Here, Yoon et al. show that translesion synthesis (TLS) opposite 1,N6-ethenodeoxyadenosine (ε dA), which disrupts Watson–Crick base pairing, occurs via Polι/Polζ- , Rev1-, and Pol θ-dependent pathways. Rev1 polymerase and Pol θ conduct TLS opposite ε dA via alternative error-prone pathways, and in contrast to extremely error-prone TLS opposite ε dA by purified Pol θ, it performs predominantly error-free TLS in human cells.
Keywords: DNA polymerase θ, εdA lesion, Hoogsteen base pairing, translesion synthesis
Abstract
Here we show that translesion synthesis (TLS) opposite 1,N6-ethenodeoxyadenosine (εdA), which disrupts Watson–Crick base pairing, occurs via Polι/Polζ-, Rev1-, and Polθ-dependent pathways. The requirement of Polι/Polζ is consistent with the ability of Polι to incorporate nucleotide opposite εdA by Hoogsteen base pairing and of Polζ to extend synthesis. Rev1 polymerase and Polθ conduct TLS opposite εdA via alternative error-prone pathways. Strikingly, in contrast to extremely error-prone TLS opposite εdA by purified Polθ, it performs predominantly error-free TLS in human cells. Reconfiguration of the active site opposite εdA would provide Polθ the proficiency for error-free TLS in human cells.
The1,N6-ethenodeoxyadenosine (εdA) adduct is formed in DNA through interaction with aldehydes derived from lipid peroxidation and by exposure to chemical carcinogens such as vinyl chloride. Lipid peroxidation is a normal chain reaction process that initiates from the oxidation of polyunsaturated fatty acids in cell membranes and results in the formation of a variety of highly reactive aldehydes, including acrolein, malonaldehyde, and trans-4-hydroxy-2-nonenal (HNE). Enals such as HNE undergo further oxidation reactions to form epoxyaldehydes and the reaction of epoxyaldehyde with DNA generates the εdA adduct (Chung et al. 1999; Luczaj and Skrzydlewska 2003).
The εdA adduct is highly inhibitory to synthesis by replicative DNA polymerases (Pols) because the exocyclic ring between the N1 and N6 positions of εdA impairs the Watson–Crick (W–C) edge of adenine (Supplemental Fig. S1). The ability of the Polι active site to push template purines into a syn conformation provides a mechanism by which this polymerase can insert nucleotides opposite DNA lesions which disrupt W–C base pairing. Biochemical studies have shown that while Polι incorporates a T correctly opposite the εdA adduct, it also incorporates a C with an about one fourth efficiency of that of T incorporation (Nair et al. 2006). Crystal structures of Polι-εdA·dTTP and Pol-εdA·dCTP ternary complexes show that similar to nonadducted A, the εdA adduct adopts a syn conformation in the Polι active site and its “Hoogsteen edge” participates in hydrogen bonding with the incoming dTTP or dCTP (Nair et al. 2006). Since Polι is highly inefficient at extending synthesis from the T or C inserted opposite εdA, the extension reaction would require another TLS Pol and biochemical studies have indicated that Polζ can extend synthesis from the εdA·dT or εdA·dC base pair (Nair et al. 2006).
Here we identify the TLS Pols that promote replication through the εdA adduct in human cells and show that replication through this adduct is mediated via three genetic pathways. As expected from biochemical and structural studies, Polι and Polζ function together in one pathway; however, contrary to biochemical and structural observations indicating a proficiency of Polι for misincorporating C opposite εdA, the Polι/Polζ pathway mediates highly error free TLS opposite this adduct in human cells. Rev1 polymerase activity contributes to a low level of TLS that is mutagenic; in this role, Rev1 functions independent of its scaffolding role with Polι in the Polι/Polζ pathway. Polθ, a member of A-family Pols, promotes replication through εdA via a third pathway. Although biochemical studies show that purified Polθ conducts highly error-prone TLS by incorporating an A opposite εdA, in human cells Polθ mediates predominantly error-free TLS opposite this adduct. We discuss the implications of this marked discrepancy and suggest that opposite DNA lesions such as εdA, Polθ active site is actively reconfigured in human cells to carry out predominantly error-free TLS.
