Abstract
The preimplantation period of life in mammals encompasses a tremendous amount of restructuring and remodeling of the embryonic genome and reprogramming of gene expression. These vast changes support metabolic activation and cellular processes that drive early cleavage divisions and enable the creation of the earliest primitive cell lineages. A major question in mammalian embryology is how such vast, sweeping changes in gene expression are orchestrated, so that changes in gene expression are exactly appropriate to meet the developmental needs of the embryo over time. Using the rhesus macaque as an experimentally tractable model species closely related to the human, we combined high quality RNA-seq libraries, in-depth sequencing and advanced systems analysis to discover the underlying mechanisms that drive major changes in gene regulation during preimplantation development. We identified the major changes in mRNA population and the biological pathways and processes impacted by those changes. Most importantly, we identified 24 key upstream regulators that are themselves modulated during development and that are associated with the regulation of over 1000 downstream genes. Through their roles in extensive gene networks, these 24 upstream regulators are situated to either drive major changes in target gene expression or modify the cellular environment in which other genes function, thereby directing major developmental transitions in the preimplantation embryo. The data presented here highlight some of the specific molecular features that likely drive preimplantation development in a nonhuman primate species and provides an extensive database for novel hypothesis-driven studies.
Keywords: transcriptome, reprogramming, genome activation, rhesus monkey, preimplantation development, embryo, pathway analysis, embryogenesis, blastocyst, cleavage stage
Introduction
The preimplantation period of life in mammals encompasses a tremendous amount of restructuring and remodeling of the embryonic genome and reprogramming of gene expression. These vast changes support metabolic activation and cellular processes that drive early cleavage divisions and enable the creation of the earliest primitive cell lineages. Global waves of DNA demethylation and methylation as well as histone modifications are associated with the activation and repression of large numbers of genes (Latham and Schultz, 2001; Zhou and Dean, 2015). In mouse embryos, the net result is a series of multiple major transcriptional activation events that anticipate major changes in cellular morphology, cellular physiology and cell lineage formation (Latham et al., 1992; Hamatani et al., 2004). Studies in cattle, human and nonhuman primate embryos indicate similarly sweeping changes in preimplantation gene expression (Misirlioglu et al., 2006; Kues et al., 2008; Vassena et al., 2011; Chitwood et al., 2017).
A major question in mammalian embryology is how such vast, sweeping changes in gene expression are orchestrated, so that the stage-by-stage changes in gene expression are exactly appropriate to meet the developmental needs of the embryo. Answering this question would significantly extend our understanding of early developmental processes and of how environmental and other factors might impact the early embryo and thereby affect later development and survival. Although previous cDNA array and RNA sequencing (RNA-seq) studies have revealed the timing and extent of major transitions in gene expression in different species, a clear understanding of what drives those changes has not been revealed, due to a combination of technical limitations and insufficient systems analysis of the expression data. Additional studies in embryonic stem cells have revealed networks of interacting genes that promote specific events related to cell lineage specification (Huang and Wang, 2014). Such studies are highly valuable, but many more essential and key events occur during the same period, and their controlling mechanisms, have yet to be explained.
Our goal in this study was to combine methods to produce high quality RNA-seq libraries, in-depth sequencing and advanced systems analysis to discover the underlying mechanisms that drive major changes in gene regulation during preimplantation development. To do this, we chose the rhesus macaque as an experimentally tractable model species that is closely related to the human and thus the most amenable to future studies to better understand early human development. Other recent reports have investigated the rhesus macaque preimplantation embryo transcriptome by RNA-seq, but have provided very low depth of coverage or variation in sequence read length employed, small numbers of biological replicates per stage, poorly described donor female information and limited systems analysis of changes associated with key developmental transitions (Chitwood et al., 2017; Wang et al., 2017). We provide here the results of an RNA-seq transcriptome study of preimplantation development in the rhesus macaque with a sufficient number of stages to detect major changes in gene expression, a greater depth of sequencing coverage and sensitivity, greater degree of biological replication and lesser opportunity for impact of maternal genotype on outcome. Additionally, our embryos were sexed and enough males and females were used to address sex as a biological variable at later stages. Using this extensive RNA-seq data set, we identified the major changes in mRNA population as well as the biological pathways and processes impacted by those changes. Most importantly, we identified 24 key upstream regulators that are themselves modulated during development and that are associated with the regulation of over 1000 downstream genes. Through their roles in extensive gene networks, these 24 upstream regulators are situated to either drive major changes in target gene expression or modify the cellular environment in which other genes function, thereby directing major developmental transitions in the preimplantation embryo. The data presented here thus highlight some of the specific molecular features that likely drive preimplantation development in a nonhuman primate species.
Materials and Methods
Oocyte collection and embryo culture
All animals were housed at the California National Primate Center in accordance with the ethics guidelines established and approved by the Institutional Animal Use and Care Administrative Advisory Committee at the University of California-Davis (Vandevoort et al., 2011). Adult female rhesus macaques (Macaca mulatta), aged 6–12 years (normal breeding age) with normal menstrual cycles, were used in this study. An ovarian stimulation protocol was used for the collection of matured oocytes (MII). This included twice daily injections of recombinant hFSH (37.5 IU; La Jolla Discount Pharmacy, La Jolla, CA) for 7 days at the onset of menstruation and was followed by a single injection hCG on Day 8 (1000 IU, La Jolla Discount Pharmacy, La Jolla, CA). Cumulus-oocyte complexes were collected by follicular aspiration approximately 30 hours post hCG injection and oocytes were assessed for maturation status. A subset of MII oocytes were collected for RNA isolation and another set of MII oocytes were used for IVF (Day 0 of culture) and subsequent embryo culture procedures for the collection of eight cell (Day 3 of culture), morula (Day 5 of culture) and expanded blastocysts (Day 6 of culture) following previously established methods (de Prada and VandeVoort, 2008; Chaffin et al., 2014). Four to five females were used as donors for each stage (MII = 5; 8-cell = 4; morula = 4; expanded blastocyst = 5). RNA-seq libraries were made with two oocytes per sample for MII oocytes and one embryo per sample for other stages.
Library preparation, sequencing, alignment and analysis
All samples were isolated for RNA following the manufacturer’s protocol using PicoPure™ RNA Extraction kit with a DNAse digestion (Qiagen; Hilden, Germany). All 52 libraries were produced for sequencing using the Ovation RNA-Seq System v2 kit, which uses Ribo-SPIA™ technology (NuGen, San Carlos, CA). Of these samples, 17 were also analyzed by constructions and sequencing libraries prepared using the Ovation® SoLo RNA-Seq System for reasons described below. The SPIA libraries included cDNA fragmentation to approximately 300 bp using a Covaris-2 sonicator and a S1 nuclease digestion as previously described (Head et al., 2011). Libraries were then purified using Agencourt AMPure XP beads (Beckman Coulter) and further processed through the Ovation Ultralow DR Multiplex Systems 1–16 (Nugen), which included end repair, adaptor ligation and library amplification following manufacture’s instruction.
Initial attempts of alignment of the libraries to the Macaca mulatta MacaM v7 genome (Zimin et al., 2014) resulted in a high level of multi-mapping and non-exonic read alignments. To rule out the possibility that this was due to faulty SPIA library preparation, 17 of the samples were processed in duplicate using the Ovation® SoLo RNA-Seq System (NuGEN, San Carlos, CA) following the manufacturer’s protocol. Library preparation included a bead purification, end repair, adaptor ligation and first round library amplification and purification. Then 20–30 ng of each library was used to for the remainder of library preparation, which included use of InDA-C primers for rRNA depletion, as well as second round library amplification and purification steps. The Solo Kit uses enzymatic shearing and all RNA-seq libraries are between 300 and 350 bp in length.
However, the library preparation method did not alter outcome, as the Pearson Correlation coefficients (0.88–0.94, P < 0.0001, Supplemental Table S1) indicated very similar results comparing SPIA and SoLo libraries. Subsequent analysis identified the MacaM v7 genome build as the source of the alignment problems, and established that alignment to the Mmul 8.1.0 (Rogers et al., 2006) alleviated the issue multi-mapping and non-exonic read alignments. Both the SoLo and SPIA libraries were used in the DESeq2 analysis, using the ‘collapseReplicates’ function to avoid inappropriate weighting of the samples in the analysis. All samples were sequenced at the Michigan State University Research Technology Support Facility, using the Illumina HiSeq 2500 (Illumina, San Diego, CA, USA) for SPIA samples or HiSeq 4000 for SoLo (Illumina, San Diego, CA, USA). The change in platform was due to an automatic upgrade at the facility. The barcoded libraries were pooled and sequenced in rapid run mode to generate 50 nucleotide unpaired end reads. Utilization of 50 nucleotide reads were selected as a means for maximizing the generation of expression data while balancing total cost. Samples were loaded at 65% of optimal loading concentration, to enhance the effectiveness of the Illumina cluster identification algorithm, along with 10% of optimal loading concentration of PhiX DNA Control library (adapter-ligated library obtained from randomly sheared PhiX DNA; Illumina), to increase initial read sequence complexity. Instead of a standard Illumina R1 sequencing primer, the SoLo Custom R1 primer was used for the sequencing of the libraries.
