Abstract
Antisense oligonucleotides (ASOs) have the potential to revolutionize medicine due to their ability to manipulate gene function for therapeutic purposes. ASOs are chemically modified and/or incorporated within nanoparticles to enhance their stability and cellular uptake, however, a major challenge is the poor understanding of their uptake mechanisms, which would facilitate improved ASO designs with enhanced activity and reduced toxicity. Here, we study the uptake mechanism of three therapeutically relevant ASOs (peptide-conjugated phosphorodiamidate morpholino (PPMO), 2’Omethyl phosphorothioate (2’OMe) and phosphorothioated tricyclo DNA (tcDNA) that have been optimized to induce exon skipping in models of Duchenne muscular dystrophy (DMD). We show that PPMO and tcDNA have high propensity to spontaneously self-assemble into nanoparticles. PPMO forms micelles of defined size and their net charge (zeta potential) is dependent on the medium and concentration. In biomimetic conditions and at low concentrations, PPMO obtains net negative charge and its uptake is mediated by class A scavenger receptor subtypes (SCARAs) as shown by competitive inhibition and RNAi silencing experiments in vitro. In vivo, the activity of PPMO was significantly decreased in SCARA1 knock-out mice compared to wild-type animals. Additionally, we show that SCARA1 is involved in the uptake of tcDNA and 2’OMe as shown by competitive inhibition and co-localization experiments. Surface plasmon resonance binding analysis to SCARA1 demonstrated that PPMO and tcDNA have higher binding profiles to the receptor compared to 2’OMe. These results demonstrate receptor-mediated uptake for a range of therapeutic ASO chemistries, a mechanism that is dependent on their self-assembly into nanoparticles.
Keywords: Antisense oligonucleotides, self-assembly, nanoparticles, scavenger receptors, uptake mechanism, delivery
Antisense oligonucleotides (ASOs) can be designed to target DNA and RNA in a sequence-specific manner to stop, alter or induce particular gene functions. Thus, they have emerged as a very promising new class of therapeutics that can target disease pathophysiology at the molecular genetic level with high specificity. One of the most advanced applications of ASOs is their use for manipulation of gene function through splice switching. ASOs can switch splicing patterns through sequence-specific targeting of pre-mRNA elements involved in exon recognition and/or consensus splice sites in a sequence-specific manner 1. This approach has been investigated as potential treatments for different types of muscular dystrophies, especially Duchenne muscular dystrophy (DMD), where ASOs have been extensively tested in disease models and are currently being evaluated in several clinical trials 2,3. DMD is caused by loss of function of the DMD gene due to deletions and/or mutations that cause the generation of premature termination codons and/or out-of-frame transcripts 4. Targeting of splice sites or putative exon splicing enhancers with ASOs can induce the removal of exons from the mature DMD transcript such that a nonsense mutation is bypassed, or alternately removal of exons around a genomic deletion can restore the mRNA reading frame.
Chemical modifications are introduced into ASOs to enhance their stability against nucleases and to prevent immune stimulation 5. Despite the promising results of the ongoing trials using ASOs, major scientific challenges remain. The principle limitation of ASOs is their poor cellular uptake due to their large molecular weight and mostly highly charged nature. Paradoxically, in practice, ASO uptake is dramatically enhanced when they are incorporated with nanoparticles that are even larger in size and much richer in charge. Most transfection reagents and ASO delivery systems rely on complexation or loading ASOs into nanoparticulate vectors 6. Moreover, under certain conditions, naked ASOs of different chemistries are taken up by cells without the need for transfection reagents in a process termed “gymnosis” via an unknown mechanism 7. Thus, understanding the uptake mechanism and resolving these seemingly paradoxical observations is very important for the clinical development of ASOs. Here we study the uptake mechanism of three ASO compounds of different chemistries that have been optimized for skipping of exon 23 in preclinical or clinical studies of DMD.
The first compound is from the peptide- morpholino family (PPMOs), which comprises a cell-penetrating peptide (CPP) attached to phosphorodiamidate morpholino oligomer; PMO 8. PMOs possess a morpholino moiety instead of the ribose moiety and the backbone phosphodiester linkages are replaced with uncharged phosphorodiamidate linkages. CPPs are short cationic peptides that enhance the uptake of the PMO into cells 8. Here, we mainly use the B-peptide conjugated PMO (B-PMO), a classical PPMO example, as our model 9. The second ASO is from the second generation of chemically modified RNAs; phosphorothioate 2’OMe. The 2’OH is replaced by a 2’OMe group and the phosphodiester linkages in the backbone are replaced by the more stable phosphorothioate linkages 5. The third ASO is from the recently developed family of tricyclo DNA (tcDNA). TcDNA has three additional C-atoms between C5’ and C3’ which increase both the affinity and hydrophobicity of the molecule together with a phosphorothioate backbone 10. Both 2’OMe and tcDNA chemistries have been shown to be taken up by cells via gymnosis7,11. All three molecules have been extensively tested in the well-established mdx mouse model for DMD, which carries a nonsense mutation in exon 23 of the Dmd gene. Of the three ASO classes, PPMOs are the most potent, achieving high levels of exon skipping in different skeletal muscles at doses as low as 6 mg/kg 9,12. TcDNA is the second most potent chemistry tested here, with a superior activity over 2’OMe when administered at a dose of 200 mg/kg demonstrating effective exon skipping even in the brain 13.
Recently, we have demonstrated that a class of pattern recognition receptors (PRRs), namely scavenger receptors (SRs), is involved in the uptake of certain CPPs when complexed with ASOs 14. SRs are a large family of PRRs that are involved in the uptake of pathogen associated molecular patterns (PAMPs) and damage associated molecular patterns (DAMPs) and play important functions in innate immunity and homeostasis 15. They are subdivided into several classes, from A to I, which are structurally diverse but functionally similar in their ability to bind polyanionic (negatively charged) particulate substrates15,16. The vast array of their ligands include: oxidized low density lipoprotein (oxLDL), acetylated LDL (acLDL), apoptotic cells, Staphylococcus aureus bacteria, HCV virus, lipopolysaccharide (LPS), PrP106–126 prion protein, viral RNA and different types of synthetic nanoparticles 15,17,18. Little is known about the structural features that are responsible for the promiscuous binding of SRs to negatively charged ligands; however, it is hypothesized that the surfaces that are engaged in ligand binding are similar in terms of shape and charge distribution, displaying clusters of cationic residues (cationic patches) 15. Additionally, due to their observed low specificity and functional overlap, they are thought to function in the form of heteromultimeric receptor complexes (signalosomes) that comprise SRs and other co-receptors 15. SRs are highly expressed in professional immune cells such as macrophages, but have also been shown to be expressed other cell types including smooth muscle cells, endothelial cells, fibroblasts, splenic dendritic cells, and epithelial cells 19. Class A SRs (SCARAs) are among the most extensively studied SRs and are characterized structurally by their collagenous domains 20. SCARAs have different subtypes, including SCARA1 (SR-AI), SCARA2 (MARCO), SCARA3, SCARA4 (Colec12), and SCARA5, among which SCARA3 and SCARA5 have been shown earlier to be involved in uptake of CPP-ASO nanocomplexes 14.
Here we study the effects of the physicochemical properties of the three different ASO compounds (PPMO, phosphorothioate 2’OMe, phosphorothioate tcDNA) used for the treatment of DMD and the role of SRs in their uptake, especially in muscle cells. We hypothesize that variation in ASO activity is due to their physicochemical properties modulated by their ability to self-assemble and to bind to SRs.
