Abstract
Purpose of Review
The environmental triggers of islet autoimmunity leading to type 1 diabetes (T1D) need to be elucidated to inform primary prevention. The Environmental Determinants of Diabetes in the Young (TEDDY) Study follows from birth 8676 children with T1D risk HLA-DR-DQ genotypes in the USA, Finland, Germany, and Sweden. Most study participants (89%) have no first-degree relative with T1D. The primary outcomes include the appearance of one or more persistent islet autoantibodies (islet autoimmunity, IA) and clinical T1D.
Recent Findings
As of February 28,2018, 769 children had developed IA and 310 have progressed to T1D. Secondary outcomes include celiac disease and autoimmune thyroid disease. While the follow-up continues, TEDDY has already evaluated a number of candidate environmental triggers, including infections, probiotics, micronutrient, and microbiome.
Summary
TEDDY results suggest that there are multiple pathways leading to the destruction of pancreatic beta-cells. Ongoing measurements of further specific exposures, gene variants, and gene-environment interactions and detailed “omics” studies will provide novel information on the pathogenesis of T1D.
Keywords: Type 1 diabetes, Autoimmunity, Children
Introduction
Autoimmunity against pancreatic islet beta-cells (islet autoimmunity, IA) leads to loss of insulin production resulting in type 1 diabetes (T1D). The incidence of childhood T1D continues to rise worldwide by 3–4% per annum [1–3]. In the USA, an estimated 1.25 million people [4] including 132,000 children and adolescents [3] are affected. Half of T1D patients are diagnosed after age 20 and the lifetime risk now exceeds 1% in North America and Europe. This pandemic is clearly environmentally induced. T1D incidence varies markedly from year to year and seasonally; outbreaks appear to be Extended author information available on the last page of the article superimposed on a steady secular increase in incidence. No genetic models can explain such a pattern and rapid increase [5]; however, the specific cause(s) remains to be determined. A number of intriguing hypotheses have been proposed and plausible environmental causes of T1D have been suggested; however, few have been consistently confirmed and none, so far, successfully modified in primary prevention.
In 2003, to overcome these limitations and to accelerate the path to primary prevention, the National Institutes of Health created a multicenter prospective cohort study—The Environmental Determinants of Diabetes in the Young (TEDDY) consortium [6, 7]. TEDDY includes three US clinical centers in Denver, Seattle, and Augusta as well as centers in Turku (Finland), Malmö (Sweden), and Munich (Germany). The Data Coordinating Center is in Tampa, FL. TEDDY aims to identify environmental factors that trigger or protect against the development of IA and T1D. The uniquely intensive follow-up of TEDDY children has already evaluated a number of environmental triggers [8•, 9-16], gene variants [17–21], and gene-environment interactions involved in the appearance of islet autoantibodies [22•, 23, 24•] preceding T1D. Models emerging from current data suggest that either a variety of exposures act on a similar pathway or that there are multiple pathways leading to the destruction of beta-cells. The progress made by TEDDY has been reflected in more than 90 peer-reviewed publications. This review provides a summary of selected TEDDY results published through June 2018.
The Goals of TEDDY
TEDDY’s overarching goal is to elucidate the etiology and pathogenesis of T1D and to inform new strategies to prevent or delay the disease. Specifically, TEDDY long-term scientific goals include:
Identify modifiable environmental factors responsible for the development of IA and T1D.
Among children with islet autoimmunity, identify predictors of progression to T1D.
Disentangle the heterogeneity of T1D by prospective characterization of endotypes [25].
To elucidate the pathogenesis of T1D trough integrative analyses of “big data” generated by “omics” study laboratories and with analytical help from the broad scientific community.
Collect and bank specimens for studies of T1D pathogenesis and development of biomarkers for T1D prediction. The specimens collected prospectively from TEDDY subjects (DNA, RNA, serum and plasma, PBMCs, stool, nasal swabs, and other samples) provide a unique resource for scientists within and outside the TEDDY consortium to test novel hypotheses.
TEDDY Study Design and Population
TEDDY’s large, intensively followed cohort provides a unique opportunity to disentangle the role of genetic and environmental factors in the complex etiology of T1D. The study design has been published [6, 7, 26]. Briefly, in 2004–2010, TEDDY screened 424,788 newborns for HLA-DR-DQ genotypes conferring increased T1D risk. In newborns without a first-degree relative with T1D (about 90% of the TEDDY cohort), the eligible haplogenotypes were HLA DR3-DQ2/DR4-DQ8, DR4-DQ8/DR4-DQ8, DR4-DQ8/ DR8-DQ4, and DR3-DQ2/DR3-DQ2. In newborns with a first-degree relative affected by T1D, an additional five haplogenotypes defined eligibility [26]. HLA DRB1*04:03 was an exclusion allele [27, 28]. Eligible enrolled children with high-risk HLA-DR-DQ (N =8676) include 922 newborn first-degree relatives (FDR) of persons with T1D and 7754 general population children (GP). TEDDY then became a closed cohort followed for development of study end points: persistent islet autoantibody, multiple persistent autoantibodies with normoglycemia (stage 1 T1D), dysglycemia (stage 2 T1D), and clinical stage 3 T1D [29].
Follow-up Schedule
Participating children completed their initial study visit by 4 months of age. Gestational and perinatal exposures were assessed; maternal serum was tested for islet autoantibodies, if the infant was positive or if the mother had diabetes. Subjects are being followed for development of study end points with meticulous assessment of environmental exposures. Detailed procedures are utilized to standardize data collection, harmonization, and management [30]. Clinic visits occur quarterly until age 4, and then every 6 months through age 15. Children with one or more persistent islet autoantibodies are followed quarterly; an OGTT is performed in these children every 6 months. Parents fill out questionnaires at regular intervals and record in the “TEDDY Book” diet, allergies, vaccinations, dietary supplements, illnesses, medication, daycare, pets, school, social groups, and significant life events. Study staff complete additional questionnaires and anthropometric measurements at each visit. Blood, stool, nasal swab, saliva, urine, toenail clippings, and drinking water are collected. Physical activity is measured by accelerometer from age 5.
Study Outcomes
The first primary outcome is the appearance of a persistent confirmed islet autoantibody. The study has a robust process of confirming outcomes in a second reference laboratory, defining persistence of autoantibodies, ruling out maternally transmitted autoantibodies, and pinpointing the timing of each islet autoantibody appearance. Autoimmune activation in response to a hypothetical trigger has been determined as the first appearing autoantibody against either insulin (IAA), GAD (GADA), or IA-2 (IA-2A) on two or more consecutive visits. Table 1 summarizes the number of TEDDY participants who have developed a single (1Ab), multiple islet autoantibodies (mAb), or T1D defined by the ADA criteria [31]. As of February 28, 2018, 769 children had developed persistent confirmed islet autoantibodies, including 447 with mAb and 310 with T1D. Secondary outcomes include celiac autoimmunity (CDA, n = 1258), celiac disease (CD, n = 471), autoimmune thyroiditis (AT, n = 373), and autoimmune thyroid disease (ATD, n = 38).
Table 1.
TEDDY enrollment and endpoints by study group, as of February 28, 2018
| Center | First-degree relatives of T1D persons (FDR) |
General population children (GP) |
||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Enrolled | IA |
T1D | Celiac |
Thyroid |
Enrolled | IA |
T1D | Celiac |
Thyroid |
|||||||
| 1Ab | mAb | CDA | CD | TA | ATD | 1Ab | mAb | CDA | CD | TA | ATD | |||||
| COL | 142 | 6 | 18 | 18 | 27 | 9 | 13 | 3 | 1231 | 41 | 48 | 29 | 174 | 67 | 59 | 10 |
| GEO | 104 | 3 | 11 | 5 | 14 | 3 | 6 | 2 | 862 | 19 | 29 | 19 | 94 | 20 | 29 | 1 |
| WAS | 125 | 4 | 11 | 7 | 14 | 1 | 7 | 0 | 1251 | 33 | 35 | 22 | 136 | 36 | 45 | 7 |
| FIN | 168 | 8 | 21 | 22 | 17 | 3 | 9 | 0 | 1666 | 67 | 98 | 70 | 247 | 84 | 84 | 6 |
| GER | 219 | 8 | 23 | 22 | 22 | 8 | 6 | 2 | 374 | 11 | 14 | 9 | 43 | 13 | 5 | 1 |
| SWE | 161 | 10 | 21 | 12 | 33 | 19 | 8 | 0 | 2364 | 112 | 118 | 75 | 436 | 207 | 102 | 6 |
| Other, USA | 9 | 0 | 0 | 0 | 1 | 1 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 | 0 |
| All | 928 | 39 | 105 | 86 | 128 | 44 | 49 | 7 | 7748 | 283 | 342 | 224 | 1130 | 427 | 324 | 31 |
lAb, single islet autoantibody; mAb, multiple islet autoantibodies; T1D, type 1 diabetes; CDA, celiac disease autoimmunity; CD, celiac disease; AT, autoimmune thyroiditis; ATD, autoimmune thyroid disease
Clinical Characteristics of Children Diagnosed with Diabetes
Despite their young age, TEDDY participants who developed T1D were often asymptomatic [32] and rarely had DKA, compared to the community cases [33]. At diagnosis, mean HbA1c was lower in TEDDY (6.8%, 51 mmol/mol) than age-matched control children diagnosed with diabetes in the community (10.5%, 91 mmol/mol) (P <0.0001). TEDDY children had significantly higher area under the curve and peak C-peptide values than the community controls throughout the first year post-diagnosis [34].