Results and Discussion
Requirement of Polι/Polζ and Polθ for Replication through the εdA adduct in human cells
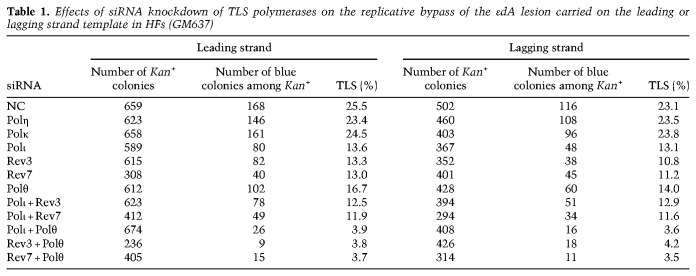
To identify the TLS Pols required for replicating through the εdA adduct, we analyzed the effects of siRNA depletion of TLS Pols individually and in combinations (Supplemental Fig. S2) on TLS frequencies resulting from replication through the lesion present on the leading or the lagging strand template of the SV40 based duplex plasmid. TLS opposite the εdA adduct carried on the leading strand template in normal human fibroblasts (HFs) and treated with control (NC) siRNA occurs with a frequency of ∼25% and TLS frequency was not affected in cells depleted for Polη or Polκ, indicating that these Pols play no significant role in TLS opposite this adduct (Table 1). In contrast, TLS frequency was reduced to ∼14% in Polι-depleted cells. A similar reduction in TLS frequency occurred upon depletion of the Rev3 or the Rev7 subunit of Polζ, and depletion of Polθ reduced TLS frequency to ∼17% (Table 1). The observation that TLS frequency remained the same in cells codepleted for Polι with Rev3 or Rev7 as that in cells depleted for any of these Pols alone (Table 1) indicated that Polι and Polζ function together in one TLS pathway. In contrast, codepletion of Polι with Polθ or of Rev3 or Rev7 with Polθ caused a drastic reduction in TLS frequency to ∼4% (Table 1), indicating that Polθ functions independently of Polι and Polζ and that TLS through the εdA adduct is mediated by two separate pathways dependent on either Polι/Polζ or Polθ. We verified this conclusion for TLS opposite εdA present on the lagging strand template (Table 1).
Table 1.
Effects of siRNA knockdown of TLS polymerases on the replicative bypass of the εdA lesion carried on the leading or lagging strand template in HFs (GM637)
Noncatalytic and catalytic roles of Rev1 in TLS opposite εdA
In human cells, Rev1 functions as an indispensable noncatalytic scaffolding component of Y-family Pols (Yoon et al. 2015). Since ∼15% of total TLS persists in HFs codepleted for Polθ with Polι or Polζ (Table 1), we considered the possibility that in addition to its role as a noncatalytic scaffolding component of Polι in the Polι/Polζ pathway, Rev1 contributes to TLS as a DNA polymerase and in this role it functions independently of Polι. To test for this possibility, we first determined whether there was evidence for Polι-dependent and Polι-independent roles of Rev1, and analyzed TLS opposite εdA in wild-type (WT), Rev1−/− (Yoon et al. 2015), and Polθ−/− (Yoon et al. 2019) mouse embryonic fibroblasts (MEFs) depleted for different TLS Pols (Supplemental Fig. S2; Supplemental Table S1). In WT MEFs, TLS opposite εdA occurs with a frequency of ∼22% and TLS is reduced to ∼8% in Rev1−/− MEFs treated with NC siRNA or depleted for Polι or Polζ. The epistasis of Rev1 over Polι in the Polι/Polζ pathway is consistent with a scaffolding role of Rev1 with Polι. To unravel the Polι-independent role of Rev1, we examined TLS in Polθ−/− MEFs depleted for Polι or Polζ, since then only the Polι/Polζ-independent role of Rev1 would remain functional. Our observation that TLS frequency is reduced to ∼3.5% in Polθ−/− MEFs depleted for Polι or Polζ strongly suggested that this residual TLS is mediated by the Polι/Polζ-independent role of Rev1. Furthermore, the almost abolition of TLS in Polθ−/− MEFs depleted for Rev1 or in Rev1−/− MEFs depleted for Polθ provided confirmatory evidence for the Polι-dependent and Polι-independent Rev1 roles.