Results and Discussion
PPMO and tcDNA have high propensity to spontaneously self-assemble into nanoparticles
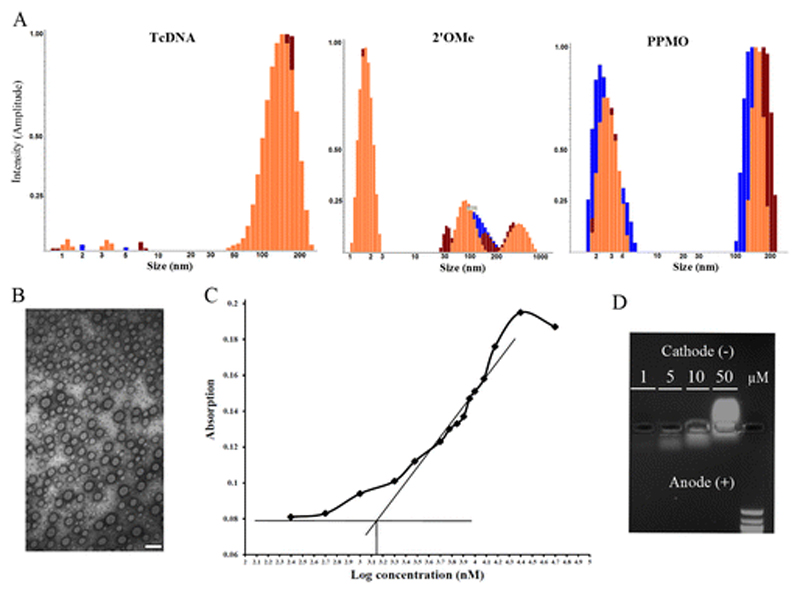
Physicochemical properties including particle size and charge are important determinants for uptake of drug delivery systems. Recently, using transmission electron microscopy (TEM) and nanoparticle tracking analysis, we found that tcDNA has a higher propensity to self-assemble into nanoparticles compared to 2’OMe 13. To investigate this further in comparison to the PPMO chemistry we used dynamic light scattering (DLS). In accordance with our previous observations, the predominant peak in the tcDNA sample was a broad particulate peak (Fig. 1A.). For PPMO, there were two peaks, one for singular molecules and one representing a particulate population. However, for 2’OMe, the predominant peak was of singular molecules at around 2 nm together with another particulate peak of much less intensity (Fig. 1A.).
Figure 1. Characterization self-assembling nanoparticles.
(A) DLS analysis of tcDNA, 2’OMe and PPMO. Each was measured 3 times (different colors) at 1 mM concentration in PBS showing the profile of the different populations present. (B) Negatively stained TEM pictures of nanoparticles formed by PPMO; Bar = 100 nm. (C) Critical micelle concentration (CMC) determination of PPMO using the dye micellization method (absorbance at 542 nm). Eosin Y concentration: 0.019 mM. The X-axis represents log the concentration in nM. CMC is the inverse log of the point at the intersection between the linear portion of the curve near the inflection point (R2=0.924) and the absorbance of the dye in the absence of any surfactant represented by the horizontal line. CMC = 1380.38 nM. (D) PPMO at different concentrations was loaded on a 1.25% agarose gel using TAE as running buffer.
Comparatively, these results show that both tcDNA and PPMO have a higher propensity to form nanoparticles than 2’OMe. In order to make sure that this property of self-assembly is retained in physiological conditions; we repeated the DLS measurements after incubation with physiological concentrations of albumin solution in PBS for 1h at 37 °C. Albumin is the most abundant serum protein representing up to 50% of total serum protein 21. While it is difficult to run DLS in the presence of full serum due to high background from diverse proteins, a simplified albumin based model generates one distinct peak of albumin around 4 nm, which also acts as an internal standard. Importantly, in this model, only tcDNA and PPMO were able to display high intensity particulate peaks, while 2’OMe failed to do so (Fig. S1 A). To test full serum conditions on these ASOs, fluorescently labelled tcDNA and PPMO were incubated with full serum for 1h at 37 °C, and subsequently fractionated using a continuous sucrose gradient. The fraction with the highest fluorescence signal was visualized via fluorescence microcopy. Densely bright particulate structures were observed for tcDNA and PPMO under these conditions, showing that the process of self-assembly into nanoparticles is maintained in physiological conditions and in the presence of serum proteins (Fig. S1 B).
Amphipathic PPMO forms micelles
To further investigate the nanoparticles formed by the PPMO, we used TEM visualization, as used previously for tcDNA and 2’OMe. PPMOs formed well-defined nanoparticles with a diameter ranging between 30 and 90 nm (Fig. 1B). We speculated that this spontaneous nanoparticle formation is due to the amphipathicitiy of the PPMO structure that leads to self-assembly into micelles. The PPMO molecule is composed of a relatively hydrophobic PMO portion and a very hydrophilic CPP that harbors multiple positive charges, a structure susceptible to micelle formation. In fact, PMOs are among the most hydrophobic ASO chemistries available. A PMO adenosine monomer has an octanol-water partition coefficient (log P) of -2.72, while the log P for an adenosine nucleotide of the 2’OMe chemistry is -4.15, -4.39 for locked nucleic acid (LNA), and -3.43 for peptide nucleic acid (PNA) 22. To confirm the micellization of PPMO, we utilized the dye micellization method 23; which is a classical method used to determine the critical micelle concentration (CMC). CMC is the concentration at which an amphipathic surface active molecule (surfactant) starts to self-assemble into micelles in the bulk of the dispersion medium 23. Dyes such as eosin, rhodamine and Sudan red are known to show a shift in the wavelength maximum (λ max) due to the presence of micelles 23. Here, we used eosin Y which has wavelength maximum at 518 nm in water; however, the presence of micelles increases its absorbance at 542 nm. CMC can be determined by plotting the change in absorbance of the micellized dye at a fixed wavelength (542 nm) against surfactant concentration. The linear portion near the inflection point is extrapolated to intersect with the absorbance of the dye in the absence of any surfactant (represented by the horizontal line in Fig. 1C.), and this concentration is the CMC 23. PPMO behaviour was typical surfactant behaviour with a CMC of about 1.4 μM. The PPMO probably behaves similarly to multi-head-group surfactants; however, the complexity of the structure due to the presence of an atypical hydrophobic tail (the PMO in this case) and up to 10 charges might require the development of specific models to understand this process.
