Abstract
Hyaluronic acid (HA) is a glycosaminoglycan of extracellular matrix related to cell surface which interacts with various cell types. To understand the role of HA during hepatocarcinogenesis, we assessed the effect of the inhibition of HA deposition and its association with heterogeneous hepatocellular carcinoma (HCC) cells. In this study, we used transgenic mice C57BL/6J-Tg(Alb1HBV)44Bri/J (HBV-TG) and normal C57BL/6 J (WT) for in vivo study, while HCC cells Huh7 and JHH6 as in vitro models. Both models were treated with an HA inhibitor 4-methylumbelliferone (4MU). We observed that 4MU treatments in animal model down-regulated the mRNA expressions of HA-related genes Has3 and Hyal2 only in HBV-TG but not in normal WT. As observed in vivo, in HCC cell lines, the HAS2 mRNA expression was down-regulated in Huh7 while HAS3 in JHH6, both with or without the presence of extrinsic HA. Interestingly, in both models, the expressions of various cancer stem cells (CD44, CD90, CD133, and EpCAM) were also decreased. Further, histological analysis showed that 4MU treatment with dose 25 mg/kg/day reduced fibrosis, inflammation, and steatosis in vivo, in addition to be pro-apoptotic. We concluded that the inhibition of HA reduced the expressions of HA-related genes and stem cells markers in both models, indicating a possible modulation of cells-to-cells and cells-to-matrix interaction.
Introduction
Hepatocellular carcinoma (HCC) is a major health problem, being the second most common cause of cancer-related death worldwide1. More than 50% HCC cases are related to chronic hepatitis B virus (HBV) infection and chronic HBV carriers have a 100-fold relative risk for developing HCC, with an annual incidence rate of 2–6% in cirrhotic patients2.
One of the main characteristics during hepatocarcinogenesis is the disturbance in the liver architecture, shown by the accumulation of extracellular matrix (ECM) in the liver parenchyma. Hyaluronic acid (hyaluronan, HA) is a linear, large and ubiquitous non-sulfated glycosaminoglycan of the ECM3. HA is found in almost all tissues but it is especially increased in those with active cell proliferation, regeneration, and repair, such as embryonic, inflamed, and tumor stroma tissues4,5. It is one of the markers of fibrosis in liver diseases6,7 and high preoperative serum HA levels can be used to predict poor prognosis in patients with HCC after hepatic resection8.
Previously published studies in various diseases models cancers had shown the relevance of HA blocking by 4-methylumbelliferone (4MU). 4MU is a derivate of 7-hydroxycoumarin (umbelliferones), substances present in many species of plants, especially umbelliferae, fabaceae, and oleaceae. It is dietary supplement since the 1990s to support liver function and detoxification, as well as in clinical trials in patients with chronic hepatitis B and C.
It had been demonstrated that a 12-weeks 4MU supplementation was also shown to ameliorate hypertriglyceridemia and hyperglycemia in mice receiving high-fat diet9. A previous study had shown that treatment of HCC cells with 4MU significantly reduced tumor cell proliferation and induced apoptosis, while primary cultured hepatocytes remained unaffected. In addition, 4MU therapy reduced hepatic and systemic levels of HA. Most important, tumors systemically treated with 4MU show extensive areas of necrosis, inflammatory infiltrates and a 2–3-fold reduction in the number of tumor satellites10.
HA links to cellular receptor CD44 and their interaction in the high-fat diet in mice promoted cancer progression and therapy resistance in different types of cancer, including in HCC11,12. CD44, an adhesion molecule, is a multifunctional cell-surface glycoprotein involved in cell-cell interactions, cell adhesion, and cells migration. It interacts with different cells populations and it is one of the phenotypical markers of cancer stem cells (CSC), either alone or in combination with other CSC markers. However, little is known about the inhibition of HA, CD44, and other cells populations during hepatocarcinogenesis. In this report, we investigate the effect of the inhibition of HA retention in the progression of HCC and its direct/indirect correlation with heterogeneous CSC populations.
Results
Down-regulation of HA genes by 4MU in HBV-TG mice
HA deposition was evident in the liver of HBV-transgenic mice (HBV-TG), especially in the hepatic periportal area while this accumulation was not noticed in wild-type mice (WT).
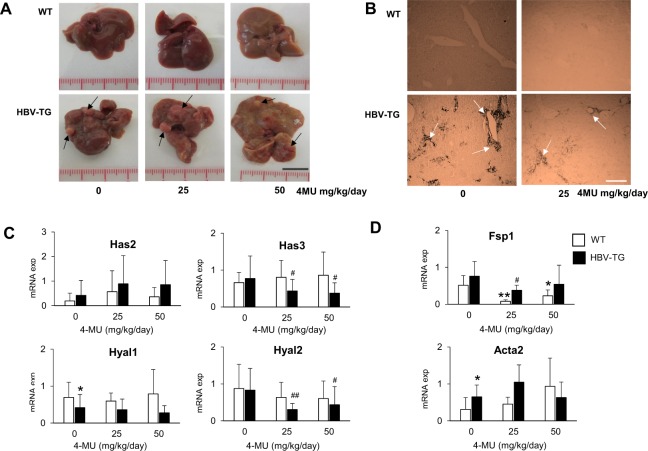
After 4MU treatment with 25 mg/kg/day (0.02%) and 50 mg/kg/day (0.04%) for 12 weeks, we still observed hepatic nodules in HBV-TG. 4MU showed a mild inhibitory effect on the growth of the tumor. Due to the fluorescence effect of 4MU, a yellowish color of the liver was noticed in all treated animals (Fig. 1A). As shown in Fig. 1B, the HA was still noticed in the liver of treated animals. No animals showed any adverse reactions during treatment; only in a group of 50 mg/kg/day HBV-TG mice, a slight increase (10%) of body weight was observed (Table 1).
Figure 1.
Effect of 4MU treatment in HBV-TG mice. (A) Macroscopic appearance of liver after 12 weeks treatment of 25 and 50 mg/kg/day 4MU (scale bar = 1 cm). (B) HA staining of the liver before and after treatment. Magnification: 200X (scale bar = 300 µm). (C) Relative mRNA expressions of hyaluronan synthases Has2 and Has3, and hyaluronidases Hyal1 and Hyal2. (D) Relative mRNA expressions of fibrosis and ECM genes Fsp1 and Acta2. Each mRNA expression of target genes was normalized to reference genes Gapdh and Actb. Statistical analysis: *p < 0.05, **p < 0.01 compared to WT 0 mg/kg/day, #p < 0.05, ##p < 0.01 compared to TG 0 mg/kg/day. WT = wild type mice, HBV-TG = HBV-transgenic mice.
Table 1.
