Abstract
Mutations in superoxide dismutase 1 (SOD1) cause 15–20% of familial amyotrophic lateral sclerosis (fALS) cases. The resulting amino acid substitutions destabilize SOD1's protein structure, leading to its self-assembly into neurotoxic oligomers and aggregates, a process hypothesized to cause the characteristic motor-neuron degeneration in affected individuals. Currently, effective disease-modifying therapy is not available for ALS. Molecular tweezers prevent formation of toxic protein assemblies, yet their protective action has not been tested previously on SOD1 or in the context of ALS. Here, we tested the molecular tweezer CLR01–a broad-spectrum inhibitor of the self-assembly and toxicity of amyloid proteins–as a potential therapeutic agent for ALS. Using recombinant WT and mutant SOD1, we found that CLR01 inhibited the aggregation of all tested SOD1 forms in vitro. Next, we examined whether CLR01 could prevent the formation of misfolded SOD1 in the G93A-SOD1 mouse model of ALS and whether such inhibition would have a beneficial therapeutic effect. CLR01 treatment decreased misfolded SOD1 in the spinal cord significantly. However, these histological findings did not correlate with improvement of the disease phenotype. A small, dose-dependent decrease in disease duration was found in CLR01-treated mice, relative to vehicle-treated animals, yet motor function did not improve in any of the treatment groups. These results demonstrate that CLR01 can inhibit SOD1 misfolding and aggregation both in vitro and in vivo, but raise the question whether such inhibition is sufficient for achieving a therapeutic effect. Additional studies in other less aggressive ALS models may be needed to determine the therapeutic potential of this approach.
Keywords: superoxide dismutase (SOD), amyotrophic lateral sclerosis (ALS) (Lou Gehrig disease), protein aggregation, mouse, inhibitor, neurodegeneration, amyloid, protein misfolding, molecular tweezer, motor neuron
Introduction
Amyotrophic lateral sclerosis (ALS)3 is a rapidly debilitating neuromuscular disorder characterized by loss of motor neuron function in the spinal cord, brain stem, and motor cortex. The resulting progressive muscle weakness and atrophy leads to death typically occurring 3–5 years after onset. Approximately 90–95% of ALS cases are sporadic, and the remaining cases are familial (fALS), caused by mutations in multiple genes (1). More than 160 mutations in the SOD1 gene, encoding the enzyme superoxide dismutase 1 (SOD1), have been described to cause amino acid substitutions that destabilize the protein leading to its aberrant self-assembly, which causes 15–20% of fALS cases (2).
SOD1 is a homodimer of a 153-amino acid polypeptide. The enzyme converts the superoxide ions produced by mitochondria into hydrogen peroxide. Some disease-associated SOD1 variants retain high activity (3), and the targeted deletion of SOD1 in transgenic mice does not induce ALS-like symptoms (4). Therefore, the mechanism by which mutant SOD1 causes ALS is believed to be not a loss of function but rather a gain of toxic function upon misfolding and self-assembly into neurotoxic oligomers and aggregates (5, 6). The latter are found in the brain and spinal cord of affected individuals as intracellular inclusions.
To date, two drugs have been approved for treatment of ALS: riluzole (7) and edaravone (8). These drugs slow disease progression mildly but do not alter substantially the disease course. Thus, there is an urgent need for a disease-modifying therapy that would halt the progression and ultimately cure ALS. Inhibition and modulation of aberrant protein self-assembly are promising strategies for therapy development, which have been explored quite extensively for other proteinopathies, such as Alzheimer's disease (AD) and Parkinson's disease (PD) (9). In contrast, to date, few studies have examined small-molecule inhibitors of SOD1 self-assembly as a potential therapy for ALS. A study using pyrimidine-2,4,6-triones showed protection from SOD1-induced cytotoxicity in cultured cells (10). Using a different mechanism, restoration of cellular protein degradation by activation of the proteasome, pyrazolones have been found to protect PC-12 cells against the toxicity of G93A-SOD1 (11). In yet a third approach, in silico screening identified azauracil- and uracil-based inhibitors, which stabilized mutant SOD1 against aggregation in human plasma (12). The therapeutic efficacy of these compounds has yet to be tested in vivo. These few examples highlight the need for further exploration of strategies directed at misfolded SOD1 self-assembly as a potential target for ALS therapy.
SOD1 was the first gene linked to familial ALS (13). A transgenic mouse model of ALS, expressing human SOD1 containing a G93A substitution, was first introduced in 1996 (14). Although this model is not ideal, as it expresses a high copy number of the transgene and consequently displays highly aggressive disease progression, it has remained not only the most thoroughly characterized ALS animal model, but also has been regarded as the gold standard in preclinical therapeutic ALS research. This mouse model mimics motor neuron loss and progressive muscle weakness (15) similar to the clinical symptoms of human ALS. The mouse model has facilitated studying mechanistic aspects of the disease, yet the rapid course of the disease may be too aggressive for many therapeutic approaches to have an effect.
Previously, we reported that molecular tweezers (MTs) are broad-spectrum inhibitors of abnormal protein self-assembly and toxicity (16–18). A lead MT, called CLR01 (Fig. 3A), has been found to inhibit the formation of toxic oligomers and aggregates of multiple disease-associated proteins (19), including those involved in AD (20, 21) and PD (22, 23). CLR01 acts as a nanochaperone by a “process-specific” mechanism, i.e. it targets the process of abnormal protein self-assembly itself rather than a particular protein. CLR01 achieves this activity by labile binding to positively charged amino acid residues, primarily Lys and to a lower extent Arg, temporarily reversing the charge from positive to negative and disrupting hydrophobic interactions in which the butylene moiety of the Lys side chain (or the propylene chain in the Arg side chain) participate. As hydrophobic and electrostatic interactions are important, particularly in the early stages of the aberrant self-assembly process in which oligomers and aggregation nuclei form, MTs effectively interfere with the formation of these early assemblies.
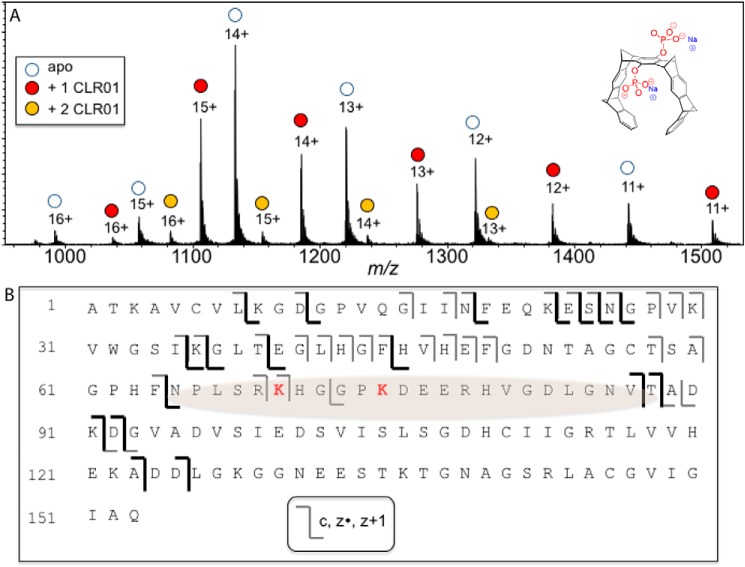
Figure 3.
Binding of CLR01 on SOD1 investigated by ESI-MS. A, ESI mass spectrum of a 10 μm SOD1/10 μm CLR01 mixture shows predominantly a 1:1 binding stoichiometry. The structure of CLR01 is shown at the top right corner. B, schematic of the ECD-MS/MS fragmentation profile for the 12+-charged 1:1 SOD1–CLR01 complex. Product ions that retained binding to CLR01 are shown in dark colored lines. From the fragmentation profile, the shaded region is suggested to be the site(s) of CLR01 binding.
CLR01 has been shown to have beneficial therapeutic effects in multiple animal models of various proteinopathies, including rodent models of AD (18, 24), PD (25), transthyretin amyloidosis (26), and desmin-related cardiomyopathy (27). However, to date, MTs have not been explored in the context of ALS in general or for inhibition of the aggregation and toxicity of SOD1 in particular. Here, we evaluated the effect of CLR01 on SOD1 aggregation in vitro and in the G93A-SOD1 mouse model of ALS. Our data reveal that CLR01 inhibits the aggregation of WT and disease-associated variant SOD1 in vitro and reduces the accumulation of misfolded SOD1 in mice. However, reduction of misfolded SOD1 did not result in significant therapeutic effects, possibly due to the fast disease progression in this model or due to other reasons discussed below.
Results
CLR01 inhibits the aggregation of wildtype (WT) and disease-associated SOD1
WT and variant SOD1 were expressed in yeast cells and purified using serial chromatography, as described previously (28). A eukaryotic yeast expression system is necessary for obtaining the correct post-translational modifications of N-terminal groups by acetylation (29). Each variant was de-metallated by incubation with excess EDTA and stored at −80 °C until use. The disease-associated variants included E21K, H46R, D76Y, and G93A. Aggregation kinetics were measured using the thioflavin-T (ThT) fluorescence assay, and the morphology of the aggregates was visualized by electron microscopy (EM). To induce misfolding and monitor aggregation, 40 μm of each variant were incubated for 48 h at 37 °C in a 96-well plate with constant agitation in the presence of 50 mm tris(2-carboxyethyl)phosphine (TCEP) to reduce disulfide bonds and increasing concentrations of CLR01. A Teflon ball was added to each well to facilitate the aggregation, as described previously (30).
