Summary
Background
C-Myc is one of the major cellular oncogenes overexpressed in non-small cell lung carcinoma (NSCLC). Its deregulated expression is necessary but not sufficient for malignant transformation. We evaluated expression of MYC gene in NSCLC patients and its association with alterations in the genes previously identified to be related to NSCLC pathogenesis, PHACTR3 and E2F4.
Methods
We analyzed MYC gene expression by qRT-PCR in 30 NSCLC patients’ samples and paired normal lung tissue. MYC expression was further statistically evaluated in relation to histopathological parameters, PHACTR3 and E2F4 gene alterations and survival. Alterations in aforementioned genes were previously detected and identified based on AP-PCR profiles of paired normal and tumor DNA samples, selection of DNA bands with altered mobility in tumor samples and their characterization by the reamplification, cloning and sequencing.
Results
MYC expression was significantly increased in NSCLC samples and its overexpression significantly associated with squamous cell carcinoma subtype. Most importantly, MYC overexpression significantly coincided with mutations in PHACTR3 and E2F4 genes, in group of all patients and in squamous cell carcinoma subtype. Moreover, patients with jointly overexpressed MYC and altered PHACTR3 or E2F4 showed trend of shorter survival.
Conclusions
Overall, MYC is frequently overexpressed in NSCLC and it is associated with mutated PHACTR3 gene, as well as mutated E2F4 gene. These joint gene alterations could be considered as potential molecular markers of NSCLC and its specific subtypes.
Keywords: E2F4, MYC, Non-small cell lung carcinoma, PHACTR3
Kratak sadržaj
Uvod
C-Myc je jedan od glavnih ćelijskih onkogena koji je prekomerno eksprimiran kod nesitnoćelijskog karcinoma pluća (NSCLC). Promene u ekspresiji ovog onkogena iako neophodne nisu dovoljne za malignu transformaciju. U ovoj studiji smo proučavali promene ekspresije MYC gena kod pacijenata sa NSCLC kao i međupovezanost sa promenama u genima PHACTR3 i E2F4, za koje je ranije pokazano da su povezani sa patogenezom NSCLC.
Metode
Ekspresija gena MYC je analizirana qRT-PCR metodom u uzorcima tumorskog i odgovarajućeg normalnog plućnog tkiva 30 pacijenata obolelih od NSCLC. Promene MYC ekspresije su dalje analizirane u odnosu na histopatološke parametre, promene u genima PHACTR3 i E2F4, kao i u odnosu na preživljavanje pacijenata. Mutacije u pomenutim genima su prethodno otkrivene i identifikovane na osnovu uporedne analize AP-PCR profila parova normalnih i tumorskih DNK uzoraka, odabira DNK traka sa izmenjenom pokretljivošcu u tumorskim uzorcima, i zatim njihovom karakterizacijom pomoću reamplifikacije, kloniranja i sekvenciranja.
Rezultati
Ekspresija gena MYC je povećana u uzorcima NSCLC i predstavlja značajnu karakteristiku karcinoma skvamoznih celija. Prekomerna ekspresija ovog gena asocira sa prisustvom mutacija u genima PHACTR3 i E2F4 u celoj grupi pacijenata, kao i u podgrupi karcinoma skvamoznih ćelija. Takođe, pacijenti sa povećanom ekspresijom MYC gena i istovremeno mutiranim PHACTR3 ili E2F4 genima su pokazali trend kraćeg preživljavanja.
Zaključak
Povećana ekspresija MYC gena je prisutna sa visokom učestalošću kod NSCLC i povezana je sa mutiranim PHACTR3 genom, kao i mutiranim E2F4 genom. Ove združene genske promene se mogu razmatrati kao potencijalni molekularni markeri NSCLC i njegovih specifičnih podtipova.
