Abstract
DSCAM and DSCAML1 are immunoglobulin and cell adhesion‐type receptors serving important neurodevelopmental functions including control of axon growth, branching, neurite self‐avoidance, and neuronal cell death. The signal transduction mechanisms or effectors of DSCAM receptors, however, remain poorly characterized. We used a human ORFeome library to perform a high‐throughput screen in mammalian cells and identified novel cytoplasmic signaling effector candidates including the Down syndrome kinase Dyrk1a, STAT3, USP21, and SH2D2A. Unexpectedly, we also found that the intracellular domains (ICDs) of DSCAM and DSCAML1 specifically and directly interact with IPO5, a nuclear import protein of the importin beta family, via a conserved nuclear localization signal. The DSCAM ICD is released by γ‐secretase‐dependent cleavage, and both the DSCAM and DSCAML1 ICDs efficiently translocate to the nucleus. Furthermore, RNA sequencing confirms that expression of the DSCAM as well as the DSCAML1 ICDs alone can profoundly alter the expression of genes associated with neuronal differentiation and apoptosis, as well as synapse formation and function. Gain‐of‐function experiments using primary cortical neurons show that increasing the levels of either the DSCAM or the DSCAML1 ICD leads to an impairment of neurite growth. Strikingly, increased expression of either full‐length DSCAM or the DSCAM ICD, but not the DSCAML1 ICD, significantly decreases synapse numbers in primary hippocampal neurons. Taken together, we identified a novel membrane‐to‐nucleus signaling mechanism by which DSCAM receptors can alter the expression of regulators of neuronal differentiation and synapse formation and function. Considering that chromosomal duplications lead to increased DSCAM expression in trisomy 21, our findings may help uncover novel mechanisms contributing to intellectual disability in Down syndrome.
Keywords: DSCAM, nuclear translocation, proteolytic cleavage, synapse formation, transcriptional regulation
Subject Categories: Neuroscience, Signal Transduction
Introduction
The precise wiring of neuronal circuits as well as the establishment of synaptic specificity majorly relies on intracellular signaling pathways downstream of diverse families of cell surface receptors. The Down syndrome cell adhesion molecule (DSCAM) receptors (Yamakawa et al, 1998) are important examples of receptors that utilize homophilic interactions during neurite growth, but can also interact with heterologous ligands (Dascenco et al, 2015). In vertebrates, there are two paralogous DSCAM genes, DSCAM and DSCAML1 (DSCAM‐Like‐1; Yamakawa et al, 1998; Agarwala et al, 2001). Human DSCAM is located in the so‐called Down syndrome critical region (DSCR) on chromosome 21 and belongs thereby to a group of genes present in three copies in Down syndrome (DS) individuals (Yamakawa et al, 1998). Notably, DSCAM levels are increased in brains of DS patients and it has been speculated that this may contribute to the cognitive disabilities observed in DS (Saito et al, 2000; Bahn et al, 2002). DSCAM is also overexpressed in brain of DS mouse models (Amano et al, 2004; Alves‐Sampaio et al, 2010), and in mice that overexpress Amyloid precursor protein (Jia et al, 2011), a causative gene in Alzheimer's disease. Studies using segmental trisomy mouse models of DS showed that triplication of genes of the DSCR including Dscam has a dose‐dependent effect on the segregation of retinal ganglion cell (RGC) axons in the lateral geniculate nucleus (LGN) in the developing retina (Blank et al, 2011). Moreover, dendrite arborization and soma spacing of subtypes of mouse RGCs have been found to be highly sensitive to both increased and decreased Dscam gene dosage (Blank et al, 2011). Overexpression of DSCAM in mice disrupts dendrite targeting and leads to increased neuronal cell death of retinal neurons (Li et al, 2015). Loss of DSCAM or DSCAML1 on the other hand results in increased neuron numbers, self‐avoidance defects, and disorganized layers in the retina (Fuerst et al, 2008, 2009), demonstrating that deregulated DSCAM levels result in altered neuronal wiring. In Drosophila, expression of three Dscam1 copies in vivo alters synaptic function at the neuromuscular junction (Lowe et al, 2018), causes synaptic targeting defects of sensory axons (Cvetkovska et al, 2013), and increased presynaptic arbor enlargement (Kim et al, 2013; Sterne et al, 2015). Thus, deregulated DSCAM levels strongly alter dendritic, axonal, and synaptic development, which may explain how DSCAM gene‐dosage imbalance could potentially contribute to the pathogenesis of neurological disorders.
Drosophila Dscam1 has been studied most extensively because of its extraordinary molecular diversity generated by alternative splicing (Schmucker et al, 2000; Hattori et al, 2008; Kise & Schmucker, 2013; Sun et al, 2013). Drosophila Dscam1 is essential for key aspects of neuronal wiring including axonal growth, guidance, targeting, and branching (Schmucker et al, 2000; Hummel et al, 2003; Chen et al, 2006; He et al, 2014; Dascenco et al, 2015), dendritic field organization (Hughes et al, 2007; Matthews et al, 2007; Soba et al, 2007), and synaptic connectivity (Millard et al, 2010). Although vertebrate DSCAMs lack extensive alternative splicing, many functions important for neuronal wiring are highly conserved from flies to mammals. Homophilic self‐recognition of DSCAMs is required in flies and mice for neurite repulsion and self‐avoidance of sister‐neurites in vivo (Hughes et al, 2007; Fuerst et al, 2008, 2010; Hattori et al, 2008; Yamagata & Sanes, 2008; Simmons et al, 2017).
Despite the conserved roles of DSCAMs in shaping neuronal circuitry, no coherent downstream signaling pathway has been identified. Vertebrate DSCAM has been implicated in the regulation of actin cytoskeleton dynamics (Ly et al, 2008; Liu et al, 2009; Purohit et al, 2012), yet in vivo evidence remains sparse. In Drosophila Dscam1 is thought to affect actin cytoskeleton dynamics through the SH2/SH3 adaptor protein Dock/Nck acting upstream of P21 activated kinase (Pak1) and Rho GTPases (Manser et al, 1994; Hall, 1998; Hing et al, 1999; Schmucker et al, 2000). However, loss of Dock or Pak1 does not lead to phenotypic defects such as dendrite self‐crossing (Hughes et al, 2007). Dscam1 also physically and genetically interacts with tubulin folding co‐factor D (TBCD), which is required for the formation of alpha‐ and beta‐tubulin heterodimers in flies. This interaction is required in vivo for glomerular targeting of olfactory neurons (Okumura et al, 2015).
Here we identify several novel cytoplasmic signaling effectors of DSCAM and DSCAML1 (DSCAM/L1). We report that Importin 5 (IPO5)‐mediated membrane‐to‐nucleus translocation of DSCAM/L1 may provide a novel signaling mode of this important receptor class. We show that the ICD of mammalian DSCAM is liberated by γ‐secretase‐mediated cleavage and that both the DSCAM and DSCAML1 ICD efficiently translocate to the nucleus. Considering that increased DSCAM levels have been proposed to contribute to intellectual disability in Down syndrome patients, we tested in gain‐of‐function experiments how increased DSCAM levels, and in particular nuclear import, might affect neurite growth and synapse formation. We show that nuclear enrichment of the DSCAM and DSCAML1 ICD in cell lines profoundly alters the transcription of genes associated with neuronal differentiation and function. Increased nuclear levels of either the DSCAM or DSCAML1 ICD strongly impair neurite outgrowth in primary mouse cortical cultures. Interestingly, only increased expression of either full‐length DSCAM or the DSCAM ICD, but not of the DSCAML1 ICD leads to a strong decrease in synapse numbers in primary mouse hippocampal neurons. Thus, these studies uncover how increased DSCAM membrane‐to‐nucleus signaling is capable of altering synaptic connectivity and suggest a novel scenario how gain of DSCAM function may contribute to neurological pathologies.
Results
Identifying human signaling effectors of DSCAM and DSCAML1
In order to identify novel cytoplasmic binding partners of vertebrate DSCAMs, we employed the mammalian protein‐protein interaction trap (MAPPIT) to monitor protein–protein interactions in mammalian cells (Fig 1A; Eyckerman et al, 2001; Lievens et al, 2009). We generated MAPPIT receptors with the intracellular domains (ICDs) of mouse DSCAM or DSCAML1 serving as baits (Fig 1A) and performed high‐throughput MAPPIT screens in which the DSCAM/L1 baits were screened against a library of about 10,000 preys (Lievens et al, 2009, 2011). From these screens, we further selected and validated IPO5, DYRK1A, DYRK1B, SH2D2A, STAT3, and USP21 as binding partners of both DSCAM and DSCAML1 in binary MAPPIT experiments including control baits and preys (Table 1; Fig 1B and G; Appendix Fig S1A and B). We also tested the interaction of the DSCAM/L1 bait receptors with preys of known binding partners including PAK1 (DSCAM) and MAGI‐1 (DSCAM and DSCAML1; Yamagata & Sanes, 2010; Purohit et al, 2012). The DSCAM/L1 baits gave rise to MAPPIT signals with MAGI‐1 and the DSCAM bait also interacted with the PAK1 prey, but these interactions caused weaker luciferase induction compared to newly identified candidates (Appendix Fig S1C and D). Recruitment of kinases and SH2‐domain adaptor proteins were expected based on previous studies on Drosophila Dscam1 (Schmucker et al, 2000); however, neither DYRK1A (also referred to as “Down syndrome kinase”) nor SH2D2A have been previously implicated in DSCAM signaling.
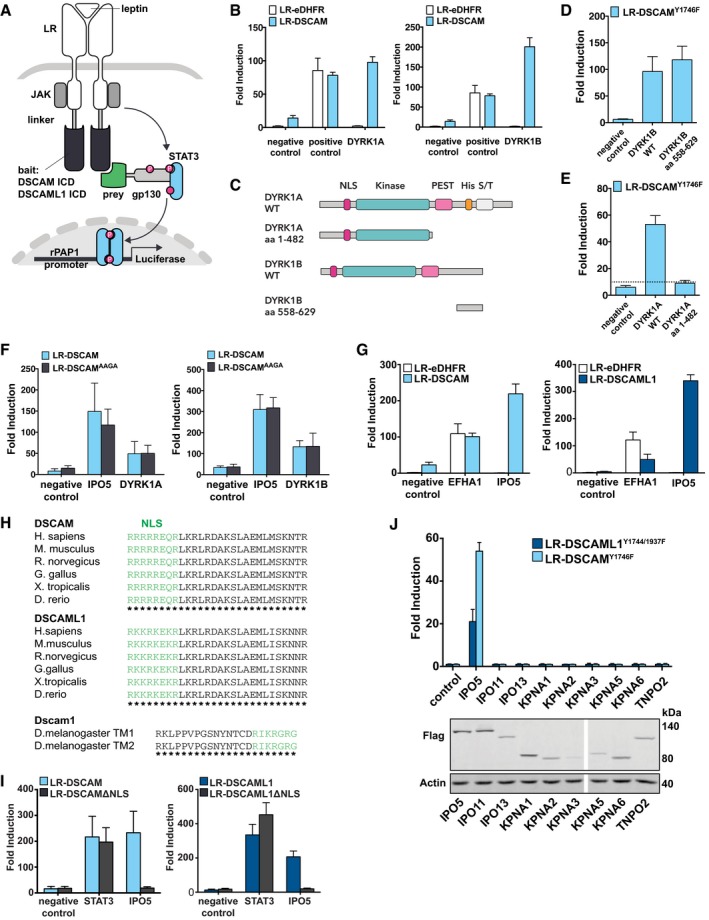
Figure 1. DSCAM and DSCAML1 interact with DYRK1A, DYRK1B, and IPO5.
-
ASchematic of the MAPPIT technique. Bait receptors consist of the mouse DSCAM or DSCAML1 ICDs fused to the C‐terminus of a signaling‐deficient leptin receptor (LR) fragment, in which the tyrosines of STAT3 recruitment motifs were mutated to phenylalanine. The preys are tethered to a gp130 cytokine receptor fragment containing functional STAT3 recruitment sites. Upon leptin stimulation, association of bait and prey restores a functional leptin receptor complex resulting in STAT3 activation, which can be monitored by a STAT3‐responsive luciferase reporter. P, phospho‐tyrosine. JAK, janus kinase.
-
BDSCAM interacts with DYRK1A and DYRK1B. LR‐DSCAM and LR‐eDHFR (negative control bait) baits were introduced in HEK cells together with a DYRK1A prey, DYRK1B prey, empty prey (negative control only consisting of the gp130 fragment), or an EFHA1 prey (positive control) and binding was tested in MAPPIT. Results for DSCAML1, see Appendix Fig S1B.
-
CSchematic of full‐length (FL) DYRK1A, DYRK1B, and truncated DYRK1A/B preys. DYRK1A contains a nuclear localization signal, a protein kinase domain, a leucine zipper motif, and a highly conserved poly‐histidine repeat; DYRK1B lacks the poly‐histidine repeat.
-
D, EThe interactions between truncated DYRK1B (aa 558–629) (D) or DYRK1A (aa 1–482) (E) preys and the DSCAM (LR‐DSCAMY1746F) bait (i.e., suppressing STAT3‐mediated background) were assayed using MAPPIT. Results for DSCAML1, see Appendix Fig S3A and B. The C‐terminus of DYRK1B is sufficient for the interaction with DSCAM, and the kinase domain is not required (D). The DYRK1A kinase domain is not sufficient to bind to DSCAM (E).
