A number of miRNAs were differentially expressed by tumor subtype and between races. These ER-subtype and race-related miRNAs may represent promising candidate biomarkers for ER−/triple negative breast cancer and may serve as new therapeutic targets for treatment.
Abstract
Breast cancer is a heterogeneous disease, characterized by molecularly and phenotypically distinct tumor subtypes, linked to disparate clinical outcomes. American women of African ancestry (AA) are more likely than those of European ancestry (EA) to be diagnosed with aggressive, estrogen receptor negative (ER−) or triple negative breast cancer, and to die of this disease. However, the underlying causes of AA predisposition to ER−/triple negative breast cancer are still largely unknown. In this study, we performed high-throughput whole-genome miRNA expression profiling in breast tissue samples from both AA and EA women. A number of differentially expressed miRNAs, i.e., DEmiRs defined as >2-fold change in expression and false discovery rate <0.05, were identified as up- or downregulated by tumor ER status or by ancestry. We found that among 102 ER-subtype-related DEmiRs identified in breast tumors, the majority of these DEmiRs were race specific, with only 23 DEmiRs shared in tumors from both AAs and EAs; this finding indicates that there are unique subsets of miRNAs differentially expressed between ER− and ER positive tumors within AAs versus EAs. Our overall results support the notion that miRNA expression patterns may differ not only by tumor subtype but by ancestry, indicating differences in tumor biology and heterogeneity of breast cancer between AAs and EAs. These results will provide the basis for further functional analysis to elucidate biological differences between AAs and EAs and to help develop targeted treatment strategies to reduce disparities in breast cancer.
Introduction
Breast cancer is a heterogeneous disease, characterized by molecularly and phenotypically distinct tumor subtypes, linked to disparate clinical outcomes. This biological heterogeneity has significant implications; as in routine clinical practice, the treatment regimens for breast cancer are still primarily determined based on hormonal receptor expression of the primary tumor, particular expression of the estrogen receptor (ER) (1). Compared with women diagnosed with ER positive (ER+) breast cancer, those with ER negative (ER−) tumors in general have a poor prognosis, partly because of their aggressive phenotype and the lack of targeted therapy. It is well established that breast cancer disproportionately affects American women of African ancestry (AA) and those of European ancestry (EA). Specifically, AA women are more likely than EAs to be diagnosed with breast cancer at younger ages; to develop more aggressive, ER− or triple negative (TN) tumors; and to die of this disease (2). One biological explanation for this mortality disparity is the higher proportion of aggressive subtypes seen in AAs. However, the underlying causes of AA predisposition to ER−/TN breast cancer are still largely unknown. Further studies are needed to investigate potential race-related tumor biological differences, and to define how such differences may drive more aggressive phenotypes in AA women.
Several studies have examined biological differences in tumors between AAs and EAs, and found a number of race-associated genomic differences on DNA methylation, genetic variants, protein and gene expression (3–5). However, studies are still limited on the potential regulatory mechanisms associated with these observed differences. MicroRNAs (miRNAs) are a family of small non-coding RNAs that function as key regulators of gene expression, through induction of mRNA degradation or interference with mRNA translation (6,7). A single miRNA can potentially regulate the expression of hundreds of genes, and as a consequence, they play essential roles in the regulation of many important biologic processes in cancer pathophysiology, including cell proliferation, differentiation, apoptosis, immunity and the stemness of the cancer cells (8,9).
Recent research has shown that miRNAs are aberrantly expressed in nearly all human cancers and that this altered miRNA expression has been associated with distinct tumor characteristics and cancer prognosis (10–13). In breast cancer, much of the miRNA research has focused on identifying changes with tumor clinicopathological factors, particularly in regards to tumor ER subtype. Some miRNAs have been shown to be up- or downregulated, and have been associated with tumor pathologic features and subtypes (11,14–16). Several miRNAs have also been found to play roles in breast cancer invasion and metastasis (17–19). However, most of these earlier studies focused on a small number of candidate miRNAs, or a defined larger subset, using real-time PCR or microarrays. In addition, most of these studies did not attempt to link miRNAs and their mRNA targets. Most importantly, few studies have examined the tumor miRNA expression changes in AA populations and their potential role in breast cancer disparities. Examining differential miRNA expression in aggressive ER− groups, which are more common in AA women, may provide insights into understanding breast cancer racial disparities. In addition, identification and quantification of miRNA changes by ER subtype within and across race may improve our understanding the biology and heterogeneity of breast cancer, e.g. how miRNA expression modulate ER, and further how these miRNAs could serve as potential therapeutic targets for novel treatments.
In this study, we aim to characterize miRNA expression patterns in breast tissue samples from both AA and EA women by performing whole-genome miRNA expression profiling using next-generation sequencing technologies (miRNA-seq). We hypothesized that miRNA expression patterns in breast cancer differ by tumor subtype and between races. Given limited data on AA women, we examined miRNA expression changes in ER− versus ER+ tumors separately for each race, and also compared the variability within and across race. For a subgroup of tumors analyzed by miRNA-seq, mRNA sequencing (RNA-seq) data were also available, which allowed us to integrate miRNA–mRNA data to examine potentially targeted pathways of miRNAs.
