Abstract
Xeroderma pigmentosum group G (XPG) protein is a pivotal element of the nucleotide excision repair pathway. XPG gene single nucleotide polymorphisms (SNPs) have been shown to confer colorectal cancer (CRC) susceptibility. In this study, we further investigated the role of Asp1104His (rs17655 G > C) in XPG on CRC risk. We genotyped the rs17655 G > C polymorphism in Chinese population comprising 1019 CRC cases and 1036 cancer-free controls. We also performed a meta-analysis to further assess the association. Overall, no significant association was detected between the rs17655 G > C and the risk of CRC. Stratified analysis also revealed no significant association. To further elucidate the association of the rs17655 with CRC susceptibility, we conducted a meta-analysis by including qualified publications and the current study. The meta-analysis results demonstrated that rs17655 G > C was associated with an increased CRC risk (CG vs. GG: OR = 1.14, 95% CI = 1.01-1.28; CC/CG vs. GG: OR = 1.12, 95% CI = 1.01-1.24; C vs. G: OR = 1.06, 95% CI = 1.01-1.11). In subgroup analysis, the significant association between the rs17655 C allele and CRC risk was found in Asians and hospital-based subgroups. Taken together, our results suggested that the XPG rs17655 G > C polymorphism is a low-penetrance susceptibility locus for CRC. Further studies are warranted to validate these findings.
Keywords: Colorectal cancer, XPG, Asp1104His, polymorphism, susceptibility
Introduction
Colorectal cancer (CRC) is considered as the third most common cancer and the fourth leading cause of cancer-related death in the world [1]. In China, CRC ranks the top five both in new cancer cases and the cancer-related cause death [2]. The etiology of CRC is highly complicated, involving the interaction between genetic and environmental factors [3]. The discovery of risk factors would help to identify high-risk individuals and develop prevention strategies. Previous epidemiological studies have led to the findings of numerous polymorphisms predisposing to CRC.
DNA repair systems play an indispensable role in protecting genome from endogenous and exogenous damages [4]. Nucleotide excision repair (NER) is the most versatile DNA repair mechanism among the five known DNA repair systems [5,6], which mainly takes the responsibility to get rid of bulky DNA adducts and UV-induced DNA damage [7]. Aberrant function of NER pathway is tightly associated with Xeroderma pigmentosum (XP), an unusual autosomal recessive disease; affected individuals are extremely vulnerable to sunlight-induced skin cancer [8]. NER pathway is composed of a number of core protein molecules, including XPA to XPG [9]. XPG [alias excision repair cross-complementation group 5 (ERCC5)] [10] is mapped to chromosome 13q22-q23 and encodes a protein of 1186-amino acid residues. XPG protein participates in the initial step of DNA repair process by recognizing the DNA damage loci [11-13]. XPG also mediates mutagenesis and cell death by influencing RNA transcription [14,15].
Single nucleotide polymorphisms (SNPs) in the XPG gene are reported to predispose to the susceptibility of several cancers, including gastric cancer [16-18], prostate cancer [19], breast cancer [20], as well as colorectal cancer [21]. Among cancer predisposing XPG SNPs, the Asp1104His polymorphism (rs17655 G > C) is most frequently investigated [22,23]. Asp1104His polymorphism is a nonsynonymous polymorphism commonly regarded as a tagger. It can result in an amino acid alteration within the protein sequence. Several studies have been performed to investigate the association between the XPG rs17655 G > C polymorphism and CRC risk, but yielded conflicting results. Therefore, further replication studies are needed to solve these discrepancies. Here, we conducted a case-control study, followed by a meta-analysis, to provide a precise evaluation of the association of interest.
Materials and methods
Study population
We recruited 1019 cases with histologically confirmed CRC in the Department of Colorectal and Anal Surgery, The First Affiliated Hospital of Zhengzhou University in the last four year. We also enrolled 1036 cancer-free controls in the same region during the same period. All the enrolled participants were unrelated ethnic Han Chinese population. Each participant provided a written informed consent. The demographic characteristics were obtained from the participants by using a self-administered questionnaire. Each participant donated 5 ml of venous blood sample on a voluntary basis. The study was approved by the Institutional Review Board of The First Affiliated Hospital of Zhengzhou University.
Genotyping
We first adopted the Qiagen Blood DNA Mini Kit (Qiagen Inc., Valencia, CA) to extract genomic DNA, according to the standard procedures. Then Taqman assay was chosen for genotyping with Applied Biosystems (Foster City, CA). We also set four duplicated positive controls and four negative controls (without DNA) in each of 384-well plates for quality control. Moreover, 10% of the samples were randomly chosen to be analyzed for a second time, and 100% concordant results were obtained.
Statistical analysis
Differences in demographic characteristics among cases and controls were tested using chi-square test. Goodness-of-fit X2 test was applied to check whether the genotype frequency distribution of rs17655 G > C in controls was deviated from Hardy-Weinberg equilibrium (HWE). Odds ratios (ORs) and 95% confidence intervals (CIs) were calculated from multivariate logistic regression, and then used to estimate the associations between rs17655 G > C and CRC risk. We also performed stratification analysis by age, gender, body mass index (BMI), smoking status, pack-years, drinking status, tumor location, and Duke stage. All statistical analysis was performed using SAS system (version 9.1; SAS Institute, Cary, NC). Statistical significance was set on the basis of two-sided P-values < 0.05.
