Abstract
We have previously demonstrated co‐receptor level‐associated functional heterogeneity in apparently homogeneous naive peripheral CD4 T cells, dependent on MHC‐mediated tonic signals. Maturation pathways can differ between naive CD4 and naive CD8 cells, so we tested whether the latter showed similar co‐receptor level‐associated functional heterogeneity. We report that, when either polyclonal and T‐cell receptor (TCR)‐transgenic monoclonal peripheral naive CD8 T cells from young mice were separated into CD8hi and CD8lo subsets, CD8lo cells responded poorly, but CD8hi and CD8lo subsets of CD8 single‐positive (SP) thymocytes responded similarly. CD8lo naive CD8 T cells were smaller and showed lower levels of some cell‐surface molecules, but higher levels of the negative regulator CD5. In addition to the expected peripheral decline in CD8 levels on transferred naive CD8 T cells in wild‐type (WT) but not in MHC class I‐deficient recipient mice, short‐duration naive T‐cell–dendritic cell (DC) co‐cultures in vitro also caused co‐receptor down‐modulation in CD8 T cells but not in CD4 T cells. Constitutive pZAP70/pSyk and pERK levels ex vivo were lower in CD8lo naive CD8 T cells and dual‐specific phosphatase inhibition partially rescued their hypo‐responsiveness. Bulk mRNA sequencing showed major differences in the transcriptional landscapes of CD8hi and CD8lo naive CD8 T cells. CD8hi naive CD8 T cells showed enrichment of genes involved in positive regulation of cell cycle and survival. Our data show that naive CD8 T cells show major differences in their signaling, transcriptional and functional landscapes associated with subtly altered CD8 levels, consistent with the possibility of peripheral cellular aging.
Keywords: CD8 T cells, cellular aging, immune aging phenotype, T‐cell transcriptome, tonic signals
Abbreviations
- APC
allophycocyanin
- BCI
(E)‐2‐benzylidene‐3‐(cyclohexylamino)‐2,3‐dihydro‐1H‐inden‐1‐one
- BMDC
bone‐marrow‐derived dendritic cells
- CFSE
carboxyfluorescein succinimidyl ester
- CTV
Cell Trace Violet
- DUSP
dual‐specific phosphatase
- ERK
extracellular signal‐regulated kinase
- FITC
fluorescein isothiocyanate
- IL‐7
interleukin‐7
- MHC
major histocompatibility complex
- Ova
ovalbumin
- PBS
phosphate‐buffered saline
- PE
phycoerythrin
- PMC
phorbol 12‐myristate 13‐acetate
- pMHC
peptide–MHC complex
- SP
single‐positive
- TCR
T‐cell receptor
- WT
wild‐type
Introduction
Functional heterogeneity within different cellular lineages, likely to be a result of stochastic processes, has been well reported.1 In the immune system, with respect to naive T cells, stochastic fluctuations in gene expressions of regulators of T‐cell receptor (TCR) signaling such as extracellular signal‐regulated kinase (ERK) kinases have been linked to the ability of T‐cell clones to sense differences in ligand affinities.2 We have recently shown that variation in baseline glucose uptake and rates of protein synthesis in naive T cells regulate their ability to respond to TCR‐mediated stimulation.3 Also, variation in expression of Src‐homology‐2 domain containing protein tyrosine phosphatases, which provides a negative feedback loop for TCR signaling, has been shown to regulate T‐cell sensitivity.2, 4
However, it is unclear how individual cells from an apparently homogeneous naive T‐cell population, which have an identical status of differentiation, show variation in molecular content. Although stochastic gene expression fluctuations may well contribute to variability in protein expression of individual cells, it is plausible that such variability may also be contributed by a variety of microenvironmental cues that naive T cells receive throughout their lifetime, thereby leading to tuning of T‐cell responsiveness and functionality.
T‐cell development in the thymus results in a steady supply of naive mature T cells, which reach the periphery. Peripheral naive T cells successfully finding their cognate peptide–major histocompatibility complex (pMHC) ligands will respond and change phenotype rapidly. However, naive T cells that do not encounter cognate pMHC will continue to circulate as naive cells in peripheral lymphoid tissues, and many may never encounter their cognate ligands until they are eventually eliminated from the peripheral pool. Hence, it is quite likely that peripheral naive T‐cell populations are heterogeneous in their post‐thymic age, even in young animals/individuals.
With age, thymic output of naive T cells decreases and the cells in the periphery undergo homeostatic proliferation,5 resulting in an increase in the number without expanding the repertoire.6 Cytokines such as interleukin‐7 (IL‐7) and IL‐15 play a major role in peripheral T‐cell homeostasis of T cells.7, 8 Self‐pMHC‐mediated signals, which play a crucial role in defining the TCR repertoire of the developing thymocytes in the thymus, are also important in the survival and maintenance of mature naive T cells in peripheral lymphoid tissues.8, 9 Homeostatic mechanisms maintain the number of peripheral naive T cells in aged mice, but the survival of individual naive T‐cell clones differs based on TCR avidity and the availability of selecting ligands.10, 11
Peripherally trafficking T cells receive multiple signals in different anatomical locations, making it possible that exposure to such signals can be non‐uniform. Further, cells that have been recirculating in the periphery for a long time would be exposed to microenvironmental signals for prolonged durations compared with recent thymic emigrants. Hence, variations are likely to exist in the patterns and frequencies of tonic and cytokine signals received by individual naive CD8 T cells depending on the time spent by them in the periphery, and the functional consequences of such heterogeneity become an interesting issue.
In this context, we have previously reported that naive CD4 T cells in aged mice show heterogeneity of hypo‐responsiveness, suggesting that the population consists of cohorts with differing functionalities, and that cellular post‐thymic aging, via MHC class II (MHCII) ‐mediated signals, may contribute to the generation of this heterogeneity of responsiveness.12 However, there is evidence that homeostatic maintenance pathways, including but not limited to the role of MHC‐mediated ‘tonic’ signals regulating the survival of naive post‐thymic T cells in the periphery, may be somewhat different for CD4 versus CD8 T cells,13, 14, 15, 16 making it interesting to examine potential co‐receptor‐associated functional heterogeneity in naive CD8 T cells.
On this background, we have demonstrated that, like naive CD4 T cells, within a naive CD8 T‐cell population with unimodal distribution of CD8 levels, lower CD8 co‐receptor levels identified cells of smaller size, poor survival, defective TCR signaling and poor proliferation, that even short‐term peripheral residence was also associated with subtle loss of CD8 levels dependent on MHCI‐mediated tonic signals. Moreover, we find that naive CD8 T cells were more sensitive than naive CD4 T cells to MHC‐mediated co‐receptor loss in vitro. Finally, we examined the global transcriptional landscape of CD8hi and CD8lo naive CD8 T cells, and show differential expression levels of cell cycle and survival‐related genes, a pattern similar to that reported from neonatal versus adult human CD8 T cells. Our data show that naive CD8 T cells show major differences in their signaling, transcriptional and functional landscapes associated with subtly altered CD8 levels, consistent with the possibility of peripheral cellular aging.
Materials and methods
Mice
Various strains of mice as indicated: C57BL/6 (B6), B6.SJL‐PtprcaPep3b/BoyJ (B6.SJL) congenic strain of C57BL/6, B6.129S2‐Tap1tm1Arp/J (Tap‐null), C57BL/6‐Tg(TcraTcrb)1100Mjb/J TCR transgenic OT1 mice specific for the H‐2Kb‐restricted OVA1 peptide SIINFEKL – were obtained from the Jackson Laboratory (Bar Harbor, ME) and maintained in the animal facility of the National Institute of Immunology. Young mice were 6–8 weeks old and aged mice were 18–20 months old at use. All mice were maintained and used, and the study was carried out, in accordance with the guidelines and recommendations of the Institutional Animal Ethics Committee. The protocol was approved by the Institutional Animal Ethics Committee.
Collection of blood and isolation of mononuclear cells from human donors
Venous blood was collected from healthy, young, adult volunteers (aged 22–35 years with equal representation of men and women) after obtaining written consent. Peripheral blood mononuclear cells were separated from heparinized blood by density‐gradient centrifugation as described previously.12 The procedures followed were approved by the Institutional Human Ethics Committee of the National Institute of Immunology. This study was carried out in accordance with the recommendations of the Institutional Human Ethics Committee of the National Institute of Immunology with written informed consent from all subjects. All subjects gave written informed consent in accordance with the Declaration of Helsinki. The protocol was approved by the Institutional Human Ethics Committee of the National Institute of Immunology.
Reagents
Antibodies for mouse CD8α (53‐6.7) [fluorescein isothiocyanate (FITC), phycoerythrin (PE), allophycocyanin (APC), APC‐eFluor 780, eFluor 450), CD8β (eBioH35‐17.2) (PE), CD3 (17A2) (APC), CD4 (RM4‐5) (FITC, PE, APC, APC‐eFluor 780), CD69 (H1.2F3) (APC, PE), CD25 (PC61.5) (PE, APC‐eFluor 780), CD62L (MEL‐14) (PE, APC, APC‐eFluor 780), CD44 (IM7) (APC, eFluorV450), CD5 (53‐7.3) (APC, eFluor 450), Qa2 (69H1‐9‐9) (FITC), CD24 (M1/69) (APC‐eFluor 780), NK1.1 (PK136) (PE, eFluor 450) and human CD8 (SK1) (FITC, eFluor 450), CD45RA (HI100) (PECY7), CD62L (DREG‐56) (FITC, eFluor 450), CD3 (OKT3) (APC) and CD56 (CMSSB) (PE) (from eBioscience, San Diego, CA); for TCRVα2 (B20.1) (PE), TCR‐β (H57‐597) (PE) CD3 (17A2) (PE), phospho‐zeta chain‐associated protein kinase 70 (ZAP70) (n3KOBUS) (PE), phospho‐ERK (MILAN8R) (PE), ZAP70 (17A/P) (PE) (BD Biosciences, San Jose, CA), and for phospho‐ZAP70 (2701), phospho‐ERK (9101), ZAP70 (2705) and ERK (9102) (Cell Signaling Technology, Danvers, MA) were used. F(abʹ)2 fragments of goat anti‐rabbit IgG1 coupled to Alexafluor 488 (Molecular Probes, Carlsbad, CA) were used where appropriate. Carboxyfluorescein succinimide ester (CFSE), Cell Trace Violet (CTV), Sytox Red (SR) (from Molecular Probes, Carlsbad, CA) were used. Functional grade purified anti‐mouse anti‐CD3 (145‐2C11) and anti‐CD28 (37.51) antibodies and anti‐human anti‐CD3 (H1T3a) and anti‐CD28 (CD28.2) antibodies (eBioscience, San Diego, CA), phorbol 12‐myristate 13‐acetate (PMA) and ionomycin (Sigma‐Aldrich, St. Loius, MO) were used for T‐cell stimulation. [3H]thymidine (Perkin Elmer, Waltham, MA) was used for radioactive assays. (E)‐2‐benzylidene‐3‐(cyclohexylamino)‐2, 3‐dihydro‐1H‐inden‐1‐one (BCI) (Sigma‐Aldrich) was used as dual‐specific phosphatase (DUSP) inhibitor. Commercially synthesized SIINFEKL (Peptron, South Korea) was used as indicated. Cells were cultured in RPMI‐1640 (Biological Industries, Beit Haemek, Israel) supplemented with 10% fetal bovine serum (Sigma‐Aldrich), 2 mm l‐glutamine and antibiotics (Sigma‐Aldrich).
