Abstract
Introduction.
Repeated measles outbreaks in countries with relatively high vaccine coverage are mainly due to failure to vaccinate and importation; however, cases in immunized individuals exist and raise questions about suboptimal measles vaccine-induced humoral immunity and/or waning immunity in a low measles-exposure environment.
Areas covered.
The plaque reduction neutralization measurement of functional measles-specific antibodies correlates with protection is the gold standard in measles serology, but it does not assess cellular-immune or other parameters that may be associated with durable and/or protective immunity after vaccination. Additional correlates of protection and long-term immunity and new determinants/signatures of vaccine responsiveness such as specific CD46 and IFI44L genetic variants associated with neutralizing antibody titers after measles vaccination, are under investigation. Current and future systems biology studies, coupled with new technology/assays and analytical approaches, will lead to an increasingly sophisticated understanding of measles vaccine-induced humoral immunity and will identify “signatures” of protective and durable immune responses.
Expert Commentary.
This will translate into the development of highly predictive assays of measles vaccine efficacy, effectiveness, and durability for prospective identification of potential low/non-responders and susceptible individuals who require additional vaccine doses. Such new advances may drive insights into the development of new/improved vaccine formulations and delivery systems.
Keywords: Measles, Measles Vaccine, Measles-Mumps-Rubella Vaccine, Genetic Association Sudies, Genetic Variation, Gene Polymorphisms, Gene Expression, Systems Biology, Antibody, Immunity, Humoral, Immunity
1. Measles in high vaccine coverage settings and measles vaccine failure.
Measles is a highly contagious disease, which has been eliminated in the United States for more than 18 years. It is still a common threat in the underdeveloped world with potentially life-threatening sequelae and over 450 known pediatric deaths each day [1]. It was estimated that over 7 million people were infected with measles worldwide in 2016 with 89,780 reported measles-related deaths [2]. Measles outbreaks have been limited in the United States during the last few years and are mainly a result of importation and predominantly involve unvaccinated individuals who travel, as well as their contacts (e.g., 86 reported measles cases in 2016; 118 measles cases in 2017; and 124 measles cases from 22 states in 2018, as of August 11, 2018) [3, 4]. In 2017, the World Health Organization announced a four-fold rise in the number of measles cases in Europe (23,927 cases), with large outbreaks affecting more than 15 European countries [5, 6]. Furthermore, over 41,000 measles cases were reported in Europe for the first half of 2018 (the most affected countries were France, Georgia, Greece, Italy, the Russian Federation, Serbia, and Ukraine), with at least 37 reported pediatric deaths [5, 6]. This resurgence of measles is primarily due to failure to vaccinate and sustain high immunization coverage with the two-dose measles vaccination schedule in the affected countries/regions. However, this is an indication that measles is and will continue to be a public health concern for both developing and developed countries for the foreseeable future.
Long-term sequelae of measles are more serious and common than previously thought. A recent study assessing national-level information from England, Wales, the United States, and Denmark, from both the pre- and the post-vaccine era, provided statistical/modeling evidence for the association of measles with the long-term increase (approximately 2- to 3-year observed impact) of non-measles infectious disease mortality in children [7]. These non-specific effects of measles on immunity to other pathogens are likely due to measles-induced immunosuppression from lymphocyte depletion of memory B and T lymphocytes and/or from measles-related functional immune impairment [8–11] . Recent research also re-estimates the rate of developing subacute sclerosing panencephalitis/SSPE, a fatal progressive inflammation of the brain resulting from persistent measles virus (MV) infection, to be 1 in 609 after measles disease occurring in infancy [12].
To prevent persisting measles endemicity and target measles for global eradication, achieving and sustaining herd immunity of at least 90–95% is required [13–17]. It is generally accepted that the live attenuated measles vaccine has a high protective efficacy, particularly after two vaccine doses (although it may be lower and not life-long compared to the wild type virus infection), and a field effectiveness of 94.1% (IQR, 88.3%-98.3%) after two doses [18]. Accordingly, measles mainly affects unvaccinated individuals. It is also indisputable that a two-dose measles vaccination program must be implemented and sustained globally to reduce measles morbidity/mortality and achieve measles eradication. What is still a subject of debate is the ability of the current measles vaccine to sustain long-term protective immunity and adequate herd immunity in settings with no wild type virus exposure (i.e., no boosting of immunity resulting from asymptomatic infection).
Primary measles vaccine failure (a failure to develop protective immunity after vaccination) is not uncommon (approximately 2 – 12% for children immunized at/around one year of age [19–21]) and can be partly managed by the administration of a second dose of vaccine and by increasing the age at first vaccination (in regions with low measles incidence) to ensure immune system maturity and loss of maternally-acquired antibodies [22]. The assessment of secondary vaccine failure (waning immunity or failure to sustain protective immunity over time) can be difficult and requires long-term monitoring of measles vaccine-induced adaptive immunity after the first and second vaccine doses and ideally vaccine efficacy data, immunogenicity data and epidemiological information on measles cases/outbreaks in the geographical area.
Annual measles outbreaks in high vaccine coverage settings also continues to occur. Although these outbreaks typically involve importation and mostly affect unvaccinated individuals, they also reveal surprisingly high numbers of vaccine failure among one- and two-dose recipients of measles-containing vaccine who were infected [15–17, 22–53]. In a recently published study of registered laboratory-confirmed measles cases in California between 2000 and 2015, Cherry and Zahn [51] report 232 measles cases with a documented vaccine history, of which 9% (20 cases) were after one dose and 11% (26 cases) were after two doses of measles-containing vaccine (median 16.7 years after last vaccination, range 6 to 23.6 years after last vaccination), indicating the likely occurrence of waning immunity [51]. Earlier studies suggest a secondary measles vaccine failure rate of ~5%, approximately 10–15 years after the second immunization [31, 54]. Previous studies have also reported a combination of laboratory methods (measles plaque reduction neutralization assay [PRN], measles IgG avidity assays, and detection of MV RNA by RT-qPCR) and criteria for classification of measles reinfection cases and secondary vaccine failure [55, 56]. Larger and more sophisticated studies are still needed to more precisely estimate secondary vaccine failure rates (waning immunity) in low measles incidence (high vaccine coverage) settings.
2. Immune measures and correlates of protection after measles vaccination.
Measles virus cell entry and infection is mediated by the known MV-specific cellular receptors SLAM/CD 150, nectin-4/NECTIN4/PVRL4 and CD46 (operational only for attenuated MV strains) [57–60]. Wild type and attenuated measles viruses elicit differential innate/inflammatory immune responses (NFκB signaling/activation of the NLRP3 inflammasome with no detectable interferon type 1 response for the wild type MV strains and detectable induction of interferon-stimulated genes [ISGs] for the attenuated MV strains, respectively), but the implications of these differences for the development of adaptive immunity are unclear [61–63]. Protective immunity to measles is accomplished by high-avidity neutralizing antibodies directed to the MV surface glycoproteins, primarily the hemagglutinin/H protein (with a modest contribution of antibodies to the fusion/F protein), which effectively neutralize SLAM-using wild type MV infection of lymphocytes [54, 61, 64–67]. Measles vaccine elicits both neutralizing and non-neutralizing antibodies against different MV proteins, as well as measles-specific cellular immunity, with limited correlation between measures of the humoral and cellular arms of immunity [68]. CD4+ T cells are not essential, but they (in particular, the follicular T-helper/Tfh cells in the lymph nodes) can facilitate protective humoral immunity by providing help for the formation of germinal centers, for the activation and differentiation of B cells, and for isotype switching and affinity maturation in antibody-secreting cells/ASCs [69]. CD8+ T cells are considered important for viral clearance [61]. Functional measles-specific neutralizing antibodies after vaccination (anti-H and anti-F) are quantified using a classical plaque reduction neutralization test (PRN) or its improved automated version, the plaque reduction microneutralization (PRMN) assay,[70–72] in specialized laboratory settings by trained personnel. Routinely, these assays rely on CD46-mediated in vitro infection of Vero cells (that do not express human SLAM) with attenuated Edmonston-based MV strains; and for these reasons, the results may not fully reflect the protective antibody efficacy upon wild virus measles exposure [54, 73].
With the above taken into consideration, the currently accepted correlate of protection against measles is a PRN titer of MV-specific neutralizing antibodies >120 (or >120 mIU/mL), which predicts protection from clinical disease [64, 71, 72, 74]. It has been repeatedly shown that serum antibodies (e.g., passively transferred immunoglobulins or transplacentally acquired antibodies) are sufficient to confer protection from measles [54, 61, 64–67]. However, it has been demonstrated that subjects with low/undetectable PRN antibody levels may still be protected from clinical measles, suggesting a role for cellular immunity in protection [54, 61, 64–67]. In addition to the PRN assay, seroprevalence studies assessing measles vaccine-induced humoral immunity apply an array of other assays (reviewed in [75–81], including a variety of automated commercial immunoassays (e.g., multiplex microsphere/bead fluorescence-based immunoassays) and microtiter-plate enzyme-linked immunoassays (EIA) reporting qualitative and/or quantitative results. Among the most commonly used are the Enzygnost® Anti-Measles Virus/IgG EIA (Siemens Health Care Diagnostics GmbH, Marburg, Germany) and the Serion Measles IgG EIA (Institut Virion\Serion GmbH, Würzburg, Germany) [75]. With few exceptions (the Enzygnost® Anti-Measles Virus/IgG), these assays were not calibrated against the 2nd WHO international measles standard (the 3rd WHO international measles standard is not currently recommended for EIA calibration[82]). More importantly, EIA assays measure antibodies to other abundant MV proteins (e.g., the N protein in addition to H and F) and have limited ability to measure antibodies to conformational epitopes; thus, their utility for categorization of individuals into immune or non-immune and assessing potential measles susceptibility is limited, particularly at the lower range of antibody titer [75]. A recently developed measles-specific assay using proteome microarray (antibody array) technology successfully detected antibodies against five MV proteins (H, F, N, P and L) in recipients of measles vaccine, and the measures/patterns were well correlated with the neutralizing antibody response [83].