The total number of pass-filter reads generated from sequencing per sample ranged from 26.7 M to 129.3 M (Supplemental Table S1). The fraction of Q30 bases ranged from 91.57% to 95.94% with and average Q of 35.80 to 39.05. Sequencing data are available at the Gene Expression Omnibus (GSE112537).
Reads were aligned the Mmul 8.1.0 genome (genome annotation release version 90) (Zerbino et al., 2018) using the Hisat2 (Kim et al., 2015) software package, excluding those aligning to ribosomal RNA (rRNA) and rRNA like genes. Exonic read counts were generated utilizing ‘featureCounts’ (Liao et al., 2014) and the differential expression calculations were conducted with the DESeq2 (Love et al., 2014) package. Although we include sequencing data for 53 libraries in GEO, we excluded from this analysis eight libraries that had <20 M aligned reads. Two samples of delayed blastocysts were also excluded as irrelevant to the purpose of the study. For this analysis, we thus used 43 libraries encompassing at least three biological replicates per sample type and samples from at least three different maternal donors (MII stage oocytes (n = 8), and embryos at the 8-cell (n = 11), morula (n = 11) and expanded blastocyst (n = 13) stages). Pre-differential expression filtering consisted of removal of genes with less than 1 FPKM (fragment counts normalized per kilobase of feature length per million mapped fragments) (Trapnell et al., 2010) present in at least five samples. This minimum of five samples was used to ensure expression at 1 FPKM or greater present in 10% or more of the samples, regardless of stage. A q-value (the P-value adjusted for multiple comparisons) below the false discovery rate of 0.05 was used as the threshold for defining DEGs. Sequencing depth achieved in this study was high, with an average of >55 M total aligned reads per sample (range 21.9 M–114.5 M). Non-rRNA exonic reads representing expressed mRNAs ranged from 4.8 M to 45.4 M with an average of 17.39 M (Supplemental Table S1). The sex of embryos was determined using the method in Midic et al. (2018). Briefly, this method uses RNA-seq data in a three step process: (1) test of Y chromosome presence by high expression of RPS4Y1/2 and low XIST, (2) test of X chromosome presence by high expression of XIST and low RPS4Y1/2, and (3) test for presences of two X chromosome copies by SNP testing (Midic et al., 2018).
All resultant DEG lists were uploaded to the Ingenuity Pathway Analysis (IPA; Qiagen, Hilden, Germany) suite of tools for exploring enrichments in Canonical Pathway (CP), Disease and Functions (DF) and Upstream Regulator (UR). IPA output consists of a P-value detailing the level of significance enrichment of the uploaded gene-set as compared to the known molecules in the database for a given CP/UR/DF entry. Those entries with a P-value less than 0.05 were classified as statistically significant. In addition, IPA provides z-score calculation allowing for the inference of directionality of the level of activity: activation/increase or inhibition/decrease. For a normal distribution, |z| > 1.96 is considered statistically significant. It should be noted that entries with a significant P-values do not necessarily result in a significant z-score; z-scores are returned on the basis of information contained within the IPA knowledge base. For a more focused analysis, we restricted IPA functional analysis to just biological function, omitting disease-based categories.
Results
RNA sequencing libraries and statistics
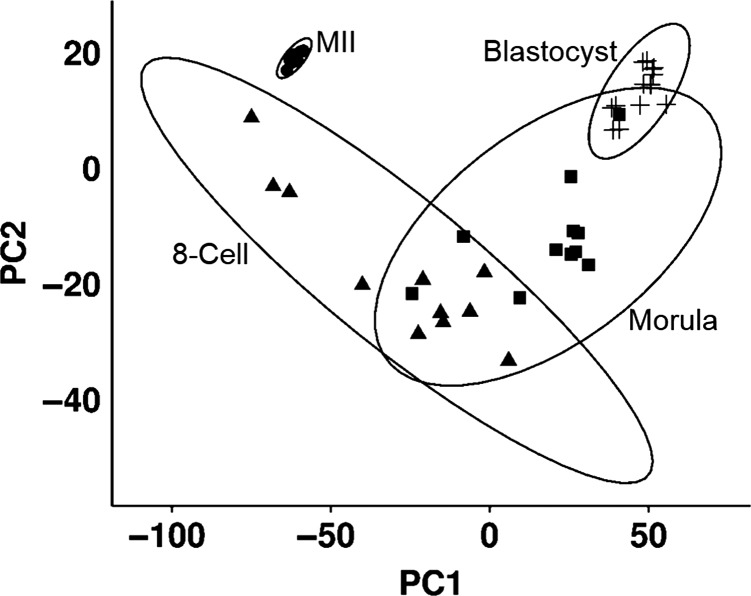
Forty-three RNA-seq libraries corresponding to MII stage oocytes (n = 8) and embryos at the 8-cell (n = 11), morula (n = 11) and expanded blastocyst (n = 13) stages were included in this study. Principal component analysis revealed that there was a high degree of clustering and extensive differences between the transcriptomes of the different developmental stages (Fig 1 and Supplemental Figure S1). Of the 8784 to 10 743 genes expressed at each stage, there were 7 349 differentially expressed genes (DEGs) observed among a total of >23 000 transcripts quantified across the developmental stages.
Figure 1.
Principle component analyses shows the relationships between the 43 samples of the metaphase II oocytes (MII), 8-cell embryos (8-cell), morula and blastocyst (Blast). Ellipses surrounding each embryo stage cluster are based on 95% confidence intervals. The MII and blastocyst samples show a tight grouping indicating high levels of homogeneity of expression while the 8-cell and morula samples have greater heterogeneity resulting in an increased spread.
Identification of trend-line groups corresponding to long-term patterns of gene regulation
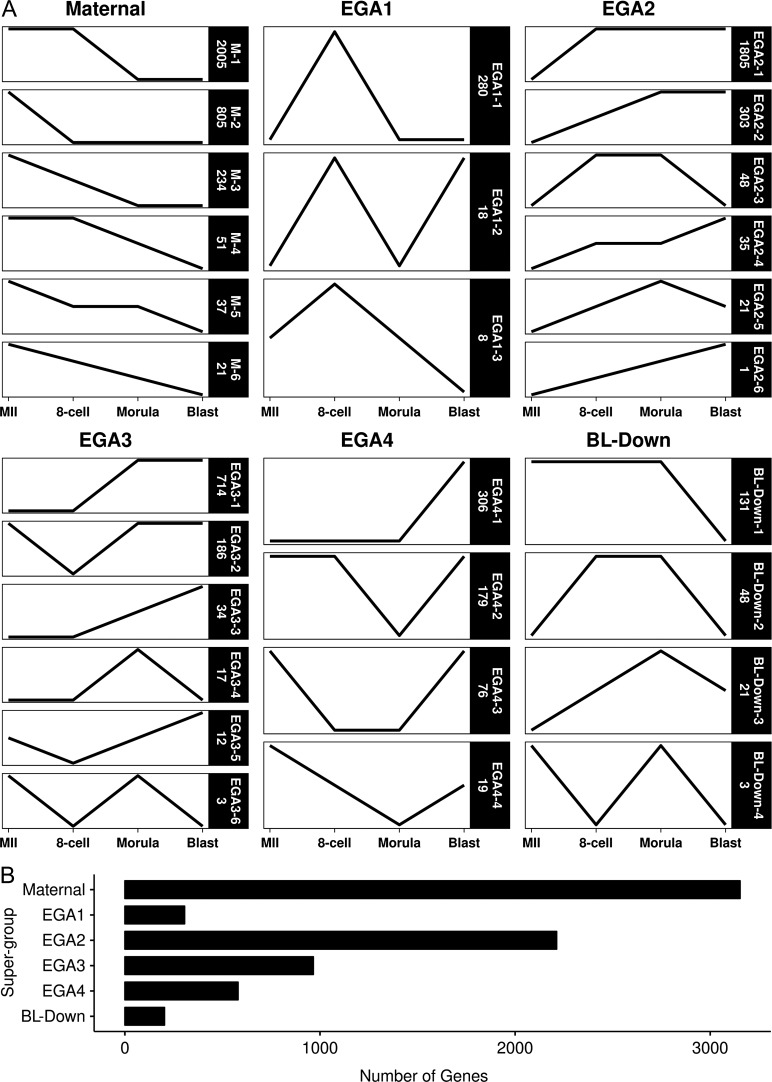
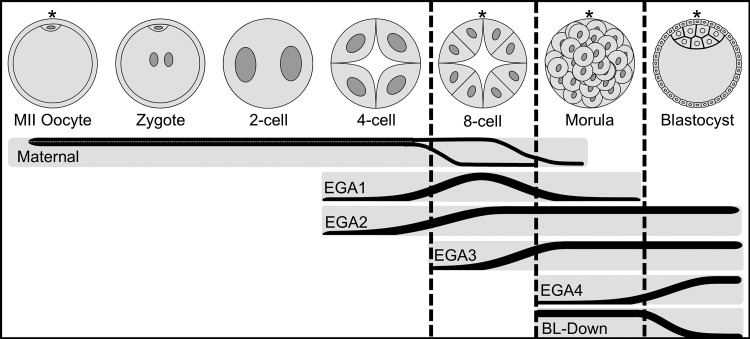
Our next step was to characterize this vast amount of change in a way that would reveal changes associated with key developmental transitions. We utilized an approach that considers long-term patterns of gene regulation and cross-stage regulatory interactions for each DEG, recognizing the temporal order of the different stages, instead of just comparing pairs of chronologically successive stages. For each gene, the statistical significance of change between successive stages was calculated and used to assign genes to one of 26 ‘trend-line’ groups (Fig. 2), which were then clustered into ‘super-groups’ consisting of oocyte-expressed mRNAs (Maternal), four major embryonic genome activation (EGA) groups (EGA1 to EGA4), and mRNAs that are repressed during the morula to blastocyst transition (BL-Down) (Fig. 3).
