Selenium is an essential element for life, with Se(IV) reduction a key step in its biogeochemical cycle. This report identifies for the first time a dissimilatory Se(IV) reductase, Srr, from a known selenite-respiring bacterium, the haloalkalophilic Bacillus selenitireducens strain MLS10. The work extends the versatility of the complex iron-sulfur molybdoenzyme (CISM) superfamily in electron transfer involving chalcogen substrates with different redox potentials. Further, it underscores the importance of biochemical and enzymological approaches in establishing the functionality of these enzymes.
KEYWORDS: complex iron-sulfur molybdoenzyme superfamily, dissimilatory selenite reductase, molybdoenzyme
ABSTRACT
The putative respiratory selenite [Se(IV)] reductase (Srr) from Bacillus selenitireducens MLS10 has been identified through a polyphasic approach involving genomics, proteomics, and enzymology. Nondenaturing gel assays were used to identify Srr in cell fractions, and the active band was shown to contain a single protein of 80 kDa. The protein was identified through liquid chromatography-tandem mass spectrometry (LC-MS/MS) as a homolog of the catalytic subunit of polysulfide reductase (PsrA). It was found to be encoded as part of an operon that contains six genes that we designated srrE, srrA, srrB, srrC, srrD, and srrF. SrrA is the catalytic subunit (80 kDa), with a twin-arginine translocation (TAT) leader sequence indicative of a periplasmic protein and one putative 4Fe-4S binding site. SrrB is a small subunit (17 kDa) with four putative 4Fe-4S binding sites, SrrC (43 kDa) is an anchoring subunit, and SrrD (24 kDa) is a chaperon protein. Both SrrE (38 kDa) and SrrF (45 kDa) were annotated as rhodanese domain-containing proteins. Phylogenetic analysis revealed that SrrA belonged to the PsrA/PhsA clade but that it did not define a distinct subgroup, based on the putative homologs that were subsequently identified from other known selenite-respiring bacteria (e.g., Desulfurispirillum indicum and Pyrobaculum aerophilum). The enzyme appeared to be specific for Se(IV), showing no activity with selenate, arsenate, or thiosulfate, with a Km of 145 ± 53 μM, a Vmax of 23 ± 2.5 μM min−1, and a kcat of 23 ± 2.68 s−1. These results further our understanding of the mechanisms of selenium biotransformation and its biogeochemical cycle.
IMPORTANCE Selenium is an essential element for life, with Se(IV) reduction a key step in its biogeochemical cycle. This report identifies for the first time a dissimilatory Se(IV) reductase, Srr, from a known selenite-respiring bacterium, the haloalkalophilic Bacillus selenitireducens strain MLS10. The work extends the versatility of the complex iron-sulfur molybdoenzyme (CISM) superfamily in electron transfer involving chalcogen substrates with different redox potentials. Further, it underscores the importance of biochemical and enzymological approaches in establishing the functionality of these enzymes.
INTRODUCTION
The assimilatory reduction and dissimilatory reduction of the selenium oxyanions selenate [Se(VI)] and selenite [Se(IV)] are key processes in the selenium biogeochemical cycle. The assimilatory reduction of the selenium oxyanion Se(IV) to selenide is integral to the synthesis of the 21st amino acid, selenocysteine (1–4), and the tRNA nucleoside 2-selenouridine (5, 6). The assimilatory reduction of Se(IV) for selenocysteine synthesis occurs in all three domains of life, but the assimilatory reduction of Se(IV) for 2-selenouridine synthesis occurs exclusively in the Bacteria and Archaea. The second major component of the selenium biogeochemical cycle is the dissimilatory reduction of the selenium oxyanions Se(VI) and Se(IV) during anaerobic respiration (1, 7). Se(VI)- and Se(IV)-respiring microorganisms have been isolated from both the bacteria and archaea, including species from the Proteobacteria (8–12), the Firmicutes (13–16), the Chrysiogenetes (17), and the Crenarchaeota (18, 19) phyla. The phylogenetic breadth represented by selenium-respiring microorganisms suggests that selenium respiration is widespread.
A stunning array of bacteria are capable of rapidly and efficiently detoxifying up to millimolar quantities of both Se(VI) and Se(IV) aerobically and anaerobically via reduction to red elemental selenium [Se(0)] via several different mechanisms (7). The phenomenon has also been observed in the Archaea (20) and the Eukarya (21, 22). This poses a significant challenge in determining whether an anaerobically grown organism reducing Se(VI) or Se(IV) is coupling this reduction with energy conservation or is merely doing this as a detoxification strategy. An additional possibility, heretofore not considered in the literature, is that Se(VI) and Se(IV) could also serve as electron sinks during fermentative growth. There are examples of organisms known to exploit elemental sulfur and polysulfides as electron sinks during fermentation (23, 24), and selenium is a stronger electrophile than sulfur (25). Thus, this possibility requires more experimental evidence to confirm whether an organism is capable of respiring oxyanions of selenium [i.e., demonstrating that Se(VI) or Se(IV) reduction is being stoichiometrically coupled to the oxidation of an electron donor].
Of the two respiratory processes, Se(VI) respiration has been studied more extensively. During anaerobic respiration on Se(VI), Se(VI) is reduced to Se(IV) in a two-electron transfer reaction (14, 26), with the Se(IV) subsequently reduced to red Se(0), presumably as a detoxification mechanism. The periplasmic Se(VI) reductase (Ser) of Thauera selenatis has been purified and fully characterized, including its genes, operon structure, and crystal structure (27–32). Se(VI) reductases have also been characterized from other bacteria, including Bacillus selenatarsenatis (33–35) and Enterobacter cloacae SLD1a-1, Escherichia coli K-12, and Citrobacter freundii strain RLS1 (36–40). These reports underscore the fact that multiple biochemical pathways and enzymes are used during Se(VI) respiration. While the known Se(VI) reductases are part of the complex iron-sulfur molybdoenzyme (CISM) superfamily (41), they differ enough to suggest that Se(VI) respiration has evolved independently multiple times in the Bacteria. Whether this is also the case for dissimilatory Se(IV) reductase was a focus of the current study.
Precious little is known about the molecular mechanisms of Se(IV) respiration, and it is uncertain whether any of the biochemical mechanisms of Se(IV) reduction identified in the literature are involved in Se(IV) respiration or other physiological processes. Genetic knockouts identified the fumarate reductase (encoded by the fccA gene) as a putative Se(IV) reductase in Shewanella oneidensis MR-1; however, only 60% of the activity was lost, and the cells were still capable of Se(IV) reduction (42). This suggests either that the fumarate reductase of S. oneidensis is involved in mediating Se(IV) reduction but is not actually the Se(IV) reductase or that the Se(IV) reduction phenotype can be partially rescued by other, unknown enzymes. Regardless, the reduction of Se(IV) via the fumarate reductase does not appear to be involved in Se(IV) respiration, given that the same mechanism has been implicated aerobically in Enterobacter cloacae Z0206 (43). A periplasmic protein possessing Se(IV) reductase activity has been purified from ER-Te-48, a strain of Shewanella frigidimarina isolated from thermal vent tube worms (44). The enzyme has been proposed to be involved in Se(IV) respiration in this organism, but its low affinity for Se(IV), with an apparent Km of 12.1 mM, makes this doubtful (44).
Current efforts to identify a respiratory Se(IV) reductase have been limited to organisms that have not been shown to be capable of using Se(IV) as a terminal electron acceptor in anaerobic respiration (45, 46). Thus far, only four microbes have been shown definitively (i.e., using mass balance) to couple oxidation of an electron donor to reduction of Se(IV) to Se(0) during a four-electron transfer reaction: Bacillus selenitireducens MLS10 (14), Pyrobaculum aerophilum IM2 (18), Desulfurispirillum indicum S5 (17), and Bacillus beveridgei MLTeJB (47). Se(IV) respiration in these organisms, in contrast to Se(VI) respiration and Se(IV) detoxification, results in the formation of black Se(0), with red Se(0) appearing as an intermediate that transitions to black Se(0) as growth approaches the stationary phase. In MLS10, this black Se(0) has been shown to form as a result of respiratory growth on red Se(0) (48), producing selenide. It is not known, however, if the black Se(0) allotropes in the other three taxa are also by-products of Se(0) respiration.
As a model for a Se(IV)-respiring organism, B. selenitireducens MLS10 is a well-suited candidate. MLS10 is a Gram-positive haloalkaliphilic firmicute isolated from Mono Lake (14). MLS10 is the first bacterium shown to be capable of Se(IV) respiration, grows readily on Se(IV) in large batch cultures, and has previously been used to provide insights into the physiology and biochemistry of both selenium and arsenic respiration (48–52). We report here on the identification and initial characterization of a putative respiratory Se(IV) reductase (Srr) from MLS10 using genomics, proteomics, and enzymology. This work provides our first insights into the biochemistry of Se(IV) respiration from an organism definitively shown to utilize selenite as a terminal electron acceptor, and it extends the catalytic versatility of a well-known family of enzymes within the larger CISM superfamily.
RESULTS
Identification of a putative Srr using nondenaturing gel enzyme assays.
