Abstract
MicroRNAs (miRNAs) control various biological processes by inducing translational repression and transcript degradation of the target genes. In mammalian development, knowledge of the timing and expression pattern of each miRNA is important to determine and predict its function in vivo. So far, no systematic analyses of the spatiotemporal expression pattern of miRNAs during mammalian neurodevelopment have been performed. Here, we isolated total RNAs from the embryonic dorsal forebrain of mice at different developmental stages and subjected these RNAs to microarray analyses. We selected 279 miRNAs that exhibited high signal intensities or ascending or descending expression dynamics. To ascertain the expression patterns of these miRNAs, we used locked nucleic acid (LNA)-modified miRNA probes in in situ hybridization experiments. Multiple miRNAs exhibited spatially restricted/enriched expression in anatomically distinct regions or in specific neuron subtypes in the embryonic brain and spinal cord, such as in the ventricular area, the striatum (and other basal ganglia), hypothalamus, choroid plexus, and the peripheral nervous system. These findings provide new insights into the expression and function of miRNAs during the development of the nervous system and could be used as a resource to facilitate studies in neurodevelopment.
Keywords: microRNA (miRNA), neurodevelopment, central nervous system (CNS), brain, mouse, embryo, gene expression, expression pattern, in situ hybridization
Introduction
The mammalian nervous system is the most complex structure of the body because of its unparalleled cellular diversity and complex connectivity, which are responsible for highly complex neural processes such as cognitive function, sensory perception, voluntary motor control, and consciousness. Numerous types of neurons and glial cells are produced with striking precision throughout the development of the nervous system, which require tight control of intrinsic signaling pathways and extrinsic factors to coordinate gene expression in a spatially and temporally specific manner (1–3). Normal development ensures that the correct complement of RNAs and proteins are present in the correct cell at the correct time. Therefore, delineation of gene expression profiles in distinct cell subtypes at different stages and characterization of the functions of these genes are critical steps in our understanding of the molecular programs that govern cell fate determination, survival, maturation, and connectivity during neural development.
Several high-throughput approaches were recently developed to identify the specific expression of genes and transcripts at cellular resolution, for example: gene expression studies by microarray or deep sequencing analysis of purified neuronal subtypes (4–6), microdissected regions of the neocortex (7–10), or single cell analysis of different regions of the brain (11–19); construction of transgenic mouse lines that express reporter genes using the promoter regions of the lineage or cell type-restricted genes and sorting the cells (20); and large-scale screening technology for RNA in situ hybridization (ISH)3 to map gene expression patterns on tissue sections (21–27). There are several publicly available databases, such as The Genepaint (23), the Brain Gene Expression Map (BGEM) (24), the Allen Brain Atlas (ABA) (25), and the Eurexpress (26), which provide comprehensive gene expression atlases for developing and adult central nervous systems. However, these databases, as well as other reports, contain large-scale in situ hybridizations of thousands of transcripts that are essentially protein-coding genes, and very few noncoding RNAs, particularly microRNAs (miRNAs), are included.
miRNAs are ∼22–nucleotide, endogenous noncoding RNAs that regulate gene expression via mediation of target mRNA degradation or translational inhibition. These molecules emerged as important post-transcriptional regulators of various aspects of nervous system development (28–30). A large amount of evidence demonstrates that miRNAs play important roles in vertebrate development and the expression of miRNAs is genetically programmed in spatially and temporally dependent patterns (31, 32). Therefore, expression profiling of miRNAs is extremely necessary to reveal the biological roles of miRNAs and miRNA-associated gene regulatory networks. However, the detection of expression patterns of miRNA in situ was technically challenging because of their small size, until development of the locked nucleic acid (LNA)-based technique (33–35).
The present study used LNA-modified probes of miRNAs to screen the expression of 279 miRNAs in E12.5 and E16.5 mouse embryos. By analyzing the spatiotemporal expression patterns and cellular localizations of these 279 miRNAs at different developmental stages, it revealed that many miRNAs exhibited specific expression in different anatomical structures, such as in different subtypes of neurons or brain regions, in the choroid plexus or arachnoids, as well as that restricted expression in non-nervous system tissues miRNAs. These data provide an entry point for subsequent investigations of the function of miRNAs in neural development processes.
Results
Analysis of the expression of miRNAs in mouse embryos using LNA probe in situ hybridization
Our initial plan was to systematically examine the expression and function of miRNAs that regulate cerebral cortex development. We first performed microarray analyses of RNAs isolated from the embryonic dorsal forebrains at different stages (E12.5 and E16.5) based on the miRBase Database Release 10.0 (http://www.mirbase.org/)4 (56) (data not shown). We repeated it again through the updated miRBase Database Release 19.0 six years later and analyzed the newly discovered miRNAs (Table S1). However, microarray analysis did not provide sufficient spatial and temporal details to determine the timing and location of the miRNAs expression. Therefore, we performed further ISH analyses to ascertain the expression of the miRNAs in the developing mouse brain. We selected 279 miRNAs that exhibited signal intensities at each stage that were higher than 500 or exhibited ascending or descending expression dynamics during the process of cerebral cortex development for further ISH analysis.
We first examined the accuracy of the LNA probes in the detection of specific miRNA expression in ISH experiments. We found the miR-128, miR-9, and several members of the let-7 family were abundantly expressed in the developing neocortex. When mutated, 2, 3, and 4 nucleotides of the probes for let-7b, miR-128, and miR-9, respectively, and the resultant probes did not detect any specific ISH signals (Fig. S1, A–D). Moreover, when transiently overexpressed, miRNAs in mouse embryos and the LNA probes could recognize their exogenous miRNAs, respectively. Also, the let-7a LNA probe only detected exogenous let-7a, but not let-7b, and vice versa, although let-7a and let-7b only differ in two nucleotides (Fig. S1, F–K). Additionally, when using probes generated by two different companies and with different LNA-modified sites of the same miRNA, their signal distributions were also consistent (Fig. S1E). These findings confirmed that the LNA probes for ISH could be used to examine the spatial and temporal expression patterns of miRNAs.
We then proceeded to perform ISH studies using LNA-modified probes (34) for different miRNAs and selected coronal head and transverse body sections (to include the brain and spinal cord, respectively) at E12.5 and coronal sections at E16.5 for the initial screening. Most miRNAs either showed ubiquitous or no detectable expression in the nervous system. For those showing specific patterns of expression, we performed a secondary screen using additional sections from other developmental stages. Overall, a total of 392 probes for 279 miRNAs and 21 control probes (randomized sequence or for mutant miRNAs) were generated and analyzed (Table S2).