Serum markers and body weight in animal model after 4MU treatment.
| mg/kg/day | WT | HBV-TG | ||||
|---|---|---|---|---|---|---|
| 0 | 25 | 50 | 0 | 25 | 50 | |
| ALT (IU/L) | 47 ± 20 | 73 ± 81 | 110 ± 136 | 186 ± 145 | 246 ± 176 | 314 ± 192 |
| AST (IU/L) | 131 ± 79 | 123 ± 40 | 141 ± 66 | 233 ± 133 | 270 ± 163 | 312 ± 177 |
| LDH (IU/L) | 925 ± 243 | 2002 ± 480** | 2129 ± 638*** | 1453 ± 339 | 2083 ± 440** | 2284 ± 992* |
| Weight (g) | 40 ± 6 | 40 ± 4 | 40 ± 5 | 39 ± 5 | 37 ± 3 | 43 ± 8 |
Statistical student’s t test: *p < 0.05, **p < 0.01, ***p < 0.001 compared to CTRL 0 mg/kg/day of each strain.
The HA-related genes were analyzed in 133 tissue samples (WT n: 20, 23, 21 and HBV-TG n: 23, 26, 20 samples, for treatment 0, 25, and 50 mg/kg/day, respectively). At basal level, HBV-TG mice had higher mRNA expression of HA synthases Has2, and lower hyaluronidase Hyal1 (p < 0.05), as compared to WT. After treatment, RTqPCR data showed that the mRNA expressions of Has3, Hyal1, and Hyal2 were decreased only in HBV-TG by around 35%, 50%, and 65%, respectively. 4MU treatment did not result in any significant effects to the Has3, Hyal1, and Hyal2 of the WT animals. However, in contrast, Has2 mRNA was up-regulated in both strains with high variability (Fig. 1C).
Further analysis of the ECM genes showed that 4MU treatment also reduced the expressions of Fsp1 (fibroblast specific protein 1) in both WT and HBV-TG mice, with the highest effect in WT (p < 0.01). However, this down-regulation was not noticed for Acta2 (alpha smooth muscle actin) (Fig. 1D).
Improvement of fibrosis stages
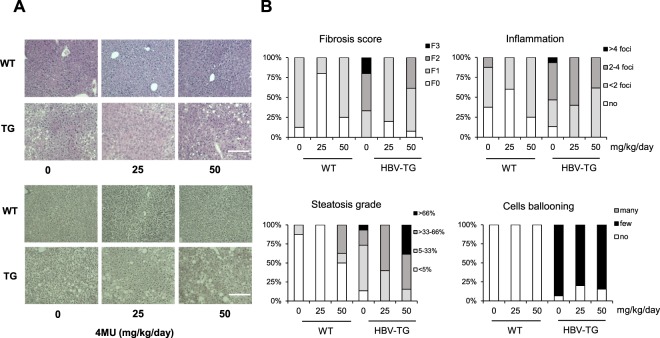
Histological analysis was performed in 21 WT (8, 5, and 8 slices for 0, 25, and 50 mg/kg/day, respectively) and 26 HBV-TG (8, 5, and 13 slices for 0, 25, and 50 mg/kg/day, respectively) as shown in Fig. 2A. The histological scoring system was based on a previous study13.
Figure 2.
4MU treatment improves histology in HBV-TG mice. (A) Representative pictures of H&E staining (upper panel) and reticulum staining (lower panel) (scale bar = 200 µm). (B) The percentage of fibrosis score, steatosis grade, and inflammation scores in the liver. WT = wild type mice, HBV-TG = transgenic mice.
As shown in Fig. 2B, at basal level the proportion of HBV-TG mice with fibrosis stages F3, F2, and F1 were 20%, 50%, and 30%, respectively. After treatment with 4MU of 25 mg/kg/day, this proportion was significantly changed as 80% of animals were F1, while F2 and F3 were not noticed. However, 40% F2 was detected in the group treated with highest 4MU concentration (50 mg/kg/day), even though none of the mice had an F3 stage. The similar beneficial effect was noticed in WT, 4MU treatment with 25 mg/kg/day showed a better result than 50 mg/kg/day, decreasing F1 from 90% to 20% and 70%, respectively.
We also observed the favorable effect of the 25 mg/kg/day treatment on both steatosis and inflammation although administration of 50 mg/kg/day increased the percentage of steatotic cells (40% vs. 10% in control group) which might be correlated with the increase of body weight. The percentage of cells ballooning was also slightly decreased (from 90% to 80%).
Quantification of serum transaminases and LDH
The quantification of serum ALT, AST, and LDH in all 56 animals was shown in Table 1. As we had previously reported14, HBV-TG animals had a significantly higher level of serum ALT (186 ± 145 vs 47 ± 20 IU/L), AST (233 ± 133 vs 131 ± 79 IU/L), and LDH (1453 ± 339 vs 925 ± 243 IU/L) compared to WT, indicating a progressive hepatic damage in these mice.
After treatment, we noticed that the level of serum ALT incresead along with the increase of 4MU concentration. This increase was more noticeable, though not statistically significant, in WT than in HBV-TG. The level of AST remained stable while LDH activity in both mouse models progressively increased, reaching for around 2-fold higher in WT (mean values: 925 to 2129 IU/L, p < 0.01) and 1.6-fold higher in HBV-TG (mean values: 1453 to 2284 IU/L, p < 0.05).
Down-regulation of HA genes by 4MU in HCC cell lines
The relative mRNA expression of HA synthases HAS2 is significantly higher in Huh7 compared to JHH6, (around 700-fold, p < 0.01), while the expression of HAS3 is high in JHH-6 (around 13-fold compared to Huh7). For hyaluronidases, the HYAL1 and HYAL2 mRNA expressions of Huh7 were around 2-fold higher than those of JHH6 (p < 0.05). Both cell lines did not express HAS1 (data not shown).
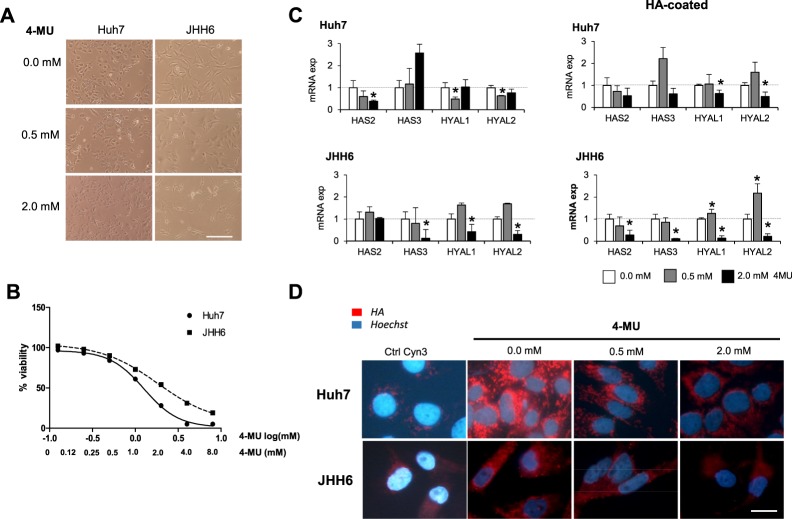
After 4MU treatment for 24 hours, by using MTT test, we noticed that the cytotoxicity of 4MU was dose-dependent, both for Huh7 and JHH6. Cells viability was gradually decreased along with the increased concentration of 4MU. In low concentration 0.5 mM (log[mM] −0.301), both cell lines showed a comparable viability for around 85%. Cells morphology remained similar with control. At high concentration 2 mM (log[mM] 0.301) the JHH6 showed higher viability compared to Huh7, for around 54% and 28% respectively. Furthermore, the cells lost their morphological origins (Fig. 3A and B).
