Abstract
O-linked β-N-acetylglucosamine (O-GlcNAc) is a single sugar modification found on many different classes of nuclear and cytoplasmic proteins. Addition of this modification, by the enzyme O-linked N-acetylglucosamine transferase (OGT), is dynamic and inducible. One major class of proteins modified by O-GlcNAc is transcription factors. O-GlcNAc regulates transcription factor properties through a variety of different mechanisms including localization, stability and transcriptional activation. Maintenance of embryonic stem (ES) cell pluripotency requires tight regulation of several key transcription factors, many of which are modified by O-GlcNAc. Octamer-binding protein 4 (Oct4) is one of the key transcription factors required for pluripotency of ES cells and more recently, the generation of induced pluripotent stem (iPS) cells. The action of Oct4 is modulated by the addition of several post-translational modifications, including O-GlcNAc. Previous studies in mice found a single site of O-GlcNAc addition responsible for transcriptional regulation. This study was designed to determine if this mechanism is conserved in humans. We mapped 10 novel sites of O-GlcNAc attachment on human Oct4, and confirmed a role for OGT in transcriptional activation of Oct4 at a site distinct from that found in mouse that allows distinction between different Oct4 target promoters. Additionally, we uncovered a potential new role for OGT that does not include its catalytic function. These results confirm that human Oct4 activity is being regulated by OGT by a mechanism that is distinct from mouse Oct4.
Keywords: Oct4, OGT, O-GlcNAc, transcription factor
Introduction
Discovered in the 1980s by Hart and coworkers, O-linked β-N-acetylglucosamine (O-GlcNAc) is found in all higher eukaryotes and is a dynamic, single sugar modification found on serine and threonine residues on many different classes of nuclear and cytoplasmic proteins (Torres and Hart 1984; Love and Hanover 2005; Zachara and Hart 2006; Teo et al. 2010a). O-GlcNAc regulates many different cellular processes such as: cell cycle control (Dehennaut et al. 2007, 2008), stress response (Zachara and Hart 2004; Ohn et al. 2008), cell signaling pathways (Wells et al. 2001; Vosseller et al. 2002a, 2002b; Gandy et al. 2006; Yang et al. 2008) and chromatin remodeling (Yang et al. 2002; Fujiki et al. 2009; Gambetta et al. 2009; Sinclair et al. 2009). The major class of proteins regulated by O-GlcNAc is transcription factors and related gene-expression modulators (Comer and Hart 1999; Vosseller et al. 2002a, 2002b; Love and Hanover 2005; Zachara and Hart 2006; Brimble et al. 2010; Teo et al. 2010b). Regulation of transcription factors via O-GlcNAc modification occurs by a variety of different mechanisms (reviewed in Brimble et al. 2010) including examples of altering protein stability (Han and Kudlow 1997), nuclear localization (Dentin et al. 2008; Sayat et al. 2008), DNA binding (Gao et al. 2003), transcriptional activation (Housley et al. 2008) and protein-protein interactions (Gewinner et al. 2004).
Unlike phosphorylation, there is only one enzyme required for the addition of O-GlcNAc, O-GlcNAc transferase (O-linked N-acetylglucosamine transferase, OGT) (Haltiwanger et al. 1992), and one for the removal, O-GlcNAcase (OGA) (Dong and Hart 1994; Gao et al. 2001). OGT is essential for embryonic and somatic cell survival in mammalian cells (Shafi et al. 2000; O'Donnell et al. 2004), Drosophila melanogaster (Ingham 1984) and Arabidopsis (Hartweck et al. 2002) but interesting not in C. elegans (Hanover et al. 2005).
During vertebrate development, the octamer-binding protein 4 (Oct4) is expressed in the oocyte and the inner cell mass (Scholer et al. 1989). Oct4 is required for early embryogenesis and maintenance of pluripotency (Nichols et al. 1998), and has been further shown to be one of the key regulatory transcription factors required for pluripotency in mammalian embryonic stem (ES) cells (Hay et al. 2004; Loh et al. 2006; Rodriguez et al. 2007). Oct4 can activate or repress multiple genes which play a role in pluripotency or early differentiation including: Sox2 (Chew et al. 2005), Nanog (Rodda et al. 2005), Fgf4 (Yuan et al. 1995), Utf1 (Nishimoto et al. 1999), cdx2 (Strumpf et al. 2005), opn (Botquin et al. 1998) as well as Oct4 itself (Chew et al. 2005). Small changes in expression level of Oct4 can induce differentiation leading to the need for tight regulation (Niwa et al. 2000; Hay et al. 2004; Rodriguez et al. 2007). The function of Oct4 protein is regulated by the addition of several post-translational modifications which can affect the protein's stability, DNA binding and transcriptional activation: SUMOylation (Tsuruzoe et al. 2006; Wei et al. 2007), ubiquitination (Xu et al. 2004, 2009; Saxe et al. 2009) and phosphorylation (Kang et al. 2009; Saxe et al. 2009; Swaney et al. 2009; Brumbaugh et al. 2012; Spelat et al. 2012). Finally, Oct4 is known to be modified with O-GlcNAc (Webster et al. 2009; Jang et al. 2012), the consequence of this modification being the focus of this paper.
Several papers have been published providing evidence that O-GlcNAc may regulate Oct4. The first came from our study involving the developmental effects of O-GlcNAc in zebrafish (Webster et al. 2009). Overexpression of OGT in zebrafish mimicked the phenotype seen in embryos deficient for the Oct4 homolog spiel ohne grenzen (spg)/pou2 (Lunde et al. 2004; Reim et al. 2004; Lachnit et al. 2008; Webster et al. 2009). Jang and colleagues mapped one site of O-GlcNAc attachment to residue T228 on Oct4 purified from mouse ES cells and showed that its transcriptional activity correlates with the level of O-GlcNAc present on the protein (Jang et al. 2012). Oct4 is conserved in both mouse and human ES cells, though its targets and function vary depending on the species suggesting a need to study the role of O-GlcNAc in human Oct4 regulation (Schnerch et al. 2010).
Human Oct4 is known to be modified by O-GlcNAc (Webster et al. 2009), although the actual site of attachment or the functional implications of this modification have not yet been determined. In this study we showed that human Oct4 is extensively modified by O-GlcNAc which can regulate transcriptional activity of a variety of promoters.
Results
hOct4 is modified beyond known mThr228 site
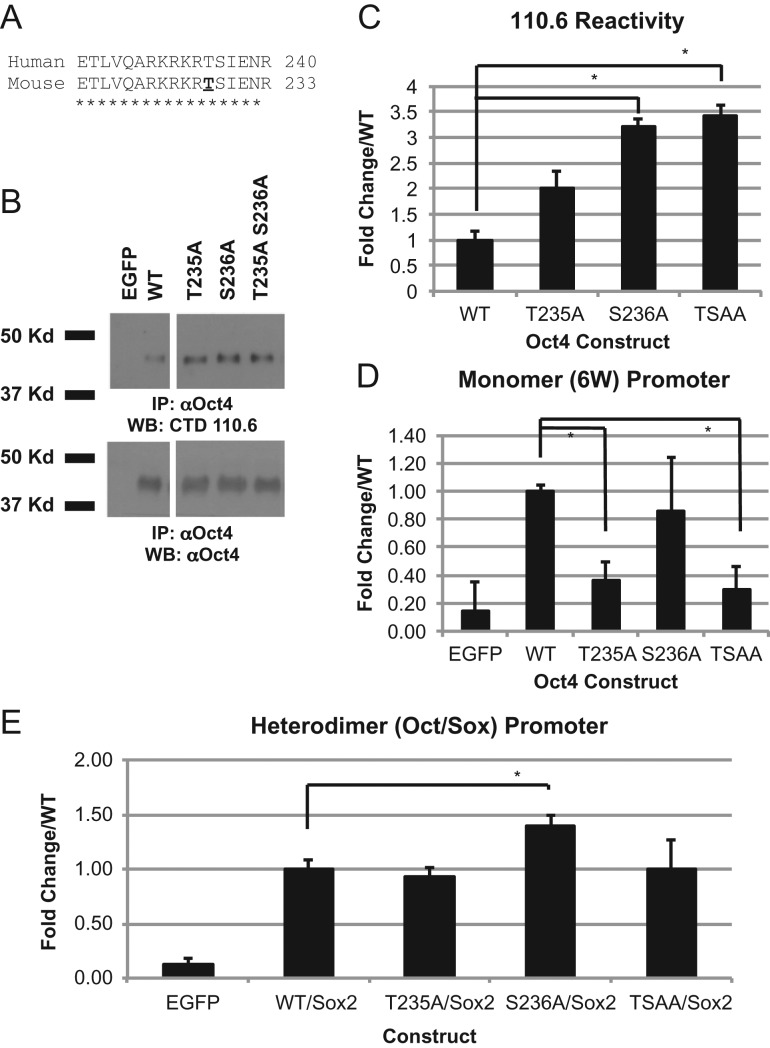
Using the online sequence comparison tool ClustalW2, human Oct4 is completely conserved in the region of the previously mapped O-GlcNAc site on mouse Oct4 responsible for transcriptional regulation (Jang et al. 2012) (Figure 1A). To determine if this site is also required in human Oct, we mutated the corresponding residue in the human sequence (T235) to an alanine using site-directed mutagenesis to prevent modification at this site. Since OGT has been shown to be promiscuous in its addition of O-GlcNAc (Cheng and Hart 2001) we also mutated the nearby residue S236 and both T235/S236 (TSAA) in combination to prevent any addition in this region. Immunoprecipitation and Western blot analysis of the Oct4 constructs expressed in human embryonic kidney (HEK293T) cells revealed that all are still reactive for O-GlcNAc as determined by the O-GlcNAc specific antibody CTD110.6 (Figure 1B). This confirms there are more sites of O-GlcNAc attachment on this protein than just the region modified. Since transcriptional activity of Oct4 was previously correlated to the amount of Oct4 O-GlcNAc modification, we quantified the amount of O-GlcNAc seen in our constructs when compared to WT. Densitometry quantification of the western blots reveals that T235 is equally modified and S236A and the TSAA double construct show higher levels of modification than WT suggesting these constructs should be more active than WT (Figure 1C). Oct family proteins can bind DNA as a monomer, or in different dimer configurations (Remenyi et al. 2001). Oct4 also frequently works in concert with Sox proteins to bind to Oct-Sox DNA elements (Remenyi et al. 2003; Rodda et al. 2005). To test our mutant Oct4 constructs we used two different Oct4 activating luciferase reporters: a promoter that contains six copies of the canonical monomer Oct binding site (6 W), and a heterodimer in which Oct4 co-operates with a Sox protein family member to activate (Oct/Sox). As was seen previously with the mouse constructs, the T235A and TSAA constructs showed a decrease in transcriptional activation of the monomer (6 W) promoter but not with the heterodimer (Oct/Sox) promoter (Figure 1D and E). The TSAA construct mimics T235A for 6 W confirming that T235 is important for activation of this promoter. Conversely, we saw an increase in the transcriptional activity for S236A construct when co-expressed with Sox2 using the Oct/Sox promotor but not with the 6 W promoter (Figure 1D and E). This suggests that modification at S236 is responsible for repression of transcriptional activation. Futhermore, TSAA construct mimics T235A construct suggesting that the modification at S236 is acting directly on the T235 site to prevent modification and subsequent increased activation seen with the S236 alone.