To determine whether Rev1 polymerase function was required for its Polι-independent role, we examined TLS in HFs expressing siRNA-sensitive WT Rev1 or the siRNA-resistant form of either the WT or the catalytically inactive Rev1 D570A, E571A mutant. TLS in cells depleted for Rev1 and carrying the vector control or the siRNA-sensitive WT Rev1 was reduced to ∼9% as compared to ∼25% in control siRNA-treated cells (Supplemental Table S2). Expression of the siRNA-resistant WT Rev1 restored TLS frequency nearly to normal levels (∼23%), while expression of siRNA-resistant D570A, E571A catalytic mutation reduced TLS to ∼20% (Supplemental Table S2). This reduction in TLS frequency closely resembles the reduction in TLS frequency that occurs in HFs codepleted for Polι and Polθ (Table 1) or in Polθ−/− MEFs depleted for Polι (Supplemental Table S1). This result suggested that Rev1 polymerase activity accounts for a small proportion of total TLS opposite εdA and that this TLS operates independently of Rev1's scaffolding role with Polι, which will remain intact in Rev1 catalytic mutant. To further ascertain the contribution of Rev1 polymerase activity to TLS, we examined the effect of Rev1 catalytic mutation on TLS opposite εdA in Rev1−/− MEFs. In Rev1−/− MEFs expressing WT Rev1, TLS occurred at a frequency of ∼19% and TLS was reduced to ∼16% in these MEFs expressing catalytic mutant Rev1 (Supplemental Table S3). This result reinforces the evidence that Rev1 polymerase activity contributes to a relatively small proportion of total TLS opposite εdA. Overall, from analyses of TLS in WT HFs, WT MEFs, Rev1−/− MEFs, and Polθ−/− MEFs, we conclude that TLS opposite εdA is mediated by two major Polι/Polζ- and Polθ-dependent pathways, respectively, and by a relatively minor Rev1 polymerase-dependent pathway (Table 1; Supplemental Tables S1–S3).
Requirement of Polθ polymerase activity for TLS opposite εdA in human cells
Human Polθ is a 290 kDa protein comprised of an N-terminal ATPase/helicase domain, a large central domain, and a C-terminal polymerase domain that shares homology with A-family DNA Pols such as Escherichia coli Pol I (Yousefzadeh and Wood 2013). We have shown previously that Polθ comprised of residues 1708–2590 performs proficient TLS opposite thymine glycol in human cells (Yoon et al. 2014) and provide evidence here that Polθ (1708–2590) is sufficient for TLS opposite εdA. Whereas TLS in control siRNA-treated HFs occurs with a frequency of ∼25%, this frequency declines to ∼15% in cells depleted for genomic Polθ and carrying either an empty vector or one expressing siRNA-sensitive WT Polθ (1708–2590) (Supplemental Table S4). Expression of an siRNA-resistant WT Polθ (1708–2590), however, restores normal TLS levels in HFs depleted for genomic Polθ, confirming that the C-terminal polymerase domain is sufficient for Polθ-dependent TLS opposite the εdA lesion. TLS was reduced to ∼15% in HFs expressing the siRNA-resistant Polθ (1708–2590) D2540A, E2541A mutant protein defective in DNA synthesis, indicating that Polθ polymerase activity is required for its role in TLS opposite εdA (Supplemental Table S4). We confirmed the requirement of Polθ polymerase activity for TLS opposite εdA in Polθ−/− MEFs (Supplemental Table S5).
The Polι/Polζ pathway conducts error-free TLS opposite εdA in human cells
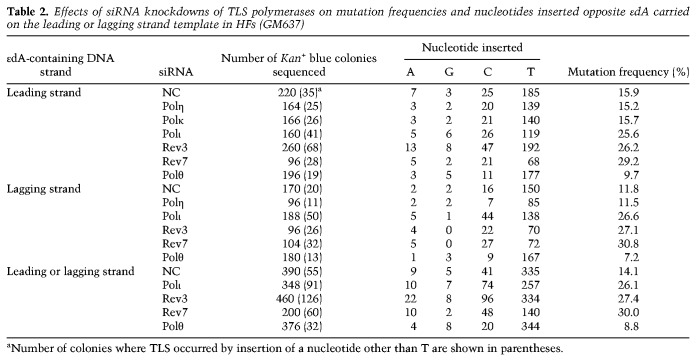
TLS opposite εdA incurs a high level of mutagenesis (Table 2). In control (NC) siRNA-treated cells, ∼16% of Kan+ blue colonies derived from TLS opposite εdA carried on the leading strand template harbored a mutation that resulted from the incorporation of primarily a C, but also of an A or a G opposite εdA. Of the incorrect nucleotides, C was incorporated at a frequency of ∼11%, whereas the A and G nucleotides were incorporated at a combined frequency of ∼5% (Table 2). As expected from the lack of requirement of Polη or Polκ for TLS opposite εdA, the spectrum and frequency of mutagenic TLS opposite this adduct was unchanged upon their depletion. Importantly, depletion of Polι, Rev3, or Rev7 raised the frequency of mutagenic TLS to ∼27% and Polθ depletion reduced mutagenic TLS to ∼10% (Table 2). These results as well as analyses of mutagenic TLS opposite εdA carried on the lagging strand template indicated that Polι/Polζ-dependent TLS operates in an error-free manner, whereas Polθ-mediated TLS is error-prone.