-
FMutation of the potential DYRK kinase substrate site does not inhibit the interaction between DSCAM and DYRK1A or DYRK1B. A potential substrate site for DYRKs was mutated in the DSCAM bait (RPGT to AAGA). The interactions of mutant or wild‐type DSCAM baits with DYRK1A, DYRK1B, and IPO5 (positive control prey) were tested in a binary MAPPIT experiments. Results for DSCAML1, see Appendix Fig S3C and D.
-
GDSCAM and DSCAML1 robustly interact with IPO5. LR‐DSCAM, LR‐DSCAML1, and LR‐eDHFR (negative control bait) baits were introduced in HEK cells together with an IPO5 prey, an empty prey (negative control), or an EFHA1 prey (positive control) and binding was tested in MAPPIT.
-
HDSCAMs exhibit a conserved NLS. Green, putative monopartite NLS motif.
-
ILoss of the NLSs of DSCAM/L1 inhibits binding of IPO5. Binding of IPO5 to wt or mutant DSCAM/L1 baits lacking the NLS (LR‐DSCAMΔNLS, LR‐DSCAML1ΔNLS) was tested in MAPPIT. Negative control, empty prey. Positive control, STAT3 prey.
-
JThe interaction between DSCAM/L1 and IPO5 is specific. The interaction between 9 importin preys and the DSCAMY1746F or the DSCAML1Y1744/1937F bait (i.e., suppressing STAT3‐mediated background signal) was evaluated in a binary MAPPIT experiment, and expression of the importin preys was confirmed by Western blot (lower panel).
Table 1.
Binding partners of DSCAM/L1 identified with MAPPIT. Further validation of the candidates can be found in Fig EV1 and Appendix Figs S1–S3
| Symbol | Name | Function | Interaction with DSCAM | Interaction with DSCAML1 |
|---|---|---|---|---|
| IPO5 | Importin 5 | Nuclear protein import. Binds to cargo NLS | Binds to NLS of DSCAM: RRRRREQR (Fig 1I) | Binds to NLS of DSCAML1: RKKRKEKR (Fig 1I) |
| STAT3 | Signal transducer and activator of transcription 3 | Transcriptional activator in response to cytokines and growth factors | Binds to YASQ motif of DSCAM (Appendix Fig S2B) | Binds to YSSQ/YHTQ motifs of DSCAML1 (Appendix Fig S2E) |
| SH2D2A | SH2 domain containing 2A | SH2 domain adaptor protein for VEGF receptor KDR. Functions in T‐cell signal transduction | N/A (Appendix Fig S2J) | Binds to YCNL motif of DSCAML1 (Appendix Fig S2I) |
| DYRK1A | Dual‐Specificity‐Tyrosine‐Phosphorylation Regulated Kinase 1A | S/T/Y‐kinase. Plays important roles in neuronal development. Down Syndrome candidate gene | Interaction through C‐terminus of DYRK1A (Fig 1E) | Interaction through C‐terminus of DYRK1A (Appendix Fig S3B) |
| DYRK1B | Dual‐Specificity‐Tyrosine‐Phosphorylation Regulated Kinase 1B | S/T/Y‐kinase. Regulates transcription in the nucleus | Interaction through C‐terminus of DYRK1B (Fig 1D) | Interaction through C‐terminus of DYRK1B (Appendix Fig S3A) |
| USP21 | Ubiquitin specific peptidase 21 | Cysteine protease with dual specificity to cleave Ubiquitin and Nedd8 | Binds to YASQ motif of DSCAM (Appendix Fig S2C) | Binds to YSSQ/YHTQ motifs of DSCAML1 (Appendix Fig S2F) |
We further validated the MAPPIT screen candidates by co‐immunoprecipitation from cell lines co‐transfected with HA‐tagged DSCAM/L1 and their novel interaction partners (Fig EV1). Additionally, we identified potential binding motifs for STAT3, SH2D2A, and USP21 (Table 1; Appendix Fig S2A–J). Mutation of those motifs in the bait receptors confirmed that specific tyrosine residues within the DSCAM and DSCAML1 ICDs function as binding sites for STAT3, USP21, and SH2D2A (Table 1; Appendix Fig S2A–J). Furthermore, we analyzed the interactions between DSCAM/L1 and DYRK1A/B in more detail. We found that a truncated DYRK1B prey, which consists of the most C‐terminal 72 amino acids and lacks the kinase domain (Fig 1C), was sufficient to interact with DSCAM and DSCAML1 baits in MAPPIT (Fig 1D; Appendix Fig S3A). Moreover, a truncated DYRK1A prey, consisting of its N‐terminal and its kinase domains but lacking its C‐terminal domains (Fig 1C), failed to bind to the DSCAM or DSCAML1 baits in contrast to full‐length (FL) DYRK1A (Fig 1E; Appendix Fig S3B). Taken together, this suggests that not the kinase domains, but the C‐terminal domains of DYRK1A and B mediate the interaction with DSCAM/L1. Both DSCAM and DSCAML1 ICDs exhibit a potential DYRK substrate motif (RPGTNP). A triple alanine substitution of the RPx(T/S/x)P motif previously shown to abolish the interaction of DYRK1A with a substrate peptide (Soundararajan et al, 2013) was generated (RPGTNP to AAGANP) for both DSCAM and DSCAML1. However, the AAGANP mutant baits could still interact with DYRKs (Fig 1F, Appendix Fig S3C and D), further suggesting that binding of DYRKs to DSCAM/L1 might be independent of their kinase domains, as previously suggested for other DYRK1A interactors (Aranda et al, 2008). In conclusion, the MAPPIT approach leads to the identification of IPO5, STAT3, DYRK1A/B, SH2D2A, and USP21 as novel potential downstream effectors of DSCAM/L1.
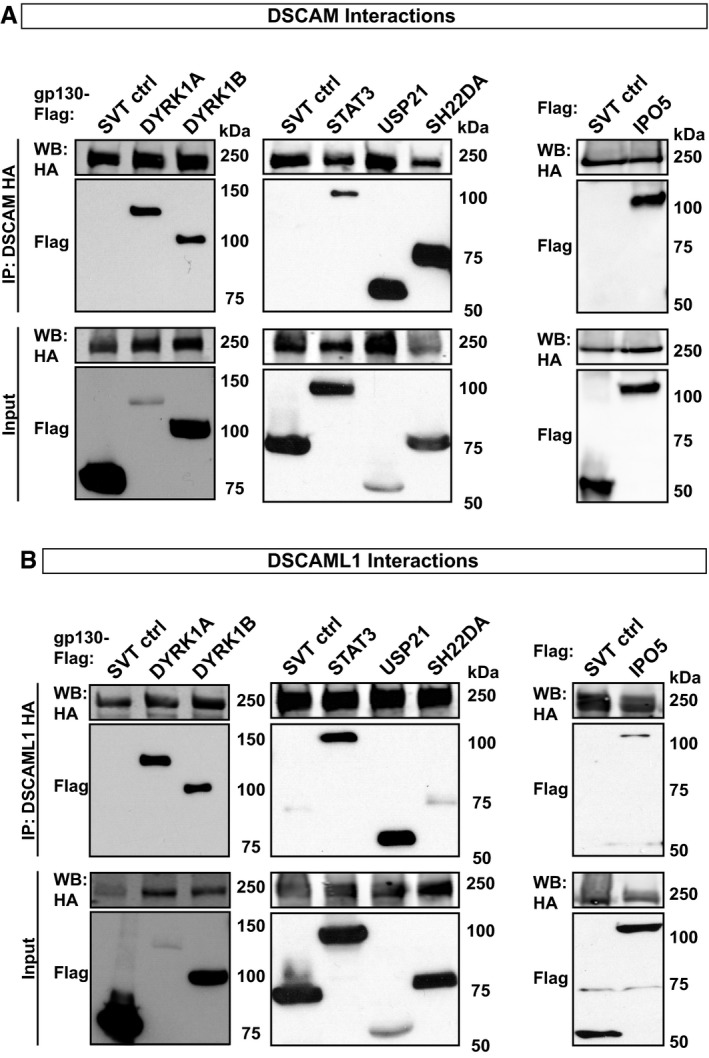
Figure EV1. Validation of MAPPIT Interactions by co‐immunoprecipitation (related to Fig 1).
-
A, BHEK293T cells or SH‐SY5Y cells were co‐transfected with either DSCAM‐HA or DSCAML1‐HA together with gp130‐Flag‐tagged SVT (SV40 large T antigen, unrelated negative control), DYRK1A, DYRK1B, STAT3, USP21, SH2D2A, or Flag‐tagged IPO5. HA‐tagged DSCAM/L1 proteins were precipitated using anti‐HA magnetic beads and co‐precipitated protein complexes were analyzed by Western blot using HA‐ and Flag‐tag specific antibodies. (A) DSCAM‐HA co‐precipitates with gp130‐Flag‐tagged DYRK1A, DYRK1B, STAT3, SH2D2A and USP21 in HEK293T cells or Flag‐tagged IPO5 in SH‐SY5Y cells. (B) DSCAML1‐HA co‐precipitates with gp130‐Flag‐tagged DYRK1A, DYRK1B, STAT3, USP21, SH2D2A, or Flag‐tagged IPO5 in HEK293T cells.
Source data are available online for this figure.
DSCAM and DSCAML1 interact with IPO5 via a nuclear localization signal
Since binary MAPPIT assays confirmed the interactions of both DSCAM and DSCAML1 with Importin 5 (IPO5; Fig 1G), we reasoned that DSCAMs might have nuclear functions. Such membrane‐to‐nucleus signaling mechanism would be similar to Neogenin and DCC, two neuronal CAMs that are closely related to vertebrate DSCAMs and cleaved by proteases resulting in the release of C‐terminal ICD fragments, which then translocate into the nucleus where they regulate gene transcription (Taniguchi et al, 2003; Goldschneider et al, 2008). IPO5 is an importin beta mediating nuclear protein import (Yaseen & Blobel, 1997) and can directly interact with the nuclear localization signal (NLS) of its cargo (Chao et al, 2012). We therefore considered that the ICDs of DSCAMs might contain an NLS serving as docking site for IPO5. Indeed, upon sequence analysis we identified a potential monopartite NLS within the membrane‐proximal regions enriched in conserved Arginine and Lysine residues (Fig 1H). Interestingly, Drosophila Dscam1 also exhibits a predicted NLS within the cytoplasmic portion that is common to all its isoforms, indicating that a membrane‐proximal NLS is highly conserved from insects to vertebrates (Fig 1H). To test its functional relevance, we generated NLS‐deficient MAPPIT bait versions (i.e., LR‐DSCAMΔNLS and LR‐DSCAML1ΔNLS), which failed to interact with the IPO5 prey but could still interact with the STAT3 control prey (Fig 1I), demonstrating that the NLSs are required for the interaction between IPO5 and DSCAM/L1. To determine specificity, we tested all full‐length importin alpha and beta preys present in the human ORFeome collection 5.1 and 8.1. Out of the 9 importins tested, only IPO5 interacted with DSCAM and DSCAML1 (Fig 1J). Together, these results show that IPO5 can bind to the membrane‐proximal NLS motifs of DSCAM and DSCAML1 with high specificity.
DSCAM is cleaved by γ‐secretase
Several neuronal transmembrane proteins including APP, DCC, Neogenin, and Notch undergo ectodomain cleavage directly followed by γ‐secretase‐mediated intra‐membrane cleavage leading to the release of their ICDs (De Strooper et al, 1999; Cupers et al, 2001; Taniguchi et al, 2003; Goldschneider et al, 2008). Shedding of the DSCAM ectodomain has been reported in flies and mice (Watson et al, 2005; Schramm et al, 2012); however, it is currently unknown whether the ICD of DSCAMs could be released by proteolytic cleavage. To investigate this, we fused HA‐epitopes to the very C‐terminus of the DSCAM/L1 ICDs as well as to FL DSCAM/L1 (Fig 2A) and expressed these fusion proteins in HEK293T cells. Immunoprecipitation with an HA‐specific antibody and Western blot (WB) analysis showed FL DSCAM migrating at approximately 250 kDA as well as a C‐terminal fragment at 55 kDa, which co‐migrates with the DSCAM ICD construct at approximately the same molecular weight (Fig 2B). Likewise, FL DSCAML1 migrated at nearly 250 kDa and we detected two additional C‐terminal fragments migrating at 65 kDa and at 60 kDa, co‐migrating with two fragments originating from the DSCAML1 ICD HA‐fusion (Fig 2B). This suggests that DSCAM and DSCAML1 are substrates for a protease and cleavage generates C‐terminal ICD fragments.
Figure 2. DSCAM family IgCAMs are cleaved releasing their ICDs.