Fig. 1.
Homologous site in human Oct4 shows similar transcriptional profile to mouse Oct4. Sequence analysis of human and mouse sequence around mapped mouse O-GlcNAc site (A). Western blot analysis to determine the presence of O-GlcNAc modification on Oct4 using O-GlcNAc specific antibody CTD110.6 on immunoprecipitated Oct4 protein expressed in HEK293T cells (B). Quantification of B using ImageJ software on 3 biological replicates. All samples were run on the same gel (see Supplementary Figure S1), lanes not pertaining to this figure were removed (C). The transcriptional activity of Oct4 determined by luciferase expression of mutant constructs in HEK293T cells showing fold change over WT with either monomer (6 W) promoter (D) or heterodimer (Oct/Sox) promoter (E). Experiments using the Oct/Sox promoter included expression of Sox2 in combination with Oct4. *P < 0.05. Student's t-test using 3 biological replicates.
OGT overexpression increases Oct4 transcriptional activation in HEK293T cells
One major issue in the field is using alanine substitution to study O-GlcNAc effects. Since both O-GlcNAc and phosphorylation occur on serine and threonine residues, the alanine mutation prevents the addition of both and thus confounding interpretation. Many sites have been shown to be both phosphorylated and O-GlcNAc modified alternatively, forming a complex interplay between these two regulatory modifications (Comer and Hart 2000; Wang et al. 2007). Many people use aspartic or glutamic acid substitution when making constructs to mimic phosphorylation, however, no such substitution is available for O-GlcNAc. To circumvent this issue, we used co-expression of OGT to distinguish between the two possibilities. If O-GlcNAc modification at the site of mutation is responsible for the decrease in transcriptional activity, then co-expression with OGT should not be able to induce transcription activation.
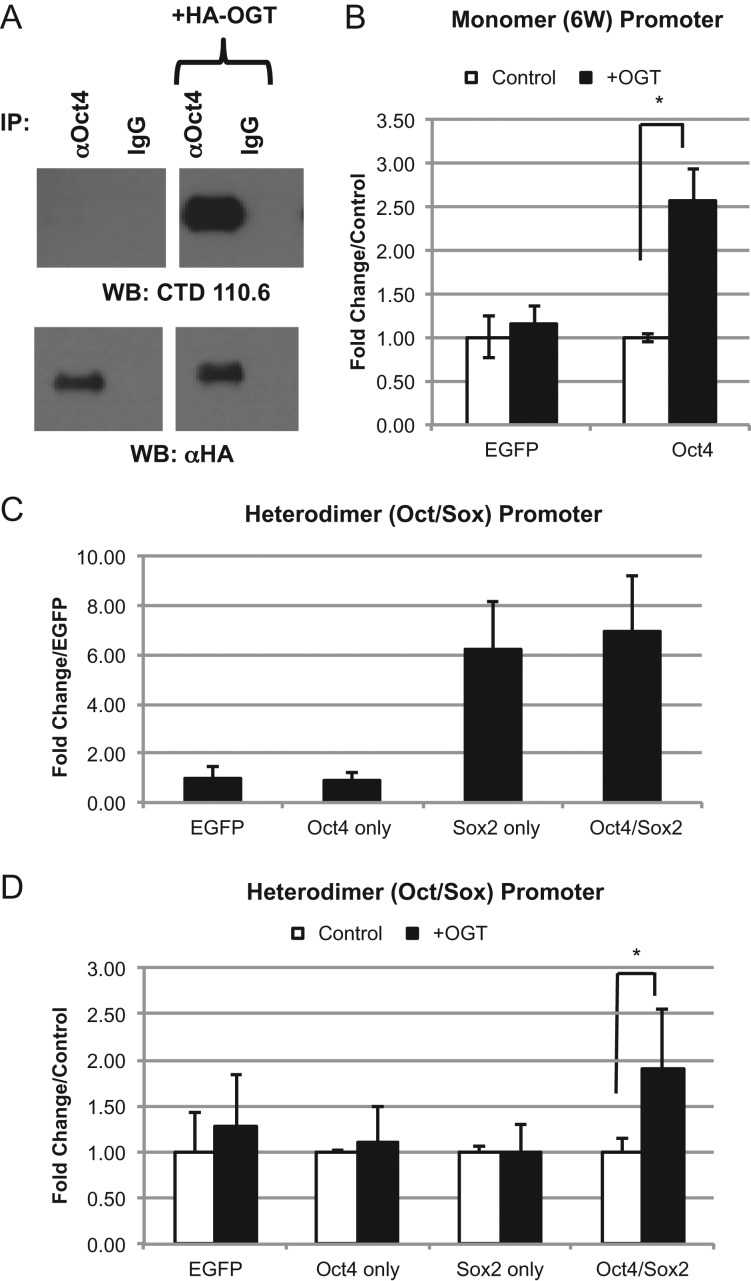
Previous studies show that the transcriptional activation of mouse Oct4 increases when OGT is co-expressed (Jang et al. 2012). This property was tested with human Oct4. First we needed to ensure that our manipulations would change the amount of O-GlcNAc present on Oct4 itself. Oct4 immunoprecipitated from HEK cells overexpressing OGT shows an increase in O-GlcNAc modification, validating our approach (Figure 2A). As expected, luciferase activity of Oct4 using the 6 W promoter was increased over control (EGFP) (Figure 1D). This activity could be enhanced 2.5-fold with co-expression of OGT. This increase in activity was specific for Oct4 since the EGFP control did not show an increase (Figure 2B). Since the use of the Oct/Sox promoter requires co-expression of Sox2, and Sox2 is also O-GlcNAc modified (Myers et al. 2011, 2016), we tested each of the components separately for activation of this promoter in the presence and absence of OGT. In the absence of OGT and Sox2, expression of Oct4 showed no change in activity and did not appear to activate the promoter above control levels (EGFP). Furthermore, this activity could not be enhanced by co-expression with OGT (Figure 2C and D). Expression of just Sox2 alone showed an increase over the control, presumably due to its interaction with Oct1 which is present in all cell types (Ryan and Rosenfeld 1997) and which has previously been shown to interact with Sox2 (Di Rocco et al. 2001). Co-expression with OGT does not increase this activity suggesting O-GlcNAc modification of Sox2 or Oct1 (Kang et al. 2013) does not play a factor in this assay. When Oct4 is co-expressed with Sox2 there is no increase over the result seen with Sox2 alone, but co-expression of OGT with both Sox2 and Oct4 increases the activity 2-fold. Taken that the condition with Sox2 alone does not increase when OGT is co-expressed, the activation seen by OGT can be attributed to the presence of Oct4 (Figure 2C and D). These results together suggest that human Oct4 transcription is activated by OGT overexpression at a variety of different promoter types.
Fig. 2.
Overexpression of OGT alters transcriptional activity of Oct4. Western blot analysis to determine the presence of O-GlcNAc modification on immunoprecipitated Oct4 protein expressed in HEK293T cells (A). WT human Oct4 was expressed in HEK293T cells with either EGFP (control) or human OGT construct. Oct4 binds promoters in either monomer (6 W) (B) or heterodimer (Oct/Sox) (C and D) configurations. *P < 0.05. Student's t-test using at least 3 biological replicates.