PPMO charge reversal in biomimetic conditions
The net charge of a nanoparticle is a function of pH and the concentration of counterions in the medium. Thus, it is important to study the properties of therapeutic compounds in biomimetic conditions in terms of pH or isotonicity where their properties at these conditions are more relevant to their biological activity. We have previously demonstrated that certain CPPs, such as PF14 which is used for ASO delivery via non-covalent complexation, change their zeta potential according to the dispersion medium 14. To determine if the same is true for the self-assembled nanoparticles of PPMO, we measured the zeta potential in different conditions. Interestingly, PPMOs displayed the same pattern, having a positive zeta potential (10 ± 2.70) in water while changing into negative values in PBS (-1.21 ± 0.63) and serum-free medium (-3.48 ±2.31) due to the change in pH and high salt concentration. Surprisingly, this charge reversal was also concentration dependent as demonstrated by gel mobility shift assay (Fig. 1D). At high concentrations (50 μM), PPMO migrates towards the cathode, indicating a net positive charge, but as the concentration gets lower, the migration pattern shifts towards the anode indicating a net negative charge in a clear visualization of the charge reversal phenomenon. These observations highlight the importance of the presence and concentration of counterions in the solution. An important parameter to consider here is ξ, which is the molar concentration ratio of counterions to surfactant 24. When ξ increases, more counterions bind to the micellar surface changing its net charge and this explains the concentration dependent charge reversal. At higher PPMO concentrations ξ is low; hence, there are not enough counterions in the running buffer to shield and reverse the exposed cationic groups on the micellar surface; and thus, migration occurs towards the cathode. As the PPMO concentration is decreased, the ξ ratio changed in favor of the concentration of counterions in the running buffer enabling shielding and reversal of the net surface charge and migration to the opposite pole. The same effect can be replicated by keeping the PPMO concentration constant while changing the concentration of the counterions. When using TBE as a running buffer with equimolar concentrations of the cationic basic species (Tris base) and the anionic acidic species (boric acid), PPMO migration pattern can be seen to be divided between both the anode and the cathode (Fig. S1 C). Interestingly, when using unbalanced TBE with excess basic species, the migration pattern shifts towards the cathode indicating net positive charge, and the opposite takes place upon using TBE with excess boric acid. This further supports our hypothesis that the net charge of the PPMO depends on the delicate balance between the concentration of the PPMO and the concentration of the counterions available in the medium.
Class A scavenger receptors are involved in PPMO uptake
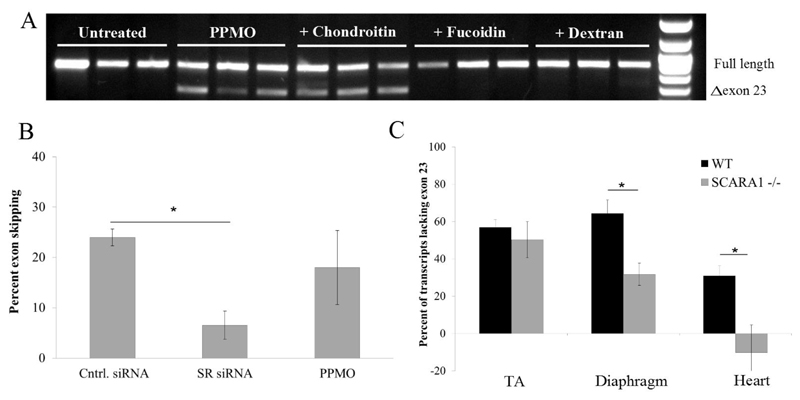
As aforementioned, we have shown previously that the uptake of certain CPP complexes with ASOs is mediated by class A scavenger receptor subtypes (SCARAs) 14. To determine whether the change in charge of the PPMO also mediates uptake through SCARAs, we tested the effects of SCARA ligands on the PPMO splice-switching activity in the C2C12 muscle cell line. Fucoidin sulfate and dextran sulfate are well-known SCARA ligands, and chondroitin sulfate is a chemically related molecule but not a specific ligand; and thus serves as a negative control. Cells were treated with the ligands or the control for 1h before treatment with the PPMO for 4 h, after which the medium was changed and cells were incubated for 20 h. Both fucoidin sulfate and dextran sulfate completely inhibited the splice-switching activity of the PPMO while chondroitin sulfate had no effect (Fig. 2A). This competitive inhibition demonstrates the involvement of SCARAs in the uptake and activity of PPMO.
Figure 2. Involvement of SCARA in PPMO uptake.
(A) Differentiated C2C12 cells (1x105) were pretreated SR ligands and control (fucoidin sulfate, dextran sulfate and chondroitin sulfate) at 50 μg/ml for 1 h then treated with PPMO (500 nM) for 4 h in Opti-MEM® before changing the medium to differentiation medium and incubation for 20 h. The products of nested reverse transcription-PCR (RT-PCR) were examined by electrophoresis on a 2% agarose gel. The top band indicates full-length transcript and the bottom band represents exon-skipped transcript (B) C2C12 myoblasts differentiated for 1 d, then treated with either siRNA cocktail targeting SACRA1,2,4 and 5 (25 nM each) using Lipofectamine RNAiMax® or scrambled control siRNA (Cntrl. siRNA, 100 nM). After 24 h, medium was changed and cells treated with PPMO (500 nm) as stated above. The products of nested RT-PCR were examined by gel electrophoresis and the percent of exon skipping was calculated using densitometry. (C) Quantitative PCR (qPCR) analysis of dystrophin exon 23-skipping in tibialis anterior muscle (TA), diaphragm and heart 1 week following intravenous injection of 10 mg/kg PPMO (Pi6a-PMO) in adult SCARA1-/- and wild-type (WT) C57 BL/6 mice (n=4). The percentage of exon 23-skipping of the Dmd transcripts was determined by normalizing exon 23-24 amplification levels to exon 20-21 levels. *P < 0.05; Student’s t-test; error bars represent mean ± SEM.
Moreover, several SCARA subtypes were expressed in the C2C12 murine myoblast cell line, including SCARA1, 3, 4 (COLEC12) and 5 (Suppl. Fig. S2A). We next tested the effect of silencing the expressed SCARA subtypes on PPMO activity. Upon using an siRNA cocktail against all the expressed SCARA subtypes (1, 3, 4 and 5), the splice-switching activity of PPMO was significantly reduced (Fig. 2B). We have recently demonstrated that the uptake and activity of PPMOs was higher in differentiated myotubes compared to undifferentiated myoblasts both in C2C1225 and H2k mdx cell-lines 26, both of which represent common models to study muscle differentiation. Interestingly, the expression of SCARA subtypes increases significantly throughout the course of differentiation of both cell lines, which correlates closely with the observed difference in PPMO uptake and activity (Fig. S2). Additionally, we performed TEM and SCARA inhibition analysis to ensure that this PPMO (Pip6a-PMO) behaves similarly to the model PPMO we are testing here (B-PMO). Pip6a peptide design is broadly similar to the B-peptide but with the inclusion of a YQFLI core motif within the arginine rich sequence 27. This change enhanced its splice-switching activity in vivo especially in the heart; however, the cause of this enhanced activity was not known. Here we show that Pip6a-PMO, similar to the B-PMO, spontaneously forms nanoparticles that can be visualized via TEM and that its uptake is significantly inhibited in the presence of SR ligands (Fig. S3). Interestingly, Pip6a-PMO nanoparticles are smaller than B-PMO nanoparticles; a property that may contribute to its enhanced biodistribution profile. Importantly, when injected in vivo in SCARA1 knock-out mice 28, the activity of Pip6a-PMO was significantly reduced in the diaphragm and heart compared to wild-type (WT) mice (Fig. 2C). This demonstrates that the SR dependent interactions observed in vitro also contribute to the biological activity of PPMOs in vivo. This does not exclude however the involvement of other receptor subtypes (SCARA3,4 and 5) and other SR classes in this process. The residual activity in the diaphragm and non-significant differences in tibialis anterior (TA) might very likely be due to compensation and differential expression of other SRs that are not altered in their expression in this model.