Figure 3.
Effect of 4MU treatment in HCC cells JHH6 and Huh7. (A) Morphological cellular changes after treatment (scale bar = 200 µm). (B) Dose-dependent cytotoxicity of 4MU with concentration ranging from 0.0–8.0 mM by MTT test. Concentration was shown as log[mM]. (C) Relative mRNA expressions of hyaluronan synthases HAS2 and HAS3, and hyaluronidases HYAL1 and HYAL2 after treatment of 0.5 and 2.0 mM of 4MU. mRNA expression was normalized to reference genes 18S and ACTB. Statistical analysis: *p < 0.05, **p < 0.01 compared to untreated control of each cell line (0.0 mM = 1.0) (D) Presence of cytoplasmic HA (scale bar = 20 µm).
Gene expression analysis showed that in both cell lines, the mRNA expressions of HYAL1 and HYAL2 were decreased at 2 mM 4MU treatment. In Huh7, a significant change was already noticed at low concentration treatment 0.5 mM. For HAS2 and HAS3, the pattern of down-regulation in each cell line was different, it found to be depended on their basal expression. In Huh7 with high HAS2, 4MU treatment down-regulated HAS2 for 60% (p < 0.05), but not for HAS3. In contrary, In JHH6 with high HAS3, 4MU significantly down-regulated HAS3 for around 85% (p < 0.05). The maximum down-regulation for both genes in both cell lines was detected at 2 mM treatment (Fig. 3C).
By using immunofluorescence against the HA-binding protein, we observed positivity of cytoplasmic HA of both cells lines. As shown in Fig. 3D, the 4-MU treatment clearly reduced the accumulation of HA.
Interestingly, when the extrinsic HA was introduced by using a HA-coated plate (8,000–15,000 MW), the expressions of HAS2 and HAS3 in both cells were even reduced after treatment. In JHH6, HAS2 was also significantly reduced. For hyaluronidases, when the HA was present, the 4MU increased the expressions of these enzymes, probably in an effort to destroy the HA (Fig. 3C).
Decrease of the CSC markers in HBV-TG mice
To explore the significance of the inhibition of HA in the profile of hepatic CSC, we further examined the expressions of CSC markers in both HCC cell lines and animal model.
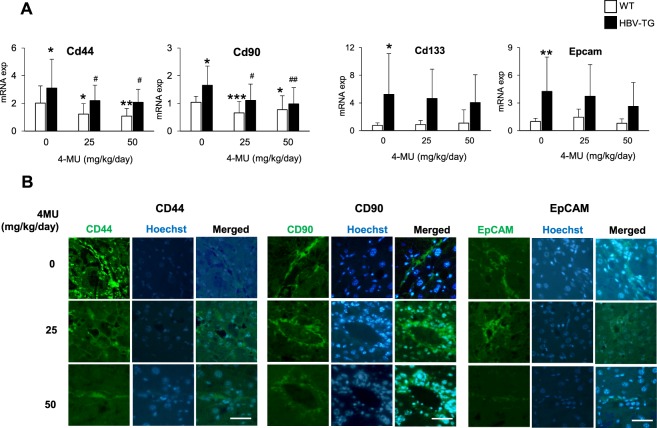
In the in vivo animal model, the CSC markers genes were analyzed in 133 tissue samples (WT n: 20, 23, 21 and HBV-TG n: 23, 26, 20 samples, for treatment 0, 25, and 50 mg/kg/day, respectively). In comparison to WT, the HBV-TG mice showed a significantly higher expressions of Cd44, Cd90, Cd133 (p < 0.05), and Epcam (p < 0.01).
The hepatic mRNA expression of Cd44 was significantly down-regulated by the 4MU treatment in both WT and HBV-TG mice. The down-regulation of Cd44 was accompanied by the decrease of Cd90 (Fig. 4A).
Figure 4.
4MU treatment reduces the expressions of CSC markers in mouse model. (A) Relative mRNA expressions of CSC markers Cd44, Cd90, Cd133, and Epcam in HBV-TG and WT mice. Statistical analysis: *p < 0.05, **p < 0.01 compared to WT 0 mg/kg/day, #p < 0.05, ##p < 0.01 compared to TG 0 mg/kg/day. (B) Representative of immunofluorescence images of protein CD44, CD90, and EpCAM in HBV-TG (scale bar = 50 µm). WT = wild type mice, HBV-TG = HBV-transgenic mice.
An interesting behavior was noticed for Cd133 and Epcam as their expressions were decreased only in TG animals, while the treatment had not effect in WT animals (Fig. 4A). Importantly, since we analyzed different portions of hepatic tissues of each TG mouse, we observed that the down-regulation of CSC markers was clearly observed in hepatic nodules compared to non-nodules. Representative images of the positivity of CSC markers CD44, CD90, and EpCAM proteins are shown in Fig. 4B.
Decrease of the CSC markers in HCC cell lines
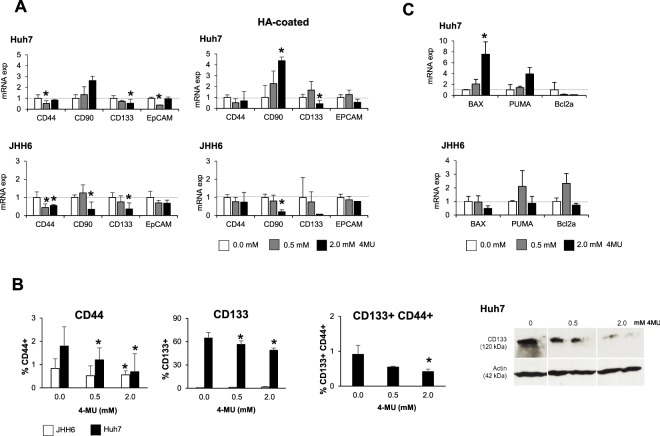
In HCC cell lines model, the expression of CD44, the receptor of HA, was significantly down-regulated (around 50%) in both cell lines after 0.5 mM 4MU treatment (p < 0.05). Interestingly, the expression of the CD133/Prominin-1, a cell surface marker of hepatic CSC, was also down-regulated (60% and 50% less than control for the JHH6 and Huh7, respectively, p < 0.05) (Fig. 5A).
Figure 5.
4MU treatment reduces the expressions of CSC markers in HCC cells JHH6 and Huh7. (A) Relative mRNA expressions of CSC markers CD44, CD90, CD133, and EpCAM after treatment. (B) Flow cytometric analysis on the number of CD44+, CD133+, and CD133+CD44+after treatment (left). Western blot representative of CD133 protein of Huh7 (right). Full-length Western blot is presented in Supplementary Fig. S1. (C) Relative mRNA expressions of apoptosis genes after treatment. mRNA expression was normalized to reference genes 18S and ACTB. Statistical analysis: *p < 0.05 vs. untreated control of each cell line (0.0 mM = 1.0).