Flow cytometry and cell sort purification
Cells were incubated with staining antibodies on ice for 30 min. Control samples were incubated in staining buffer alone [phosphate‐buffered saline (PBS) containing 1% fetal calf serum and 0·05% sodium azide] or with an appropriate isotype‐matched control antibody. The cells were then washed with PBS. For detecting intracellular proteins (pZAP, pERK, ZAP and ERK), cells were fixed with BD Cytofix buffer (BD Biosciences) for 30 min on ice and permeabilized with BD Phosphoflow Perm Buffer III (BD Biosciences) for 30 min on ice, followed by staining according to the manufacturer's instructions (BD Bioscience, Cell Signaling Technology). Samples were analyzed on a flow cytometer (FACSVerse or FACSARIA III, BD Biosciences) and data were analyzed with flowjo software (Treestar, Ashland, OR). All flow cytometry data followed a normal distribution, and mean fluorescence intensity was calculated with flowjo software. Scaling in histograms was calculated by normalizing to the peak height at the mode of the distribution so that the maximum y‐axis value in the absolute count histogram was 100%.
Naive CD8 T cells were flow cytometrically sort‐purified as CD8+ CD62Lhi CD44lo. Naive CD8 T cells were further sorted on the basis of surface CD8 expression by gating 10% brightest and 10% dimmest cells to yield ‘CD8hi’ and ‘CD8lo’ sub‐populations, respectively. Sorted cells were subjected to RNA‐sequencing analysis. For sorting human CD8hi and CD8lo naive CD8 T cells, peripheral blood mono nuclear cells were isolated from the blood of healthy donors by Ficoll–Hypaque density gradient centrifugation (Ficoll‐Paque; GE Healthcare Biosciences, Little Chalfont, UK) and naive (CD8+ CD45RA+ CD62L+) CD8 T cells were sort purified into CD8hi and CD8lo subsets. For sorting CD8hi and CD8lo single‐positive (SP) thymocytes, thymic single‐cell suspensions were made. CD8+ CD4− CD24− Qa2+ cells, identified as CD8SP cells, were further gated as CD8lo and CD8hi and flow cytometrically sorted as described for peripheral cells (FACSAria III, BD Bioscience).
In all experiments where comparisons of CD8 levels were made, aliquots of cell groups being compared were frozen, and all aliquots were thawed, stained and analyzed together, so as to avoid flow cytometric artifacts.
Staining for dead cells
Cells were first stained for various surface markers and washed with PBS or RPMI‐1640. Cells were then incubated with 5 nm of Sytox‐green or Sytox‐red dye in saline for ~15 min at room temperature. The samples were then directly analyzed on a flow cytometer.
Bone‐marrow‐derived dendritic cell culture
Bone‐marrow‐derived dendritic cells (BMDCs) were cultured as described.17 Briefly, BMDCs were grown by culturing mouse bone marrow cells with recombinant granulocyte–monocyte colony‐stimulating factor (PeproTech, Rocky Hill, NJ) for 7 days, with periodic growth factor replenishment. Semi‐adherent cells obtained on day 7 were used for experiments.
Adoptive transfer assays
Naive CD8 T cells were sort‐purified from spleen and lymph nodes of C57BL/6 mice and labeled with CFSE or CTV and intravenously transferred into anesthetized recipient mice. Donor cells were identified by the CFSE or CTV labels on various days after transfer. For testing the in vivo stability of CD8hi and CD8lo subsets, sort‐purified naive CD8hi and CD8lo cells were labeled with CFSE and CTV respectively, mixed in a 1 : 1 ratio and 4 × 106 cells were intravenously transferred into anesthetized recipient mice. B6.SJL (CD45.1) and B6Thy1.1 (CD90.1) mice were used for congenic transfers and cell origins were identified by surface expressions of CD45.1/CD45.2 or CD90.1/CD90.2 markers.
[3H]thymidine incorporation assay and CFSE/CTV dilution
For estimating DNA synthesis, sorted naive CD8hi and CD8lo naive CD8 T cells were cultured at a density of 50 000 cells/200 μl medium per well of 96‐well flat‐bottomed plates pre‐coated with various concentrations of anti‐CD3 as indicated and anti‐CD28 (3 μg/ml) mixed together. PMA (100 ng/ml) and various concentrations of ionomycin were used for stimulation where indicated. Sort‐purified OT1 CD8hi and CD8lo naive CD8 T cells were stimulated with irradiated splenic cells pulsed with various concentrations of SIINFEKL peptide. Both sets of stimuli were optimized by titration on total CD8/OT1 T cells, although not for the sorted CD8hi or CD8lo cells. After 48–60 hr of culture, cells were pulsed with 0·5 μCi of [3H]thymidine for 12 hr and plates were frozen followed by harvesting (Molecular Devices, CA, USA) onto glass‐fiber filter‐mats (Wallac, Turku, Finland) and counting (Microbeta plate counter, Perkin Elmer, Waltham, MA).
For assessing cell proliferation, spleen and lymph node cell suspensions were labeled with 10 μm CFSE or 5 μm CTV at a density of 100 × 106 cells/ml in serum‐free RPMI media for 5 min at room temperature. Labeled cells were washed with serum containing RPMI and sort purified into CD8hi and CD8lo naive CD8 T cells. Sort‐purified cells were stimulated as indicated above and CFSE/CTV dilution was assessed 48–60 hr later.
RNA isolation, library construction and high‐throughput sequencing
Total RNA was extracted using TRIzol (Invitrogen, Carlsbad, CA) and RNA was co‐precipitated in the presence of Glycoblue (Thermo Fisher Scientific, Waltham, MA). Isolated RNA samples from CD8hi and CD8lo naive CD8 T cells were analyzed for integrity (RIN) using RNA 6000 Nano kit and 2100 Bioanalyzer (Agilent Technologies, Santa Clara, CA). RNA samples were further used for library construction using Truseq polyA sample prep kit V2 (Illumina, San Diego, CA) following the manufacturer's instructions. Briefly, the PolyA RNA fraction was enriched using oligo‐dT‐magnetic beads and heat‐denatured. Fragmented RNA was end‐repaired and ligated using specific barcode adapters. Enriched libraries were analyzed on the bioanalyzer using high‐sensitivity DNA kit and pooled before cluster generation. Samples were sequenced on a Hiseq 2500 platform using V4 SBS chemistry.
Differential gene expression analysis
Differential gene expression analysis was performed using edger 18 and the new Tuxedo protocol.19 Specifically, processed RNA sequencing reads were aligned using hisat2 (v2.0.5). Aligned reads were assembled using stringtie (v1.3.2d). A count matrix prepared using prepDE.py script was used for normalization and differential expression in edger (v3.12.1). Gene ontology analysis was carried out using clusterprofiler (v2.4.3).20 Heatmaps were plotted using normalized CPM values in R using heatmap3 function (v1.1.1). The raw data have been uploaded on the NCBI SRA Database under Accession number SRP115539.
A publicly available expression array data set (GSE61570) 21 for human adult and neonatal CD8 T cells was analyzed using GEO2R. Genes significantly differentially expressed in neonatal and adult CD8 T cells (adv. P value < 0·05, log2FC of 1) were shortlisted. Genes enriched in both neonatal CD8 T cells and CD8hi naive CD8 T cells were subjected to enrichment analysis using panther 22 and revigo.23 Statistical significance of the overlap was tested using the overlap_stats (v0.011) program (http://nemates.org/MA/progs/overlap_stats_prog.html) considering a total of 26 083 genes.
Quantitative RT‐PCR analysis
For validation of RNA‐Seq results from CD8hi and CD8lo naive CD8 T cells, 100 ng of total RNA was subjected to cDNA synthesis (iScript, Bio‐Rad, Hercules, CA). Some of the genes that were found to be differentially expressed in CD8hi and CD8lo naive CD8 T cells in the RNA‐Seq analysis were subjected to quantitative RT‐PCR analysis (FastStart, Roche, Basel, Switzerland). Relative quantification was performed using the ΔCt method with 18s rRNA as control (primer details in the Supplementary material, Table S1).
Statistical analysis
For comparison of means between two groups, Student's t‐test was performed. For multi‐group comparisons, an analysis of variance test with Bonferroni's multiple comparison correction was used. Values of P < 0·05 were considered significant.
Results
Relative levels of CD8 on naive CD8 T cells correlate with responsiveness
We have previously reported functional heterogeneity in naive CD4 T cells associated with subtly differing CD4 levels.12 We therefore began to examine if naive CD8 T cells showed similar heterogeneity. First, we sorted splenic naive CD8 T cells from young mice, which showed a unimodal and normal distribution of CD8 expression, into CD8hi and CD8lo decile subsets (Fig. 1a) and had no contamination of natural killer cells or CD3− cells (Fig. 1b). These sort‐purified cells were stimulated with plate‐coated anti‐CD3 + anti‐CD28 monoclonal antibodies (3 μg/ml each) for 12 hr and the frequencies of live cells expressing the activation marker CD69 were estimated by flow cytometry. Significantly lower proportions of CD8lo cells expressed CD69 compared with CD8hi cells (Fig. 1c). This difference broadly persisted even when CD69 and CD25 expression was tested at later times post‐activation (see Supplementary material, Fig. S1). CD25 (IL‐2Rα) expression was also tested on these CD8hi and CD8lo cells 24 hr after activation. Again, lower frequencies of CD8lo cells expressed CD25 than CD8hi cells (Fig. 1d), indicating poor early activation in CD8lo cells. Naive CD8hi cells continued to have higher CD8 levels than CD8lo cells upon stimulation for 24 hr with anti‐CD3 + anti‐CD28 (Fig. 1e). In fact, it is noteworthy that CD8 levels on CD8hi cells decline measurably upon activation as expected, whereas CD8lo cells do not show such a decline (Fig. 1e), possibly indicating that they have already reached the lowest permissible level of CD8.
