated by the absence of a neighboring protomer at the 3′ end of the NCLP whose NTDARM would normally displace P. The presence of either or both of these interactions would be able to place L in the vicinity of the 3′ end of the genomic or antigenomic RNA during transcription and replication, as illustrated in the hypothetical proposition shown in Fig. 7 (see also ref. 34).
Fig. 7.
The 3′ RNA is accessible to the RNA-dependent RNA polymerase complex. Representation of the possible positioning of the polymerase complex relative to the solvent accessible 3′ end of the RNA genome (orange surface) at the terminus of the NC (adjacent N protomers are shown in different shades of blue). Only NTAIL of the terminal N protomer is shown for simplicity. The protomer can bind P via two possible mechanisms, via an interaction between the NTAIL molecular recognition element and the XD domain of P, and via the N-terminal peptide of P that could bind to N, due to the absence of adjacent N protomers that displace all other P peptides in the NC. The unfolded domains PNTD and NTAIL were generated using the flexible-meccano algorithm (56). The conformation of the polymerase (L) is taken from the structure of the homologous enzyme from VSV (57). The binding of L to P is unknown and is shown figuratively here.
Comparison with the recently published N0P structures from NiV and MeV shows significant conformational changes as a result of NCLP assembly. The first involves the previously observed reorientation of the N- and C-terminal lobes of NCORE (20), facilitated by concerted conformational changes on both sides of the RNA binding pocket. In particular helix α6, situated on the lower rim of the RNA-binding groove, exhibits significant degrees of conformational flexibility, enabling modulation of the dimensions of the groove as a function of the RNA sequence, as reflected in the observed correlation between RNA and helical position in MeV and PIV5 N–RNA structures (SI Appendix, Fig. S8). Our structures show that upon assembly the α6 helix extends significantly to constitute almost the entire lower edge of the binding pocket, allowing for optimal positioning of basic and polar sidechains that contribute to stabilization of the bound RNA (Figs. 3 and 4).
The high-resolution structures also allow us to investigate protein–RNA interactions that may be important for NCLP assembly and stability. We observe RNA-backbone:protein interactions lining the bottom (N-terminal lobe of NCORE) of the groove (K180, T183, R194, and R195) that were identified in previous MeV and PIV5 NCLP structures, and a notable lack of interactions with the top of the groove (C-terminal lobe of NCORE) related to the presence of the cavity. There are few interactions between nucleotide bases and protein moieties, principally because the three bases pointing into the RNA binding groove form a surprisingly large internal cavity, together with adjacent protomers (Fig. 4). Note that a recent study of the N–RNA complex of VSV showed that polyamines can bind in a similar pocket formed within the NC, and that this binding inhibits viral replication (35).
Four putative nucleobase:protein interactions were nevertheless identified (Fig. 4D), involving bases on the edge of this pocket. Two of these (R195, Q202) exhibit favorable geometry for the formation of stabilizing hydrogen bonds with bases in positions A5/G5 and A6, respectively, and are conserved between Paramyxoviridae. The sidechain conformation of E263 is lacking, but in the case of favorable sidechain rotamers could form hydrogen–bond interactions with N6 of A4. N351 also lacks sidechain density, but is potentially able to form cation–π interactions with the A2/C2 base. Both E263 and N351 are conserved through Morbilli- and Henipaviruses.
Using NMR we are able to follow the process of NCLP assembly in real time (Figs. 5 and 6). Mutation of the four sites identified as potentially involved in nucleobase–protein interactions all resulted in slower NCLP assembly (Fig. 6), suggesting that N:RNA stability relies on the concerted effect of multiple intermolecular interactions along the entire binding groove. Mutations of three different amino acids in helix α6 of the related mumps virus resulted in reduced polymerase activity in minigenome assays (26). Putative interactions between Q202 and an RNA base were recently proposed on the basis of the crystal structure of PIV5 rings (27, 36). On this basis, Q202 was suggested to play a role in initiation at replication promotors in PIV2, where Q202A shows increased luciferase activity, but no effect on RNA binding or editing. The present study shows that Q202 of MeV stabilizes the 3′ terminus of the RNA (position 6). We can speculate whether disruption of this mechanism could render the RNA more flexible and possibly more accessible to the polymerase.
