Abstract
p53 mutants can form amyloid-like structures that accumulate in cells. p53 reactivation with induction of massive apoptosis-1 (PRIMA-1) and its primary active metabolite, 2-methylene-3-quinuclidinone (MQ), can restore unfolded p53 mutants to a native conformation that induces apoptosis and activates several p53 target genes. However, whether PRIMA-1 can clear p53 aggregates is unclear. In this study, we investigated whether PRIMA-1 can restore aggregated mutant p53 to a native form. We observed that the p53 mutant protein is more sensitive to both PRIMA-1 and MQ aggregation inhibition than WT p53. The results of anti-amyloid oligomer antibody assays revealed that PRIMA-1 reverses mutant p53 aggregate accumulation in cancer cells. Size-exclusion chromatography of the lysates from mutant p53-containing breast cancer and ovarian cell lines confirmed that PRIMA-1 substantially decreases p53 aggregates. We also show that MDA-MB-231 cell lysates can “seed” aggregation of the central core domain of recombinant WT p53, corroborating the prion-like behavior of mutant p53. We also noted that this aggregation effect was inhibited by MQ and PRIMA-1. This study provides the first demonstration that PRIMA-1 can rescue amyloid-state p53 mutants, a strategy that could be further explored as a cancer treatment.
Keywords: amyloid, protein aggregation, p53, anticancer drug, protein misfolding
Introduction
Cancer is a major public health problem worldwide. Among cancer cases, more than 50% present p53 mutations. Of these, missense mutations are associated with the expression of functional proteins that have distinct activity compared with the WT form (1) and are associated with increased susceptibility to cancer and worse prognosis. p53 is a transcription factor involved in cell fate decisions after DNA damage; in the presence of abundant damage, p53 can activate cell cycle arrest, apoptosis, or senescence (2). There is mounting evidence that mutant p53 is an extremely important drug target (3–5).
Several p53 mutants form amyloid structures that accumulate in the cell (6–10). In the past few years, our group has shown that the dominant-negative effect of mutant p53 involves its ability to promote WT p53 aggregation by inducing a conformational change in a prion-like manner (6, 7, 11). Gain-of-function (GoF)4 effects, including increased invasion, altered migration (12, 13), and drug resistance, have been reported for several mutants (14–16). GoF effects have also been related to mutant p53 aggregation and its interaction with other proteins, such as the paralogous p63 and p73 proteins (8, 11, 17–19).
PRIMA-1 (p53 reactivation with induction of massive apoptosis-1) was the first p53 reactivator compound described (20). PRIMA-1 restores several unfolded p53 mutants to a WT-like conformation, which induces apoptosis and activates a number of p53 target genes (21–23), thereby reducing xenograft tumor size in mice (24). Phase I studies of PRIMA-1MET, a PRIMA-1 analog associated with cisplatin, have been concluded, and phase II trials are now in development (25, 26). The mechanisms through which PRIMA-1 and its analog PRIMA-1MET function are controversial. The effects of PRIMA-1 appear to depend on the cell background, i.e. different protein–protein interactions occur according to the oncogenes found in cancer cells. Hypoxia is one of the factors that increase the sensitivity of p53 mutants to PRIMA-1MET (27, 28). Lambert et al. (29) reported that PRIMA-1 and other analogues are pro-drugs that are metabolized in vivo and converted into a common major active metabolite, 2-methylene 3-quinuclidinone (MQ), which is responsible for the structural stabilization of p53 mutants. MQ is a nucleophile acceptor that reacts with thiol groups, and cysteine residues in proteins are covalently modified via a Michael addition reaction. Lambert et al. (29) showed that the PRIMA-1/MQ concentration is crucial for the reactivity of an increasing number of cysteines. One cysteine residue appears to have the most importance in the conformational shift promoted by MQ: Cys-124 (30).This specific residue is located in a transiently open binding pocket between loop L1 and sheet S3 of the p53 core domain and is surrounded by the hotspots where the missense mutations are located. Recently, Zhang et al. (31) identified two cysteine residues as the main targets for MQ: Cys-124 and Cys-277. As described by the authors, these appear to be key residues in the functional stabilization of mutant R175H. However, the mechanism through which PRIMA-1 reactivates mutant p53 or its structural features before and after reaction with MQ have not yet been elucidated. Moreover, the implications of p53 aggregation for cancer need to be further explored.
The Michael reaction of MQ with cysteine is not exclusive to p53; other cellular proteins are also susceptible. The extent to which MQ reacts with other proteins and might thereby alter their function and cause toxicity to cancer cells is not completely known. An example of a protein that reacts with MQ is thioredoxin reductase 1 (TrxR1), which is transformed from a reductase to an NADPH oxidase that can produce reactive oxygen species and cause cytotoxicity in p53-null cell lines (28, 32). However, the importance of a drug targeting mutant p53 is undeniable because the number of candidate cases for possible treatment is very large. Additionally, these off-target effects appear to be positive because they seem to cooperate in the reactivation of mutant p53 and thus boost the anticancer effect of PRIMA-1 and its analogs (26).
In this study, we asked whether PRIMA-1 is capable of clearing p53 aggregation. We also investigated whether aggregated p53 is reactivated by PRIMA-1. Through immunoprecipitation assays using the anti-amyloid oligomer antibody A11, we show that PRIMA-1 mobilizes amyloid-state p53, promoting its partial reactivation and de-aggregation, thus leading to apoptosis. Size-exclusion chromatography (SEC) of the lysates from the cancer cell lines containing mutant p53 corroborated that PRIMA-1 led to a substantial decrease in p53 aggregates. Additionally, we show in vitro that PRIMA-1/MQ can inhibit the ability of mutant p53 to act as a seed, in a prion-like manner, to accelerate WT p53 aggregation. We provide the first demonstration of the molecular mechanism through which PRIMA-1 rescues amyloid mutant p53 and thereby decreases dominant-negative and GoF effects. Our findings reinforce the notion that mutant p53 aggregation is an excellent target for the development of new antineoplastic drugs.
Results
PRIMA-1 and its active metabolite, MQ, inhibit in vitro p53 aggregation
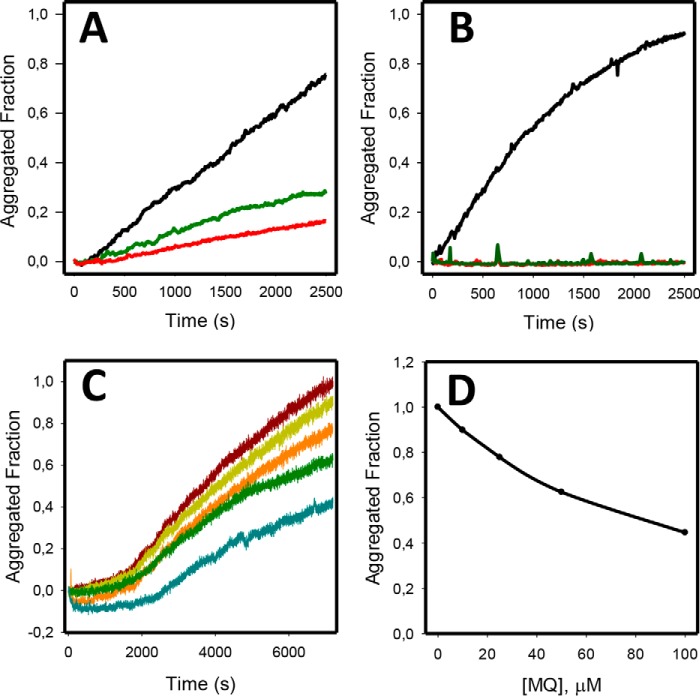
PRIMA-1 is known to stabilize mutant p53 and restore a folded, active state. However, the effect of PRIMA-1 on amyloid-state mutant p53 has never been investigated. To determine whether p53 amyloid aggregation could be affected by either PRIMA-1 or MQ, we incubated the DNA-binding, central core domain of WT p53 (WTp53C) or its mutant R248Q (R248Qp53C) at 37 °C for 1 h. As observed in Fig. 1, A and B, although WTp53C aggregation was inhibited by PRIMA-1 and MQ at 100 μm, the mutant form was more strongly inhibited. The concentration of the compounds for this experiment was selected according to the number of cysteines present in the central core domain of p53, which corresponds to 10, bringing us to a protein-to-PRIMA-1/MQ ratio of 1:10. Lower MQ concentrations (50, 25, and 10 μm) also abolished R248Qp53C aggregation (Fig. 1C). Fig. 1D shows a concentration-dependent inhibition of the R248Q mutant after 2 h of aggregation at 37 °C.
Figure 1.
PRIMA-1 and MQ inhibit both R248Qp53C and WTp53C aggregation at 37 °C. The samples (5 μm) were incubated at 37 °C for 1 h in the presence of PRIMA-1, MQ, or 0.1% DMSO (final concentration) as the control. A, WTp53C aggregation in the absence (black) or presence of 100 μm PRIMA-1 (green line) or 100 μm MQ (red line). B, R248Qp53C aggregation in the absence (black line) or presence of 100 μm PRIMA-1 (red) or 100 μm MQ (green line). C, lower concentrations of MQ inhibited R248Qp53C aggregation: control R248Qp53C (dark red line), 10 μm MQ (dark yellow line), 25 μm MQ (orange line), 50 μm (green line), and 100 μm MQ (blue line). D, MQ dose-dependent inhibition of R248Q mutant p53 aggregation at 37 °C after 2 h. Protein aggregation was assessed by monitoring the increase in the ThT fluorescence intensity (excitation, 440 nm; emission, 482 nm) at a ThT:protein ratio of 5:1. All samples were diluted in 50 mm Tris (pH 7.2), 150 mm NaCl, 5 mm DTT, and 5% glycerol. This figure shows one representative experiment; n = 4.
