Abstract
In translation initiation, a 43S preinitiation complex (PIC) containing eIF1 and a ternary complex (TC) of GTP-bound eIF2 and Met-RNAi scans the mRNA for the start codon. AUG recognition triggers eIF1 release and rearrangement from an open PIC conformation to a closed state with more tightly-bound Met-tRNAi (PIN state). Cryo-EM models reveal eIF2β contacts with eIF1 and Met-tRNAi exclusive to the open complex that should destabilize the closed state. eIF2β or eIF1 substitutions disrupting these contacts increase initiation at UUG codons, and compound substitutions also derepress translation of GCN4, indicating slower TC recruitment. The latter substitutions slow TC loading while stabilizing TC binding at UUG codons in reconstituted PICs, indicating a destabilized open complex and shift to the closed/PIN state. An eIF1 substitution that should strengthen the eIF2β:eIF1 interface has the opposite genetic and biochemical phenotypes. eIF2β is also predicted to restrict Met-tRNAi movement into the closed/PIN state, and substitutions that should diminish this clash increase UUG initiation in vivo and stabilize Met-tRNAi binding at UUG codons in vitro with little effect on TC loading. Thus, eIF2β anchors eIF1 and TC to the open complex, enhancing PIC assembly and scanning, while impeding rearrangement to the closed conformation at non-AUG codons.
INTRODUCTION
The process of translation initiation selects the start codon for protein synthesis, determining the reading frame for elongation and N-terminus of the polypeptide, and also influencing the rate at which the protein is synthesized. Accordingly, accurate identification of the translation start codon is critical to ensure the synthesis of the correct cellular proteins in the proper amounts. In eukaryotes, translation initiation occurs via a scanning mechanism, wherein the small (40S) subunit of the ribosome recruits methionyl initiator tRNA (Met-tRNAi) in a ternary complex (TC) with GTP-bound eukaryotic initiation factor 2 (eIF2), in a reaction stimulated by factors eIF1, eIF1A and eIF3. The resulting 43S preinitiation complex (PIC) attaches to the 5′ end of mRNA and scans the mRNA leader for an AUG start codon. The nucleotide sequence immediately surrounding the start codon—the AUG context—particularly at the −3 and +4 positions (numbered from the A of AUG (+1)) also influences the efficiency of start codon selection. In the scanning PIC, eIF1 and eIF1A promote an open, scanning-conducive conformation of the 40S subunit with TC bound in a relatively unstable conformation, ‘POUT’, which facilitates the inspection of successive triplets in the peptidyl (P) decoding site for complementarity with the anticodon of Met-tRNAi. The GTP bound to eIF2 can be hydrolyzed, stimulated by GTPase activating protein eIF5, but eIF1 blocks release of inorganic phosphate (Pi) at non-AUG codons. Start codon recognition triggers dissociation of eIF1 from the 40S subunit, enabling both Pi release from eIF2-GDP·Pi and more stable TC binding to the PIC, with Met-tRNAi more fully accommodated in the ‘PIN’ state. Subsequent dissociation of eIF2-GDP and other eIFs from the 48S PIC enables eIF5B-catalyzed subunit joining and formation of an 80S initiation complex ready to commence protein synthesis (1).
eIF1 plays a dual role in translation initiation. It promotes the open conformation of the PIC, to which TC rapidly loads in the POUT conformation, and it ensures initiation fidelity by blocking Pi release and impeding isomerization to the closed/PIN state at non-AUG codons or start codons in poor context (Figure 1A). Structural analyses of the PIC reveal that eIF1 and eIF1A promote rotation of the 40S head relative to the body, which likely enhances TC binding, while eIF1 clashes with Met-tRNAi in the PIN state. Hence, eIF1 dissociation from the 40S subunit is required for start codon recognition, and mutations that weaken eIF1 binding to 40S subunits confer dual defects in vivo: (i) they reduce the rate of TC loading, since eIF1 promotes TC binding to the open conformation of the PIC; and (ii) they increase initiation at near cognate codons or AUG codons in poor context, by destabilizing the open/POUT state and favoring rearrangement to the closed/PIN state (1,2). A reduced rate of TC loading resulting from such eIF1 mutations confers derepressed translation of GCN4 mRNA in vivo (the Gcd− phenotype), as slower TC binding to PICs scanning the GCN4 mRNA leader allows inhibitory upstream open reading frames (uORFs) to be bypassed in favor of reinititation further downstream at the GCN4 coding sequence (3). Increased initiation at near-cognate codons in such eIF1 mutants also restores translation of his4-303 mRNA, lacking the AUG start codon, by elevating initiation at an in-frame UUG triplet at the third codon (the Sui− phenotype) (4) (Figure 1B). The AUG codon of the eIF1 gene itself (SUI1 in yeast) occurs in suboptimal context and the frequency of its recognition is inversely related to eIF1 abundance, establishing a negative feedback loop that maintains proper eIF1 levels (5,6). Whereas overexpressing WT eIF1 suppresses initiation at its own suboptimal AUG codon, eIF1 mutants defective for 40S binding relax discrimination against poor context and increase the translational efficiency of SUI1 mRNA, elevating expression of such eIF1 variants. These effects have been attributed to the altered rates of eIF1 dissociation from the scanning PIC, impeding or enhancing, respectively, inappropriate isomerization to the closed state at non-AUG codons or at AUG codons in poor context (7,8).
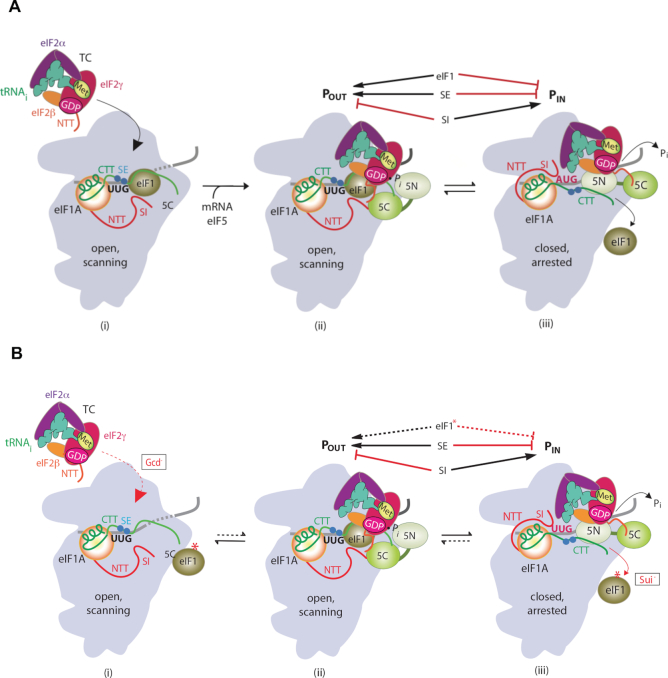
Figure 1.
Model describing conformational rearrangements of the PIC during scanning and start codon recognition and the consequences of Sui− substitutions in eIF1. (A) Assembly of the PIC, scanning and start codon selection in WT cells. (i) eIF1 and the scanning enhancer (SE) elements in the CTT of eIF1A stabilize an open conformation of the 40S subunit to which TC rapidly loads. (ii) The 43S PIC in the open conformation scans the mRNA for the start codon with Met-tRNAi bound in the POUT state. The GAP domain in the N-terminal domain of eIF5 (5N) stimulates GTP hydrolysis by the TC to produce GDP-Pi, but release of Pi is blocked. The unstructured NTT of eIF2β interacts with eIF1 to stabilize eIF1•40S association and the open conformation. (iii) On AUG recognition, the Met-tRNAi moves from the POUT to PIN state, clashing with eIF1. Movement of eIF1 away from the P site disrupts its interaction with the eIF2β-NTT, and the latter interacts with the eIF5-CTD instead. eIF1 dissociates from the 40S subunit, and the eIF1A SE elements move away from the P site. The eIF5-NTD dissociates from eIF2 and interacts with the 40S subunit and the eIF1A CTT, facilitating Pi release and blocking reassociation of eIF1 with the 40S subunit. (Above) The arrows summarize that eIF1 and the eIF1A SE elements promote POUT and block the transition to the PIN state, whereas the scanning inhibitor (SI) elements in the NTT of eIF1A stabilize the PIN state. (Adapted from 7,28,29.) (B) An eIF1 substitution that weakens that factor's binding to the 40S subunit destabilizes the open/POUT conformation, reducing the rate of TC loading and increasing selection of near-cognate (UUG) start codons. (i) Aberrant dissociation of mutant eIF1 from the 40S subunit reduces the prevalence of the open/POUT conformation, decreasing the rate of TC loading and conferring the Gcd− phenotype (red dotted arrow). (ii, iii) Once TC eventually binds to the PIC and scanning commences, an increased frequency of mutant eIF1 dissociation from the open/POUT conformation enables more frequent rearrangement to the PIN state at UUG codons, conferring the Sui− phenotype (red solid arrow).
Many mechanistic aspects of the translation initiation process remain unclear, including the exact molecular mechanism by which recognition of a start codon triggers isomerization of the PIC from the open/POUT state to the closed/PIN state and the cessation of scanning. Cryo-electron microscopy (cryo-EM) structures of two distinct py48S complexes, both containing eIF1, eIF1A, mRNA and TC, but appearing to represent distinct intermediates in scanning and start codon recognition (py48S-open and py48S-closed), shed light on this process (9). The py48S-open complex exhibits an upward movement of the 40S head from the body that widens both the mRNA binding cleft and the P site, eliminating certain 40S contacts with the mRNA and Met-tRNAi that occur only in py48S-closed. Conversely, the closed structure shows a constricted mRNA channel and narrowed P site that encloses Met-tRNAi. Comparing these two structures enables predictions about the factors and specific residues preferentially stabilizing either the open or the closed state of the PIC. Further, these structures show clear density for the globular portion of the β-subunit of eIF2, unresolved in previous PIC structures, allowing predictions about the molecular functions of eIF2β in TC loading and start codon recognition.
Evidence from the cryo-EM structures indicates that the helix-turn-helix (HTH) domain of eIF2β forms contacts with eIF1 that are specific to the open conformation of the PIC, as the HTH domain moves away from eIF1 during rearrangement to the closed complex (9) (Figure 2A). We reasoned that substitutions in the eIF2β HTH domain that perturb these contacts would promote premature eIF1 release, and thereby increase the likelihood of isomerization to the closed state at UUG codons or AUG codons in suboptimal context, conferring the Sui− phenotype and increasing eIF1 expression from SUI1 mRNA containing native, poor context. The loss of contacts between eIF1 and eIF2β specific to the open state is also expected to reduce eIF1 occupancy of the open PIC and reduce the rate of TC loading to confer the Gcd− phenotype. This is the same collection of phenotypes noted above for eIF1 mutations that weaken its 40S binding.
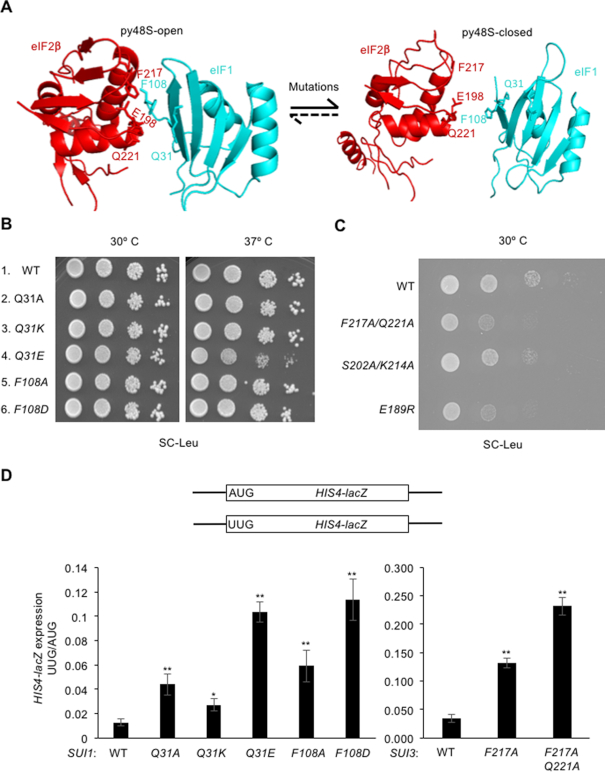
Figure 2.
