Abstract
Background
The role of exosomal circular RNAs (circRNAs) in Hepatocellular carcinoma (HCC) cells with high metastatic potential has been little studied.
Methods
Exosomal circRNA from cells with non-metastatic (HepG2), low metastatic (97L), and high metastatic (LM3) potential were sequencing. Metastatic-related circRNAs in serum from HCC patients were measured and their association with clinical prognosis was evaluated. Furthermore, candidate functional circRNAs in LM3-derived exosomes was assessed.
Findings
LM3 exosomes enhanced the cell migration and invasion potential of HepG2 and 97 L cells. CircPTGR1, a circRNA with three isoforms, was specifically expressed in exosomes from 97 L and LM3 cells, upregulated in serum exosomes from HCC patients and was associated with the clinical stage and prognosis. Knockdown of circPTGR1 expression suppressed the migration and invasion of HepG2 and 97L cells induced by co-culturing with LM3 exosomes. Bioinformatics, co-expression analysis, and a luciferase assay indicated that circPTGR1 competed with MET to target miR449a.
Interpretation
Higher metastatic HCC cells can confer this potential on those with lower or no metastatic potential via exosomes with circPTGR1, resulting in increased migratory and invasive abilities in those cells.
Fund
National Natural Science Foundation of China (No. 81470870, 81670601, 81570593), Guangdong Natural Science Foundation (No. 2015A030312013, 2015A030313038), Sci-tech Research Development Program of Guangdong Province (2014B020228003), Sci-tech Research Development Program of Guangzhou City (No. 201508020262, 201400000001-3, 201604020001, 201607010024), Innovative Funds for Small and Medium-Sized Enterprises of Guangdong Province (2016A010119103), Pearl River S&T Nova Program of Guangzhou (201710010178), and National 13th Five-Year Science and Technology Plan Major Projects of China (No. 2017ZX10203205-006-001).
Keywords: circPTGR1, Hepatocellular carcinoma, Metastasis, miR449a
Research in context.
Evidence before this study
Exosomes originating from various types of hepatocellular carcinoma (HCC) cells have been characterized, and their effects on cell growth, metastasis, and drug resistance have been determined. Circular RNA (circRNA) is a class of RNA derived from precursor mRNA, which has been reported to play roles in tumors, including HCC. However, exosomal circRNA and its biological function in HCC cells, especially those with high metastatic potential, have been little studied.
Added value of this study
This study has provided information about exosomal circRNA in HCC cells with different metastatic potentials. In particular, we identified three isoforms of circPTGR1 that are preferentially located in exosomes derived from LM3, an HCC cell line with high metastatic potential. These were found to be associated with the clinical stage and prognosis of HCC patients, indicating their prognostic value in the clinical setting. In addition, knockdown of LM3 exosomal circPTGR1 significantly reduced tumor progression in non- and low-metastatic cell lines both in vivo and in vitro, suggesting the involvement of the miR449a/MET pathway in this effect.
Implications of all the available evidence
HCC cells with higher metastatic potential may communicate with less metastatic and non-metastatic cells via exosomal cargo such as circPTGR1, thereby affecting cell fate. Exosomes from highly metastatic cells with a high abundance of circPTGR1 may influence cells with lower malignancy by downregulating miR449a-MET interactions in the recipient cells, leading to the disruption of tumor microenvironment homeostasis and the promotion of HCC progression. Because circPTGR1 is highly abundant and is aberrantly expressed in malignant cells and in cells from patients with metastases, it could function as a prognostic biomarker and therapeutic target for HCC.
Alt-text: Unlabelled Box
1. Introduction
Hepatocellular carcinoma (HCC) is the fifth most commonly diagnosed cancer in China and the third leading cause of cancer-related death worldwide [1,2], responsible for >600,000 deaths annually [3]. Its high incidence (16 cases per 100,000 inhabitants) and poor prognosis have led to it being an increasing financial burden [4]. There is therefore an urgent need for a better understanding of the pathology of HCC and for candidate biomarkers to allow its early detection and prognosis and for developing therapeutic strategies.
Exosomes are small extracellular vesicles (30–150 nm in diameter) with an intact lipid bilayer; they are endocytic in origin are able to encapsulate cargo such as lipids, RNA, DNA, and proteins from the parent cell. There is evidence that exosomes exist in almost all mammalian cells, including tumor cells [5]. RNA and proteomics analyses have clarified the role played by tumor-derived exosomes in tumor development and progression [6]. Kogure et al., identified exosomal microRNAs (miRNAs) in HCC that were differentially expressed between a donor H3B cell and the recipient HepG2 cell and found that HCC cell-derived exosomes downregulated hepatocarcinogenesis-related transforming growth factor β activated kinase-1 (TAK1), as well as its associated signaling pathway, resulting in the promotion of HepG2 cell growth [7]. A study that characterized the RNA and proteome contents of exosomes revealed that exosomes derived from metastatic HCC cell lines could enhance hepatocyte motility [8].
Circular RNAs (circRNAs) are a class of endogenous noncoding RNAs with cell type-specific expression, which function as miRNA sponges to regulate gene expression. It is thought they are generated by the backsplicing of exons and/or introns during the process of precursor mRNA splicing. However, unlike the linear transcripts from which they are generated, circRNAs lack the 5′ cap and 3′ polyadenylated tail, making them more resistant than the linear mRNAs to RNAase [9]. A recent study suggested that circRNAs are abundant and stable in exosomes derived from liver cancer cells [10], and it has been suggested that their expression is modulated by endonucleic activity and exosomes [11]. Together, these findings suggest a possible therapeutic role of exosomal circRNAs in controlling cancer progression, with a potential application as promising biomarkers in the diagnosis of HCC. However, most previous studies of HCC exosomes have focused on miRNAs and proteins, and research into the functional roles of exosomal circRNAs derived from HCC cells is limited.
The aim of this study was to investigate the RNA profile of HCC-derived exosomes and the potential role of an HCC exosomal circRNA in tumor cell migration and invasion, and to clarify the underlying mechanisms.
2. Materials and methods
2.1. Cell lines and culture
The human hepatocellular carcinoma cell lines HepG2, L-O2, SMCC-7721, HEP3B and HUH7 were purchased from the Cell Bank of the Chinese Academy of Sciences (Shanghai, China). MHCC97-L (97 L), MHCC 97H (97H) and HCC-LM3 (LM3) cells were kindly provided by Prof. Zhaoyou Tang (Liver Cancer Institute, Zhongshan Hospital, Fudan University, Shanghai, China). The cells were grown in high-glucose Dulbecco's Modified Eagle Medium (DMEM; Invitrogen, CA, USA) supplemented with 10% fetal bovine serum (FBS; Invitrogen) and 1% penicillin/streptomycin. All the cultures were incubated at 37 °C with 5% CO2.
To construct both the circPTGR1 and the corresponding parent linear PTGR1 stable knockdown cell lines, short hairpin RNAs (shRNAs) containing the si-circPTGR1 (GAAGAAAGCGTCTCCTGAT), si-PTGR1(GGACCCTGAAGAAGCACTT) sequence were cloned into lentiviral vectors (Forevergen, Guangzhou, China). A vector containing the si-NC (CTTTCTCCGAACGTG TCAC) sequence was used as a negative control. All constructed lentivirus vectors were transfected with the packaging plasmids pGag/Pol, pRev, and pVSV-G into 293 T cells with Lipofectamine 2000. Viruses in cell supernatant were collected at 48 h and 72 h post transfection and transduced to LM3 cells, in order to generate the shcircPTGR1, shPTGR1, and control NC cell lines.
2.2. Patients and specimens
A total of 82 HCC patients and 47 healthy people at the Third Affiliated Hospital of Sun Yat-sen University (Guangzhou, China), were enrolled in this study between November 2015 and April 2017. All the patients had been diagnosed with primary HCC, and none had received any preoperative treatment. The patients underwent surgical resection, and serum samples were collected on the day of surgery. Their clinicopathological characteristics are presented in Supplementary Table 1. The protocol for collecting clinical samples was approved by the Ethics Committee of the Third Affiliated Hospital of Sun Yat-sen University (Guangzhou, China), and the patients provided informed consent before samples were collected.
2.3. Isolation and identification of cellular exosomes
Cells cultured on 15 cm plates with DMEM containing 10% FBS were replenished with serum-free medium when they reached 80% confluence and were maintained in culture in a 37 °C incubator with 5% CO2 until the cell medium was collected for exosome isolation after 48 h. The cell medium was centrifuged at 600 ×g for 5 min, followed by at 12,000 ×g for 25 min to remove any cell debris and possible apoptotic bodies. The supernatants were then incubated overnight with ExoQuick-TC exosome precipitation solution (System Biosciences, CA, USA) at 4 °C and were then centrifuged at 1500 ×g for 30 min to harvest the exosome pellet. The exosomes were resuspended in 100 μl 1× phosphate-buffered saline (PBS) and verified with electron microscopy JEM-1400 (JEOL, Tokyo, Japan). A NanoSight LM10 instrument (Nanosight, Malvern, UK) was used to analyze the size and number of exosomes, following the manufacturers' instructions.
2.4. Blood preparation and exosome isolation
After centrifugation of the whole blood at 1600 ×g for 10 min at 4 °C, the aspirated serum was stored at −80 °C until use. Exosomes were isolated from the serum sample using ExoQuick Exosome Precipitation Solution (System Biosciences). Briefly, the serum sample was centrifuged at 3000 ×g for 15 min to remove cell debris. Next, 63 μl of ExoQuick Exosome Precipitation Solution was added to 250 μl of the serum sample and mixed well. Then, 125 μL ExoQuick Exosome Precipitation Solution was added to 500 μL serum and mixed well. After incubating at 4 °C for 30 min, the mixture was centrifuged at 1500 ×g for 30 min. The supernatant was then removed, and the tubes were centrifuged for another 5 min. Finally, the exosomes were resuspended with PBS.
2.5. Exosome fluorescence assay
An exosome fluorescence assay was used to validate the internalization of labeled LM3-Exosome (0 ng, 10 ng, and 25 ng) by HepG2 cells. Firstly, we resuspended LM3-Exosome in 500 μl PBS in a 1.5 ml Eppendorf tube, and added 50 μl of 10× labeling dye Exo-Green to the LM3-Exosome preparation. The mixture was then incubated at 37 °C for 10 min without shaking. 100ul ExoQuick-TC was added to the solution and incubated at 4 °C for 30 min. The Eppendorf tube was spun at 14,000 rpm for 3 min and the supernatant was carefully aspirated from the corner of the tube and the LM3-Exosome was resuspended in 500 μl PBS for downstream applications. Cells were seeded at a density of 5 × 10 [3] cells/well in 24-well plates and co-cultured with different concentrations of labeled LM3-Exosome for 2–24 h. Finally, the cells were observed under a fluorescence microscope (Excitation: 494 nm; Emission: 521 nm (green), Filter setting: Typical GFP filter set).