Remarkably, the R195A mutation resulted in almost complete stalling of assembly (Figs. 5 and 6). Although R195 has previously been proposed to interact with the phosphate group of A5/G5 (11), an additional, potentially stronger interaction is observed in both NC6A and NC5′ between the R195 sidechain and the A5/G5 base. Such interactions have been commonly observed in protein–RNA complexes, although they are more common for guanines than adenines (37, 38). This tight bipartite interaction implicating both sidechain and backbone of the same nucleotide provides a strong constraint on the conformation of the 3-4-5 stacking motif, and is potentially important for the stability of the 5:6 in–out transition and the N–RNA interaction. This additional stability may be related to the reduced conformational disorder compared with the 2:3 transition. Both polyA and 5′ RNA has a purine at this position, and it is interesting to speculate whether this interaction can be maintained with a pyrimidine in position 5. By inspection, it would appear that such an interaction could only be maintained if the base adopted the less energetically favorable syn conformation (39). Further structural studies of these specific sequences will be necessary to determine how they may be accommodated. The sidechains of R195 and Y260 pack closely to form the interior of the RNA binding pocket, and it is possible that mutation of R195 perturbs Y260 and its associated stabilization of the stacked bases in positions 3-4-5. The impact of the R195A mutation on NCLP formation demonstrated by NMR establishes that it plays an important role in the assembly process.
In conclusion, the ability to form well-ordered NCLPs assembled on specific RNA sequences has allowed us to determine the structures of N–RNA complexes comprising polyA and viral genomic 5′ RNA sequences. Two aspects of this study are unique: first, the known RNA sequences allow us to establish the 3′ and 5′ binding modes in assembled NCs; and second, they afford increased resolution that reveals a wealth of information concerning the structure and stability of assembled NCLPs. These structures thus unambiguously identify the initial binding site and register of genomic RNA binding, revealing that the three nucleotides at the 3′ end of the RNA genome are largely exposed in the assembled NCLP, making them accessible to the RNA-dependent RNA polymerase complex, and suggesting a role for N–P interactions in positioning L during RNA processing. Local conformational flexibility is also observed in the bound RNA, in particular at one of the two switch junctions between bases pointing into and out of the binding pocket. Local conformational changes in N are induced by NCLP formation, in particular, helix α6, constituting the lower edge of the RNA binding pocket, exhibits substantial conformational freedom that appears to be important in modulating the binding of different RNA sequences. Finally, stabilizing interactions between RNA bases and protein sidechains were identified and four putative nucleobase interaction sites in the binding groove of N were mutated, resulting in systematic changes in assembly kinetics, with one specific amino acid, R195, proving essential for formation of NCLPs.
Materials and Methods
Expression and Purification.
MeV N0P constructs were expressed and purified essentially as previously described (24). Briefly, the plasmids encoding for P1–50-TEV-N1–405-8His (pET41c) were transformed into Escherichia coli Rosetta (λDE3)/pRARE (Novagen), grown in lysogeny broth (LB) or M9 medium supplied with stable isotopes for NMR (15N) at 37 °C until an OD between 0.6 and 1. Isopropyl β-d-1thiogalactopyranoside (IPTG) was added to a final concentration of 1 mM and expression was continued at 20 °C overnight. The cells were lysed by sonication in 20 mM Tris pH 8, 150 mM NaCl, and the cleared lysate was subjected to standard Ni-affinity purification. The protein was eluted from the Ni-beads with an imidazole concentration of 400 mM. N0P was cleaved using tobacco etch virus (TEV) protease in an overnight dialysis as described elsewhere (24) and the protein was finally transferred into 50 mM Na-phosphate pH 6, 150 mM NaCl, 2 mM DTT by size-exclusion chromatography (Superdex 200 column; GE Healthcare). All experiments were conducted in this final buffer.
Electron Microscopy.
The NCLPs used for EM negative stain and cryo-EM were formed by adding 50 μM RNA (HO-ACCAGA-OH or HO-AAAAAA-OH) to 20 μM of N0P in a final volume of 20 μL. The samples were incubated overnight at 25 °C.
Negative staining.