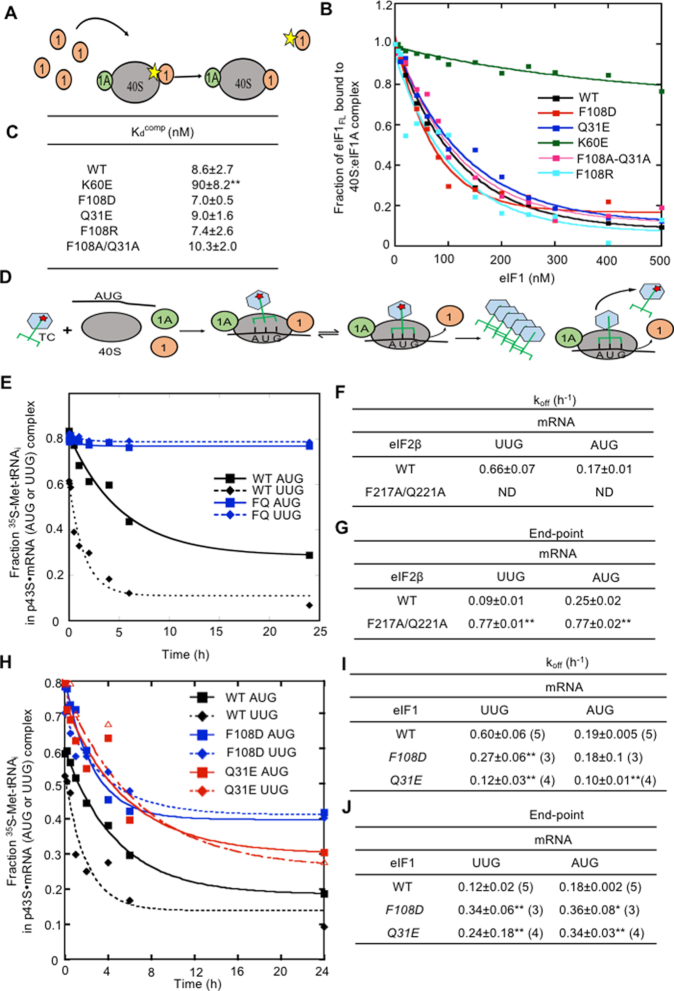
Substitutions at the eIF2β:eIF1 interface of the open complex decrease discrimination against UUG initiation codons in vivo. (A) Interactions between eIF2β and eIF1 in the py48S-open and py48S-closed states of the PIC (9). eIF2β is shown in red, eIF1 in cyan. Residues substituted in this study are shown as sticks and labeled. Arrows indicate that substitutions disrupting these contacts favor the closed state by allowing eIF1 release. (B) Ten-fold serial dilutions of JCY03 derivatives with the indicated SUI1 alleles were spotted on synthetic complete medium lacking leucine (SC-Leu) and incubated at 30°C or 37°C for 2–3 days. (C) Ten-fold serial dilutions of KAY18 derivatives with the indicated SUI3 alleles were spotted on SC-Leu and incubated at 30°C for 2 days. (D) HIS4-lacZ reporters with AUG or UUG start codons assayed to calculate UUG:AUG initiation ratios. Derivatives of sui1Δ his4-301 strain JCY03 containing the indicated SUI1 alleles or derivatives of KAY18 containing the indicated SUI3 alleles, each harboring the appropriate HIS4-lacZ reporter, were cultured in synthetic dextrose minimal medium (SD) supplemented with His and Trp at 30°C to A600 of ∼1.0, and β-galactosidase activities (in units of nanomoles of o-nitrophenyl-β-d-galactopyranoside cleaved per min per mg) were measured in whole cell extracts (WCEs). The ratio of expression of the UUG to AUG reporter was calculated from four to six different transformants, and the mean and SEMs were plotted. Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (*P < 0.05; **P < 0.01).
The HTH domain of eIF2β also forms distinct contacts with the Met-tRNAi in the py48S-open and py48S-closed structures. Downward movement of the 40S head towards the body in the open-to-closed transition is associated with movement of the eIF2β-HTH away from the anticodon stem-loop (ASL) of Met-tRNAi (9) (Supplementary Figure S1A and B). We hypothesized that eIF2β-HTH: Met-tRNAi-ASL interactions specific to the open state help to stabilize the Met-tRNAi in the open conformation in the absence of a perfect codon-anticodon duplex and other canonical P site tRNA contacts restricted to the closed complex. They might also enhance the rate of TC loading to the open conformation. If so, then eIF2β substitutions that perturb these eIF2β:Met-tRNAi contacts should destabilize the open PIC conformation and confer the dual Sui− and Gcd− phenotypes predicted above for disruptions at the eIF1:eIF2β interface.
In addition to contacting the Met-tRNAi-ASL in the open conformation, the eIF2β-HTH is predicted to clash with the D-loop of Met-tRNAi bound in the PIN state when eIF2β is bound in the open conformation, as revealed by an overlay of the py48S-open and py48S-closed structures (Supplementary Figure S1C). This clash would occur as a result of Met-tRNAi movement relative to eIF2β in the downward movement of the 40S head toward the body during isomerization to the closed state and is avoided by displacement of eIF2β in py48S-closed relative to py48S-open (9). This structural evidence suggests that eIF2β might act to sterically oppose transition of the PIC to the closed state before an appropriate start codon is recognized by impeding accommodation of Met-tRNAi in the PIN state. Hence, we predicted that substitutions in the eIF2β HTH domain that diminish the predicted clash would favor isomerization to the closed state and increase its likelihood at non-AUG or suboptimal AUG codons. However, these substitutions are not predicted to destabilize the open complex, and should therefore have a minimal effect on the rate of TC loading. As such, they should confer Sui− but not Gcd− phenotypes. This phenotypic signature is rare among known Sui− mutations, most of which act through destabilization of the open PIC, but it has been observed in Sui− substitutions of eIF1 Loop 2, which act by a similar mechanism to alleviate the clash between eIF1 in the open conformation and Met-tRNAi in the closed/PIN state (10).
In an effort to support the physiological relevance of the py48S-open and py48S-closed structures reported by Llacer et al., we previously generated mutations in eIF2β and eIF1 at the eIF1:eIF2β interface and found that they reduced stringent selection of AUG start codons in the manner expected from a role for this interface in promoting the open conformation of the PIC (9). Here, we expanded on these genetic findings with analysis of additional substitutions perturbing the eIF1:eIF2β interface in the open complex, and observed opposing effects on initiation fidelity for substitutions that either weaken this mutual interface (promoting eIF1 release and reducing the stringency of start codon selection) or strengthen it (stabilizing eIF1 binding and increasing the requirement for an AUG start codon in preferred context). We further demonstrated that eIF1:eIF2β interactions promote recognition of optimal Kozak context in addition to stringent AUG selection, as well as the rapid recruitment of TC to the open complex in vivo. Importantly, we recapitulated the effects of exemplar substitutions that weaken eIF1:eIF2β interactions in reducing TC loading and increasing near-cognate start codon selection in a fully purified translation initiation system. A similar in vitro reconstitution was achieved for substitutions at the eIF2β:Met-tRNAi interface found exclusively in the open conformation that both impair TC loading and stringent AUG selection in vivo. Finally, we provide genetic and biochemical evidence that a predicted clash between eIF2β in the open complex with Met-tRNAi in the closed state helps to restrict transition to the closed conformation to AUG start codons, without perturbing the open conformation of the PIC. Together, these findings firmly establish three critical roles for eIF2β in accurate start codon recognition: (i) eIF2β is involved in anchoring eIF1 specifically to the open PIC, promoting rapid TC loading and helping to prevent premature release of eIF1 at non-AUG codons; (ii) eIF2β stabilizes binding of Met-tRNAi specifically to the open conformation of the PIC, when many contacts between Met-tRNAi and the 40S subunit are not yet formed; and (iii) eIF2β functions similarly to eIF1 by impeding rearrangement of Met-tRNAi to the PIN state via a steric clash with Met-tRNAi D-loop, thus helping to limit this transition to AUG start codons in vivo. Hence, eIF2β emerges as a key component of the complex network of physical interactions within the PIC that promote ribosomal scanning and restrict initiation to AUG codons in optimum context.
MATERIALS AND METHODS
Plasmid and yeast strain constructions
Yeast strains used in this study are listed in Supplementary Table S2. Strains ATY100 and ATY122 to ATY128 harboring mutant SUI1 alleles on single-copy (sc) LEU2 plasmids were derived from strain JCY03 (MATa ura3-52 leu2-3, leu2-112 trp1-Δ63 his4-301(ACG) sui1Δ::hisG p1200 (sc URA3 SUI1) by plasmid shuffling, using growth on 5-fluoorotic acid (5-FOA) medium to select for loss of URA3 plasmid p1200 containing WT SUI1 (plasmid-shuffling). The QuikChange site-directed mutagenesis system (Stratagene) was employed with the primers indicated in Supplemental Table S3 to generate all mutant SUI1 plasmids listed in Supplemental Table S1, using as templates plasmids pJCB101 for yeast plasmids, or pTYB2-eIF1 for plasmids used to express recombinant eIF1 proteins in bacteria for purification. The high-copy (hc) plasmid p4385 (hc TC, TRP1) was generated from p1780-IMT (hc TC, URA3) by the ‘marker swap’ approach using the URA3-to-TRP1 converter plasmid pUT11 (11).
To generate strains LMY103-LMY117, LMY130-LMY134 and LMY142 expressing eIF2β variants, strain KAY18 was transformed to Leu+ with single-copy (sc) LEU2 plasmids harboring WT or mutant SUI3 alleles constructed from plasmid p920 (Supplemental Table S1) using the appropriate primers (Supplemental Table S3) and the QuikChange site-directed mutagenesis system (Stratagene) according to the manufacturer's directions. The S202A/K214A double substitution was made sequentially by introducing the S202A mutation into a plasmid containing K214A in a second round of site-directed mutagenesis. The resident WT SUI3, URA3 plasmid p921 was evicted by counter-selection on 5-FOA medium. Strains LMY128, LMY129, and LMY137, used for purification of eIF2 variants containing eIF2β-S202A/K214A, -F217A/Q221A or -E189R, respectively, were constructed from H3840 by plasmid shuffling to replace pAV1089 (containing URA3 and WT SUI3) with high-copy LEU2 plasmids containing the appropriate SUI3 alleles derived from pAV1726 by site-directed mutagenesis, as described above (Supplemental Table S1).
Biochemical assays using yeast cell extracts
Assays of β-galactosidase activity in whole cell extracts (WCEs) were performed as described previously (12). For Western analysis, WCEs were prepared by trichloroacetic acid extraction as previously described (13) and immunoblot analysis was conducted as previously described (14) using antibodies against eIF1/Sui1 (15) and Hcr1 (15). Enhanced chemiluminiscence (Amersham) was used to visualize immune complexes, and signal intensities were quantified by densitometry using NIH ImageJ software.
Biochemical analysis in the reconstituted yeast system
WT eIF1 and eIF1 variants Q31E, F108D, F108R, Q31A/F108A and K60E were expressed in bacterial strain BL21(DE3) Codon Plus cells (Agilent Technologies) and purified using the IMPACT system (New England Biolabs) as described previously (16). His6-tagged WT eIF2 and eIF2 variants were overexpressed in yeast and purified as described (16). 40S subunits were purified as described previously from strain YAS2488 (16). Model mRNAs with sequences 5′-GGAA[UC]7UAUG[CU]10C-3′ and 5′-GGAA[UC]7UUUG[CU]10C-3′ were purchased from Thermo Scientific. Yeast tRNAiMet was synthesized from a hammerhead fusion template using T7 RNA polymerase, charged with [35S]-methionine, and used to prepare radiolabeled eIF2·GDPNP·[35S]-Met-tRNAi ternary complexes ([35S]-TC), all as previously described (16). Charged, unlabeled yeast Met-tRNAiMet was purchased from tRNA Probes, LLC. For eIF1 binding competition experiments, WT eIF1 protein was labeled at its C-terminus with Cys-Lys-ϵ-fluorescein dipeptide, using the Expressed Protein Ligation system, as previously described (17).
TC dissociation rate constants (koff) were measured by monitoring the amount of [35S]-TC that remains bound to 40S·eIF1·eIF1A·mRNA (43S·mRNA) complexes over time, in the presence of excess unlabeled TC (chase), using a native gel shift assay to separate 40S-bound from unbound [35S]-TC. 43S·mRNA complexes were preassembled for 2 h at 26°C in reactions containing 40S subunits (20 nM), eIF1A (1 μM), eIF1 (WT or mutant variants, 1 μM), eIF5-G31R (1 μM) (where indicated), mRNA (10 μM), and [35S]-TC (pre-assembled from 0.25 μM eIF2 (WT or mutant variants), 0.1 mM GDPNP, and 1 nM [35S]-Met-tRNAi) in 60 μl of reaction buffer (30 mM HEPES–KOH (pH 7.4), 100 mM potassium acetate (pH 7.4), 3 mM magnesium acetate, and 2 mM dithiothreitol). To initiate each dissociation reaction, a 6 μl-aliquot of the preassembled 43S·mRNA complexes was mixed with 3 μl of 3-fold concentrated unlabeled TC chase (comprised of 2 μM WT eIF2, 0.3 mM GDPNP and 0.9 μM Met-tRNAi), representing a 300-fold excess over labeled TC in the final dissociation reaction, and incubated for the prescribed period of time. A converging time course was employed so that all dissociation reactions were terminated simultaneously by the addition of native-gel dye and loaded directly on a running native gel. The fraction of [35S]-Met-tRNAi remaining in 43S complexes at each time point was determined by quantifying the 40S-bound and unbound signals by Phosphor Imaging, normalized to the ratio observed at the earliest time-point; and the data were fit with a single exponential equation (18).