Meta-analysis
We further evaluated the association between rs17655 G > C and CRC risk using meta-analysis. PubMed, EMBASE, and MEDLINE databases were used to conduct systematic literature searches. The search terms were as follows: “colorectal cancer or colorectal tumor or colorectal carcinoma or colorectal neoplasm or CRC”, “Xeroderma pigmentosum group G or XPG or rs17655 or Asp1104His”, and “polymorphism or SNP or variant or variation”. Literature searches were updated to July 1, 2018. Between-study heterogeneity was determined by a chi-square-based Q-Test. The random-effects model (the DerSimonian and Laird method) would be performed in the presence of heterogeneity, whereas the fixed-effects model (the Mantel-Haenszel method) would be performed [23-25]. The funnel plot and the Egger’s linear regression test were used to assess publication bias. In addition, sensitivity analysis was also applied to assess the strength of the study. The meta-analysis was conducted using STATA version 11.0 (Stata Corporation, College Station, TX, USA).
Results
Population characteristics
The demographic characteristics of 1019 CRC patients and 1036 cancer-free controls were shown in Table 1. No significant difference was observed in the distributions of age (P = 0.508) and gender (P = 0.230) between the cases and controls. The percentage of ever smokers (28.75%) were significantly lower in cases than in controls (45.46%). Significant difference was also detected in pack-years between cases and controls. Moreover, cases were less likely to be drinkers than controls. As to the location of tumor, 46.81% of lesions (477 cases) occurred in colon, while 53.19% of lesions (542 cases) in rectum. In term of tumor stage, 46 (4.51%), 314 (30.81%), 380 (37.29%), and 279 cases (27.38%) were diagnosed with Duke’s stage A, B, C, and D diseases, respectively.
Table 1.
Demographic characteristics of the colorectal cancer patients and controls
| Variables | Cases (n = 1019) | Controls (n = 1036) | Pa | ||
|---|---|---|---|---|---|
|
| |||||
| No. | % | No. | % | ||
| Age range, year | 23-87 | 24-85 | 0.508 | ||
| Mean ± SD | 56.58 ± 12.69 | 57.25 ± 11.82 | |||
| ≤ 58 | 546 | 53.58 | 650 | 52.12 | |
| > 58 | 473 | 46.42 | 496 | 47.88 | |
| Gender | 0.230 | ||||
| Female | 389 | 38.17 | 369 | 35.62 | |
| Male | 630 | 61.83 | 667 | 64.38 | |
| BMI | < 0.0001 | ||||
| < 18.0 | 90 | 8.83 | 9 | 0.87 | |
| 18-24.9 | 717 | 70.36 | 606 | 58.49 | |
| 25.0-29.9 | 193 | 18.94 | 362 | 34.94 | |
| > 30.0 | 19 | 1.86 | 59 | 5.69 | |
| Smoking status | < 0.0001 | ||||
| Never | 726 | 71.25 | 565 | 54.54 | |
| Ever | 293 | 28.75 | 471 | 45.46 | |
| Pack-year | < 0.0001 | ||||
| 0 | 726 | 71.25 | 565 | 54.54 | |
| ≤ 30 | 151 | 14.82 | 294 | 28.38 | |
| > 30 | 142 | 13.94 | 177 | 17.08 | |
| Drinking status | < 0.0001 | ||||
| No | 847 | 83.12 | 763 | 73.65 | |
| Yes | 172 | 16.88 | 273 | 26.35 | |
| Tumor locations | |||||
| Colon | 477 | 46.81 | / | / | |
| Rectal | 542 | 53.19 | / | / | |
| Duke stages | |||||
| A | 46 | 4.51 | / | / | |
| B | 314 | 30.81 | / | / | |
| C | 380 | 37.29 | / | / | |
| D | 279 | 27.38 | / | / | |
SD, standard deviation; BMI, body mass index.
Two-sided Chi-square test for the distributions between patients and controls.
XPG gene rs17655 G > C polymorphism and colorectal cancer risk
The genotype distribution of the XPG gene rs17655 G > C and the association results were summarized in Table 2. The frequency distribution of rs17655 G > C was consistent with HWE in the control subjects (P = 0.854). We observed no significant association between rs17655 G > C and CRC risk.
Table 2.
Association between XPG rs17655 G > C polymorphism and colorectal cancer risk
| Genotype | Cases | Controls | P a | OR (95% CI) | P | AOR (95% CI)b | P b | ||
|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||
| No. | % | No. | % | ||||||
| rs17655 (HWE = 0.854) | |||||||||
| GG | 248 | 24.34 | 265 | 25.58 | 1.00 | 1.00 | |||
| CG | 510 | 50.05 | 515 | 49.71 | 1.06 (0.86-1.31) | 0.601 | 0.99 (0.79-1.24) | 0.947 | |
| CC | 261 | 25.61 | 256 | 24.71 | 1.09 (0.85-1.39) | 0.492 | 1.10 (0.85-1.43) | 0.461 | |
| Additive | 0.781 | 1.04 (0.92-1.18) | 0.493 | 1.05 (0.92-1.20) | 0.459 | ||||
| Dominant | 771 | 75.66 | 771 | 74.42 | 0.515 | 1.07 (0.88-1.31) | 0.516 | 1.03 (0.83-1.27) | 0.797 |
| Recessive | 758 | 74.39 | 780 | 75.29 | 0.637 | 1.05 (0.86-1.28) | 0.637 | 1.11 (0.90-1.37) | 0.342 |
OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio; HWE, Hardy-Weinberg equilibrium.
Chi-square test for genotype distributions between patients and controls.
Adjusted for age, gender, BMI, smoking and drinking status.
Stratification analysis
The stratified study was performed to explore the association between rs17655 G > C polymorphism and CRC risk by age, gender, BMI, smoking status, pack-year, drinking status, tumor location, and Duke stage. However, we did not find any significant association (Table 3).
Table 3.