Materials and methods
Tissue sample collection
Fresh frozen tissue samples were obtained from 58 breast cancer patients (29 AA and 29 EA women) and 10 (5 AA and 5 EA) healthy women. The protocol was approved by the Roswell Park Institutional Review Board. Breast tumor tissues were collected from patients who were treated at Roswell Park, after patient consent for use of remnant tissue for research, snap frozen and stored by the Roswell Park Pathology Network Shared Resource. Normal breast tissue samples were from women undergoing reduction mammoplasty, with pathology determined free of any abnormalities, as described previously (20). Total RNA was extracted and stored at −80°C until use.
miRNA sequencing and data processing
miRNA sequencing was performed at the University at Buffalo New York State Center of Excellence in Bioinformatics and Life Sciences. A total of 1 μg of total RNA was used for small RNA cDNA library preparation using the NEBNext Multiplex Small RNA kit, which has been optimized to minimize adaptor-dimers contamination while producing high-yield, high-diversity libraries. The Blue Pippin Prep from Sage Sciences was used for size selection. The samples were sequenced on the Illumina HiSeq 2500 platform, a powerful and efficient ultra-high-throughput sequencing system, using High Output 50-Cycle Single Read sequencing. We ran 12 samples in each lane, which could generate a good sequencing depth, an average of ~12.5 million reads per sample. All samples were randomized across the six lanes with respect to patient’s race for sequencing. According to Illumina guidelines for small RNA sequencing, 1–2 million reads is an accepted range for expression profiling experiments, whereas 2–5 million reads is the accepted range for discovery applications.
The 3′ adapter (AGATCGGAAGAGCACACGTCTGAACTCCAGTCA) in the reads was trimmed by cutadapt (version 1.12). Only the reads with at least 6 nt perfect match against the adapter sequences and longer than 15 nt were kept for the downstream analysis. The trimmed reads were aligned to human reference genome (hg19) by Burrows−Wheeler Aligner (version 0.7.15) with no mismatches allowed. The miRNA profiling pipeline developed by the British Columbia Genome Sciences Centre was implemented to quantify miRNA expression. The-Cancer-Genome-Atlas-formatted miRNA quantification data were generated including expression levels for the known pre-miRNAs and mature miRNAs for all samples. The variance stabilizing transformation was applied to raw count data by DESeq2 package. Unsupervised hierarchical clustering analysis was performed on miRNA expression patterns of breast tissue samples.
Detection of differentially expressed miRNAs
The R package DESeq2 (21) was used to identify differentially expressed miRNAs (DEmiRs) between groups. Specifically, DESeq2 normalizes read counts using size factors, which is the median ratio of a sample’s observed counts to the geometric mean of counts across all samples. Differences in read counts for each individual miRNA were evaluated in breast tissues between groups, i.e. normal versus patients, ER− versus ER+ tumors, and AAs versus EAs. DESeq2 estimates dispersion using the Cox Reid-adjusted profile likelihood method. The negative binomial generalized linear model was further used to obtain maximum likelihood estimates for each miRNA’s log-fold change between two groups, and then the Wald test was used to determine DEmiRs. The false discovery rate (FDR) was computed using the Benjamini and Hochberg approach to adjust for multiple comparisons. The statistically significant DEmiRs were defined as fold change (FC) >2.0 and FDR<0.05.
The Cancer Genome Atlas (TCGA) data processing analysis
TCGA miRNA expression data were downloaded from GDAC Broad Institute (http://gdac.broadinstitute.org/runs/stddata__2016_01_28/data/BRCA/20160128/), and only data generated by the same sequencing platform as this study (i.e. Illumina HiSeq) were included. Only samples annotated as ‘primary solid tumor’, with race ‘white’ or ‘black or African American’ and ‘Positive’ or ‘Negative’ ER status were included. This filtering process retained 648 samples for the downstream analysis, with tumor characteristics described in Supplementary Table 1, available at Carcinogenesis Online. All data were processed and analyzed following the same pipeline as used in the current study. The DEmiRs were defined the same parameters as our data for comparison.
RNA-seq and data processing
Among the 58 breast tumors included in the miRNA-seq, RNA-seq was performed on 50 tumors, as described previously (22). Briefly, a 1 μg’s aliquot of RNA quantified by Quant-iT RiboGreen RNA assay Kit (Invitrogen #R11490) was used for library preparation using TruSeq(r) Stranded Total RNA LT with Ribo-Zero TM Gold (Illumina #RS1222301). Library preparations were run on HiSeq2500, generating 100 bp stranded paired-end reads. In this analysis, the RNA-seq reads were aligned to human reference genome (hg19) by STAR (version 2.5.2b) with the GENCODE gene annotation (version 19). Then RSEM (version 1.2.31) was applied for the gene quantification, the expression level for each gene was measured in counts per million. Similar to miRNA-seq data analysis, the significant differentially expressed mRNAs were defined by the DESeq2 package with FC>2.0 and FDR<0.05. We further excluded 19 samples with relatively low useable sequencing reads (i.e. <25% reads), leaving 31 tumor samples in the following miRNA–mRNA integrated analysis, which include 17 ER+ and 14 ER− tumors (Supplementary Table 2, available at Carcinogenesis Online).
miRNA target gene prediction and pathway analysis
The Ingenuity Pathway Analysis (IPA) microRNA Target Filter was used to identify mRNA targets for the DEmiRs. IPA incorporates experimentally validated targets from TarBase and miRecords, predicted targets from TargetScan (Human), and manually collected miRNA–mRNA interactions from the peer-reviewed literature, which allows for the integration of miRNA with both validated and predicted mRNA targets. After prioritizing the miRNA–mRNA relationships in IPA, RNA-seq data were incorporated into the target filter analysis pipeline. Then, the expression-pairing filter was applied to specifically identify mRNA targets inversely correlated in expression with corresponding miRNAs. Finally, the Core Analysis in IPA was performed to understand the functions of these target genes. A right-tailed Fisher’s exact test was run through IPA software and functions with P-value <0.05 were considered significant. In this analysis, we focused on miRNAs and mRNA targets with inverse relationships based on the hypothesis that miRNAs bind to their target mRNA and negatively regulate its expression.