Ključne reči: E2F4, MYC, nesitnoćelijski karcinom pluća, PHACTR3
Introduction
For more than 3 decades lung cancer has been the most common type of human malignancies, representing approximately 12–13% of all newly diagnosed cancer cases worldwide (1, 2). The estimated number of lung cancer related deaths has been constantly rising during this period (2). Although there have been improvements in lung cancer patient survival, it is still very poor with less than 18% 5-year survival rate (3). Therefore, scientists are persistently looking for novel and reliable molecular markers to improve diagnosis of lung cancer, optimize treatments and enable monitoring disease progression and response to therapy. Molecules that regulate cellular processes essential for maintaining malignant phenotype, such as c-Myc oncogene, stand out as promising candidates to serve as molecular markers. This nuclear phosphoprotein functions as transcriptional regulator of multiple genes and controls cell proliferation, induces apoptosis, and regulates stemness, senescence, metabolism and genome stability (4). Its expression is deregulated in number of human cancers including breast cancer, Burkitt’s lymphoma, medulloblastoma, neuroblastoma and lung cancer (5). In most studies on non-small cell lung carcinoma (NSCLC), a predominant clinical form of lung cancer, c-Myc protein is overexpressed in 40–75% of cases (6, 7, 8, 9). Even though it is highly expressed in NSCLC, it does not associate with histopathological parameters and its prognostic value for this type of lung cancer is not clear (7, 9, 10). These data implicate that c-Myc is important for NSCLC pathogenesis but not sufficient to individually drive this process. Accordingly, we evaluated expression of MYC gene in 30 NSCLC patients’ samples and analyzed its association with histopathological parameters and patients survival. Our main focus was to investigate the possible association of MYC expression with alterations in the genes previously identified to be related with NSCLC pathogenesis, PHACTR3 and E2F4 (11). The aim was to determine their mutual importance for NSCLC pathogenesis and potential to serve as molecular markers of this disease. Association analyses of MYC overexpression and aforementioned gene alterations were performed on whole set of patients’ samples and samples stratified according to histopathological parameters. To evaluate their joint prognostic signifcance we analyzed their effects on patients’ survival.
Materials and Methods
Tissue samples
We analyzed paired tumor samples and adjacent normal lung tissue from 30 patients with NSCLC who underwent surgery at the Clinic for Thoracic Surgery (Belgrade, Serbia). The specimens were frozen in liquid nitrogen, where they were kept until DNA and RNA extraction. The samples were collected and used after obtaining patients’ informed consents and approval from the Ethics Committee, in accordance with the ethical standards laid down in the 1964 Declaration of Helsinki and its later amendments. Diagnosis of NSCLC and its classification according to histological subtype, grade stage, and lymph node invasion were established by histological analyses of the surgical specimens. Distribution of all specimens by relevant histopathological parameters was presented previously (11). Overall, 9 adenocarcinomas and 21 squamous cell carcinomas were included in the study. 26 NSCLC samples were well to moderately differentiated (grade 1/2), while 4 were poorly differentiated (grade 3). 16 samples were in early pathological stages (stages I/II), and 14 cases were in late stage (stage III). Lymph node invasion was observed in 25 samples. Neither pre-surgical chemotherapy nor radiotherapy had been administered.
RNA extraction and Reverse Transcription
Total RNA was isolated from tumor and corresponding normal tissues samples of all 30 NSCLC patients. The isolation was carried out using Trizol® reagent (Invitrogen Life Technologies, USA) according to the manufacturer’s instructions. RNA was quantified by spectrophotometry and quality was verified by agarose gel electrophoresis. Subsequently, reverse transcription reactions were performed using 2 mg of total RNA, with a High-capacity cDNA reverse transcription kit (Applied Biosystems), following the manufacturer’s protocol.
Quantitative real-time PCR
Quantitative real time PCR (qRT-PCR) was performed to evaluate the mRNA expression level of MYC gene. PCR reactions with 100 ng of cDNA were performed using pair of primers specific for MYC transcripts (12). For each sample ACTB was used as a reference gene for the normalization of target gene expression (13). The primer sequences were as follows: 5’-CAA GAG GCG AAC ACA CAA CGT CT-3’ (sense) and 5’-AAC TGT TCT CGT CGT TTC CGC AA-3’ (antisense) for MYC, and 5 -TGG ACA TCC GCAAAG ACC TGT AC-3 (sense) and 5-TCA GGA GGA GCA ATG ATC TTG A-3 (antisense) for the ACTB. PCR reactions were performed using a Maxima SYBR Green/ROX qPCR Master Mix (Thermo Fisher Scientific, USA) on a QuantStudio 3 Real-Time PCR system (Thermo Fisher Scientific, USA) according to the manufacturer’s recommendations. Each sample was tested in triplicate and relative gene expression levels were analyzed by the 2–ΔΔCt method (14).
Identification of NSCLC associated genes
In our preceding study we identified several genes not previously related to NSCLC pathogenesis (11). Overall procedure of identifying those genes included: DNA isolation, generation and comparison of AP-PCR profiles of paired normal and tumor DNA samples, selection of DNA bands with altered mobility in tumor versus matching normal samples and their further characterization through the reamplification, cloning and sequencing.
