Summary
Salmonella Typhimurium induces inflammatory diarrhea and uptake into intestinal epithelial cells using the Salmonella pathogenicity island 1 (SPI1) type III secretion system (T3SS). Three AraC-like regulators, HilD, HilC, and RtsA, form a feed-forward regulatory loop that activates transcription of hilA, encoding the activator of the T3SS structural genes. Many environmental signals and regulatory systems are integrated into this circuit to precisely regulate SPI1 expression. A subset of these regulatory factors affect translation of hilD, but the mechanisms are poorly understood. Here, we identified two sRNAs, FnrS and ArcZ, which repress hilD translation, leading to decreased production of HilA. FnrS and ArcZ are oppositely regulated in response to oxygen, one of the key environmental signals affecting expression of SPI1. Mutational analysis demonstrates that FnrS and ArcZ bind to the hilD mRNA 5’ UTR, resulting in translational repression. Deletion of fnrS led to increased HilD production under low aeration conditions, whereas deletion of arcZ abolished the regulatory effect on hilD translation aerobically. The fnrS arcZ double mutant has phenotypes in a mouse oral infection model consistent with increased expression of SPI1. Together, these results suggest that coordinated regulation by these two sRNAs maximizes HilD production at an intermediate level of oxygen.
Keywords: Salmonella infection, SPI1, HilD, FnrS, ArcZ
Graphical Abstract
Salmonella is a leading cause of gastrointestinal disease worldwide. Proper temporal and spatial expression of the Salmonella SPI1 type-three secretion system is critical for invasion of the host intestinal epithelium. Here, we show that two oxygen-dependent sRNAs, FnrS and ArcZ, regulate production of the invasion machinery, tuning SPI1 expression to a particular oxygen level consistent with that at the epithelial surface.
Introduction
Bacterial pathogens encounter various host environments and adapt their physiology and virulence traits accordingly. Salmonella enterica serovar Typhimurium preferentially infects epithelial cells in the distal ileum of the small intestine, causing inflammatory diarrhea and initiating systemic infection (Savidge et al., 1991; Clark et al., 1994; Penheiter et al., 1997; Jepson and Clark, 2001). Both invasion and induction of inflammation is dependent on the type three secretion system (T3SS) encoded on Salmonella pathogenicity island 1 (SPI1), which directly injects bacterial effectors into the host cell cytosol to initiate cytoskeletal rearrangement and alter signal transduction (Penheiter et al., 1997; Galan and Collmer, 1999; Galan, 2001; Ellermeier and Slauch, 2007).
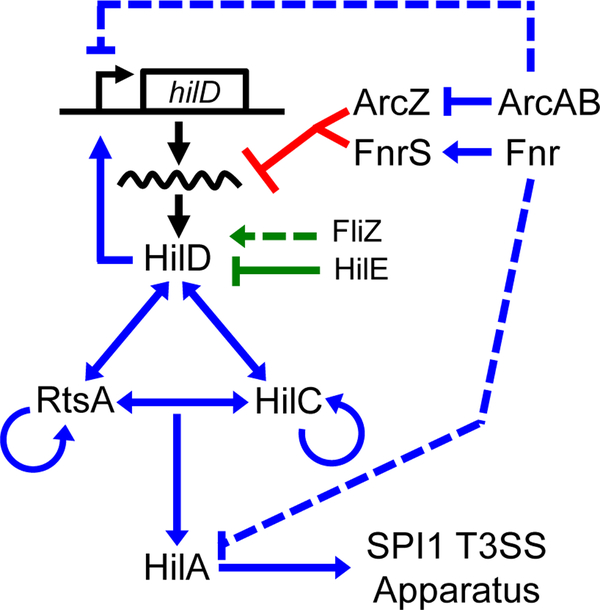
Production of the SPI1 T3SS is tightly regulated in response to numerous environmental cues (Jones, 2005; Ellermeier and Slauch, 2007; Golubeva et al., 2012). HilA (hyperinvasion locus A), encoded within the SPI1 locus, directly activates the T3SS structural genes (Bajaj et al., 1996; Darwin and Miller, 1999; Eichelberg and Galan, 1999). Transcription of hilA is activated by three AraC-like proteins, HilD, HilC, and RtsA, which each bind the promoter of hilA to directly enhance transcription. In addition, HilD, HilC, and RtsA activate their own transcription and the transcription of each other, forming a complex feed-forward regulatory loop (Olekhnovich and Kadner, 2002; Ellermeier et al., 2005; Olekhnovich and Kadner, 2006; Olekhnovich and Kadner, 2007) (Fig 1).
Figure 1. Simplified model of the SPI1 T3SS regulatory circuit.
Blue lines indicate transcriptional regulation, green lines indicate regulation of HilD at the protein level, and red lines indicate regulation of hilD translation. Dotted lines indicate that the exact mechanism of regulation is not known and is likely indirect.
A number of environmental signals and regulatory systems are integrated into the SPI1 regulatory circuit primarily at the level of hilD translation or HilD protein activity (Golubeva et al., 2012), while HilC and RtsA act to amplify these inducing signals (Ellermeier and Slauch, 2007; Saini et al., 2010; Golubeva et al., 2012) (Fig. 1). For example, FliZ and HilE control HilD protein activity (Baxter et al., 2003; Chubiz et al., 2010; Golubeva et al., 2012; Grenz et al., 2018). Several systems have been shown to control hilD mRNA translation or stability, but the underlying mechanism is understood in only a couple of cases (Martinez et al., 2011). Given the role of HilD as signal integrator and dominant activator of hilA transcription, characterizing the direct regulation of hilD translation is crucial to understanding SPI1 regulation at a systems level.
Small regulatory RNAs (sRNAs) are increasingly recognized as key regulators of bacterial physiology and virulence (Frohlich and Vogel, 2009; De Lay et al., 2013; Desnoyers et al., 2013; Jagodnik et al., 2017). Ranging from 50 to 500 nucleotides in length, sRNAs control gene expression by base pairing with target mRNAs. The RNA chaperones Hfq or ProQ facilitate base pairing-dependent regulation by sRNAs on mRNA targets (Moller et al., 2002; Geissmann and Touati, 2004; Mandin and Gottesman, 2009; Vogel and Luisi, 2011; Henderson et al., 2013; Smirnov et al., 2016). The outcomes of sRNA-mRNA base pairing vary, but usually the translation and/or stability of the mRNA is affected either positively or negatively (Majdalani et al., 1998; Majdalani et al., 2001; Vanderpool and Gottesman, 2004; Masse et al., 2005; McCullen et al., 2010). In Salmonella, both experimental and bioinformatic approaches have been used to identify more than 325 sRNAs, but few of them are characterized (Kroger et al., 2012; Kroger et al., 2013; Srikumar et al., 2015; Colgan et al., 2016; Smirnov et al., 2016). Dramatic changes in expression of all SPI1 genes in hfq mutants compared to wild-type Salmonella implicate sRNAs in direct or indirect control of SPI1 gene expression (Sittka et al., 2007; Storz et al., 2011).
Low oxygen tension and high osmolarity are conditions traditionally used to induce the SPI1 T3SS (Ni Bhriain et al., 1989; Lee and Falkow, 1990; Tartera and Metcalf, 1993; Ellermeier and Slauch, 2007; Jennewein et al., 2015). Salmonella primarily utilizes two major regulators, Fnr and the ArcAB two-component system, to adapt to oxygen-related environmental changes (Fink et al., 2007; Lim et al., 2013; Troxell and Hassan, 2016). Fnr is directly inactivated by oxygen via oxidation of its 4Fe-4S cluster (Khoroshilova et al., 1997). Fnr is known to affect expression of the SPI1 T3SS (Fink et al., 2007; Golubeva et al., 2012). Our previous data suggest that Fnr negatively regulates SPI1 by repressing hilA expression, but the mechanism is unclear (Golubeva et al., 2012). In the mouse model, fnr mutants show a mild loss of virulence (Craig et al., 2013).
The activity of the ArcAB two-component system is influenced by oxygen availability (Georgellis et al., 2001; Lu et al., 2002). ArcB is a histidine sensor kinase, and controls the phosphorylation of ArcA, the response regulator. ArcB activity responds to the oxidation state of the quinone pool of the respiratory chain, thus acting as an indirect oxygen sensor (Bauer et al., 1999). ArcB phosphorylates ArcA under anaerobic conditions and dephosphorylates ArcA-P under aerobic conditions. ArcA-P controls expression of various genes including some involved in aerobic metabolism. ArcA also affects hilA expression under aerobic growth conditions, but the mechanism is unclear (Lim et al., 2013). Despite several links between oxygen sensing and regulation of SPI1 gene expression, the overall mechanisms of oxygen-mediated control of SPI1 remain enigmatic.
Two sRNAs, FnrS and ArcZ, are produced in response to changes in oxygen tension. Data from E. coli and Salmonella show that FnrS is produced under anaerobic conditions due to Fnr-mediated activation of fnrS transcription, whereas ArcZ is produced primarily under aerobic conditions; its transcription is repressed anaerobically by ArcA-P (Papenfort et al., 2009; Durand and Storz, 2010; Mandin and Gottesman, 2010). FnrS represses genes related to aerobic metabolism and genes encoding products that protect cells from reactive oxygen species (Boysen et al., 2010; Durand and Storz, 2010). ArcZ regulates rpoS, csgD and eptB mRNAs along with several others encoding products related to aerobic metabolism (Papenfort et al., 2009; Mandin and Gottesman, 2010; Moon et al., 2013; Mika and Hengge, 2014).
Here, we show that these two sRNAs, FnrS and ArcZ, provide missing links between environmental oxygen changes and changes in SPI1 gene expression. FnrS and ArcZ both inhibit hilD translation, thus leading to decreased expression of hilA and the SPI1 T3SS (Fig 1). We suggest that this dual repression of hilD by two sRNAs produced at different oxygen concentrations optimizes production of the SPI1 T3SS to intermediate oxygen tensions. This may be one mechanism by which Salmonella cells precisely control the production of the SPI1 T3SS at the right time and place within the host GI tract.
Results
Screening an E. coli sRNA library for regulators of hilD translation initiation
We previously showed that multiple regulatory signals feed into the SPI1 T3SS system by affecting hilD at the post-transcriptional level (Golubeva et al., 2012). We hypothesized that some of this regulation is mediated through sRNAs. To identify direct sRNA regulators of hilD translation, we took advantage of an E. coli sRNA library that includes key sRNA regulators of cellular physiology (Mandin and Gottesman, 2010), and virulence gene control (Vogel, 2009). The sRNAs included in this library are highly conserved between E. coli and Salmonella. We created a PBAD-hilD’-’lacZ translational fusion in E. coli under the control of an arabinose-inducible promoter (Mandin and Gottesman, 2009). The hilD translational fusion contains the 35-nt 5’ UTR and the first 11 codons of hilD fused in-frame to lacZ (Fig S1). This is analogous to the in-locus lacZ fusion that we have used in Salmonella, and our previous studies showed that this region of hilD is sufficient for the observed post-transcriptional regulation (Golubeva et al., 2012). Performing our initial screening in E. coli reduced the complications imposed by the complex feed-forward loop controlling SPI1 gene expression in Salmonella (Ellermeier et al., 2005; Golubeva et al., 2012).
Plasmids encoding one of 23 sRNAs from E. coli under control of a lac promoter (Mandin and Gottesman, 2009) were each introduced into an E. coli PBAD-hilD’-’lacZ fusion strain. Strains were cultured in low salt LB (LSLB) with 100 μM IPTG to induce expression of the sRNA and 0.001% arabinose to induce the reporter fusion and β-galactosidase activity was measured. Five of the sRNAs in the library, RydC, GcvB, MicC, ArcZ, and FnrS, caused an ~2-fold or greater decrease in expression of the hilD’-’lacZ fusion (Fig S2). Overexpression of either FnrS or ArcZ had the greatest effect and we have focused on these sRNAs.