Antibody avidity assays are also emerging as useful tests for distinguishing primary from secondary humoral immune response during measles outbreaks in high vaccine coverage settings. Avidity is defined as the cumulative strength of attachment/binding of multivalent antibodies to multivalent antigens. A new MV-specific IgG avidity assay was developed by the CDC using a modified commercial EIA assay with the use of the denaturant diethylamine (DEA), and validated with a panel of reference serum samples [84]. This assay provides useful thresholds for classification of antibodies into high/low or intermediate avidity and can supplement IgM antibody assays in the serological assessment of measles cases, as well as facilitate the classification of secondary vaccine failures [84].
Measurement of other markers of measles-specific humoral immunity are also currently being introduced and used, but their outputs (immune outcomes) are not generally accepted as correlates or surrogates of protection against measles. The memory B cell ELISPOT assay uses peripheral blood mononuclear cells (PBMCs) or purified B cells to provide a quantitative measure of the frequencies of pre-existing measles-specific memory B cells after non-specific polyclonal B cell stimulation followed by enumeration of antigen-specific ASCs [85–89]. Similarly, a plasmablast ELISPOT assay (without non-specific B cell stimulation) directly measures the frequencies of the antigen-specific circulating plasmablasts that peak around day 7 after vaccination [90]. There has been some controversy in the literature over the correlation of measles-specific antibody titers with the frequencies of measles-specific memory B cells and the use of the latter as a predictor of the duration of antibody response/antibody waning [87–89]. Memory B cells are important for a prompt humoral response upon antigen re-exposure, but most likely long-term measles-specific antibody production is maintained by antigen-specific long-lived plasma cells in the bone marrow rather than reactivation/differentiation of memory B cells [91–95]. No feasible assay for large-scale studies exists today to reliably measure the quantity and characteristics of antigen-specific long-lived plasma cells (as well as the antigen-specific Tfh cells) in humans due to their specific niche/homing (ideally assessment requires bone marrow and lymph node biopsies). Assessment of measles vaccine-induced humoral immunity in vulnerable populations (particularly in pediatric patients suffering from chronic infections and/or immunosuppression) is also important for the maintenance of measles control and eradication/elimination efforts for the general population. A study assessing B cell compartment immunity in 70 HIV-1-infected children established the importance of early antiretroviral therapy for the maintenance of long-term immunity (measles-specific memory B cell frequencies and protective measles antibody titers) after routine vaccinations [96]. Similarly, other conditions associated with immunosuppression (e.g., transplantation and primary immunodeficiencies) often lead to impaired development and/or waning of measles vaccine-induced immunity [97–99]. Monitoring of measles immunity in such cases is critical for the optimization of population vaccination strategies and maintenance of long-term protection against measles. Addressing the current knowledge and public health gap, as well as barriers to measles elimination activities, requires new approaches in assessing and predicting humoral immune response after measles vaccination in order to prospectively identify vaccine responders and non- or low-responders and/or potentially measles susceptible individuals whose protective titers wane over time. Such approaches and perspectives are in line with the WHO 2012–2020 Global Measles and Rubella Strategic Elimination Plan and the Midterm Review which recommended “research on susceptibility profiles for measles, and research related to outbreaks in high vaccine coverage settings [100], and are reviewed in the sections below with a focus on identifying determinants/signatures of measles vaccine-induced humoral immunity.
3. Contribution of HLA and candidate immune response genes to measles vaccine-induced variations in humoral immunity.
Over the last 23 years, our group has investigated the wide inter-individual variation in circulating humoral antibody responses after routine measles vaccination in highly immunized healthy populations and has systematically defined genetic contributions to inter-individual immune response variations and vaccine failure (reviewed in [17, 49]).
Both host genetics and environmental factors contribute to variability in immune responses to vaccines. Among the host genetic determinants that are involved in protective immunity against measles are the highly polymorphic HLA and non-HLA genetic variants. In this regard, HLA allelic associations with humoral immune responses after measles vaccination have been studied in detail. Measles vaccine-induced immunity can involve strong HLA class I- and class II-restricted CD8+ and CD4+ T cell immune responses. Some key HLA class I (B*57:01, B*35:03) and class II (DQB1*06:02, DQB1*03:03, DB1*07:01 and DRB1:15:01) alleles have also shown confirmed associations with inter-individual variations in measles antibody responses after two doses of measles vaccine [101]. Furthermore, in a large cohort of 2,506 healthy immunized subjects (age 11 to 41 years), specific class I and class II HLA types—such as B*57:01, DQB1*06:02, and DRB1*15:05—have been clearly associated with measles vaccine-specific neutralizing antibody titers [102]. These reproducible associations between HLA molecules and immune response outcomes have led to the identification of measles virus epitopes presented by HLA [103–105]. Such synthetic peptides/epitopes can be used to design personalized measles vaccines [106, 107].
Multiple population-based vaccine studies have also demonstrated associations between candidate genes/SNPs and variations in measles vaccine-induced immune responses, including vaccine non-response and vaccine failure [17, 26, 101, 106, 108–124]. One study included healthy children (n=745) who received 2 doses of measles vaccine and were genotyped for a panel of innate SNP markers, such as vitamin A (RARA, RARB, and RARG), and vitamin D receptor (RXRA) genes, a transmembrane receptor CD46, CD209 (DC-SIGN), host antiviral sensor and effector (VISA, DDX58, OAS1-3, MX2, ADAR), TRIM (TRIM 5, 22, 25) and TLR (TLR2,3,4,7,8) genes. Multiple polymorphisms and haplotypes in these genes have been found to be significantly associated with humoral and/or cellular immune response markers [109, 112–114, 125], and some were subsequently replicated (e.g., the CD46 rs2724384 genetic variant [126, 127]). These findings point to additional non-HLA genetic variants/genes as being critical determinants modulating the adaptive immune responses to measles vaccine. Likewise, multigenic effects on measles vaccine-induced immunity have been examined using a large collection of SNPs (n=1,912) that tag 126 candidate genes. Combined analyses of all these SNPs provided evidence that a multigenic model may explain variations in antibody levels (p=0.05) and in cell-mediated IFN-g ELISPOT response (p=0.02) variance [110]. The genetic studies reported above have identified important and informative genetic determinants of measles vaccine response heterogeneity and led to a large-scale state-of-the-art genome-wide association study (GWAS) that allowed for identification of additional genetic determinants (SNPs and immune response pathways) of measles vaccine-induced immunity [126, 128].
4. Genome-wide genetic association studies and measles vaccination: from genetic association to function of genetic determinants of humoral immunity.
The advances in technology and statistical analysis during the last decade have allowed for enhanced genome-wide interrogation of the human genome (i.e., GWAS studies) for identification of determinants of host response to measles vaccination in an unbiased (by prior knowledge) way [126, 129–131].
In a sophisticated GWAS in children, Feenstra et al. [129] identified two genetic loci on chromosome 1 that were associated with febrile seizures after measles-mumps-rubella (MMR) vaccination but not with unrelated febrile seizures. These two genetic loci harbor the interferon-stimulated gene IFI44L and the measles virus receptor gene (for attenuated MV strains) CD46.
In a population-based study of 2,872 healthy subjects (age 11–41 years) who had received 2 doses of MMR vaccine, we performed measles neutralizing antibody titer assays after a second vaccination (median 3.4 years). After correcting for multiple confounding variables (e.g., age, time since last MMR vaccination, etc.) and excluding subjects with conditions affecting immune response, we documented a wide range of MV-specific humoral immunity, with a median Ab titer of 845 mIU/mL (IQR: 394 to 1,683). Among these subjects, 94 subjects (3.3%) had non-protective levels of circulating neutralizing antibody (<120 mIU/mL), and 338 subjects (11.8%) had neutralizing antibody <210 mIU/mL, which corresponds to a PRN neutralizing dose/ND50 titer of 120 [132]. These data and other recent reports from the literature [22, 51, 52, 133] raise questions about suboptimal measles vaccine-induced humoral immunity and/or waning immunity among highly vaccinated populations in a low measles exposure environment.
The above cohort was used to assess for genetic factors contributing to MV-specific neutralizing antibody response in the first GWAS study of measles vaccine-induced immunity, which estimated the heritability of vaccine-induced measles antibody titers to be ~49% [126, 127]. This GWAS (unrelated to the GWAS by Feenstra B. et al., on febrile seizures [129]) independently identified the two chromosome 1 genetic regions (the 1q31.1 region harboring the IFI44L gene with 9 significant SNPs; and the 1q32 region harboring the CD46 gene with 20 significant non-coding SNPs) to be associated with the measured MV-specific neutralizing antibody titer after MMR vaccination. Several overlapping SNP associations were found between the two studies (i.e., the intergenic SNP rs1318653 near the CD46 locus and the coding IFI44L His73Arg SNP 273259) in concert with multiple SNP associations with immune responses not reported previously [126]. The top SNP association in subjects of European ancestry is an intronic genetic variant rs2724374 (p-value = 4.88×10−09), located near the CD46 intron 8-exon 8 boundary, which is demonstrated to affect/cause genetic splicing (the skipping of the CD46 serine/threonine/proline-rich/STP B exon), resulting in differential abundance of CD46 isoforms associated with different genotypes [126, 134, 135]. The minor allele of this SNP (G) was significantly associated in a dose-response dependent manner with ~50% reduction in MV neutralizing antibody titer after vaccination [126]. This allele is likely responsible for the splicing of exon B to favor the generation of CD46 isoforms with a shorter (and less O-glycosylated) STP region, as demonstrated by a genotype-specific RT-PCR isoform analysis and a DEXSeq analysis of NGS data [126]. We and others have suggested that MV binding and fusion capacity in cells expressing the CD46 C1/C2 (shorter) isoforms vs. the BC1/BC2 (longer) isoforms differ, and this variance may translate to differences in both viral replication and triggering of immune response pathways after live virus vaccination [126, 136–138]. The functional effects of the CD46 rs2724374 genetic variant are summarized in Fig. 1. Ongoing functional studies in human cells expressing/overexpressing different CD46 isoforms demonstrate clear difference in MV infection/replication and innate immune pathway activation depending on the prevalence of specific CD46 isoforms (Haralambieva and Poland, unpublished data). Lastly, the presence/absence of STP exons (associated also with difference in the O-glycosylation) in the extracellular portion of CD46 can result in altered processing, altered CD46 shedding/cleavage by metalloproteinases, altered downregulation and cell surface expression, and profound differences in T cell function and TCR signaling [139, 140].