2.6. Cell proliferation, migration, invasion assays, and flow cytometry
Cells were incubated with exosomes (10 μg/ml) or PBS for the indicated time periods. For the valuation of cell proliferation, the [3-(4,5-dimethylthiazol-2-yl)-5-(3-carboxymethoxyphenyl)-2-(4-sulfophenyl)-2H-tetrazolium] MTS assays were conducted according to a previous report [12], but briefly, the cells (2 × 104 per well) were seeded into 96-well plates and cultured for 3 days. MTS reagents were added to assess cell proliferation at days 1, 2, and 3. For the cell invasion and migration analysis, 1 × 106 cells were seeded in the upper chambers that were pre-coated with or without Matrigel (Matrigel BD biosciences, NY, USA). Cells in the upper chambers were maintained in serum-free DMEM, whereas those in the lower chambers were maintained in DMEM with 10% FBS. At 48 h, all cells that had transferred to the lower chambers were stained with 0.5% crystal violet. Positive staining cells from six to eight fields of chambers in each group were counted under a microscope (Olympus, Tokyo, Japan).
Cells were seeded into 6-well plates at a density of 1 × 106 per well and cultured for 12 h prior to exosome incubation. Cells were incubated with exosomes (10 μg/ml) for 48 h, collected by trypsin treatment, and resuspended in cold PBS. For measurement of the cell cycle, cells were fixed overnight in cold 70% ethanol and stained with propidium iodide (PI) in staining buffer (50 μg/ml PtdIns (Sigma, CA, USA) and 20 μg/ml RNase in PBS for 2 h at 4 °C. In order to detect apoptosis, cells were stained with Annexin V-APC and 7-AAD (BioLegend, CA, USA) according to the manufacturer's instructions. The nuclear DNA content at each phase of the cell cycle and the apoptotic cell proportion were assessed with flow cytometry (Becton-Dickinson, CA, USA).
2.7. Western blot analysis
Exosome pellets were lysed in lysis buffer (8 M Urea/2.5% SDS containing 5 μg.ml−1 leupeptin, 1 μg.ml−1 pepstatin, and 1 mM phenylmethylsulphonyl fluoride) as previously described [13]. The Bradford method was used for protein quantification and Western blot was conducted as previously reported [12]. The following primary antibodies were used: Alix, HSPA8, MET, and GAPDH (Cell Signaling Technology, Inc., MA, USA), Tsg101, CD63 (Abcam, CA, UK), and PTGR1 (Sigma-Aldrich, St. Louis). Blots were visualized with enhanced chemiluminescent agents (Forevergen biosciences, GZ, China).
2.8. RNA isolation and qRT-PCR detection
Trizol reagent (Life Technologies, CA, USA) was used to isolate the total RNA from cells, following the standard protocol. Exosomal RNA was extracted using a SeraMir™ Exosome RNA Amplification Kit (System Biosciences). RNA quantification was performed with Qubit 3.0 (Thermo Fisher, MA, USA). To compare expression levels between groups, quantitative real-time PCR (qRT-PCR) was performed using an RNA-direct SYBR Realtime PCR Master Mix (TOYOBO, Osaka, Japan) and an ABI 7500 real-time PCR system (Applied Biosystems, CA, USA). The qRT-PCR results were analyzed with the 2-ΔΔCt method. To validate the backspliced circRNAs predicted by RNA-seq, RT-PCR was carried out using circRNA-specific divergent primers. The primers used are listed in Supplementary Table 2.
2.9. RNA library preparation and sequencing
RNA integrity and size distributions were assessed using Agilent 2100 Bioanalyzer pico-RNA chips (Agilent, CA, USA). RNA libraries were prepared according to the instructions for the VAHTS Total RNA-seq (H/M/R) Library Prep Kit from Illumina® (VAZYME, Nanjing, China). RNA-seq was performed as previously described [12], and the libraries were created and sequenced using the Illumina HiSeq 2500 platform. The RNA-seq data were uploaded to NCBI database (https://trace.ncbi.nlm.nih.gov/Traces/sra/?study=SRP165940). The accession number was NCBI: SRP165940.
2.10. Bioinformatics analysis
The reads from the Illumina sequencer were subjected to mRNA, lncRNA, and circRNA analyses and the data were aligned with the human reference genome GRCh37/hg19 using TOPHAT v2.1.0. Counts of mapped reads for each gene were normalized by the number of reads per kilobase per million mapped reads, which allowed the comparison of expression levels between genes. A differential expression analysis of two samples was performed using a previous statistical model [14]. The P-values were adjusted using Benjamini and Hochberg's approach for controlling the false discovery rate and an adjusted P-value of <0.05 was considered to indicate differential expression. To construct the hierarchical clustering analysis, the heatmap package in R version 1.0.8 (https://cran.r-project.org/web/packages/pheatmap/) was used to cluster the differentially expressed circRNAs that overlapped between the 97 L and LM3 cells.
The biological processes involving these genes were obtained from the Kyoto Encyclopedia of Genes and Genomes (KEGG) pathways database (http://www.genome.jp/kegg/) (adjusted P-value <.05; gene count ≥2). The miRanda (http://www.microrna.org/) and TargetScan (http://www.targetscan.org/) software packages were used to predict the circPRTG1 miRNA targets and the potential mRNA targets of the miRNAs. The circRNAs–miRNAs–mRNA interaction network was constructed by merging the common targets of the circRNAs and mRNAs, as previously described [15]. Finally, the network obtained was visualized with Cytoscape software (V 2.8.3, http://www.cytoscape.org/).
2.11. In vivo metastasis assay
An experimental metastasis model was developed using athymic nude mice and the LM3 HCC cell line. The mice were anesthetized with pentobarbital and a small transverse incision was made below the sternum to expose the liver. After carefully exposing the liver, 2 × 106 viable cells were preincubated with PBS, shcircPTGR1, or negative control exosomes and were slowly injected into the upper left lobe of the liver with a 28-gage needle. Four weeks after the injection, the mice were sacrificed under anesthesia and the tumor metastasis was examined with a stereo microscope.
2.12. Statistical analysis
The statistical analyses were performed with SPSS version 16.0 (SPSS, Inc.) and GraphPad Prism version 6.0 (GraphPad Software). Data from three or more independent experiments are presented as means ± standard error of the mean. Data that were not distributed normally were transformed into a normal distribution before analysis. The relationship between the circRNAs and the clinicopathological parameters of the HCC patients was analyzed using one-way ANOVA. Differences between two groups were evaluated using Student's t-test. P < .05 was considered to indicate a statistically significant difference. The expression in liver tissues of PTGR1 mRNA and the patient survival data were obtained from The Cancer Genome Atlas Liver Hepatocellular Carcinoma (TCGA LIHC) dataset (https://tcga-data.nci.nih.gov/) and the OncoLnc Anaya dataset [16]. Kaplan–Meier survival curves were created with GraphPad Prism and compared using the log-rank test.
3. Results
3.1. LM3-derived exosomes promoted migration and invasion in HepG2 and 97 L cells
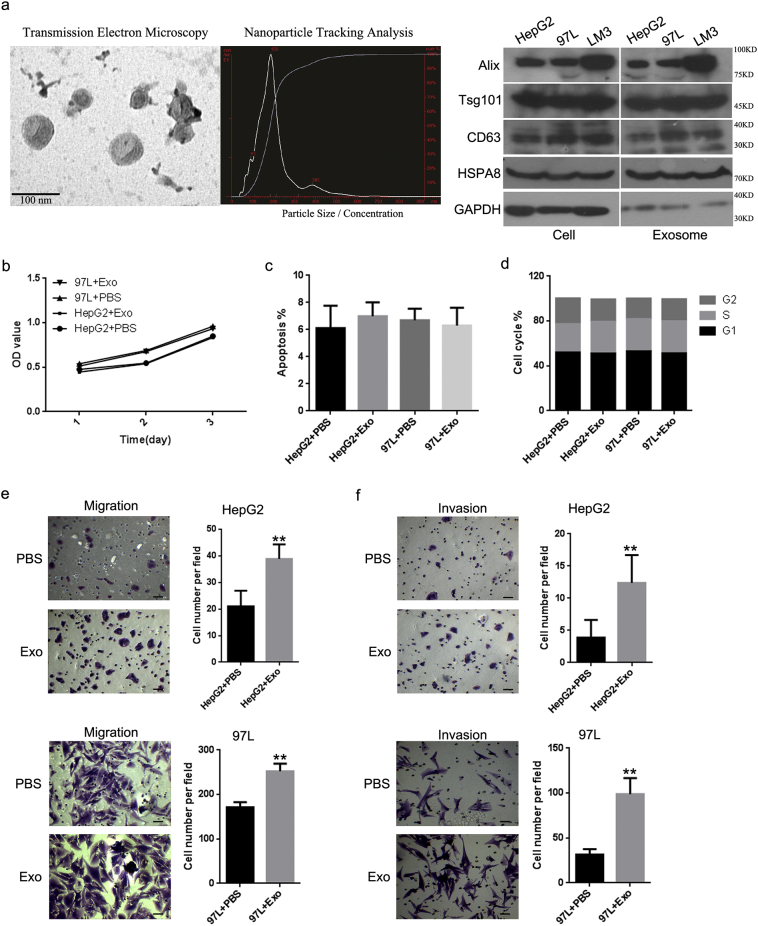
Exosomes isolated from cells with metastatic potential were verified and used to evaluate whether such cells could affect the biological functions of cells with little or no metastatic potential. The identity of the exosomes was confirmed by electron microscopy, which revealed that exosomes with cup-shape morphology were 50–100 nm in diameter (Fig. 1a). Nanoparticle Tracking Analysis showed a size range of 50–250 nm. There were no obvious differences in the shape or size of exosomes secreted by the HepG2, 97 L, and LM3 cells. In addition, western blot analysis confirmed the presence of the exosome-associated markers Alix, Tsg101, and CD63 in isolated exosomes (Fig. 1a). We labeled LM3-derived exosomes with ExoGlow-Protein and followed their uptake by HepG2 cells. The punctate fluorescence signal confirmed the internalization of the labeled LM3 exosomes by HepG2 cells (Supplementary Fig. 1a).
Fig. 1.
LM3-derived exosomes promoted the cell migratory and invasive ability of HepG2 and 97 L cells. (a) Exosome morphology viewed by transmission electron microscopy. Scale bar, 100 nm. NanoSight tracking analysis was used to measure the diameters of exosomes isolated from LM3 cells. The exosomal markers Alix, Tsg101, CD63, and HSPA8 were detected by western blot. An MTS assay, cytometry, and transwell assays were used to analyze the effects of LM3-derived exosomes, compared with treatment with phosphate-buffered saline (PBS), on HepG2 and 97 L cells for the following characteristics: (b) cell proliferation; (c) apoptosis; (d) cycle distribution; (e) cell migration (HepG2, P < .0001; 97 L, P < .0001); and (f) invasion (HepG2, P = .0004; 97 L, P < .0001). Scale bars: 100 μm. Error bars indicate standard deviation. **P < .01 vs. PBS.