Figure 1.
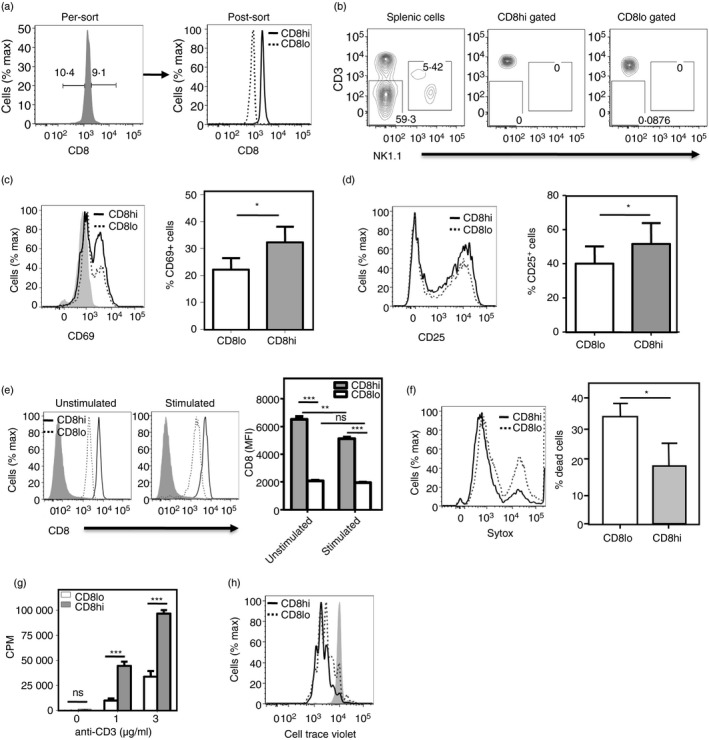
Relative levels of CD8 on naive CD8 T cells correlate with responsiveness. (a) Sort profiles are shown for naive (CD62Lhi CD44lo) CD8 T cells sort‐purified on the basis of the highest and lowest deciles of CD8 expression as naive CD8hi (~10% cells with highest CD8 expression) and naive CD8lo (~10% cells with lowest CD8 expression). (b) Frequencies of CD3− and NK + cells in total splenic cells, and CD8hi and CD8lo gated naive CD8 T cells. (c,d) CD69 (c) and CD25 (d) expression is shown (left panels) on sort‐purified CD8hi and CD8lo naive CD8 T cells stimulated with anti‐CD3 + anti‐CD28 for 12 hr (CD69) or 24 hr (CD25). Frequencies of CD69/CD25‐expressing cells are shown (n = 3; right panels; *P < 0·05). (e) CD8 levels on CD8hi and CD8lo naive CD8 T cells either left unstimulated or stimulated for 24 hr with anti‐CD3 + anti‐CD28 (3 μg/ml each). (f) Frequencies of dead cells as estimated by Sytox dye staining 12 hr after stimulation are shown as histograms (left panel) and as analyzed (n = 4; right panel; ***P < 0·001). (g) [3H]Thymidine incorporation by sort‐purified naive CD8hi and naive CD8lo cells 60 hr after stimulation with titrating concentrations of anti‐CD3 + anti‐CD28 (3 μg/ml). Mean ± SE of triplicate cultures (of three independent experiments). CPM, counts per minute; ***P < 0·001; ns, not significant. (h) Cell Trace Violet dilution by sort‐purified CD8hi and CD8lo naive CD8 T cells stimulated with anti‐CD3 + anti‐CD28 for 48 hr. Three plots represent data from three independent experiments
The extent of cell death upon activation was also estimated in these purified CD8hi and CD8lo naive CD8 T cells using Sytox dye staining. CD8lo cells showed significantly greater death than CD8hi cells did at 12 hr after stimulation (Fig. 1f), although it must be noted that the stimulation conditions had been optimized for total CD8 T cells and not for CD8lo cells. Further, CD8hi cells showed more DNA synthesis than CD8lo cells did upon activation by graded concentrations of anti‐CD3 with anti‐CD28, as assayed by [3H]thymidine incorporation 60–72 hr after activation (Fig. 1g). Proliferation of CD8hi and CD8lo cells was measured using progressive halving of CTV fluorescence within daughter cells, following each cell division. CTV‐labeled CD8hi and CD8lo cells were stimulated with plate‐coated anti‐CD3 + anti‐CD28 for 48 hr and Sytox‐negative cells were gated as live cells and used for analysis of CTV dilution. CD8hi cells proliferated better than CD8lo cells did upon activation (Fig. 1h). Hence, CD8lo naive CD8 T cells showed poor activation and survival in response to TCR‐mediated stimulation.
Phenotypic differences between CD8hi and CD8lo naive CD8 T cells
The CD8‐brightest and CD8‐dullest deciles of naive CD8 T cells from young mice were gated (Fig. 2a), and the relative cell sizes and levels of CD5, CD127 (IL‐7Rα) and TCR on them were examined. While CD8lo cells were smaller and had lower levels of both TCR and CD127, they showed equivalent levels of CD5, a negative regulator of T‐cell activation 24, 25 (Fig. 2b–e). As CD8lo cells were smaller than CD8hi cells, it was possible that the difference in, say, TCR levels between them was simply a correlate of cell size. We therefore gated CD8hi and CD8lo cells of equal size and then examined their TCR levels. Notably, even on CD8hi and CD8lo cells of equal size, TCR levels remain lower on the CD8lo cells (see Supplementary material, Fig. S2).
Figure 2.
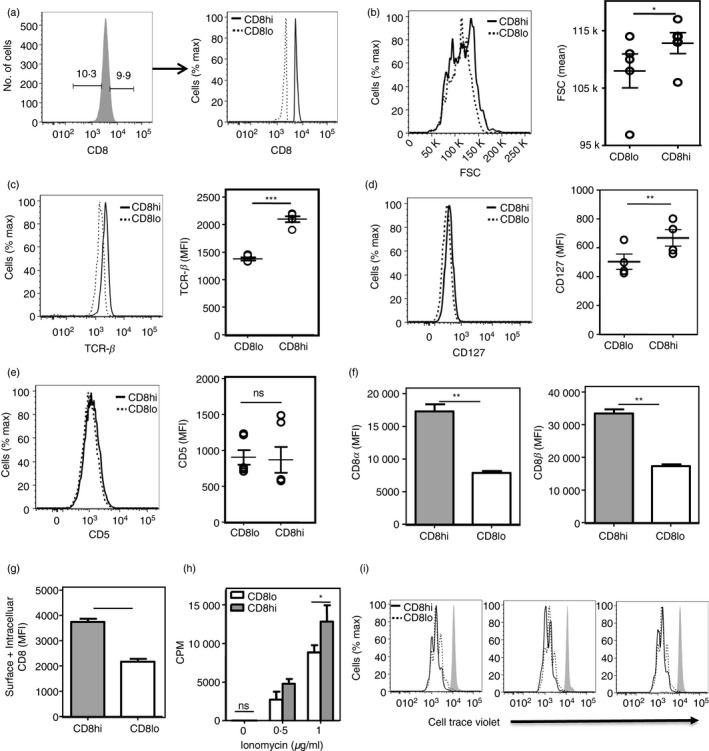
Phenotypic differences between CD8hi and CD8lo naive CD8 T cells. (a) Strategy for gating naive (CD62Lhi CD44lo) CD8T cells into CD8hi‐ and CD8lo‐decile subsets. (b–e) Comparison of cell size (forward scatter; FSC; (b), and expression of T‐cell receptor‐β (TCR‐β) (c), CD127 [interleukin‐17 receptor α (IL‐7Rα)] (d) and CD5 (e) levels between naive CD8hi and naive CD8lo cells (n = 5). ***P < 0·001, **P < 0·01, *P < 0·05, ns, non‐significant; MFI, mean fluorescence intensity. (f) Levels of CD8α and CD8β on naive CD8 T cells gated into CD8hi‐ and CD8lo‐decile subsets based on CD8α expression. (g) Total (surface + intracellular) CD8 levels in CD8hi and CD8lo naive CD8 T cells (P <0·001). (h) [3H]Thymidine incorporation assay on CD8hi and CD8lo cells, stimulated using PMA (100 ng/ml) and titrating concentrations of ionomycin, for 60 hr. Mean ± SE of triplicate cultures (of three independent experiments). (i) Cell Trace Violet dilution by sort‐purified CD8hi and CD8lo naive CD8 T cells stimulated with PMA (100 ng/ml) and ionomycin (1 μg/ml) for 48 hr. Three plots represent data from three independent experiments
We observed that the different levels of cell‐surface CD8α expression in CD8hi and CD8lo naive CD8 T cells were accompanied by a similar difference in cell‐surface CD8β levels (Fig. 2f). We next addressed if only CD8 surface display was different between the CD8hi and CD8lo naive CD8 T cells or were the total CD8 levels higher in CD8hi cells. For this, the CD8hi and CD8lo naive CD8 T cells were sort‐purified and total (cell surface and intracellular) CD8 levels were determined. We observed that the total (cell‐surface and intracellular) CD8 levels were indeed higher in CD8hi naive CD8 T cells (Fig. 2g).
The CD8lo naive CD8 T cells had lower CD8 and TCR levels, so we next asked if the relatively poor responsiveness of these cells was restricted to and explicable by lower TCR levels. When TCR‐independent triggers, namely PMA and titrating concentrations of the calcium ionophore ionomycin which together bypass the TCR signaling cascade and directly activate secondary messengers, were used to activate CD8hi and CD8lo cells, we found that CD8lo cells still showed poorer proliferative responses as assayed by [3H]thymidine incorporation and CTV dilution 60 hr after activation compared with CD8hi cells (Fig. 2h,i). Hence, the hypo‐responsiveness associated with CD8lo naive CD8 T cells is a global cellular characteristic.