Table 2.
Effects of siRNA knockdowns of TLS polymerases on mutation frequencies and nucleotides inserted opposite εdA carried on the leading or lagging strand template in HFs (GM637)
To get an estimate of the overall contributions of the Polι/Polζ and Polθ pathways to error-free and mutagenic TLS, respectively, we pooled the mutation data for the two DNA strands. Overall, mutagenic TLS opposite εdA in WT HFs occurs with a frequency of ∼14%, this frequency rises to ∼27% in cells depleted for Polι or Polζ, and declines to ∼9% in Polθ-depleted cells (Table 2). Whereas incorporation of C occurs with a frequency of ∼10% in control siRNA-treated cells, it rises to ∼20% in cells depleted for Polι or Polζ and declines to ∼5% in Polθ-depleted cells. These data suggested that Polθ-mediated TLS contributes to C misincorporation opposite εdA and raised the possibility that Rev1 polymerase activity contributes to mutagenic TLS that persists in Polθ-depleted cells.
The DNA polymerase activities of Polθ and Rev1 account for error-prone TLS opposite εdA
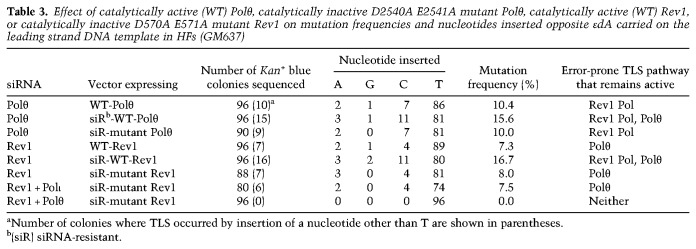
To test whether the Rev1 polymerase activity contributes to mutagenic TLS and to determine whether Rev1 and Polθ polymerase activities provide alternative routes of mutagenic TLS, we analyzed the effects of mutations in active site residues in their polymerase domains on mutagenic TLS opposite εdA. As shown in Table 3, in Polθ-depleted cells expressing siRNA-sensitive WT Polθ, ∼10% of TLS products harbor mutations, and the frequency of mutagenic TLS rises to ∼16% in cells expressing siRNA-resistant WT Polθ, suggesting that ∼6% of mutations result from Polθ-mediated TLS. Expression of the siRNA-resistant catalytic mutant of Polθ also reduced mutation frequency to ∼10%, indicating that Polθ’s polymerase activity is required for its role in mutagenic TLS. In Rev1-depleted cells expressing siRNA-sensitive WT Rev1, mutagenic TLS occurs at a frequency of ∼7%, and expression of the siRNA-resistant WT Rev1 raises the frequency of mutagenic TLS to ∼17% (Table 3). This result indicated the requirement of Rev1 for mutagenic TLS. To establish that Rev1 polymerase activity was required for mutagenic TLS and that Rev1 functions in this role independent of Polι, we expressed siRNA-resistant Rev1 catalytic mutant in Rev1-depleted cells or in cells codepleted for Rev1 and Polι (Table 3, items 6 and 7). In both cases, mutagenic TLS was reduced to 8%, confirming the Polι-independent role of Rev1 polymerase in mutagenic TLS. The observation that codepletion of Rev1 and Polθ in cells expressing siRNA-resistant Rev1 catalytic mutant causes a complete inhibition of mutagenic TLS (Table 3, last item) supports the conclusion that Rev1 polymerase and Polθ polymerase activities provide alternative routes for mutagenic TLS opposite εdA (Table 3). Since no mutations occur in cells lacking Polθ and Rev1 polymerase activity but retaining the Rev1 scaffolding function, where only the Polι/Polζ pathway would remain functional, Polι/Polζ-dependent TLS must operate in an error-free manner (Table 3, last item).
Table 3.