TC association rates were measured by mixing [35S]-TC with 40S·eIF1·eIF1A·mRNA complexes and quenching the binding reaction at various times by adding a 300-fold excess of unlabeled WT TC. Reactions were assembled as described above using 6 μl of sample and 3 μl of chase, and completed reactions were mixed with 2 μl of native gel dye before resolving 10 μl by native gel electrophoresis. As above, samples were loaded within minutes of one another on a running native gel. The kobs values were calculated by plotting the fraction of [35S]-Met-tRNAi bound to 40S·eIF1·eIF1A·mRNA complexes against time and fitting the data with a single exponential equation. The resulting kobs values were plotted against the 40S subunit concentrations used in different experiments and the data were fit to a straight line. The slopes of these lines correspond to the second-order rate constants (kon) for TC binding.
Fluorescence anisotropy measurements of equilibrium binding constants (Kd) for eIF1 binding to 40S·eIF1A complexes were performed using a T-format Spex Fluorolog-3 (J.Y. Horiba) as described previously (17). The excitation and emission wavelengths were 497 and 520 nm, respectively. The data were fit with a quadratic equation describing the competitive binding of two ligands to a receptor, as previously described (8).
RESULTS
Substitutions at the eIF2β:eIF1 interface decrease discrimination against suboptimal codons in vivo
Comparing the cryo-EM structures of the py48S-open and py48S-closed complexes reveals contacts between eIF2β and eIF1 restricted to the open complex (9) (Figure 2A) involving a potential stacking interaction of the side chains of Phe residues eIF2β-F217 and eIF1-F108. eIF2β-E198 and -Q221 appear to facilitate this interaction by stabilizing the position of eIF1-F108 via interactions with the protein backbone, and eIF2β-K216 and -G200 interact with eIF1-Q31. In an effort to disrupt the eIF2β-eIF1 interaction from each side of the interface, we introduced substitutions into eIF2β residues E198 and F217, and into eIF1 residues F108 and Q31. We predicted that perturbing the eIF2β:eIF1 interaction at each of these positions would destabilize eIF1 binding to the open complex, thus favoring isomerization to the closed state and increasing initiation at UUG codons.
Mutations introducing eIF2β substitutions were generated in a SUI3 allele under its native promoter on a single-copy LEU2 plasmid, and the resultant mutant alleles were used to replace wild type SUI3 on a URA3 plasmid (plasmid shuffling) by counter-selection on medium containing 5-fluoroorotic acid (5-FOA). Similarly, eIF1 mutations were generated in a SUI1 allele under its native promoter on a single-copy LEU2 plasmid, and the mutant sui1 alleles were used to replace WT SUI1 by plasmid-shuffling. Among the eIF1 substitutions, only eIF1-Q31E conferred a Slg− phenotype, which was restricted to 37°C (Figure 2B, row 4 versus row 1). The eIF2β-E198A substitution was lethal, preventing growth on 5-FOA medium, whereas the double substitution eIF2β-F217A/Q221A produced a slow-growth (Slg−) phenotype (Figure 2C, row 2 versus row 1) at 30°C. Previously, we established that eIF2β-F217A/Q221A is expressed at WT levels and forms a complex with eIF2α and eIF2γ comparably to WT eIF2β (9).
To examine the effects of the eIF2β and eIF1 substitutions on start codon selection, we first measured expression of matched HIS4-lacZ reporters containing either an AUG or UUG start codon. The eIF2β-F217A, eIF1-Q31A, eIF1-Q31K and eIF1-F108A substitutions all produced an ∼2–4 -fold increase of the UUG:AUG ratio, while the double mutant eIF2β-F217A/Q221A and single mutants eIF1-Q31E and eIF1-F108D produced approximately order-of-magnitude increases compared to the WT strains (Figure 2D). The increased UUG:AUG ratios observed for eIF1-F108A, eIF1-F108D, and eIF2β-F217A/Q221A are in accordance with our previous observations on these variants (9). These hypoaccuracy phenotypes of multiple substitutions at the eIF2β:eIF1 interface support the notion that physical contacts between these factors observed exclusively in py48S-open help to stabilize eIF1 binding to the open, scanning conformation of the PIC. The greater increases in the HIS4-lacZ UUG:AUG initiation ratio conferred by replacing Q31 and F108 with acidic residues E or D versus Ala (Figure 2D) might result from electrostatic repulsion with acidic residue E198 of eIF2β beyond the loss of eIF1:eIF2β contacts conferred by the corresponding Ala replacements. The much weaker hypoaccuracy phenotype of the eIF1-Q31K substitution might result from a new salt-bridge that can be formed between this altered residue and eIF2β-E198 (Figure 2D).
Substitutions at the eIF2β:eIF1 interface reduce discrimination against AUG codons in suboptimal context
There is evidence that the presence of eIF1 on the PIC disfavors initiation at AUG codons present in poor Kozak context as well as at non-AUG codons. Thus, eIF1 discriminates against the suboptimal sequence context of its own start codon in SUI1 mRNA to autoregulate its cellular abundance (5,6). Moreover, Sui− eIF1 mutations that elevate UUG initiation also reduce discrimination against the poor context of the SUI1 AUG codon and increase eIF1 abundance (5,8). Consistent with these previous findings, the eIF1-Q31A, Q31E, F108A, F108D and eIF2β-F217A/Q221A substitutions, which elevate UUG initiation on HIS4-lacZ mRNA (Figure 2D), all increase eIF1 abundance to some extent (Figure 3A). Importantly, the acidic substitutions eIF1-Q31E and eIF1-F108D with the strongest hypoaccuracy phenotypes (Figure 2D) also confer the largest increases in eIF1 levels, whereas Q31K, showing the smallest increase in UUG initiation, confers no significant increase in eIF1 abundance (Figure 3A). Furthermore, all of these substitutions except Q31A and Q31K increase expression of a SUI1-lacZ fusion containing the native, poor context of the SUI1 AUG codon (-3CGU-1), but not that of a modified SUI1-lacZ fusion containing optimum context (-3AAA-1) (Figure 3B). In WT cells, the latter SUI1opt-lacZ fusion is expressed at ∼2-fold higher levels than the native SUI1-lacZ fusion. Accordingly, the SUI1opt-lacZ:SUI1-lacZ expression ratio is significantly diminished by eIF1-Q31A, Q31E, F108A, F108D and eIF2β-F217A/Q221A (Figure 3B). It is noteworthy that the strongest reductions in the SUI1opt-lacZ:SUI-lacZ expression ratio resulted from the eIF1-Q31E, eIF1-F108D, and eIF2β-F217A/Q221A substitutions (Figure 3B), which also conferred the greatest increases in UUG initiation among this group of mutants (Figure 2D).
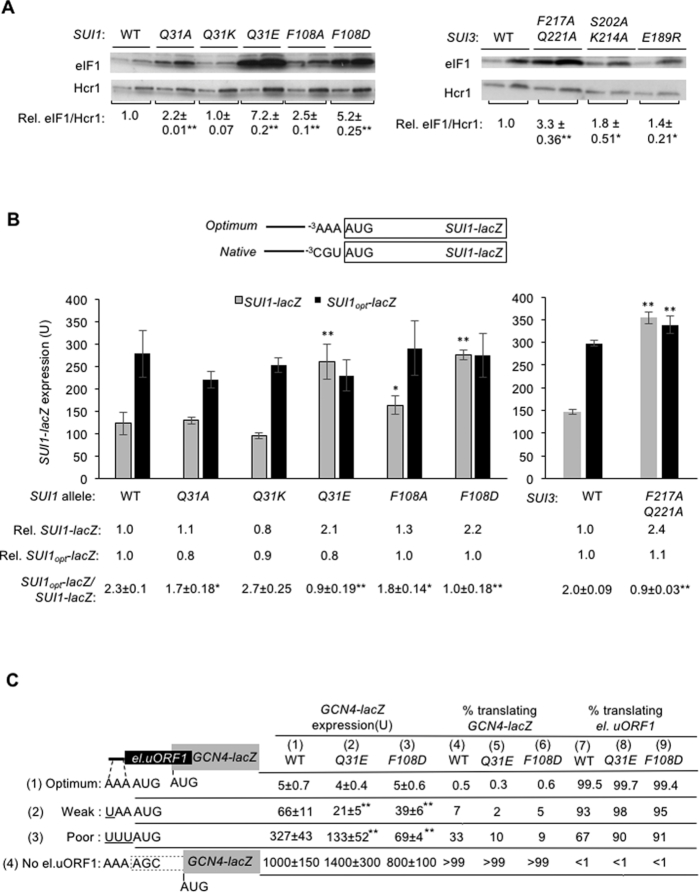
Figure 3.
Substitutions at the eIF2β:eIF1 interface of the open complex decrease discrimination against AUG codons in suboptimal contexts in vivo. (A) Derivatives of sui1Δ his4-301 strain JCY03 containing the indicated SUI1 alleles or derivatives of KAY18 containing the indicated SUI3 alleles were cultured in SD supplemented with His, Trp and Ura at 30°C to A600 of ∼1.0, and WCEs were subjected to western blot analysis using antibodies against eIF1 and Hcr1 (loading control). Two amounts of each extract differing by a factor of two were loaded in successive lanes. eIF1 western signals were normalized to those for Hcr1 and mean values (±SEM) were calculated from seven biological replicates. (B) The same strains as in (A) but harboring sc plasmids (pPMB24 or pPMB25) with SUI1-lacZ fusions with native suboptimal (-3CGU-1) or optimum (-3AAA-1) AUG contexts were cultured and assayed for β-galactosidase activities as in Figure 2B. Mean expression levels and SEMs were calculated from three transformants, and relative (Rel.) mean expression levels normalized to that of the WT strain are listed below, along with expression ratios for the SUI1-lacZ versus SUI1-opt-lacZ reporters. (C) Transformants of JCY03 harboring WT SUI1, sui1-Q31E or sui1-F108D and el.uORF1 GCN4-lacZ reporters (pC3502, pC3503 or pC4466) containing the depicted optimum (row 1), weak (row 2) or poor (row 3) context of uAUG-1, or an uORF-less GCN4-lacZ reporter with a mutated uAUG-1 (pC3505, row 4), were assayed for β-galactosidase activities as in Figure 2B. Mean expression values with SEMs were determined from six transformants (columns 1, 2 and 3). The percentages of scanning ribosomes that translate el.uORF1 (columns 7, 8 and 9) or leaky-scan uAUG-1 and translate GCN4-lacZ instead (columns 4, 5 and 6) were calculated from results in columns 1, 2 and 3 by comparing the amount of expression observed for each uORF-containing reporter to the uORF-less construct, as described in Supplementary Figure S2B and C. (A–C) Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (*P < 0.05; **P < 0.01).
To support the conclusion that these substitutions reduce discrimination against AUGs in poor context, we asked whether they increase recognition of the suboptimal AUG codon of an upstream ORF, and thereby decrease expression of the downstream ORF encoded on the same mRNA. To this end, we assayed expression of GCN4-lacZ reporters containing a single, modified version of upstream ORF1 elongated to overlap the GCN4 main ORF (el.uORF1). With the WT (optimal) context -3AAA-1 for el.uORF1, virtually all scanning ribosomes recognize its AUG codon (uAUG-1) and, because reinitiation at the downstream GCN4 ORF following el.uORF1 translation is nearly non-existent, GCN4-lacZ expression is extremely low (19). In WT cells, replacing only the optimal A with U at the –3 position of el.uORF1 increases leaky scanning of uAUG-1 to produce an ∼7-fold increase in GCN4-lacZ translation. Introducing the poor context of -3UUU-1 further increases leaky scanning, for an ∼33-fold increase in GCN4-lacZ expression, and eliminating uAUG-1 altogether increases GCN4-lacZ expression by >100-fold (Figure 3C, column 1 (WT), rows 1–4 and Supplementary Figure S2A and C). From these results, the percentages of scanning ribosomes that either translate el.uORF1 or bypass (leaky-scan) uAUG-1 and translate GCN4-lacZ instead can be calculated, revealing that >99.5%, ∼93% and ∼67% of scanning ribosomes recognize uAUG-1 in optimum, weak, and poor context, respectively, in WT cells (Figure 3C, columns 4 and 7; see legend, Supplementary Figure S2B).
