Abstract
Cancer is a complex collection of diseases that are to some degree unique to each patient. Precision oncology aims to identify the best drug treatment regime using molecular data on tumor samples. While omics-level data is becoming more widely available for tumor specimens, the datasets upon which co learning methods can be trained vary in coverage from sample to sample and from data type to data type. Methods that can ‘connect the dots’ to leverage more of the information provided by these studies could offer major advantages for maximizing predictive potential. We introduce a multi-view machine-learning strategy called PLATYPUS that builds ‘views’ from multiple data sources that are all used as features for predicting patient outcomes. We show that a learning strategy that finds agreement across the views on unlabeled data increases the performance of the learning methods over any single view. We illustrate the power of the approach by deriving signatures for drug sensitivity in a large cancer cell line database. Code and additional information are available from the PLATYPUS website https://sysbiowiki.soe.ucsc.edu/platypus.
Keywords: Pattern Recognition, Machine Learning, Multiple View Learning, Cancer, Drug Sensitivity, Incompleteness, Unlabeled Data, Semi-Supervised, Co-Training, Integrative Genomics, Systems Biology, Multidimensional, Multi-Omic
1. Introduction
Predicting whether a tumor will respond to a particular treatment strategy remains a challenging and important task. However, the availability and cost of screening compound libraries for a tumor sample remains prohibitive. At the same time, the use of genomic assays, such as DNA and RNA sequencing, for clinical decision making are on the rise. As the costs for these high-throughput assays drop, applying ‘genomic signatures’ from machine-learning trained on external data in place of the more expensive direct drug assay becomes an option.
One obstacle to achieving this goal is the ability to find training sets for machine-learning classifiers for which comprehensive clinical outcomes are available, e.g. survival or drug sensitivity. Non–uniformity of large composite datasets such as The Cancer Genome Atlas (TCGA, cancergenome.nih.gov) forces many existing approaches to ignore data unless it is available for all samples. At the same time, many studies have samples that would be useful to analyze beyond their original purpose, yet cannot be included because they lack outcome data.
The large number of variables compared to far fewer samples can often result in biologically irrelevant solutions.12 However, issues related to the over-determined nature of the problem sets can be minimized by using prior knowledge to inform feature selection techniques. Incorporating this information can guide learning methods to both more generalizable and interpretable solutions. For example, several approaches that include database-mined gene–gene interaction information have shown promise for interpreting cancer genomics data and utilizing it to predict outcomes.1,10,16,18 In addition, ensembles can reduce error caused by small sample sizes.17
We present a multiple view learning (MVL) framework called PLATYPUS (Progressive LAbel Training bY Predicting Unlabeled Samples) that combines the advantages of the knowledge-driven and ensemble approaches. ‘Views’ are feature extractions of particular data platforms that encode specific prior knowledge and are each allowed to vote on the predicted outcome, providing a more complete and diverse glimpse into the underlying biology. The framework infers outcome labels for unlabeled samples by maximizing prediction agreement between multiple views, thus including more of the data in the classifiers. It reduces overfitting caused by small sample sizes both by predicting labels for unlabeled samples and by incorporating prior knowledge.8
A typical approach in machine learning is to train classifiers on a subset of samples containing all of the data, impute missing data, or train ensembles based on data availability, but are generally restricted to samples with the majority of the data for each sample.20 The semi–supervised MVL approach learns missing patient outcome labels, thus allowing the use of all available labeled and unlabeled datasets. PLATYPUS trains on one or more views and then co-trains on the unlabeled samples. By doing this, PLATYPUS can make predictions on any patient regardless of data availability. This increases overall classifier accuracy while also finding solutions that generalize to the entire population– which has proven extremely difficult in high–feature, low–sample problems.2 A comparison of PLATYPUS to other related methods is provided in Supplemental Section S1.