Cohort Retention
TEDDY is a demanding protocol requiring frequent visits, blood draws, extensive interviews, and athome tasks. Retention of study participants is our central focus with the goals of minimizing withdrawal and assessing possible bias. Of the original cohort, 5731 (66%) participants are actively engaged in the protocol 13 years since enrollment began; 31% have withdrawn from in-person visits, and only 3% have been lost to follow-up. Withdrawal was lower among FDR (24%) compared to GP (32%). Importantly, 71% of withdrawals occurred in the first 2 years of life and have steadily declined as the cohort grew older (Fig. 2). In the past 3 years, combined withdrawal and loss has been < 1% annually and even less among those with autoantibodies. Withdrawn subjects are contacted annually to update contact information and disease status and to offer re-enrollment; on average, 1853 (69%) of withdrawn participants complete the update annually. This surveillance has identified 23 subjects diagnosed with T1D and 27 with celiac disease. Additionally, 738 (40% of those contacted) have expressed interest in reengaging, and 403 subjects re-joined the study by completing another visit. A long-distance protocol, allowing for remote data and sample collection, includes 13% of the cohort. Local and national diabetes registries [3, 35–37] have been successfully engaged in surveillance for T1D among withdrawn and lost-to-follow-up participants.
Fig. 2.
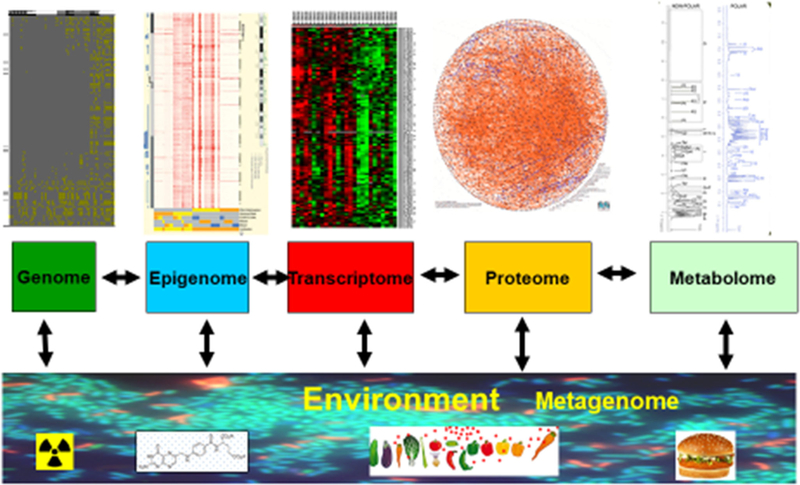
TEDDY nested case-control analyses [38]. All cases of persistent confirmed IA (N = 418) including childrenwho developed T1D (N = 114) ascertained by May 31, 2012 were selected. A total 1253 controls were matched to the cases of persistent confirmed IA (generally 3:1) on clinical center, sex, and family history of T1D. All the controls were included in studies for the dietary biomarkers and metabolomics and 418 controls (1:1) were included in metagenomics and gene expression studies. For the 114 T1D cases, 342 controls were selected for dietary biomarkers and metabolomics and 114 controls for the other studies. Number of samples tested include the following: dietary biomarkers (n = 23,594), metabolomics (n = 12,959), gene expression (n = 5200), microbiome (n = 13,403), viral metagenomics (n = 6380), proteomics (n = 5500)
The Nested Case-Control Study (NCCS)
Children who reached the study outcomes by 2012 (418 with autoantibodies and 114 with T1D) and matched controls (N = 1595; up to 3 controls were selected for each case) formed a nested case-control study—a multidimensional omic analysis of serial samples [38] (Fig. 1). Cases of persistent confirmed IA seroconverted at the median age 1.8 years (IQR 1.0–2.8). Children who developed T1D had median age of diagnosis 2.4 years (1.6–3.4). A total of 1253 controls were matched to the cases of persistent confirmed IA (generally 3:1) on clinical center, sex, and family history of T1D (Fig. 1). All the controls were included in studies for the dietary biomarkers and metabolomics and 418 controls (1:1) were included in metagenomics and gene expression studies. For the 114 T1D cases, 342 controls were selected for dietary biomarkers and metabolomics and 114 controls for the other studies.
Fig. 1.
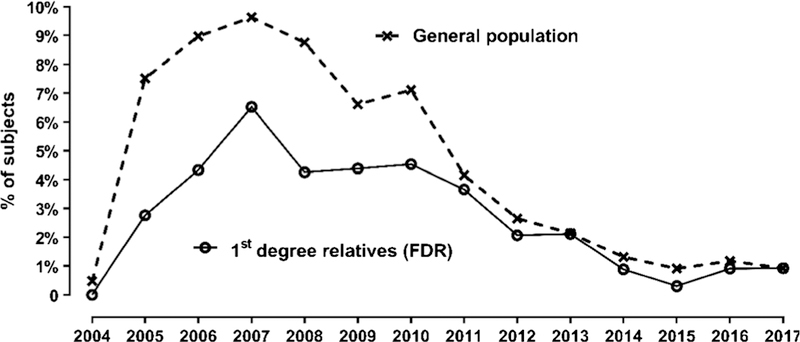
TEDDY cohort retention. Annual loss to follow-up (%)
Results in Major Study Areas
Cumulative Incidence of Islet Autoimmunity and T1D
The incidence of IA peaks at 2 years of life and decreases steadily thereafter. While additional children develop IA throughout childhood, their progression to T1D is slower than in those who developed IA as infants and toddlers. Either IAA first or GADA first was observed in 82% of the children (Table 2); a second autoantibody appeared within 1 year in 60% of the subjects who have developed either IAA first or GADA first [20]. By age 10 years, the cumulative incidence of any persistent confirmed islet autoantibody was 12.2% (95%CI, 11.4–13.1%) and for multiple islet autoantibodies it was 7.1% (95%CI, 6.5–7.8%). TEDDY has observed that 24% of children with single islet autoantibodies, particularly those with IAA (29%), lost their autoantibody without developing other islet autoantibodies [39]. It is important to determine whether such early transient autoimmunity can reappear in adolescents, and what factors are associated with remission compared to relapse of autoimmunity. TEDDY is also exploring profiles of autoantibodies that are associated with rapid or slow progression to diabetes [20, 40], and methods that can distinguish IgM from IgG islet autoantibodies [41, 42]. In the 5 years after the appearance of IA, T1D developed in 47% with three, 36% with two, and 11% with one autoantibody [40]. Higher IAA and IA-2A levels, but not GADA levels, predicted progression to T1D among persistently autoantibody-positive TEDDY children [40, 43].
Table 2.
First appearing islet autoantibody in 536 TEDDY children who seroconverted up to the age of 6 years
 |
Heterogeneity of Islet Autoimmunity and T1D
Previous studies have suggested a dichotomy in the pattern of islet autoantibodies: an early appearance of IAA, distinct from a later appearance of predominantly GADA-positive children [44–47]. TEDDY has confirmed this dichotomy in a large international population of children with and without T1D relatives [20, 23, 48, 49] and has discovered novel determinants of these IA phenotypes [23, 24•]. Children who present initially only with IAA carry usually one or two copies of the HLA DR4-DQ8 haplotype. This “IAA-first” phenotype peaks at 1 year of life and its incidence rapidly declines thereafter (Fig. 3). In contrast, a “GADA-first” phenotype presents later, persists at a steadier incidence, and is associated with HLA DR3-DQ2 [24•, 47].
Fig.3.
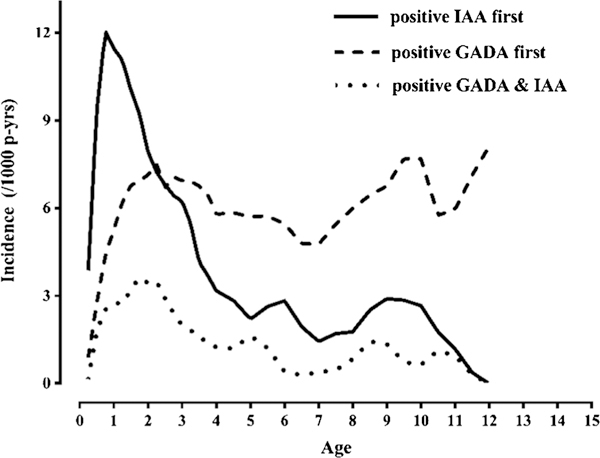
The incidence of islet autoantibodies. Early “IAA-first” phenotype followed by “GADA-first” phenotype
Cellular Immune Markers
TEDDY has regularly collected and frozen peripheral blood mononuclear cells (PBMC) from 5066 of the participants, including 554 who have developed islet autoantibodies. This provides opportunities to determine how the peripheral immune repertoire matures in childhood and how genetics and environment modify this development, and further characterize the immune cells that are associated with islet autoimmunity. Clinical Centers isolate and freeze sterile PBMC from TEDDY subjects in triplicate vials. Recent development in single-cell analysis and RNA sequencing will allow analysis of circulating blood cell subsets in relation to the appearance of the first islet autoantibody, progression to multiple autoantibodies, and clinical onset of diabetes. In order to fully capitalize on these opportunities, TEDDY has recruited experts in measuring and understanding innate and adaptive immune responses for workshops and ongoing pilot studies. The results of these experiments will guide the effort to comprehensively profile the immune development and immune repertoire before and during progression of islet autoimmunity. The analyses will focus on how environmental exposures that are associated with islet autoimmunity modify these immune responses.