Figure 2.
Classification and quantification of the 7349 DEGs into 26 trend-line groups, categorized within each super-group. Trend-line groups were defined by mRNAs showing the same patterned in changes of expression. An increase or decrease in the slope of a line indicates a significant change in expression, while a horizontal line represents a significant change. Panel A has the individual trend-lines for each trend-line groups, collated by their respective super-group. Maternal trend-lines have maximal expression at MII with no subsequent increase, limited to decreases or no changes. EGA1 through EGA4 trend-lines are defined by their shared first significant increases at 8-cell, morula and blastocyst stages, respectively. BL-Down are trend-lines with a decrease at the blastocyst stage. Each trend-line group in Panel A is labeled on the right with its designation (e.g. EGA1-1) and number of mRNAs in the group (e.g. 280). Summation of all genes classified by super-group are plotted in Panel B. Note that BL-Down groups 2, 3 and 4 are the same as EGA2-3, EGA2-5 and EGA3-6.
Figure 3.
Depiction of the major stages of preimplantation development and the corresponding relationship of the super-groups. Stages with sequenced data are denoted with an asterisk and bounded by vertical dashed lines representing transitions between the represented samples. The six super-groups are plotted in relation to development stage, increase or decrease in slope indicates change in mRNA abundances during a transition. Maternal = mRNAs of maternal origin with decrease in mRNA abundance, EGA1 through EGA4 = embryonic genome activation, BL-Down = mRNAs with decreasing expression at blastocyst.
The Maternal group was the largest with 3153 genes total (M1–M6 trend-line groups; Figs 2 and 3 and Supplemental Table S2). Starting with the transition from MII to 8-cell stage, we first defined two super-groups with increased mRNA expression, one transiently increased at the 8-cell stage (EGA1) and a second in which expression either remained elevated or increased further past the 8-cell stage (EGA2) (Figs 2 and 3, Supplemental Tables S3 and S4). EGA1 contained 306 genes and encompassed two known markers of the initial embryonic genome activation state reported in mice and humans (DUXA and ZSCAN4) (Falco et al., 2007; De Iaco et al., 2017). EGA2 contained another 2213 genes. The next two super-groups in temporal sequence were EGA3, with 966 genes displaying their first significant increase in expression at the 8-cell to morula transition, and EGA4, containing 580 mRNAs that displayed significant increases between the morula and expanded blastocyst stages (Figs 2 and 3, Supplemental Tables S5 and S6). The last super-group contained 203 mRNAs that decreased in abundance between morula and expanded blastocyst stages (BL-Down) (Figs 2 and 3, Supplemental Table S7). (Note that BL-Down groups 2, 3 and 4 are the same as EGA2-3, EGA2-5 and EGA3-6.)
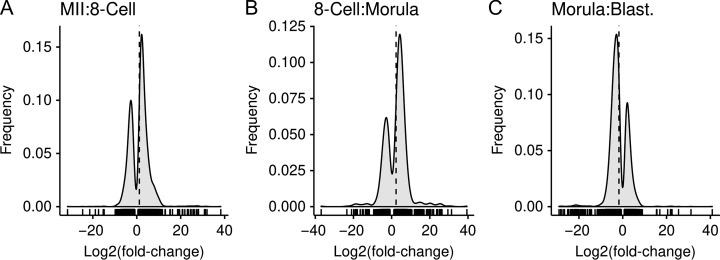
The magnitude and importance of the EGA1 and EGA2 super-groups was evident in the log2(fold-change) plot comparing MII and 8-cell stages, which shows a prevalence of mRNAs undergoing increased expression (Fig. 4 panel A). Interestingly, the log2(fold-change) plot comparing 8-cell and morula stages (Fig. 4, panel B) revealed many more decreases in expression than increases, marking this interval as the largest period of maternal mRNA destruction. Thereafter, the log2(fold-change) plot comparing morula and blastocyst stages (Fig. 4 panel C) revealed that mRNAs decreasing in expression substantially outnumber mRNAs undergoing increasing expression.
Figure 4.
Frequency distribution of the log2(fold-change) for the differentially expressed genes between the two consecutive stages. These graphs illustrate that the increasing and decreasing in expression distributions over time. The x-axis plots the log2(fold-change) and the frequency on the y-axis. A vertical dashed line denotes the mean log2(fold-change). Short vertical lines between the x-axis and the frequency curve denote individual data points. There was a marked number of genes with an increase in expression for the MII to 8-cell and 8-cell to morula transitions, while the morula to blastocyst had majority decreases.
IPA analysis of super-groups
The foregoing analysis revealed thousands of gene expression changes but a high degree of co-regulation amongst these mRNAs expressed, with 94% of the >7000 DEGs encompassed by just 10 of the 26 trend-line groups described above. To understand the biological consequence of this vast amount of change, and to identify potential regulators that drive this change, we applied IPA analysis in two complementary approaches.
First, we examined pathways, functions and regulators associated with specific super-groups in order to discover the specific roles of those cohorts of genes in development. This approach was applied to Maternal and EGA1 to EGA3 super-groups. IPA analysis of these super-groups identified multiple different canonical pathways and biological functions, including key signaling pathways, pathways related to cell cycle, DNA repair and gene expression, and other essential cellular functions (Fig. 5; Supplemental Tables S8–S15). Because EGA1 and EGA2 occurred simultaneously, we also applied IPA analysis to the EGA1 and EGA2 groups combined. This revealed additional biological effects not observed by examining EGA1 and EGA2 separately, and overall indicated a predominant effect in promoting cell division and survival (Fig. 6; Supplemental Tables S16 and S17). IPA analysis of EGA4 revealed significant activation of the planar cell polarity (PCP) pathway, along with functions involved in metabolism of carbohydrates and fatty acids (Fig. 6; Supplemental Tables S18 and S19). IPA analysis of the BL-Down super-group yielded limited results (Supplemental Table S20).
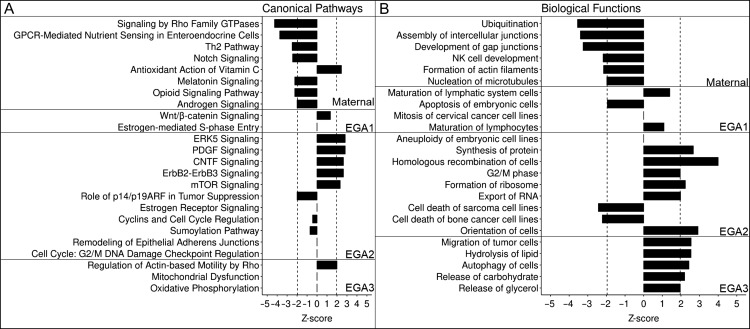
Figure 5.