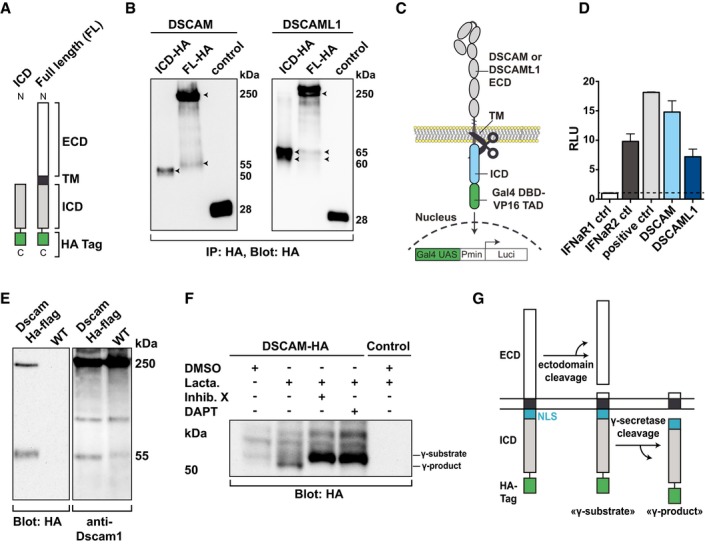
- Schematic of C‐terminally HA‐tagged DSCAM and DSCAML1 constructs. ECD, extracellular domain. TM, transmembrane domain. ICD, intracellular domain. HA, human influenza hemagglutinin epitope.
- Immunoblot showing C‐terminal ICD fragments resulting from cleavage of DSCAM and DSCAML1. HA‐tagged constructs shown in (A) were expressed in HEK293 cells, immunoprecipitated, and subsequently immunoblotted. Control, HA‐tagged YFP. Arrowheads indicate full‐length DSCAM/L1 and cleavage products.
- Schematic of the cleavage reporter assay. DSCAM or DSCAML1 was fused to the DNA‐binding domain of Gal4 (Gal4DBD) and the transcriptional activation domain of VP16 (VP16TAD). Intra‐membrane cleavage results in nuclear translocation of cleaved fragments and activation of a Luciferase reporter in the nucleus.
- Cleavage reporter assay confirming cleavage of DSCAM/L1. HEK293 cells were transfected with DSCAM‐, DSCAML1‐, IFNaR1‐, or IFNaR2‐Gal4DBD‐VP16 or Gal4DBD‐VP16 alone and co‐transfected with Gal50hIL6luc and pRL‐TK and luciferase activity was quantified after 24 h. Results are expressed as relative luciferase units (RLU), which are calculated as the ratio of firefly (Gal50hIL6luc) and Renilla (pRL‐TK) luciferase activities.
- Immunoblot showing Drosophila Dscam 1 cleavage in vivo. Brain lysates from wt and from transgenic Dscam1‐HA‐Flag tagged flies (i.e., endogenous tag within the ICD of the Dscam1 gene) were immunoblotted and probed with HA‐specific and Dscam1‐ICD‐specific antibodies.
- DSCAM is cleaved by γ‐secretase. HA‐immunoblot from lysates of DSCAM‐HA expressing stable HEK293 cells treated over night with DMSO, or Lactacystin (10 μM) in the presence or absence of the γ‐secretase inhibitors DAPT (10 μM) or inhibitor X (10 μM).
- Model of secretase‐mediated DSCAM cleavage according to our results and literature on related secretase‐cleaved receptors.
We further employed a cleavage luciferase reporter assay in which DSCAM and DSCAML1 were C‐terminally fused to a Gal4DBD–VP16 transactivation domain (TAD; Karlstrom et al, 2002; May et al, 2002). Cleavage of these constructs releases the Gal4DBD–VP16 fusion protein fragment, which can transcriptionally activate a co‐transfected luciferase gene (Fig 2C). As compared to control cells expressing a non‐cleavable IFNaR1‐Gal4DBD‐VP16 receptor, cells expressing the DSCAM or DSCAML1 fusion proteins showed a strong increase in luciferase activity similar to cells expressing a constitutively cleaved (IFNaR2‐Gal4bd‐VP16) or a constitutively active control (Gal4bd‐VP16‐empty; Fig 2D). These data confirm that the ICDs of DSCAM and DSCAML1 can be released upon proteolytic cleavage. The apparent MW of the fragment detected by WB suggests that likely the entire ICD is shuttled to the nucleus.
We also investigated a potential in vivo cleavage of Drosophila Dscam1. In brain lysates of wild type (wt) as well as flies harboring a BAC‐based genomic transgene where the full‐length Dscam1 gene is HA‐tagged in exon 22 (i.e., cytoplasmic domain; see Appendix Supplementary Methods), we detected FL Dscam1 at 250 kDa and an additional band at 55 kDa, while these bands were absent in control lysates from wt flies (Fig 2E). Using a previously validated antibody directed against the ICD of fly Dscam1 ICD (Watson et al, 2005), we also detected FL Dscam1 as well as a 55 kDa fragment in WT samples (Fig 2E), suggesting that endogenous Dscam1 is cleaved as well. This suggests that Drosophila Dscam1 is processed by proteolysis in vivo and that the cleavage of DSCAM family IgCAMs is conserved from vertebrates to flies.
To determine whether vertebrate DSCAM is a substrate of γ‐secretase, we treated HEK cells stably expressing C‐terminally HA‐tagged DSCAM with or without the proteasome inhibitor Lactacystin in the presence or absence of two different γ‐secretase inhibitors (DAPT and Inhibitor X). Lactacystin was added to stabilize intracellular fragments generated by γ‐secretase as these are rapidly degraded by the proteasome (Cupers et al, 2001). WB analysis of lysates in the presence of Lactacystin and in the absence of Inhibitor X or DAPT revealed a stabilized fragment of about 50 kDa (Fig 2F). This fragment was absent in the presence of either Inhibitor X or DAPT, and instead, a prominent higher‐molecular‐mass fragment of 55 kDa was detected (Fig 2F). Taken together, this suggests that DSCAM is a novel γ‐secretase substrate and its cleavage releases an apparent 50 kDa “γ‐product” (Fig 2G).
The ICDs of DSCAM and DSCAML1 translocate into the nucleus
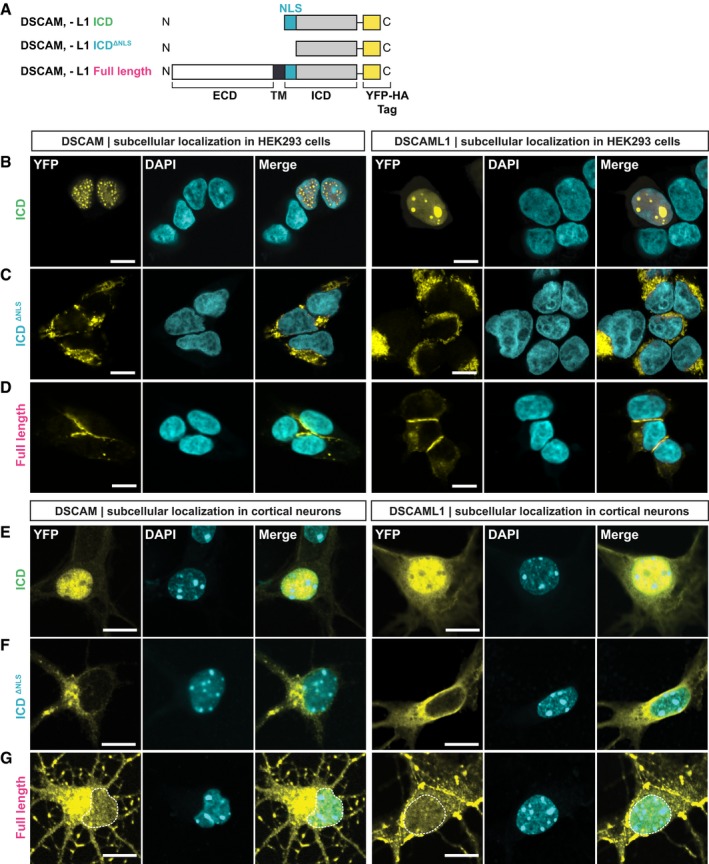
The predicted NLS motif in DSCAM as well as DSCAML1 is highly conserved in vertebrates where an Arginine‐ or Lysine‐rich eight amino acid peptide in DSCAM or DSCAML1, respectively, is followed by an invariant 20 amino acid sequence (Fig 1H). Moreover, DSCAM or DSCAML1 interactions with the nuclear import factor IPO5 are eliminated when the putative NLS motifs are missing in MAPPIT baits (Fig 1I). To further validate that these conserved NLS motifs are indeed necessary for nuclear import of the DSCAM/L1 ICDs, we added C‐terminal YFP‐tags to wt and NLS‐deficient protein forms (Fig 3A). We expressed these YFP‐tagged constructs in HEK293T cells and analyzed their subcellular localization. Both wt DSCAM and DSCAML1 ICD were predominantly located in the nucleus (Fig 3B), whereas in cells expressing the NLS‐deficient ICDs YFP fluorescence was absent from the nucleus but present throughout the cytoplasm or in perinuclear compartments (Fig 3C). On the other hand, FL YFP‐tagged DSCAM and DSCAML1 were enriched at the membrane between cell‐cell contacts and also present in the cytoplasm, yet undetectable in the nucleus even upon Lactacystin treatment (Fig 3D). This suggests that in HEK cells, where DSCAM and DSCAML1 are not endogenously expressed, cleavage of full‐length proteins is scarce, although it can be detected in cell lysates (Fig 2F). This however is different in developing cortical neurons (see below), which endogenously express both DSCAM and DSCAML1 (Barlow et al, 2002; Cui et al, 2013). We nucleofected E14.5 cortical neurons and analyzed the subcellular localization of the same YFP‐tagged proteins (Fig 3A) after 4 days in vitro (DIV4). The YFP‐tagged ICDs, similarly to our findings in HEK cells, were enriched in the nucleus of cortical neurons (Fig 3E), whereas the NLS‐deficient ICDs were predominantly localized in the cytoplasm of cell bodies and neurites (Fig 3F). In line with their function as neuronal CAMs, FL DSCAM/L1 YFP‐fusions were localized at the neuronal membrane and enriched at neurite‐neurite contacts (Fig 3G). Strikingly, in neurons expressing YFP‐tagged FL DSCAM/L1 we also detected substantial YFP‐signal in the nucleus upon Lactacystin treatment (Fig 3G) most likely resulting from cleavage of DSCAM receptors. Hence, there is a striking difference between HEK cells and primary neurons, suggesting that cleavage or nuclear import of DSCAM is facilitated in neurons. We also investigated the potential effect of γ‐secretase inhibition on DSCAM nuclear levels in primary mouse neurons and human neuroblastoma SH‐SY5Y cells transfected with FL DSCAM‐YFP. In both SH‐SY5Y cells (Fig EV2A and B) and primary neurons (Fig EV2C and D), simultaneous treatment with Lactacystin and inhibitor X strongly and significantly diminished nuclear YFP levels as compared to Lactacystin treatment alone (Fig EV2), showing that γ‐secretase cleavage products of DSCAM enrich in the nucleus. Taken together, our results show that in mammalian neurons, overexpression of DSCAM/L1 leads to significant proteolytic cleavage of FL receptors and that a conserved membrane‐proximal NLS motif is essential for translocation of the entire ICD to the nucleus.
Figure 3. The NLSs of DSCAM and DSCAML1 are required for nuclear translocation of their ICDs.
-
ASchematic of YFP‐tagged DSCAM/L1 constructs expressed in (B–G). A C‐terminal YFP‐HA tag was fused to the ICDs of DSCAM/L1, to ICD constructs lacking the NLS (ICDΔNLS), and to full‐length DSCAM/L1.
-
B–DYFP‐HA‐tagged DSCAM/L1 constructs (yellow) shown in (A) were expressed in HEK293 cells and immuno‐stained for YFP. DAPI was used to visualize the nuclei (cyan). The ICDs of DSCAM and DSCAML1 localize to the nucleus of HEK293 cells (B). Deletion of the NLSs (DSCAM/L1 ICDΔNLS) impairs nuclear localization of the DSCAM and DSCAML1 ICDs (C). Full‐length DSCAM and DSCAML1 are not detected in the nucleus of HEK293 cells treated with 10 μM Lactacystin (D).
-
E–GYFP‐tagged DSCAM/L1 constructs shown in (A) were expressed in primary mouse cortical neurons at E14.5 until DIV4. Neurons were treated with 10 μM Lactacystin and then immuno‐stained for YFP (yellow) and treated with DAPI (cyan).The ICDs of DSCAM/L1 enrich in the nucleus of primary neurons (E). DSCAM/L1 ICDΔNLS constructs (i.e., lacking the NLS) are localized in the cytoplasm (F). Full‐length DSCAM and DSCAML1 can be detected in the nucleus (G).
Figure EV2. γ‐Secretase regulates nuclear enrichment of the DSCAM ICD (related to Fig 3).
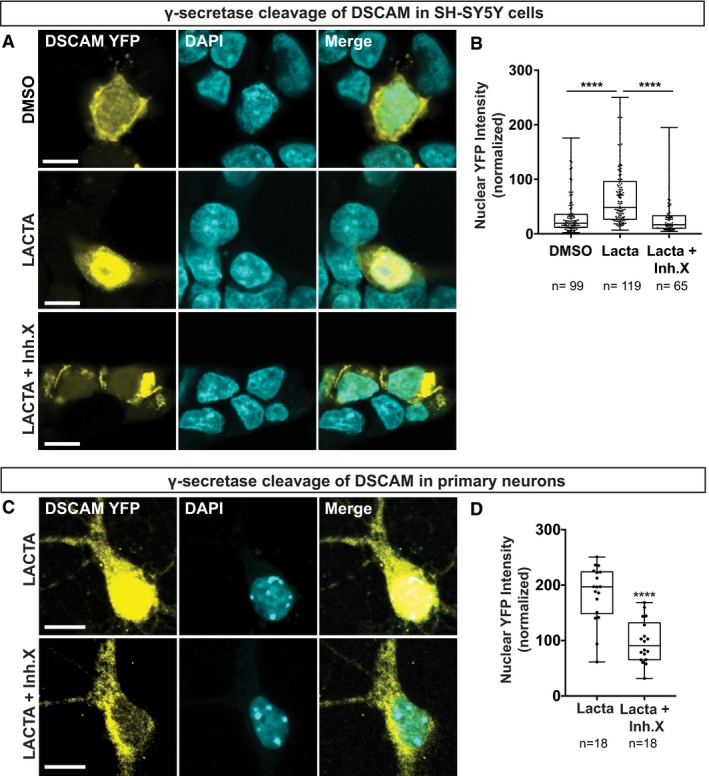
-
A, B24 h post‐transfection SH‐SY5Y cells were incubated over night with DMSO (control) or Lactacystin (10 μM) in the presence or absence of γ‐secretase inhibitor X (10 μM). (A) Cells were immuno‐stained for YFP and treated with DAPI. (B) Quantification of DSCAM nuclear localization, which is expressed as nuclear YFP intensity normalized to the nuclear area.