Subjecting the mutants to this analysis revealed that eIF1 substitution Q31E reduces leaky scanning of uAUG-1, as indicated by significantly reduced GCN4-lacZ expression for the two reporters containing el.uORF1 with weak or poor context, but not for the el.uORF1 reporter with optimum context nor the uORF-less reporter (Figure 3C, cf. columns 1–2, rows 1–4). However, calculating the percentages of ribosomes that recognize uAUG-1 revealed that eIF1-Q31E confers a substantial increase in recognition of uAUG-1 only for the poor-context (UUU) reporter, from ∼67% to ∼90%, while producing only a small increase for the weak-context (UAA) reporter, from ∼93% to ∼98% (Figure 3C, cf. columns 7–8, rows 2–3; Supplementary Figure S2B, Q31E versus WT, Weak & Poor). Similar results were obtained for the eIF1-F108D substitution, increasing recognition of uAUG-1 in poor-context (UUU) from ∼67% to ∼91%; while producing only a slight increase for weak-context (UAA), from ∼93% to ∼95%, and none for optimal context (AAA) (Figure 3C, cf. columns 7 & 9, rows 1–3; Supplementary Figure S2B). Concordant results were also obtained for the eIF1-Q31A and eIF1-F108A substitutions (Supplementary Figure S3A-D). Together, the results show that these eIF1 substitutions reduce discrimination against poor context, both for the SUI1 AUG codon and uAUG-1 of GCN4 mRNA, to an extent that generally parallels their effects in relaxing discrimination against UUG start codons at HIS4. We conclude that the eIF2β:eIF1 substitutions reduce discrimination against AUG codons in poor context and UUG start codons to similar extents.
Substitutions at the eIF2β:eIF1 interface reduce the rate of TC loading in vivo
Mutations that destabilize the open conformation of the PIC typically confer dual Sui− and Gcd− phenotypes, such as the K60E substitution in eIF1 Loop 1 that reduces eIF1 affinity for the 40S subunit (8). Because eIF1 promotes TC loading on the open conformation of the PIC (20), the reduced 40S occupancy of eIF1-K60E slows TC binding and confers derepression of GCN4 translation in vivo (Gcd− phenotype) as revealed using a GCN4-lacZ reporter (Figure 4A, columns 1–2). The 40S binding defect also enables inappropriate dissociation of eIF1-K60E from the scanning PIC at non-AUG codons to elevate UUG initiation (8). In contrast, substitutions in eIF1 Loop 2 that remove steric or electrostatic clashes with Met-tRNAi that impede transition to the closed/PIN state, but do not affect eIF1:40S association, increase UUG initiation without reducing the rate of TC loading in vivo (Gcd+) (10).
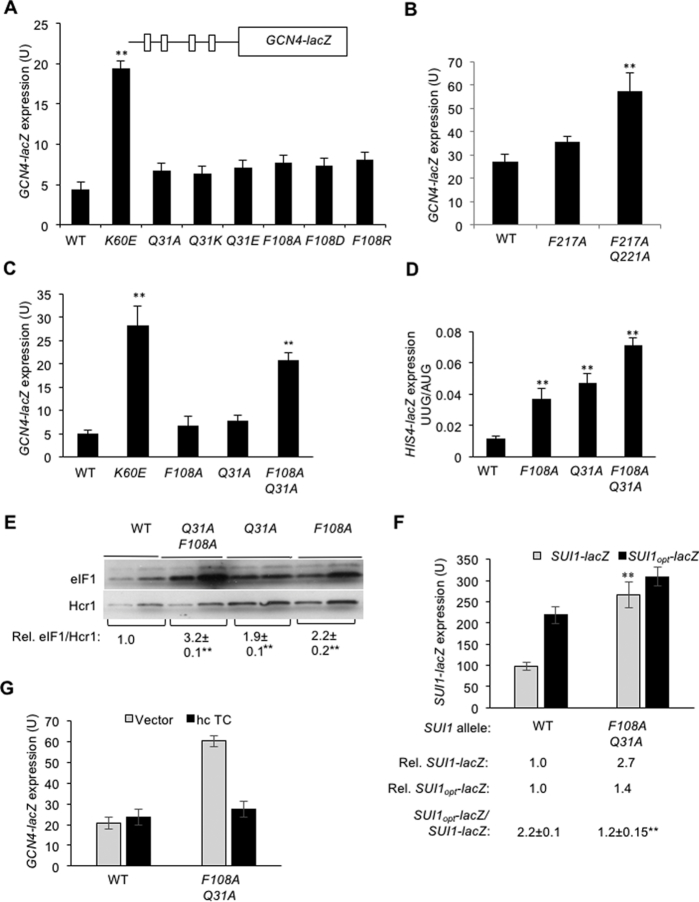
Figure 4.
Double substitutions at the eIF2β:eIF1 interface of the open complex derepress GCN4-lacZ expression and decrease discrimination against the poor context of the eIF1 AUG codon in vivo. (A) Transformants of JCY03 containing the indicated SUI1 alleles and the WT GCN4-lacZ reporter (depicted schematically) on plasmid p180 were assayed for β-galactosidase activities as in Figure 2B. Mean expression levels and SEMs calculated from six to eight transformants of each strain are plotted. (B) Derivatives of KAY18 containing the indicated SUI3 alleles and the WT GCN4-lacZ reporter were assayed for β-galactosidase activities as in Figure 2B. (C) Derivatives of JCY03 containing the indicated SUI1 alleles and WT GCN4-lacZ reporter were assayed for β-galactosidase activities as in Figure 2B. (D) Transformants of JCY03 containing the indicated SUI1 alleles and HIS4-lacZ reporters were assayed for β-galactosidase activities as in Figure 2B. (E) Derivatives of sui1Δ his4-301 strain JCY03 containing the indicated SUI1 alleles were subjected to western blot analysis as in Figure 3A. The eIF1 western signals were normalized to those for Hcr1 and mean values (±SEM) were calculated from three biological replicates. (F) Derivatives of sui1Δ his4-301 strain JCY03 containing the indicated SUI1 alleles and harboring plasmids (pPMB24 or -25) with SUI1-lacZ fusions with native suboptimal (-3CGU-1) or optimum (-3AAA-1) AUG contexts were cultured and assayed for β-galactosidase activities as in Figure 2B. Mean expression levels and SEMs were calculated from six transformants, and relative (Rel.) mean expression levels normalized to that of the WT strain are listed below, along with expression ratios for the SUI1-lacZ versus SUI1opt-lacZ reporters. (G) Transformants of JCY03 containing the indicated SUI1 alleles on a sc plasmid, either hc TC plasmid (p4835) or empty vector (YCplac112), and the WT GCN4-lacZ reporter were assayed for β-galactosidase activities as in Figure 2B. (A–G) Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (*P < 0.05; **P < 0.01).
None of the single eIF1 substitutions at the eIF2β:eIF1 interface significantly derepresses GCN4-lacZ expression (Figure 4A), and eIF2β-F217A increases expression only slightly (Figure 4B). However, substantial derepression of GCN4-lacZ occurred when eIF2β-F217A was combined with Q221A in the eIF2β-F217A/Q221A double substitution (Figure 4B), as reported previously (9). Derepression of GCN4-lacZ expression also was conferred when the eIF1 substitutions F108A and Q31A were combined (Figure 4C). The eIF1-F108A/Q31A double substitution also conferred a larger increase in the UUG:AUG initiation ratio compared to the two single substitutions (Figure 4D), as shown above for eIF2β-F217A/Q221A versus eIF2β-F217A alone (Figure 2D). Moreover, eIF1-Q31A/F108A conferred a greater increase in eIF1 abundance compared to the corresponding single substitutions (Figure 4E), and it elevated expression of the native SUI1-lacZ fusion and reduced the SUI1opt-lacZ:SUI1-lacZ expression ratio (Figure 4F) considerably more than did the eIF1-Q31A or eIF1-F108A single substitutions (Figure 3B). Together, these results indicate that combining the F108A and Q31A substitutions in eIF1 produces a synthetic reduction in discrimination against UUG codons and poor-context AUG codons in parallel with a derepression of GCN4-lacZ expression not observed in the corresponding single substitution mutants. Importantly, the latter defect was reversed by co-overexpressing all components of TC (Met-tRNAi, eIF2α, eIF2β and eIF2γ) from a high-copy plasmid (hc TC) (Figure 4G) as expected if the Gcd− phenotype of the eIF1-Q31A/F108A mutant results from slower TC loading that can be overcome by mass action at elevated TC concentrations. These genetic findings support the idea that substantially impairing the eIF1:eIF2β interface with multiple substitutions in either protein reduces the rate of TC loading to the open PIC conformation in addition to facilitating rearrangement from the open to closed conformation at poor start codons.
Because the Gcd− phenotype is conferred by eIF1 substitutions that weaken its binding to 40S subunits, we purified recombinant forms of the eIF1 variants and measured their 40S binding affinity by determining their ability to compete with fluorescently labeled WT eIF1 (eIF1FL) for binding to purified 40S·eIF1A complexes in vitro. Pre-assembled 40S·eIF1A· eIF1FL complexes were challenged with increasing concentrations of unlabeled mutant or WT eIF1 proteins, and the fraction of bound eIF1FL remaining at each competitor concentration was determined by monitoring changes in fluorescence anisotropy (Figure 5A and B). In accordance with previous results (8,10), the helix α1 variant K60E competed poorly with WT eIF1FL for binding to 40S·eIF1A complexes, increasing the eIF1 dissociation constant (Kd) by ∼10-fold. By contrast, the eIF1-Q31E, F108D and eIF1-Q31A/F108A mutants competed with WT eIF1FL indistinguishably from WT unlabeled eIF1, indicating no significant change in their affinity for 40S·eIF1A complexes (Figure 5B and C). These findings indicate that the decreased rate of TC binding inferred from the Gcd− phenotype of the F108A/Q31A double substitution, and the relaxed discrimination against poor initiation sites observed for this variant and the eIF1-F108D and eIF1-Q31E variants with single acidic substitutions, do not involve a weaker interaction of eIF1 with the 40S subunit.
Figure 5.
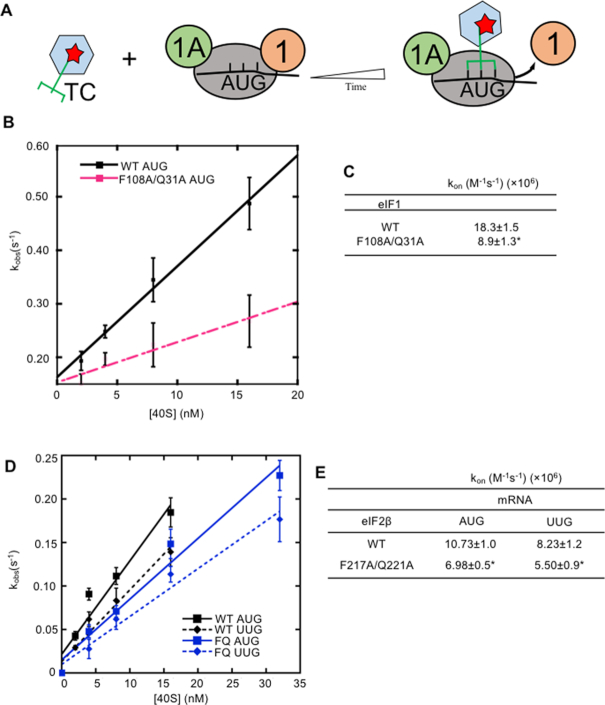
Substitutions at the eIF2β:eIF1 interface of the open complex do not impair eIF1:40S interaction but enhance the closed/PIN conformation of py48S PICs at UUG codons in vitro. (A–C) Measurement of eIF1 binding constants. Fluorescein-labeled WT eIF1 (5 nM) was pre-bound to 40S subunits (15 nM) in the presence of eIF1A (1 μM), mixed with increasing concentrations of unlabeled WT eIF1, eIF1-K60E, eIF1-F108D, eIF1-Q31E, eIF1-F108R or eIF1-F108D/Q31E, and the change in fluorescence anisotropy was measured (A). One of two replicate experiments is shown (B), from which mean Kdcomp values and average deviations were calculated (C). (D–J) Measurement of TC dissociation kinetics. Partial 48S complexes were assembled with radiolabeled TC containing WT or mutant eIF2β, eIF1A, model mRNA containing an AUG or UUG start codon, and WT or mutant eIF1 proteins; chased with excess unlabeled TC for increasing periods of time; and the fraction of labeled Met-tRNAi bound to the PIC at each time-point was determined by EMSA (D). A representative plot of the fraction of [35S]-Met-tRNAi incorporated into p43S·mRNA complexes is plotted as a function of time for eIF2β-F217A/Q221A (E) from which mean rate constants (F) and end points (G) (with SEMs) were calculated. ND, not determined. Similar plots were created for eIF1 variants F108D and Q31E (H) and were used to calculate mean rate constants (I) and end points (with SEMs) (J) from between three and five replicate experiments (numbers in parentheses). (F-J) Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (*P< 0.05; **P < 0.01).