2. System and methods
2.1. Data
At the time of download the Cancer Cell Line Encyclopedia (CCLE) contained genomic, phenotype, clinical, and other annotation data for 1,037 cancer cell lines,7 described in Section S2. Of these, drug sensitivity data was available for 504 cell lines and 24 drugs. Drug response was converted to a binary label in order to transform the regression problem into a classification problem. For each compound, cell lines were divided into quartiles ranked by ActArea; The bottom 25% were assigned to the ‘non–sensitive’ class and the top 25% to the ‘sensitive’ class. Cell lines lying in the middle were marked with ‘intermediate’ and considered unlabeled in this test (Fig. S2). Note that these samples are often the most difficult to classify as they represent those with a range of sensitivities that may span orders of magnitude where the growth inhibition curve has its steepest changes as a function of drug concentration. Thus, the ability to input a binary designation for the growth inhibition using a co-training strategy could in itself have advantages over approaches that identify cutoffs in the drug response curves that are more-or-less arbitrary, without the use of a clear optimization criteria, and without the ability to make use of genomic signatures.
2.2. Single views and co-training
PLATYPUS uses co–training (Fig. 1) between single views to learn labels for unlabeled samples. Single views are based on different feature sets. Genomic or clinical features can be used directly (baseline views), or transformed using a biological prior (interpreted views). We built four baseline views from the CCLE data: expression, CNV, mutation, Sample- and Patient-Specific (SPS) information; and many interpreted views (Section S3). Each view can be set up with the best suited machine learning algorithm and optimized parameters for its task, e.g. a random forest or an elastic net (Section S5.1).
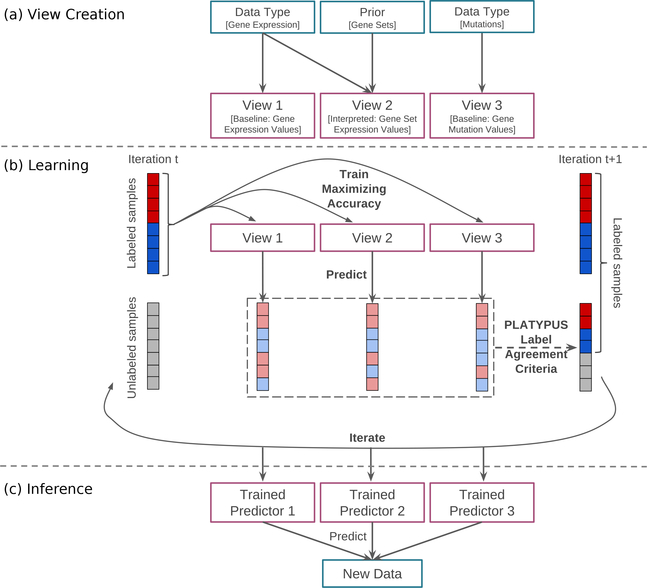
Fig. 1.
PLATYPUS framework illustrated with three views. (a) Creation of single views using sample data and optional prior knowledge. (b) Iterative Learning: Each view maximizes prediction accuracy on the labeled samples; unlabeled samples predicted with high confidence are added to the known sample set; repeat until no new samples are labeled. (c) Models from the final iteration of PLATYPUS training applied to new data.
Co–training works by training a separate classifier using each view as a separate feature set to make independent predictions, then incorporating disagreement into the loss function. Each view trains on the labeled data then predicts labels for the common unlabeled set. High confidence labels are passed as truth in the next iteration. Co–training methods iterate until either convergence, some threshold (a minimal change in label definition on the unlabeled samples) is attained, or a maximum number of iterations is reached.
After co–training, each view can be used as a standalone classifier that incorporates learning from one or more data platforms without relying solely on that data platform. Since views are trained in conjunction, the trained models will incorporate the perspectives of all views. This also provides a measure of influence from all views when applying any of the classifiers to new data, without requiring data for those views when making predictions.
2.3. Maximizing agreement across views through label assignment
The key step in the PLATYPUS approach is the inference of outcome labels for a set of unlabeled data. Each training iteration seeks to improve the agreement of the assignments given to the unlabeled data across all views. Views are first created by applying machine learning methods using either the features directly, or from gene set summaries or subsetting (Section S3). Fig. 1 shows an overview of PLATYPUS using three views. Any number of views may be used– in this paper, up to 10 views are used per experiment.
PLATYPUS searches iteratively for a label assignment that improves the agreement on unlabeled data (Fig. 1(b)). At each iteration t, the views are trained on labeled data and the labels for unlabeled samples are inferred. Because the set of labels can change across iterations, we denote the training data with sensitive labels as T+(t) and those with non–sensitive labels as T−(t) at iteration t. T+(0) and T−(0) are the given sets of sensitive and non–sensitive training samples before learning labels, respectively. The set of unlabeled samples is denoted U(t), with all unlabeled samples before learning labels as U(0).