DNA extraction
DNA was extracted using the phenol/chloroform/isoamyl alcohol method (15). The quality of the DNA was verified by electrophoresis on 0.8% agarose gel and staining with ethidium bromide. The DNA concentration was established spectrophotometrically.
AP-PCR DNA fingerprinting
AP-PCR (Arbitrarily Primed Polymerase Chain Reaction) was used to generate DNA fingerprint profiles of paired DNA samples, normal and tumor, of the same patient (16). In short, six different, randomly chosen, primers were used to amplify DNA, initially under low-stringency conditions and then at high-stringency conditions, to generate informative fingerprints of moderate complexity that could distinguish tumor from normal tissue. The reactions were performed in a GeneAmp® PCR System 9700 (Applied Biosystems, Foster City, CA, USA) using PCR reaction kits supplied by Fermentas Life Sciences (Lithuania). Each reaction consisted of an initial denaturation step (95 °C for 5 min), 4 cycles at low-stringency conditions (specific for each primer), 35 cycles at high-stringency conditions (specific for each primer), and a final extension (72 °C for 7 min). Five out of six tested primers (p53 A, MDR A, E5A p53, GAPDH A, and GAPDH S) produced informative sequence alterations differentiating normal from tumor tissue. Primer sequences, AP-PCR amplification profiles and reaction conditions are described in our previous publications (16, 11) and summarized in Table I. The AP-PCR products were separated on 6-8% non-denaturing polyacrylamide gels and visualized by silver staining.
Table I.
AP-PCR primer sequences, amplification profiles and reaction conditions.
| Primers | Sequences | Amplification profiles | Reaction conditions | |
|---|---|---|---|---|
| Low-stringency conditions | High-stringency conditions | |||
| p53A | 5’-TTG GGC AGT GCT CGC TTA GT-3’ | 95 °C 30”; 40 °C 2’; 72 °C 1’ | 95 °C 30”; 60 °C 2’; 72 °C 1’ | 0.2 mmol/L each dNTP 3.5 mmol/L MgCl2 5 μmol/L primer 1U Taq DNA Polymerase |
| MDR A | 5’-GTT CAA ACT TCT GCT CCT GA-3’ | 95 °C 30”; 40 °C 2’; 72 °C 1’ | 95 °C 30”; 58 °C 2’; 72 °C 1’ | 0.4 mmol/L each dNTP 2.5 mmol/L MgCl2 5 μmol/L primer 1U Taq DNA Polymerase |
| E5A p53 | 5’-CAG CCC TGT CGT CTC TCC AG-3’ | 95 °C 30”; 40 °C 2’; 72 °C 1’ | 95 °C 30”; 55 °C 2’; 72 °C 1’ | 0.6 mmol/L each dNTP 3.5 mmol/L MgCl2 3 μmol/L primer 1U Taq DNA Polymerase |
| GAPDH A | 5’-AGC CTT CTC CAT GGT GGT GAA GAC-3’ | 95 °C 30”; 50 °C 2’; 72 °C 1’ | 95 °C 30”; 72 °C 2’; 72 °C 1’ | 0.2 mmol/L each dNTP 2.5 mmol/L MgCl2 3 μmol/L primer 1U Taq DNA Polymerase |
| GAPDH S | 5’-CGG AGT CAA CGG ATT TGG TCG TAT-3’ | 95 °C 30”; 50 °C 2’; 72 °C 1’ | 95 °C 30”; 70 °C 2’; 72 °C 1’ | 0.4 mmol/L each dNTP 2.5 mmol/L MgCl2 5 μmol/L primer 1U Taq DNA polymerase |
Isolation, cloning and sequencing of altered bands
Some of the DNA bands with altered mobility, in tumor compared to normal samples, were selected for further characterization. Briefly, PCR amplicons of chosen bands were scrapped from silver stained gels with a hypodermic 22-gauge needle previously soaked in the PCR master mix solution. The needle was dipped in the PCR master mix for 2 min and then discarded. Further reamplification of the PCR products was performed with the same primers used for AP-PCR reactions at high-stringency conditions specific for each particular primer. The obtained products were cloned and sequenced as described (11). In short, the reamplified material was administrated on 1.5% agarose gels, than purified using DNA Extraction Kit (Fermentas Life Sciences, Lithuania) and cloned with GeneJetTM PCR Cloning Kit (Fermentas Life Sciences, Lithuania) according to manufacturers’ instructions. After cloning procedure, plasmids were purified using GeneJetTM Plasmid Miniprep Kit (Fermentas Life Sciences, Lithuania). Further, sequencing was performed in both directions on several clones for each selected DNA band on an ABI Prism 3130 Genetic Analyzer automated sequencer (Applied Biosystems, Foster City, CA, USA) using BigDye Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems, Foster City, CA, USA). The obtained sequences were analyzed using BLAST software in the NCBI GenBank and EBI (Sanger Institute) database to identify selected alterations.