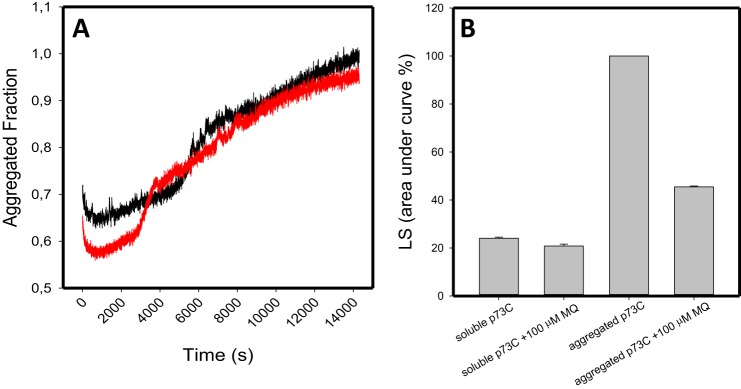
We also show that p73, a p53 paralog, is not as sensitive to MQ as p53 because we see a very modest effect of MQ on its DNA-binding domain aggregation when followed by ThT fluorescence and light scattering (Fig. 2). The effects on the reduction of the light scattering were significant but smaller than those found with p53 (Fig. 2B). p73C has higher stability, aggregates much more slowly, and has less exposure of backbone hydrogen bonds than p53C. This is probably the reason why the thiol groups of p73C are much less susceptible to reaction with MQ through the Michael addition reaction
Figure 2.
Human p73 central core domain (p73C) aggregation in the presence of MQ. MQ shows a small effect on p73C aggregation after 4 h at 37 °C. A, p73C at 5 μm was submitted to aggregation at 37 °C for 4 h, monitored by ThT binding at 25 μm. Black line, p73C; red line, p73C treated with 100 μm MQ. B, p73C light scattering (LS), before (soluble p73C and soluble p73C + 100 μm MQ) and after the 4-h kinetics (aggregated p73C and aggregated p73C + 100 μm MQ). All samples were diluted in 50 mm Tris (pH 7.2), 150 mm NaCl, 5 mm DTT, and 5% glycerol. This figure shows one representative experiment; n = 2.
PRIMA-1 lowers amyloid oligomers levels in mutant p53 cancer cells
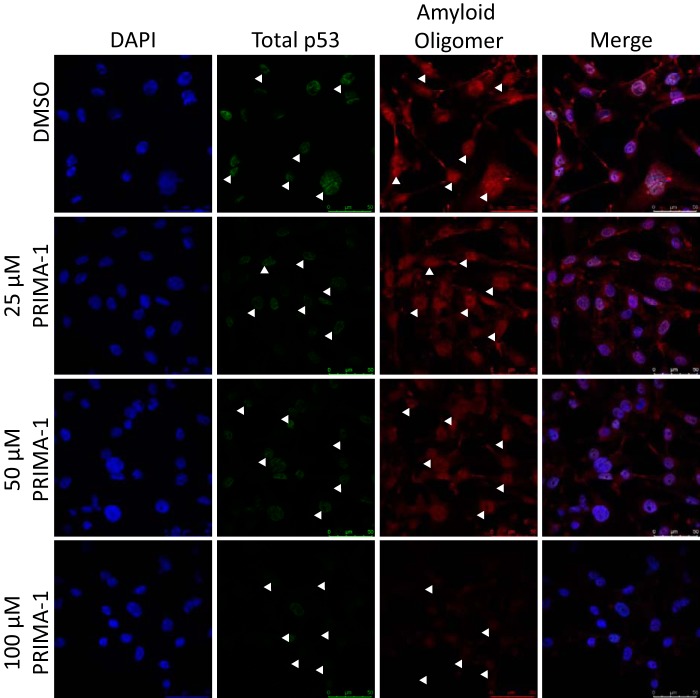
Through immunofluorescence and confocal microscopy, we previously demonstrated that the p53 mutant R280K strongly aggregates in MDA-MB-231 cells (7) in a pattern similar to those observed in biopsies of breast cancer tissues (33). In a similar assay, in which we treated MDA-MB-231 cells (Fig. 3, mutant p53) or MCF-7 cells (Fig. 4A, WT p53), we observed that, when MDA-MB-231 cells were treated with PRIMA-1 at increasing concentrations (25–100 μm), the levels of amyloid oligomers detected in the samples were reduced in a dose-dependent manner (Fig. 3). Moreover, a significant reduction in p53 levels was observed for all PRIMA-1 concentrations tested (Fig. 3B). Additionally, although reported previously (34, 35) with the time of treatment employed here (16 h), we did not observe a representative number of cells with p53 nucleolar staining upon PRIMA-1 treatment.
Figure 3.
Mutant p53 and amyloid oligomer staining are reduced in the MDA-MB-231 cell line after PRIMA-1 treatment in a concentration-dependent manner. A, MDA-MB-231 cells, treated with 25, 50, or 100 μm PRIMA-1 or 0.1% DMSO as the control for 16 h, were labeled with anti-p53 (DO-1) and anti-oligomer (A11) antibodies. Nuclei were stained with DAPI. Columns from left to right show nuclear staining with DAPI, p53 labeling, labeling of amyloid oligomers, and merged images. White arrowheads indicate cells stained with either DO-1 or A11 antibodies in their correspondent panels. Magnification, ×400. Scale bar = 50 μm.
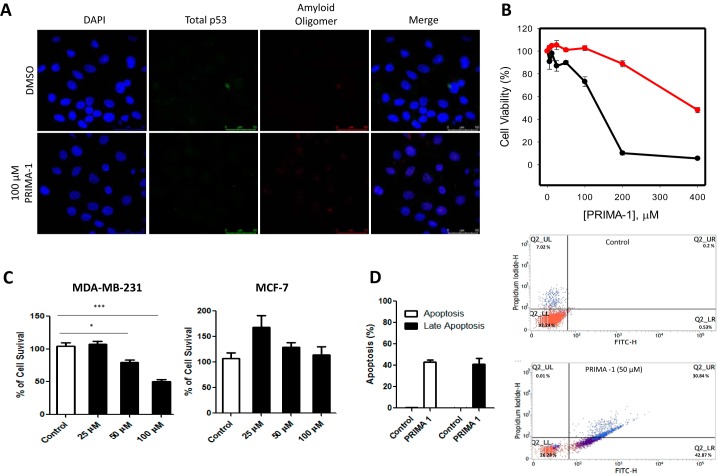
Figure 4.
A, MCF-7 cells were treated for 16 h with either 100 μm PRIMA-1 or 0. 1% DMSO as a control. The cells were labeled with anti-p53 (DO-1) and anti-oligomer (A11) antibodies, and nuclei were stained with DAPI. Columns from left to right show DAPI, p53 labeling, labeling of amyloid oligomers, and merged images. Magnification, ×400. Scale bar = 50 μm. B, MTT assay showing MDA-MB-231 (black line) and MCF-7 (red line) cell viability after treatment for 24 h with a serial dilution of PRIMA-1 from 0 (0.2% DMSO) to 200 μm. n = 4. C, cell proliferation assay using trypan blue showing a dose-dependent inhibition of cell proliferation after 24 h of treatment with PRIMA-1. n = 3 and significance marked with *(p < 0.05) and with *** (p < 0.001). D, AnnexinV/PI assay showing that PRIMA-1 induces apoptosis of MDA-MB-231 cells at 50 μm PRIMA-1, showing its effectiveness in this cell line in the concentration range used in this study.
MCF-7 cells present a markedly lower level of amyloid oligomer and p53 labeling, as reported previously (7) (Fig. 4A). The concentrations used for this and subsequent experiments were selected according to a 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide (MTT) cell viability assay (Fig. 4B), in which we observed a minimum of 80% cell viability for MDA-MB-231 cells after treatment with 100 μm PRIMA-1 and an even smaller reduction in MCF-7 cell viability. Also, Fig. 4, C and D, indicates that the concentrations used in this study are sufficient to inhibit cell proliferation and trigger apoptosis in the cell line used.
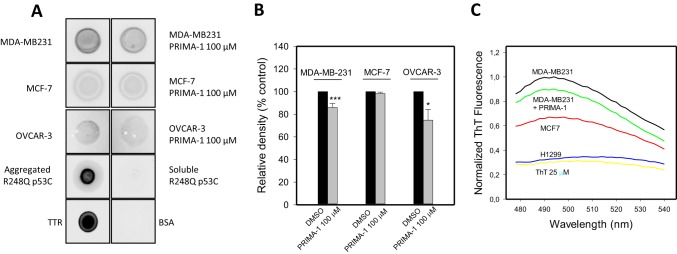
To further understand the results obtained in the immunofluorescence assay and to further understand their correlation with in vitro assays using WTp53C or R248Qp53C, we used a dot blot assay to evaluate the interaction between p53 aggregates formed by the R280K mutant in the MDA-MB-231 and the R248Q mutant in OVCAR-3 cell lines and the anti-amyloid oligomer antibody A11 (Fig. 5). We observed a significant reduction in A11 antibody binding when mutant p53 cells were treated with PRIMA-1 (Fig. 5A). The quantification is shown in Fig. 5B. We also observed that the levels of A11 antibody binding in MCF-7 cells treated with 100 μm PRIMA-1 did not differ significantly from those in control cells. Another probe for amyloid structures, thioflavin T (ThT), was used to estimate the levels of amyloid-state p53 in MDA-MB-231 (mutant p53), MCF-7 (WTp53), and H1299 (p53-null) cell lysates. As shown in Fig. 5C, the mutant p53 cell line (MDA-MB-231) developed the highest ThT fluorescence peak at ∼492 nm compared with the 25 μm ThT spectrum background. When treated with PRIMA-1 at 100 μm, the levels were reduced by 33.4%. As a control, we used MCF-7 (WT p53, lower aggregation, although a basal level is detected (see Fig. 4A and Ref. 7) and H1299 (p53-null, no amyloid oligomers detected) cell lysates. As expected, the H1299 cell line presented the lowest level of amyloid structures bound to ThT.