Fig. 1.
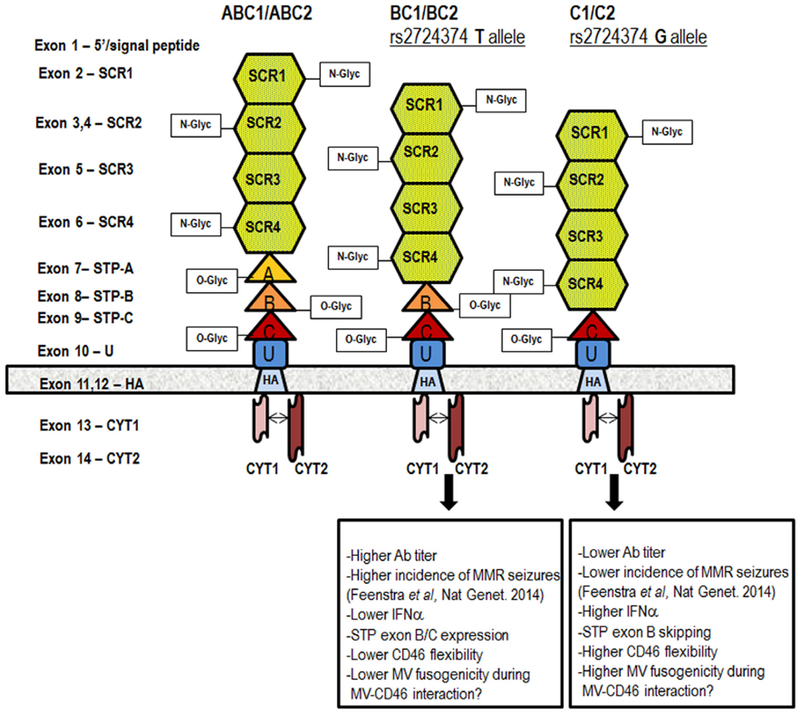
Measles virus receptor CD46 and functional effects of CD46 rs2724374
The figure above is published with permission from Human Genetics. [126, 127] The extracellular portion of CD46 consists of four N-glycosylated conserved short consensus repeats SCR1-4; a STP region that is O-glycosylated (encoded by exons 7/A, 8/B and 9/C); and a region of unknown function (U). The four most common CD46 isoforms are defined based on the present STP exon/exons and the cytoplasmic tail (CYT1 or CYT2): BC1 and BC2 (with B and C exons/domains in the STP and with either CYT1 or CYT2), and C1 and C2 (with C exon/domain in the STP and with either CYT1 or CYT2). The effect of CD46 rs2724374 on CD46 isoform prevalence (exon B expression or skipping), interaction between CD46 and MV, and immune response following measles vaccination is also summarized for the different genotypes.
The discovery of CD46 and IFI44L genetic variants as determinants of measles vaccine-induced humoral immunity (and adverse events) after measles vaccination could translate into the development of inexpensive chips/platforms for prospective identification of potential non-responders and susceptible individuals who will eventually need additional vaccine doses, as well as improved vaccines capable of overcoming any genetic restrictions.
5. Statistical challenges and solutions for analyzing high-dimensional data (genetic association studies, GWAS).
It is well known that immune response is strongly influenced by both genetic and environmental factors. Because the immune system is fine-tuned [141], individual genetic factors tend to have small effects on immune response, making it statistically challenging to uncover the main causes. To overcome this challenge, careful study design and large sample sizes are crucial. The design of studies should control for factors known to influence response, such as age at immunization, sex, prior vaccinations, etc. It is also critical to control for laboratory batch effects of assay measurements, and to normalize response to make unbiased comparisons [142]. Although there are multiple genes known to influence immune response, such as genes in the HLA region, the agnostic approach of GWAS offer the advantage of new discoveries, albeit at the requirement of large sample sizes.
GWAS studies [143] have achieved enormous success at identifying the genomic regions that harbor genetic determinants of complex traits [144–150], with over 2,000 traits registered in the Catalog of published genome-wide association studies with association p-values less than 10−5 with single nucleotide polymorphisms [151]. This tremendous success can be attributed to the large sample sizes required to have sufficient power to detect small effects of genes, as well as inexpensive genotyping microarrays that contain a large number of SNPs. Furthermore, large-scale reference panels provide a way to reliably impute SNPs that are close to the SNPs that are measured in microarrays [152], resulting in approximately 10 million SNPs frequently available for analyses.
To summarize GWAS p-values that measure the marginal association of one SNP at a time with a trait, Manhattan plots of p-values are frequently used, followed with LocusZoom plots for regions of interest [153]. This provides a way to focus on the SNPs with the smallest (i.e., most significant) p-values in distinct regions. The SNPs with the smallest p-values are sometimes called the lead or index SNPs. Because there are many SNPs tested for their association with a trait, GWAS results are most reliable when SNP associations achieve the accepted genome-wide statistical significance threshold of p-value<5×10−8 [154, 155].
A caution about the lead SNP is that there is a reasonable chance that it does not have a direct causal effect on the trait [156]. Rather, the lead SNP is often in linkage disequilibrium/LD with an unmeasured causal variant [157]. This is because the SNPs on microarrays, called tag-SNPs, are chosen because they serve as surrogates for large genomic regions. Their ability to be faithful surrogates stems from their high correlation with neighboring unmeasured SNPs (i.e., high LD) [158, 159]. This means that the association between a tag-SNP and a trait can be indirect, resulting from a tag-SNP statistically associated with an unmeasured causal SNP and the causal SNP having a direct effect on the trait. To increase the density of SNPs, hopefully capturing the causal variant or at least refining its location, statistical imputation of neighboring SNPs is a widely accepted technique [160, 161].
The patterns of LD among SNPs can be complex. This makes it challenging to determine the underlying causal variants by inspecting the marginal association of a trait with one SNP at a time. Statistical methods that jointly analyze all the SNPs in a region, particularly methods that are designed for fine-mapping [162, 163], are useful to prioritize SNPs for subsequent functional studies. Additional insights can be gained by genomic annotation of SNPs that assign biological function based on publicly available resources [164–168].
Many of the trait-associated SNPs discovered by GWAS are not in gene-coding regions; rather, they map to non-coding regions, often in areas enriched for regulatory elements, such as enhancers, promoters, insulators, and silencers [169]. This suggests that SNPs discovered by GWAS influence the amount of expression of nearby genes (referred to as expression quantitative trait loci; eQTL), and this altered expression ultimately influences the trait. Statistical methods can be useful to integrate eQTL data (i.e., genes whose RNA levels are associated with specific SNPs) with GWAS data (i.e., traits associated with specific SNPs) in order to quantify the evidence of a causal pathway from SNP to gene-expression to a complex trait. Some methods are based on testing causal models [170], some are based on Mendelian randomization [171], and some are Bayesian approaches [172, 173], but they all have the common strategy of evaluating whether mRNA is a mediator between a SNP and a trait. A variety of approaches are provided elsewhere [162].
6. Gene expression and systems biology-based approaches for the discovery of determinants/signatures of vaccine-induced immunity.
The application of high-dimensional gene expression and/or other omics technologies to reliably identify the determinants/signatures reflecting the development and maintenance of measles vaccine immunity is still in its infancy. Several gene expression studies during the course of measles infection, or after measles vaccination and/or in vitro infection, have provided useful but limited information about the role of specific genes and pathways in the development of measles immunity [62, 174, 175]. Two recent mRNA-Seq studies profiling gene and miRNA expression in the cells of high and low antibody measles vaccine responders have identified essential plasma cell survival and homing factors (e.g., CD93, a key factor for the preservation of plasma cells in the bone marrow and for sustained production of antibodies [176]) and B cell-specific miRNA expression patterns [177] that were associated with neutralizing antibody titer after vaccination.
Systems biology approaches have also yielded important insights into the development of humoral immunity following vaccination. Most systems vaccinology studies focus on the high dimensional analysis of the transcriptome, proteome, metabolome, and/or microbiome. It is equally important to comprehensively evaluate the immune response both in terms of the cell subsets involved and the effector functions produced. This type of approach can be especially useful for pathogens without clearly defined correlates of protection [71], as humoral immunity can be conferred by antigen-specific antibodies with myriad effector functions including: neutralization, complement fixation, opsonization, and enhancement of cellular responses. Each of these effector functions can then be linked to specific gene expression patterns, or “signatures,” necessary for their development. Querec et al. were able to identify a gene signature that predicted neutralizing antibody responses to the yellow fever vaccine with 100% accuracy [178]. This signature involved expression of TNFRSF17, a gene producing the receptor for the BLyS-BAFF B cell growth factor. A similar study across three influenza seasons found that expression levels of CaMKIV were inversely proportional to the titer of hemagglutination inhibiting antibodies [179]. Genomic signatures have also been found to be associated with protection following vaccination with the RTS, S malaria vaccine [180]. Collectively, these and similar studies identify critical pathways necessary for the development of humoral immunity. Further research is needed to identify how these and/or other genes/pathways contribute to measles immunity. The resulting signatures may serve as important predictive biomarkers of immunogenicity or vaccine efficacy.
Novel technologies are allowing researchers to investigate immune responses at an unprecedented level of detail. Mass cytometry (CyTOF®) using heavy-metal conjugated antibodies has expanded the number of parameters one can measure from 12–18 to 40+. This increase in capability enables more comprehensive immune profiling of leukocyte phenotype and higher resolution of functional characteristics (Fig. 2). Similarly, single-cell sequencing (scRNA-Seq) allows us to evaluate individual cellular transcriptomes rather than the average gene expression of all cells in a biological specimen (Fig. 2). One can observe the expression level of transcription factors, cellular receptors, signaling molecules, and other immunologically important genes within relevant cell types [181]. Sequencing of individual T cell and B cell receptors provides information on T/B cell diversity, clonal expansion, and somatic hypermutation [182]. An exciting next step in the field of systems vaccinology will be the combination of multiple single-cell resolution technologies into multi-omics approaches [183]. One example is CITE-Seq (or REAP-Seq), which can be used to simultaneously characterize both surface expression and transcriptome in individual cells, thereby combining the advantages of next-generation sequencing and flow cytometry. One might use these techniques to assess the individual transcriptomes of tetramer-positive cells in order to gain insights into transcriptional activity in antigen-specific cells [184, 185]. Similarly, scMT-Seq or scTrio-Seq allow for the simultaneous analysis of the genome, transcriptome, and epigenome of individual cells for a comprehensive cell-specific snapshot of genetic landscape and activity [186, 187].