In the absence of CLR01, all the variants showed a typical time-dependent sigmoidal increase in ThT fluorescence consisting of an initial lag phase indicating misfolding and nucleation, followed by an exponential increase in fluorescence, and finally a plateau when most of the monomers have been exhausted (Fig. 1, A–E, red curve). In the presence of CLR01, dose-dependent inhibition of the aggregation was observed in all cases. The dose dependence of the inhibition varied for the different isoforms but was complete or nearly complete at 5-fold excess CLR01 in all cases (Fig. 1, A–E, green curve).
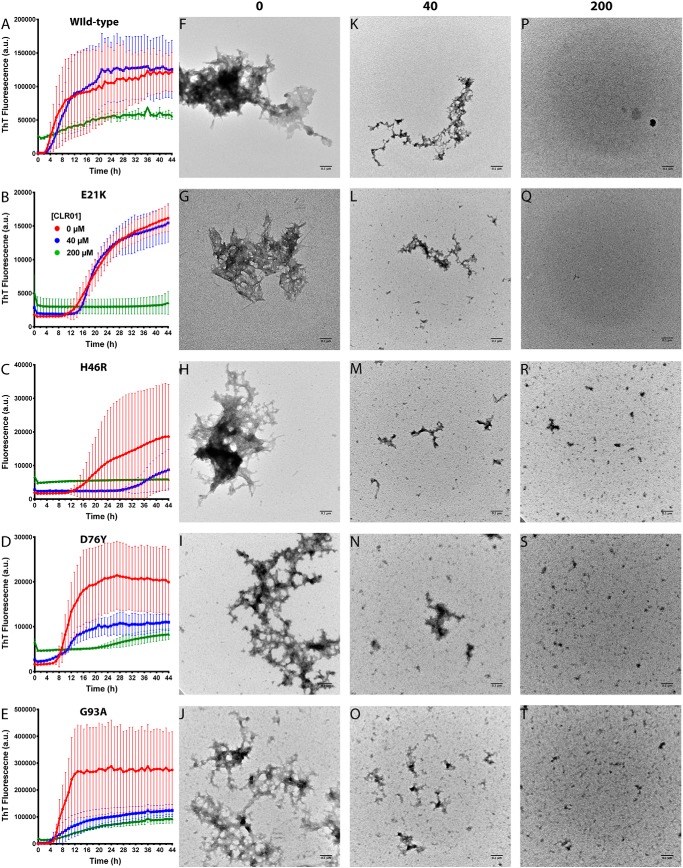
Figure 1.
CLR01 inhibits SOD1 aggregation in vitro. A–E, aggregation kinetics was measured using the ThT fluorescence assay. WT SOD1 (A) or the variants E21K (B), H46R (C), D76Y (D), and G93A (E) were each incubated at 40 μm in the absence or presence of 40 or 200 μm CLR01. F–T, the morphology of the protein was examined at the end of each aggregation reaction by EM. Scale bars, 0.1 μm.
Examination of the ThT curves showed that CLR01 affected all the stages of the aggregation process, including the lag phase (Fig. S1A), the slope of the exponential phase (Fig. S1B), and the final plateau (Fig. S1C), although the dose dependence of the inhibition varied among the different variants. As shown in Fig. 1 and Fig. S1, in some cases, e.g. WT SOD1, little inhibition was observed at a 1:1 SOD1/CLR01 ratio, whereas in other cases, such as H46R-SOD1, substantial inhibition was found already with equimolar CLR01. The additional binding sites for CLR01 at Lys-21 in E21K-SOD1 and Arg-46 in H46R-SOD1 did not appear to play a major role in the activity of the compound. The exact mechanism underlying the differences among the SOD1 variants is not known and will require further investigation.
Morphological examination by EM showed that in the absence of CLR01, all the SOD1 variants formed primarily amorphous aggregates (Fig. 1, F–J). After a long incubation (30–60 days), typical amyloid fibrils also were apparent (Fig. 2). In the presence of equimolar concentrations of CLR01, smaller aggregates were observed (Fig. 1, K–O). This reduction of aggregate size was consistent in all cases, but it did not correlate well with the ThT results. For example, in the cases of WT SOD1 or E21K-SOD1, the ThT change in the presence of equimolar CLR01 was similar to the control reactions in the absence of CLR01, yet the morphological examination showed substantially smaller aggregates, suggesting that these aggregates contained a high quotient of cross-β-structures. At a 5-fold molar excess of CLR01, only small amorphous structures were apparent, in agreement with a strong inhibition of the ThT fluorescence increase (Fig. 1, P–T).
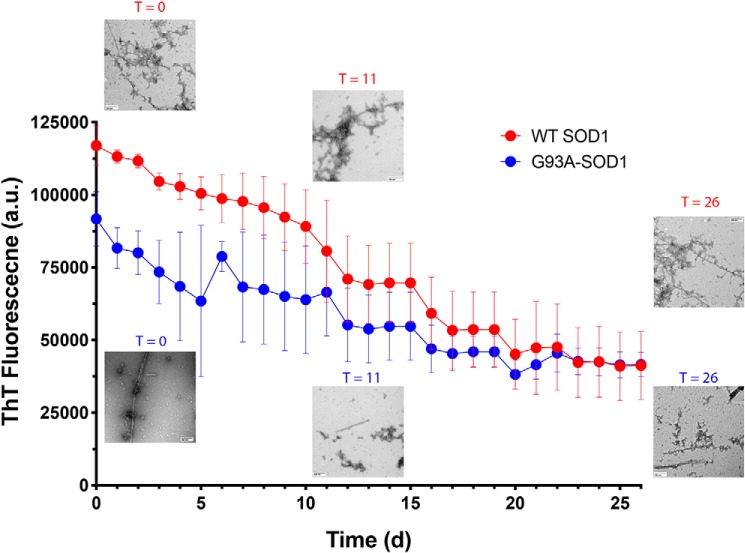
Figure 2.
CLR01 partially disrupts the cross-β-structure in pre-formed SOD1 aggregates. Forty μm WT SOD1 or G93A-SOD1 were aged for 6 months and then incubated with 400 μm CLR01 in the presence of ThT, and the fluorescence was monitored every day for 26 days. Aliquots were taken in the beginning of the reaction (day 0), after 11 days, and at the end of the reaction (day 26) and examined by EM.
Previously, CLR01 was shown not only to inhibit the aggregation of amyloidogenic proteins, such as Aβ40, Aβ42 (19), and α-synuclein (22), but also to disaggregate pre-formed fibrils over several weeks when added at a 10-fold excess. Here, we asked whether it could also dissociate pre-formed fibrils or other aggregates of SOD1. To answer the question, we added a 10-fold excess of CLR01 and an equimolar concentration of ThT to WT SOD1 or G93A-SOD1 that had been previously agitated at 37 °C for 48 h and then left at room temperature for 6 months. The mixtures were incubated without agitation at 37 °C for 26 days, and ThT fluorescence was measured every 24 h. Aliquots from each reaction were taken on days 0, 11, and 26 for morphological examination by EM. The initial ThT fluorescence in the WT SOD1 reactions was ∼20% higher than in the G93A-SOD1 reactions (Fig. 2).
In both cases, the ThT fluorescence gradually decreased to about 50% of the original fluorescence value after ∼20 days, at which point the value did not change anymore, suggesting that CLR01 dissociated some but not all of the SOD1 aggregates. The morphological examination showed that at all time points, a mixture of fibrils and large and small amorphous aggregates was present, which did not show a clear trend of change during the experiment, possibly due to the high morphological variability of the aggregates. This disagreement between the change observed by ThT but not by EM suggests that CLR01 can disrupt some but not all of the cross-β-structure of pre-formed SOD1 fibrils and/or amorphous aggregates under the experimental conditions we used.
Identification of the binding region of CLR01 on SOD1
SOD1 contains 11 Lys residues (12 in the E21K variant), each of which is a potential binding site for CLR01. Previous studies with amyloid β-protein (19), α-synuclein (23), huntingtin exon 1 (31), and tau (32) have shown that not all of these binding sites are equal, and the molecular tweezer prefers certain Lys residues over others. Thus, we asked whether a preferred binding site for CLR01 also existed in SOD1. To answer the question, we used native mass spectrometry (MS) with electrospray ionization (ESI). These experiments showed that when SOD1 and CLR01 were mixed at a 1:1 ratio, the 1:1 complex was the predominant CLR01-bound stoichiometry; from the summed intensities of the peaks in the native ESI mass spectrum (Fig. 3A), the relative ratios of the apo-, 1-ligand bound, and 2-ligand bound forms were 0.55:0.40:0.05. However, this does not preclude multiple binding sites for CLR01 on SOD1. By measuring the normalized signal intensities for each such complex, equilibrium dissociation constants were calculated using a Scatchard plot, as was done previously for α-synuclein (23). The first and second dissociation constants were measured to be 2.93 and 44.9 μm, respectively.