Samples were applied to the clean side of carbon on mica (carbon/mica interface) and negatively stained with 2% sodium silicotungstate (pH 7.0). Micrographs were taken with a T12 FEI microscope at 120 kV and a magnification of 30,000×.
Cryo-EM.
For cryo-EM, 3.5 μL of sample at about 1 mg/mL were applied to glow discharged (25 mA, 40 s) R2/1 quantifoil grids (Quantifoil Micro Tools). A Vitrobot (20°, 100% humidity, 2-s blotting time, and force 1) was used to flash-freeze the grid. Grids were transferred to a Polara electron microscope working at 300 kV. Special care has been taken to correct for the coma. Movies (40 frames of 0.1 s and a dose of 1 electron/Å2 per frame) were recorded in an automatic mode on a K2 summit direct electron detector in superresolution mode using the Latitude S (Gatan) software at a nominal magnification of 30,000× (0.816 Å per pixel at the image level) with defocus values between 1.2 μm and 3.5 μm. A total number of 186 and 211 movies were acquired for NCLP6A and NCLP5′, respectively.
Helical NCLP5′ Reconstruction.
For initial reconstruction, movies were motion-corrected with unblur (40) and aligned movie sums obtained from all 40 aligned frames were used. Only micrographs showing Thon rings exceeding 8 Å and no notable astigmatism were retained. Helices were manually selected from each micrograph using e2helixboxer.py from EMAN2 (41) and 40,267 overlapping 768 × 768-pixel2 segments were extracted with a step size of 30 Å. The resulting stack of segments was convolved by the corresponding contrast transfer functions (CTFs) determined using CTFFIND3 and CTFTILT (42). We used our previous helical reconstruction of the truncated MeV NCs (11) filtered to 30-Å resolution as an initial model, and first applied the previously determined symmetry parameters (pitch of 49.54 Å and 12.34 subunits per turn) during refinement with SPRING (43). This procedure converged to a 3.9-Å resolution structure as estimated based on the 0.143 cut-off of the Fourier shell correlation (FSC). Helical parameters were then refined using SPRING segrefine3dgrid module by setting up progressively narrower and finer grid searches with the initial grid spanning between a pitch of 47 and 50 Å and units per turn between 12.3 and 12.4, and a final grid with a step of 0.01 Å and 0.01 units per turn. This resulted into a pitch of 48.6 Å and 12.35 subunits per turn, further improving the map resolution to 3.6 Å. Reducing the dose to 11 e−/Å2 (15 frames) for further processing resulted into a map with a resolution of 3.4 Å, obtained from 38,820 segments (310,560 asymmetric units after symmetrisation). The dataset size was then increased by performing automatic picking in RELION (44), using 2D class-averages obtained from the manually picked dataset as templates, resulting in 102,353 segments of 660 × 660 pixel2 with a distance between segments of 31 Å. In addition, per particle CTF determination was done with Gctf (45). A final 3D refinement in RELION using these segments and the per particle defocus values r resulted into an improved map with a pitch of 48.58 Å and 12.346 subunits per turn, to a global resolution of 3.3 Å (FSC0.143 cut-off) with an automatically determined B-factor of −115 Å−2 (SI Appendix, Fig. S2).
Helical NCLP6A Reconstruction.
The frames 2–20 (total dose 16 e−/Å2) were aligned and averaged with motioncor2 (46), and the dose-weighted sums were used for further processing. For initial reconstruction, the data were binned by a factor of 2 to a pixel size of 1.632 Å. Helices were manually selected from each micrograph using e2helixboxer.py and 28,990 overlapping 360 × 360-pixel2 segments were extracted with a step size of 30 Å. The resulting stack of segments was convolved by the corresponding CTFs determined using CTFFIND3 and CTFTILT (42), and a phase-flipped version was created for initial symmetry refinement. Using the NCLP5′ reconstruction filtered to 10 Å as initial model, 15 iterations of refinement were computed using an in-house implementation of the IHRSR method (47) in SPIDER (48), resulting in refined symmetry parameters of 48.35-Å pitch and 12.34 units per turn. With these values, a first reconstruction was computed using SPRING to a resolution of 3.7 Å (FSC0.143 cut-off), from 28,886 segments (231,088 asymmetric units after symmetrization). The dataset size was then increased by performing automatic picking in RELION, using 2D class-averages obtained from the manually picked dataset as templates, resulting in 111,582 segments with a distance between segments of 31 Å. Using these segments, 3D refinement in RELION (with helical symmetry refinement) gave a reconstruction of 3.4-Å resolution (FSC0.143 cut-off) with a pitch of 48.38 Å and 12.341 subunits per turn. To further improve the map, segments were reextracted from the unbinned dataset (pixel size 0.816 Å) with a segment size of 720 × 720 pixel2, and Gctf was used to calculate per particle CTF parameters. A final 3D refinement in RELION resulted into an improved map with a global resolution of 3.3 Å (FSC0.143 cut-off) with an automatically determined B-factor of −122 Å−2 (SI Appendix, Fig. S2).