V is the set of views used in the PLATYPUS run. In iteration t, each view v ∈ V is trained to maximize its prediction accuracy on the labeled samples T+(t) and T−(t). The accuracy of view v at iteration t is determined using cross–validation of the training samples and is written here as a(v, t), where a(v,0) is the single view accuracy before learning labels. A prediction is then made by the trained models for each unlabeled sample s. Let l(v, s, t) be the prediction of sample s by view v in iteration t where it is 1 if predicted sensitive and 0 otherwise. The single view votes are summarized to a sensitive ensemble vote L+(s, t) and non–sensitive ensemble vote L−(s, t) for each sample (Eq. 1 and 2).
| (1) |
| (2) |
Only views with data to predict sample s are taken into account: V s = {v ∈ V : v has data for s}; and the different views are weighted by w(v,t) (Eq. 3). View accuracies within [0.5,1] are rescaled to [0,1] and log–scaled. Views with an accuracy lower than 0.5 are given a weight of 0 since it indicates worse than random predictions.
| (3) |
To determine, which unlabeled samples are added to the training data for the next iteration, we define Lmax(t), the strongest vote found between all samples in iteration t (Eq. 4), and Ψ(t), the set of samples reaching the strongest vote (Eq. 5).
| (4) |
| (5) |
In order to favor missing data for a sample over conflicting predictions, we define Lmin(t) as mins∈Ψ(t){min{L+(s, t),L−(s, t)}}, the weakest contrary vote that is found between all samples in Ψ(t).
All samples meeting both the strongest vote and the weakest contrary vote conditions (Label Agreement Criteria) build the set of new training samples , which are added to T+(t) and T−(t) for the next iteration’s training data:
| (6) |
| (7) |
| (8) |
To avoid adding predictions with low confidence, Lmax(t) needs to stay above a certain value, otherwise no labels are added to the training data in iteration t. This can be adjusted by the learning threshold λ, which represents the fraction of the maximal reachable vote, i.e. when all views agree. By default λ is 75%.
The training process continues until a convergence criterion is met: either all labels have been learned, no new labels have been learned in the last iteration, or a maximum number of iterations has been reached. After termination of the learning process, the trained single–view predictors can be used independently or as an ensemble via PLATYPUS (Fig. 1(c)).
3. Results
3.1. Preliminary experiments to optimize PLATYPUS performance
We ran 120 different PLATYPUS variants to predict drug sensitivity in the CCLE cell lines to identify the best way to combine the views for this application. As mentioned in the Data Section (Section S2), samples with intermediate levels of sensitivity for a particular drug were treated as unlabeled and used by the co-training to maximize agreement across views. The conversion of this regression problem into a classification problem in which drug sensitivities arbitrarily are discretized into sensitive versus insensitive (top and bottom 25%), reflects the reality of the clinical setting in which a decision must be made to either treat or not treat a particular patient. The test measures the co-training strategy’s ability to infer sensitivities for cell lines that are the most difficult to classify.
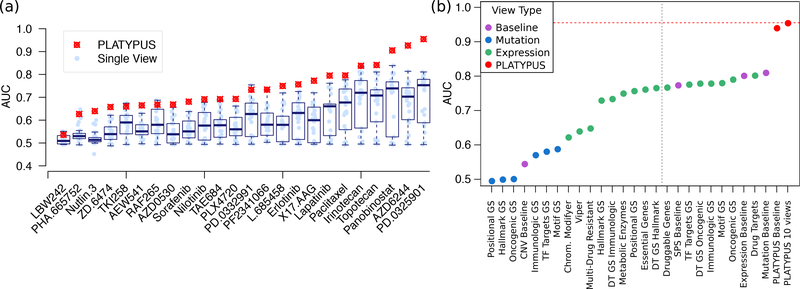
We first asked whether the interpretive views that use gene set information provide benefit over using only the baseline views (Section 2.2). We then determined a weighting scheme for the ensemble to achieve better performance. We ran PLATYPUS using the 4 baseline views and the 3, 5, 7, and 10 best–performing single views for each of the 24 CCLE drugs at a λ = 75% learning threshold, for a total of 120 different PLATYPUS variants (5 per drug). Fig. 2(a) shows the highest accuracy PLATYPUS models as well as each of the single view scores. In almost all cases PLATYPUS significantly outperforms single view models, most notably for the MEK inhibitors AZD6244 and PD-0325901, and HDAC inhibitor Panobinostat. Adding interpreted views to PLATYPUS increased PD-0325901 AUC from 0.94 to 0.99 (Fig. 2(b)), motivating their continued inclusion in PLATYPUS models. Furthermore, within 10 iterations, most PLATYPUS runs added 90% or more of the unlabeled cell lines to the labeled set, effectively doubling the number of samples on which the models trained. We look more closely at the results from the best overall performing PLATYPUS model, PD-0325901, as well as important features from each of its models, in Section 3.2.