Genetic Determinants of Islet Autoimmunity
The entire TEDDY cohort was genotyped using the ImmunoChip array [50] incorporating SNPs associated with one of 12 autoimmune diseases, including ~ 20,000 “wild card” SNPs, to comprise 176,586 SNPs in 186 loci. TEDDY first analyzed 5164 Caucasian children for 41 non-HLA SNPs that achieved genome-wide significance for association with T1D in the genome-wide association scan meta-analysis conducted by the Type 1 Diabetes Genetics Consortium [51]. In a time-to-event analysis, eight SNPs achieved significant association to the appearance of a first islet autoantibody and the following four remained significant after adjustment for multiple testing: rs2476601 in PTPN22, rs2292239 in ERBB3, rs3184504 in SH2B3, and rs1004446 in INS [17]. These SNPs were also significantly associated with T1D, in particular, rs2476601. Although genes in the HLA region remain the most important genetic risk factors for T1D, other non-HLA genetic factors contribute to a first appearing autoantibody, the first step in the pathogenesis of T1D.
A multivariate proportional hazards model of TEDDY made it possible to further dissect the genetic association with the first appearing autoantibody; rs2476601 in PTPN22 and rs2292239 in ERBB3 were related to both IAA first and GADA first [24•]. PTPN22 is a tyrosine phosphatase that inhibits T cell receptor signaling and selectively promote type I interferon responses [52]. ERBB3 is a tyrosine kinase cell surface growth factor receptor overexpressed in many cancers and viewed as a therapeutic target [53]. The common allele in rs689 in INS was the only non-HLA genetic factor that increased the risk for IAA first [24•]. The mechanism is thought to be explained by a reduced expression of insulin on thymocytes which lessens the efficiency of tolerance induction. The minor alleles in rs3184504 in SH2B3 and in rs3757247 in BACH2 increased the risk for GADA first but not for IAA first [24•]. SH2B adapter protein 3 (SH2B3) is commonly expressed in hematopoietic cells. BACH2, on the other hand, is a transcription factor essential for T and B lymphocytes. IA was also associated with a novel region near PPIL2 with suggestive evidence [19] for this recently reported minor autoantigen in newly diagnosed T1D [54, 55]. PPIL2 is a member of the cyclophilin family of peptidylprolyl isomerases; the protein is involved in protein folding and immunosuppression by cyclosporin A. TTC34/PRDM16 (tetratricopeptide repeat domain 34) was associated with IAA as the first appearing autoantibody and RBFOX1 with GADA first [19]. SNPs within the complement genes are also associated with IA [18].
TEDDY also tested if gestational infections in interactions with child’s HLA and non-HLA genes affected the appearance of IA [23]. Gestational infections were not associated with the first appearing islet autoantibodies overall; however, they showed a protective influence on IAA-first among CTLA4-(AG, GG) children [23]. The predominant associations of HLA-DR-DQ 4–8/4–8 with IAA and HLA-DR-DQ 3–2/3–2 with GADA were not observed if a gestation respiratory infection was reported. The role of gestational respiratory infection may depend on offspring HLA and CTLA-4 alleles and supports a bidirectional trigger for IAA or GADA as a first appearing beta-cell autoantibody in early life [23].
Genetic Determinants of Multiple Autoantibodies and T1D
PTPN22 and INS as well as a novel region, PXK/PDHB, were associated with the risk for multiple islet autoantibodies. Interestingly, the appearance of a second autoantibody was not related to HLA [20, 24•]. The extensive genetic analyses in the TEDDY study made it possible to exploit a genetic risk score (GRS) to stratify risk of developing multiple islet autoantibodies and type 1 diabetes [21]. The positive predictive value for multiple islet autoantibodies and T1D was determined in 3498 children in whom genetic scores were calculated from 41 SNPs. These children of mainly European descent had a greater than 10% risk for multiple islet autoantibodies, and a nearly twofold higher risk than children identified by high-risk HLA genotypes alone [21]. This GRS based on TEDDY genetics is currently used to screen newborn children to be enrolled in a primary prevention study with oral insulin in Germany [56, 57].
Infectious Triggers of Islet Autoimmunity and Promoters of Progression to T1D
TEDDY has used a unique exploratory approach to study the role of microbial exposures in the development of IA and T1D without any pathogen-specific a priori hypothesis. Comprehensive questionnaires (TEDDY book) are used to monitor the occurrence of infections, antibiotic treatments, and vaccinations. Omics-based laboratory technologies then identify specific various microbes and to characterize in detail the complete microbiome (including virome and parasitome) in longitudinal series of stool and blood samples and nasal swabs.
TEDDY has first developed methods to identify and classify > 113,000 parent-reported infectious episodes [58]. Importantly, respiratory infections in early childhood were temporally associated with the appearance of islet autoantibodies [8•]. In further analyses, respiratory infections predicted development of both the “IAA-first” and “GADA-first” phenotypes, while acute gastroenteritis predicted only the later-onset “GADA-first” phenotype (M. Lönnrot, unpublished). Acute gastroenteritis also predicted the appearance of transglutaminase autoantibodies—a marker of celiac disease [59]. In contrast to previous smaller studies, TEDDY found no association between the use of antibiotics and development of IA or celiac disease-associated autoimmunity [60].
Vaccinations
Vaccinations received by TEDDY participants have been classified and validated against medical records. Pandemrix® flu vaccination that triggered narcolepsy in Finland and Sweden did not increase the risk of IA or T1D in Finnish or Swedish TEDDY participants. In contrast, a negative association was found between the vaccine and IA or T1D [15] in Finland, but this was not seen in Swedish children.
Serum Virome
The association between viruses and IA/T1D has been analyzed using nested case-control design and nextgeneration sequencing to characterize the complete virome in stool, serum, white blood cell, and nasal swab samples. The first such study was carried out in a small subgroup of TEDDY infants, who rapidly developed T1D. They did not have more viruses detectable in serum than matched controls [12]. The virome analyses of stool, serum, “buffy coats,” and nasal swab specimens in a larger cohort of case and control children are in progress.
Gut Microbiome
The gut bacteriome in serial stool samples collected between 3 and 40 months of life from 903 children was characterized using 16S rRNA gene sequencing (n =11,717) and metagenomic sequencing (n = 10,602). TEDDY demonstrated the feasibility to detect lactobacilli in mail-in infant stools [61]. In all TEDDY geographical locations, the gut microbiome showed three distinct developmental phases, characterized primarily by reducing Bifidobacterium and increasing diversity. These phases correlated with diet, especially with breast-feeding which was the most significant factor associated with the microbiome structure. While a number of factors were identified to determine the development of gut bacteriome in early childhood, no association was found with IA [62, 63]. However, genes mapping to bacterial fermentation pathways, including the production of short-chain fatty acids (SCFA), were decreased in cases of IA, supporting previously postulated protective effects of SCFA on early-onset human T1D.
Gut Virome
Enteroviruses, which are among the main T1D- associated viruses, were found frequently by RT-PCR in stool samples but strikingly less often in Finland than in other TEDDY countries [64], consistent with the polio hypothesis [65]. Poliovirus infection led to paralysis most frequently in countries with a high standard of hygiene, lower frequency of poliovirus infections, and lower prevalence of maternal antibodies. Gut virome was also analyzed using mass sequencing. Preliminary results suggest an association between enteroviral infections and IA (R. Lloyd et al., unpublished).
Dietary Triggers of Islet Autoimmunity and Promoters of Progression to T1D
TEDDY has harmonized the food composition databases across the four participating countries for nutrient data [12, 66] and for food group data [67]. Exposure to probiotics in early infancy was found to decrease the risk of IA in the highest risk children [9], while use of hydrolyzed infant formula increased the risk of IA [13].Intake of soluble fiber is not associated with IA [16]. We have reported differences in supplement use, diet, and feeding patterns by country, sociodemographic status, and diabetes status of the parents [68–71].
Dietary Biomarkers
Vitamin D sufficiency was associated with a lower risk of IA overall, and particularly in children with at least one minor allele at a SNP in the vitamin D receptor gene [22•]. Higher levels of fish-derived fatty acids (EPA, DPA) in erythrocytes during infancy were also associated with lower risk of IA [72].
