Abstract
Background
Transcriptomic research of blood cell lineages supports the understanding of distinct features of the immunopathology in human malaria.
Methods
We used microarray hybridization, validated by real-time RT-PCR to analyze whole blood gene expression in healthy Gabonese children and children with various conditions of Plasmodium falciparum infection, including i) asymptomatic infection, ii) uncomplicated malaria, iii) malaria associated with severe anemia and iv) cerebral malaria.
Findings
Our data indicate that the expression profile of 22 genes significantly differed among the investigated groups. Immunoglobulin production, complement regulation and IFN beta signaling, in particular IRF7 and ISRE binding signatures in the corresponding genes, were most conspicuous. Down-regulation in cerebral malaria seems to rely on AhRF, GABP and HIF1 hypoxia transcription factors. ARG1, BPI, CD163, IFI27, HP and TNFAIP6 transcript levels correlated positively with lactatemia, and negatively with hemoglobin concentrations.
Interpretation
Differences in gene expression profile reflect distinct immunopathological mechanisms of P. falciparum infection. They emerge as potential prognostic markers for early therapeutic measures and need to be validated further.
Fund
This work was supported by a grant of the NGFN (Nationales Genomforschungsnetz 01GS0114) and by a CNPq (Conselho Nacional de Desenvolvimento Científico e Tecnológico, Brazil) PhD scholarship for A. B. W. Boldt. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
Keywords: Uncomplicated malaria, Severe malaria, Cerebral malaria, Transcriptome, Plasmodium falciparum, Microarray, Gabon
Research in context.
Evidence before this study
Based on microarray technology, several studies have tried to more precisely describe particular features of the host's immune response against the Plasmodium parasite in both naturally and experimentally infected humans. Here, we extend previous studies on the expression of candidate genes in whole blood of Gabonese children with malaria and describe the whole blood signature of gene expression in relevant clinical presentations of childhood malaria.
Added value of this study
Modifications in gene expression when different blood cells interact with the parasite during P. falciparum infection were demonstrated. Immunoglobulin production, complement regulation and IFN beta signaling emerged as most discrepant features between uncomplicated malaria and other clinical presentations of P. falciparum infections. Especially, 22 genes significantly differed among the investigated groups. Importantly, the expression of genes were correlated with clinical parameters of malaria, such as lactatemia and hemoglobin concentrations.
Implication of all the available evidence
This work is an attempt to describe the whole blood signature of gene expression in the most important clinical manifestations of childhood malaria. This study provides specific signatures for gene expression profiles and could differentiate among uncomplicated malaria, severe malarial anemia and cerebral malaria phenotypes.
Alt-text: Unlabelled Box
1. Introduction
Although treatable and significantly declining over the last decade, malaria is still a major health problem in many tropical regions, especially in sub-Saharan Africa. According to the World Health Organization (WHO), there were an estimated 216 million clinical cases and 445,000 fatalities in 2016 [1], representing a 21% reduction in new cases and a 29% decline in fatalities, compared to 2010 estimates. Among a plethora of different factors, the clinical spectrum of malaria manifestations depends on various host and parasite factors [2]. In Gabon, malaria is hyperendemic, and transmission is generally intense and perennial [3]. Children younger than 36 months suffer most often from severe anemia (59% of cases), followed by malaria-associated hypoglycemia (32%), cerebral malaria (21%) and severe respiratory distress (19%) [4]. The overall case fatality rate is approximately 3%, and cerebral malaria, together with respiratory distress, is the highest risk for fatal outcomes [4].
Functional genomics allows for unraveling pathological mechanisms, biomarkers and new drug targets. Based on microarray technology, several studies have tried to more precisely describe particular features of the host's immune response against the Plasmodium parasite in both naturally and experimentally infected humans [5,6]. Here, we extend previous studies on the expression of candidate genes in whole blood of Gabonese children with malaria [7,8] and describe the whole blood signature of gene expression in relevant clinical presentations of childhood malaria. The various presentations of P. falciparum infection were discriminated, and discrete signatures for transcription factor binding sites in promoters of genes were identified, either up-regulated across all disease manifestations, specifically up-regulated in uncomplicated malaria or specifically down-regulated in cerebral malaria. Similar gene expression profiles in each of these clinical presentations likely result from common regulation at the transcriptional level.
2. Materials and methods
2.1. Subjects and samples
Febrile children between 0.5 and 6 years of age presenting to the Centre Hôspitalier de Libreville and to the Albert Schweitzer Hospital in Lambaréné, Gabon, were recruited. Criteria qualifying for the present study were i) confirmed P. falciparum parasitemia by microscopic examination of thick and thin blood smears; ii) no evidence of other severe diseases according to WHO's definition of severe malaria (World Health Organization and Communicable Diseases Cluster 1–90) [9]; iii) hemoglobin level < 5 g/dL for severe malarial anemia patients; and iv) Blantyre Coma Score (BCS) ≤ 2 for patients with cerebral malaria. Thick and thin smears were prepared for parasite counts. Patients presenting with acute malaria and anemic normochromic and normocytic red blood cell counts were hospitalized and treated with a combination therapy (Clindamycin in combination with chloroquine or quinine) according to local standards [10].
Patients were classified into different subgroups, namely i) children with asymptomatic P. falciparum infection (A), ii) uncomplicated malaria (U), iii) severe malarial anemia (S) and iv) cerebral malaria (Ce). The only difference between U and A groups was the absence of clinical symptoms in the asymptomatic group, for at least 5 consecutive days. Healthy control children (Co) were recruited from neighbourhood compounds in the vicinity of the Centre Hôspitalier de Libreville and had negative results for parasite counts in thick and thin smears. With the exception of parasitemia, the Co, U and S groups differed significantly with respect to age, respiration rate, degree of spleen enlargement, white blood cell count, glycemia, hemoglobin and hematocrit levels, lactatemia and temperature (Suppl. Table 1b). Approximately 5 mL of whole blood were obtained by venipuncture, transferred into two PaxGene RNA tubes (Qiagen, Hilden, Germany) of 2.5 mL each and stored at −80 °C prior to transportation to the Institute of Tropical Medicine, University of Tübingen, Germany. Written informed consent was obtained by parents/guardians of children after explaining the nature and possible consequences of the study. The Ethics Committees of the International Foundation for the Albert Schweitzer Hospital in Lambaréné, Gabon, and the Medical Faculty of the University of Tübingen, Germany, approved the study. All experiments were performed in accordance with relevant guidelines and regulations.
2.2. Experimental design
In order to investigate gene expression in whole blood cells of the different groups (A, U, S, Ce, Co), we hybridized the Human Genome oligonucleotide array chip (Affymetrix, Santa Clara, CA) with pooled cRNA. To this end, we used 20 samples per group (A, U, S, Ce and Co). For each pool, we used 4 individual samples, ending up with 5 pools per group (20/4 = 5) and 25 pools in total (5 Co + 5 A + 5 U + 5 S + 5 Ce = 25). We hybridized two different microarray chips: U133A and U133B with each pool. These chips complement each other: U133A probing well-annotated genes and U133B, putative genes in the genome. They probe whole human genome-derived transcripts through a set of 11 perfectly-matched “PM” probes and 11 mismatched “MP” probes, designed to be complementary to different parts of the transcript sequence (“MP” differ by only one nucleotide from “PM”). A high signal intensity from PM cells is as important, as a low signal intensity in MP cells, in order to consider the hybridization results for further analysis. This approach greatly reduces the chance of false positives or miscalling due to cross-matching parasite-derived RNA, reassuring the quality of signal calling. The pools were statistically homogeneous within each group, with respect to age, body weight, temperature, respiration rate, BCS, hemoglobin concentration, hematocrit, white cell count, glycemia and lactatemia average values, except glycemia within A and hemoglobin/hematocrit within U (Suplementary Table 1a). We compared the gene expression profiles obtained from the microarrays of patient groups with those of healthy control children. Potential genes with up- or down-regulation were validated by TaqMan real-time RT-PCR assay.