Fig. 2.
PLATYPUS Performance. (a) Boxplot showing performance (in AUC) sorted by PLATY-PUS score, of all single views and the best PLATYPUS score. PLATYPUS score for each drug is the highest from the 3,5,7, and 10 view runs. (b) AUC for PD-0325901 sensitivity predictions for each single view, colored by view type. The 10 views to the right of the gray line are used in the PLATYPUS ensemble. See Fig. S3 for single view AUCs for all drugs. DT = Drug Target; GS = Gene Set.
We next investigated how to combine the ensemble of different views to improve the PLATYPUS method’s accuracy. Previous studies show that combining multiple weak but independent models will result in much higher model accuracy.17,20 Similarly, previous work has shown that using biological priors can reduce the influence of noise present in biological data.10,11,18 However, it is not clear how models can be combined in an ensemble to achieve the best results. First, we tested a weighting scheme where each view contributed equally to the final prediction, however this made the model sensitive to information–poor views (data not shown). We then tested an AUC–weighted voting scheme, which derives view weights for the current iteration based on the AUC obtained from the previous iteration (Eq. 3). Doing so allows the PLATYPUS ensemble to incorporate a large number of views, without the need for a pre-selection step, where each view has the opportunity to either become more accurate, and contribute more to the prediction outcome, during label learning, or is effectively ignored if it never reaches a high accuracy.
Figs. S5 and S6 show the effectiveness of label learning validation (LLV) for each of the 24 drugs in CCLE. Most of the drug models learn labels correctly, however model AUC decreases once a model starts to learn labels incorrectly. Over many iterations this can lead to a model where the majority of labels are learned incorrectly (e.g. Nutlin-3, Fig. S6). We found that this risk can be minimized by setting a high confidence threshold for label learning and by using many information–independent views. In our experiments, LLV consistently helps identify optimal parameters to run PLATYPUS on a given dataset.
Without missing data, PLATYPUS is equivalent to a classic ensemble classifier and often outperforms any single view model. In order to understand the benefits of using additional unlabeled data, we compared the ‘ensemble’ (first) iteration of PLATYPUS to the final and the ‘best’ iterations. We define ‘best’ as the iteration with the highest AUC. Interestingly, in almost all cases, the PLATYPUS AUC is higher than the ensemble AUC (Fig. S4). The use of more samples by PLATYPUS helps ensure a more generalizable model. For the experiments in this paper, we intentionally set a high number for maximum iterations to show how label learning can degrade over time, and therefore the final iteration often scores poorly. Label learning degradation is avoidable by using high label learning thresholds and an appropriate number of iterations.
3.2. Predicting drug sensitivity in cell lines
Our analysis focuses on the full CCLE dataset, composed of 36 tumor types. For most drugs, the Sample- and Patient-Specific (SPS) view has the highest starting view performance with AUCs ranging from 0.6 to 0.8, and expression baseline views often performed similarly. The mutation view is effective for some drugs (e.g. MEK inhibitors). Three of the four baseline views are top performers for predicting cancer cell line sensitivity to PD-0325901 (Fig. 2(b)), a MEK1/2 inhibitor. CNV view performance was never high enough to warrant inclusion in PLATYPUS models except as the ‘aggregated copy number changes’ feature in the SPS view.