Stratification analysis for the association between XPG rs17655 G > C polymorphism and colorectal cancer risk
| Variables | GG | CG/CC | OR (95% CI) | P | AOR (95% CI)a | P a |
|---|---|---|---|---|---|---|
|
| ||||||
| Cases/controls | ||||||
| Age, median | ||||||
| ≤ 58 | 124/125 | 422/415 | 1.03 (0.77-1.36) | 0.864 | 1.01 (0.75-1.35) | 0.964 |
| > 58 | 124/140 | 349/356 | 1.11 (0.83-1.47) | 0.483 | 1.04 (0.77-1.42) | 0.789 |
| Gender | ||||||
| Females | 92/90 | 297/279 | 1.04 (0.75-1.45) | 0.812 | 1.06 (0.74-1.51) | 0.764 |
| Males | 156/175 | 474/492 | 1.08 (0.84-1.39) | 0.543 | 1.02 (0.78-1.32) | 0.910 |
| BMI | ||||||
| < 18.0 | 19/6 | 71/3 | 7.47 (1.71-32.68) | 0.008 | 13.58 (2.33-79.11) | 0.004 |
| 18-24.9 | 171/150 | 546/456 | 1.05 (0.82-1.35) | 0.702 | 1.04 (0.80-1.34) | 0.788 |
| 25.0-29.9 | 53/93 | 140/269 | 0.91 (0.62-1.36) | 0.652 | 0.87 (0.58-1.32) | 0.512 |
| > 30.0 | 5/16 | 14/43 | 1.04 (0.32-3.36) | 0.946 | 1.09 (0.33-3.66) | 0.887 |
| Smoking status | ||||||
| Never | 174/134 | 552/431 | 0.99 (0.76-1.28) | 0.917 | 0.93 (0.71-1.22) | 0.579 |
| Ever | 74/131 | 219/340 | 1.14 (0.82-1.59) | 0.438 | 1.15 (0.81-1.64) | 0.425 |
| Pack-year | ||||||
| 0 | 174/134 | 552/431 | 0.99 (0.76-1.28) | 0.917 | 0.94 (0.72-1.24) | 0.676 |
| ≤ 30 | 34/82 | 117/212 | 1.33 (0.84-2.11) | 0.222 | 1.26 (0.77-2.05) | 0.361 |
| > 30 | 40/49 | 102/128 | 0.98 (0.60-1.60) | 0.923 | 0.75 (0.43-1.33) | 0.324 |
| Drinking status | ||||||
| Never | 205/180 | 642/583 | 0.97 (0.77-1.22) | 0.774 | 0.95 (0.75-1.21) | 0.677 |
| Ever | 43/85 | 129/188 | 1.36 (0.88-2.09) | 0.165 | 1.29 (0.82-2.02) | 0.271 |
| Tumor locations | ||||||
| Colon | 117/265 | 360/771 | 1.06 (0.82-1.36) | 0.664 | 1.00 (0.76-1.30) | 0.969 |
| Rectal | 131/265 | 411/771 | 1.08 (0.85-1.37) | 0.540 | 1.07 (0.83-1.38) | 0.607 |
| Duke stages | ||||||
| A + B | 90/265 | 270/771 | 1.03 (0.78-1.36) | 0.829 | 1.04 (0.78-1.39) | 0.783 |
| C + D | 158/265 | 501/771 | 1.09 (0.87-1.37) | 0.457 | 1.02 (0.80-1.30) | 0.859 |
OR, odds ratio; CI, confidence interval; AOR, adjusted odds ratio; BMI, body mass index.
Adjusted for age, gender, BMI, smoking and drinking status.
Meta-analysis results
Meta-analysis was also carried out to further explore the association of rs17655 G > C polymorphism with CRC risk by combining qualified publications and our data. Overall, 14 eligible case-control studies were pooled together to evaluate such association [26-37] (Table 4). As shown in Table 5 and Figure 1, pooled results indicated that rs17655 G > C polymorphism was associated with an increased CRC susceptibility (CG vs. GG: OR = 1.14, 95% CI = 1.01-1.28; CC/CG vs. GG: OR = 1.12, 95% CI = 1.01-1.24; C vs. G: OR = 1.06, 95% CI = 1.01-1.11). Stratified analysis by ethnicity revealed significant association between rs17655 G > C genotype and CRC risk among Asian (CC vs. GG: OR = 1.18, 95% CI = 1.04-1.35; CG vs. GG: OR = 1.25, 95% CI = 1.00-1.54; CC/CG vs. GG: OR = 1.23, 95% CI = 1.03-1.47; C vs. G: OR = 1.10, 95% CI = 1.03-1.17), but not among Caucasians or Africans (Figure 2). Regarding source of controls (Figure 3), significant association was detected between rs17655 G > C and an increased CRC risk in hospital-based studies (CC vs. GG: OR = 1.14, 95% CI = 1.01-1.29; CG vs. GG: OR = 1.26, 95% CI = 1.11-1.44; CC/CG vs. GG: OR = 1.24, 95% CI = 1.11-1.37; C vs. G: OR = 1.11, 95% CI = 1.05-1.17). Regarding HWE (Figure 4), significant association was only detected between rs17655 G > C and an increased CRC risk in HWE ≤ 0.05 studies (CC/CG vs. GG: OR = 1.13, 95% CI = 1.00-1.28). Leave-one-out sensitivity analysis result demonstrated that no removal of any single study could lead to substantial change in pooled results. Moreover, no evidence of obvious asymmetry in Begg’s funnel plots was found.
Table 4.