Top canonical pathways and biological functions for the Maternal and EGA1 through EGA3 super-groups. Pathways (Panel A) and biological functions (Panel B) are grouped by super-group, magnitude of graph denotes z-score. Significance was set at |z| > 1.96 and is denoted by the vertical dashed lines. All entries were found to have significant P-values (P < 0.05).
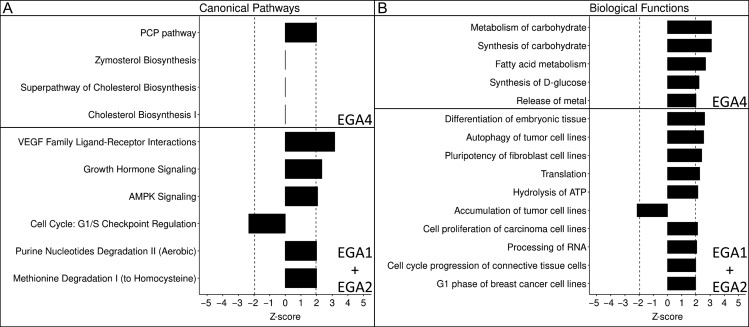
Figure 6.
Top canonical pathways and biological functions for EGA4 and the EGA1+EGA2 groups. Pathways (Panel A) and biological functions (Panel B) are grouped by super-group, magnitude of graph denotes z-score. Significance was set at |z| > 1.96 and is denoted by the vertical dashed lines. All entries were found to have significant P-values (P < 0.05).
IPA analysis of pathways and biological functions associated with major developmental transitions
Having identified the major gene regulatory groups and their likely impacts on embryo development, we next sought to discover the combined impacts of these groups on the embryo during the major developmental transitions. To do this, we employed a second approach to IPA, by merging combinations of multiple trend-line groups into ‘mega-groups’ to capture effects driven by the entire spectrum of changes occurring during three major developmental transitions: oocyte-to-8-cell, 8-cell-to-morula and morula-to-blastocyst.
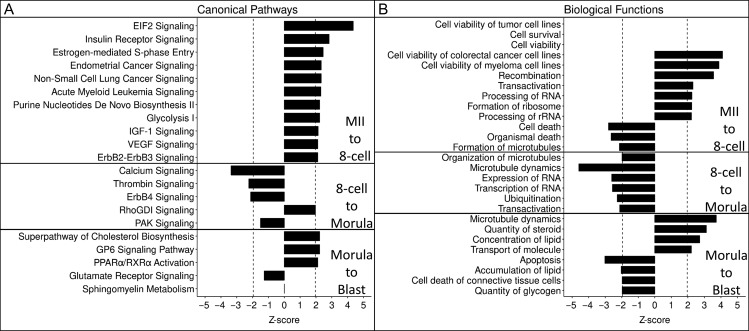
The switch from maternal to embryonic control of development is a key event during preimplantation development of all mammals and combines translation and degradation of maternally derived transcripts with the transcriptional activation of the embryonic genome. Our results above revealed that this transition occurs in part from MII to 8-cell stage and in part from the 8-cell to morula stage. IPA on the combination of 2938 genes undergoing significant increases or decreases between MII and 8-cell stage (M-2, EGA1-1, EGA2-1, EGA2-3) garnered 179 significant results, 37 with a significant z-score (32 activated, 5 inhibited). A number of activated functions were identified involving cell survival/viability and recombination. Inhibited functions consisted of cell/organismal death and the formation and organization of microtubules (Fig. 7, panel B, Supplemental Table S21). Canonical pathway analysis revealed activation of EIF2 signaling, estrogen-mediated S-phase entry and other pathways (Fig. 7, panel A, Supplemental Table S22). IPA applied to the eight cell to morula stage genes (M-1, M-3 and EGA3-1) revealed activation of RhoGDI signaling and inhibition of sphingosine-1-phosphate, PAK signaling, microtubule dynamics as well as expression and transcription of RNA (Fig. 7, Supplemental Table S23).
Figure 7.
Top canonical pathways (Panel A) and biological functions (Panel B) for mega-groups. Significance was set at |z| > 1.96 and is denoted by the vertical dashed lines. All entries were found to have significant P-values (P < 0.05).
IPA of the gene modulations associated with the transition from morula to blastocyst (EGA3, EGA4, BL-Down, M-4, M-5, M-6, and EGA1-2) revealed a total of 25 pathways (2 activated, 1 inhibited), and 206 biological functions (28 activated and 8 inhibited). Compared to the individual analysis of EGA4 and BL-Down, superpathway of cholesterol biosynthesis emerged as activated, along with GP6 signaling and inhibition of RhoGDI signaling, and there was activation of microtubule dynamics, quantity of steroid and concentration of lipid (Fig. 7, Supplemental Tables S24 and S25). There were 10 additional significant affected canonical pathways revealed (Supplemental Table S25).
Effect of embryo sex on pathways and biological functions
Once the embryo expresses its own genome, it is possible to determine the sex of the embryo from which the library was made (Midic et al., 2018). We tested whether embryo sex had an impact on IPA results for morulae and blastocysts, using sets of DEGs identified by comparing male and female embryos at morula and blastocyst stages (Supplemental Tables S26 and S27). There were eight significantly affected canonical pathways at the morula stage and 14 at the blastocyst stage (Supplemental Table S28). None of the pathways had a significant z-score. Each stage had a different affected biological function; quantity of cells at morula (inhibited in females, z = −2.274) and conversion of lipid at blastocyst (inhibited in females, z = −1.968).
Upstream regulators as novel drivers of major developmental transitions
A powerful tool afforded by IPA is the ability to identify upstream regulators (URs) that may drive significant changes in biological pathways and processes, and thus play a role in the progressive acquisition of embryo characteristics from one stage to the next. We note that, because we are measuring mRNA expression, and some UR-effector relationships in IPA may be assigned on the basis of protein–protein interactions, interpretation of possible biological meaning must account for UR functional class (transcription factors, kinases, etc.).
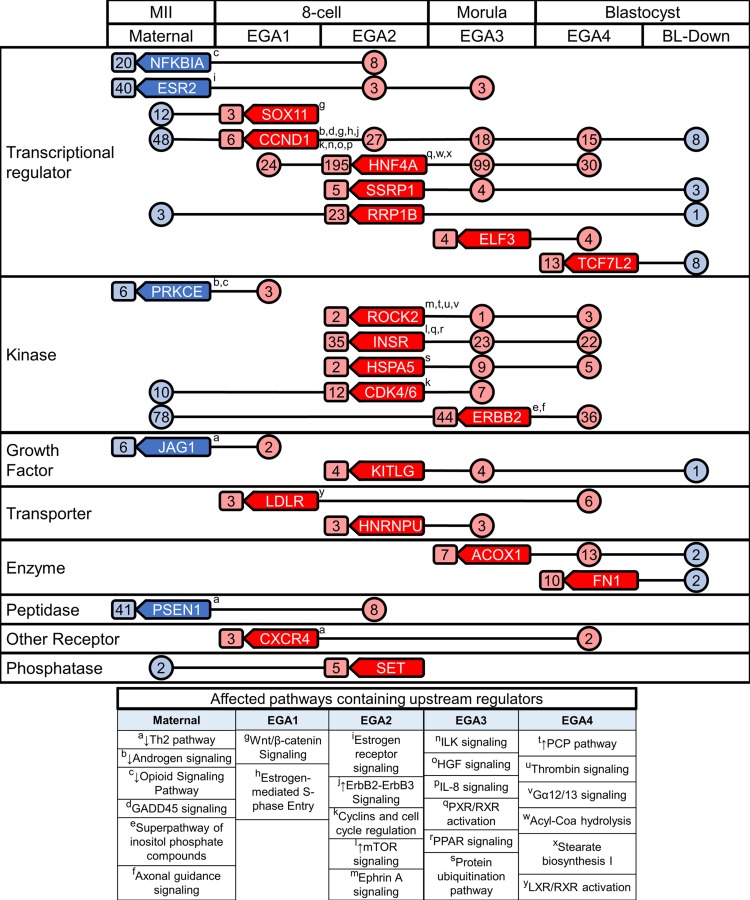
We identified 83 significant URs that were differentially expressed; 23 from the maternal super-group, 9 from EGA1, 24 from EGA2, 16 from EGA3, and 11 from EGA4 (Supplemental Table S29). Of these 83 URs, 24 were associated with target molecules that were regulated within other super-groups (Fig. 8). These 24 URs have diverse functions, including nine transcription factors, six kinases and nine others (receptor, peptidase, growth factor, transporter and enzyme) (Fig. 8; Supplemental Table S30). Interestingly, of these 24 URs, nine were observed to be regulated in equivalent stages when compared to either of the two previously published rhesus monkey preimplantation embryo RNA-seq studies (overlap of six in each study; Supplemental Table S31) (Chitwood et al., 2017; Wang et al., 2017). The other members of this 24 UR set were either not detected or were not observed to be differentially regulated, most likely due to lower sensitivity of mRNA detection smaller numbers of replicate samples, and/or slight differences in relative developmental timing of embryos within morphological stages. Two URs (CDK4/6 and HSPA5) were returned for all three studies.