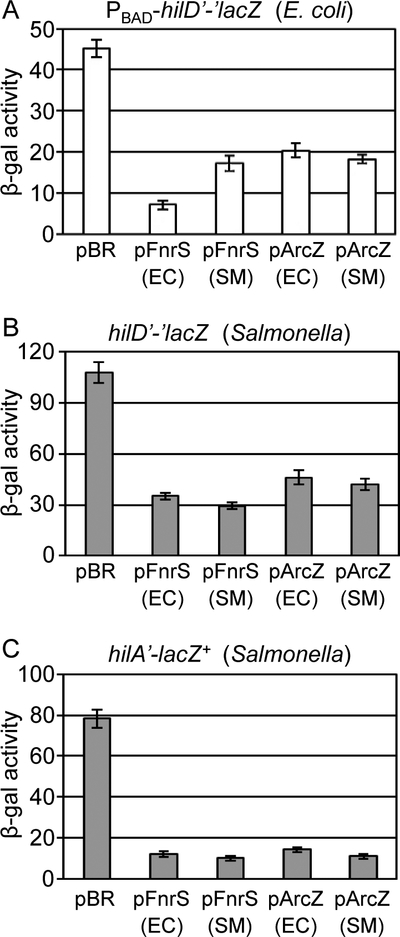
Both FnrS (122 nt) and the processed ArcZ (57 nt) are highly conserved between E.coli and Salmonella, each differing by only 2 nucleotides in the stem-loop of the terminators (Papenfort et al., 2009; Durand and Storz, 2010; Chao et al., 2017). Despite this conservation, we cloned fnrS and arcZ from Salmonella Typhimurium strain 14028 into the pBRpLac. Overexpression of either the E.coli or Salmonella FnrS and ArcZ sRNAs led to decreased expression of the hilD’-’lacZ fusion in E. coli (Fig 2A). We then introduced the plasmids producing the Salmonella and E. coli FnrS and ArcZ sRNAs into a Salmonella strain containing the in locus hilD’-’lacZ fusion (similar fusion joint but under the hilD promoter; Fig S1). Overexpression of each of the sRNAs caused a >2-fold decrease in activity of the reporter fusion (Fig 2B). These data show that FnrS and ArcZ negatively regulate hilD translation.
Figure 2. FnrS and ArcZ downregulate SPI1 expression by repressing hilD translation.
(A) β-galactosidase activity in E. coli strains containing the hilD’-’lacZ translational fusion and plasmids overexpressing FnrS or ArcZ from either E. coli (EC) and Salmonella (SM) grown in the presence of 100 μM IPTG and 0.001% arabinose to induce the sRNA expression and the fusion lacZ protein expression, respectively. β-galactosidase activity in Salmonella strains containing (B) a hilD’-’lacZ translational fusion, or (C) a hilA’-lacZ+ transcriptional fusion and plasmids overexpressing FnrS or ArcZ from either E. coli (EC) or Salmonella (SM) grown in SPI1 inducing conditions. β-galactosidase activity units are defined as (μmol of ONP formed min−1) x 106/(OD600 x ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JMS6500, JS892, or JS749, each with plasmid pBRplac, pFnrS-EC, pFnrS-SM, pArcZ-EC, or pArcZ-SM.
We then tested whether overexpression of FnrS or ArcZ also affected the expression of the SPI1 regulators HilC and RtsA, which act with HilD in the feed-forward loop controlling hilA expression. Translational lacZ fusions to hilC and rtsA were created in E. coli. Overexpression of neither sRNA affected hilC or rtsA translation in E. coli (Fig S3A and B), suggesting that these sRNAs only regulate hilD translation.
To further examine effects of the sRNAs on SPI1 expression, we overexpressed FnrS or ArcZ in a Salmonella strain carrying a hilA’-lacZ+ transcriptional fusion (Fig S1). Both FnrS and ArcZ dramatically repressed hilA expression (Fig 2C). This enhanced repression is consistent with a direct regulatory effect of each sRNA on hilD translation, which, due to the feed forward regulatory loop (Fig. 1), is expected to decrease expression of rtsA, hilC and dampen hilD autoregulation, all of which would lead to decreased transcription of hilA. Importantly, neither FnrS nor ArcZ affect expression of a hilA’-’lacZ translational fusion in E coli (Fig S3C), showing that these sRNAs do not directly control hilA expression. These data suggest that FnrS and ArcZ are direct sRNA regulators of hilD translation in Salmonella.
FnrS and ArcZ basepair directly with the hilD mRNA
We hypothesized that FnrS and ArcZ regulate SPI1 via base pairing interactions with the hilD mRNA. Computational prediction suggested that both FnrS and ArcZ bind to the hilD mRNA near the ribosome binding site (RBS) (Fig 3A). To test these predictions, we performed mutational analyses using the PBAD-hilD’-’lacZ fusion in E. coli. FnrS nts 37 to 49 are predicted to basepair with hilD mRNA from nts −8 to +5 relative to the initiation AUG (Fig 3A). Consistent with this prediction, a mutant FnrS G47C was not able to regulate wild type hilD’-’lacZ (Fig 3B). A compensatory mutation at position −6 in the hilD fusion restored the regulation by mutant FnrS (Fig 3B). These genetic data support the model that FnrS requires this base pairing interaction to regulate hilD translation.
Figure 3. FnrS and ArcZ regulate hilD translation by direct base-pairing interactions.
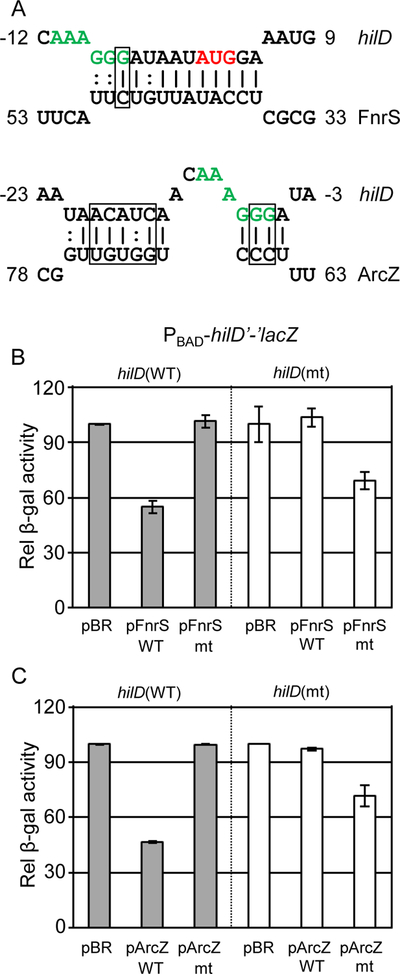
(A) Predicted base-pairing interactions between each sRNA and hilD mRNA. For hilD, nucleotides are numbered from the translational start site. The ribosome binding site of hilD mRNA is highlighted in green, and the translational start site of hilD is highlighted in red. Boxes mark nucleotides for which complementary mutations were created in hilD and each sRNA. (B) Relative β-galactosidase activity in E. coli strains containing the wild type or mutated hilD’-’lacZ translational fusion and plasmids overexpressing either the wild type (pFnrS) or mutated (pFnrS mt) sRNA grown as indicated in Figure 2A. (C) Relative β-galactosidase activity in E. coli strains containing the wild type or mutated hilD’-’lacZ translational fusion and plasmids overexpressing either the wild type (pArcZ) or mutated (pArcZ mt) sRNA grown as indicated in Figure 2A. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JMS6500, JMS6501, and JMS6502 with plasmid pBRplac, pFnrS-SM, pFnrS-mt, pArcZ-SM, or pArcZ-mt.
ArcZ nts 65–76 are predicted to interact with hilD nts −5 to −21 relative to the AUG (Fig 3A). Although several single mutations in this region failed to disrupt the interaction, mutating the boxed ArcZ nts disrupted the interaction with hilD mRNA and the resulting mutant ArcZ failed to regulate wild-type hilD translation (Fig 3C). Introduction of compensatory mutations in the hilD fusion restored the regulation by the mutant ArcZ, suggesting that ArcZ requires this base pairing interaction to regulate hilD translation.
Oxygen-dependent regulation of sRNAs
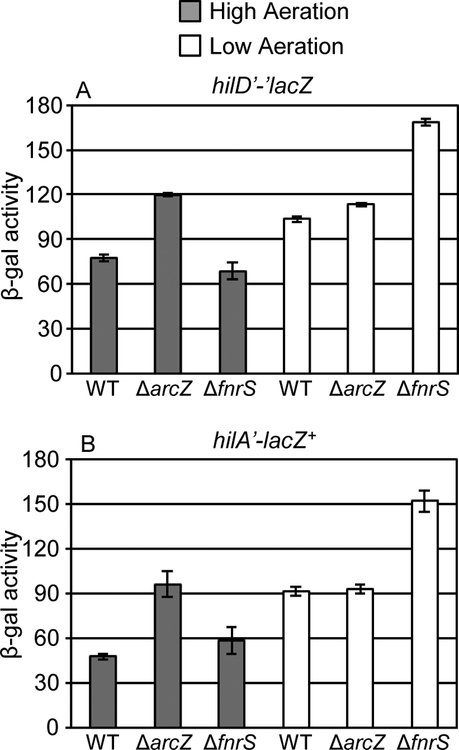
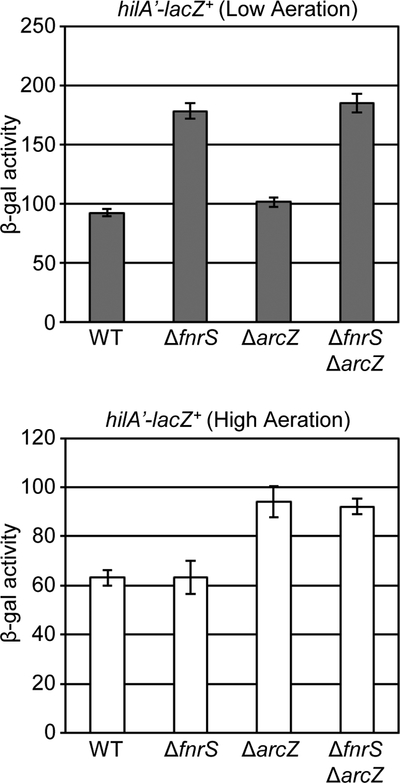
Oxygen is regarded as one of the key regulatory signals for control of the SPI1 system (Ni Bhriain et al., 1989; Lee and Falkow, 1990; Tartera and Metcalf, 1993; Ellermeier and Slauch, 2007; Jennewein et al., 2015). Since FnrS and ArcZ are known to be differentially produced in response to oxygen availability (Papenfort et al., 2009; Boysen et al., 2010; Durand and Storz, 2010; Mandin and Gottesman, 2010), we hypothesized that these sRNAs provide one link between changes in environmental oxygen concentrations and control of SPI1 through regulation of hilD translation. To test this, we created fnrS or arcZ deletion mutations in both the hilD’-’lacZ translational and the hilA’-lacZ+ transcriptional fusion backgrounds, and measured β-galactosidase activity under high-aeration and low-aeration conditions. We observed higher activity from both the hilD’-’lacZ and hilA’-lacZ+ fusions under low aeration conditions (Fig 4), in agreement with previous data (Ellermeier et al., 2005). Deletion of fnrS led to increased hilD translation when strains were grown in low aeration (Fig 4A); the effect on hilA transcription was slightly greater than that on hilD (Fig 4B). However, in high aeration, wildtype and fnrS mutants showed similar levels of activity for both the hilD’-’lacZ and hilA’-lacZ+ fusions (Fig 4). In contrast, deletion of arcZ caused increased levels of hilD translation only when the strains were incubated under high aeration conditions, with a concomitant increase in hilA expression (Fig 4). There was no phenotype conferred by the arcZ deletion in low aeration (Fig 4). These data show that both FnrS and ArcZ affect hilD translation under conditions known to induce their expression, high aeration for ArcZ and low aeration for FnrS. The phenotypes are consistent with a model in which these sRNAs act to optimize hilD translation, and thus SPI1 expression, in response to some intermediate oxygen level.