Effect of catalytically active (WT) Polθ, catalytically inactive D2540A E2541A mutant Polθ, catalytically active (WT) Rev1, or catalytically inactive D570A E571A mutant Rev1 on mutation frequencies and nucleotides inserted opposite εdA carried on the leading strand DNA template in HFs (GM637)
To further ascertain the contributions of Rev1 polymerase-dependent and Polθ-dependent pathways to mutagenic TLS and to verify the requirement of Polι for error-free TLS in the Polι/Polζ pathway, we analyzed the mutation spectrum of TLS products in WT and Polθ−/− MEFs, and in Rev1−/− MEFS lacking or expressing the catalytic mutant Rev1 protein (Supplemental Table S6). The effects of Polθ and Rev1 knockouts on the frequency and mutation spectrum of TLS products are remarkably similar to that in HFs depleted for these Pols (Table 3) and the results that no mutations are recovered in Rev1−/− MEFs expressing the catalytic mutant Rev1 protein and depleted for Polθ (Supplemental Table S6) provide confirmatory evidence that Rev1 polymerase and Polθ provide alternate pathways of mutagenic TLS, and that the Polι/Polζ pathway, which would remain functional in these MEFs, operates in an error-free manner.
Biochemical analysis of Polθ polymerase activity for TLS opposite εdA
For these studies, we examined DNA synthesis by Pol θ across from either an undamaged A or εdA. Whereas opposite an undamaged A template Polθ inserts a T but also a C or a G (Fig. 1, lanes 1–5), opposite εdA Polθ primarily inserts an A (Fig. 1, lanes 9,4,19). Although Polθ can insert each of the four dNTPs opposite εdA, importantly, in the presence of all four dNTPs, dATP is preferentially inserted (Fig. 1, lane 10). Since dATP was preferentially incorporated opposite εdA in a template in which the downstream 5′ template nucleotide was a T residue, we determined whether Polθ used a frameshift mechanism to incorporate dATP opposite the downstream template T rather than opposite εdA. To examine this, we used templates containing εdA that harbor either an A or a G as the next templating residue downstream from the lesion (Fig. 1, lanes 11–20). In each case, an A was the best incorporated nucleotide opposite εdA, suggesting that Polθ is not frameshifting the template and using the downstream nucleotide, for if it were, then the pattern of insertion would be different for each substrate and a preference for the W–C partner of the downstream nucleotide would be observed.
Figure 1.
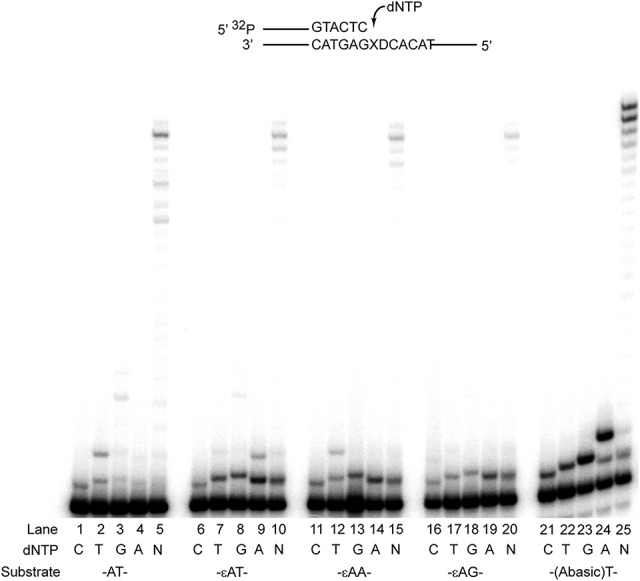
Nucleotide incorporation opposite A or εdA by Polθ. 0.5 or 5 nM Polθ was incubated with 10 nM DNA substrate and 25 µM of either dGTP, dATP, dTTP, dCTP, or all four dNTPs for 5 min at 37°C. Reactions containing a single or all four dNTPs (N) are indicated. The DNA substrate used in lanes 1–5 harbor undamaged template A at the primer terminus, and reactions contained 0.5 nM Polθ. The substrate in lanes 6–20 harbored εdA at the primer terminus followed by either T (lanes 6–10), A (lanes 11–15), or G (lanes 16–20) residue, indicated by D in the template sequence. Synthesis opposite an abasic site is shown in lanes 21–25. Reactions in lanes 6–25 were all carried out with 5 nM Polθ. X in the template sequence indicates the position of undamaged A, εdA, or an abasic site.
Polθ exhibits a high proficiency for replicating through an abasic site by inserting an A opposite it and by extending synthesis (Fig. 1, lanes 24,25; Seki et al. 2004). Although Polθ is more efficient in performing TLS opposite an abasic site than opposite εdA, the similar pattern of nucleotide incorporation opposite the two DNA lesions (Fig. 1) suggests that the εdA adduct becomes extrahelical and Polθ incorporates an A opposite an abasic site-like mode.