Fig. 2.
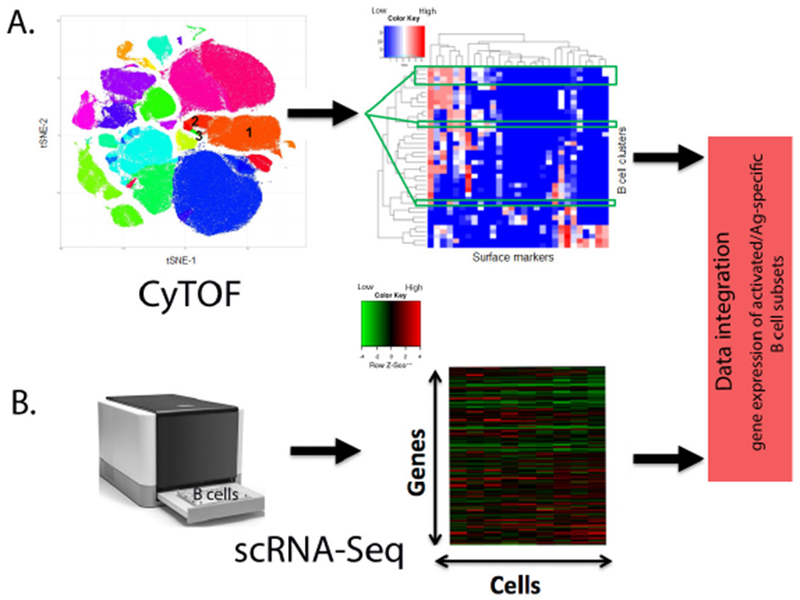
CyTOF and scRNA-Seq analysis of B cell subsets after vaccination.
A) Schematic representation of CyTOF. t-SNE plot of cell clusters defined by cellular markers. The annotation of the numbered cell clusters is as follows: 1. naïve B cells; 2. memory B and 3. plasmablasts. For clarity only three of the relevant B cell clusters are shown. Heat map displaying the expression levels (blue=low, red=high) of each cell surface marker in columns and each B cell cluster of interest in rows. B) Schematic representation of scRNA-Seq. Heat map displaying the gene expression levels (green=low, red=high) within single B cells (assay cell input is purified B cells). Data integration allows for the identification of gene expression signatures within activated and/or antigen-specific B cell subsets after vaccination.
7. Expert commentary.
Given the morbidity and mortality of measles, repeated importations, continuing outbreaks, contraindications to live attenuated measles vaccines for an increasingly immunocompromised population, and unmerited concerns over the safety of the current measles vaccine, it is apparent that new vaccine types are needed.
Unique among vaccine-preventable human pathogens, measles is the most transmissible human disease—requiring at least 90–95% herd immunity for disease control. The current vaccine licensed in the United States has a measurable primary and secondary failure rate that leads to population-level immunity that is often less than that required for herd immunity [17, 49]. This is especially true in regions of the world where seroconversion rates for MMR (and other measles-containing vaccines) are lower than in the United States and Europe.
The ideal measles vaccine, even if parenteral, should induce lifelong immunity after one dose, have little or no contraindications, be manufactured inexpensively, not require a cold chain and be safely stored for long periods of time, and be administered by a variety of health care personnel—particularly in low income countries.
For many of the above reasons, even more appealing would be vaccines that do not require parenteral administration and could be given to infants below the age of 12 months (i.e., no maternal antibody interference), ideally without the need for highly trained health care personnel and suitable for use in low resource settings. In this regard, oral or skin patch vaccines would be ideal candidates if they could be inexpensively made.
To this end, possibilities currently being investigated include protein and peptide-based vaccines using one or more measles viral proteins (H, F, and N proteins and peptides), recombinant protein vaccines, DNA-based vaccine constructs, and virus-vectored vaccines, as well as different delivery systems, such as microneedle skin patch vaccines and aerosol vaccines [107, 117, 188–195]. A unique recently developed administration method is via oral disintegrating films, which have been loaded with measles virus nanoparticles. Early studies in pigs have been promising [196].
8. Five-year view.
Over the next five years, we will continue to see an increasingly sophisticated understanding of measles vaccine-induced humoral immunity. Currently, humoral “measles immunity” is assessed most commonly using either EIA assays or fluorescence-based immunoassays. In research settings, measles neutralizing antibody may also be measured. The former two assays are not direct functional antibody assays and hence can be misleading. Neutralizing antibody on the other hand, is a direct measure of functional antibody responses, that has been correlated with protection against disease, and therefore, is a better measure of humoral immunity. However, the assay is laborious, expensive, and has lower throughput; therefore, it is not generally used clinically. In addition, our current correlate of immunity/protection is incomplete, as it does not evaluate cellular immune or other parameters that may also be important for durable and/or protective immunity. Systems biology studies are beginning to reveal genetic and molecular “signatures” of protective immune response. In time, it may be possible to narrow such signatures to highly predictive assays of vaccine responsiveness and efficacy/effectiveness, and to identify precise correlates of protection. In addition, protein and antibody array assays may provide a more holistic view of humoral immune response; in other words, they will allow direct measures of antibody response to specific proteins within a pathogen. In the case of measles, for example, we may be able to measure anti-H, anti-F, anti-N, and anti-P antibodies or specific antibody patterns as better correlates of immunity in a high-throughput, low-cost manner. Finally, measures of immune durability are critically needed, and much research is warranted on this topic.
9. Key issues.
Measles outbreaks continue to occur in high-vaccine coverage and low-measles exposure settings.
To address the barriers to measles elimination activities and the current knowledge gap, new/additional correlates of protection and new approaches for evaluating and predicting humoral immune response after measles vaccination are needed.
Genetic determinants of measles vaccine-induced neutralizing antibody response (e.g., CD46 and IFI44L genetic variants, other genetic markers) are under investigation.
Systems biology and/or other “omics” studies are likely to identify “signatures” of protective and durable immune response after measles vaccination.
The discovery of predictive “signatures” of measles vaccine immunogenicity, efficacy, and long-term effectiveness will identify individuals in need of additional vaccine doses and/or new improved measles vaccines.
Acknowledgments
Research reported in this publication was supported by the National Institute of Allergy And Infectious Diseases of the National Institutes of Health under award numbers U01AI089859 and R37AI048793. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Footnotes
References
*References of interest
**References of considerable interest
- 1.Progress towards regional measles elimination - worldwide, 2000-2016. Releve epidemiologique hebdomadaire. 2017. October 27;92(43):649–59. [PubMed] [Google Scholar]
- 2.Dabbagh A, Patel MK, Dumolard L, et al. Progress Toward Regional Measles Elimination - Worldwide, 2000-2016. MMWR. 2017. October 27;66(42):1148–1153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Centers for Disease Control and Prevention. Measles Cases and Outbreaks. http://www.cdc.gov/measles/cases-outbreaks.html Date accessed: September 30, 2018.
- 4.Hall V, Banerjee E, Kenyon C, et al. Measles Outbreak - Minnesota April-May 2017. MMWR. 2017. July 14;66(27):713–717. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.World Health Organization. Europe observes a 4-fold increase in measles cases in 2017 compared to previous year. http://www.euro.who.int/en/media-centre/sections/press-releases/2018/europe-observes-a-4-fold-increase-in-measles-cases-in-2017-compared-to-previous-year Date accessed: September 30, 2018.
- 6.Williams S European Measles Cases Quadrupled in 2017. https://www.the-scientist.com/the-nutshell/european-measles-cases-quadrupled-in-2017-30261 The Scientist. 2018
- *7.Mina MJ, Metcalf CJ, de Swart RL, et al. Long-term measles-induced immunomodulation increases overall childhood infectious disease mortality. Science. 2015. May 8;348(6235):694–9. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Provides statistical evidence for the non-specific effects of measles on host resistance/immunity to other pathogens
- **8.de Vries RD, McQuaid S, van AG, et al. Measles immune suppression: lessons from the macaque model. PLoS Pathog. 2012;8(8):e1002885. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Explains measles-induced immunosuppression from infection and immune-mediated lymphocyte depletion of follicular B and CD45RA(−) memory T-lymphocytes
- **9.de Vries RD, de Swart RL. Measles immune suppression: functional impairment or numbers game? PLoS Pathogens. 2014. December;10(12):e1004482. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Explains measles-induced immune suppression resulting from depletion of immune cell subsets, which is masked by the proliferation of MV-specific and bystander lymphocytes (explains the “measles paradox”).
- *10.Laksono BM, Grosserichter-Wagener C, de Vries RD, et al. In Vitro Measles Virus Infection of Human Lymphocyte Subsets Demonstrates High Susceptibility and Permissiveness of both Naive and Memory B Cells. Journal of Virology. 2018. April 15;92(8). [DOI] [PMC free article] [PubMed] [Google Scholar]; *Demonstrates the high susceptibility/permissiveness of human memory B cells to measles virus.
- 11.Laksono BM, de Vries RD, McQuaid S, et al. Measles Virus Host Invasion and Pathogenesis. Viruses. 2016. July 28;8(8). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Wendorf KA, Winter K, Zipprich J, et al. Subacute Sclerosing Panencephalitis: The Devastating Measles Complication That Might Be More Common Than Previously Estimated. Clinical infectious diseases. 2017. July 15;65(2):226–232. [DOI] [PubMed] [Google Scholar]
- 13.Muller CP, Kremer JR, Best JM, et al. Reducing global disease burden of measles and rubella: report of the WHO Steering Committee on research related to measles and rubella vaccines and vaccination, 2005. Vaccine. 2007;25(1):1–9. [DOI] [PubMed] [Google Scholar]
- 14.Patel MK, Gacic-Dobo M, Strebel PM, et al. Progress Toward Regional Measles Elimination - Worldwide, 2000-2015. MMWR. 2016. November 11;65(44): 1228–1233. [DOI] [PubMed] [Google Scholar]
- 15.Meissner HC, Strebel PM, Orenstein WA. Measles vaccines and the potential for worldwide eradication of measles. Pediatrics. 2004;114(4):1065–1069. [DOI] [PubMed] [Google Scholar]
- 16.Elliman D, Sengupta N. Measles. Curr OpinInfect Dis. 2005;18(3):229–234. [DOI] [PubMed] [Google Scholar]
- *17.Haralambieva IH, Ovsyannikova IG, Pankratz VS, et al. The genetic basis for interindividual immune response variation to measles vaccine: new understanding and new vaccine approaches. Expert Review of Vaccines. 2013. January;12(1):57–70. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Summarizes the immunogenetic findings underlying the interindividual immune response variation to measles vaccine.