-
C, DPrimary mouse hippocampal neurons were isolated at E18 and nucleofected at DIV0. At DIV9, neurons were incubated over night with Lactacystin (5 μM) in the presence or absence of inhibitor X (10 μM) and immuno‐stained on DIV10 for YFP and DAPI (C). (D) Quantification of DSCAM nuclear localization, which is expressed as nuclear YFP intensity normalized to the nuclear area.
Nuclear localization of the DSCAM/L1 ICDs alters the expression of genes involved in neuronal wiring
Cleavage and nuclear import has been described for a number of membrane receptors, yet this mechanism has previously not been implicated in DSCAM signaling. We therefore investigated the possibility that nuclear translocation of the DSCAM/L1 ICDs could alter gene transcription. To test this, we generated inducible isogenic HEK293 t‐Rex‐Flp‐In cell lines stably expressing the YFP‐tagged DSCAM and DSCAML1 ICDs as well as cytoplasmic and nuclear YFP controls and subsequently performed RNA deep sequencing (RNA‐seq). We then determined the global transcriptome changes of cells expressing the YFP‐tagged DSCAM/L1 ICDs versus cells expressing a nuclear YFP as control (Fig 4A). We excluded from our analysis genes that were already differentially expressed in cytoplasmic versus nuclear YFP expressing cells (P‐value ≤ 0.1), as these genes are unlikely affected by DSCAM ICD expression (Fig 4A). Notably, choosing this stringent cutoff may have led to the elimination of some positive candidates of differentially expressed genes (DEGs), which may, nevertheless, be relevant candidates rather than false positives. Statistical analysis (P‐value ≤ 0.0005; log2 fold change ≥ 0.58, ≤ −0.58) revealed a total of 1,028 DEGs upon DSCAM ICD expression (i.e., DSCAM data set; Figs 4B and EV3A) and 860 DEGs upon DSCAML1 ICD expression (i.e., DSCAML1 data set; Figs 4B and EV3B), with 552 DEGs that are in common to the two data sets (Fig 4C). Functional categorization of those DEGs using Ingenuity Pathway Analysis (IPA) identified “nervous system development and function” as a statistically highly enriched biological function (P < 0.005) and “axonal guidance signaling” as the most enriched canonical pathway (P = 2.45E‐05) in both data sets (Fig EV3C and D; Appendix Tables S1 and S2) encompassing about 240 DEGs for DSCAM and 190 DEGs for DSCAML1 altogether. To illustrate the remarkable enrichment for neuronal differentiation genes, we narrowed the DSCAM and DSCAML1 data sets down to about 90 candidate genes based on their well‐known functions in axon guidance, neurite repulsion, branching, synaptogenesis, and neuronal cell death (Fig 4D and E). For simplicity, they were further grouped based on subcellular localization (membrane, cytoplasmic, extracellular, and nuclear; Fig 4D and E).
Figure 4. The ICDs of DSCAM and DSCAML1 alter the expression of genes involved in neuronal circuit formation.
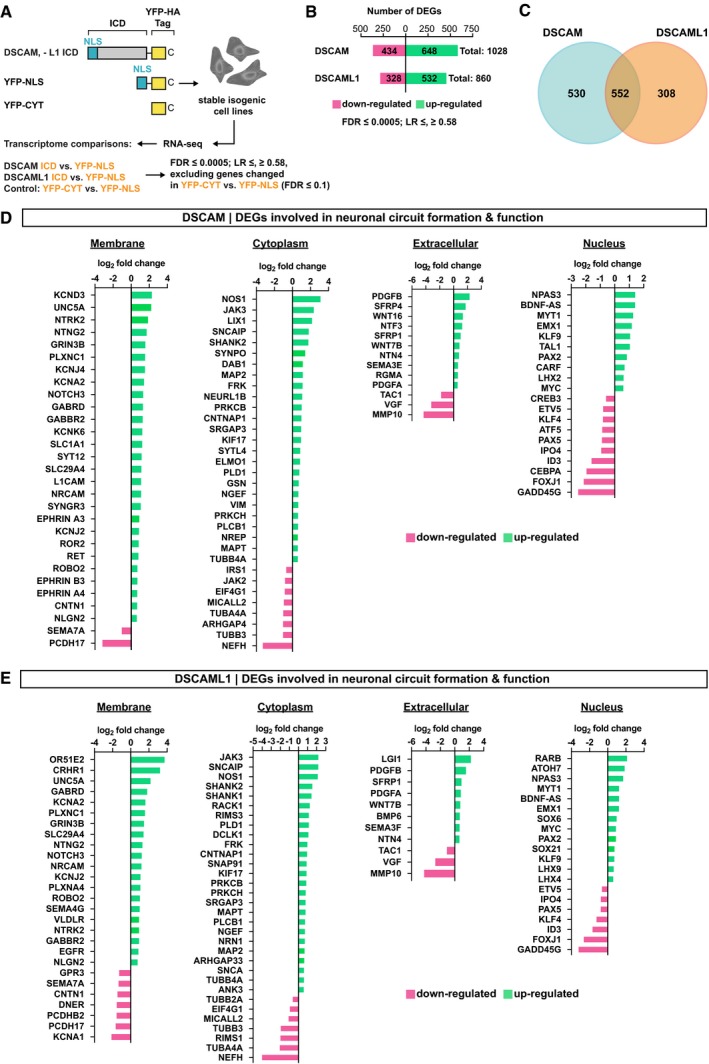
-
AOutline of RNA‐seq experiment. FDR, false discovery rate (corrected P‐value). LR, log ratio (log2 fold change).
-
B–EResults of Ingenuity Pathway Core Analysis (FDR ≤ 0.0005; LR ≥ 0.58, ≤ −0.58) comparing the transcriptomes of stable cell lines expressing the DSCAM ICD versus YFP‐NLS, or DSCAML1 ICD versus YFP‐NLS. Genes that changed expression in nuclear versus cytoplasmic YFP controls (YFP‐NLS versus YFP‐CYT) were excluded from the analysis (FDR ≤ 0.1). DEGs, differentially expressed genes. (B) Bar graph showing the number of up‐ and down‐regulated genes in the DSCAM/L1 data sets. (C) Venn diagram showing the number of overlapping and individual DEGs between the DSCAM and DSCAML1 data sets. (D, E) Selection of DEGs involved in neuronal circuit formation and function in DSCAM ICD (D) or DSCAML1 ICD (E) expressing cells relative to YFP‐NLS‐expressing cells. For simplicity, DEGs were grouped according to their subcellular localization. Up‐ and down‐regulation is expressed as log ratio (i.e., log2 fold change). Positive log ratio, up‐regulation (green). Negative log ratio, down‐regulation (magenta). LR ≥ 0.58, ≤ −0.58 corresponds to an approximate gene expression fold change ≥ 1.5, ≤ −1.5.
Figure EV3. Enriched Biological Functions in DSCAM and DSCAML1 RNA‐seq datasets (related to Fig 4).

-
A, BVolcano plots showing all profiled up‐ and down‐regulated genes as log2 fold change versus −log10 of the false discovery rate (FDR) corrected P‐value in the DSCAM (A) and DSCAML1 (B) data sets. Statistically relevant DEGs are separated from statistically non‐relevant DEGs by a horizontal striped line by FDR ≤ 0.0005 cutoff. Vertical striped lines indicate the log ratio (LR) cutoff (LR ≥ 0.58, ≤ −0.58; fold change ≥ 1.5, ≤ 1.5). For simplicity, genes differentially expressed in nuclear versus cytoplasmic YFP control cell lines (FDR ≥ 0.1) are not shown. (A) Volcano plot showing all profiled genes in DSCAM ICD versus nuclear YFP expressing cell lines. Note that DSCAM is highly up‐regulated (log2 fold change, 6.0345; −log10 of P‐value, 124.678) due to experimental overexpression of the DSCAM ICD. (B) Volcano plot showing all profiled genes in DSCAML1 ICD versus nuclear YFP expressing cell lines. DSCAML1 is highly up‐regulated (log2 fold change, 7.553) due to experimental overexpression of the DSCAML1 ICD. The arrow indicates that due to a FDR P‐value of 0, this data point cannot be plotted on a −log10 scale.
-
C, DResults of Ingenuity Pathway Core Analysis (FDR ≤ 0.0005; LR ≥ 0.58, ≤ −0.58). Enriched biological functions related to physiological system development and function and canonical pathways. X‐axis shows likelihood of association between a set of genes in our dataset and a biological function, expressed as −log (P‐value). Enriched biological functions and pathways were scored according to their P‐value, calculated with the Fischer's exact test. The y‐axis shows the annotations of the enriched biological functions or canonical pathways of differentially expressed genes. (C) Functional classification of DEGs in DSCAM ICD relative to YFP‐NLS‐expressing cells (P‐value ≤ 0.05). (D) Functional classification of DEGs in DSCAML1 ICD relative to YFP‐NLS‐expressing cells (P‐value ≤ 0.05).
-
EValidation of DEGs of the DSCAM Dataset in Neuro2A cells. Mouse Neuro 2A cells were transfected with the YFP‐tagged DSCAM ICD (DSCAM ICD), a nuclear YFP control (YFP Ctrl), or left non‐transfected (NT Ctrl). 48 h post‐transfection total mRNA was isolated and reverse transcribed into cDNA. mRNA transcription levels were quantified by semi‐quantitative real‐time PCR (qRT–PCR) using gene specific primers. mRNA levels between different samples were normalized using GAPDH and actin as reference genes.
Multiple receptors and ligands involved in axon guidance and branching as well as structural components of the tubulin cytoskeleton were differentially expressed in both the DSCAM and DSCAML1 datasets; for example, UNC5A (Unc‐5 Netrin Receptor A), TUBB3 (Tubulin Beta 3 Class III), Robo2 (Roundabout Guidance Receptor 2), PLXNC1 (Plexin C1), NTN4 (Netrin 4), and NTNG2 (Netrin G2; Fig 4D and E). Moreover, several neurotrophin‐type signaling molecules were differentially expressed, including NTRK2 (Neurotrophic Tyrosine Kinase Receptor 2, also known as TrkB) and NTF3 (neurotrophin 3, DSCAM only). Despite the overlap between DEGs in the DSCAM/DSCAML1 datasets (Fig 4C), several gene expression changes were specific for DSCAM including a cluster of Ephrins, namely EFNA3 (Ephrin A3), EFNA4 (Ephrin A4), EFNB3 (Ephrin B3), as well as L1CAM (L1 Cell Adhesion Molecule), SEMA3E (Semaphorin 3E), RGMA (Repulsive Guidance Molecule Family Member A), and SFRP4 (Secreted Frizzled Related Protein 4; Fig 4D).
To validate this striking enrichment of nervous system specific genes, we analyzed gene expression changes of some DEGs in the neuronal crest‐derived mouse Neuro 2A (N2A) cell line. We isolated RNA from N2A cells transfected with DSCAM‐ICD‐YFP or YFP alone, as well as from non‐transfected cells and analyzed quantitative gene expression changes of mouse Unc5a, Tubb3, Pcdh17, and Nos1 by semi‐quantitative real‐time PCR (qRT–PCR). We found that all four genes were up‐regulated in N2A cells expressing the DSCAM ICD compared to control conditions (Fig EV3E). To determine whether nuclear enrichment of the DSCAM ICD could affect gene expression levels in developing neurons, we then performed single‐molecule fluorescent in situ hybridization (smFISH) on hippocampal cultures transfected with DSCAM‐ICD‐YFP or YFP alone using probes against Unc5a, Pcdh17, and Ntrk2 (TrkB) mRNAs. We focused on these three candidates since they have crucial functions in developing neurons. Unc5a is a Netrin receptor mediating axon repulsion (Leonardo et al, 1997; Hong et al, 1999) and acts as dependence receptor required for neuronal survival when associated with Netrin (Llambi et al, 2001). Pcdh17, a member of the non‐clustered protocadherins, regulates axon extension as well as synaptic vesicle assembly and transmission (Hoshina et al, 2013; Hayashi et al, 2014). Ntrk2 belongs to the Trk family of receptor tyrosine kinases, which are key regulators of neural cell death and survival (Lu et al, 2005), and also have important functions in neurite branching, as well as synapse formation and function (Park & Poo, 2013). Multiplexing of each candidate probe with a probe against YFP mRNA allowed us to detect single YFP‐positive neurons (Fig 5A, C and E). Quantification of single mRNA puncta in individual YFP‐positive neurons revealed that Unc5a was up‐regulated and Pcdh17 and Ntrk2 (TrkB) were down‐regulated in DSCAM ICD expressing hippocampal neurons (Fig 5B, D and F).