We performed nondenaturing gel enzyme assays using reduced methyl viologen as an electron donor and Se(IV) as the electron acceptor to identify candidates for the putative Srr. Periplasmic, particulate (i.e., membrane) and cytoplasmic protein fractions from MLS10 cells grown with Se(IV) were tested. All three fractions showed activity at the same location in the gel, but the bulk of the activity was in the periplasmic fraction (as determined by protein quantification and gel staining). The band of activity from the periplasmic fraction was excised and subjected to both SDS-PAGE and liquid chromatography-tandem mass spectrometry (LC-MS/MS) to determine the identities of the eluted proteins (Fig. 1). SDS-polyacrylamide gels indicated that the active band consisted primarily of an approximately 80-kDa protein (Fig. 1, lane C). The LC-MS/MS results revealed that the active band that eluted directly from the nondenaturing gel contained 3 proteins identified with high confidence as a trimethylamine-N-oxide (TMAO) reductase, a 4Fe-4S ferredoxin iron-sulfur protein, and a rhodanese domain-containing protein corresponding to proteins encoded by the genes Bsel_1476, Bsel_1477, and Bsel_1475, respectively, from the MLS10 genome as annotated by the Joint Genome Institute (Table 1).
FIG 1.
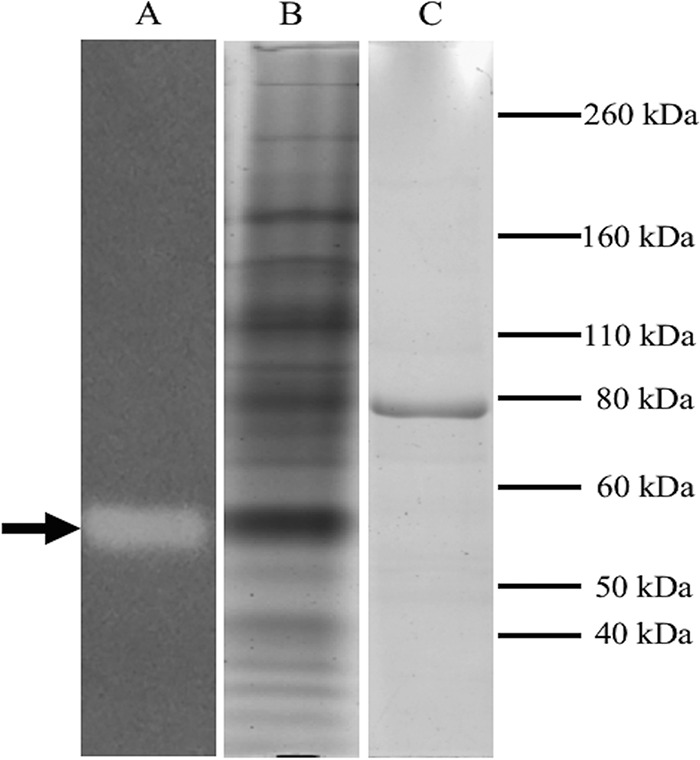
Identification of the respiratory Se(IV) reductase (Srr) in the periplasmic fraction of Bacillus selenitireducens strain MLS10. Lane A, nondenaturing gel developed for Se(IV) reductase activity with reduced methyl viologen. The clearing indicates Srr activity (arrow). Lane B, lane A after staining with Coomassie brilliant blue, showing a protein band at the same location as the Srr activity. Lane C, SDS-polyacrylamide gel of the excised band of Srr activity from lane A visualized by staining with Coomassie brilliant blue.
TABLE 1.
LC-MS/MS results for proteins eluted from nondenaturing and SDS-polyacrylamide gels
| Sample | Locus tag, GenBank accession no. | Protein annotation | Score | No. of unique peptides |
|---|---|---|---|---|
| Se(IV) reductase active band eluted from nondenaturing gel shown in Fig. 1A | Bsel_1476, WP_013172412 | Trimethylamine-N-oxide reductase | 256.3 | 16 |
| Bsel_1477, WP_013172413 | 4Fe-4S ferredoxin iron-sulfur binding domain protein | 30.7 | 5 | |
| Bsel_1475, WP_013172411 | Rhodanese domain protein | 19.3 | 3 | |
| 80-kDa protein from SDS-PAGE shown in Fig. 1B | Bsel_1476, WP_013172412 | Trimethylamine-N-oxide reductase | 191.7 | 21 |
| 80-kDa band from second anion-exchange column shown in Fig. 2 | Bsel_1476, WP_013172412 | Trimethylamine-N-oxide reductase | 139.8 | 15 |
Kinetics of Srr.
Efforts to measure the kinetics of Srr were complicated by several factors. We found that Se(IV) by itself could oxidize methyl viologen, benzyl viologen, flavin adenine dinucleotide (FAD), flavin mononucleotide (FMN), and NADPH at pH 7.0 and below. Methyl viologen, however, was stable at pH 7.5 and above. Thus, we were able to test Se(IV) reduction over a pH range between 7.5 and 10. The enzyme, however, showed no activity above pH 8.0. Further, any trace of Se(0) would also result in spurious methyl viologen oxidation, as was the case for the whole-cell and cell lysate fractions. Hence, a complete purification table was not possible. Nonetheless, starting with the periplasmic fraction, we were able to obtain fractions highly enriched in Srr (Fig. 2) after two sequential steps of anion-exchange chromatography, as determined by LC-MS/MS analysis of the 80-kDa protein (Table 1). The enzyme was found to have a high affinity for Se(IV), with an apparent Km of 145 ± 53 μM, a Vmax of 23 ± 2.5 μM min−1, and a kcat of 23 ± 2.68 s−1 (Fig. 3). It also appeared to be specific to Se(IV), as no activity was observed using arsenate, selenate, or thiosulfate as an electron acceptor in the assay. Inductively coupled plasma MS (ICP-MS) found evidence for both Mo and Fe, with a ratio of about 1 mol Mo to 10 mol Fe, suggesting that metal loss had likely occurred during purification.
FIG 2.
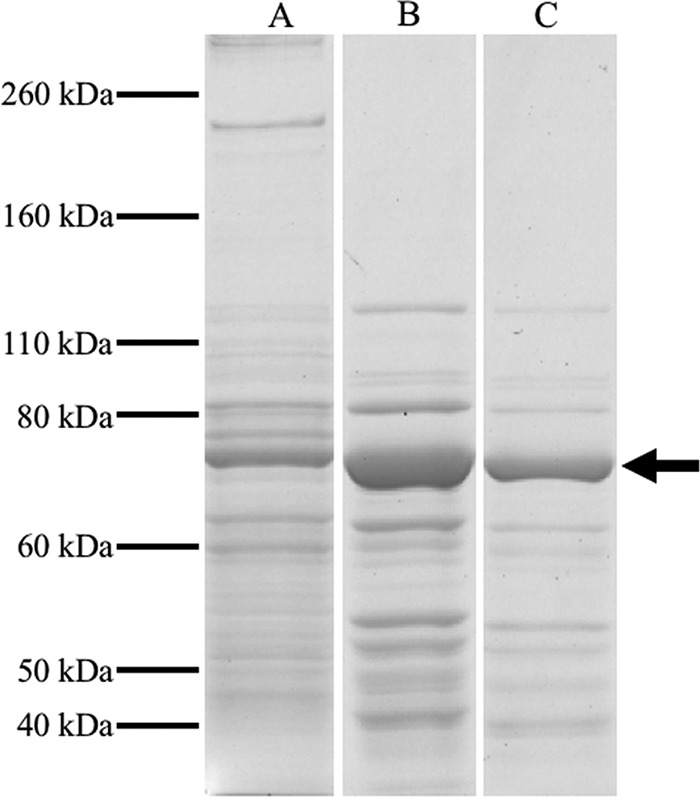
SDS-polyacrylamide gels of the Bacillus selenitireducens strain MLS10 periplasmic fraction and Srr-active fractions from ion-exchange columns. Lane A, periplasmic fraction. Lane B, Srr-active fraction eluted from the first anion-exchange column loaded with the periplasmic fraction. Lane C, Srr-active fraction obtained from a second anion-exchange column. The 80-kDa band (arrow) corresponds to the putative SrrA.
FIG 3.
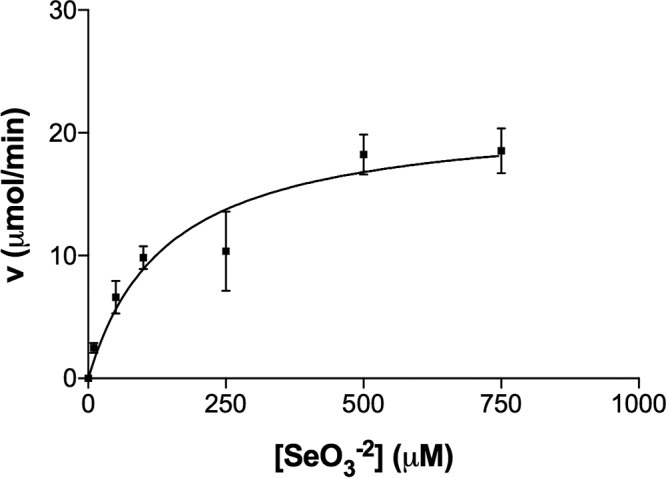
Kinetics of Srr from Bacillus selenitireducens strain MLS10. The kinetics of the enzyme are expressed in terms of methyl viologen oxidized.