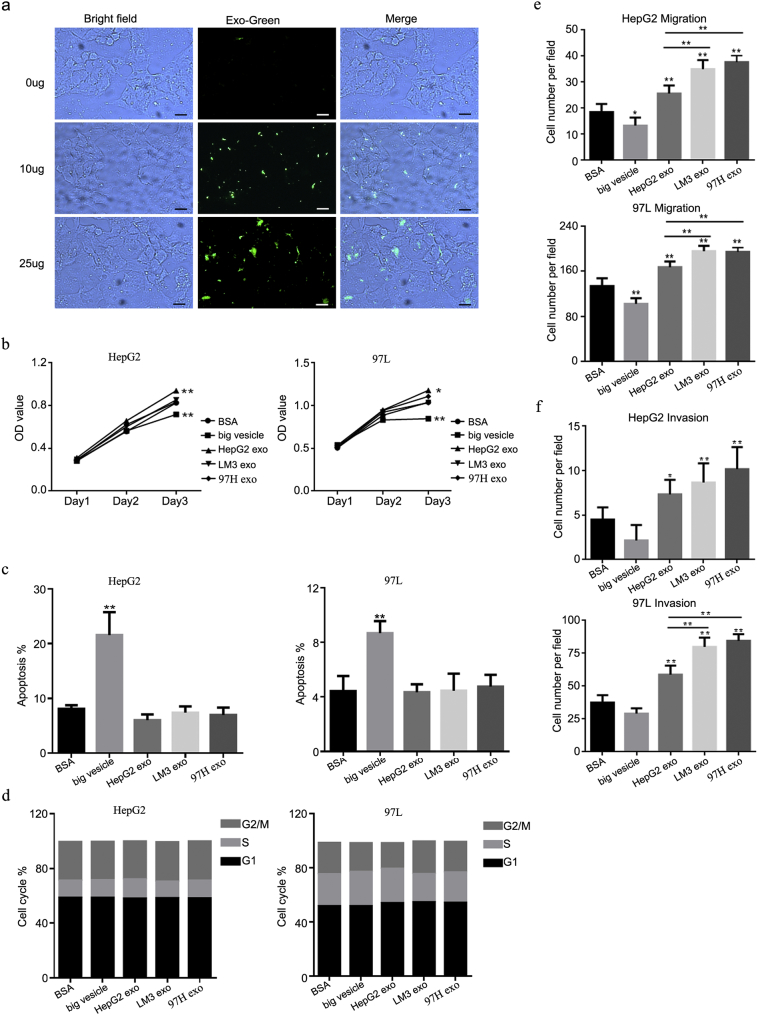
Supplementary Fig. 1.
(a) ExoGlow-Protein provided a clear visualization of the internalization by HepG2 cells of labeled LM3 exosomes. Scale bars: 100 μm. (b) MTS assay (HepG2: BSA vs. bigger vesicle, P = .0074; BSA vs. HepG2 exosomes (HepG2 exo), P = .0034; 97 L: BSA vs. bigger VC, P = .0042; BSA vs. HepG2 exo, P = .0107). (c) Flow cytometry (P < .0001 vs. BSA). (d) Flow cytometry (P = .001 vs. BSA). (e) Migration assay (HepG2: BSA vs. HepG2 exo, P = .0037; BSA vs. LM3 exo, P < .0001; BSA vs. 97H exo, P < .0001; HepG2 exo vs. LM3 exo, P = .0002; HepG2 exo vs. 97H exo, P < .0001; 97 L: BSA vs. big vesicle, P = .0002; BSA vs. HepG2 exo, P < .0001; BSA vs. LM3 exo, P < .0001; BSA vs. 97H exo, P < .0001; HepG2 exo vs. LM3 exo, P = .0006; HepG2 exo vs. 97H exo, P = .0011). (f) Invasion assay (HepG2: BSA vs. LM3 exo, P = .0073; BSA vs. 97H exo, P = .0002; 97 L: BSA vs. HepG2 exo, P < .0001; BSA vs. LM3 exo, P < .0001; BSA vs. 97H exo, P < .0001; HepG2 exo vs. LM3 exo, P < .0001; HepG2 exo vs. 97H exo, P < .0001). Transwell assays were used to evaluate the effects of LM3-derived exosomes on cell proliferation, apoptosis, cycle distribution, cell migration, and invasion in HepG2 and 97 L cells treated with BSA, big vesicles, HepG2 exosomes, LM3 exosomes, and 97H exosomes. Error bars indicated standard deviations. * P < .05 vs. BSA; ** P < .01 vs. BSA.
Next, HepG2 and 97 L cells were incubated with LM3 exosomes and subjected to MTS, flow cytometry, and transwell assays to determine cell proliferation, apoptosis, the cell cycle, migration, and invasion. Cells treated with phosphate-buffered saline (PBS) were used as a control. The MTS assay showed no significant differences between the LM3 exosomes and the PBS treatment in either the HepG2 or the 97 L cell lines (Fig. 1b), and there were no statistically significant differences in the apoptosis levels or cycle distribution between the exosomes and PBS-treated HepG2 and 97 L cells (Fig. 1c, d). However, there were significant differences in cell numbers based on the transwell migration and invasion assays. The number of migrated HepG2 cells was more than two-fold higher in the group incubated with LM3 exosomes compared to the PBS-treated control (P < .0001) (Fig. 1e). A similar increase was also clearly detected in the 97 L cells incubated with LM3 exosomes (P < .0001). Similar results were obtained in the invasion assay (P < .0001), in which, compared with the controls, more than double the number of cells in both of the cell lines treated with LM3 exosomes migrated into the lower compartment (HepG2, P = .0004; 97 L, P < .0001; Fig. 1f). In addition, bovine serum albumin, big vesicles, and exosomes derived from HepG2 were used as further controls to rule out contamination from the culture conditions or the exosome isolation process (Supplementary Fig. 1b–f). Exosomes were also isolated from another highly metastatic cell line, 97H, with similar results; there were no significant differences in cell viability, apoptosis level, or cycle distribution between the LM3 or 97H exosomes incubated with the HepG2 or 97 L cell lines. However, there were significant differences in cell numbers based on the transwell migration and invasion assays (Supplementary Fig. 1b–f).
3.2. RNA deep sequencing revealed different RNA profiles for exosomes from HepG2, 97 L, and LM3 cells
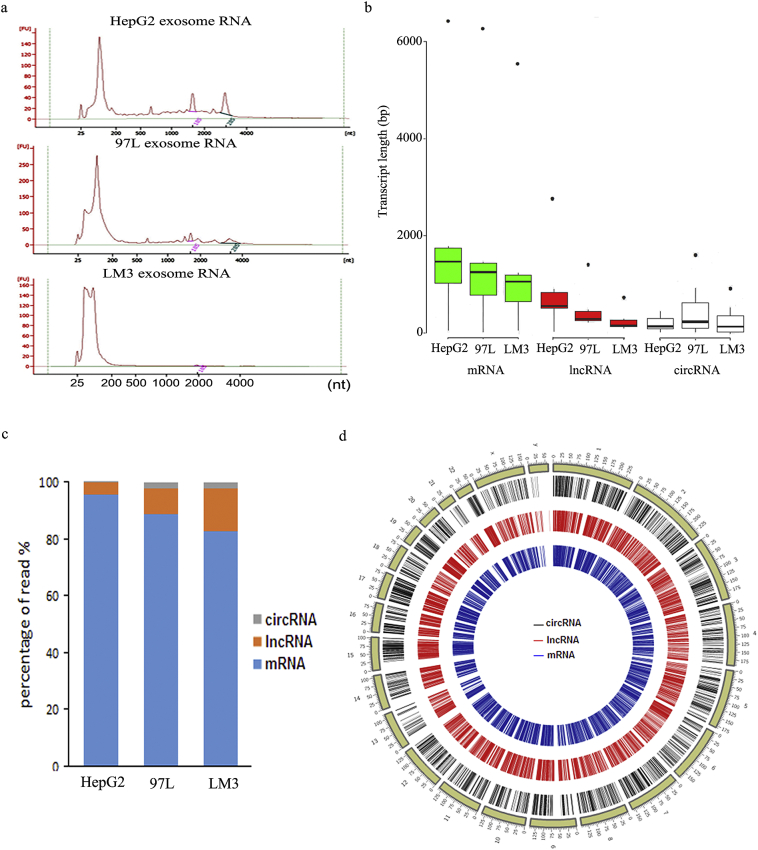
An Agilent Bioanalyzer was used to analyze the exosomal total RNA profiles of HepG2, 97 L, and LM3 cells. This demonstrated that exosomes derived from LM3 and 97 L cells had relatively lower levels of 28S and 18S ribosomal RNA (rRNA) compared with those from HepG2 cells. Compared with HepG2 exosomal RNA, 97 L and LM3 exosomal RNA included a greater proportion of short-length RNAs (Fig. 2a).
Fig. 2.
RNA profiles of HepG2, 97 L, and LM3-derived exosomes revealed by RNA sequencing. (a) Bioanalyzer analyses of RNA isolated from HepG2, 97 L, and LM3-derived exosomes. (b) Transcript length distribution of mRNAs, lncRNAs, and circRNAs in HepG2, 97 L, and LM3-derived exosomes. The dots indicate discrete values. (c) Abundance of mRNAs, lncRNAs, and circRNAs in HepG2, 97 L, and LM3-derived exosomes. (d) Circos plots representing the distribution of all the mRNAs (black), lncRNAs (red), and circRNAs (blue) from HepG2, 97 L, and LM3-derived exosomes on different chromosomes. The outermost track (yellow) shows the different chromosomes. (For interpretation of the references to colour in this figure legend, the reader is referred to the web version of this article).
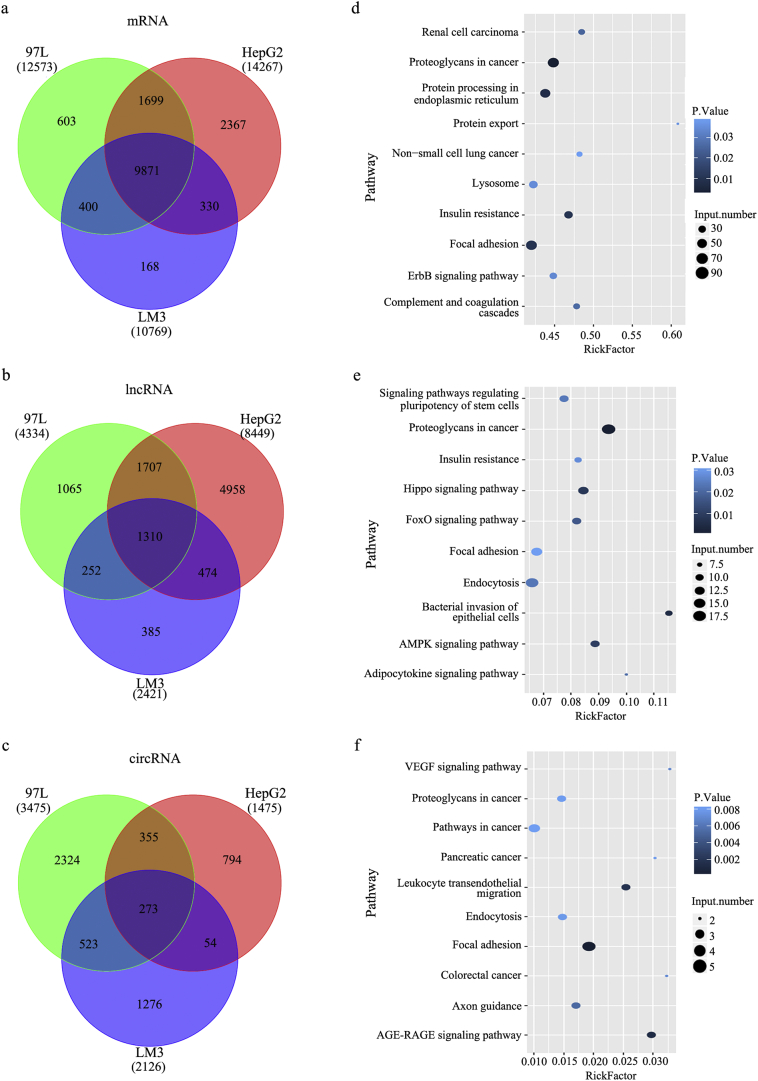
To investigate whether RNA was involved in the observed effects of LM3 cells on HepG2 and 97 L cells, differences in the RNA profiles of HCC cell lines with different metastatic potentials were analyzed with RNA sequencing (RNA-seq). The transcript length of mRNA and long noncoding RNA (lncRNA) tended to decrease gradually in all three cell lines, whereas the transcript length of circRNA was homogeneous in the HepG2, 97 L, and LM3 cells (Fig. 2b). The analysis of the percentage reads of circRNA, lncRNA, and mRNA in the derived exosomes of the three cell lines revealed different trends. A higher abundance of circRNA and lncRNA and a lower abundance of mRNA were observed in the 97 L and LM3 cells compared with the HepG2 cells (Fig. 2c). Although the three kinds of RNA were homogeneously distributed on chromosomes (Fig. 2d), many genes were differentially expressed between the three cell lines (Fig. 3a, b, and c). The differentially expressed RNAs were subjected to KEGG pathway enrichment analysis to capture the relevant biological processes. This showed enrichment in cancer-associated pathways, with the mRNAs enriched mainly in cancer proteoglycan and focal adhesion pathways (Fig. 3d) and the lncRNAs in cancer proteoglycan, endocytosis, and focal adhesion pathways (Fig. 3e). In particular, the circRNAs were primarily enriched in cell motility-related leukocyte transendothelial migration and focal adhesion pathways (Fig. 3f), indicating their role in moderated cell migration and invasion. Their role in HCC cell metastasis was investigated further.