It is important to note that these observations for PPMOs could have more general implications on understanding the mechanism of action of both CPPs and antimicrobial peptides (AMPs). Several secondary amphipathic CPPs are known to self-assemble into nanoparticles 29. Furthermore, Kohno et al. 30 have recently shown that the uptake of an antimicrobial peptide (K8L9) at subcytotoxic concentrations is mediated by neuropilin-1 and low-density lipoprotein-related protein receptor 1 (LRP1), a receptor with scavenger like properties. Moreover, using siRNA screens, Kondo et al. 31 demonstrated the involvement of M160 (CD163L1; scavenger receptor cysteine-rich type I) in the uptake of a tumor homing CPP (CPP44). This shows that the phenomena of self-assembly and SR interaction are more general and that several SR receptor classes might be involved in the CPP/AMP mechanism of action; however, the details of their complimentary, synergistic or compensatory mechanisms require further studies. Additionally, the observation that net charge is dependent on concentration might explain different mechanisms of activity for CPPs and/or AMPs. At low concentrations, when there are sufficient counterions to shield and reverse the positive charge, receptor-mediated uptake might be the predominant mechanism of action. However, at high concentrations, when the net charge is positive, direct membrane interactions might predominate; which could explain lytic and toxic effects at such concentrations.
Varied involvement of SRs in the uptake of PPMO, tcDNA and 2’OMe chemistries
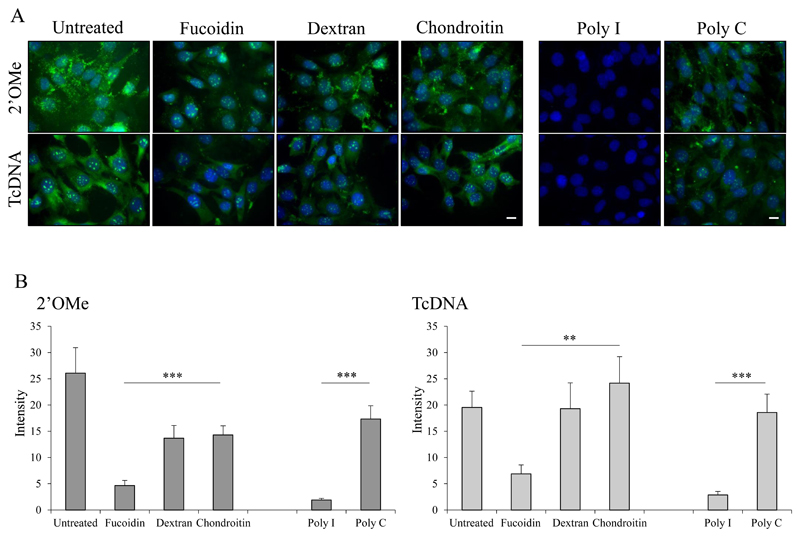
To determine the involvement of SRs in the gymnotic uptake of the other ASO chemistries we tested the effect of several SR ligands on the uptake of naked tcDNA and 2’OMe. The uptake of FITC-tcDNA and FITC-2’OMe was monitored in the presence of polyinosinic acid (poly I), polycytidylic acid (poly C), fucoidin sulfate, dextran sulfate and chondroitin sulfate. While fucoidin and dextran sulfates are more specific for SCARAs, poly I is more general as it also targets class C (SR-CI), class E (ORL-1/Lox-1) and class F (SREC) SRs 32 in addition to other receptors with scavenger properties such as Mac133 and nucleolin which binds quadruplex DNA structures 34,35. Poly C serves as the control for poly I. The uptake of tcDNA and 2’OMe was only partially inhibited in the presence of fucoidin and dextran sulfates, but almost completely inhibited in the presence of poly I (93.6% and 89.0% respectively) (Fig. 3) demonstrating the involvement of SCARAs together with other poly I sensitive SRs in the uptake, probably in heteromultimeric signalosome complexes. SRs have been previously suggested to be involved in the uptake of negatively charged ASOs 36–39, and it has been shown that the formation of multimers or the capacity to form G-quadruplexes enhance the uptake and activity of naked ASOs 40–42. This aggregation was also shown to enhance binding to SRs 43,44. Thus, it is feasible to speculate that the superior activity of tcDNA is related to its propensity to self-assemble into nanoparticles mediating better SR interaction and subsequent gymnotic uptake. Indeed, we observed that the extent of uptake of fluorescently labelled tcDNA is significantly higher than that of 2’OMe despite having similar biological activity upon lipofection (Fig. S4). Equivalent biological activity using lipofection was also demonstrated using sequences targeting exon 51 (Fig. S4D). This shows that despite other factors that might contribute to the superior activity of tcDNA (mRNA binding for example), a higher uptake profile due to higher propensity to form nanoparticles significantly contributes to this increased activity. This is more evident in the case of PPMO, which displays several folds higher uptake than the other two chemistries (Fig. S4).
Figure 3. Role of SRs in the uptake of tcDNA and 2’OMe.
(A) Differentiated C2C12 cells (2.5x104/cm2) were pretreated with SR ligands including fucoidin sulfate or polyinosinic acid (poly I), and controls including chondroitin sulfate or polycytidylic acid (poly C), at 100 μg/ml for 1 h. Then cells were incubated with FITC-2’OMePS (200 nM) or FITC-tcDNA (200 nM) for 4 h and visualized by florescence microscopy. Scale bar, 20 μm. (B) Quantitative fluorescence image analysis, mean fluorescence intensity was quantified using ImageJ software. *P < 0.05, **P < 0.01, ***P < 0.001; one-way ANOVA, error bars represent mean ± SEM.
Furthermore, we studied the effect of other factors on the uptake of the ASOs. Incubation at 4 °C significantly reduced the uptake of all the chemistries indicating the involvement of an active, energy-dependent mechanism of uptake (Fig S4). Incubation in the presence of serum on the other hand had only a small negative effect on uptake (Fig. S4), which might be due to the high stability of these heavily chemically modified ASOs.
The mechanism of spontaneous tcDNA self-assembly is not well understood. While tcDNA lacks the dichotomy of structure of the PPMO, the three additional C-atoms increase the hydrophobicity of the molecule. We speculate that this increased hydrophobicity might impart secondary amphipathic properties on the structure enabling self-assembly via mechanisms resembling the self-assembly of secondary amphipathic peptides 29. Alternatively, the extra rings in the tcDNA might facilitate stacking and generation of structures resembling poly-G aggregates that were seen to enhance the uptake of ASOs and binding to SRs 41–44. Structural and molecular modeling studies are underway to unravel the mechanism of tcDNA self-assembly.