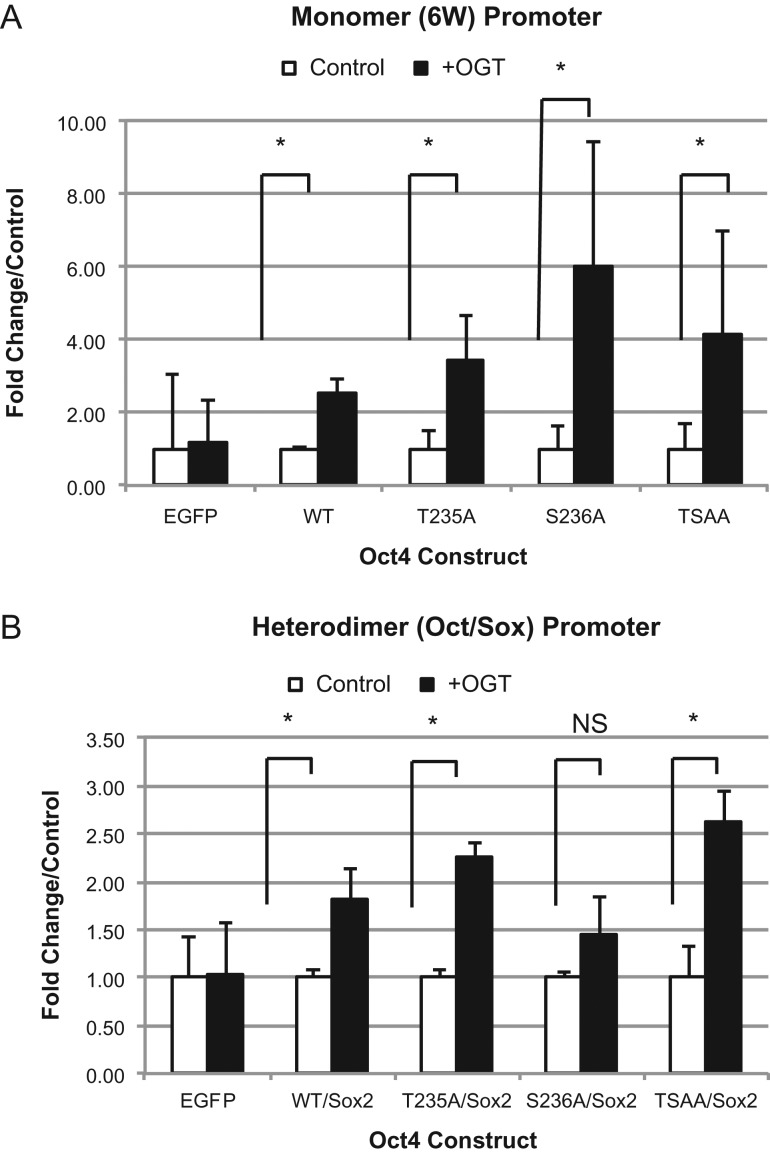
When we used this principle in our system, co-expression of OGT increased transcriptional activity of all the constructs tested (Figure 3A). Although T235A showed a decrease in transcriptional activation using the monomer promoter, we saw an increase in activity when the construct was co-expressed with OGT. This suggests that O-GlcNAc is acting at a site other than the sites tested to activate transcription. When we looked at the Oct/Sox promoter we saw an increase in all constructs when co-expressed with OGT except for S236A (Figure 3B) suggesting this site is important for OGT activation of Oct/Sox. Curiously, we saw the TSAA construct could be induced using both promoters suggesting that there is regulation of transcriptional activation at another site on the protein.
Fig. 3.
hOct4 is Modified Beyond Known mT228 Site. Luciferase expression of mutant constructs alone using monomer (6 W) promoter (A) or the heterodimer (Oct/Sox) promoter (with Sox2) (B) in the presence of EGFP (control) or OGT. Data are plotted as fold change over control for each mutant. *P < 0.05. Student's t-test using at least 3 biological replicates.
Oct4 is modified by O-GlcNAc at multiple sites
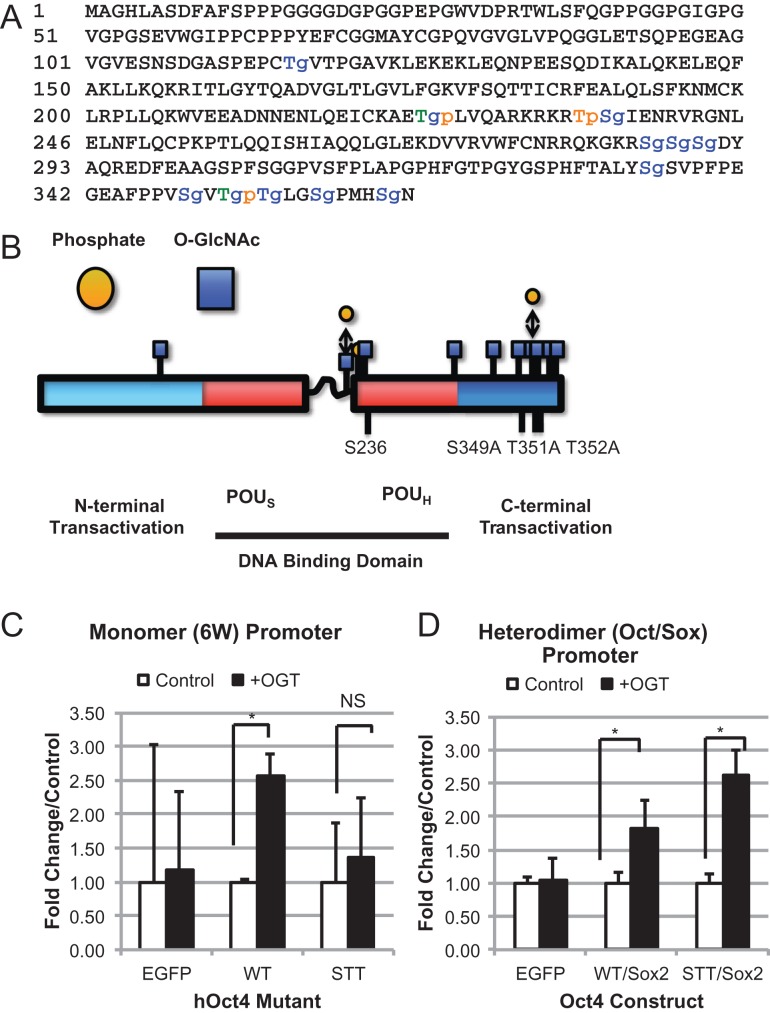
The ability of OGT to induce transcriptional activation with our TSAA mutant human Oct4 and the increased O-GlcNAc reactivity of our constructs prompted us to determine the other sites of O-GlcNAc attachment. Site mapping O-GlcNAc sites on proteins such as transcription factors is extremely difficult due to the low stoichiometry and low abundance of the protein in the cell. Initial mass spectrometry analysis undertaken on Oct4 protein immunopurified from H9 human ES cells showed evidence of several peptides modified by O-GlcNAc as evidenced by neutral loss. However, definitive sites of attachment could not be determined from this data (unpublished data). Instead, our study used immunoprecipitated proteins from HEK293T cells expressing Oct4 protein co-expressed with OGT to increase the abundance of O-GlcNAc modification on Oct4. Previous studies have shown that co-expression of OGT can increase the stoichiometry and will add O-GlcNAc to bona fide O-GlcNAc sites (Yuzwa et al. 2011). Peptides containing 10 novel O-GlcNAc sites and 3 phosphorylation sites, one of which is novel, were found using a mixture of collision-induced dissociation (CID) and electron-transfer dissociation (ETD) techniques (Figure 4A and B, Table I). Representative spectra from all modified peptides are shown: Full MS showing parent mass, CID spectra showing neutral loss of the HexNAc ion(s), and ETD spectra showing the peptide sequence with O-GlcNAc assignment (Figure 4A and B, Table 1, and Supplementary data). One site mapped as O-GlcNAc modified was S236. This site is adjacent to the homologous site in the mouse (Jang et al. 2012) and is a known site of phosphorylation (Swaney et al. 2009; Brumbaugh et al. 2012). The majority of O-GlcNAc residues were mapped to the C-terminal peptide, GEAFPPVSVTTLGSPMHSN (see Supplementary Table SI). Of note, three O-GlcNAc residues were mapped simultaneously to a single peptide which is unusual but not unprecedented (Capotosti et al. 2011). The tentative assignments of T351, T352 and S359 were made though additional hydroxyl-containing amino acids nearby may in fact harbor the O-GlcNAc sites (Supplementary data).
Fig. 4.
O-GlcNAc modification of C-terminal peptide of Oct4 regulates transcriptional activity of monomer promoter. Full human Oct4 sequence showing mapped O-GlcNAc sites (denoted in red with a lower case g) and phosphorylation sites (denoted in blue with a lower case p). Sites that were mapped with both O-GlcNAc and phosphate are shown in green (A). Graphical representation of A (B). Luciferase expression of C-terminal mutant construct using 6 W promoter (C) or the Oct/Sox promoter (D) in the presence of EGFP (control) or OGT, showing fold change over control for each mutant. *P < 0.05. Student's t-test using at least 3 biological replicates. This figure is available in black and white in print and in color at Glycobiology online.
Table I.
O-GlcNAc and phosphorylation sites mapped on oct4
| Sequence | Modifications |
|---|---|
| GASPEPCTVTPGAVKLE | T116-HexNAc |
| TLVQARKRKRTSIE | T225-HexNAc, S236-HexNAc |
| T225-Phospho, T235-Phospho | |
| KDVVRVWFCNRRQKGKRSSSD | S288 or S289 or S290 |
| AAGSPFSGGPVSFPLAPGPHFGTPGY | S335-HexNAc |
| GSPHFTALYSSVPFPEGE | |
| GEAFPPVSVTTLGSPMHSN | S349-HexNAc, |
| S349-HexNAc/S355-HexNAc, | |
| S349-HexNAc/T351-Phospho, | |
| T351-HexNAc/S359-HexNAc, | |
| T351-HexNAc/T352-HexNAc/S359-HexNAc |
Residues modified with HexNAc are bold and underlined. Commas denote separate modifications on the same peptide, forward slash denotes modifications found on the same peptide.