Results
miRNA expression profiling in normal and tumor breast tissue
miRNA-seq was performed to profile miRNA expression in breast tissue samples from 58 breast cancer patients (29 AA and 29 EA) and 10 (5 AA and 5 EA) women undergoing reduction mammoplasty. Overall patient and tumor characteristics were comparable between AA and EA patients (Table 1).
Table 1.
Tumor characteristics of 58 breast cancer patients.
| AA | EA | ||
|---|---|---|---|
| Characteristics | N (29) | N (29) | P a |
| Age | 0.57 | ||
| <50 | 8 | 10 | |
| ≥50 | 21 | 19 | |
| ER status | 0.60 | ||
| Positive | 15 | 17 | |
| Negative | 14 | 12 | |
| PR status | 0.79 | ||
| Positive | 13 | 14 | |
| Negative | 16 | 15 | |
| HER2 | 0.01 | ||
| Positive | 4 | 12 | |
| Negative | 25 | 16 | |
| Grade | 0.79 | ||
| Moderate-differentiated | 6 | 4 | |
| Poorly differentiated | 23 | 25 | |
| Node status | 1.00 | ||
| Negative | 10 | 10 | |
| Positive | 19 | 19 | |
| Tumor stage | 1.00 | ||
| Stage I and II | 18 | 18 | |
| Stage III and IV | 11 | 11 |
aChi-square or Fisher’s exact test, as appropriate.
We detected a total of 2113 mature miRNAs in this sample set. Of the 1333 miRNAs detected in normal breast, all but 16 were also expressed in breast tumors along with an additional 780 miRNAs identified only in cancer samples (total of 2097). Unsupervised hierarchical clustering based on the top most 500 highly expressed miRNAs identified two distinct clusters of miRNA expression patterns (Supplementary Figure 1, available at Carcinogenesis Online). Cluster 1 contains normal breast samples (yellow bar), cluster 2 includes tumor samples (green/purple), with cluster 2a enriched for ER− cancers (green bar) and cluster 2b enriched for ER+ subtypes (purple bar). We profiled only a small number of normal breast samples because our study focused on differences in tumors by race and ER status; nevertheless, our data showed that miRNA expression patterns clearly distinguish tumor from normal breast tissue. No significant clustering was observed between tumor samples from the two races. As shown in Supplementary Figure 2, available at Carcinogenesis Online, of the 500 miRNAs identified as differentially expressed (DEmiRs, FC>2; FDR<0.05) between tumor and normal tissue, 106 were specific to AAs, 106 were unique to EAs and 288 were observed for both AAs and EAs.
Identification of DEmiRs between ER− and ER+ tumors in AAs and EAs
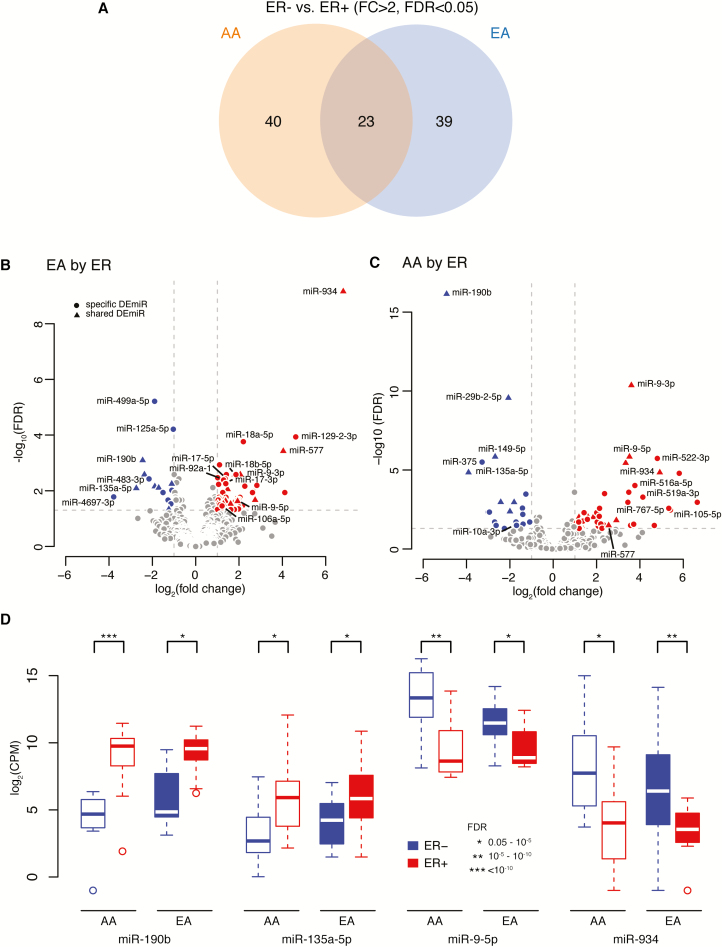
As data are limited in tumor miRNA expression profiling in AA populations and there is a clear pattern of miRNA differences by ER subtype, in the current study, we first performed analyses to identify miRNAs differentially expressed by ER status for each race, separately. In cancers from AA women, we identified 63 DEmiRs between ER− and ER+ breast tumors (43 upregulated and 20 downregulated). Similarly, in tumors from EA women, we identified 62 DEmiRs (47 miRs upregulated and 15 downregulated). Among the 102 unique DEmiRs by ER status identified in breast tumors from either AA or EA women, 23 DEmiRs were identified in both races with the same direction in expression change, 40 were specific to AAs and 39 were specific to EAs (Figure 1A). As shown in volcano and box plots (Figure 1B–D), in breast tumors from both AA and EA women, miR-934 and miR-9 (-3p, -5p) were among the most highly expressed and upregulated miRNAs in ER− versus ER+ tumors, whereas miR-190b and miR-135a-5p were among the highly downregulated. In contrast, multiple members of the miR-17-92 cluster (e.g. miR-17, -18a, -18b and -92a-1) and paralogues, miR-106a-363 and miR-106b-25, were all upregulated specifically in EA tumors, whereas miR-4697-3p and miR-483-3p were downregulated. On the other hand, in AA tumors, multiple members of the miR-515 family (e.g. miR-522, -516a, -519a) were upregulated, whereas miR-375 and miR-10a-3p/5p were downregulated in these samples. The full list of these significant DEmiRs by ER subtype for each group is presented in Supplementary Table 3a–c, available at Carcinogenesis Online.