To elucidate the main binding site(s) of CLR01 on SOD1, we used tandem MS (MS/MS) or “top-down” MS of the protein–ligand complex. Using electron capture dissociation (ECD), covalent backbone bonds of the polypeptide could be cleaved, whereas noncovalent forces holding the ligand bound to the macromolecule were maintained. ECD-MS/MS of protein–ligand complexes yielded product ions from dissociation of the protein chain, some of which were still bound to the ligand (Table S1). The sequence of these ligand-bound fragments was mapped onto the full-length protein sequence to determine the ligand-binding region(s). The 12+-charged ion for the 1:1 SOD1–CLR01 complex was analyzed using this approach; protein fragments of the c- (retaining the N terminus) and z- (retaining the C terminus) product-ion series were detected. Some product ions from dissociation of the polypeptide backbone corresponded to unbound protein, whereas other product ions retained binding to CLR01 (Fig. 3B, dark lines). The data were interpreted in a similar fashion as in our previous work (19, 23, 32). CLR01 is bound to both the C terminus retaining product ion z89 (cleavage between Phe-64 and Asn-65) and the N terminus retaining product ion c87 (cleavage between Val-87 and Thr-88, see Table 1). These data allowed localizing the binding of CLR01 to the region SOD1(65–88), which contains two likely binding sites at Lys-70 and Lys-75. Arg-69 and Arg-79 also may be minor contributors to the binding in this region. The region is on the surface of the protein in the crystal structure (34) and is thus readily available for CLR01 binding even before misfolding.
Examination of the impact of CLR01 on weight, muscle weakness, and respiration in the G93A-SOD1 transgenic mouse model of ALS
In view of the ability of CLR01 to inhibit the aggregation of SOD1 in vitro, we wanted to determine its impact on SOD1 aggregation in the G93A-SOD1 mouse model (14). Mice were treated with 0.5 or 5.0 mg/kg CLR01 administered by a daily subcutaneous (s.c.) injection. A control group was treated with vehicle (sterile saline). According to the standardized instructions of Prize4Life, which provided the mice for the study, the treatment started at 50 days of age and continued until the mice met the criteria for euthanasia due to advanced weakness. Each group consisted of 12 female and 12 male mice.
Most previous studies in models of neurodegenerative diseases used s.c. administration via osmotic minipumps (24–26). This approach offers continuous drug administration but is limited to 5–6 weeks. In addition, although the aqueous solubility of CLR01 is high (>10 mm), at the limit of solubility, the highest dose possible using the model 1004 minipump used previously is 1.2 mg/kg/day. The terms of the Prize4Life-supported study mandated that extension of life must be the primary outcome measure. As the G93A-SOD1 mice tend to develop disease at 110–120 days of age and die at 130–140 days of age, using the same pumps for this study was impractical. Therefore, we decided to administer CLR01 by a s.c. daily injection, as was done in a recent study using a desmin-related cardiomyopathy mouse model (27). In using this method of administration, we took into account that the plasma half-life of CLR01, 2–3 h, and its first-pass blood–brain barrier penetration, 2–3% (35), may limit the exposure of the target, G93A-SOD1, to the compound. However, we reasoned that the much slower clearance of CLR01 from the CNS relative to clearance from the plasma (35) might allow gradual accumulation of the compound in the CNS and increase target engagement.
In addition to recording disease onset and disease duration before mice reached the criteria for euthanasia, every week of the study we weighed the mice, tested their strength using the grip-strength and rotarod (starting on week 13) tests, and measured unchallenged and challenged pulmonary function, as described previously (36, 37). Loss of body weight also can reflect motor impairment that occurs during disease progression. All the animals consistently gained weight up to 13 weeks (males) or 14 weeks (females) of age, after which the weight began to decline (Fig. 4, A and B). During the entire treatment period, there were no significant differences among the treatment groups in weight, morbidity, mortality, or signs of distress, suggesting that the treatment did not cause overt adverse effects in the mice.
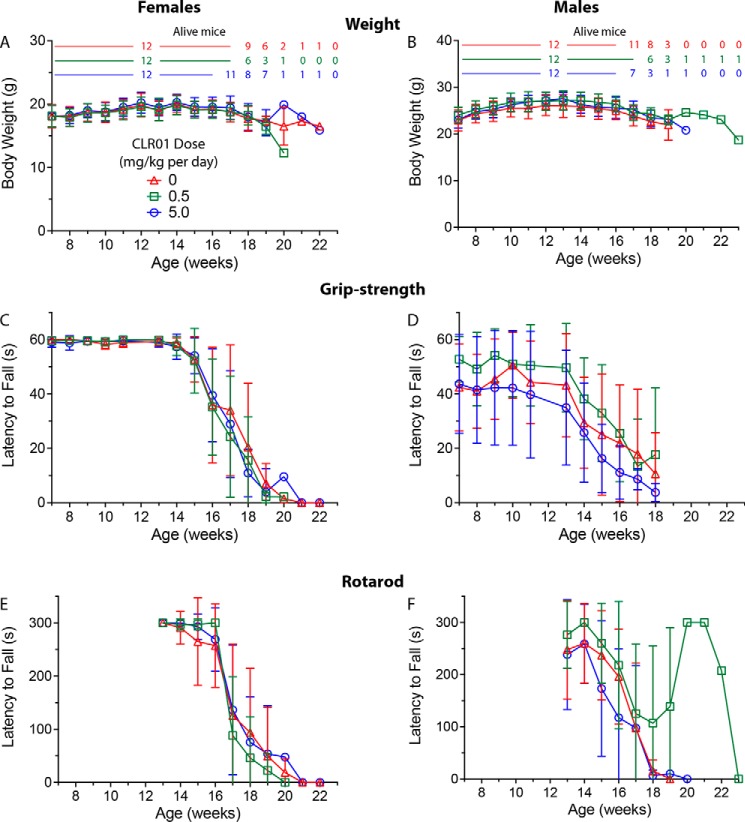
Figure 4.
CLR01 treatment did not affect mouse weight or motor performance significantly. Male and female (n = 12 per group each) G93A-SOD1 transgenic mice were administered a daily s.c. dose of saline vehicle or CLR01 (0.5 or 5 mg/kg) from 50 days of age until meeting criteria for euthanasia. Body weight (A and B) and grip-strength (C and D) were monitored weekly starting on week 7, and rotarod performance (E and F) was tested weekly starting on week 13 (E and F). The key in A is applicable to all the other panels. The number of animals used to calculate the average and S.D. at each time point is shown above A and B using the same color code and are applicable also to C–F. To facilitate comparison among the three measurements, all the graphs are shown with the same time scale. Missing data points in D were not recorded.
In the grip-strength test, female mice began to show weakness at week 15 and males at week 14, in agreement with the known and more aggressive course of the disease in male mice in this model (38, 39). There were no differences in weakness progression among the females in the three treatment groups (Fig. 4C). In the males, a trend toward faster progression in the high-dose group and slower progression in the low-dose group was observed, yet the differences were not statistically significant. The rotarod test was performed starting on week 13 and showed a similar trend to the grip-strength test. There was a slight delay in onset of weakness in the females in the low-dose group and a trend toward slightly better performance of the males in this group, but these differences were not statistically significant.
Patients with ALS invariably develop respiratory muscle weakness, and most die from pulmonary complications (40). Therefore, we used plethysmography to test whether the treatment would affect the pulmonary function of the mice. In this test, mice are placed in a chamber that allows them to breathe naturally, unrestrained and untethered. The system measures the small changes in the air that is exchanged in and out of the entire chamber because of the animal's respiration (36). Mice were given a hypercapnic challenge to increase tidal volume and frequency. Measurements were performed every week between week 13 and week 18. Similar to the grip-strength and rotarod tests, the values of both unchallenged and challenged expiratory and inspiratory volume were similar in the all three treatment groups and did not show significant differences (supporting Respiratory function text and Fig. S2).
Effect of CLR01 on disease onset, disease duration, and survival
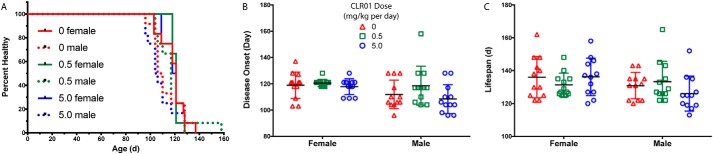
A Kaplan-Meier analysis of timing of disease onset did not show statistically significant differences among the groups (Fig. 5A). A small trend toward delayed onset was observed in both female and male mice in the low-dose treatment group, but it did not reach statistical significance. Disease onset, determined by the first detectable sign of leg weakness, scissoring of the legs when lifted by the tail, was earlier by 6 days on average in male mice than in female mice (Fig. 5B, F(1, 66) = 6.794, p = 0.011) but was not significantly different among the treatment groups. There was no statistically significant difference in overall lifespan among any of the groups (Fig. 5C).
Figure 5.
CLR01 did not increase G93A-SOD1 mouse survival. A, Kaplan-Meier analysis of disease onset in male and female (n = 12 per group each) G93A-SOD1 mice. B, disease onset was determined on the day of the first sign of weakness, crossing of hind legs when mice were lifted by the tail. C, life span represents the day the mouse either died or met criteria for euthanasia. The key in B is applicable also to C.