Statistical analysis
Significant differences between the data sets were determined by GraphPad Prism 6 software (La Jolla, CA USA) and statistical package R (version 3.1.3 (2015-03-09) Copyright (C) 2015 The R Foundation for Statistical Computing). Paired t-test was used to assess statistical differences in expression level of MYC gene between paired tumor and normal sample of each patient. On the other hand, difference in MYC relative expression level between total tumor and normal samples was evaluated using unpaired t-test. Fisher exact test was used to examine the relationship between histopathological parameters (histological subtype, grade stage, and lymph node invasion) and MYC overexpression. The same statistical analysis was used to study the possible associations between frequencies of MYC overexpression and alterations in PHACTR3 and E2F4. Survival analyses were performed by Kaplan & Meier product-limit method. The log rank test was used to evaluate the significance of the difference between pairs of survival probabilities. Overall survival rate was calculated from day one after surgery to the last follow-up examination or death of the patient. The total follow up period was 117 months. Statistical differences were considered significant when p value was < 0.05.
Results
MYC gene expression analysis
Alterations in MYC gene expression between tumor and matching normal samples were analyzed by qRT-PCR. MYC gene expression was significantly higher in NSCLC samples (p = 0.018) compared with normal lung samples (Figure 1). Specifically, MYC gene was overexpressed in 11 out of 30 tumor samples (37%).
Figure 1.
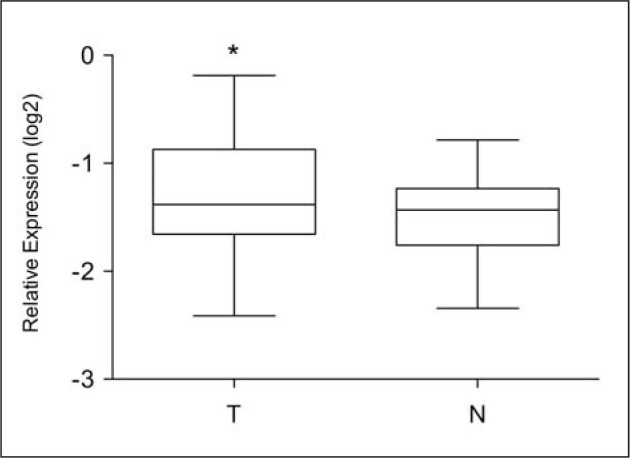
MYC gene expression. Box plot analysis of MYC expression level in primary tumor (T) and normal (N) samples. Results are presented as log2 of MYC gene expression relative to ACTB gene level. ∗ indicates p<0.05 as significant gene overexpression in tumor samples compared to normal samples.
Increased MYC gene expression in relation to histopathological parameters
Significance of MYC overexpression was initially evaluated in relation to major histopathological parameters: NSCLC subtype, histological grade, stage and lymph node invasion (Table II). Statistical analyses revealed that MYC overexpression was significantly associated with histological subtype (p=0.01). In adenocarcinoma increased MYC expression was completely absent (0/9), while in squamous cell carcinoma MYC overexpression was more frequently present (11/21, 52.4%).
Table II.
Association between MYC overexpression and histopathological parameters.
| Parameter | Total NPb | MYC overexpression | ||
|---|---|---|---|---|
| Yes NP (%) | No NP (%) | p value | ||
| Total | 30 | 11 | 19 | |
| NSCLC subtype | ||||
| Adenocarcinoma | 9 | 0 (0.0) | 9 (100.0) | 0.01c |
| Squamous cell carcinoma | 21 | 11 (52.4) | 10 (47.6) | |
| Histological gradea | ||||
| g1/2 | 26 | 11 (42.3) | 15 (57.7) | 0.36 |
| g3 | 4 | 0 (0.0) | 4 (100.0) | |
| Stage | ||||
| I/II | 16 | 6 (37.5) | 10 (62.5) | 1 |
| III | 14 | 5 (35.7) | 9 (64.3) | |
| Lymph node invasion | ||||
| Positive | 25 | 9 (36.0) | 16 (64.0) | 1 |
| Negative | 5 | 2 (40.0) | 3 (60.0) | |
NP number of patients per group;
bold indicates statistically significant value, p<0.05.