Co-localization and SCARA1binding
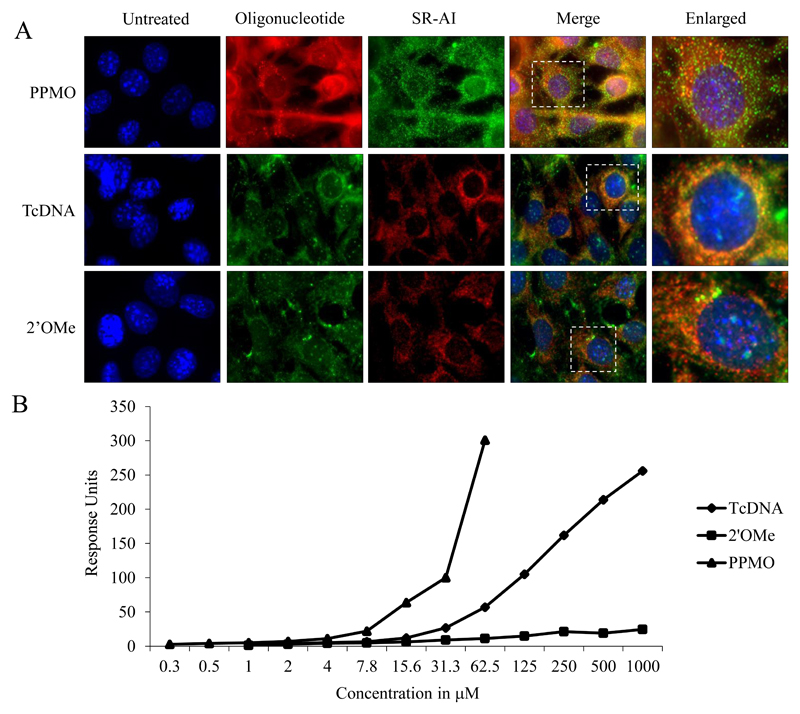
To demonstrate direct association between ASOs and receptor we performed co-localization experiments and Surface Plasmon Resonance (SPR) binding analysis. Immunocytochemical analysis using an anti-SCARA1 antibody showed co-localization of all three ASO compounds with the receptor (Fig. 4A). Furthermore, SPR was used to comparatively study the relative binding of the different chemistries to SCARA1. His-tagged SCARA-1 receptors were immobilized on the chip using an anti-His-tag antibody. Response units were calculated for specific binding after subtraction of non-specific binding to the chip or the antibody. PPMO demonstrated the highest binding followed by tcDNA, while 2’OMe displayed minimal binding (Fig. 4B). It was evident that the chemistries that are able to form nanoparticles demonstrated more efficient binding to the receptor, which is in accordance with the mode of action of SRs.
Figure 4. Interaction with SCARA1.
(A) Representative images of the cellular co-localization of Cy5-PPMO or FITC-2’OMePS and FITC-tcDNA with rat anti-mouse SCARA1 antibody in differentiated C2C12 myotubes at 4 h as measured by fluorescence microscopy (Cy5- PPMO, FITC-tcDNA and FITC- 2’OMePS were used at 200, 500 and 500 nM, respectively). Scale bar, 20 μm. (B) Binding experiments were performed using a Biacore 3000 system. His-tagged SCARA1 receptor was immobilized on the chip using an anti-his-tag antibody to give < 420 RU. Different ASOs in PBS were injected at 10 μL/min at 25 °C. Data traces were zeroed in the x and y axis after subtraction of non-specific binding.
Conclusions
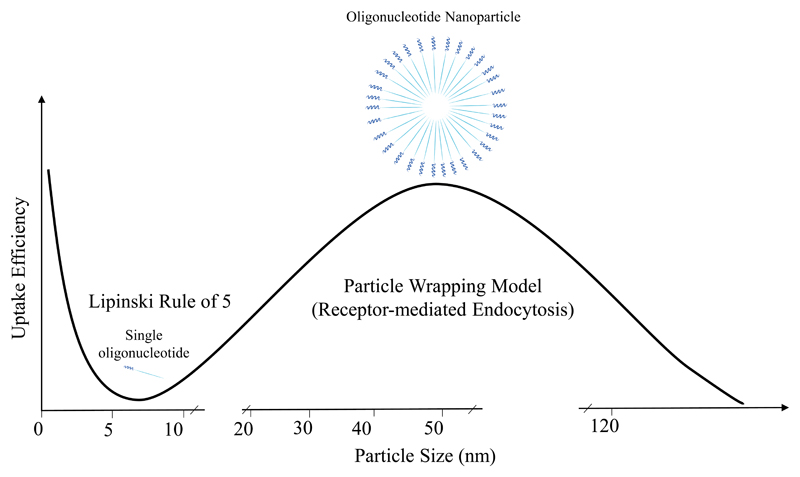
Our data shows that two potent ASOs, PPMO and tcDNA, have a higher propensity to self-assemble into nanoparticles and better binding to SCARA1 compared to the less potent 2’OMe. We hypothesize that nanoparticle assembly enhances ASO uptake based on the particle-wrapping model for receptor-mediated uptake of nanoparticles 45,46. In this model, optimal uptake requires a certain threshold of particle size. Below this threshold uptake will be impeded by the high energy cost required for a high curvature of the membrane to wrap the particle, and above it the uptake will be limited by the number of the receptors available for efficient particle wrapping 47. An optimal radius for uptake has been predicted theoretically to be around 25-30 nm and has also been validated experimentally 48. Interestingly, TEM pictures show that both PPMO and tcDNA13 nanoparticles are within this size range. Thus, the self-assembly process is important for mediating this type of interaction through SRs. The importance of the self-assembly process for uptake can be depicted in a model were single ASO molecules fail the criteria required for passive uptake into cells due to charge and size restrictions (Lipinski rule of 5 49), however, they can gain access via receptor mediated endocytosis only if they reach a certain size threshold (particle-wrapping model) (Fig. 5).
Figure 5. An illustration describing the relation between two different models of cellular uptake.
Singular molecules follow the Lipinski rule of 5 49 (molecular weight is less than 500 Da, lipophilicity-expressed as is less than 5, the number of hydrogen bond donators is less than 5 and the number of hydrogen bond acceptors is less than 10). This leads to sharp decrease in uptake with increasing size and charge of a molecule. That is why singular naked ASOs fail to cross cell-membranes. However, when they reach certain size, either by self-assembly or incorporation into nanoparticle-based delivery vectors, they lie within the scope of the receptor-mediated uptake process, especially through pattern recognition receptors of the innate immune system such as scavenger receptors. For this process to take place the nanoparticles have to possess a minimum radius (rminimum, around 22 nm) 45. Beyond an upper limit (rmaximum, around 60 nm) uptake starts to drop once more. Below the minimum radius, uptake is impeded by the high energy cost required for high curvature of the membrane for particle wrapping, and above the maximum radius uptake will be limited by the number of the receptors available for efficient particle interaction 47.
Unlike preformed synthetic nanoparticles, this process is spontaneous and dynamic, which means that the particles form and deform under different conditions. However, the apparent high propensity to self-assemble increases the probability of PPMO and tcDNA to form nanoparticles in proximity to the cell surface compared to other chemistries. In this regard, PPMOs and tcDNA resemble in vivo nanoparticle-based delivery systems and in vitro transfection reagents without the need for exogenous delivery or complexing agents. Furthermore, the spontaneous and reversible nature of the self-assembly process can explain the enhanced biodistribution of PPMOs and tcDNAs to tissues that are inaccessible to conventional nanoparticles, like skeletal muscle for example. While preformed nanoparticles are unable to extravasate into most tissues except liver and spleen due to the size restriction of capillary fenestrations, we speculate that self-assembling chemistries are able to extravasate through capillary fenestrations as single molecules and reform nanoparticles upon accumulation and reaching high local concentrations in situ. However, more in vivo pharmacokinetic and biodistribution studies are needed to elucidate these mechanisms in more detail as a limitation of in vitro cell systems is that they differentially express proteins on their surface when compared to in vivo conditions.
We believe that such models are crucial for understanding the pharmacokinetics and dynamics of current ASO therapeutics for better design and development of new ASO chemistries and delivery vehicles. Based on the findings discussed above, novel drug delivery platforms can be designed to enhance the propensity of self-assembly or to target SCARAs. These findings also highlight the importance of understanding the uptake mechanism for the clinical development of ASOs and pave the way for successfully applying ASOs to the treatment of genetic diseases.