Figure 4. Loss of FnrS or ArcZ results in the increased level of hilA expression due to the abolished repression of hilD translation.
β-galactosidase activity in Salmonella strains containing a (A) hilD’-’lacZ translational or (B) hilA’-lacZ+ transcriptional fusion in the wildtype, ΔarcZ, or ΔfnrS background grown under either high aeration or low aeration conditions as described in Experimental procedures. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JS892, JS2123, JS2124, JS749, JS2125 and JS2126.
Mechanism of FnrS or ArcZ mediated regulation of hilD translation in Salmonella
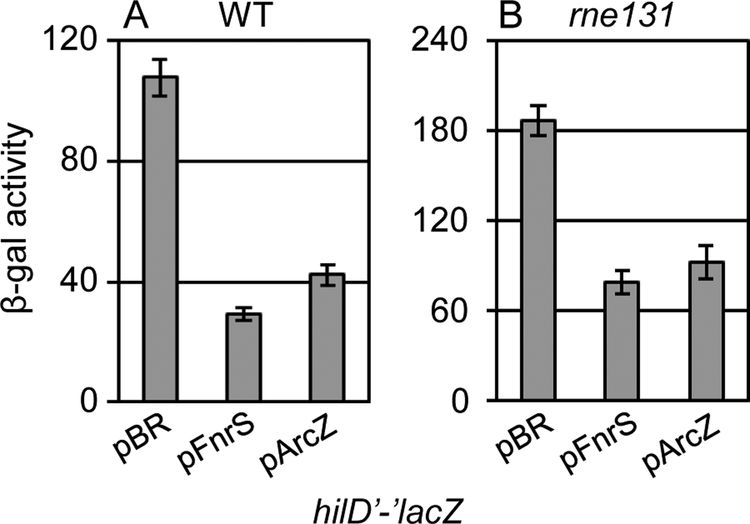
sRNA-mRNA base pairing interactions can induce mRNA degradation by RNase E (Masse et al., 2003; Prevost et al., 2011; Lalaouna et al., 2013). To determine whether FnrS or ArcZ promote hilD mRNA turnover via RNase E, we created an rne131 mutation in Salmonella (Vanzo et al., 1998; Lopez et al., 1999; Viegas et al., 2007). The rne131 mutation truncates RNase E, maintaining enzymatic activity, but preventing assembly of the degradosome, which in many cases also eliminates sRNA-dependent mRNA turnover (Lopez et al., 1999; Masse et al., 2003; Rice and Vanderpool, 2011). RNaseE enzymatic activity is required for processing of the ArcZ sRNA (Chao et al., 2017). Significant levels of processed ArcZ were present in the mutant background (Fig S4). The rne131 mutation caused a moderate increase in the basal level of hilD’-’lacZ fusion activity in Salmonella. However, both FnrS and ArcZ still repressed hilD translation in the rne131 mutant background (Fig 5). These data support the idea that both FnrS and ArcZ directly block translation of the hilD mRNA and that repression does not require mRNA degradation.
Figure 5. Mechanism of FnrS and ArcZ regulation of hilD mRNA translation in Salmonella.
β-galactosidase activity in Salmonella strains containing the hilD’-’lacZ translational fusion and either the empty vector or plasmids overexpressing FnrS or ArcZ from Salmonella in a (A) wildtype or (B) rne131 background grown in SPI1 inducing conditions. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JS892, JS2118 or JS2119, each with plasmid pBRplac, pFnrS-SM, or pArcZ-SM.
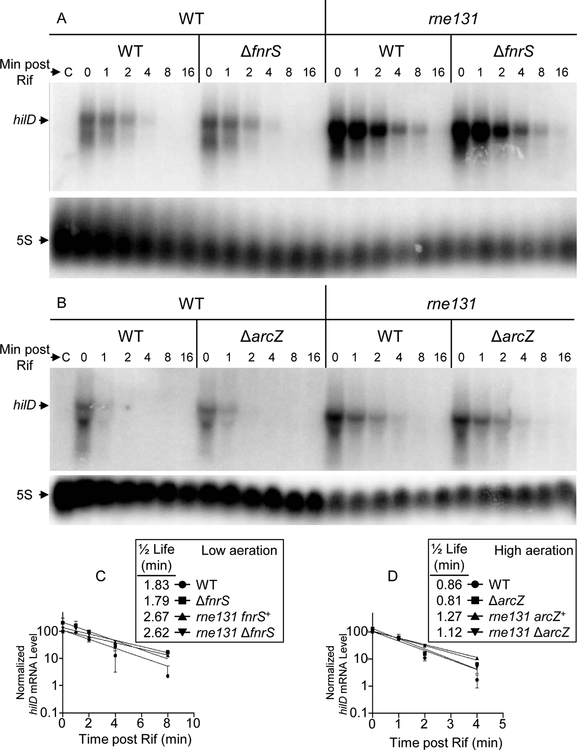
To further examine the mechanisms of regulation, we determined the hilD mRNA half-life in wild type, arcZ, fnrS, or rne131 mutant backgrounds under high aeration and low aeration conditions. We monitored expression of the hilD’-’lacZ fusion indicating that the mutations conferred the expected phenotype under the exact conditions in which we isolated RNA (High aeration: WT, 50±4; arcZ, 69±4; Low aeration: WT, 57±3; fnrS, 94±8 β-gal units). The hilD mRNA had a half-life of approximately 1.8 min under low aeration conditions (Fig 6). Interestingly, the half-life decreased to 0.85 min under high aeration conditions, suggesting that control of mRNA degradation is one mechanism of regulation in response to oxygen. However, the half-life was essentially unaffected by loss of either ArcZ or FnrS (Fig 6), consistent with interpretation above that these sRNAs act by directly blocking translation rather than inducing degradation of the message. The half-life of the hilD message was significantly increased in the rne131 background, showing the involvement of the degradosome in overall mRNA stability (Fig 6).
Figure 6. The half-life of hilD mRNA.
Cells were grown under (A) low aeration or (B) high aeration conditions and RNA was isolated at various time points after addition of rifampicin, and processed as described in Experimental procedures. (“C” indicates a sample from a ΔhilD strain.) The northern blots are representative of two independent experiments. The intensities of the hilD mRNA bands were quantified and normalized to the 5S bands. The WT bands at 0 min was considered 100%. mRNA decay curves represent the means and the standard errors (SEM) for the two experiments Strains used: 14028, JS481, JS2117, JS2121, JS2165, JS2166, and JS2167.
FnrS acts independently of Fur to regulate SPI1 expression
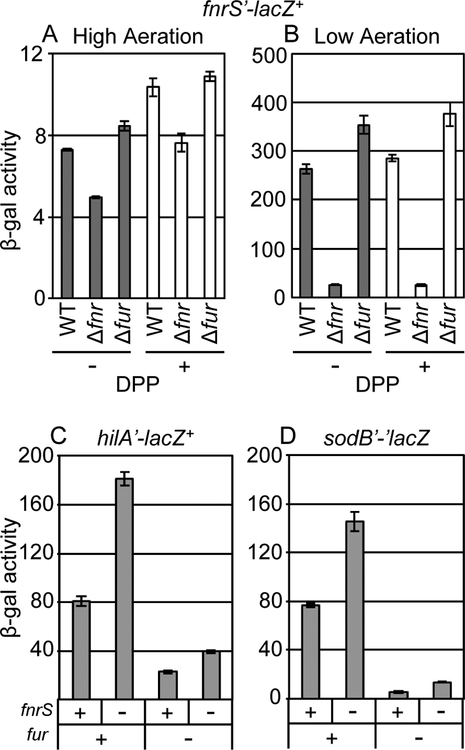
Anaerobic activation of fnrS is mainly dependent on Fnr, but is also affected by ArcA and CRP (Durand and Storz, 2010). It has been reported that Fur, the ferric uptake regulator, also affects fnrS expression (Colgan et al., 2016). Fur is known to positively regulate HilD expression. This regulation requires both the hilD promoter and HilD protein and is likely the result of Fur affecting H-NS dependent repression of hilD transcription (Lavrrar et al., 2002; Olekhnovich and Kadner, 2006; Olekhnovich and Kadner, 2007; Ellermeier and Slauch, 2008; Teixido et al., 2011; Troxell et al., 2011). To better understand the relationship between these global regulators and the potential link to SPI1 gene expression via FnrS, we tested whether the expression of fnrS depends on Fnr or Fur in response to oxygen concentration and iron availability in Salmonella. We created a transcriptional fusion to fnrS, and deleted either fnr or fur. All strains were tested under high and low aeration and in the presence or absence of dipyridyl, an iron chelator that induces iron starvation and causes Fur derepression of target genes (Ikeda et al., 2005; Ellermeier and Slauch, 2008). Under high aeration conditions, transcription of fnrS was very low (Fig 7A, note scale). Expression of fnrS was strongly induced under low aeration conditions (Fig 7B), and this activation was predominantly dependent on Fnr. Neither addition of dipyridyl nor deletion of fur had a substantial impact on fnrS transcription (Fig 7B). These data suggest that the production of FnrS is oxygen-dependent and controlled primarily by Fnr in Salmonella under our experimental conditions.
Figure 7. FnrS regulation of SPI1 expression is independent of Fur.
β-galactosidase activity in Salmonella strains containing an fnrS’-lacZ+ transcriptional fusion in the wild type, Δfnr, or Δfur background grown in (A) high aeration or (B) low aeration conditions in the presence or absence of 200 μM dipyridyl (DPP). β-galactosidase activity in Salmonella strains containing (C) hilA’-lacZ+ transcriptional fusion or (D) sodB’-’lacZ translational fusion in the wild type or the indicated mutant background grown in SPI1 inducing conditions. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JS2129, JS2130, JS2131, JS749, JS2125, JS583, JS2132, JS619, JS2133, JS620, and JS2134.
We then examined the regulation of hilA by FnrS and Fur. We constructed fnrS and fur mutations in Salmonella strains with a hilA’-lacZ+ transcriptional fusion. For a control, we examined expression of a sodB’-’lacZ translational fusion. Fur regulates sodB indirectly via repressing transcription of the two paralogous sRNAs, RyhB-1 and RyhB-2 in Salmonella (Troxell et al., 2011). FnrS also regulates sodB in E. coli (Durand and Storz, 2010). When we measured β-galactosidase activity of these fusions in SPI1-inducing conditions, the sodB fusion behaved as expected; sodB is independently regulated by Fur and FnrS (Fig 7D). Likewise, we observed increased levels of hilA’-lacZ+ in the fnrS mutant compared to wild-type. Deletion of fur in these strains reduced the expression of the hilA’-lacZ+ fusion. However, the effect of fnrS was still observed in the fur background (Fig 7C). These data suggest that the regulation of hilA by FnrS is independent of the regulation by Fur.