2.3. RNA extraction and microarray hybridization
Total RNA was extracted from whole blood using the PAXgene Blood RNA System (Qiagen, Hilden, Germany), pooled in equal amounts to a total of 5 μg from four individuals and the first- and second-strand cDNA was synthesized using the SuperScript Double-Stranded cDNA Synthesis Kit (Life Technologies Inc., Rockville, MD, USA) and an oligo-dT24-T7 primer. cRNA was prepared and labeled with biotinylated UTP and CTP by in vitro transcription using a T7 promoter-coupled double-stranded cDNA as template and the T7 RNA Transcript Labeling Kit (ENZO Diagnostics Inc., Farmingdale, NY, USA). After purification and precipitation at −20 °C, 20 μg of this cRNA were fragmented by heat and ion-mediated hydrolysis at 94 °C [200 mM Tris-acetate (pH 8.1), 500 mM KOAc, 150 mM MgOAc] and hybridized to the microarray. Increasing concentrations of spiked controls were added, expected to hybridize to the 5′ and 3′ transcript regions of the Escherichia coli genes bioB, bioC and bioD and of the cre gene of the P1 bacteriophage. The arrays were hybridized with the targets at 45 °C for 16 h, washed at 25 °C with 6× saline sodium phosphate-EDTA (0.9 M NaCl, 60 mM NaH2PO4, 6 mM EDTA+0.01% Tween 20) for 10 min, followed by a stringent washing step at 50 °C with 100 mM MES, 0.1 M [Na+], 0.01% Tween 20 for 12 min. Subsequently, the arrays were stained with 10 μg/mL phycoerythrin-conjugated streptavidin (Molecular Probes Europe BV, Leiden, Netherlands) for 10 min. Hybridization, washing and staining procedures were performed using the GeneChip® Fluidics Station 400 (Affymetrix, Santa Clara, CA, USA). Fluorescence intensities were assessed using a laser confocal scanner (Agilent GeneArray ® Scanner) with excitation of 488 nm, spot size 4 μm and pixel space 3 μm and the scanned images were analysed using Affymetrix® Microarray Suite version 5.0 (Affymetrix, Santa Clara, CA, USA). Sample loading and variations in staining were standardized by scaling the signals on all arrays to constant target intensity (TGT 150). The results of each microarray hybridization are publicly available in the GEO series GSE1124.
2.4. Microarray data filtering
The investigation was done in accordance with MIAME (minimal information about a microarray experiment) guidelines (www.mged.org). All with the exception of three U133B microarrays were homogeneous with respect to average background, noise (Raw Q) and percentage of genes called present; had spiked control signals within the expected range, and adequate 3′/5′ ratio values (under 3) of the signals of the housekeeping gene transcripts (Glyceraldehyde 3-phosphate dehydrogenase - GAPDH, 18SrRNA). The scaling factors of all arrays (except those three) compared to each other and did not differ by a factor higher than 3.3. Therefore, signals were compared between the microarrays of two groups, totalizing 25 AxCo, 25 UxA, 25 UxCo, 25 SxCo, 25 SxA, 25 SxU, 25 CexCo, 25 CexA, 25 CexU, and 25 SxU comparisons for U133A and U133B (for the U133B, there were 15 AxCo and 20 CexCo, CexA, CexU and CexS comparisons).
Signals on the arrays, signal ratios among them and the corresponding measure of likelihood of change and direction (“change P-value”) were calculated with the Affymetrix® Microarray Suite version 5.0 (Affymetrix, Santa Clara, CA, USA). Probe sets whose signal intensities reached values under 50 in all the investigated groups were excluded. The expression of a certain gene was considered increased or decreased if it varied positively by a factor ≥ 1.9 or negatively by a factor ≤ −1.9, and the corresponding change in p value was <0.15 or > 0.85, respectively, in at least 15 comparisons. The one class analysis of the Significance Analysis of Microarrays (SAM) program [11] was applied for the signal ratios (logged on base 2) to identify those genes having significant changes at a false discovery rate of 0.004%. In order to build the heat maps, the Genesis 1.6.0 Beta 1 software (http://genome.tugraz.at/) was used. We performed unsupervised biclustering with the medians of AxCo, UxCo, UxA, SxCo, SxA, SxU, CexCo, CexA, CexU and CexS comparisons using SAMBA (Statistical-Algorithmic Method for Bicluster Analysis) and unsupervised clustering with the AxCo, UxA, SxU and CexU comparisons using CLICK (CLuster Identification via Connectivity Kernels). Clusters were further evaluated for significant enrichment in gene ontology terms (with TANGO - Tool for ANalysis of GO enrichments), transcription factor binding sites (with PRIMA - PRomoter Integration in Microarray Analysis) and micro RNA regulation (with FAME - Functional Assignment of MicroRNAs via Enrichment). All these algorithms are embedded in the EXPANDER 5.2 (EXPression ANalyzer and DisplayER) software package (http://acgt.cs.tau.ac.il/expander/). For each software, we applied default parameters.
We extracted normalized data from the Stanford University Microarray Database (http://genome-www5.stanford.edu/) referring to microarray results of individual samples of a Kenyan cohort with six healthy children, six children with uncomplicated malaria, three children with severe malarial anemia and six children with cerebral malaria [5]. Samples of these children were evaluated with the Stanford University cDNA lymphochip microarray platform and analysed with an unsupervised clustering approach that included samples of children with bacterial infections. We re-analysed all samples, except those of bacteria-infected children in order to find genes similarly regulated in the UxCo, SxU and CexU comparisons in both studies. We extracted and reanalysed the Affymetrix U133A data sets from 1AGSE5418 (http://www.ncbi.nlm.nih.gov/geo) with gene expression changes in peripheral blood mononuclear cells of Cameroonian adults with acute malaria before and after treatment with chloroquine [6].
The DAVID (Database for Annotation, Visualization and Integrated Discovery) Bioinformatics Resources (v6.7; https://david.ncifcrf.gov/) was used with default parameters in order to evaluate functional clusters of genes with significant EASE (Expression Analysis Systematic Explorer) scores. The DAVID Functional Annotation Clustering algorithm measures relationships among the Gene Ontology (GO) and other annotation terms based on the degrees of their co-associated genes to group similar, redundant, and heterogeneous annotation contents from identical or different resources into annotation groups (clustering stringency was set at medium). Canonical pathways were identified using Ingenuity Pathway Analysis software (Ingenuity Systems, Redwood City, CA) with default parameters. Networks were scored based on the number of network eligible molecules that the networks contained. The higher the score, the lower was the probability of finding the observed number of network eligible molecules in a given network by random chance.
2.5. TaqMan real-time RT-PCR
We used a real-time RT-PCR assay (Assay-on-Demand Gene Expression Products, Applied Biosystems, Foster City, CA, USA) to measure the expression of 29 selected genes on 25 Co, 33 U and 30 S samples (Suppl. Table 1). These 29 genes are listed in the supplementary data (Suppl. Table 2). Samples were assayed in 20 μL reactions consisting of 1 ng/μL cDNA, 1× TaqMan Universal Master Mix (2×) and 1× of the Assays on Demand™ Gene Expression Mix (Applied Biosystems, Foster City, CA, USA). TaqMan probes were labeled with the 5′ reporter dye (6-FAM) and the nonfluorescent quencher dye with a minor groove binder (MGB). We performed PCR amplification and mRNA detection using a GeneAmp 5700 Sequence Detection System (Applied Biosystems, Foster City, CA, USA) under the following conditions: 10 min at 95 °C, 40 cycles of 15 s at 95 °C and 60 s at 60 °C. After using the ΔΔCt method to normalize the real-time results according to the gene expression of HPRT1 (hypoxanthine phosphoribosyltransferase 1) as a housekeeping gene and reference samples, resulting values were converted into logarithms (base 2).
2.6. Statistical analysis
Differences between groups were analysed using ANOVA and student's t-tests where appropriate, and P values were corrected for multiple comparisons using the Bonferroni/Dunn method. All possible correlations between expression levels of the genes as well as between clinical data and the gene expression values of the individuals using a multivariate correlation test in JMP 9 software (SAS Campus Drive, Cary, NC, USA) were investigated. A double-hierarchical cluster analysis of the genes indicating significant differences between groups was performed.
3. Results
3.1. General malaria transcriptome
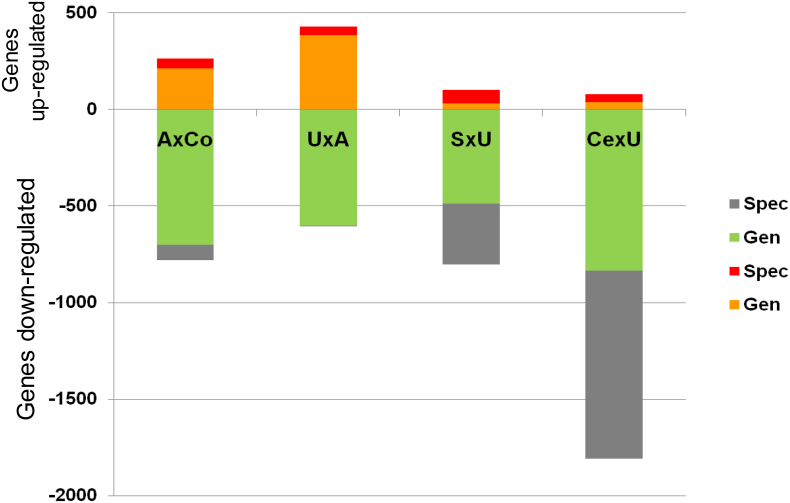
A total of 4643 transcripts were significantly regulated in all comparisons made among controls (Co), asymptomatic P. falciparum infection (A), uncomplicated malaria (U), severe malarial anemia (S) and cerebral malaria (Ce). The comparisons A versus Co (AxCo), U versus A (UxA), S versus U (SxU) and Ce versus U (CexU) were further investigated in detail. Except for UxA, where the number of up- and down-regulated genes was almost equal, the majority of genes was down-regulated. Genes were classified into 55 functional annotation clusters, of which seven in U and two in Ce had up-regulated genes. Seventeen clusters were shared among AxCo, UxA, SxU or CexU. The largest of the shared clusters included only down-regulated genes and showed annotation terms relative to distinct locations (e.g. membrane-enclosed), transcription regulation at the DNA and RNA level, cell activation and death (Suppl. Table 3). We also observed more genes specifically regulated in cerebral malaria (1017/1889; 53.8%) compared to the other clinical conditions (130/1042; 12.5% in A; 45/1033; 4.4% in U; and 386/906; 42.6% in S) (P < 10E−7 for each comparison) (Fig. 1).