OGT regulates Oct4 transcription at S236 and S349A T351A T352A (STT)
To determine the impact of O-GlcNAc on transcriptional activation of the sites mapped, we undertook site-directed mutagenesis to make the constructs summarized in Table II. Again, due to the promiscuous nature of OGT, we mutated serine or threonine residues that are adjacent or close to the mapped site. For one peptide, we could not assign the exact site of O-GlcNAc attachment, however, this peptide only has three possible sites of attachment, S288, S289 or S290, so all were mutated together for analysis (Table II). Due to the large number of modifications present on the C-terminal peptide, we looked at the online prediction software Yin-Yang 1.2 Server (Gupta and Brunak 2002) to narrow down the candidates. S349 had the highest predicted score for modification and was also seen to be modified in the most number of peptides (Supplementary Table SI). T351 and T352 are adjacent to S349 and are also seen to be modified in several peptides so all three were modified to create STT construct. When expressed in HEK cells, all the Oct4 constructs showed O-GlcNAc reactivity confirming Oct4 has multiple sites of O-GlcNAc modification (Supplementary Figure S1). Transcriptional analysis using both 6 W and Oct/Sox reporters was undertaken as previously described in Figure 2. OGT failed to induce the STT construct when using the 6 W reporter but not the Oct/Sox reporter suggesting that this residue is important for activation of Oct/Sox genes but not monomer genes (Figure 4C and D). The remaining constructs showed an increase in activity when co-expressed with OGT using the 6 W and Oct/Sox promoter (Supplementary Figures S2 and S3). Although the constructs TT and T235 did not show a significant increase due to the amount of variability seen in the assay we did not consider these since they were trending in that direction. When looking at the Oct/Sox reporter all the constructs showed increased activity (Supplementary Figure S3) except for the S236A construct described earlier (Figure 3B). Transcriptional activity of all the constructs used in this study are summarized in Table II.
Table II.
Summary of transcriptional activity of Oct4 constructs used in this study
| hOct4 Construct | 6 W | Oct/Sox |
|---|---|---|
| WT | Increase with OGT | Increase with OGT |
| T116A T118A | Increase with OGT | Increase with OGT |
| T225A | Increase with OGT | Increase with OGT |
| T235A | Increase with OGT | Increase with OGT |
| S236A | Increase with OGT | No increase with OGT |
| T235A S236A | Increase with OGT | Increase with OGT |
| S288A S289A S290A | Increase with OGT | Increase with OGT |
| S349A T351A T352A | No increase with OGT | Increase with OGT |
OGA inhibitor GlcNAcstatin does not regulate Oct4 transcription
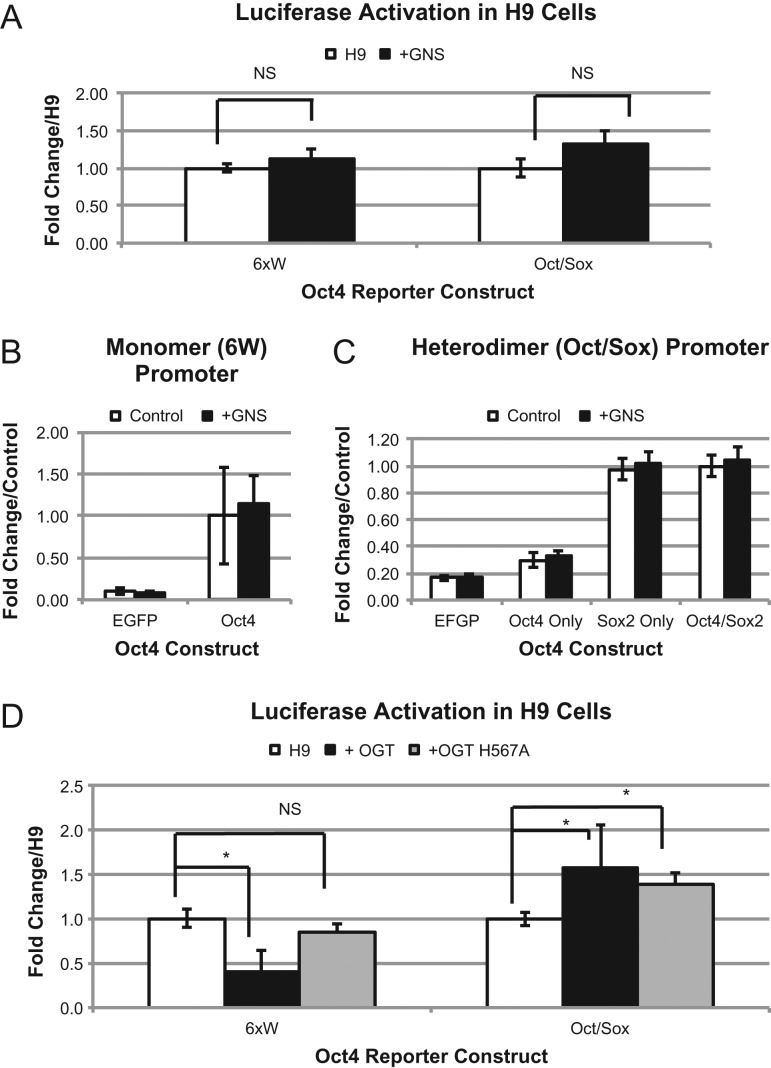
Since Oct4 is central in regulating pluripotency in ES cells, modulation of transcriptional activity would likely alter pluripotency and differentiation properties. Previous studies in mouse ES cells has shown that increased O-GlcNAc levels due to treatment with the O-GlcNAcase inhibitor STZ (Roos et al. 1998) and the more specific inhibitor GlcNAcstatin (GNS) (Dorfmueller et al. 2006, 2009) leads to stabilized pluripotency and delayed differentiation (Jang et al. 2012; Speakman et al. 2014). We wanted to determine if this observation was due to altered Oct4 transcriptional activity so we moved our analysis into H9 human ES cells to see if there is regulation of endogenous Oct4 by O-GlcNAc. After 24 h of GNS treatment, global O-GlcNAc levels on total cell proteins increased as well as levels of O-GlcNAc specifically on Oct4 (Supplementary Figure S4). To our surprise, treatment of H9 cells with GNS does not increase either Oct4 promoters tested (Figure 5A). To rule out any cell type specific difference, we repeated our experiments in HEK293T cells with GNS but still saw no transcriptional induction (Figure 5B and C). This suggested to us that OGT increases transcriptional activation of Oct4 by: inducing a normally unmodified O-GlcNAc site to become modified, using an indirect method of activation (modifies another protein not normally glycosylated) or by a mechanism that is OGT dependent but O-GlcNAc independent. To distinguish between these possibilities, we repeated the experiment in H9 cells using a catalytically inactivated mutant, OGT H567A, described previously (Martinez-Fleites et al. 2008) (Supplementary Figure S5). The 6 W promoter shows a significant decrease in activity when WT-OGT is expressed. When OGT H567A is expressed, there is no change from the control (Figure 5D). This suggests that catalytic activity of OGT is important for this promoter. The Oct/Sox promoter showed an increase in activity regardless of which OGT was co-expressed suggesting that the catalytic activity of this enzyme is not important for promoting activation of this promoter. Together these results demonstrate that OGT and O-GlcNAc modification can alter the transcriptional ability of Oct4 via different mechanisms for different promoters.
Fig. 5.
Association with OGT alters transcriptional activation of Oct4 specific luciferase constructs in H9 hES cells. Luciferase activity with both promoters of endogenous Oct4 in H9 cells (A) or expressed WT-Oct4 in HEK293T cells (B and C) treated with or without 100 nM GNS. Luciferase activity of endogenous Oct4 in H9 cells expressing either EGFP (control), OGT or catalytically dead OGT (OGT H567A) (D). *P < 0.05. Student's t-test using at least 3 biological replicates.
Discussion
Human Oct4 transcriptional activity is regulated by O-GlcNAc at two different regions
In our study, we found two regions on human Oct4 that are sensitive to OGT induced activation: T235A S236 (TSAA) and S349A T351A T352A (STT). In concurrence with the mouse study, we saw a decrease in transcriptional activity with T235A. However, we saw an increase in transcriptional activity of our T235A mutant when OGT is co-expressed suggesting that there is at least one other site responsible. In our study, we mapped an O-GlcNAc residue to S236, and a phosphate to T235. With our S236A mutant, we saw an increase in the Oct/Sox reporter activity over WT protein, which could not be increased by OGT. Surprisingly, when both T235 and S236 were mutated the induction ability returned suggesting yet another site is involved in activation. This suggests that sites other than T235 and S236 are important in the induction of activation by OGT but both clearly play a role in regulating the Oct/Sox promoter.
Our data set showed an abundance of O-GlcNAc modifications on the C-terminal peptide (Supplementary data). Previous studies have shown that the C-terminal transactivation domain of Oct4 is required for full activation (Niwa et al. 2002), so it is no surprise to find the majority of the sites here nor the effect seen with our STT mutant construct. About 5 of the 10 sites mapped lie within the terminal 12 residues, with 1 peptide being modified by as many as 3 O-GlcNAc residues. Although unusual it is not unprecedented. Recent papers have shown peptides containing three O-GlcNAc sites in close proximity on Host cell factor 1 (HCF1) and histone 2B (Capotosti et al. 2011; Hahne et al. 2012). HCF and C/EBPb both have O-GlcNAc sites mapped on adjacent residues (Li et al. 2009; Myers et al. 2011).
Both our mutant constructs showed a response to different promoters. S236 was important for activation of the heterodimer (Oct/Sox) promoter, and the STT mutant was important for the monomer (6 W) activation. As a master regulator of pluripotency, Oct4 binds to many different types of promoters to regulate multiple genes involved in both maintenance of pluripotency and differentiation of ES cells (Boyer et al. 2005; Loh et al. 2006). These results suggest that OGT is acting at different parts of the protein to regulate which promoter is being activated.
Oct4 is modified with multiple O-GlcNAc modifications on or close to other known post-translational modifications
Site mapping of human Oct4 revealed 10 unique O-GlcNAc sites that have the potential to fine-tune Oct4 function as they are involved in, or are in proximity to, other characterized modifications. Except for S335 and S349, all other residues mapped in this study are also modified by phosphorylation, shown either in this study or a previously published study (Brumbaugh et al. 2012). The reciprocal nature of O-GlcNAc with phosphorylation has been well documented as these two modifications form a complex interplay (Comer and Hart 2000; Wang et al. 2007). Our approach increases the likelihood of deciphering the difference between these two possibilities by manipulating O-GlcNAc levels by overexpression of OGT with the assumption that transcriptional activity will not change if the altered site is an OGT target. Combining these results with O-GlcNAc site mapping, increases our confidence of the correct assumption. However, this cannot rule out completely the possibility of altered phosphorylation. Phosphorylation mimics were not included in this study since modifications by phosphorylation at one site can influence the O-GlcNAc modification status at an adjacent or nearby site (Comer and Hart 2001; Hu et al. 2010), complicating the interpretation of the results.