Figure 5. Expression of the DSCAM ICD changes mRNA levels of neuronal genes in primary neurons.
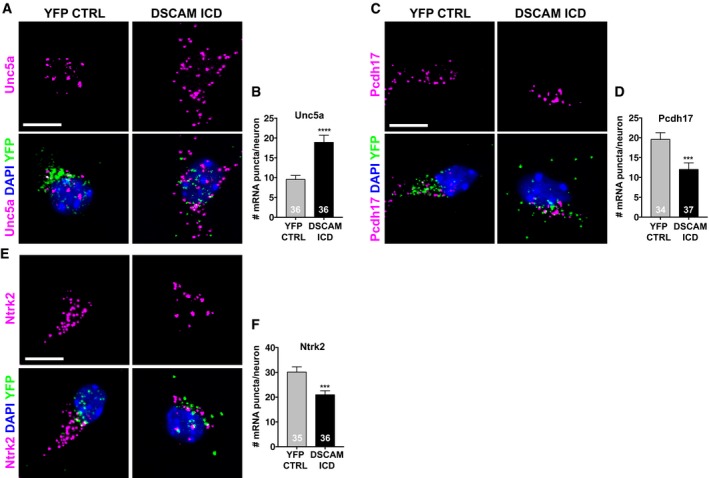
-
A–F(A, C, E) Primary hippocampal neurons were isolated at E18 and nucleofected with YFP‐HA‐tagged DSCAM ICD or a nuclear YFP‐HA control. Neurons were fixed at DIV5, and single mRNA molecules were detected using RNA FISH probes against mouse Unc5a (A), Pcdh17 (C), and Ntrk2 (E) (magenta). Single transfected neurons were identified by using an RNA FISH probe against eYFP (green), and nuclei were visualized with DAPI (blue). Scale bars 10 μm. Partial confocal z‐stacks are shown. (B, D, F) Quantification Unc5a (B), Pcdh17 (D), and Ntrk2 (F) mRNA levels in DSCAM‐ICD‐YFP‐HA expressing versus YFP‐HA expressing control neurons.
Thus, increased nuclear levels of the DSCAM ICD resulted in up‐regulation of Unc5a in human and mouse cell lines as well as in primary neurons, whereas for Pcdh17, Ntrk2, and Tubb3, we observed different directions of transcriptional changes depending on the cellular system studied. This can be explained by the fact that different cellular systems have distinct transcriptional profiles and thus varying sets of key transcriptional regulators potentially involved in the gene expression changes induced by the DSCAM ICD.
Taken together, we show that nuclear enrichment of the DSCAM/DSCAML1 ICDs triggers transcriptional changes of genes involved in neurite repulsion, axon guidance, branching, synapse formation, and neuronal cell death and survival in cell lines. Moreover, increased nuclear DSCAM ICD levels lead to specific up‐regulation of the repulsive Netrin receptor Unc5a in primary neurons and cell lines, suggesting that transcriptional regulation via membrane‐to‐nucleus signaling of DSCAM might play a role in mediating its repulsive functions during neuronal circuit formation.
Nuclear DSCAM and DSCAML1 impair neurite outgrowth
Since our transcriptome analysis suggests that overexpression of DSCAM/L1 ICDs alters the expression of genes involved in neuronal differentiation and synapse formation, we next investigated the role of these ICDs in primary neurons. It has been shown that overexpression of full‐length DSCAM in cortical pyramidal neurons significantly inhibits axon growth and branching in a dosage‐dependent manner (Jain & Welshhans, 2016). Furthermore, DSCAM overexpression in mouse hippocampal neurons inhibits dendritic branching and leads to a reduction of total dendrite length (Alves‐Sampaio et al, 2010). In order to test whether a membrane‐to‐nucleus signaling loop might contribute to the regulation of neurite growth, we first transduced low‐density cortical cultures at DIV1 with lentiviral constructs expressing YFP‐tagged wt and NLS‐deficient DSCAM/L1 ICDs, as well as a nuclear YPF control (Fig 6A). We immuno‐stained the cultures for neuron‐specific tubulin (Tuj1) at DIV4 and subsequently traced the tubulin cytoskeleton from randomly selected single YFP‐positive neurons (Fig 6B; Appendix Fig S4A–C). Nuclear localization of the DSCAM or DSCAML1 ICDs resulted in a significant reduction of total neurite length as compared to controls with YFP expression (DSCAM, 39% shorter; DSCAML1 47% shorter) or NLS‐deficient ICDs (DSCAM, 44% shorter; DSCAML1, 44% shorter; Fig 6C and D). Moreover, we observed a significant decrease in the length of the longest neurite in neurons expressing the DSCAM ICD (49% shorter) or DSCAML1 ICDs (48% shorter; Fig 6C and D). Notably, we did not observe significant differences in total neurite length or length of the longest neurite when comparing cultures expressing the YFP control and the NLS‐deficient ICDs (Fig 6C and D). Taken together, this demonstrates that enhanced nuclear translocation of the DSCAM and DSCAML1 ICDs profoundly impairs neurite outgrowth and development of primary cortical neurons.
Figure 6. Nuclear translocation of the DSCAM/L1 ICDs impairs neurite outgrowth in primary cortical neurons.
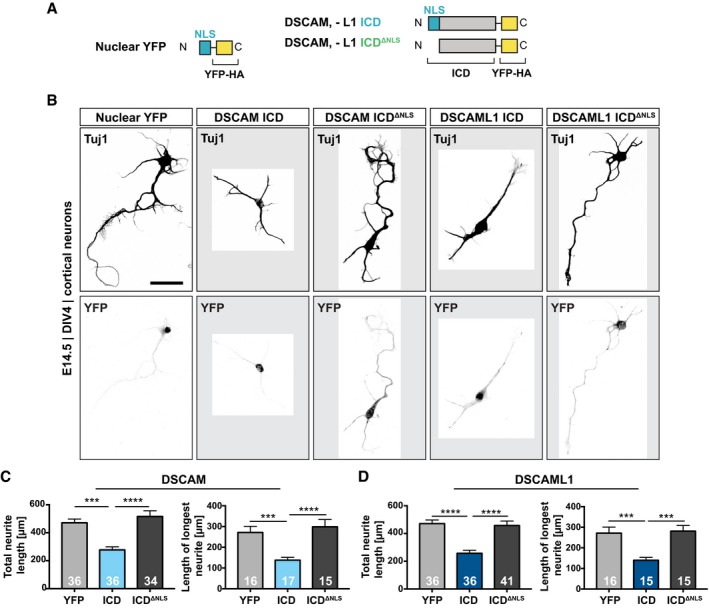
-
ASchematic of C‐terminally YFP‐HA‐tagged lentiviral DSCAM/L1 constructs expressed in (B).
-
BNeuron‐specific tubulin (Tuj1) staining showing the morphology of primary cortical neurons transduced with lentiviral expression constructs shown in A. Primary cortical cultures from E14.5 mouse embryos were transduced with lentivirus at DIV1. At DIV4, cultures were treated for 6 h with Mg132 (1 μM) and immuno‐stained for Tuj1 and YFP, and nuclei were visualized with DAPI. Images were converted to grayscale, and ROIs were cropped and displayed on a gray background. Single confocal projections of YFP‐positive neurons are shown. Additional examples can be found in Appendix Fig S4A. Scale bar, 50 μm.
-
C, DQuantification of total neurite length and of the length of the longest neurite. The neuronal tubulin cytoskeleton was traced, and the neurite length was determined using ImageJ. Total neurite length was calculated as the sum of length of all neurites of a given neuron in μm. The longest neurite of each neuron was measured in μm using the simple neurite tracing tool of ImageJ. See also Appendix Fig S4B and C. Total neurite length and the length of the longest neurite are decreased in neurons expressing the DSCAM ICD as compared to control neurons expressing YFP or DSCAM‐ICDΔNLS (C). Total neurite length and the length of the longest neurite are decreased in neurons expressing the DSCAML1 ICD as compared to control neurons expressing YFP or DSCAML1‐ICDΔNLS (D).
Increased nuclear DSCAM levels inhibit synapse formation
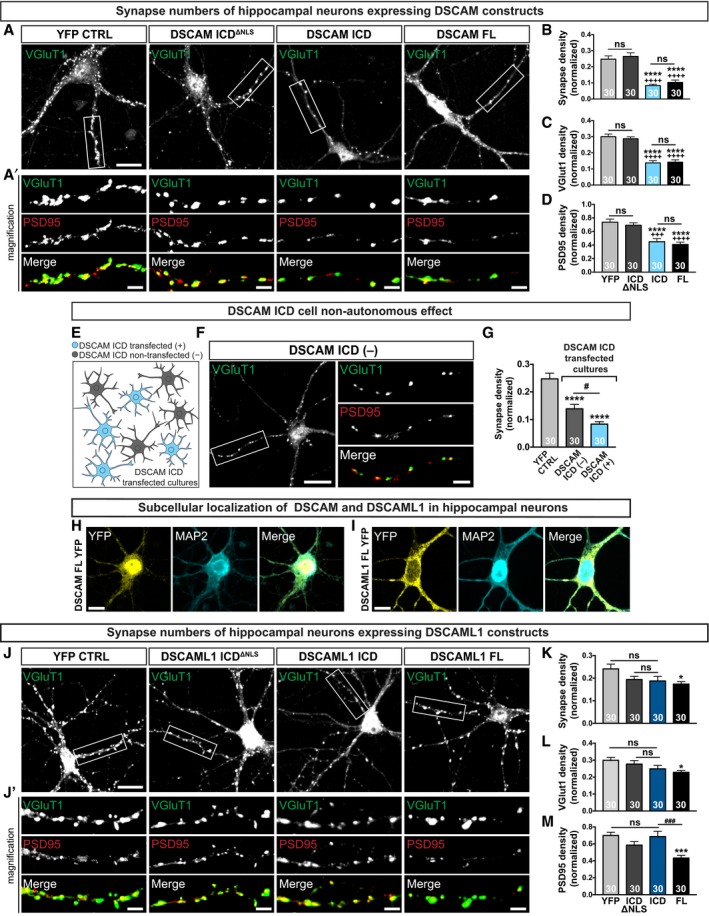
The DSCAM/L1 transcriptome data sets also contained a significant number of genes known to regulate synapse formation or function. Yet a role of mammalian DSCAM at the synapse or during synapse development has not been investigated directly. This may in part be due to the fact that early neurodevelopmental defects in DSCAM mutant mice likely mask later roles at synapses. We therefore set out to test whether nuclear translocation of the DSCAM/L1 ICDs could affect synapse density in developing neurons. Primary mouse hippocampal neurons isolated from E18 embryos were electroporated with wt and NLS‐deficient YFP‐fusions of the DSCAM/L1 ICDs, YFP‐tagged full‐length DSCAM/L1, and a nuclear YPF control. At DIV10, we determined the density of excitatory synapses defined as puncta double‐positive for the postsynaptic marker PSD95 and the presynaptic marker VGlut1 normalized over dendrite length as described previously (Ko et al, 2009; Savas et al, 2015). Expression of full‐length DSCAM or of the DSCAM ICD strongly and significantly decreased synapse density as compared to control cultures expressing YFP or the NLS‐deficient mutant (Fig 7A and B). This reduction of synapse density was characterized by a decrease in puncta positive for both PSD95 and VGlut1 (Fig 7C and D; Appendix Fig S5A). This suggests that nuclear enrichment of the DSCAM ICD is sufficient to recapitulate the effect of full‐length DSCAM on synapse density.
Figure 7. The DSCAM but not the DSCAML1 ICD inhibits synapse development in hippocampal neuron cultures.
-
A–DAnalysis of synapse density in E18 hippocampal neuron cultures electroporated with YFP‐tagged DSCAM ICD or full‐length DSCAM (DSCAM‐FL) in comparison with control neurons electroporated with YFP alone or YFP‐tagged DSCAM‐ICDΔNLS. VGlut1‐, PSD95‐, and double‐positive puncta density are decreased in DSCAM ICD and DSCAM‐FL‐expressing neurons as compared to control neurons expressing YFP (****P ≤ 0.0001) or DSCAM‐ICDΔNLS (++++ P ≤ 0.0001, +++ P ≤ 0.001). (B) VGlut1 and PSD95 double‐positive puncta normalized to dendritic length (synapse density). (C) VGlut1‐positive puncta normalized to dendritic length. (D) PSD95‐positive puncta normalized to dendritic length.
-
E–GThe DSCAM ICD has a cell non‐autonomous effect. Reduced synapse density in cultures electroporated with the DSCAM ICD in both transfected (DSCAM ICD TF) and neighboring non‐transfected neurons (DSCAM ICD NTF) as compared to control neurons expressing YFP (****P ≤ 0.0001, # P ≤ 0.05).
-
H, ISubcellular localization of recombinant YFP‐tagged full‐length DSCAM or DSCAML1 in hippocampal neurons. Scale bars, 10 μm.