Sequence analysis of Srr.
The gene sequences inferred from the LC-MS/MS data for the three subunits were used to locate the putative srr operon in the genome of B. selenitireducens. Annotated as Bsel_1475 to Bsel_1480, it was found to contain six genes, srrE, srrA, srrB, srrC, srrD, and srrF, with SrrA the catalytic subunit (80 kDa), SrrB a small electron transfer subunit (17.7 kDa), SrrC (43 kDa) an anchoring subunit, and SrrD (24 kDa) a chaperon protein (Fig. 4). Both SrrE (38 kDa) and SrrF (45.6 kDa) were annotated as rhodanese domain-containing proteins (see Fig. S1 in the supplemental material). Features of SrrA include a twin-arginine translocation (TAT) motif for export to the periplasm (confirming the original observation that Srr is periplasmic), a motif associated with a 4Fe-4S cluster, and a molybdopterin guanine dinucleotide coordination motif coordinated by a cysteine residue (Fig. S1A). The annotation by the Joint Genome Institute (who sequenced the genome) of SrrA as a TMAO reductase, however, is most likely incorrect for several reasons. First, the molybdopterin cofactor of TMAO reductase is coordinated by a serine residue; second, the TMAO reductase catalytic subunit lacks a 4Fe-4S cluster (27). The primary sequence of the annotated 4Fe-4S ferredoxin iron-sulfur binding domain protein (SrrB) suggests that this protein contains four 4Fe-4S clusters (Fig. S1B). SrrC is a homolog of the polysulfide reductase NrfD membrane-anchoring subunit and has no apparent binding motifs for either a c- or b-type heme. SrrD, encoded by Bsel_1479, is most likely a homolog of the NarJ protein, which functions to facilitate incorporation of the molybdopterin cofactor into the active site of the respiratory nitrate reductase of E. coli (53). The PROSITE tool (54) identified two structural motifs in the rhodanese domain proteins annotated by Bsel_1475 (SrrE) and Bsel_1480 (SrrF). Both SrrE and SrrF contain a lipoprotein motif for attachment to the cytoplasmic membrane at the N termini of the proteins and two rhodanese domains, with the rhodanese domain closest to the N terminus containing an active-site cysteine residue (Fig. S1E and F).
FIG 4.
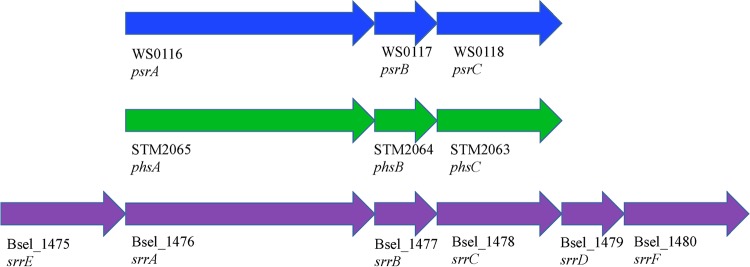
Operon organization of the Wolinella succinogenes (WS) polysulfide reductase, Salmonella enterica serovar Typhimurium (STM) thiosulfate reductase, and putative Bacillus selenitireducens MLS10 (Bsel) selenite reductase. The genes psrA, phsA, and srrA encode the catalytic subunits of the Psr/Phs/Srr reductase complex; psrB, phsB, and srrB encode the electron transfer subunits; and psrC, phsC, and srrC encode the membrane anchor subunits. The gene srrD encodes a protein with strong sequence homology to the NarJ protein, which is involved in facilitating the incorporation of the molybdopterin cofactor into the catalytic subunit NarG. The genes srrE and srrF encode rhodanese domain-containing proteins. The arrows refer to the direction of transcription.
Phylogenetic analysis of SrrA.
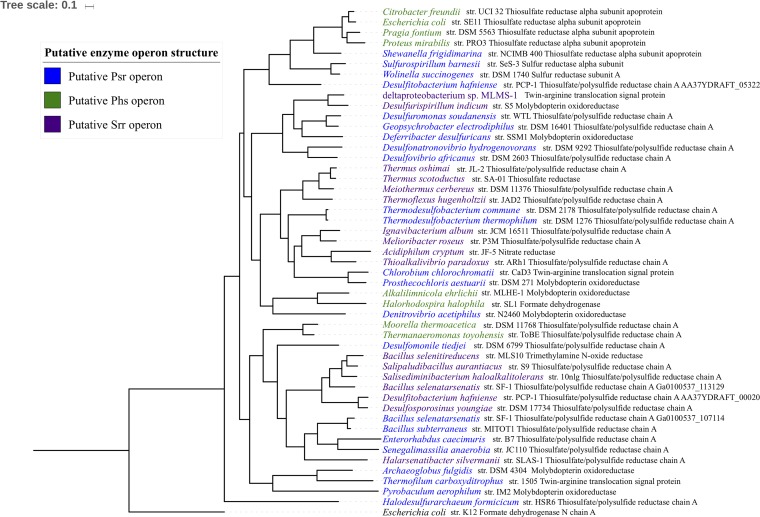
In order to place SrrA more accurately within the CISM superfamily, Bayesian phylogenies of SrrA homologs were constructed using the catalytic subunit of formate dehydrogenase N from E. coli as the outgroup (Fig. 5). The ProtTest tool (55) and MrBayes (56) program indicated the appropriate amino acid substitution and rate variation models. The ProtTest tool showed that the LG amino acid substitution model (57) and the WAG amino acid substitution model (58) (each with a gamma distribution [59] or with a gamma distribution with a portion of invariant sites) best described the observed differences in the amino acid sequences of the SrrA homologs. MrBayes also concurred that the WAG amino acid substitution model best fit our data with a confidence of 1.0.
FIG 5.
Bayesian phylogenetic tree of SrrA from MLS10 and related homologs. The phylogenetic analysis was done using MrBayes, using the LG amino acid substitution model with a gamma distribution. The protein annotation after each species name is the annotation assigned by the Joint Genome Institute Integrated Microbial Genomes database. All but two bootstrap values were 100 (as shown). The tree scale bar refers to 0.1 amino acid substitution per site.
All four models (i.e., LG with a gamma distribution, LG with a gamma distribution with a portion of invariant sites, WAG with a gamma distribution, and WAG with a gamma distribution with a portion of invariant sites), strongly supported the positioning of SrrA within the polysulfide/thiosulfate reductase (Psr/Phs) family of CISM molybdoenzymes. They also yielded identical topologies, with each node showing robust support (posterior probabilities greater than or equal to 0.9). When the Tracer program (60) was used to assess convergence in the log likelihood of Markov chain Monte Carlo (MCMC) runs and compare the likelihoods of the four evolutionary models, it suggested that the LG amino acid substitution model best fit our data, with either a gamma or a gamma with a proportion of invariant sites being equally likely (Fig. 5).
The Psr/Phs family mediates polysulfide and thiosulfate respiration in bacteria, both of which are crucial components of the sulfur biogeochemical cycle (53). The involvement of Psr in polysulfide respiration has been studied most extensively in Wolinella succinogenes (61–63), whereas Salmonella enterica serovar Typhimurium has been used to elucidate the involvement of Phs in thiosulfate respiration (64, 65). The Psr operon of W. succinogenes encodes a catalytic subunit (PsrA), a four-4Fe-4S electron transfer subunit (PsrB), and a membrane anchor subunit that lacks heme groups (PsrC). The Phs operon of S. enterica serovar Typhimurium has an identical organization, with the exception that the membrane anchor subunit (PhsC) harbors two heme b groups. PsrA/PhsA catalytic subunits and the PsrB/PhsB electron transfer subunits are closely related, while PsrC is closely related to the membrane anchor of nitrite reductases and PhsC is closely related to the membrane anchor of formate dehydrogenases (27). Thus, genome structure was used to suggest other potential Srr candidates. In examining the genomic context of srrA homologs, three operon types were apparent. Several SrrA homologs were found to be encoded in putative Psr operons (Fig. 4). Other SrrA homologs were encoded in putative Phs operons (Fig. 5). The third operon organization encoded a catalytic subunit, a four-4Fe-4S electron transfer subunit, a membrane anchor subunit that lacks heme groups, and one or two additional rhodanese domain proteins. We propose that this operon organization is typical for the Srr operon. When these operon organizations were mapped onto the phylogenies, we found that the Psr, Phs, and Srr operons did not form monophyletic clades. This suggests that the differences in the physiological functions of these enzymes do not stem from evolutionarily conserved differences in the primary sequences of the putative PsrA/PhsA/SrrA homologs.