Fnr regulates SPI1 expression independent of FnrS
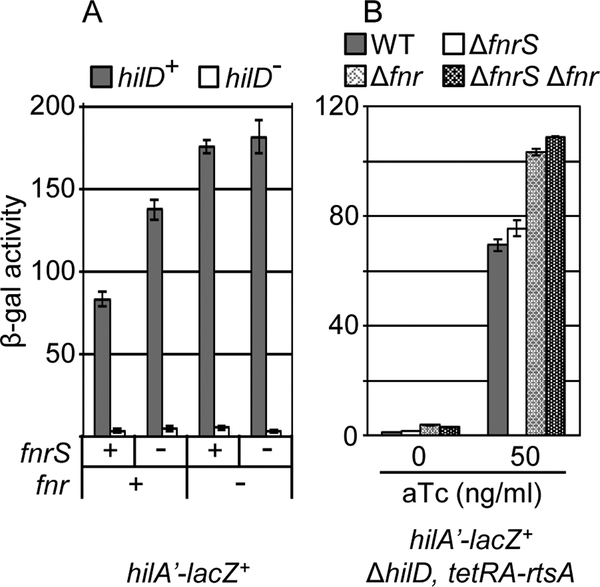
We have previously shown that Fnr represses hilA expression independent of HilD (Golubeva et al., 2012), in seeming contrast to repression of hilD translation by FnrS. This suggests that Fnr controls expression of SPI1 via more than one mechanism. To distinguish between these mechanisms, we determined the activity of the hilA’-lacZ+ transcriptional fusion in strains containing fnr and fnrS deletions. As shown in figure 8A, deletion of fnrS increased hilA transcription only in the fnr+ background. This is as expected, given that FnrS expression is dependent on Fnr. However, deletion of fnr increased hilA-lacZ transcription in both fnrS+ and fnrS− backgrounds. This suggests that Fnr has an additional FnrS-independent regulatory effect on hilA transcription.
Figure 8. Fnr regulates SPI1 expression independent of FnrS.
(A) β-galactosidase activity in Salmonella strains containing the hilA’-lacZ+ transcriptional fusion in the wild type, ΔfnrS, or Δfnr background in the presence or absence of HilD grown in low aeration conditions. (B) β-galactosidase activity in Salmonella strains containing hilA’-lacZ+ transcriptional fusion and the indicated mutations in the ΔhilD background with RtsA protein produced under a tetracycline regulated promoter grown under low aeration conditions with the indicated a-tetracycline concentrations. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JS749, JS2125, JS2135, JS2136, JS2137, JS2138, JS2139, and JS2140.
Expression of hilA in all of these backgrounds was dependent on HilD, making it impossible to distinguish how Fnr and FnrS feed into the regulatory circuit. To separate the effects of FnrS on hilD expression, we placed rtsA under control of a tetracycline inducible promoter. In this background, we can activate expression of hilA in the absence of HilD. As shown in figure 8B, adding 50 ng anhydrotetracycline in this background restored hilA expression to approximately wild type levels in the absence of HilD. Deletion of fnr in this background increased hilA expression, showing that this effect is independent of HilD. Deletion of fnrS in these strains had no significant effect, proving that the sRNA acts solely by controlling hilD translation. These data are consistent with a model in which Fnr controls hilA via both FnrS (translation of hilD) and some mechanism that affects hilA transcription independent of HilD.
The ArcAB two-component system feeds into the system via multiple pathways
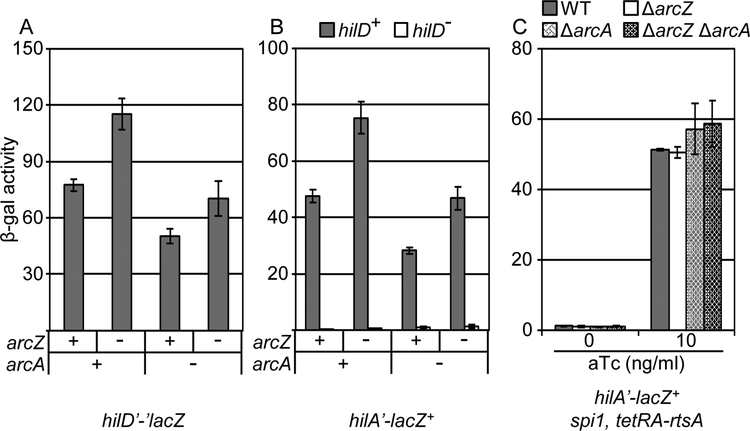
The expression of ArcZ is repressed by ArcA under anaerobic conditions (Mandin and Gottesman, 2010). Inhibition of ArcAB by the oxidized quinone pool in the respiratory chain leads to derepression of arcZ under aerobic growth conditions (Georgellis et al., 2001; Mandin and Gottesman, 2010). In a recent study, ArcA was reported to have a regulatory effect on hilA expression (Lim et al., 2013). We tested how ArcAB and ArcZ each feed into the SPI1 regulatory circuit. We deleted arcZ and/or arcA in either hilD’-’lacZ or hilA’-lacZ+ backgrounds and performed β-galactosidase assays under high aeration conditions. The data (Fig 9A) indicate that, as above, deletion of arcZ increased hilD’-’lacZ expression. In contrast, deletion of arcA decreased expression of the hilD’-’lacZ fusion. Further deletion of arcZ in the arcA background led to the expected increase in hilD’-’lacZ expression. We also observed a similar decrease of a hilD’-lacZ+ transcriptional fusion in absence of ArcA (Fig S5), showing that this effect is at the transcriptional level. (Note that these fusion strains are hilD null, negating any effects of autoregulation.) These data suggest that ArcAB positively controls hilD via two separate pathways: repressing arcZ expression and activating hilD transcription by some unknown mechanism. These effects on hilD are reflected in hilA expression, which mirrors expression of HilD. Both effects are HilD dependent as reflected by the absence of any significant phenotype in a hilD null strain where hilA is activated by RtsA (Fig 9C).
Figure 9. ArcA regulates SPI1 expression independent of ArcZ.
β-galactosidase activity in Salmonella strains containing the (A) hilD’-’lacZ translational fusion or the (B) hilA’-lacZ transcriptional fusion in a wild type, ΔarcZ, and/or ΔarcA background grown in high aeration conditions. (C) β-galactosidase activity in Salmonella strains containing hilA’-lacZ+ transcriptional fusion and the indicated mutations in the spi1 (Δspi1 ΔrtsA) background with RtsA protein produced under a tetracycline regulated promoter grown under high aeration conditions with the indicated a-tetracycline concentrations. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JS892, JS2124, JS2143, JS2144, JS749, JS2126, JS2145, JS2146, JS2147, JS2148, JS2149, JS2150, JS2077, JS2151, JS2152, and JS2153.
The ArcAB system was reported to control hilD transcription through activation of LoiA, a LysR type regulator encoded in SPI14 that presumably binds directly to the hilD promoter (Jiang et al., 2017). To test this possibility, we deleted loiA or the entire SPI14 island in both hilD’-lacZ+ and hilA’-lacZ+ fusion strains. We confirmed these deletions by PCR analysis and performed β-galactosidase assays under both low and high aeration conditions. We observed no effect on either hilD’-lacZ+ or hilA’-lacZ+ expression (Fig S6). Therefore, we conclude that phenotypes that we observe are independent of LoiA.
Impact of sRNA regulation of SPI1 in mouse models of infection
Both FnrS and ArcZ repress translation of hilD, resulting in decreased hilA expression under low or high aeration conditions. We suppose that this regulation defines an oxygen “window” for optimal activation of SPI1. Mouse competition assays were used to ask if this regulation is relevant in the animal model of infection. We created a mutant deleted for both fnrS and arcZ in a hilA’-lacZ+ background. (The fusion has no significant effect on virulence, Table S3, and was present in both strain backgrounds.) Before testing the strain in mice, we measured the phenotype conferred by the double deletion in low aeration and high aeration conditions. Compared to wild type and individual deletion mutants, the double deletion of both sRNAs increased hilA expression independent of aeration levels (Fig 10).
Figure 10. Loss of both FnrS and ArcZ abolishes oxygen-mediated regulation of hilA expression.
β-galactosidase activity in Salmonella strains containing the hilA’-lacZ+ transcriptional fusion in the wild type, ΔfnrS, ΔarcZ, or ΔfnrS ΔarcZ background grown in either high aeration or low aeration conditions. β-galactosidase activity units are defined as (μmol of ONP formed min−1) × 106/(OD600 × ml of cell suspension) and are reported as mean ± standard deviation where n=3. Strains used: JS749, JS2125, JS2126. and JS2160.
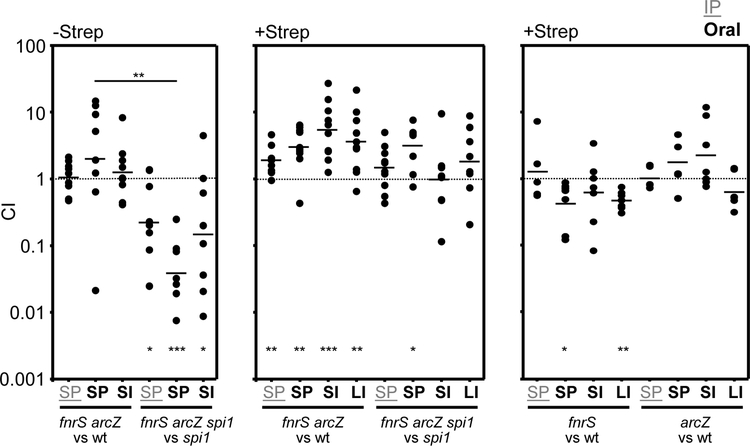
To address the contribution of the FnrS- and ArcZ-mediated regulation to virulence in the host, we used oral mouse competition assays (dependent on the SPI1 T3SS), and intraperitoneal (IP) infection (bypassing the need for SPI1). Streptomycin treatment of mice leads to availability of anaerobic terminal electron acceptors and as well as oxygen (Clark and Barrett, 1987; Stecher and Hardt, 2008; Winter et al., 2010; Thiennimitr et al., 2011; Lopez et al., 2012; Winter et al., 2013; Faber et al., 2017). Given the role of these sRNAs in regulating aspects of respiration (Papenfort et al., 2009; Durand and Storz, 2010; Mandin and Gottesman, 2010), we tested the effects of removing FnrS and ArcZ in in normal (Strep−) or streptomycin-treated (Strep+) mice. To determine whether any observed effects were due to changes in SPI1 expression, we also performed competition assays in a spi1 null background (Δspi1 ΔrtsA).
There was marked variability in the competitive indices between individual mice, making it difficult to make significant conclusions. However, there was a trend that was particularly evident in the normal (Strep−) mice. Given the fact that SPI1 is not required systemically after invasion, the ratio of strains in the spleen after oral infection reflects the ratio of invasion (Ellermeier et al., 2005). The fnrS arcZ double mutant competed equally with the wild-type strain in both oral and IP infections (Fig 11). In contrast, in the Δspi1 mutant background, fnrS arcZ mutants were at a competitive disadvantage compared to the fnrS+arcZ+ strain in orally infected mice. We interpret these data to suggest that, whereas loss of FnrS and ArcZ confers a SPI1-independent competitive disadvantage, the increased expression of SPI1 in the sRNA mutant leads to an overall increase in invasion. These phenotypes are largely negated in Strep+ mice, consistent with significant changes in the intestinal environment. Interestingly, deletion of each of the sRNAs alone did not confer a phenotype suggesting, perhaps, that it is the requirement to adapt to subtly different oxygen concentrations that is critical. This could also explain the large variability in individual mice, possibly reflecting variations in oxygen concentrations in different niches in the intestine.
Figure 11. Mouse competition assays.