We interpret the findings described thus far to indicate that the eIF1:eIF2β interface, found exclusively in the open conformation of the PIC, serves as a barrier to achieving the closed state, and that single substitutions at this interface that increase UUG initiation facilitate transition from the open to closed states at poor start codons by weakening this interface/barrier, without materially destabilizing the open conformation. The eIF1:eIF2β interface also faciliates TC binding during assembly of the open complex, and the more substantial weakening of the interface by multiple eIF1 or eIF2β substitutions reduces the rate of TC loading to the open complex in addition to conferring an even greater frequency of inappropriate transition to the closed state at poor start codons during scanning.
Substitutions at the eIF2β:eIF1 interface promote Met-tRNAi accommodation in the PIN conformation of the closed 48S PIC in vitro
To provide biochemical evidence that substitutions at the eIF2β:eIF1 interface facilitate isomerization to the closed/PIN state, we measured their effects on the rate of TC dissociation from PICs reconstituted from purified components in vitro. Partial 43S·mRNA PICs were reconstituted by incubating TCs pre-assembled with eIF2 (purified from yeast containing WT or mutant eIF2β subunits), [35S]-Met-tRNAi and nonhydrolyzable GTP analog (GDPNP), with 40S subunits, saturating concentrations of WT or mutant eIF1, WT eIF1A and an uncapped, unstructured model mRNA containing either an AUG or a UUG start codon [mRNA(AUG) or mRNA(UUG)]. The pre-assembled 43S·mRNA PICs were incubated with a chase of excess unlabeled WT TC for increasing time-periods, and the fraction of [35S]-labeled TC remaining bound to the PIC was quantified after resolving 40S-bound and unbound fractions via native gel electrophoresis, from which the rate of dissociation (koff) was calculated (Figure 5D). Previous work has indicated that TC bound in the open/POUT conformation is unstable during electrophoresis and cannot be visualized. Thus, the measured rate of TC dissociation in these assays largely reflects the proportion of complexes in the PIN state and the stability of that state with either an AUG or UUG in the P site (18,21).
In agreement with previous findings (7,18,21,22), we observed that TC dissociates more rapidly from the mRNA(UUG) versus mRNA(AUG) PICs in reactions containing all WT components (Figure 5E and F, WT; Supplementary Figure S4; see representative data in Supplementary Figure S5B), reflecting the reduced formation and relative instability of the PIN state at near-cognate UUG versus AUG codons. The endpoint of the dissociation reaction is also seen to vary with start codon identity. At AUG codons, TC has dissociated from ∼75% of PICs during the time course of our assay while the remaining ∼25% of complexes are completely stable over this period (Figure 5E and G, WT). Those complexes from which no dissociation occurs are thought to reflect a shift to a hyperstable state distinct from the PIN state (21). The lower endpoint for complexes assembled on mRNA(UUG), of only ∼9% of starting complexes (Figure 5E and G), presumably reflects the lower stability of the closed/PIN complex at near-cognate UUG codons, from which fewer complexes may enter the hyperstable state.
Interestingly, the double substitution eIF2β-F217A/Q221A results in dramatic stabilization of PICs at both AUG and UUG start codons. The degree of stabilization was such that off-rates could not be determined, because there was very little dissociation from these complexes on the time-scale of our assay (Figure 5E and F). The endpoints for both AUG and UUG start codons reflect almost no dissociation over 24 h as compared to complexes before the addition of chase. This is similar to the effect seen previously for double substitutions of Watson-Crick base pairs in the ASL of Met-tRNAi (maintaining W:C pairing), which confer Sui− phenotypes (21). These data are consistent with the Sui− phenotype of eIF2β-F217A/Q221A, and suggest that this double eIF2β substitution shifts the equilibrium in favor of the closed/PIN state, which can be accessed with an elevated frequency at UUG codons. The fact that TC does not dissociate from ∼80% of complexes containing eIF2β-F217A/Q221A further indicates that most complexes are able to enter the highly stable state even with a near-cognate UUG codon in the P site (Figure 5E and G).
Related results were obtained for the purified eIF1 Sui− variants F108D and Q31E, as these substitutions decreased the rate of TC dissociation (koff) by ∼2.3- to 5.0-fold from PICs assembled on mRNA(UUG) (Figure 5H–I; see representative data in Supplementary Figure S5C). They also decreased the extent of TC dissociation from these PICs as indicated by ∼1.5- to 2.7-fold increases in the fraction of TC remaining bound to the PIC at the reaction end-points (Figure 5H & J), thus indicating increased formation of the hyper-stable complex at UUG codons. Using mRNA(AUG), eIF1-Q31E decreased the koff by ∼2-fold, considerably smaller than the 5-fold reduction observed for mRNA(UUG); while eIF1-F108D had no significant effect on the rate of dissociation at AUG codons (Figure 5H–I), and both substitutions decreased the extent of TC dissociation at AUG codons (Figure 5H and J). These findings support the conclusion that substitutions in eIF1 or eIF2β that perturb the eIF2β:eIF1 interface in the open complex reduce a barrier to isomerization to the closed state, particularly at near-cognate UUG codons, in accordance with the increased initiation at UUG codons such mutations produce in vivo.
Substitutions at the eIF2β:eIF1 interface reduce the rate of Met-tRNAi loading to the open complex in vitro
As noted above, the eIF1-Q31A/F108A and eIF2β-F217A/Q221A double substitutions appear to reduce the rate of TC loading to the open conformation of the PIC, as indicated by elevated GCN4-lacZ reporter expression (Figure 4B and C). To support this interpretation, we measured the kinetics of TC binding to 40S·eIF1A·eIF1·mRNA complexes assembled with either WT eIF1 or eIF1-Q31A/F108A using the gel mobility shift assay described above. WT TC preassembled with [35S]-Met-tRNAi was added to initiate the reactions, which were terminated at each time point with excess unlabeled TC (Figure 6A). The pseudo-first-order rate constant (kobs) was measured at different 40S concentrations to obtain the second-order rate constant (kon) (18). Compared to WT eIF1, eIF1-Q31A/F108A confers an ∼2-fold decrease in kon for mRNA(AUG) (Figure 6B and C, Representative data in Supplementary Figure S6B). Using radiolabeled TCs assembled with eIF2 containing the eIF2β-F217A/Q221A mutant subunit, nearly identical ∼1.5-fold decreases in kon were measured for both mRNA(AUG) and mRNA(UUG) compared to WT TCs (Figure 6D and E). The relatively smaller reduction in kon for eIF2β-F217A/Q221A versus eIF1-Q31A/F108A (Figure 6C and E) is in keeping with the weaker Gcd− phenotype observed for the former mutant in vivo (Figure 4B and C). The similar reduction in kon conferred by eIF2β-F217A/Q221A for PICs assembled with mRNA(AUG) or mRNA(UUG) fits with the notion that this eIF2β variant reduces the initial step of TC binding to the open conformation of the PIC, which is relatively insensitive to the nature of the start codon (18). These findings support the conclusion that the eIF1-Q31A/F108A and eIF2β-F217A/Q221A substitutions destabilize the open complex, reducing the rate of TC loading to this state while increasing rearrangement to the closed state at UUG codons during scanning.
Figure 6.
Double substitutions at the eIF2β:eIF1 interface of the open complex reduce rates of TC binding in vitro. (A) Schematic for measurement of TC association kinetics. WT TC preassembled with [35S]-Met-tRNAi was mixed with pre-formed 40S·eIF1A·eIF1·mRNA complexes, incubated for increasing times, and reactions were terminated at each time-point with a chase of excess unlabeled TC. The fraction of labeled Met-tRNAi bound to the PIC at each time-point was determined by EMSA. The pseudo-first-order rate constant (kobs) was measured at different 40S concentrations to obtain the second order rate constant (kon). (B, C) Determination of TC kon values as described in (A) for WT or eIF1-F108A/Q31A mutant and mRNA(AUG). The mean kobs values were determined from at least three independent experiments at each 40S concentration. The resulting mean kobs values are plotted against each 40S concentration with error bars representing SEMs (B). kobs values from each of three independent experiments were plotted against 40S concentration in order to calculate kon, and mean kon values (with SEMs) are shown (C). (D, E) Determination of TC kon values as described in (A–C) for WT eIF2 or eIF2 containing the eIF2β-F217A/Q221A (FQ) variant for partial 43S·mRNA complexes containing mRNA(AUG) or mRNA(UUG). The mean kobs values determined from three independent experiments at each 40S concentration were plotted versus the 40S concentration (D), and the mean calculated kon values determined from three independent experiments are shown (E). Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (*P< 0.05).
eIF1-F108R increases discrimination against near-cognate UUG codons in vivo and favors the open/POUT conformation of the PIC in vitro
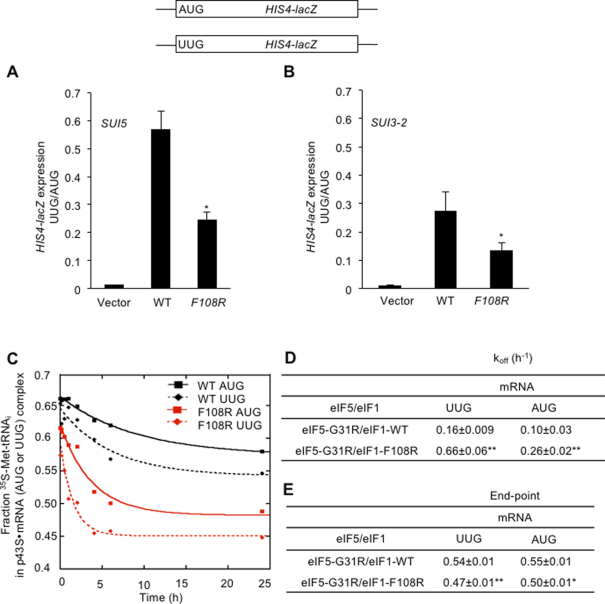
Having found that the acidic substitution eIF1-F108D shifts the system to the closed complex and increases UUG initiation, and noting the presence of the essential acidic residue eIF2β E198 at the eIF1:eIF2β interface (Figure 2A, left), we asked whether substituting eIF1 F108 with the basic residue arginine would produce the opposite effect on the system versus eIF1-F108D by strengthening the eIF1:eIF2β interface through a new salt-bridge between eIF1 R108 and native eIF2β E198. Indeed, eIF1-F108R was found to partially suppress the elevated UUG initiation conferred by the dominant Sui− alleles SUI5 and SUI3-2, encoding the eIF5-G31R and eIF2β-S264Y variants, respectively (23), thus conferring an Ssu− phenotype (Figure 7A and B, WT versus F108R).
Figure 7.
eIF1 substitution F108R at the eIF2β:eIF1 interface of the open complex increases discrimination against UUG start codons in vivo and disfavors PIN at UUG start codons in vitro. (A, B) HIS4-lacZ reporters with AUG or UUG start codons were assayed as in Figure 2D (except that Trp was omitted from the medium) to calculate the UUG:AUG initiation ratio in the presence of the dominant Sui− allele SUI5 on plasmid p4281 (A) or SUI3-2 on plasmid p4280 (B). (C–E) TC dissociation kinetics were assayed as in Figure 5D–G for partial 43S·mRNA complexes containing mRNA(AUG) or mRNA(UUG) and assembled with WT eIF1 or eIF1-F108R in the presence of the eIF5-G31R variant (SUI5). Representative curves selected from three independent experiments are shown (C) from which mean rate constants (D) and end points (E) (with SEMs) were calculated. (A–E) Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (*P < 0.05; **P < 0.01).
The eIF1-F108R substitution does not detectably alter eIF1 binding to 40S·eIF1A complexes in vitro (Figure 5B and C), suggesting that it reduces UUG initiation by impeding transition from the open to closed conformation of the PIC during scanning. Supporting this interpretation, eIF1-F108R increases the rate of TC dissociation from PICs reconstituted in vitro in the presence of the SUI5 eIF5 variant, eIF5-G31R (Figure 7C and D). Consistent with our previous results (7), in reactions with WT eIF1 and eIF5-G31R, TC dissociates very little over the time course of the experiment from complexes containing an AUG or UUG start codon, yielding rate constants of only 0.10 or 0.16 h−1, respectively (Figure 7C and D), reflecting stabilization of the closed/PIN state by this eIF5 variant. Importantly, the TC dissociation rates for the complexes assembled with eIF1-F108R and eIF5-G31R were increased by ∼2.5-fold for mRNA(AUG) and ∼4-fold for mRNA(UUG) compared to the koff values observed with WT eIF1 and eIF5-G31R (Figure 7C and D). The extent of TC dissociation was also elevated somewhat by eIF1-F108R, as reflected in lower reaction end-points (Figure 7E). These findings provide biochemical evidence that eIF1-F108R destabilizes the closed/PIN state at both AUG and UUG start codons, with a relatively stronger effect on the near-cognate triplet, thereby overriding the opposing effect of the eIF5-G31R substitution in preferentially enhancing the stability of the UUG complex (7). These findings help to account for the decreased utilization of UUG codons conferred by the F108R substitution in vivo.