Figure 8.
Diagram of the key 24 upstream regulators (URs) across the preimplantation period. Upstream regulators are shown in rounded boxes with a left facing point in their super-group origin. The numbers of downstream effectors regulated by each UR are shown within circles or rounded boxes. Boxed numbers adjacent to URs denote regulation within the stage of origin, and circled numbers connected by lines indicate transient regulation. Superscripts (a–y) denote that the UR was a member of a canonical pathway. Red coloring indicates an upregulation of URs and downstream targets, while blue indicates downregulation. These 24 URs target/interact with 1053 of the differentially expressed genes during development.
Some of these 24 differentially regulated URs displayed significant (P < 0.05) effects on target molecules at multiple stages. Significant z-scores were obtained for some effects, but not for others; for the latter situations, the possible direction of effect could be inferred from up- or downregulation of the target molecules. Some affected URs were associated with DEGs within the same super-group in which the URs were regulated, or with DEGs within the same embryo stage (i.e. EGA1 and EGA2 for 8-cell stage), and thus may exert short-term actions. URs could also be associated with DEGs in other super-groups/embryo stages, indicating possible long-term actions (Fig. 8).
As an example of short-term regulation, ESR2 was associated with 40 downregulated DEGs in the maternal super-group, HNF4A was associated with 195 upregulated DEGs within EGA2, and ERBB2 was associated with 44 upregulated DEGs within EGA3 (Fig. 8). Short-term interactions were also seen within the 8-cell stage but between super-groups. For example, CCND1 increases in expression during EGA1, and its downstream effectors include members of the concurrently activated EGA2 super-group and its affected CPs and biological functions.
An example of the utility of the UR analysis for detecting long-term UR actions can be seen in INSR. This gene is upregulated at the 8-celll stage as part of the EGA2 super-group and displays significant positive z-scores for effects at the morula and blastocyst stages (EGA3 and EGA4) indicating activation of its downstream effects, with over 20 downstream DEGs at each stage (Fig. 8). CCND1 provides another interesting example, being itself upregulated during the 8-cell stage as part of EGA1 and displaying a negative z-score (z = −1.2). Although the z-score was below the significance threshold, this effect was accompanied by significant downregulation of 48 downstream effector mRNAs as part of the M-1 trend-line group. Thereafter, distinct groups of many CCND1 target molecules were upregulated as part of EGA3 or EGA4, or downregulated as part of the BL-Down super-group, and CCND1 was significantly associated with these later changes (P < 0.05) even though it displayed either no z-score, or a positive z-score below the significance threshold. Other URs (ERBB2, RRP1B, CDK4/6, SET) with increased expression at EGA2 or EGA3 were associated with degraded maternal mRNAs. Reciprocally, URs with diminishing expression in the maternal super-group (PRKCE, JAG1) were associated with increased target molecule expression at EGA1. Interestingly, the largest number of developmentally modulated URs were those upregulated as part of EGA2.
Additional insight into long-term effects of developmentally regulate URs was obtained through a novel analysis linking affected URs to IPA canonical pathways that are modulated within other super-groups and embryonic stages. First, we identified the canonical pathways that contain the URs themselves (Fig. 8, Supplemental Table S30). Second, we identified affected canonical pathways that contained affected target molecules for each UR (Supplemental Tables S32 and S33). Third, we identified the biological functions associated with the target molecules for each differentially regulated UR (Supplemental Tables S34–S36). These combined approaches provided an estimate of the degree to which the 24 URs acting across stages contributed to the overall range of gene expression changes throughout preimplantation development. Of the 7349 DEGs identified among the 26 trend-line groups, 4423 were matched with a molecule definition in the IPA database. Affected canonical pathways encompassed 1051 of these DEGs, and affected biological functions encompassed 2906 DEGs, for a total of 3048 DEGs. The 24 URs acting across super-groups regulate 1053 (34.5%) of these 3 048 DEGs as downstream effectors. Analysis of the canonical pathways and biological functions affected by these 24 URs revealed a striking collection of both short-term and long-term actions (Fig. 8).
Long-term modes of action include cases where URs are upregulated during EGA1, EGA2 or EGA3 and modulate pathways and processes at later embryonic stages. One example is ROCK2, a UR that is upregulated during EGA2 and affects members of the EGA4 super-group (Fig. 8). Interestingly, ROCK2 is also a member of the planar cell polarity pathway, which is activated in the EGA4 super-group. The EGA4 super-group also included DEGs in the planar cell polarity pathway that are targets of the EGA1-1 URs: CCND1 and LDLR, and the EGA2-1 UR: ERBB2. Thus, URs associated with EGA1 and EGA2 were associated with regulation of planar cell polarity as part of EGA4 regulation. Similarly, two of the most significant EGA3-associated pathways (mitochondrial dysfunction and oxidative phosphorylation) had more than six of the affected molecules targeted by the HNF4A and INSR (two EGA2-1 URs). HNF4A also targeted more than 50% of the affected molecules in an EGA4 biological function (synthesis and metabolism of carbohydrate). Thus, HNF4A was activated as part of EGA2 (8-cell stage) but may contribute to modulation of pathways and processes at both morula (EGA3) and blastocyst (EGA4) stages.
We observed downregulation of effector genes as part of the Maternal super-group that were also associated with upregulation of some URs during EGA1, EGA2 or even EGA3 (Fig. 8). This may reflect either a causal role for the URs in promoting mRNAs degradation or a reduction in the ability of URs to function as a consequence of maternal mRNA degradation. For example, ERBB2 (an EGA3-1 UR) is associated with downregulated effectors included in M-1 trend-line group and related to the Th2 pathway, assembly of intercellular junctions, formation of intercellular junctions and development of gap junctions. Because M-1 and EGA-3 genes are concurrently modulated, this may reflect a causal role. Alternatively, maternal mRNA downregulation associated with transcription factors that are upregulated during EGA (e.g. CCND1) could reflect a causal role of the UR if it regulates maternal mRNA degradation, or could indicate a reduction in expression of downstream effectors that limits the UR actions. URs from the EGA super-groups may have exerted inhibitory effects on the BL-Down groups. This included short-term and long-term modes of action. A long-term inhibitory affect was seen with CCND1 and SOX11 (EGA1-1 URs) targeting downregulated (BL-Down) DEGs associated with formation of actin filaments (significantly inhibited).
Because the rhesus monkey is expected to be an excellent model for studying human reproduction, we examined the patterns of expression of these same 24 key URs with data reported for the same morphological stages of human embryos (Dang et al., 2016). Eleven of the 24 URs displayed equivalent patterns of regulation (Table I). Six additional URs displayed conservative differences (EGA1 vs. EGA2; EGA3 vs. EGA2; EGA3 vs. EGA4), which may reflect slight differences in relative embryo developmental timing within morphology at time of embryo lysis. Three maternally expressed URs displayed either no expression or no difference in expression in the human data, and one UR (FN1) displayed disparate patterns of regulation between human and monkey. Thus, overall, the results for human and monkey were very similar.
Table I.
Comparison of upstream regulator expression patterns between rhesus monkey and human embryos.
| This Study | Dang et al. (2016) | |
|---|---|---|
| GEO | GSE112537 | GSE71318 |
| Median Exon Reads | 16 513 169 | 16 648 486 |
| No. Samples | ||
| MII | 8 | 4 |
| 8-Cell | 11 | 5 |
| Morula | 11 | 4 |
| Blast. | 13 | 9 |
| Total Expressed Genes | 23 677 | 17 764 |
| Upstream Regulator Super-Groups | ||
| PSEN1 | Maternal | IS/NSa |
| PRKCE | Maternal | NED |
| NFKBIA | Maternal | Maternal |
| JAG1 | Maternal | Maternal |
| ESR2 | Maternal | IS/NSa |
| SOX11 | EGA1 | EGA2 |
| LDLR | EGA1 | EGA4 |
| CXCR4 | EGA1 | EGA2 |
| CCND1 | EGA1 | EGA2 |
| SSRP1 | EGA2 | IS/NSa |
| SET | EGA2 | EGA2 |
| RRP1B | EGA2 | EGA2 |
| ROCK2 | EGA2 | EGA2 |
| KITLG | EGA2 | EGA2 |
| INSR | EGA2 | EGA2 |
| HSPA5 | EGA2 | NED |
| HNRNPU | EGA2 | EGA2 |
| HNF4A | EGA2 | EGA2 |
| CDK4/6 | EGA2 | EGA2 |
| ERBB2 | EGA3 | EGA2 |
| ELF3 | EGA3 | EGA2 |
| ACOX1 | EGA3 | EGA4 |
| TCF7L2 | EGA4 | EGA4 |
| FN1 | EGA4 | Maternal |
aInsuficient sampling/not significant.