This mRNA result was confirmed by flow cytometry where we observed that the percentage of CD44+ cells decreased from 0.8% to 0.5% in JHH6 and from 1.8% to 0.7% in Huh7 after treatment (p < 0.05). The reduction of CD133 was also noticed. The percentage of CD133+ in Huh7 was significantly decreased from 65% to 49% (p < 0.05). The reduction of double-positive CD133+ CD44+ was also observed in Huh7, the percentage of this cell population decreased for around 40% and 50% for 0.5 mM and 2.0 mM (p < 0.05). The protein expression of the CD133 was confirmed by Western Blot (Fig. 5B). The mRNA expression of EpCAM was significantly decreased in Huh7 and in lower extent in JHH6; mRNA expression of CD90 was decreased only in JHH6.
Interestingly, in Huh7, the decrease of CD133 and EpCAM after 4MU treatment was also accompanied by the increase of pro-apoptotic genes PUMA and BAX and the decrease of anti-apoptotic gene Bcl2a (Fig. 5C).
Discussion
In this study we report the effect of 4MU, an HA inhibitor, in the down-regulation of HA-related genes and the reduction of HA accumulation in two sets of experimental models, an HCC-transgenic mouse model and human HCC cell lines. For the animal model, we employed the HBV-transgenic mouse model C57BL/6J-Tg(Alb1HBV)44Bri/J (HBV-TG). This strain contains the HBV fragment codifies the envelope protein (HBsAg) and its intrahepatic concentration corresponds with the incidence of HCC15. Our previous study showed early liver inflammation caused by the HBsAg accumulation together with an increased expression of the markers of hematopoietic stem cells markers in the liver. The progression of liver damage up-regulated different CSC populations, such as EpCAM and CD13314.
Regardless of the etiologies, the main risk factor of HCC development is liver cirrhosis. It is preceded by a progressive fibrosis, characterized by the scarring of fibrous connective tissue as a reparative response to injury or damage. The level of HA has been used as a non-invasive biomarker to assess liver fibrosis (reviewed in16). In chronic hepatitis B, various studies had demonstrated that HA level increased with the increase of the stages of liver fibrosis, to be highest in liver cirrhosis (F4)17–23. It was reported that low serum HA level was associated with spontaneous HBsAg clearance24 while in HCC, high serum HA levels (100 ng/ml or above) were associated with shorter recurrence-free survival. Interestingly, also in patients with HCC without severe liver fibrosis, serum HA level was correlated with the presence of multiple tumors8. In tissues, by using immunostaining, higher expression of HA was significantly exhibited in HCC tissues compared with paired normal liver tissues. High HA expression was also correlated with poor overall survival, large tumor size, and poor tumor differentiation12.
In our animal study, we started the 12 weeks treatment in 9-month old mice, an age when in HBV-TG mice begin the initial development of liver dysplasia leading to HCC at 12-month of age. The 12-month old HBV-TG mice had higher expression of Has2 and lower expression of Hyal1 compared to WT mice. And as expected, HBV-TG mice showed a high incidence of liver fibrosis stages F2 and F3 and the deposition of HA in the liver. They also had higher expression of fibrosis marker Acta2 and Fsp1. After treatment with 4MU we observed the decrease of HA deposition in the liver. Importantly, it was accompanied by a significant reduction in the incidences of F2 and F3 in HBV-TG mice, especially in 25 mg/kg/day treatment. It is also noteworthy to notice that the reversal of hepatic histological states was also observed by the improvement of steatosis and inflammation, as well as cells ballooning.
Our gene expression analysis showed that 4MU treatment reduced the mRNA expressions of HA-synthesis (HAS) Has3 and HA-degradation (hyaluronidase) enzyme Hyal2, while it slightly down-regulated Hyal1. Has3 codifies for the production of low molecular weight HA that has been shown to induce inflammatory cytokines25. Our data on the down-regulation of Has3 mRNA and the reduction of fibrosis and inflammation is consistent with previous animal studies in lung and colitis model25–27. Previously, it was shown that Has3 mRNA in the liver was increased after carbon tetrachloride exposure for fibrosis induction in normal mice28.
One of the important findings was that the down-regulation of HA-related mRNAs were observed only in treated HBV-TG mice, but not in WT mice, suggesting that 4MU does not affect the HA synthesis in normal hepatic cells. This conclusion is in line with previous reports showing the anticancer effect of 4MU in HCC. In xenograft HCC models, 4MU significantly reduced tumor cell proliferation while primary cultured hepatocytes remained unaffected10. 4MU also reduced the expression of CD31 and inhibited systemic level of VEGF and IL-6 production was dramatically reduced in isolated Kupffer cells29.
To better understand the data obtained in vivo, we performed in vitro experiments using two human HCC cell lines with different degree of differentiation, Huh7 and JHH6. We observed that HAS2 mRNA was highly expressed in Huh7, a well-differentiated HCC cell line, while on the contrary HAS3 was highly expressed in JHH6, a poor differentiated HCC cell line. As exhibited in mouse models, 4MU treatment reduced the expressions of HAS in vitro: it down-regulated the high basal level of HAS2 mRNA in Huh7, and the high level of HAS3 mRNA in JHH6. This finding suggests that the effect of 4MU in targeting HA-enzymes depends on the initial level of HAS in the cells.
As in mouse model, 4MU also down-regulated the mRNA of hyaluronidases HYAL1 and HYAL2 in both cell lines. The results in HCC are in line with previous reports in vitro in breast, melanoma, ovarian, squamous carcinoma cell lines30,31 and in vivo mouse models of bladder, malignant pleural mesothelioma, malignant peripheral nerve sheath, and mostly in pancreatic cancer32–37.
To observe the effect of 4MU in the deposition of extracellular HA, we added gene expression analysis of HA-related enzymes in the presence of extrinsic HA. By treating the cells on the HA-coated plate (8,000–15,000 MW), we observed a higher expressions hyaluronidases compared to cells grown in non-coated plates, probably in an effort to destroy the presence of HA. In addition, the reduction of both HAS2 and HAS3 were also noticed in JHH6.
Further, we checked whether the reduction of HA enzymes mRNA was also accompanied by the decrease of its cell receptor CD44. The CD44 molecule (Indian blood group) is a multifunctional cell-surface glycoprotein involved in cell-cell interactions, cell adhesion, and cells migration. It serves as an adhesion/homing protein receptor for the HA in the ECM38. The expression of CD44 in HCC is associated with poor prognosis and tumor aggressiveness39,40. Our data in human samples showed that CD44 mRNA was increased in HCC as compared to normal liver tissues (data not shown). Further, CD44 has been implicated as one of CSC markers in HCC, as a single marker as well as in combination with other CSC markers.