Fig. 1.
Number of genes up- and down-regulated during malaria. Spec: specifically regulated (not significantly regulated in any of the other clinical conditions examined in this study). Gen: genes found similarly regulated in other comparisons. Orange and red: genes up-regulated; light and dark green: genes down-regulated. AxCo: comparison between children with asymptomatic P. falciparum infection and healthy children. UxA: comparison between children with uncomplicated malaria and children with asymptomatic infection. SxU: comparison between children with severe malarial anemia and children with uncomplicated malaria. CexU: comparison between children with cerebral malaria and children uncomplicated malaria.
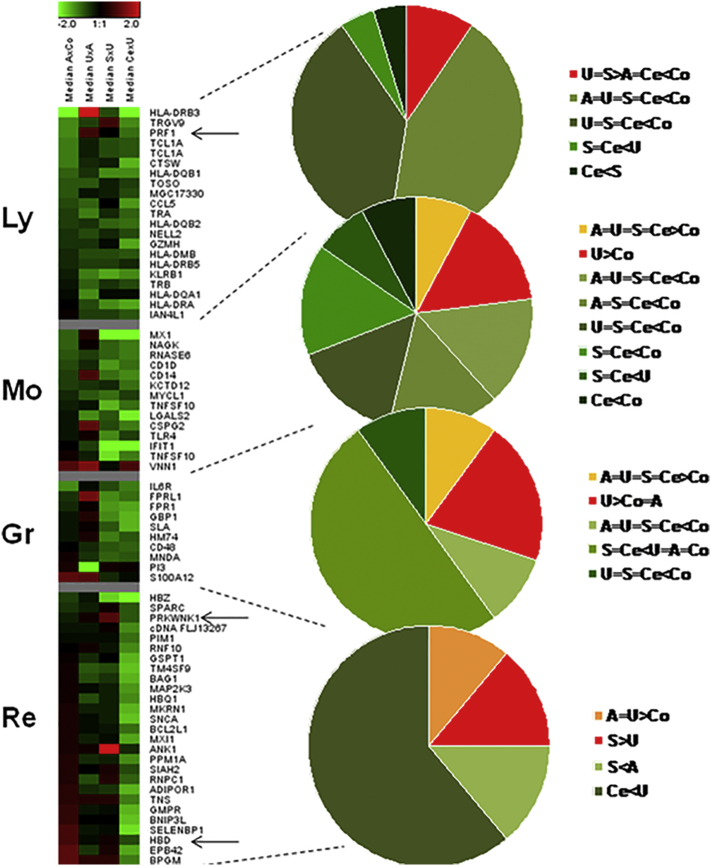
The considerable down-regulation of genes observed in P. falciparum infected individuals was also reflected in the expression signature of the major blood cell lineages (Fig. 2). This could result from cell sequestration in the tissues and/or cell apoptosis or cell destruction. We found genes involved in cytotoxic T-lymphocyte mediated apoptosis of target cells and glucocorticoid receptor signaling, whose anti-inflammatory effects lead to apoptosis and were specifically up-regulated in the U group. Interestingly, gene repression was significantly more pronounced in lymphocytic than in monocytic and granulocytic lineages. Some genes (MHC classes I and II, T cell receptor gene, CD3G) are included in the calcium-induced T lymphocyte apoptosis pathway. In addition, other pathways significantly implicated in all infected children modulate B lymphocyte activation and antibody production, namely Primary Immunodeficiency Signaling, PI3K Signaling in B Lymphocytes and B Cell Development (Fig. 3).
Fig. 2.
Distribution of up- and down- regulated genes according to cell source. Red: genes up-regulated; green: genes down regulated. The medians of 5 logged fold change values (base 2) between children with asymptomatic P. falciparum infection compared with healthy children (AxCo), children with uncomplicated malaria compared with children having asymptomatic infection (UxA), children with malaria associated with severe anemia compared with children having uncomplicated malaria (SxU) and children with cerebral malaria compared with children having uncomplicated malaria (CexU) were colorimetrically represented in the columns. The expression pattern of the genes is represented in the horizontal strips. The grey strips define the limits between the cell-specific groups of genes for lymphocytes (Ly), monocytes (Mo), granulocytes (Gr) and reticulocytes (Re). Arrows: gene expression patterns verified by real-time RT-PCR in samples of children with uncomplicated malaria and severe malaria associated with severe anemia. Gene expression profiles were allocated to specific cell types according to previous research done on the same platform (www.affymetrix.com/support/technical/technotes/blood_technote.pdf). Due to thrombocytopenia, a common finding in malarial disease, RNA amounts of platelets were too low to compare with those obtained from other blood cell sources (lymphocyte, monocyte, granulocyte and reticulocytes), and were thus not shown.
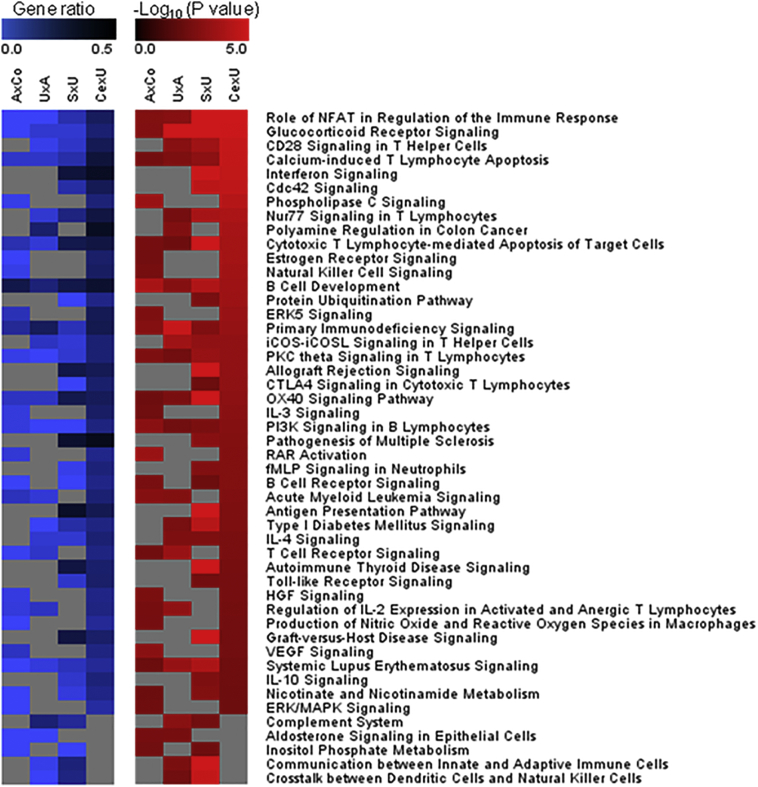
Fig. 3.
Pathways predicted to be affected by gene regulation during P. falciparum infection. Using Ingenuity software, 114 canonical pathways were predicted to be affected by transcriptional regulation. Of these pathways, 42% were shared by at least two of the AxCo, UxA, SxU and CexU comparisons. Values were colorimetrically shown in the columns for pathways with significant P values (<0.05) in at least two comparisons: children with asymptomatic P. falciparum infection compared with healthy children (AxCo), children with uncomplicated malaria compared with children having asymptomatic infection (UxA), children with malaria associated with severe anemia compared with children having uncomplicated malaria (SxU) and children with cerebral malaria compared with children having uncomplicated malaria (CexU). At the left side, colorimetric representation of the gene ratio (percentage of transcriptionally regulated gene products in the pathway - maximally 50%: darker blue means more enriched). At the right side, colorimetric representation of the P values (−log P values on base 10, maximally P = .00001: brighter red means lower P values). Grey cells stand for non significant results.