Discussion
The major findings of this study are that (1) there is a vast amount of change in gene expression (>7000 DEGs total) during preimplantation development in the rhesus monkey that can be summarized in just 26 trend-line groups, and (2) over one-third of the eligible DEGs analyzed by IPA are associated with just 24 URs that are themselves developmentally regulated and display predicted activation or inhibition states, on the basis of downstream effector mRNA expression. This novel identification of these 24 URs as key developmentally regulated factors playing controlling roles in large networks of genes provides new mechanistic insight into the control of preimplantation development.
In some cases, modulation of downstream DEGs may enable or disable upstream regulator function. In other cases, downstream DEG modulation in association with UR modulation likely reflects a mechanistic effect wherein a UR is controlling the associated DEGs. Downregulation of an inhibitory UR may allow downstream DEGs to be upregulated, and increased expression of an activating UR may direct the downstream DEG activation. In either case, such connections suggest important modes of regulation in the early embryo and a major controlling influence of the 24 URs identified here. Additional URs may regulate other DEGs that could not be analyzed in IPA, and obviously the identification of these 24 URs here using IPA does not discount previously identified key regulators in early embryos.
These 24 URs include transcription factors, kinases, receptors, peptidases, growth factors, transporters and enzymes. This diversity of function indicates that some of these key URs can act directly to mediate changes in gene expression (‘instructive’ effects), while others modify the cellular environment to potentiate key developmental events (‘permissive’ effects), and yet others may have their actions constrained by down-modulation of effectors at a prior stage.
One example of a long-range ‘instructive’ function is seen for HNF4A, a transcription factor that is upregulated during EGA2 and is associated with increased expression of downstream effectors during EGA3 and EGA4. Another interesting example of an instructive function is a group of transcription regulators expressed during EGA1, EGA2 or EGA3 (ERBB2, SOX11, SET) that may down-regulate maternal mRNAs as part of genome reprogramming and help establish the embryonic cell cycle. A possible example of a ‘permissive’ function is the downregulation of NFKBIA maternal mRNA, which may release of NFKB from cytoplasmic sequestration to allow its nuclear action during the 8-cell stage. Other examples of permissive actions include kinases or kinase regulators, such as increased expression of CCND1 mRNA during EGA1, which is then available to drive both positive and negative effects through diverse downstream effectors at later times.
In addition to identifying the 24 upstream regulators, our approach to defining long-term modes of regulation and long-term gene interactions across 8 cell stages yielded new insight into regulatory mechanisms driving preimplantation development, including major developmental transitions (oocyte–embryo, oocyte–morula and morula–blastocyst mega-groups) and provided a greater understanding of the pathways and processes associated with those transitions. When taking this long-range view, we identified metabolism-related and growth factor-related signaling and biological process related to microtubules, transcription, translation, cell survival and ubiquitination as key processes, as well as associated DEGs as key molecules driving preimplantation development. Activation of growth factor signaling via VEGF and IGF-1 were prominent features of the oocyte to morula transition. Contribution of VEGF to preimplantation development is conserved, as it is also developmentally regulated in bovine and porcine embryos and improves cleavage and blastocyst rates in bovine embryos (Einspanier et al., 2002; Biswas et al., 2011). IGF-1, which is secreted by the preimplantation embryo and co-expressed with its receptor in oocytes and embryos, promotes cleavage, differentiation, transcriptional activation and prevention of apoptosis, as well as implantation (Heyner et al., 1993; Inzunza et al., 2010). Interestingly, we observed relaxin receptor (RXFP1) mRNA expressed in oocytes as well as embryos, with a higher abundance in 8-cell and morula stage embryos. This suggests both maternal and embryonic origins of the RXFP mRNA, and the potential for relaxin signaling in oocytes as well as embryos. Relaxin stimulates production of VEGF and other growth factors and is produced in primate ovaries during the luteal phase of the menstrual cycle. Relaxin also enhances rhesus monkey blastocyst quality (Vandevoort et al., 2011). This suggests a possible early role of ovarian relaxin and oocyte-, as well as embryo-, expressed receptor in contributing to embryo quality.
Ribosome biogenesis is another key function that emerged in our analysis of the oocyte to morula transition, reminiscent of nucleolus reformation and the key role for nucleoli in preimplantation development (Maddox-Hyttel et al., 2007; Golomb et al., 2014; Fulka and Aoki, 2016). We observed both DEGs and biological functions related to ribosome biogenesis during EGA2, including EMG1 (essential for mitotic growth 1) and RBM19 (RNA binding motif Protein 19), which plays a critical role in ribosome biogenesis and is essential for mouse preimplantation development (Zhang et al., 2008; Wu et al., 2010).
The next critical transition that we wanted to highlight was the morula to blastocyst transition, allowing us to assess the important processes involved in blastocyst formation. Key features of the morula to blastocyst transition that emerged in our data set included increased biosynthesis of lipid, cholesterol and steroids, increased PPARα/RXRα signaling and increased carbohydrate metabolism. The importance of these processes was also reported for blastocyst formation in other species (rodents, pig and bovine) (Mohan et al., 2002; Huang, 2008; Abbott, 2009; Blitek and Szymanska, 2017). The role of cholesterol and lipids in preimplantation embryos are still being discovered. Our results indicate that lipids, cholesterol and PPAR/RxR signaling are likely especially important during the morula to blastocyst development. Another novel finding for the morula to blastocyst transition was the activation of the GP6 or GPVI signaling in blastocysts in relation to morula embryos. GP6 signaling is involved in platelet activation and collagen binding (Semeniak et al., 2016). Platelet-activating factor (PAF) plays a role in embryo viability and pregnancy (Ripps et al., 1993; Roudebush et al., 2002).
Another aspect of the transition to blastocyst is planar cell polarity (PCP) signaling. The PCP pathway is increased during EGA4. PCP is a branch of the WNT signaling pathway and is required for embryonic development (Gao, 2012). PCP plays a key role in embryo compaction and creation of inner and outer cell lineages (Tao et al., 2009; Sokol, 2015). It is also known for its role in cell polarization, cell migration, axon guidance and chemotaxis, and is dependent on the RhoA-ROCK1 and ROCK2 cascade (Sebbagh and Borg, 2014). ROCK2 is a PCP upstream regulator (Zhang et al., 2014) and is required for proper inner cell mass morphology and lineage formation, cleavage and blastocyst formation (Laeno et al., 2013; Duan et al., 2014; Kwon et al., 2016). We find that ROCK2 mRNA first increases in abundance during EGA2, indicating that events as early as the 8-cell stage in the monkey likely facilitate PCP, in support of cavitation and cell lineage formation during subsequent stages.
Next, we identified key processes or signaling pathways that were modulated throughout the preimplantation period. One prominent process that emerged from our analysis is the modulation of mitochondrial function and metabolism-related signaling during development. Changes in mitochondria-related functions (mitochondrial dysfunction, OXPHOS, TCA cycle II, and Mito L-carnitine shuttle pathway) were major elements associated with EGA3. The activation of mitochondrial functions after the 8-cell stage sets the stage for mitochondria increasing in number and undergoing structural changes to provide for increased oxidative metabolism, oxygen consumption, ATP production and respiratory function, as the embryo develops (Dumollard et al., 2007; Cecchino et al., 2018). The oocyte and embryo mitochondrial activity is also directly related to embryo viability (Dumollard et al., 2007; Kaneko, 2016). The UR analyses highlights HNF4A and INSR as potential key players in this transition, consistent with regulatory functions identified in other cell types (Louvi et al., 1997; Stoffel and Duncan, 1997; Barry and Thummel, 2016; Logan et al., 2018). Our transcriptome data thus highlight the potential key contribution of increasing mitochondrial function during the morula stage to continued preimplantation development.