In bone metastasis cell lines, it had been reported that CD44 cell acts as the CSC and promotes bone metastases by enhancing tumorigenicity, cell motility, and HA production41. Accordingly, we also investigated whether other CD44-associated CSC markers, CD90, CD133, and EpCAM42–45, were affected by 4MU treatment. We observed that both in mice model and in human HCC cell lines, CD44 mRNA was decreased after 4MU treatment. The decrease of CD44 mRNA in cells Huh7 and JHH6 was confirmed by the reduction of CD44+ cells using flow cytometry where the CD44-positivity was reduced by about 50% after treatment in both cell lines.
Besides the decrease of CD44 cells, we found the 4MU treatment also reduced various HCC CSC markers. In mouse models, 4MU significantly decreased the expression of Cd90 in both HBV-TG and in WT mice in a concentration-dependent manner, as for Cd44. Interestingly, even though we did not reach significant value due to the high heterogeneity of mRNA expressions, we observed the mRNA expressions of Cd133 and Epcam were also down-regulated.
In line with the in vivo model, 4MU reduced the expressions of CD133 and EpCAM in both Huh7 and JHH6, while the CD90 mRNA expression was reduced only in JHH6. As the down-regulation of CD133 was found to be the most significant, we performed a quantification of CD133+ cells and CD133 protein expression in Huh7 by using flow cytometry and Western blot, respectively. Both analyses confirmed the down-regulation of CD133 molecule after 4MU treatment.
The mechanisms by which 4MU inhibits the properties of CSC is still unclear. In oral squamous cell carcinoma cell lines, a population identified as rapid adherent cells (RAC) to collagen-IV was enriched for its high tumorigenic ability. Using scanning electron microscopy, 4MU was shown to altered membrane morphology of the RAC and significantly decreased their membrane capacitance46.
In the aforementioned bone metastasis study, 4MU decreased CD44 together with CSC tumor sphere and osteoclast-like cell formation in vitro41. A study in colorectal, osteosarcoma, and lung cancer cell lines showed that irradiation induced the expression of HAS2, resulting in their radioresistance, one of the hallmarks of the CSC47. Here, we observed that 4MU increased the pro-apoptosis genes Bax and Puma, while decreased the anti-apoptosis gene Bcl2a in Huh7. It was known that CSC CD133 is correlated with anti-apoptosis property48,49. However, the effect of 4MU in the epithelial mesenchymal transition (EMT), another hallmark of CSC, was unclear. Perhaps, a treatment for 24 hours was not sufficient to block the EMT (supplementary material).
Our study in a transgenic model is in line with a recent data on the 4MU treatment in an orthotopic HCC model established in fibrotic livers. 4MU, in combination with an adenovirus encoding interleukin-12 genes elicited a potent antitumor effect accompanied by the reduction of mRNA levels of CSC markers50.
In this study, we found that 4MU therapy in this in vivo model was acceptable for both experimental doses of 25 and 50 mg/kg/day. Even though we did not see the disappearance of liver nodules, a clear improvement in liver histology and the marked decrease of CSC markers in nodules was observed supporting the anticancer properties of 25 mg/kg/day of 4MU. A 4MU study in osteosarcoma mice model showed only a mild inhibitory effect on the growth of the primary tumor but a marked inhibition (75% reduction) in lung metastasis51.
In conclusion, we demonstrated that the reduction of HA by 4MU lowered HA-related genes that reduced deposition of HA in HCC. The inhibition of HA was accompanied by a reduction of CSC markers CD44 and CD133, as well as CD90 and EpCAM cells, indicating a possible mechanism of HA in cells-to-cells and cells-to-matrix interaction.
Materials and Methods
Animal model
Fifty-six male Hepatitis B Virus (HBV)-transgenic mouse C57BL/6J-Tg(Alb1HBV)44Bri/J (HBV-TG, n = 28) and its wild-type counterpart C57BL/6 J (WT, n = 28) were maintained in the animal facility of the University of Trieste. Mice were randomly selected for the treatment group. The animal procedure was performed in accordance with the proper care and use of laboratory animals. The study protocol was approved by the Ethical Committee of the University of Trieste and the Ministry of Health of the Republic of Italy (Decreto no: 57/2012 – B).
HCC cell lines
Human HCC cell lines Huh7 (JCRB0403) and JHH6 (JCRB1030) were obtained from the Japan Health Science Research Resources Bank (HSRRB, Tokyo, Japan). Huh7 cells were grown in DMEM medium (high glucose) supplemented with 10% (v/v) FBS, 1% L-glutamine and 1% antibiotics. JHH6 cells were grown in Williams’ E medium supplemented with 10% (v/v) FBS, 1% L-glutamine and 1% antibiotics. The cultures were maintained at 37 °C in a humidified 5% CO2 incubator and when they reached 80–90% confluence they were routinely expanded by 0.05% trypsin detachment.
Inhibition of HA deposition by 4MU
For in vivo study, 9 month old mice, for both HBV-TG (initial development of liver dysplasia) and WT, were given 4MU (Sigma Aldrich, Milan, Italy) with concentration 25 mg/kg/day (0.02%) and 50 mg/kg/day (0.04%) by oral administration in drinking water for 12 weeks. Mice without 4MU administration were used as control group (0 mg/kg/day). At the end of treatment, animals were weighted and euthanized in deep anesthesia. Blood and liver tissues were collected immediately after sacrifice. Three different portions of liver were sliced and collected in sterile tubes for subsequent gene, protein, and histochemical analysis. Body and liver weight were measured.
For in vitro study, HCC cell line Huh7 and JHH6 cells were cultured in a 6-well plate with or without 0.2 mg/ml HA (8,000–15,000 MW) (Sigma) with an initial concentration of 100,000 cell/ml and 50,000 cell/ml, respectively, for 24 hours. 4MU was dissolved in stock solution and subsequently in the growth medium. 4MU concentration ranging from 0 to 8 mM was given for treatment for 24 hours. After treatment, the cells were collected for flow cytometry analysis, total RNA extraction, and total protein extraction. The protocol of HA-coating was based on a previous study52.
Quantification of serum transaminases and lactate dehydrogenase
After collection, mice sera were separated from whole blood and they were stored at −80 °C until analyzed. Serum alanine aminotransferase (ALT), aspartate aminotransferase (AST), and lactate dehydrogenase (LDH) were quantified with the standard enzymatic methods (Roche Cobas Analyzer) at the IRCCS Burlo Garofalo children hospital.
Histology
Liver tissues were fixed in 4% formalin for at least 24 hours. The samples were included in paraffin block with the automated Sakura method at the Santa Maria della Misericordia Hospital. The fixed slices were subjected to hematoxylin & eosin (HE) and reticulum staining to assess the staging of liver fibrosis. Histological analysis was examined by a pathologist. Diagnostic and the scoring system in histological analysis was referred to the validated consensus of non-alcoholic steatohepatitis and fibrosis13.