Main characteristics of included studies for the final meta-analysis
| Name | Year | Region | Ethnicity | Design | Genotype | Case | Control | MAF | HWE | ||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| |||||||||||||||
| Method | GG | CG | CC | All | GG | CG | CC | All | |||||||
| Bigler | 2005 | USA | Caucasian | PB | Taqman | 440 | 237 | 36 | 713 | 353 | 226 | 37 | 616 | 0.24 | 0.917 |
| Huang | 2006 | USA | Caucasian | PB | Sequencing | 407 | 243 | 29 | 679 | 403 | 265 | 29 | 697 | 0.23 | 0.073 |
| Pardini | 2008 | Czech | Caucasian | HB | PCR-RFLP | 334 | 177 | 21 | 532 | 356 | 153 | 23 | 532 | 0.19 | 0.211 |
| Joshi | 2009 | USA | Caucasian | FB | Taqman | 183 | 114 | 11 | 308 | 213 | 137 | 11 | 361 | 0.22 | 0.046 |
| Canbay | 2011 | Turkey | Caucasian | PB | PCR-RFLP | 43 | 34 | 2 | 79 | 148 | 83 | 16 | 247 | 0.23 | 0.352 |
| Gil | 2012 | Poland | Caucasian | PB | PCR-RFLP | 86 | 35 | 11 | 132 | 64 | 31 | 5 | 100 | 0.21 | 0.625 |
| Liu | 2012 | China | Asian | HB | PCR-RFLP | 233 | 603 | 192 | 1028 | 329 | 537 | 219 | 1085 | 0.45 | 0.996 |
| Du | 2014 | China | Asian | HB | TaqMan | 286 | 459 | 133 | 878 | 355 | 405 | 124 | 884 | 0.37 | 0.623 |
| Steck | 2014 | USA | Caucasian | PB | MassARRAY | 183 | 100 | 15 | 298 | 335 | 170 | 27 | 532 | 0.21 | 0.372 |
| Steck | 2014 | USA | African | PB | MassARRAY | 65 | 120 | 39 | 224 | 100 | 151 | 66 | 317 | 0.45 | 0.519 |
| Paszkowska-Szczur | 2015 | Poland | Caucasian | HB | Taqman | 429 | 272 | 32 | 733 | 869 | 404 | 85 | 1358 | 0.21 | 0.0001 |
| Sun | 2015 | China | Asian | HB | PCR-RFLP | 216 | 476 | 198 | 890 | 227 | 497 | 186 | 910 | 0.48 | 0.004 |
| Kabzinski | 2015 | Poland | Caucasian | HB | QPCR | 36 | 171 | 27 | 234 | 43 | 175 | 20 | 238 | 0.45 | < 0.001 |
| Su | Current | China | Asian | HB | Taqman | 248 | 510 | 261 | 1019 | 265 | 515 | 256 | 1036 | 0.50 | 0.854 |
MAF, minor allele frequency; HWE, Hardy-Weinberg equilibrium; PB, population based; HB, hospital based; FB, family based; PCR-RFLP, polymerase chain reaction-restriction fragment length polymorphism.
Table 5.
Meta-analysis of the association between XPG rs17655 G > C polymorphism and colorectal cancer risk
| Variables | No. of studies | Cases/controls | Homozygous | Heterozygous | Recessive | Dominant | Allele comparing | |||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|
|
| ||||||||||||
| CC vs. GG | CG vs. GG | CC vs. CG/GG | CC/CG vs. GG | C vs. G | ||||||||
|
| ||||||||||||
| OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | OR (95% CI) | P het | |||
| All | 14 | 7747/8913 | 1.09 (0.98-1.22) | 0.584 | 1.14 (1.01-1.28) | 0.002 | 0.99 (0.90-1.09) | 0.593 | 1.12 (1.01-1.24) | 0.013 | 1.06 (1.01-1.11) | 0.477 |
| Ethnicity | ||||||||||||
| Caucasian | 9 | 3708/4681 | 0.93 (0.76-1.15) | 0.651 | 1.07 (0.93-1.23) | 0.046 | 0.91 (0.75-1.12) | 0.503 | 1.05 (0.94-1.18) | 0.163 | 1.02 (0.95-1.10) | 0.557 |
| Asian | 4 | 3815/3915 | 1.18 (1.04-1.35) | 0.716 | 1.25 (1.00-1.54) | 0.006 | 1.03 (0.92-1.15) | 0.572 | 1.23 (1.03-1.47) | 0.025 | 1.10 (1.03-1.17) | 0.394 |
| African | 1 | 224/317 | 0.91 (0.55-1.51) | / | 1.22 (0.83-1.81) | / | 0.80 (0.52-1.24) | / | 1.13 (0.78-1.64) | / | 0.98 (0.77-1.25) | / |
| Source of control | ||||||||||||
| PB | 6 | 2125/2509 | 0.91 (0.71-1.17) | 0.757 | 0.97 (0.85-1.12) | 0.330 | 0.89 (0.70-1.13) | 0.639 | 0.95 (0.84-1.07) | 0.544 | 0.95 (0.86-1.05) | 0.806 |
| HB | 7 | 5314/6043 | 1.14 (1.01-1.29) | 0.395 | 1.26 (1.11-1.44) | 0.041 | 1.01 (0.91-1.12) | 0.336 | 1.24 (1.11-1.37) | 0.148 | 1.11 (1.05-1.17) | 0.804 |
| FB | 1 | 308/361 | 1.16 (0.49-2.75) | / | 0.97 (0.71-1.33) | / | 1.18 (0.50-2.76) | / | 0.98 (0.72-1.34) | / | 1.00 (0.77-1.30) | / |
| HWE | ||||||||||||
| > 0.05 | 10 | 5582/6046 | 1.11 (0.98-1.26) | 0.100 | 1.14 (0.98-1.33) | 0.090 | 0.98 (0.88-1.09) | 0.709 | 1.20 (0.98-1.28) | 0.105 | 1.05 (0.98-1.13) | 0.141 |
| ≤ 0.05 | 4 | 2165/2867 | 1.05 (0.81-1.37) | 0.722 | 1.13 (0.94-1.36) | 0.196 | 1.02 (0.75-1.38) | 0.895 | 1.13 (1.00-1.28) | 0.047 | 1.07 (0.98-1.17) | 0.116 |
OR, odds ratio; CI, confidence interval; PB, population based; HB, hospital based; FB, family based; HWE, Hardy-Weinberg equilibrium.