The miRNAs enriched in embryonic neural progenitor cells
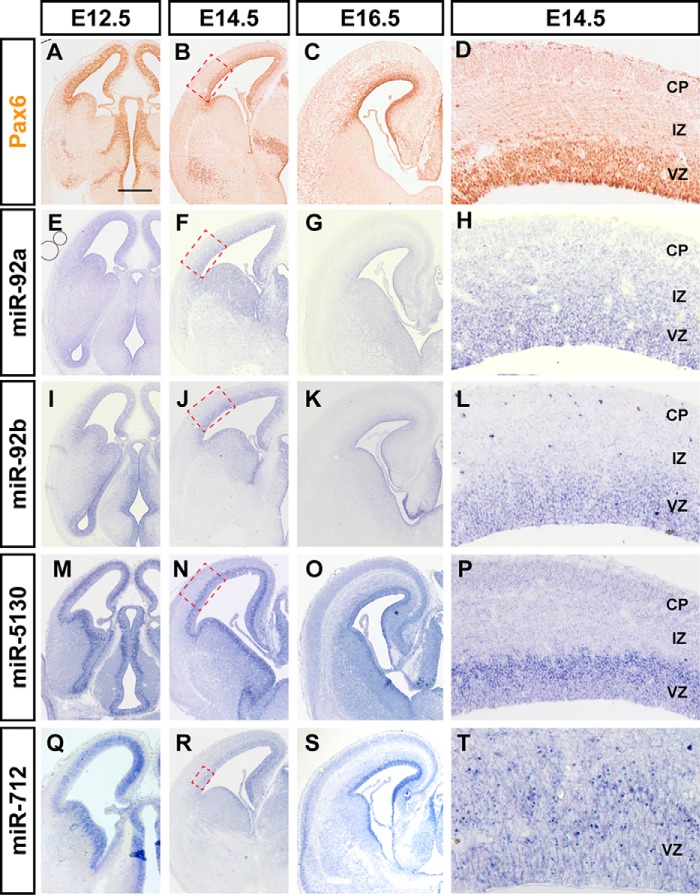
Neural progenitor cells in the embryonic nervous system, which include stem cells and precursors with more limited self-renewal capability and differentiation potential, mostly reside in the ventricular zones (VZ). Radial glial cells are stem cells responsible for producing most neurons for the neocortex, which arise from the dorsal telencephalon, and express a transcription factor called Pax6 (Fig. 1). We also used miR-124, a neuron-specific miRNA not expressed in the progenitor cells, to mark both the migrating neurons in the subventricular zone (SVZ) and intermediate zone (IZ) and the mature neurons in the cortical plate (CP) (Fig. S2, A and B). Multiple miRNAs were highly expressed in the VZ of the dorsal forebrain during neurogenesis (E12.5–E16.5), including the miRNAs that were exclusively expressed in the VZ, and miRNAs that were expressed in the VZ and the CP (Fig. 1 and data not shown). Particularly, some miRNAs exhibited temporal-gradient expression patterns during neocortical development, such as miR-92a and miR-92b, which exhibited similar patterns in the microarray data (Table S1).
Figure 1.
miRNAs exhibit enriched expression in the ventricular zone in the developing mouse brain. The dynamics of Pax6 (A–D), miR-92a (E–H), miR-92b (I–L), miR-5130 (M–P), and miR-712 (Q–T) expression in coronal sections of mouse brains at E12.5 (A, E, I, M, and Q), E14.5 (B, D, F, H, J, L, N, P, R, and T), and E16.5 (C, G, K, O, and S). D, H, L, P, and T are the magnified images of the red dashed boxes in B, F, J, N, and R. The expression pattern of Pax6 was performed by immunohistochemistry and miRNAs performed by in situ hybridization. All images show the same magnification. Scale bar: 500 μm.
Notably, miR-5130 was specifically expressed in the VZ. As expected, the ISH signal of miR-5130 is co-localized with Sox2, a marker for VZ neural progenitor cells, but not with intermediate progenitor cell marker Tbr2 in VZ and SVZ (Fig. S2, F–K). The neural progenitors in the VZ exhibit a remarkable feature termed interkinetic nuclear migration, in which their nuclei migrate between the apical surface and the basal part of the VZ as cell cycle progresses. S phase (DNA synthesis) nuclei are located in the basal half of the VZ, and M phase (mitotic) nuclei are largely restricted to the apical surface (36, 37). miR-5130 expressing cells include S-phase nuclei, which could be labeled by a short (30 min) pulse of BrdU (Fig. S2, C–E). These results indicate that the miR-5130 is specifically expressed in the VZ progenitor cells.
miR-712 is another miRNA that is enriched in the germinal region throughout the entire central nervous system, and it is highly expressed in precursor cells from E11.5 to E16.5 (Fig. 1, M–O, and Fig. S2, L and M). Unlike most other miRNAs, miR-712 is derived from pre-rRNA (38), and its intracellular localization is concentrated in dots, which likely exist in the nuclei rather than throughout the cytoplasm (Fig. 1P). miR-712 signals also existed in cortical neurons, with enrichment in layer-V of the neocortex, at E18.5 and postnatal day 1 (P1), after comparing with the markers of each layer of the neocortex (Fig. S2, L and M, and data not shown).
The miRNAs exhibiting spatially restricted expression in the brain
Among the 279 screened miRNAs exhibited, differential regional enrichment in expression across the brain could be categorized using anatomical regions.
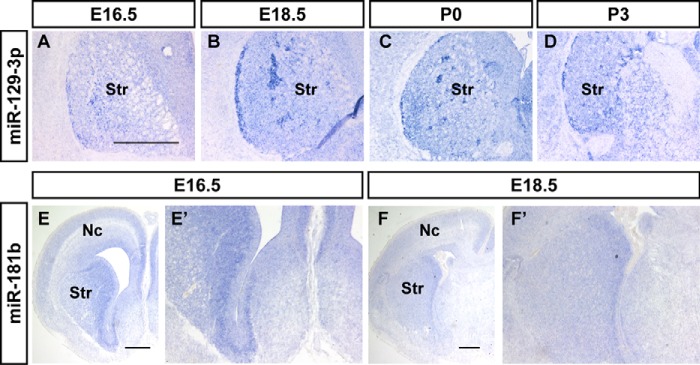
miR-129-3p and miR-181b were enriched in the subpallial structures of the brain. miR-129-3p was abundantly expressed in the caudate-putamen complex (CPu) of the basal ganglia in the coronal section of E16.5 to P3 mouse telencephalon (Fig. 2, A–D). Notably, miR-129-3p was not expressed in all of the cells in the area, but rather in bundles of subpopulation cells during perinatal or early postnatal stages (Fig. 2, B and C). This result indicates the presence of cytoarchitectural differences or subdivisions in the caudate-putamen domain. miR-129-3p was also expressed in the cerebral cortex, but it was relatively weak during cortical development. miR-181b was primarily expressed in the ventral SVZ and weakly expressed in the dorsal germinal area (Fig. 2, E and F, and E′ and F′). miR-541 is another miRNA that was majorly expressed in the ventral subpallial structures, predominantly in the basal ganglia, thalamus, and hypothalamus of the diencephalon during E12.5 to E18.5 (Fig. 4, O and P).
Figure 2.
The restricted expression pattern of miRNAs in the mouse ventral forebrain. In situ hybridization patterns of miR-129–3p (A–D) and miR-181b (E, F, E′, and F′) on sections during the brain development. A–D, the expression patterns of miR-129-3p in coronal sections of mouse brain at E16.5 (A), E18.5 (B), P0 (C), and P3 (D). E, F, E′, and F′, the miR-181b signals in the coronal section of mouse brains at E16.5 (E and E′) and E18.5 (F and F′). The indicated areas of E and F are magnified in E′ and F′. Nc, neocortex; Str, striatum. Scale bar in A is 500 nm for A–D, scale bars in E and F are 500 μm.