Interpreted views often outperform the SPS view (Fig. S3). We found several examples in which a biological prior view outperformed the data–specific view, e.g. Metabolic Enzymes, Drug Targets, and Chromatin Modifying Enzymes are better at predicting Lapatinib sensitivity than the baseline expression predictor. The Drug Target Gene Set Hallmark view outperforms data–specific views in Irinotecan and Panobinostat sensitivity predictions. Such examples can be found for all compounds except for the MEK inhibitors, for which the baseline mutations view is always the top performer.
In general, views incorporating expression data have high accuracy (Fig. S3), whereas mutation views are comparable to a random prediction in most cases. This could be due to the presence of many passenger mutations that have little bearing on cell fitness and drug response. In one notable exception, AZD6244, the Drug Target Mutation view is more accurate than the Drug Target Expression view. Generally, interpreted mutation views outperform their baseline counterpart. For example, the Drug Target Mutation view is more accurate than the baseline mutation view in both Irinotecan and Topotecan. Furthermore, the Drug Target Mutation view trained on PD-0325901 increases the relative feature weights for RAS genes, suggesting that it identifies the exclusivity of RAS/BRAF mutations described in Section S6. However overall, mutation views have low accuracy despite mutations being key to drug sensitivity, indicating that other representations that increase the signal-to-noise ratio of this data should be explored in future work.
The Drug Target Gene Set views created from Molecular Signatures Database (MSigDB) gene set collections perform well overall, especially on Irinotecan, Topotecan, and Panobinostat (Fig. S3). For most compounds the Drug Target Gene Set Hallmark is more accurate than the Oncogenic and Immunologic. A possible reason is that these gene sets are from the Hallmark collection, which are re–occurring, highly reliable gene sets built from combinations of other gene set collections. Their similar performance could also be due to overlap in the gene sets. We recommend that users test for and subsequently remove highly correlated views before running label learning, and intend to incorporate this into future versions of PLATYPUS. One approach to handling correlated views is to extend the ensemble vote step to use stacked learning instead of the current agreement formula. By training a model on the predictions from each view, PLATYPUS may be better able to handle correlated views by treating them with less weight than more independent views.
In addition to the MSigDB gene set views, master regulator–based predictors via Virtual Inference of Protein activity by Enriched Regulon analysis (VIPER)13 were tested but are not among the top performing ones for any drug. This could be due to use of a generic regulon as VIPER input rather than tissue–specific versions for each cell line.13
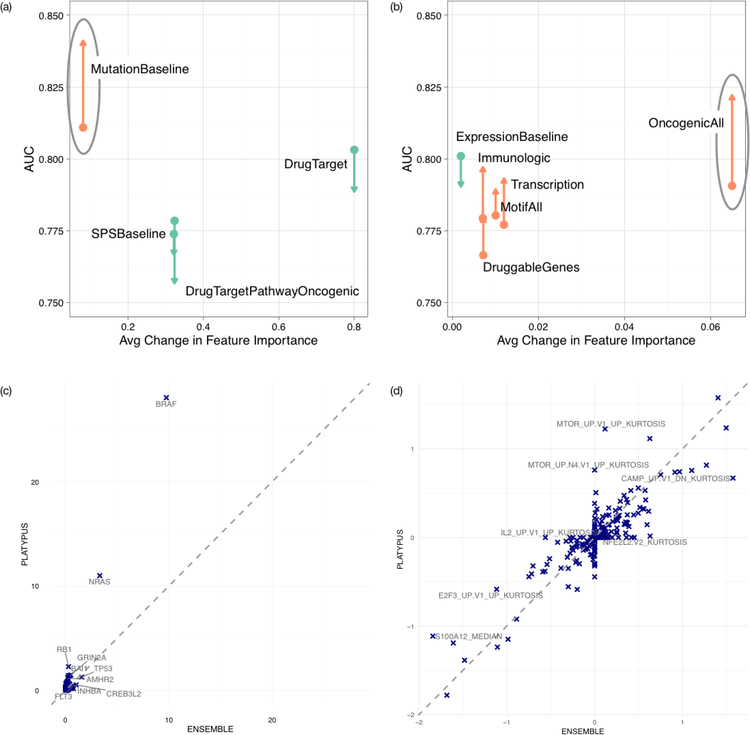
The PLATYPUS model for the drug PD-0325901 achieved the highest accuracy of all experiments, with a near perfect AUC. We therefore chose to further investigate the results of this drug to identify the nature by which the MVL approach finds an improved classification. PD-0325901 was initially tested in papillary thyroid carcinoma cell lines and is known to be especially effective in cell lines with BRAF mutations.14 Since these are frequent in the CCLE data, the high accuracy of the single view models is expected. Fig. 3 shows changes from the ensemble to the ‘best’ PLATYPUS PD-0325901 models. Single view AUCs mostly increase after several iterations, and feature weights within the models also shift to varying degrees. In the baseline mutations view, RAS gene mutations have higher Gini coefficient changes in the PLATYPUS model than in the ensemble (Fig. 3(c)), indicating increased model importance of those genes. Past studies of the CCLE data7 and our analysis (Section S6 and Table S3) have found RAS and BRAF mutations in the data tend to be mutually exclusive, both of which are linked to PD-0325901 sensitivity (Fig. S1). Thus, PLATYPUS is better able to identify the dual importance of RAS/BRAF mutations than the single view and ensemble models.