Biochemical analysis of Rev1 polymerase activity for TLS opposite εdA
Rev1 specifically incorporates dCTP opposite template G (Nelson et al. 1996; Haracska et al. 2002). In the Rev1 active site, template G, and the incoming dCTP do not pair with each other; instead, the template G residue is evicted from the DNA helix and makes hydrogen bonds via its Hoogsteen edge with the amino acids in the G loop of Rev1, while an Arg residue in Rev1 forms hydrogen bonds with the incoming dCTP (Nair et al. 2005; Swan et al. 2009). Rev1 favors nucleotide incorporation opposite template G over other nucleotides because the G loop makes specific hydrogen bonds with the Hoogsteen edge of templating G. Incorporation of dCTP opposite template A is much less favorable, likely due to the presence of the N6 H-bond donor on adenine which would sterically hinder binding by the G loop. Since εdA lacks the N6 amino group (Supplemental Fig. S1A), it would not sterically clash with the G loop. Accordingly, we found that under identical conditions, nucleotide incorporation opposite εdA is better than opposite template A (Supplemental Fig. S3). In fact, yeast Rev1 incorporates dCTP opposite εdA with a catalytic efficiency only approximately fivefold less than template G, whereas opposite template A, yeast Rev1 is ∼200-fold less efficient (data not shown). This proposed better binding of εdA by the Rev1 G loop over template A would also account for the incorporation of dGTP and dTTP mispairs opposite εdA that is not observed opposite template A (Supplemental Fig. S3, lanes 1,3 vs. 6,8,11,13,16,18). Since in the presence of all four dNTPs (Supplemental Fig. S3, lanes 10,15,20), Rev1 preferentially incorporates dCTP, dCTP is incorporated opposite εdA with the highest catalytic efficiency.
Pathways for replicating through εdA in human cells
Based upon our observations in HFs, and in WT, Polθ−/−, Rev1−/− MEFs, we conclude that replication through the εdA adduct is mediated via two Rev1-dependent TLS pathways in which Rev1 functions as a noncatalytic component of Polι in the Polι/Polζ pathway and as a DNA polymerase that functions independently of the Polι/Polζ pathway and via a third pathway dependent on Polθ (Supplemental Fig. S4). Polι/Polζ-dependent TLS operates in an error-free manner, Rev1 polymerase activity makes a relatively minor contribution to TLS and it acts in an error-prone manner, and Polθ polymerase activity provides the alternative route of mutagenic TLS.
Role of Polι/Polζ-dependent TLS in error-free replication through εdA in human cells
Although structural studies have shown that Polι can accommodate dTTP or dCTP opposite εdA via Hoogsteen base pairing, and biochemical studies have shown that Polι incorporates a T opposite εdA with only an approximately fourfold higher catalytic efficiency than a C (Nair et al. 2006), our genetic studies reveal that Polι-dependent TLS opposite this lesion operates in an error-free manner in human cells. In the Polι active site, εdA is pushed into a syn conformation and it forms a Hoogsteen base pair with the incoming T or C residue (Nair et al. 2006). While the ability of Polι to push the adduct into the syn conformation explains its proficiency for TLS opposite εdA, it does not explain how Polι avoids C incorporation in human cells. To explain the high fidelity of Polι opposite εdA in human cells, we suggest that TLS Pols function as a component of a multiprotein ensemble and that their fidelity is actively regulated in such an ensemble by protein–protein interactions and posttranslational modifications. A similar mechanism would account for predominantly error-free replication by TLS Pols through other DNA lesions (Yoon et al. 2009, 2010, 2017, 2018; Conde et al. 2015).
Reconfiguration of Polθ active site opposite εdA in human cells
Similar to other A family Pols, Polθ incorporates nucleotides opposite undamaged A, G, C, or T templates via W–C base pairing. Since εdA disrupts W–C base pairing, purified Polθ could replicate through this adduct by pushing it out of the DNA helix into an abasic-like mode, then inserting an A opposite the adduct site and extending synthesis. The feasibility of this scenario is suggested from the proficient ability of Polθ to replicate through an abasic site where it inserts an A opposite the abasic site and then extends synthesis. Structural studies with Polθ have verified its ability to insert an A opposite an abasic site (Zahn et al. 2015).