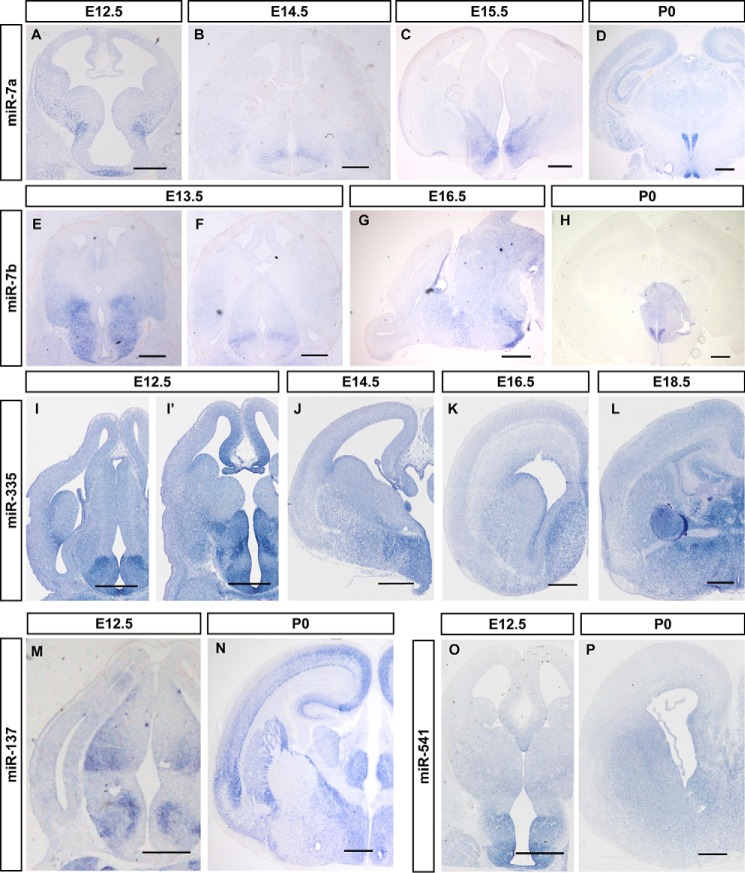
Figure 4.
miRNAs are enriched in the developing mouse hypothalamus. In situ hybridization patterns for miR-7a (A–D), miR-7b (E–H), miR-335 (I–L), miR-137 (M and N), and miR-541 (O and P) in sections during brain development. A–D, the expression patterns of miR-7a in coronal sections of mouse brain at E12.5 (A), E14.5 (B), E16.5 (C), and E18.5 (D). E–H, miR-7b expression in coronal (E, F, and H) and sagittal sections (G) of mouse brain at E13.5, E16.5 and P0. I–L, O, and P, frontal sections through the telencephalon of E12.5-P0 mice, at a rostral or middle level of the pallidum, showing the dynamic expression of the miR-335 (I–L) and miR-541 (O and P) in the hypothalamus. The material shown corresponds to E12.5 (I, I′, and O), E14.5 (J), E16.5 (K and N), E18.5 (L), and P0 (P). M and N, in situ hybridization of miR-137 shows its expression pattern in the brain at E12.5 (M) and P0 (N). Scale bars in A–P are 500 μm.
miR-708, miR-135a, miR-135b, and miR-335 exhibited enriched expression in the thalamus and hypothalamus in the P0 brain. miR-135a and miR-135b were primarily expressed in the epithalamus, dorsal thalamus, and hypothalamus of the diencephalon at E14.5, E16.5, and P0, and especially in the habenular nucleus of the epithalamus (Fig. 3, A–F, and Fig. S3). miR-135a and miR-135b were more enriched in the habenular nucleus (Fig. 3, C and F, and Fig. S3, C and F). miR-708 was primarily expressed in the hippocampus and thalamus at E12.5, and ISH signals were heavily detected in the hippocampus, cingulate cortex, as well as dorsal thalamus at E14.5 to E18.5, with special abundance in the ventral lateral thalamic nucleus (Fig. 3, H and I). Weak ISH signals of miR-708 were detected in layer VI of the neocortex (Fig. 3I). Layer VI of the cerebral cortex sends out connections to dorsal thalamus nuclei, and these results may reflect information flow within the brain.
Figure 3.
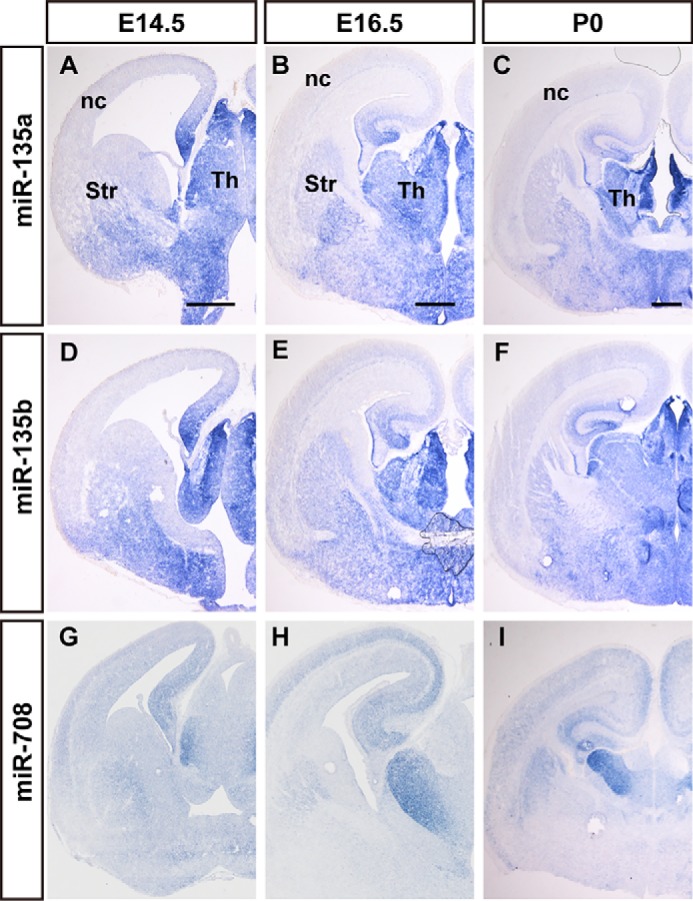
The expression of thalamus-enriched miRNAs at E14.5 to E18.5 mouse brain. The dynamics of miR-135a (A–C), miR-135b (D–F), and miR-708 (G–I) expression in coronal sections of mouse brains during E14.5 (A, D, and G), E16.5 (B, E, and H), and E18.5 (C, F, and I), were performed by in situ hybridization. nc, neocortex; Str, striatum; Th, thalamus. Scale bars in A, D, and G are 500 nm, scale bars in B, E, and H are 500 nm, scale bars in C, F, and I are 500 μm.
Some miRNAs were abundantly expressed in the developing thalamus, and some miRNAs were abundantly and selectively expressed in the developing hypothalamus. Coronal sections of P0 brains revealed the specific expression of miR-7a and miR-7b in the paraventricular nucleus (PVN) and the suprachiasmatic nucleus (SCN) in the mouse hypothalamus (Fig. 4, D and H, and Fig. S4). miR-7a was expressed in the pallidal differentiating zone and anterobasal nucleus at E12.5 (Fig. 4A). Transverse sections of E14.5 embryos revealed limited expression of miR-7a in the ventral part of the hypothalamus (Fig. 4B). Frontal and horizontal sections through the telencephalon of E16.5 mouse embryos revealed the selective expression of miR-7a in supraventricular and paraventricular nuclei (Fig. 4C and Fig. S4), according to the updated anatomic reference atlas redrawn from the Allen Developing Mouse Brain Atlas (http://developingmouse.brain-map.org/).4miR-7b exhibited an expression pattern similar to miR-7a. Sagittal sections of E16.5 mouse brains revealed heavily enriched miR-7b in the hypothalamus region (Fig. 4G). miR-335 was primarily expressed in the posterior hypothalamic and paraventricular nucleus in E12.5, E14.5 and E16.5, and it is more obvious in the paraventricular nucleus at E18.5 (Fig. 4, I–L).