g1/2, well to moderately differentiated; g3, poorly differentiated;
Increased MYC gene expression in relation to gene alterations
We further examined MYC expression in relation to the presence of alterations in PHACTR3 and E2F4. In previous study (11), mutations in these two genes were identified according to sequence data and BLAST search results. Specifically, multiple nucleotide substitutions were detected in PHACTR3 and E2F4.
Association between MYC overexpression and altered PHACTR3 gene
Patients with altered PHACTR3 gene had more frequently increased MYC gene expression (Table III; p=0.015). Among six patients with altered PHACTR3 gene, 5 (83.3%) had increased MYC mRNA level, while in 1 patient (16.7%) MYC was not overexpressed. Similar association was present in subgroup of patients with squamous cell carcinoma (Table III; p=0.035). In this group all patients with PHACTR3 alterations had overexpressed MYC.
Table III.
Association of overexpressed MYC oncogene with altered PHACTR3 and E2F4 genes.
| MYC overexpression | Squamous cell carcinoma | ||||
|---|---|---|---|---|---|
| MYC overexpression | |||||
| Yes | No | Yes | No | ||
| NPa (%) | NP (%) | NPa (%) | NP (%) | ||
| PHACTR3 alteration | Yes | 5 (83.3) | 1 (16.7) | 5 (100.0) | 0 (0.0) |
| No | 6 (25.0) | 18 (75.0) | 6 (37.5) | 10 (62.5) | |
| p value | 0.015b | 0.035 | |||
| E2F4 alteration | Yes | 5 (83.3) | 1 (16.7) | 5 (100.0) | 0 (0.0) |
| No | 6 (25.0) | 18 (75.0) | 6 (37.5) | 10 (62.5) | |
| p value | 0.015b | 0.035 | |||
NP number of patients per group;
bold indicates statistically significant values, p<0.05.
Association between MYC overexpression and altered E2F4 gene
MYC overexpression in the presence of E2F4 gene alterations showed the same pattern of conjunction as with the incidence of altered PHACTR3 gene (Table III). Namely, 83.3% of patients with abnormal E2F4 had overexpressed MYC compared to 16.7% of patients that did not show overexpression of this oncogene (p=0.015). In squamous cell carcinoma subtype altered E2F4 exclusively coincided with MYC overexpression (100%) while unchanged E2F4 was predominantly associated with non-overexpressed MYC (62.5% versus 37.5%; p=0.035).
Effects of gene alterations and MYC overexpression on survival rate
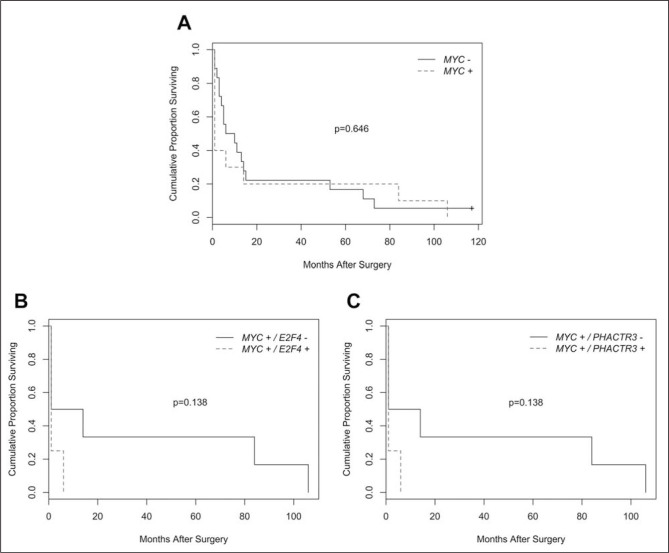
Kaplan-Meier survival curves were analyzed to evaluate the effect of MYC gene overexpression in the presence of aforementioned gene alterations on NSCLC patients’ survival. When individually examined, MYC overexpression did not significantly affect the survival rate (Figure 2A; p=0.646). However, when considered in combination with altered PHACTR3 and E2F4, MYC overexpression showed tendency towards shorter survival although did not reach statistical significance. The median survival time was 1 month for patients with overexpressed MYC and altered PHACTR3 or E2F4 compared to 7.5 months for those with overexpressed MYC only (Figure 2B and C; p=0.138).