Figure 1.
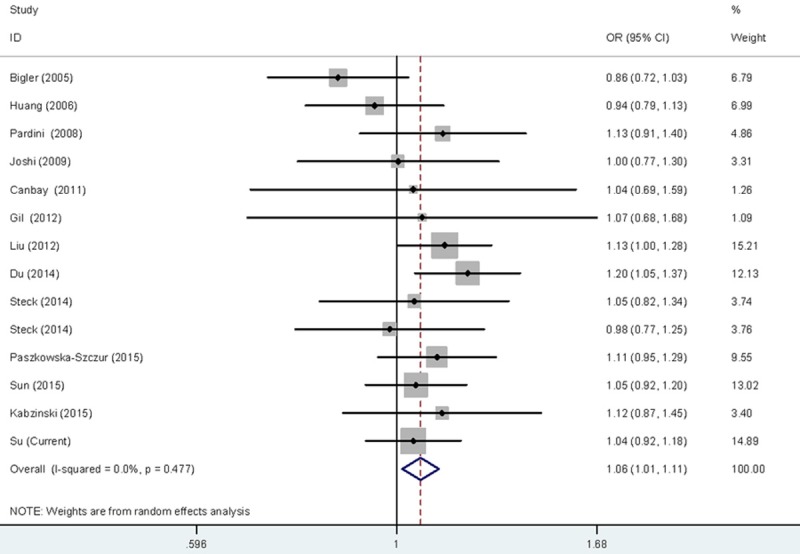
Forest plot for the CRC susceptibility associated with the rs17655 G > C polymorphism under allele comparison model. The horizontal lines represent the study-specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI.
Figure 2.
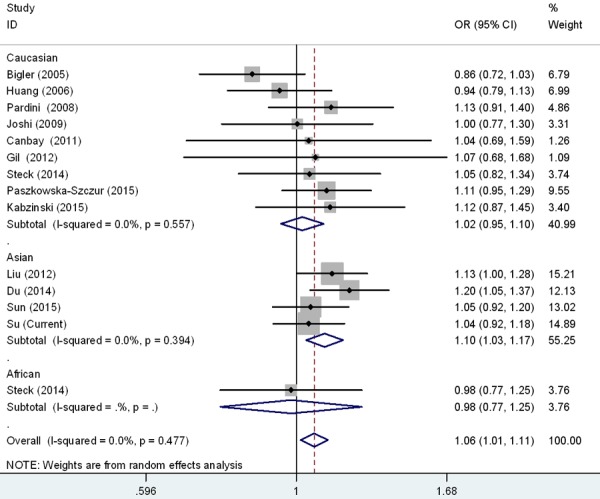
Forest plot for the CRC susceptibility associated with the rs17655 G > C polymorphism stratified by ethnicities under allele comparison model. The horizontal lines represent the study-specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI.
Figure 3.
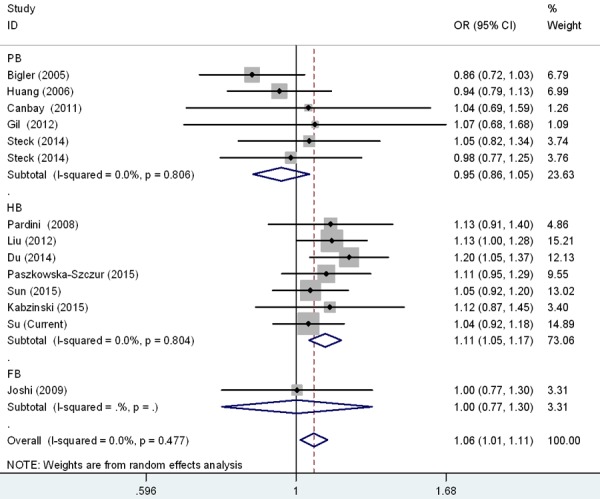
Forest plot for the CRC susceptibility associated with the rs17655 G > C polymorphism stratified by design under allele comparison model. The horizontal lines represent the study-specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI.
Figure 4.
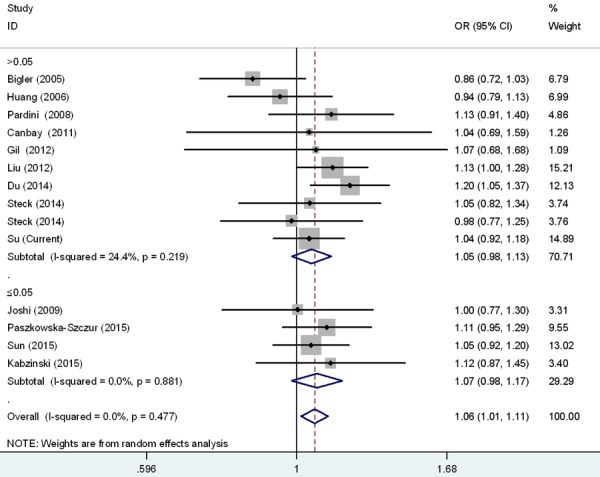
Forest plot for the CRC susceptibility associated with the rs17655 G > C polymorphism stratified by HWE under allele comparison model. The horizontal lines represent the study-specific ORs and 95% CIs, respectively. The diamond represents the pooled results of OR and 95% CI.
Discussion
In the present study, we further explored the predisposing role of XPG rs17655 G > C polymorphism in CRC. The results of our case-control study failed to provide supportive evidence of the association between the XPG gene rs17655 G > C polymorphism and CRC risk. However, the following meta-analysis demonstrated that the XPG rs17655 G > C polymorphism confers increased CRC risk.