Effect of CLR01 on SOD1 accumulation and inflammation in mouse spinal cord
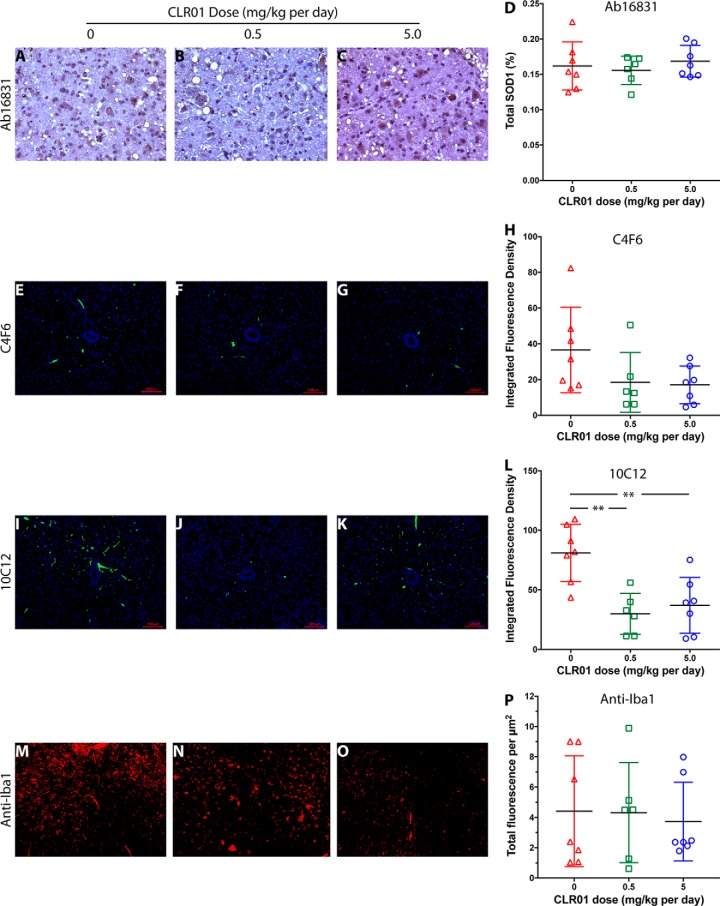
Following treatment and euthanasia, spinal cords were harvested. However, because of the aggressive course of the disease in this model, most of the mice still did not fulfill the criteria for euthanasia on the day before their death and were found dead the next day, when their tissues had begun undergoing necrosis and therefore could not be used for immunohistochemistry (IHC) analysis. Therefore, only euthanized mice (n = 7 (vehicle), 6 (low-dose group), 7 (high-dose group)) were included in the IHC analysis. The anterior horn in the lumbar region of the spinal cord of these mice was analyzed by IHC for the presence of motor neurons (anti-NeuN antibody), total SOD1 (Ab16831), G93A-SOD1 (C4F6), misfolded SOD1 (10C12), and microglia (anti-Iba1). Monoclonal antibody (mAb) C4F6 was reported previously to recognize misfolded SOD1 (41, 42). However, our own characterization has shown that this mAb recognizes specifically G93A-SOD1 regardless of misfolding (43). In contrast, we found that mAb 10C12 was highly sensitive for misfolded SOD1 (both WT and the G93A variant) (43), and therefore, we used it to analyze this form of the protein.
NeuN staining showed that in most of the sections no motor neurons were left. In a few sections, we found 1–2 surviving motor neurons, but their low frequency indicated that in the vast majority of the mice the motor neurons degenerated completely or almost completely by the time the mice reached the criteria for euthanasia, precluding quantitative analysis. IHC analysis of total SOD1 using polyclonal antibody Ab16831, which recognizes both mouse and human SOD1, showed little difference among the treatment groups (Fig. 6, A–D), in agreement with the expectation that normal SOD1 is not affected by CLR01 treatment. C4F6 staining showed a dose-dependent trend toward reduction in G93A-SOD1, which did not reach statistical significance (Fig. 6, E–H). This trend may reflect a decrease in misfolded G93A-SOD1 in the treated mice, likely due to increased clearance of the misfolded protein, as was reported previously for α-synuclein in zebrafish treated with CLR01 (22). C4F6 does not recognize metallated G93A-SOD1 (43), which could suggest that the decreased signal in treated mice might reflect stabilization of the native, metallated form of the protein. However, based on the known mechanism of action of CLR01 (17), this is an unlikely explanation of the data. A significant, ∼3-fold reduction was found in 10C12-reactive misfolded SOD1 (Fig. 6, I–L). The integrated fluorescence density in the vehicle-treated mice, was 81.9, compared with 29.8 in the low-dose group and 37.0 in the high-dose group, demonstrating that CLR01 engaged the target and reduced misfolded G93A-SOD1 in the spinal cord of the mice, in agreement with the inhibition of aggregation observed in vitro (Fig. 1, E, J, O, and T). Finally, staining with anti-Iba1 suggested a trend toward a decrease in microglial density (Fig. 6, M–O, quantified as total fluorescence/μm2) in the high-dose group, but due to high variability, this effect was not statistically significant (Fig. 6P).
Figure 6.
Analysis of SOD1 and microglia in mouse spinal cord. Lumbar spinal cord sections from euthanized mice were analyzed by IHC. A–C, representative images of pAb 16831, a pan-SOD1 antibody, staining for total mouse and human SOD1 in vehicle-treated (A), 0.5 mg/kg (B), and 5.0 mg/kg (C) CLR01-treated mice. D, quantitative analysis of pAb 16831 staining. E–H, representative images of mAb C4F6, which stains specifically apo G93A-SOD1 in vehicle-treated (E), 0.5 mg/kg (F), and 5.0 mg/kg (G) CLR01-treated mice. H, quantitative analysis of mAb C4F6 staining. I–L, representative images of mAb 10C12, which stains specifically early-forming species of misfolded SOD1 in vehicle-treated (I), 0.5 mg/kg (J), and 5.0 mg/kg (K) CLR01-treated mice. L, quantitative analysis of mAb 10C12 staining. M–P, representative images of anti-Iba1 pAb, which stains microglia in vehicle-treated (M), 0.5 mg/kg (N), and 5.0 mg/kg (O) CLR01-treated mice. P, quantitative analysis of anti-Iba1 staining. The scale bars in E–G and I–K denote 100 μm and are applicable also to M–O. **, p < 0.01, one-way ANOVA with post hoc Tukey test.
Discussion
To date, the two Federal Drug Administration-approved drugs targeting ALS potentially have considerable side effects. Thus, developing disease-modifying agents for ALS is an urgent need, and testing such therapy in transgenic mice is a crucial step in the drug-development process. Here, we tested whether the molecular tweezer CLR01, which has been shown to inhibit the formation of toxic oligomers and aggregates in other diseases, would be effective against aberrant SOD1 self-assembly and therefore offer potential therapy for ALS. For initial in vitro testing, in addition to WT SOD1, we examined the variants E21K, H46R, D76Y, and G93A, in which the disease-associated amino acid substitutions are located in different regions along the SOD1 amino acid sequence. As expected, based on the broad-spectrum mechanism of CLR01, in all cases, we found that the molecular tweezer inhibited the aggregation of SOD1 dose-dependently, with complete or near complete inhibition at 5-fold excess CLR01 (Fig. 1, A–E). Interestingly, the two cases in which the aggregation was not inhibited at a 1:1 SOD1/CLR01 concentration ratio were the WT and E21K isoforms. E21K may require a higher concentration of CLR01 for inhibition of aggregation due to the presence of the extra binding site at Lys-21 relative to the other isoforms. However, the observation that WT SOD1 required more CLR01 for inhibition than the aggregation-prone variants H46R, D76Y, and G93A is difficult to explain and will require further study. Morphological examination showed that inhibition of the ThT fluorescence increase correlated with inhibition of aggregation (Fig. 1). As SOD1 contains 11 Lys residues, the Lys/CLR01 concentration ratio at the highest CLR01 concentration corresponds to 2.2:1 (2.4:1 for E21K-SOD1), respectively, suggesting that when approximately half of the Lys residues are bound by CLR01, complete inhibition is achieved. It is important to remember, however, that the binding of CLR01 is highly labile, which means that at this concentration ratio, most of the Lys residues likely are bound part of the time.
To gain further insight into which Lys residues are most likely to be engaged by CLR01, we used top-down MS and determined that CLR01's main binding site was in the region comprising residues 65–88, which contains two Lys and two Arg residues. These experiments suggest that CLR01 binds preferentially to the Lys residues at positions 70 and 75. Previously, several regions in SOD1 have been reported to be important for the aggregation process, including residues 14–21, 30–38, 101–107, and 147–153 (44) and 10–23, 92–122, and 137–153 (45). In addition, residues 28–38 have been suggested to be involved in mediating SOD1 toxicity, presumably in an amyloidogenic oligomer structure. The main binding sites of CLR01, Lys-70, and/or Lys-75 are not involved in any of these segments, which may explain why a 5-fold excess of CLR01 is needed for complete or near-complete inhibition of SOD1 aggregation (Fig. 1). Of the segments listed above, SOD1(30–38) contains two Lys residues, at positions 30 and 36. These residues obviously are also in the segment implicated in toxicity, SOD1(28–38). In addition, Lys-23 is at the C terminus of the fragment SOD1(10–23), and Lys-122 is at the C terminus of SOD1(92–122). The other amyloidogenic segments do not contains Lys residues and therefore are not affected directly by CLR01. It is thus reasonable to hypothesize that the inhibitory activity of CLR01 is mediated by binding to Lys-30 and/or Lys-36, but binding to these residues likely occurs secondary to the binding of CLR01 to Lys-70/Lys-75, diminishing the effectivity of the molecular tweezer against SOD1.