Atomic Coordinate Refinement.
Initially, we placed into the helical EM density our previous atomic model of the truncated nucleoprotein monomer bound to random RNA from insect cells and modeled with polyC-RNA6 (PDB ID code 4UFT), using the rigid body option implemented in Chimera (49). Because the synthetic ACCAGA-RNA6 used in this study to mimic the 5′ sequence of the MeV genome possessed an OH group at the 5′ end instead of a phosphate group, corresponding breaks in the backbone density of the RNA in the EM map could be clearly detected. These breaks enabled us to: (i) identify the beginning and the end of each RNA6 strand, and (ii) numerically mutate the original polyC-RNA6 into HO-ACCAGA-OH in COOT (50). The resulting monomer model was replaced into the helical NC density in Chimera. The fitted monomer model was then superimposed with the atomic model of the MeV nucleoprotein core crystallized in an open N0P-like conformation (PDB ID code 5E4V) (21) and solved at 2.7-Å resolution, that we had to cut into NTD and CTD between amino acids 266 and 267 for individual rigid-body fitting to simulate the nucleoprotein closure upon RNA binding and helical polymerization. In addition, we overlaid the atomic model of the truncated PIV5 nucleoprotein as extracted from the structure of a tridecameric PIV5 NCORE -RNA ring (PDB ID code 4XJN) solved at 3.11 Å by X-ray crystallography (15). Comparison of these three atomic models allowed us to improve the visual match between the model and the EM density and to create a composite model of the monomer.
We then generated its symmetry-related copies using the determined pitch and number of subunits per turn, and extracted a segment of the helical EM map corresponding to three consecutive monomers, to take into account intersubunit interfaces during model refinement. The central subunit of the atomic model corresponding to this trimer map was then manually rebuilt with COOT by improving the main-chain position and fitting all visible side chains possible and a new atomic model of the trimer created based on this improved monomer model. This trimeric model was refined against the EM map using Refmac5 (51). Only Fourier coefficients up to 3.2-Å resolution were taken into account, and the map was used either directly or blurred by applying a B-factor of +50 Å−2. These values of resolution cut-off and B-factor were estimated from a plot of average amplitudes of Fourier coefficients (<|F|>) as a function of resolution and for a series of different positive and negative B-factors between −150 Å−2 and +150 Å−2. Jelly-body refinement (51) was performed to reduce overfitting. In addition, various sets of external restraints were used at different stages of refinement. Specifically, LibG (52) was used to maintain stacking between bases, and ProSMART (53) supplied additional restraints on interatomic distances based on either the 5E4V or the 4XJN models, or using a library of standard backbone conformations corresponding to different secondary-structure elements. The model was improved by iterative cycles of manual model rebuilding of the central subunit, reassembly of an improved trimeric model, and Refmac5 refinement. Noncrystallographic symmetry was taken into account to improve interfaces and minimize clashes between adjacent subunits. Final stages of refinement were performed using the modular workflow for refinement of atomic models rsref (54) by iterative cycles of automatic refinement of the previous trimeric model, extraction of the central subunit and reassembly into a new trimeric model by symmetrization.
The final atomic model of the central subunit was reexpanded into a helical structure and an FSC between the model and the cryo-EM map (SI Appendix, Fig. S2) estimates a resolution as about 3.4 Å at the 0.5 criterion. The NCLP6A model was refined from the NCLP5′ model resulting in an almost identical structure except in the region of the RNA bases.