- 18.Uzicanin A, Zimmerman L. Field effectiveness of live attenuated measles-containing vaccines: a review of published literature. J Infect Dis. 2011;204 Suppl 1: S133–S148. [DOI] [PubMed] [Google Scholar]
- 19.Boulianne N, De Serres G, Ratnam S, et al. Measles, mumps, and rubella antibodies in children 5-6 years after immunization: effect of vaccine type and age at vaccination. Vaccine. 1995;13:1611–1616. [DOI] [PubMed] [Google Scholar]
- 20.Christenson B, B”ttiger M Measles antibody: comparison of long-term vaccination titres, early vaccination titres and naturally acquired immunity to and booster effects on the measles virus. Vaccine. 1994;12:129–133. [DOI] [PubMed] [Google Scholar]
- 21.Mitchell LA, Tingle AJ, D cD, et al. Serologic responses to measles, mumps, and rubella (MMR) vaccine in healthy infants: Failure to respond to measles and mumps components may influence decisions on timing of the second dose of MMR. Canadian Journal of Public Health. 1998;89(5):325–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Moss W Measles in Vaccinated Individuals and the Future of Measles Elimination. Clin Infect Dis. 2018; 67: 1320–1321. [DOI] [PubMed] [Google Scholar]
- 23.Moss WJ. Measles control and the prospect of eradication. Current Topics in Microbiology and Immunology. 2009;330:173–189. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Measles Fact Sheet No. 286. http://www.who.int/en/news-room/fact-sheets/detail/measles 2018.
- 25.Parker Fiebelkorn A, Redd SB, Gallagher K, et al. Measles in the United States during the postelimination era. J Infect Dis. 2010;202(10):1520–1528. [DOI] [PubMed] [Google Scholar]
- 26.Poland GA, Jacobson RM, Thampy AM, et al. Measles re-immunization in children seronegative after initial immunization. JAMA. 1997;277:1156–1158. [PubMed] [Google Scholar]
- 27.Seward JF, Orenstein WA. A rare event: A measles outbreak in a population with high 2-dose measles vaccine coverage. Clinical Infectious Diseases. 2012;55(3):403–5. [DOI] [PubMed] [Google Scholar]
- 28.De Serres G, Boulianne N, Defay F, et al. Higher risk of measles when the first dose of a 2-dose schedule of measles vaccine is given at 12-14 months versus 15 months of age. Clin Infect Dis. 2012;55(3):394–402. [DOI] [PubMed] [Google Scholar]
- 29.Poland GA, Jacobson RM. Failure to reach the goal of measles elimination. Apparent paradox of measles infections in immunized persons. Arch In Med. 1994;154:1815–1820. [PubMed] [Google Scholar]
- 30.Paunio M, Peltola H, Valle M, et al. Explosive school-based measles outbreak: intense exposure may have resulted in high risk, even among revaccinees. Am J Epidemiol. 1998; 148(11): 1103–1110. [DOI] [PubMed] [Google Scholar]
- 31.Mathias RG, Meekison WG, Arcand TA, et al. The role of secondary vaccine failures in measles outbreaks. Am J Public Health. 1989;79:475–478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Sugerman DE, Barskey AE, Delea MG, et al. Measles outbreak in a highly vaccinated population, San Diego, 2008: role of the intentionally undervaccinated. Pediatrics. 2010;125(4):747–755. [DOI] [PubMed] [Google Scholar]
- 33.Pannuti CS, Morello RJ, Moraes JC, et al. Identification of primary and secondary measles vaccine failures by measurement of immunoglobulin G avidity in measles cases during the 1997 Sao Paulo epidemic. Clin Diagn Lab Immunol. 2004; 11(1):119–122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Hickman CJ, Hyde TB, Sowers SB, et al. Laboratory characterization of measles virus infection in previously vaccinated and unvaccinated individuals. J Infect Dis. 2011;204 Suppl 1:S549–S558. [DOI] [PubMed] [Google Scholar]
- 35.Glass K, Grenfell BT. Waning immunity and subclinical measles infections in England. Vaccine. 2004;22(29-30):4110–4116. [DOI] [PubMed] [Google Scholar]
- 36.Mossong J, Nokes DJ, Edmunds WJ, et al. Modeling the impact of subclinical measles transmission in vaccinated populations with waning immunity. Am J Epidemiol. 1999; 150(11): 1238–1249. [DOI] [PubMed] [Google Scholar]
- 37.Whitaker JA, Poland GA. Measles and mumps outbreaks in the United States: Think globally, vaccinate locally [Editorial], Vaccine. 2014. August 20;32(37):4703–4. [DOI] [PubMed] [Google Scholar]
- 38.Papania MJ, Wallace GS, Rota PA, et al. Elimination of endemic measles, rubella, and congenital rubella syndrome from the Western hemisphere: the US experience [Historical Article], JAMA Pediatr. 2014. February;168(2):148–55. [DOI] [PubMed] [Google Scholar]
- 39.Notes from the field: measles outbreak associated with a traveler returning from India - North Carolina, April-May 2013. MMWR. 2013. September 13;62(36):753. [PMC free article] [PubMed] [Google Scholar]
- 40.Notes from the field: measles outbreak among members of a religious community - Brooklyn, New York, March-June 2013. MMWR. 2013. September 13;62(36):752–3. [PMC free article] [PubMed] [Google Scholar]
- 41.Zipprich J, Hacker JK, Murray EL, et al. Notes from the field: measles - California, January 1-April 18, 2014. MMWR. 2014. April 25;63(16):362–3. [PMC free article] [PubMed] [Google Scholar]
- 42.Notes from the field: measles outbreak—Indiana, June-July 2011. Morbidity and Mortality Weekly Report. 2011;34:1169. [PubMed] [Google Scholar]
- 43.Notes from the field: Multiple cases of measles after exposure during air travel--Australia and New Zealand, January 2011. MMWR. 2011; 60(25):851. [PubMed] [Google Scholar]
- 44.Notes from the field: measles outbreak--Hennepin county, Minnesota, February--March 2011. MMWR. 2011;60(13):421. [PubMed] [Google Scholar]
- 45.Katz SL, Hinman AR. Summary and conclusions: measles elimination meeting, 16-17 March 2000. J Infect Dis. 2004; 189 Suppl ES43–S47. [DOI] [PubMed] [Google Scholar]
- 46.Measles once again endemic in the United Kingdom [News], Euro Surveill. 2008. July 3; 13(27). [PubMed] [Google Scholar]
- 47.Measles - United States, 2011. MMWR. 2012;61:253–257. [PubMed] [Google Scholar]
- 48.Rosen JB, Rota JS, Hickman CJ, et al. Outbreak of measles among persons with prior evidence of immunity, New York City, 2011. Clinical infectious diseases : an official publication of the Infectious Diseases Society of America. 2014. May;58(9): 1205–10. doi: 10.1093/cid/ciul05. [DOI] [PubMed] [Google Scholar]
- **49.Haralambieva IH, Kennedy RB, Ovsyannikova IG, et al. Variability in Humoral Immunity to Measles Vaccine: New Developments. Trends Mol Med. 2015. December;21(12):789–801. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Summarizes new studies/findings underlying variability in humoral immunity to measles vaccine.
- 50.Markowitz LE, Preblud SR, Fine PEM, et al. Duration of live measles vaccine-induced immunity. Pediatr Infect Dis J. 1990;9:101–110. [DOI] [PubMed] [Google Scholar]
- *51.Cherry JD, Zahn M Clinical Characteristics of Measles in Previously Vaccinated and Unvaccinated Patients in California. Clin Infect Dis. 2018;67(9): 1315–1319. [DOI] [PubMed] [Google Scholar]; *Confirms measles in subjects with ≥2 doses of measles vaccine (11% of 232 US measles cases with verified vaccination history)
- 52.Clemmons NS, Wallace GS, Patel M, et al. Incidence of Measles in the United States, 2001-2015. Jama. 2017. October 3;318(13): 1279–1281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Increased transmission and outbreaks of measles--European Region, 2011. MMWR. 2011. December 2;60(47): 1605–10. [PubMed] [Google Scholar]
- 54.Griffin DE. Measles Vaccine. Viral Immunology. 2018. March;31(2):86–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Sowers SB, Rota JS, Hickman CJ, et al. High Concentrations of Measles Neutralizing Antibodies and High-Avidity Measles IgG Accurately Identify Measles Reinfection Cases. Clin Vaccine Immunol. 2016. August;23(8):707–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Hahne SJ, Nic Lochlainn LM, van Burgel ND, et al. Measles Outbreak Among Previously Immunized Healthcare Workers, the Netherlands, 2014. J Infect Dis. 2016. December 15;214( 12): 1980–1986. [DOI] [PubMed] [Google Scholar]
- 57.Tatsuo H, Ono N, Tanaka K, et al. SLAM (CDwl50) is a cellular receptor for measles virus. Nature. 2000;406(6798): 893–897. [DOI] [PubMed] [Google Scholar]
- **58.Noyce RS, Bondre DG, Ha MN, et al. Tumor Cell Marker PVRL4 (Nectin 4) Is an Epithelial Cell Receptor for Measles Virus. PLoS Pathog. 2011;7(8):el002240. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Discovers PVRL4 (Nectin 4) as a cell receptor for measles virus.
- **59.Muhlebach MD, Mateo M, Sinn PL, et al. Adherens junction protein nectin-4 is the epithelial receptor for measles virus. Nature. 2011;480(7378):530–533. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Discovers PVRL4 (Nectin 4) as a cell receptor for measles virus.