In view of the ability of CLR01 to inhibit SOD1 aggregation in vitro, we hypothesized that it could be used therapeutically for fALS cases caused by mutant SOD1 and took the first step toward testing this hypothesis by treating the G93A-SOD1 mouse model with CLR01. This model is the most widely used and characterized mouse model for ALS, displaying typical symptoms of ALS, including progressive muscle weakness, weight loss, and difficulty with mobility, and has a 50% survival rate at 128.9 ± 9.1 days (14).
As expected, misfolded SOD1 detected by mAb 10C12, which we have found recently to be highly sensitive to early SOD1 misfolding, was reduced by ∼3-fold (p < 0.01) in the spinal cord of the CLR01-treated mice compared with vehicle-treated mice (Fig. 6, I–L). Interestingly, the reduction was similar in both treatment groups, suggesting that 0.5 mg/kg daily injection of CLR01 achieved maximal target engagement. The reduction in misfolded SOD1 correlated with a decrease in G93A-SOD1 measured using mAb C4F6 (Fig. 6, E–H), but not total SOD1 (Fig. 5, A–D).
Despite the significant decrease in misfolded SOD1, the treatment did not affect significantly the loss of motor neurons and the ensuing rapidly progressive weakness leading to death of the mice (Fig. 4, A and C). Interestingly, one male mouse in the 0.5 mg/kg group deviated significantly from the rest, lived until 165 days of age, and started showing a decline in rotarod performance only on week 22 (Fig. 4F). However, it is not known whether the treatment contributed to the extension of life and slower decline in this mouse. Most mice showed initial signs of weakness starting on day 110–120 (Fig. 4, C–F), and subsequently their disease progressed for ∼1–4 weeks until they either fulfilled the criteria for euthanasia or died. There was a small delay of the disease onset in the mice receiving 0.5 mg/kg CLR01. In contrast, male mice receiving 5.0 mg/kg CLR01 appeared to have an earlier onset and a more severe progression (Fig. 5, A–C).
Male mice consistently showed earlier disease onset than female mice, as reported previously (14). This was readily observed in the grip-strength (Fig. 2, C and D) and rotarod (Fig. 2, E and F) tests, in which the males showed earlier signs of motor difficulty than the females. Consequently, the male survival rate was shorter than that of the females (Fig. 5A). 5.0 mg/kg CLR01 caused a more rapid decline in the performance of the male mice in the grip-strength and rotarod tests, whereas 0.5 mg/kg caused a small delay (Fig. 2, D and F). These trends did not reach statistical significance and were not observed in female mice. Nonetheless, because they were observed consistently in a number of tests, they do suggest that CLR01 treatment may be beneficial at lower doses and serve as a cautionary note for the use of higher doses. It is not clear at this point whether these effects are specific to the G93A-SOD1 mice or are more general.
Toxicity studies in WT mice showed that CLR01 had a high-safety margin. A single dose of 100 mg/kg CLR01 intraperitoneally caused temporary distress due to liver injury (35), as would be expected for such a high dose, but the symptoms resolved completely within 2 h without any mortality (n = 8). Daily administration of 10 mg/kg CLR01 i.p. for 30 days had a single significant effect of ∼40% reduction of plasma cholesterol, but no other effects on mortality, morbidity, general behavior, histology, or serology (35). In view of these results, it is unlikely that the slight adverse effects observed upon daily administration of 5.0 mg/kg s.c. reflected general toxicity.
The discrepancy between the observed clearance of misfolded SOD1 (Fig. 4, I–L) and lack of significant effect on the motor tests or the disease course was surprising. One possible explanation of the results is that misfolded SOD1 is not the cause of the disease in fALS or at least in this mouse model. Although we cannot rule out this possibility, it would contradict a large body of literature without providing an explanation for the cause of the disease (46). Alternative explanations are as follows: 1) the high-copy overexpression of the SOD1 transgene in the G93A mouse caused a rapid disease progression, which cannot be counteracted by aggregation inhibitors, such as CLR01; 2) insufficient Cu2+ levels in the brain due to the high-copy overexpression of the transgene, which is unaffected by CLR01; 3) the sub-optimal route of administration, which might have limited target exposure; and 4) formation of toxic SOD1 structures that are unaffected by CLR01. If build-up of CLR01 in the CNS was slower with daily injection than using continuous administration via pumps, as was done previously, the amount of compound available for preventing formation of toxic SOD1 assemblies might have not been sufficient in the high-copy number, aggressive G93A-SOD1 model. The nondiscriminatory nature of CLR01's mechanism of action, which is based on binding to exposed Lys and Arg residues regardless of assembly state, and the presence of 11 Lys and 4 Arg residues in SOD1, make the existence of putative, specific forms that are unaffected by CLR01 unlikely. The successful improvement of the disease phenotype in this mouse model following treatment with CuII(SP-4-2) (47) supports the possibility that Cu2+ depletion, rather than misfolded G93A-SOD1, was a major contributor to the pathological process in the mice. Finally, it is not possible to formally exclude the possibility that CLR01 treatment converted SOD1 into toxic structures that are not recognized by mAb 10C12. If this is the case, it would contradict all the studies reported so far using CLR01 in cell culture or in vivo, which showed beneficial effects of the compound. We hypothesize that examination of CLR01 or other molecular-tweezer derivatives with improved pharmacokinetic characteristics in a model with slower progression may lead to a better outcome of disease onset and mouse survival.
Experimental procedures
Materials
ThT, TCEP, EDTA, sodium acetate, sodium chloride, and other high-purity reagents and buffers were obtained from Sigma or ThermoFisher Scientific. The Na+ salt of CLR01 was prepared and purified as described previously (48). The compound was dissolved in a sterile-saline solution for administration to animals.
Yeast expression system
EG118 yeast cells, a deletion mutant for yeast sod1 (sod1Δ) (49), were transformed with YEp351, a yeast multicopy shuttle vector containing a LEU2-selectable marker and WT or a mutant human SOD1 gene (50). Plasmids containing sequences for WT and SOD1 variants G93A, E21K, H46R, and D76Y were a kind gift from Dr. Joan Valentine, UCLA.
Mouse model
Transgenic B6SJL-Tg mice hemizygous for G93A-SOD1 as used in this study were obtained through Prize4Life from The Jackson Laboratory. Animal care was provided in accordance with the United States Public Health Service Guide for the Care and Use of Laboratory Animals. All procedures were approved by the Institutional Animal Care and Use Committee of UCLA.
Antibodies
For antibodies and dilutions, rabbit pAb ab16831 (1:1,000) was obtained from Abcam. Mouse mAb C4F6 (1:250) was from MédiMabs, Montréal, Québec, Canada. Mouse mAb 10C12 (1:500) was a generous gift from Dr. Neil Cashman, University of British Columbia, Canada. Guinea pig pAb anti-Iba1 (1:250) was from Synaptic Systems, Göttingen, Germany. Biotinylated anti-rabbit IgG (1:500, Vector Laboratories), Alexa Fluor 488–labeled anti-mouse IgG (1:500), and Alexa Fluor 594–conjugated anti-guinea pig IgG (1:2500, ThermoFisher Scientific) were used as secondary antibodies.
Protein purification and demetallation
EG118 Saccharomyces cerevisiae cells were transformed with each plasmid. Yeast cells were grown at 30 °C with shaking at 220 rpm in yeast extract/peptone or synthetic dropout without Leu, containing 2% glucose. The culture was grown for 5–7 days until reaching A600 = 0.6–0.9. Then the cells were resuspended in a double volume of lysis buffer containing 250 mm Tris-HCl, 150 mm NaCl, and 0.1 mm EDTA, pH 8.0. Cells were lysed by adding 0.5-mm glass beads and vortexing in a blender at high speed. Proteins were purified using sequential hydrophobic interaction, ion-exchange, and size-exclusion chromatographies, as described previously (51). The de-metallated (apo) form of each variant was obtained by dialyzing first against 50 mm EDTA, 100 mm sodium acetate, pH 3.8, then against 100 mm NaCl, 100 mm sodium acetate, pH 3.8, and finally against 10 mm potassium phosphate, pH 7.0. The concentration of the proteins was determined by measuring absorbance at 280 nm using ϵ = 10,800 m−1 cm−1.
ThT assay
De-metallated variants of SOD1 were incubated at 40 μm in the absence or presence of 40 or 200 μm CLR01 in a buffer containing 10 mm potassium phosphate, 40 μm ThT, and 50 mm TCEP, pH 7.4. The aggregation reactions were carried out in 96-well plates, each containing a final volume of 100 μl and a Teflon ball to assist with agitation. Triplicate samples were incubated at 37 °C with agitation at 300 rpm in a plate reader (Synergy HTX, BioTek Instruments) in three independent experiments. ThT fluorescence was measured every 10 min using λex = 420 nm and λem = 485 nm (52). For clarity, data points are presented only every 30 min in Fig. 1.