Final figures were generated with University of California, San Francisco Chimera (49) and PyMol (Schrödinger).
NMR Spectroscopy.
Spectral assignment of N0P in the pH 6 buffer was transferred from the assignment described previously (24). All NMR experiments were performed at a temperature of 25 °C and 1H frequencies of 700 or 850 MHz. The assembly of NCLPs were followed by recording a series of SOFAST (55) 1H-15N HMQC spectra over time before and after initiation of NCLP assembly by HO-ACCAGA-OH. The spectra were recorded with 100 complex points in the indirect dimension, 200-ms recycling delay, and eight transients at a protein concentration of 100 μM and an RNA concentration of 200 μM. This yielded a time resolution of about 8 min per experiment. These conditions were kept identical for all N0P mutants.
The time traces for the peaks corresponding to P (residues 4, 5, 10, 14, 16, 17, 22, 25, 29, 40) were extracted from the respective peak intensities throughout the kinetics. Residues 16, 17, and 40 of P were excluded from fitting of the R195A mutant assembly, as no intensity was observable for these residues throughout the whole kinetics. To avoid cross compensation between different exponential rates and to guarantee easy comparison of assembly speed between the different N0P mutants, only a single exponential growth rate was used to fit the data. The intensity build-up curves were fit individually for every peak and were then averaged per N0P mutant (Fig. 6A). Error bars were plotted from the SD.
Supplementary Material
Acknowledgments
The authors thank Daphna Fenel and Leandro Estrozi for invaluable help with computing and image processing. I.G. thanks Garib Mushrudov for initial help with building of atomic models into the cryoelectron microscopy density using REFMAC. This work was supported by Grenoble Research for Integrated Cellular and Structural Biology (GRAL) (Grant ANR-10-LABX-49-01), Finovi (Fondation innovations en infectiologie) and the Fondation Recherche Médicale (Equipe DEQ20170336754). The work used the platforms of the Grenoble Instruct-ERIC centre (ISBG; UMS 3518 CNRS-CEA-UGA-EMBL) with support from FRISBI (Grant ANR-10-INSB-05-02) and GRAL (Grant ANR-10-LABX-49-01) within the Grenoble Partnership for Structural Biology. S.M. acknowledges funding from a European Molecular Biology Organization long-term fellowship (ALTF 468-2014) and European Commission (Grant EMBOCOFUND2012 GA-2012-600394) via Marie Curie Action. The electron microscope facility is supported by the Rhône-Alpes Region, the Fondation Recherche Medicale, the Fonds Européen de Développement Régional, the Centre National de la Recherche Scientifique, the Commissariat à l’Energie Atomique, the University of Grenoble, European Molecular Biology Laboratory, and the GIS-Infrastrutures en Biologie Sante et Agronomie. A.D. is supported by the Fondation Recherche Medicale via Grant ARF20160936266.
Footnotes
The authors declare no conflict of interest.
This article is a PNAS Direct Submission. O.P. is a guest editor invited by the Editorial Board.
Data deposition: The atomic coordinates have been deposited in the Protein Data Bank, https://www.wwpdb.org/ (PDB ID code 6H5Q) and the Electron Microscopy Data Bank, https://www.ebi.ac.uk/pdbe/emdb/ (entry ID EMD-0141).
This article contains supporting information online at www.pnas.org/lookup/suppl/doi:10.1073/pnas.1816417116/-/DCSupplemental.