- 60.Cattaneo R Four viruses, two bacteria, and one receptor: membrane cofactor protein (CD46) as pathogens’ magnet. J Virol. 2004;78(9):4385–4388. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Griffin DE. The Immune Response in Measles: Virus Control, Clearance and Protective Immunity. Viruses. 2016. October 12;8(10). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Zilliox MJ, Parmigiani G, Griffin DE. Gene expression patterns in dendritic cells infected with measles virus compared with other pathogens. Proc Natl Acad Sci USA. 2006;103(9):3363–3368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Haralambieva IH, Ovsyannikova IG, Dhiman N, et al. Differential cellular immune responses to wild-type and attenuated edmonston tag measles virus strains are primarily defined by the viral phosphoprotein gene. J Med Virol. 2010;82(11):1966–1975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chen RT, Markowitz LE, Albrecht P, et al. Measles antibody: reevaluation of protective titers. J Infect Dis. 1990; 162(5): 1036–1042. [DOI] [PubMed] [Google Scholar]
- 65.de Swart RL, Yuksel S, Osterhaus AD. Relative contributions of measles virus hemagglutinin- and fusion protein-specific serum antibodies to virus neutralization. J Virol. 2005;79(17): 11547–11551. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Polack FP, Lee SH, Permar S, et al. Successful DNA immunization against measles: Neutralizing antibody against either the hemagglutinin or fusion glycoprotein protects rhesus macaques without evidence of atypical measles. Nat Med. 2000;6(7):776–781. [DOI] [PubMed] [Google Scholar]
- **67.Bouche FB, Ertl OT, Muller CP. Neutralizing B cell response in measles. Viral Immunol. 2002;15(3):451–471. [DOI] [PubMed] [Google Scholar]; **Reviews and summarises findings on mealses-specific B cell responses.
- 68.de Vries RD, de Swart RL. Evaluating measles vaccines: can we assess cellular immunity? Exp Rev Vaccines. 2012. July;11(7):779–82. [DOI] [PubMed] [Google Scholar]
- 69.Crotty S A brief history of T cell help to B cells. Nat Rev Immunol. 2015. March;15(3):185–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Haralambieva IH, Ovsyannikova IG, Vierkant RA, et al. Development of a novel efficient fluorescence-based plaque reduction microneutralization assay for measles immunity. Clin Vaccine Immunol. 2008;15(7):1054–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *71.Plotkin SA. Correlates of protection induced by vaccination. Clin Vaccine Immunol. 2010;17(7):1055–1065. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Summarizes correlates of protection after vaccination with different vaccines.
- 72.Haralambieva IH, Ovsyannikova IG, O’Byrne M, et al. A large observational study to concurrently assess persistence of measles specific B-cell and T-cell immunity in individuals following two doses of MMR vaccine. Vaccine. 2011;29(27):4485–4491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Polack FP, Hoffman SJ, Crujeiras G, et al. A role for nonprotective complement-fixing antibodies with low avidity for measles virus in atypical measles. Nat Med. 2003;9(9):1209–1213. [DOI] [PubMed] [Google Scholar]
- 74.World Health Organization. Correlates of vaccine-induced protection: methods and implications. http://apps.who.int/iris/bitstream/handle/10665/84288/WHO_IVB_13.01_eng.pdf;jsessionid=F944A52651CB43AEC79D14568617D67D?sequence=1 Geneva, Switzerland: 2013. [Google Scholar]
- 75.Dimech W, Mulders MN. A review of testing used in seroprevalence studies on measles and rubella. Vaccine. 2016. July 29;34(35):4119–22. [DOI] [PubMed] [Google Scholar]
- 76.Dimech W, Mulders MN. A 16-year review of seroprevalence studies on measles and rubella. Vaccine. 2016. July 29;34(35):4110–4118. [DOI] [PubMed] [Google Scholar]
- 77.Thompson KM, Odahowski CL. Systematic Review of Measles and Rubella Serology Studies. Risk Anal. 2016. July;36(7):1459–86. [DOI] [PubMed] [Google Scholar]
- 78.Binnicker MJ, Jespersen DJ, Rollins LO. Evaluation of the Bio-Rad BioPlex Measles, Mumps, Rubella, and Varicella-Zoster Virus IgG multiplex bead immunoassay. Clin Vaccine Immunol 2011. September;18(9):1524–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Smits GP, van Gageldonk PG, Schouls LM, et al. Development of a bead-based multiplex immunoassay for simultaneous quantitative detection of IgG serum antibodies against measles, mumps, rubella, and varicella-zoster virus. Clin Vaccine Immunol 2012. March;19(3):396–400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Tischer A, Andrews N, Kafatos G, et al. Standardization of measles, mumps and rubella assays to enable comparisons of seroprevalence data across 21 European countries and Australia. Epidemiol Infect. 2007. July;135(5):787–97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hubschen JM, Bork SM, Brown KE, et al. Challenges of measles and rubella laboratory diagnostic in the era of elimination. Clin Microbiol Infect 2017. August;23(8):511–515. [DOI] [PubMed] [Google Scholar]
- 82.World Health Organization. Expert committee on Biological standardization, October 8-12, 2007: report on the collaborative study to investigate the relationship between the 1st IRP and the 2nd and 3rd international standards for anti-measles serum/plasma in both ELISA and PRNT. http://www.who.int/biologicals/BS07%202076anti-measles.pdf Geneva, Switzerland: 2007. [Google Scholar]
- *83.Haralambieva IH, Simon WL, Kennedy RB, et al. Profiling of Measles-Specific Humoral Immunity in Individuals Following Two Doses of MMR Vaccine Using Proteome Microarrays. Viruses. 2015;7(3): 1113–33. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Provides data on measles-specific antibody levels to different measles virus proteins after vaccination.
- *84.Mercader S, Garcia P, Bellini WJ. Measles virus IgG avidity assay for use in classification of measles vaccine failure in measles elimination settings. Clin Vaccine Immunol. 2012. November;19(11):1810–7. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Provides methodology (measles-specific IgG avidity assay) for classification of measles vaccine failure in settings with high vaccine coverage.
- 85.Latner DR, McGrew M, Williams N, et al. Enzyme-linked immunospot assay detection of mumps-specific antibody-secreting B cells as an alternative method of laboratory diagnosis. Clin Vaccine Immunol. 2011;18(1):35–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Carter MJ, Mitchell RM, Meyer Sauteur PM, et al. The Antibody-Secreting Cell Response to Infection: Kinetics and Clinical Applications. Front Immunol. 2017;8:630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Kakoulidou M, Ingelman-Sundberg H, Johansson E, et al. Kinetics of antibody and memory B cell responses after MMR immunization in children and young adults. Vaccine. 2013. January 11;31(4):711–7. [DOI] [PubMed] [Google Scholar]
- 88.Buisman AM, de Rond CG, Ozturk K, et al. Long-term presence of memory B-cells specific for different vaccine components. Vaccine. 2009. December 10;28(1):179–86. [DOI] [PubMed] [Google Scholar]
- 89.Titanji K, De Milito A, Cagigi A, et al. Loss of memory B cells impairs maintenance of long-term serologic memory during HIV-1 infection. Blood. 2006. September 1;108(5):1580–7. [DOI] [PubMed] [Google Scholar]
- 90.Ellebedy AH, Jackson KJ, Kissick HT, et al. Defining antigen-specific plasmablast and memory B cell subsets in human blood after viral infection or vaccination. Nat Immunol 2016. October;17(10): 1226–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **91.Hammarlund E, Thomas A, Amanna IJ, et al. Plasma cell survival in the absence of B cell memory. Nat Commun. 2017. November 24;8(1):1781. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Provides evidence that long-lived plasma cells represent a key cell population responsible for long-term antibody production.
- 92.Amanna IJ, Carlson NE, Slifka MK. Duration of humoral immunity to common viral and vaccine antigens. N Engl J Med. 2007;357(19):1903–1915. [DOI] [PubMed] [Google Scholar]
- **93.Amanna IJ, Slifka MK. Mechanisms that determine plasma cell lifespan and the duration of humoral immunity. Immunol Rev. 2010. July;236:125–38. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Reviews findings and hypotheses on plasma cells with regard to the duration of humoral immunity (antibody response).
- 94.Fairfax KA, Kallies A, Nutt SL, et al. Plasma cell development: from B-cell subsets to long-term survival niches. Semin Immunol 2008. February;20(1):49–58. [DOI] [PubMed] [Google Scholar]
- **95.Radbruch A, Muehlinghaus G, Luger EO, et al. Competence and competition: the challenge of becoming a long-lived plasma cell. Nat Reviews Immunol. 2006. October;6(10):741–50. [DOI] [PubMed] [Google Scholar]; **Reviews findings on long-lived plasma cells with regard to the duration of humoral immunity (antibody response).