Figure 1.
The differentially expressed miRNAs [DEmiRs: fold change (FC)>2; false discovery rate (FDR)<0.05] between estrogen receptor negative (ER−) and ER positive (ER+) tumors in African ancestry (AA) and European ancestry (EA) women. (A) Venn diagram showing the number of DEmiRs by ER status specific to AA women (n = 40), common in both AA and EA women (n = 23) and specific to EA women (n = 39). (B and C) Volcano plots showing selected up- or downregulated DEmiRs by ER status in EA (B) and in AA women (C). Closed circles represent DEmiRs specific to either AA or EA women; triangles represents DEmiRs common in both AA and EA women; red represents for upregulated and blue for downregulated DEmiRs. The log2 fold change of miRNA expression in ER− versus ER+ tumors is on the x-axis, and the y-axis shows the log10 of FDR (adjusted P-value). (D) Box plots of log2 of counts per million reads (CPM) of miR-190b, miR-135a-5p, miR-9-5p and miR-934 in ER− and ER+ tumors among AA and EA women. Blue represents ER− and red represents ER+ tumor.
Identification of DEmiRs between breast tumors from AA and EA women
We then compared miRNA expression patterns between tumors from the two race groups, stratified by tumor ER status. Our analysis identified eight miRs differentially expressed between AAs and EAs within ER+ tumors (Table 2). Of these, miR-105-5p, miR-767-5p and miR-122-5p, were among the most downregulated, whereas miR-187-3p and miR-937-3p were the most upregulated in AAs compared with EAs. Four DEmiRs by race were identified within ER− tumors (Table 2), all four of which (miR-10a-3p, miR-10a-5p, miR-3176 and miR-190b) were downregulated in AAs compared with EAs.
Table 2.
MiRNAs significantly differentially expressed (FC > 2, FDR < 0.05) in breast tumors between AA and EA women.
| miRNA | ER+ tumors (AA versus EA) | miRNA | ER− tumors (AA versus EA) | ||||||
|---|---|---|---|---|---|---|---|---|---|
| Regulation | Log2 FC | P-value | FDR | Regulation | Log2 FC | P-value | FDR | ||
| hsa-miR-105-5p | Down | 5.49 | 3.06E-05 | 0.014 | hsa-miR-3176 | Down | 2.79 | 2.48E-06 | 0.002 |
| hsa-miR-767-5p | Down | 5.26 | 3.69E-05 | 0.014 | hsa-miR-10a-3p | Down | 2.73 | 2.62E-06 | 0.001 |
| hsa-miR-122-5p | Down | 3.16 | 4.70E-05 | 0.014 | hsa-miR-10a-5p | Down | 2.54 | 5.20E-06 | 0.007 |
| hsa-miR-3607-5p | Down | 1.99 | 0.0003 | 0.036 | hsa-miR-190b | Down | 2.25 | 8.32E-05 | 0.02 |
| hsa-miR-499a-5p | Down | 1.23 | 8.28E-05 | 0.019 | |||||
| Has-miR-4482-5p | Up | 2.21 | 0.0002 | 0.036 | |||||
| Has-miR-937-3p | Up | 2.21 | 0.0002 | 0.036 | |||||
| Has-miR-187-3p | Up | 2.52 | 0.0004 | 0.044 | |||||
Validation of DEmiRs by analysis of TCGA
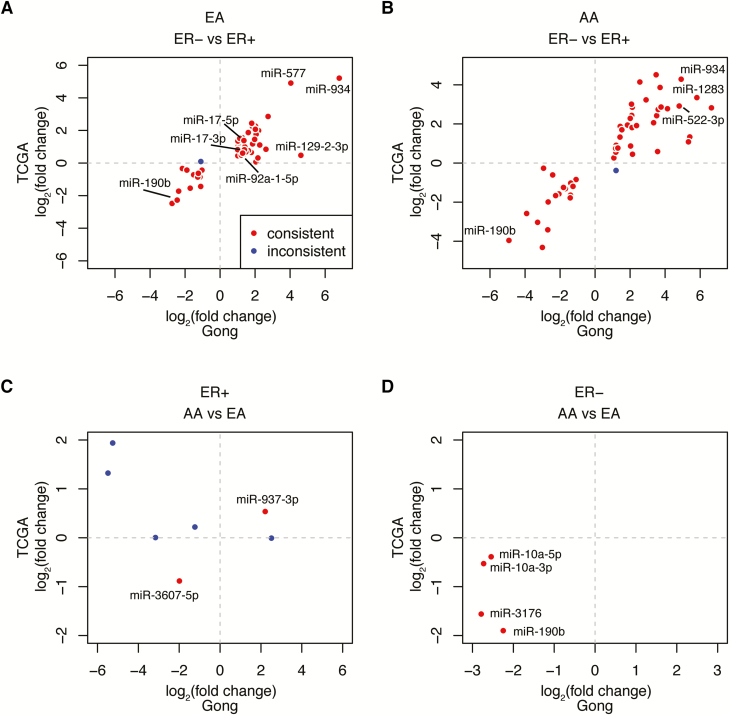
The same analysis pipeline was used to identify DEmiRs in 648 breast cancer samples in TCGA. When comparing changes in miRNA expression by ER status, we observed good concordance in the ‘direction’ (i.e. up- or downregulated in ER− tumors compared with ER+ tumors) of expression change in our dataset with those identified in TCGA; the large majority of DEmiRs identified in our study showed the same direction of expression differences between ER-subtypes as in the TCGA dataset (Figure 2A and B, plotted in red). For example, miR-934 was among the most upregulated, whereas miR-190b was among the most downregulated in ER− versus ER+ tumors in both races in both datasets. Consistent with our data, in EAs, multiple members of the miR-17-92 cluster, such as miR-17a-5p/-3p, were upregulated, whereas in AAs, several members of miR-515 family, such as miR-522 and miR-1283, were significantly upregulated. There were only a few DEmiRs that were inconsistent with direction of expression differences (plotted in blue). In contrast, the concordance was lower among the few DEmiRs identified by race, especially in ER+ tumors (Figure 2C and D). We found that in ER− tumors, two (miR-3176 and miR-190b) showed consistent expression changes in both datasets, whereas the other two (miR-10a-3p, -5p) showed consistent direction of expression differences but did not reach statistical significance in TCGA data to be identified as a DEmiRs. In ER+ tumors, two DEmiRs (miR-937 and miR-3607) showed consistent changes in the direction of expression whereas the others did not (Figure 2C). The FC and P-values of these miRNAs are included in Supplementary Tables 4a–c and 5a and b, available at Carcinogenesis Online.