T235/S236, S355 and S289/S290 are modified by AKT, ERK and PKA/PIM1, respectively (Brumbaugh et al. 2012; Lin et al. 2012), although the actual consequence of phosphorylation at these sites has not yet been fully investigated. Although our study did not show any consequence of these sites in transcriptional activation, considering the importance these kinases play in signaling pathways responsible for maintaining pluripotency/differentiation (Dalton 2013), blocking phosphorylation by modification with O-GlcNAc at these sites is likely to play an important regulatory role.
Differences between human ES cells, mouse ES cells and HEK cells
Once we moved our analysis into human ES cells, we saw a decrease in transcriptional activation of the monomer (6 W) reporter by OGT in H9 cells (Figure 5), as opposed to the increase seen in HEK293T cells using 6 W reporter (Figure 2). The difference shown here may be due to a difference in signaling pathways activated in the two different cell types. It has been shown previously that the C-terminal transactivation domain of Oct4 is differentially phosphorylated in different cell types, which correlates to its activity (Brehm et al. 1997). Futhermore, there are discrepancies between mouse and human Oct4. Unlike mouse Oct4, we found no correlation between the amount of O-GlcNAc modification and transactivation ability, and a large amount of modifications on the C-terminus (Jang et al. 2012). It is possible it is due to differences in detection and the use of different promoters, however, it is well known that there are major differences between mouse and human ES cells in genes expression and signaling pathways(Schnerch et al. 2010). This points to the importance of further investigation into the exact site of attachment. Taken together, the differences of modification in different cell types should be investigated further in future studies and will make selection of a system for studying function important.
OGA inhibitor GlcNacstatin did not recapitulate OGT overexpression results
Most surprisingly, although the use of GlcNAcstatin in our cells yielded increased O-GlcNAc levels on Oct4, it did not yield the same results as OGT overexpression in our transcriptional activation assays (Figure 5). OGA inhibitors are widespread in the field and are often used interchangeably with OGT overexpression to modulate O-GlcNAc levels. Originally streptozotocin and PUGNAc were used to inhibit OGA, but more recently their use is limited due to the off-target effects seen (Szkudelski 2001; Macauley et al. 2005). More recently it was shown that PUGNAc can inhibit the pro-survival action of insulin in a manner that is independent of O-GlcNAc levels (Teo et al. 2016) cautioning the use of the assumption that the effect seen is due to increased O-GlcNAc levels. Currently, newly designed inhibitors such as GlcNAcstatin and Thiamet G (Yuzwa et al. 2008) have taken over as the inhibitors of choice as they are much more specific (Dorfmueller et al. 2006). There is also the potential that cycling time of O-GlcNAc is essential for proper function and that overexpression of OGT, that would shift the equilibrium towards modified but not inhibit cycling, is not identical to OGA inhibition, that would also shift the equilibrium towards modified but would also inhibit cycling.
Enzymatic activity of OGT is not always required for transcriptional activation of Oct4
Since GNS failed to alter transcriptional activity of Oct4 it suggests there may be another function of OGT separate from its ability to modify Oct4 with O-GlcNAc. Although both methods lead to an increase of O-GlcNAc, they do so by very different mechanisms. The inhibition of OGA increases O-GlcNAc levels by breaking the cycle and preventing removal of O-GlcNAc. Overexpression of OGT increases O-GlcNAc levels by changing the ratio of OGT to OGA in the cell. The use of an inactive OGT mutant in our transcription experiments points to another function of OGT that does not rely on its catalytic activity. Two independent groups found OGT bound to Oct4 when looking for interaction partners suggesting these proteins form a complex (Pardo et al. 2010; van den Berg et al. 2010). Oct4 has been shown to require a bridging factor for full activation (Scholer et al. 1991). It is entirely possible that OGT acts as a bridging protein between Oct4 and Sox2 to bring them to this promoter. However, we cannot rule out the possibility that the inactive OGT is forming a complex with other functional OGT proteins in the cell (Haltiwanger et al. 1992; Jinek et al. 2004), or acting as a lectin by binding existing O-GlcNAc residues. Further investigation into the non-enzymatic functions of OGT and defining its interactome will be required in future studies.
OGT differentially regulates Oct4 transcriptional activation
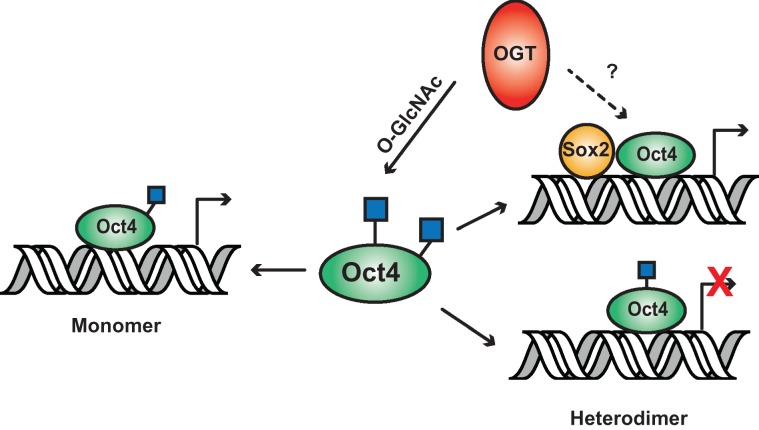
Taken together, we believe that OGT is responsible for regulating Oct transcriptional activation by using several different mechanisms (Figure 6). Human Oct4 is abundantly modified by OGT on the C-terminal transactivation domain which regulates its ability to activate the monomer promoter. Addition of GNS does not activate this promoter although catalytic activity of OGT is required. This suggests that attachment of O-GlcNAc is necessary for activation and only occurs when OGT is in high abundance. Modification of S236 leads to repression of activation of the heterodimer (Oct/Sox) promoter through possible interaction with T235 O-GlcNAc modification. Since both these sites are also phosphorylated, this allows O-GlcNAc to interact with signaling pathways, including ERK signaling, that currently regulate stem cell pluripotency and more specifically Oct4 (Brumbaugh et al. 2012). Finally, OGT could possibly activate the heterodimer promoter by acting as a bridging protein at the promoter.
Fig. 6.
Model of how OGT regulates Oct4 transcriptional activation of two different promoters. OGT regulates transcriptional activity of Oct4 by two different mechanisms. OGT transfers O-GlcNAc to Oct4 C-terminal domain to activate the monomer promoter. Modification of Oct4 at S236 represses activation of the heterodimer (Oct/Sox) promoter. OGT is hypothesized to participate in activation of heterodimer promoter independently of modification by directly interacting with the Oct/Sox complex (dotted line). This figure is available in black and white in print and in color at Glycobiology online.
Conclusion
This study revealed that human Oct4 is highly modified by O-GlcNAc and the majority of these modifications are located in the C-terminal activation domain. We found two regions that were sensitive to transcriptional activation by OGT, one that has been previously described and a novel region, which are responsible for activation of different Oct4 promoter types. These results show the importance of post-translational modifications in regulating transcription factors such as Oct4 which activate many different promoters. Full characterization of the genes associated with these promoters will need to be focused on in the future. We also discovered that OGT does not always require its catalytic function and hypothesize that it plays a previously undescribed role of acting as a bridging protein between Oct4 and Sox2 to activate transcription at these promoters.
Materials and methods
Cell culture and transfections
H9 human ES cells were maintained on Matrigel™ (BD biosciences) in StemPro® hESC media (Life Technologies) using Accutase™ passaging (ICT). HEK293T cells were maintained in 10% FBS/DMEM. Transfections were carried out using X-tremeGENE HP DNA Transfection Reagent (Roche) or JetPRIME (Polyplus) as per manufacture instructions. Cells were treated with GlcNAcstatin (GNS) (Gift from Daan van Aalten, University of Dundee, UK) by adding 100 nM directly to the appropriate media every 24 h.
Immunoprecipitation, western blotting and quantification
Immunoprecipitations were carried out using 1 mg of protein in Tris buffers containing 1%NP40, 0.1%SDS. Western blotting was carried out using standard conditions. Antibodies used in this study: Oct4 (Santa Cruz), 110.6 and HA (gift from Gerald Hart). ImageJ software (NIH) was used for the quantification of film exposures. Amount of O-GlcNAc was determined by dividing the value for the 110.6 antibody by the amount of HA or Oct4 measured. P-values were determined using standard Student's t-test undertaken on at least 3 biological replicates.
Luciferase assays
About 6 W luciferase constructs were kindly donated by Dr Jonathan Saxe et al. (2009). Oct/Sox promoter was obtained from Addgene (plasmid 15,686) (Tokuzawa et al. 2003). Luciferase expression was detected using Promega Dual Glo® Luciferase Assay System per manufacture instructions. All luciferase values were normalized to Renilla luciferase expression used as an internal transfection control. Student's t-test was carried out in excel on a minimum of biological triplicate samples.
Sample preparation for analysis of mass spectrometry
Human Oct4 was co-expressed in HEK293T cells with human OGT. Immunoprecipitation of ten 10 cm plates was carried out as described above, eluted with 0.1 M Glycine pH 2.5 and neutralized to pH 8.0 with Tris. The eluted samples were reduced with 10 mM dithiothreitol (DTT) for 1 h at 56°C, carboxyamidomethylated with 55 mM iodoacetamide (ICH2CONH2, Sigma) in the dark for 45 min, and then digested with 3 μg of sequence grade Glu-C (Promega) in 100 mM phosphate buffer at pH 7.0 overnight at 37°C. After digestion, the peptides were acidified with 1% trifluoroacetic acid (TFA). Desalting was subsequently performed with C18 spin columns (Vydac Silica C18, The Nest Group, Inc.) and the resulting peptides were dried down in a Speed Vac and stored at −20°C until analysis.