Some miRNAs were expressed in special types of neurons. For example, miR-137 was enriched in the dorsolateral nucleus and the ventral posterolateral nucleus of the thalamus, the striatum, and nucleus accumbens of the telencephalon, and cerebral peduncle of the midbrain. A layer distribution pattern of miR-137 was observed for the ISH signals in the neocortex at E16.5 (Fig. 4, M and N, and data not shown). These structural distributions suggest that miR-137 is related to cholinergic neurotransmission neurons (39). The spatially restricted expression of miRNAs in anatomically distinct brain regions or specific neuron subtypes reveals the highly localized enrichment of the miRNAs that are associated with the developmental or functional of related neurons.
miRNAs exhibit restricted expression in motor neurons of the developing spinal cord
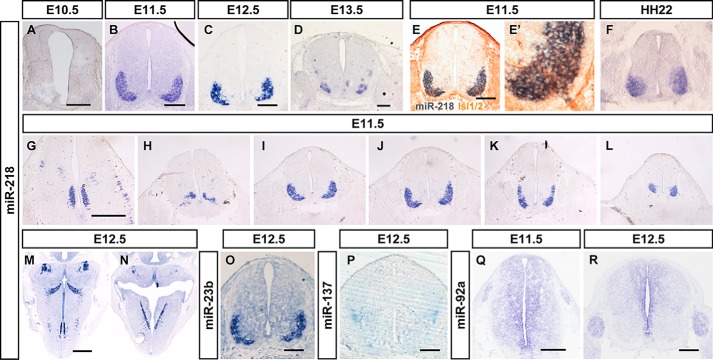
Multiple miRNAs also showed region-specific expression in the developing spinal cord. miR-218 and miR-23a were expressed in the ventrolateral spinal cord, where motor neurons reside, in E12.5 thoracic spinal slices (Fig. 5, C and O). Weaker miR-218 signals were observed in the motoneurons of the spinal cord at E10 during the onset of motor neuron differentiation. miR-218 maintained its exclusive motor neuron expression pattern throughout embryonic spinal cord development (Fig. 5, A–D), and it was detected in all subtypes of motor neurons from the medulla to the spinal cord, and in lateral motor column and medial motor column spinal motor neurons from cervical to lumbar regions (Fig. 5, G–L). miR-218 was also selectively expressed in the brainstem motor nucleus to the spinal cord motor neurons in cross-sections of E12.5 embryos (Fig. 5, M and N). miR-218 is conserved in vertebrates, and it exhibits similar expression patterns in chicken embryos. miR-218 was specifically expressed in motor neuron regions at Hamburger and Hamilton (HH) stage 22 (Fig. 5F), which was further confirmed using the generic motor neuron markers Islet-1/2. Co-labeling experiments in the spinal cords of E11.5 mice revealed that most miR-218–positive cells co-expressed Islet-1/2 (Fig. 5, E and E′, and Fig. S5, A and B).
Figure 5.
The dynamics expression of miRNAs in mouse spinal cord and brainstem. The expression patterns of miR-218 (A–N), miR-23b (O), miR-137 (P), and miR-92a (Q and R) in the mouse spinal cord and brainstem during E10.5 (A), E11.5 (B, E–E′, G–L, and Q), E12.5 (C, M–P, and R), and E13.5 (D), using in situ hybridization. E and E′, a section of a WT E11.5 mouse spinal cord double-labeled for Islet-1/2 protein and miR-218 miRNA. F, the expression patterns of miR-218 in the HH22 stage of chicken spinal cord. G–L, show the selected images of serial sections of miR-218 expression from the cervical segment to lumbar segments. Scale bars in A–E and O–R are 200 nm, scale bars in G and M and are 500 μm, the same for H–L and N, respectively.
miRNAs exhibit choroid plexus-enriched expression in the developing mouse brain
The choroid plexus is a highly vascularized secretory tissue in the brain ventricles. The choroid plexus is responsible for regulation of brain homeostasis via the production of cerebrospinal fluid (CSF), and it acts as the blood-CSF barrier (40). miR-204 and miR-449 were specifically expressed in the choroid plexuses of the lateral and 3rd ventricles at E12.5 and E16.5 (Fig. 6). The expression of miR-204 in the developing mouse embryo was primarily localized to the choroid plexus in the coronal section of E12.5 to E16.5 embryonic mouse brains (Fig. 6, D–F). miR-449 was also specifically expressed in the choroid plexus from E12.5 to E16.5, as well as miR-204 (Fig. 6). Higher magnification analyses of the distribution of miR-449 in the choroid plexus revealed that the signal primarily localized to the ependyma of the choroid plexus (Fig. 6, A–C, and data not shown).
Figure 6.
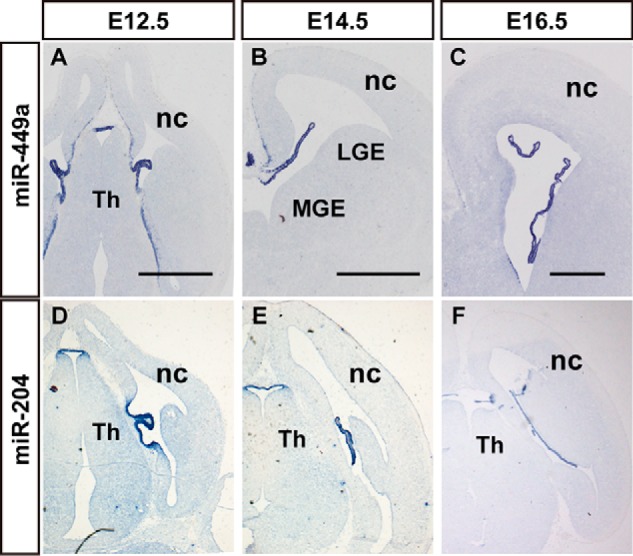
miRNAs exhibit choroid plexus-enriched expression in the developing mouse brain. In situ hybridization patterns of miR-449a (A–C) and miR-204 (D–F) in sections during the brain development. A–C, the expression patterns of miR-449a in the coronal section of mouse brain at E12.5 (A), E14.5 (B), and E16.5 (C). D–F, the expression of miR-204 during brain development at E12.5 (D), E14.5 (E), and E16.5 (F). nc, neocortex; LGE, lateral ganglionic eminence; MGE, medial ganglionic eminence; Th, thalamus. Scale bar in A–C and D–F is 500 μm.
miRNAs exhibit enriched expression in the ganglion cells of the peripheral nervous system
Early embryos (E11.5–E13.5) were frozen as whole-mount, and our analyses included the entire head, the upper cervical segment, and the lumbar region. Therefore, our data provide spatial expression information on miRNAs in developing cranial facial tissues and spinal cord. Some miRNAs were enriched in sensory organs. At E12.5, miR-96 and miR-182 were prominently expressed in ganglion cells of the dorsal root ganglia (DRG), which are located peripherally alongside the spinal cord, and trigeminal ganglia, which are in the roof of the neopallial cortex in horizontal views of the brain (Fig. 7). The DRG and TG are agglomerate-formed tissue that contain thousands of primary afferent neurons, including the nociceptive, mechanoreceptive, and proprioceptive sensory neurons. However, it was difficult to determine the positive cells of miR-96 and miR-182 because of one of the subcategories or all of them.
Figure 7.