Figure 5.
Dot blot of cancer cell lysates showing that the amyloid oligomer levels are reduced by PRIMA-1 treatment. A, dot blots for A11 were performed with equal protein amounts from lysates of MDA-MB-231, MCF-7, or OVCAR-3 cells treated with 100 μm PRIMA-1 or 0.1% DMSO. Aggregated R248Qp53C at 20 μm and transthyretin (TTR) at 10 μm were used as positive controls, and soluble R248Qp53C at 20 μm and 0.1% BSA were used as negative controls. B, quantification of dot blots from a total of four independent experiments. A significant reduction in amyloid oligomers levels in cells treated with PRIMA-1 is indicated by asterisks (***, p < 0.001; *, p < 0.05). Quantification was performed using ImageJ software (version 1.43r, National Institutes of Health). C, ThT fluorescence in the presence of lysates from cells with different p53 statuses. ThT spectra were obtained after incubation of 15 μg of protein from different cell lysates with 25 μm ThT. The result is representative of three independent measurements.
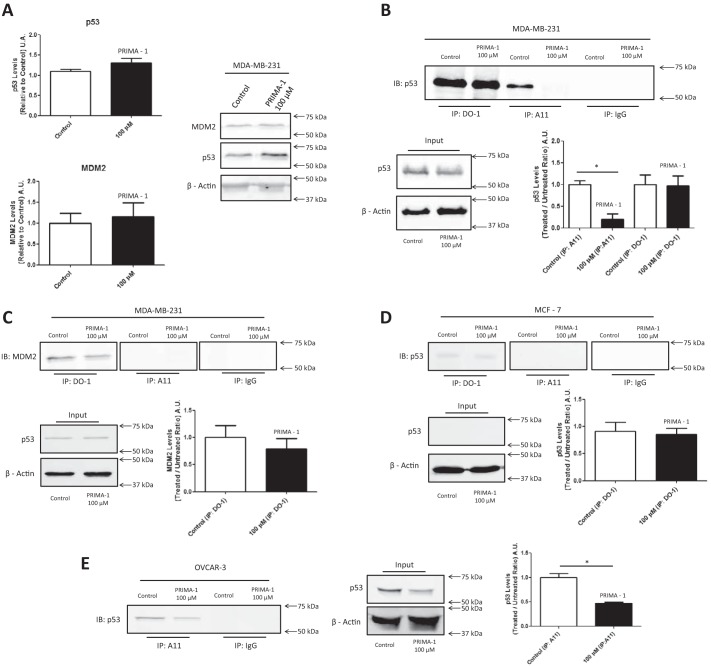
As shown in Fig. 6, A and B, lysates from 100 μm PRIMA-1–treated and DMSO-treated MDA-MB-231 cells were analyzed by Western blotting using the anti-p53 antibody DO-1. In this assay, we did not observe a significant difference in the p53 levels after PRIMA-1 treatment or in MDM2 levels. This result, together with the dot blot assay, prompted us to develop an immunoprecipitation assay using A11 and to then analyze the p53 levels in the amyloid fractions to quantify p53 mobilization by PRIMA-1 from the total amyloid content in the cell (Fig. 6B). We noticed a stark difference in p53 levels when the samples were immunoprecipitated with A11 (Fig. 6B); this change appears to be related to the rescue of p53 folding promoted by PRIMA-1, which reduces the level of aggregated p53. We found no difference when the proteins were immunoprecipitated with DO-1, which indicated the total amount of p53 in the cell and is in accordance with the input p53 levels.
Figure 6.
The p53 amyloid fraction of mutant p53 cancer cells is mobilized by PRIMA-1 treatment. A, Western blotting of lysates of MDA-MB-231 control cells treated with 0.1% DMSO or 100 μm PRIMA-1 for 18 h and quantification of p53 and MDM2 levels. n = 5. β-Actin was used as the reference. B, MDA-MB-231 cell lysates treated with PRIMA-1 or DMSO were immunoprecipitated with DO-1 and A11 and immunoblotted for p53 (anti-p53 rabbit polyclonal antibody). n = 3. C, detection and quantification of MDM2 co-immunoprecipitation. n = 3. D, immunoprecipitation assays using MCF-7 cells and their quantification. n = 3. E, immunoprecipitation assay using OVCAR-3 cells and their quantification. n = 3. *, p < 0.05. Overall, mouse IgG was used as an antibody control.
Immunoprecipitates were also probed for MDM2 (Fig. 6C), the most important p53 regulator in the cell. Although MDM2 was not detected in the cell lysates because of its low content in mutant-p53 cell lines (Fig. 6A), differences were detected in the immunoprecipitation results. MDM2 was found at similar amounts in the DO-1 immunoprecipitates, but aggregated p53 does not seem to interact with MDM2 (Fig. 6C). Moreover, the PRIMA-1-mediated stabilization of mutant p53 does not appear to induce MDM2 expression in this cell line, which is supposed to occur upon WT p53 activation (36–38). Fig. 6D shows the same immunoprecipitation reactions using A11 and DO-1 with MCF-7 cells. In this case, in the DO-1 immunoprecipitate, we observed a slight, nonsignificant reduction in p53 levels upon PRIMA-1 treatment, and p53 was not co-immunoprecipitated with A11, which indicates that the A11 staining detected in both the immunofluorescence and dot blot assays might include other amyloid proteins present in this cell line (39–41). Fig. 6E shows that the effect observed for the R280K mutant in the MDA-MB-231 cell line is reproduced with mutant R248Q in OVCAR-3, which has been shown to be susceptible to PRIMA-1 or PRIMA-1MET (24, 42). The amount of p53 in the amyloid fraction is lower when cells are treated with PRIMA-1.
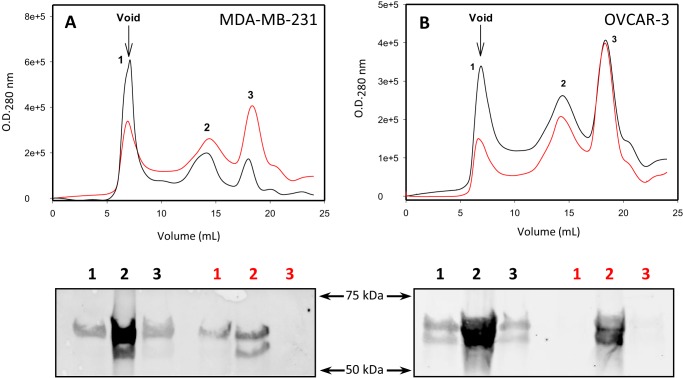
To confirm the results seen in the immunoprecipitation assay with A11, we performed SEC using both MDA-MB-231 (Fig. 7A) cells and OVCAR-3 (Fig. 7B) cells treated with 0.1% DMSO or 100 μm PRIMA-1 for 18 h. We observed three major peaks in all chromatograms, from which we precipitated proteins, separated through SDS-PAGE, and analyzed through Western blotting. In Fig. 7, we observe a reduction in peak 1 for both cell lines upon PRIMA-1 treatment. This peak corresponds to the column void volume, in which aggregated p53 is eluted, as shown by Western blotting (Fig. 7, A and B).
Figure 7.
Size-exclusion chromatography of cell lysates. A and B, MDA-MB-231 (A) and OVCAR3 (B) cell lysates were subjected to size-exclusion chromatography for detection of p53 aggregates in cells treated with either 100 μm PRIMA-1 (red line) or DMSO (black line). Aggregates eluted in the column void volume. Fractions eluted were separated through SDS-PAGE and immunoblotted for p53 (DO1). n = 3.