DISCUSSION
Our work substantially advances our knowledge of the biology of selenium by providing the first report of a putative respiratory Se(IV) reductase in a bona fide Se(IV)-respiring organism. It is especially significant that we have strong evidence that SrrA is a member of the PsrA/PhsA clade of the CISM superfamily. The PsrA/PhsA clade of the CISM superfamily has representatives from both Bacteria and Archaea and is deeply branched in phylogenies of CISM family enzymes, suggesting that this protein family is ancient (27). These enzymes mediate crucial reactions involving transient sulfur species in the sulfur biogeochemical cycle. While little is known about the environmental distribution and availability of polysulfide and thiosulfate, a number of reports suggest that polysulfide respiration is an important component of the sulfur biogeochemical cycle in a number of environments, including soda lakes and oceans (66–68). Our identification of SrrA as a member of the PsrA/PhsA clade thus not only expands our knowledge of the catalytic diversity of this clade but also implicates these enzymes in a crucial step of the selenium biogeochemical cycle.
Phylogenetic evidence suggests that the CISM oxidases and terminal reductases for several toxic oxyanions, including the T. selenatis selenate reductase (SerA), the Ideonella dechloratans chlorate reductase (ClrA), and the anaerobic arsenite oxidase (ArxA) of Alkalilimnicola ehrlichii, diverged from more deeply branched clades within the CISM superfamily, namely, the respiratory nitrate reductase (NarG) and arsenate reductase (ArrA) families (1, 69). Further, it appears that SrrA defines neither a monophyletic group nor a distinct branch of the PsrA/PhsA clade. This is very surprising given that members of the Psr/Phs enzyme family mediate electron transfer of substrates with remarkably different redox potentials. Both polysulfide and thiosulfate are highly reduced substrates, with redox potentials of −260 mV and −402 mV, respectively (23, 70). Se(IV), however, has a redox potential of 216 mV (7). While it is possible that the oxidation state of the Srr involves the rhodanese domain-containing proteins, these proteins are found in other enzymes of the clade. Alternatively, the modulation could be due to features unique to the catalytic pocket and the ligand binding site.
The affinity of SrrA for Se(IV) is much higher than that reported for other selenium oxidoreductases. Previously reported Km values have ranged from high micromolar to millimolar (44). A curious feature shared by this enzyme family is that the electron transfer subunit does not appear to be necessary for enzyme activity. This contrasts strongly with previous work on CISM superfamily enzymes, where the electron transport subunit is necessary for activity (28, 52, 71). The observable pH optimum for the enzyme was also unexpected. As a haloalkaliphile, MLS10 grows at a pH of 9.8, and the previously published purification of the MLS10 respiratory arsenate reductase found that that enzyme had an optimal pH of 9.5 (52). No Se(IV) reductase activity was observed above pH 8.0, despite the fact that Srr should also have a periplasmic topology (e.g., presence of a TAT motif). More puzzling still was the apparent specificity of SrrA for Se(IV). Mono Lake, where MLS10 was isolated, is particularly rich in arsenic (72), with lake water containing 200 μM arsenic. This is decidedly not the case for selenium, as previous work estimated the concentration of selenium in Mono Lake water at 38 nM (47). Further, the arsenate reductase (Arr) from MLS10 can use Se(VI) and Se(IV) to oxidize reduced methyl viologen in vitro (52). It seems that MLS10 cells express SrrA specifically for Se(IV) respiration, despite the fact that Se(IV) is not readily available as an energy source in the environment from which it was isolated. Regardless, both Se(IV) and Se(VI) are rich sources of energy compared to many other terminal electron acceptors (73), and this may offer an important selective advantage to microorganisms that can respire selenium oxyanions, even in environments not particularly rich in selenium.
Neither SrrE nor SrrF appears to be essential for Se(IV) respiration in vitro, yet our nondenaturing in-gel enzyme assays suggest that SrrE at least functions in vivo with SrrA and SrrB to reduce Se(IV) to Se(0). The use of rhodanese domain-containing proteins during anaerobic respiration has previously been documented in W. succinogenes during polysulfide respiration (74). The Sud protein, while not part of the Psr operon, is a single-active-site rhodanese domain-containing protein that increases the affinity of PsrA in W. succinogenes by nearly an order of magnitude when W. succinogenes is grown on polysulfide. Curiously, there appears to be no relation between the Sud protein and SrrE and SrrF, as both Srr rhodanese proteins contain two active sites rather than one, and BLAST searches for the Sud protein in the MLS10 genome did not identify any homologs. Nonetheless, it is possible that SrrE and SrrF perform an analogous role in MLS10 in increasing the affinity of Srr for Se(IV). It should be noted that a putative Srr operon has been reported previously in Desulfitobacterium hafniense TCE1; it encodes a protein homologous to the SrrF from MLS10 and likely is involved in sulfide detoxification (75). Curiously, the authors could not stimulate expression of any other components of the putative Srr operon on any growth substrates tested. While the authors did not test for expression of this operon when growing TCE1 on Se(IV), previous attempts to cultivate D. hafniense strains on Se(IV) have not been successful (76).
This report is part of a growing body of literature that emphasizes the extraordinary catalytic diversity of CISM superfamily enzymes (36). It extends these findings by offering evidence that dissimilatory Se(IV) reduction in B. selenitireducens is mediated by a Psr/Phs homolog. Although gene knockouts will provide definitive proof that Srr is the respiratory Se(IV) reductase in MLS10, this work underscores the importance of, establishing that reduction of selenium is coupled to energy conservation. Our phylogenetic analyses suggest that there may be more species capable of respiring Se(IV); thus, including it as a possible electron acceptor when characterizing new isolates is warranted. It remains to be seen if Srr-mediated Se(IV) respiration is conserved in other Se(IV)-respiring organisms or if multiple pathways for Se(IV) respiration have evolved. Lastly, it is appears that Srr may mediate the reduction of Se(IV) to Se(0), an overall four-electron transfer reaction. This would be unique among CISM superfamily members, as they typically catalyze oxygen atom transfers involving two electrons (41). Whether an intermediate step involving a transient selenium species, two cycles of reduction, or additional components are involved remains to be determined.
MATERIALS AND METHODS
Cultivation and fractionation of MLS10 cells and determination of protein content.
Cells of MLS10 were cultivated in the medium described by Switzer Blum et al. (14). The medium consisted of (grams liter−1) (NH4)SO4 (0.1), MgSO4·7H2O (0.025), K2HPO4 (0.15), KH2PO4 (0.08), NaCl (40), Na2CO3 (10.6), NaHCO3 (4.2), and yeast extract (0.2) with 5 ml liter−1 of SL-10 trace element solution. The pH of the medium was adjusted to 9.8, and the medium was made anaerobic by degassing under a 100% N2 atmosphere. The medium was amended with sodium lactate (10 mM) and sodium selenite (10 mM) as the electron donor and acceptor, respectively, and with cysteine-HCl (0.025%) as a reducing agent. Cells were cultivated in 2-liter Pyrex bottles with lids modified to accommodate black rubber septa to prevent O2 contamination. The cells were harvested via centrifugation at 9,000 rpm for 20 min at 4°C as described previously (52). Periplasmic protein fractions were obtained by suspending the harvested cells in a buffer containing 50 mM Tris-HCl (pH 8.0), 5 mM EDTA, and 500 mM sucrose. The cells were treated with lysozyme (0.6 mg ml−1) and incubated at 37°C for 15 min and subsequently at 4°C for 30 min. The cells were centrifuged at 7,232 × g for 15 min using an Eppendorf 5810R refrigerated centrifuge (Eppendorf AG, Hamburg, Germany). The supernatant was the periplasmic fraction. The spheroplasts were suspended in a buffer containing 20 mM Tris-HCl (pH 8.0) and 1 mM EDTA and sonicated with a Microson XL-2000 sonicator (Misonix Incorporated, Farmingdale, NY) three times at 12 W. The spheroplasts were centrifuged at 7,232 × g for 15 min. The sonicated cells were treated with DNase (0.1 mg ml−1) for approximately 15 min and centrifuged at 7,232 × g for 15 min. The resulting cell lysate was ultracentrifuged (Optima XE-90; Beckman Coulter Inc., Brea, CA) at 100,000 × g for 1 h to obtain the cytoplasmic (the supernatant) and particulate fractions. The particulate fractions were suspended in the buffer of 20 mM Tris-HCl (pH 8.0) and 1 mM EDTA. The concentration of protein in the different fractions was determined using the Lowry method (77).
Nondenaturing gel enzyme assays for Se(IV) reductase activity.
Nondenaturing gels were made according to standard protocols (78), with the exception that CHAPS [3-(3-cholamidopropyl)dimethylammonio)-1-propanesulfonate] was used in lieu of SDS (sodium dodecyl sulfate). Nondenaturing gels consisted of an 8% acrylamide running gel and a 4% acrylamide stacking gel. Approximately 125 μg each of the periplasmic, particulate, and cytoplasmic fractions was incubated overnight in a nondenaturing 4× Laemmli buffer without both reducing agents and SDS and with 1% CHAPS. The nondenaturing gels were run for 6 h at 4°C and 60 V in a reservoir that contained a nondenaturing Tris-glycine buffer with 0.05% CHAPS substituted for 0.1% SDS.