HA staining
HA staining of mouse liver paraffinated slices was performed as described previously10. Briefly, paraffin liver sections were incubated with 3% H2O2–methanol for 30 min at room temperature to block endogenous peroxidase, followed by avidin, biotin and protein-blocking solution (Vector, Buenos Aires, Argentina), and bHA-BP (# 385911, Calbiochem) in BSA–PBS. Negative controls was stained with bHA-BP and pretreated with 100 U/mL of Streptomyces hyaluronidase (Calbiochem). Peroxidase complex (Sigma-Aldrich, Buenos Aires, Argentina) was used as a revealing system.
In human HCC cell lines, the cytoplasmic HA positivity were detected by hyaluronic acid binding protein (HABP). After 4MU treatment, cells grown in cover slip were fixed in 3% paraformaldehyde. Cells were then blocked, permeabilized and quenched by 1% BSA, 0.1% Triton X, 50Mm Glycine in PBS. Incubation with HABP-biotin labeled (5 ug/ml) (Merck, Milan, Italy) that binds specifically and strongly to HA ≥ 2000 MW was done overnight in 4 °C in a humid chamber. The day after, cells were washed and incubated with streptavidin-Cyn3 for 1 hour in room temperature. The nuclei were stained by Hoechst 33258. Controls without HABP (ctrl Cyn3) were included. Images were generated using fluorescence microscope Leica DM2000 (Leica, Wetzlar, Germany).
Total RNA isolation
Total RNA from HCC cell lines and mouse liver tissues was extracted using the TriReagent (Sigma Aldrich) according to the manufacture’s protocol. RNA was quantified at 260 nm in a DU®730 spectrophotometer (Beckman Coulter, Fullertone, CA, USA). The RNA purity was evaluated according to MIQE guidelines53 by measuring the ratio A260/A280 with appropriate purity values between 1.8 and 2.0.
Reverse Transcription quantitative real time PCR (RT-qPCR)
cDNA synthesis was obtained by reverse transcription (RT) of 1 µg of total RNA using High Capacity cDNA Reverse Transcription Kits (Applied Biosystems, Foster City, CA, USA) according to the manufacture’s suggestions.
RT-qPCR was performed using iQ SYBR Green Supermix protocol (Bio-Rad Laboratories, Milan, Italy). Each PCR amplification was carried out in 15 µL reaction volume containing 25 ng of cDNA, 1x iQ SYBR Green Supermix, and 250 nM gene-specific sense and anti-sense primers. Reactions were run on a Bio-Rad iQ5 real-time PCR detection system (iCycler IQ5 software, version 3.1; Bio-Rad) together with two reference genes. The list of the primers description and sequences were shown in Supplementary Materials.
Flow cytometry of CSC markers
The presence of CSC surface marker antigens was detected using antibodies CD90/THY1 (Stem Cell Technologies, Vancouver, BC, Canada), CD133/PROM1 (Miltenyi Biotec GmbH, Bergisch Gladbach, Germany), and CD44 (Abcam, Cambridge, UK). Flow cytometric analysis was performed immediately on a FACSCalibur flow cytometer (Becton Dickinson, NJ, USA). A total of 10,000 events were analyzed per sample. Data were presented as a mean for at least three independent experiments.
Immunofluorescence of CSC markers
For mouse liver tissues, after de-paraffinization with xylene and rehydration with gradual concentration of ethanol, antigen retrieval was performed by microwave heat in sodium citrate pH 6.0. The autofluorescence background of paraffin tissues was quenched by combination of photo bleaching and Sudan Black B dye54. Direct immunostaining was performed with fluorochromes-conjugated antibodies against mouse CD44 (Miltenyi Biotec) CD90 (Santa Cruz Biotechnology, Californa, USA) and CD326/EpCAM, Miltenyi Biotec). Proteins positivity was observed by using a fluorescence microscope Leica DM2000.
Protein extraction and Western Blot
Western Blot was performed for CD133. Total protein of Huh7 was extracted by using cell lysis buffer. Twenty ug of total protein of Huh7 were size-separated, together with molecular weight standards (Fermentas) by (SDS–PAGE) on polyacrylamide gel, using a Mini Protein III Cell (Bio-Rad). After SDS–PAGE, proteins were electro-transferred with a semi-dry blotting system onto immune-blot PVDF membranes (Bio-Rad) using a Mini Trans-Blot Cell (Bio-Rad). Membrane was incubated overnight with anti-CD133 (clone W6B3C1) and with peroxidase-conjugated secondary antibody. Actin was used as a housekeeping protein. The peroxidase reaction was obtained by exposure of membrane in the ECL-Plus Western blot detection system solutions (ECL Plus Western blotting Detection Reagents, GE-Healthcare Bio-Sciences, Italia).
Statistical analysis
Statistical analysis was performed by one-way ANOVA with post-hoc Bonferroni’s multiple comparison sets. Students’t test was performed for statistical comparison between groups (cells and strains) using software InStat Version 3.05 (GraphPad Software, Inc., La Jolla, CA, USA). Statistical significance was set to p < 0.05.
Supplementary information
Acknowledgements
The authors thank the support of Dr. Stebel, Dr. Barbetta, and Dr. Zarattini (animal facility, University of Trieste) and the group of Prof. Bulla (Department of Life Sciences, University of Trieste) for the reagents for HABP staining. BA was supported by a fellowship the Italian Association for the Study of the Liver (AISF), SI by a fellowship from the Istituto Italiano di Cultura, Jakarta and Fondazione Italiana Fegato, CHCS by the University of Trieste. This study was supported by the project 297 Nutrizione e Salute (2012–2014) ‘Dalla nutrigenetica alla nutraceutical’ of the Italian Ministro dell’ Istruzione, dell’ Università e della Ricerca (Art. 13 D.LGS 297/99) and in part by an in-house grant from Fondazione Italiana Fegato.
Author Contributions
C.H.C.S., B.A., C.T. and G.M. designed the study. C.H.C.S. and B.A. performed animal and cell lines experiments. C.H.C.S., B.A. and S.I.I. performed gene expression, flow cytometry, immunofluorescence, and W.B. experiments. E.F. performed H.A. staining. C.A. performed histopathological analysis. F.V. and A.R. quantified serum markers. C.H.C.S., C.T. and G.M. wrote the manuscript. The Ms was read and approved by all authors.
Data Availability
The data and images generated during the current study are available from the corresponding author on reasonable request.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-40436-6.