PRIMA-1 and MQ inhibit p53 aggregation seeding promoted by the R280K mutant in MDA-MB-231 cell lysates
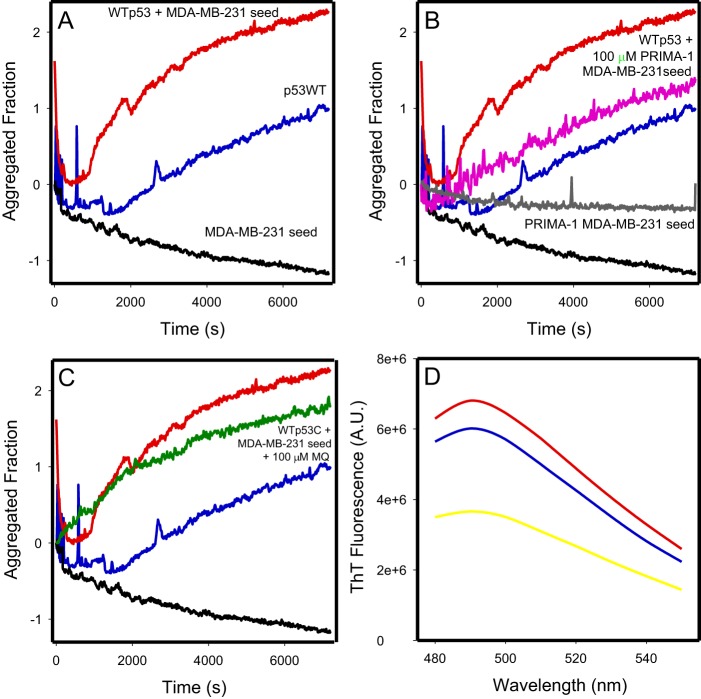
A seeding experiment was performed using MDA-MB-231 cell lysate to accelerate the conversion of WTp53C into the amyloid form. This experiment is similar to those used for prion conversion with mammalian PrP (RT Quick) (43), in which a misfolded template of the protein, called a seed, catalyzes the conversion of the correctly folded protein, speeding up the amyloid formation reaction. We observed increments in ThT fluorescence over time. In Fig. 8A, the blue line represents WTp53C aggregation at 37 °C for 2 h. Addition of the MDA-MB-231 total cell lysate at a concentration of 3 μg/ml showed a clear seeding effect with a reduction in the lag phase of the WTp53C curve (Fig. 8A, red line). When the MDA-MB-231 lysate is prepared from cells treated with 100 μm PRIMA-1, an equal protein quantity is much less effective in seeding aggregation (Fig. 8B, magenta line). Moreover, when the seeding experiment was performed in the presence of 100 μm MQ, which was added at time zero of the reaction (Fig. 8C, green line), inhibition of both p53 aggregation and MDA-MB-231 extract aggregation seeding was observed. Fig. 8D represents the ThT wavelength spectra of the samples presented in Fig. 8A at the end of the assay.
Figure 8.
Seeding of WT p53 aggregation by MDA-MB-231 protein extract. A–C, WTp53C at 5 μm (blue lines), protein extract at 3 μg/ml incubated with WTp53C at 5 μm (red lines), and control of protein extract alone used as the seed at 3 μg/ml (black lines). B, MDA-MB-231 cell extract treated with 3 μg/ml PRIMA-1 was incubated with 5 μm WTp53C (magenta line). MDA-MB-231 cell extract treated with PRIMA-1 was used as the seed at 3 μg/ml (gray line). The traces for WTp53C (blue line), protein extract incubated with WTp53C (red line), and control of protein extract alone used as the seed (black line) from A are repeated in the panel. C, MDA-MB-231 protein extract at 3 μg/ml was incubated with 5 μm WTp53C and 100 μm MQ (green line); the traces for WTp53C (blue line), protein extract incubated with WTp53C (red line), and control of protein extract alone used as the seed (black line) from A are repeated in the panel. D, ThT spectra (excitation, 450 nm; emission, 480 to 550 nm) after the 2-h kinetics experiment in A: WTp53C at 5 mm (blue line), protein extract at 3 μg/ml incubated with WTp53C at 5 μm (red line), and control of protein extract alone used as the seed at 3 μg/ml (yellow line). Aggregation was monitored by thioflavin T fluorescence emission (excitation, 440 nm; emission, 482 nm) over time at 37 °C. The ThT concentration was 25 μm, and measurements were performed at pH 7.2 in 50 mm Tris buffer, 150 mm NaCl, 5% glycerol, and 5 mm DTT. Aggregated fraction = (Fobs − FI)/(FF − FI), where F is the ThT fluorescence emission, Fobs is the observed fluorescence emission, FI is the initial fluorescence, and FF is the final fluorescence. The figure shows an experiment representative of three independent experiments.
Discussion
p53 aggregation has been the subject of study for several different groups in the past few years (7, 11, 44–48). These groups, including ours, have been working on discovering novel molecules that affect mutant p53 aggregates and have been attempting to obtain a deeper understanding of its aggregation process. We realize that p53 does not behave as a classic amyloid, forming arranged fibers with a unique pattern of organization, as discussed by Wang and Fersht (49) and Silva et al. (48). p53 has different sites available for aggregation, which leads to less organized amyloid structures because of its variability and weakens any attempt to block only one aggregation-prone site. In contrast, PRIMA-1, the lead compound that paved the way to APR-246 (PRIMA-1MET), the only compound targeting mutant p53 in phase II clinical trials (26), is a Michael acceptor with the capacity to promote p53 refolding. Thus, we propose that several of the mutant-exposed aggregation sites are hidden by PRIMA-1, which inhibits mutant p53 aggregation. These reasons led us to use PRIMA-1 as a model for studying p53 aggregation modulation.
Here we aimed to investigate the role of PRIMA-1 and MQ in the amyloid fraction of p53 found in cancer cells and to determine whether PRIMA-1 can reactivate amyloid-state p53. We observed that the aggregation of mutant p53 was more sensitive to both PRIMA-1 and MQ inhibition than WT p53 (Fig. 1). This finding is consistent with data presented by Lambert et al. (29), who showed that the R175H p53 mutant is more prone to covalent MQ binding than the WT protein because of the number of cysteines accessible by MQ. However, our study provides the first demonstration of a role of PRIMA-1 in p53 aggregation. In agreement with the findings that the aggregation propensity of mutant p53 is due to a network of multiple aggregation-prone sites in the protein (49), we propose that mutant p53 has a greater chance to expose SH groups from different regions that can react with MQ, which decreases the predisposition to interact with other p53 molecules. This finding would also explain the relatively low specificity of PRIMA-1/MQ for p53. In fact, we found that p73 aggregation is only partially inhibited by MQ (Fig. 2). This is related to results obtained with H1299 cells expressing p73α submitted to PRIMA-1MET treatment (50), in which very little effect is seen on this isoform. On the other hand, p73β isoform and p63γ have shown a capacity to induce gene transactivation. Although mutations in these p53 homologs are very rare in cancers, they have a crucial role in animal development (51, 52). Also, the overexpression of their isoforms in tumors, which appear to block the transactivation activity of the full-length protein (53), is related to bad prognosis (54). Finally, PRIMA-1 has been suggested to release p73 bound to mutant p53 (55). This might have additional effects on attenuating the gain-of-function effects of mutant p53 aggregates that interact with p73 (8, 17, 18).
The effects of MQ on cellular proteins other than mutant p53 are related to the GSH and thioredoxin reductase system. It is known to react with thiol groups from cysteines present in TrxR1, converting it to an active oxidase and on GSH and free cysteine itself, thus impairing GSH synthesis (28, 42). Also, more recently, the binding of mutant p53 to Nrf2, an antioxidant transcription factor, has been described to lead to decreased expression of SLC7A11 (56). These alterations result in a cellular environment that favors apoptosis through different mechanisms independent of p53 reactivation. On the other hand, these same alterations make cells more sensitive to reactivated MQ-p53 effects. In a general way, it is a consensus that several factors contribute to make PRIMA-1/PRIMA-1MET compounds more valuable compared with other Michael acceptors that have high toxicity in vivo (21, 42), including the advantage of being a prodrug, only converted to MQ within the cell. Also, the favorable molecular orbital properties from the MQ structure (57), leading to mild potency of MQ as a Michael acceptor, added to its effects on redox mechanisms of the cell that positively modify the cancer environment, are additional advantageous effects. As has been shown for MDA-MB-231 in Fig. 4, the PRIMA-1 ability to induce apoptosis and impair cell growth and proliferation has been described for different mutant p53 cell lines (20, 23, 42), including p53-null cell lines with insertion of p53 mutants (20, 21).
We also observed that the amyloid content was reduced in MDA-MB-231 cells treated with PRIMA-1 (Figs. 3 and 5). This finding supports the previous suggestion that the amyloid content in these cells was reduced and that PRIMA-1 acts on the amyloid p53 fraction in these cells. This result was further confirmed through immunoprecipitation assays (Fig. 6, B and E). ThT, an amyloid probe, was also used to assess the amyloid content in different cell lines lysates (Fig. 5C). We observed that cancer cells expressing mutant p53 had higher levels of amyloid structures compared with control cells. After treatment with PRIMA-1, the level of amyloid aggregation was decreased.
Immunoprecipitation assays detected a notably lower p53 level in the A11 precipitates of PRIMA-1-treated cells compared with DMSO-treated cells (Fig. 6, B and E). A11 is not a specific antibody for p53, and other proteins can exist in an amyloid state in cancer cells (39, 58–60). Thus, A11 might bind to amyloid proteins other than mutant p53 (11, 58). However, in the A11 immunoprecipitation assay, mutant p53 present in the amyloid oligomer immunoprecipitate was detected by Western blotting, which makes this assay more specific to the amyloid p53 fraction than the dot blot assay and is further confirmed by the reduction of p53 levels in the fractions eluted in the void volume in SEC for both mutant p53 cell lines used in this work (Fig. 7).
Because MDM2 is a well-known regulator of p53, MDM2 levels were also assessed in immunoprecipitates. The low MDM2 level observed in the mutant p53 cell line is acceptable because of the previously reported lower affinity of MDM2 for mutant p53 (and the lack of difference in MDM2 levels after PRIMA-1MET administration, which has been reported previously) (23). MDM2 was not found in A11 precipitates but was only associated with DO-1 precipitates, which indicated that, at a low level, MDM2 binds to R280K mutant p53 but not in its amyloid state (Fig. 6C). Curiously, up-regulation of MDM2 by activated p53 (36, 37) did not occur in PRIMA-1-treated MDA-MB-231cells. This finding was also observed for p53 mutants in bladder cancer cells (61) and indicates that the mutant p53 recovery promoted by PRIMA-1 might not recover all of WTp53 functions. Wiech et al. (62) report the formation of “pseudo-aggregates” of mutant p53, MDM2, and HSP70 that form amyloid-like structures. With respect to MDM2, this effect was not observed under the conditions used in our experiments.