Figure 2.
DEmiRs identified in the current (Gong) study and in The Cancer Genome Atlas (TCGA). (A and B) Scatter plot of fold-change by estrogen receptor (ER) subtype for DEmiRs from Gong (x-axis) versus TCGA (y-axis) in European ancestries (EAs) (A) and in African ancestries (AAs) (B). (C and D) Scatter plot of fold-change by race for DEmiRs from Gong (x-axis) versus TCGA (y-axis) in ER+ (C) and in ER− tumors (D). DEmiRs with consistent fold change direction in both data are plotted in red; and DEmiRs with inconsistent fold-change direction are plotted in blue.
We also similarly present DEmiRs identified either in our dataset or in TCGA in Supplementary Figure 3, available at Carcinogenesis Online. For DEmiRs by ER status (Supplementary Figure 3A and B, available at Carcinogenesis Online), we observed, overall, that DEmiRs identified in both datasets showed consistent changes in expression direction (plotted in orange). A number of miRNAs were identified as DEmiRs (i.e. FC>2, FDR<0.05) only in our study or in the TCGA dataset (plotted in red and blue, respectively); nevertheless, most of these miRNAs showed the same direction of expression change in both datasets. In contrast, fewer DEmiRs by race were identified in both data, and the concordance was lower, especially in ER+ tumors (Supplementary Figures 3C and D, available at Carcinogenesis Online).
mRNA target identification of DEmiRs and pathway analysis
To investigate the biological relevance of the DEmiRs, their target genes and corresponding expression values were uploaded into the Dataset Files of IPA software. The pathway analysis in IPA was then performed to identify biological functions and predominant canonical pathways from the IPA library based on their significance and enrichment in these pathways. Our findings suggest that these miRNAs play a role in various key molecular processes related to and beyond ER biology. The top canonical pathways related to DemiRs by ER status in EAs and AAs are shown in Figure 3. Several ER-subtype associated pathways showed similar activities in both races, such as p53 signaling, Wnt/β-catenin and estrogen-dependent breast cancer signaling. In contrast, a number of pathways showed different levels of activities between EAs and AAs. For example, several G protein related pathways, including G-proteins, Gαi, Gαs, Gβγ and cAMP-mediated signaling, were upregulated in AAs, but downregulated in EAs. As a case in point, among targets within the cAMP-mediated signaling pathway, our mRNAseq analysis showed that several G-protein coupled receptors (GPCR; e.g. FFAR3, GLP1R and NPR3) were upregulated in ER− compared with ER+ tumors in AA women, but not in EA women. We also found a small number of gene targets for the few DEmiRs identified by race. The gene targets for several DEmiRs are listed in Supplementary Table 6a and b, available at Carcinogenesis Online. For example, we found in ER− tumors, several target genes of miR-3176 (e.g. ADIPOQ, SCRG1, USH1G and PLA2G2A) were upregulated in AA versus EA women.
Figure 3.
Top canonical pathways from IPA on by-ER DEmiR-target genes in AA and EA women. The heatmap shows the top pathways enriched from target genes of DEmiRs in EA and in AA women, respectively. The calculated activation Z score indicates a pathway with genes exhibiting overall increased mRNA levels (positive Z score: red bars) or decreased mRNA levels (negative Z score: blue bars).