References
- 1.Finch JT, Gibbs AJ. Observations on the structure of the nucleocapsids of some paramyxoviruses. J Gen Virol. 1970;6:141–150. doi: 10.1099/0022-1317-6-1-141. [DOI] [PubMed] [Google Scholar]
- 2.Egelman EH, Wu SS, Amrein M, Portner A, Murti G. The Sendai virus nucleocapsid exists in at least four different helical states. J Virol. 1989;63:2233–2243. doi: 10.1128/jvi.63.5.2233-2243.1989. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Tarbouriech N, Curran J, Ruigrok RW, Burmeister WP. Tetrameric coiled coil domain of Sendai virus phosphoprotein. Nat Struct Biol. 2000;7:777–781. doi: 10.1038/79013. [DOI] [PubMed] [Google Scholar]
- 4.Communie G, et al. Structure of the tetramerization domain of measles virus phosphoprotein. J Virol. 2013;87:7166–7169. doi: 10.1128/JVI.00487-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bruhn JF, et al. Crystal structure of the nipah virus phosphoprotein tetramerization domain. J Virol. 2014;88:758–762. doi: 10.1128/JVI.02294-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Huber M, et al. Measles virus phosphoprotein retains the nucleocapsid protein in the cytoplasm. Virology. 1991;185:299–308. doi: 10.1016/0042-6822(91)90777-9. [DOI] [PubMed] [Google Scholar]
- 7.Curran J, Marq JB, Kolakofsky D. An N-terminal domain of the Sendai paramyxovirus P protein acts as a chaperone for the NP protein during the nascent chain assembly step of genome replication. J Virol. 1995;69:849–855. doi: 10.1128/jvi.69.2.849-855.1995. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Schoehn G, et al. The 12 Å structure of trypsin-treated measles virus N-RNA. J Mol Biol. 2004;339:301–312. doi: 10.1016/j.jmb.2004.03.073. [DOI] [PubMed] [Google Scholar]
- 9.Bhella D, Ralph A, Yeo RP. Conformational flexibility in recombinant measles virus nucleocapsids visualised by cryo-negative stain electron microscopy and real-space helical reconstruction. J Mol Biol. 2004;340:319–331. doi: 10.1016/j.jmb.2004.05.015. [DOI] [PubMed] [Google Scholar]
- 10.Desfosses A, Goret G, Farias Estrozi L, Ruigrok RWH, Gutsche I. Nucleoprotein-RNA orientation in the measles virus nucleocapsid by three-dimensional electron microscopy. J Virol. 2011;85:1391–1395. doi: 10.1128/JVI.01459-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gutsche I, et al. Structural virology. Near-atomic cryo-EM structure of the helical measles virus nucleocapsid. Science. 2015;348:704–707. doi: 10.1126/science.aaa5137. [DOI] [PubMed] [Google Scholar]
- 12.Albertini AAV, et al. Crystal structure of the rabies virus nucleoprotein-RNA complex. Science. 2006;313:360–363. doi: 10.1126/science.1125280. [DOI] [PubMed] [Google Scholar]
- 13.Green TJ, Zhang X, Wertz GW, Luo M. Structure of the vesicular stomatitis virus nucleoprotein-RNA complex. Science. 2006;313:357–360. doi: 10.1126/science.1126953. [DOI] [PubMed] [Google Scholar]
- 14.Tawar RG, et al. Crystal structure of a nucleocapsid-like nucleoprotein-RNA complex of respiratory syncytial virus. Science. 2009;326:1279–1283. doi: 10.1126/science.1177634. [DOI] [PubMed] [Google Scholar]
- 15.Alayyoubi M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus parainfluenza virus 5 nucleoprotein-RNA complex. Proc Natl Acad Sci USA. 2015;112:E1792–E1799. doi: 10.1073/pnas.1503941112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Su Z, et al. Electron cryo-microscopy structure of Ebola virus nucleoprotein reveals a mechanism for nucleocapsid-like assembly. Cell. 2018;172:966–978.e12. doi: 10.1016/j.cell.2018.02.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sugita Y, Matsunami H, Kawaoka Y, Noda T, Wolf M. Cryo-EM structure of the Ebola virus nucleoprotein-RNA complex at 3.6 Å resolution. Nature. 2018;563:137–140. doi: 10.1038/s41586-018-0630-0. [DOI] [PubMed] [Google Scholar]
- 18.Calain P, Roux L. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol. 1993;67:4822–4830. doi: 10.1128/jvi.67.8.4822-4830.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Kolakofsky D, et al. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: The rule of six revisited. J Virol. 1998;72:891–899. doi: 10.1128/jvi.72.2.891-899.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yabukarski F, et al. Structure of Nipah virus unassembled nucleoprotein in complex with its viral chaperone. Nat Struct Mol Biol. 2014;21:754–759. doi: 10.1038/nsmb.2868. [DOI] [PubMed] [Google Scholar]