Fig. 3.
Performance and feature weight changes for single views between ensemble and PLATYPUS in predicting sensitivity to PD-0325901. (a) For each random forest view, the average Gini change for all features between the ensemble and the best PLATYPUS iteration, plotted against the view AUC for the ensemble (arrow tail) and PLATYPUS (arrow head). Circled view is shown in detail in (c). (b) Same as (a), but showing the elastic net views and their average change in feature weights. (c) Scatter plot where each point is a feature in the Baseline Mutations view. Plot shows the ensemble feature weight versus the PLATYPUS feature weight. (d) Same as (c), but showing feature weight changes in the Oncogenic (OncogenicAll in (b)) view.
We also chose to look at a case where PLATYPUS failed to achieve an improvement. LBW242 is one such case. The single views for this drug all have near random scores. However, instead of identifying an improvement through view combination as is the usual case in our experiments (e.g. PHA-665752 and Nutlin-3), the PLATYPUS models also achieved near random performance (Fig. 2(a)). Further investigation reveals that the performance may not be the fault of PLATYPUS. Instead, little signal may be available in the drug sensitivity labels for this case due to our quantization strategy (i.e. using the upper- and lower-quartiles for the resistant and sensitive classes). The dose-response curve for LBW242 shows very few of the CCLE cell lines may be truly sensitive. While our approach creates balanced class sizes and ensures continuity between experiments, finding a more nuanced per–drug cutoff would likely improve model performance. Suboptimal label cutoffs lead to a low signal–to–noise ratio in the labels for a few of the drugs, which in general leads to low classifier performance.19 It is also possible that the metric for drug sensitivity for some drugs is ineffective. Traditional methods to quantify sensitivity are dependent on population growth and thus slow–growing cell lines may appear to be resistant to all drugs.6
These results are consistent with previous findings that have shown sensitivity to some compounds is easier to predict than others.9 For example, the two MEK inhibitors (PD-0325901, AZD6244) and Panobinostat have higher overall accuracy in the single view models (Fig. S3). Interestingly, in the case of Panobinostat, the ‘Chromatin Modifiers’ and ‘Positional Gene Set’ PLATYPUS views have higher single view accuracy than the baseline expression view, which could indicate that there is an epigenetic effect from chromatin modifiers. We postulate that a small region of the genome has been unwound, lending sensitivity to Panobinostat. PLATYPUS captures this interaction, whereas single view models do not.
3.3. Key features from PLATYPUS models
Each machine-learning algorithm used by a view has its own internal feature selection. We extracted features from these models to evaluate the most informative features. Fig. 3(a-b) show changes in single view model performance and average feature importance within those models, before and after PLATYPUS training. Fig. 3(c-d) show feature changes and enrichment of those features within one of the views. Fig. 3(c) highlights how PLATYPUS is able to remove feature weights of spurious correlations between cell line mutations and the true mutation features of importance, NRAS and BRAF. While the overall feature weights in the single view model do not have large changes from the ensemble to PLATYPUS frameworks, there is a large shift in 2 key features which are known to be significantly associated with sensitivity to this particular drug. PLATYPUS is able to avoid overfitting the model whereas the ensemble is unable to draw from external information. In Fig. 3(d), the model has significantly changed both in AUC and in feature weights between the ensemble and PLATYPUS experiments.