TCR‐transgenic monoclonal CD8lo naive CD8 T cells also show hypo‐responsiveness
We next tested whether the hypo‐responsiveness of CD8lo naive CD8 T cells was dependent on their polyclonal nature, with intrinsic differences between different clones leading to enrichment of poor responders in the CD8lo subset. To address this, CD8hi and CD8lo naive CD8 T cells were sort‐purified from TCR‐transgenic OT1 mice and the absence of any contaminating natural killer cells or CD3– cells was confirmed (Fig. 3a,b). The sort‐purified CD8hi and CD8lo naive CD8 OT1 cells were characterized functionally by stimulation with antigen‐presenting cells pulsed with the cognate SIINFEKL peptide. As with polyclonal CD8lo naive CD8 T cells, CD8lo OT1 cells showed poorer survival 12 hr after stimulation as estimated by Sytox dye staining (Fig. 3c), although it must be noted again that the stimulation conditions had been optimized for total OT1 CD8 T cells and not for CD8lo cells in particular. Lower frequencies of CD8lo OT1 naive CD8 T cells expressed CD69 12 hr after stimulation (Fig. 3d) and CD25 at 24 hr after stimulation (Fig. 3e). [3H]Thymidine incorporation as well as CTV dilution were poorer in activated CD8lo OT1 cells after 60 hr (Fig. 3f,g). These data indicated that the hypo‐responsiveness of CD8lo naive CD8 T cells was independent of clonal diversity.
Figure 3.
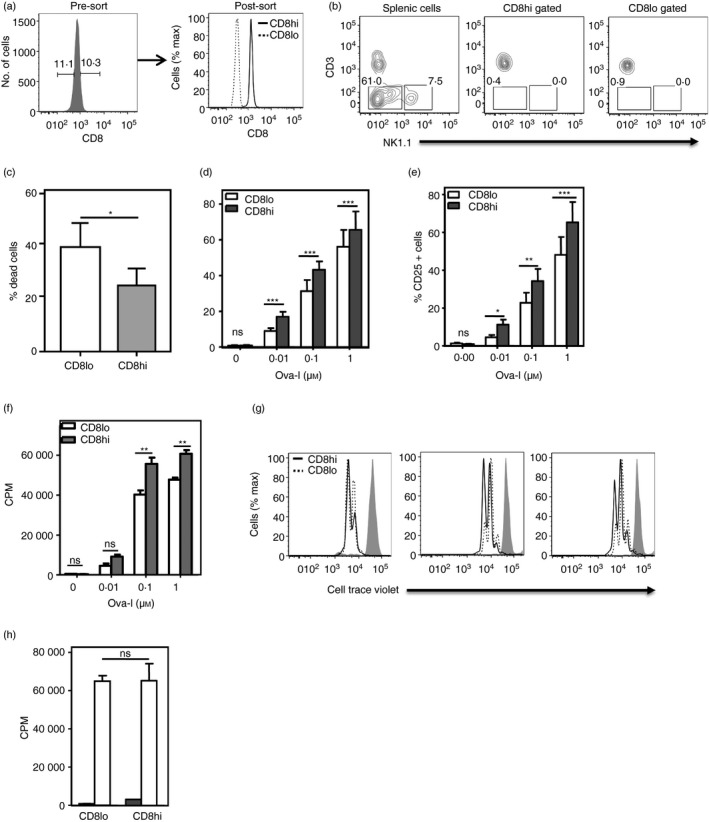
Naive CD8lo cells from T‐cell receptor (TCR) ‐transgenic OT1 mice are hypo‐responsive to cognate ligand. (a) Sort‐purification profiles of CD8hi and CD8lo naive CD8 T cells from TCR‐transgenic OT1 mice. (b) Frequencies of CD3− and NK + cells in total splenic cells, and CD8hi and CD8lo gated naive CD8 T cells from OT1 mice. (c) Frequencies of dead cells as estimated by Sytox dye staining 12 hr after stimulation are shown (n = 3; *P < 0·05). (d, e) Frequencies of CD69 (d; shown as % CD69+ cells) and CD25 (e) expression on sort‐purified CD8hi and CD8lo naive CD8 T cells stimulated with graded concentrations of OVA‐I peptide as indicated for 12 hr (CD69) or 24 hr (CD25). (n = 3; ***P < 0·001, **P < 0·01, *P < 0·05; ns, not significant; two‐way analysis of variance with Bonferroni correction). (f, g) [3H]Thymidine incorporation (f) and Cell Trace Violet dilution assay (g) to measure proliferation of sort‐purified CD8hi and CD8lo naive CD8 OT1 T cells 60 hr after stimulation with OVA‐I peptide as shown. Mean ± SE of triplicate cultures (of four independent experiments). CPM, counts per minute; **P < 0·01. Three plots of Cell Trace Violet dilution represent data from three independent experiments. (h) CD8hi‐ and CD8lo‐decile subsets were sort‐purified from mature CD8+ CD4− Qa2+ CD24− thymocytes and [3H]thymidine incorporation was measured 60 hr after stimulation with anti‐CD3 + anti‐CD28 (3 μg/ml each). Mean ± SE of triplicate cultures (one of four independent experiments); CPM, counts per minute; ns, not significant.
Lack of association between the CD8lo phenotype and hypo‐responsiveness in CD8SP thymocytes
Mature CD8SP thymocytes (CD8+ CD4− Qa2+ CD24−) in the lower and higher deciles of the CD8 distribution were sort‐purified as CD8lo and CD8hi cells, respectively. When these CD8hi and CD8lo CD8SP thymocytes were stimulated with plate‐coated anti‐CD3 and anti‐CD28 (3 μg/ml each) for 60–72 hr and proliferation was measured as [3H]thymidine incorporation, both subsets showed equivalent responses (Fig. 3h), unlike the peripheral CD8hi and CD8lo naive CD8 T cells. This finding indicated that the hypo‐responsiveness of peripheral CD8lo naive CD8 T cells was not thymic in origin and likely developed in the periphery.
Human CD8hi and CD8lo naive CD8 T cells show properties similar to those from mice
We tested if the distinctions observed between CD8hi and CD8lo naive CD8 T from mice were also found in human CD8 T cells. To address this, we isolated naive CD8 T cells (CD8+ CD45RA+ CD62L+) from human peripheral blood mononuclear cells of healthy donors. Naive CD8 T cells in the lower and higher deciles of the CD8 distribution were sort‐purified as CD8lo and CD8hi cells, respectively (Fig. 4a) and the absence of any contaminating natural killer cells or CD3− cells was confirmed (Fig. 4b). Human CD8lo naive CD8 T cells were found to be smaller in size (Fig. 4c), consistent with the mouse data. The sort‐purified human CD8hi and CD8lo cells were then stimulated with plate‐coated anti‐CD3 and anti‐CD28 monoclonal antibodies and proliferation was measured by [3H]thymidine incorporation. Like CD8hi cells from mice, human CD8hi cells proliferated better in response to TCR‐mediated stimulation than CD8lo cells (Fig. 4d).
Figure 4.
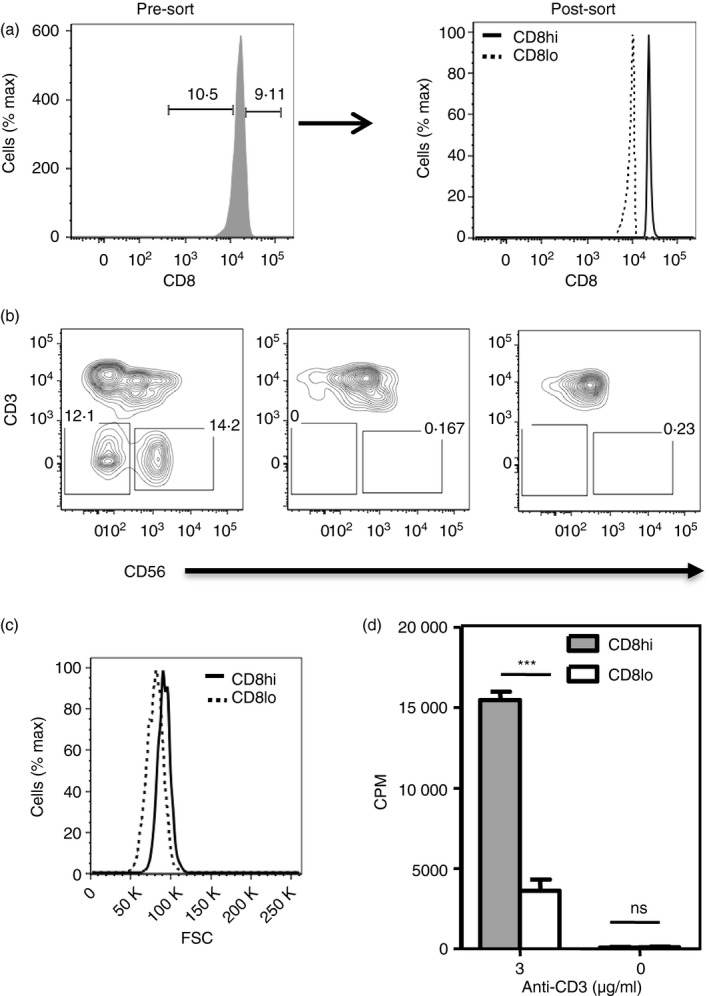
Human CD8hi and CD8lo naive CD8 T cells show properties similar to those from mice. (a) Sort‐purification profiles of naive (CD8+ CD45RA + CD62L+) CD8hi‐ and CD8lo‐decile subsets from human peripheral blood mononuclear cells (PBMCs). (b) Frequencies of CD3− and NK + cells in total PBMCs, and CD8hi and CD8lo gated naive CD8 T cells from human PBMCs. (c) Comparison of cell size (forward scatter; FSC) between human naive CD8hi and naive CD8lo cells. (d) [3H]Thymidine incorporation assay to measure proliferation of sort‐purified human naive CD8hi and naive CD8lo cells 96‐hr after stimulation with anti‐CD3 + anti‐CD28.