Body Fat and Physical Activity
Accelerated weight gain may increase the risk for T1D [73–76]. TEDDY monitors factors associated with insulin resistance and beta-cell overload including BMI and body composition as well as exercise and fitness. TEDDY subjects aged ≥ 5 years wear the Actigraph® GT3X+ accelerometer around the waist for 1 week per year to generate objective physical activity data. Over 16,000 measurements have been completed, so far. Preliminary data demonstrated significant variability in the activity levels by country after adjusting for sex, race/ethnicity, and mother’s education. Body composition is measured at each visit, using bioelectrical impedance assessment (BIA) device (Tanita® DC-430U).
Celiac Disease and Autoimmune Thyroid Disease
Annual transglutaminase autoantibody screening for CD began at 2 years of age. TEDDY also screens for autoantibodies to thyroid peroxidase and thyroglobulin starting at 8 years of age. Islet and transglutaminase autoantibodies co-occurred in 1.5% of children—more often than that expected by chance alone. Islet autoantibodies usually, but not always, appeared earlier than transglutaminase autoantibodies [77]. TEDDY has reported major country differences in the incidence of celiac disease autoimmunity [78], the clinical features [79], genetic predictors [80, 81], and the role of the early infant diet [82, 83], and infectious factors [59, 60] in risk of celiac disease. Interestingly, gluten introduction before 17 weeks or later than 26 weeks was not associated with increased risk for CDA or CD, controlling for country, HLA genotype, sex, and family history of CD [83].
Psychosocial Studies and Support
TEDDY has sought to determine the cognitive, emotional, and behavioral impact of the study participation on the child and parent [84–87]. Families that experience maladjustment to the stress of a child developing islet autoimmunity are offered professional counseling partially supported by the study. TEDDY has documented the following: (i) heightened parent anxiety associated with IAs in the child; (ii) the substantial number of parents who underestimate the child’s T1D risk despite multiple educational efforts; (iii) the substantial number of parents who report efforts to prevent T1D even when there is no known proven means to prevent the disease; and (iv) psychological manifestations of celiac disease autoimmunity in young children. We have also identified factors associated with study recruitment, retention, and compliance and developed/tested methods to improve study retention and compliance [88–94].
In the next 5 years, we will examine the long-term cognitive, emotional, and behavioral impact of TEDDY on participants. We will expand our studies to include the impact on children—who are now old enough to provide responses themselves—as well as parents. In addition, we are pursuing the extent to which parent report of dietary changes to prevent T1D—which are common—are associated with actual differences in the child’s diet.
As the participants get older, we have developed and put in place age-appropriate measures of primary study variables (stress, cognitive, emotional, and behavioral impact). In addition, we are evaluating tools (e.g., child-focused books, hand-outs) to help children understand TEDDY. We will use this information to develop additional tools and approaches to improve child and parent understanding of TEDDY, the child consent/assent process, and child/parent engagement in TEDDY, as well as assessing the impact of TEDDY on those families whose children do develop T1D. These assessments will also permit us to examine the role of stress together with other environmental triggers on the initiation and progression of islet autoimmunity.
TEDDY Study Mechanisms for Inclusion of New Hypotheses and Technologies
TEDDY utilizes multiple mechanisms to enrich the study scope and to optimize the protocol as new discoveries and technologies become available. Examples include:
TEDDY Science Workshops on Immune Markers (March 04, 2013), Systems Biology (March 05, 2013), Application of New Technology (October 07, 2014 and May 19, 2015), Infectious Agents (November 13, 2017), Immune Markers Analysis (November 14, 2017), and Epigenetics (November 16, 2017) which gathered investigators and international experts to discuss innovative approaches and methodology. Recommendations from this workshop led to the design of TEDDY pilot studies and the development of RFPs (requests for proposals).
Children’s Health Exposure Analysis Resource (CHEAR) Project in collaboration with CHEAR, approved by NIEHS in Feb. 2018, TEDDY will assess whether environmental exposures and toxicants (phthalates, phenols, metals) affect inflammatory markers and lead to the initiation of islet autoimmunity or progression to T1D.
Ancillary Study applications are encouraged from the research community at large for studies on samples or data collected by TEDDY to answer research questions that complementthe study objectives and thereby enhance the value of the project (for policy, see http://teddy.epi.usf.edu/research). They are evaluated for merit, feasibility, competition for samples, and potential impact. Over the last 5 years, 11 proposals have been reviewed and six approved. The NIDDK Data Repository periodically receives data from the TEDDY study and encourages researches to request access to these data (www.niddkrepository.org).
TEDDY Data Sharing Policy has been developed to structure the external release of TEDDY data not available through the NIDDK repository (http://teddy.epi.usf.edu/research). Access to TEDDY data has been approved for 62 proposals, including 38 from TEDDY investigators and 24 from outside researchers.
Discussion/Future Directions
TEDDY cohort’s size, population diversity, intensive followup, and application of unbiased “omics” technologies position the study well to discover the environmental cause(s) of T1D and the mechanisms by which they act. In addition to studies reviewed above, TEDDY is carrying out analyses of transcriptomic, proteomic, and metabolomics profiles of a large number of cases and controls. Epigenetic analyses are planned for the near future. TEDDY can evaluate both highly prevalent and rare exposures that require a larger sample size and assess a wide range of environmental factors prospectively from early age and repeatedly as they evolve during childhood and adolescence. Extended follow-up is critical as the effects of the exposures may have a long lag time. The TEDDY population covers a wide spectrum of genetic variation and environmental exposures by including diverse populations: Finnish, Swedish, German, southeast US with representation of African Americans, southwest US with Hispanics, and northwest US with Asian Americans. The study participants are genotyped using high-density SNPs and whole-genome sequencing and their environmental exposures are assessed prospectively allowing novel geneenvironment analyses. In addition to IA and T1D, TEDDY defines celiac and thyroid autoimmunity phenotypes. One of the main goals of TEDDY moving forward is integration of “big data” generated by our omics laboratories with analytical help from the broad scientific community. The large-scale meticulous banking of study specimens in the NIDDK repository will facilitate access to the samples by the research community for years to come.
TEDDY has already collected an extraordinary amount of prospective data and biological samples from high-risk children followed through the initial 8 years of life—the critical period for triggering islet autoimmunity. However, islet autoimmunity begins frequently also after age 8 and may be an important determinant of T1D diagnosed in adults. Little is known about environmental triggers of islet autoimmunity in older children and TEDDY will now address this gap in our understanding of T1D etiology and heterogeneity.
Acknowledgements
The TEDDY Study Group (see Appendix)
Funding Information The TEDDY Study is funded by U01 DK63829, U01 DK63861, U01 DK63821, U01 DK63865, U01 DK63863, U01 DK63836, U01 DK63790, UC4 DK63829, UC4 DK63861, UC4 DK63821, UC4 DK63865, UC4 DK63863, UC4 DK63836, UC4 DK95300, UC4 DK100238, UC4 DK106955, UC4 DK112243, UC4 DK117483, and Contract No. HHSN267200700014C from the National Institute of Diabetes and Digestive and Kidney Diseases (NIDDK), National Institute of Allergy and Infectious Diseases (NIAID), National Institute of Child Health and Human Development (NICHD), National Institute of Environmental Health Sciences (NIEHS), Centers for Disease Control and Prevention (CDC), and JDRF. This work supported in part by the NIH/NCATS Clinical and Translational Science Awards to the University of Florida (UL1 TR000064) and the University of Colorado (UL1 TR001082).
Appendix: TEDDY Study Acknowledgments
The TEDDY Study Group
Colorado Clinical Center:
Marian Rewers, M.D., Ph.D., PI1,4,5,6,10,11, Kimberly Bautista12, Judith Baxter9,10,12,15, Daniel Felipe-Morales, Kimberly Driscoll, Ph.D.9, Brigitte I. Frohnert, M.D.2,14, Marisa Gallant, M.D.13, Patricia Gesualdo2,6,12,14,15, Michelle Hoffman12,13,14, Rachel Karban12, Edwin Liu, M.D.13, Jill Norris, Ph.D.2,3,12, Adela Samper-Imaz, Andrea Steck, M.D.3,14, Kathleen Waugh6,7,12,15, Hali Wright12. University of Colorado, Anschutz Medical Campus, Barbara Davis Center for Childhood Diabetes.