- 21.Guryanov SG, Liljeroos L, Kasaragod P, Kajander T, Butcher SJ. Crystal structure of the measles virus nucleoprotein core in complex with an N-terminal region of phosphoprotein. J Virol. 2015;90:2849–2857. doi: 10.1128/JVI.02865-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Renner M, et al. Nucleocapsid assembly in pneumoviruses is regulated by conformational switching of the N protein. eLife. 2016;5:e12627. doi: 10.7554/eLife.12627. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Aggarwal M, Leser GP, Kors CA, Lamb RA. Structure of the paramyxovirus PIV5 nucleoprotein in complex with an amino-terminal peptide of the phosphoprotein. J Virol. 2017;92:e01304–e01317. doi: 10.1128/JVI.01304-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Milles S, et al. Self-assembly of measles virus nucleocapsid-like particles: Kinetics and RNA sequence dependence. Angew Chem Int Ed Engl. 2016;55:9356–9360. doi: 10.1002/anie.201602619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Green TJ, et al. Access to RNA encapsidated in the nucleocapsid of vesicular stomatitis virus. J Virol. 2011;85:2714–2722. doi: 10.1128/JVI.01927-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Severin C, et al. Releasing the genomic RNA sequestered in the mumps virus nucleocapsid. J Virol. 2016;90:10113–10119. doi: 10.1128/JVI.01422-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Matsumoto Y, Ohta K, Kolakofsky D, Nishio M. A point mutation in the RNA-binding domain of human parainfluenza virus type 2 nucleoprotein elicits abnormally enhanced polymerase activity. J Virol. 2017;91:e02203–e02216. doi: 10.1128/JVI.02203-16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Shapovalov MV, Dunbrack RL., Jr A smoothed backbone-dependent rotamer library for proteins derived from adaptive kernel density estimates and regressions. Structure. 2011;19:844–858. doi: 10.1016/j.str.2011.03.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Meyer EA, Castellano RK, Diederich F. Interactions with aromatic rings in chemical and biological recognition. Angew Chem Int Ed Engl. 2003;42:1210–1250. doi: 10.1002/anie.200390319. [DOI] [PubMed] [Google Scholar]
- 30.Cox R, et al. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci USA. 2014;111:15208–15213. doi: 10.1073/pnas.1413268111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Tubbs JD, et al. The nuclear magnetic resonance of CCCC RNA reveals a right-handed helix, and revised parameters for AMBER force field torsions improve structural predictions from molecular dynamics. Biochemistry. 2013;52:996–1010. doi: 10.1021/bi3010347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Iseni F, et al. Chemical modification of nucleotide bases and mRNA editing depend on hexamer or nucleoprotein phase in Sendai virus nucleocapsids. RNA. 2002;8:1056–1067. doi: 10.1017/s1355838202029977. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Milles S, et al. An ultraweak interaction in the intrinsically disordered replication machinery is essential for measles virus function. Sci Adv. 2018;4:eaat7778. doi: 10.1126/sciadv.aat7778. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sourimant J, Plemper RK. Organization, function, and therapeutic targeting of the morbillivirus RNA-dependent RNA polymerase complex. Viruses. 2016;8:E251. doi: 10.3390/v8090251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gumpper RH, et al. A polyamide inhibits replication of vesicular stomatitis virus by targeting RNA in the nucleocapsid. J Virol. 2018;92:e00146-18. doi: 10.1128/JVI.00146-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Matsumoto Y, Ohta K, Kolakofsky D, Nishio M. The control of paramyxovirus genome hexamer length and mRNA editing. RNA. 2018;24:461–467. doi: 10.1261/rna.065243.117. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nobeli I, Laskowski RA, Valdar WSJ, Thornton JM. On the molecular discrimination between adenine and guanine by proteins. Nucleic Acids Res. 2001;29:4294–4309. doi: 10.1093/nar/29.21.4294. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Morozova N, Allers J, Myers J, Shamoo Y. Protein-RNA interactions: Exploring binding patterns with a three-dimensional superposition analysis of high resolution structures. Bioinformatics. 2006;22:2746–2752. doi: 10.1093/bioinformatics/btl470. [DOI] [PubMed] [Google Scholar]