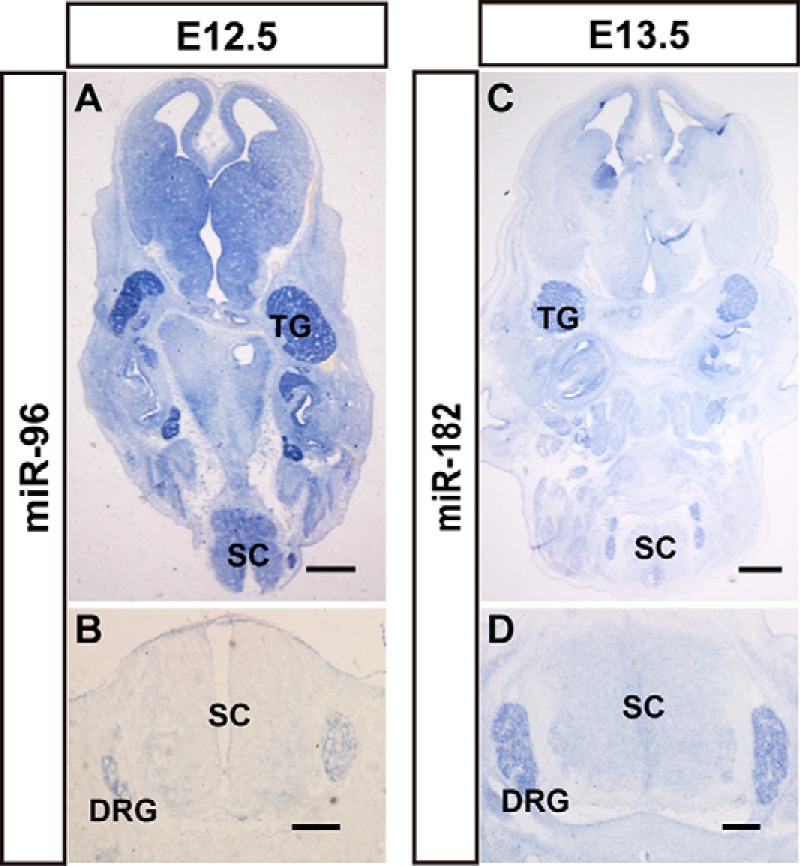
miRNAs show enriched expression in the ganglion cells of the peripheral nervous system. The expression patterns of miR-96 (A and B) and miR-182 (C and D) in mouse brains, nerve ganglia, and spinal cord. The signal of miR-96 expression in horizontal sections of whole body (A) and spinal cord (B) at E12.5 is shown. The expression patterns of miR-182 in horizontal sections of whole body (C) and spinal cord (D) at E13.5 is shown. TG, trigeminal ganglia; SC, spinal cord; Scale bars in A and C are 500 μm, scale bars in B and D are 200 μm.
miRNAs exhibit restricted expression in non-nervous system tissues
Several nervous system-specific miRNAs were identified, such as miR-9 and miR-128. We also found that some miRNAs were not expressed in the nervous system but exhibited tissue-specific expression. The in situ hybridization signals of miR-199a-5p, miR-199a-3p, and miR-199b* were highly restricted to epithelial tissues in E12.5 embryos (Fig. S6, A, E, and I) and in the meningeal tissue outside of nerve tissue (Fig. S6, A–L). miR-451 was primarily expressed in the liver, but it was also found in cerebrovascular structures during development (Fig. S6, M and S-V). miR-154 was not expressed in the liver, but it was specifically expressed in the inferior vena cava (Fig. S6P). Frontal sections exhibited miR-205 robust expression in the nasal lumen epithelium (Fig. S6, N and O), and miR-429 was primarily expressed in vomeronasal and olfactory epithelium (Fig. S6, Q–R).
Discussion
Ascertaining the spatiotemporal dynamics of transcriptomes is important to understand the gene regulation networks of organ development and function, and the elucidation of the timing and location of each gene, including miRNAs, is crucial to predict the physiological function in biological processes. In this study, we systematically analyzed the expression patterns of 279 miRNAs during nervous system development of embryonic mice and found tissue or region-specific expression of some miRNAs.
As mentioned earlier, due to the difficulty of in situ hybridization of miRNAs, we carefully examined various aspects of the validity and reliability of our miRNA probes by mutating nucleotides, detecting exogenous miRNAs, and showing a loss of signal in Dicer mutants (data not shown). miRNA ISH experiments were repeated using at least 3 sets of embryos. We also used different companies to synthesize probes for the same miRNAs since we started the project in 2007, and we purchased LNA-modified probes from companies such as Takara before Exiqon had applied for an LNA patent. We also confirmed the consistency and reliability of the results.
Our findings are consistent with previous sporadic reports. Pfaff and colleagues (41) and Lee and colleagues (42) found miR-218 expression in spinal motor neurons. miR-449 was also found that specifically expressed in the choroid plexuses of the lateral and 3rd ventricles at E13.5 and E15.5 (43). miR-451 exhibited strong hybridization signals in the embryonic liver, which is consistent with previous reports that miR-451 is required for erythroid development (44, 45) and the maintenance of hepatic gluconeogenesis (46). These results further confirm the reliability of our in situ hybridization results.
Many miRNAs are frequently arranged into clusters that are transcribed as a single polycistronic primary transcript and processed into multiple individual mature miRNAs by RNA-binding proteins. miR-96 and miR-182 belong to the miR-183 cluster, which consists of miR-183, -96, and -182. miR-96 and miR-182 show similar expression patterns in mouse embryos during development, and these miRNAs are highly enriched in ganglion cells of the peripheral nervous system. Our results indicate that the miR-183 cluster should be transcribed together and processed similarly. Kitamoto and colleagues (47) used quantitative real-time PCR analyses and confirmed miR-183 cluster enrichment in DRG in adult rats. Other studies demonstrated that miR-183 cluster miRNAs regulated sensory perception, including the development of the retina and inner ear (48, 49), and point mutations in the seed region of miRNA-96 caused hearing loss in mice (50) and human (51). miR-183/96/182 null mice display severe deficits in vision, hearing, balance, and smell (49). Peng et al. (52) revealed that the miR-183 cluster controlled the gene networks of physiological and dysfunctional pain via targeting the majority of pain-regulated genes in mice.
Therefore, the specific expression patterns of these miRNAs reveal their exact roles in development or functional maintenance. For example, miR-92b could regulate the differentiation of neural progenitor cells and the precise development of cortical neurons, which when overexpression of miR-92b reduce the proportion of precursor cells, especially Tbr2+ cells, and lead the cells distributed in the superficial layer of the neocortex (Fig. S7). miR-7a and miR-7b were specifically expressed in the PVN and the SCN in the hypothalamus of mice at different developmental stages. PVN is an important neurosecretory nucleus in the hypothalamus, and it is the main secretor of oxytocin. SCN is the most important circadian pacemaker in mammals, and it regulates a series of physiological behaviors and activities. Loss of a mammalian circular RNA locus causes a neuropsychiatric disorder phenotype that may be the results of miR-7 deregulation (53).
In conclusion, several miRNAs exhibited spatially restricted/enriched expression in anatomically distinct regions or in specific neuron subtypes in the embryonic brain and spinal cord, such as in the ventricular area, the striatum (and other basal ganglia), hypothalamus, choroid plexus, and the peripheral nervous system. There are also miRNAs exhibiting restricted/enriched expression outside the nervous system. These findings will facilitate further studies to understand the development and function of these neurons and to insights for treating neurodegenerative diseases.
Experimental procedures
Animals
All animals used for experiments were 8–12-week-old CD1 mice from the Beijing Vital River Laboratory Animal Limited Company (Beijing, China). Mice were maintained in the Animal Centre of Peking Union Medical College. All animal care and experiments were approved by the Institutional Animal Care and Use Committee of the Chinese Academy of Medical Sciences and Peking Union Medical College with all procedures in compliance with the Experimental Animal Regulations (China Science and Technology Commission Order No. 2).
Tissue preparation
Timed pregnant mouse embryonic and postnatal tissues were harvest for the day of the vaginal plug was defined as embryonic day (E) 0.5, and the day of birth as postnatal day (P) 0. The embryonic brain tissues were directly fixed at 4 °C overnight in 4% paraformaldehyde (PFA) in 100 mm phosphate-buffered saline (PBS, pH 7.4). For postnatal brain, the pups were transcardially perfused with 4% PFA (100 mm PBS), followed by brain dissection and further PFA fixation at 4 °C overnight. Fixed tissues were cryoprotected in 25% sucrose in PBS and equilibrated in the O.C.T. Compound (Sakura, Finetek, Japan) for 15–30 min before freezing. Sixteen-μm thick cryosections were generated and stored at −80 °C.