3.2. Asymptomatic P. falciparum infection
We observed 36 genes specifically regulated during asymptomatic infection with at least three times higher intensity of expression (Suppl. Table 4). Most of these genes are involved in RNA processing and nucleotide binding (Suppl. Table 3). In fact, unsupervised clustering yielded a large cluster enriched for RNA-binding and ribosomal protein coding genes in the chromosomal region 5p (1.6%, P = 7.3E−5) and Xq15 (0.4%, P = 5.4E−5) (Suppl. Fig. 1). Three pathways with >15% modulated genes were uniquely regulated in asymptomatic P. falciparum infection, namely the biosynthesis of valine, leucine and isoleucine (with 5/12 or 42% modulated genes - BCAT1, IARS2, VARS, PDHB, IARS), DNA Methylation and Transcriptional Repression Signaling (with 5/19 or 26% - MTA1, RBBP7, SAP30, DNMT1, ARID4B) and Thrombopoietin Signaling (with 9/53 or 17% - MYC, FOS, PRKCQ, PIK3R1, SOS1, PRKCE, PRKCH, MAP2K1, PRKCB) (Fig. 3). This suggests that active gene regulation through chromatin remodelling may contribute to maintaining and preserving the asymptomatic status during P. falciparum infection.
3.3. Uncomplicated malaria
Corroborating the findings in asymptomatic infection, DAVID clusters up-regulated in uncomplicated malaria and down-regulated in the two severe malaria sub-groups contained annotations relative to ‘immunoglobulin’, ‘immunoglobulin-like fold’, ‘lysosome’ and ‘defense response’ (Suppl. Table 3). The up-regulated genes on chromosome 12 and chromosome 14q32 encode immune proteins (5.4%, P = 3.3E−5) as well as immunoglobulins (1.3%, P = 4.4E−5), respectively, and plasma B cell-related transcription factors and proteins, namely EGR1 (early growth response 1), POU2F2 (POU class 2 homeobox 2, also addressed as OCT2), FOS (v-fos FBJ murine osteosarcoma viral oncogene homolog), FOSL2 (FOS-like antigen 2) and DUSP2 (dual specificity phosphatase 2). They were enriched with binding sites for ATF (PLAU; plasminogen activator urokinase), Tel-2 (ETV7 or ETS translocation variant 7), AP2 alpha (TFAP2A; transcription factor activating enhancer binding protein 2 alpha), IRF-7 (interferon regulatory factor 7), ETF (TEAD2; transcriptional enhancer factor domain family member 2) and STAT1 and STAT2 (recognizing interferon-stimulated response elements or ISRE, in response to IFN type I) (Suppl. Table 5). Interestingly, the genes encoding Tel-2 (ETV7), IRF-7 and the receptor for ATF (PLAUR) were up-regulated in uncomplicated malaria and down-regulated in both severe forms of malaria (anemia and cerebral malaria). This follows the expression pattern of 7 out of 12 immunoglobulin (Ig) transcripts that were repressed in group A, became expressed in U, and mostly down-regulated again in group S. Two of them, IGHG1 (immunoglobulin heavy constant gamma 1) and IGHV (immunoglobulin heavy chain variable region) were up-regulated as high as FOS, an important Ig gene transcription factor, and were found uniquely up-regulated in the UxA comparison (Suppl. Table 6). There was a corresponding regulation of IgM, IgD, IgE and IgA in AxCo and UxA comparisons, observed in the Primary Immunodeficiency Signaling pathway (7/45 or 16% modulated in A - BLNK, IGH@, IGL@, IGHM, IGHA1, CD79A, IGHD and 10/45 or 22% modulated genes in U - IL7R, CD40LG, CD3E, IGH@, CIITA, IGHM, IGHA1, JAK3, CD3D, IGHD) and ‘B cell development’ canonical pathways.
Consistent with our findings, Kenyan children [5] showed a similar regulation of 99 genes in uncomplicated malaria. Several of these genes were represented by more than one probeset of Affymetrix arrays, e.g. interferon-responsive and MHC genes (Suppl. Fig. 3). In contrast, only one gene encoding the nuclear histone component (HIST2H2AA) and the heat shock protein Hsp70 gene showed a similar expression pattern before and after chloroquine treatment in acute malaria of adult patients from Cameroon, compared with Gabonese children. Interestingly, hemoglobin delta had a completely opposite regulation in the adult Cameroonian cohort [6] (Suppl. Fig. 3D). A dynamic gene regulation pattern was also revealed in certain genes encoding critical proteins of the complement canonical pathway. While the C1q encoding genes C1QA and C1QB were up-regulated in children of the A and U groups, C4 up-regulation was unique to the A group, and the activation of complement regulatory genes as SERPING1 (C1 inhibitor) and CR1 (complement receptor 1 or CD35) was specific for the U group.
3.4. Severe malaria - severe malarial anemia
Genes specifically regulated with a more than three-fold expression intensity in severe malarial anemia encode proteins involved in amino acid transport, phospholipid metabolic processes and positive regulation of nitrogen compound metabolic processes (Suppl. Table 7). Unique pathways with >15% of gene products affected were the Lipid Antigen Presentation by CD1 (24%), the Role of Pattern Recognition Receptors in Recognition of Bacteria and Viruses (17%), Altered T Cell and B Cell Signaling in Rheumatoid Arthritis (17%), MIF Regulation of Innate Immunity (17%), MIF-mediated Glucocorticoid Regulation (16%), Dendritic Cell Maturation (16%) and April Mediated Signaling (16%) in the SxU comparison. A set of up-regulated genes was also found in the reticulocytic lineage restricted to the subgroup of malaria associated with severe anemia: PRKWNK1 (WNK lysine deficient protein kinase 1; WNK1), ANK1 (ankyrin 1), SIAH2 (seven in absentia homolog 2), RNPC1 (RNA binding motif protein 38; RBM38), tensin (TNS), hemoglobin delta (HBD), EPB42 (erythrocyte membrane protein band 4.2) and BPGM (2,3-bisphosphoglycerate mutase) (Fig. 2). The gene encoding clusterin (CLU), which prevents formation of the terminal complement attack complex, and the alternative pathway factor D, were repressed in children with either kind of severe malaria, and C1QBP (C1q binding protein), which inhibits C1q activation, was specifically down-regulated in cerebral malaria. Children with severe malarial anemia and cerebral malaria presented genes significantly enriched for specific transcription factor binding sites, namely PEA3 (or ETV4, polyoma enhancing activator-3) and C/EBP delta (CCAAT/enhancer binding protein), as well as for the microRNA recognition sites mir15/16/195/4 (Suppl. Fig. 2). Kenyan and Gabonese children showed similar regulation of 49 genes in malaria associated with severe anemia, and several of the 49 genes were represented by more than one probeset of the Affymetrix arrays, e.g. interferon-responsive and MHC genes (Suppl. Fig. 3). This result reinforces the predicted down-regulation of antigen presentation and interferon signaling by the canonical pathways in severe malaria.
3.5. Severe malaria - cerebral malaria
Genes specifically regulated with at least three times higher expression intensity in cerebral malaria encode proteins mostly involved in ubiquitin-dependent metabolic processes (CexU comparison) (Suppl. Table 8). Genes specifically repressed in cerebral malaria (CexU comparison) were further enriched with AhR-HIF (complex of two helix-loop-helix proteins: aryl hydrocarbon receptor and hypoxia inducible factor), GABP (GA binding protein transcription factor), AhR and HIF-1 transcription factor binding sites (Suppl. Fig. 1, Suppl. Table 9). Unique pathways with >15% of gene products affected were the Nucleotide Excision Repair Pathway (27%), the Glutamate Metabolism (25%), VDR/RXR Activation (24%), PI3K/AKT Signaling (22%), EIF2 Signaling (22%), Cell Cycle: G2/M DNA Damage Checkpoint Regulation (22%), Mitotic Roles of Polo-Like Kinase (21%), CCR5 Signaling in Macrophages (20%), EGF Signaling (20%), GM-CSF Signaling (20%), Regulation of Actin-based Motility by Rho (19%), Regulation of eIF4 and p70S6K Signaling (18%), Cholecystokinin/Gastrin-mediated Signaling (18%), PDGF Signaling (18%), mTOR Signaling (17%), Androgen Signaling (17%), Rac Signaling (17%) and Insulin Receptor Signaling (16%) in the CexU comparison. We re-evaluated individual microarray data of Kenyan children (5) and found that both cohorts showed similar regulation patterns of 150 genes in Ce. Several of them were present in more than one probe set, e.g. interferon-responsive and MHC genes (Suppl. Fig. 3). Moreover, INFB1 (interferon beta 1) and a constellation of IFNB1-regulated genes were observed in six networks, including one with 32 down-regulated genes connected to INFB1 in Ce (Suppl. Fig. 4).