Figure 2.
Kaplan–Meier overall survival analyses. No difference in survival was observed between patients with overexpressed (MYC+) and non-overexpressed MYC gene (MYC-) (A). Patients with overexpressed MYC and mutated E2F4 (MYC+/ E2F4+) (B) or PHACTR3 (MYC+/ PHACTR3+) (C) showed trend of decreased survival compared to those with overexpressed MYC only (MYC+/ E2F4-, MYC+/ PHACTR3-).
Discussion
C-Myc is one of the most important cellular oncogenes necessary for malignant transformation (17). Deregulation of its expression is critical for the pathogenesis of various cancer types, including lung cancer (5). C-Myc protein is overexpressed in 40–75% of NSCLC patients (6, 7, 8, 9). However, data obtained by RT-PCR are not consistent. Wu et al. have reported only 18% NSCLC cases with MYC overexpression (18), while Hsu and colleagues have found overexpressed MYC in 91% of NSCLC patients’ samples (19). In our study, we evaluated MYC gene expression by qRT-PCR analysis in 30 NSCLC patients’ samples and observed its overexpression in 37% of cases. This result corresponds to most studies on c-Myc protein expression (7, 20). Moreover, we have observed for the first time that MYC overexpression is a significant characteristic of squamous cell carcinoma subtype rather than adenocarcinoma. C-Myc overexpression was previously shown to be more frequently present in squamous cell carcinoma but without statistical significance (9), while Xu et al. (8) reported it to be significant characteristic of adenocarcinoma.
Major focus of this study was to analyze MYC gene overexpression in relation to alterations in genes associated with NSCLC pathogenesis (11). Tested gene alterations, PHACTR3 and E2F4 were shown to be associated with MYC overexpression. Moreover, these alterations significantly coincide with MYC overexpression in squamous cell carcinoma. PHACTR3 (Phosphatase and actin regulator 3/ scaffold-associated PP1 inhibiting protein), also known as scapinin, is a protein phosphatase 1-binding protein associated with nuclear non-chromatin structures (the nuclear matrix or nucleoskeleton) (21). It directly interacts with actin and regulates actin cytoskeleton structures influencing cell spreading and motility (22). At the same time, nuclear F-actin activates transcription of c-Myc through Wnt/β-catenin signaling (23). We speculate that actin could be one of potential interconnections between PHACTR3 alterations and MYC overexpression. In our previous study PHACTR3 was shown to be altered in 6 out of 30 patients, mostly with squamous cell carcinoma (11). Recently, significance of PHACTR3 as parameter for non-invasive lung cancer diagnosis was evaluated along with other diagnostic and risk biomarkers (24).
Considering interrelationship between MYC overexpression and mutational status of E2F4 the observed positive association is not surprising. E2F4 is a part of complex involved in repression of MYC transcription in response to different signals (25). This transcriptional regulator was shown to be necessary for normal development of lung tissue (26). However, its importance and role in pathogenesis of NSCLC was examined only in our earlier publication (11). In that study, altered E2F4 was detected in 20% of patients, predominantly with squamous cell carcinoma.
The influence of c-Myc overexpression on NSCLC patient survival is inconclusive. Some authors showed c-Myc association with shorter survival (9), while others reported no effects on NSCLC prognosis (7). We observed no significant difference in life span between NSCLC patients with overexpressed and non-overexpressed MYC gene. However, the presence of altered PHACTR3 or E2F4 along with overexpressed MYC contributed to shorter survival but without prognostic significance. Considering our previous results this joint effect of MYC and altered genes could be mainly attributed to the significant negative effects of altered PHACTR3 and E2F4 genes on NSCLC patients’ survival (11).
Overall this study implicates frequent presence of overexpressed MYC gene in NSCLC patients and its applicative potential as molecular marker of squamous cell carcinoma. Additionally, the results point out the relevance of overexpressed MYC in association with altered PHACTR3 and E2F4 genes as joint molecular markers of NSCLC and its specific subtypes. Most importantly, we identified positive associations between MYC overexpression and aforementioned gene alterations not previously considered in NSCLC nor other cancer types, therefore opening a novel research perspective in studying c-Myc role in carcinogenesis. In this context, functional interconnection of MYC and altered genes will be of particular interest in our further studies.