Substitutions in eIF2β that disrupt contacts with the Met-tRNAi anticodon stem-loop (ASL) favor the closed/PIN complex and perturb TC binding to the open complex
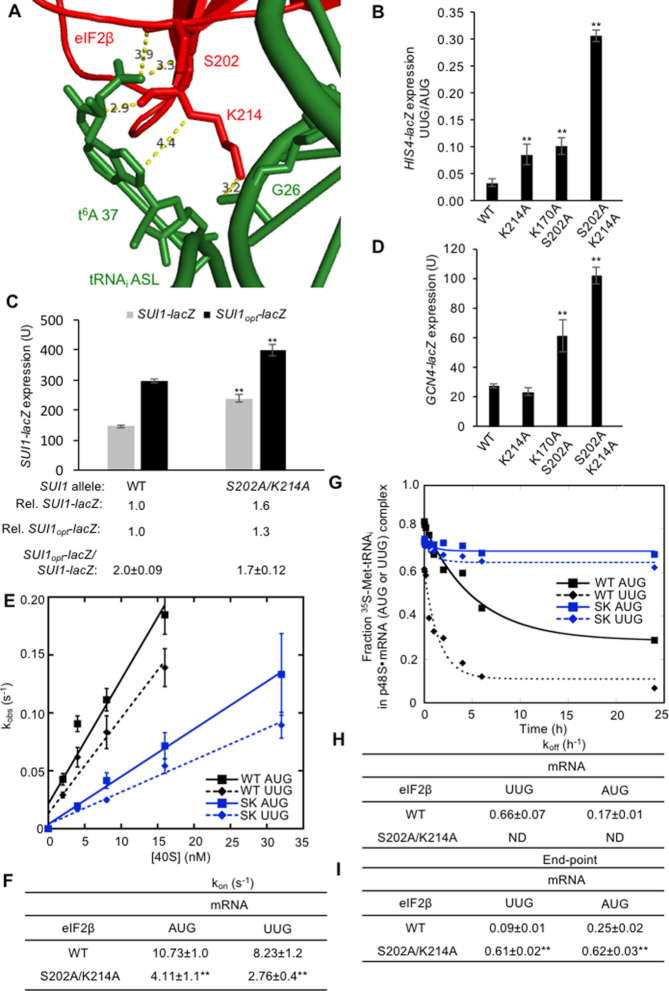
Comparing the cryo-EM structures of py48S-open and py48S-closed complexes revealed contacts between eIF2β and the ASL of Met-tRNAi that are unique to the open state (Supplementary Figure S1A and B). These include eIF2β residues S202, K170 and K214, which lie in proximity to the modified anticodon loop nucleotide t6A37 (Figure 8A). In the closed complex, eIF2β is more compact and forms an alternative interface with the acceptor stem and D-loop, but not the ASL of Met-tRNAi (Supplementary Figure S1B). The structures also reveal that the open conformation has a widened P site in which many of the canonical contacts between the ASL and the 40S body are missing (9). We therefore hypothesized that in the absence of these latter contacts, eIF2β residues interacting with the ASL have a role in stabilizing Met-tRNAi binding to the PIC in the open conformation, leading to the prediction that substituting these residues would increase initiation at poor initiation codons in vivo.
Figure 8.
Genetic and biochemical evidence that eIF2β substitutions at the eIF2β:Met-tRNAi interface of the open complex decrease the rate of TC binding and enhance the closed/PIN conformation at UUG codons. (A) Contacts between eIF2β and the Met-tRNAi ASL in the py48S-open complex. In py48S-closed, eIF2β has moved away and instead contacts the Met-tRNAi D-loop (Supplementary Figure S1B). eIF2β is shown in red, Met-tRNAi in green. Measurements are in angstroms. (B) Derivatives of strain KAY18 containing the indicated SUI3 alleles and harboring HIS4-lacZ reporters with AUG or UUG start codons were assayed for β-galactosidase activities as in Figure 2D. The ratio of expression of the UUG to AUG reporter was calculated from at least four different measurements, and the mean and S.E.M.s were plotted. (C) Derivatives of strain KAY18 containing the indicated SUI3 alleles and harboring plasmids (pPMB24 or pPMB25) with SUI1-lacZ fusions with either the native suboptimal (-3CGU-1) or optimized (-3AAA-1) AUG context were cultured and assayed for β-galactosidase activities as in Figure 2D. Mean expression levels and SEMs were calculated from four transformants, and relative (Rel.) mean expression levels normalized to that of the WT strain are listed, along with expression ratios for the SUI1-lacZ versus SUI1opt-lacZ reporters. (D) Derivatives of strain KAY18 containing the indicated SUI3 alleles and harboring a GCN4-lacZ reporter on plasmid p180 were cultured in synthetic complete medium lacking leucine and uracil (SC-L-U) at 30°C to A600 of ∼0.8, and β-galactosidase activities were measured as in Figure 2D. Mean GCN4-lacZ expression (± SEM) in units of β-galactosidase activity was calculated from four transformants. (E, F) The kon values for TC binding to partial 43S·mRNA(AUG) or 43S·mRNA(UUG) complexes were determined as in Figure 6A–C for WT eIF2 and eIF2 containing eIF2β-S202A/K214A from three independent experiments for each eIF2/mRNA combination. (G–I) TC dissociation rates were measured as in Figure 5D–G for WT eIF2 or eIF2 containing eIF2β-S202A/K214A for partial 43S·mRNA(AUG) or 43S·mRNA(UUG) complexes. One of three replicate experiments is shown for each eIF2/mRNA combination (G), from which mean rate constants (H) and end points (I) with SEMs were calculated. ND, not determined. (B–I) Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (**P < 0.01).
A single alanine substitution at K170 did not significantly increase the HIS4-lacZ UUG:AUG initiation ratio (data not shown), whereas the eIF2β-K214A substitution elevated the UUG:AUG ratio 2.5-fold. However, combining Ala substitutions of either K170 or K214 with S202A resulted in stronger phenotypes, with eIF2β-K170A/S202A increasing the UUG:AUG ratio 3.0-fold and eIF2β-S202A/K214A producing a 9.1-fold increase (Figure 8B), in accordance with our earlier findings on the -S202A/K214A variant (9). These results indicate that the loss of a positive charge at eIF2β residue 214 relaxes discrimination against near cognate start codons, particularly when a hydroxyl group is absent at residue 202, whereas loss of the basic side-chain of K170 has a similar but weaker effect on the system. The eIF2β-S202A/K214A variant also slightly increases eIF1 abundance (Figure 3A, right), and elevates expression of a SUI1-lacZ reporter containing the native, suboptimal AUG context while decreasing the SUI1opt-lacZ:SUI1-lacZ ratio (Figure 8C), suggesting a modest reduction in discrimination against AUG codons in poor sequence context in addition to increasing UUG initiation. These results support the prediction that perturbing the eIF2β:Met-tRNAi interface specific to the open complex would increase the probability of rearrangement to the closed state at non-AUG codons and AUGs in poor context. The eIF2β-S202A/K214A variant is both expressed and forms a complex with eIF2α and eIF2γ at WT levels (9).
We also examined whether these eIF2β substitutions impair TC loading using the GCN4-lacZ reporter described above. While no Gcd− phenotype was seen for the single substitutions eIF2β-K214A or eIF2β-K170A (data not shown), the double substitutions eIF2β-K170A/S202A and eIF2β-S202A/K214A derepressed GCN4-lacZ expression by 2.2- and 3.7-fold, respectively (Figure 8D). Because TC loads onto the open state of the PIC, this TC loading defect, combined with the Sui− phenotypes described above, provide in vivo evidence that contacts between eIF2β residues and the ASL contribute to TC binding specifically to the open complex.
This last conclusion was further supported by biochemical analysis. Measuring the kinetics of TC dissociation revealed that eIF2β-S202A/K214A stabilized TC binding at both AUG and UUG codons, such that no dissociation was seen over the time-scale of our experiments (Figure 8G–I). This defect was coupled with a ∼2.6- to 3-fold reduction in the rate of TC binding to PICs (kon) with AUG or UUG start codons (Figure 8E and F). These results signify a destabilization of the open conformation, with attendant reduction in the rate of TC loading and a shift toward the closed conformation and subsequent rearrangement to the hyper-stable complex, on perturbing the eIF2β:Met-tRNAi ASL interface unique to the open complex.
eIF2β substitutions at the interface with the Met-tRNAi D-loop favor rearrangement to the closed/PIN complex with minimal perturbation of TC binding to the open complex
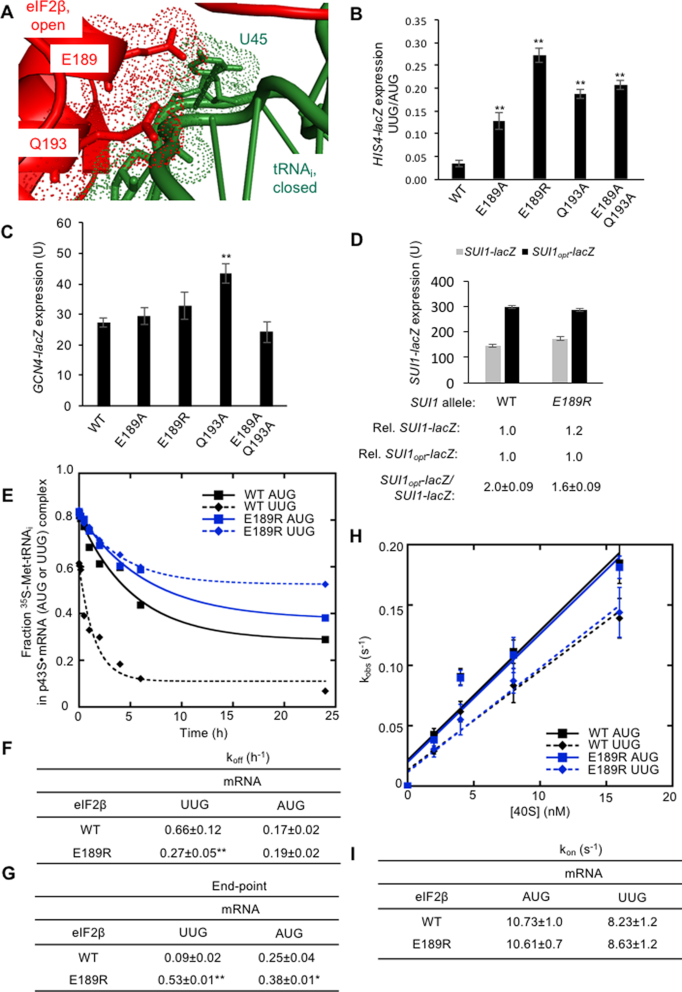
In addition to its contacts with the ASL described above, eIF2β forms differential contacts with the D-loop of Met-tRNAi in the open and closed conformations of the PIC. Interestingly, overlaying the two cryo-EM structures revealed extensive steric and electrostatic clashes between eIF2β in py48S-open and Met-tRNAi in py48S-closed, particularly involving eIF2β residues E189 and Q193 (Figure 9A, Figure S1C). These predicted clashes might indicate that eIF2β functions analogously to eIF1 during scanning to impede transition of Met-tRNAi to the PIN state of the closed complex before an AUG codon enters the P site. If so, then alanine substitutions of these residues should alleviate the clash, allowing Met-tRNAi to move into the closed/PIN state inappropriately at non-AUG codons. Indeed, we found that eIF2β-E189A, eIF2β-Q193A, and the double substitution eIF2β-E189A/Q193A elevated the UUG:AUG ratio between ∼3.8- and ∼6.1-fold (Figure 9B). Interestingly, substituting E189 with positively charged Arg conferred a considerably larger increase in UUG initiation compared to E189A (Figure 9B), whereas the eIF2β-Q193R substitution was lethal. eIF2β-E189R conferred a slow-growth phenotype (Figure 2C). Increased UUG initiation in response to the Ala substitutions of these eIF2β residues supports the notion that relieving a clash with Met-tRNAi at these positions removes a barrier to PIN. That UUG initiation was further elevated by introducing a positive charge at position 189 suggests that introducing electrostatic attraction with Met-tRNAi can stabilize the closed/PIN state at UUG codons. Previously, we made similar findings for Ala and Arg substitutions in Loop-2 of eIF1, which is also predicted to clash with the Met-tRNAi D-loop as a means of restricting transition to the closed complex (10). eIF2β-E189R slightly increased eIF1 abundance (Figure 3A, right) and modestly reduced the SUI1opt-lacZ:SUI1-lacZ expression ratio (Figure 9D), indicating that eIF2β-E189R has a relatively small effect in promoting transition to the closed/PIN state at AUGs in poor context compared to its effect at UUG codons, suggesting that a codon:anticodon mismatch in the P-site is required for its effect on the system.