Discussion
Unlike previously published studies that focused on EA women, we evaluated miRNA expression patterns in breast tissues of both AA and EA women. Our results show that genome-wide miRNA profiling can clearly distinguish normal from tumor tissue, as well as most ER− tumors from ER+ tumors. We identified a number of miRNAs differentially expressed by tumor ER subtype within both races, and found that a majority were race specific, with only 23 DEmiRs shared in tumors from both AA and EA women. These data indicate that there are unique subsets of miRNAs significantly associated with breast cancer ER phenotypes within AA and EA samples. Further, in the direct comparison of tumor miRNA expression levels between tumor samples from AAs and EAs, we observed modest differences, as eight and four miRNAs were identified within ER+ and within ER− tumors, respectively, further suggesting potential racial differences in tumor miRNA expression.
A subset of identified miRNAs was differentially regulated in ER− versus ER+ tumors, regardless of race. Top DEmiRs include miR-934, miR-9-3p/5p, miR-18a/18b, miR-135b and miR-190b, which showed expression changes consistent with results from our analysis of the TCGA dataset and from several previous studies (15,16,23–26). These observations are in line with the fact that most published studies have focused exclusively on EAs. Interestingly, in both races, miR-934 was highly upregulated in ER−/TN compared with ER+ tumors, and was previously found to be associated with downregulated ESR1 expression (25,26). Consistent with these reports, ESR1 was one of the most negatively correlated target genes for miR-934 in our dataset and in TCGA. These findings raise the intriguing possibility that such miRNAs may be functionally involved in the development or maintenance of the ER− tumor phenotype, through repression of pro-luminal gene expression (e.g. ESR1). Thus, miR-934 may represent a promising biomarker for ER−/TN breast cancer and possible future therapeutic target for aggressive types of breast cancer for both AA and EA women.
Interestingly, we identified a number of ER-subtype specific miRNAs unique to tumors from either EAs or AAs. Within EA tumors, multiple members of the miR-17-92 cluster and its two paralogues were significantly upregulated in ER− compared with ER+ tumors in our dataset and in TCGA. This was also similarly observed in the study by Enerly et al.(25), which found these were the most significantly differentially expressed clusters, and several of these miRNAs were overexpressed in highly proliferative, basal-like tumors. The deregulation of members of these clusters has been frequently reported in several cancer types (27–30), and they have been found to target genes such as PTEN and TGF-β signaling, with important functions in cell cycle progression, apoptosis and angiogenesis (31–33). Within AA tumors in contrast, we identified multiple members of the miR-515 family that were significantly upregulated in ER− versus ER+ tumors. Members of the miR-515 family belong to the chromosome 19 miRNA cluster, which recent studies have implicated in cancer and stem cell biology (34). In the single previous study that examined differences in miRNA expression by tumor subtype in AAs, miR-522 was found to be upregulated in TN versus non-TN tumors (35). Overexpression of miR-522 has been shown to promote breast cancer cell detachment, invasion, and mesenchymal gene expression, properties associated with metastasis (36). Collectively, our results suggest that miRNAs in different clusters may be specifically involved in the biology of the aggressive, ER−/TN breast cancer in EAs or in AAs. The unique expression changes of these miRNAs by ER status of EA or AA women could be used as additional diagnostic and prognostic biomarkers to stratify patients and to better assess treatment responses. More importantly, it is feasible to design novel, effective therapeutics that directly target these miRNAs and/or their target genes for treatment of aggressive, ER− breast cancer, specifically for AA or EA women.
As for overall functions of these ER-subtype associated DEmiRs, a number of signaling pathways were enriched in either AAs or EAs. Particularly, we found several G-protein-related pathways were among the most upregulated pathways in tumors from AAs compared with those from EAs. G-proteins are important cell-surface molecules and responsible for cellular signal transduction (37). G-protein activation through GPCR stimulation triggers the production of other second messengers (e.g. cAMP), which further regulate critical intracellular pathways and drive the migration, invasion and metastasis of cancer cells (38,39). A recent study showed that GPCR 161 (GPR161) is overexpressed specifically in TN breast cancer and associated with poor prognosis (40). Our findings that DEmiR target genes enriched specifically for G-protein pathways in AA women who are often diagnosed with aggressive types of breast cancer are intriguing, as emerging evidence suggests that GPCRs and downstream signaling networks have a crucial role in tumor growth, angiogenesis and metastasis (39). It is likely that G-proteins and GPCRs may be suitable targets for cancer prevention and treatment, particularly for AA women (41).
Compared with differences by ER status, we observed fewer DEmiRs when directly comparing tumors by race within a specific ER subtype. This may be explained by our limited sample size, and the fact that race-based differences in tumor miRNA expression may be moderate in magnitude. However, several by-race DEmiRs within ER− tumors were consistent with results from TCGA. For example, miR-190b and miR-3176 were significantly downregulated in AAs compared with EAs. Lower expression of miR-190b and miR-3176 has been associated with reduced breast cancer metastasis-free survival (16) and prostate cancer chemoresistance (42), respectively. Given the small number of significant mRNA targets identified for the small number of race-associated DEmiRs, we did not carry out a formal pathway analysis. However, we observed that several target genes of miR-3176, such as ADIPOQ and PLA2G2A, were associated with breast cancer progression (43) and poor therapeutic response and survival in rectal cancer (44). Further larger studies will be needed to validate these intriguing findings.