Another interesting result that emerged from our analysis is the potential role of estrogen signaling in the early embryo. A loss of estrogen during maturation can block embryo development and additional supplement of maturation media with estrogen has shown to improve blastocyst formation (Bondesson et al., 2015; Kubo et al., 2015). However, there is still limited knowledge on the importance of estrogen-mediated signaling within the embryo itself during preimplantation development. We identified estrogen-mediated S phase in our mega group for the oocyte-to-embryo (8-cell) transition and super-group EGA1, as well as estrogen receptor signaling within EGA2. The estrogen-mediated S phase pathway also includes three DEGs related to cell cycle progression (CCNA1, CCND1, and MYC), which were also associated with key affected biological functions (mitosis, cell survival, cell viability and G2/M phase) associated with the oocyte-to-embryo transition. Although the role of estrogen-mediated signaling during preimplantation development is not fully understood, this finding suggests this may be a worthwhile line of investigation.
One of the strengths of this dataset is the use of the rhesus monkey as a model. This species has highly similar reproductive physiology to humans, especially in comparison to rodent models. There is also evidence that both nuclear and cytoplasmic maturation of primate oocytes resemble that of humans (Chaffin and Vandevoort, 2013; Ruebel et al., 2018) and that primate embryos require key regulatory mechanisms not observed in rodents (Vandevoort et al., 2011). Genetically, there is a high degree of similarity between rhesus and human genomes (Schramm and Bavister, 1999), and an accompanying similarity in gene regulation was evident in the expression patterns of the 24 URs examined in detail here. This similarity further highlights the importance of the rhesus monkey as a model for conducting studies that would not be acceptable using human embryos.
The dataset presented here stands as a major new resource for primate embryology. Our data set provides greater depth, sensitivity and biological replication within oocyte or embryo stage than any previous rhesus monkey study, and is the first to address sex as a biological variable in the early rhesus embryo. Most importantly, our novel method of analysis provides a detailed view of affected pathways, processes and functions that are associated with overt histological and biological changes during the preimplantation period. This study is the first in any species to provide a long-range analysis to illuminate regulatory effects of key genes across multiple stages. The extensive database presented here should help promote novel hypothesis-driven studies. The focused group of 24 key upstream regulators in particular provides a new framework for hypothesis development and mechanistic studies of the early embryo, how those pathways dictate embryo quality and viability, and how they may be affected by exogenous factors. The ability to design and execute such studies in a superior nonhuman primate model of human embryos may facilitate the development of improved assisted reproduction methods in humans.
Supplementary Material
Acknowledgements
The authors thank Dana Dagget for her technical assistance and Kailey Vincent and Ben Goheen for help with library preparation.
Authors’ roles
PZS processed and analyzed data, prepared figures and tables and wrote the manuscript; MLR analyzed and interpreted data and wrote the manuscript; UM processed, analyzed and interpreted data; CAV produced samples, provided funding and edited the manuscript; KEL conceived the study, provided funding, interpreted data and wrote the manuscript.
Funding
This work was supported by: grants from the National Institutes of Health, Office of Research Infrastructure Programs Division of Comparative Medicine Grants R24 (OD012221 to K.E.L., OD011107/RR00169 (California National Primate Research Center), and OD010967/RR025880 to C.A.V.); the Eunice Kennedy Shiver National Institute of Child Health and Human Development of the National Institutes of Health (under the award number T32HD087166); AgBioResearch, Michigan State University; and Michigan State University. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
Competing Interests
The authors have nothing to disclose.
References
- Abbott BD. Review of the expression of peroxisome proliferator-activated receptors alpha (PPAR alpha), beta (PPAR beta), and gamma (PPAR gamma) in rodent and human development. Reprod Toxicol 2009;27:246–257. [DOI] [PubMed] [Google Scholar]
- Barry WE, Thummel CS. The Drosophila HNF4 nuclear receptor promotes glucose-stimulated insulin secretion and mitochondrial function in adults. Elife 2016;5:e11183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biswas D, Jung EM, Jeung EB, Hyun SH. Effects of vascular endothelial growth factor on porcine preimplantation embryos produced by in vitro fertilization and somatic cell nuclear transfer. Theriogenology 2011;75:256–267. [DOI] [PubMed] [Google Scholar]
- Blitek A, Szymanska M. Peroxisome proliferator-activated receptor (PPAR) isoforms are differentially expressed in peri-implantation porcine conceptuses. Theriogenology 2017;101:53–61. [DOI] [PubMed] [Google Scholar]
- Bondesson M, Hao R, Lin CY, Williams C, Gustafsson JA. Estrogen receptor signaling during vertebrate development. Biochim Biophys Acta 2015;1849:142–151. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cecchino GN, Seli E, Alves da Motta EL, Garcia-Velasco JA. The role of mitochondrial activity in female fertility and assisted reproductive technologies: overview and current insights. Reprod Biomed Online 2018;36:686–697. [DOI] [PubMed] [Google Scholar]
- Chaffin CL, Latham KE, Mtango NR, Midic U, VandeVoort CA. Dietary sugar in healthy female primates perturbs oocyte maturation and in vitro preimplantation embryo development. Endocrinology 2014;155:2688–2695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chaffin CL, Vandevoort CA. Follicle growth, ovulation, and luteal formation in primates and rodents: a comparative perspective. Exp Biol Med (Maywood) 2013;238:539–548. [DOI] [PubMed] [Google Scholar]
- Chitwood JL, Burruel VR, Halstead MM, Meyers SA, Ross PJ. Transcriptome profiling of individual rhesus macaque oocytes and preimplantation embryos. Biol Reprod 2017;97:353–364. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dang Y, Yan L, Hu B, Fan X, Ren Y, Li R, Lian Y, Yan J, Li Q, Zhang Y et al. Tracing the expression of circular RNAs in human pre-implantation embryos. Genome Biol 2016;17:130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Iaco A, Planet E, Coluccio A, Verp S, Duc J, Trono D. DUX-family transcription factors regulate zygotic genome activation in placental mammals. Nat Genet 2017;49:941–945. [DOI] [PMC free article] [PubMed] [Google Scholar]
- de Prada JK, VandeVoort CA. Growth hormone and in vitro maturation of rhesus macaque oocytes and subsequent embryo development. J Assist Reprod Genet 2008;25:145–158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Duan X, Chen KL, Zhang Y, Cui XS, Kim NH, Sun SC. ROCK inhibition prevents early mouse embryo development. Histochem Cell Biol 2014;142:227–233. [DOI] [PubMed] [Google Scholar]
- Dumollard R, Duchen M, Carroll J. The role of mitochondrial function in the oocyte and embryo. Curr Top Dev Biol 2007;77:21–49. [DOI] [PubMed] [Google Scholar]
- Einspanier R, Schonfelder M, Muller K, Stojkovic M, Kosmann M, Wolf E, Schams D. Expression of the vascular endothelial growth factor and its receptors and effects of VEGF during in vitro maturation of bovine cumulus-oocyte complexes (COC). Mol Reprod Dev 2002;62:29–36. [DOI] [PubMed] [Google Scholar]
- Falco G, Lee SL, Stanghellini I, Bassey UC, Hamatani T, Ko MS. Zscan4: a novel gene expressed exclusively in late 2-cell embryos and embryonic stem cells. Dev Biol 2007;307:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fulka H, Aoki F. Nucleolus precursor bodies and ribosome biogenesis in early mammalian embryos: old theories and new discoveries. Biol Reprod 2016;94:143. [DOI] [PubMed] [Google Scholar]
- Gao B. Wnt regulation of planar cell polarity (PCP). Curr Top Dev Biol 2012;101:263–295. [DOI] [PubMed] [Google Scholar]
- Golomb L, Volarevic S, Oren M. p53 and ribosome biogenesis stress: the essentials. FEBS Lett 2014;588:2571–2579. [DOI] [PubMed] [Google Scholar]
- Hamatani T, Carter MG, Sharov AA, Ko MS. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell 2004;6:117–131. [DOI] [PubMed] [Google Scholar]
- Head SR, Komori HK, Hart GT, Shimashita J, Schaffer L, Salomon DR, Ordoukhanian PT. Method for improved Illumina sequencing library preparation using NuGEN Ovation RNA-Seq system. Biotechniques 2011;50:177–181. [DOI] [PubMed] [Google Scholar]