-
J–MSynapse density in hippocampal neuron cultures electroporated with YFP‐tagged DSCAML1 ICD or full‐length DSCAML1 (DSCAML1‐FL) in comparison with control neurons electroporated with YFP alone or YFP‐tagged DSCAML1‐ICDΔNLS. (K) Decreased synapse density in DSCAML1‐FL expressing neurons as compared to control neurons expressing YFP (*P ≤ 0.05). (L) Decreased VGlut1‐positive puncta density in DSCAML1‐FL expressing neurons as compared to control neurons expressing YFP (*P ≤ 0.05). (M) Decreased PSD95‐positive puncta density in DSCAML1‐FL expressing neurons as compared to control neurons expressing YFP (***P ≤ 0.001) or DSCAML1 ICD (### P ≤ 0.001).
Figure 8. Model of nuclear DSCAM signaling.
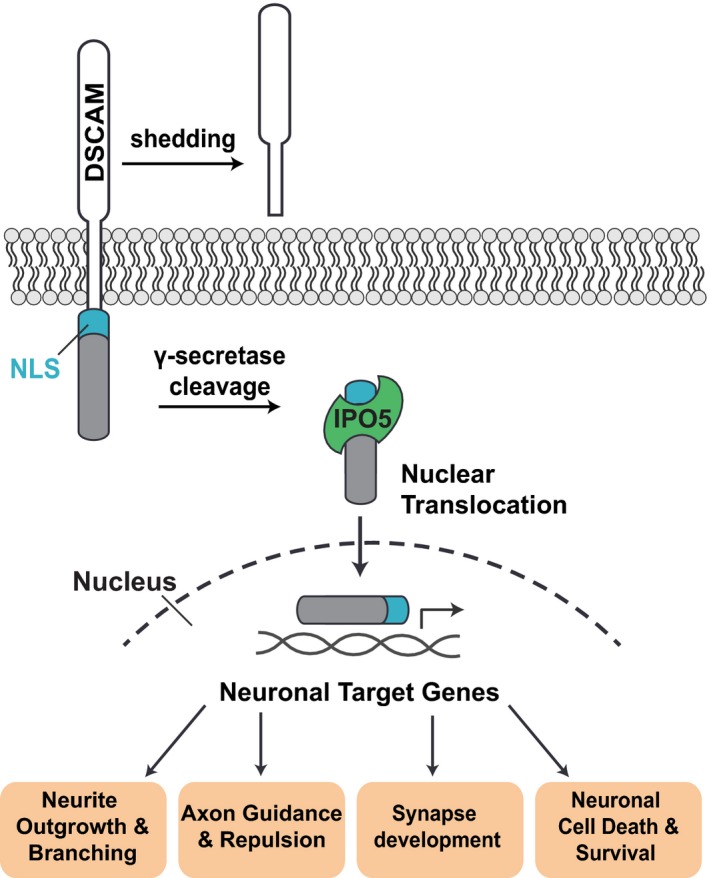
γ‐secretase‐mediated intra‐membrane cleavage of DSCAM receptors results in the release of the DSCAM ICD, which is likely proceeded by shedding of the DSCAM ectodomain. Interaction of IPO5 with the NLS of DSCAM then leads to importin‐mediated nuclear import of the DSCAM ICD. In the nucleus, the DSCAM ICD may regulate the transcription of genes involved in neuronal development and function, thereby regulating processes such as neurite outgrowth, branching, and repulsion, as well as synapse formation, axon guidance, and neuronal cell death and survival.
Given the transcriptional changes in numerous secreted factors, we also examined the potential cell non‐autonomy of defects in synapse formation in DIV10 hippocampal cultures electroporated with the DSCAM ICD (Fig 7E). Strikingly, in cultures electroporated with the DSCAM ICD, neighboring non‐transfected neurons also exhibited significantly reduced synapse density characterized by a reduction of both PSD95 and VGlut1 puncta (Fig 7F and G; Appendix Fig S5B–D). However, this cell non‐autonomous effect on synapse density was weaker as compared to neurons transfected with the DSCAM ICD (Fig 7G). We therefor conclude that nuclear translocation of the DSCAM ICD has a cell autonomous and a cell non‐autonomous effect on the development of synapses in hippocampal neurons. A cell non‐autonomous function would be consistent with the scenario that a DSCAM ICD‐dependent nuclear signaling loop alters expression and levels of (likely secreted) factors that inhibit synapse formation in neighboring neurons.
We further observed that in primary DIV10 hippocampal neurons full‐length DSCAM and DSCAML1 expression resulted in a different pattern of subcellular localization. While for DSCAM a substantial nuclear localization was apparent, DSCAML1 was predominantly located at the cell membrane (Fig 7H and I). Moreover, expression of full‐length DSCAML1 in hippocampal neurons resulted in only a slight reduction of synapse density as compared to YFP expressing control cultures, and expression of the DSCAML1 ICD alone did not have an effect on synapse density (Fig 7J–M, Appendix Fig S6). The small effect of full‐length DSCAML1 on synapse formation in contrast likely does not depend on a nuclear feedback loop in line with the distinct subcellular distribution we observed for DSCAM and DSCAML1 in these neurons. Taken together, this suggests that the effect of full‐length DSCAM on synapse formation in hippocampal neurons depends to a significant degree on cleavage and nuclear translocation of its ICD.
Discussion
Novel membrane‐to‐nucleus signaling of DSCAMs
Previous studies emphasized the importance of local cytoskeletal regulation as the central modus of DSCAM‐dependent signaling (Schmucker et al, 2000; Liu et al, 2009; Purohit et al, 2012; Huang et al, 2015; Okumura et al, 2015). Several findings presented in this study, however, reveal a previously overlooked yet potentially conserved membrane‐to‐nucleus signaling mechanism of DSCAM receptors. The top candidates of our newly identified DSCAM/L1 binding partners are IPO5, STAT3, SH2D2A, DYRK1A, DYRK1B, and USP21.
All of these factors show a surprisingly close link to nuclear functions, the most prominent one being the transcription factor STAT3. It is well known that STAT3 dimers directly bind to DNA and get activated downstream of cytokine receptors, through phosphorylation by receptor tyrosine kinases such as the epidermal growth factor receptor (EGFR), and non‐receptor tyrosine kinases like Src kinases (David et al, 1996; Reich & Liu, 2006). Moreover, the kinase DYRK1A can get recruited to promoters of genes actively transcribed by RNA polymerase II (RNAPII), where it phosphorylates RNAPII, thereby activating gene transcription (Di Vona et al, 2015). DYRK1B, SH2D2A, and USP21 have also been shown to be involved in transcriptional regulation (Lim et al, 2002; Marti et al, 2006; Nakagawa et al, 2008).
These findings together with our studies on proteolytic processing and nuclear import of DSCAM suggest a potentially important membrane‐to‐nucleus signaling mechanism of DSCAM receptors. Such a mechanism resembles for example the signaling mode of the Notch receptor (Bray, 2006). Ligand binding to Notch (i.e., Jagged and Delta) promotes shedding of the Notch ectodomain by ADAM‐family metalloproteases, followed by a second intra‐membrane cleavage mediated by the γ‐secretase complex (De Strooper et al, 1999; Bray, 2006). The second cleavage releases the Notch intracellular domain (Nicd), which in turn translocates to the nucleus where it acts as transcriptional co‐factor to modulate gene expression of Notch target genes (De Strooper et al, 1999; Bray, 2006). While Schramm and colleagues previously reported shedding of the DSCAM ectodomain (Schramm et al, 2012), and ectodomain cleavage has also been shown for Drosophila Dscam1 (Watson et al, 2005), the shedding protease of DSCAM receptors has not yet been identified. It is possible, however, that the reported DSCAM shedding product leaves an ectodomain stub, which serves as γ‐secretase substrate. In the case of Notch, metalloprotease cleavage depends on ligand binding, whereas subsequent intra‐membrane cleavage by γ‐secretase is constitutive (Struhl & Adachi, 2000). This raises the question of whether cleavage and subsequent nuclear localization of the DSCAM/L1 ICDs might be ligand dependent. However, we observed γ‐secretase cleavage of DSCAM in the absence of ligand treatment in cell cultures. Thus, it is possible that also extracellular factors alone or in combination with homophilic DSCAM interactions in cis or trans may regulate DSCAM cleavage.
Functions of DSCAM‐dependent membrane‐to‐nucleus signaling in neural development
Accumulating evidence supports the concept that specific aspects of neuronal morphogenesis may be controlled by dynamic transcriptional feedback programs in response to transmembrane receptor signaling at growth cones or synapses (Bao et al, 2004; Yang et al, 2009; Neuhaus‐Follini & Bashaw, 2015). For example, the axon guidance receptor Frazzled (Fra), which is the Drosophila homologue of DCC and expressed in neuronal growth cones, was shown to be cleaved by γ‐secretase releasing its intracellular domain (Neuhaus‐Follini & Bashaw, 2015). The Fra ICD translocates to the nucleus where it functions as a transcriptional activator regulating the expression of commissureless in vivo (Yang et al, 2009; Neuhaus‐Follini & Bashaw, 2015). Commissureless on the other hand is a factor controlling midline crossing by negatively regulating the amount of repulsive Robo receptors at the cell surface of pre‐crossing commissural axons (Yang et al, 2009; Neuhaus‐Follini & Bashaw, 2015). Here we show that nuclear enrichment of the DSCAM ICD leads to transcriptional up‐regulation of the repulsive axon guidance receptor Unc5a as well as down‐regulation of the neurotrophin receptor Ntrk2 in primary neuron cultures. Thus, it is tempting to speculate that a potential transcriptional regulation mediated by DSCAM receptors might serve to selectively regulate the responsiveness of neurons to repulsive axon guidance and/or death and survival promoting cues and thereby contribute to the spatial, temporal, and cell type‐specific fine‐tuning of neural circuit assembly. Importantly, the surprisingly strong impact of the DSCAM ICD on synapse formation and the observed non‐autonomous inhibition of synapse formation are certainly consistent with a DSCAM‐dependent retrograde signaling via nuclear import.
DSCAM and DSCAML1 specifically interacted with the importin beta IPO5, whereas deletion of the identified NLSs abolished this specific interaction and suppressed nuclear translocation of the DSCAM/L1 ICDs in cell lines and cultured neurons. This suggests a direct role of IPO5 in the nuclear import of the DSCAM/L1 ICDs. Nuclear protein import can be accomplished by a complex consisting of importin alpha and beta or by importin beta acting alone (Freitas & Cunha, 2009; Xu et al, 2010). IPO5 was shown to interact directly with the NLS of the cytoplasmic polyadenylation element‐binding protein (CPEB) 3, mediating its NMDA‐induced nuclear translocation in neurons (Chao et al, 2012). Considering that we tested five out of seven human importin alphas (Miyamoto et al, 2016) and none of them interacted with DSCAM/L1, it seems also possible that IPO5 binds directly to DSCAM/L1 without involvement of an importin alpha. Interestingly, we noted that successful co‐immunoprecipitation (Co‐IP) of DSCAM/L1 and IPO5 required the strict presence of Tyrosine‐, as well as Serine‐, and Threonine‐Phosphatase inhibitors. This suggests that the interaction between DSCAM receptors and IPO5 is likely regulated by phosphorylation, potentially involving multiple classes of kinases. Moreover, the interaction between DSCAM and IPO5 was confirmed by Co‐IP in SH‐SY5Y human neuroblastoma cells (also in the presence of phosphatase inhibitors). Notably, as opposed to HEK293T cells we could observe low levels of nuclear localization of DSCAM‐YFP in SH‐SY5Y cells even in the absence of proteasomal inhibitor, suggesting that kinases and/or phosphatases regulating the interaction between DSCAM and IPO5 might be expressed at different levels in those cells.
Even though intra‐membrane cleavage and nuclear translocation has been shown for several neuronal receptors (Kopan et al, 1996; Taniguchi et al, 2003; Goldschneider et al, 2008; Neuhaus‐Follini & Bashaw, 2015), very little is known about the molecular mechanisms of their nuclear translocation from distant axon terminals or synapses. A study in Drosophila showed that nuclear import of the C‐terminus of the Wnt receptor Frizzled2 requires importins to promote postsynaptic development (Mosca & Schwarz, 2010). Importins are localized in dendrites, axons, and synapses (Hanz et al, 2003; Thompson et al, 2004; Mosca & Schwarz, 2010; Perry et al, 2012), and their molecular interactions and subcellular localization at synapses can be altered by neuronal activity during neuronal plasticity (Thompson et al, 2004; Jeffrey et al, 2009). Furthermore, importins are involved in synapse‐to‐nucleus transport as well as nuclear import of synaptic proteins (Lever et al, 2015; Panayotis et al, 2015), and have been reported to enable retrograde signaling in regenerating axons after injury (Hanz et al, 2003; Perry et al, 2012). Our data indicate that importin‐mediated membrane‐to‐nucleus signaling may also play a role in neurite and synapse development downstream of DSCAMs. Taken together, we speculate that importin‐mediated membrane‐to‐nucleus communication from distant growth cones and synapses may represent a more general attribute of neuronal receptor signaling.
Potential gain‐of‐function effects of DSCAM by gene dosage increase in trisomy 21
Interestingly, mouse models of Down syndrome (e.g., Ts1Cje) as well as studies on DS patients reveal abnormal patterns of gene expression in the brain during embryonic development and adulthood (Guedj et al, 2015a,b). These studies suggest that overexpression of certain proteins in DS brains may cause long‐term transcriptional changes during brain development. Interestingly, in addition to DSCAM also DYRK1A is located in the DS critical region (Shindoh et al, 1996; Yamakawa et al, 1998) and both are overexpressed in DS patients (Saito et al, 2000; Liu et al, 2008). It seems therefore possible that co‐expression of these two signaling factors may lead to a synergistic enhancement of this signaling unit. Also, the recruitment of SH2D2A, which is specifically expressed in activated T cells (Spurkland et al, 1998), may be a potentially interesting lead to test for its contribution in DS given that DS patients suffer from immune system deficiencies including decreased levels of T cells (Ram & Chinen, 2011).