Fig. 3.
Comparison of the exosomal RNA profiles of HepG2, 97 L, and LM3 cells and the top 10 enriched KEGG pathways for differentially expressed RNAs. (a–c) Venn diagrams of overlapping differentially expressed exosomal RNAs from HepG2, 97 L, and LM3 cells: (a) mRNAs; (b) lncRNAs; and (c) circRNAs. (d–f) Diagrams of the top 10 enriched KEGG pathways for differentially expressed exosomal RNAs: (d) mRNAs; (e) lncRNAs; and (f) circRNAs.
3.3. Profiling revealed differences in circRNA between the HCC cells
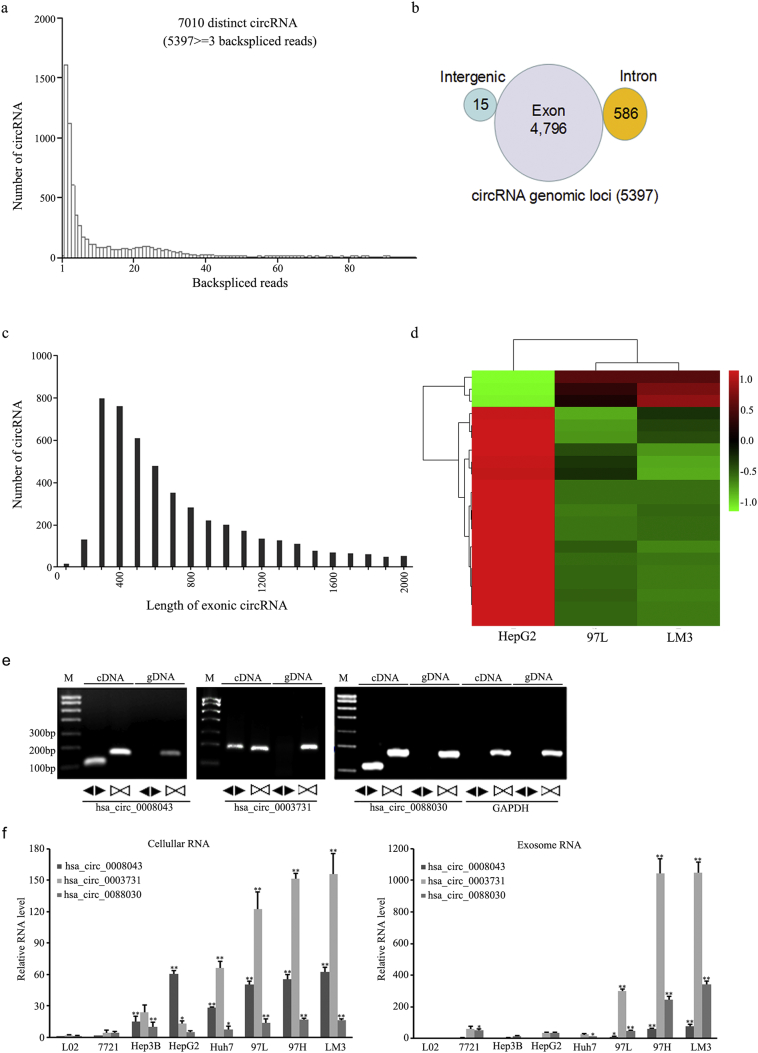
RNA-seq was used to characterize circRNA transcripts, which were annotated by using the RefSeq database. A total of 5397 distinct circRNA candidates containing at least three unique backspliced reads were predicted for the HepG2, 97 L, and LM3 cells (Fig. 4a). Of these, 4796 candidates were mapped to exon region of known transcripts, and 586 were mapped to intron regions. Only 15 circRNAs from gene intergenic regions were identified (Fig. 4b). The length distribution showed that most exonic circRNAs were <700 nt in length, with a median length of ~500 nt (Fig. 4c). The cluster heatmap for circRNAs differentially expressed in 97 L and LM3 cells revealed distinct expression patterns in these cells with different metastatic potential. Compared with HepG2 cells, several circRNAs were downregulated in 97 L and LM3 cells, and a smaller number were upregulated. The expression pattern of circRNAs in 97 L cells was very similar to that in LM3 cells but quite different from that of HepG2 cells (Fig. 4d).
Fig. 4.
Identification of circRNAs in RNA sequencing data and the determination of differentially expressed circRNAs in HepG2, 97 L, and LM3 cell-derived exosomes. (a) The length distributions of backspliced circRNAs in HepG2, 97 L, and LM3 cells. (b) The genomic origins of HepG2, 97 L, and LM3 cell-derived circRNAs. (c) The length distributions of exonic circRNAs in HepG2, 97 L, and LM3 cells. (d) Heat map showing the expression profiles of exosomal circRNA in HepG2, 97 L, and LM3 cells. (e) The identification of three isoforms of circPTGR1 by RT-PCR, using divergent primers. (f) Fold enrichment of the three isoforms of circPTGR1 detected by qRT-PCR compared to the linear PTGR1 mRNAs in exosomes and their parent cells in eight hepatocellular carcinoma cell lines. Error bars indicate standard deviations. * P < .05 vs. L-O2; ** P < .01 vs. L-O2. (For cellular RNA, Hsa_circ_0008043: L-O2 vs. Hep3B, P = .0004; L-O2 vs. HepG2, huh7, 97 L, 97H, and LM3, P < .0001; Hsa_circ_0003731: L-O2 vs. huh, 97 L, 97H, hand LM3, P < .0001; Hsa_circ_0088030, L-O2 vs. Hep3B, P = .0038; L-O2 vs. huh7, P = .0397; L-O2 vs. 97 L, 97H, and LM3, P < .0001. For exosomal RNA, Hsa_circ_0008043: L-O2 vs. 97 L, P = .0435; L-O2 vs. 97H and LM3, P < .0001; Hsa_circ_0003731: L-O2 vs. huh, 97 L, 97H, and LM3, P < .0001; Hsa_circ_0088030, L-O2 vs. Hep3B, P = .0169; L-O2 vs. huh7, P = .0003; L-O2 vs. 97 L, P = .0007; L-O2 vs. 97H, LM3, P < .0001).
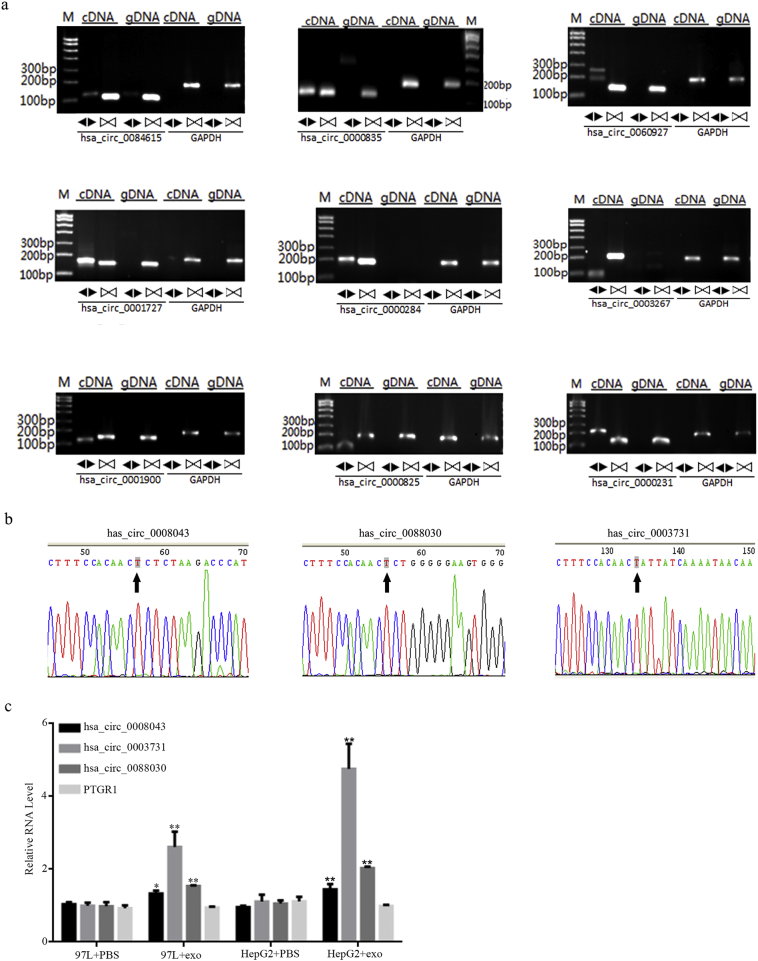
To validate the candidate circRNAs for backsplicing, RT-PCR was performed using divergent primers for the top 12 most differentially expressed circRNAs (Supplementary Table 2). As expected, PCR products of the expected size were amplified from complementary DNA by the divergent primers. All 12 candidates exhibited absent bands in the genomic DNA based on gel electrophoresis after staining with ethidium bromide (three are shown in Fig. 4e and nine in Supplementary Fig. 2a).
Supplementary Fig. 2.
(a) Nine of the top 12 differentially expressed circRNAs identified by RT-PCR. Identification of the remaining three circRNAs (hsa_circ_0008043, hsa_circ_0003731, and hsa_circ_0088030) is shown in Fig. 4 in the main text. (b) Sanger sequencing confirmation of the divergent amplicons of hsa_circ_0008043, hsa_circ_0003731, and hsa_circ_0088030. (c) A qRT-PCR assay analyzing the effects of LM3 exosomes on the expression of circPTGR1 and PTGR1 in 97 L and HepG2 P < .05 vs. PBS; ** P < .01 vs. PBS.