O-GlcNAc site mapping of Oct4 in HEK by LC-MS/MS
The peptides resuspended with 19.5 μL of mobile phase A (0.1% formic acid, FA, in water) and 0.5 μL of mobile phase B (80% acetonitrile, ACN, and 0.1% formic acid in water) and filtered with 0.2 μm filters (Nanosep, PALL). The samples were loaded off-line onto a nanospray tapered capillary column/emitter (360 × 75 × 15 μm, PicoFrit, New Objective, 15 cm column) that was self-packed with C18 reverse phase (RP) resin (Waters) in a nitrogen pressure bomb for 10 min at 1000 psi (∼5 μL load). The peptides were separated using the Dionex UltiMate 3000 nano-LC system (ThermoFisher) with a 180 min linear gradient of increasing mobile phase B at a flow rate of 120 nL/min. The LC-MS/MS analysis was performed using the Orbitrap Fusion Tribrid MS (ThermoFisher) equipped with a Nanospray Flex Ion Source at 2.2 kV spray voltage and 280°C ion transfer tube temperature. The full FTMS (Fourier transform mass spectrometry) spectrum, typically recorded at 120,000 of resolution in positive ion and profile mode, was acquired at 300–2000 m/z followed by the MS/MS spectra of ITMS (ion trap mass spectrometry) on the 15 most intense ions from the targeted mass lists or data dependent MS/MS spectra on the most intense ion with dynamic exclusion at 30 s duration time. The targeted ions were isolated by the quadruple at 1.5 m/z isolation window for CID and 3.0 m/z for ETD and fragmented by decision-tree algorithm by alternating between CID at 38% normalized collision energy and ETD at 80 ms of reaction time for above triply charged and 150 ms of reaction time with 40% of supplemental activation for doubly charged ions.
Detection of O-linked glycosylation
The raw files were searched against the Oct4 database including contaminant database (along with reversed proteins as decoys) using Proteomic Discoverer (Thermo Scientific) with a peptide tolerance of 30 ppm; a MS/MS tolerance of 0.8 Da; the carbamidomethylated cysteine; oxidation of methionine and phosphorylation and O-linked glycosylation (HexNAc) of serine and threonine as variable modifications. The peptide sequences were identified by Proteomic Discoverer from the CID and ETD spectra and verified manually. The glycosylations and phosphorylations on the peptides were verified by the presence of corresponding neutral loss fragment ions of sugar and phosphate such as the HexNAc at 203.08 Da and phosphate at 79.97 and 97.98 Da calculating charge states in CID spectra. In total, 34O-GlcNAc or phosphorylation sites were observed via multiple LC-MS/MS runs in each experiment. The best scored glyco- and phospho-peptides based on XCorr value that were manually validated for neutral loss peaks are listed in Supplementary Table SI. Representative MS and MS/MS spectra are shown in Figure 4, remaining spectra can be found in the Supplementary data.
Supplementary Material
Acknowledgments
Thank you to Daan van Aalten for the GlcNAcstatin, Gerald W. Hart for the CTD110.6 and HA antibody, Jonathan Saxe for the 6 W luciferase construct and Chin Fen Teo for testing the OGT H567A construct.
Supplementary data
Funding
A portion of this research was supported by NIH/NIGMS (National Center for Biomedical Glycomics, P41GM103490, LW Senior Investigator). This research was also supported in part by Basic Science Research Program and the Priority Research Centers Program through the National Research Foundation of Korea (NRF) funded by the Ministry of Education, Science and Technology (grant number: NRF 2011-0,013,961 and NRF 2010-0,029,634, JML).
Conflict of interest statement
None declared.
References
- Botquin V, Hess H, Fuhrmann G, Anastassiadis C, Gross MK, Vriend G, Scholer HR. 1998. New POU dimer configuration mediates antagonistic control of an osteopontin preimplantation enhancer by Oct-4 and Sox-2. Genes Dev. 12:2073–2090. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer LA, Lee TI, Cole MF, Johnstone SE, Levine SS, Zucker JP, Guenther MG, Kumar RM, Murray HL, Jenner RG et al. . 2005. Core transcriptional regulatory circuitry in human embryonic stem cells. Cell. 122:947–956. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brehm A, Ohbo K, Scholer H. 1997. The carboxy-terminal transactivation domain of Oct-4 acquires cell specificity through the POU domain. Mol Cell Biol. 17:154–162. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brimble SN, Wollaston-Hayden EE, Teo CF, Morris AC, Wells L. 2010. The role of the O-GlcNAc modification in regulating eukaryotic gene expression. Curr Signal Transduct Ther. 5:12–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbaugh J, Hou Z, Russell JD, Howden SE, Yu P, Ledvina AR, Coon JJ, Thomson JA. 2012. Phosphorylation regulates human OCT4. Proc Natl Acad Sci USA. 109:7162–7168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Capotosti F, Guernier S, Lammers F, Waridel P, Cai Y, Jin J, Conaway JW, Conaway RC, Herr W. 2011. O-GlcNAc transferase catalyzes site-specific proteolysis of HCF-1. Cell. 144:376–388. [DOI] [PubMed] [Google Scholar]
- Cheng X, Hart GW. 2001. Alternative O-glycosylation/O-phosphorylation of serine-16 in murine estrogen receptor beta: Post-translational regulation of turnover and transactivation activity. J Biol Chem. 276:10570–10575. [DOI] [PubMed] [Google Scholar]
- Chew JL, Loh YH, Zhang W, Chen X, Tam WL, Yeap LS, Li P, Ang YS, Lim B, Robson P et al. . 2005. Reciprocal transcriptional regulation of Pou5f1 and Sox2 via the Oct4/Sox2 complex in embryonic stem cells. Mol Cell Biol. 25:6031–6046. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Comer FI, Hart GW. 1999. O-GlcNAc and the control of gene expression. Biochim Biophys Acta. 1473:161–171. [DOI] [PubMed] [Google Scholar]
- Comer FI, Hart GW. 2000. O-Glycosylation of nuclear and cytosolic proteins. Dynamic interplay between O-GlcNAc and O-phosphate. J Biol Chem. 275:29179–29182. [DOI] [PubMed] [Google Scholar]
- Comer FI, Hart GW. 2001. Reciprocity between O-GlcNAc and O-phosphate on the carboxyl terminal domain of RNA polymerase II. Biochemistry. 40:7845–7852. [DOI] [PubMed] [Google Scholar]
- Dalton S. 2013. Signaling networks in human pluripotent stem cells. Curr Opin Cell Biol. 25:241–246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dehennaut V, Hanoulle X, Bodart JF, Vilain JP, Michalski JC, Landrieu I, Lippens G, Lefebvre T. 2008. Microinjection of recombinant O-GlcNAc transferase potentiates Xenopus oocytes M-phase entry. Biochem Biophys Res Commun. 369:539–546. [DOI] [PubMed] [Google Scholar]
- Dehennaut V, Lefebvre T, Sellier C, Leroy Y, Gross B, Walker S, Cacan R, Michalski JC, Vilain JP, Bodart JF. 2007. O-linked N-acetylglucosaminyltransferase inhibition prevents G2/M transition in Xenopus laevis oocytes. J Biol Chem. 282:12527–12536. [DOI] [PubMed] [Google Scholar]
- Dentin R, Hedrick S, Xie J, Yates J 3rd, Montminy M. 2008. Hepatic glucose sensing via the CREB coactivator CRTC2. Science. 319:1402–1405. [DOI] [PubMed] [Google Scholar]
- Di Rocco G, Gavalas A, Popperl H, Krumlauf R, Mavilio F, Zappavigna V. 2001. The recruitment of SOX/OCT complexes and the differential activity of HOXA1 and HOXB1 modulate the Hoxb1 auto-regulatory enhancer function. J Biol Chem. 276:20506–20515. [DOI] [PubMed] [Google Scholar]
- Dong DL, Hart GW. 1994. Purification and characterization of an O-GlcNAc selective N-acetyl-beta-D-glucosaminidase from rat spleen cytosol. J Biol Chem. 269:19321–19330. [PubMed] [Google Scholar]
- Dorfmueller HC, Borodkin VS, Schimpl M, Shepherd SM, Shpiro NA, van Aalten DM. 2006. GlcNAcstatin: A picomolar, selective O-GlcNAcase inhibitor that modulates intracellular O-glcNAcylation levels. J Am Chem Soc. 128:16484–16485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dorfmueller HC, Borodkin VS, Schimpl M, van Aalten DM. 2009. GlcNAcstatins are nanomolar inhibitors of human O-GlcNAcase inducing cellular hyper-O-GlcNAcylation. Biochem J. 420:221–227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fujiki R, Chikanishi T, Hashiba W, Ito H, Takada I, Roeder RG, Kitagawa H, Kato S. 2009. GlcNAcylation of a histone methyltransferase in retinoic-acid-induced granulopoiesis. Nature. 459:455–459. [DOI] [PubMed] [Google Scholar]
- Gambetta MC, Oktaba K, Muller J. 2009. Essential role of the glycosyltransferase sxc/Ogt in polycomb repression. Science. 325:93–96. [DOI] [PubMed] [Google Scholar]
- Gandy JC, Rountree AE, Bijur GN. 2006. Akt1 is dynamically modified with O-GlcNAc following treatments with PUGNAc and insulin-like growth factor-1. FEBS Lett. 580:3051–3058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gao Y, Miyazaki J, Hart GW. 2003. The transcription factor PDX-1 is post-translationally modified by O-linked N-acetylglucosamine and this modification is correlated with its DNA binding activity and insulin secretion in min6 beta-cells. Arch Biochem Biophys. 415:155–163. [DOI] [PubMed] [Google Scholar]