3.6. Validation of gene expression
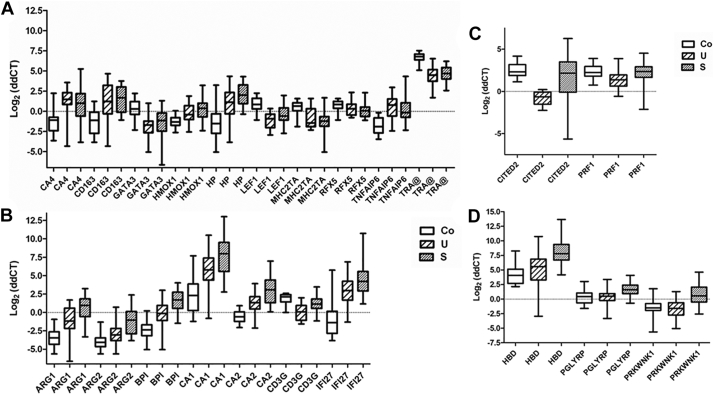
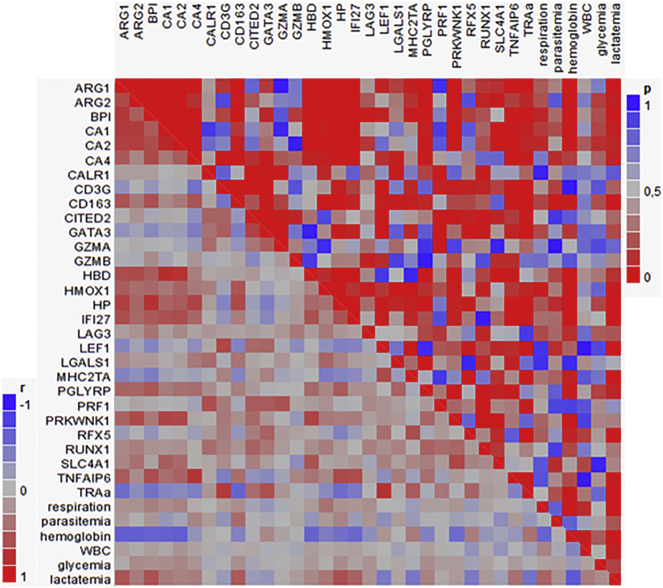
We selected a panel of 29 genes (seven of which were regulated in Kenyan children (5) (Suppl. Fig. 3) to validate their expression profiles by Taqman real-time RT-PCR in samples from Gabonese children with uncomplicated malaria and severe malarial anemia. Out of 29 genes, 22 showed significant difference in their expression between uncomplicated malaria and severe malarial anemia (Fig. 4). There was complete agreement in the expression pattern of 14 genes and small differences only in the others. Genes with no relevant alteration of expression (CALR1, GZMA, GZMB, LAG3, LGALS1 and SLC4A1) were excluded from further analyses. Ten genes showed no expression difference between uncomplicated and severe malaria and were, thus, considered as part of the “malaria gene expression signature” (Fig. 4A). Seven genes were expressed higher in severe malarial anemia (Fig. 4B). RUNX1 had a particular profile with no difference between malaria cases and controls, but a difference between uncomplicated (down-regulated) and severe malarial anemia (up-regulated) (P < .001). CITED2 and PRF1 were specifically down-regulated in uncomplicated malaria, whereas HBD, PGLYRP and PRKWNK1 were up-regulated in severe malarial anemia (Fig. 4C and D). The highest number of strong positive intergenic correlations occurred between functionally related genes: TRA@, CD3G, GATA3, LEF1, MHC2TA and RFX5; and genes whose transcript numbers correlated negatively with hemoglobin concentrations: ARG1, ARG2, BPI, CA1, CA2, CA4, CD163, HBD, HMOX1, HP, IFI27, PGLYRP, PRKWNK1 and TNFAIP6. Among these, CD163 was the only gene that correlated positively with parasitemia. CD163, ARG1, BPI and HP were correlated strongly and positively with lactatemia, followed by IFI27 and TNFAIP6. Weaker negative correlations of lactatemia with TRA@ gene, the transcription factor genes LEF1, MHC2TA and RFX5 were observed (Fig. 5).
Fig. 4.
Expression pattern of selected genes validated by real-time RT-PCR. The expression pattern of selected genes was verified by Taqman real-time RT-PCR. Box-plots illustrate medians with 25 and 75 percentiles and whiskers from minimum to maximum. Groups were compared with the unpaired student's t-test and the P values corrected with the Bonferroni method. Genes are listed on the X-axis in alphabetical order. Co = controls; U = uncomplicated malaria; S = malaria associated with severe anemia. Genes similarly regulated in symptomatic malaria (A) include: CA4 (carbonic anhydrase 4), CD163 (CD163 scavenger receptor), GATA3 (GATA binding protein 3), HMOX1 (heme oxygenase 1), HP (haptoglobin), LEF1 (lymphoid enhancer-binding factor 1), MHC2TA (major histocompatibility complex class 2, transactivator), RFX5 (regulatory factor X 5), TNFAIP6 (tumor necrosis factor alpha induced protein 6), TRA@ (T cell receptor alpha). P < .0001 for UxCo, except for HMOX1 (P < .001), HP (P < .01) and RFX5 (not significant, but P < .01 for SxCo). Differences between S and U were not significant. Genes with strong up-regulation (except CD3G) in severe malaria (B) include: ARG1 (arginase 1), ARG2 (arginase 2), BPI (bactericidal permeability increasing protein), CA1 (carbonic anhydrase1), CA2 (carbonic anhydrase 2), CD3G (CD3 gamma), IFI27 (interferon-alpha inducible protein 27). P < .0001 for UxCo, except for ARG2 (P < .01). P < .0001 for SxU except for ARG1 and BPI (P < .001); CA1, CA2 and IFI27 (P < .01). Genes with unique regulation in uncomplicated malaria (C) include: CITED2 (Cbp/p300-interacting transactivator, with Glu/Asp-rich carboxy-terminal domain, 2), PRF1 (perforin 1). P < .0001 for U ≠ Co, except for PRF1 (P < .01). Differences between S and Co were not significant. Genes uniquely expressed in severe malaria (D) include: HBD (hemoglobin delta), PGLYRP (peptidoglycan recognition protein 1), PRKWNK1 (lysine-deficient protein kinase 1). P < .0001 for SxU, except for PGLYRP (P < .001). Differences between U and Co were not significant.
Fig. 5.
Correlation between gene expression patterns determined by real-time RT-PCR and clinical data. The microarray expression pattern of selected genes was verified by Taqman real time RT-PCR and correlated with respiration rate, parasitemia, hemoglobin concentration, white blood cell count (WBC), glycemia and lactatemia in a multivariate analysis. R and P values were colorimetrically represented in the left and right halves of the square, respectively, e.g. hemoglobin concentration was negatively correlated with the expression of at least 12 genes (blue colored squares of the “hemoglobin” row, r value near −1), being this correlation highly significant (red colored squares of the “hemoglobin” column, p value near 0). On the other hand, most of the same genes were significantly but positively correlated with lactatemia (red for “lactatemia” row and column).
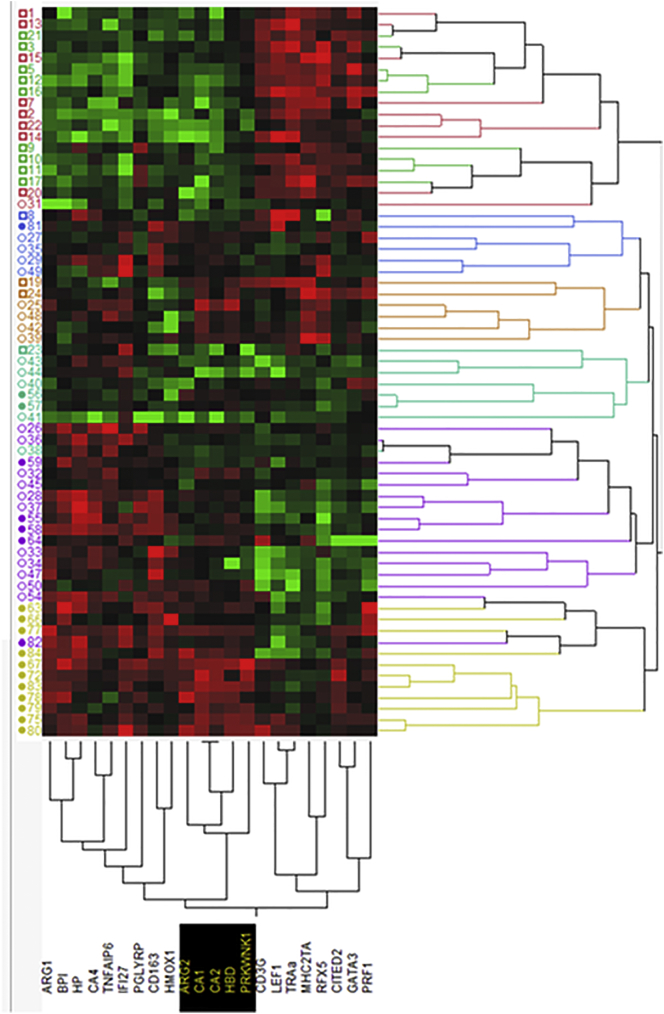
We used 22 genes with significant P values (Fig. 4) to cluster the samples according to the gene expression profiles and to the gene signatures of the individual samples. The expression profiles of healthy control children, however with four exceptions, clustered together with down-regulation of the ARG1, BPI, HP, CA4, TNFAIP6, IFI27, PGLYRP, CD163, HMOX1, ARG2, CA1, CA2, HBD and PRKWNK1 genes and up-regulation of the CD3G, LEF1, TRA@, MHC2TA, RFX5, CITED2, GATA3 and PRF1 genes. Children with uncomplicated malaria shared four different patterns of expression profiles, showing that this clinical condition has rather heterogeneous gene regulation signatures. Three of the patterns were shared by seven children with severe malarial anemia, and six of these children shared low expression of those genes that were up-regulated in healthy children. The severe malarial anemia cluster showed a unique type of gene expression profile with either overexpression of ARG1, BPI, HP, IFI27 and/or TNFAIP6, all of which were positively correlated with lactatemia. Five genes predictive of severe malarial anemia are ARG2, CA1, CA2, HBD and PRKWNK1 (Fig. 6). These genes were positively correlated with hemoglobin concentration, but not with lactatemia, and the last four are typically expressed in reticulocytes.