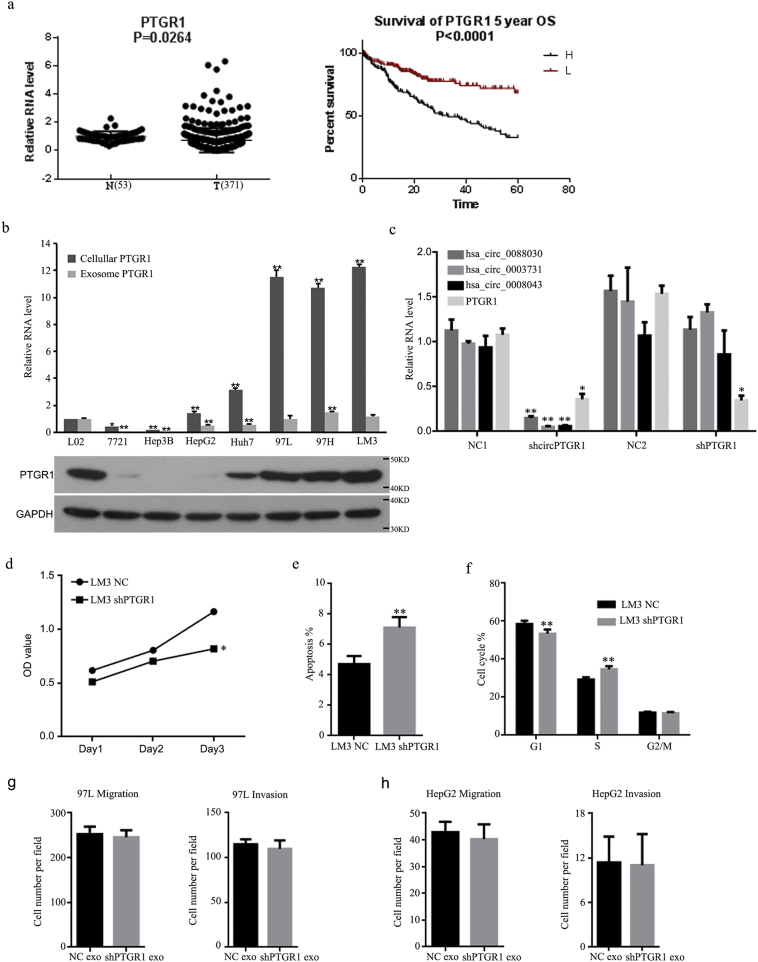
3.4. The expression of exosomal circPTGR1 was associated with poor outcomes in HCC patients
Of the 12 candidate circRNAs, hsa_circ_0008043, hsa_circ_0003731, and hsa_circ_0088030 were all transcribed from the same gene (prostaglandin reductase 1, PTGR1) and were therefore collectively named circPTGR1. Sanger sequencing confirmed the existence of circPTGR1 (Supplementary Fig. 2b). Interestingly, the RNA-seq showed the three isoforms were exclusively expressed in 97 L and LM3 cells, but not in HepG2 cells (Supplementary Table 3). The three circRNAs and their parent linear PTGR1 were selected for further qPCR analysis in both HCC cells and serum-derived exosomes. The relative expressions of hsa_circ_0008043, hsa_circ_0003731, and hsa_circ_0088030 were significantly upregulated in both the exosomes and their parent cells, with cell invasion enhanced in eight cell lines, particularly the 97H and LM3 cell lines (Fig. 4f).
To confirm that circPTGR1 from LM3 exosomes could be transferred to recipient cell lines, 97 L and HepG2 cells were incubated with PBS and with LM3 exosomes for 24 h. Compared with the PBS, incubation with LM3 exosomes resulted in upregulated expression of circPTGR1 but no significant change in PTGR1 mRNA level (Supplementary Fig. 2c).
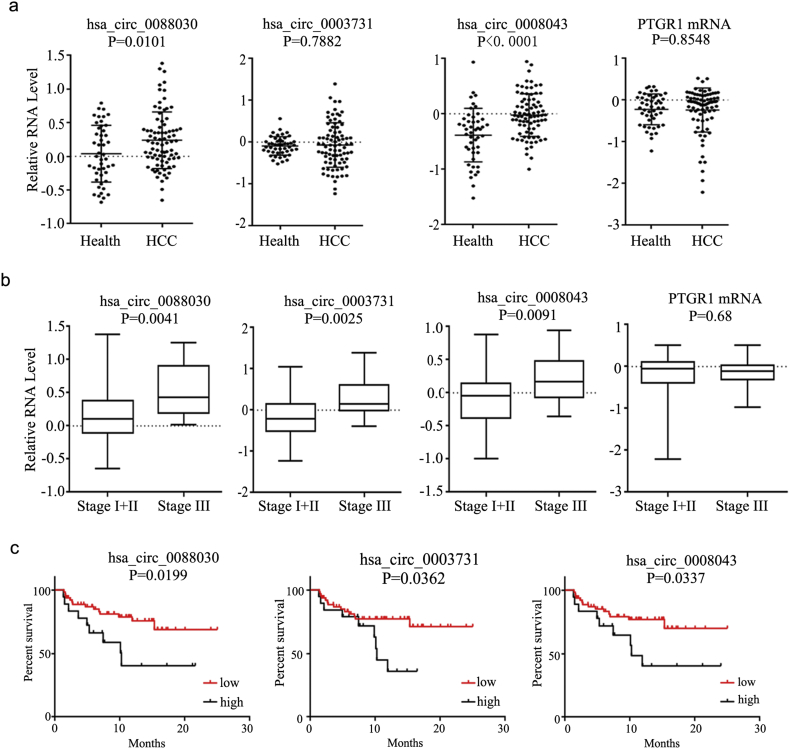
Data from The Cancer Genome Atlas (TCGA) showed slightly upregulated PTGR1 mRNA in HCC tissue. A survival analysis showed better survival rates in patients with lower PTGR1 levels than in those with high PTGR1 levels (Supplementary Fig. 3a). However, higher linear PTGR1 levels were found in more-invasive HCC cells but not in exosomes (Supplementary Fig. 3b). Compared with exosomes from healthy controls, the serum exosomes of tumor patients showed increased expression of circRNAs, but not of PTGR1 mRNA (Fig. 5a). Interestingly, the expression of circPTGR1 differed significantly between the different clinical stages of HCC. In contrast, no significant difference between HCC stages was found in serum linear PTGR1 exosome levels (Fig. 5b). The HCC patients were divided into two groups according to the expression level of circPTGR1, the low and high expression groups. Survival curves of disease-free survival for the two groups were used to analyze the relationship between the expression of circPTGR1 and the prognosis. The survival curves differed significantly between the two groups (Fig. 5c), showing that the prognosis for HCC patients with low expression of circPTGR1 was better than that for those with high circPTGR1 expression.
Supplementary Fig. 3.
(a) The relative expression of PTGR1 in liver tissue based on data from the TCGA database. (b) PTGR1 mRNA and protein levels in the eight tumor cells lines. *P < .05 or ** P < .01 vs. L-O2 cells. (For cellular PTGR1 mRNA: L-O2 vs. 7721, P = .0320;LO2 vs. Hep3B, P = .0015; L-O2 vs. HepG2, P = .1057; L-O2 vs. huh7, P < .0001; L-O2 vs. 97 L, P < .0001; L-O2 vs. 97H, P < .0001; and L-O2 vs. LM3, P < .0001. For exosome PTGR1 mRNA: L-O2 vs. 7721, P < .0001; L-O2 vs. Hep3B, P = .0001; L-O2 vs. HepG2, P = .0005; L-O2 vs. huh7, P < .0001; L-O2 vs. 97 L, P = .9979; L-O2 vs. 97H, P = .0002; and L-O2 vs. LM3, P = .2167.) (c) The knockdown efficiency of circPTGR1 and its counterpart linear PTGR1 in LM3 exosomes. (Compared with NC1: hsa_circ_0088030, P = .0171; hsa_circ_0003731, P = .0001; hsa_circ_0008043, P = .0180; and linear PTGR1, P = .0131. Compared with NC2: hsa_circ_0088030, P = .1109; hsa_circ_0003731, P = .9373; hsa_circ_0008043, P = .4131; and linear PTGR1, P = .0116). * P < .05 vs. negative control (NC); ** P < .01 vs. NC. (d–f) An MTS assay (d) and cytometry (e, f) were used to analyze the effects of knockdown PTGR1 expression on cell proliferation (P < .0001), apoptosis (P = .0086), and cycle distribution (G1, P = .0046; S, P = .0005). * P < .05 vs. NC; ** P < .01 vs. NC. (g, h) Transwell assays were used to analyze cell migration and invasion in HepG2 and 97 L cells treated with shNC or shPTGR1 exosomes. Error bars indicate standard deviations. ** P < .01 vs. NC.
Fig. 5.
The expression of circPTGR1 in clinical specimens. (a) The relative expression of PTGR1 and circPTGR1 in serum exosomes from HCC patients and healthy controls. (b) The relative expression of PTGR1 and circPTGR1 in serum exosomes from HCC patients at different clinical stages. (c) The relationship between the expression of circPTGR1 and the prognosis of HCC patients.
3.5. LM3 exosome-derived circPTGR1 promoted the progression of HCC in vitro and in vivo
To evaluate the effects of exosomal LM3 circPTGR1 on HepG2 and 97 L cells, circPTGR1 was knocked down in LM3 cells with the shcircPTGR1 lentivirus. Exosomes from the circPTGR1-knockdown cells (shcircPTRG1) and the corresponding negative control (shNC) were isolated. The knockdown efficiency of shcircPTGR1 and shPTGR1 in exosomes was confirmed by qPCR (Supplementary Fig. 3c).
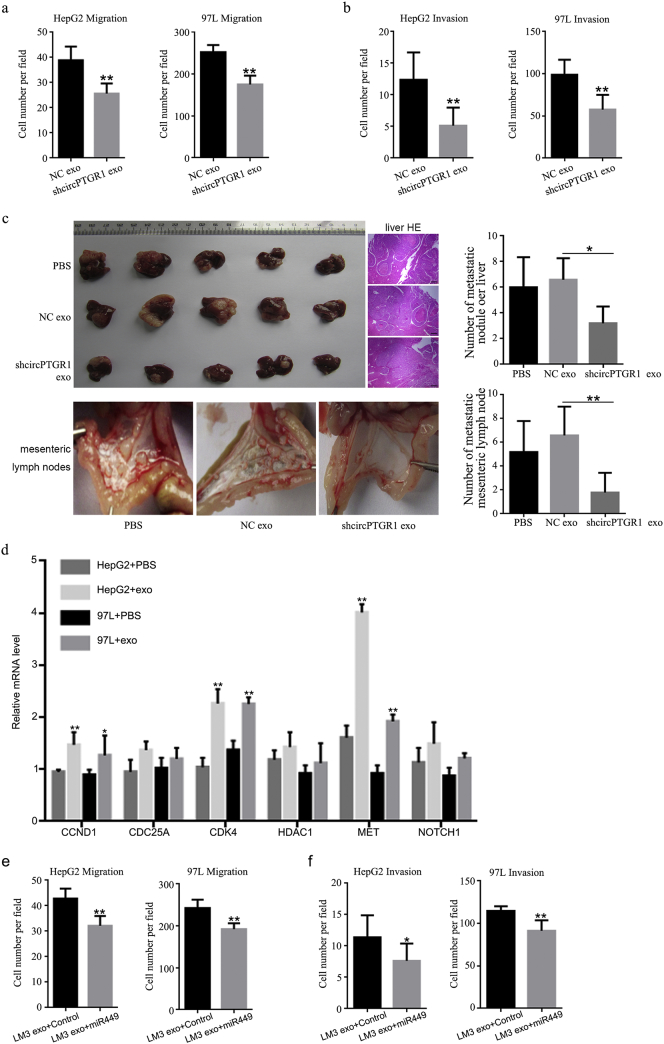
The resulting LM3 exosomes with low circPTGR1 expression were co-cultured with HepG2 and 97 L cells. Cell migration and invasion assays revealed significantly lower numbers of migrated cells in the HepG2 and 97 L cells with shcircPTGR1 exosomes than for the cells incubated with shNC exosomes (migration: HepG2, P < .0001; 97 L, P < .0001; Fig. 6a; invasion: HepG2, P = .0015; 97 L, P = .0024; Fig. 6b). The knockdown of PTGR1 resulted in lower cell viability, higher levels of apoptosis, and cell arrest at the S phase in LM3 cells; this was consistent with the survival analysis based on the TCGA data (Supplementary Fig. 3d–f). In contrast, co-culture with exosomes derived from shPTGR1 cells did not influence the migration and invasion of 97 L and HepG2 cells (Supplementary Fig. 3g and h). Thus, the effects of exosomes with low circPTGR1 expression on tumor metastasis were due to the downregulation of circPTGR1.