- 96.Pensieroso S, Cagigi A, Palma P, et al. Timing of HAART defines the integrity of memory B cells and the longevity of humoral responses in HIV-1 vertically-infected children. Proc Natl Acad Sci USA. 2009. May 12;106(19):7939–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Rocca S, Santilli V, Cotugno N, et al. Waning of vaccine-induced immunity to measles in kidney transplanted children. Medicine. 2016. September;95(37):e4738. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Kotton CN. Immunization after kidney transplantation-what is necessary and what is safe? Nat Rev Nephrol. 2014. Oct;10(10):555–62. [DOI] [PubMed] [Google Scholar]
- 99.Cotugno N, Finocchi A, Cagigi A, et al. Defective B-cell proliferation and maintenance of long-term memory in patients with chronic granulomatous disease. J Allergy Clin Immunol. 2015. March;135(3):753–61 e2. [DOI] [PubMed] [Google Scholar]
- 100.World Health Organization. Global Measles & Rubella Strategic Plan 2012-2020. http://apps.who.int/iris/bitstream/10665/44855/1/9789241503396_eng.pdf Geneva: 2012. [Google Scholar]
- 101.Ovsyannikova IG, Pankratz VS, Vierkant RA, et al. Consistency of HLA associations between two independent measles vaccine cohorts: a replication study. Vaccine. 2012;30(12):2146–2152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Ovsyannikova IG, Schaid DJ, Larrabee BR, et al. A large population-based association study between HLA and KIR genotypes and measles vaccine antibody responses. PLos ONE. 2017;12(2):e0171261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Schellens IM, Meiring HD, Hoof I, et al. Measles Virus Epitope Presentation by HLA: Novel Insights into Epitope Selection, Dominance, and Microvariation. Front Immunol. 2015;6:546. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Ovsyannikova IG, Johnson KL, Muddiman DC, et al. Identification and characterization of novel, naturally processed measles virus class II HLA-DRB1 peptides. J Virol 2004;78(1):42–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Ovsyannikova IG, Johnson KL, Naylor S, et al. Naturally processed measles virus peptide eluted from class II HLA-DRB1*03 recognized by T lymphocytes from human blood. Virology. 2003;312(2):495–506. [DOI] [PubMed] [Google Scholar]
- 106.Poland GA, Ovsyannikova IG, Kennedy RB, et al. Vaccinomics and a new paradigm for the development of preventive vaccines against viral infections. Omics. 2011;15(9):625–636. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Ovsyannikova IG, Johnson KL, Bergen HR III, et al. Mass spectrometry and peptide-based vaccine development. Clin PharmacolTher. 2007;82(6):644–652. [DOI] [PubMed] [Google Scholar]
- 108.Clifford HD, Hayden CM, Khoo SK, et al. Polymorphisms in key innate immune genes and their effects on measles vaccine responses and vaccine failure in children from Mozambique. Vaccine. 2012. September 21;30(43):6180–5. [DOI] [PubMed] [Google Scholar]
- 109.Ovsyannikova IG, Haralambieva IH, Vierkant RA, et al. The role of polymorphisms in toll-like receptors and their associated intracellular signaling genes in measles vaccine immunity. Human Genetics. 2011;130(4):547–561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Kennedy RB, Ovsyannikova IG, Haralambieva IH, et al. Multigenic control of measles vaccine immunity mediated by polymorphisms in measles receptor, innate pathway, and cytokine genes. Vaccine. 2012;30(12):2159–2167. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Poland GA, Ovsyannikova IG, Jacobson RM, et al. Heterogeneity in vaccine immune response: the role of immunogenetics and the emerging field of vaccinomics. Clin Pharmacol Ther. 2007;82(6):653–664. [DOI] [PubMed] [Google Scholar]
- 112.Ovsyannikova IG, Haralambieva IH, Vierkant RA, et al. Effects of vitamin A and D receptor gene polymorphisms/haplotypes on immune responses to measles vaccine. Pharmacogenet Genomics. 2012;22(1):20–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ovsyannikova IG, Haralambieva IH, Vierkant RA, et al. The association of CD46, SLAM, and CD209 cellular receptor gene SNPs with variations in measles vaccine-induced immune responses--a replication study and examination of novel polymorphisms. Hum Hered. 2011;72(3):206–223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Haralambieva IH, Ovsyannikova IG, Umlauf BJ, et al. Genetic polymorphisms in host antiviral genes: associations with humoral and cellular immunity to measles vaccine. Vaccine. 2011;29(48):8988–8997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Haralambieva IH, Ovsyannikova IG, Kennedy RB, et al. Associations between single nucleotide polymorphisms and haplotypes in cytokine and cytokine receptor genes and immunity to measles vaccination. Vaccine. 2011;29(45):7883–7895. PubMed Central PMCID: PMCPMCID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116.Yoo KH, Agarwal K, Butterfield M, et al. Assessment of humoral and cell-mediated immune response to measles-mumps-rubella vaccine viruses among patients with asthma. Allergy Asthma Proc. 2010;31(6):499–506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Poland GA, Ovsyannikova IG, Jacobson RM. Application of pharmacogenomics to vaccines. Pharmacogenomics. 2009;10(5):837–852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Dhiman N, Ovsyannikova IG, Vierkant RA, et al. Associations between SNPs in toll-like receptors and related intracellular signaling molecules and immune responses to measles vaccine: preliminary results. Vaccine. 2008;26 (14):1731–1736. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Poland GA, Ovsyannikova IG, Jacobson RM. Genetics and immune response to vaccines In: Kaslow RA, McNicholl JM, Hill AVS, editors. Genetic Susceptibility to Infectious Diseases. New York, New York: Oxford University Press; 2008. p. 1–447. [Google Scholar]
- 120.Clifford HD, Yerkovich ST, Khoo SK, et al. TLR3 and RIG-I gene variants: Associations with functional effects on receptor expression and responses to measles virus and vaccine in vaccinated infants. Hum Immunol. 2012;73(6):677–685. [DOI] [PubMed] [Google Scholar]
- 121.Clifford HD, Hayden CM, Khoo SK, et al. CD46 measles virus receptor polymorphisms influence receptor protein expression and primary measles vaccine responses in naive Australian children. Clin Vaccine Immunol. 2012;19(5):704–710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Clifford HD, Yerkovich ST, Khoo SK, et al. Toll-like receptor 7 and 8 polymorphisms: associations with functional effects and cellular and antibody responses to measles virus and vaccine. Immunogenetics. 2012;64(3):219–228. [DOI] [PubMed] [Google Scholar]
- 123.Clifford HD, Richmond P, Khoo SK, et al. SLAM and DC-SIGN measles receptor polymorphisms and their impact on antibody and cytokine responses to measles vaccine. Vaccine. 2011;29(33):5407–5413. [DOI] [PubMed] [Google Scholar]
- 124.Newport MJ. The genetic regulation of infant immune responses to vaccination. Front Immunol. 2015;6:18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Ovsyannikova IG, Haralambieva IH, Vierkant RA, et al. Associations between polymorphisms in the antiviral TRIM genes and measles vaccine immunity. Hum Immunol. 2013. June;74(6):768–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- **126.Haralambieva IH, Ovsyannikova IG, Kennedy RB, et al. Genome-Wide Associations of CD46 and IFI44L Genetic Variants with Neutralizing Antibody Response to Measles Vaccine. Hum Genet. 2017;136(4):421–435. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Discovers CD46 and IFI44L genetic variants as important determinants of neutralizing antibody response to measles vaccination.
- 127.Schaid DJ, Haralambieva IH, Larrabee BR, et al. Heritability of vaccine-induced measles neutralizing antibody titers. Vaccine 2017;35(10):1390–1394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Marchini J, Donnelly P, Cardon LR. Genome-wide strategies for detecting multiple loci that influence complex diseases. Nat Genet. 2005;37(4):413–417. [DOI] [PubMed] [Google Scholar]
- **129.Feenstra B, Pasternak B, Geller F, et al. Common variants associated with general and MMR vaccine-related febrile seizures. Nat Genet. 2014. December;46(12):1274–82. [DOI] [PMC free article] [PubMed] [Google Scholar]; **Discovers genetic drivers of MMR vaccine-associated febrile seizures.
- 130.Pasternak B, Feenstra B, Melbye M, et al. Improving vaccine safety through a better understanding of vaccine adverse events. Clin Infect Dis. 2015. May 15;60(10): 1586–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Hammer C, Begemann M, McLaren PJ, et al. Amino Acid Variation in HLA Class II Proteins Is a Major Determinant of Humoral Response to Common Viruses. Am J Hum Genet. 2015. November 5;97(5):738–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 132.Voigt EA, Ovsyannikova IG, Haralambieva IH, et al. Genetically defined race, but not sex, is associated with higher humoral and cellular immune responses to measles vaccination. Vaccine. 2016. August 30;34(41):4913–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Moss WJ. Measles. Lancet. 2017. December 2;390(10111):2490–2502. [DOI] [PubMed] [Google Scholar]
- 134.Hull J, Campino S, Rowlands K, et al. Identification of common genetic variation that modulates alternative splicing. PLoS Genet. 2007. June;3(6):e99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Zhao K, Lu ZX, Park JW, et al. GLiMMPS: robust statistical model for regulatory variation of alternative splicing using RNA-seq data. Genome Biol. 2013;14(7):R74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Buchholz CJ, Gerlier D, Hu A, et al. Selective expression of a subset of measles virus receptor-competent CD46 isoforms in human brain. Virology 1996;217(1):349–355. [DOI] [PubMed] [Google Scholar]
- 137.Iwata K, Seya T, Ueda S, et al. Modulation of complement regulatory function and measles virus receptor function by the serine-threonine-rich domains of membrane cofactor protein (CD46). Biochem J. 1994. November 15;304 ( Pt 1):169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Buchholz CJ, Schneider U, Devaux P, et al. Cell entry by measles virus: Long hybrid receptors uncouple binding from membrane fusion. Virology 1996;70:3716–3723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 139.Ni Choileain S, Astier AL. CD46 processing: a means of expression. Immunobiol 2012. February;217(2):169–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Ni Choileain S, Hay J, Thomas J, et al. TCR-stimulated changes in cell surface CD46 expression generate type 1 regulatory T cells. Sci Signal. 2017. October 24;10(502). [DOI] [PubMed] [Google Scholar]
- 141.Flajnik MF, Kasahara M. Origin and evolution of the adaptive immune system: genetic events and selective pressures. Nat Rev Genet 2010. January;11(1):47–59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Oberg AL, McKinney BA, Schaid DJ, et al. Lessons learned in the analysis of high-dimensional data in vaccinomics. Vaccine. 2015;S0264-410X:574-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Hardy J, Singleton A. Genomewide association studies and human disease. N Engl J Med. 2009. April 23;360(17):1759–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 144.Wellcome Trust Case Control C. Genome-wide association study of 14,000 cases of seven common diseases and 3,000 shared controls. Nature. 2007;447(7145):661–678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Yang J, Benyamin B, McEvoy BP, et al. Common SNPs explain a large proportion of the heritability for human height. Nat Genet 2010. July;42(7):565–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Willer CJ, Schmidt EM, Sengupta S, et al. Discovery and refinement of loci associated with lipid levels. Nature Genetics. 2013. November;45(11):1274–1283. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Nikpay M, Goel A, Won HH, et al. A comprehensive 1,000 Genomes-based genome-wide association meta-analysis of coronary artery disease. Nat Genet. 2015. October;47(10):1121–1130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Al Olama AA, Kote-Jarai Z, Berndt SI, et al. A meta-analysis of 87,040 individuals identifies 23 new susceptibility loci for prostate cancer. Nat Genet. 2014. October;46(10): 1103–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Fuchsberger C, Flannick J, Teslovich TM, et al. The genetic architecture of type 2 diabetes. Nature. 2016. August 4;536(7614):41–47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 150.Biological insights from 108 schizophrenia-associated genetic loci. Nature. 2014. July 24;511(7510):421–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *151.MacArthur J, Bowler E, Cerezo M, et al. The new NHGRI-EBI Catalog of published genome-wide association studies (GWAS Catalog). Nucleic Acids Res. 2017. January 4;45(D1):D896–D901. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Describes the NHGRI-EBI Catalog of published genome-wide association studies.