The gel was transferred to a Bactron IV anaerobic chamber (Sheldon Manufacturing Inc., Cornelius, OR), and the nondenaturing gel enzyme assay was performed under anoxic conditions. The nondenaturing gel was incubated in a solution containing 10 mM methyl viologen reduced with 10 mM sodium dithionite, 20 mM Tris-HCl (pH 8.0), and 1 mM EDTA for several minutes. The gel was incubated in a solution containing 20 mM sodium selenite, 20 mM Tris-HCl (pH 8.0), and 1 mM EDTA until a band of Se(IV) reductase activity was observed. The nondenaturing gel was stained overnight in a solution of 0.1% Coomassie brilliant blue R-250, 40% methanol, and 10% glacial acetic acid and was destained the following day in a solution of 50% methanol and 10% glacial acetic acid. The destained nondenaturing gels were visualized using a GS-800 densitometer (Bio-Rad, Hercules, CA). Se(IV) reductase active bands were excised from the nondenaturing gels and soaked overnight in 4× Laemmli buffer. The Laemmli buffer supernatant was used in SDS-PAGE to evaluate the protein composition and estimate the molecular weights of the proteins. SDS-polyacrylamide gels were made following standard protocols (78) and run at 160 V for approximately 1.5 h.
Phylogenetic analysis of the MLS10 SrrA.
Phylogenetic trees were constructed using homologs of the MLS10 SrrA protein subunit obtained using the DELTA-BLAST tool (79). The formate dehydrogenase N alpha subunit of E. coli was selected as an outgroup (80). The genomic context of each putative MLS10 SrrA homolog was then evaluated using the Integrated Microbial Genomes tool (81). The homologs were aligned using the MUSCLE tool (82) available in the MEGA 7 program (83). The alignments were trimmed using the trimAl tool (84). The sequence alignments were trimmed such that all columns with gaps in more than 20% of the SrrA homolog sequences or columns with a similarity score below 0.001 were omitted, with the caveat that 60% of the columns were to be conserved. This resulted in a trimmed sequence alignment of 722 amino acids for phylogenetic analysis. The alignment was inspected manually to ensure that the putative 4Fe-4S sequence motif at the N termini of the SrrA homologs and the E. coli formate dehydrogenase N alpha subunit was aligned, as was the cysteine residue predicted to coordinate the molybdopterin guanine dinucleotide cofactor.
Bayesian phylogenies were constructed using the MrBayes 3.2.6 phylogenetic program (56). The phylogenetic analysis consisted of 2 parallel Metropolis-coupled Markov chain Monte Carlo (MCMC) analyses (85) with 4 chains each. Both the ProtTest software program (55) and MrBayes (56) were used to evaluate probable amino acid substitution models and rate variation models. The four most probable combinations of amino acid substitution and rate variation models were chosen from the ProtTest program, and the most probable amino acid substitution model was chosen using MrBayes by specifying a mixed amino acid substitution model prior to the analysis when analyzing the data using rate variation models with a gamma distribution and with a gamma distribution with a proportion of invariant sites. All tested models were run in triplicates and yielded identical topologies, with consistent levels of posterior probability support at each node. Convergence of the MCMC runs was tested using the Tracer 1.6.0 software program (60). The resulting trees were visualized with the iTOL program (86).
Enrichment of MLS10 SrrA.
Protein fractions highly enriched in Se(IV) reductase activity were obtained using a Toyopearl GigaCap DEAE 650M anion-exchange resin (Tosoh Bioscience, Tokyo, Japan) for anion-exchange chromatography. Chromatographic separation of proteins was performed using the Bio-Rad (Hercules, CA) BioLogic LP chromatography system. Periplasmic protein fractions were loaded onto the resin for the first anion-exchange chromatography run, and the proteins were eluted in a 20 mM Tris-HCl (pH 8.0) buffer using an NaCl gradient (0 to 500 mM NaCl). Elution of proteins from the column was monitored using a UV detector at 280 nm. Se(IV) reductase activity was assessed in a 100 mM phosphate (KH2PO4-NaOH) buffer (pH 7.5) with 500 μM methyl viologen and 500 μM sodium selenite. Assays were performed in 2-ml volumes with approximately 50 μg total protein, and the solution was made anaerobic by degassing in a 100% N2 atmosphere (oxygen was removed by passage through a heated copper oven). The reaction was initiated by adding sodium dithionite to a final concentration of 50 μM. Oxidation of methyl viologen was monitored at 600 nm using a T3 single-beam spectrophotometer (Persee, Auburn, CA). After screening of the first Se(IV) reductase active fractions, two fractions showing the most Se(IV) reductase activity were run on a smaller column using the identical anion-exchange resin. The fractions were diluted in an equal volume of 20 mM Tris-HCl–1 mM EDTA (pH 8.0) buffer to reduce the salt concentration. The NaCl gradient was identical to that in the first run, as was the procedure for screening Se(IV) reductase activity. The purity of the fractions demonstrating Se(IV) reductase activity was assessed using SDS-PAGE. SDS-polyacrylamide gels were made according to standard protocols (78). Running gels consisted of 8% acrylamide, and stacking gels consisted of 4% acrylamide. Approximately 20 μg total protein was suspended in 4× Laemmli sample buffer and then incubated at 90°C for 5 min. Gels were run at 160 V for approximately 1.5 h. The gels were stained overnight, destained, and visualized as described above for nondenaturing gels.
Kinetic analysis of SrrA.
The Km of the MLS10 SrrA was obtained using the Se(IV) reductase assay described above, with the exception that the concentration of Se(IV) was adjusted to include the following concentrations: 10 μM, 50 μM, 100 μM, 250 μM, 500 μM, and 750 μM. Each Se(IV) concentration was assessed in triplicate. All Se(IV) reductase assay mixtures contained approximately 150 μg total protein from enriched fractions, and reactions were initiated with the addition of sodium dithionite to a final concentration of 50 μM and monitored at 600 nm using a T3 single-beam spectrophotometer (Persee, Auburn, CA). Se(IV) reductase activity was determined by calculating the amount of methyl viologen oxidized (μM min−1) using the extinction coefficient for methyl viologen of 13 mM−1 cm−1 (52). The Km, Vmax, and kcat of SrrA were calculated with the Prism 7 (GraphPad Software, La Jolla, CA) statistical analysis software package using the Michaelis-Menten equation with a nonlinear regression fit. The kcat was calculated using the concentration of SrrA in the 2-ml cuvette as a constant. The substrate specificity of SrrA was assessed using the same assay but with a 500 μM concentration of the following alternative electron acceptors: arsenate, selenate, and thiosulfate.
UV-visible spectroscopy analysis of enriched fractions of SrrA.
Fractions enriched with SrrA were analyzed with a UV-1800 dual-beam spectrophotometer (Shimadzu Scientific Instruments, Columbia, MD). A 1-ml aliquot of the enriched fraction was analyzed in a cuvette over a wavelength range of 800 to 300 nm to obtain a spectrum of the enzyme in its oxidized state. A spectrum of the enzyme in its reduced was obtained by the addition of crystals of sodium dithionite. In both cases, 1 ml of blank Tris-HCl (pH 8.0) containing 20 mM Tris-HCl, 1 mM EDTA, and 500 mM NaCl was used as a control in the reference cuvette.
Analysis of metals associated with SrrA.
Approximately 3.6 mg total protein (∼800-μl volume) from active fractions after elution from the first anion-exchange column was filtered through a 0.22-μm PES filter (VWR, Bridgeport, NJ) to remove particles and diluted 1:100 in a 2% nitric acid solution. The samples were analyzed on a NexION X300 ICP-MS with S10 autosampler and the NexION 300× ICP-MS software (Perkin-Elmer, San Jose, CA). CPI International single-element standards were used to create multielement standards for ICP-MS. All solutions were stabilized in 2% nitric acid (trace metal grade, sub-boil distilled). All blanks, standards, and samples were spiked in-line with an internal standard composed as a mixture of beryllium, germanium, and thallium mix solution single-element standards (CPI International, Santa Rosa, CA), and 2% nitric acid was used as the rinse solution.
LC-MS/MS analysis of protein bands.
Bands displaying Se(IV) reductase activity in nondenaturing gels and protein bands in SDS-polyacrylamide gels of fractions enriched in SrrA were excised and diced into approximately 1-mm-long pieces. The Coomassie brilliant blue R-250 stain removed by several washes in a 50% acetonitrile–25 mM ammonium bicarbonate buffer. The gel pieces were stored in a 25 mM ammonium bicarbonate buffer. Samples were digested with trypsin and loaded onto a 100-μm-by-2-cm Acclaim PepMap 100 C18 Nanotrap column (5 μm, 100 Å) with an Ultimate 3000 liquid chromatograph (Thermo Fisher, Pittsburgh, PA) at 3 μl/min. The peptides were separated on a custom silica capillary column packed with C18 reverse-phase material (Halo peptide; 2.7 μM, 160 Å). The samples were run at 0.3 μl/min starting with 96% solvent A (100% water, 0.1% formic acid) and 4% solvent B (100% acetonitrile, 0.1% formic acid) over 25 min, followed by a 45-minute gradient from 10% to 90% solvent B. The column effluent was directly coupled to the Q-exactive orbitrap mass spectrometer using a Flex ion source (Thermo Fisher, Pittsburgh, PA). The mass spectrometer was controlled by Xcalibur 2.2 software and operated in data-dependent acquisition mode. Spectra of peptide mass were acquired from an m/z range of 300 to 2,000 at a high mass-resolving power. The top 25 most abundant charged ions were subjected to higher-energy collisional dissociation (HCD) with an m/z range of 300 to 2,000 with an intensity threshold of 3.3e4, with charge states of less than 7 and unassigned charge states excluded.