Consequences of peripheral residence for naive CD8 T cells
We next tested the stability of the CD8hi and CD8lo phenotype of these naive CD8 T‐cell subsets, by purifying and labeling them followed by adoptive co‐transfer into congenic recipient mice. An aliquot of these cells was cryopreserved so that analysis of cells before and after transfer could be performed in parallel. Recipient mice were killed 4 days later and their splenic cells were similarly cryopreserved. Parallel comparison of CD8 levels on naive CD8hi and CD8lo cells before and 4 days after transfer showed that adoptively transferred CD8lo cells retained their CD8 levels, but CD8hi cells lost some CD8 intensity 4 days after transfer (Fig. 5a,b). These data indicated that peripheral cellular aging of naive CD8 T cells was correlated to the decline of CD8 levels on them.
Figure 5.
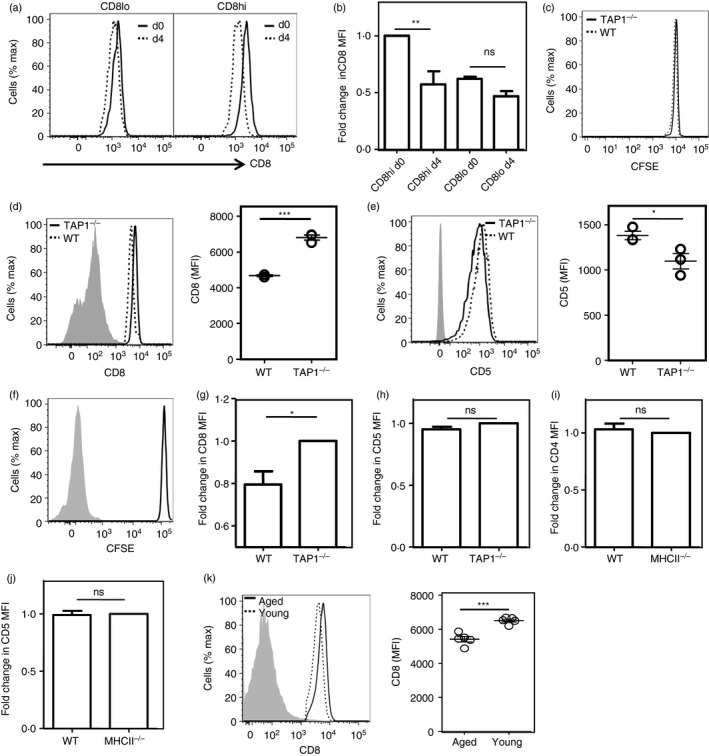
Consequences of peripheral residence and tonic signals on co‐receptor levels on naive T cells. (a,b) CD8 levels on transferred CD8hi and CD8lo naive CD8 T cells before (day 0) and 4 days after transfer (day 4) into wild‐type (WT) recipient mice. Representative histograms (a) and analyzed data (b; n = 3; **P < 0·01) are shown. (c) CFSE profiles of donor naive OT1 cells assayed 4 days after transfer in wild‐type (WT) and TAP1‐null mice. (d) CD8 levels on donor naive CD8 OT1 cells 4 days after transfer in wild‐type (WT) and TAP1‐null mice. Representative histograms (left panel; gray histogram, negative control) and analyzed data (right panel; n = 3; *** P < 0·001) are shown. (E) CD5 levels of donor naive CD8 OT1 cells 4 days after transfer in WT and TAP1‐null mice. Representative histograms (left panel; gray histogram, negative control) and analyzed data (right panel; n = 3; *P < 0·05) are shown. (f) CFSE dilution assay on naive CD8 OT1 cells co‐cultured with bone‐marrow‐derived dendritic cells (BMDCs) for 3 days. Unstained control is shown as gray‐shaded histogram. (g, h) CD8 levels (g) and CD5 levels (h) on naive OT1 cells co‐cultured with either WT or TAP1‐null BMDCs. Unstained control is shown as gray shaded histogram. (i, j) CD4 levels (i) and CD5 levels (j) on naive OT2 cells co‐cultured with either WT or MHCII‐null BMDCs. Unstained control is shown as gray‐shaded histogram. (k) CD8 levels on aged (18–20 months old) and young (4–8 weeks old) naive CD8 T cells stained as a mixture in a single well are shown (left panel), and mean fluorescence intensity (MFI) values are shown (n = 5; right panel; ***P < 0·001). Gray histogram represents negative control
MHCI‐mediated tonic signals modulate co‐receptor levels on naive CD8 T cells
After emigration from the thymus, naive T cells recirculate in the periphery, where they encounter ‘tonic’ signals mediated by non‐cognate peptide/s bound to the restricting MHC molecule. Cells with long post‐thymic cellular ages would therefore be expected to have encountered more of these signals. There is evidence that MHCI‐mediated tonic signals have a role in modulating co‐receptor levels on peripheral naive CD8 T cells.16 We confirmed this by CFSE‐labeling naive CD8 T OT1 cells and adoptively transferring them into either WT or TAP1‐null mice, which express little MHCI on cell surfaces. CFSE bright donor cells were analyzed 4 days after transfer (Fig. 5c). CD8 levels were lower, but CD5 levels were higher in naive CD8 T cells parked in WT mice (Fig. 5d,e). These data were consistent with the possibility that MHCI‐mediated tonic signals to naive CD8 T cells, as indicated by higher CD5 levels, contributed to reduction of cell surface CD8 levels.
These findings were tested by in vitro recapitulation of the cognate stimulus‐independent ‘tonic’ signaling in vivo. Naive CD8 OT1 cells were co‐cultured with BMDCs of WT and TAP1‐null mice for 72 hr in the presence of IL‐7 (10 ng/ml) to ensure survival. Cells did not proliferate during that time, as shown by CFSE dilution data (Fig. 5f). We observed that naive CD8 OT1 cells co‐cultured with WT BMDCs showed lower CD8 levels than those cultured with TAP1‐null BMDCs (Fig. 5g), although CD5 levels were similar (Fig. 5h). Interestingly, although MHCII‐mediated tonic signals cause co‐receptor down‐modulation in naive CD4 T cells in vivo,12 CD4 OT2 cells co‐cultured with WT and MHCII‐null BMDCs showed equivalent CD4 and CD5 levels (Fig. 5i,j). In addition to confirming that MHCI‐mediated tonic signals cause co‐receptor down‐modulation in naive CD8 T cells,16 these observations suggest that tonic signaling may well be subtly different between naive CD4 and naive CD8 T cells.
Our data are consistent with the possibility that that CD8lo cells from young mice are likely to be those with longer peripheral residence, so are likely to have encountered more MHCI‐mediated tonic signals. Naive T cells from aged animals have a longer average peripheral residence time than those from young animals.5, 26 We therefore tested splenic naive CD8 T cells from young or aged mice, and found that CD8 levels were subtly but reproducibly lower in the latter (Fig. 5k).
Poorer baseline signaling in CD8lo cells
We further looked for evidence of differential signaling in the CD8hi and CD8lo subsets of naive CD8 T cells, by examining the baseline ex vivo phosphorylation status of signaling molecules in TCR signaling cascades such as Zap70/Syk and ERK. Baseline ZAP70/Syk and ERK phosphorylation and total ERK, but not total ZAP70/Syk levels, were found to be significantly lower in CD8lo cells than CD8hi cells (Figs 6a–d). Although reproducible, these differences were modest. Unfortunately, our efforts to obtain sufficient numbers of CD8hi and CD8lo cells to confirm these results with titrated Western blot analyses have been unsuccessful. We therefore attempted to confirm these findings, made using direct PE‐conjugates of the relevant antibodies (BD; dilutions as per manufacturer's recommendations), by using different antibodies for the flow cytometric analysis for pZAP70/pSyk and pERK in a two‐step protocol using relevant non‐labeled rabbit antibodies (Cell Signaling Technologies; dilutions as recommended by the manufacturer), followed by fluorochrome‐coupled secondary F(abʹ)2 goat anti‐rabbit IgG1 (0·5 μg/100 μl). We found similar differences in pZAP‐79/p‐Syk and pERK in these experiments (see Supplementary material, Fig. S3). Phosphorylation of ERK has been shown to be negatively regulated by DUSP activity.27 Therefore, we tested if treatment with a DUSP inhibitor, BCI, could rescue the hypo‐responsiveness of CD8lo naive CD8 T cells. BCI partially rescued the proliferative response of CD8lo, but not of CD8hi, naive CD8 T cells to anti‐CD3 + anti‐CD28 (Fig. 6e), indicating that the DUSP‐ERK axis is likely to be one, but not the only, pathway involved in the hypo‐responsiveness of CD8lo naive CD8 T cells.
Figure 6.
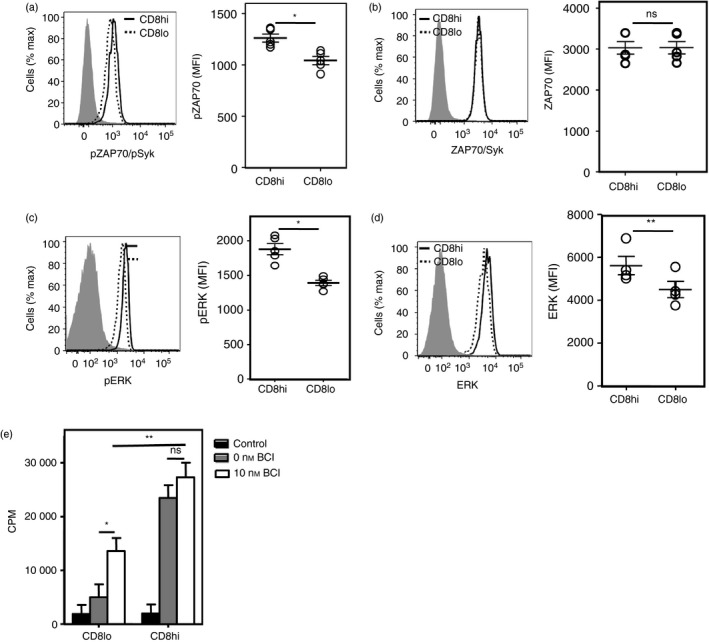
Compromised baseline signaling in CD8lo cells. (a–d) Baseline levels of pZAP‐70/pSyk (a), pERK (b), total ZAP70/Syk (c) and total ERK (d) on CD8hi and CD8lo naive CD8 T cells. Isotype controls are shown as gray‐shaded histograms. (e) [3H]Thymidine incorporation on sort‐purified CD8hi and CD8lo naive CD8 T cells either treated or not with DUSP inhibitor (BCI) for 1 hr before activation with anti‐CD3 + anti‐CD28 for 60 hr. Mean ± SE of triplicate cultures. Data representative of three independent experiments. CPM, counts per minute; *P < 0·05, ns = non‐significant.