Several previous studies demonstrated that tyrosine‐phosphorylation levels of DSCAMs may play an important functional role (Liu et al, 2009; Purohit et al, 2012; Dascenco et al, 2015) and fly Dscam1 interacts with the SH2/SH3 adaptor protein Dock/Nck (Schmucker et al, 2000). Consistent with this, our findings now further suggest that STAT3 and the adaptor protein SH2D2A interact with tyrosine‐containing motifs within the DSCAM/L1 ICDs. The SH2 domains of both STAT3 and SH2D2A are known to bind to phosphorylated tyrosine residues in the context of such motifs (Stahl et al, 1995; Shao et al, 2004; Tinti et al, 2013). Thus, the interactions between DSCAMs and SH2‐domain containing proteins seem to play a central and conserved role in Dscam signaling in the context of dynamic changes of tyrosine‐phosphorylation levels.
Taken together, investigating potentially deleterious DSCAM‐dependent gain‐of‐function effects and in particular the transcriptional alterations in response to activity of the DSCAM ICD may greatly increase our understanding of the molecular mechanisms that contribute to the emergence of neurodevelopmental disorders and disabilities associated with Down syndrome.
Materials and Methods
Additional details and methods are provided in the Appendix Supplementary Methods including a list of primer sequences (Appendix Table S3) and a list of antibodies (Appendix Table S4).
MAPPIT
MAPPIT screens were performed as described previously (Lievens et al, 2009; Simicek et al, 2013). The prey collection screened consisted of a subset of 9,870 human ORF preys selected from the human ORFeome collection version 5.1 (http://horfdb.dfci.harvard.edu/hv5/). We would like to note that DSCAM/L1 bait receptors gave rise to background signals in binary MAPPIT experiments, i.e., generating a signal in the absence of an interaction (Appendix Fig S2D and G). This can be explained by activation of endogenous STAT3 signaling upon binding to the DSCAM/L1 bait receptors. Consistently, the STAT3 binding deficient mutant DSCAM (i.e., LR‐DSCAMY1746F) and DSCAML (i.e., LR‐DSCAML1Y1744F/Y1937F) baits did not generate background signals and were therefore used in follow‐up binding experiments as indicated (Appendix Fig S2D and G).
Neuronal cultures and transfections
Low‐density neuronal cultures were established as described in Kaech and Banker (2006). In brief, neurons were cultured from E14‐15 cortices or E18 hippocampi of C57BL/6J mice and plated on poly‐D‐lysine (Millipore)‐ and laminin (Invitrogen)‐coated glass coverslips (Marienfeld GmbH) and grown on top of a glial feeder layer. Neurons were maintained in Neurobasal medium (Invitrogen) supplemented with B27, glucose, glutamax, penicillin/streptomycin (Invitrogen), 20 mg/ml insulin (Sigma), and 25 mM b‐mercaptoethanol. Hippocampal neurons were electroporated with 3 μg of plasmid DNA just before plating with the mouse neuron AMAXA Nucleofector kit (Lonza) and immuno‐stained at DIV10 or processed for RNAscope at DIV5. Lentiviral infections of cortical neurons were performed at DIV1, and neuronal morphology was analyzed at DIV4. To stabilize intracellular DSCAM/L1 fragments in IHC experiments, hippocampal cultures were treated at DIV9 with the proteasomal inhibitor Mg132 (1 μM, Invivogen) for 12 h until DIV10 and cortical cultures were treated with Mg132 (1 μM) or Lactacystin (10 μM, Calbiochem) for 6 h prior to fixation and immuno‐staining (see figure legends). For γ‐secretase inhibition experiments, primary hippocampal neurons were treated at DIV9 over night with 5 μM Lactacystin (Calbiochem) alone or in combination with 10 μM Inhibitor X (Calbiochem) and then fixed and immuno‐stained at DIV10.
Image acquisition and analysis
Images were taken with a Zeiss LSM710 confocal microscope using a 40×/1.2NA water immersion or a 20×/0.8NA objective (Zeiss, Jena, Germany). Confocal images were processed by the use of the Fiji (Schindelin et al, 2012) software. Images were cropped, and brightness and contrast were adjusted on sample and control images. Analysis of neurite length was performed on YFP‐positive neurons using the skeletonize/analyze skeleton and simple neurite tracer plug‐ins in Fiji. Analysis of synaptic density was performed on transfected YFP‐positive neurons as described in Savas et al (2015). Following RNA FISH, mRNA molecules of single YFP‐positive neurons were counted using the “find maxima” plugin in Fiji. For more details, see Appendix Supplementary Methods.
RNA sequencing
Libraries were prepared from RNA of three 3 biologically independent experiments. Sequence‐libraries of each sample were equimolarly pooled and sequenced on an Illumina NextSeq 500 instrument (High Output, 75 bp, Single Reads, v2) at the VIB Nucleomics core (www.nucleomics.be). Differentially expressed genes were uploaded into the IPA software (Ingenuity Systems, http://www.ingenuity.com). An IPA core analysis was performed focusing on both up‐ and down‐regulated molecules and setting the log ratio (LR) cutoff ≥ 0.58 (fold change ≥ 1.5) and the FDR cutoff ≤ 0.0005 (range 0.0–0.0005).
Single‐molecule fluorescent in situ hybridization
Hippocampal neuron cultures nucleofected with DSCAM‐YFP‐Ha or YFP‐Ha plasmid DNA were fixed at DIV5 in 4% PFA/sucrose (w/v) in PBS for 30 min at RT. Single‐molecule FISH was performed using the RNAScope® Fluorescent Multiplex Reagent kit (Advanced Cell Diagnostics). Coverslips were immobilized on glass slides and pretreated with the RNAscope® Protease III reagent (Advanced Cell Diagnostics) for 15 min at RT. Probe hybridization was achieved with an EYFP probe (312131‐C3) multiplexed with either Mm‐Unc5a (429301), or Mm‐Pcdh17 (489901‐C2), or Mm‐Ntrk2 (423611) followed by amplifying hybridization probes. Heating steps were performed using a HybEZ™ oven. DAPI was used to visualize nuclei, and samples were mounted with ProLong Gold Antifade (Thermo Fisher).
Author contributions
SMS, DS, SL, JT, designed research; SMS, KH, AM, LFR, DM, DD, ASDS, and NV performed research; SL, JT, DD, BDS, ES, M‐LE, SN, YK, and JDW contributed reagents/analytical tools; SMS, SL, WVD, DD, SN and SP analyzed data; SMS and DS wrote the paper.
Conflict of interest
The authors declare that they have no conflict of interest.
Supporting information
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1J
Source Data for Figure 2F
Acknowledgements
RNA‐seq library preparation, sequencing and statistical analysis were performed by VIB Nucleomics Core (www.nucleomics.be). S.M.S. and M.L.E. were supported by the Boehringer Ingelheim Fonds. LFR. and DD. are supported by an FWO postdoctoral fellowship. JDW. is supported by ERC Starting Grant (#311083), FWO Odysseus grant, and KU Leuven Methusalem Grant (3M140280). This work was supported by KU Leuven C1 (C14/16/049) grant (DS), the Fonds voor Wetenschappelijk Onderzoek (FWO; G098615N, G0E0717N (DS); G0H2818N (DS and JDW)), the KU Leuven and VIB, a Methusalem grant from the KU Leuven/Flemish Government to BDS (3M140280). BDS is supported by the Bax‐Vanluffelen Chair for Alzheimer's Disease and “Opening the Future” of the Leuven Universiteit Fonds (LUF). BDS is supported by Vlaams Initiatief voor Netwerken voor Dementie Onderzoek (VIND, Strategic Basic Research Grant IWT 135043).
The EMBO Journal (2019) 38:e99669
Data availability
RNA‐seq raw data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO Series accession number GSE122568 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122568).
References
- Agarwala KL, Ganesh S, Tsutsumi Y, Suzuki T, Amano K, Yamakawa K (2001) Cloning and functional characterization of DSCAML1, a novel DSCAM‐like cell adhesion molecule that mediates homophilic intercellular adhesion. Biochem Biophys Res Commun 285: 760–772 [DOI] [PubMed] [Google Scholar]
- Alves‐Sampaio A, Troca‐Marin JA, Montesinos ML (2010) NMDA‐mediated regulation of DSCAM dendritic local translation is lost in a mouse model of Down's syndrome. J Neurosci 30: 13537–13548 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amano K, Sago H, Uchikawa C, Suzuki T, Kotliarova SE, Nukina N, Epstein CJ, Yamakawa K (2004) Dosage‐dependent over‐expression of genes in the trisomic region of Ts1Cje mouse model for Down syndrome. Hum Mol Genet 13: 1333–1340 [DOI] [PubMed] [Google Scholar]
- Aranda S, Alvarez M, Turro S, Laguna A, de la Luna S (2008) Sprouty2‐mediated inhibition of fibroblast growth factor signaling is modulated by the protein kinase DYRK1A. Mol Cell Biol 28: 5899–5911 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bahn S, Mimmack M, Ryan M, Caldwell MA, Jauniaux E, Starkey M, Svendsen CN, Emson P (2002) Neuronal target genes of the neuron‐restrictive silencer factor in neurospheres derived from fetuses with Down's syndrome: a gene expression study. Lancet 359: 310–315 [DOI] [PubMed] [Google Scholar]
- Bao J, Lin H, Ouyang Y, Lei D, Osman A, Kim TW, Mei L, Dai P, Ohlemiller KK, Ambron RT (2004) Activity‐dependent transcription regulation of PSD‐95 by neuregulin‐1 and Eos. Nat Neurosci 7: 1250–1258 [DOI] [PubMed] [Google Scholar]
- Barlow GM, Lyons GE, Richardson JA, Sarnat HB, Korenberg JR (2002) DSCAM: an endogenous promoter drives expression in the developing CNS and neural crest. Biochem Biophys Res Commun 299: 1–6 [DOI] [PubMed] [Google Scholar]
- Blank M, Fuerst PG, Stevens B, Nouri N, Kirkby L, Warrier D, Barres BA, Feller MB, Huberman AD, Burgess RW, Garner CC (2011) The Down syndrome critical region regulates retinogeniculate refinement. J Neurosci 31: 5764–5776 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bray SJ (2006) Notch signalling: a simple pathway becomes complex. Nat Rev Mol Cell Biol 7: 678–689 [DOI] [PubMed] [Google Scholar]
- Chao HW, Lai YT, Lu YL, Lin CL, Mai W, Huang YS (2012) NMDAR signaling facilitates the IPO5‐mediated nuclear import of CPEB3. Nucleic Acids Res 40: 8484–8498 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen BE, Kondo M, Garnier A, Watson FL, Puettmann‐Holgado R, Lamar DR, Schmucker D (2006) The molecular diversity of Dscam is functionally required for neuronal wiring specificity in Drosophila . Cell 125: 607–620 [DOI] [PubMed] [Google Scholar]
- Cui S, Lao L, Duan J, Jin G, Hou X (2013) Tyrosine phosphorylation is essential for DSCAML1 to promote dendrite arborization of mouse cortical neurons. Neurosci Lett 555: 193–197 [DOI] [PubMed] [Google Scholar]
- Cupers P, Orlans I, Craessaerts K, Annaert W, De Strooper B (2001) The amyloid precursor protein (APP)‐cytoplasmic fragment generated by gamma‐secretase is rapidly degraded but distributes partially in a nuclear fraction of neurones in culture. J Neurochem 78: 1168–1178 [DOI] [PubMed] [Google Scholar]
- Cvetkovska V, Hibbert AD, Emran F, Chen BE (2013) Overexpression of Down syndrome cell adhesion molecule impairs precise synaptic targeting. Nat Neurosci 16: 677–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dascenco D, Erfurth ML, Izadifar A, Song M, Sachse S, Bortnick R, Urwyler O, Petrovic M, Ayaz D, He H, Kise Y, Thomas F, Kidd T, Schmucker D (2015) Slit and receptor tyrosine phosphatase 69D confer spatial specificity to axon branching via Dscam1. Cell 162: 1140–1154 [DOI] [PMC free article] [PubMed] [Google Scholar]
- David M, Wong L, Flavell R, Thompson SA, Wells A, Larner AC, Johnson GR (1996) STAT activation by epidermal growth factor (EGF) and amphiregulin. Requirement for the EGF receptor kinase but not for tyrosine phosphorylation sites or JAK1. J Biol Chem 271: 9185–9188 [DOI] [PubMed] [Google Scholar]
- De Strooper B, Annaert W, Cupers P, Saftig P, Craessaerts K, Mumm JS, Schroeter EH, Schrijvers V, Wolfe MS, Ray WJ, Goate A, Kopan R (1999) A presenilin‐1‐dependent gamma‐secretase‐like protease mediates release of Notch intracellular domain. Nature 398: 518–522 [DOI] [PubMed] [Google Scholar]
- Di Vona C, Bezdan D, Islam AB, Salichs E, Lopez‐Bigas N, Ossowski S, de la Luna S (2015) Chromatin‐wide profiling of DYRK1A reveals a role as a gene‐specific RNA polymerase II CTD kinase. Mol Cell 57: 506–520 [DOI] [PubMed] [Google Scholar]