Our study has several strengths. We performed miRNA expression profiling using miRNA-seq, which not only enables quantitation of miRNA expression differences between groups, but also characterizes, globally, the presence and abundance of all miRNAs across the miRome. Unlike most previous studies, we examined tumor miRNA expression changes in tumors from both AA and EA women and used TCGA data as a validation of our findings. Although our primary focus was not to provide mechanistic data, we integrated miRNA–mRNA expression data and investigated potential biological functions and pathways for altered miRNAs and their target genes. This study also has limitations. Our sample size is relatively small, though we used comprehensive high-throughput technology with adequate depth and matched patients between groups. In contrast to miRNA expression differences between tumor subtypes within a given race, we observed smaller differences between races within a given tumor subtype. This could be explained by limited statistical power, and the fact that absolute race-based differences in tumor miRNA expression may be moderate. In addition, although the major purpose of this current study is to examine expression patterns of mature miRNAs, recent work suggested that isoforms of miRNAs (i.e. isomiRs) differ by breast cancer subtypes and between races (45,46). We will consider integrating expression of isomiRs in our future analyses.
In summary, limited studies to date have evaluated the potential role of miRNAs in breast cancer disparities. Our study identified a number of dysregulated miRNAs not only by tumor subtype within a given race, but between races within a given subtype, indicating differences in tumor biology and heterogeneity of breast cancer between EAs and AAs. Although exact functional roles in breast carcinogenesis remain to be further elucidated, these ER– and race-related miRNAs may represent promising candidate biomarkers for ER−/TN cancers and may serve as new therapeutic targets for treatment.
Funding
Breast Cancer Research Foundation (to C.B.A.); National Institutes of Health/National Cancer Institute [K07 CA178293 (to Z.G.), U24CA232979, R01 CA1332641 and P01 CA151135]; The Roswell Park Pathology Network Shared Resource, DataBank and BioRepository, and Bioinformatics are Roswell Park CCSG Shared Resources (NIH P30 CA016056).
Conflict of Interest Statement: None declared.
Supplementary Material
Abbreviations
- AA
African ancestry
- DEmiRs
differentially expressed miRNAs
- EA
European ancestry
- ER
estrogen receptor
- FC
fold change
- FDR
false discovery rate
- GPCR
G-protein-coupled receptor
- miRNA
microRNA
- TCGA
The Cancer Genome Atlas
- IPA
Ingenuity Pathway Analysis
References
- 1. Yersal O., et al. (2014)Biological subtypes of breast cancer: prognostic and therapeutic implications. World J. Clin. Oncol., 5, 412–424. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Troester M.A., et al. (2018)Racial differences in PAM50 subtypes in the Carolina breast cancer study. J. Natl. Cancer Inst., 110, 176–182. doi: 10.1093/jnci/djx135 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Ambrosone C.A., et al. (2014)Genome-wide methylation patterns provide insight into differences in breast tumor biology between American women of African and European ancestry. Oncotarget, 5, 237–248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Parada H., Jr, et al. (2017)Race-associated biological differences among luminal A and basal-like breast cancers in the Carolina breast cancer study. Breast Cancer Res., 19, 131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Huo D., et al. (2017)Comparison of breast cancer molecular features and survival by African and European ancestry in The Cancer Genome Atlas. JAMA Oncol., 3, 1654–1662. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Mattick J.S., et al. (2006)Non-coding RNA. Hum. Mol. Genet., 15 Spec No 1, R17–R29. [DOI] [PubMed] [Google Scholar]
- 7. He L., et al. (2004)MicroRNAs: small RNAs with a big role in gene regulation. Nat. Rev. Genet., 5, 522–531. [DOI] [PubMed] [Google Scholar]
- 8. Bartel D.P. (2004)MicroRNAs: genomics, biogenesis, mechanism, and function. Cell, 116, 281–297. [DOI] [PubMed] [Google Scholar]
- 9. Friedman R.C., et al. (2009)Most mammalian mRNAs are conserved targets of microRNAs. Genome Res., 19, 92–105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Calin G.A., et al. (2005)A MicroRNA signature associated with prognosis and progression in chronic lymphocytic leukemia. N. Engl. J. Med., 353, 1793–1801. [DOI] [PubMed] [Google Scholar]
- 11. Iorio M.V., et al. (2005)MicroRNA gene expression deregulation in human breast cancer. Cancer Res., 65, 7065–7070. [DOI] [PubMed] [Google Scholar]
- 12. Schetter A.J., et al. (2008)MicroRNA expression profiles associated with prognosis and therapeutic outcome in colon adenocarcinoma. JAMA, 299, 425–436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Di Leva G., et al. (2013)miRNA profiling of cancer. Curr. Opin. Genet. Dev., 23, 3–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Blenkiron C., et al. (2007)MicroRNA expression profiling of human breast cancer identifies new markers of tumor subtype. Genome Biol., 8, R214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. van Schooneveld E., et al. (2015)Dysregulation of microRNAs in breast cancer and their potential role as prognostic and predictive biomarkers in patient management. Breast Cancer Res., 17, 21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Cizeron-Clairac G., et al. (2015)MiR-190b, the highest up-regulated miRNA in ERα-positive compared to ERα-negative breast tumors, a new biomarker in breast cancers?BMC Cancer, 15, 499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Zhu S., et al. (2008)MicroRNA-21 targets tumor suppressor genes in invasion and metastasis. Cell Res., 18, 350–359. [DOI] [PubMed] [Google Scholar]