Major differences in transcriptional landscapes of CD8hi and CD8lo naive CD8 T cells
We next looked for evidence of any changes in the transcriptional landscapes in the CD8hi and CD8lo subsets of naive CD8 T cells by sort‐purifying the two subsets and performing RNA sequencing. The CD8α transcript levels were higher in CD8hi cells than CD8lo naive CD8 T cells, as estimated by RNA sequencing, suggesting quantitative transcriptional modulation (Fig. 7a). We also identified 315 genes with a statistically significant difference in expression of over twofold between CD8hi and CD8lo naive CD8 T cells (Fig. 7b), indicating heterogeneity within the naive CD8 T‐cell compartment. Pathway analysis identified ‘cell cycle’ as the molecular network most significantly associated with transcripts showing differential expression in CD8hi and CD8lo naive CD8 T cells (Fig. 7c,d). As expected, Mki67 (which encodes Ki‐67), and other genes encoding key regulators involved in the proliferation of CD8 T cells, such as Ccbn2, Uhrf1, Melk and Mcm10 showed transcript abundance in the CD8hi cells (Fig. 7c); for some of these, we confirmed the difference by quantitative RT‐PCR (Fig. 7e) with a 100% confirmation rate of seven selected genes from the RNAseq results by real‐time RT‐PCR. Gene enrichment analysis suggested enrichment of genes involved in the cell cycle in CD8hi cells, but many pro‐apoptotic genes including FasL, mmp9, ccl5, sox4 and itgb were enriched in the CD8lo cells (Fig. 7f,g), which was consistent with our observation that CD8lo cells have poor survival in in vitro cultures. Levels of Nr4a1 (which encodes Nur77) were also higher in the CD8lo naive CD8 T cells (Fig. 7f,g). This also correlated with the possibility of differential tonic signaling, although Nur77 expression may also be modulated by other signals.
Figure 7.
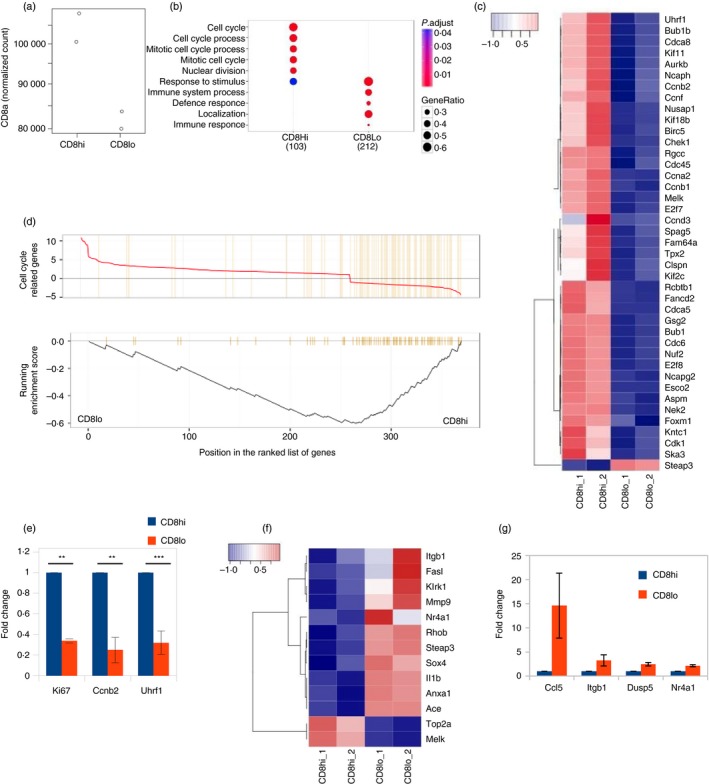
Major changes in transcriptional landscapes of naive CD8 T cells as a result of cellular aging. (a) Comparison of CD8 transcript levels in sort‐purified CD8hi and CD8lo naive CD8 T cells by RNA sequencing. (b) Comparison of gene cluster functions of genes deferentially expressed between CD8hi and CD8lo cells. (c) Gene Set Enrichment analysis (GSEA) of cell cycle genes (GO:0007049). (d, f) Expression heatmaps of cell cycle genes and genes involved in apoptosis. Plotted using normalized counts per minute values (key = Z‐score). (e, g) Quantitative RT‐PCR validation of genes identified in combined RNA‐Seq microarray analysis.
We further compared our RNA‐Seq data on CD8hi and CD8lo naive CD8 T cells with a published data set on human microarray analysis GSE6157021 on neonatal and adult CD8 T‐cell samples using Geo2R, under the assumption that neonatal CD8 T cells will show less cellular post‐thymic aging than adult CD8 T cells. A number of genes that were differentially expressed between neonatal and adult human microarray samples were also found to be differentially expressed between CD8hi and CD8lo naive CD8 T cells (see Supplementary material, Fig. S4). This analysis yielded 82 hits, of which those that showed higher expression in both mouse CD8hi cells and human neonatal samples were plotted. We observed a significant overlap of 19 genes between CD8hi and neonatal samples; however, only one gene overlapped between CD8hi cells and adult human CD8 T cells. When a similar analysis was performed for mouse CD8lo cells and adult human CD8 T cells, we found that there were 20 genes that showed higher expression in mouse both CD8lo cells and adult human CD8 T cells, indicating striking similarities between CD8lo and adult human CD8 T cells. Although the overlap appears quite modest, it is considerably different between CD8hi and CD8lo cells, especially as the comparison is with bulk neonatal/adult CD8 T cells, cross‐species and cross‐platform. We have performed a GO analysis that is also included for information (see Supplementary material, Fig. S4).
In summary, our findings from the RNA‐Seq data further validated the existence of heterogeneity in the naive CD8 T‐cell compartment as well as suggesting that post‐thymic aging leads to major changes in the transcriptional and functional program of naive CD8 T cells.
Discussion
The mechanisms causing variability in homogeneous naive T‐cell populations are likely to be a mixture of the stochastic and the environmentally induced. We have previously reported that cell population variation in glucose metabolism and rate of protein synthesis could be involved in regulating the variation in the ability of CD8 T cells to respond to TCR‐mediated stimulation.3 Recent reports have traced the in vivo CD8 T‐cell response at single‐cell level, demonstrating that there is a strong heterogeneity in the numerical output of clonal expansion from individual T cells.28 Moreover, the behavior of individual T cells varies markedly with respect to differentiation and recall capacity.28 However, in addition to stochastic factors, functional heterogeneity in a T‐cell population can also result due to differential life histories of individual cells. Within a young naive T‐cell compartment, cells are not synchronized for age because it is composed of cells that have either recently emigrated from the thymus or those that have been continuously recirculating in the peripheral lymphoid organs without having encountered a cognate ligand. Hence, there is a diversity of post‐thymic age among peripheral naive T cells. We have previously reported that this diversity contributes to the functional and phenotypic heterogeneity of naive CD4 T cells, at least partly dependent on MHCII‐mediated tonic signals.12
A corollary of these findings would be that naive CD8 T cells behave in the same way; and that diversity of post‐thymic age among peripheral naive CD8 T cells contributes to their functional and phenotypic heterogeneity in an MHCI‐dependent fashion. However, there are substantial differences between naive CD4 and naive CD8 T cells with regard to their interactions with self‐pMHC. Naive CD8 T cells in MHCI‐deficient environments in vivo tend to die rapidly, but naive CD4 T cells do not do so in lymphoreplete situations,14, 15 identifying a major potential difference between them. Also, the periodicity of pMHC contact is likely to differ widely between naive CD4 T and naive CD8 T cells, as MHCII is normally expressed on select cell subsets of bone marrow origin, whereas MHCI is far more widely expressed in most cellular lineages.29
It was therefore important to examine whether the functional and phenotypic heterogeneity correlated with CD4 levels we had observed in naive CD4 T cells were also detectable in naive CD8 T cells. We have now shown that, although there is indeed broad similarity between naive CD4 and naive CD8 T cells in that, like naive CD4 T cells, co‐receptor CD8 levels correlate with phenotypic and functional differences in naive CD8 T cells so identifying hidden heterogeneity in them, there are also subtle differences between them. Subtle yet consistent differences in co‐receptor levels, either CD4 or CD8, mark naive T cells of different functionality and correlate with potentially peripheral ages, whereas an in vitro MHC‐deficient environment is sufficient to induce co‐receptor level reduction in naive CD8 T cells but not in naive CD4 T cells. Further, we also report a comprehensive transcriptomic analysis of CD8lo and CD8hi naive CD8 T cells, with identification of resultant differences consistent with the observed functional differences between them.
When naive CD8 T cells were purified into CD8hi and CD8lo subsets, the CD8lo cells, which had lower TCR‐β and IL‐7Rα expression and were smaller in size, were hypo‐responsive, like CD4lo naive CD4 T cells, to TCR‐mediated activation by showing poor viability and hypo‐responsiveness, indicating that subtle differences in CD8 levels in an apparently homogeneous naive CD8 T‐cell population did indeed correlate with functional heterogeneity. As CD8lo cells not only responded poorly to TCR‐mediated activation but also showed lower TCR levels, it was possible that lower TCR as well as CD8 levels were contributing to poor activation. However, even when TCR‐mediated signaling was bypassed by using PMA and ionomycin as the stimulus, CD8lo cells proliferated relatively poorly. Hence, CD8hi and CD8lo naive CD8 T cells showed functional differences independent of TCR or co‐receptor levels or avidity. The observation that even among monoclonal naive CD8 T cells from TCR‐transgenic mice, CD8lo cells responded more poorly than CD8hi cells ruled out the possibility that functional variation between CD8hi and CD8lo naive CD8 T cells was due to intrinsic differences between different T‐cell clones within a polyclonal population. Hence, within a naive CD8 T‐cell population with unimodally distributed co‐receptor levels, there exists distinct phenotypic as well as functional heterogeneity. Strikingly, our RNA sequencing data on CD8hi and CD8lo naive CD8 T cells showed that genes encoding key regulators of either proliferation or survival were differentially expressed between the two subsets, indicating that the transcriptional program of naive CD8 T cells is also correlated with subtle differences in CD8 levels. This functional variation in naive CD8 T cells may well have real‐life consequences. Hence, CD8 T‐cell responses to infections may show heterogeneity as a consequence of the relative preponderance of CD8hi and CD8lo cells in the naive CD8 T‐cell population. Such interesting possibilities remain to be investigated.