A reduction in p53 and amyloid oligomer levels in the MCF-7 cell line was not seen in the dot blot, Western blotting for p53, or immunoprecipitation with A11, which indicates that MCF-7 cells are not affected by PRIMA-1 treatment under normoxic conditions, which has been reported previously (63, 64). In contrast, because PRIMA-1 affects more than one target in cancer cells, we cannot eliminate the possibility that the reduction in p53 levels or amyloid content is due to the activation of other regulatory proteins (26).
We tested the ability of MDA-MB-231 cell lysates containing mutant p53 to induce aggregation of recombinant WT p53C, and we observed a clear seeding effect indicative of aggregates in the cell extract promoting WTp53C aggregation in a prion-like manner. This result is in agreement with the evidence for a prion-like behavior of aggregates of p53 (7, 11, 19, 65). Lysates obtained from cells treated with PRIMA-1 had a weaker seeding capacity, which corroborates our findings of a reduction in the cellular amyloid-state p53 content after PRIMA-1 treatment. Finally, by adding MQ to the reaction medium, both WTp53C aggregation (Fig. 1A) and mutant p53 seeding (Fig. 8C) were diminished, which confirms that PRIMA-1/MQ inhibits WTp53C aggregation and mobilizes amyloid-state mutant p53 to reactivate p53. In this case, we must observe that p53 aggregation was not reduced to very low levels as seen in Fig. 1. This effect could be due to a difference in the speed of two different reactions happening at the same time: the seeding reaction versus MQ Michael addition and p53C stabilization. Further experiments would be necessary to elucidate this.
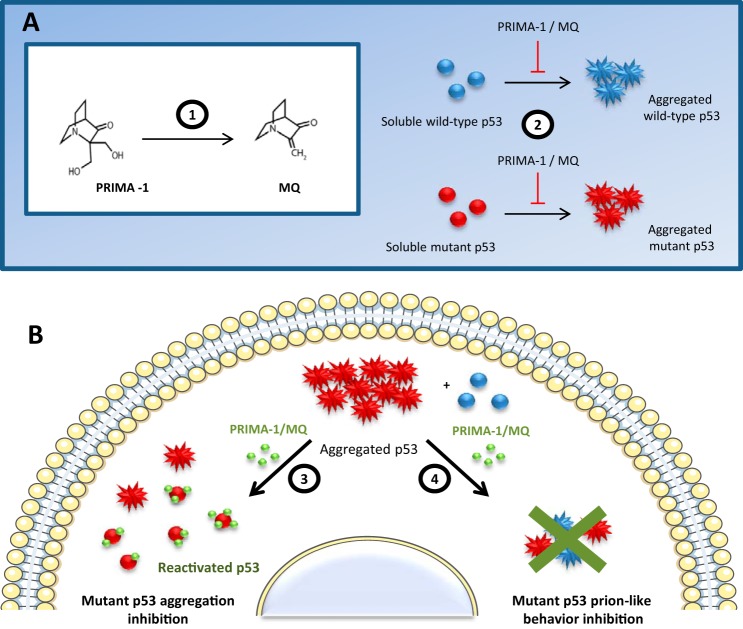
The scheme shown in Fig. 9 summarizes our results concerning the effects of PRIMA-1/MQ on p53 aggregation. Clearly, mutant p53 aggregation is intensely decreased when mutant p53 is modified by these compounds, and the scheme outlines how the rescue effects of PRIMA-1 might be mediated through a combination of aggregation reduction and functional recovery of aggregated p53. Another striking effect is the inhibition of seeding by the treated extract, characterized by the PRIMA-1 property of disassembling p53 amyloid aggregates, which impairs their prion-like features.
Figure 9.
Summary of PRIMA-1/MQ effects on amyloid-state p53. 1, PRIMA-1 is converted to MQ in biological systems, as described by Lambert et al. (29). 2, PRIMA-1 and MQ inhibit p53C aggregation in vitro. 3, PRIMA-1 mobilizes the aggregated p53 pool and stabilizes mutant p53, resulting in recovery of WT p53 functions and a reduction in cellular amyloid content. 4, PRIMA-1/MQ blocks the seeding of WT p53C aggregation by mutant p53 present in cell lysates, which might be part of the general effect of PRIMA-1/MQ on p53 seen in mutant p53 cancer cells.
Further studies are necessary to reveal the cellular conditions that lead to the formation of these aggregates and to identify the intermediate species that might produce deleterious effects. Additionally, other p53 mutations leading to aggregation should be investigated. Although revealing the mechanism of action of PRIMA-1 has been the goal of several different studies, our work demonstrates the origin of PRIMA-1 stabilized mutant p53 and highlights a novel target in cancer cells. This study provides the first demonstration that amyloid-state p53 can be rescued by a small compound, and this strategy warrants further exploration as a cancer treatment.
Experimental procedures
Chemicals
All reagents were of analytical grade. Distilled water was filtered and deionized through a Millipore water purification system.
Cell lines
Cell lines were obtained from the Rio de Janeiro Cell Bank. H1299 and OVCAR-3 cells were maintained in RPMI supplemented with 10% fetal bovine serum and 10 units/ml penicillin/streptomycin (Gibco). MDA-MB-231 and MCF-7 cells were maintained in Dulbecco's modified Eagle's medium supplemented with 10% fetal bovine serum and 10 units/ml penicillin/streptomycin (Gibco). All cell lines were maintained at 37 °C with 5% CO2.
Protein purification
WT and R248Q mutant human p53C (residues 94–312) and p73C were overexpressed in Escherichia coli BL21 (DE3) from plasmid clones using the pET11a vectors (Novagen) (6, 66). Overexpression was induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at an A600 of 1.2. Cell suspensions were kept overnight at 15 °C before centrifugation at 10,000 × g for 15 min. The cell pellets were resuspended in 50 mm Tris-HCl (pH 7.2), 50 mm NaCl, 2.5 mm DTT, and 5% glycerol (v/v) with 1 mm phenylmethylsulfonyl fluoride. The suspension was sonicated on ice, and the samples were purified using SP-Sepharose in a Shimadzu ultra-fast liquid chromatograph. Briefly, the samples were loaded onto a Superdex S200 HR10/30 column (GE Healthcare) equilibrated in buffer containing 50 mm Tris-HCl (pH 7.2), 150 mm NaCl, 5 mm DTT, and 5% glycerol (v/v) and analyzed at a flow rate of 0.7 ml/min. The protein samples were stored at −80 °C.
Thioflavin T fluorescence
ThT fluorescence spectra were acquired using an ISS-PC1 spectrofluorometer (ISS, Champaign, IL) with an excitation wavelength of 440 nm, and the emitted light in the 480- 510-nm range was collected. All measurements were performed using p53 assay buffer containing 50 mm Tris-HCl (pH 7.2), 150 mm NaCl, 5 mm DTT, and 5% glycerol (v/v). For the recombinant protein aggregation procedures, 5 μm p53C (WT and R248Q mutant) was incubated with 25 μm ThT at 37 °C for 2 h. ThT emission was collected over time at 482 nm. To obtain the ThT spectra of the cell extract, 15 μg of protein lysates was diluted in p53 assay buffer and incubated with 25 μm ThT for 5 min before the ThT spectra were collected.
Light scattering measurements
Light scattering measurements were carried out in an ISS-PC1 spectrofluorometer. Light scattering excitation wavelength was fixed at 420 nm, and the emission spectrum was recorded from 400 to 440 nm.
Immunofluorescence co-localization assays
Cells were grown to 70–80% confluence and treated with PRIMA-1 or an equivalent volume of DMSO for 16 h. The cells were then washed twice with PBS and incubated with ice-cold 1:1 methanol:acetone solution for 15 min at −20 °C. The cells were simultaneously labeled with a mouse monoclonal anti-human p53 DO-1 primary antibody (1:200) and an anti-oligomer A11 primary antibody (1:1,000) for 2 h at room temperature. The cells were subsequently incubated with Alexa 568–conjugated goat anti-mouse and Alexa 647–conjugated goat anti-rabbit (Life Technologies) secondary antibodies (1:2,000) for 1 h at room temperature in the dark. The cells were washed twice with PBS, and cover slips were mounted with Prolong Diamond with DAPI (Life Technologies) and analyzed by confocal microscopy (Leica TCS SPE confocal microscope) at ×40,000 magnification.
Annexin V-FITC/propidium iodide apoptosis detection
The apoptosis detection assay was performed using the Annexin V-FITC apoptosis detection kit (APOAF-50TST, Sigma-Aldrich). 106 cells were plated onto 24-well plates and, after 24 h, treated with DMSO as a control or PRIMA-1 at 50 μm for 18 h and treated according to the kit directions. The samples were analyzed by flow cytometry (BD FACSVerseTM).
Cell lysate preparation
Cells treated with either 0.1% DMSO or 100 μm PRIMA-1 for 18 h were washed three times with PBS, lysed with liquid nitrogen in a buffer containing 10 mm Tris-HCl (pH 7.5), 150 mm NaCl, 1 mm EDTA, 1% Triton, 1 mm Na3VO4, 5 mm NaF, and protease inhibitor mixture (Sigma), and then centrifuged at 3,000 × g for 5 min. The protein content of the samples was quantified according to Lowry et al. (67) and stored at −80 °C.