Fig. 6.
Double-hierarchical clustering of samples and selected genes using gene expression profiles. The expression profiles of 22 genes measured by real-time RT-PCR and showing significant differences between groups were used to cluster samples of healthy children (open squares), children with uncomplicated malaria (open circles) and with malaria associated with severe anemia (filled circles) on the left, as well as the genes at bottom of the Figure. The log2∆∆CT values were colorimetrically represented in the columns, the expression profiles per sample, in the horizontal strips. Common expression profiles are shown in the same color. The gene cluster highlighted in black contains genes whose expression pattern was unique to malaria associated with severe anemia. The numbers on the left side of the figure are sample codes. The red squares indicate upregulated genes in their expression profile and the green squares represent those genes whicgh are down regulated.
4. Discussion
This work is an attempt to describe the whole blood signature of gene expression in the most important clinical manifestations of childhood malaria, and validated in 22 genes known to be expressed in the principal blood cell lineages. A previous study did not differentiate among uncomplicated malaria, severe malarial anemia and cerebral malaria, considering them as overlapping ‘acute malaria syndromes’. In that study, patients were divided in two unsupervised clusters based on differences in neutrophil-gene expression, but the patients could not be clearly distinguished by any clinical parameter [5]. We re-evaluated these previous data and observed a considerable overlap between genes regulated in Kenyan and Gabonese children with symptomatic malaria. That not withstanding, we have to take into account, and address as potential limitations of our study, i) the small number of patients we studied, ii) differences in geographic locations and epidemiological particularities (holo- to mesoendemic in coastal Kenya vs. hyperendemic in Gabon) and iii) distinct microarray platforms (Stanford lymphochips for the Kenyan and Affymetrix microarrays for the Gabonese study). Therefore, although using the same method of RNA extraction, the list of commonly modulated genes generated through our comparisons cannot yet be considered definitive. In contrast to the results in children, adults with acute malaria from Cameroon shared a small number of genes before and after treatment, of which two were also modulated in childhood malaria [6].
In asymptomatic malaria, genes involved in RNA processing and nucleotide binding are specifically regulated, indicating that although without overt disease, asymptomatic children are rather able to temporarily maintain their status by orchestrating active gene regulation through chromatin remodelling. This probably affects the production of immunoglobulin chain transcripts found repressed in asymptomatic children, but specifically activated in paediatric uncomplicated malaria. Another study from The Gambia found that asymptomatic children had a more limited antibody repertoire than children with uncomplicated malaria, suggesting that the accumulation of antibody specificities against a large fraction of parasite antigens may not be protective against symptoms and may impair the function of protective antibody specificities [12].
In symptomatic malaria, basophil and eosinophil transcripts remained repressed (e.g. CLC; Charcot-Leyden crystal protein), whereas other genes encoding components of the granulocyte and myeloid lineages become activated. Levels of CD163, HP, BPI, ARG1, IFI27 and TNFAIP6 are correlated with clinical parameters and thus represent potential indicators of parasitemia, hemoglobin levels and lactatemia. On the phagocytic surface, CD163 scavenges hemoglobin-haptoglobin complexes generated after rupture of infected erythrocytes, whereas the soluble form has potential anti-inflammatory properties and is found at higher levels in symptomatic malaria [13], thus, it correlates weakly with parasite biomass. CD163 is in fact the only marker associated with the degree of parasitemia. Therefore, increased CD163 consumption may lead to CD163 up-regulation, paralleling an increase in parasitemia. Expression of CD163 and all other aforementioned genes is positively correlated with lactate and inversely with hemoglobin levels. Lactatemia occurs because persistent oxygen deficit leads to a replacement of intracellular aerobic respiration with anaerobic glycolysis and excessive production of lactic acid. Lactic acidosis lowers the tissue pH and blunts the vasomotor response, ultimately causing widespread tissue hypoxia, acidosis and an increased risk of mortality [14]. BPI haplotypes are associated with a higher risk of developing rapid airflow decline after hematopoietic cell transplantation [15]. ARG1 competes with nitric oxide synthase for L-arginine, hydrolyzing this substrate to ornithine and urea. Higher amounts of urea are produced by activated ARG1 in erythrocytes pre-exposed to free heme released through rupture of infected erythrocytes [16]. Higher levels of urea parallels the decrease of hemoglobin and the increase of lactate levels in children affected with severe malaria [17]. IFI27 associates with or inserts into the mitochondrial membrane and its transient expression leads to decreased numbers of viable cells and enhanced sensitivity to DNA-damage induced apoptosis [18], suggesting involvement of IFI27 in the apoptosis mechanisms that generate lymphopenia during severe malarial anemia [7]. In addition, expression of TNFAIP6, along with CA1, CA2 and CA4, is correlated positively with upsA var gene expression in P. falciparum infected Gabonese children [8].
In contrast to the granulocyte, myeloid and erythroid lineages, more than half of the transcripts known to be expressed in lymphocytes were down-regulated. Among these transcripts, we found CD3G, TRA@ and their transcription factors GATA3 and LEF1, which is in line with our previous results indicating lower membrane expression of the CD3-TCR complex as well as CD8+ and CD4+ T cell apoptosis in Gabonese children with symptomatic malaria [7]. A higher apoptotic rate found in CD85j + B cells [19] is supported by our study showing down-regulation of LILRB1 (encoding CD85j) in Gabonese and Kenyan children with severe malarial anemia. Furthermore, MHC class II genes and two of their most important transcription factors, RFX5 and MHC2TA, are significantly down-regulated in symptomatic malaria and may hamper antigen presentation. This down-regulation has been associated with a higher apoptotic rate of HLA class II positive cells [7] and correlates with the hemozoin load [20]. Up-regulation of HMOX1, SOD2 (superoxide dismutase 2), MMP9 (matrix metalloproteinase 9), LY96 (lymphocyte antigen 96) and CD14 (monocyte differentiation antigen CD14) is evident in both Gabonese and Kenyan children with uncomplicated malaria as well as in murine macrophage-like cells with hemozoin-derived lipid peroxidation components [20]. In contrast to this study, the up-regulation of MMP9 is counter-regulated by the activation of TIMP1 (tissue inhibitor of metalloproteinase 1). Some genes up-regulated in symptomatic disease shared mir-15/16/195/4 regulation. These miRNAs regulate the expression of genes involved in cell division, metabolism, stress response, and angiogenesis and are associated with human cancers as well as with cardiovascular and neurodegenerative diseases [21]. Some genes share transcription binding sites for PEA3 (linked to enhanced growth factor/ERK signaling) and C/EBP delta (acts as an amplifier of NF-κB responses, discriminating between transient and persistent TLR4 signals, and leading to enhanced TLR8 transcription) [22,23]. In the present study, TLR4 and TLR8 are the only Toll-like receptor genes up-regulated in symptomatic malaria.