Finland Clinical Center:
Jorma Toppari, M.D., Ph.D., PI¥˄1,,4,11,14, oni g. simell, M.D., Ph.D., Annika Adamsson, Ph.D.˄12, Suvi Ahonen*±§, Heikki Hyöty, M.D., Ph.D.*±6, Jorma Ilonen, M.D., Ph.D.¥¶3, Sanna Jokipuu˄, Leena Karlsson˄, Miia Kähönenμ¤, Mikael Knip, M.D., Ph.D.*±5, Mirva Koreasalo*±§2, Kalle Kurppa, M.D., Ph.D.*±13, Tiina Latva-ahoμ¤, Maria Lönnrot, M.D., Ph.D.*±6, Markus Mattila*, Elina Mäntymäki˄, Katja Multasuoμ¤, Tiina Niininen±*12, Sari Niinistö±§2, Mia Nyblom*±, Sami Oikarinen, Ph.D.*±, Paula Ollikainenμ¤, Petra Rajala˄, Jenna Rautanen±§, Anne Riikonen*±§, Minna Romo˄, Suvi Ruohonen˄, Juulia Rönkäμ¤, Satu Simell, M.D., Ph.D.¥13, Tuula Simell, Ph.D.¥12, Maija Sjöberg¥˄12,14, Aino Steniusμ¤12, Sini Vainionpää˄, Eeva Varjonen¥˄12, Riitta Veijola, M.D., Ph.D.μ¤14, Suvi M. Virtanen, M.D., Ph.D.*±§2, Mari Vähä-Mäkilä˄, Mari Åkerlund*±§, Katri Lindfors, Ph.D.*13 ¥University of Turku, *University of Tampere, μUniversity of Oulu, ˄Turku University Hospital, Hospital District of Southwest Finland, ±Tampere University Hospital, ¤Oulu University Hospital, §National Institute for Health and Welfare, Finland, ¶University of Kuopio.
Georgia/Florida Clinical Center:
Jin-Xiong She, Ph.D., PI1,3,4,11, Desmond Schatz, M.D.*4,5,7,8, Diane Hopkins12, Leigh Steed12,13,14,15, Jennifer Bryant, Janey Adams*12, Katherine Silvis2, Michael Haller, M.D.*14, Melissa Gardiner, Richard McIndoe, Ph.D., Ashok Sharma, Stephen W. Anderson, M.D.˄, Laura Jacobsen, M.D.*14, John Marks, DHSc.*14, P.D. Towe*. Center for Biotechnology and Genomic Medicine, Augusta University. *University of Florida, ˄Pediatric Endocrine Associates, Atlanta.
Germany Clinical Center:
Anette G. Ziegler, M.D., PI1,3,4,11, Andreas Beyerlein, Ph.D.2, Ezio Bonifacio Ph.D.*5, Anita Gavrisan, Cigdem Gezginci, Anja Heublein, Michael Hummel, M.D.13, Sandra Hummel, Ph.D.2, Andrea Keimer2, Annette Knopff7, Charlotte Koch, Sibylle Koletzko, M.D.¶13, Claudia Ramminger, Roswith Roth, Ph.D.9, Marlon Scholz, Joanna Stock9,12,14, Katharina Warncke, M.D.14, Lorena Wendel, Christiane Winkler, Ph.D.2,12,15. Forschergruppe Diabetes e.V. and Institute of Diabetes Research, Helmholtz Zentrum München, Forschergruppe Diabetes, and Klinikum rechts der Isar, Technische Universität München. *Center for Regenerative Therapies, TU Dresden, ¶Dr. von Hauner Children’s Hospital, Department of Gastroenterology, Ludwig Maximillians University Munich.
Sweden Clinical Center:
Åke Lernmark, Ph.D., PI1,3,4,5,6,8,10,11,15, Daniel Agardh, M.D., Ph.D.13, Carin Andrén Aronsson, Ph.D.2,12,13, Maria Ask, Jenny Bremer, Ulla-Marie Carlsson, Corrado Cilio, Ph.D., M.D.5, Emelie Ericson-Hallström, Annika Fors, Lina Fransson, Thomas Gard, Rasmus Bennet, Carina Hansson, Susanne Hyberg, Hanna Jisser, Fredrik Johansen, Berglind Jonsdottir, M.D., Ph.D., Silvija Jovic, Helena Elding Larsson, M.D., Ph.D. 6,14, Marielle Lindström, Markus Lundgren, M.D., Ph.D.14, Maria Månsson-Martinez, Maria Markan, Jessica Melin12, Zeliha Mestan, Caroline Nilsson, Karin Ottosson, Kobra Rahmati, Anita Ramelius, Falastin Salami, Sara Sibthorpe, Anette Sjöberg, Birgitta Sjöberg, Carina Törn, Ph.D. 3,15, Anne Wallin, Åsa Wimar14, Sofie Åberg. Lund University.
Washington Clinical Center:
William A. Hagopian, M.D., Ph.D., PI1,3,4, 5 6,7,11,13 14, Michael Killian6,7,12,13, Claire Cowen Crouch12,14,15, Jennifer Skidmore2, Ashley Akramoff, Jana Banjanin, Masumeh Chavoshi, Kayleen Dunson, Rachel Hervey, Rachel Lyons, Arlene Meyer, Denise Mulenga, Jared Radtke, Davey Schmitt, Julie Schwabe, Sarah Zink. Pacific Northwest Research Institute.
Pennsylvania Satellite Center:
Dorothy Becker, M.D., Margaret Franciscus, MaryEllen Dalmagro-Elias Smith2, Ashi Daftary, M.D., Mary Beth Klein, Chrystal Yates. Children’s Hospital of Pittsburgh of UPMC.
Data Coordinating Center:
Jeffrey P. Krischer, Ph.D.,PI14,5,10,11, Sarah Austin-Gonzalez, Maryouri Avendano, Sandra Baethke, Rasheedah Brown12,15, Brant Burkhardt, Ph.D.5,6, Martha Butterworth2, Joanna Clasen, David Cuthbertson, Christopher Eberhard, Steven Fiske9, Dena Garcia, Jennifer Garmeson, Veena Gowda, Kathleen Heyman, Belinda Hsiao, Francisco Perez Laras, Hye-Seung Lee, Ph.D.1,2,13,15, Shu Liu, Xiang Liu, Ph.D.2,3,9,14, Kristian Lynch, Ph.D. 5,6,9,15 Colleen Maguire, Jamie Malloy, Cristina McCarthy12,15, Aubrie Merrell, Steven Meulemans, Hemang Parikh, Ph.D.3, Ryan Quigley, Cassandra Remedios, Chris Shaffer, Laura Smith, Ph.D.9,12, Susan Smith12,15, Noah Sulman, Ph.D., Roy Tamura, Ph.D.1,2,13, Ulla Uusitalo, Ph.D.2,15, Kendra Vehik, Ph.D.4,5,6,14,15, Ponni Vijayakandipan, Keith Wood, Jimin Yang, Ph.D., R.D.2,15. Past staff: Michael Abbondondolo, Lori Ballard, David Hadley, Ph.D., Wendy McLeod. University of South Florida.
Project scientist:
Beena Akolkar, Ph.D.1,3,4,5,6,7,10,11. National Institutes of Diabetes and Digestive and Kidney Diseases.
Other contributors:
Kasia Bourcier, Ph.D.5, National Institutes of Allergy and Infectious Diseases. Thomas Briese, Ph.D.6,15, Columbia University. Suzanne Bennett Johnson, Ph.D.9,12, Florida State University. Eric Triplett, Ph.D.6, University of Florida.
Teddy Study Group: Affiliated Laboratories
Laboratory personnel should be copied-and-pasted for inclusion in the TEDDY Study Group Appendix on relevant manuscripts. Please review the labs below to determine which locations should be included in your manuscript’s Acknowledgements. (The Repository should be included for any manuscript referring to lab data.)
Autoantibody Reference Laboratories:
Liping Yu, M.D. ˄5, Dongmei Miao, M.D.˄, Polly Bingley, M.D., FRCP*5, Alistair Williams*, Kyla Chandler*, Olivia Ball*, Ilana Kelland*, Sian Grace*, Ben Gillard*. ˄Barbara Davis Center for Childhood Diabetes, University of Colorado Denver, *Bristol Medical School, University of Bristol UK.
Cortisol Laboratory:
Elisabeth Aardal Eriksson, M.D., Ph.D., Ing-Marie Lundgren, Ewa Lönn Karlsson, Dzeneta Nezirevic Dernroth, Ph.D. Department of Clinical Chemistry, Linköping University Hospital, Linköping, Sweden.
Dietary Biomarkers Laboratory:
Iris Erlund, Ph.D.2, Irma Salminen, Jouko Sundvall, Nina Kangas, Petra Arohonka. National Institute for Health and Welfare, Helsinki, Finland.
HbA1c Laboratory:
Randie R. Little, Ph.D., Curt Rohlfing. Diabetes Diagnostic Laboratory, Dept. of Pathology, University of Missouri School of Medicine.
HLA Reference Laboratory:
William Hagopian3, MD, PhD, Masumeh Chavoshi, Jared Radtke, Julie Schwabe. Pacific Northwest Research Institute, Seattle WA. (Previously Henry Erlich, Ph.D.3, Steven J. Mack, Ph.D., Anna Lisa Fear. Center for Genetics, Children’s Hospital Oakland Research Institute.)
Metabolomics Laboratory:
Oliver Fiehn, Ph.D., Bill Wikoff, Ph.D., Brian Defelice, Dmitry Grapov, Ph.D., Tobias Kind, Ph.D., Mine Palazoglu, Luis Valdiviez, Benjamin Wancewicz, Gert Wohlgemuth, Joyce Wong. UC Davis Metabolomics Center.