Even though Polθ contributes to error-prone TLS opposite εdA in human/mouse cells, in both HFs and MEFs, it incorporates a T at a frequency of ∼92%, a C at ∼5%, and an A at ∼3% (Table 3; Supplemental Table S6). Thus in spite of the propensity of purified Polθ for incorporating an A opposite εdA, Polθ-mediated TLS opposite this adduct is predominantly error-free in human cells. Polθ could accomplish this by pushing εdA into a syn conformation and by forming a Hoogsteen base pair with the incoming T. The low frequency of C insertion could also occur by similar means. Although in its potential to form a Hoogsteen base pair between εdA in syn conformation and an incoming T or C in anti conformation Polθ would resemble Polι, the two Pols would differ in a number of ways. Polι has a preformed active site which enables it to similarly position an undamaged A or an εdA in the syn configuration; hence, Polι would adopt a similar mechanism for performing TLS in human cells as it portrays in biochemical assays. In striking contrast, Polθ would need to adopt entirely different means for conducting TLS opposite εdA in human cells than those adopted by purified Polθ.
To explain the adoption of different TLS mechanisms by Polθ in human cells vs in biochemical assays with purified enzyme, we suggest that Polθ functions in TLS in human cells as a component of a multiprotein ensemble and that protein–protein interactions and posttranslational modifications in that ensemble actively modulate Polθ active site such that the εdA adduct is pushed and stabilized in a syn conformation, allowing for Hoogsteen base pairing primarily with the T. Thus, whereas protein–protein interactions in the Polθ-containing multiprotein ensemble would actively reconfigure Polθ active site for Hoogsteen base pairing opposite εdA enabling it to conduct relatively high fidelity TLS in human cells, in Polι with its preformed active site, the overall mechanism of Hoogsteen base pairing would stay the same in the multiprotein ensemble in human cells as with purified Pol but its fidelity opposite εdA will be greatly enhanced in human cells.
Materials and methods
Construction of εdA plasmid vectors and translesion synthesis assays
The 16-mer oligonucleotides containing εdA, purchased from the Midland Certified Reagent Company, Inc., and the in-frame target sequence of lacZ′ gene in the resulting vectors are shown in Supplemental Figure S1B. The WT kanamycin gene (Kan+) was placed on the same DNA strand as εdA and lacz′ in this DNA strand is in-frame and functional for β-gal. The opposite DNA strand harbors an SpeI restriction site containing a +1 frameshift, which makes it nonfunctional for β-gal. This DNA strand carries the Kan− gene. The detailed methods for construction of lesion containing SV40 duplex plasmid, for assays for translesion synthesis, and for mutation analyses of TLS products have been published previously (Yoon et al. 2009, 2010).
Protein expression and purification
Rev1 core (amino acid residues 330–883) and Polθ core (amino acid residues 1708–2590) proteins were each expressed and purified from yeast as fusion proteins harboring an N-terminal Glutathione-S transferase (GST) tag as described (Johnson et al. 2006). Rev1 and Polθ proteins were expressed from plasmids pBJ1228 and pPOL507, respectively. The GST tags were removed by prescisson protease during purification.
DNA polymerase assays
Proteins were assayed using the standard DNA polymerase reaction conditions (Johnson et al. 2006). The DNA substrate consisted of the 52mer temple 5′-TTCGTATAATGCCTACACDXGAGTACCGGAGCATCGTCGTGACTGGGAAAAC-3′, where D indicates either G, A, or T and X indicates either an undamaged A or εdA annealed to the 32mer 32P radiolabeled primer 5′-GTTTTCCCAGTCACGACGATGCTCCGGTACTC-3′. Reactions (5 µL), carried out for 5 min at 37°C, contained 10 nM DNA substrate, 25 µM dNTP and protein concentrations as indicated in the figure legends. Reaction products were separated by 15% TBE PAGE containing 8 M urea as described, and visualized by phosphorimaging on a Typhoon FLA 7000 (GE Healthcare).
Supplementary Material
Acknowledgments
These studies were supported by National Institutes of Health grants ES022948 and GM126087.
Author contributions: J.H.Y. performed the genetic experiments and analyzed the data; R.E.J. performed biochemical experiments and analyzed the data; L.P. and S.P. designed and coordinated the study; J.H.Y., R.E.J., L.P., and S.P. wrote the paper.
Footnotes
Supplemental material is available for this article.
Article published online ahead of print. Article and publication date are online at http://www.genesdev.org/cgi/doi/10.1101/gad.320531.118.