In uncomplicated malaria, we identified specific up-regulation of immunoglobulin transcripts and plasma B cell-related transcription factors and proteins (POU2F2, EGR1, FOS, FOSL2, DUSP2) [24]. This up-regulation is particularly evident in the comparison of uncomplicated malaria with the group of asymptomatic children. Several of these immunoglobulin transcripts are down-regulated or up-regulated less efficiently in severe malaria, in agreement with the finding that children with uncomplicated malaria have a different antibody repertoire compared to those with severe malaria [25,26]. Children with uncomplicated malaria shared the induction of interferon and interferon-responsive genes, as found in experimental and natural malaria [5,6]. We found a specific induction of IFN type I regulated genes, indicated by up-regulation of IRF7 (interferon regulatory factor 7) and IRF7- and ISRE-regulated genes in uncomplicated malaria, but strong down-regulation in both Gabonese and Kenyan children with severe malaria. IRF7 and ISRE-binding transcription factors respond to IFN beta signaling through STAT1 and STAT2 (signal transducer and activator of transcription 1 and 2) activation, which are highly up-regulated in uncomplicated malaria. In addition, we found a similar expression profile for IRF2BP2 (interferon regulatory factor 2 binding protein 2), which inhibits IRF2 activity in response to IFN beta [27].
In severe malarial anemia, our data suggest i) increased erythrocyte production measured as higher reticulocyte transcript levels (e.g. PRKWNK1 and HBD), ii) increased intra-vascular hemolysis and raised parasitemia levels reflected by up-regulation and positive correlation of genes encoding proteins removing free hemoglobin (e.g. HP and CD163); and iii) enhanced extra-vascular hemolysis due to down-regulation of genes encoding complement regulatory proteins (e.g. SERPING1 or C1inh and CR1 or CD35). Similarly, a previous study indicated diminished CD35 and CD55 levels expressed on erythrocyte membranes and enhanced erythrophagocytosis in Kenyan children with severe malarial anemia, compared to children with uncomplicated malaria [28]. Several markers, including low haptoglobin (Hp) levels due to higher Hp consumption, indicate increased intravascular hemolysis in these children [29]. Also in accordance with out findings, raised CD163 transcripts were correlated with increasing parasitemia levels in a recent study done with Indonesian adults [30]. Decreased erythropoiesis could be caused by expression of the Cbp/p300-interacting transactivator 2 (CITED2), which suppresses erythropoietin production [31]. Similarly, the homologous mouse gene is up-regulated in the spleen of P. berghei-infected mice, whereas typical reticulocytic genes encoding the hemoglobins and ankyrin are down-regulated [32].
The clinical course of cerebral malaria is characterized by encephalopathy and loss of consciousness, focal accumulation of cytokines in the brain parenchyma, immunologic dysfunction, and endothelial cell activation [33]. We found MHC class II genes, ferro chelatase (FECH), pre-synaptic synuclein alpha protein (SNCA), ankyrin 1 (ANK1), STAT1, the ubiquitin mediated proteolytic pathway RAD23A protein, hydroxyacylglutathione hydrolase (HAGH) and of 2,3-bisphosphoglycerate mutase (BPGM) all to be down-regulated. Down-regulation of HAGH and of other genes involved in glutamate metabolism is correlated with acidosis and neuronal cell death [34]. In contrast, we observed an up-regulation for defensin alpha DEFA1 and down-regulation for the fraktalkine receptor CX3CR1 (also recently found by others [35], to be modulated in Gambian children with cerebral malaria) and myelin and lymphocyte protein MAL. Furthermore, we found strong repression of IFN beta-regulated genes and of genes with key-roles in IFN signaling, of which IFN beta has emerged as a strong candidate for the treatment of cerebral malaria [36,37]. Down-regulation of several genes in cerebral malaria seems to be a response to hypoxia orchestrated by AhRF, GABP and HIF1 transcription factors. This is in agreement with hypoxia effects due to sequestration of infected erythrocytes and vessel occlusion in children with cerebral malaria. Thus, improvements of perfusion to diminish hypoxic injury may be beneficial in children with cerebral malaria [38].
A limitation of this study is that, a quantitative PCR comparison with arrays could not be validated by plots comparing qPCR mRNA levels with array density PCR mRNA expression. Another limitation is that, this particular work was carried out in 2005, when methodologies such as RNA Seq and NGS platforms were not available. Embarking on these new methodologies could have additionally provided an in depth understanding on the expression profile of different clinical phenotypes.
In conclusion, modifications in gene expression when different blood cells interact with the parasite during P. falciparum infection were demonstrated. Immunoglobulin production, complement regulation and IFN beta signaling emerged as most discrepant features between uncomplicated malaria and other clinical presentations of P. falciparum infections. Although several associations in the inferred transcriptional network should be validated further, ARG1, BPI, CD163, IFI27, HP and TNFAIP6 have a potential to serve as prognostic markers, directing early therapeutic measures to prevent malarial disease evolution and death.
Acknowledgments
Acknowledgements
The authors thank all children and their families for their participation in this study. We are also grateful to Stephen Müller, who helped with the SAM analysis and to Adrian Luty and Kim Bucci for helpful discussions, Cristina Tena-Tomás, Velia Grummes, Andrea Weierich and Sven Poths for excellent technical assistance.
.
Competing interests
None of the authors has any financial or non-financial competing interests, and none has served or currently serve on the editorial board of this journal, have acted as an expert witness in relevant legal proceedings, or have sat or currently sit on a committee for an organization that may benefit from publication of the paper.
Author's contributions
JK designed the study. MPG involved in the study design. AB, YK, AD and MK performed the experiments. TPV, PGK supervised the study procedures and contributed to materials. AB, HVT TPV, and CGM analysed the data and wrote the manuscript. All authors agreed with the results and conclusions.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ebiom.2018.12.055.
Contributor Information
Angelica B.W. Boldt, Email: angelicaboldt@gmail.com.
Thirumalaisamy P. Velavan, Email: velavan@medizin.uni-tuebingen.de.
Appendix A. Supplementary data
Supplementary material
References
- 1.WHO . World Health Organization; Geneva: 2017. World Malaria Report 2017. [Google Scholar]
- 2.Remme J.H., Binka F., Nabarro D. Toward a framework and indicators for monitoring roll back malaria. Am J Trop Med Hyg. 2001 January;64(1–2 Suppl):76–84. doi: 10.4269/ajtmh.2001.64.76. [DOI] [PubMed] [Google Scholar]
- 3.Wildling E., Winkler S., Kremsner P.G., Brandts C., Jenne L., Wernsdorfer W.H. Malaria epidemiology in the province of Moyen Ogoov, Gabon. Trop Med Parasitol. 1995 June;46(2):77–82. [PubMed] [Google Scholar]
- 4.Issifou S., Kendjo E., Missinou M.A., Matsiegui P.B., Dzeing-Ella A., Dissanami F.A. Differences in presentation of severe malaria in urban and rural Gabon. Am J Trop Med Hyg. 2007 December;77(6):1015–1019. [PubMed] [Google Scholar]
- 5.Griffiths M.J., Shafi M.J., Popper S.J., Hemingway C.A., Kortok M.M., Wathen A. Genomewide analysis of the host response to malaria in Kenyan children. J Infect Dis. 2005 May 15;191(10):1599–1611. doi: 10.1086/429297. [DOI] [PubMed] [Google Scholar]
- 6.Ockenhouse C.F., Hu W.C., Kester K.E., Cummings J.F., Stewart A., Heppner D.G. Common and divergent immune response signaling pathways discovered in peripheral blood mononuclear cell gene expression patterns in presymptomatic and clinically apparent malaria. Infect Immun. 2006 October;74(10):5561–5573. doi: 10.1128/IAI.00408-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kalmbach Y., Boldt A.B., Mordmuller B., Kombila M., Grobusch M.P., Kremsner P.G. Reduced CD3/TCR complex expression leads to immunosuppression during Plasmodium falciparum malaria. Parasitol Res. 2009 February;104(3):575–582. doi: 10.1007/s00436-008-1232-9. [DOI] [PubMed] [Google Scholar]
- 8.Kalmbach Y., Rottmann M., Kombila M., Kremsner P.G., Beck H.P., Kun J.F. Differential var. gene expression in children with malaria and antidromic effects on host gene expression. J Infect Dis. 2010 July 15;202(2):313–317. doi: 10.1086/653586. [DOI] [PubMed] [Google Scholar]