Fig. S8 shows a closeup of the changes within the Fig. 3(d) view between PLATYPUS and a general ensemble. It focuses on one feature from the view, MTOR up V1 up kurtosis, which had the biggest increase in feature weight from ensemble to PLATYPUS. At a glance, this gene set is not associated with cancer– it describes genes that are regulated by an inhibitor used to prevent graft rejection by blocking cell proliferation signals via mTOR. However, the gene set kurtosis correlates with ActArea and with our binary drug sensitivity labels (Fig. S8(a-b)). A closer look shows that this is because of gene-gene correlations within the gene set. Kurtosis features are intended to capture large changes within the gene set. Mean and median gene set correlation values do not capture cell line differences in the co-correlated gene clusters, whereas kurtosis highlights extreme values. No one gene expression correlates strongly with the kurtosis of the whole set (Fig. S8(c, e)), and so the set cannot be replaced with a single gene expression value. Clusters within the gene set are linked to EGFR signaling (cluster IV, genes marked E), metastasis and Basal vs Mesenchymal BRCA (cluster V, genes marked M and B respectively), and resistance to several cancer drugs (clusters II and V, genes marked R). Gene-gene correlations shown in Fig. S8(d) combine to form the overall kurtosis score. As shown in Fig. S8(e), many genes related to cancer processes are the driving force in the gene set kurtosis score. This highlights how small overall changes combine to improve PLATYPUS accuracy over the ensemble.
Many of the highly ranked features from other models (Fig. S7 shows expression view for PD-0325901, other data not shown) are known oncogenes, for example ETV4 was previously found to be correlated with MEK inhibitor sensitivity.15 SPRY2, a kinase inhibitor, correlates with BRAF mutation status, both of which are predictive of sensitivity to PD-0325901, AZD6244, and PLX4720. DUSP6 has been named as a marker of FGFR inhibitor sensitivity4 and a previous study shows a weak inverse correlation between DUSP6 expression and sensitivity to MEK1/2 inhibitors.3 Thus PLATYPUS recapitulates several known markers of drug sensitivity.
4. Conclusions
When compared to a traditional ensemble and to single view predictors, PLATYPUS often has higher AUC (Fig. 2). The multi-view approach uses the set of unlabeled samples as links between different views to find agreement in the different feature spaces. Since label learning validation shows that labels are learned correctly in most cases, the increase in improve model performance may be due to doubling the number of samples that can be considered while training. In 96% of our experiments, PLATYPUS outperforms an ensemble (Fig. S4). Furthermore, PLATYPUS outperforms 85% of the single views and has higher AUC than all of the single views for 17 of the 24 drugs. No one single view consistently outperforms any of the PLATYPUS models In order to retain such high performance without PLATYPUS, a user would need to test all single view models.
Important features from PLATYPUS views (both baseline and interpreted) have previously been linked to drug sensitivity. The approach generally improves AUC while incorporating significantly more data and allowing uncertainty– a necessity in medical research. By combining extracted features from each of the MVL model views, the user is provided a clearer picture of the key facets of sensitivity to each drug. We also investigated the generality of PLATYPUS by applying it to the prediction of an aggressive subtype of prostate cancer and found it generalized to an external validation set not used during training (see Supplemental Section S7). Overall, PLATYPUS enables the use of samples with missing data, benefits from views without high correlation, and is a flexible form of MVL amenable to biological problems.
The PLATYPUS co-training approach has several important advantages. First, it is ideal when samples have missing data, a common scenario in bioinformatics. Imagine a new patient entering a clinic for whom not all of the same data is available as was collected for a large drug trial. A PLATYPUS model trained on the drug trial data is able to predict drug response for this patient without retraining, simply by restricting to views for which there is patient data. For example, a sample with only expression data could be provided predictions using the expression–based views. Predicted label confidence for that sample will be much lower since there are no scores from the missing views, ensuring that labels for samples with complete data will be inferred in earlier iterations than those with missing data. PLATYPUS automatically sets weights for view predictions, implicitly accounting for missing data, and ensuring future predictions are not constrained by limited data. Second, co-training allows for the use of different classification methods for each data type, capturing the strengths of each data type and increasing flexibility in the framework. Third, PLATYPUS is effective when using information–divergent views. Fourth, co–training combines predictions at a later stage in the algorithm, so that views are trained independently. This is ideal for ensemble learning, which has shown to be highly effective when models/views are independent, even with low individual model accuracy.5,17
It is worth mentioning some distinct limitations of the approach as a pointer toward future work. First, if missing data correspond to cases that are more difficult to classify, rather than missing at random, the poorer performance of individual views may result in appreciably lower agreement, and thus little benefit in combining views. Second, combining multiple views introduces the need for setting additional parameters (e.g. the agreement threshold). This requires a user to gain familiarity with the performance of newly incorporated views in test runs before final results can be obtained. Finally, highly correlated views can inflate the agreement voting and down-weigh other, uncorrelated views. A future adjustment could incorporate prediction correlation on the labeled samples for the voting of unlabeled samples.