ISH
The ISH procedure was performed as described (54). Probes of miRNAs labeled with digoxigenin for in situ hybridization were purchased from Exiqon (Denmark), Takara (Japan), and Sangon (China). A total of 392 probes containing 279 miRNAs and 21 control or mutant miRNAs were generated (Table S1).
Immunohistochemistry
Immunohistochemical analyses of the cryosections and cells were performed as previously described (55). 4′,6-diamidino-2-phenylindole (ZLI-9557, ZSGB-Bio, China) was used for DNA staining to reveal the nuclei. The following primary antibodies used for immunohistochemical analyses were as follows: rabbit anti-Pax6 (PRB-278P, Covance), rabbit anti-Sox2 (ab97959, Abcam), rabbit anti-Tbr2 (ab23345, Abcam), mouse anti-HB9 and anti-Islet-1/2 (kindly provided by Mengsheng Qiu). Secondary antibodies included horseradish peroxidase-labeled goat anti-mouse IgG (PV-6002, ZSGB-Bio, China) and Alexa Fluor 594 (CA-11005, Molecular Probes).
For pulse labeling of cells in the S phase, the pregnant mice were intraperitoneally injected with BrdU (5-bromo-2′-deoxyuridine, 75 mg/kg of body weight) at E13.5 and sacrificed 30 min (for proliferation) after injection. Immunofluorescence for BrdU staining was 2 n HCl pretreated brain sections, followed by standard immunofluorescence processes.
Author contributions
P. S., C. W., W. L., L. H., B. Y., B. Q., and X. P. data curation; P. S., W. L., X. R., and X. P. formal analysis; P. S., B. Q., and X. P. funding acquisition; P. S., C. W., W. L., X. R., L. H., Y. Z., P. C., X. Z., and B. Y. validation; P. S., C. W., W. L., X. R., C. L., L. H., Y. Z., H. F., and M. W. investigation; P. S., C. W., X. R., and C. L. methodology; P. S. and X. P. writing-original draft; J. Y., B. Q., and X. P. supervision; J. Y., B. Q., and X. P. project administration.
Supplementary Material
Acknowledgments
We thank Prof. Weimin Zhong (Yale University) for very helpful discussions and guidance, and Dr. Hao Huang and Prof. Mengsheng Qiu (Hangzhou Normal University) for technical assistance. We also thank the State Key Laboratory of Medical Molecular Biology for support throughout this study, as well as members of our laboratory for discussions.
This work was supported by National Key Research and Development Program of China Grants 2016YFA0100702 and 2016YFC0902502, National Natural Science Foundation of China Grant 31670789, and CAMS Innovation Fund for Medical Sciences (CIFMS) Grants 2016-I2M-2-001, 2016-I2M-1-004, and 2017-I2M-1-004. The authors declare that they have no conflicts of interest with the contents of this article.
This article contains Figs. S1–S7 and Tables S1 and S2.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- ISH
- in situ hybridization
- LNA
- locked nucleic acid
- mRNA
- microRNA
- VZ
- ventricular zones
- SVZ
- subventricular zone
- CP
- cortical plate
- PVN
- paraventricular nucleus
- SCN
- suprachiasmatic nucleus
- CSF
- cerebrospinal fluid
- DRG
- dorsal root ganglia
- PFA
- paraformaldehyde.
References
- 1. Gao P., Postiglione M. P., Krieger T. G., Hernandez L., Wang C., Han Z., Streicher C., Papusheva E., Insolera R., Chugh K., Kodish O., Huang K., Simons B. D., Luo L., Hippenmeyer S., and Shi S. H. (2014) Deterministic progenitor behavior and unitary production of neurons in the neocortex. Cell 159, 775–788 10.1016/j.cell.2014.10.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Rowitch D. H., and Kriegstein A. R. (2010) Developmental genetics of vertebrate glial-cell specification. Nature 468, 214–222 10.1038/nature09611 [DOI] [PubMed] [Google Scholar]
- 3. Miller F. D., and Gauthier A. S. (2007) Timing is everything: making neurons versus glia in the developing cortex. Neuron 54, 357–369 10.1016/j.neuron.2007.04.019 [DOI] [PubMed] [Google Scholar]
- 4. Arlotta P., Molyneaux B. J., Chen J., Inoue J., Kominami R., and Macklis J. D. (2005) Neuronal subtype-specific genes that control corticospinal motor neuron development in vivo. Neuron 45, 207–221 10.1016/j.neuron.2004.12.036 [DOI] [PubMed] [Google Scholar]
- 5. Molyneaux B. J., Goff L. A., Brettler A. C., Chen H. H., Hrvatin S., Rinn J. L., and Arlotta P. (2015) DeCoN: genome-wide analysis of in vivo transcriptional dynamics during pyramidal neuron fate selection in neocortex. Neuron 85, 275–288 10.1016/j.neuron.2014.12.024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Sugino K., Hempel C. M., Miller M. N., Hattox A. M., Shapiro P., Wu C., Huang Z. J., and Nelson S. B. (2006) Molecular taxonomy of major neuronal classes in the adult mouse forebrain. Nat. Neurosci. 9, 99–107 10.1038/nn1618,10.1080/10284150600772148 [DOI] [PubMed] [Google Scholar]
- 7. Zhong Y., Takemoto M., Fukuda T., Hattori Y., Murakami F., Nakajima D., Nakayama M., and Yamamoto N. (2004) Identification of the genes that are expressed in the upper layers of the neocortex. Cereb. Cortex 14, 1144–1152 10.1093/cercor/bhh074 [DOI] [PubMed] [Google Scholar]
- 8. Bernard A., Lubbers L. S., Tanis K. Q., Luo R., Podtelezhnikov A. A., Finney E. M., McWhorter M. M., Serikawa K., Lemon T., Morgan R., Copeland C., Smith K., Cullen V., Davis-Turak J., Lee C. K., et al. (2012) Transcriptional architecture of the primate neocortex. Neuron 73, 1083–1099 10.1016/j.neuron.2012.03.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Bakken T. E., Miller J. A., Ding S. L., Sunkin S. M., Smith K. A., Ng L., Szafer A., Dalley R. A., Royall J. J., Lemon T., Shapouri S., Aiona K., Arnold J., Bennett J. L., Bertagnolli D., et al. (2016) A comprehensive transcriptional map of primate brain development. Nature 535, 367–375 10.1038/nature18637 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Fertuzinhos S., Li M., Kawasawa Y. I., Ivic V., Franjic D., Singh D., Crair M., and Sestan N. (2014) Laminar and temporal expression dynamics of coding and noncoding RNAs in the mouse neocortex. Cell Rep. 6, 938–950 10.1016/j.celrep.2014.01.036 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Lein E., Borm L. E., and Linnarsson S. (2017) The promise of spatial transcriptomics for neuroscience in the era of molecular cell typing. Science 358, 64–69 10.1126/science.aan6827 [DOI] [PubMed] [Google Scholar]
- 12. Johnson M. B., Wang P. P., Atabay K. D., Murphy E. A., Doan R. N., Hecht J. L., and Walsh C. A. (2015) Single-cell analysis reveals transcriptional heterogeneity of neural progenitors in human cortex. Nat. Neurosci. 18, 637–646 10.1038/nn.3980 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Poulin J. F., Tasic B., Hjerling-Leffler J., Trimarchi J. M., and Awatramani R. (2016) Disentangling neural cell diversity using single-cell transcriptomics. Nat. Neurosci. 19, 1131–1141 10.1038/nn.4366 [DOI] [PubMed] [Google Scholar]