- Heyner S, Shi CZ, Garside WT, Smith RM. Functions of the IGFs in early mammalian development. Mol Reprod Dev 1993;35:421–425; discussion 425–426. [DOI] [PubMed] [Google Scholar]
- Huang JC. The role of peroxisome proliferator-activated receptors in the development and physiology of gametes and preimplantation embryos. PPAR Res 2008;2008:732303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huang X, Wang J. The extended pluripotency protein interactome and its links to reprogramming. Curr Opin Genet Dev 2014;28:16–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Inzunza J, Danielsson O, Lalitkumar PG, Larsson O, Axelson M, Tohonen V, Danielsson KG, Stavreus-Evers A. Selective insulin-like growth factor-I antagonist inhibits mouse embryo development in a dose-dependent manner. Fertil Steril 2010;93:2621–2626. [DOI] [PubMed] [Google Scholar]
- Kaneko KJ. Metabolism of preimplantation embryo development: a bystander or an active participant? Curr Top Dev Biol 2016;120:259–310. [DOI] [PubMed] [Google Scholar]
- Kim D, Langmead B, Salzberg SL. HISAT: a fast spliced aligner with low memory requirements. Nat Methods 2015;12:357–360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kubo N, Cayo-Colca IS, Miyano T. Effect of estradiol-17beta during in vitro growth culture on the growth, maturation, cumulus expansion and development of porcine oocytes from early antral follicles. Anim Sci J 2015;86:251–259. [DOI] [PubMed] [Google Scholar]
- Kues WA, Sudheer S, Herrmann D, Carnwath JW, Havlicek V, Besenfelder U, Lehrach H, Adjaye J, Niemann H. Genome-wide expression profiling reveals distinct clusters of transcriptional regulation during bovine preimplantation development in vivo. Proc Natl Acad Sci USA 2008;105:19768–19773. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon J, Kim NH, Choi I. ROCK activity regulates functional tight junction assembly during blastocyst formation in porcine parthenogenetic embryos. PeerJ 2016;4:e1914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Laeno AM, Tamashiro DA, Alarcon VB. Rho-associated kinase activity is required for proper morphogenesis of the inner cell mass in the mouse blastocyst. Biol Reprod 2013;89:122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Latham KE, Garrels JI, Chang C, Solter D. Analysis of embryonic mouse development: construction of a high-resolution, two-dimensional gel protein database. Appl Theor Electrophor 1992;2:163–170. [PubMed] [Google Scholar]
- Latham KE, Schultz RM. Embryonic genome activation. Front Biosci 2001;6:D748–D759. [DOI] [PubMed] [Google Scholar]
- Liao Y, Smyth GK, Shi W. featureCounts: an efficient general purpose program for assigning sequence reads to genomic features. Bioinformatics 2014;30:923–930. [DOI] [PubMed] [Google Scholar]
- Logan S, Pharaoh GA, Marlin MC, Masser DR, Matsuzaki S, Wronowski B, Yeganeh A, Parks EE, Premkumar P, Farley JA et al. Insulin-like growth factor receptor signaling regulates working memory, mitochondrial metabolism, and amyloid-beta uptake in astrocytes. Mol Metab 2018;9:141–155. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Louvi A, Accili D, Efstratiadis A. Growth-promoting interaction of IGF-II with the insulin receptor during mouse embryonic development. Dev Biol 1997;189:33–48. [DOI] [PubMed] [Google Scholar]
- Love MI, Huber W, Anders S. Moderated estimation of fold change and dispersion for RNA-seq data with DESeq2. Genome Biol 2014;15:550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maddox-Hyttel P, Svarcova O, Laurincik J. Ribosomal RNA and nucleolar proteins from the oocyte are to some degree used for embryonic nucleolar formation in cattle and pig. Theriogenology 2007;68:S63–S70. [DOI] [PubMed] [Google Scholar]
- Midic U, VandeVoort CA, Latham KE. Determination of single embryo sex in Macaca mulatta and Mus musculus RNAseq transcriptome profiles. Physiol Genomics 2018;50:628–635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Misirlioglu M, Page GP, Sagirkaya H, Kaya A, Parrish JJ, First NL, Memili E. Dynamics of global transcriptome in bovine matured oocytes and preimplantation embryos. Proc Natl Acad Sci USA 2006;103:18905–18910. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mohan M, Malayer JR, Geisert RD, Morgan GL. Expression patterns of retinoid X receptors, retinaldehyde dehydrogenase, and peroxisome proliferator activated receptor gamma in bovine preattachment embryos. Biol Reprod 2002;66:692–700. [DOI] [PubMed] [Google Scholar]
- Ripps BA, Zhu YP, Burwinkel TH, Kim HN, Buster JE, Minhas BS. Platelet-activating factor production from in vitro and in vivo fertilized murine embryos is similar. Am J Reprod Immunol 1993;30:101–104. [DOI] [PubMed] [Google Scholar]
- Rogers J, Garcia R, Shelledy W, Kaplan J, Arya A, Johnson Z, Bergstrom M, Novakowski L, Nair P, Vinson A et al. An initial genetic linkage map of the rhesus macaque (Macaca mulatta) genome using human microsatellite loci. Genomics 2006;87:30–38. [DOI] [PubMed] [Google Scholar]
- Roudebush WE, Wininger JD, Jones AE, Wright G, Toledo AA, Kort HI, Massey JB, Shapiro DB. Embryonic platelet-activating factor: an indicator of embryo viability. Hum Reprod 2002;17:1306–1310. [DOI] [PubMed] [Google Scholar]
- Ruebel ML, Schall PZ, Midic U, Vincent KA, Goheen B, VandeVoort CA, Latham KE. Transcriptome analysis of rhesus monkey failed-to-mature oocytes: deficiencies in transcriptional regulation and cytoplasmic maturation of the oocyte mRNA population. Mol Hum Reprod 2018;24:478–494. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schramm RD, Bavister BD. A macaque model for studying mechanisms controlling oocyte development and maturation in human and non-human primates. Hum Reprod 1999;14:2544–2555. [DOI] [PubMed] [Google Scholar]
- Sebbagh M, Borg JP. Insight into planar cell polarity. Exp Cell Res 2014;328:284–295. [DOI] [PubMed] [Google Scholar]
- Semeniak D, Kulawig R, Stegner D, Meyer I, Schwiebert S, Bosing H, Eckes B, Nieswandt B, Schulze H. Proplatelet formation is selectively inhibited by collagen type I through Syk-independent GPVI signaling. J Cell Sci 2016;129:3473–3484. [DOI] [PubMed] [Google Scholar]
- Sokol SY. Spatial and temporal aspects of Wnt signaling and planar cell polarity during vertebrate embryonic development. Semin Cell Dev Biol 2015;42:78–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stoffel M, Duncan SA. The maturity-onset diabetes of the young (MODY1) transcription factor HNF4alpha regulates expression of genes required for glucose transport and metabolism. Proc Natl Acad Sci USA 1997;94:13209–13214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tao H, Suzuki M, Kiyonari H, Abe T, Sasaoka T, Ueno N. Mouse prickle1, the homolog of a PCP gene, is essential for epiblast apical-basal polarity. Proc Natl Acad Sci USA 2009;106:14426–14431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trapnell C, Williams BA, Pertea G, Mortazavi A, Kwan G, van Baren MJ, Salzberg SL, Wold BJ, Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol 2010;28:511–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandevoort CA, Mtango NR, Latham KE, Stewart DR. Primate preimplantation embryo is a target for relaxin during early pregnancy. Fertil Steril 2011;96:203–207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vassena R, Boue S, Gonzalez-Roca E, Aran B, Auer H, Veiga A, Izpisua Belmonte JC. Waves of early transcriptional activation and pluripotency program initiation during human preimplantation development. Development 2011;138:3699–3709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang X, Liu D, He D, Suo S, Xia X, He X, Han JJ, Zheng P. Transcriptome analyses of rhesus monkey preimplantation embryos reveal a reduced capacity for DNA double-strand break repair in primate oocytes and early embryos. Genome Res 2017;27:567–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu X, Sandhu S, Patel N, Triggs-Raine B, Ding H. EMG1 is essential for mouse pre-implantation embryo development. BMC Dev Biol 2010;10:99. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Giron CG et al. Ensembl 2018. Nucleic Acids Res 2018;46:D754–D761. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang JY, Dong HS, Oqani RK, Lin T, Kang JW, Jin DI. Distinct roles of ROCK1 and ROCK2 during development of porcine preimplantation embryos. Reproduction 2014;148:99–107. [DOI] [PubMed] [Google Scholar]
- Zhang J, Tomasini AJ, Mayer AN. RBM19 is essential for preimplantation development in the mouse. BMC Dev Biol 2008;8:115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhou LQ, Dean J. Reprogramming the genome to totipotency in mouse embryos. Trends Cell Biol 2015;25:82–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zimin AV, Cornish AS, Maudhoo MD, Gibbs RM, Zhang X, Pandey S, Meehan DT, Wipfler K, Bosinger SE, Johnson ZP et al. A new rhesus macaque assembly and annotation for next-generation sequencing analyses. Biol Direct 2014;9:20. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.