Dot blot assay
Cell lysates (15 μg of protein) were placed onto a nitrocellulose membrane in a final volume of 2 μl. The membrane was blocked with Odyssey blocking buffer (LI-COR) for 1 h at room temperature and incubated for 18 h at 4 °C with the primary antibody A11 at 1:5,000 dilution (Millipore). The membrane was then washed with TBS-T (20 mm Tris-HCl, pH 7.4, 150 mm NaCl, 0.1% Tween 20) and incubated with IR-Dye® 800CW goat anti-rabbit secondary antibody at 1:15,000 dilution for 1 h at room temperature in Odyssey blocking buffer. After four 5-min washes with TBS-T and two washes with TBS, the membrane was analyzed in an Odyssey detection system (LI-COR). We used BSA and soluble p53CR248Q at 20 μm as a negative control and R248Q p53C aggregates at 20 μm and aggregated transthyretin TTR as positive controls.
MTT assay
Cells were plated onto 96-well plates to a confluence between 80% and 90% at the time of the assay. After 24 h, 100 μl of serial PRIMA-1 dilutions ranging from 0 (0.2% DMSO) to 200 μm were added to the wells, and the cells were treated for 24 h. The next day, 0.5 mg/ml MTT in PBS was added to the cells for 2 to 4 h. The crystals formed were then solubilized in DMSO, and the plate was analyzed with a SpectraMax Paradigm multi-mode microplate reader (Molecular Devices) at 570 nm and 650 nm.
Cell proliferation
MDA-MB-231 and MCF-7 cells were cultured in a 24-well plate. The next day, cells were treated with 25, 50, or 100 μm PRIMA-1 for 18 h. Then cells were washed with PBS and detached with 100 μl of trypsin, which was neutralized with 300 μl of Dulbecco's modified Eagle's medium high-glucose with 10% fetal calf serum. A 10-μl aliquot was mixed with trypan blue dye (1:1), and the viable cells were counted in a Neubauer's chamber using an optical microscope.
SEC of cell lysates
To analyze the degree of p53 aggregation inhibition by PRIMA-1 in mutant-p53 cell lines, MDA-MB-231 or OVCAR-3 cells lysates treated with either 0.1% DMSO or 100 μm PRIMA-1 were loaded onto a Superdex S200 HR10/30 column (GE Healthcare) equilibrated in a buffer containing 50 mm Tris-HCl, 150 mm NaCl, 5 mm DTT, and 5% glycerol (pH 7.5)) using a 0.4 ml/min flow in a Shimadzu ultra-fast liquid chromatograph. The eluted fractions were precipitated with TCA, washed with acetone, and resuspended in sample buffer for SDS-PAGE and Western blot analysis.
Western blotting
Cell lysates (100 μg of the control and treated samples) were resolved by SDS-PAGE (12.5%), and the separated proteins were transferred to polyvinylidene difluoride low-fluorescence membranes (Millipore). The membranes were blocked for 1 h at 4 °C in LI-COR blocking buffer and then incubated overnight with primary antibodies (1:10,000). The following antibodies were used: anti-p53 (DO-1), anti-MDM2 (D-7) (both from Santa Cruz Biotechnology), and anti-β-actin (Sigma-Aldrich). The membranes were then incubated with IR-Dye® 800CW goat anti-rabbit or anti-mouse antibody (LI-COR) (1:15,000) for 1 h at room temperature. Immunoreactive bands were visualized in an Odyssey detection system (LI-COR) according to the manufacturer's instructions. The β-actin levels in the cell lysates were used as a loading control. Densitometric quantification of the bands was performed using ImageJ software (version 1.43r, National Institutes of Health).
Immunoprecipitation assays
Cell lysates (500 μg), which were obtained as described above, were incubated with the antibodies (A11, 1:1,000 dilution; DO-1, 1:100 dilution, according to the antibody datasheets) in a total volume of 1 ml of PBS for 1 h at 4 °C. Then, 20 μl of protein A/G PLUs–agarose (Santa Cruz Biotechnology) was added, and the samples were incubated overnight at 4 °C on a rocker platform. Immunoprecipitates were collected by centrifugation at 1,000 × g and 4 °C for 5 min. The supernatants were discarded, and the pellets were washed four times with 1 ml of cold PBS with centrifugation at 1,000 × g and 4 °C for 5 min between each wash. The final pellet was resuspended in 40 μl of electrophoresis sample buffer and boiled for 3 min. After pelleting the beads, 20- or 40-μl aliquots were analyzed by SDS-PAGE and Western blotting.
Statistical analyses
Statistical analyses were performed using SigmaPlot for Windows version 12.0 (Systat Software, Inc.). According to the parametric or nonparametric distribution of the values, Student's t test or Mann–Whitney rank sum test was used for statistical analyses between groups, respectively.
Author contributions
L. P. R. and J. L. S. conceptualization; L. P. R. and J. L. S. data curation; L. P. R., G. D. S. F., and J. L. S. formal analysis; L. P. R. and J. L. S. funding acquisition; L. P. R., G. D. S. F., C. L. C., S. M. M. V. A., and D. C. F. C. investigation; L. P. R., G. D. S. F., D. C. F. C., and J. L. S. visualization; L. P. R., D. C. F. C., and J. L. S. writing-original draft; L. P. R. and J. L. S. writing-review and editing; G. D. S. F., D. C. F. C., and J. L. S. validation; C. L. C., S. M. M. V. A., R. S. C., and D. C. F. C. methodology; J. L. S. supervision; J. L. S. project administration.
This work was supported by Serrapilheira Institute Grant Serra-1708-15204 (to L. P. R.), Fundação do Câncer, Conselho Nacional de Desenvolvimento Científico e Tecnológico Grants 465395/2014-7 and 402321/2016-2 (to J. L. S.), Fundação Carlos Chagas Filho de Amparo à Pesquisa do Estado do Rio de Janeiro Grant 202955/2015 (to J. L. S.), Ministério da Saúde, and Financiadora de Estudos e Projetos of Brazil. The authors declare that they have no conflicts of interest with the contents of this article.
- GoF
- gain-of-function
- MQ
- 2-methylene-3-quinuclidinone
- SEC
- size-exclusion chromatography
- ThT
- thioflavin T
- MTT
- 3-(4,5-dimethylthiazol-2-yl)-2,5-diphenyltetrazolium bromide
- DAPI
- 4′,6-diamidino-2-phenylindole.
References
- 1. Freed-Pastor W. A., and Prives C. (2012) Mutant p53: One name, many proteins. Genes Dev. 26, 1268–1286 10.1101/gad.190678.112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Vousden K. H., and Lane D. P. (2007) p53 in health and disease. Nat. Rev. Mol. Cell Biol. 8, 275–283 10.1038/nrm2147 [DOI] [PubMed] [Google Scholar]
- 3. Alexandrova E. M., Yallowitz A. R., Li D., Xu S., Schulz R., Proia D. A., Lozano G., Dobbelstein M., and Moll U. M. (2015) Improving survival by exploiting tumour dependence on stabilized mutant p53 for treatment. Nature 523, 352–356 10.1038/nature14430 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Zhu J., Sammons M. A., Donahue G., Dou Z., Vedadi M., Getlik M., Barsyte-Lovejoy D., Al-awar R., Katona B. W., Shilatifard A., Huang J., Hua X., Arrowsmith C. H., and Berger S. L. (2015) Gain-of-function p53 mutants co-opt chromatin pathways to drive cancer growth. Nature 525, 206–211 10.1038/nature15251 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Muller P. A., and Vousden K. H. (2014) Mutant p53 in cancer: new functions and therapeutic opportunities. Cancer Cell 25, 304–317 10.1016/j.ccr.2014.01.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Ishimaru D., Andrade L. R., Teixeira L. S., Quesado P. A., Maiolino L. M., Lopez P. M., Cordeiro Y., Costa L. T., Heckl W. M., Weissmüller G., Foguel D., and Silva J. L. (2003) Fibrillar aggregates of the tumor suppressor p53 core domain. Biochemistry 42, 9022–9027 10.1021/bi034218k [DOI] [PubMed] [Google Scholar]
- 7. Ano Bom A. P., Rangel L. P., Costa D. C., de Oliveira G. A., Sanches D., Braga C. A., Gava L. M., Ramos C. H., Cepeda A. O., Stumbo A. C., De Moura Gallo C. V., Cordeiro Y., and Silva J. L. (2012) Mutant p53 aggregates into prion-like amyloid oligomers and fibrils: Implications for cancer. J. Biol. Chem. 287, 28152–28162 10.1074/jbc.M112.340638 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Xu J., Reumers J., Couceiro J. R., De Smet F., Gallardo R., Rudyak S., Cornelis A., Rozenski J., Zwolinska A., Marine J. C., Lambrechts D., Suh Y. A., Rousseau F., and Schymkowitz J. (2011) Gain of function of mutant p53 by coaggregation with multiple tumor suppressors. Nat. Chem. Biol. 7, 285–295 10.1038/nchembio.546 [DOI] [PubMed] [Google Scholar]
- 9. Lasagna-Reeves C. A., Clos A. L., Castillo-Carranza D., Sengupta U., Guerrero-Muñoz M., Kelly B., Wagner R., and Kayed R. (2013) Dual role of p53 amyloid formation in cancer; loss of function and gain of toxicity. Biochem. Biophys. Res. Commun. 430, 963–968 10.1016/j.bbrc.2012.11.130 [DOI] [PubMed] [Google Scholar]