- 18. Tavazoie S.F., et al. (2008)Endogenous human microRNAs that suppress breast cancer metastasis. Nature, 451, 147–152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Ma L., et al. (2010)miR-9, a MYC/MYCN-activated microRNA, regulates E-cadherin and cancer metastasis. Nat. Cell Biol., 12, 247–256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Dumitrescu R.G., et al. (2010)Familial and racial determinants of tumour suppressor genes promoter hypermethylation in breast tissues from healthy women. J. Cell. Mol. Med., 14, 1468–1475. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Love M.I., et al. (2014)Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol., 15, 550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Espinal A.C., et al. (2017)FOXA1 hypermethylation: link between parity and ER-negative breast cancer in African American women?Breast Cancer Res. Treat., 166, 559–568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Volinia S., et al. (2012)Breast cancer signatures for invasiveness and prognosis defined by deep sequencing of microRNA. Proc. Natl. Acad. Sci. USA., 109, 3024–3029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Barbano R., et al. (2017)Stepwise analysis of MIR9 loci identifies miR-9-5p to be involved in Oestrogen regulated pathways in breast cancer patients. Sci. Rep., 7, 45283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Enerly E., et al. (2011)miRNA-mRNA integrated analysis reveals roles for miRNAs in primary breast tumors. PLoS One, 6, e16915. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Castilla M.A., et al. (2014)VGLL1 expression is associated with a triple-negative basal-like phenotype in breast cancer. Endocr. Relat. Cancer, 21, 587–599. [DOI] [PubMed] [Google Scholar]
- 27. Matsubara H., et al. (2007)Apoptosis induction by antisense oligonucleotides against miR-17-5p and miR-20a in lung cancers overexpressing miR-17-92. Oncogene, 26, 6099–6105. [DOI] [PubMed] [Google Scholar]
- 28. Olive V., et al. (2010)mir-17-92, a cluster of miRNAs in the midst of the cancer network. Int. J. Biochem. Cell Biol., 42, 1348–1354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Petrocca F., et al. (2008)Emerging role of miR-106b-25/miR-17-92 clusters in the control of transforming growth factor beta signaling. Cancer Res., 68, 8191–8194. [DOI] [PubMed] [Google Scholar]
- 30. Navon R., et al. (2009)Novel rank-based statistical methods reveal microRNAs with differential expression in multiple cancer types. PLoS One, 4, e8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. O’Donnell K.A., et al. (2005)c-Myc-regulated microRNAs modulate E2F1 expression. Nature, 435, 839–843. [DOI] [PubMed] [Google Scholar]
- 32. Ivanovska I., et al. (2008)MicroRNAs in the miR-106b family regulate p21/CDKN1A and promote cell cycle progression. Mol. Cell. Biol., 28, 2167–2174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Tsuchida A., et al. (2011)miR-92 is a key oncogenic component of the miR-17-92 cluster in colon cancer. Cancer Sci., 102, 2264–2271. [DOI] [PubMed] [Google Scholar]
- 34. Nguyen P.N., et al. (2017)Selective activation of miRNAs of the primate-specific chromosome 19 miRNA cluster (C19MC) in cancer and stem cells and possible contribution to regulation of apoptosis. J. Biomed. Sci., 24, 20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sugita B., et al. (2016)Differentially expressed miRNAs in triple negative breast cancer between African-American and non-Hispanic white women. Oncotarget, 7, 79274–79291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Tan S.M., et al. (2014)Sequencing of captive target transcripts identifies the network of regulated genes and functions of primate-specific miR-522. Cell Rep., 8, 1225–1239. [DOI] [PubMed] [Google Scholar]
- 37. Neves S.R., et al. (2002)G protein pathways. Science, 296, 1636–1639. [DOI] [PubMed] [Google Scholar]
- 38. Dorsam R.T., et al. (2007)G-protein-coupled receptors and cancer. Nat. Rev. Cancer, 7, 79–94. [DOI] [PubMed] [Google Scholar]
- 39. O’Hayre M., et al. (2014)Novel insights into G protein and G protein-coupled receptor signaling in cancer. Curr. Opin. Cell Biol., 27, 126–135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Feigin M.E., et al. (2014)G-protein-coupled receptor GPR161 is overexpressed in breast cancer and is a promoter of cell proliferation and invasion. Proc. Natl. Acad. Sci. USA, 111, 4191–4196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Lappano R., et al. (2011)G protein-coupled receptors: novel targets for drug discovery in cancer. Nat. Rev. Drug Discov., 10, 47–60. [DOI] [PubMed] [Google Scholar]
- 42. Li J., et al. (2016)Exosome-derived microRNAs contribute to prostate cancer chemoresistance. Int. J. Oncol., 49, 838–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Libby E.F., et al. (2016)Globular adiponectin enhances invasion in human breast cancer cells. Oncol Lett., 11, 633–641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. He H.L., et al. (2015)PLA2G2A overexpression is associated with poor therapeutic response and inferior outcome in rectal cancer patients receiving neoadjuvant concurrent chemoradiotherapy. Histopathology, 66, 991–1002. [DOI] [PubMed] [Google Scholar]
- 45. Telonis A.G., et al. (2015)Beyond the one-locus-one-miRNA paradigm: microRNA isoforms enable deeper insights into breast cancer heterogeneity. Nucleic Acids Res., 43, 9158–9175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Telonis A.G., et al. (2018)Race disparities in the contribution of miRNA isoforms and tRNA-derived fragments to triple-negative breast cancer. Cancer Res., 78, 1140–1154. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.