Transmission electron microscopy
Ten-μl aliquots were taken at the end of each aggregation reaction and applied onto freshly glow-discharged, carbon-coated 400-mesh copper grids, rinsed with deionized water, and stained with 1% uranyl acetate, as described previously (28). Images were obtained using a JEOL JEM1200-EX transmission electron microscope at 80 kV.
Remodeling of SOD1 aggregates by CLR01
Forty μm WT SOD1 or G93A-SOD1 solutions were aged for 6 months. The pre-formed aggregates were mixed with 10-fold excess CLR01 in a 96-well plate and covered with a sealing film to avoid evaporation. Triplicate samples were incubated at 37 °C without agitation. ThT fluorescence was measured every 24 h using λex = 420 nm, λem = 485 nm (52). The samples were shaken for 20 s in the plate reader (Synergy HTX, BioTek Instruments) each time before taking the reading. Ten-μl aliquots were taken on days 0, 11, and 26 for morphological analysis by EM.
Native MS
Native electrospray ionization-MS was performed using a Bruker Solarix 15-Tesla FT-ICR mass spectrometer (Billerica, MA), equipped with an Infinity ICR cell. Protein solutions were prepared at a concentration of 5–10 μm in 20 mm ammonium acetate, pH 6.8. Samples were loaded into a borosilicate glass capillary coated with Au/Pd (Proxeon) for nanoelectrospray. The analyte solution flow rate was 50–100 nl/min. The ESI voltage was set to 1.1 kV. Drying gas was applied at 0.5 liters/min at 160 °C in the ESI source region to aid desolvation. Transfer ion optics were tuned to the following settings: capillary 200 V, deflector plate at 180 V, funnel 120 V, and skimmer 40 V. For top-down MS experiments, the ECD parameters were as follows: 10-ms electron pulse, a 1-V bias, and 15 V applied to the lens. Product ions were manually identified using the Bruker DataAnalysis software and assigned by a home-built Matlab program using a ≤10 ppm mass accuracy tolerance.
Mouse treatment
G93A-SOD1 transgenic mice were kept in standard mouse housing with 12-h light/dark cycles and ad libitum chow and water. Daily s.c. injections of CLR01 started at 50 days of age. Treatment ended when the mice met either one of the criteria for euthanasia: either loss of ≥10% of body weight or the inability of the mouse to right itself within 30 s when placed on either of its sides.
Experimental groups
The cohort comprised 72 mice, 36 males and 36 females, that were randomized into three treatment groups receiving 0, 0.5, or 5.0 mg/kg CLR01 in 0.9% (w/v) sterile saline by daily s.c. injection. Each group included 12 females and 12 males. The mice were weighed and tested for strength using the grip-strength and rotarod tests weekly. In addition, the respiratory volumes of the mice under normal and challenging conditions (elevated CO2) were measured weekly.
Grip-strength test
The test started on week 7. Each mouse was placed on a wire lid of a conventional housing cage, and the lid was turned upside down. The latency from the beginning of the test until the mouse stood with at least two limbs on the lid was timed. The animals had three attempts to stand for a maximum of 60 s each for five trials, and the longest latency was recorded.
Rotarod test
Mice started training on the rotarod apparatus daily 1 week before data recording began on week 13. Animals were placed onto the cylinder at a constant speed of 15 rpm for 5 min. The longest latency to fall in three trials was recorded for up to 300 s per trial.
Pulmonary function
The inspiratory and expiratory flow were measured in conscious, unrestrained mice using a whole-body plethysmograph (Buxco, Data Sciences International). The system measures the small changes in the air that are exchanged in and out of the entire chamber due to the animal's respiration. The chamber was calibrated before every experiment, selecting the most appropriate response observed in FinePoint Software (Buxco, Data Sciences International) to the injection of 1 ml of air into the chamber. After calibration, mice were placed into the chamber and acclimated for 55 min. CO2 concentration was maintained during the acclimation and room-air breathing periods by using a bias-flow regulator. After acclimation, baseline respiratory function was measured for 5 min. After baseline correction, bias flow was turned down for the first CO2 exposure. The bias flow was turned back on for 10 min and again turned off for the second CO2 exposure. Both exposures used 4 ml of CO2 for 5 min. All data were collected and analyzed using FinePoint, which uses algorithms that include variables affecting respiratory function, such as humidity and temperature, to calculate the measured box flow and the physiological values of the respiratory parameters from the animal.
Immunohistochemistry
Euthanized animals were perfused with 4% paraformaldehyde. Spinal cords were removed and post-fixed for 2 h in 4% paraformaldehyde. Thereafter, the tissue was dehydrated, paraffinized, and sectioned. To prepare the lumbar spinal cord sections for immunohistochemistry analysis, all slides were incubated at 50–60 °C for 60 min. Slides then were deparaffinized by soaking the sections in xylene four times and rehydrated sequentially in 100, 95, and 70% ethanol. Antigens were retrieved by placing sections in 10 mm sodium citrate buffer, pH 6.0, and heating in a pressure cooker for 10 min and allowed to cool back down to 20 °C. Sections then were washed in 0.05% (v/v) Tween 20 in PBS, blocked in 5% (v/v) normal goat serum (NGS) in PBS, and incubated with each primary antibody in 2.5% (v/v) NGS in PBS at the dilutions mentioned above. Sections stained with Ab16831 were subsequently washed and incubated with a biotinylated anti-rabbit secondary antibody in wash buffer. Slides were then incubated with an avidin-biotinylated enzyme complex (ABC Elite Vectastain kit, Vector Labs) using a peroxidase detection system for 80 min at 37 °C as described previously (24). Antigen was visualized by incubation for 2 min with metal-enhanced 3,3′-diaminobenzidine tetrahydrochloride (ThermoFisher Scientific). Finally, sections were counterstained with hematoxylin for 30 s. After staining, sections were dehydrated sequentially in 70, 95, and 100% ethanol and then treated with xylene. All Ab16831 sections were coverslipped using Permount and dried in a chemical fume hood for 3 days. Quantitation of Ab16831-positive staining was performed using a StereoInvestigator system. Regions of interest were drawn around the individual cells. Cells containing SOD1 within the gray matter were normalized to the total area quantified. Each section was quantified twice, and the average of the two counts was used. Other antibodies were visualized by fluorescence microscopy.
Fluorescence microscopy
Both primary antibodies C4F6 and 10C12 were visualized using an Alexa Fluor 488–conjugated fluorescent anti-mouse secondary antibody. Anti-Iba1 was visualized using an Alexa Fluor 594 anti-guinea pig secondary antibody. Nuclei were counterstained with 4′,6-diamidino-2-phenylindole. Images were acquired using a Keyence BZ9000 fluorescence microscope. The central aspect of the spinal cord was used to define the area analyzed, and background fluorescence was eliminated using the background–reduction function in the software controlling the microscope. Quantification of integrated fluorescence per unit area (in μm2) was done using ImageJ (33).
Statistical analysis
All statistical analyses were performed using Prism 7.0d (GraphPad Software). To identify differences among groups, Student's t test was used for female versus male mouse group analysis, and one-way analysis of variance (ANOVA) was used for comparison among treatment groups. p < 0.05 was considered statistically significant.
Author contributions
R. M., H. M., and P. W. data curation; R. M., H. M., P. W., J. A. L., M. W., and G. B. formal analysis; R. M., H. M., P. W., and G. B. investigation; R. M., H. M., and J. A. L. writing-original draft; H. M., M. J. S., M.W., and G. B. methodology; C. I. C., N. S., R. S. A., F.-G. K., and T. S. resources; M. J. S., M. W., and G. B. conceptualization; M. J. S., J. A. L., M. W., and G. B. supervision; M. J. S., J. A. L., M. W., and G. B. funding acquisition; M. W. and G. B. project administration; M. W. and G. B. writing-review and editing; G. B. validation.
Supplementary Material
Acknowledgments
We acknowledge the contribution of Dr. Christine Fontanilla to performing experiments and collecting data. We are grateful to Dr. Joan Valentine (UCLA) for the SOD1-containing plasmids; Dr. Marie-Françoise Chesselet (UCLA) for the use of her StereoInvestigator system; Dr. Neil Cashman (University of British Columbia) for mAb 10C12; Nihal Satyadev for help with immunohistochemistry; and Prize4Life for providing the mice for the study.
This work was supported by a Judith and Jean Pape Adams Charitable Foundation grant (to G. B.), the David Vickter Foundation (to M. W.), Prize4Life (to G. B. and M. W.), RGK Foundation Grant 20143057 (G. B.), National Institutes of Health/NINDS F32 Fellowship NS087858 (to C. F.), National Institutes of Health/NIAMS Grant P30 AR057230 (to M. J. S.), National Institutes of Health/NIGMS Grant R01 GM103479 (to J. A. L.), and National Institutes of Health/NCRR Grant S10 RR028893 (to J. A. L.). The authors declare that they have no conflicts of interest with the contents of this article. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S2, Table S1 and supporting Ref. 1.