Fig. 6.
The knockdown of circPTGR1 suppressed HCC tumor metastasis, and MET was one of the target genes of circPTGR1. A transwell assay was performed to determine the knockdown effects of circPTGR1 in LM3-derived exosomes on HepG2 and 97 L cells for (a) cell migration (HepG2, P < .0001; 97 L, P < .0001), and (b) invasion (HepG2, P = .0015; 97 L, P = .0024). ** P < .01 vs. negative control (NC). (c) The in vivo metastasis model. After the intrahepatic injection of cells treated with shcircPTGR1 or NC exosomes, the mice developed tumors in the liver, and hematoxylin and eosin staining revealed metastatic nodules in the liver tissue. Scale bars: 100 μm. The metastatic nodules were counted manually and the number of metastases per mouse presented as the mean ± SD (liver, P = .0306; mesenteric lymph nodes, P = .0004). * P < .05 vs. NC; ** P < .01 vs. NC. (d) The relative expression levels of candidate downstream mRNA targets of circPTGR1 in the network were detected with qRT-PCR in HepG2 and 97 L cells incubated with LM3-derived exosomes. * P < .05 vs. PBS; ** P < .01 vs. PBS (CCND1: HepG2, P = .0004; 97 L, P = .0105; CDK4: HepG2, P < .0001; 97 L, P = .0006; MET: HepG2, P < .0001; 97 L, P = .0001). (e) Overexpression of miR449a inhibited LM3 exosome-mediated cell migration. (HepG2, P = .0002; 97 L, P = .0001.) ** P < .01 vs. control. (f) Overexpression of miR449a inhibited LM3 exosome-mediated cell invasion. (HepG2, P = .0325; 97 L, P = .0006.) * P < .05 vs. control; ** P < .01 vs. control.
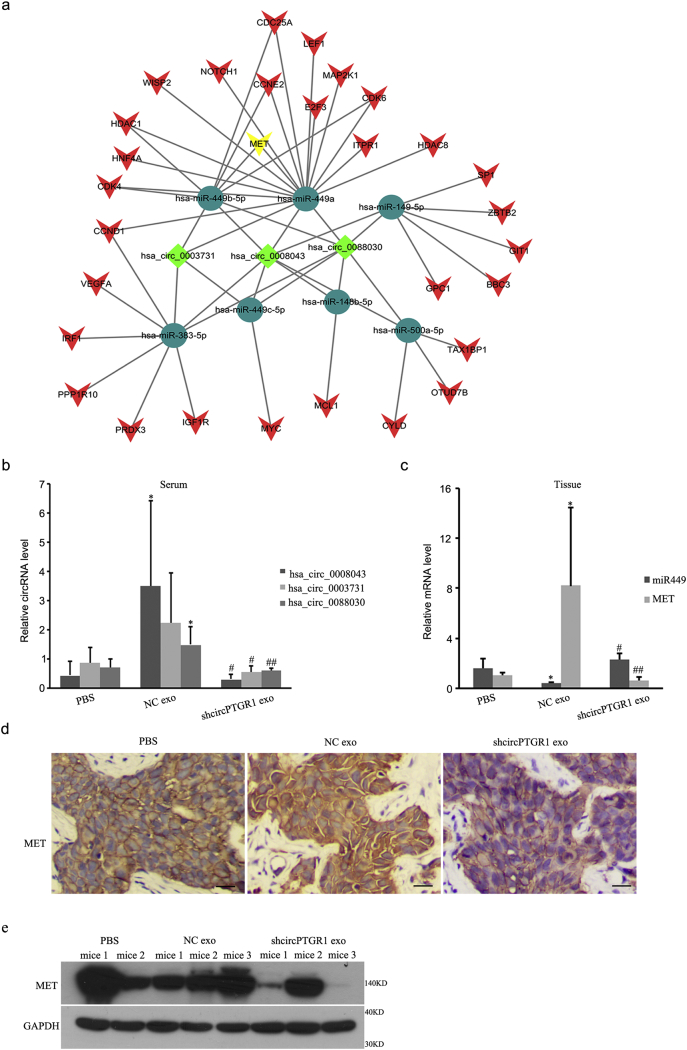
An in vivo metastatic model was created by injecting mice intrahepatically with LM3 cells preincubated with the different exosomes or PBS as control. Four weeks after cell implantation, >90% of the treated animals developed liver tumors, and hematoxylin and eosin (HE) staining revealed that the mice injected with LM3 cells treated with shcircPTGR1 exosomes had fewer metastatic nodules in their liver tissue compare with cells treated with NC exosomes (P = .0306; Fig. 6c, upper panel). In addition, the mice injected with shcircPTGR1 exosome-treated cells had fewer metastatic tumor nodules in the mesentery, whereas more tumors were visible in the mesenteric lymph nodes of the mice treated with NC exosomes and PBS (P = .0004; Fig. 6c, lower panel), indicating that shcircPTGR1 suppressed HCC metastasis. Exosomes isolated from the serum of mice injected with shcircPTGR1 also showed significantly decreased circPTGR1 expression levels compared with the NC exosome group, whereas there was no significant difference between the mice injected with shcircPTGR1 exosome-treated cells and those injected with PBS-treated cells (Supplementary Fig. 4b).
Supplementary Fig. 4.
(a) The predicted circPTGR1–miRNA–mRNA interaction network, generated by Cytoscape. (b) circPTGR1 mRNA expression level in exosomes isolated from serum obtained from the in vivo metastasis model mice (hsa_circ_0088030: NC exo vs. PBS, P = .0102; sh exo vs. NC exo, P = .0043; hsa_circ_0003731: sh exo vs. NC exo, P = .0142; hsa_circ_0008043: NC exo vs. PBS, P = .0193; sh exo vs. NC exo, P = .0240). (c, d) MET expression levels in tissues from the in vivo metastasis model mice. (For miR449: NC exo vs. PBS, P = .0108; sh exo vs. NC exo, P = .0003. For MET: NC exo vs. PBS, P = .0230; sh exo vs. NC exo, P = .0163.) Scale bars: 100 μm. * P < .05 vs. PBS treatment. #P < .05 or ## P < .01 vs. NC group. (e) MET protein level in tissues from the in vivo metastasis model mice.
3.6. CircPTGR1 regulated HCC progression through the miR449a/MET pathway
To explore the underlying mechanism by which circPTGR1 regulated cell migration and invasion in HCC cells, we performed an RNA-seq analysis to identify changes in gene expression in HepG2 and 97 L cells after incubation with LM3-derived exosomes. This detected hundreds of genes that were differentially expressed in the exosome-treated cells compared with the control PBS-treated cells. Interactions between the circPTGR1 and its target miRNAs were theoretically predicted by conserved seed-matching sequencing using Arraystar software for miRNA target prediction based on the TargetScan and miRanda databases. An entire network of circPTGR1–miRNA–mRNA interactions was delineated using Cytoscape (Supplementary Fig. 4a). The three circPTGR1 isoforms were at the center of the network, connected to seven miRNAs, which were further connected to their respective potential complementary binding mRNAs. Among these, miR449a, miR449b, and miR449c were highly homologous [17]. In particular, miR449a had the most candidate mRNA targets and was assumed to be one of the specific targets (Supplementary Fig. 4a). Confirming the involvement of these targets in the effects of HCC progression mediated by LM3 exosomes, six mRNAs that contained miR449a and/or miR449b binding sites were detected by qPCR in HepG2 and 97 L cells treated with either LM3 exosomes or PBS; these were CCND1, CDC25A, CDK4, HDAC1, MET, and NOTCH1. The qPCR results confirmed that CCND1, CDK4, and MET were significantly upregulated compared with the PBS-treated controls (CCND1: HepG2, P = .0004; 97 L, P = .0105; CDK4: HepG2, P < .0001; 97 L, P = .0006; MET: HepG2, P < .0001; 97 L, P = .0001; Fig. 6d), whereas the expression levels of CDC25A, HDAC1 and NOTCH1 were not significantly increased in the cells incubated with LM3 exosomes. Of these mRNAs, the qPCR results showed that MET, a candidate target, was the most closely related to miR449a (Fig. 6d).
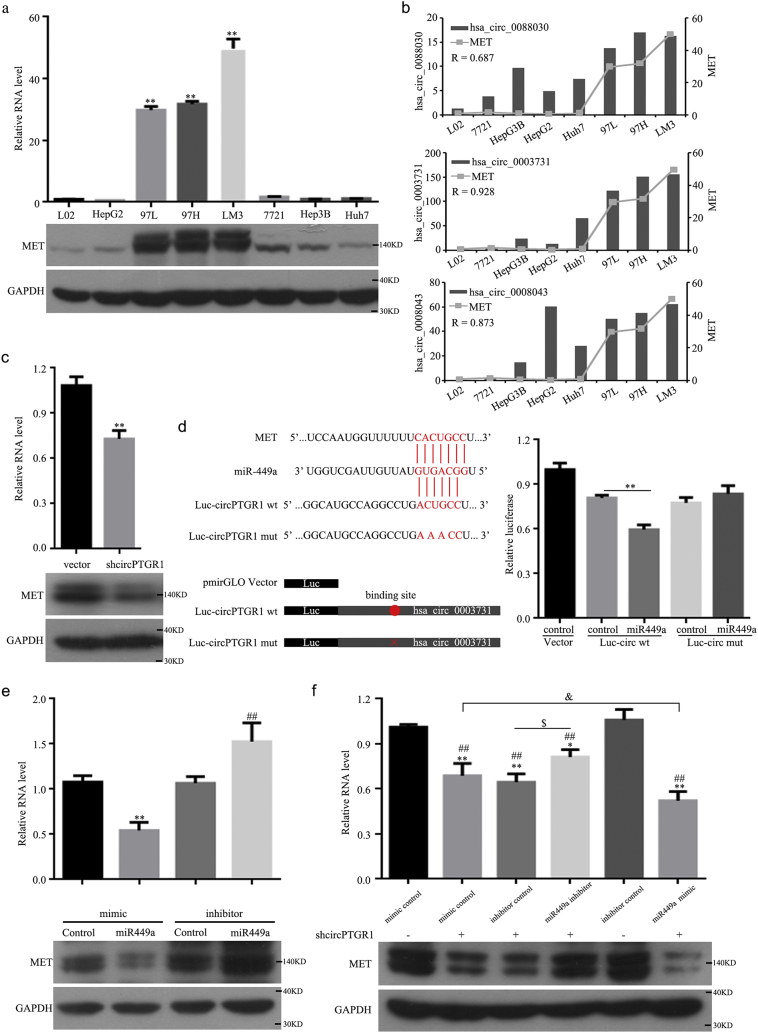
Because MET was the most differentially expressed target after exosome treatment, we measured its expression level in eight HCC cell lines (Fig. 7a). This showed that MET was upregulated in the 97 L, 97H, and LM3 metastatic cell lines compared to the other lines. The correlation coefficients for the correlations of has_circ_0088030, hsa_circ_0003731, and hsa_circ_0088043 with MET mRNA were 0.687, 0.928, and 0.873, respectively, suggesting a close relationship between circPTGR1 and MET (Fig. 7b). MET expression significantly decreased after the knockdown of circPTGR1 (P = .0016; Fig. 7c). These results indicated a positive correlation between circPTGR1 and MET.