References
- 1.Bray F, Ren JS, Masuyer E, Ferlay J. Global estimates of cancer prevalence for 27 sites in the adult population in 2008. Int. J. Cancer. 2013;132:1133–1145. doi: 10.1002/ijc.27711. [DOI] [PubMed] [Google Scholar]
- 2.Llovet JM, Burroughs A, Bruix J. Hepatocellular carcinoma. Lancet. 2003;362:1907–1917. doi: 10.1016/S0140-6736(03)14964-1. [DOI] [PubMed] [Google Scholar]
- 3.Toole BP. Hyaluronan: from extracellular glue to pericellular cue. Nat. Rev. Cancer. 2004;4:528–539. doi: 10.1038/nrc1391. [DOI] [PubMed] [Google Scholar]
- 4.Toole BP. Hyaluronan promotes the malignant phenotype. Glycobiology. 2002;12:37R–42R. doi: 10.1093/glycob/12.3.37R. [DOI] [PubMed] [Google Scholar]
- 5.Alaniz L, Garcia M, Rizzo M, Piccioni F, Mazzolini G. Altered hyaluronan biosynthesis and cancer progression: an immunological perspective. Mini Rev. Med. Chem. 2009;9:1538–1546. doi: 10.2174/138955709790361485. [DOI] [PubMed] [Google Scholar]
- 6.Parés A, et al. Serum hyaluronate reflects hepatic fibrogenesis in alcoholic liver disease and is useful as a marker of fibrosis. Hepatology. 1996;24:1399–1403. doi: 10.1002/hep.510240615. [DOI] [PubMed] [Google Scholar]
- 7.Guha IN, Parkes J, Roderick PR, Harris S, Rosenberg WM. Non-invasive markers associated with liver fibrosis in non-alcoholic fatty liver disease. Gut. 2006;55:1650–1660. doi: 10.1136/gut.2006.091454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mima K, et al. Preoperative serum hyaluronic acid level as a prognostic factor in patients undergoing hepatic resection for hepatocellular carcinoma. Br. J. Surg. 2014;101:269–276. doi: 10.1002/bjs.9343. [DOI] [PubMed] [Google Scholar]
- 9.Sim MO, Ham JR, Lee H-I, Seo KI, Lee MK. Long-term supplementation of umbelliferone and 4-methylumbelliferone alleviates high-fat diet induced hypertriglyceridemia and hyperglycemia in mice. Chem. Biol. Interact. 2014;216:9–16. doi: 10.1016/j.cbi.2014.03.003. [DOI] [PubMed] [Google Scholar]
- 10.Piccioni F, et al. Antitumor effects of hyaluronic acid inhibitor 4-methylumbelliferone in an orthotopic hepatocellular carcinoma model in mice. Glycobiology. 2012;22:400–410. doi: 10.1093/glycob/cwr158. [DOI] [PubMed] [Google Scholar]
- 11.Henry JC, et al. miR-199a-3p targets CD44 and reduces proliferation of CD44 positive hepatocellular carcinoma cell lines. Biochem. Biophys. Res. Commun. 2010;403:120–125. doi: 10.1016/j.bbrc.2010.10.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Li JH, et al. Over expression of hyaluronan promotes progression of HCC via CD44-mediated pyruvate kinase M2 nuclear translocation. Am. J. Cancer Res. 2016;6:509–521. [PMC free article] [PubMed] [Google Scholar]
- 13.Kleiner DE, et al. Design and validation of a histological scoring system for nonalcoholic fatty liver disease. Hepatology. 2005;41:1313–1321. doi: 10.1002/hep.20701. [DOI] [PubMed] [Google Scholar]
- 14.Anfuso B, et al. Activation of hepatic stem cells compartment during hepatocarcinogenesis in a HBsAg HBV-transgenic mouse model. Sci. Rep. 2018;8:13168. doi: 10.1038/s41598-018-31406-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Dunsford HA, Sell S, Chisari FV. Hepatocarcinogenesis due to chronic liver cell injury in hepatitis B virus transgenic mice. Cancer Res. 1990;50:3400–3407. [PubMed] [Google Scholar]
- 16.Neuman MG, Cohen LB, Nanau RM. Hyaluronic acid as a non-invasive biomarker of liver fibrosis. Clin. Biochem. 2016;49:302–315. doi: 10.1016/j.clinbiochem.2015.07.019. [DOI] [PubMed] [Google Scholar]
- 17.Stibbe KJM, et al. Comparison of non-invasive assessment to diagnose liver fibrosis in chronic hepatitis B and C patients. Scand. J. Gastroenterol. 2011;46:962–972. doi: 10.3109/00365521.2011.574725. [DOI] [PubMed] [Google Scholar]
- 18.Lee KG, et al. Usefulness of non‐invasive markers for predicting liver cirrhosis in patients with chronic hepatitis B. J. Gastroenterol. Hepatol. 2009;25:94–100. doi: 10.1111/j.1440-1746.2009.05953.x. [DOI] [PubMed] [Google Scholar]
- 19.Park SH, et al. Usefulness of multiple biomarkers for the prediction of significant fibrosis in chronic hepatitis B. J. Clin. Gastroenterol. 2011;45:361–365. doi: 10.1097/MCG.0b013e31820d3458. [DOI] [PubMed] [Google Scholar]
- 20.Kim BK, et al. Prospective validation of ELF test in comparison with Fibroscan and FibroTest to predict liver fibrosis in Asian subjects with chronic hepatitis B. Plos One. 2012;7:e41964. doi: 10.1371/journal.pone.0041964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Li F, et al. Role of hyaluronic acid and laminin as serum markers for predicting significant fibrosis in patients with chronic hepatitis B. Braz. J. Infect. Dis. 2012;16:9–14. doi: 10.1016/s1413-8670(12)70267-2. [DOI] [PubMed] [Google Scholar]
- 22.Chen J, et al. Study on noninvasive laboratory tests for fibrosis in chronic HBV infection and their evaluation. J. Clin. Lab. Anal. 2013;27:5–11. doi: 10.1002/jcla.21554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Montazeri G, et al. Serum hyaluronate as a non-invasive marker of hepatic fibrosis and inflammation in HBeAg-negative chronic hepatitis B. BMC Gastroenterol. 2005;5:32. doi: 10.1186/1471-230X-5-32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Harkisoen S, et al. Low serum hyaluronic acid levels associated with spontaneous HBsAg clearance. Eur. J. Clin. Microbiol. Infect. Dis. 2015;34:2183–2189. doi: 10.1007/s10096-015-2467-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mascarenhas MM, et al. Low molecular weight hyaluronan from stretched lung enhances interleukin-8 expression. Am. J. Respir. Cell Mol. Biol. 2004;30:51–60. doi: 10.1165/rcmb.2002-0167OC. [DOI] [PubMed] [Google Scholar]
- 26.Bai KJ, et al. The role of hyaluronan synthase 3 in ventilator-induced lung injury. Am. J. Respir. Crit. Care Med. 2005;172:92–98. doi: 10.1164/rccm.200405-652OC. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Kessler SP, Obery DR, de la Motte C. Hyaluronan synthase 3 null mice exhibit decreased intestinal inflammation and tissue damage in the DSS-induced colitis model. Int. J. Cell Biol. 2015;2015:745237. doi: 10.1155/2015/745237. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.McCracken JM, Jiang L, Deshpande KT, O’Neil MF, Pritchard MT. Differential effects of hyaluronan synthase 3 deficiency after acute vs chronic liver injury in mice. Fibrogenesis Tissue Repair. 2016;9:4. doi: 10.1186/s13069-016-0041-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Piccioni F, et al. 4-methylumbelliferone inhibits hepatocellular carcinoma growth by decreasing IL-6 production and angiogenesis. Glycobiology. 2015;25:825–835. doi: 10.1093/glycob/cwv023. [DOI] [PubMed] [Google Scholar]