XPG is an endonuclease responsible for a dual incision in NER pathway. XPG cut the DNA strand at the 3’ end of the lesion, and maintain the DNA repair complex in the damaged site with ERCC1/XPF complex by generating 5’ incision [38-41]. Genetic variations of XPG may impair the DNA repair capacity and genome integrity, consequently leading to the initiation of carcinogenesis. The association of XPG rs17655 G > C (Asp1104His) polymorphism with colorectal cancer risk has been widely investigated, and results are controversial. Paszkowska-Szczur et al. [35] failed to detect significant associations between XPG rs17655 G > C and CRC risk. Such null associations were also presented in a study conducted by Canbay et al. [31] in Turkish population with 79 CRC cases and 247 healthy controls. Opposite results regarding the association were also reported. In a Czech hospital-based case-control study including 532 cases and 532 controls, the XPG rs17655 G > C was shown to increase the risk of CRC [29]. Liu et al. [33] observed that heterozygotes and homozygotes of this variant were more likely to have CRC than wild controls, in a Chinese population study including 1028 CRC cases and 1085 controls. More recently, Du et al. [26] also verified the risk effect of XPG rs17655 G > C polymorphism on CRC in a Chinese population.
Replication study is a golden standard to validate a association. We performed this case-control study to further elucidate the contribution of XPG rs17655 G > C polymorphism to CRC susceptibility. We found that the XPG rs17655 G > C polymorphism was not significantly associated with CRC risk, either in the overall analysis or stratification analysis. The null association may be attributed to therelatively small sample size or the low-penetrance of this SNP. Therefore, we next conducted a meta-analysis to comprehensively evaluate this association. Our meta-analysis indicated that individuals with CG and CC/CG genotype were more likely to be susceptible to CRC. Stratified analysis by ethnicity showed that significant association was observed among Asians, but not Caucasians. A variety of reasons may help to explain the discrepant results, such as differences in linkage disequilibrium structure, allele frequency, and lifestyles as well as diversities of geography and living environments [42]. Moreover, different results from the current study and meta-analysis regarding the association between rs17655 G > C and CRC risk might be due to different sample size, ethnicity, allele frequency and histological type of tumor.
The sample size of this study is moderate with 1019 cases and 1036 controls. Moreover, this meta-analysis is by far the largest pooled study to investigate the association of interest. Therefore, the conclusion obtained is convincing. However, several limitations still exist. First, we only analyzed one SNP in this study, more potentially functional SNPs in the XPG gene should be explored in the future. Second, the environmental variables were not included, which might also affect the risk of CRC. Third, selection bias and information bias could not be ruled out since all the participants were enrolled from the same hospital. Fourth, the moderate sample size of this study might have no sufficient power to detect the weak impact of SNP. Fifth, our study was a case-control study with subjects from north China. The current findings may not well represent other nationality and ethnicities. Finally, functional studies should be performed to elucidate the mechanism underlying this association.
In conclusion, we found that XPG rs17655 G > C polymorphism was associated with CRC susceptibility in Asian populations. More case-control studies with larger sample size are warranted to confirm our findings.
Acknowledgements
This study was funded by the Institutional Fund of the First Affiliated Hospital of Zhengzhou University.
Disclosure of conflict of interest
None.
References
- 1.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2017. CA Cancer J Clin. 2017;67:7–30. doi: 10.3322/caac.21387. [DOI] [PubMed] [Google Scholar]
- 2.Chen W, Zheng R, Baade PD, Zhang S, Zeng H, Bray F, Jemal A, Yu XQ, He J. Cancer statistics in China, 2015. CA Cancer J Clin. 2016;66:115–132. doi: 10.3322/caac.21338. [DOI] [PubMed] [Google Scholar]
- 3.Fahy B, Bold RJ. Epidemiology and molecular genetics of colorectal cancer. Surg Oncol. 1998;7:115–123. doi: 10.1016/s0960-7404(99)00021-3. [DOI] [PubMed] [Google Scholar]
- 4.Ishikawa T, Zhang SS, Qin X, Takahashi Y, Oda H, Nakatsuru Y, Ide F. DNA repair and cancer: lessons from mutant mouse models. Cancer Sci. 2004;95:112–117. doi: 10.1111/j.1349-7006.2004.tb03190.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hoeijmakers JH. Genome maintenance mechanisms for preventing cancer. Nature. 2001;411:366–374. doi: 10.1038/35077232. [DOI] [PubMed] [Google Scholar]
- 6.Wood RD, Mitchell M, Sgouros J, Lindahl T. Human DNA repair genes. Science. 2001;291:1284–1289. doi: 10.1126/science.1056154. [DOI] [PubMed] [Google Scholar]