- 152.McCarthy S, Das S, Kretzschmar W, et al. A reference panel of 64,976 haplotypes for genotype imputation. Nat Genet. 2016. October;48(10):1279–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Pruim RJ, Welch RP, Sanna S, et al. LocusZoom: regional visualization of genome-wide association scan results. Bioinformatics. 2010. September 15;26(18):2336–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154.Manolio TA. Genomewide association studies and assessment of the risk of disease. N Engl J Med. 2010. July 8;363(2):166–76. [DOI] [PubMed] [Google Scholar]
- 155.Pe’er I, Yelensky R, Altshuler D, et al. Estimation of the multiple testing burden for genomewide association studies of nearly all common variants. Genet Epidemiol. 2008. May;32(4):381–5. [DOI] [PubMed] [Google Scholar]
- 156.MacArthur DG, Manolio TA, Dimmock DP, et al. Guidelines for investigating causality of sequence variants in human disease. Nature. 2014. April 24;508(7497):469–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157.van de Bunt M, Cortes A, Brown MA, et al. Evaluating the Performance of Fine-Mapping Strategies at Common Variant GWAS Loci. PLoS Genet. 2015;11(9):e1005535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158.Ding K, Kullo IJ. Methods for the selection of tagging SNPs: a comparison of tagging efficiency and performance. Eur J Hum Genet 2007. February;15(2):228–36. [DOI] [PubMed] [Google Scholar]
- 159.Stram DO. Tag SNP selection for association studies. Genet Epidemiol 2004. December;27(4):365–74. [DOI] [PubMed] [Google Scholar]
- 160.Marchini J, Howie B. Genotype imputation for genome-wide association studies. Nat Rev Genet. 2010. July; 11(7):499–511. [DOI] [PubMed] [Google Scholar]
- 161.Li Y, Willer C, Sanna S, et al. Genotype imputation [10.1146/annurev.genom.9.081307.164242 doi] AnnuRevGenomics HumGenet. 2009;10:387–406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162.Schaid DJ, Chen W, Larson NB. From genome-wide associations to candidate causal variants by statistical fine-mapping. Nat Rev Genet. 2018. August;19(8):491–504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163.Spain SL, Barrett JC. Strategies for fine-mapping complex traits. Hum Molec Genet 2015. October 15;24(R1):R111–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164.Rhee SY, Wood V, Dolinski K, et al. Use and misuse of the gene ontology annotations. Nat Rev Genet. 2008. July;9(7):509–15. [DOI] [PubMed] [Google Scholar]
- 165.Harrow J, Frankish A, Gonzalez JM, et al. GENCODE: the reference human genome annotation for The ENCODE Project. Genome Res. 2012. September;22(9):1760–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166.The ENCODE (ENCyclopedia Of DNA Elements) Project. Science. 2004. October 22;306(5696):636–40. [DOI] [PubMed] [Google Scholar]
- 167.Andersson R, Gebhard C, Miguel-Escalada I, et al. An atlas of active enhancers across human cell types and tissues. Nature. 2014. March 27;507(7493):455–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168.Kundaje A, Meuleman W, Ernst J, et al. Integrative analysis of 111 reference human epigenome. Nature. 2015. February 19;518(7539):317–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 169.Maurano MT, Humbert R, Rynes E, et al. Systematic localization of common disease-associated variation in regulatory DNA. Science. 2012. September 7;337(6099):1190–5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 170.Millstein J, Zhang B, Zhu J, et al. Disentangling molecular relationships with a causal inference test. BMC genetics. 2009. May 27;10:23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171.Zhu Z, Zhang F, Hu H, et al. Integration of summary data from GWAS and eQTL studies predicts complex trait gene targets. Nature Genetics. 2016. May;48(5):481–7. [DOI] [PubMed] [Google Scholar]
- 172.Giambartolomei C, Vukcevic D, Schadt EE, et al. Bayesian test for colocalisation between pairs of genetic association studies using summary statistics. PLoS Genet. 2014. May;10(5):e1004383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 173.Hormozdiari F, van de Bunt M, Segre AV, et al. Colocalization of GWAS and eQTL Signals Detects Target Genes. Am J Hum Genet. 2016. December 1;99(6):1245–1260. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174.Zilliox MJ, Moss WJ, Griffin DE. Gene expression changes in peripheral blood mononuclear cells during measles virus infection. Clin Vaccine Immunol. 2007;14(7):918–923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 175.Dhiman N, Ovsyannikova IG, Oberg AL, et al. Immune activation at effector and gene expression levels after measles vaccination in healthy individuals: a pilot study. Hum Immunol. 2005;66:1125–1136. [DOI] [PubMed] [Google Scholar]
- *176.Haralambieva IH, Zimmermann MT, Ovsyannikova IG, et al. Whole Transcriptome Profiling Identifies CD93 and Other Plasma Cell Survival Factor Genes Associated with Measles-Specific Antibody Response after Vaccination. PLos ONE. 2016;11(8):e0160970. [DOI] [PMC free article] [PubMed] [Google Scholar]; *Assesses the gene expression changes in peripheral blood mononuclear cells during measles virus infection. Provides insights into important genes involved in the host response to measles.
- 177.Haralambieva IH, Kennedy RB, Simon WL, et al. Differential miRNA expression in B cells is associated with inter-individual differences in humoral immune response to measles vaccination. PLos ONE. 2018;13(1):e0191812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- *178.Querec TD, Akondy RS, Lee EK, et al. Systems biology approach predicts immunogenicity of the yellow fever vaccine in humans. Nat Immunol. 2009; 10(1):116–125. [DOI] [PMC free article] [PubMed] [Google Scholar]; **A landmark paper: one of the first reports applying a systems biology approach to predict response to vaccination (with yellow fever vaccine).
- 179.Nakaya HI, Wrammert J, Lee EK, et al. Systems biology of seasonal influenza vaccination in humans. Nat Immunol. 2011;12(8):786–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 180.Kazmin D, Nakaya HI, Lee EK, et al. Systems analysis of protective immune responses to RTS,S malaria vaccination in humans. Proc Natl Acad Sci USA. 2017. February 28;114(9):2425–2430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181.Papalexi E, Satija R. Single-cell RNA sequencing to explore immune cell heterogeneity. Nat Rev Immunol. 2018. January;18(1):35–45. [DOI] [PubMed] [Google Scholar]
- 182.Friedensohn S, Khan TA, Reddy ST. Advanced Methodologies in High-Throughput Sequencing of Immune Repertoires. Trends Biotechnol. 2017. March;35(3):203–214. [DOI] [PubMed] [Google Scholar]
- 183.Hu Y, An Q, Sheu K, et al. Single Cell Multi-Omics Technology: Methodology and Application. Front Cell Dev Biol. 2018;6:28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184.Stoeckius M, Hafemeister C, Stephenson W, et al. Simultaneous epitope and transcriptome measurement in single cells. Nat Methods. 2017. September;14(9):865–868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 185.Peterson VM, Zhang KX, Kumar N, et al. Multiplexed quantification of proteins and transcripts in single cells. Nature Biotechnology. 2017. October;35(10):936–939. [DOI] [PubMed] [Google Scholar]
- 186.Hou Y, Guo H, Cao C, et al. Single-cell triple omics sequencing reveals genetic, epigenetic, and transcriptomic heterogeneity in hepatocellular carcinomas. Cell Res. 2016. March;26(3):304–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 187.Hu Y, Huang K, An Q, et al. Simultaneous profiling of transcriptome and DNA methylome from a single cell. Genome Biol. 2016. May 5;17:88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188.Frantz PN, Teeravechyan S, Tangy F. Measles-derived vaccines to prevent emerging viral diseases. Microbes and infection. 2018. Oct-Nov;20(9-10):493–500 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 189.Griffin DE. Current progress in pulmonary delivery of measles vaccine. Exp Rev Vaccines. 2014. June;13(6):751–9. [DOI] [PubMed] [Google Scholar]
- 190.Griffin DE, Pan CH. Measles: old vaccines, new vaccines. Curr Topics Microbiol Immunol. 2009;330:191–212. [DOI] [PubMed] [Google Scholar]
- 191.de Swart RL, de Vries RD, Rennick LJ, et al. Needle-free delivery of measles virus vaccine to the lower respiratory tract of non-human primates elicits optimal immunity and protection. NPJ Vaccines. 2017;2:22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192.de Vries RD, Stittelaar KJ, Osterhaus AD, et al. Measles vaccination: new strategies and formulations. Expert RevVaccines. 2008;7(8):1215–1223. [DOI] [PubMed] [Google Scholar]
- 193.Joyce JC, Carroll TD, Collins ML, et al. A Microneedle Patch for Measles and Rubella Vaccination Is Immunogenic and Protective in Infant Rhesus Macaques. J Infect Dis. 2018. June 5;218(1): 124–132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194.Edens C, Collins ML, Goodson JL, et al. A microneedle patch containing measles vaccine is immunogenic in non-human primates. Vaccine. 2015. March 12;S0264-410X(15):00285-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 195.Putz MM, Muller CP. The rationale of a peptide-conjugate vaccine against measles. Vaccine. 2003. January 30;21(7-8):663–6. [DOI] [PubMed] [Google Scholar]
- 196.Gala RP, Popescu C, Knipp GT, et al. Physicochemical and Preclinical Evaluation of a Novel Buccal Measles Vaccine. AAPS Pharm Sci Tech. 2017. February;18(2):283–292. [DOI] [PubMed] [Google Scholar]