Methods
Antisense oligonucleotides (ASOs)
All ASOs target the donor splice site of exon 23 of the mouse dystrophin pre-mRNA. The most efficient sequence was chosen for all chemistries from previously reported studies. TcDNA-PS (5’-AACCTCGGCTTACCT-3’) was synthesized by SYNTHENA, Bern. 2’OMePS (5’- GGCCAAACCUCGGCUUACCU-3’) was synthesized by Integrated DNA Technologies (IDT, USA) and PMO (5’-GGCCAAACCTCGGCTTACCTGAAAT-3’) was ordered from Gene Tools LLC. Conjugations of peptide (B-peptide: RXRRBRRXRRBRXB, Pip6a: RXRRBRRXR YQFLI RXRBRXRB, X, 6-aminohexanoic acid; B, b-alanine) with PMO were synthesized through use of a stable amide linker as described elsewhere 50.
DLS, TEM, CMC and zeta potential
DLS measurements were performed on Viscotek 802 instrument (Malvern, USA) using 30 μl of 1 mM ASO in PBS. DLS measurements in simulated physiologic conditions were performed by mixing equal volumes of the ASOs with filtered albumin (BSA, Sigma-Aldrich, Germany) solution in PBS (4.25 g/dL) at 37 °C for 1h then measuring on the DLS machine after appropriate dilution (10x). For TEM visualization, PPMO was mounted on formvar/carbon coated 200 mesh nickel grids (Agar Scientific, UK), then negatively stained using an aqueous solution of uranyl acetate and visualized using a JEOL 1010 transmission electron microscope (JEOL, Peabody, MA, USA) at 50,000 x magnification. For CMC measurements, eosin Y at a final concentration of 0.019 mM was mixed with different concentrations of PPMO (50, 25, 15, 12, 10, 9, 8, 7, 6, 5, 4, 3, 2, 1, 0.5, 0.25, 0.125 μM). UV absorption was measured at 542 nm using Biotek Synergy HT spectrophotometer (Biotek, USA). Zeta potential measurements were carried out on a Zetasizer instrument (Malvern, USA). Charge reversal gel experiments were run on 1.25% agarose gel with ethidium bromide using different buffers including: TAE, TBE, TBE with 4x Tris and TBE with 4x boric acid and visualized using UV. For visualization nanoparticles in serum, FITC-labelled ASOs were incubated in 100 μl of whole serum at a final concentration of 10μM (Fetal Bovine Serum, Heat inactivated (FBS)) for 1 h at 37°C. After incubation, ASOs were layered on top of a sucrose gradient. The gradient was composed of seven 1.25 ml fractions of sucrose dissolved in PBS. The concentrations of the fractions from the top to the bottom were 15%-45% in 5% increments. Once layered, the gradients’ interfaces were smoothened through diffusion by vertical incubation at 4°C overnight. The loaded gradients were spun in an ultracentrifuge (Beckman Coulter OptimaLE 80K) using a swinging bucket rotor (Beckman Coulter, SW 41Ti) at 200000 xg for 4h at 4°C. The gradients were then retrieved and 1ml fractions from top to bottom were collected (10 fractions in total per ASO). 100 μl per fraction were transferred to a clear bottom black plate and screened for fluorescence signal. The fractions with highest signal were loaded on a sandwiched coverslip-slide (two spacers made of double sided tape were used to fix the coverslip on the slide) treated with 50 μl of a 5mg/mL BSA in PBS solution for 10 minutes and subsequently imaged using an oil immersion 100x objective in a Nikon Eclipse TE2000-U equipped with a Watec WAT-902H camera.
Cell culture and PCR
Murine C2C12 cells were grown in DMEM (Dulbecco's Modified Eagle Medium), high glucose, GlutaMAX media (Life Technologies, USA) with 10% fetal bovine serum (FBS, Life Technologies) and 1% penicillin/streptomycin (Life Technologies). Murine H2k mdx myoblasts were cultured in gelatin (0.01%)-coated flasks at 33 °C, under 10% CO2, in Dulbecco’s modified Eagles medium (DMEM PAA laboratories) supplemented with 20% heat-inactivated fetal bovine serum (FBS Gold, PAA laboratories), 2% chicken embryo extract (Seralab), 1% penicillin/streptomycin- neomycin antibiotic mixture (Life Technologies) and 3 pg/ml g-interferon (PeproTech). For differentiation cells (1x105) were seeded into wells of a 24-well plate and the medium was changed after 24 h into differentiation medium consisting of DMEM containing 2% horse serum (Life Technologies) and differentiated for 3-4 days before experimentation. For PPMO treatment, cells were treated with a concentration of 500 nM in serum-free Opti-MEM® medium for 4 h, the medium was then changed for differentiation medium and incubation continued for a further 20 h. For RT-PCR detection of exon skipping, cells were lysed and RNA harvested using RNeasy kit (Qiagen, Germany) and quantified, then used for nested PCR procedure. Briefly, RNA was amplified on 2 steps with gene-specific primers (Ex 20-26, Fwd: CAG AAT TCT GCC AAT TGC TGA G-, Rev: TTC TTC AGC TTG TGT CAT CC) using Gene Amp PCR core kit (Life Technologies). Then cDNA was further amplified using Amplitaq Gold polymerase (Life Technologies, USA) with primers: Ex 20-26: Fwd: CCC AGT CTA CCA CCC TAT CAG AGC, Rev: CCT GCC TTT AAG GCT TCC TT).
PCR products were examined by electrophoresis on a 2% agarose gel. qPCR analysis was performed on cDNA from C2C12 and H2k mdx cells using 25 ng cDNA template and amplified with Taqman Gene Expression Master Mix (Applied Biosystems, Warrington, UK) on a StepOne Plus Thermocycler (Applied Biosystems, Warrington, UK). Taqman probes targeting SCARA1/3/4/5 (Life Technologies) were used and murine Glyceraldehyde 3-phosphate dehydrogenase (GAPDH) probes were used as an internal control for cDNA levels.
Scavenger receptor (SR) inhibition
Differentiated C2C12 or H2k mdx (1x105) were treated with SR ligands and controls: fucoidin sulfate, dextran sulfate, chondroitin sulfate, polyinosinic acid, polycytidylic acid (Sigma-Aldrich, Germany) for 1 h before adding the ASOs. Cells were then treated with the ASOs and analyzed as stated above. For siRNA treatment, C2C12 myoblasts differentiated for 24 h were treated with either a cocktail of siRNAs, SACRA1,2,4 and 5, 25 nM each (siGenome SMART pool, Dharmacon, USA) or scrambled control siRNA (100 nM) using Lipofectamine RNAiMax® (Life Technologies) according to the manufacturer’s protocol. After 24 h, medium was changed and cells were treated with PPMO as explained earlier.
Animal experiments
Experiments were carried out in the Biomedical Sciences Unit, University of Oxford according to procedures authorized by the UK Home Office. SCARA1 -/- and C57BL/6 mice (14 month old, n = 4) were used. SCARA1-/- mice were generated by Professor Tatsuhiko Kodama 28, and a colony has been maintained in our lab since then. Pip6-PMO conjugates were prepared in 0.9% saline solution at a final dose of 10 mg/kg and administered via the tail vein. One week later mice were sacrificed by CO2 inhalation, and tissues harvested and snap-frozen in cooled isopentane before storage at –80 °C. Total RNA was extracted from tissues using TRIzol reagent (Life Technologies) following manufacturer’s instructions. For quantitative analysis of exon skipping levels, 1 μg of RNA was reverse transcribed using the High Capacity cDNA RT Kit (Applied Biosystems, Warrington, UK) according to manufacturer’s instructions. qPCR analysis was performed using 25 ng cDNA template and amplified with Taqman Gene Expression Master Mix (Applied Biosystems, Warrington, UK) on a StepOne Plus Thermocycler (Applied Biosystems, Warrington, UK). Levels of Dmd exon 23 skipping was determined by multiplex qPCR of FAM-labelled primers spanning Exon 20-21 (Assay Mm.PT.47.9564450, Integrated DNA Technologies, Leuven, Belgium) and HEX-labelled primers spanning Exon 23-24 (Mm.PT.47.7668824, Integrated DNA Technologies). The percentage of Dmd transcripts skipping exon 23 was determined by normalizing Dmd exon 23-24 amplification levels to Dmd exon 20-21 levels.