- Gao Y, Wells L, Comer FI, Parker GJ, Hart GW. 2001. Dynamic O-glycosylation of nuclear and cytosolic proteins: Cloning and characterization of a neutral, cytosolic beta-N-acetylglucosaminidase from human brain. J Biol Chem. 276:9838–9845. [DOI] [PubMed] [Google Scholar]
- Gewinner C, Hart G, Zachara N, Cole R, Beisenherz-Huss C, Groner B. 2004. The coactivator of transcription CREB-binding protein interacts preferentially with the glycosylated form of Stat5. J Biol Chem. 279:3563–3572. [DOI] [PubMed] [Google Scholar]
- Gupta R, Brunak S. 2002. Prediction of glycosylation across the human proteome and the correlation to protein function. Pac Symp Biocomput. 7:310–322. [PubMed] [Google Scholar]
- Hahne H, Moghaddas Gholami A, Kuster B. 2012. Discovery of O-GlcNAc-modified proteins in published large-scale proteome data. Mol Cell Proteomics. 11:843–850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haltiwanger RS, Blomberg MA, Hart GW. 1992. Glycosylation of nuclear and cytoplasmic proteins. Purification and characterization of a uridine diphospho-N-acetylglucosamine:polypeptide beta-N-acetylglucosaminyltransferase. J Biol Chem. 267:9005–9013. [PubMed] [Google Scholar]
- Han I, Kudlow JE. 1997. Reduced O glycosylation of Sp1 is associated with increased proteasome susceptibility. Mol Cell Biol. 17:2550–2558. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanover JA, Forsythe ME, Hennessey PT, Brodigan TM, Love DC, Ashwell G, Krause M. 2005. A Caenorhabditis elegans model of insulin resistance: Altered macronutrient storage and dauer formation in an OGT-1 knockout. Proc Natl Acad Sci U S A. 102:11266–11271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hartweck LM, Scott CL, Olszewski NE. 2002. Two O-linked N-acetylglucosamine transferase genes of Arabidopsis thaliana L. Heynh. have overlapping functions necessary for gamete and seed development. Genetics. 161:1279–1291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hay DC, Sutherland L, Clark J, Burdon T. 2004. Oct-4 knockdown induces similar patterns of endoderm and trophoblast differentiation markers in human and mouse embryonic stem cells. Stem Cells. 22:225–235. [DOI] [PubMed] [Google Scholar]
- Housley MP, Rodgers JT, Udeshi ND, Kelly TJ, Shabanowitz J, Hunt DF, Puigserver P, Hart GW. 2008. O-GlcNAc regulates FoxO activation in response to glucose. J Biol Chem. 283:16283–16292. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hu P, Shimoji S, Hart GW. 2010. Site-specific interplay between O-GlcNAcylation and phosphorylation in cellular regulation. FEBS Lett. 584:2526–2538. [DOI] [PubMed] [Google Scholar]
- Ingham PW. 1984. A gene that regulates the bithorax complex differentially in larval and adult cells of Drosophila. Cell. 37:815–823. [DOI] [PubMed] [Google Scholar]
- Jang H, Kim TW, Yoon S, Choi SY, Kang TW, Kim SY, Kwon YW, Cho EJ, Youn HD. 2012. O-GlcNAc regulates pluripotency and reprogramming by directly acting on core components of the pluripotency network. Cell Stem Cell. 11:62–74. [DOI] [PubMed] [Google Scholar]
- Jinek M, Rehwinkel J, Lazarus BD, Izaurralde E, Hanover JA, Conti E. 2004. The superhelical TPR-repeat domain of O-linked GlcNAc transferase exhibits structural similarities to importin alpha. Nat Struct Mol Biol. 11:1001–1007. [DOI] [PubMed] [Google Scholar]
- Kang J, Gemberling M, Nakamura M, Whitby FG, Handa H, Fairbrother WG, Tantin D. 2009. A general mechanism for transcription regulation by Oct1 and Oct4 in response to genotoxic and oxidative stress. Genes Dev. 23:208–222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kang J, Shen Z, Lim JM, Handa H, Wells L, Tantin D. 2013. Regulation of Oct1/Pou2f1 transcription activity by O-GlcNAcylation. FASEB J. 27:2807–2817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lachnit M, Kur E, Driever W. 2008. Alterations of the cytoskeleton in all three embryonic lineages contribute to the epiboly defect of Pou5f1/Oct4 deficient MZspg zebrafish embryos. Dev Biol. 315:1–17. [DOI] [PubMed] [Google Scholar]
- Li X, Molina H, Huang H, Zhang YY, Liu M, Qian SW, Slawson C, Dias WB, Pandey A, Hart GW et al. . 2009. O-linked N-acetylglucosamine modification on CCAAT enhancer-binding protein beta: role during adipocyte differentiation. J Biol Chem. 284:19248–19254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin Y, Yang Y, Li W, Chen Q, Li J, Pan X, Zhou L, Liu C, Chen C, He J et al. . 2012. Reciprocal regulation of Akt and Oct4 promotes the self-renewal and survival of embryonal carcinoma cells. Mol Cell. 48:627–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loh YH, Wu Q, Chew JL, Vega VB, Zhang W, Chen X, Bourque G, George J, Leong B, Liu J et al. . 2006. The Oct4 and Nanog transcription network regulates pluripotency in mouse embryonic stem cells. Nat Genet. 38:431–440. [DOI] [PubMed] [Google Scholar]
- Love DC, Hanover JA. 2005. The hexosamine signaling pathway: Deciphering the “O-GlcNAc code”. Sci STKE. 2005:re13. [DOI] [PubMed] [Google Scholar]
- Lunde K, Belting HG, Driever W. 2004. Zebrafish pou5f1/pou2, homolog of mammalian Oct4, functions in the endoderm specification cascade. Curr Biol. 14:48–55. [DOI] [PubMed] [Google Scholar]
- Macauley MS, Whitworth GE, Debowski AW, Chin D, Vocadlo DJ. 2005. O-GlcNAcase uses substrate-assisted catalysis: Kinetic analysis and development of highly selective mechanism-inspired inhibitors. J Biol Chem. 280:25313–25322. [DOI] [PubMed] [Google Scholar]
- Martinez-Fleites C, Macauley MS, He Y, Shen DL, Vocadlo DJ, Davies GJ. 2008. Structure of an O-GlcNAc transferase homolog provides insight into intracellular glycosylation. Nat Struct Mol Biol. 15:764–765. [DOI] [PubMed] [Google Scholar]
- Myers SA, Panning B, Burlingame AL. 2011. Polycomb repressive complex 2 is necessary for the normal site-specific O-GlcNAc distribution in mouse embryonic stem cells. Proc Natl Acad Sci U S A. 108:9490–9495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Myers SA, Peddada S, Chatterjee N, Friedrich T, Tomoda K, Krings G, Thomas S, Maynard J, Broeker M, Thomson M et al. . 2016. SOX2 O-GlcNAcylation alters its protein-protein interactions and genomic occupancy to modulate gene expression in pluripotent cells. Elife. 5:e10647. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nichols J, Zevnik B, Anastassiadis K, Niwa H, Klewe-Nebenius D, Chambers I, Scholer H, Smith A. 1998. Formation of pluripotent stem cells in the mammalian embryo depends on the POU transcription factor Oct4. Cell. 95:379–391. [DOI] [PubMed] [Google Scholar]
- Nishimoto M, Fukushima A, Okuda A, Muramatsu M. 1999. The gene for the embryonic stem cell coactivator UTF1 carries a regulatory element which selectively interacts with a complex composed of Oct-3/4 and Sox-2. Mol Cell Biol. 19:5453–5465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Masui S, Chambers I, Smith AG, Miyazaki J. 2002. Phenotypic complementation establishes requirements for specific POU domain and generic transactivation function of Oct-3/4 in embryonic stem cells. Mol Cell Biol. 22:1526–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Niwa H, Miyazaki J, Smith AG. 2000. Quantitative expression of Oct-3/4 defines differentiation, dedifferentiation or self-renewal of ES cells. Nat Genet. 24:372–376. [DOI] [PubMed] [Google Scholar]
- O'Donnell N, Zachara NE, Hart GW, Marth JD. 2004. Ogt-dependent X-chromosome-linked protein glycosylation is a requisite modification in somatic cell function and embryo viability. Mol Cell Biol. 24:1680–1690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ohn T, Kedersha N, Hickman T, Tisdale S, Anderson P. 2008. A functional RNAi screen links O-GlcNAc modification of ribosomal proteins to stress granule and processing body assembly. Nat Cell Biol. 10:1224–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pardo M, Lang B, Yu L, Prosser H, Bradley A, Babu MM, Choudhary J. 2010. An expanded Oct4 interaction network: Implications for stem cell biology, development, and disease. Cell Stem Cell. 6:382–395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reim G, Mizoguchi T, Stainier DY, Kikuchi Y, Brand M. 2004. The POU domain protein spg (pou2/Oct4) is essential for endoderm formation in cooperation with the HMG domain protein casanova. Dev Cell. 6:91–101. [DOI] [PubMed] [Google Scholar]
- Remenyi A, Lins K, Nissen LJ, Reinbold R, Scholer HR, Wilmanns M. 2003. Crystal structure of a POU/HMG/DNA ternary complex suggests differential assembly of Oct4 and Sox2 on two enhancers. Genes Dev. 17:2048–2059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Remenyi A, Tomilin A, Pohl E, Lins K, Philippsen A, Reinbold R, Scholer HR, Wilmanns M. 2001. Differential dimer activities of the transcription factor Oct-1 by DNA-induced interface swapping. Mol Cell. 8:569–580. [DOI] [PubMed] [Google Scholar]