It was notable that CD8lo naive CD8 T cells showed lower IL‐7Rα (CD127) expression. Interleukin‐7 is a critical survival factor for naive T cells,7, 30 so lower CD127 levels may suggest decreased survival potential, but this interpretation is complicated by the finding that CD127 expression is reduced after its engagement by IL‐7,31 so low CD127 levels may also be a result of efficient IL‐7 encounters.
The observation that mature thymocytes about to enter the periphery do not show functional distinctions between their CD8lo and CD8hi sub‐populations, although preliminary, is consistent with the possibility that variation in CD8 levels in CD8SP thymocytes is likely to be stochastic and without functional consequences, and that hypo‐responsiveness of peripheral CD8lo cells is post‐thymic in origin. In keeping with this, transfer of sort‐purified peripheral CD8hi cells in vivo showed that their CD8 levels went down within 4 days of peripheral residence. Interestingly, CD8 levels remained unchanged in CD8lo cells, possibly suggesting regulatory mechanisms preventing decline in CD8 levels below this point.
We have confirmed previous data that naive CD8 T‐cell transfer into MHCI‐deficient TAP1‐null and WT recipients identifies MHCI‐mediated decline in CD8 levels.16 However, the in vitro equivalents of such experiments show down‐modulation of CD8 on naive CD8 T cells but not of CD4 on naive CD4 T cells in our hands. Consistent with a role for peripheral microenvironmental influences in regulating co‐receptor levels, it has been previously shown that CD8 expression is transcriptionally enhanced by IL‐7 and other common γ‐chain cytokine signals.32 In the steady state, survival of naive CD8 T cells in vivo is known to depend on their ability to get tonic signals from self pMHC complexes 33 and absence of such complexes leads to a steady loss of the population.34 Notably, although this is true for both NCD4 and naive CD8 T cells in mostly non‐physiological lymphopenic situations,14, 15, 16 it is only true for naive CD8 but not for naive CD4 T cells in the physiologically more common lymphoreplete situations.14, 15, 35 The patterns, periodicity and duration of MHC‐mediated tonic signals would quite likely differ between MHCI and MHCII for naive CD8 and naive CD4 T cells, respectively, because of differences in the cellular lineage specificity of MHCI and MHCII expression. So it appears possible that, compared with naive CD4 T cells, naive CD8 T cells are more sensitive to as well as more dependent on MHC‐mediated signals. It has also been reported previously that removal from MHCI‐sufficient environments enhanced naive CD8 T‐cell functionality,16 indicating that tonic signaling may induce hypo‐responsiveness while maintaining cell survival.
CD5 is described as a negative regulator of TCR signaling via self pMHC complexes.24, 25, 36 However, the correlation between CD5 levels on naive T cells and their responsiveness has been controversial. Higher CD5 levels have been shown to be correlated with better responsiveness of naive T cells.37 On the other hand, studies in lymphopenic mice have shown that naive CD8 T cells with high CD5 levels tend to have lower TCR levels, which in turn indicate a requirement for higher avidity self pMHC interaction for survival.38 Also, tonic MHC‐mediated signals lead to up‐regulation of CD5 levels and to induction of hypo‐responsiveness in both CD4 and CD8 T cells.16, 39 Our data showed that CD5 levels were higher on naive CD8 T cells transferred into MHCI‐sufficient recipients. This may indicate that CD5 levels reflect the duration of tonic signaling and might be inversely correlated with responsiveness as previously reported. However, it remains possible that, between naive T cells that have received tonic signals for comparable durations, higher CD5 levels may be correlated with higher responsiveness. It is also noteworthy that the differences in functionality between the naive CD8lo and CD8hi cells persist even in a monoclonal TCR‐transgenic T‐cell population, in which the quality of tonic signaling would be expected to be relatively invariant, implicating the duration of tonic signaling as a major determinant of heterogeneity of responsiveness.
Naive T cells from aged mice have been reported to show higher susceptibility to death and proliferate poorly than naive T cells from young mice. However, it has also been demonstrated that aged T cells that survive primary activation behave similar to young cells in terms of proliferation and death, suggesting that only a subset of cells in the aged T‐cell population is hypo‐responsive.40 In this context, as age‐related decline in thymic output appears to lead to longer average peripheral residence time of naive T cells in the periphery,6, 41 our observation that naive CD8 T cells from aged mice have lower CD8 levels than naive CD8 T cells from young mice may be consistent with the interpretation that prolonged peripheral residence leads to tonic signal‐induced hypo‐responsiveness marked by a subtle decline in CD8 levels.
Phosphorylation of ERK has been shown to be poor in aged T cells due to DUSP6, whose expression increases with age. The DUSP inhibitor BCI has been shown to rescue functional defects in aged human T cells.42 Our previous data with naive CD4 T cells showed that the ERK–DUSP axis was also important in the hypo‐responsiveness of CD4lo naive CD4 T cells,12 and that finding is consistent for CD8lo naive CD8 T cells in the present data, although the differences in basal phosphorylation of signal transduction intermediates we find between CD8hi and CD8lo cells are modest and based on tentative flow cytometric assays.
Compared with naive CD4 T cells, naive CD8 T cells appeared to be more sensitive to, as well as more dependent on, MHC‐mediated signals, so it was reasonable to look for major transcriptional program modifications in CD8hi versus CD8lo naive CD8 T cells. Our RNA‐Seq analysis, supported by quantitative RT‐PCR data, indicates that CD8hi naive CD8 T cells show higher expression of genes that are positive regulators of cell cycle, suggesting that they may be more poised to enter cycle upon activation, whereas CD8lo naive CD8 T cells show higher expression of pro‐apoptotic genes, suggesting that they may be more poised for cell death upon activation, and both expectations are consistent with our functional data. Finally, we not only made similar functional findings in the human system, but a comparative analysis of reported transcriptomes from neonatal versus adult CD8 T cells and our CD8hi/lo naive CD8 T‐cell transcriptomes indicated that CD8hi naive CD8 T cells indeed resemble neonatal, presumably ‘young’ human CD8 T cells. Hence, from a number of different perspectives, our data reveal functional heterogeneity in a seemingly homogeneous naive CD8 T‐cell population. The genesis and consequences of this heterogeneity are clearly issues of great future interest.
Funding
The study was supported in part by grants from the Department of Biotechnology (to AG # BT/PR12849/MED/15/35/2009; to VB # BT/PR14420/Med/29/213/2010; to SG # BT/COE/34/SP17426/2016 and to SRath # BT/PR‐14592/BRB/10/858/2010), and from the Department of Science and Technology, Government of India (to VB # SR/SO/HS‐0005/2011 and #EMR/2015/001074; to SRath # SB/SO/HS/210/2013), and from the Arkansas Biosciences Institute (to JD). SK is supported by an intramural postdoctoral fellowship from the Indian Institute of Science Education and Research (IISER), Pune, and SP is supported by a senior research fellowship from the Council of Scientific and Industrial Research, India. The National Institute of Immunology is supported by the Department of Biotechnology, Government of India. IISER Pune is supported by the Ministry of Human Resource Development, Government of India.
Author contributions
RB, RG, ASC, SJP, SPK and SRane performed experiments and analyzed data. RB, SJP and SPK plotted figures. RB wrote the manuscript draft. SG, JD, AG, VB and SRath conceptualized the project, designed and supervised experiments and edited the manuscript. All authors read and approved the final manuscript.
Disclosures
SRath is a non‐executive director of Ahammune Biosciences Private Limited, Pune, India, and a member of the scientific advisory boards of Curadev Pharma Private Limited, NOIDA, India, and Mynvax Private Limited, Bangalore, India. Other authors have no financial interests to declare.
Supporting information
Table S1. Quantitative RT‐PCR analysis: primer details
Figure S1. Induction of CD69 on CD8hi versus CD8lo naive CD8 T cells
Figure S2. Cell‐surface TCR levels on CD8hi versus CD8lo naive CD8 T cells of equal size
Figure S3. Compromised baseline signaling in CD8lo cells
Figure S4. Comparison of mouse CD8hi and CD8lo RNA‐Seq with human neonatal and adult CD8 T‐cell expression array (GSE61570).
Acknowledgments
We thank Inderjit Singh and Dr. P. Nagarajan for help in animal breeding and maintenance and K. Rajesh Kumar for assistance in flow cytometric sorting, and Dr. P. Chandramouli Reddy for discussions.