- 7.Leibeling D, Laspe P, Emmert S. Nucleotide excision repair and cancer. J Mol Histol. 2006;37:225–238. doi: 10.1007/s10735-006-9041-x. [DOI] [PubMed] [Google Scholar]
- 8.Constantinou A, Gunz D, Evans E, Lalle P, Bates PA, Wood RD, Clarkson SG. Conserved residues of human XPG protein important for nuclease activity and function in nucleotide excision repair. J Biol Chem. 1999;274:5637–5648. doi: 10.1074/jbc.274.9.5637. [DOI] [PubMed] [Google Scholar]
- 9.De Weerd-Kastelein EA, Keijzer W, Bootsma D. Genetic heterogeneity of xeroderma pigmentosum demonstrated by somatic cell hybridization. Nat New Biol. 1972;238:80–83. doi: 10.1038/newbio238080a0. [DOI] [PubMed] [Google Scholar]
- 10.Emmert S, Schneider TD, Khan SG, Kraemer KH. The human XPG gene: gene architecture, alternative splicing and single nucleotide polymorphisms. Nucleic Acids Res. 2001;29:1443–1452. doi: 10.1093/nar/29.7.1443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gillet LC, Scharer OD. Molecular mechanisms of mammalian global genome nucleotide excision repair. Chem Rev. 2006;106:253–276. doi: 10.1021/cr040483f. [DOI] [PubMed] [Google Scholar]
- 12.Lin J, Swan GE, Shields PG, Benowitz NL, Gu J, Amos CI, de Andrade M, Spitz MR, Wu X. Mutagen sensitivity and genetic variants in nucleotide excision repair pathway: genotype-phenotype correlation. Cancer Epidemiol Biomarkers Prev. 2007;16:2065–2071. doi: 10.1158/1055-9965.EPI-06-1041. [DOI] [PubMed] [Google Scholar]
- 13.Scherly D, Nouspikel T, Corlet J, Ucla C, Bairoch A, Clarkson SG. Complementation of the DNA repair defect in xeroderma pigmentosum group G cells by a human cDNA related to yeast RAD2. Nature. 1993;363:182–185. doi: 10.1038/363182a0. [DOI] [PubMed] [Google Scholar]
- 14.Lee SK, Yu SL, Prakash L, Prakash S. Requirement of yeast RAD2, a homolog of human XPG gene, for efficient RNA polymerase II transcription. implications for Cockayne syndrome. Cell. 2002;109:823–834. doi: 10.1016/s0092-8674(02)00795-x. [DOI] [PubMed] [Google Scholar]
- 15.Barreto G, Schafer A, Marhold J, Stach D, Swaminathan SK, Handa V, Doderlein G, Maltry N, Wu W, Lyko F, Niehrs C. Gadd45a promotes epigenetic gene activation by repair-mediated DNA demethylation. Nature. 2007;445:671–675. doi: 10.1038/nature05515. [DOI] [PubMed] [Google Scholar]
- 16.Zhou RM, Niu CX, Wang N, Liu L, Huang X, Chen ZF, Huo XR, Hao YL, Li Y. XPG gene polymorphisms and the risk of gastric cardia adenocarcinoma. Genet Test Mol Biomarkers. 2016;20:432–437. doi: 10.1089/gtmb.2015.0333. [DOI] [PubMed] [Google Scholar]
- 17.He J, Qiu LX, Wang MY, Hua RX, Zhang RX, Yu HP, Wang YN, Sun MH, Zhou XY, Yang YJ, Wang JC, Jin L, Wei QY, Li J. Polymorphisms in the XPG gene and risk of gastric cancer in Chinese populations. Hum Genet. 2012;131:1235–1244. doi: 10.1007/s00439-012-1152-8. [DOI] [PubMed] [Google Scholar]
- 18.Hua RX, Zhuo ZJ, Zhu J, Jiang DH, Xue WQ, Zhang SD, Zhang JB, Li XZ, Zhang PF, Jia WH, Shen GP, He J. Association between genetic variants in the XPG gene and gastric cancer risk in a southern Chinese population. Aging (Albany NY) 2016;8:3311–3320. doi: 10.18632/aging.101119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mirecka A, Paszkowska-Szczur K, Scott RJ, Gorski B, van de Wetering T, Wokolorczyk D, Gromowski T, Serrano-Fernandez P, Cybulski C, Kashyap A, Gupta S, Golab A, Slojewski M, Sikorski A, Lubinski J, Debniak T. Common variants of xeroderma pigmentosum genes and prostate cancer risk. Gene. 2014;546:156–161. doi: 10.1016/j.gene.2014.06.026. [DOI] [PubMed] [Google Scholar]
- 20.Ma SH, Ling FH, Sun YX, Chen SF, Li Z. Investigation on the role of XPG gene polymorphisms in breast cancer risk in a Chinese population. Genet Mol Res. 2016;15 doi: 10.4238/gmr.15028066. [DOI] [PubMed] [Google Scholar]
- 21.Hua RX, Zhuo ZJ, Zhu J, Zhang SD, Xue WQ, Zhang JB, Xu HM, Li XZ, Zhang PF, He J, Jia WH. XPG gene polymorphisms contribute to colorectal cancer susceptibility: a two-stage case-control study. J Cancer. 2016;7:1731–1739. doi: 10.7150/jca.15602. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhu ML, Wang M, Cao ZG, He J, Shi TY, Xia KQ, Qiu LX, Wei QY. Association between the ERCC5 Asp1104His polymorphism and cancer risk: a meta-analysis. PLoS One. 2012;7:e36293. doi: 10.1371/journal.pone.0036293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Zhao J, Chen S, Zhou H, Zhang T, Liu Y, He J, Zhu J, Ruan J. XPG rs17655 G > C polymorphism associated with cancer risk: evidence from 60 studies. Aging (Albany NY) 2018;10:1073–1088. doi: 10.18632/aging.101448. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.He J, Liao XY, Zhu JH, Xue WQ, Shen GP, Huang SY, Chen W, Jia WH. Association of MTHFR C677T and A1298C polymorphisms with non-Hodgkin lymphoma susceptibility: evidence from a meta-analysis. Sci Rep. 2014;4:6159. doi: 10.1038/srep06159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.He J, Wang F, Zhu JH, Chen W, Cui Z, Jia WH. No association between MTR rs1805087 A > G polymorphism and non-Hodgkin lymphoma susceptibility: evidence from 11 486 subjects. Leuk Lymphoma. 2015;56:763–767. doi: 10.3109/10428194.2014.935370. [DOI] [PubMed] [Google Scholar]