- 10. Bullock A. N., Henckel J., DeDecker B. S., Johnson C. M., Nikolova P. V., Proctor M. R., Lane D. P., and Fersht A. R. (1997) Thermodynamic stability of wild-type and mutant p53 core domain. Proc. Natl. Acad. Sci. 94, 14338–14342 10.1073/pnas.94.26.14338 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Silva J. L., De Moura Gallo C. V., Costa D. C., and Rangel L. P. (2014) Prion-like aggregation of mutant p53 in cancer. Trends Biochem. Sci. 39, 260–267 10.1016/j.tibs.2014.04.001 [DOI] [PubMed] [Google Scholar]
- 12. Muller P. A., Caswell P. T., Doyle B., Iwanicki M. P., Tan E. H., Karim S., Lukashchuk N., Gillespie D. A., Ludwig R. L., Gosselin P., Cromer A., Brugge J. S., Sansom O. J., Norman J. C., and Vousden K. H. (2009) Mutant p53 drives invasion by promoting integrin recycling. Cell 139, 1327–1341 10.1016/j.cell.2009.11.026 [DOI] [PubMed] [Google Scholar]
- 13. Adorno M., Cordenonsi M., Montagner M., Dupont S., Wong C., Hann B., Solari A., Bobisse S., Rondina M. B., Guzzardo V., Parenti A. R., Rosato A., Bicciato S., Balmain A., and Piccolo S. (2009) A nutant-p53/Smad complex opposes p63 to empower TGFβ-induced metastasis. Cell 137, 87–98 10.1016/j.cell.2009.01.039 [DOI] [PubMed] [Google Scholar]
- 14. Blandino G., Levine A. J., and Oren M. (1999) Mutant p53 gain of function: differential effects of different p53 mutants on resistance of cultured cells to chemotherapy. Oncogene 18, 477–485 10.1038/sj.onc.1202314 [DOI] [PubMed] [Google Scholar]
- 15. Wong R. P., Tsang W. P., Chau P. Y., Co N. N., Tsang T. Y., and Kwok T. T. (2007) p53-R273H gains new function in induction of drug resistance through down-regulation of procaspase-3. Mol. Cancer Ther. 6, 1054–1061 10.1158/1535-7163.MCT-06-0336 [DOI] [PubMed] [Google Scholar]
- 16. Schilling T., Kairat A., Melino G., Krammer P. H., Stremmel W., Oren M., and Müller M. (2010) Interference with the p53 family network contributes to the gain of oncogenic function of mutant p53 in hepatocellular carcinoma. Biochem. Biophys. Res. Commun. 394, 817–823 10.1016/j.bbrc.2010.03.082 [DOI] [PubMed] [Google Scholar]
- 17. Kehrloesser S., Osterburg C., Tuppi M., Schäfer B., Vousden K. H., and Dötsch V. (2016) Intrinsic aggregation propensity of the p63 and p73 TI domains correlates with p53R175H interaction and suggests further significance of aggregation events in the p53 family. Cell Death Differ. 23, 1952–1960 10.1038/cdd.2016.75 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Cino E. A., Soares I. N., Pedrote M. M., De Oliveira G. A. P., and Silva J. L. (2016) Aggregation tendencies in the p53 family are modulated by backbone hydrogen bonds. Sci. Rep. 10.1038/srep32535 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Costa D. C. F., de Oliveira G. A. P., Cino E. A., Soares I. N., Rangel L. P., and Silva J. L. (2016) Aggregation and prion-like properties of misfolded tumor suppressors: is cancer a prion disease? Cold Spring Harb. Perspect. Biol. 8, 1–22 10.1101/cshperspect.a023614 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Bykov V. J., Issaeva N., Shilov A., Hultcrantz M., Pugacheva E., Chumakov P., Bergman J., Wiman K. G., and Selivanova G. (2002) Restoration of the tumor suppressor function to mutant p53 by a low-molecular-weight compound. Nat. Med. 8, 282–288 10.1038/nm0302-282 [DOI] [PubMed] [Google Scholar]
- 21. Bykov V. J., Issaeva N., Zache N., Shilov A., Hultcrantz M., Bergman J., Selivanova G., and Wiman K. G. (2005) Reactivation of mutant p53 and induction of apoptosis in human tumor cells by maleimide analogs. J. Biol. Chem. 280, 30384–30391 10.1074/jbc.M501664200 [DOI] [PubMed] [Google Scholar]
- 22. Bykov V. J., Zache N., Stridh H., Westman J., Bergman J., Selivanova G., and Wiman K. G. (2005) PRIMA-1MET synergizes with cisplatin to induce tumor cell apoptosis. Oncogene 24, 3484–3491 10.1038/sj.onc.1208419 [DOI] [PubMed] [Google Scholar]
- 23. Lambert J. M., Moshfegh A., Hainaut P., Wiman K. G., and Bykov V. J. (2010) Mutant p53 reactivation by PRIMA-1MET induces multiple signaling pathways converging on apoptosis. Oncogene 29, 1329–1338 10.1038/onc.2009.425 [DOI] [PubMed] [Google Scholar]
- 24. Bykov V. J., Issaeva N., Selivanova G., and Wiman K. G. (2002) Mutant p53-dependent growth suppression distinguishes PRIMA-1 from known anticancer drugs: a statistical analysis of information in the National Cancer Institute database. Carcinogenesis 23, 2011–2018 10.1093/carcin/23.12.2011 [DOI] [PubMed] [Google Scholar]
- 25. Lehmann S., Bykov V. J., Ali D., Andrén O., Cherif H., Tidefelt U., Uggla B., Yachnin J., Juliusson G., Moshfegh A., Paul C., Wiman K. G., and Andersson P. O. (2012) Targeting p53 in vivo: a first-in-human study with p53-targeting compound APR-246 in refractory hematologic malignancies and prostate cancer. J. Clin. Oncol. 30, 3633–3639 10.1200/JCO.2011.40.7783 [DOI] [PubMed] [Google Scholar]
- 26. Bykov V. J. N., Eriksson S. E., Bianchi J., and Wiman K. G. (2017) Targeting mutant p53 for efficient cancer therapy. Nat. Rev. Cancer. 18, 89–102 10.1038/nrc.2017.109 [DOI] [PubMed] [Google Scholar]
- 27. Rieber M., and Strasberg-Rieber M. (2012) Hypoxia, Mn-SOD and H2O2 regulate p53 reactivation and PRIMA-1 toxicity irrespective of p53 status in human breast cancer cells. Biochem. Pharmacol. 84, 1563–1570 10.1016/j.bcp.2012.09.003 [DOI] [PubMed] [Google Scholar]
- 28. Peng X., Zhang M. Q. Z., Conserva F., Hosny G., Selivanova G., Bykov V. J. N., Arnér E. S. J., and Wiman K. G. (2013) APR-246/PRIMA-1MET inhibits thioredoxin reductase 1 and converts the enzyme to a dedicated NADPH oxidase. Cell Death Dis. 4, e881 10.1038/cddis.2013.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Lambert J. M., Gorzov P., Veprintsev D. B., Söderqvist M., Segerbäck D., Bergman J., Fersht A. R., Hainaut P., Wiman K. G., and Bykov V. J. (2009) PRIMA-1 reactivates mutant p53 by covalent binding to the core domain. Cancer Cell 15, 376–388 10.1016/j.ccr.2009.03.003 [DOI] [PubMed] [Google Scholar]
- 30. Wassman C. D., Baronio R., Demir Ö., Wallentine B. D., Chen C. K., Hall L. V., Salehi F., Lin D. W., Chung B. P., Hatfield G. W., Richard Chamberlin A., Luecke H., Lathrop R. H., Kaiser P., and Amaro R. E. (2013) Computational identification of a transiently open L1/S3 pocket for reactivation of mutant p53. Nat. Commun. 4, 1407 10.1038/ncomms2361 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Zhang Q., Bykov V. J. N., Wiman K. G., and Zawacka-Pankau J. (2018) APR-246 reactivates mutant p53 by targeting cysteines 124 and 277. Cell Death Dis. 9, 439 10.1038/s41419-018-0463-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Bykov V. J., and Wiman K. G. (2014) Mutant p53 reactivation by small molecules makes its way to the clinic. FEBS Lett. 588, 2622–2627 10.1016/j.febslet.2014.04.017 [DOI] [PubMed] [Google Scholar]
- 33. Levy C. B., Stumbo A. C., Ano Bom A. P., Portari E. A., Cordeiro Y., Carneiro Y., Silva J. L., and De Moura-Gallo C. V. (2011) Co-localization of mutant p53 and amyloid-like protein aggregates in breast tumors. Int. J. Biochem. Cell Biol. 43, 60–64 10.1016/j.biocel.2010.10.017 [DOI] [PubMed] [Google Scholar]
- 34. Rökaeus N., Klein G., Wiman K. G., Szekely L., and Mattsson K. (2007) PRIMA-1MET induces nucleolar accumulation of mutant p53 and PML nuclear body-associated proteins. Oncogene 26, 982–992 10.1038/sj.onc.1209858 [DOI] [PubMed] [Google Scholar]
- 35. Russo D., Ottaggio L., Penna I., Foggetti G., Fronza G., Inga A., and Menichini P. (2010) PRIMA-1 cytotoxicity correlates with nucleolar localization and degradation of mutant p53 in breast cancer cells. Biochem. Biophys. Res. Commun. 402, 345–350 10.1016/j.bbrc.2010.10.031 [DOI] [PubMed] [Google Scholar]
- 36. Barak Y., Juven T., Haffner R., and Oren M. (1993) Mdm2 expression is induced by wild type p53 activity. EMBO J. 12, 461–468 10.1002/j.1460-2075.1993.tb05678.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Juven T., Barak Y., Zauberman A., George D. L., and Oren M. (1993) Wild type p53 can mediate sequence-specific transactivation of an internal promoter within the mdm2 gene. Oncogene 8, 3411–3416 [PubMed] [Google Scholar]