Mice were infected (intraperitoneally) IP or orally (as noted) with a 50:50 mix of the indicated strains. Competition assays were performed in streptomycin-treated (+Strep) or untreated (−Strep) mice (see Experimental procedures). Bacteria were recovered from the spleen (designated SP) in the case of IP competition assays or from the spleen (SP), small intestine (SI), and large intestine (LI) in oral competitions. The competitive index (CI) was calculated as described in experimental procedures and is shown for each mouse. The line indicates the geometric mean for each set. The Student t test was used to compare the CIs to the inocula or between groups. p=<0.05 (*), p=<0.005 (**), p=<0.0005 (***). The strains used were JS749, JS2125, JS2164, JS2160, JS2162, JS2075.
Discussion
Multiple environmental signals influence expression of the SPI1 T3SS and the integration of these signals presumably allows the cell to determine the appropriate time and place to produce the invasion machinery. Our long-term goal is to understand the relative impact of these various environmental factors and how these signals are mechanistically integrated into the SPI1 regulatory circuit (Ellermeier and Slauch, 2007; Saini et al., 2010; Golubeva et al., 2012). Much of the regulatory input is integrated at HilD, affecting either the translation or activity of the HilD protein (Saini et al., 2010; Golubeva et al., 2012). Consistent with this dominant role, we identified two sRNAs, FnrS and ArcZ, that directly affect SPI1 T3SS activation via regulating hilD translation. Both FnrS and ArcZ repress hilD translation resulting in significant inhibition of hilA transcription (Fig 1). These sRNAs have no direct effects on hilC, rtsA, or hilA translation. Bioinformatic prediction and genetic analyses proved that both FnrS and ArcZ sRNAs basepair near the ribosome binding site of the hilD message (Fig 4) leading to direct translational inhibition; the RNase E degradosome was not required for hilD translational regulation by these sRNAs. The hilD mRNA also has an unusually long ~300 nt 3’ UTR, which apparently affects the stability of the message. Several mechanisms of regulation at the 3’ UTR have been characterized (Lopez-Garrido et al., 2014; Gaviria-Cantin et al., 2017; El Mouali et al., 2018).
Oxygen and osmolarity have been regarded as key environmental signals that control SPI1 T3SS activation, but it is clear that oxygen concentrations impact the SPI1 regulatory circuit through several mechanisms. For example, we have previously shown that the rtsA, hilC, and hilD promoters each respond to oxygen levels in the absence of all known regulators (Ellermeier et al., 2005). Our results here show that the half-life of the hilD mRNA is decreased under high aeration by an unknown mechanism (Fig 6). It is also clear from our results that Fnr and ArcAB affect SPI1 expression in complex ways that are independent of the sRNAs (see below). Available iron, the levels of which are significantly affected by oxygen (Ikeda et al., 2005) affect SPI1 expression via Fur (Ellermeier and Slauch, 2008). Further investigations are required to completely understand how oxygen influences SPI1 gene expression.
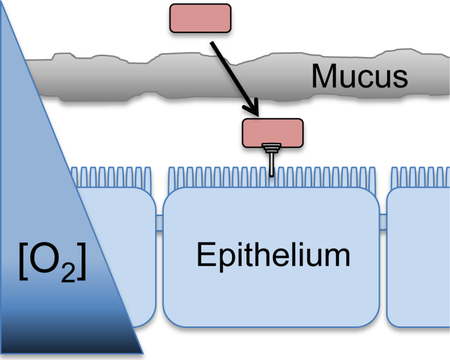
Both FnrS and ArcZ repress hilD translation and the their production is differentially controlled by oxygen levels. We propose that these sRNAs tune expression of SPI1 to be maximally produced at some optimal intermediate oxygen concentration. Studies in E. coli suggest that Fnr is active at O2 concentrations below ~20 μM, whereas the ArcA shifts toward the dephosphorylated state at about this same concentration or above (Tseng et al., 1996). The intestine has both longitudinal and radial gradients of oxygen. Recent studies show that the oxygen concentration in the lumen of conventional mice drops from >50 μM in the duodenum to ~9 μM in the terminal ileum (Friedman et al., 2018). But there is also a considerable radial gradient. The midpoint of the lumen is the most anoxic, with any available oxygen being quickly reduced by resident bacteria or other chemical reactions (Friedman et al., 2018). The concentration rises steeply below the mucus and at the surface of the intestinal epithelial cells. Interestingly, data suggest that the oxygen concentration in this region is on the order of 20 μM (Espey, 2013; Albenberg et al., 2014), the concentration at which neither FnrS nor ArcZ should be produced.
There are also chemical gradients along the intestine that act as signals to optimize SPI1 expression. For example, we and others have provided data showing that short and long chain fatty acids control SPI1 gene expression. Diet-derived long chain fatty acids negatively regulate expression by directly binding to HilD to prevent DNA binding (Golubeva et al., 2016). Presumably, these fatty acids are absorbed along the small intestine, being at their lowest concentration in the distal small intestine. Both acetate and proprionate activate SPI1 gene expression (Lawhon et al., 2002; Hung et al., 2013). These short chain fatty acids are at their highest concentration in the distal small intestine (Argenzio et al., 1974; Cummings et al., 1987; Macfarlane et al., 1992). In contrast, butyrate, produced by the strict anaerobes in the colon, negatively regulates SPI1 expression (Gantois et al., 2006; Bronner et al., 2018; Gillis et al., 2018). We believe that these concentration gradients are such that SPI1 is optimally expressed in the distal small intestine.
Pretreatment of mice with oral streptomycin and subsequent infection with Salmonella leads to SPI1-mediated induction of inflammatory diarrhea (Que and Hentges, 1985; Barthel et al., 2003). Salmonella benefits from the inflammatory response by taking advantage of newly available carbon sources and terminal electron acceptors, including tetrathionate and nitrate, thereby out-competing fermenting bacteria (Clark and Barrett, 1987; Stecher and Hardt, 2008; Winter et al., 2010; Thiennimitr et al., 2011; Lopez et al., 2012; Winter et al., 2013; Faber et al., 2017). FnrS and ArcZ both affect expression of numerous genes, including those encoding central metabolic and respiratory enzymes (Papenfort et al., 2009; Durand and Storz, 2010; Mandin and Gottesman, 2010). Therefore, it is not surprising that there were significant differences observed between normal and streptomycin-treated mice infected with the fnrS arcZ double mutant. To separate the pleiotropic effects of the sRNAs from their specific role in SPI1 regulation, we tested loss of the sRNAs in a spi1 mutant background. Although normally such a test is straightforward, in this case interpretation is complicated by the fact that the inflammatory response in the streptomycin-treated mice is largely SPI1 dependent (Que and Hentges, 1985; Barthel et al., 2003).
FnrS is anaerobically induced by the major anaerobic transcriptional regulator, Fnr (Boysen et al., 2010; Durand and Storz, 2010). However, it was reported that Fur also activates the expression of FnrS in exponential growth (Colgan et al., 2016). Because oxygen affects iron availability, Fur-mediated regulation is oxygen dependent (Kehres et al., 2002). However, under the conditions that we tested, FnrS expression is dominantly regulated by environmental oxygen and Fnr. Iron depletion or deletion of fur did not affect the FnrS expression (Fig 6-C).
In addition to activating fnrS expression leading to translational inhibition of hilD, Fnr decreases hilA transcription independently of FnrS and HilD (Fig 7). Of the two effects, FnrS plays a slightly greater role. We could not identify any obvious Fnr binding sites in the hilA promoter, suggesting that this regulation may be indirect. Such indirect effects would not be surprising given the response of the SPI1 system to numerous physiological factors and the pleiotropic role of Fnr in central metabolism (Fink et al., 2007; Golubeva et al., 2012). But direct or indirect, Fnr induction in low oxygen leads to a decrease in hilA transcription and hence SPI1 expression through both HilD-dependent and -independent mechanisms.
Like Fnr, the ArcAB two-component system regulates SPI1 through multiple mechanisms. Above 20 μM, ArcB begins to shift ArcA to the de-phosphorylated state (Tseng et al., 1996). But this is not a sharp cutoff and ArcA-P levels are significant under a wide range of oxygen concentrations (Rolfe et al., 2011). Different genes will require varying levels of ArcA-P to be regulated. Deletion of arcA leads to decreased hilD transcription independent of ArcZ, suggesting that there is still significant ArcA-P under our aerobic conditions, even though arcZ is not being significantly repressed. As with Fnr, this is almost certainly an indirect effect on hilD transcription. It was recently proposed that the transcriptional regulator LoiA is transcriptionally repressed by ArcA in low oxygen and that LoiA directly activates hilD transcription (Jiang et al., 2017). However, in our hands, loss of LoiA had no effect on hilD or hilA expression under either low- or high-aeration conditions (Fig S3). Therefore, ArcA is controlling hilD transcription by an unknown mechanism. It is also interesting that the two effects are seemingly in opposition.
FnrS and ArcZ play an important role in the regulation of SPI1 in response to oxygen, adding to our overall understanding of the mechanisms by which environmental signals relevant in the intestine feed into the regulatory network. Although oxygen levels have long been considered an important parameter controlling SPI1, the system responds to oxygen via multiple pathways and more studies will be required to fully understand how these various mechanisms are integrated.
Experimental procedures
Strain construction
Bacterial strains and plasmids are described in Table S1. All Salmonella enterica serovar Typhimurium strains created for this study are isogenic derivatives of strain 14028 [American Type Culture Collection (ATCC)] and were constructed using P22 HT105/1 int-201 (P22)-mediated transduction (Maloy et al., 1996). Deletion of various genes and concomitant insertion of an antibiotic resistance cassette was carried out using lambda Red-mediated recombination (Datsenko and Wanner, 2000; Yu et al., 2000; Ellermeier et al., 2002). The end-points of each deletion are indicated in Table S1. In all cases, the appropriate insertion of the antibiotic resistance marker was confirmed by polymerase chain reaction analysis. In each case, the constructs resulting from this procedure were moved into an unmutagenized background by P22 transduction. When appropriate, antibiotic resistance cassettes were removed using the temperature sensitive plasmid pCP20 carrying the FLP recombinase (Cherepanov and Wackernagel, 1995). To create transcriptional lacZ fusions of FnrS or ArcZ, the insertion mutations in FnrS and ArcZ were converted to transcriptional lac+ fusions using an FLP/FRT-mediated site-specific recombination method as previously described (Ellermeier et al., 2002).
Construction of translational lacZ reporter fusions in E. coli
The translational lacZ reporter fusions were constructed using lambda Red recombination in the E. coli strain PM1205 as described previously (Mandin and Gottesman, 2009). All fusions are under PBAD control. The 5’ UTR and early coding regions of hilD, hilC, rtsA, or hilA were fused in-frame to lacZ to create translational fusions (Fig S1). The corresponding DNA fragments were amplified from purified genomic DNA of Salmonella using the primers in Table S2 with homology to the PBAD promoter or to lacZ. The PCR fragments were purified using a PCR purification Kit (Qiagen) and competent cells were prepared as described (Mandin and Gottesman, 2009). Recombinants were selected on sucrose minimal plates (M63 salts, 0.2% glycerol, 5% sucrose) containing 40 μg ml−1 of 5-bromo-4-chloro-3-indolyl-β-D-galactopyranoside (X-Gal). To create fusions with mutations in the hilD 5’ UTR, the nucleotide changes were encoded in the amplifying primers (Table S2).
Media, reagents, enzymatic assays, and growth conditions
Lysogeny Broth medium containing 10 g tryptone, 5 g yeast extract, and 0 g NaCl per liter (designated no salt LB, NSLB), 5 g NaCl per liter (low salt LB, LSLB) or 10 g NaCl per liter (high salt LB, HSLB) were used as indicated. Superoptimal broth with catabolite repression (SOC) was used for the recovery of transformants (Maloy et al., 1996). Bacterial strains were normally grown at 37°C except for the strains containing the temperature sensitive plasmids, pCP20 or pKD46, which were grown at 30°C. When required, antibiotics were used at the following concentrations: 100 μg ml−1 ampicillin (Ap), 20 μg ml−1 chloramphenicol (Cm), 50 μg ml−1 kanamycin (Kn), 25 μg ml−1 tetracycline (Tet), and 50 μg ml−1 apramycin (Apr). Primers were purchased from IDT. Enzymes were purchased from New England Biolabs or Invitrogen.