- 14. Shekhar K., Lapan S. W., Whitney I. E., Tran N. M., Macosko E. Z., Kowalczyk M., Adiconis X., Levin J. Z., Nemesh J., Goldman M., McCarroll S. A., Cepko C. L., Regev A., and Sanes J. R. (2016) Comprehensive classification of retinal bipolar neurons by single-cell transcriptomics. Cell 166, 1308–1323.e1330 10.1016/j.cell.2016.07.054 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Pollen A. A., Nowakowski T. J., Shuga J., Wang X., Leyrat A. A., Lui J. H., Li N., Szpankowski L., Fowler B., Chen P., Ramalingam N., Sun G., Thu M., Norris M., Lebofsky R., et al. (2014) Low-coverage single-cell mRNA sequencing reveals cellular heterogeneity and activated signaling pathways in developing cerebral cortex. Nat. Biotechnol. 32, 1053–1058 10.1038/nbt.2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Usoskin D., Furlan A., Islam S., Abdo H., Lönnerberg P., Lou D., Hjerling-Leffler J., Haeggström J., Kharchenko O., Kharchenko P. V., Linnarsson S., and Ernfors P. (2015) Unbiased classification of sensory neuron types by large-scale single-cell RNA sequencing. Nat. Neurosci. 18, 145–153 10.1038/nn.3881 [DOI] [PubMed] [Google Scholar]
- 17. Hu P., Fabyanic E., Kwon D. Y., Tang S., Zhou Z., and Wu H. (2017) Dissecting cell-type composition and activity-dependent transcriptional state in mammalian brains by massively parallel single-nucleus RNA-Seq. Mol. Cell 68, 1006–1015.e1007 10.1016/j.molcel.2017.11.017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Liu S. J., Nowakowski T. J., Pollen A. A., Lui J. H., Horlbeck M. A., Attenello F. J., He D., Weissman J. S., Kriegstein A. R., Diaz A. A., and Lim D. A. (2016) Single-cell analysis of long non-coding RNAs in the developing human neocortex. Genome Biol. 17, 67 10.1186/s13059-016-0932-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Nowakowski T. J., Bhaduri A., Pollen A. A., Alvarado B., Mostajo-Radji M. A., Di Lullo E., Haeussler M., Sandoval-Espinosa C., Liu S. J., Velmeshev D., Ounadjela J. R., Shuga J., Wang X., Lim D. A., West J. A., et al. (2017) Spatiotemporal gene expression trajectories reveal developmental hierarchies of the human cortex. Science 358, 1318–1323 10.1126/science.aap8809 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Gong S., Zheng C., Doughty M. L., Losos K., Didkovsky N., Schambra U. B., Nowak N. J., Joyner A., Leblanc G., Hatten M. E., and Heintz N. (2003) A gene expression atlas of the central nervous system based on bacterial artificial chromosomes. Nature 425, 917–925 10.1038/nature02033 [DOI] [PubMed] [Google Scholar]
- 21. Hawrylycz M. J., Lein E. S., Guillozet-Bongaarts A. L., Shen E. H., Ng L., Miller J. A., van de Lagemaat L. N., Smith K. A., Ebbert A., Riley Z. L., Abajian C., Beckmann C. F., Bernard A., Bertagnolli D., Boe A. F., et al. (2012) An anatomically comprehensive atlas of the adult human brain transcriptome. Nature 489, 391–399 10.1038/nature11405 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Gray P. A., Fu H., Luo P., Zhao Q., Yu J., Ferrari A., Tenzen T., Yuk D. I., Tsung E. F., Cai Z., Alberta J. A., Cheng L. P., Liu Y., Stenman J. M., Valerius M. T., et al. (2004) Mouse brain organization revealed through direct genome-scale TF expression analysis. Science 306, 2255–2257 10.1126/science.1104935 [DOI] [PubMed] [Google Scholar]
- 23. Visel A., Thaller C., and Eichele G. (2004) GenePaint.org: an atlas of gene expression patterns in the mouse embryo. Nucleic Acids Res. 32, D552–556 10.1093/nar/gkh029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Magdaleno S., Jensen P., Brumwell C. L., Seal A., Lehman K., Asbury A., Cheung T., Cornelius T., Batten D. M., Eden C., Norland S. M., Rice D. S., Dosooye N., Shakya S., Mehta P., and Curran T. (2006) BGEM: an in situ hybridization database of gene expression in the embryonic and adult mouse nervous system. PLoS Biol. 4, e86 10.1371/journal.pbio.0040086 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Lein E. S., Hawrylycz M. J., Ao N., Ayres M., Bensinger A., Bernard A., Boe A. F., Boguski M. S., Brockway K. S., Byrnes E. J., Chen L., Chen L., Chen T. M., Chin M. C., Chong J., et al. (2007) Genome-wide atlas of gene expression in the adult mouse brain. Nature 445, 168–176 10.1038/nature05453 [DOI] [PubMed] [Google Scholar]
- 26. Diez-Roux G., Banfi S., Sultan M., Geffers L., Anand S., Rozado D., Magen A., Canidio E., Pagani M., Peluso I., Lin-Marq N., Koch M., Bilio M., Cantiello I., Verde R., et al. (2011) A high-resolution anatomical atlas of the transcriptome in the mouse embryo. PLoS Biol. 9, e1000582 10.1371/journal.pbio.1000582 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. McKee A. E., Minet E., Stern C., Riahi S., Stiles C. D., and Silver P. A. (2005) A genome-wide in situ hybridization map of RNA-binding proteins reveals anatomically restricted expression in the developing mouse brain. BMC Dev. Biol. 5, 14 10.1186/1471-213X-5-14 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Ebert M. S., and Sharp P. A. (2012) Roles for microRNAs in conferring robustness to biological processes. Cell 149, 515–524 10.1016/j.cell.2012.04.005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Jonas S., and Izaurralde E. (2015) Towards a molecular understanding of microRNA-mediated gene silencing. Nat. Rev. Genet. 16, 421–433 10.1038/nrg3965 [DOI] [PubMed] [Google Scholar]
- 30. Rajman M., and Schratt G. (2017) MicroRNAs in neural development: from master regulators to fine-tuners. Development 144, 2310–2322 10.1242/dev.144337 [DOI] [PubMed] [Google Scholar]
- 31. Sokol N. S. (2012) Small temporal RNAs in animal development. Curr. Opin. Genet. Dev. 22, 368–373 10.1016/j.gde.2012.04.001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Pauli A., Rinn J. L., and Schier A. F. (2011) Non-coding RNAs as regulators of embryogenesis. Nat. Rev. Genet. 12, 136–149 10.1038/nrg2904 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Kloosterman W. P., Wienholds E., de Bruijn E., Kauppinen S., and Plasterk R. H. (2006) In situ detection of miRNAs in animal embryos using LNA-modified oligonucleotide probes. Nat. Methods 3, 27–29 10.1038/nmeth843 [DOI] [PubMed] [Google Scholar]
- 34. Obernosterer G., Martinez J., and Alenius M. (2007) Locked nucleic acid-based in situ detection of microRNAs in mouse tissue sections. Nat. Protoc. 2, 1508–1514 10.1038/nprot.2007.153 [DOI] [PubMed] [Google Scholar]
- 35. Silahtaroglu A. N., Nolting D., Dyrskjøt L., Berezikov E., Møller M., Tommerup N., and Kauppinen S. (2007) Detection of microRNAs in frozen tissue sections by fluorescence in situ hybridization using locked nucleic acid probes and tyramide signal amplification. Nat. Protoc. 2, 2520–2528 10.1038/nprot.2007.313 [DOI] [PubMed] [Google Scholar]