References
- 1. Altschuler SJ, Wu LF, Bakal C, Li F, Sun Y, Perrimon N et al Cellular heterogeneity: do differences make a difference? Cell 2010; 141:559–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Feinerman O, Veiga J, Dorfman JR, Germain RN, Altan‐Bonnet G. Variability and robustness in T cell activation from regulated heterogeneity in protein levels. Science 2008; 321:1081–4. 10.1126/science.1158013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Balyan R, Gund R, Ebenezer C, Khalsa JK, Verghese DA, Krishnamurthy T et al Modulation of naive CD8 T cell response features by ligand density, affinity, and continued signaling via internalized TCRs. J Immunol (2017) Available at: http://www.jimmunol.org/content/early/2017/01/18/jimmunol.1600083 [Accessed August 7, 2017] [DOI] [PubMed] [Google Scholar]
- 4. Gascoigne NR, Acuto O. THEMIS: a critical TCR signal regulator for ligand discrimination. Curr Opin Immunol 2015; 33:86–92. [DOI] [PubMed] [Google Scholar]
- 5. Den Braber I, Mugwagwa T, Vrisekoop N, Westera L, Mögling R, Bregje de Boer A et al Maintenance of peripheral naive T cells is sustained by thymus output in mice but not humans. Immunity 2012; 36:288–97. [DOI] [PubMed] [Google Scholar]
- 6. Nikolich‐Žugich J. Ageing and life‐long maintenance of T‐cell subsets in the face of latent persistent infections. Nat Rev Immunol 2008; 8:512–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Rathmell JC, Farkash EA, Gao W, Thompson CB. IL‐7 enhances the survival and maintains the size of naive T cells. J Immunol 2001; 167:6869–76. Available at: http://www.ncbi.nlm.nih.gov/pubmed/11739504 [Accessed August 8, 2017] [DOI] [PubMed] [Google Scholar]
- 8. Surh CD, Sprent J. Regulation of mature T cell homeostasis. Semin Immunol 2005; 17:183–91. [DOI] [PubMed] [Google Scholar]
- 9. Kirberg J, Berns A, Boehmer HV. Peripheral T cell survival requires continual ligation of the T cell receptor to major histocompatibility complex‐encoded molecules. J Exp Med 1997; 186:1269–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Barthlott T, Wright RJ, Stockinger B. Normal thymic selection of TCR transgenic CD4 T cells, but impaired survival in the periphery despite the presence of selecting MHC molecules. J Immunol (1998) 161: Available at: http://www.jimmunol.org/content/161/8/3992.short [Accessed August 15, 2017] [PubMed] [Google Scholar]
- 11. Boursalian TE, Bottomly K. Survival of naive CD4 T cells: roles of restricting versus selecting MHC class II and cytokine milieu. J Immunol (1999) 162: Available at: http://www.jimmunol.org/content/162/7/3795 [Accessed August 15, 2017] [PubMed] [Google Scholar]
- 12. Rane S, Das R, Ranganathan V, Prabhu S, Das A, Mattoo H et al Peripheral residence of naive CD4 T cells induces MHC class II‐dependent alterations in phenotype and function. BMC Biol 2014; 12:106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Brocker T. Survival of mature CD4 T lymphocytes is dependent on major histocompatibility complex class II‐expressing dendritic cells. J Exp Med (1997) 186: Available at: http://jem.rupress.org/content/186/8/1223 [Accessed May 14, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Clarke SRM, Rudensky AY. Survival and homeostatic proliferation of naive peripheral CD4+ T cells in the absence of self peptide: MHC complexes. J Immunol (2000) 165: Available at: http://www.jimmunol.org/content/165/5/2458?ijkey=b009b8b5bcc04110232c640806fc309ab86cb0ed&keytype2=tf_ipsecsha [Accessed August 8, 2017] [DOI] [PubMed] [Google Scholar]
- 15. Dorfman JR, Štefanová I, Yasutomo K, Germain RN. CD4+ T cell survival is not directly linked to self–MHC‐induced TCR signaling. Nat Immunol 2000; 1:329–35. [DOI] [PubMed] [Google Scholar]
- 16. Takada K, Jameson SC. Self–class I MHC molecules support survival of naive CD8 T cells, but depress their functional sensitivity through regulation of CD8 expression levels. J Exp Med 2009; 206:2253–69. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17. Inaba K, Inaba M, Romani N, Aya H, Deguchi M, Ikehara S et al Generation of large numbers of dendritic cells from mouse bone marrow cultures supplemented with granulocyte/macrophage colony‐stimulating factor. J Exp Med (1992) 176:1693–702. Available at: http://www.ncbi.nlm.nih.gov/pubmed/1460426 [Accessed February 19, 2018] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Anders S, McCarthy DJ, Chen Y, Okoniewski M, Smyth GK, Huber W et al Count‐based differential expression analysis of RNA sequencing data using R and Bioconductor. Nat Protoc 2013; 8:1765–86. [DOI] [PubMed] [Google Scholar]
- 19. Pertea M, Kim D, Pertea GM, Leek JT, Salzberg SL. Transcript‐level expression analysis of RNA‐seq experiments with HISAT. StringTie and Ballgown. Nat Protoc 2016; 11:1650–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Yu G, Wang L‐G, Han Y, He Q‐Y. clusterProfiler: an R package for comparing biological themes among gene clusters. OMICS 2012; 16:284–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Galindo‐Albarran AO, Lopez‐Portales OH, Gutierrez‐Reyna DY, Rodriguez‐Jorge O, Sanchez‐Villanueva JA, Ramirez‐Pliego O et al CD8+ T cells from human neonates are biased toward an innate immune response. Cell Rep 2016; 17:2151–60. [DOI] [PubMed] [Google Scholar]
- 22. Mi H, Dong Q, Muruganujan A, Gaudet P, Lewis S, Thomas PD. PANTHER version 7: improved phylogenetic trees, orthologs and collaboration with the Gene Ontology Consortium. Nucleic Acids Res 2010; 38:D204–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Supek F, Bošnjak M, Škunca N, Šmuc T. REVIGO summarizes and visualizes long lists of gene ontology terms. PLoS ONE 2011; 6:e21800. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Azzam HS, Grinberg A, Lui K, Shen H, Shores EW, Love PE. CD5 expression is developmentally regulated by T cell receptor (TCR) signals and TCR avidity. J Exp Med (1998) 188:2301–11. Available at: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC2212429/pdf/98-1130.pdf [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Perez‐Villar JJ, Whitney GS, Bowen MA, Hewgill DH, Aruffo AA, Kanner SB. CD5 negatively regulates the T‐cell antigen receptor signal transduction pathway: involvement of SH2‐containing phosphotyrosine phosphatase SHP‐1. Mol Cell Biol 1999; 19:2903–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Tsukamoto H, Clise‐Dwyer K, Huston GE, Duso DK, Buck AL, Johnson LL et al Age‐associated increase in lifespan of naive CD4 T cells contributes to T‐cell homeostasis but facilitates development of functional defects. Proc Natl Acad Sci USA 2009; 106:18333–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Kondoh K, Nishida E. Regulation of MAP kinases by MAP kinase phosphatases. Biochim Biophys Acta ‐ Mol Cell Res 2007; 1773:1227–37. [DOI] [PubMed] [Google Scholar]
- 28. Gerlach C, Rohr JC, Perie L, van Rooij N, van Heijst JWJ, Velds A et al Heterogeneous differentiation patterns of individual CD8+ T cells. Science 2013; 340:635–9. [DOI] [PubMed] [Google Scholar]
- 29. Neefjes J, Jongsma MLM, Paul P, Bakke O. Towards a systems understanding of MHC class I and MHC class II antigen presentation. Nat Rev Immunol 2011; 11:823–36. [DOI] [PubMed] [Google Scholar]
- 30. Thompson JC, Rathmell EA, Farkash W, Gao CB, Rathmell JC, Farkash EA et al Size of naive T cells IL‐7 enhances the survival and maintains the IL‐7 enhances the survival and maintains the size of naive T cells. J Immunol Ref 2001; 167:6869–76. [DOI] [PubMed] [Google Scholar]
- 31. Park J‐H, Yu Q, Erman B, Appelbaum JS, Montoya‐Durango D, Grimes HL et al Suppression of IL7Rα transcription by IL‐7 and other prosurvival cytokines. Immunity 2004; 21:289–302. [DOI] [PubMed] [Google Scholar]
- 32. Park J‐H, Adoro S, Lucas PJ, Sarafova SD, Alag AS, Doan LL et al“Coreceptor tuning”: cytokine signals transcriptionally tailor CD8 coreceptor expression to the self‐specificity of the TCR. Nat Immunol 2007; 8:1049–59. [DOI] [PubMed] [Google Scholar]
- 33. Garbi N, Hämmerling G, Tanaka S. Interaction of ERp57 and tapasin in the generation of MHC class I–peptide complexes. Curr Opin Immunol 2007; 19:99–105. [DOI] [PubMed] [Google Scholar]
- 34. Freitas AA, Rocha B. Peripheral T cell survival. Curr Opin Immunol 1999; 11:152–6. [DOI] [PubMed] [Google Scholar]
- 35. Marrack P, Kappler J. Control of T cell viability. Annu Rev Immunol 2004; 22:765–87. [DOI] [PubMed] [Google Scholar]
- 36. Tarakhovsky A, Kanner SB, Hombach J, Ledbetter JA, Muller W, Killeen N et al A role for CD5 in TCR‐mediated signal transduction and thymocyte selection. Science 1995; 269:535–7. Available at: http://www.ncbi.nlm.nih.gov/pubmed/7542801 [DOI] [PubMed] [Google Scholar]
- 37. Mandl JN, Monteiro JP, Vrisekoop N, Germain RN. T cell‐positive selection uses self‐ligand binding strength to optimize repertoire recognition of foreign antigens. Immunity 2013; 38:263–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Kieper WC, Burghardt JT, Surh CD. A role for TCR affinity in regulating naive T cell homeostasis. J Immunol (2003) 172: Available at: http://www.jimmunol.org/content/172/1/40.short [Accessed August 17, 2017] [DOI] [PubMed] [Google Scholar]
- 39. Smith K, Seddon B, Purbhoo MA, Zamoyska R, Fisher AG, Merkenschlager M. Sensory adaptation in naive peripheral CD4 T cells. J Exp Med (2001) 194: Available at: http://jem.rupress.org/content/194/9/1253 [Accessed May 14, 2017] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Mattoo H, Faulkner M, Kandpal U, Das R, Lewis V, George A et al Naive CD4 T cells from aged mice show enhanced death upon primary activation. Int Immunol 2009; 21:1277–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Tsukamoto H, Huston GE, Dibble J, Duso DK, Swain SL. Bim dictates naive CD4 T cell lifespan and the development of age‐associated functional defects. J Immunol 2010; 185:4535–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Li G, Yu M, Lee W‐W, Tsang M, Krishnan E, Weyand CM et al Decline in miR‐181a expression with age impairs T cell receptor sensitivity by increasing DUSP6 activity. Nat Med 2012; 18:1518–24. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Table S1. Quantitative RT‐PCR analysis: primer details
Figure S1. Induction of CD69 on CD8hi versus CD8lo naive CD8 T cells
Figure S2. Cell‐surface TCR levels on CD8hi versus CD8lo naive CD8 T cells of equal size
Figure S3. Compromised baseline signaling in CD8lo cells
Figure S4. Comparison of mouse CD8hi and CD8lo RNA‐Seq with human neonatal and adult CD8 T‐cell expression array (GSE61570).