- 39.Sokoloski JE, Godfrey SA, Dombrowski SE, Bevilacqua PC. Prevalence of syn nucleobases in the active sites of functional RNAs. RNA. 2011;17:1775–1787. doi: 10.1261/rna.2759911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Brilot AF, et al. Beam-induced motion of vitrified specimen on holey carbon film. J Struct Biol. 2012;177:630–637. doi: 10.1016/j.jsb.2012.02.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Tang G, et al. EMAN2: An extensible image processing suite for electron microscopy. J Struct Biol. 2007;157:38–46. doi: 10.1016/j.jsb.2006.05.009. [DOI] [PubMed] [Google Scholar]
- 42.Mindell JA, Grigorieff N. Accurate determination of local defocus and specimen tilt in electron microscopy. J Struct Biol. 2003;142:334–347. doi: 10.1016/s1047-8477(03)00069-8. [DOI] [PubMed] [Google Scholar]
- 43.Desfosses A, Ciuffa R, Gutsche I, Sachse C. SPRING—An image processing package for single-particle based helical reconstruction from electron cryomicrographs. J Struct Biol. 2014;185:15–26. doi: 10.1016/j.jsb.2013.11.003. [DOI] [PubMed] [Google Scholar]
- 44.Scheres SHW. A Bayesian view on cryo-EM structure determination. J Mol Biol. 2012;415:406–418. doi: 10.1016/j.jmb.2011.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Zhang K. Gctf: Real-time CTF determination and correction. J Struct Biol. 2016;193:1–12. doi: 10.1016/j.jsb.2015.11.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng SQ, et al. MotionCor2: Anisotropic correction of beam-induced motion for improved cryo-electron microscopy. Nat Methods. 2017;14:331–332. doi: 10.1038/nmeth.4193. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Egelman EH. Reconstruction of helical filaments and tubes. Methods Enzymol. 2010;482:167–183. doi: 10.1016/S0076-6879(10)82006-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Shaikh TR, et al. SPIDER image processing for single-particle reconstruction of biological macromolecules from electron micrographs. Nat Protoc. 2008;3:1941–1974. doi: 10.1038/nprot.2008.156. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Pettersen EF, et al. UCSF Chimera—A visualization system for exploratory research and analysis. J Comput Chem. 2004;25:1605–1612. doi: 10.1002/jcc.20084. [DOI] [PubMed] [Google Scholar]
- 50.Emsley P, Cowtan K. Coot: Model-building tools for molecular graphics. Acta Crystallogr D Biol Crystallogr. 2004;60:2126–2132. doi: 10.1107/S0907444904019158. [DOI] [PubMed] [Google Scholar]
- 51.Murshudov GN, et al. REFMAC5 for the refinement of macromolecular crystal structures. Acta Crystallogr D Biol Crystallogr. 2011;67:355–367. doi: 10.1107/S0907444911001314. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Brown A, et al. Tools for macromolecular model building and refinement into electron cryo-microscopy reconstructions. Acta Crystallogr D Biol Crystallogr. 2015;71:136–153. doi: 10.1107/S1399004714021683. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Nicholls RA, Fischer M, McNicholas S, Murshudov GN. Conformation-independent structural comparison of macromolecules with ProSMART. Acta Crystallogr D Biol Crystallogr. 2014;70:2487–2499. doi: 10.1107/S1399004714016241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Hoffmann NA, Jakobi AJ, Vorländer MK, Sachse C, Müller CW. Transcribing RNA polymerase III observed by electron cryomicroscopy. FEBS J. 2016;283:2811–2819. doi: 10.1111/febs.13732. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Schanda P, Brutscher B. Very fast two-dimensional NMR spectroscopy for real-time investigation of dynamic events in proteins on the time scale of seconds. J Am Chem Soc. 2005;127:8014–8015. doi: 10.1021/ja051306e. [DOI] [PubMed] [Google Scholar]
- 56.Ozenne V, et al. Flexible-meccano: A tool for the generation of explicit ensemble descriptions of intrinsically disordered proteins and their associated experimental observables. Bioinformatics. 2012;28:1463–1470. doi: 10.1093/bioinformatics/bts172. [DOI] [PubMed] [Google Scholar]
- 57.Liang B, et al. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell. 2015;162:314–327. doi: 10.1016/j.cell.2015.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.