β-Galactosidase assays were performed using a microtiter plate assay as previously described on strains grown under the indicated conditions (Slauch and Silhavy, 1991). β-Galactosidase activity units are defined as (mmol of ortho-nitrophenyl-β-galactoside formed min-1) x106/(OD600 X ml of cell suspension) and are reported as mean ± standard deviation where n = 4. Cultures used to measure β-galactosidase activity in Salmonella were initially inoculated into NSLB and grown overnight, then subcultured 1/100 and grown under one of the following conditions: (i) statically overnight in 3 ml of HSLB in a 13 × 100 mm tube, referred to as either Low Aeration or SPI1 inducing; (ii) on a platform shaker at 225 r.p.m. in 4 ml of HSLB in a 125 ml baffled flask to an OD600 of 0.8, referred to as High Aeration. Cultures used to measure β-galactosidase activity in E. coli were initially inoculated into LSLB, grown overnight, and subcultured 1/100 into 2 ml of LSLB with 100 μM IPTG and 0.001% Arabinose in a 13 × 100 mm tube and incubated on a roller drum to an OD600 of 0.5.
Plasmid construction and site-directed mutagenesis of sRNA constructs
The pFnrS and pArcZ plasmids were constructed by PCR amplifying fnrS or arcZ from strain 14028 using primers F-AatII-FnrS and R-EcoRI-FnrS or F-AatII-ArcZ and R-EcoRI-ArcZ, respectively (Table S2). The PCR products were subsequently cloned into the pBR-plac vector after digestion with AatII and EcoRI (Mandin and Gottesman, 2009).
Various bioinformatics tools (Zuker, 2003; Kruger and Rehmsmeier, 2006; Busch et al., 2008) were used to predict the region of FnrS or ArcZ that base pairs with the hilD mRNA. The Quick Change Lightening Site Directed Mutagenesis Kit (Stratagene) was used to create the corresponding mutant constructs with the primers listed in Table S2.
Northern analysis
High aeration cultures were grown as described above. For low aeration conditions, overnight NSLB cultures were subcultured 1/100 into 16 ml of HSLB in a 20 × 150 mm tube and incubated for 3 hours on a platform shaker at 225 rpm. For the measurement of the half-life of hilD mRNA, 500 μg of rifampicin was added into the bacterial cultures (Holmqvist et al., 2018). Immediately upon addition of rifampicin (time 0) and at 1, 2, 4, 8 and 16 minutes after the treatment, 800 μl aliquots of bacterial cells were collected. Bacterial cells were immediately suspended in 915 μl of 65°C phenol solution (15–594-047, Invitrogen) with 120 μL of Lysis buffer (0.3M NaOAc (pH 5.2), 8% SDS, and 0.02 M EDTA (pH 8.0)) and then incubated at 65°C for 10 minutes while shaking (Ares, 2012). After centrifugation at 13,000 rpm for 10 min, the aqueous fraction was collected and added to 500 μl of a phenol: chloroform: isoamylalchol (P:C:I, 25:24:1) solution (pH 6.6, AM9732, Invitrogen) for further purification. After centrifugation, the subsequent aqueous portion were transferred into 1.3 ml of ice-chilled ethanol and incubated at −80°C for at least 2 hours. After centrifugation at 13,000 rpm for 10 min, the supernatant was carefully removed and the pellets were washed with 1 ml 70% ethanol. The RNA pellets were allowed to air-dry and suspended in 25 μl DEPC-treated H2O.
For each sample, 20 μg total RNA was denatured in 3X volume of Formaldehyde Loading Dye (AM8552, Ambion) at 95°C for 3 min, and separated on a 1.2% agarose gel with 1X MOPS buffer and 7% formaldehyde for 1 h at 85 V. RNA was transferred to a BrightStar™-Plus Positively Charged Nylon Membrane (AM10104, Ambion) by capillary transfer with Northern MAX transfer buffer (AM8672, Ambion)(2005). To probe hilD mRNA, a radiolabeled random-primed probe was generated from 25 ng of PCR fragment corresponding to the hilD orf following the manufacturer’s instructions (18187–013, Life Technologies).
To examine ArcZ processing, we used aliquots from the 0 time point wild type and rne131 samples grown under high aeration. Total RNA (20 μg) was denatured in 1× RNA loading buffer II (AM8546G, Ambion) at 95°C for 3 min, and separated by 6% polyacrylamide gel with 7 M urea for 2 h at 300 V (Chao et al., 2017). RNA was transferred to BrightStar™-Plus Positively Charged Nylon Membrane (AM10104, Ambion) by electro- blotting (1 h, 50 V, 4°C) in 1× TBE buffer. To produce a radiolabeled oligo probe antisense to ArcZ , 10 pmol of ArcZ oligonucleotide was incubated with 25 μCi of [γ−32P]-ATP and 1 U T4 polynucleotide kinase (M0236S, NEB) at 37°C for 1 hour.
After crosslinking by 0.12 J/cm2 UV light, the membranes were hybridized with radiolabeled DNA probes at 42°C overnight in ULTRAhyb hybridization buffer (AM8670, Invitrogen). In all cases, as a loading control, 5S RNA was detected by radio-labeled oligo probe, synthesized as for the ArcZ probe. Signal was visualized on a phosphorimager (Fuji FLA-3000) and quantified using the Image Quant image analyzer (ImageGauge V4.22). Decay curves corresponding to rifampicin chase experiments were generated by using GraphPad Prism version 8.0 (Sinha et al., 2018).
Mouse and in vitro competition assays
Bacteria were initially inoculated into 2 ml LSLB, grown overnight, then subcultured 1/35 in 4 ml HSLB (1% NaCl) in 125 mL flasks and grown for 4 h with aeration at 200 rpm. BALB/c mice (Harlan) (10 to 13 weeks old) were inoculated either orally or intraperitoneally (i.p.) with 0.2 ml of a bacterial suspension. For oral infections of normal mice, the bacteria were washed and suspended at 5 × 108 (wt background) or 109 (spi1 background) cells per 0.2 ml in sterile 0.1 M sodium phosphate buffer, pH 8.0. Before infection, food and water were withheld for 4 hours and mice were orally inoculated with the indicated number of bacteria, after which the food and water were provided immediately. For oral infections of streptomycin-treated mice, the bacteria were washed and suspended at 5 × 107 (wt background) or 5×108 (spi1 background) cells per 0.2 ml in sterile 0.1 M sodium phosphate buffer, pH 8.0. For streptomycin treatment, food and water were withheld for 4 hours; then mice were treated with 20 mg of streptomycin delivered intragastrically, after which the food and water were provided immediately. At 20 hours after the streptomycin treatment, food and water were withheld for 4 hours and mice were orally inoculated with the indicated number of bacteria, after which the water was provided immediately and food was provided at 2 hours post infection. For intraperitoneal infections, the cells were diluted to 103 cells per 0.2 ml in sterile PBS. For oral infections, mice were sacrificed by CO2 asphyxiation at 3.5 days after inoculation and the spleens, small intestines, and large intestines were harvested. For i.p. infections, the mice were sacrificed by CO2 asphyxiation between 4 and 5 days after inoculation and spleens were harvested. These organs were homogenized, and serial dilutions of the homogenates were plated on the appropriate medium to determine the number of CFU per organ. The relative percentage of each strain recovered was determined by replica plating to the appropriate antibiotic-containing medium. In all competition assays, the inoculum consisted of a 1:1 mix of two bacterial strains. The actual CFU and relative percentage represented by each strain was determined by direct plating of the inoculum. The competitive index (CI) was calculated as (percentage of strain A recovered/percentage of strain B recovered)/(percentage of strain A inoculated/percentage of strain B inoculated). All strains were independently reconstructed and the competition assays were repeated to ensure that any phenotypes were the result of the designated mutations. To measure competitive growth in vitro, strains were initially inoculated into 2 ml LSLB, grown overnight, then mixed 1:1, diluted and inoculated into indicated medium/condition at 103 bacteria per tube/flask, then grown for 16h in indicated conditions. In all cases, the Student t test was used to determine whether the output ratio was significantly different from the input ratio or to compare groups of mice.
Supplementary Material
Acknowledgements
We thank members of the Slauch lab, Dhriti Sinha and Nick DeLay for helpful discussions. This work was supported by the National Institutes of Health grant R01 GM120182 to CKV and JMS.
Footnotes
Ethics statement
All animal work was reviewed and approved by the University of Illinois Institutional Animal Care and Use Committee (IACUC). Procedures were performed in our AAALAC accredited facility in accordance with University and PHS guidelines under protocol 15214. All efforts were made to minimize animal suffering.
Conflict of interest
The authors declare that they have no conflict of interest with the contents of this article.