Supplementary Material
Acknowledgments
We thank Evan Paull, Dan Carlin, Yulia Newton, Pablo Cordero, Anya Tsalenko, Robert Kincaid, and Artem Sokolov for feedback during PLATYPUS implementation. K.G. was supported by an Agilent Fellowship. V.F. was supported by a PROMOS scholarship of the Technical University of Munich (TUM). J.S. was supported by grants from NCI (U24-CA143858, 1R01CA180778, NHGRI (5U54HG006097), and NIGMS (5R01GM109031).
References
- 1.Turki T and Wei Z, BMC systems biology 11, p. 94 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Airola A, Pahikkala T, Waegeman W, De Baets B and Salakoski T, in Machine learning in systems biology, 2009. [Google Scholar]
- 3.Gupta S, Chaudhary K, Kumar R, Gautam A, Nanda JS, Dhanda SK, Brahmachari SK and Raghava GP, Scientific reports 6, p. 23857 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Nakanishi Y, Mizuno H, Sase H, Fujii T, Sakata K, Akiyama N, Aoki Y, Aoki M and Ishii N, Molecular cancer therapeutics, molcanther (2015). [DOI] [PubMed] [Google Scholar]
- 5.Bagnall A, Lines J, Bostrom A, Large J and Keogh E, Data Mining and Knowledge Discovery 31, 606 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hafner M, Niepel M, Chung M and Sorger PK, Nature methods 13, p. 521 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Barretina J, Caponigro G, Stransky N, Venkatesan K, Margolin AA, Kim S, Wilson CJ, Lehár J, Kryukov GV, Sonkin D et al. , Nature 483, p. 603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Cheng W-Y, Yang T-HO and Anastassiou D, PLoS computational biology 9, p. e1002920 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Costello JC, Heiser LM, Georgii E, Gönen M, Menden MP, Wang NJ, Bansal M, Hintsanen P, Khan SA, Mpindi J-P et al. , Nature biotechnology 32, p. 1202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Seoane JA, Day IN, Gaunt TR and Campbell C, Bioinformatics 30, 838 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Vaske CJ, Benz SC, Sanborn JZ, Earl D, Szeto C, Zhu J, Haussler D and Stuart JM, Bioinformatics 26, i237 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Venet D, Dumont JE and Detours V, PLoS computational biology 7, p. e1002240 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Alvarez MJ, Shen Y, Giorgi FM, Lachmann A, Ding BB, Ye BH and Califano A, Nature genetics 48, p. 838 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Henderson YC, Chen Y, Frederick MJ, Lai SY and Clayman GL, Molecular cancer therapeutics, 1535 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Leow CC-Y, Gerondakis S and Spencer A, Blood cancer journal 3, p. e105 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jang IS, Dienstmann R, Margolin AA and Guinney J, in Pacific Symposium on Biocomputing Co-Chairs, 2014. [PMC free article] [PubMed] [Google Scholar]
- 17.Rokach L, Artificial Intelligence Review 33, 1 (2010). [Google Scholar]
- 18.Iorio F, Knijnenburg TA, Vis DJ, Bignell GR, Menden MP, Schubert M, Aben N, Gonçalves E, Barthorpe S, Lightfoot H et al. , Cell 166, 740 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Frénay B and Verleysen M, IEEE transactions on neural networks and learning systems 25, 845 (2014). [DOI] [PubMed] [Google Scholar]
- 20.Kriegel H-P and Zimek A, Proceedings of MultiClustKDD (2010).
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.