- 38. Perry M. E., Piette J., Zawadzki J. A., Harvey D., and Levine A. J. (1993) The mdm-2 gene is induced in response to UV light in a p53-dependent manner. Proc. Natl. Acad. Sci. U.S.A. 90, 11623–11627 10.1073/pnas.90.24.11623 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Antony H., Wiegmans A. P., Wei M. Q., Chernoff Y. O., Khanna K. K., and Munn A. L. (2012) Potential roles for prions and protein-only inheritance in cancer. Cancer Metastasis Rev. 31, 1–19 10.1007/s10555-011-9325-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mizejewski G. J. (2017) Breast cancer and amyloid bodies: is there a role for amyloidosis in cancer-cell dormancy? Breast Cancer 9, 287–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Takagi K., Ito S., Miyazaki T., Miki Y., Shibahara Y., Ishida T., Watanabe M., Inoue S., Sasano H., and Suzuki T. (2013) Amyloid precursor protein in human breast cancer: an androgen-induced gene associated with cell proliferation. Cancer Sci. 104, 1532–1538 10.1111/cas.12239 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Mohell N., Alfredsson J., Fransson A., Uustalu M., Byström S., Gullbo J., Hallberg A., Bykov V. J. N., Björklund U., and Wiman K. G. (2015) APR-246 overcomes resistance to cisplatin and doxorubicin in ovarian cancer cells. Cell Death Dis. 6, e1794 10.1038/cddis.2015.143 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Vieira T. C., Cordeiro Y., Caughey B., and Silva J. L. (2014) Heparin binding confers prion stability and impairs its aggregation. FASEB J. 28, 2667–2676 10.1096/fj.13-246777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Chen Z., Chen J., Keshamouni V. G., and Kanapathipillai M. (2017) Polyarginine and its analogues inhibit p53 mutant aggregation and cancer cell proliferation in vitro. Biochem. Biophys. Res. Commun. 489, 130–134 10.1016/j.bbrc.2017.05.111 [DOI] [PubMed] [Google Scholar]
- 45. Soragni A., Janzen D. M., Johnson L. M., Lindgren A. G., Thai-Quynh Nguyen A., Tiourin E., Soriaga A. B., Lu J., Jiang L., Faull K. F., Pellegrini M., Memarzadeh S., and Eisenberg D. S. (2016) A designed inhibitor of p53 aggregation rescues p53 tumor suppression in ovarian carcinomas. Cancer Cell 29, 90–103 10.1016/j.ccell.2015.12.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Herzog G., Shmueli M. D., Levy L., Engel L., Gazit E., Klärner F. G., Schrader T., Bitan G., and Segal D. (2015) The Lys-specific molecular tweezer, CLR01, modulates aggregation of the mutant p53 DNA binding domain and inhibits its toxicity. Biochemistry 54, 3729–3738 10.1021/bi501092p [DOI] [PubMed] [Google Scholar]
- 47. Wilcken R., Wang G., Boeckler F. M., and Fersht A. R. (2012) Kinetic mechanism of p53 oncogenic mutant aggregation and its inhibition. Proc. Natl. Acad. Sci. U.S.A. 109, 13584–13589 10.1073/pnas.1211550109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Silva J. L., Cino E. A., Soares I. N., Ferreira V. F., and A. P. de Oliveira G. (2018) Targeting the Prion-like aggregation of mutant p53 to combat cancer. Acc. Chem. Res. 51, 181–190 10.1021/acs.accounts.7b00473 [DOI] [PubMed] [Google Scholar]
- 49. Wang G., and Fersht A. R. (2017) Multisite aggregation of p53 and implications for drug rescue. Proc. Natl. Acad. Sci. 10.1073/pnas.1700308114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Rökaeus N., Shen J., Eckhardt I., Bykov V. J. N., Wiman K. G., and Wilhelm M. T. (2010) PRIMA-1 MET /APR-246 targets mutant forms of p53 family members p63 and p73. Oncogene 29, 6442–6451 10.1038/onc.2010.382 [DOI] [PubMed] [Google Scholar]
- 51. Mills A. A., Zheng B., Wang X. J., Vogel H., Roop D. R., and Bradley A. (1999) p63 is a p53 homologue required for limb and epidermal morphogenesis. Nature 398, 708–713 10.1038/19531 [DOI] [PubMed] [Google Scholar]
- 52. Pozniak C. D., Radinovic S., Yang A., McKeon F., Kaplan D. R., and Miller F. D. (2000) An anti-apoptotic role for the p53 family member, p73, during developmental neuron death. Science 289, 304–306 10.1126/science.289.5477.304 [DOI] [PubMed] [Google Scholar]
- 53. Grob T. J., Novak U., Maisse C., Barcaroli D., Lüthi A. U., Pirnia F., Hügli B., Graber H. U., De Laurenzi V., Fey M. F., Melino G., and Tobler A. (2001) Human ΔNp73 regulates a dominant negative feedback loop for TAp73 and p53. Cell Death Differ. 8, 1213–1223 10.1038/sj.cdd.4400962 [DOI] [PubMed] [Google Scholar]
- 54. Buhlmann S., and Pützer B. M. (2008) DNp73 a matter of cancer: mechanisms and clinical implications. Biochim. Biophys. Acta 1785, 207–216 10.1016/j.bbcan.2008.01.002 [DOI] [PubMed] [Google Scholar]
- 55. Bykov V. J. N., Selivanova G., and Wiman K. G. (2003) Small molecules that reactivate mutant p53. Eur. J. Cancer 39, 1828–1834 10.1016/S0959-8049(03)00454-4 [DOI] [PubMed] [Google Scholar]
- 56. Liu D. S., Duong C. P., Haupt S., Montgomery K. G., House C. M., Azar W. J., Pearson H. B., Fisher O. M., Read M., Guerra G. R., Haupt Y., Cullinane C., Wiman K. G., Abrahmsen L., Phillips W. A., and Clemons N. J. (2017) Inhibiting the system xC-/glutathione axis selectively targets cancers with mutant-p53 accumulation. Nat. Commun. 8, 14844 10.1038/ncomms14844 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Kaar J. L., Basse N., Joerger A. C., Stephens E., Rutherford T. J., and Fersht A. R. (2010) Stabilization of mutant p53 via alkylation of cysteines and effects on DNA binding. Protein Sci. 19, 2267–2278 10.1002/pro.507 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Chemes L. B., Noval M. G., Sánchez I. E., and de Prat-Gay G. (2013) Folding of a cyclin box: linking multitarget binding to marginal stability, oligomerization, and aggregation of the retinoblastoma tumor suppressor ab pocket domain. J. Biol. Chem. 288, 18923–18938 10.1074/jbc.M113.467316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Kirilyuk A., Shimoji M., Catania J., Sahu G., Pattabiraman N., Giordano A., Albanese C., Mocchetti I., Toretsky J. A., Uversky V. N., and Avantaggiati M. L. (2012) An intrinsically disordered region of the acetyltransferase p300 with Similarity to prion-like domains plays a role in aggregation. PLoS ONE 7, e48243 10.1371/journal.pone.0048243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Latonen L., Moore H. M., Bai B., Jäämaa S., and Laiho M. (2011) Proteasome inhibitors induce nucleolar aggregation of proteasome target proteins and polyadenylated RNA by altering ubiquitin availability. Oncogene 30, 790–805 10.1038/onc.2010.469 [DOI] [PubMed] [Google Scholar]
- 61. Piantino C. B., Reis S. T., Viana N. I., Silva I. A., Morais D. R., Antunes A. A., Dip N., Srougi M., and Leite K. R. (2013) Prima-1 induces apoptosis in bladder cancer cell lines by activating p53. Clinics 68, 297–303 10.6061/clinics/2013(03)OA03 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62. Wiech M., Olszewski M. B., Tracz-Gaszewska Z., Wawrzynow B., Zylicz M., and Zylicz A. (2012) Molecular mechanism of mutant p53 stabilization: the role of HSP70 and MDM2. PLoS ONE 7, e51426 10.1371/journal.pone.0051426 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63. Liang Y., Besch-Williford C., and Hyder S. M. (2009) PRIMA-1 inhibits growth of breast cancer cells by re-activating mutant p53 protein. Int. J. Oncol. 35, 1015–1023 [DOI] [PubMed] [Google Scholar]
- 64. Synnott N. C., Murray A., McGowan P. M., Kiely M., Kiely P. A., O'Donovan N., O'Connor D. P., Gallagher W. M., Crown J., and Duffy M. J. (2017) Mutant p53: a novel target for the treatment of patients with triple-negative breast cancer? Int. J. Cancer. 140, 234–246 10.1002/ijc.30425 [DOI] [PubMed] [Google Scholar]
- 65. Ghosh S., Salot S., Sengupta S., Navalkar A., Ghosh D., Jacob R., Das S., Kumar R., Jha N. N., Sahay S., Mehra S., Mohite G. M., Ghosh S. K., Kombrabail M., Krishnamoorthy G., et al. (2017) P53 amyloid formation leading to its loss of function: implications in cancer pathogenesis. Cell Death Differ. 24, 1784–1798 10.1038/cdd.2017.105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Ishimaru D., Ano Bom A. P., Lima L. M., Quesado P. A., Oyama M. F., de Moura Gallo C. V., Cordeiro Y., and Silva J. L. (2009) Cognate DNA stabilizes the tumor suppressor p53 and prevents misfolding and aggregation. Biochemistry 48, 6126–6135 10.1021/bi9003028 [DOI] [PubMed] [Google Scholar]
- 67. Lowry O. H., Rosebrough N. J., Farr A. L., and Randall R. J. (1951) Protein measurement with the folin-phenol reagent. J. Biol. Chem. 193, 265–275 [PubMed] [Google Scholar]