Figure 9.
Genetic and biochemical evidence that eIF2β substitutions designed to alleviate a predicted clash between eIF2β and Met-tRNAi enhance transition to the closed state at UUG codons but do not alter the rate of TC loading. (A) Overlaying the structure of eIF2β in py48S-open with Met-tRNAi in py48S-closed reveals predicted clashes between eIF2β (including residues E189 and Q193) and Met-tRNAi (including residue U45) that should impede rearrangement of Met-tRNAi to the closed state before AUG recognition. eIF2β is shown in red, Met-tRNAi in green. (B) Derivatives of strain KAY18 containing the indicated SUI3 alleles and harboring HIS4-lacZ reporters were analyzed as in Figure 2D. The ratio of expression of the UUG to AUG reporter was calculated from at least four independent transformants, and the mean and S.E.M.s were plotted. (C) Derivatives of strain KAY18 containing the indicated SUI3 alleles and harboring a GCN4-lacZ reporter were analyzed as in Figure 2D. Mean GCN4-lacZ (±SEM) was determined from at least four independent transformants. (D) Derivatives of strain KAY18 containing the indicated SUI3 alleles and harboring plasmids (pPMB24 or pPMB25) with SUI1-lacZ fusions with either native suboptimal (-3CGU-1) or optimized (-3AAA-1) AUG context were analyzed as in Figure 2D. Mean expression levels and SEMs were calculated from four transformants, and relative (Rel.) mean expression levels normalized to that of the WT strain are listed, along with expression ratios for the SUI1-lacZ versus SUI1opt-lacZ reporters. (E–G) TC dissociation rates were measured as in Figure 5D–G. One of three replicate experiments is shown for WT eIF2 and eIF2 containing the variant eIF2β-E189R (E), from which mean rate constants (F) and end points (G) with SEMs were calculated. (H, I) TC association rates were measured as in Figure 6A–C for WT eIF2 and eIF2 containing eIF2β-E189R and partial 43S·mRNA(AUG) or 43S·mRNA(UUG) complexes. The kobs values are plotted against the concentration of 40S subunits (H), and the calculated TC on-rates (kon) are shown (I). Values are the averages of three independent experiments. (B–I) Asterisks indicate significant differences between mutant and WT as judged by a two-tailed, unpaired Student's t-test (**P< 0.01).
Of the eIF2β substitutions in this region, only Q193A produced a significant but modest 1.6-fold derepression of GCN4-lacZ expression. It is noteworthy that no derepression was observed for eIF2β-E189A/Q193A nor –E189R (Figure 9C) that conferred equal or greater increases in UUG initiation compared to eIF2β-Q193A (Figure 9B). The general lack of Gcd− phenotypes for substitutions at this interface suggests that the rate of TC loading is not impaired, supporting our hypothesis that these substitutions remove a clash with Met-tRNAi that impedes transition to the closed state during scanning without affecting the stability of the open state of the PIC.
Our interpretation of the in vivo phenotypes of the E189R variant is supported by biochemical analysis. Similar to the eIF1-F108D mutation, we observed that eIF2β-E189R lowered the TC off-rate (koff) for UUG complexes by 2-fold while dramatically increasing the number of complexes from which Met-tRNAi does not dissociate (∼5-fold) (Figure 9E–G). These results indicate a shift toward the closed conformation and subsequent rearrangement to the hyperstable state. The TC on-rates (kon) for mRNA(AUG) and mRNA(UUG) were unaffected by the E189R substitution (Figure 9H and I), indicating that this mutation does not confer a TC loading defect. This is consistent with the lack of Gcd− phenotype seen in vivo for this variant (Figure 9C) and supports our interpretation that the E189R substitution does not affect the stability of the open complex to which TC binds but only facilitates transition to the closed/PIN state.
DISCUSSION
In this report, we provide a combination of genetic and biochemical evidence that eIF2β plays a direct role in stimulating TC loading in the assembly of 43S PICs, and that it supports eIF1 function both in promoting scanning of the mRNA for AUG codons and in suppressing initiation at non-AUG start codons. These functions of eIF2β are mediated by its separate interactions with eIF1 and the ASL of Met-tRNAi found specifically in the open conformation of the PIC. We also provide evidence that eIF2β cooperates with eIF1 in preventing stable binding of Met-tRNAi in the P site at near cognate start codons through physical clashes predicted between both eIF1 and eIF2β in their locations found in the open complex and Met-tRNAi in the closed state. Hence, as demonstrated previously for eIF1 (1), eIF2β has a dual role in promoting the open scanning conformation of the PIC while impeding rearrangement to the closed conformation, which helps to restrict the transition from open to closed conformations of the PIC to AUG codons in optimum context.
eIF2β:eIF1 and eIF2β:Met-tRNAi interactions restricted to the PIC open conformation reduce the rate of TC loading and increase initiation at poor initiation sites
Interactions of eIF2β with eIF1 and Met-tRNAi were first observed in the py48S-open complex, thought to represent the scanning complex with Met-tRNAi not tightly locked into the P site, and these contacts were absent in the py48S-closed complex, thought to represent the PIC following AUG recognition (9). In the downward movement of the 40S head towards the 40S body in the transition from py48S-open to py48S-closed, the TC moves in conjunction with the 40S head, and the HTH domain of eIF2β is displaced from its interface with eIF1. At the same time, the eIF2β-HTH domain moves along the Met-tRNAi and is no longer engaged with the ASL in the closed complex. Because the eIF2β:eIF1 and eIF2β:ASL interactions occur exclusively in the open complex (Supplementary Figure S1D), we hypothesized that they help to prevent eIF1 dissociation from the scanning PIC, and that they also anchor eIF1 in the position needed for its predicted clash with Met-tRNAi in the closed conformation to help impede rearrangement to the PIN state at non-AUG codons (9,10).
We previously provided genetic support for this hypothesis (9), and have confirmed and extended the evidence here that substitutions in either eIF1 or eIF2β that perturb their interface in the open complex increase aberrant transition to the closed conformation at near-cognate UUG codons in vivo, elevating the UUG:AUG initiation ratio for a HIS4-lacZ reporter in vivo. We obtained biochemical evidence indicating that such substitutions reduce rates of TC dissociation from py48S PICs reconstituted at UUG codons in vitro, indicating a shift to the closed conformation (and subsequent transition to a hyperstable state) that exhibits stable Met-tRNAi binding in the P site. Additional strong support for our hypothesis came from the finding that the eIF1-F108R substitution, designed to strengthen the eIF1:eIF2β interface, affected the system in ways opposite to the substitutions expected to weaken the interface, decreasing UUG initiation in a mutant rendered hypoaccurate by the eIF5-G31R variant (SUI5) and increasing the TC off-rate at UUG codons from py48S PICs assembled in the presence of eIF5-G31R.
An additional in vivo manifestation of eIF1 substitutions that favor the open-to-closed transition is the reduced leaky scanning of AUG start codons in poor context (5). We observed this phenotype here for both eIF1 and eIF2β substitutions that perturb their interface in py48S-open, as indicated by increased initiation at the eIF1 AUG present in the native, poor context, but not when context was optimized. In addition, they increased initiation at the AUG for the GCN4 uORF1 when it resides in poor context, with attendant reduced initiation at the downstream GCN4-lacZ coding sequences. Thus, we have demonstrated that the eIF2β:eIF1 interface in the open complex is required for discrimination against poor context at AUG codons in addition to suppressing initiation at near-cognate triplets.
Substitutions at the eIF1:eIF2β interface also impair assembly of the PIC by reducing the rate of TC loading. This was revealed previously for the eIF2β-F217A/Q221A double mutant by its derepression of a GCN4-lacZ reporter (9), and was also observed here for the eIF1-F108A/Q31A double mutant. Translation of GCN4 mRNA is regulated by a delayed reinitiation mechanism wherein a decreased rate of TC loading blocks reinitiation at the uORFs and allows it to occur downstream at the GCN4-lacZ coding sequences instead (3). Here, we provided biochemical support for this interpretation by demonstrating a reduced rate of TC binding to py43S⋅mRNA complexes reconstituted in vitro using the eIF1-F108A/Q31A and eIF2β-F217A/Q221A variants. For the eIF1 mutant, we further established that the substitutions do not decrease eIF1 affinity for reconstituted 40S⋅eIF1A complexes in vitro, supporting a specific destabilization of the eIF1: eIF2β interface as the defect underlying both the assembly and accuracy phenotypes of these substitutions.
The fact that eIF1 and eIF2β substitutions that destabilize the open conformation simultaneously reduce the on-rate and decrease the off-rate for TC in reconstituted py48S complexes is consistent with previous biochemical results indicating that eIF1 in the open complex accelerates TC binding, but that TC is more tightly bound to closed complexes lacking eIF1 (that would exist following eIF1 release at the start codon) (20). The dual biochemical effects of the eIF1 and eIF2β substitutions in decreasing the rate of TC binding to the open conformation and reducing TC dissociation from the closed complex are paralleled by their dual in vivo phenotypes of derepressing GCN4-lacZ expression and elevating the HIS4-lacZ UUG:AUG initiation ratio. Hence, our combined genetic and biochemical analyses provide strong evidence that the eIF1:eIF2β interface both stabilizes the open conformation and opposes transition to the closed state during scanning (Supplementary Figure S7). These data further identify a direct molecular mechanism for the enhancement of TC recruitment by eIF1. Whereas single substitutions in eIF1 or eIF2β conferred elevated UUG initiation, multiple substitutions were required to derepress GCN4-lacZ expression, suggesting that only a moderate perturbation of the eIF1:eIF2β interface is sufficient to increase the probability that eIF1 dissociates from the PIC during scanning, or to diminish eIF1′s predicted clash with Met-tRNAi at non-AUG codons. However, a larger perturbation conferred by multiple substitutions in eIF1 or eIF2β is required to impair eIF1′s enhancement of TC recruitment during PIC assembly.
Interactions between eIF2β and the ASL of Met-tRNAi were also revealed in the py48S-open complex that are absent in py48S-closed (9), suggesting that they too might specifically stabilize the open, scanning conformation of the PIC. Supporting this hypothesis, we demonstrated that the eIF2β-S202A/K214A double substitution, designed to perturb the eIF2β:ASL interface, confers the dual in vivo phenotypes of derepressed GCN4-lacZ expression and elevated UUG initiation, and produced the corresponding biochemical defects of a reduced TC on-rate and decreased TC off-rate, respectively, in reconstituted py48S PICs. Thus, the eIF2β:ASL interactions, like eIF1:eIF2β contacts, appear to enhance TC loading and scanning in the open conformation of the PIC, and disfavor rearrangement to the closed state, at near cognate start codons (Supplementary Figure S7).
It is noteworthy that the eIF2β substitutions at the eIF2β:ASL interface have a smaller stimulatory effect on initiation at the poor-context AUG codon of eIF1 compared to initiation at the UUG start codon of the HIS4-lacZ reporter, whereas the eIF1 substitutions at the eIF1:eIF2β interface, and other eIF1 substitutions at the 40S interface described previously (8), confer more comparable increases in initiation at poor-context AUG and UUG codons. The enhanced recognition of poor-context AUGs produced by eIF1 substitutions could result from a decreased rate of scanning that increases the dwell-time at each triplet and provides more time to complete downstream reactions required for start codon selection (‘slow-scanning’ mechanism). Alternatively, it could result from an increased probability of rearrangement to the closed conformation at each triplet that elevates the likelihood of initiation at AUGs in poor context (‘shifted equilibrium’ mechanism). We have invoked the latter mechanism to account for the increased UUG initiation conferred by eIF1 substitutions that shift the equilibrium from the open to closed conformation of reconstituted PICs at UUG codons (Figure 1B & Supplementary Figure S6).
While our data do not directly address the mechanism by which the recognition of suboptimal start codons is enhanced, one way to account for our findings is to propose that both the eIF1 substitutions and eIF2β substitutions perturbing the eIF1:eIF2β interface increase UUG initiation by the ‘shifted equilibrium’ mechanism, whereas they enhance initiation at poor-context AUG codons by the ‘slow scanning’ mechanism mentioned above. The eIF2β substitutions at the interface with the Met-tRNAi ASL would also enhance UUG initiation by the ‘shifted equilibrium’ mechanism, as they confer reduced TC off-rates in reconstituted PICs. However, because they do not reduce the occupancy of eIF1 in the scanning PIC, they cannot increase initiation at poor-context AUGs by the slow-scanning mechanism and can do so only by shifting the equilibrium between open and closed conformations of the PIC.