Fig. 7.
CircPTGR1 regulated hepatocellular carcinoma (HCC) metastasis via the miR449a–MET pathway. (a) MET expression was detected by qRT-PCR and western blotting in eight HCC cell lines. P < .01 vs. L-O2 cells (L-O2 vs. 97 L, 97H, and LM3, P < .0001). (b) Expression levels of MET and circPTGR1 in eight HCC cell lines were evaluated with qRT-PCR and correlation analysis. (c) The knockdown efficiency of circPTGR1 in LM3 exosomesP <0.01 vs. vector (P = .0016). (d) The predicted binding site of miRNAs shared by circPTGR1 and MET was confirmed with a luciferase assay by the cotransfection of mutated circPTGR1 sequences that targeted the seed region of miR449a with miR449a mimics. ** P < .01 vs. Luc-circ-wt control (P = .0004). Error bars indicate standard deviations. (e) Overexpression or inhibition of miR449a, as shown by changes in the MET expression level (mimic control vs. miR449 mimic, P = .0034; mimic control vs. miR449 inhibitor, P = .0094; inhibitor control vs. miR449 inhibitor, P = .0083). (f) The expression of MET was downregulated by circPTGR1 and reversed by the miR449a inhibitor. ** P < .01 vs. mimic control (−). (Mimic control (+), P = .0002; inhibitor control (+), P < .0001; miR449a inhibitor (+), P = .0134; miR449a mimic (+), P < .0001P < 0.01 vs. inhibitor control (−). (Mimic control P < (+), P < .0001; inhibitor control (+), P < .0001; miR449a inhibitor (+), P < .0001; miR449a mimic (+), P < .0001.) $, P < .05 (miR449a inhibitor vs. inhibitor control (+), P ≤ 0.04429; and miR449a mimic vs. mimic control P < (+), P = .04429.) (+) cotransfected with shcircPTGR1; (−) cotransfected with shNC.
A bioinformatics analysis suggested that there was a complementary binding site of 5-nt nucleic acids between circPTGR1 and the 5′ end of miR449a, a region that was competitively matched by the 3′ untranslated region of MET (P = .0004; Fig. 7d). To confirm the regulatory relationship between circPTGR1 and miR449a, the predicted binding site sequence of the 5-nt nucleic acids of circPTGR1 was mutated. A dual luciferase assay confirmed that plasmids carrying wild-type circPTGR1 (Luc-circ-wt) and mutant circPTGR1 (Luc-circ-mut) were cotransfected with a miR449a or negative control mimic into HepG2 cells. The Luc-circ-wt luciferase activity was significantly reduced in the cells cotransfected with miR449a compared with those cotransfected with the negative control. However, the Luc-circ-mut activity did not change, indicating that circPTGR1 specifically interacts with miR449a in HCC cells (Fig. 7d).
To confirm the relationship between miRNA449a and MET, we synthesized miR449a mimic/inhibitor sequences and observed the MET expression levels. The MET levels decreased with miR449a overexpression but increased with miR449a inhibition (mimic control vs. miR449 mimic, P = .0034; inhibitor control vs. miR449 inhibitor, P = .0083; Fig. 7e). Analyzing the co-expression relationship between miR449a, MET, and circPTGR1 showed that the level of MET was decreased with shcircPTGR1 alone, but the co-expression of shcircPTGR1 and miR449a inhibitor reversed this decrease in MET expression, and miR449a mimic and shcircPTGR1 further downregulated MET expression compared with shcircPTGR1 alone (Fig. 7f). In addition, circPTGR1 knockdown abolished the decreased miR449a and increased MET expression levels due to negative control exosomes in vivo (Supplementary Fig. 4c–e). These results suggest that circPTGR1 can promote migration and metastasis in HCC through a circPTGR1–miR449a–MET pathway.
4. Discussion
Emerging evidence has suggested a role of exosomes in intercellular communication, thereby contributing to a wide variety of diseases [18]. Exosomes released from donor cells are taken up by recipient cells through fusion with the plasma membrane or through endocytosis, affecting biological events and cellular activity [6,19]. Studies of HCC have characterized exosomes originating from different types of donor cells and revealed their effects on cell growth, metastasis, and drug resistance, indicating material exchange between donor and receipt cells [8,[20], [21], [22]]. However, the interplay between HCC cell lines with different metastatic potential is not yet sufficiently understood. In this study, we investigated the differences between exosomes derived from cells with different metastatic properties, and we observed the effects of exosomes derived from LM3 cells, which are known to have high metastatic properties, on HepG2 (a non-metastatic cell line) and 97 L cells (a low-metastatic cell line). A functional analysis revealed that incubating HepG2 and 97 L cells with LM3-derived exosomes enhanced the cell migration and invasion abilities of both cell lines, but did not affect their cell proliferation, apoptosis, or cell cycle distribution. These findings suggest that LM3 exosomes have a primary function of regulating the metastatic potential of HCC cells, but not their tumorigenicity.
Exosomal cargo, such as RNA, DNA, and proteins, may reflect specific disease-related characteristics of the origin of the exosomes. It has been suggested that exosomes communicate with neighboring or distant cells by the horizontal transfer of their cargo molecules to recipient cells, thereby influencing cancer progression and metastasis [6,23]. Among these cargo molecules, RNAs, particularly noncoding RNAs such as lncRNAs, miRNAs, and circRNAs, have been identified as being specifically expressed under different physiological and pathological conditions [24,25]. In this study, we compared the divergent exosomal RNAs between HepG2, 97 L, and LM3 exosomes. RNA-seq data showed similar patterns of RNA distribution in the highly and low metastatic cells compared with the HepG2 cells. The KEGG pathway enrichment analysis results suggested that the lncRNAs which were differentially expressed between 97 L and LM3 were enriched in tumor-related pathways, with circRNA showing especially high enrichment in metastasis-related pathways. To the best of our knowledge, no previous study has analyzed the function of exosomal circRNA in HCC. We therefore paid particular attention to the circRNAs derived from HCC cell exosomes that were differentially expressed. Interestingly, we found three isoforms of circPTGR1 that were selectively expressed in exosomes derived from highly metastatic cells and were preferentially located in LM3 exosomes. In patients, the expression of circPTGR1 was associated with the clinical stage of their HCC, indicating its prognostic value in the clinical setting. A previous study investigated the interaction between HCC tumor cells and liver parenchyma cells, characterizing the RNA and protein contents of exosomes using RNA-seq and mass spectrometry in HCC cell lines (HKCI-C3, 97 L, and HKCI-8) and hepatocyte cells (MIHA) [8]. This identified several RNAs that were highly expressed in MHCC97L cells. Specifically, S100A4 was uniquely expressed in the MHCC97L and HKCI-8 cell lines. Our study adds new information about the exosomal content, particularly the circRNAs profile, derived from highly metastatic cells.
The role of circRNAs in HCC has been reported in several studies [12,26], and their existence within exosomes has been confirmed. Li et al. discovered abundant stable circRNAs in LM3-derived cells [10] and Dou et al. detected exosomal circRNAs in colon cancer cells [27]. However, little is known about their functions. Using exosomes with low circPTGR1 expression, we revealed the role of circPTGR1 in HCC progression, both in vivo and in vitro. We identified three isoforms of circPTGR1 as the transcription products of linear PTGR1. The knockdown of circPTGR1 resulted in significant reductions in the expression of both circPTGR1 and linear PTGR1. It has been reported that PTGR1 is upregulated in HCC [28]; however, the mechanism has largely remained unexplored. A recent study reported that NRF2 plays an important role in the upregulation of PTGR1, thereby promoting HCC proliferation in response to oxidative stress [29]. In our study, the knockdown of PTGR1 mRNA inhibited cell proliferation, but it did not influence metastasis in LM3 cells; this was consistent with the findings of previous studies and with TCGA data. In addition, the expression of PTGR1 in serum exosomes did not differ between patients at different clinical stages, and exosomes derived from shPTGR1 cells did not influence the migration and invasion of 97 L and HepG2 cells. Thus, we conclude that the observed effects were attributable mainly to circPTGR1 and not to PTGR1. This finding may provide new insights into the previously reported functions of PTGR1 in tumor development [[30], [31], [32]], namely that the circular forms of PTGR1 can regulate HCC tumor metastasis and play a role unique from that of the parent mRNA.
In this study, we characterized the facilitation of MET expression by circPTGR1 competing with the seed sequence of miR449a. Previously, miR449a was described as having roles in cell differentiation [33] and tumor suppression [34,35], as well as involvement in the inhibition of tumor growth and metastasis in HCC [[35], [36], [37]]. Consistent with the findings of these previous studies, we confirmed that miR449a overexpression inhibited HCC cell migration and invasion. Among the identified targets of miR449a [17], MET is thought to promote HCC progression [22,38]. Our study provided further evidence of the regulatory relationship between miR449a and MET in HCC progression. Interestingly, miR449a and MET have been reported to be involved in lncARSR-mediated sunitinib drug resistance in renal cancer; in this process, lncARSR from the resistant cells is packed into exosomes and effectively transfers the resistant phenotype to drug-sensitive cells [15]. It is likely that circPTGR1 derived from LM3 cells enhanced the metastatic abilities of 97 L and HepG2 cells in a similar way by affecting the miR449a–MET pathway of the recipient cells.
In summary, our study provided evidence that cells with higher metastatic potential could confer this potential on low-metastatic and non-metastatic cells via exosomes, resulting in an increase in the migratory and invasive abilities of these cells. In addition, the study revealed the role in this effect played by circPTGR1. It is possible that exosomes from highly metastatic cells with a high abundance of circPTGR1 may influence cells with lower metastatic potential by downregulating miR449a–MET interactions in the recipient cells, leading to a disruption in tumor microenvironment homeostasis and promoting HCC progression. Because circPTGR1 is highly abundant and is aberrantly expressed in malignant cells and in cells from patients with metastases, it could function as a prognostic biomarker and therapeutic target in HCC.
The following are the supplementary data related to this article.
Supplementary material
Acknowledgments/funding sources
This study was supported by the National Natural Science Foundation of China (No. 81470870, 81670601, 81570593), Guangdong Natural Science Foundation(No. 2015A030312013, 2015A030313038), Sci-tech Research Development Program of Guangdong Province (2014B020228003), Sci-tech Research Development Program of Guangzhou City (No. 201508020262, 201400000001-3, 201604020001, 201607010024), Innovative Funds for Small and Medium-Sized Enterprises of Guangdong Province (2016A010119103), Pearl River S&T Nova Program of Guangzhou(201710010178), and National 13th Five-Year Science and Technology Plan Major Projects of China (No. 2017ZX10203205-006-001).