- ALS
- amyotrophic lateral sclerosis
- ANOVA
- analysis of variance
- ESI
- electrospray ionization
- fALS
- familial ALS
- IHC
- immunohistochemistry
- NGS
- normal goat serum
- SOD1
- Cu-Zn superoxide dismutase
- TCEP
- tris(2-carboxyethyl)phosphine
- ThT
- thioflavin T
- MT
- molecular tweezer
- AD
- Alzheimer's disease
- PD
- Parkinson's disease
- ECD
- electron capture dissociation
- CNS
- central nervous system.
References
- 1. Chiò A., Logroscino G., Traynor B. J., Collins J., Simeone J. C., Goldstein L. A., and White L. A. (2013) Global epidemiology of amyotrophic lateral sclerosis: a systematic review of the published literature. Neuroepidemiology 41, 118–130 10.1159/000351153 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Sreedharan J., and Brown R. H. Jr. (2013) Amyotrophic lateral sclerosis: problems and prospects. Ann. Neurol. 74, 309–316 10.1002/ana.24012 [DOI] [PubMed] [Google Scholar]
- 3. Ratovitski T., Corson L. B., Strain J., Wong P., Cleveland D. W., Culotta V. C., and Borchelt D. R. (1999) Variation in the biochemical/biophysical properties of mutant superoxide dismutase 1 enzymes and the rate of disease progression in familial amyotrophic lateral sclerosis kindreds. Hum. Mol. Genet. 8, 1451–1460 10.1093/hmg/8.8.1451 [DOI] [PubMed] [Google Scholar]
- 4. Reaume A. G., Elliott J. L., Hoffman E. K., Kowall N. W., Ferrante R. J., Siwek D. F., Wilcox H. M., Flood D. G., Beal M. F., Brown R. H. Jr., Scott R. W., and Snider W. D. (1996) Motor neurons in Cu/Zn superoxide dismutase-deficient mice develop normally but exhibit enhanced cell death after axonal injury. Nat. Genet. 13, 43–47 10.1038/ng0596-43 [DOI] [PubMed] [Google Scholar]
- 5. Johnston J. A., Dalton M. J., Gurney M. E., and Kopito R. R. (2000) Formation of high molecular weight complexes of mutant Cu, Zn-superoxide dismutase in a mouse model for familial amyotrophic lateral sclerosis. Proc. Natl. Acad. Sci. U.S.A. 97, 12571–12576 10.1073/pnas.220417997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Siddique T., and Ajroud-Driss S. (2011) Familial amyotrophic lateral sclerosis, a historical perspective. Acta Myol. 30, 117–120 [PMC free article] [PubMed] [Google Scholar]
- 7. Martin D., Thompson M. A., and Nadler J. V. (1993) The neuroprotective agent riluzole inhibits release of glutamate and aspartate from slices of hippocampal area CA1. Eur. J. Pharmacol. 250, 473–476 10.1016/0014-2999(93)90037-I [DOI] [PubMed] [Google Scholar]
- 8. Sawada H. (2017) Clinical efficacy of edaravone for the treatment of amyotrophic lateral sclerosis. Expert Opin. Pharmacother. 18, 735–738 10.1080/14656566.2017.1319937 [DOI] [PubMed] [Google Scholar]
- 9. Liu T., and Bitan G. (2012) Modulating self-assembly of amyloidogenic proteins as a therapeutic approach for neurodegenerative diseases: strategies and mechanisms. Chem. Med. Chem. 7, 359–374 10.1002/cmdc.201100585 [DOI] [PubMed] [Google Scholar]
- 10. Xia G., Benmohamed R., Kim J., Arvanites A. C., Morimoto R. I., Ferrante R. J., Kirsch D. R., and Silverman R. B. (2011) Pyrimidine-2,4,6-trione derivatives and their inhibition of mutant SOD1-dependent protein aggregation. Toward a treatment for amyotrophic lateral sclerosis. J. Med. Chem. 54, 2409–2421 10.1021/jm101549k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Trippier P. C., Zhao K. T., Fox S. G., Schiefer I. T., Benmohamed R., Moran J., Kirsch D. R., Morimoto R. I., and Silverman R. B. (2014) Proteasome activation is a mechanism for pyrazolone small molecules displaying therapeutic potential in amyotrophic lateral sclerosis. ACS Chem. Neurosci. 5, 823–829 10.1021/cn500147v [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Nowak R. J., Cuny G. D., Choi S., Lansbury P. T., and Ray S. S. (2010) Improving binding specificity of pharmacological chaperones that target mutant superoxide dismutase-1 linked to familial amyotrophic lateral sclerosis using computational methods. J. Med. Chem. 53, 2709–2718 10.1021/jm901062p [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Rosen D. R., Siddique T., Patterson D., Figlewicz D. A., Sapp P., Hentati A., Donaldson D., Goto J., O'Regan J. P., and Deng H. X. (1993) Mutations in Cu/Zn superoxide dismutase gene are associated with familial amyotrophic lateral sclerosis. Nature 362, 59–62 10.1038/362059a0 [DOI] [PubMed] [Google Scholar]
- 14. Gurney M. E., Pu H., Chiu A. Y., Dal Canto M. C., Polchow C. Y., Alexander D. D., Caliendo J., Hentati A., Kwon Y. W., and Deng H. X. (1994) Motor neuron degeneration in mice that express a human Cu,Zn superoxide dismutase mutation. Science 264, 1772–1775 10.1126/science.8209258 [DOI] [PubMed] [Google Scholar]
- 15. Kim R. B., Irvin C. W., Tilva K. R., and Mitchell C. S. (2015) State of the field: an informatics-based systematic review of the SOD1-G93A amyotrophic lateral sclerosis transgenic mouse model. Amyotroph Lateral Scler Frontotemporal Degener. 17, 1–14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Attar A., and Bitan G. (2014) Disrupting self-assembly and toxicity of amyloidogenic protein oligomers by “molecular tweezers”–from the test tube to animal models. Curr. Pharm. Des. 20, 2469–2483 10.2174/13816128113199990496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Schrader T., Bitan G., and Klärner F. G. (2016) Molecular tweezers for lysine and arginine-powerful inhibitors of pathologic protein aggregation. Chem. Commun. 52, 11318–11334 10.1039/C6CC04640A [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Malik R., Di J., Nair G., Attar A., Taylor K., Teng E., Klärner F. G., Schrader T., and Bitan G. (2018) Using molecular tweezers to remodel abnormal protein self-assembly and inhibit the toxicity of amyloidogenic proteins. Methods Mol. Biol. 1777, 369–386 10.1007/978-1-4939-7811-3_24 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Sinha S., Lopes D. H., Du Z., Pang E. S., Shanmugam A., Lomakin A., Talbiersky P., Tennstaedt A., McDaniel K., Bakshi R., Kuo P. Y., Ehrmann M., Benedek G. B., Loo J. A., Klärner F. G., et al. (2011) Lysine-specific molecular tweezers are broad-spectrum inhibitors of assembly and toxicity of amyloid proteins. J. Am. Chem. Soc. 133, 16958–16969 10.1021/ja206279b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Sinha S., Du Z., Maiti P., Klärner F. G., Schrader T., Wang C., and Bitan G. (2012) Comparison of three amyloid assembly inhibitors: the sugar scyllo-inositol, the polyphenol epigallocatechin gallate, and the molecular tweezer CLR01. ACS Chem. Neurosci. 3, 451–458 10.1021/cn200133x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Zheng X., Liu D., Klärner F. G., Schrader T., Bitan G., and Bowers M. T. (2015) Amyloid β-protein assembly: the effect of molecular tweezers CLR01 and CLR03. J. Phys. Chem. B 119, 4831–4841 10.1021/acs.jpcb.5b00692 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Prabhudesai S., Sinha S., Attar A., Kotagiri A., Fitzmaurice A. G., Lakshmanan R., Lakshmanan R., Ivanova M. I., Loo J. A., Klärner F. G., Schrader T., Stahl M., Bitan G., and Bronstein J. M. (2012) A novel “molecular tweezer” inhibitor of α-synuclein neurotoxicity in vitro and in vivo. Neurotherapeutics 9, 464–476 10.1007/s13311-012-0105-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Acharya S., Safaie B. M., Wongkongkathep P., Ivanova M. I., Attar A., Klärner F. G., Schrader T., Loo J. A., Bitan G., and Lapidus L. J. (2014) Molecular basis for preventing α-synuclein aggregation by a molecular tweezer. J. Biol. Chem. 289, 10727–10737 10.1074/jbc.M113.524520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Attar A., Ripoli C., Riccardi E., Maiti P., Li Puma D. D., Liu T., Hayes J., Jones M. R., Lichti-Kaiser K., Yang F., Gale G. D., Tseng C. H., Tan M., Xie C. W., Straudinger J. L., et al. (2012) Protection of primary neurons and mouse brain from Alzheimer's pathology by molecular tweezers. Brain 135, 3735–3748 10.1093/brain/aws289 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Richter F., Subramaniam S. R., Magen I., Lee P., Hayes J., Attar A., Zhu C., Franich N. R., Bove N., De La Rosa K., Kwong J., Klärner F. G., Schrader T., Chesselet M. F., and Bitan G. (2017) A molecular tweezer ameliorates motor deficits in mice overexpressing α-synuclein. Neurotherapeutics 14, 1107–1119 10.1007/s13311-017-0544-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Ferreira N., Pereira-Henriques A., Attar A., Klärner F. G., Schrader T., Bitan G., Gales L., Saraiva M. J., and Almeida M. R. (2014) Molecular tweezers targeting transthyretin amyloidosis. Neurotherapeutics 11, 450–461 10.1007/s13311-013-0256-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Xu N., Bitan G., Schrader T., Klärner F. G., Osinska H., and Robbins J. (2017) Inhibition of mutant αB crystallin-induced protein aggregation by a molecular tweezer. J. Am. Heart Assoc. 