- 30.Kultti A, et al. 4-Methylumbelliferone inhibits hyaluronan synthesis by depletion of cellular UDP-glucuronic acid and downregulation of hyaluronan synthase 2 and 3. Exp. Cell Res. 2009;315:1914–1923. doi: 10.1016/j.yexcr.2009.03.002. [DOI] [PubMed] [Google Scholar]
- 31.Urakawa H, et al. Inhibition of hyaluronan synthesis in breast cancer cells by 4-methylumbelliferone suppresses tumorigenicity in vitro and metastatic lesions of bone in vivo. Int. J. Cancer. 2012;130:454–466. doi: 10.1002/ijc.26014. [DOI] [PubMed] [Google Scholar]
- 32.Morera DS, et al. Hyaluronic acid family in bladder cancer: potential prognostic biomarkers and therapeutic targets. Br. J. Cancer. 2017;117:1507–1517. doi: 10.1038/bjc.2017.318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Nagase H, et al. 4-Methylumbelliferone suppresses hyaluronan synthesis and tumor progression in SCID mice intra-abdominally inoculated with pancreatic cancer cells. Pancreas. 2017;46:190–197. doi: 10.1097/MPA.0000000000000741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Cho H, et al. Trametinib plus 4-Methylumbelliferone exhibits antitumor effects by ERK blockade and CD44 downregulation and affects PD-1 and PD-L1 in malignant pleural mesothelioma. J. Thorac. Oncol. 2017;12:477–490. doi: 10.1016/j.jtho.2016.10.023. [DOI] [PubMed] [Google Scholar]
- 35.Yoshida E, et al. Antitumor effects of the hyaluronan inhibitor 4-methylumbelliferone on pancreatic cancer. Oncol. Lett. 2016;12:2337–2344. doi: 10.3892/ol.2016.4930. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kudo, D., Suto, A. & Hakamada, K. The development of a novel therapeutic strategy to target hyaluronan in the extracellular matrix of pancreatic ductal adenocarcinoma. Int. J. Mol. Sci. 18, (2017). [DOI] [PMC free article] [PubMed]
- 37.Ikuta K, et al. Antitumor effects of 4-methylumbelliferone, a hyaluronan synthesis inhibitor, on malignant peripheral nerve sheath tumor. Int. J. Cancer. 2017;140:469–479. doi: 10.1002/ijc.30460. [DOI] [PubMed] [Google Scholar]
- 38.Misra S, et al. Hyaluronan-CD44 interactions as potential targets for cancer therapy. FEBS J. 2011;278:1429–1443. doi: 10.1111/j.1742-4658.2011.08071.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Gao Y, et al. Knockdown of CD44 inhibits the invasion and metastasis of hepatocellular carcinoma both in vitro and in vivo by reversing epithelial-mesenchymal transition. Oncotarget. 2015;6:7828–7837. doi: 10.18632/oncotarget.3488. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Luo Y, Tan Y. Prognostic value of CD44 expression in patients with hepatocellular carcinoma: meta-analysis. Cancer Cell Int. 2016;16:47. doi: 10.1186/s12935-016-0325-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hiraga T, Ito S, Nakamura H. Cancer stem-like cell marker CD44 promotes bone metastases by enhancing tumorigenicity, cell motility, and hyaluronan production. Cancer Res. 2013;73:4112–4122. doi: 10.1158/0008-5472.CAN-12-3801. [DOI] [PubMed] [Google Scholar]
- 42.Yang ZF, et al. Significance of CD90+cancer stem cells in human liver cancer. Cancer Cell. 2008;13:153–166. doi: 10.1016/j.ccr.2008.01.013. [DOI] [PubMed] [Google Scholar]
- 43.Zhu Z, et al. Cancer stem/progenitor cells are highly enriched in CD133+CD44+population in hepatocellular carcinoma. Int. J. Cancer. 2010;126:2067–2078. doi: 10.1002/ijc.24728. [DOI] [PubMed] [Google Scholar]
- 44.Cao L, et al. Sphere-forming cell subpopulations with cancer stem cell properties in human hepatoma cell lines. BMC Gastroenterol. 2011;11:71. doi: 10.1186/1471-230X-11-71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Seto K, et al. A novel small-molecule WNT inhibitor, IC-2, has the potential to suppress liver cancer stem cells. Anticancer Res. 2017;37:3569–3579. doi: 10.21873/anticanres.11727. [DOI] [PubMed] [Google Scholar]
- 46.Liang X, Graham KA, Johannessen AC, Costea DE, Labeed FH. Human oral cancer cells with increasing tumorigenic abilities exhibit higher effective membrane capacitance. Integr. Biol. 2014;6:545–554. doi: 10.1039/C3IB40255J. [DOI] [PubMed] [Google Scholar]
- 47.Shen YN, et al. Inhibition of HAS2 induction enhances the radiosensitivity of cancer cells via persistent DNA damage. Biochem. Biophys. Res. Commun. 2014;443:796–801. doi: 10.1016/j.bbrc.2013.12.026. [DOI] [PubMed] [Google Scholar]
- 48.Piao LS, et al. CD133+liver cancer stem cells modulate radioresistance in human hepatocellular carcinoma. Cancer Lett. 2012;315:129–137. doi: 10.1016/j.canlet.2011.10.012. [DOI] [PubMed] [Google Scholar]
- 49.Zhang X, Shao J, Li X, Cui L, Tan Z. Docetaxel promotes cell apoptosis and decreases SOX2 expression in CD133-expressing hepatocellular carcinoma stem cells by suppressing the PI3K/AKT signaling pathway. Oncol. Rep. 2018;41:1067–1074. doi: 10.3892/or.2018.6891. [DOI] [PubMed] [Google Scholar]
- 50.Rodríguez MM, et al. 4Mu decreases CD47 expression on hepatic cancer stem cells and primes a potent antitumor T cell response induced by interleukin-12. Mol. Ther. 2018;26:2738–2750. doi: 10.1016/j.ymthe.2018.09.012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Arai E, et al. Inhibition of hyaluronan retention by 4-methylumbelliferone suppresses osteosarcoma cells in vitro and lung metastasis in vivo. Br. J. Cancer. 2011;105:1839–1849. doi: 10.1038/bjc.2011.459. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Corradetti B, et al. Hyaluronic acid coatings as a simple and efficient approach to improve MSC homing toward the site of inflammation. Sci. Rep. 2017;7:7991. doi: 10.1038/s41598-017-08687-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Bustin SA, et al. The MIQE guidelines: minimum information for publication of quantitative real-time PCR experiments. Clin. Chem. 2009;55:611–622. doi: 10.1373/clinchem.2008.112797. [DOI] [PubMed] [Google Scholar]
- 54.Viegas MS, Martins TC, Seco F, do Carmo A. An improved and cost-effective methodology for the reduction of autofluorescence in direct immunofluorescence studies on formalin-fixed paraffin-embedded tissues. Eur. J. Histochem. 2007;51:59–66. [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The data and images generated during the current study are available from the corresponding author on reasonable request.