Data analysis.
The higher-energy collisional dissociated data were extracted using Proteome Discoverer implemented in Sequest version 1.4.1.14 (Thermo Fisher, Pittsburgh, PA). The files were searched against the UniProt Bacillus selenitireducens MLS10 database, which contains 3,231 proteins. Sequest was searched with a fragment ion tolerance of 0.020 Da and a parent ion tolerance of 10 ppm. Carbamidomethylation of cysteine was specified as a fixed modification. The enzyme specificity was set to trypsin with missed cleavage of 3. A target decoy database search was performed. Peptide identifications were accepted with a target false-discovery rate (FDR) between 0.05 (relaxed, moderate confidence) and 0.01 (strict, high confidence). The validation was based on the q value, which is the minimal FDR at which the identification is considered correct. A post-database search filter was applied with an Xcorr value greater than 2 for high peptide confidence.
Supplementary Material
ACKNOWLEDGMENTS
This work was supported in part by the Bayer School of Natural and Environmental Sciences.
We thank J. Switzer Blum (U.S. Geological Survey) for aiding in the culture work and T. Cantley (Duquesne University) and D. J. Bain (University of Pittsburgh) for metal analysis.
Mention of brand name products does not constitute an endorsement by the U.S. Geological Survey.
Footnotes
Supplemental material for this article may be found at https://doi.org/10.1128/JB.00614-18.
REFERENCES
- 1.Stolz JF, Basu P, Santini JM, Oremland RS. 2006. Arsenic and selenium in microbial metabolism. Annu Rev Microbiol 60:107–130. doi: 10.1146/annurev.micro.60.080805.142053. [DOI] [PubMed] [Google Scholar]
- 2.Kryukov GV, Gladyshev VN. 2004. The prokaryotic selenoproteome. EMBO Rep 5:538–543. doi: 10.1038/sj.embor.7400126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Carlson BA, Xu X-M, Kryukov GV, Rao M, Berry MJ, Gladyshev VN, Hatfield DL. 2004. Identification and characterization of phosphoseryl-tRNA[Ser]Sec kinase. Proc Natl Acad Sci U S A 101:12848–12853. doi: 10.1073/pnas.0402636101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kryukov GV, Castellano S, Novoselov SV, Lobanov AV, Zehtab O, Guigó R, Gladyshev VN. 2003. Characterization of mammalian selenoproteomes. Science 300:1439–1443. doi: 10.1126/science.1083516. [DOI] [PubMed] [Google Scholar]
- 5.Veres Z, Tsai L, Scholz TD, Politino M, Balaban RS, Stadtman TC. 1992. Synthesis of 5-methylaminomethyl-2-selenouridine in tRNAs: 31P NMR studies show the labile selenium donor synthesized by the selD gene product contains selenium bonded to phosphorus. Proc Natl Acad Sci U S A 89:2975–2979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Wolfe MD, Ahmed F, Lacourciere GM, Lauhon CT, Stadtman TC, Larson TJ. 2004. Functional diversity of the rhodanese homology domain: the Escherichia coli ybbB gene encodes a selenophosphate-dependent tRNA 2-selenouridine synthase. J Biol Chem 279:1801–1809. doi: 10.1074/jbc.M310442200. [DOI] [PubMed] [Google Scholar]
- 7.Nancharaiah YV, Lens PNL. 2015. Ecology and biotechnology of selenium-respiring bacteria. Microbiol Mol Biol Rev 79:61–80. doi: 10.1128/MMBR.00037-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Oremland RS, Switzer Blum JS, Culbertson CW, Visscher PT, Miller LG, Dowdle P, Strohmaier FE. 1994. Isolation, growth, and metabolism of an obligately anaerobic, selenate-respiring bacterium, strain SES-3. Appl Environ Microbiol 60:3011–3019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Macy JM, Rech S, Auling G, Dorsch M, Stackebrandt E, Sly LI. 1993. Thauera selenatis gen. nov., sp. nov., a member of the beta subclass of Proteobacteria with a novel type of anaerobic respiration. Int J Syst Bacteriol 43:135–142. doi: 10.1099/00207713-43-1-135. [DOI] [PubMed] [Google Scholar]
- 10.Narasingarao P, Häggblom MM. 2007. Pelobacter seleniigenes sp. nov., a selenate-respiring bacterium. Int J Syst Evol Microbiol 57:1937–1942. doi: 10.1099/ijs.0.64980-0. [DOI] [PubMed] [Google Scholar]
- 11.Nakagawa T, Iino T, Suzuki K, Harayama S. 2006. Ferrimonas futtsuensis sp. nov. and Ferrimonas kyonanensis sp. nov., selenate-reducing bacteria belonging to the Gammaproteobacteria isolated from Tokyo Bay. Int J Syst Evol Microbiol 56:2639–2645. doi: 10.1099/ijs.0.64399-0. [DOI] [PubMed] [Google Scholar]
- 12.Narasingarao P, Häggblom MM. 2006. Sedimenticola selenatireducens, gen. nov., sp. nov., an anaerobic selenate-respiring bacterium isolated from estuarine sediment. Syst Appl Microbiol 29:382–388. doi: 10.1016/j.syapm.2005.12.011. [DOI] [PubMed] [Google Scholar]
- 13.Blum JS, Stolz JF, Oren A, Oremland RS. 2001. Selenihalanaerobacter shriftii gen. nov., sp. nov., a halophilic anaerobe from Dead Sea sediments that respires selenate. Arch Microbiol 175:208–219. [DOI] [PubMed] [Google Scholar]
- 14.Switzer Blum J, Burns Bindi A, Buzzelli J, Stolz JF, Oremland RS. 1998. Bacillus arsenicoselenatis, sp. nov., and Bacillus selenitireducens, sp. nov.: two haloalkaliphiles from Mono Lake, California that respire oxyanions of selenium and arsenic. Arch Microbiol 171:19–30. doi: 10.1007/s002030050673. [DOI] [PubMed] [Google Scholar]
- 15.Yamamura S, Yamashita M, Fujimoto N, Kuroda M, Kashiwa M, Sei K, Fujita M, Ike M. 2007. Bacillus selenatarsenatis sp. nov., a selenate- and arsenate-reducing bacterium isolated from the effluent drain of a glass-manufacturing plant. Int J Syst Evol Microbiol 57:1060–1064. doi: 10.1099/ijs.0.64667-0. [DOI] [PubMed] [Google Scholar]
- 16.Abin CA, Hollibaugh JT. 2017. Desulfuribacillus stibiiarsenatis sp. nov., an obligately anaerobic, dissimilatory antimonate- and arsenate-reducing bacterium isolated from anoxic sediments, and emended description of the genus Desulfuribacillus. Int J Syst Evol Microbiol 67:1011–1017. doi: 10.1099/ijsem.0.001732. [DOI] [PubMed] [Google Scholar]
- 17.Rauschenbach I, Narasingarao P, Häggblom MM. 2011. Desulfurispirillum indicum sp. nov., a selenate- and selenite-respiring bacterium isolated from an estuarine canal. Int J Syst Evol Microbiol 61:654–658. doi: 10.1099/ijs.0.022392-0. [DOI] [PubMed] [Google Scholar]
- 18.Huber R, Sacher M, Vollmann A, Huber H, Rose D. 2000. Respiration of arsenate and selenate by hyperthermophilic Archaea. Syst Appl Microbiol 23:305–314. doi: 10.1016/S0723-2020(00)80058-2. [DOI] [PubMed] [Google Scholar]
- 19.Slobodkina GB, Lebedinsky AV, Chernyh NA, Bonch-Osmolovskaya EA, Slobodkin AI. 2015. Pyrobaculum ferrireducens sp. nov., a hyperthermophilic Fe(III)-, selenate- and arsenate-reducing crenarchaeon isolated from a hot spring. Int J Syst Evol Microbiol 65:851–856. doi: 10.1099/ijs.0.000027. [DOI] [PubMed] [Google Scholar]
- 20.Güven K, Mutlu MB, Çırpan C, Kutlu HM. 2013. Isolation and identification of selenite reducing Archaea from Tuz (salt) Lake In Turkey. J Basic Microbiol 53:397–401. doi: 10.1002/jobm.201200008. [DOI] [PubMed] [Google Scholar]
- 21.Nickerson WJ, Falcone G. 1963. Enzymatic reduction of selenite. J Bacteriol 85:763–771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Rosenfeld CE, Kenyon JA, James BR, Santelli CM. 2017. Selenium (IV, VI) reduction and tolerance by fungi in an oxic environment. Geobiology 15:441–452. doi: 10.1111/gbi.12224. [DOI] [PubMed] [Google Scholar]
- 23.Hedderich R, Klimmek O, Kröger A, Dirmeier R, Keller M, Stetter KO. 1998. Anaerobic respiration with elemental sulfur and with disulfides. FEMS Microbiol Rev 22:353–381. doi: 10.1111/j.1574-6976.1998.tb00376.x. [DOI] [Google Scholar]
- 24.Schönheit P, Schäfer T. 1995. Metabolism of hyperthermophiles. World J Microbiol Biotechnol 11:26–57. [DOI] [PubMed] [Google Scholar]