6, e006182 10.1161/JAHA.117.006182 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chattopadhyay M., Durazo A., Sohn S. H., Strong C. D., Gralla E. B., Whitelegge J. P., and Valentine J. S. (2008) Initiation and elongation in fibrillation of ALS-linked superoxide dismutase. Proc. Natl. Acad. Sci. U.S.A. 105, 18663–18668 10.1073/pnas.0807058105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Hallewell A. R., Mills R., Tekamp-Olson P., Blacher R., Rosenberg S., Ötting, Masiarz F. F., and Scandella C. J. (1987) Amino terminal acetylation of authentic human Cu,Zn superoxide dismutase produced in yeast. Nature Biotechnol. 5, 363–366 [Google Scholar]
- 30. Giehm L., and Otzen D. E. (2010) Strategies to increase the reproducibility of protein fibrillization in plate reader assays. Anal. Biochem. 400, 270–281 10.1016/j.ab.2010.02.001 [DOI] [PubMed] [Google Scholar]
- 31. Vöpel T., Bravo-Rodriguez K., Mittal S., Vachharajani S., Gnutt D., Sharma A., Steinhof A., Fatoba O., Ellrichmann G., Nshanian M., Heid C., Loo J. A., Klärner F. G., Schrader T., Bitan G., et al. (2017) Inhibition of Huntingtin exon-1 aggregation by the molecular tweezer CLR01. J. Am. Chem. Soc. 139, 5640–5643 10.1021/jacs.6b11039 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Nshanian M., Lantz C., Wongkongkathep P., Schrader T., Klärner F. G., Blümke A., Despres C., Ehrmann M., Smet-Nocca C., Bitan G., and Loo J. A. (2019) Native top-down mass spectrometry and ion mobility spectrometry of the interaction of tau protein with a molecular tweezer assembly modulator. J. Am. Soc. Mass Spectrom. 30, 16–23 10.1007/s13361-018-2027-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Abramoff M. D., Magelhaes P. J., and Ram S. J. (2004) Image processing with ImageJ. Biophotonics Intl. 11, 36–42 [Google Scholar]
- 34. Rakhit R., and Chakrabartty A. (2006) Structure, folding, and misfolding of Cu,Zn superoxide dismutase in amyotrophic lateral sclerosis. Biochim. Biophys. Acta 1762, 1025–1037 10.1016/j.bbadis.2006.05.004 [DOI] [PubMed] [Google Scholar]
- 35. Attar A., Chan W. T., Klärner F. G., Schrader T., and Bitan G. (2014) Safety and pharmacological characterization of the molecular tweezer CLR01–a broad-spectrum inhibitor of amyloid proteins' toxicity. BMC Pharmacol. Toxicol. 15, 23 10.1186/2050-6511-15-23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Capote J., Kramerova I., Martinez L., Vetrone S., Barton E. R., Sweeney H. L., Miceli M. C., and Spencer M. J. (2016) Osteopontin ablation ameliorates muscular dystrophy by shifting macrophages to a pro-regenerative phenotype. J. Cell Biol. 213, 275–288 10.1083/jcb.201510086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Brooks S. P., and Dunnett S. B. (2009) Tests to assess motor phenotype in mice: a user's guide. Nat. Rev. Neurosci. 10, 519–529 10.1038/nrn2652 [DOI] [PubMed] [Google Scholar]
- 38. Heiman-Patterson T. D., Deitch J. S., Blankenhorn E. P., Erwin K. L., Perreault M. J., Alexander B. K., Byers N., Toman I., and Alexander G. M. (2005) Background and gender effects on survival in the TgN(SOD1-G93A)1Gur mouse model of ALS. J. Neurol. Sci. 236, 1–7 10.1016/j.jns.2005.02.006 [DOI] [PubMed] [Google Scholar]
- 39. Scott S., Kranz J. E., Cole J., Lincecum J. M., Thompson K., Kelly N., Bostrom A., Theodoss J., Al-Nakhala B. M., Vieira F. G., Ramasubbu J., and Heywood J. A. (2008) Design, power, and interpretation of studies in the standard murine model of ALS. Amyotrophic Lateral Sclerosis 9, 4–15 10.1080/17482960701856300 [DOI] [PubMed] [Google Scholar]
- 40. Nichols N. L., Van Dyke J., Nashold L., Satriotomo I., Suzuki M., and Mitchell G. S. (2013) Ventilatory control in ALS. Respir. Physiol. Neurobiol. 189, 429–437 10.1016/j.resp.2013.05.016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Bosco D. A., Morfini G., Karabacak N. M., Song Y., Gros-Louis F., Pasinelli P., Goolsby H., Fontaine B. A., Lemay N., McKenna-Yasek D., Frosch M. P., Agar J. N., Julien J. P., Brady S. T., and Brown R. H. Jr. (2010) Wild-type and mutant SOD1 share an aberrant conformation and a common pathogenic pathway in ALS. Nat. Neurosci. 13, 1396–1403 10.1038/nn.2660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Brotherton T. E., Li Y., Cooper D., Gearing M., Julien J. P., Rothstein J. D., Boylan K., and Glass J. D. (2012) Localization of a toxic form of superoxide dismutase 1 protein to pathologically affected tissues in familial ALS. Proc. Natl. Acad. Sci. U.S.A. 109, 5505–5510 10.1073/pnas.1115009109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Atlasi R. S., Malik R., Corrales C. I., Tzeplaeff L., Whitelegge J. P., Cashman N. R., and Bitan G. (2018) Investigation of anti-SOD1 antibodies yields new structural insight into SOD1 misfolding and surprising behavior of the antibodies themselves. ACS Chem. Biol. 13, 2794–2807 10.1021/acschembio.8b00729 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Ivanova M. I., Sievers S. A., Guenther E. L., Johnson L. M., Winkler D. D., Galaleldeen A., Sawaya M. R., Hart P. J., and Eisenberg D. S. (2014) Aggregation-triggering segments of SOD1 fibril formation support a common pathway for familial and sporadic ALS. Proc. Natl. Acad. Sci. U.S.A. 111, 197–201 10.1073/pnas.1320786110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ida M., Ando M., Adachi M., Tanaka A., Machida K., Hongo K., Mizobata T., Yamakawa M. Y., Watanabe Y., Nakashima K., and Kawata Y. (2016) Structural basis of Cu,Zn-superoxide dismutase amyloid fibril formation involves interaction of multiple peptide core regions. J. Biochem. 159, 247–260 10.1093/jb/mvv091 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. van Es M. A., Dahlberg C., Birve A., Veldink J. H., van den Berg L. H., and Andersen P. M. (2010) Large-scale SOD1 mutation screening provides evidence for genetic heterogeneity in amyotrophic lateral sclerosis. J. Neurol. Neurosurg. Psychiatry 81, 562–566 10.1136/jnnp.2009.181453 [DOI] [PubMed] [Google Scholar]
- 47. Hilton J. B., Mercer S. W., Lim N. K., Faux N. G., Buncic G., Beckman J. S., Roberts B. R., Donnelly P. S., White A. R., and Crouch P. J. (2017) Cu(II)(atsm) improves the neurological phenotype and survival of SOD1(G93A) mice and selectively increases enzymatically active SOD1 in the spinal cord. Sci. Rep. 7, 42292 10.1038/srep42292 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Talbiersky P., Bastkowski F., Klärner F. G., and Schrader T. (2008) Molecular clip and tweezer introduce new mechanisms of enzyme inhibition. J. Am. Chem. Soc. 130, 9824–9828 10.1021/ja801441j [DOI] [PubMed] [Google Scholar]
- 49. Gralla E. B., and Valentine J. S. (1991) Null mutants of Saccharomyces cerevisiae Cu,Zn superoxide dismutase: characterization and spontaneous mutation rates. J. Bacteriol. 173, 5918–5920 10.1128/jb.173.18.5918-5920.1991 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Wei J. P., Srinivasan C., Han H., Valentine J. S., and Gralla E. B. (2001) Evidence for a novel role of copper-zinc superoxide dismutase in zinc metabolism. J. Biol. Chem. 276, 44798–44803 10.1074/jbc.M104708200 [DOI] [PubMed] [Google Scholar]
- 51. Hayward L. J., Rodriguez J. A., Kim J. W., Tiwari A., Goto J. J., Cabelli D. E., Valentine J. S., and Brown R. H. Jr. (2002) Decreased metallation and activity in subsets of mutant superoxide dismutases associated with familial amyotrophic lateral sclerosis. J. Biol. Chem. 277, 15923–15931 10.1074/jbc.M112087200 [DOI] [PubMed] [Google Scholar]
- 52. Rogers D. R. (1965) Screening for amyloid with the thioflavin-T fluorescent method. Am. J. Clin. Pathol. 44, 59–61 10.1093/ajcp/44.1.59 [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.