Acknowledgements
This study was supported by the Ministry of Education, Science and Technological Development of the Republic of Serbia (Grant No III41031).
Glossary
List of abbreviations
- NSCLC
non-small cell lung carcinoma
- qRT-PCR
quantitative real time polymerase chain reaction
- AP-PCR
arbitrarily primed polymerase chain reaction
- ACTB
beta-actin
- PHACTR3
phosphatase and actin regulator 3/ scaffold-associated PP1 inhibiting protein.
Footnotes
Conflict of interest
Conflict of interest statement: The authors stated that they have no conflicts of interest regarding the publication of this article.
References
- 1.Parkin DM, Pisani P, Ferlay J.. Estimates of the worldwide incidence of eighteen major cancers in 1985. Int J Cancer. 1993;54:594–606. doi: 10.1002/ijc.2910540413. [DOI] [PubMed] [Google Scholar]
- 2.Torre LA, Bray F, Siegel RL, Ferlay J, Lortet-Tieulent J, Jemal A.. Global cancer statistics, 2012. CA Cancer J Clin. 2015;65:87–108. doi: 10.3322/caac.21262. [DOI] [PubMed] [Google Scholar]
- 3.Torre LA, Siegel RL, Jemal A.. Lung Cancer Statistics. Adv Exp Med Biol. 2016;893:1–19. doi: 10.1007/978-3-319-24223-1_1. [DOI] [PubMed] [Google Scholar]
- 4.Chen BJ, Wu YL, Tanaka Y, Zhang W.. Small molecules targeting c-Myc oncogene: promising anti-cancer therapeutics. Int J Biol Sci. 2014;10:1084–96. doi: 10.7150/ijbs.10190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Albihn A, Johnsen JI, Henriksson MA.. MYC in oncogenesis and as a target for cancer therapies. Adv Cancer Res. 2010;107:163–224. doi: 10.1016/S0065-230X(10)07006-5. [DOI] [PubMed] [Google Scholar]
- 6.Dong QZ, Wang Y, Dong XJ, Li ZX, Tang ZP, Cui QZ. et al. CIP2A is overexpressed in non-small cell lung cancer and correlates with poor prognosis. Ann Surg Oncol. 2011;18:857–65. doi: 10.1245/s10434-010-1313-8. [DOI] [PubMed] [Google Scholar]
- 7.Jiang R, Wang X, Jin Z, Li K.. Association of Nuclear PIM1 Expression with Lymph Node Metastasis and Poor Prognosis in Patients with Lung Adenocarcinoma and Squamous Cell Carcinoma. J Cancer. 2016;7:324–34. doi: 10.7150/jca.13422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Xu X, Sun PL, Li JZ, Jheon S, Lee CT, Chung JH.. Aberrant Wntl/beta-catenin expression is an independent poor prognostic marker of non-small cell lung cancer after surgery. J Thorac Oncol. 2011;6:716–24. doi: 10.1097/JTO.0b013e31820c5189. [DOI] [PubMed] [Google Scholar]
- 9.Kiani A, Abedini A, Adcock IM, Mirenayat M, Taghavi K, Mortaz E, Kazempour-Dizaji M.. Association between vitamin D deficiencies in sarcoidosis with disease activity, course of disease and stages of lung involvements. J Med Biochem. 2018;37:103–9. doi: 10.1515/jomb-2017-0041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Volm M, Koomagi R.. Prognostic relevance of c-Myc and caspase-3 for patients with non-small cell lung cancer. Oncol Rep. 2000;7:95–8. doi: 10.3892/or.7.1.95. [DOI] [PubMed] [Google Scholar]
- 11.Bankovic J, Stojsic J, Jovanovic D, Andjelkovic T, Milinkovic V, Ruzdijic S. et al. Identification of genes associated with non-small-cell lung cancer promotion and progression. Lung Cancer. 2010;67:151–9. doi: 10.1016/j.lungcan.2009.04.010. [DOI] [PubMed] [Google Scholar]
- 12.Wong H, Anderson WD, Cheng T, Riabowol KT.. Monitoring mRNA expression by polymerase chain reaction: the »primer-dropping« method. Anal Biochem. 1994;223:251–8. doi: 10.1006/abio.1994.1581. [DOI] [PubMed] [Google Scholar]