- 26.Du H, Zhang X, Du M, Guo N, Chen Z, Shu Y, Zhang Z, Wang M, Zhu L. Association study between XPG Asp1104His polymorphism and colorectal cancer risk in a Chinese population. Sci Rep. 2014;4:6700. doi: 10.1038/srep06700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Bigler J, Ulrich CM, Kawashima T, Whitton J, Potter JD. DNA repair polymorphisms and risk of colorectal adenomatous or hyperplastic polyps. Cancer Epidemiol Biomarkers Prev. 2005;14:2501–2508. doi: 10.1158/1055-9965.EPI-05-0270. [DOI] [PubMed] [Google Scholar]
- 28.Huang WY, Berndt SI, Kang D, Chatterjee N, Chanock SJ, Yeager M, Welch R, Bresalier RS, Weissfeld JL, Hayes RB. Nucleotide excision repair gene polymorphisms and risk of advanced colorectal adenoma: XPC polymorphisms modify smoking-related risk. Cancer Epidemiol Biomarkers Prev. 2006;15:306–311. doi: 10.1158/1055-9965.EPI-05-0751. [DOI] [PubMed] [Google Scholar]
- 29.Pardini B, Naccarati A, Novotny J, Smerhovsky Z, Vodickova L, Polakova V, Hanova M, Slyskova J, Tulupova E, Kumar R, Bortlik M, Barale R, Hemminki K, Vodicka P. DNA repair genetic polymorphisms and risk of colorectal cancer in the Czech republic. Mutat Res. 2008;638:146–153. doi: 10.1016/j.mrfmmm.2007.09.008. [DOI] [PubMed] [Google Scholar]
- 30.Joshi AD, Corral R, Siegmund KD, Haile RW, Le Marchand L, Martinez ME, Ahnen DJ, Sandler RS, Lance P, Stern MC. Red meat and poultry intake, polymorphisms in the nucleotide excision repair and mismatch repair pathways and colorectal cancer risk. Carcinogenesis. 2009;30:472–479. doi: 10.1093/carcin/bgn260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Canbay E, Cakmakoglu B, Zeybek U, Sozen S, Cacina C, Gulluoglu M, Balik E, Bulut T, Yamaner S, Bugra D. Association of APE1 and hOGG1 polymorphisms with colorectal cancer risk in a Turkish population. Curr Med Res Opin. 2011;27:1295–1302. doi: 10.1185/03007995.2011.573544. [DOI] [PubMed] [Google Scholar]
- 32.Gil J, Ramsey D, Stembalska A, Karpinski P, Pesz KA, Laczmanska I, Leszczynski P, Grzebieniak Z, Sasiadek MM. The C/A polymorphism in intron 11 of the XPC gene plays a crucial role in the modulation of an individual’s susceptibility to sporadic colorectal cancer. Mol Biol Rep. 2012;39:527–534. doi: 10.1007/s11033-011-0767-5. [DOI] [PubMed] [Google Scholar]
- 33.Liu D, Wu HZ, Zhang YN, Kang H, Sun MJ, Wang EH, Yang XL, Lian MQ, Yu ZJ, Zhao L, Olopade OI, Wei MJ. DNA repair genes XPC, XPG polymorphisms: relation to the risk of colorectal carcinoma and therapeutic outcome with Oxaliplatin-based adjuvant chemotherapy. Mol Carcinog. 2012;51(Suppl 1):E83–93. doi: 10.1002/mc.21862. [DOI] [PubMed] [Google Scholar]
- 34.Steck SE, Butler LM, Keku T, Antwi S, Galanko J, Sandler RS, Hu JJ. Nucleotide excision repair gene polymorphisms, meat intake and colon cancer risk. Mutat Res. 2014;762:24–31. doi: 10.1016/j.mrfmmm.2014.02.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paszkowska-Szczur K, Scott RJ, Gorski B, Cybulski C, Kurzawski G, Dymerska D, Gupta S, van de Wetering T, Masojc B, Kashyap A, Gapska P, Gromowski T, Kladny J, Lubinski J, Debniak T. Polymorphisms in nucleotide excision repair genes and susceptibility to colorectal cancer in the Polish population. Mol Biol Rep. 2015;42:755–764. doi: 10.1007/s11033-014-3824-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Sun K, Gong A, Liang P. Predictive impact of genetic polymorphisms in DNA repair genes on susceptibility and therapeutic outcomes to colorectal cancer patients. Tumour Biol. 2015;36:1549–1559. doi: 10.1007/s13277-014-2721-3. [DOI] [PubMed] [Google Scholar]
- 37.Kabzinski J, Przybylowska K, Dziki L, Dziki A, Majsterek I. An association of selected ERCC2 and ERCC5 genes polymorphisms, the level of oxidative DNA damage and its repair efficiency with a risk of colorectal cancer in Polish population. Cancer Biomark. 2015;15:413–423. doi: 10.3233/CBM-150488. [DOI] [PubMed] [Google Scholar]
- 38.Friedberg EC, Bond JP, Burns DK, Cheo DL, Greenblatt MS, Meira LB, Nahari D, Reis AM. Defective nucleotide excision repair in xpc mutant mice and its association with cancer predisposition. Mutat Res. 2000;459:99–108. doi: 10.1016/s0921-8777(99)00068-3. [DOI] [PubMed] [Google Scholar]
- 39.Friedberg EC. DNA damage and repair. Nature. 2003;421:436–440. doi: 10.1038/nature01408. [DOI] [PubMed] [Google Scholar]
- 40.O’Donovan A, Davies AA, Moggs JG, West SC, Wood RD. XPG endonuclease makes the 3’ incision in human DNA nucleotide excision repair. Nature. 1994;371:432–435. doi: 10.1038/371432a0. [DOI] [PubMed] [Google Scholar]
- 41.Wakasugi M, Reardon JT, Sancar A. The non-catalytic function of XPG protein during dual incision in human nucleotide excision repair. J Biol Chem. 1997;272:16030–16034. doi: 10.1074/jbc.272.25.16030. [DOI] [PubMed] [Google Scholar]
- 42.Sawyer SL, Mukherjee N, Pakstis AJ, Feuk L, Kidd JR, Brookes AJ, Kidd KK. Linkage disequilibrium patterns vary substantially among populations. Eur J Hum Genet. 2005;13:677–686. doi: 10.1038/sj.ejhg.5201368. [DOI] [PubMed] [Google Scholar]