References
- Chung F-L, Zhang L, Ocando JE, Nath RG. 1999. Role of 1,N2-propanodeoxygunosine adducts as endogenous DNA lesions in rodents and humans. IARC Sci Publ 150: 45–53. [PubMed] [Google Scholar]
- Conde J, Yoon JH, Roy Choudhury J, Prakash L, Prakash S. 2015. Genetic control of replication through N1-methyladenine in human cells. J Biol Chem 290: 29794–29800. 10.1074/jbc.M115.693010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Haracska L, Prakash S, Prakash L. 2002. Yeast Rev1 protein is a G template-specific DNA polymerase. J Biol Chem 277: 15546–15551. 10.1074/jbc.M112146200 [DOI] [PubMed] [Google Scholar]
- Johnson RE, Prakash L, Prakash S. 2006. Yeast and human translesion DNA synthesis polymerases: expression, purification, and biochemical characterization. Methods Enzymol 408: 390–407. 10.1016/S0076-6879(06)08024-4 [DOI] [PubMed] [Google Scholar]
- Luczaj W, Skrzydlewska E. 2003. DNA damage caused by lipid peroxidation products. Cell Mol Biol Lett 8: 391–413. [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. 2005. Rev1 employs a novel mechanism of DNA synthesis using a protein template. Science 309: 2219–2222. 10.1126/science.1116336 [DOI] [PubMed] [Google Scholar]
- Nair DT, Johnson RE, Prakash L, Prakash S, Aggarwal AK. 2006. Hoogsteen base pair formation promotes synthesis opposite the 1,N6-ethenodeoxyadenosine lesion by human DNA polymerase ι. Nat Struct Mol Biol 13: 619–625. 10.1038/nsmb1118 [DOI] [PubMed] [Google Scholar]
- Nelson JR, Lawrence CW, Hinkle DC. 1996. Deoxycytidyl transferase activity of yeast REV1 protein. Nature 382: 729–731. 10.1038/382729a0 [DOI] [PubMed] [Google Scholar]
- Seki M, Masutani C, Yang LW, Schuffert A, Shigenori I, Bahar I, Wood RD. 2004. High-efficiency bypass of DNA damage by human DNA polymerase Q. EMBO J 23: 4484–4494. 10.1038/sj.emboj.7600424 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Swan MK, Johnson RE, Prakash L, Prakash S, Aggarwal AK. 2009. Structure of the human REV1-DNA-dNTP ternary complex. J Mol Biol 390: 699–709. 10.1016/j.jmb.2009.05.026 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Prakash L, Prakash S. 2009. Highly error-free role of DNA polymerase η in the replicative bypass of UV-induced pyrimidine dimers in mouse and human cells. Proc Natl Acad Sci 106: 18219–18224. 10.1073/pnas.0910121106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, Prakash L, Prakash S. 2010. Error-free replicative bypass of (6-4) photoproducts by DNA polymerase ζ in mouse and human cells. Genes Dev 24: 123–128. 10.1101/gad.1872810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. 2014. A role for DNA polymerase θ in promoting replication through oxidative DNA lesion, thymine glycol, in human cells. J Biol Chem 289: 13177–13185. 10.1074/jbc.M114.556977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Park J, Conde J, Wakamiya M, Prakash L, Prakash S. 2015. Rev1 promotes replication through UV lesions in conjunction with DNA polymerases η, ι, and κ but not DNA polymerase ζ. Genes Dev 29: 2588–2662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Roy Choudhury J, Park J, Prakash S, Prakash L. 2017. Translesion synthesis DNA polymerases promote error-free replication through the minor-groove DNA adduct 3-deaza-3-methyladenine. J Biol Chem 292: 18682–18688. 10.1074/jbc.M117.808659 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon JH, Hodge RP, Hackfeld LC, Park J, Roy Choudhury J, Prakash S, Prakash L. 2018. Genetic control of predominantly error-free replication through an acrolein-derived minor-groove DNA adduct. J Biol Chem 293: 2949–2958. 10.1074/jbc.RA117.000962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J-H, McArthur MJ, Park J, Basu D, Wakamiya M, Prakash L, Prakash S. 2019. Error-prone replication through UV lesions by DNA polymerase θ protects against skin cancers. Cell 10.1016/j.cell.2019.01.023 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yousefzadeh MJ, Wood RD. 2013. DNA polymerase POLQ and cellular defense against DNA damage. DNA Repair (Amst) 12: 1–9. 10.1016/j.dnarep.2012.10.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zahn KE, Averill AM, Aller P, Wood RD, Doublié S. 2015. Human DNA polymerase θ grasps the primer terminus to mediate DNA repair. Nat Struct Mol Biol 22: 304–311. 10.1038/nsmb.2993 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.