- 25.Reich HJ, Hondal RJ. 2016. Why nature chose selenium. ACS Chem Biol 11:821–841. doi: 10.1021/acschembio.6b00031. [DOI] [PubMed] [Google Scholar]
- 26.Macy JM, Michel TA, Kirsch DG. 1989. Selenate reduction by a Pseudomonas species: a new mode of anaerobic respiration. FEMS Microbiol Lett 52:195–198. [DOI] [PubMed] [Google Scholar]
- 27.Schröder I, Rech S, Krafft T, Macy JM. 1997. Purification and characterization of the selenate reductase from Thauera selenatis. J Biol Chem 272:23765–23768. [DOI] [PubMed] [Google Scholar]
- 28.Krafft T, Bowen A, Theis F, Macy JM. 2000. Cloning and sequencing of the genes encoding the periplasmic-cytochrome b-containing selenate reductase of Thauera selenatis. DNA Seq J DNA Seq Mapp 10:365–377. doi: 10.3109/10425170009015604. [DOI] [PubMed] [Google Scholar]
- 29.Harel A, Häggblom MM, Falkowski PG, Yee N. 2016. Evolution of prokaryotic respiratory molybdoenzymes and the frequency of their genomic co-occurrence. FEMS Microbiol Ecol 92:fiw187. doi: 10.1093/femsec/fiw187. [DOI] [PubMed] [Google Scholar]
- 30.Maher MJ, Macy JM. 2002. Crystallization and preliminary X-ray analysis of the selenate reductase from Thauera selenatis. Acta Crystallogr D Biol Crystallogr 58:706–708. [DOI] [PubMed] [Google Scholar]
- 31.Maher MJ, Santini J, Pickering IJ, Prince RC, Macy JM, George GN. 2004. X-ray absorption spectroscopy of selenate reductase. Inorg Chem 43:402–404. doi: 10.1021/ic035136n. [DOI] [PubMed] [Google Scholar]
- 32.Dridge EJ, Watts CA, Jepson BJN, Line K, Santini JM, Richardson DJ, Butler CS. 2007. Investigation of the redox centres of periplasmic selenate reductase from Thauera selenatis by EPR spectroscopy. Biochem J 408:19–28. doi: 10.1042/BJ20070669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fujita M, Ike M, Nishimoto S, Takahashi K, Kashiwa M. 1997. Isolation and characterization of a novel selenate-reducing bacterium, Bacillus sp. SF-1. J Ferment Bioeng 83:517–522. doi: 10.1016/S0922-338X(97)81130-0. [DOI] [Google Scholar]
- 34.Kuroda M, Yamashita M, Miwa E, Imao K, Fujimoto N, Ono H, Nagano K, Sei K, Ike M. 2011. Molecular cloning and characterization of the srdBCA operon, encoding the respiratory selenate reductase complex, from the selenate-reducing bacterium Bacillus selenatarsenatis SF-1. J Bacteriol 193:2141–2148. doi: 10.1128/JB.01197-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Grimaldi S, Schoepp-Cothenet B, Ceccaldi P, Guigliarelli B, Magalon A. 2013. The prokaryotic Mo/W-bisPGD enzymes family: a catalytic workhorse in bioenergetic. Biochim Biophys Acta 1827:1048–1085. doi: 10.1016/j.bbabio.2013.01.011. [DOI] [PubMed] [Google Scholar]
- 36.Watts CA, Ridley H, Condie KL, Leaver JT, Richardson DJ, Butler CS. 2003. Selenate reduction by Enterobacter cloacae SLD1a-1 is catalysed by a molybdenum-dependent membrane-bound enzyme that is distinct from the membrane-bound nitrate reductase. FEMS Microbiol Lett 228:273–279. doi: 10.1016/S0378-1097(03)00782-1. [DOI] [PubMed] [Google Scholar]
- 37.Ridley H, Watts CA, Richardson DJ, Butler CS. 2006. Resolution of distinct membrane-bound enzymes from Enterobacter cloacae SLD1a-1 that are responsible for selective reduction of nitrate and selenate oxyanions. Appl Environ Microbiol 72:5173–5180. doi: 10.1128/AEM.00568-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Losi ME, Frankenberger WT. 1997. Reduction of selenium oxyanions by Enterobacter cloacae SLD1a-1: Isolation and growth of the bacterium and its expulsion of selenium particles. Appl Environ Microbiol 63:3079–3084. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bébien M, Kirsch J, Méjean V, Verméglio A. 2002. Involvement of a putative molybdenum enzyme in the reduction of selenate by Escherichia coli. Microbiology 148:3865–3872. doi: 10.1099/00221287-148-12-3865. [DOI] [PubMed] [Google Scholar]
- 40.Theisen J, Yee N. 2014. The molecular basis for selenate reduction in Citrobacter freundii. Geomicrobiol J 31:875–883. doi: 10.1080/01490451.2014.907377. [DOI] [Google Scholar]
- 41.Rothery RA, Workun GJ, Weiner JH. 2008. The prokaryotic complex iron-sulfur molybdoenzyme family. Biochim Biophys Acta 1778:1897–1929. doi: 10.1016/j.bbamem.2007.09.002. [DOI] [PubMed] [Google Scholar]
- 42.Li D-B, Cheng Y-Y, Wu C, Li W-W, Li N, Yang Z-C, Tong Z-H, Yu H-Q. 2014. Selenite reduction by Shewanella oneidensis MR-1 is mediated by fumarate reductase in periplasm. Sci Rep 4:3735. doi: 10.1038/srep03735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Song D, Li X, Cheng Y, Xiao X, Lu Z, Wang Y, Wang F. 2017. Aerobic biogenesis of selenium nanoparticles by Enterobacter cloacae Z0206 as a consequence of fumarate reductase mediated selenite reduction. Sci Rep 7:3239. doi: 10.1038/s41598-017-03558-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Maltman C, Donald LJ, Yurkov V. 2017. Two distinct periplasmic enzymes are responsible for tellurite/tellurate and selenite reduction by strain ER-Te-48 associated with the deep sea hydrothermal vent tube worms at the Juan de Fuca Ridge black smokers. Arch Microbiol 199:1113–1120. doi: 10.1007/s00203-017-1382-1. [DOI] [PubMed] [Google Scholar]
- 45.Klonowska A, Heulin T, Vermeglio A. 2005. Selenite and tellurite reduction by Shewanella oneidensis. Appl Environ Microbiol 71:5607–5609. doi: 10.1128/AEM.71.9.5607-5609.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Maltman C, Walter G, Yurkov V. 2016. A diverse community of metal(loid) oxide respiring bacteria is associated with tube worms in the vicinity of the Juan de Fuca Ridge black smoker field. PLoS One 11:e0149812. doi: 10.1371/journal.pone.0149812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Baesman SM, Stolz JF, Kulp TR, Oremland RS. 2009. Enrichment and isolation of Bacillus beveridgei sp. nov., a facultative anaerobic haloalkaliphile from Mono Lake, California, that respires oxyanions of tellurium, selenium, and arsenic. Extrem Life Extreme Cond 13:695–705. doi: 10.1007/s00792-009-0257-z. [DOI] [PubMed] [Google Scholar]
- 48.Herbel MJ, Blum JS, Oremland RS, Borglin SE. 2003. Reduction of elemental selenium to selenide: experiments with anoxic sediments and bacteria that respire Se-oxyanions. Geomicrobiol J 20:587–602. doi: 10.1080/713851163. [DOI] [Google Scholar]
- 49.Herbel MJ, Johnson TM, Oremland RS, Bullen TD. 2000. Fractionation of selenium isotopes during bacterial respiratory reduction of selenium oxyanions. Geochim Cosmochim Acta 64:3701–3709. doi: 10.1016/S0016-7037(00)00456-7. [DOI] [Google Scholar]
- 50.Oremland RS, Herbel MJ, Blum JS, Langley S, Beveridge TJ, Ajayan PM, Sutto T, Ellis AV, Curran S. 2004. Structural and spectral features of selenium nanospheres produced by Se-respiring bacteria. Appl Environ Microbiol 70:52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Baesman SM, Bullen TD, Dewald J, Zhang D, Curran S, Islam FS, Beveridge TJ, Oremland RS. 2007. Formation of tellurium nanocrystals during anaerobic growth of bacteria that use Te oxyanions as respiratory electron acceptors. Appl Environ Microbiol 73:2135–2143. doi: 10.1128/AEM.02558-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Afkar E, Lisak J, Saltikov C, Basu P, Oremland RS, Stolz JF. 2003. The respiratory arsenate reductase from Bacillus selenitireducens strain MLS10. FEMS Microbiol Lett 226:107–112. doi: 10.1016/S0378-1097(03)00609-8. [DOI] [PubMed] [Google Scholar]
- 53.Blasco F, Dos Santos JP, Magalon A, Frixon C, Guigliarelli B, Santini CL, Giordano G. 1998. NarJ is a specific chaperone required for molybdenum cofactor assembly in nitrate reductase A of Escherichia coli. Mol Microbiol 28:435–447. [DOI] [PubMed] [Google Scholar]