Fluorescence microscopy and spectrophotometry
For immunofluorescence, cells were treated with either Cy5 conjugated PPMO 51, or FITC conjugated 2′OMePS or tcDNA oligonucleotides for 4 hours then washed 3 times with PBS plus (PBS containing Ca2+ and Mg2+) solution, and fixed with methanol at -20° C for 10 minutes.
Cells were then washed and stored in PBS at 4° C for future immunofluorescence. For colocalization, cells were treated with 0.1% Triton-X100 (Sigma-Aldrich) in PBS for 10 minutes, and washed three times with PBS plus, then blocked with 1% bovine serum albumin (BSA, Sigma Aldrich) containing PBS for 1 h. Cells were then incubated with rat anti-mouse SCARA1 (1:200 dilution, Bio-Rad, Hercules, CA), then washed three times with PBS plus and treated with 1:500 Alexa Fluor 488 goat anti-rat (Life Technologies) for 1 h. DAPI (1:5000 dilution, Sigma Aldrich) staining was then completed for 2 minutes, after which cells were washed and mounted with fluorescent mounting medium S3023 (Dako, Tokyo, Japan) onto glass slides. Visualization was carried out on a Leica fluorescent microscope with pictures taken by Axiovision fluorescent camera and Axiovision software (Zeiss, Oberkochen, Germany). Spectrophotometric measurements for the uptake of FITC-labeleld ASOs were performed in tissue-culture coated black plates with clear optical bottom (Corning, USA) using Victor3 spectrophotometer (Perkin Elmer, USA) 24 h after ASO addition.
SPR
Binding experiments were performed with a Biacore 3000 system (GE Healthcare) using CM5 chip. Anti-his-tag antibody was immobilized using amine coupling. Subsequently, His-tagged recombinant mouse SR-AI (SCARA1) (R&D Systems, USA) receptor was immobilized on a CM5 chip to give < 420 RU. For the binding assay, ASOs in PBS were injected at 10 μL/min at 25 °C. Data traces were zeroed in the x and y axis after subtraction of non-specific binding.
Supplementary Material
Supporting Information Available: [Self-assembly into nanoparticles is essential for receptor mediated uptake of therapeutic antisense oligonucleotides.] This material is available free of charge via the Internet at http://pubs.acs.org.
Acknowledgements
Work in the laboratory of MJG was supported by the Medical Research Council (MRC programme number U105178803). SELA is supported by Swedish Medical Research Council (VR-Med and EuroNanoMED2) and the Swedish Society of Medical Research (SSMF). AGC was supported by the European Commission under the Seventh Framework Programme (FP7), as part of the Marie Curie Initial Training Network, EScoDNA (GA no. 317110). We would like to thank Dr. David Staunton at the Biophysical Instrumentation Facility, Department of Biochemistry and Mr. Marcus Bridge at the Dunn School of Pathology for help with physicochemical characterization and SPR.
Footnotes
Conflict of Interest
The authors declare no competing financial interest.
References
- 1.Kole R, Krainer AR, Altman S. Nat Rev Drug Discov. 2012;11:125–140. doi: 10.1038/nrd3625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wood MJ, Gait MJ, Yin H. Brain. 2010;133:957–972. doi: 10.1093/brain/awq002. [DOI] [PubMed] [Google Scholar]
- 3.Koo T, Wood MJ. Hum Gene Ther. 2013;24:479–488. doi: 10.1089/hum.2012.234. [DOI] [PubMed] [Google Scholar]
- 4.Douglas AG, Wood MJ. Mol Cell Neurosci. 2013;56:169–185. doi: 10.1016/j.mcn.2013.04.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Deleavey GF, Damha MJ. Chem Biol. 2012;19:937–954. doi: 10.1016/j.chembiol.2012.07.011. [DOI] [PubMed] [Google Scholar]
- 6.Mastrobattista E, van der Aa MA, Hennink WE, Crommelin DJ. Nat Rev Drug Discov. 2006;5:115–121. doi: 10.1038/nrd1960. [DOI] [PubMed] [Google Scholar]
- 7.Soifer HS, Koch T, Lai J, Hansen B, Hoeg A, Oerum H, Stein CA. Methods Mol Biol. 2012;815:333–346. doi: 10.1007/978-1-61779-424-7_25. [DOI] [PubMed] [Google Scholar]
- 8.El Andaloussi SA, Hammond SM, Mager I, Wood MJ. Curr Gene Ther. 2012;12:161–178. doi: 10.2174/156652312800840612. [DOI] [PubMed] [Google Scholar]
- 9.Yin H, Moulton HM, Seow Y, Boyd C, Boutilier J, Iverson P, Wood MJ. Hum Mol Genet. 2008;17:3909–3918. doi: 10.1093/hmg/ddn293. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Leumann CJ. Nucleic Acids Symp Ser (Oxf) 2006:55–56. doi: 10.1093/nass/nrl167. [DOI] [PubMed] [Google Scholar]
- 11.Murray S, Ittig D, Koller E, Berdeja A, Chappell A, Prakash TP, Norrbom M, Swayze EE, Leumann CJ, Seth PP. Nucleic Acids Res. 2012;40:6135–6143. doi: 10.1093/nar/gks273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yin H, Moulton HM, Betts C, Merritt T, Seow Y, Ashraf S, Wang Q, Boutilier J, Wood MJ. Mol Ther. 2010;18:1822–1829. doi: 10.1038/mt.2010.151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Goyenvalle A, Griffith G, Babbs A, Andaloussi SE, Ezzat K, Avril A, Dugovic B, Chaussenot R, Ferry A, Voit T, Amthor H, et al. Nat Med. 2015 doi: 10.1038/nm.3765. [DOI] [PubMed] [Google Scholar]
- 14.Ezzat K, Helmfors H, Tudoran O, Juks C, Lindberg S, Padari K, El Andaloussi S, Pooga M, Langel Ü. FASEB J. 2012;3:1172–80. doi: 10.1096/fj.11-191536. [DOI] [PubMed] [Google Scholar]
- 15.Canton J, Neculai D, Grinstein S. Scavenger receptors in homeostasis and immunity. Nat Rev Immunol. 2013;13:621–634. doi: 10.1038/nri3515. [DOI] [PubMed] [Google Scholar]
- 16.Platt N, Gordon S. Chem Biol. 1998;5:R193–203. doi: 10.1016/s1074-5521(98)90156-9. [DOI] [PubMed] [Google Scholar]
- 17.Pluddemann A, Neyen C, Gordon S. Methods. 2007;43:207–217. doi: 10.1016/j.ymeth.2007.06.004. [DOI] [PubMed] [Google Scholar]