References
- (2005) Northern blotting: transfer of denatured RNA to membranes. Nature Methods 2: 997. [Google Scholar]
- Albenberg L, Esipova TV, Judge CP, Bittinger K, Chen J, Laughlin A, et al. (2014) Correlation between intraluminal oxygen gradient and radial partitioning of intestinal microbiota. Gastroenterology 147: 1055–1063 e1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ares M (2012) Bacterial RNA Isolation. Cold Spring Harbor Protocols 2012: pdb.prot071068. [DOI] [PubMed] [Google Scholar]
- Argenzio RA, Southworth M and Stevens CE (1974) Sites of organic acid production and absorption in the equine gastrointestinal tract. Am J Physiol 226: 1043–1050. [DOI] [PubMed] [Google Scholar]
- Bajaj V, Lucas RL, Hwang C and Lee CA (1996) Co-ordinate regulation of Salmonella typhimurium invasion genes by environmental and regulatory factors is mediated by control of hilA expression. Mol Microbiol 22: 703–714. [DOI] [PubMed] [Google Scholar]
- Barthel M, Hapfelmeier S, Quintanilla-Martinez L, Kremer M, Rohde M, Hogardt M, et al. (2003) Pretreatment of mice with streptomycin provides a Salmonella enterica serovar Typhimurium colitis model that allows analysis of both pathogen and host. Infect Immun 71: 2839–2858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bauer CE, Elsen S and Bird TH (1999) Mechanisms for redox control of gene expression. Annu Rev Microbiol 53: 495–523. [DOI] [PubMed] [Google Scholar]
- Baxter MA, Fahlen TF, Wilson RL and Jones BD (2003) HilE interacts with HilD and negatively regulates hilA transcription and expression of the Salmonella enterica serovar Typhimurium invasive phenotype. Infect Immun 71: 1295–1305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boysen A, Moller-Jensen J, Kallipolitis B, Valentin-Hansen P and Overgaard M (2010) Translational regulation of gene expression by an anaerobically induced small non-coding RNA in Escherichia coli. J Biol Chem 285: 10690–10702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bronner DN, Faber F, Olsan EE, Byndloss MX, Sayed NA, Xu G, et al. (2018) Genetic ablation of butyrate utilization attenuates gastrointestinal Salmonella disease. Cell Host Microbe 23: 266–273 e264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Busch A, Richter AS and Backofen R (2008) IntaRNA: efficient prediction of bacterial sRNA targets incorporating target site accessibility and seed regions. Bioinformatics 24: 2849–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chao Y, Li L, Girodat D, Forstner KU, Said N, Corcoran C, et al. (2017) In vivo cleavage map illuminates the central role of rnase e in coding and non-coding RNA pathways. Mol Cell 65: 39–51. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cherepanov PP and Wackernagel W (1995) Gene disruption in Escherichia coli: TcR and KmR cassettes with the option of Flp-catalyzed excision of the antibiotic-resistance determinant. Gene 158: 9–14. [DOI] [PubMed] [Google Scholar]
- Chubiz JE, Golubeva YA, Lin D, Miller LD and Slauch JM (2010) FliZ regulates expression of the Salmonella pathogenicity island 1 invasion locus by controlling HilD protein activity in Salmonella enterica serovar Typhimurium. J Bacteriol 192: 6261–6270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA and Barrett EL (1987) The phs gene and hydrogen sulfide production by Salmonella typhimurium. J Bacteriol 169: 2391–2397. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark MA, Jepson MA, Simmons NL and Hirst BH (1994) Preferential interaction of Salmonella typhimurium with mouse Peyer’s patch M cells. Res Microbiol 145: 543–552. [DOI] [PubMed] [Google Scholar]
- Colgan AM, Kroger C, Diard M, Hardt WD, Puente JL, Sivasankaran SK, et al. (2016) The impact of 18 ancestral and horizontally-acquired regulatory proteins upon the transcriptome and sRNA landscape of Salmonella enterica serovar Typhimurium. PLoS Genet 12: e1006258. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Craig M, Sadik AY, Golubeva YA, Tidhar A and Slauch JM (2013) Twin-arginine translocation system (tat) mutants of Salmonella are attenuated due to envelope defects, not respiratory defects. Mol Microbiol 89: 887–902. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cummings JH, Pomare EW, Branch WJ, Naylor CP and Macfarlane GT (1987) Short chain fatty acids in human large intestine, portal, hepatic and venous blood. Gut 28: 1221–1227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Darwin KH and Miller VL (1999) InvF is required for expression of genes encoding proteins secreted by the SPI1 type III secretion apparatus in Salmonella typhimurium. J Bacteriol 181: 4949–4954. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Datsenko KA and Wanner BL (2000) One-step inactivation of chromosomal genes in Escherichia coli K-12 using PCR products. Proc Natl Acad Sci U S A 97: 6640–6645. [DOI] [PMC free article] [PubMed] [Google Scholar]
- De Lay N, Schu DJ and Gottesman S (2013) Bacterial small RNA-based negative regulation: Hfq and its accomplices. J Biol Chem 288: 7996–8003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desnoyers G, Bouchard MP and Masse E (2013) New insights into small RNA-dependent translational regulation in prokaryotes. Trends Genet 29: 92–98. [DOI] [PubMed] [Google Scholar]
- Durand S and Storz G (2010) Reprogramming of anaerobic metabolism by the FnrS small RNA. Mol Microbiol 75: 1215–1231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eichelberg K and Galan JE (1999) Differential regulation of Salmonella typhimurium type III secreted proteins by pathogenicity island 1 (SPI-1)-encoded transcriptional activators InvF and HilA. Infect Immun 67: 4099–4105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- El Mouali Y, Gaviria-Cantin T, Sanchez-Romero MA, Gibert M, Westermann AJ, Vogel J, et al. (2018) CRP-cAMP mediates silencing of Salmonella virulence at the post-transcriptional level. PLoS Genet 14: e1007401. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ellermeier CD, Ellermeier JR and Slauch JM (2005) HilD, HilC and RtsA constitute a feed forward loop that controls expression of the SPI1 type three secretion system regulator hilA in Salmonella enterica serovar Typhimurium. Mol Microbiol 57: 691–705. [DOI] [PubMed] [Google Scholar]
- Ellermeier CD, Janakiraman A and Slauch JM (2002) Construction of targeted single copy lac fusions using lambda Red and FLP-mediated site-specific recombination in bacteria. Gene 290: 153–161. [DOI] [PubMed] [Google Scholar]
- Ellermeier JR and Slauch JM (2007) Adaptation to the host environment: regulation of the SPI1 type III secretion system in Salmonella enterica serovar Typhimurium. Curr Opin Microbiol 10: 24–29. [DOI] [PubMed] [Google Scholar]
- Ellermeier JR and Slauch JM (2008) Fur regulates expression of the Salmonella pathogenicity island 1 type III secretion system through HilD. J Bacteriol 190: 476–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Espey MG (2013) Role of oxygen gradients in shaping redox relationships between the human intestine and its microbiota. Free Radic Biol Med 55: 130–140. [DOI] [PubMed] [Google Scholar]
- Faber F, Thiennimitr P, Spiga L, Byndloss MX, Litvak Y, Lawhon S, et al. (2017) Respiration of microbiota-derived 1,2-propanediol drives Salmonella expansion during colitis. PLoS Pathog 13: e1006129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fink RC, Evans MR, Porwollik S, Vazquez-Torres A, Jones-Carson J, Troxell B, et al. (2007) FNR is a global regulator of virulence and anaerobic metabolism in Salmonella enterica serovar Typhimurium (ATCC 14028s). J Bacteriol 189: 2262–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman ES, Bittinger K, Esipova TV, Hou L, Chau L, Jiang J, et al. (2018) Microbes vs. chemistry in the origin of the anaerobic gut lumen. Proc Natl Acad Sci U S A 115: 4170–4175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frohlich KS and Vogel J (2009) Activation of gene expression by small RNA. Curr Opin Microbiol 12: 674–682. [DOI] [PubMed] [Google Scholar]
- Galan JE (2001) Salmonella interactions with host cells: type III secretion at work. Annu Rev Cell Dev Biol 17: 53–86. [DOI] [PubMed] [Google Scholar]
- Galan JE and Collmer A (1999) Type III secretion machines: Bacterial devices for protein delivery into host cells. Science 284: 1322–1328. [DOI] [PubMed] [Google Scholar]
- Gantois I, Ducatelle R, Pasmans F, Haesebrouck F, Hautefort I, Thompson A, et al. (2006) Butyrate specifically down-regulates Salmonella pathogenicity island 1 gene expression. Appl Environ Microbiol 72: 946–949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gaviria-Cantin T, El Mouali Y, Le Guyon S, Romling U and Balsalobre C (2017) Gre factors-mediated control of hilD transcription is essential for the invasion of epithelial cells by Salmonella enterica serovar Typhimurium. PLoS Pathog 13: e1006312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Geissmann TA and Touati D (2004) Hfq, a new chaperoning role: binding to messenger RNA determines access for small RNA regulator. Embo j 23: 396–405. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Georgellis D, Kwon O and Lin EC (2001) Quinones as the redox signal for the arc two-component system of bacteria. Science 292: 2314–2316. [DOI] [PubMed] [Google Scholar]
- Gillis CC, Hughes ER, Spiga L, Winter MG, Zhu W, Furtado de Carvalho T, et al. (2018) Dysbiosis-associated change in host metabolism generates lactate to support Salmonella growth. Cell Host Microbe 23: 54–64 e56. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubeva YA, Ellermeier JR, Cott Chubiz JE and Slauch JM (2016) Intestinal long-chain fatty acids act as a direct signal to modulate expression of the Salmonella pathogenicity island 1 type III secretion system. mBio 7: e02170–02115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Golubeva YA, Sadik AY, Ellermeier JR and Slauch JM (2012) Integrating global regulatory input into the Salmonella pathogenicity island 1 type III secretion system. Genetics 190: 79–90. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grenz JR, Cott Chubiz JE, Thaprawat P and Slauch JM (2018) HilE regulates HilD by blocking dna binding in Salmonella enterica serovar Typhimurium. J Bacteriol 200. doi: 10.1128/JB.00750-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Henderson CA, Vincent HA, Casamento A, Stone CM, Phillips JO, Cary PD, et al. (2013) Hfq binding changes the structure of Escherichia coli small noncoding RNAs OxyS and RprA, which are involved in the riboregulation of rpoS. RNA 19: 1089–1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Holmqvist E, Li L, Bischler T, Barquist L and Vogel J (2018) Global maps of ProQ binding in vivo reveal target recognition via rna structure and stability control at mRNA 3′ ends. Molecular Cell 70: 971–982.e976. [DOI] [PubMed] [Google Scholar]
- Hung CC, Garner CD, Slauch JM, Dwyer ZW, Lawhon SD, Frye JG, et al. (2013) The intestinal fatty acid propionate inhibits Salmonella invasion through the post-translational control of HilD. Mol Microbiol 87: 1045–1060. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ikeda JS, Janakiraman A, Kehres DG, Maguire ME and Slauch JM (2005) Transcriptional regulation of sitABCD of Salmonella enterica serovar Typhimurium by MntR and Fur. J Bacteriol 187: 912–922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jagodnik J, Brosse A, Le Lam TN, Chiaruttini C and Guillier M (2017) Mechanistic study of base-pairing small regulatory RNAs in bacteria. Methods 117: 67–76. [DOI] [PubMed] [Google Scholar]
- Jennewein J, Matuszak J, Walter S, Felmy B, Gendera K, Schatz V, et al. (2015) Low-oxygen tensions found in Salmonella-infected gut tissue boost Salmonella replication in macrophages by impairing antimicrobial activity and augmenting Salmonella virulence. Cell Microbiol 17: 1833–1847. [DOI] [PubMed] [Google Scholar]
- Jepson MA and Clark MA (2001) The role of M cells in Salmonella infection. Microbes Infect 3: 1183–1190. [DOI] [PubMed] [Google Scholar]
- Jiang L, Feng L, Yang B, Zhang W, Wang P, Jiang X, et al. (2017) Signal transduction pathway mediated by the novel regulator LoiA for low oxygen tension induced Salmonella typhimurium invasion. PLoS Pathog 13: e1006429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones BD (2005) Salmonella invasion gene regulation: a story of environmental awareness. J Microbiol 43 Spec No: 110–117. [PubMed] [Google Scholar]
- Kehres DG, Janakiraman A, Slauch JM and Maguire ME (2002) SitABCD is the alkaline Mn(2+) transporter of Salmonella enterica serovar Typhimurium. J Bacteriol 184: 3159–3166. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khoroshilova N, Popescu C, Munck E, Beinert H and Kiley PJ (1997) Iron-sulfur cluster disassembly in the FNR protein of Escherichia coli by O2: [4Fe-4S] to [2Fe-2S] conversion with loss of biological activity. Proc Natl Acad Sci U S A 94: 6087–6092. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kroger C, Colgan A, Srikumar S, Handler K, Sivasankaran SK, Hammarlof DL, et al. (2013) An infection-relevant transcriptomic compendium for Salmonella enterica Serovar Typhimurium. Cell Host Microbe 14: 683–695. [DOI] [PubMed] [Google Scholar]