Metagenomics and Microbiome Laboratory:
Joseph F. Petrosino, Ph.D.6*, Nadim J. Ajami, Ph.D.*, Richard E. Lloyd, Ph.D.6*, Matthew C. Ross, Ph.D.*, Jacqueline L. O’Brien, Ph.D.*, Diane S. Hutchinson, Ph.D.*, Daniel P. Smith, Ph.D.*, Matthew C. Wong*, Xianjun Tian, Ph.D.*, Tulin Ayvaz*, Auriole Tamegnon*, Nguyen Truong*, Hannah Moreno*, Lauren Riley*, Eduardo Moreno*, Tonya Bauch*, Lenka Kusic*, Ginger Metcalf˄, Donna Muzny˄, HarshaVArdhan Doddapaneni, Ph.D.˄, Richard Gibbs, Ph.D.˄. *Alkek Center for Metagenomics and Microbiome Research, Department of Molecular Virology and Microbiology, Baylor College of Medicine, ˄Human Genome Sequencing Center, Baylor College of Medicine.
OGTT Laboratory:
Santica M. Marcovina, Ph.D., Sc.D., Vinod P. Gaur, Ph.D., Northwest Lipid Metabolism and Diabetes Research Laboratories, University of Washington.
Proteomics Laboratory:
Richard D. Smith, Ph.D., Thomas O. Metz, Ph.D., Charles Ansong, Ph.D., Bobbie-Jo Webb- Robertson, Ph.D., Hugh D. Mitchell, Ph.D., Ernesto S. Nakayasu, Ph.D., and Wei-Jun Qian, Ph.D. Pacific Northwest National Laboratory.
Repository:
Sandra Ke, Niveen Mulholland, Ph.D. NIDDK Biosample Repository at Fisher BioServices.
RNA Laboratory and Gene Expression Laboratory:
Jin- Xiong She, Ph.D., PI1,3,4,11, Richard Mclndoe, Ph.D., Haitao Liu, M.D., John Nechtman, Yansheng Zhao, Na Jiang, M.D., Yanna Tian, M.S. Guangkuo Dong, M.S. Jinfiniti Biotech, LLC.
SNP Laboratory:
Stephen S. Rich, Ph.D.3, Wei-Min Chen, Ph.D.3, Suna Onengut-Gumuscu, Ph.D.3, Emily Farber, Rebecca Roche Pickin, Ph.D., Jonathan Davis, Jordan Davis, Dan Gallo, Jessica Bonnie, Paul Campolieto. Center for Public Health Genomics, University of Virginia.
Thyroid Laboratory:
William E. Winter, M.D., David L. Pittman. UF Health Pathology Laboratories’ Endocrinology Laboratory, University of Florida.
Committees:
1Ancillary Studies, 2Diet, 3Genetics, 4Human Subjects/ Publicity/Publications, 5Immune Markers, 6Infectious Agents, 7Laboratory Implementation, 8Maternal Studies, 9Psychosocial, 10Quality Assurance, 11Steering, 12Study Coordinators, 13Celiac Disease, 14Clinical Implementation, 15Quality Assurance Subcommittee on Data Quality.
Footnotes
Compliance with Ethical Standards
Conflict of Interest Marian Rewers, William Hagopian, Jin-Xiong She, Desmond Schatz, Anette-G Ziegler, Beena Akolkar, and Jeffrey Krischer declare that they have no conflict of interest.
Heikki Hyöty reports grants from National Institute of Health (NIH) to carry out the TEDDY study; and being a Shareholder and member of the board of Vactech Ltd., which develops vaccines against picornaviruses.
Ake Lernmark is a Member of the Scientific Advisory Board of Diamyd Medical AB, Stockholm, Sweden.
Jorma Toppari reports grants from NIH/NIDDK.
Human and Animal Rights and Informed Consent This article does not contain any studies with human or animal subjects performed by any of the authors. The TEDDY study was approved by local institutional review or ethics boards at each site (University of Washington, Seattle; University of Colorado; Medical College of Georgia, Augusta; University of South Florida, Tampa; University of Turku, Finland; Technische Universitat, Munich, Germany; Lund University, Malmo, Sweden) and is monitored by an External Evaluation Committee formed by the National Institutes of Health.
References
Papers of particular interest, published recently, have been highlighted as:
• Of importance
- 1.Group DERI. Secular trends in incidence of childhood IDDM in 10 countries. Diabetes. 1990;39(7):858–64. [PubMed] [Google Scholar]
- 2.Patterson CC, Dahlquist GG, Gyurus E, Green A, Soltesz G. Incidence trends for childhood type 1 diabetes in Europe during 1989–2003 and predicted new cases 2005–20: a multicentre prospective registration study. Lancet. 2009;373(9680):2027419–2933. [DOI] [PubMed] [Google Scholar]
- 3.Mayer-Davis EJ, Lawrence JM, Dabelea D, Divers J, Isom S, Dolan L, et al. Incidence trends of type 1 and type 2 diabetes among youths, 2002–2012. N Engl J Med. 2017;376(15):1419–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.National Center for Chronic Disease Prevention and Health Promotion DoDT. National diabetes statistics report, 2017. Estimates of diabetes and its burden in the United States. 2017. http://www.diabetes.org/assets/pdfs/basics/cdc-statistics-report-2017.pdf.
- 5.Rewers M, Ludvigsson J. Environmental risk factors for type 1 diabetes. Lancet. 2016;387(10035):2340419–28. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) study: study design. Pediatr Diabetes. 2007;8(5):286–98. [DOI] [PubMed] [Google Scholar]
- 7.TEDDY Study Group. The Environmental Determinants of Diabetes in the Young (TEDDY) study. Ann N Y Acad Sci. 2008;1150:1–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.• Lonnrot M, Lynch KF, Elding Larsson H, et al. Respiratory infections are temporally associated with initiation of type 1 diabetes autoimmunity: the TEDDY study. Diabetologia. 2017;60(10): 1931–40.TEDDY reports that respiratory infections increase the risk of islet autoimmunity in young children. Islet autoimmunity developed in 454 out of 7869 high-risk children followed from birth. Each infection in a 9-month period increased the subsequent risk of autoimmunity by 6%.
- 9.Uusitalo U, Liu X, Yang J, Aronsson CA, Hummel S, Butterworth M, et al. Association of early exposure of probiotics and islet autoimmunity in the TEDDY study. JAMA Pediatr. 2016;170(1):20–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Uusitalo U, Lee HS, Andren Aronsson C, et al. Early infant diet and islet autoimmunity in the TEDDY study. Diabetes Care. 2018;41(3):522–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Lundgren M, Steed LJ, Tamura R, et al. Analgesic antipyretic use among young children in the TEDDY study: no association with islet autoimmunity. BMC Pediatr. 2017;17(1):127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Ziegler AG, Pflueger M, Winkler C, Achenbach P, Akolkar B, Krischer JP, et al. Accelerated progression from islet autoimmunity to diabetes is causing the escalating incidence of type 1 diabetes in young children. J Autoimmun. 2011;37(1):3–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Hummel S, Beyerlein A, Tamura R, Uusitalo U, Andrén Aronsson C, Yang J, et al. First infant formula type and risk of islet autoimmunity in The Environmental Determinants of Diabetes in the Young (TEDDY) study. Diabetes Care. 2017;40(3):398–404. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Elding Larsson H, Vehik K, Haller MJ, Liu X, Akolkar B, Hagopian W, et al. Growth and risk for islet autoimmunity and progression to type 1 diabetes in early childhood: The Environmental Determinants of Diabetes in the Young study. Diabetes. 2016;65(7):1988–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Elding Larsson H, Lynch KF, Lonnrot M, et al. Pandemrix(R) vaccination is not associated with increased risk of islet autoimmunity or type 1 diabetes in the TEDDY study children. Diabetologia. 2018;61(1):193–202. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Beyerlein A, Liu X, Uusitalo UM, Harsunen M, Norris JM, Foterek K, et al. Dietary intake of soluble fiber and risk of islet autoimmunity by 5 y of age: results from the TEDDY study. Am J Clin Nutr. 2015;102(2):345–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Torn C, Hadley D, Lee HS, et al. Role of type 1 diabetes-associated SNPs on risk of autoantibody positivity in the TEDDY study. Diabetes. 2015;64(5): 1818–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Torn C, Liu X, Hagopian W, et al. Complement gene variants in relation to autoantibodies to beta cell specific antigens and type 1 diabetes in the TEDDY study Sci Rep. 2016;6:27887. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sharma A, Liu X, Hadley D, Hagopian W, Chen WM, Onengut-Gumuscu S, et al. Identification of non-HLA genes associated with development of islet autoimmunity and type 1 diabetes in the prospective TEDDY cohort. J Autoimmun. 2018;89:90–100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Krischer JP, Liu X, Lernmark A, Hagopian WA, Rewers MJ, She JX, et al. The influence of type 1 diabetes genetic susceptibility regions, age, sex, and family history on the progression from multiple autoantibodies to type 1 diabetes: a TEDDY study report. Diabetes. 2017;66(12):3122–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bonifacio E, Beyerlein A, Hippich M, Winkler C, Vehik K. Genetic scores to stratify risk of developing multiple islet autoantibodies and type 1 diabetes: a prospective study in children. PLoS Med. 2018;15(4):e1002548. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.• Norris JM, Lee HS, Frederiksen B, Erlund I, Uusitalo U, Yang J, et al. Plasma 25-hydroxyvitamin D concentration and risk of islet autoimmunity. Diabetes. 2018;67(1):146–54.TEDDY provides evidence for a protective effect of higher plasma vitamin D levels for development of islet autoimmunity in high-risk children. However, this protective effect is limited to children who carry specific vitamin D receptor genotypes.