- 54.de Castro E, Sigrist CJA, Gattiker A, Bulliard V, Langendijk-Genevaux PS, Gasteiger E, Bairoch A, Hulo N. 2006. ScanProsite: detection of PROSITE signature matches and ProRule-associated functional and structural residues in proteins. Nucleic Acids Res 34:W362–W365. doi: 10.1093/nar/gkl124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Darriba D, Taboada GL, Doallo R, Posada D. 2011. ProtTest 3: fast selection of best-fit models of protein evolution. Bioinformatics 27:1164–1165. doi: 10.1093/bioinformatics/btr088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Ronquist F, Teslenko M, van der Mark P, Ayres DL, Darling A, Höhna S, Larget B, Liu L, Suchard MA, Huelsenbeck JP. 2012. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol 61:539–542. doi: 10.1093/sysbio/sys029. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Le SQ, Gascuel O. 2008. An improved general amino acid replacement matrix. Mol Biol Evol 25:1307–1320. doi: 10.1093/molbev/msn067. [DOI] [PubMed] [Google Scholar]
- 58.Whelan S, Goldman N. 2001. A general empirical model of protein evolution derived from multiple protein families using a maximum-likelihood approach. Mol Biol Evol 18:691–699. doi: 10.1093/oxfordjournals.molbev.a003851. [DOI] [PubMed] [Google Scholar]
- 59.Yang Z. 1994. Maximum likelihood phylogenetic estimation from DNA sequences with variable rates over sites: approximate methods. J Mol Evol 39:306–314. [DOI] [PubMed] [Google Scholar]
- 60.Rambaut A, Drummond AJ, Xie D, Baele G, Suchard MA. 2018. Posterior summarisation in Bayesian phylogenetics using Tracer 1.7. Syst Biol 67:901–904. doi: 10.1093/sysbio/syy032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Schröder I, Kröger A, Macy JM. 1988. Isolation of the sulphur reductase and reconstitution of the sulphur respiration of Wolinella succinogenes. Arch Microbiol 149:572–579. doi: 10.1007/BF00446763. [DOI] [Google Scholar]
- 62.Krafft T, Bokranz M, Klimmek O, Schröder I, Fahrenholz F, Kojro E, Kröger A. 1992. Cloning and nucleotide sequence of the psrA gene of Wolinella succinogenes polysulphide reductase. Eur J Biochem 206:503–510. [DOI] [PubMed] [Google Scholar]
- 63.Dietrich W, Klimmek O. 2002. The function of methyl-menaquinone-6 and polysulfide reductase membrane anchor (PsrC) in polysulfide respiration of Wolinella succinogenes. Eur J Biochem 269:1086–1095. [DOI] [PubMed] [Google Scholar]
- 64.Heinzinger NK, Fujimoto SY, Clark MA, Moreno MS, Barrett EL. 1995. Sequence analysis of the phs operon in Salmonella typhimurium and the contribution of thiosulfate reduction to anaerobic energy metabolism. J Bacteriol 177:2813–2820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Stoffels L, Krehenbrink M, Berks BC, Unden G. 2012. Thiosulfate reduction in Salmonella enterica is driven by the proton motive force. J Bacteriol 194:475–485. doi: 10.1128/JB.06014-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Findlay AJ. 2016. Microbial impact on polysulfide dynamics in the environment. FEMS Microbiol Lett 363 [DOI] [PubMed] [Google Scholar]
- 67.Vavourakis CD, Ghai R, Rodriguez-Valera F, Sorokin DY, Tringe SG, Hugenholtz P, Muyzer G. 2016. Metagenomic insights into the uncultured diversity and physiology of microbes in four hypersaline soda lake brines. Front Microbiol 7:211. doi: 10.3389/fmicb.2016.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Wright JJ, Mewis K, Hanson NW, Konwar KM, Maas KR, Hallam SJ. 2014. Genomic properties of marine group A bacteria indicate a role in the marine sulfur cycle. ISME J 8:455–468. doi: 10.1038/ismej.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Zargar K, Conrad A, Bernick DL, Lowe TM, Stolc V, Hoeft S, Oremland RS, Stolz J, Saltikov CW. 2012. ArxA, a new clade of arsenite oxidase within the DMSO reductase family of molybdenum oxidoreductases. Environ Microbiol 14:1635–1645. doi: 10.1111/j.1462-2920.2012.02722.x. [DOI] [PubMed] [Google Scholar]
- 70.Thauer RK, Jungermann K, Decker K. 1977. Energy conservation in chemotrophic anaerobic bacteria. Bacteriol Rev 41:100–180. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Krafft T, Macy JM. 1998. Purification and characterization of the respiratory arsenate reductase of Chrysiogenes arsenatis. Eur J Biochem 255:647–653. [DOI] [PubMed] [Google Scholar]
- 72.Oremland RS, Dowdle PR, Hoeft S, Sharp JO, Schaefer JK, Miller LG, Switzer Blum J, Smith RL, Bloom NS, Wallschlaeger D. 2000. Bacterial dissimilatory reduction of arsenate and sulfate in meromictic Mono Lake, California. Geochim Cosmochim Acta 64:3073–3084. doi: 10.1016/S0016-7037(00)00422-1. [DOI] [Google Scholar]
- 73.Newman DK, Ahmann D, Morel FMM. 1998. A brief review of microbial arsenate respiration. Geomicrobiol J 15:255–268. doi: 10.1080/01490459809378082. [DOI] [Google Scholar]
- 74.Klimmek O, Kreis V, Klein C, Simon J, Wittershagen A, Kröger A. 1998. The function of the periplasmic Sud protein in polysulfide respiration of Wolinella succinogenes. Eur J Biochem 253:263–269. [DOI] [PubMed] [Google Scholar]
- 75.Prat L, Maillard J, Rohrbach-Brandt E, Holliger C. 2012. An unusual tandem-domain rhodanese harbouring two active sites identified in Desulfitobacterium hafniense. FEBS J 279:2754–2767. doi: 10.1111/j.1742-4658.2012.08660.x. [DOI] [PubMed] [Google Scholar]
- 76.Niggemyer A, Spring S, Stackebrandt E, Rosenzweig RF. 2001. Isolation and characterization of a novel As(V)-reducing bacterium: implications for arsenic mobilization and the genus Desulfitobacterium. Appl Environ Microbiol 67:5568–5580. doi: 10.1128/AEM.67.12.5568-5580.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Lowry OH, Rosebrough NJ, Farr AL, Randall RJ. 1951. Protein measurement with the Folin phenol reagent. J Biol Chem 193:265–275. [PubMed] [Google Scholar]
- 78.Sambrook J, Russell DW. 2006. SDS-polyacrylamide gel electrophoresis of proteins. Cold Spring Harb Protoc 2006:pdb.prot4540. doi: 10.1101/pdb.prot4540. [DOI] [PubMed] [Google Scholar]
- 79.Altschul SF, Madden TL, Schäffer AA, Zhang J, Zhang Z, Miller W, Lipman DJ. 1997. Gapped BLAST and PSI-BLAST: a new generation of protein database search programs. Nucleic Acids Res 25:3389–3402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Jormakka M, Törnroth S, Byrne B, Iwata S. 2002. Molecular basis of proton motive force generation: structure of formate dehydrogenase-N. Science 295:1863–1868. doi: 10.1126/science.1068186. [DOI] [PubMed] [Google Scholar]
- 81.Markowitz VM, Korzeniewski F, Palaniappan K, Szeto E, Werner G, Padki A, Zhao X, Dubchak I, Hugenholtz P, Anderson I, Lykidis A, Mavromatis K, Ivanova N, Kyrpides NC. 2006. The integrated microbial genomes (IMG) system. Nucleic Acids Res 34:D344–D348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Edgar RC. 2004. MUSCLE: multiple sequence alignment with high accuracy and high throughput. Nucleic Acids Res 32:1792–1797. doi: 10.1093/nar/gkh340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Kumar S, Stecher G, Tamura K. 2016. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Capella-Gutiérrez S, Silla-Martínez JM, Gabaldón T. 2009. trimAl: a tool for automated alignment trimming in large-scale phylogenetic analyses. Bioinforma Oxf Engl 25:1972–1973. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Altekar G, Dwarkadas S, Huelsenbeck JP, Ronquist F. 2004. Parallel Metropolis coupled Markov chain Monte Carlo for Bayesian phylogenetic inference. Bioinforma Oxf Engl 20:407–415. doi: 10.1093/bioinformatics/btg427. [DOI] [PubMed] [Google Scholar]
- 86.Letunic I, Bork P. 2016. Interactive tree of life (iTOL) v3: an online tool for the display and annotation of phylogenetic and other trees. Nucleic Acids Res 44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.