- Rodda DJ, Chew JL, Lim LH, Loh YH, Wang B, Ng HH, Robson P. 2005. Transcriptional regulation of nanog by OCT4 and SOX2. J Biol Chem. 280:24731–24737. [DOI] [PubMed] [Google Scholar]
- Rodriguez RT, Velkey JM, Lutzko C, Seerke R, Kohn DB, O'shea KS, Firpo MT. 2007. Manipulation of OCT4 levels in human embryonic stem cells results in induction of differential cell types. Exp Biol Med (Maywood). 232:1368–1380. [DOI] [PubMed] [Google Scholar]
- Roos MD, Xie W, Su K, Clark JA, Yang X, Chin E, Paterson AJ, Kudlow JE. 1998. Streptozotocin, an analog of N-acetylglucosamine, blocks the removal of O-GlcNAc from intracellular proteins. Proc Assoc Am Physicians. 110:422–432. [PubMed] [Google Scholar]
- Ryan AK, Rosenfeld MG. 1997. POU domain family values: Flexibility, partnerships, and developmental codes. Genes Dev. 11:1207–1225. [DOI] [PubMed] [Google Scholar]
- Saxe JP, Tomilin A, Scholer HR, Plath K, Huang J. 2009. Post-translational regulation of Oct4 transcriptional activity. PLoS ONE. 4:e4467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sayat R, Leber B, Grubac V, Wiltshire L, Persad S. 2008. O-GlcNAc-glycosylation of beta-catenin regulates its nuclear localization and transcriptional activity. Exp Cell Res. 314:2774–2787. [DOI] [PubMed] [Google Scholar]
- Schnerch A, Cerdan C, Bhatia M. 2010. Distinguishing between mouse and human pluripotent stem cell regulation: The best laid plans of mice and men. Stem Cells. 28:419–430. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Ciesiolka T, Gruss P. 1991. A nexus between Oct-4 and E1A: Implications for gene regulation in embryonic stem cells. Cell. 66:291–304. [DOI] [PubMed] [Google Scholar]
- Scholer HR, Hatzopoulos AK, Balling R, Suzuki N, Gruss P. 1989. A family of octamer-specific proteins present during mouse embryogenesis: Evidence for germline-specific expression of an Oct factor. EMBO J. 8:2543–2550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shafi R, Iyer SP, Ellies LG, O'Donnell N, Marek KW, Chui D, Hart GW, Marth JD. 2000. The O-GlcNAc transferase gene resides on the X chromosome and is essential for embryonic stem cell viability and mouse ontogeny. Proc Natl Acad Sci U S A. 97:5735–5739. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sinclair DA, Syrzycka M, Macauley MS, Rastgardani T, Komljenovic I, Vocadlo DJ, Brock HW, Honda BM. 2009. Drosophila O-GlcNAc transferase (OGT) is encoded by the Polycomb group (PcG) gene, super sex combs (sxc). Proc Natl Acad Sci U S A. 106:13427–13432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Speakman CM, Domke TC, Wongpaiboonwattana W, Sanders K, Mudaliar M, van Aalten DM, Barton GJ, Stavridis MP. 2014. Elevated O-GlcNAc levels activate epigenetically repressed genes and delay mouse ES cell differentiation without affecting naive to primed cell transition. Stem Cells. 459:455–459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spelat R, Ferro F, Curcio F. 2012. Serine 111 phosphorylation regulates OCT4A protein subcellular distribution and degradation. J Biol Chem. 287:38279–38288. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Strumpf D, Mao CA, Yamanaka Y, Ralston A, Chawengsaksophak K, Beck F, Rossant J. 2005. Cdx2 is required for correct cell fate specification and differentiation of trophectoderm in the mouse blastocyst. Development. 132:2093–2102. [DOI] [PubMed] [Google Scholar]
- Swaney DL, Wenger CD, Thomson JA, Coon JJ. 2009. Human embryonic stem cell phosphoproteome revealed by electron transfer dissociation tandem mass spectrometry. Proc Natl Acad Sci U S A. 106:995–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szkudelski T. 2001. The mechanism of alloxan and streptozotocin action in B cells of the rat pancreas. Physiol Res. 50:537–546. [PubMed] [Google Scholar]
- Teo CF, El-Karim EG, Wells L. 2016. Dissecting PUGNAc-mediated inhibition of the pro-survival action of insulin. Glycobiology. 26:1198–1208. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ. 2010. a. Generation of O-GlcNAc specific monoclonal antibodies using a novel synthetic immunogen. FASEB J. 24:904–907. [Google Scholar]
- Teo CF, Ingale S, Wolfert MA, Elsayed GA, Not LG, Chatham JC, Wells L, Boons GJ. 2010. b. Glycopeptide-specific monoclonal antibodies suggest new roles for O-GlcNAc. Nat Chem Biol. 6:338–343. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tokuzawa Y, Kaiho E, Maruyama M, Takahashi K, Mitsui K, Maeda M, Niwa H, Yamanaka S. 2003. Fbx15 is a novel target of Oct3/4 but is dispensable for embryonic stem cell self-renewal and mouse development. Mol Cell Biol. 23:2699–2708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Torres CR, Hart GW. 1984. Topography and polypeptide distribution of terminal N-acetylglucosamine residues on the surfaces of intact lymphocytes. Evidence for O-linked GlcNAc. J Biol Chem. 259:3308–3317. [PubMed] [Google Scholar]
- Tsuruzoe S, Ishihara K, Uchimura Y, Watanabe S, Sekita Y, Aoto T, Saitoh H, Yuasa Y, Niwa H, Kawasuji M et al. . 2006. Inhibition of DNA binding of Sox2 by the SUMO conjugation. Biochem Biophys Res Commun. 351:920–926. [DOI] [PubMed] [Google Scholar]
- van den Berg DL, Snoek T, Mullin NP, Yates A, Bezstarosti K, Demmers J, Chambers I, Poot RA. 2010. An Oct4-centered protein interaction network in embryonic stem cells. Cell Stem Cell. 6:369–381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vosseller K, Sakabe K, Wells L, Hart GW. 2002. a. Diverse regulation of protein function by O-GlcNAc: A nuclear and cytoplasmic carbohydrate post-translational modification. Curr Opin Chem Biol. 6:851–857. [DOI] [PubMed] [Google Scholar]
- Vosseller K, Wells L, Lane MD, Hart GW. 2002. b. Elevated nucleocytoplasmic glycosylation by O-GlcNAc results in insulin resistance associated with defects in Akt activation in 3T3-L1 adipocytes. Proc Natl Acad Sci U S A. 99:5313–5318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang Z, Pandey A, Hart GW. 2007. Dynamic interplay between O-linked N-acetylglucosaminylation and glycogen synthase kinase-3-dependent phosphorylation. Mol Cell Proteomics. 6:1365–1379. [DOI] [PubMed] [Google Scholar]
- Webster DM, Teo CF, Sun Y, Wloga D, Gay S, Klonowski KD, Wells L, Dougan ST. 2009. O-GlcNAc modifications regulate cell survival and epiboly during zebrafish development. BMC Dev Biol. 9:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wei F, Scholer HR, Atchison ML. 2007. Sumoylation of Oct4 enhances its stability, DNA binding, and transactivation. J Biol Chem. 282:21551–21560. [DOI] [PubMed] [Google Scholar]
- Wells L, Vosseller K, Hart GW. 2001. Glycosylation of nucleocytoplasmic proteins: Signal transduction and O-GlcNAc. Science. 291:2376–2378. [DOI] [PubMed] [Google Scholar]
- Xu H, Wang W, Li C, Yu H, Yang A, Wang B, Jin Y. 2009. WWP2 promotes degradation of transcription factor OCT4 in human embryonic stem cells. Cell Res. 19:561–573. [DOI] [PubMed] [Google Scholar]
- Xu HM, Liao B, Zhang QJ, Wang BB, Li H, Zhong XM, Sheng HZ, Zhao YX, Zhao YM, Jin Y. 2004. Wwp2, an E3 ubiquitin ligase that targets transcription factor Oct-4 for ubiquitination. J Biol Chem. 279:23495–23503. [DOI] [PubMed] [Google Scholar]
- Yang X, Ongusaha PP, Miles PD, Havstad JC, Zhang F, So WV, Kudlow JE, Michell RH, Olefsky JM, Field SJ et al. . 2008. Phosphoinositide signalling links O-GlcNAc transferase to insulin resistance. Nature. 451:964–969. [DOI] [PubMed] [Google Scholar]
- Yang X, Zhang F, Kudlow JE. 2002. Recruitment of O-GlcNAc transferase to promoters by corepressor mSin3A: Coupling protein O-GlcNAcylation to transcriptional repression. Cell. 110:69–80. [DOI] [PubMed] [Google Scholar]
- Yuan H, Corbi N, Basilico C, Dailey L. 1995. Developmental-specific activity of the FGF-4 enhancer requires the synergistic action of Sox2 and Oct-3. Genes Dev. 9:2635–2645. [DOI] [PubMed] [Google Scholar]
- Yuzwa SA, Macauley MS, Heinonen JE, Shan X, Dennis RJ, He Y, Whitworth GE, Stubbs KA, McEachern EJ, Davies GJ et al. . 2008. A potent mechanism-inspired O-GlcNAcase inhibitor that blocks phosphorylation of tau in vivo. Nat Chem Biol. 4:483–490. [DOI] [PubMed] [Google Scholar]
- Yuzwa SA, Yadav AK, Skorobogatko Y, Clark T, Vosseller K, Vocadlo DJ. 2011. Mapping O-GlcNAc modification sites on tau and generation of a site-specific O-GlcNAc tau antibody. Amino acids. 40:857–868. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. 2004. O-GlcNAc a sensor of cellular state: The role of nucleocytoplasmic glycosylation in modulating cellular function in response to nutrition and stress. Biochim Biophys Acta. 1673:13–28. [DOI] [PubMed] [Google Scholar]
- Zachara NE, Hart GW. 2006. Cell signaling, the essential role of O-GlcNAc. Biochim Biophys Acta. 1761:599–617. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.