- Kroger C, Dillon SC, Cameron AD, Papenfort K, Sivasankaran SK, Hokamp K, et al. (2012) The transcriptional landscape and small RNAs of Salmonella enterica serovar Typhimurium. Proc Natl Acad Sci U S A 109: E1277–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruger J and Rehmsmeier M (2006) RNAhybrid: microRNA target prediction easy, fast and flexible. Nucleic Acids Res 34: W451–454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lalaouna D, Simoneau-Roy M, Lafontaine D and Masse E (2013) Regulatory RNAs and target mRNA decay in prokaryotes. Biochim Biophys Acta 1829: 742–747. [DOI] [PubMed] [Google Scholar]
- Lavrrar JL, Christoffersen CA and McIntosh MA (2002) Fur-DNA interactions at the bidirectional fepDGC-entS promoter region in Escherichia coli. J Mol Biol 322: 983–995. [DOI] [PubMed] [Google Scholar]
- Lawhon SD, Maurer R, Suyemoto M and Altier C (2002) Intestinal short-chain fatty acids alter Salmonella typhimurium invasion gene expression and virulence through BarA/SirA. Mol Microbiol 46: 1451–1464. [DOI] [PubMed] [Google Scholar]
- Lee CA and Falkow S (1990) The ability of Salmonella to enter mammalian-cells is affected by bacterial-growth state. Proc Natl Acad Sci U S A 87: 4304–4308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lim S, Yoon H, Kim M, Han A, Choi J, Choi J, et al. (2013) Hfq and ArcA are involved in the stationary phase-dependent activation of Salmonella pathogenicity island 1 (SPI1) under shaking culture conditions. J Microbiol Biotechnol 23: 1664–1672. [DOI] [PubMed] [Google Scholar]
- Lopez-Garrido J, Puerta-Fernandez E and Casadesus J (2014) A eukaryotic-like 3’ untranslated region in Salmonella enterica hilD mRNA. Nucleic Acids Res 42: 5894–5906. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez CA, Winter SE, Rivera-Chavez F, Xavier MN, Poon V, Nuccio SP, et al. (2012) Phage-mediated acquisition of a type III secreted effector protein boosts growth of Salmonella by nitrate respiration. mBio 3. doi: 10.1128/mBio.00143-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopez PJ, Marchand I, Joyce SA and Dreyfus M (1999) The C-terminal half of RNase E, which organizes the Escherichia coli degradosome, participates in mRNA degradation but not rRNA processing in vivo. Mol Microbiol 33: 188–199. [DOI] [PubMed] [Google Scholar]
- Lu S, Killoran PB, Fang FC and Riley LW (2002) The global regulator ArcA controls resistance to reactive nitrogen and oxygen intermediates in Salmonella enterica serovar Enteritidis. Infect Immun 70: 451–461. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Macfarlane GT, Gibson GR and Cummings JH (1992) Comparison of fermentation reactions in different regions of the human colon. J Appl Bacteriol 72: 57–64. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Chen S, Murrow J, St John K and Gottesman S (2001) Regulation of RpoS by a novel small RNA: the characterization of RprA. Mol Microbiol 39: 1382–1394. [DOI] [PubMed] [Google Scholar]
- Majdalani N, Cunning C, Sledjeski D, Elliott T and Gottesman S (1998) DsrA RNA regulates translation of RpoS message by an anti-antisense mechanism, independent of its action as an antisilencer of transcription. Proc Natl Acad Sci U S A 95: 12462–12467. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maloy SR, Stewart VJ and Taylor RK (1996) Genetic analysis of pathogenic bacteria: a laboratory manual, p. xix + 603 pp. Cold Spring Harbor Laboratory Press, Plainville, NY 11803–2500. [Google Scholar]
- Mandin P and Gottesman S (2009) A genetic approach for finding small RNAs regulators of genes of interest identifies RybC as regulating the DpiA/DpiB two-component system. Mol Microbiol 72: 551–565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mandin P and Gottesman S (2010) Integrating anaerobic/aerobic sensing and the general stress response through the ArcZ small RNA. Embo j 29: 3094–3107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martinez LC, Yakhnin H, Camacho MI, Georgellis D, Babitzke P, Puente JL, et al. (2011) Integration of a complex regulatory cascade involving the SirA/BarA and Csr global regulatory systems that controls expression of the Salmonella SPI-1 and SPI-2 virulence regulons through HilD. Mol Microbiol 80: 1637–1656. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Escorcia FE and Gottesman S (2003) Coupled degradation of a small regulatory RNA and its mRNA targets in Escherichia coli. Genes Dev 17: 2374–2383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masse E, Vanderpool CK and Gottesman S (2005) Effect of RyhB small RNA on global iron use in Escherichia coli. J Bacteriol 187: 6962–6971. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McCullen CA, Benhammou JN, Majdalani N and Gottesman S (2010) Mechanism of positive regulation by DsrA and RprA small noncoding RNAs: pairing increases translation and protects rpoS mRNA from degradation. J Bacteriol 192: 5559–5571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mika F and Hengge R (2014) Small RNAs in the control of RpoS, CsgD, and biofilm architecture of Escherichia coli. RNA Biol 11: 494–507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moller T, Franch T, Hojrup P, Keene DR, Bachinger HP, Brennan RG, et al. (2002) Hfq: a bacterial Sm-like protein that mediates RNA-RNA interaction. Mol Cell 9: 23–30. [DOI] [PubMed] [Google Scholar]
- Moon K, Six DA, Lee HJ, Raetz CR and Gottesman S (2013) Complex transcriptional and post-transcriptional regulation of an enzyme for lipopolysaccharide modification. Mol Microbiol 89: 52–64. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ni Bhriain N, Dorman CJ and Higgins CF (1989) An overlap between osmotic and anaerobic stress responses: a potential role for DNA supercoiling in the coordinate regulation of gene expression. Mol Microbiol 3: 933–942. [DOI] [PubMed] [Google Scholar]
- Olekhnovich IN and Kadner RJ (2002) DNA-binding activities of the HilC and HilD virulence regulatory proteins of Salmonella enterica serovar Typhimurium. J Bacteriol 184: 4148–4160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Olekhnovich IN and Kadner RJ (2006) Crucial roles of both flanking sequences in silencing of the hilA promoter in Salmonella enterica. J Mol Biol 357: 373–386. [DOI] [PubMed] [Google Scholar]
- Olekhnovich IN and Kadner RJ (2007) Role of nucleoid-associated proteins Hha and H-NS in expression of Salmonella enterica activators HilD, HilC, and RtsA required for cell invasion. J Bacteriol 189: 6882–6890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Papenfort K, Said N, Welsink T, Lucchini S, Hinton JC and Vogel J (2009) Specific and pleiotropic patterns of mRNA regulation by ArcZ, a conserved, Hfq-dependent small RNA. Mol Microbiol 74: 139–158. [DOI] [PubMed] [Google Scholar]
- Penheiter KL, Mathur N, Giles D, Fahlen T and Jones BD (1997) Non-invasive Salmonella typhimurium mutants are avirulent because of an inability to enter and destroy M cells of ileal Peyer’s patches. Mol Microbiol 24: 697–709. [DOI] [PubMed] [Google Scholar]
- Prevost K, Desnoyers G, Jacques JF, Lavoie F and Masse E (2011) Small RNA-induced mRNA degradation achieved through both translation block and activated cleavage. Genes Dev 25: 385–396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Que JU and Hentges DJ (1985) Effect of streptomycin administration on colonization resistance to Salmonella typhimurium in mice. Infect Immun 48: 169–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rice JB and Vanderpool CK (2011) The small RNA SgrS controls sugar-phosphate accumulation by regulating multiple PTS genes. Nucleic Acids Res 39: 3806–3819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rolfe MD, Ter Beek A, Graham AI, Trotter EW, Asif HM, Sanguinetti G, et al. (2011) Transcript profiling and inference of Escherichia coli K-12 ArcA activity across the range of physiologically relevant oxygen concentrations. J Biol Chem 286: 10147–10154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saini S, Ellermeier JR, Slauch JM and Rao CV (2010) The role of coupled positive feedback in the expression of the SPI1 type three secretion system in Salmonella. PLoS Pathog 6: e1001025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Savidge TC, Smith MW, James PS and Aldred P (1991) Salmonella-induced M-cell formation in germ-free mouse Peyer’s patch tissue. Am J Pathol 139: 177–184. [PMC free article] [PubMed] [Google Scholar]
- Sinha D, Matz LM, Cameron TA and De Lay NR (2018) Poly(A) polymerase is required for RyhB sRNA stability and function in Escherichia coli. Rna 24: 1496–1511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sittka A, Pfeiffer V, Tedin K and Vogel J (2007) The RNA chaperone Hfq is essential for the virulence of Salmonella typhimurium. Mol Microbiol 63: 193–217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Slauch JM and Silhavy TJ (1991) Cis-acting ompF mutations that result in OmpR-dependent constitutive expression. J Bacteriol 173: 4039–4048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smirnov A, Forstner KU, Holmqvist E, Otto A, Gunster R, Becher D, et al. (2016) Grad-seq guides the discovery of ProQ as a major small RNA-binding protein. Proc Natl Acad Sci U S A 113: 11591–11596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Srikumar S, Kroger C, Hebrard M, Colgan A, Owen SV, Sivasankaran SK, et al. (2015) RNA-seq brings new insights to the intra-macrophage transcriptome of Salmonella typhimurium. PLoS Pathog 11: e1005262. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stecher B and Hardt WD (2008) The role of microbiota in infectious disease. Trends Microbiol 16: 107–114. [DOI] [PubMed] [Google Scholar]
- Storz G, Vogel J and Wassarman KM (2011) Regulation by small RNAs in bacteria: expanding frontiers. Mol Cell 43: 880–891. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tartera C and Metcalf ES (1993) Osmolarity and growth phase overlap in regulation of Salmonella typhi adherence to and invasion of human intestinal cells. Infect Immun 61: 3084–3089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Teixido L, Carrasco B, Alonso JC, Barbe J and Campoy S (2011) Fur activates the expression of Salmonella enterica pathogenicity island 1 by directly interacting with the hilD operator in vivo and in vitro. PLoS One 6: e19711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thiennimitr P, Winter SE, Winter MG, Xavier MN, Tolstikov V, Huseby DL, et al. (2011) Intestinal inflammation allows Salmonella to use ethanolamine to compete with the microbiota. Proc Natl Acad Sci U S A 108: 17480–17485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Troxell B and Hassan HM (2016) Interplay between O2 and iron in gene expression: environmental sensing by FNR, ArcA, and Fur in bacteria In: Stress and Environmental Regulation of Gene Expression and Adaptation in Bacteria. John Wiley & Sons, Inc., pp. 1079–1089. [Google Scholar]
- Troxell B, Sikes ML, Fink RC, Vazquez-Torres A, Jones-Carson J and Hassan HM (2011) Fur negatively regulates hns and is required for the expression of HilA and virulence in Salmonella enterica serovar Typhimurium. J Bacteriol 193: 497–505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tseng CP, Albrecht J and Gunsalus RP (1996) Effect of microaerophilic cell growth conditions on expression of the aerobic (cyoABCDE and cydAB) and anaerobic (narGHJI, frdABCD, and dmsABC) respiratory pathway genes in Escherichia coli. J Bacteriol 178: 1094–1098. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vanderpool CK and Gottesman S (2004) Involvement of a novel transcriptional activator and small RNA in post-transcriptional regulation of the glucose phosphoenolpyruvate phosphotransferase system. Mol Microbiol 54: 1076–1089. [DOI] [PubMed] [Google Scholar]
- Vanzo NF, Li YS, Py B, Blum E, Higgins CF, Raynal LC, et al. (1998) Ribonuclease E organizes the protein interactions in the Escherichia coli RNA degradosome. Genes Dev 12: 2770–2781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Viegas SC, Pfeiffer V, Sittka A, Silva IJ, Vogel J and Arraiano CM (2007) Characterization of the role of ribonucleases in Salmonella small RNA decay. Nucleic Acids Res 35: 7651–7664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vogel J (2009) A rough guide to the non-coding RNA world of Salmonella. Mol Microbiol 71: 1–11. [DOI] [PubMed] [Google Scholar]
- Vogel J and Luisi BF (2011) Hfq and its constellation of RNA. Nat Rev Microbiol 9: 578–589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Thiennimitr P, Winter MG, Butler BP, Huseby DL, Crawford RW, et al. (2010) Gut inflammation provides a respiratory electron acceptor for Salmonella. Nature 467: 426–429. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, et al. (2013) Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339: 708–711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu D, Ellis HM, Lee EC, Jenkins NA, Copeland NG and Court DL (2000) An efficient recombination system for chromosome engineering in Escherichia coli. Proc Natl Acad Sci U S A 97: 5978–5983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuker M (2003) Mfold web server for nucleic acid folding and hybridization prediction. Nucleic Acids Res 31: 3406–3415. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.