Declaration of interests
The authors declare no competing interests.
Author contributions
Guoying Wang, Wei Liu and Yong Zou wrote the paper; Genshu Wang, Yinan Deng and Jingyan Luo performed the experiments; Yingcai Zhang and Hua Li analyzed the data; Qi Zhang, Yang Yang and Guihua Chen conducted the experiments. All authors approved the final manuscript.
Contributor Information
Qi Zhang, Email: keekee77@126.com.
Yang Yang, Email: yyang1971@126.com.
Guihua Chen, Email: pxkpzj@163.com, chgh1955@126.com.
References
- 1.Chen W., Zheng R., Baade P.D., Zhang S., Zeng H., Bray F. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66(2):115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 2.Forner A., Llovet J.M., Bruix J. Hepatocellular carcinoma. Lancet (London, England) 2012;379(9822):1245–1255. doi: 10.1016/S0140-6736(11)61347-0. [DOI] [PubMed] [Google Scholar]
- 3.Parkin D.M., Bray F., Ferlay J., Pisani P. Global cancer statistics, 2002. CA Cancer J Clin. 2005;55(2):74–108. doi: 10.3322/canjclin.55.2.74. [DOI] [PubMed] [Google Scholar]
- 4.Ferlay J., Shin H.R., Bray F., Forman D., Mathers C., Parkin D.M. Estimates of worldwide burden of cancer in 2008: GLOBOCAN 2008. Int J Cancer. 2010;127(12):2893–2917. doi: 10.1002/ijc.25516. [DOI] [PubMed] [Google Scholar]
- 5.Shahabipour F., Barati N., Johnston T.P., Derosa G., Maffioli P., Sahebkar A. Exosomes: nanoparticulate tools for RNA interference and drug delivery. J Cell Physiol. 2017;232(7):1660–1668. doi: 10.1002/jcp.25766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Azmi A.S., Bao B., Sarkar F.H. Exosomes in cancer development, metastasis, and drug resistance: a comprehensive review. Cancer Metastasis Rev. 2013;32(3–4):623–642. doi: 10.1007/s10555-013-9441-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Kogure T., Lin W.L., Yan I.K., Braconi C., Patel T. Intercellular nanovesicle-mediated microRNA transfer: a mechanism of environmental modulation of hepatocellular cancer cell growth. Hepatology. 2011;54(4):1237–1248. doi: 10.1002/hep.24504. (Baltimore, Md) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.He M., Qin H., Poon T.C., Sze S.C., Ding X., Co N.N. Hepatocellular carcinoma-derived exosomes promote motility of immortalized hepatocyte through transfer of oncogenic proteins and RNAs. Carcinogenesis. 2015;36(9):1008–1018. doi: 10.1093/carcin/bgv081. [DOI] [PubMed] [Google Scholar]
- 9.Greene J., Baird A.M., Brady L., Lim M., Gray S.G., McDermott R. Circular RNAs: biogenesis, function and role in human diseases. Front Mol Biosci. 2017;4:38. doi: 10.3389/fmolb.2017.00038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Li Y., Zheng Q., Bao C., Li S., Guo W., Zhao J. Circular RNA is enriched and stable in exosomes: a promising biomarker for cancer diagnosis. Cell Res. 2015;25(8):981–984. doi: 10.1038/cr.2015.82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lasda E., Parker R. Circular RNAs co-precipitate with extracellular vesicles: a possible mechanism for circRNA clearance. PloS one. 2016;11(2) doi: 10.1371/journal.pone.0148407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yao Z., Luo J., Hu K., Lin J., Huang H., Wang Q. ZKSCAN1 gene and its related circular RNA (circZKSCAN1) both inhibit hepatocellular carcinoma cell growth, migration, and invasion but through different signaling pathways. Mol Oncol. 2017;11(4):422–437. doi: 10.1002/1878-0261.12045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Melo S.A., Luecke L.B., Kahlert C., Fernandez A.F., Gammon S.T., Kaye J. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature. 2015;523(7559):177–182. doi: 10.1038/nature14581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Audic S., Claverie J.M. The significance of digital gene expression profiles. Genome Res. 1997;7(10):986–995. doi: 10.1101/gr.7.10.986. [DOI] [PubMed] [Google Scholar]
- 15.Liu Q., Zhang X., Hu X., Dai L., Fu X., Zhang J. Circular RNA related to the chondrocyte ECM regulates MMP13 expression by functioning as a MiR-136 'Sponge' in human cartilage degradation. Sci Rep. 2016;6 doi: 10.1038/srep22572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Anaya J. OncoLnc: linking TCGA survival data to mRNAs, miRNAs, and lncRNAs. PeerJ Comput Sci. 2016;2 [Google Scholar]
- 17.Lize M., Klimke A., Dobbelstein M. MicroRNA-449 in cell fate determination. Cell Cycle. 2011;10(17):2874–2882. doi: 10.4161/cc.10.17.17181. (Georgetown, Tex) [DOI] [PubMed] [Google Scholar]
- 18.Abels E.R., Breakefield X.O. Introduction to extracellular vesicles: biogenesis, RNA cargo selection, content, release, and uptake. Cell Mol Neurobiol. 2016;36(3):301–312. doi: 10.1007/s10571-016-0366-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mulcahy L.A., Pink R.C., Carter D.R. Routes and mechanisms of extracellular vesicle uptake. J Extracell Vesicles. 2014;3 doi: 10.3402/jev.v3.24641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Pan Q., Ramakrishnaiah V., Henry S., Fouraschen S., de Ruiter P.E., Kwekkeboom J. Hepatic cell-to-cell transmission of small silencing RNA can extend the therapeutic reach of RNA interference (RNAi) Gut. 2012;61(9):1330–1339. doi: 10.1136/gutjnl-2011-300449. [DOI] [PubMed] [Google Scholar]
- 21.Qu Z., Wu J., Wu J., Luo D., Jiang C., Ding Y. Exosomes derived from HCC cells induce sorafenib resistance in hepatocellular carcinoma both in vivo and in vitro. J Exp Clin Cancer Res: CR. 2016;35(1):159. doi: 10.1186/s13046-016-0430-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Tey S.K., Mao X., Yam J.W.P. Role of nuclear Met-derived exosomes in hepatocellular carcinoma metastasis and lung premetastatic niche formation. Eur J Cancer. 2016;61:S63–S. [Google Scholar]
- 23.Suchorska W.M., Lach M.S. The role of exosomes in tumor progression and metastasis (Review) Oncol Rep. 2016;35(3):1237–1244. doi: 10.3892/or.2015.4507. [DOI] [PubMed] [Google Scholar]
- 24.Cai S., Cheng X., Pan X., Li J. Emerging role of exosomes in liver physiology and pathology. Hepatol Res. 2017;47(2):194–203. doi: 10.1111/hepr.12794. [DOI] [PubMed] [Google Scholar]
- 25.Kahlert C., Kalluri R. Exosomes in tumor microenvironment influence cancer progression and metastasis. J Mol Med. 2013;91(4):431–437. doi: 10.1007/s00109-013-1020-6. (Berlin, Germany) [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zheng Q., Bao C., Guo W., Li S., Chen J., Chen B. Circular RNA profiling reveals an abundant circHIPK3 that regulates cell growth by sponging multiple miRNAs. Nat Commun. 2016;7 doi: 10.1038/ncomms11215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Dou Y., Cha D.J., Franklin J.L., Higginbotham J.N., Jeppesen D.K., Weaver A.M. Circular RNAs are down-regulated in KRAS mutant colon cancer cells and can be transferred to exosomes. Sci Rep. 2016;6 doi: 10.1038/srep37982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Sanchez-Rodriguez R., Torres-Mena J.E., De-la-Luz-Cruz M., Bernal-Ramos G.A., Villa-Trevino S., Chagoya-Hazas V. Increased expression of prostaglandin reductase 1 in hepatocellular carcinomas from clinical cases and experimental tumors in rats. Int J Biochem Cell Biol. 2014;53:186–194. doi: 10.1016/j.biocel.2014.05.017. [DOI] [PubMed] [Google Scholar]
- 29.Sanchez-Rodriguez R., Torres-Mena J.E., Quintanar-Jurado V., Chagoya-Hazas V., Rojas Del Castillo E., Del Pozo Yauner L. Ptgr1 expression is regulated by NRF2 in rat hepatocarcinogenesis and promotes cell proliferation and resistance to oxidative stress. Free Radic Biol Med. 2017;102:87–99. doi: 10.1016/j.freeradbiomed.2016.11.027. [DOI] [PubMed] [Google Scholar]
- 30.Huang X., Zhou W., Zhang Y., Liu Y. High expression of PTGR1 promotes NSCLC cell growth via positive regulation of cyclin-dependent protein kinase complex. Biomed Res Int. 2016;2016 doi: 10.1155/2016/5230642. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 31.Xue L., Zhu Z., Wang Z., Li H., Zhang P., Wang Z. Knockdown of prostaglandin reductase 1 (PTGR1) suppresses prostate cancer cell proliferation by inducing cell cycle arrest and apoptosis. Biosci Trends. 2016;10(2):133–139. doi: 10.5582/bst.2016.01045. [DOI] [PubMed] [Google Scholar]
- 32.Yang S., Luo F., Wang J., Mao X., Chen Z., Wang Z. Effect of prostaglandin reductase 1 (PTGR1) on gastric carcinoma using lentivirus-mediated system. Int J Clin Exp Pathol. 2015;8(11):14493–14499. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 33.Wu J., Bao J., Kim M., Yuan S., Tang C., Zheng H. Two miRNA clusters, miR-34b/c and miR-449, are essential for normal brain development, motile ciliogenesis, and spermatogenesis. Proc Natl Acad Sci U S A. 2014;111(28):E2851–E2857. doi: 10.1073/pnas.1407777111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Chen H., Lin Y.W., Mao Y.Q., Wu J., Liu Y.F., Zheng X.Y. MicroRNA-449a acts as a tumor suppressor in human bladder cancer through the regulation of pocket proteins. Cancer Lett. 2012;320(1):40–47. doi: 10.1016/j.canlet.2012.01.027. [DOI] [PubMed] [Google Scholar]
- 35.You J., Zhang Y., Li Y., Fang N., Liu B., Zu L. MiR-449a suppresses cell invasion by inhibiting MAP2K1 in non-small cell lung cancer. Am J Cancer Res. 2015;5(9):2730–2744. [PMC free article] [PubMed] [Google Scholar]
- 36.Chen S.P., Liu B.X., Xu J., Pei X.F., Liao Y.J., Yuan F. MiR-449a suppresses the epithelial-mesenchymal transition and metastasis of hepatocellular carcinoma by multiple targets. BMC Cancer. 2015;15:706. doi: 10.1186/s12885-015-1738-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Liu S., Liu K., Zhang W., Wang Y., Jin Z., Jia B. miR-449a inhibits proliferation and invasion by regulating ADAM10 in hepatocellular carcinoma. Am J Transl Res. 2016;8(6):2609–2619. [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 38.Giordano S., Columbano A. Met as a therapeutic target in HCC: facts and hopes. J Hepatol. 2014;60(2):442–452. doi: 10.1016/j.jhep.2013.09.009. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Supplementary material