- 36. Spear P. C., and Erickson C. A. (2012) Interkinetic nuclear migration: a mysterious process in search of a function. Dev. Growth Differ. 54, 306–316 10.1111/j.1440-169X.2012.01342.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Kosodo Y., Suetsugu T., Suda M., Mimori-Kiyosue Y., Toida K., Baba S. A., Kimura A., and Matsuzaki F. (2011) Regulation of interkinetic nuclear migration by cell cycle-coupled active and passive mechanisms in the developing brain. EMBO J. 30, 1690–1704 10.1038/emboj.2011.81 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Son D. J., Kumar S., Takabe W., Kim C. W., Ni C. W., Alberts-Grill N., Jang I. H., Kim S., Kim W., Won Kang S., Baker A. H., Woong Seo J., Ferrara K. W., and Jo H. (2013) The atypical mechanosensitive microRNA-712 derived from pre-ribosomal RNA induces endothelial inflammation and atherosclerosis. Nat. Commun. 4, 3000 10.1038/ncomms4000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Allaway K. C., and Machold R. (2017) Developmental specification of forebrain cholinergic neurons. Dev. Biol. 421, 1–7 10.1016/j.ydbio.2016.11.007 [DOI] [PubMed] [Google Scholar]
- 40. Lun M. P., Monuki E. S., and Lehtinen M. K. (2015) Development and functions of the choroid plexus-cerebrospinal fluid system. Nat. Rev. Neurosci. 16, 445–457 10.1038/nrn3921 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Amin N. D., Bai G., Klug J. R., Bonanomi D., Pankratz M. T., Gifford W. D., Hinckley C. A., Sternfeld M. J., Driscoll S. P., Dominguez B., Lee K. F., Jin X., and Pfaff S. L. (2015) Loss of motoneuron-specific microRNA-218 causes systemic neuromuscular failure. Science 350, 1525–1529 10.1126/science.aad2509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Thiebes K. P., Nam H., Cambronne X. A., Shen R., Glasgow S. M., Cho H. H., Kwon J. S., Goodman R. H., Lee J. W., Lee S., and Lee S. K. (2015) miR-218 is essential to establish motor neuron fate as a downstream effector of Isl1-Lhx3. Nat. Commun. 6, 7718 10.1038/ncomms8718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Redshaw N., Wheeler G., Hajihosseini M. K., and Dalmay T. (2009) microRNA-449 is a putative regulator of choroid plexus development and function. Brain Res. 1250, 20–26 10.1016/j.brainres.2008.11.020 [DOI] [PubMed] [Google Scholar]
- 44. Rasmussen K. D., Simmini S., Abreu-Goodger C., Bartonicek N., Di Giacomo M., Bilbao-Cortes D., Horos R., Von Lindern M., Enright A. J., and O'Carroll D. (2010) The miR-144/451 locus is required for erythroid homeostasis. J. Exp. Med. 207, 1351–1358 10.1084/jem.20100458 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Dore L. C., Amigo J. D., Dos Santos C. O., Zhang Z., Gai X., Tobias J. W., Yu D., Klein A. M., Dorman C., Wu W., Hardison R. C., Paw B. H., and Weiss M. J. (2008) A GATA-1-regulated microRNA locus essential for erythropoiesis. Proc. Natl. Acad. Sci. U.S.A. 105, 3333–3338 10.1073/pnas.0712312105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Zhuo S., Yang M., Zhao Y., Chen X., Zhang F., Li N., Yao P., Zhu T., Mei H., Wang S., Li Y., Chen S., and Le Y. (2016) MicroRNA-451 negatively regulates hepatic glucose production and glucose homeostasis by targeting glycerol kinase-mediated gluconeogenesis. Diabetes 65, 3276–3288 10.2337/db16-0166 [DOI] [PubMed] [Google Scholar]
- 47. Aldrich B. T., Frakes E. P., Kasuya J., Hammond D. L., and Kitamoto T. (2009) Changes in expression of sensory organ-specific microRNAs in rat dorsal root ganglia in association with mechanical hypersensitivity induced by spinal nerve ligation. Neuroscience 164, 711–723 10.1016/j.neuroscience.2009.08.033 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Busskamp V., Krol J., Nelidova D., Daum J., Szikra T., Tsuda B., Jüttner J., Farrow K., Scherf B. G., Alvarez C. P., Genoud C., Sothilingam V., Tanimoto N., Stadler M., Seeliger M., et al. (2014) miRNAs 182 and 183 are necessary to maintain adult cone photoreceptor outer segments and visual function. Neuron 83, 586–600 10.1016/j.neuron.2014.06.020 [DOI] [PubMed] [Google Scholar]
- 49. Fan J., Jia L., Li Y., Ebrahim S., May-Simera H., Wood A., Morell R. J., Liu P., Lei J., Kachar B., Belluscio L., Qian H., Li T., Li W., Wistow G., and Dong L. (2017) Maturation arrest in early postnatal sensory receptors by deletion of the miR-183/96/182 cluster in mouse. Proc. Natl. Acad. Sci. U.S.A. 114, E4271–E4280 10.1073/pnas.1619442114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Lewis M. A., Quint E., Glazier A. M., Fuchs H., De Angelis M. H., Langford C., van Dongen S., Abreu-Goodger C., Piipari M., Redshaw N., Dalmay T., Moreno-Pelayo M. A., Enright A. J., and Steel K. P. (2009) An ENU-induced mutation of miR-96 associated with progressive hearing loss in mice. Nat. Genet. 41, 614–618 10.1038/ng.369 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Mencía A., Modamio-Hoybjør S., Redshaw N., Morín M., Mayo-Merino F., Olavarrieta L., Aguirre L. A., del Castillo I., Steel K. P., Dalmay T., Moreno F., and Moreno-Pelayo M. A. (2009) Mutations in the seed region of human miR-96 are responsible for nonsyndromic progressive hearing loss. Nat. Genet. 41, 609–613 10.1038/ng.355 [DOI] [PubMed] [Google Scholar]
- 52. Peng C., Li L., Zhang M. D., Bengtsson Gonzales C., Parisien M., Belfer I., Usoskin D., Abdo H., Furlan A., Häring M., Lallemend F., Harkany T., Diatchenko L., Hökfelt T., Hjerling-Leffler J., and Ernfors P. (2017) miR-183 cluster scales mechanical pain sensitivity by regulating basal and neuropathic pain genes. Science 356, 1168–1171 10.1126/science.aam7671 [DOI] [PubMed] [Google Scholar]
- 53. Piwecka M., Glazar P., Hernandez-Miranda L. R., Memczak S., Wolf S. A., Rybak-Wolf A., Filipchyk A., Klironomos F., Cerda Jara C. A., Fenske P., Trimbuch T., Zywitza V., Plass M., Schreyer L., Ayoub S., et al. (2017) Loss of a mammalian circular RNA locus causes miRNA deregulation and affects brain function. Science 357, eaam8526 10.1126/science.aam8526 [DOI] [PubMed] [Google Scholar]
- 54. Zhou J., Wang R., Zhang J., Zhu L., Liu W., Lu S., Chen P., Li H., Yin B., Yuan J., Qiang B., Shu P., and Peng X. (2017) Conserved expression of ultra-conserved noncoding RNA in mammalian nervous system. Biochim. Biophys. Acta 1860, 1159–1168 10.1016/j.bbagrm.2017.10.002 [DOI] [PubMed] [Google Scholar]
- 55. Shu P., Fu H., Zhao X., Wu C., Ruan X., Zeng Y., Liu W., Wang M., Hou L., Chen P., Yin B., Yuan J., Qiang B., and Peng X. (2017) MicroRNA-214 modulates neural progenitor cell differentiation by targeting Quaking during cerebral cortex development. Sci. Rep. 7, 8014 10.1038/s41598-017-08450-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Kozomara A., and Griffiths-Jones S. (2014) miRBase: annotating high confidence microRNAs using deep sequencing data. Nucleic Acids Res. 42, D68–D73 10.1093/nar/gkt1181 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.