- 9.Severe falciparum malaria. World Health Organization, communicable diseases clusterTrans R Soc Trop Med Hyg. 2000 April;94(Suppl. 1):S1–90. [PubMed] [Google Scholar]
- 10.Kremsner P.G., Winkler S., Brandts C., Neifer S., Bienzle U., Graninger W. Clindamycin in combination with chloroquine or quinine is an effective therapy for uncomplicated Plasmodium falciparum malaria in children from Gabon. J Infect Dis. 1994 February;169(2):467–470. doi: 10.1093/infdis/169.2.467. [DOI] [PubMed] [Google Scholar]
- 11.Tusher V.G., Tibshirani R., Chu G. Significance analysis of microarrays applied to the ionizing radiation response. Proc Natl Acad Sci U S A. 2001 April 24;98(9):5116–5121. doi: 10.1073/pnas.091062498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gray J.C., Corran P.H., Mangia E., Gaunt M.W., Li Q., Tetteh K.K. Profiling the antibody immune response against blood stage malaria vaccine candidates. Clin Chem. 2007 July;53(7):1244–1253. doi: 10.1373/clinchem.2006.081695. [DOI] [PubMed] [Google Scholar]
- 13.Kusi K.A., Gyan B.A., Goka B.Q., Dodoo D., Obeng-Adjei G., Troye-Blomberg M. Levels of soluble CD163 and severity of malaria in children in Ghana. Clin Vaccine Immunol. 2008 September;15(9):1456–1460. doi: 10.1128/CVI.00506-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Jarvis J.N., Planche T., Bicanic T., Dzeing-Ella A., Kombila M., Issifou S. Lactic acidosis in Gabonese children with severe malaria is unrelated to dehydration. Clin Infect Dis. 2006 June 15;42(12):1719–1725. doi: 10.1086/504329. [DOI] [PubMed] [Google Scholar]
- 15.Chien J.W., Zhao L.P., Hansen J.A., Fan W.H., Parimon T., Clark J.G. Genetic variation in bactericidal/permeability-increasing protein influences the risk of developing rapid airflow decline after hematopoietic cell transplantation. Blood. 2006 March 1;107(5):2200–2207. doi: 10.1182/blood-2005-06-2338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Omodeo-Sale F., Cortelezzi L., Vommaro Z., Scaccabarozzi D., Dondorp A.M. Dysregulation of l-arginine metabolism and bioavailability associated to free plasma heme. Am J Physiol Cell Physiol. 2010 July;299(1):C148–C154. doi: 10.1152/ajpcell.00405.2009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gouado I., Pankoui M.J., Fotso K.H., Zambou O., Nguele S., Combes V. Physiopathologic factors resulting in poor outcome in childhood severe malaria in Cameroon. Pediatr Infect Dis J. 2009 December;28(12):1081–1084. doi: 10.1097/INF.0b013e3181ab489d. [DOI] [PubMed] [Google Scholar]
- 18.Rosebeck S., Leaman D.W. Mitochondrial localization and pro-apoptotic effects of the interferon-inducible protein ISG12a. Apoptosis. 2008 April;13(4):562–572. doi: 10.1007/s10495-008-0190-0. [DOI] [PubMed] [Google Scholar]
- 19.Kalmbach Y., Boldt A.B., Fendel R., Mordmuller B., Kremsner P.G., Kun J.F. Increase in annexin V-positive B cells expressing LILRB1/ILT2/CD85j in malaria. Eur Cytokine Netw. 2006 September;17(3):175–180. [PubMed] [Google Scholar]
- 20.Schrimpe A.C., Wright D.W. Comparative analysis of gene expression changes mediated by individual constituents of hemozoin. Chem Res Toxicol. 2009 March 16;22(3):433–445. doi: 10.1021/tx8002752. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Finnerty J.R., Wang W.X., Hebert S.S., Wilfred B.R., Mao G., Nelson P.T. The miR-15/107 group of microRNA genes: evolutionary biology, cellular functions, and roles in human diseases. J Mol Biol. 2010 September 24;402(3):491–509. doi: 10.1016/j.jmb.2010.07.051. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Litvak V., Ramsey S.A., Rust A.G., Zak D.E., Kennedy K.A., Lampano A.E. Function of C/EBPdelta in a regulatory circuit that discriminates between transient and persistent TLR4-induced signals. Nat Immunol. 2009 April;10(4):437–443. doi: 10.1038/ni.1721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Dissanayake K., Toth R., Blakey J., Olsson O., Campbell D.G., Prescott A.R. ERK/p90(RSK)/14-3-3 signalling has an impact on expression of PEA3 Ets transcription factors via the transcriptional repressor capicua. Biochem J. 2011 February 1;433(3):515–525. doi: 10.1042/BJ20101562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Segel G.B., Woodlock T.J., Xu J., Li L., Felgar R.E., Ryan D.H. Early gene activation in chronic leukemic B lymphocytes induced toward a plasma cell phenotype. Blood Cells Mol Dis. 2003 May;30(3):277–287. doi: 10.1016/s1079-9796(03)00035-4. [DOI] [PubMed] [Google Scholar]
- 25.Tamborrini M., Liu X., Mugasa J.P., Kwon Y.U., Kamena F., Seeberger P.H. Synthetic glycosylphosphatidylinositol microarray reveals differential antibody levels and fine specificities in children with mild and severe malaria. Bioorg Med Chem. 2010 June 1;18(11):3747–3752. doi: 10.1016/j.bmc.2010.04.059. [DOI] [PubMed] [Google Scholar]
- 26.Tebo A.E., Kremsner P.G., Piper K.P., Luty A.J. Low antibody responses to variant surface antigens of Plasmodium falciparum are associated with severe malaria and increased susceptibility to malaria attacks in Gabonese children. Am J Trop Med Hyg. 2002 December;67(6):597–603. doi: 10.4269/ajtmh.2002.67.597. [DOI] [PubMed] [Google Scholar]
- 27.Childs K.S., Goodbourn S. Identification of novel co-repressor molecules for Interferon regulatory factor-2. Nucleic Acids Res. 2003 June 15;31(12):3016–3026. doi: 10.1093/nar/gkg431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Waitumbi J.N., Donvito B., Kisserli A., Cohen J.H., Stoute J.A. Age-related changes in red blood cell complement regulatory proteins and susceptibility to severe malaria. J Infect Dis. 2004 September 15;190(6):1183–1191. doi: 10.1086/423140. [DOI] [PubMed] [Google Scholar]
- 29.Fendel R., Brandts C., Rudat A., Kreidenweiss A., Steur C., Appelmann I. Hemolysis is associated with low reticulocyte production index and predicts blood transfusion in severe malarial anemia. PLoS One. 2010;5(4) doi: 10.1371/journal.pone.0010038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Yamagishi J., Natori A., Tolba M.E., Mongan A.E., Sugimoto C., Katayama T. Interactive transcriptome analysis of malaria patients and infecting Plasmodium falciparum. Genome Res. 2014 September;24(9):1433–1444. doi: 10.1101/gr.158980.113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Freedman S.J., Sun Z.Y., Kung A.L., France D.S., Wagner G., Eck M.J. Structural basis for negative regulation of hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol. 2003 July;10(7):504–512. doi: 10.1038/nsb936. [DOI] [PubMed] [Google Scholar]
- 32.Sexton A.C., Good R.T., Hansen D.S., D'Ombrain M.C., Buckingham L., Simpson K. Transcriptional profiling reveals suppressed erythropoiesis, up-regulated glycolysis, and interferon-associated responses in murine malaria. J Infect Dis. 2004 April 1;189(7):1245–1256. doi: 10.1086/382596. [DOI] [PubMed] [Google Scholar]
- 33.Deininger M.H., Kremsner P.G., Meyermann R., Schluesener H.J. Focal accumulation of cyclooxygenase-1 (COX-1) and COX-2 expressing cells in cerebral malaria. J Neuroimmunol. 2000 July 1;106(1–2):198–205. doi: 10.1016/s0165-5728(00)00187-9. [DOI] [PubMed] [Google Scholar]
- 34.Lewerenz J., Dargusch R., Maher P. Lactacidosis modulates glutathione metabolism and oxidative glutamate toxicity. J Neurochem. 2010 April;113(2):502–514. doi: 10.1111/j.1471-4159.2010.06621.x. [DOI] [PubMed] [Google Scholar]
- 35.Lee H.J., Georgiadou A., Walther M., Nwakanma D., Stewart L.B., Levin M. Integrated pathogen load and dual transcriptome analysis of systemic host-pathogen interactions in severe malaria. Sci Transl Med. 2018 June 27;10(447) doi: 10.1126/scitranslmed.aar3619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Franklin B.S., Ishizaka S.T., Lamphier M., Gusovsky F., Hansen H., Rose J. Therapeutical targeting of nucleic acid-sensing Toll-like receptors prevents experimental cerebral malaria. Proc Natl Acad Sci U S A. 2011 March 1;108(9):3689–3694. doi: 10.1073/pnas.1015406108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Morrell C.N., Srivastava K., Swaim A., Lee M.T., Chen J., Nagineni C. Beta interferon suppresses the development of experimental cerebral malaria. Infect Immun. 2011 April;79(4):1750–1758. doi: 10.1128/IAI.00810-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Beare N.A., Harding S.P., Taylor T.E., Lewallen S., Molyneux M.E. Perfusion abnormalities in children with cerebral malaria and malarial retinopathy. J Infect Dis. 2009 January 15;199(2):263–271. doi: 10.1086/595735. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material