- 18.Arnida, Janat-Amsbury MM, Ray A, Peterson CM, Ghandehari H. Eur J Pharm Biopharm. 2011;77:417–423. doi: 10.1016/j.ejpb.2010.11.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.DeWitte-Orr SJ, Collins SE, Bauer CM, Bowdish DM, Mossman KL. PLoS Pathog. 2010;6:e1000829. doi: 10.1371/journal.ppat.1000829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Bowdish DM, Gordon S. Immunol Rev. 2009;227:19–31. doi: 10.1111/j.1600-065X.2008.00728.x. [DOI] [PubMed] [Google Scholar]
- 21.Zaias J, Mineau M, Cray C, Yoon D, Altman NH. J Am Assoc Lab Anim Sci. 2009;48:387–390. [PMC free article] [PubMed] [Google Scholar]
- 22.Cheng CJ, Saltzman WM, Slack FJ. Curr Med Chem. 2013;20:3582–3593. doi: 10.2174/0929867311320290004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Patist A, Bhagwat SS, Penfield KW, Aikens P, Shah DO. J Surfactants Deterg. 2000;3:53–58. [Google Scholar]
- 24.Ge W, Kesselman E, Talmon Y, Hart DJ, Zakin JL. J Non Newtonian Fluid Mech. 2008;154:1–12. [Google Scholar]
- 25.Aoki Y, Nagata T, Yokota T, Nakamura A, Wood MJ, Partridge T, Takeda S. Hum Mol Genet. 2013;22:4914–4928. doi: 10.1093/hmg/ddt341. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lehto T, Castillo Alvarez A, Gauck S, Gait MJ, Coursindel T, Wood MJ, Lebleu B, Boisguerin P. Nucleic Acids Res. 2014;42:3207–3217. doi: 10.1093/nar/gkt1220. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betts C, Saleh AF, Arzumanov AA, Hammond SM, Godfrey C, Coursindel T, Gait MJ, Wood MJ. Mol Ther Nucleic Acids. 2012;1:e38. doi: 10.1038/mtna.2012.30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Suzuki H, Kurihara Y, Takeya M, Kamada N, Kataoka M, Jishage K, Ueda O, Sakaguchi H, Higashi T, Suzuki T, Takashima Y, et al. Nature. 1997;386:292–296. doi: 10.1038/386292a0. [DOI] [PubMed] [Google Scholar]
- 29.Pujals S, Fernandez-Carneado J, Lopez-Iglesias C, Kogan MJ, Giralt E. Biochim Biophys Acta. 2006;1758:264–279. doi: 10.1016/j.bbamem.2006.01.006. [DOI] [PubMed] [Google Scholar]
- 30.Kohno M, Horibe T, Ohara K, Ito S, Kawakami K. Chem Biol. 2014;21:1522–1532. doi: 10.1016/j.chembiol.2014.09.008. [DOI] [PubMed] [Google Scholar]
- 31.Kondo E, Saito K, Tashiro Y, Kamide K, Uno S, Furuya T, Mashita M, Nakajima K, Tsumuraya T, Kobayashi N, Nishibori M, et al. Nat Commun. 2012;3:951. doi: 10.1038/ncomms1952. [DOI] [PubMed] [Google Scholar]
- 32.Peiser L, Gordon S. Microbes Infect. 2001;3:149–159. doi: 10.1016/s1286-4579(00)01362-9. [DOI] [PubMed] [Google Scholar]
- 33.Zhou H, Liao J, Aloor J, Nie H, Wilson BC, Fessler MB, Gao HM, Hong JS. J Immunol. 2013;190:115–125. doi: 10.4049/jimmunol.1202136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Bates PJ, Kahlon JB, Thomas SD, Trent JO, Miller DM. J Biol Chem. 1999;274:26369–26377. doi: 10.1074/jbc.274.37.26369. [DOI] [PubMed] [Google Scholar]
- 35.Burge S, Parkinson GN, Hazel P, Todd AK, Neidle S. Nucleic Acids Res. 2006;34:5402–5415. doi: 10.1093/nar/gkl655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bijsterbosch MK, Manoharan M, Rump ET, De Vrueh RL, van Veghel R, Tivel KL, Biessen EA, Bennett CF, Cook PD, van Berkel TJ. Nucleic Acids Res. 1997;25:3290–3296. doi: 10.1093/nar/25.16.3290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Biessen EA, Vietsch H, Kuiper J, Bijsterbosch MK, Berkel TJ. Mol Pharmacol. 1998;53:262–269. doi: 10.1124/mol.53.2.262. [DOI] [PubMed] [Google Scholar]
- 38.de Diesbach P, N'Kuli F, Berens C, Sonveaux E, Monsigny M, Roche AC, Courtoy PJ. Nucleic Acids Res. 2002;30:1512–1521. doi: 10.1093/nar/30.7.1512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sawai K, Mahato RI, Oka Y, Takakura Y, Hashida M. J Pharmacol Exp Ther. 1996;279:284–290. [PubMed] [Google Scholar]
- 40.Benimetskaya L, Berton M, Kolbanovsky A, Benimetsky S, Stein CA. Nucleic Acids Res. 1997;25:2648–2656. doi: 10.1093/nar/25.13.2648. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Wu CC, Castro JE, Motta M, Cottam HB, Kyburz D, Kipps TJ, Corr M, Carson DA. Hum Gene Ther. 2003;14:849–860. doi: 10.1089/104303403765701141. [DOI] [PubMed] [Google Scholar]
- 42.Bishop JS, Guy-Caffey JK, Ojwang JO, Smith SR, Hogan ME, Cossum PA, Rando RF, Chaudhary N. J Biol Chem. 1996;271:5698–5703. doi: 10.1074/jbc.271.10.5698. [DOI] [PubMed] [Google Scholar]
- 43.Suzuki K, Doi T, Imanishi T, Kodama T, Tanaka T. Eur J Biochem. 1999;260:855–860. doi: 10.1046/j.1432-1327.1999.00233.x. [DOI] [PubMed] [Google Scholar]
- 44.Pearson AM, Rich A, Krieger M. J Biol Chem. 1993;268:3546–3554. [PubMed] [Google Scholar]
- 45.Gao H, Shi W, Freund LB. Proc Natl Acad Sci U S A. 2005;102:9469–9474. doi: 10.1073/pnas.0503879102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Nel AE, Madler L, Velegol D, Xia T, Hoek EM, Somasundaran P, Klaessig F, Castranova V, Thompson M. Nat Mater. 2009;8:543–557. doi: 10.1038/nmat2442. [DOI] [PubMed] [Google Scholar]
- 47.Shi W, Wang J, Fan X, Gao H. Phys Rev E Stat Nonlin Soft Matter Phys. 2008;78 doi: 10.1103/PhysRevE.78.061914. 061914. [DOI] [PubMed] [Google Scholar]
- 48.Jiang W, Kim BY, Rutka JT, Chan WC. Nat Nanotechnol. 2008;3:145–150. doi: 10.1038/nnano.2008.30. [DOI] [PubMed] [Google Scholar]
- 49.Lipinski CA, Lombardo F, Dominy BW, Feeney PJ. Adv Drug Deliv Rev. 2001;46:3–26. doi: 10.1016/s0169-409x(00)00129-0. [DOI] [PubMed] [Google Scholar]
- 50.Wu RP, Youngblood DS, Hassinger JN, Lovejoy CE, Nelson MH, Iversen PL, Moulton HM. Nucleic Acids Res. 2007;35:5182–5191. doi: 10.1093/nar/gkm478. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Shabanpoor F, Gait MJ. Chem Commun (Camb) 2013;49:10260–10262. doi: 10.1039/c3cc46067c. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.