The predicted clash of eIF2β in the open conformation with Met-tRNAi in the closed state impedes initiation at near-cognate start codons
Previous studies have shown that eIF1 physically restricts full accommodation of Met-tRNAi in the P-site of the scanning complex in a manner that restricts rearrangement to the closed state until a perfect AUG:anticodon duplex is formed in the P site (10). The eIF1: Met-tRNAi clash was revealed by superimposing eIF1 in py48S-open with Met-tRNAi in py48S-closed. Similar modeling here revealed that the HTH domain of eIF2β in py48S-open clashes with Met-tRNAi in py48S-closed (Supplementary Figure S1C), and we hypothesized that eIF2β cooperates with eIF1 in opposing Met-tRNAi binding in the PIN state prior to AUG recognition. Supporting this idea, Ala substitutions in eIF2β residues E189 and Q193 designed to diminish the predicted clash with the D-loop of Met-tRNAi, increased UUG initiation in vivo. Interestingly, the E189R substitution conferred a stronger phenotype compared to eIF2β-E189A, suggesting that replacing electrostatic repulsion with attraction allows eIF2β to stabilize rather than impede Met-tRNAi binding in the closed complex and thereby overcome the destabilizing effect of a UUG:anticodon mismatch more effectively than is achieved by merely eliminating the acidic side chain of eIF2β E189. Consistent with this interpretation, the eIF2β-E189R substitution shifted the equilibrium to the closed state in vitro, reducing the TC off-rate from UUG codons in reconstituted py48S complexes. Importantly, however, the E189R substitution had no effect on GCN4-lacZ expression in vivo, nor on the on-rate of TC in reconstituted PICs. These findings can be explained by proposing that diminishing the predicted electrostatic clash of the eIF2β HTH domain with Met-tRNAi (or replacing it with electrostatic attraction) removes a barrier to the PIN state of the closed complex that increases UUG initiation, but it does not destabilize the open complex and, hence, does not slow TC loading to the open conformation of the PIC. We recently obtained similar results for substitutions in Loop-2 of eIF1 that were designed to diminish its predicted clash with Met-tRNAi (10). Hence, we propose that the eIF2β HTH cooperates with eIF1 Loop-2 in the scanning PIC to establish a physical barrier to isomerization of Met-tRNAi to the PIN state of the closed complex (Supplementary Figure S7). Both eIF1 and the eIF2β HTH in their respective positions in the open complex are predicted to clash with different surfaces of the Met-tRNAi in the closed complex, and their contacts with one another in the open complex should mutually reinforce their distinct inhibitory interactions with Met-tRNAi (Supplementary Figure S1D).
The eIF2β-E189R substitution does not increase initiation at the poor-context AUG start codon of eIF1 mRNA and in this respect resembles the eIF2β substitutions at the interface with Met-tRNAi-ASL in the open complex in selectively increasing initiation at UUG codons. Because eIF2β-E189R is not expected to perturb eIF1 binding to the open complex, it should not reduce the rate of scanning and overcome poor AUG context by the ‘slow-scanning’ mechanism proposed above. The same reasoning should apply to the eIF1 loop-2 mutations that diminish the predicted clash with Met-tRNAi. However, these substitutions were found to increase initiation at poor-context AUGs in addition to UUG codons (10). As a possible explanation for this discrepancy, it could be proposed that eIF1 loop-2 substitutions additionally perturb an interaction of eIF1 with the eIF3c-NTD (24) required for a WT rate of scanning. More work will be required to determine whether decreased discrimination against poor context can be achieved simply by removing a barrier to the PIN state, which is sufficient to increase UUG initiation, or whether a kinetic defect in scanning is also required.
In summary, through a combination of genetics and biochemistry, we have identified three distinct interactions involving the eIF2β HTH domain that increase the accuracy of initiation. These involve separate eIF2β contacts with eIF1 and the Met-tRNAi ASL, which occur specifically in the open conformation of the PIC, and the predicted clash of the eIF2β HTH domain in its position in the open complex with Met-tRNAi in its location in the closed state—all of which impede the open-to-closed transition of the PIC and help limit initiation to AUG codons. These functions of eIF2β, and the analogous regulatory roles of eIF1, join a growing list of molecular interactions visualized in the py48S-open or py48S-closed PICs (9) that regulate scanning and start codon recognition, including interaction of the eIF1A N-terminal tail with the codon:anticodon duplex (25), interactions of Rps5/uS7 with eIF2α (22,26), and Rps3-mRNA interactions at the mRNA entry channel of the 40S subunit (27). Our results further establish a direct role for eIF1 in promoting TC binding to the open complex through its interactions with the eIF2β-HTH, and we show that this aspect of PIC assembly is also stimulated by eIF2β-HTH interactions with the Met-tRNAi ASL.
Supplementary Material
ACKNOWLEDGEMENTS
We thank Jinsheng Dong, Fan Zhang, Jagpreet Nanda and Jon Lorsch for advice and assistance in using the yeast reconstituted system, and all members of our laboratory and those of Jon Lorsch, Tom Dever and Nick Guydosh for invaluable suggestions.
SUPPLEMENTARY DATA
Supplementary Data are available at NAR Online.
FUNDING
Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health. Funding for open access charge: Intramural Research Program of the Eunice Kennedy Shriver National Institute of Child Health and Human Development, National Institutes of Health.
Conflict of interest statement. None declared.
REFERENCES
- 1. Hinnebusch A.G. The scanning mechanism of eukaryotic translation initiation. Annu. Rev. Biochem. 2014; 83:779–812. [DOI] [PubMed] [Google Scholar]
- 2. Hinnebusch A.G. Structural insights into the mechanism of scanning and start codon recognition in eukaryotic translation initiation. Trends Biochem. Sci. 2017; 42:589–611. [DOI] [PubMed] [Google Scholar]
- 3. Hinnebusch A.G. Translational regulation of GCN4 and the general amino acid control of yeast. Annu. Rev. Microbiol. 2005; 59:407–450. [DOI] [PubMed] [Google Scholar]
- 4. Donahue T. Sonenberg N, Hershey JWB, Mathews MB. Translational Control of Gene Expression. 2000; Cold Spring Harbor: Cold Spring Harbor Laboratory Press; 487–502. [Google Scholar]
- 5. Martin-Marcos P., Cheung Y.N., Hinnebusch A.G.. Functional elements in initiation factors 1, 1A, and 2beta discriminate against poor AUG context and non-AUG start codons. Mol. Cell. Biol. 2011; 31:4814–4831. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ivanov I.P., Loughran G., Sachs M.S., Atkins J.F.. Initiation context modulates autoregulation of eukaryotic translation initiation factor 1 (eIF1). Proc. Natl. Acad. Sci. U.S.A. 2010; 107:18056–18060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Martin-Marcos P., Nanda J.S., Luna R.E., Zhang F., Saini A.K., Cherkasova V.A., Wagner G., Lorsch J.R., Hinnebusch A.G.. Enhanced eIF1 binding to the 40S ribosome impedes conformational rearrangements of the preinitiation complex and elevates initiation accuracy. RNA. 2014; 20:150–167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Martin-Marcos P., Nanda J., Luna R.E., Wagner G., Lorsch J.R., Hinnebusch A.G.. beta-hairpin loop of eIF1 mediates 40S ribosome binding to regulate initiator tRNAMet recruitment and accuracy of AUG selection in vivo. J. Biol. Chem. 2013; 288:27546–27562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Llacer J.L., Hussain T., Marler L., Aitken C.E., Thakur A., Lorsch J.R., Hinnebusch A.G., Ramakrishnan V.. Conformational differences between open and closed states of the eukaryotic translation initiation complex. Mol. Cell. 2015; 59:399–412. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Thakur A., Hinnebusch A.G.. eIF1 Loop 2 interactions with Met-tRNAi control the accuracy of start codon selection by the scanning preinitiation complex. Proc. Natl. Acad. Sci. U.S.A. 2018; 115:E4159–E4168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Cross F.R. ‘Marker swap’ plasmids: convenient tools for budding yeast molecular genetics. Yeast. 1997; 13:647–653. [DOI] [PubMed] [Google Scholar]
- 12. Moehle C.M., Hinnebusch A.G.. Association of RAP1 binding sites with stringent control of ribosomal protein gene transcription in Saccharomyces cerevisiae. Mol. Cell. Biol. 1991; 11:2723–2735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Reid G.A., Schatz G.. Import of proteins into mitochondria. Yeast cells grown in the presence of carbonyl cyanide m-chlorophenylhydrazone accumulate massive amounts of some mitochondrial precursor polypeptides. J. Biol. Chem. 1982; 257:13056–13061. [PubMed] [Google Scholar]
- 14. Nanda J.S., Cheung Y.N., Takacs J.E., Martin-Marcos P., Saini A.K., Hinnebusch A.G., Lorsch J.R.. eIF1 controls multiple steps in start codon recognition during eukaryotic translation initiation. J. Mol. Biol. 2009; 394:268–285. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Olsen D.S., E.M. S., Mathew A., Zhang F., Krishnamoorthy T., Phan L., Hinnebusch A.G.. Domains of eIF1A that mediate binding to eIF2, eIF3 and eIF5B and promote ternary complex recruitment in vivo. EMBO J. 2003; 22:193–204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Acker M.G., Kolitz S.E., Mitchell S.F., Nanda J.S., Lorsch J.R.. Reconstitution of yeast translation initiation. Methods Enzymol. 2007; 430:111–145. [DOI] [PubMed] [Google Scholar]
- 17. Maag D., Lorsch J.R.. Communication between eukaryotic translation initiation factors 1 and 1A on the yeast small ribosomal subunit. J. Mol. Biol. 2003; 330:917–924. [DOI] [PubMed] [Google Scholar]
- 18. Kolitz S.E., Takacs J.E., Lorsch J.R.. Kinetic and thermodynamic analysis of the role of start codon/anticodon base pairing during eukaryotic translation initiation. RNA. 2009; 15:138–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Grant C.M., Miller P.F., Hinnebusch A.G.. Requirements for intercistronic distance and level of eIF-2 activity in reinitiation on GCN4 mRNA varies with the downstream cistron. Mol. Cell. Biol. 1994; 14:2616–2628. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Passmore L.A., Schmeing T.M., Maag D., Applefield D.J., Acker M.G., Algire M.A., Lorsch J.R., Ramakrishnan V.. The eukaryotic translation initiation factors eIF1 and eIF1A induce an open conformation of the 40S ribosome. Mol. Cell. 2007; 26:41–50. [DOI] [PubMed] [Google Scholar]
- 21. Dong J., Munoz A., Kolitz S.E., Saini A.K., Chiu W.L., Rahman H., Lorsch J.R., Hinnebusch A.G.. Conserved residues in yeast initiator tRNA calibrate initiation accuracy by regulating preinitiation complex stability at the start codon. Genes Dev. 2014; 28:502–520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Visweswaraiah J., Pittman Y., Dever T.E., Hinnebusch A.G.. The beta-hairpin of 40S exit channel protein Rps5/uS7 promotes efficient and accurate translation initiation in vivo. Elife. 2015; 4:e07939. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Huang H., Yoon H., Hannig E.M., Donahue T.F.. GTP hydrolysis controls stringent selection of the AUG start codon during translation initiation in Saccharomyces cerevisiae. Genes Dev. 1997; 11:2396–2413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Obayashi E., Luna R.E., Nagata T., Martin-Marcos P., Hiraishi H., Singh C.R., Erzberger J.P., Zhang F., Arthanari H., Morris J. et al.. Molecular landscape of the ribosome Pre-initiation complex during mRNA Scanning: Structural role for eIF3c and its control by eIF5. Cell Rep. 2017; 18:2651–2663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Martin-Marcos P., Zhou F., Karunasiri C., Zhang F., Dong J., Nanda J., Kulkarni S.D., Sen N.D., Tamame M., Zeschnigk M. et al.. eIF1A residues implicated in cancer stabilize translation preinitiation complexes and favor suboptimal initiation sites in yeast. Elife. 2017; 6:e31250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Visweswaraiah J., Hinnebusch A.G.. Interface between 40S exit channel protein uS7/Rps5 and eIF2alpha modulates start codon recognition in vivo. Elife. 2017; 6:e22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Dong J., Aitken C.E., Thakur A., Shin B.S., Lorsch J.R., Hinnebusch A.G.. Rps3/uS3 promotes mRNA binding at the 40S ribosome entry channel and stabilizes preinitiation complexes at start codons. Proc. Natl. Acad. Sci. U.S.A. 2017; 114:E2126–E2135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Hinnebusch A.G., Lorsch J.R.. The mechanism of eukaryotic translation initiation: new insights and challenges. Cold Spring Harb. Perspect. Biol. 2012; 4:a011544. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Nanda J.S., Saini A.K., Muñoz A.M., Hinnebusch A.G., Lorsch J.R.. Coordinated movements of eukaryotic translation initiation factors eIF1, eIF1A, and eIF5 trigger phosphate release from eIF2 in response to start codon recognition by the ribosomal preinitiation complex. J. Biol. Chem. 2013; 288:5316–5329. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.