- 23.Lynch KF, Lee HS, Torn C, et al. Gestational respiratory infections interacting with offspring HLA and CTLA-4 modifies incident beta-cell autoantibodies. J Autoimmun. 2018;86:93–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.• Krischer JP, Lynch KF, Lernmark A, Hagopian WA, Rewers MJ, She JX, et al. Genetic and environmental interactions modify the risk of diabetes-related autoimmunity by 6 years of age: the TEDDY study. Diabetes Care. 2017;40(9):1194–202.TEDDY demonstrates an apparent heterogeneity of type 1 diabetes and the underlying genetic and environmental determinants.
- 25.Russell CD, Baillie JK. Treatable traits and therapeutic targets: goals for systems biology in infectious disease. Curr Opin Syst Biol. 2017;2:140–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Hagopian WA, Erlich H, Lernmark A, Rewers M, Ziegler AG, Simell O, et al. The environmental determinants of diabetes in the young (TEDDY): genetic criteria and international diabetes risk screening of 421 000 infants. Pediatr Diabetes. 2011;12(8):733–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Cucca F, Lampis R, Frau F, Macis D, Angius E, Masile P, et al. The distribution of DR4 haplotypes in Sardinia suggests a primary association of type I diabetes with DRB1 and DQB1 loci. Hum Immunol. 1995;43(4):301–8. [DOI] [PubMed] [Google Scholar]
- 28.Erlich HA, Valdes AM, Noble JA. Prediction of type 1 diabetes. Diabetes. 2013;62(4):1020–1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Insel RA, Dunne JL, Atkinson MA, Chiang JL, Dabelea D, Gottlieb PA, et al. Staging presymptomatic type 1 diabetes: a scientific statement of JDRF, the Endocrine Society, and the American Diabetes Association. Diabetes Care. 2015;38(10):1964–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Vehik K, Fiske SW, Logan CA, Agardh D, Cilio CM, Hagopian W, et al. Methods, quality control and specimen management in an international multicentre investigation of type 1 diabetes: TEDDY Diabetes Metab Res Rev. 2013;29(7):557–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.American Diabetes Association. Classification and diagnosis of diabetes. Diabetes Care. 2017;40(Suppl 1):S11–24.27979889 [Google Scholar]
- 32.Elding Larsson H, Vehik K, Gesualdo P, Akolkar B, Hagopian W, Krischer J, et al. Children followed in the TEDDY study are diagnosed with type 1 diabetes at an early stage of disease. Pediatr Diabetes. 2014;15(2):118–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Elding Larsson H, Vehik K, Bell R, Dabelea D, Dolan L, Pihoker C, et al. Reduced prevalence of diabetic ketoacidosis at diagnosis of type 1 diabetes in young children participating in longitudinal follow-up. Diabetes Care. 2011;34(11):2347–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Steck AK, Larsson HE, Liu X, Veijola R, Toppari J, Hagopian WA, et al. Residual beta-cell function in diabetes children followed and diagnosed in the TEDDY study compared to community controls. Pediatr Diabetes. 2017;18(8):794–802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Harjutsalo V, Sund R, Knip M, Groop PH. Incidence of type 1 diabetes in Finland. JAMA. 2013;310(4):427–8. [DOI] [PubMed] [Google Scholar]
- 36.Bendas A, Rothe U, Kiess W, Kapellen TM, Stange T, Manuwald U, et al. Trends in incidence rates during 1999–2008 and prevalence in 2008 of childhood type 1 diabetes mellitus in Germany—model- based national estimates. PLoS One. 2015;10(7):e0132716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Hanberger L, Birkebaek N, Bjarnason R, Drivvoll AK, Johansen A, Skrivarhaug T, et al. Childhood diabetes in the Nordic countries: a comparison of quality registries. J Diabetes Sci Technol. 2014;8(4): 738–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lee HS, Briese T, Winkler C, et al. Next-generation sequencing for viruses in children with rapid-onset type 1 diabetes. Diabetologia. 2013;56(8):1705–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Vehik K, Lynch KF, Schatz DA, Akolkar B, Hagopian W, Rewers M, et al. Reversion of beta-cell autoimmunity changes risk of type 1 diabetes: TEDDY study. Diabetes Care. 2016;39(9):1535–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Steck AK, Vehik K, Bonifacio E, Lernmark A, Ziegler AG, Hagopian WA, et al. Predictors of progression from the appearance of islet autoantibodies to early childhood diabetes: The Environmental Determinants of Diabetes in the Young (TEDDY). Diabetes Care. 2015;38(5):808–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yu L, Dong F, Miao D, Fouts AR, Wenzlau JM, Steck AK. Proinsulin/insulin autoantibodies measured with electrochemiluminescent assay are the earliest indicator of predia¬betic islet autoimmunity. Diabetes Care. 2013;36(8):2266–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Yu LZ, Miao D, Waugh K, Jiang L, Steck A, Liu L, et al. Electrochemiluminescence-based IAA and GADA assays detect the appearance of islet autoimmunity earlier than radioimmunoassay in a significant proportion of children. San Francisco: Immunology of Diabetes Society; 2017. [Google Scholar]
- 43.Kohler M, Beyerlein A, Vehik K, et al. Joint modeling of longitudinal autoantibody patterns and progression to type 1 diabetes: results from the TEDDY study. Acta Diabetol. 2017;54(11):1009–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Ilonen J, Hammais A, Laine AP, Lempainen J, Vaarala O, Veijola R, et al. Patterns of beta-cell autoantibody appearance and genetic associations during the first years of life. Diabetes. 2013;62(10): 3636–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ziegler AG, Bonifacio E. Age-related islet autoantibody incidence in offspring of patients with type 1 diabetes. Diabetologia. 2012;55(7):1937–43. [DOI] [PubMed] [Google Scholar]
- 46.Ziegler AG, Hummel M, Schenker M, Bonifacio E. Autoantibody appearance and risk for development of childhood diabetes in offspring of parents with type 1 diabetes: the 2-year analysis of the German BABYDIAB study. Diabetes. 1999;48(3):460–8. [DOI] [PubMed] [Google Scholar]
- 47.Hagopian WA, Sanjeevi CB, Kockum I, Landin-Olsson M, Karlsen AE, Sundkvist G, et al. Glutamate decarboxylase-, insulin-, and islet cell-antibodies and HLA typing to detect diabetes in a general population-based study of Swedish children. J Clin Invest. 1995;95(4):1505–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Krischer JP, Lynch KF, Schatz DA, et al. The 6 year incidence of diabetes-associated autoantibodies in genetically at-risk children: the TEDDY study. Diabetologia. 2015;58(5):980–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.de Goffau MC, Luopajarvi K, Knip M, Ilonen J, Ruohtula T, Harkonen T, et al. Fecal microbiota composition differs between children with beta-cell autoimmunity and those without. Diabetes. 2013;62:1238–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Parkes M, Cortes A, van Heel DA, Brown MA. Genetic insights into common pathways and complex relationships among immunemediated diseases. Nat Rev Genet. 2013;14(9):661–73. [DOI] [PubMed] [Google Scholar]
- 51.Cooper JD, Howson JM, Smyth D, et al. Confirmation of novel type 1 diabetes risk loci in families. Diabetologia. 2012;55(4): 996–1000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Stanford SM, Bottini N. PTPN22: the archetypal non-HLA autoimmunity gene. Nat Rev Rheumatol. 2014;10(10):602–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Roepstorff K, Grovdal L, Grandal M, Lerdrup M, van Deurs B. Endocytic downregulation of ErbB receptors: mechanisms and relevance in cancer. Histochem Cell Biol. 2008;129(5):563–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Muller D, Telieps T, Eugster A, et al. Novel minor HLA DR associated antigens in type 1 diabetes. Clin Immunol. 2018;194:87–91. [DOI] [PubMed] [Google Scholar]
- 55.Bian X, Wasserfall C, Wallstrom G, Wang J, Wang H, Barker K, et al. Tracking the antibody immunome in type 1 diabetes using protein arrays. J Proteome Res. 2017;16(1):195–203. [DOI] [PubMed] [Google Scholar]
- 56.Ziegler AG, Danne T, Dunger DB, Berner R, Puff R, Kiess W, et al. Primary prevention of beta-cell autoimmunity and type 1 diabetes—the Global Platform for the Prevention of Autoimmune Diabetes (GPPAD) perspectives. Mol Metab. 2016;5(4):255–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hommel A, Haupt F, Delivani P, Winkler C, Stopsack M, Wimberger P, et al. Screening for type 1 diabetes risk in newborns: the Freder1k pilot study in Saxony. Horm Metab Res. 2018;50(1): 44–9. [DOI] [PubMed] [Google Scholar]