- 13.NicAmhlaoibh R, Heenan M, Cleary I, Touhey S, O’Loughlin C, Daly C. et al. Altered expression of mRNAs for apoptosis-modulating proteins in a low level multidrug resistant variant of a human lung carcinoma cell line that also expresses mdr1 mRNA. Int J Cancer. 1999;82:368–76. doi: 10.1002/(sici)1097-0215(19990730)82:3<368::aid-ijc10>3.0.co;2-e. [DOI] [PubMed] [Google Scholar]
- 14.Livak KJ, Schmittgen TD.. Analysis of relative gene expression data using real-time quantitative PCR and the 2(-Delta Delta C(T)) Method. Methods. 2001;25:40–28. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 15.Sambrook J, Fritch EF, Maniatis T. Molecular cloning: a laboratory manual. Second. New York: Cold Spring Harbor: Laboratory Press; 1989. pp. E.3–E.4. [Google Scholar]
- 16.Markovic J, Stojsic J, Zunic S, Ruzdijic S, Tanic N.. Genomic instability in patients with non-small cell lung cancer assessed by the arbitrarily primed polymerase chain reaction. Cancer Invest. 2008;26:262–8. doi: 10.1080/07357900701708385. [DOI] [PubMed] [Google Scholar]
- 17.Boehm JS, Hession MT, Bulmer SE, Hahn WC.. Transformation of human and murine fibroblasts without viral oncoproteins. Mol Cell Biol. 2005;25:6464–74. doi: 10.1128/MCB.25.15.6464-6474.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Wu W, Fan YH, Kemp BL, Walsh G, Mao L.. Overexpression of cdc25A and cdc25B is frequent in primary non-small cell lung cancer but is not associated with overexpression of c-myc. Cancer Res. 1998;58:4082–5. [PubMed] [Google Scholar]
- 19.Cooks T, Miaw J, Hsia JY, Shai SE, Chen CY.. Concordant expression of the telomerase-associated genes in nonsmall cell lung cancer. Eur J Surg Oncol. 2003;29:59–49. doi: 10.1016/s0748-7983(03)00108-2. [DOI] [PubMed] [Google Scholar]
- 20.Cooks T, Dong QZ, Cui QZ, Papavassiliou P, Wang ED, Wang EH.. Ataxia-telangiectasia group D complementing gene (ATDC) promotes lung cancer cell proliferation by activating NF-kappaB pathway. PLoS One. 2013;8:e63676. doi: 10.1371/journal.pone.0063676. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Sagara J, Higuchi T, Hattori Y, Moriya M, Sarvotham H, Shima H. et al. Scapinin, a putative protein phosphatase-1 regulatory subunit associated with the nuclear non-chromatin structure. J Biol Chem. 2003;278:45611–9. doi: 10.1074/jbc.M305227200. [DOI] [PubMed] [Google Scholar]
- 22.Sagara J, Arata T, Taniguchi S.. Scapinin, the protein phosphatase 1 binding protein, enhances cell spreading and motility by interacting with the actin cytoskeleton. PLoS One. 2009;4:e4247. doi: 10.1371/journal.pone.0004247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yamazaki S, Yamamoto K, de Lanerolle P, Harata M.. Nuclear F-actin enhances the transcriptional activity of beta-catenin by increasing its nuclear localization and binding to chromatin. Histochem Cell Biol. 2016;145:389–99. doi: 10.1007/s00418-016-1416-9. [DOI] [PubMed] [Google Scholar]
- 24.Hubers AJ, Brinkman P, Boksem RJ, Rhodius RJ, Witte BI, Zwinderman AH. et al. Combined sputum hypermethylation and eNose analysis for lung cancer diagnosis. J Clin Pathol. 2014;67:707–11. doi: 10.1136/jclinpath-2014-202414. [DOI] [PubMed] [Google Scholar]
- 25.Chen CR, Kang Y, Siegel PM, Massague J.. E2F4/5 and p107 as Smad cofactors linking the TGFbeta receptor to c-myc repression. Cell. 2002;110:19–32. doi: 10.1016/s0092-8674(02)00801-2. [DOI] [PubMed] [Google Scholar]
- 26.Danielian PS, Bender Kim CF, Caron AM, Vasile E, Bronson RT, Lees JA.. E2f4 is required for normal development of the airway epithelium. Dev Biol. 2007;305:564–76. doi: 10.1016/j.ydbio.2007.02.037. [DOI] [PMC free article] [PubMed] [Google Scholar]