- 58.Lonnrot M, Lynch K, Larsson HE, et al. A method for reporting and classifying acute infectious diseases in a prospective study of young children: TEDDY. BMC Pediatr. 2015;15:24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Kemppainen KM, Lynch KF, Liu E, Lönnrot M, Simell V, Briese T, et al. Factors that increase risk of celiac disease autoimmunity after a gastrointestinal infection in early life. Clin Gastroenterol Hepatol. 2017;15(5):694–702.e695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kemppainen KM, Vehik K, Lynch KF, Larsson HE, Canepa RJ, Simell V, et al. Association between early-life antibiotic use and the risk of islet or celiac disease autoimmunity. JAMA Pediatr. 2017;171 (12):1217–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Salami F, Abels M, Hyoty H, et al. Detection of Latobacilli in monthly mail-in stool samples from 3–18 months old infants at genetic risk for type 1 diabetes. Int Journal of Probiotics Prebiotics. 2012;7(3–4):135–44. [PMC free article] [PubMed] [Google Scholar]
- 62.Stewart CJ, Ajami NJ, O’Brien JL, et al. Temporal development of the gut microbiome in early childhood—the TEDDY study. Nature. 2018;563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Vatanen T, Franzosa EA, Schwager R, et al. The human gut microbiome of early onset type 1 diabetes in the TEDDY study. Nature. 2018;563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Sioofy-Khojine A-B, Oikarinen S, Honkanen H, Huhtala H, Lehtonen JP, Briese T, et al. Molecular epidemiology of enteroviruses in young children at increased risk of type 1 diabetes. PLoS One. 2018;13:e0201959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Nurminen N, Oikarinen S, Hyoty H. Virus infections as potential targets of preventive treatments for type 1 diabetes. Rev Diabet Stud. 2012;9(4):260–71. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Uusitalo U, Kronberg-Kippila C, Aronsson CA, Schakel S, Schoen S, Mattisson I, et al. Food composition database harmonization for between-country comparisons of nutrient data in the TEDDY study. J Food Compost Anal. 2011;24(4–5):494–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Joslowski G, Yang J, Aronsson CA, Ahonen S, Butterworth M, Rautanen J, et al. Development of a harmonized food grouping system for between-country comparisons in the TEDDY study. J Food Compost Anal. 2017;63:79–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Aronsson CA, Vehik K, Yang J, Uusitalo U, Hay K, Joslowski G, et al. Use of dietary supplements in pregnant women in relation to sociodemographic factors—a report from The Environmental Determinants of Diabetes in the Young (TEDDY) study. Public Health Nutr. 2013;16(8):1390–402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Andren Aronsson C, Uusitalo U, Vehik K, et al. Age at first introduction to complementary foods is associated with sociodemographic factors in children with increased genetic risk of developing type 1 diabetes. Matern Child Nutr. 2015;11(4): 803–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Hummel S, Vehik K, Uusitalo U, McLeod W, Aronsson CA, Frank N, et al. Infant feeding patterns in families with a diabetes history— observations from The Environmental Determinants of Diabetes in the Young (TEDDY) birth cohort study. Public Health Nutr. 2014;17(12):2853–62. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Yang J, Tamura RN, Uusitalo UM, Aronsson CA, Silvis K, Riikonen A, et al. Vitamin D and probiotics supplement use in young children with genetic risk for type 1 diabetes. Eur J Clin Nutr. 2017;71(12):1449–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Niinistö SEI, Lee H-S, Uusitalo U, Salminen I, Aronsson CA, Hummel S, et al. Higher EPA and DPA status in erythrocyte during infancy is associated with reduced risk of islet autoimmunity. San Francisco: Immunologu of Diabetes Society; 2017. [Google Scholar]
- 73.Hypponen E, Virtanen SM, Kenward MG, Knip M, Akerblom HK, Childhood Diabetes in Finland Study Group. Obesity, increased linear growth, and risk of type 1 diabetes in children. Diabetes Care. 2000;23(12):1755–60. [DOI] [PubMed] [Google Scholar]
- 74.Knip M, Reunanen A, Virtanen SM, Nuutinen M, Viikari J, Akerblom HK. Does the secular increase in body mass in children contribute to the increasing incidence of type 1 diabetes? Pediatr Diabetes. 2008;9(3 Pt 2):46–9. [DOI] [PubMed] [Google Scholar]
- 75.Wilkin TJ. The accelerator hypothesis: weight gain as the missing link between type I and type II diabetes. Diabetologia. 2001;44(7): 914–22. [DOI] [PubMed] [Google Scholar]
- 76.Dahlquist G Can we slow the rising incidence of childhood-onset autoimmune diabetes? The overload hypothesis. Diabetologia. 2006;49(1):20–4. [DOI] [PubMed] [Google Scholar]
- 77.Hagopian W, Lee HS, Liu E, Rewers M, She JX, Ziegler AG, et al. Co-occurrence of type 1 diabetes and celiac disease autoimmunity. Pediatrics. 2017;140(5):e20171305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Liu E, Lee HS, Aronsson CA, Hagopian WA, Koletzko S, Rewers MJ, et al. Risk of pediatric celiac disease according to HLA haplotype and country. N Engl J Med. 2014;371(1):42–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Agardh D, Lee HS, Kurppa K, Simell V, Aronsson CA, Jorneus O, et al. Clinical features of celiac disease: a prospective birth cohort. Pediatrics. 2015;135(4):627–34. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Sharma A, Liu X, Hadley D, Hagopian W, Liu E, Chen WM, et al. Identification of non-HLA genes associated with celiac disease and country-specific differences in a large, international pediatric cohort. PLoS One. 2016;11(3):e0152476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Hadley D, Hagopian W, Liu E, et al. HLA-DPB1*04:01 protects genetically susceptible children from celiac disease autoimmunity in the TEDDY study. Am J Gastroenterol. 2015;110(6):915–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Andren Aronsson C, Lee HS, Koletzko S, et al. Effects of gluten intake on risk of celiac disease: a case-control study on a Swedish birth cohort. Clin Gastroenterol Hepatol. 2016;14(3):403–409.e403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Aronsson CA, Lee HS, Liu E, Uusitalo U, Hummel S, Yang J, et al. Age at gluten introduction and risk of celiac disease. Pediatrics. 2015;135(2):239–45. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Johnson SB, Lynch KF, Roth R, Schatz D. My child is islet autoantibody positive: impact on parental anxiety. Diabetes Care. 2017;40(9):1167–72. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Swartling U, Lynch K, Smith L, Johnson SB. Parental estimation of their child’s increased type 1 diabetes risk during the first 2 years of participation in an international observational study: results from the TEDDY study. J Empir Res Hum Res Ethics. 2016;11 (2): 106–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Roth R, Lynch K, Lernmark B, Baxter J, Simell T, Smith L, et al. Maternal anxiety about a child’s diabetes risk in the TEDDY study: the potential role of life stress, postpartum depression, and risk perception. Pediatr Diabetes. 2015;16(4):287–98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Smith LB, Lynch KF, Baxter J, Lernmark B, Roth R, Simell T, et al. Factors associated with maternal-reported actions to prevent type 1 diabetes in the first year of the TEDDY study. Diabetes Care. 2014;37(2):325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Johnson SB, Lynch KF, Baxter J, Lernmark B, Roth R, Simell T, et al. Predicting later study withdrawal in participants active in a longitudinal birth cohort study for 1 year: the TEDDY study. J Pediatr Psychol. 2016;41(3):373–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Lernmark B, Lynch K, Baxter J, et al. Participant experiences in the environmental determinants of diabetes in the young study: common reasons for withdrawing. J Diabetes Res. 2016;2720650:2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Haghighi M, Johnson SB, Qian X, Lynch KF, Vehik K, Huang S. A comparison of rule-based analysis with regression methods in understanding the risk factors for study withdrawal in a pediatric study. Sci Rep. 2016;6:30828. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Yang J, Lynch KF, Uusitalo UM, Foterek K, Hummel S, Silvis K, et al. Factors associated with longitudinal food record compliance in a paediatric cohort study. Public Health Nutr. 2016;19(5):804–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Johnson SB, Lynch KF, Lee HS, Smith L, Baxter J, Lernmark B, et al. At high risk for early withdrawal: using a cumulative risk model to increase retention in the first year of the TEDDY study. J Clin Epidemiol. 2014;67(6):609–11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Lernmark B, Lynch K, Ballard L, et al. Reasons for staying as a participant in the environmental determinants of diabetes in the young (TEDDY) longitudinal study. J Clin Trials. 2012;2(2). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Baxter J, Vehik K, Johnson SB, Lernmark B, Roth R, Simell T. Differences in recruitment and early retention among ethnic minority participants in a large pediatric cohort: the TEDDY study. Contemp Clin Trials. 2012;33(4):633–40. [DOI] [PMC free article] [PubMed] [Google Scholar]