- Edgar R, Domrachev M, Lash AE (2002) Gene Expression Omnibus: NCBI gene expression and hybridization array data repository. Nucleic Acids Res 30: 207–210 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eyckerman S, Verhee A, der Heyden JV, Lemmens I, Ostade XV, Vandekerckhove J, Tavernier J (2001) Design and application of a cytokine‐receptor‐based interaction trap. Nat Cell Biol 3: 1114–1119 [DOI] [PubMed] [Google Scholar]
- Freitas N, Cunha C (2009) Mechanisms and signals for the nuclear import of proteins. Curr Genomics 10: 550–557 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Koizumi A, Masland RH, Burgess RW (2008) Neurite arborization and mosaic spacing in the mouse retina require DSCAM. Nature 451: 470–474 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Bruce F, Tian M, Wei W, Elstrott J, Feller MB, Erskine L, Singer JH, Burgess RW (2009) DSCAM and DSCAML1 function in self‐avoidance in multiple cell types in the developing mouse retina. Neuron 64: 484–497 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuerst PG, Harris BS, Johnson KR, Burgess RW (2010) A novel null allele of mouse DSCAM survives to adulthood on an inbred C3H background with reduced phenotypic variability. Genesis 48: 578–584 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Goldschneider D, Rama N, Guix C, Mehlen P (2008) The neogenin intracellular domain regulates gene transcription via nuclear translocation. Mol Cell Biol 28: 4068–4079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj F, Pennings JL, Ferres MA, Graham LC, Wick HC, Miczek KA, Slonim DK, Bianchi DW (2015a) The fetal brain transcriptome and neonatal behavioral phenotype in the Ts1Cje mouse model of Down syndrome. Am J Med Genet A 167: 1993–2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guedj F, Pennings JL, Wick HC, Bianchi DW (2015b) Analysis of adult cerebral cortex and hippocampus transcriptomes reveals unique molecular changes in the Ts1Cje mouse model of down syndrome. Brain Pathol 25: 11–23 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hall A (1998) Rho GTPases and the actin cytoskeleton. Science 279: 509–514 [DOI] [PubMed] [Google Scholar]
- Hanz S, Perlson E, Willis D, Zheng JQ, Massarwa R, Huerta JJ, Koltzenburg M, Kohler M, van‐Minnen J, Twiss JL, Fainzilber M (2003) Axoplasmic importins enable retrograde injury signaling in lesioned nerve. Neuron 40: 1095–1104 [DOI] [PubMed] [Google Scholar]
- Hattori D, Millard SS, Wojtowicz WM, Zipursky SL (2008) Dscam‐mediated cell recognition regulates neural circuit formation. Annu Rev Cell Dev Biol 24: 597–620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hayashi S, Inoue Y, Kiyonari H, Abe T, Misaki K, Moriguchi H, Tanaka Y, Takeichi M (2014) Protocadherin‐17 mediates collective axon extension by recruiting actin regulator complexes to interaxonal contacts. Dev Cell 30: 673–687 [DOI] [PubMed] [Google Scholar]
- He H, Kise Y, Izadifar A, Urwyler O, Ayaz D, Parthasarthy A, Yan B, Erfurth ML, Dascenco D, Schmucker D (2014) Cell‐intrinsic requirement of Dscam1 isoform diversity for axon collateral formation. Science 344: 1182–1186 [DOI] [PubMed] [Google Scholar]
- Hing H, Xiao J, Harden N, Lim L, Zipursky SL (1999) Pak functions downstream of Dock to regulate photoreceptor axon guidance in Drosophila . Cell 97: 853–863 [DOI] [PubMed] [Google Scholar]
- Hong K, Hinck L, Nishiyama M, Poo MM, Tessier‐Lavigne M, Stein E (1999) A ligand‐gated association between cytoplasmic domains of UNC5 and DCC family receptors converts netrin‐induced growth cone attraction to repulsion. Cell 97: 927–941 [DOI] [PubMed] [Google Scholar]
- Hoshina N, Tanimura A, Yamasaki M, Inoue T, Fukabori R, Kuroda T, Yokoyama K, Tezuka T, Sagara H, Hirano S, Kiyonari H, Takada M, Kobayashi K, Watanabe M, Kano M, Nakazawa T, Yamamoto T (2013) Protocadherin 17 regulates presynaptic assembly in topographic corticobasal Ganglia circuits. Neuron 78: 839–854 [DOI] [PubMed] [Google Scholar]
- Huang H, Shao Q, Qu C, Yang T, Dwyer T, Liu G (2015) Coordinated interaction of Down syndrome cell adhesion molecule and deleted in colorectal cancer with dynamic TUBB3 mediates Netrin‐1‐induced axon branching. Neuroscience 293: 109–122 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hughes ME, Bortnick R, Tsubouchi A, Baumer P, Kondo M, Uemura T, Schmucker D (2007) Homophilic Dscam interactions control complex dendrite morphogenesis. Neuron 54: 417–427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hummel T, Vasconcelos ML, Clemens JC, Fishilevich Y, Vosshall LB, Zipursky SL (2003) Axonal targeting of olfactory receptor neurons in Drosophila is controlled by Dscam. Neuron 37: 221–231 [DOI] [PubMed] [Google Scholar]
- Jain S, Welshhans K (2016) Netrin‐1 induces local translation of down syndrome cell adhesion molecule in axonal growth cones. Dev Neurobiol 76: 799–816 [DOI] [PubMed] [Google Scholar]
- Jeffrey RA, Ch'ng TH, O'Dell TJ, Martin KC (2009) Activity‐dependent anchoring of importin alpha at the synapse involves regulated binding to the cytoplasmic tail of the NR1‐1a subunit of the NMDA receptor. J Neurosci 29: 15613–15620 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jia YL, Jing LJ, Li JY, Lu JJ, Han R, Wang SY, Peng T, Jia YJ (2011) Expression and significance of DSCAM in the cerebral cortex of APP transgenic mice. Neurosci Lett 491: 153–157 [DOI] [PubMed] [Google Scholar]
- Kaech S, Banker G (2006) Culturing hippocampal neurons. Nat Protoc 1: 2406–2415 [DOI] [PubMed] [Google Scholar]
- Karlstrom H, Bergman A, Lendahl U, Naslund J, Lundkvist J (2002) A sensitive and quantitative assay for measuring cleavage of presenilin substrates. J Biol Chem 277: 6763–6766 [DOI] [PubMed] [Google Scholar]
- Kim JH, Wang X, Coolon R, Ye B (2013) Dscam expression levels determine presynaptic arbor sizes in Drosophila sensory neurons. Neuron 78: 827–838 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kise Y, Schmucker D (2013) Role of self‐avoidance in neuronal wiring. Curr Opin Neurobiol 23: 983–989 [DOI] [PubMed] [Google Scholar]
- Ko J, Fuccillo MV, Malenka RC, Sudhof TC (2009) LRRTM2 functions as a neurexin ligand in promoting excitatory synapse formation. Neuron 64: 791–798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kopan R, Schroeter EH, Weintraub H, Nye JS (1996) Signal transduction by activated mNotch: importance of proteolytic processing and its regulation by the extracellular domain. Proc Natl Acad Sci USA 93: 1683–1688 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Leonardo ED, Hinck L, Masu M, Keino‐Masu K, Ackerman SL, Tessier‐Lavigne M (1997) Vertebrate homologues of C. elegans UNC‐5 are candidate netrin receptors. Nature 386: 833–838 [DOI] [PubMed] [Google Scholar]
- Lever MB, Karpova A, Kreutz MR (2015) An importin code in neuronal transport from synapse‐to‐nucleus? Front Mol Neurosci 8: 33 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li S, Sukeena JM, Simmons AB, Hansen EJ, Nuhn RE, Samuels IS, Fuerst PG (2015) DSCAM promotes refinement in the mouse retina through cell death and restriction of exploring dendrites. J Neurosci 35: 5640–5654 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lievens S, Vanderroost N, Van der Heyden J, Gesellchen V, Vidal M, Tavernier J (2009) Array MAPPIT: high‐throughput interactome analysis in mammalian cells. J Proteome Res 8: 877–886 [DOI] [PubMed] [Google Scholar]
- Lievens S, Peelman F, De Bosscher K, Lemmens I, Tavernier J (2011) MAPPIT: a protein interaction toolbox built on insights in cytokine receptor signaling. Cytokine Growth Factor Rev 22: 321–329 [DOI] [PubMed] [Google Scholar]
- Lim S, Jin K, Friedman E (2002) Mirk protein kinase is activated by MKK3 and functions as a transcriptional activator of HNF1alpha. J Biol Chem 277: 25040–25046 [DOI] [PubMed] [Google Scholar]
- Liu F, Liang Z, Wegiel J, Hwang YW, Iqbal K, Grundke‐Iqbal I, Ramakrishna N, Gong CX (2008) Overexpression of Dyrk1A contributes to neurofibrillary degeneration in Down syndrome. FASEB J 22: 3224–3233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu G, Li W, Wang L, Kar A, Guan KL, Rao Y, Wu JY (2009) DSCAM functions as a netrin receptor in commissural axon pathfinding. Proc Natl Acad Sci USA 106: 2951–2956 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Llambi F, Causeret F, Bloch‐Gallego E, Mehlen P (2001) Netrin‐1 acts as a survival factor via its receptors UNC5H and DCC. EMBO J 20: 2715–2722 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lowe SA, Hodge JJL, Usowicz MM (2018) A third copy of the Down syndrome cell adhesion molecule (Dscam) causes synaptic and locomotor dysfunction in Drosophila . Neurobiol Dis 110: 93–101 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lu B, Pang PT, Woo NH (2005) The yin and yang of neurotrophin action. Nat Rev Neurosci 6: 603–614 [DOI] [PubMed] [Google Scholar]
- Ly A, Nikolaev A, Suresh G, Zheng Y, Tessier‐Lavigne M, Stein E (2008) DSCAM is a netrin receptor that collaborates with DCC in mediating turning responses to netrin‐1. Cell 133: 1241–1254 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Manser E, Leung T, Salihuddin H, Zhao ZS, Lim L (1994) A brain serine/threonine protein kinase activated by Cdc42 and Rac1. Nature 367: 40–46 [DOI] [PubMed] [Google Scholar]
- Marti F, Garcia GG, Lapinski PE, MacGregor JN, King PD (2006) Essential role of the T cell‐specific adapter protein in the activation of LCK in peripheral T cells. J Exp Med 203: 281–287 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews BJ, Kim ME, Flanagan JJ, Hattori D, Clemens JC, Zipursky SL, Grueber WB (2007) Dendrite self‐avoidance is controlled by Dscam. Cell 129: 593–604 [DOI] [PubMed] [Google Scholar]
- May P, Reddy YK, Herz J (2002) Proteolytic processing of low density lipoprotein receptor‐related protein mediates regulated release of its intracellular domain. J Biol Chem 277: 18736–18743 [DOI] [PubMed] [Google Scholar]
- Millard SS, Lu Z, Zipursky SL, Meinertzhagen IA (2010) Drosophila dscam proteins regulate postsynaptic specificity at multiple‐contact synapses. Neuron 67: 761–768 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyamoto Y, Yamada K, Yoneda Y (2016) Importin alpha: a key molecule in nuclear transport and non‐transport functions. J Biochem 160: 69–75 [DOI] [PubMed] [Google Scholar]
- Mosca TJ, Schwarz TL (2010) The nuclear import of Frizzled2‐C by Importins‐beta11 and alpha2 promotes postsynaptic development. Nat Neurosci 13: 935–943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nakagawa T, Kajitani T, Togo S, Masuko N, Ohdan H, Hishikawa Y, Koji T, Matsuyama T, Ikura T, Muramatsu M, Ito T (2008) Deubiquitylation of histone H2A activates transcriptional initiation via trans‐histone cross‐talk with H3K4 di‐ and trimethylation. Genes Dev 22: 37–49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neuhaus‐Follini A, Bashaw GJ (2015) The intracellular domain of the frazzled/DCC receptor is a transcription factor required for commissural axon guidance. Neuron 87: 751–763 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Okumura M, Sakuma C, Miura M, Chihara T (2015) Linking cell surface receptors to microtubules: tubulin folding cofactor D mediates Dscam functions during neuronal morphogenesis. J Neurosci 35: 1979–1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panayotis N, Karpova A, Kreutz MR, Fainzilber M (2015) Macromolecular transport in synapse to nucleus communication. Trends Neurosci 38: 108–116 [DOI] [PubMed] [Google Scholar]
- Park H, Poo MM (2013) Neurotrophin regulation of neural circuit development and function. Nat Rev Neurosci 14: 7–23 [DOI] [PubMed] [Google Scholar]
- Perry RB, Doron‐Mandel E, Iavnilovitch E, Rishal I, Dagan SY, Tsoory M, Coppola G, McDonald MK, Gomes C, Geschwind DH, Twiss JL, Yaron A, Fainzilber M (2012) Subcellular knockout of importin beta1 perturbs axonal retrograde signaling. Neuron 75: 294–305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Purohit AA, Li W, Qu C, Dwyer T, Shao Q, Guan KL, Liu G (2012) Down syndrome cell adhesion molecule (DSCAM) associates with uncoordinated‐5C (UNC5C) in netrin‐1‐mediated growth cone collapse. J Biol Chem 287: 27126–27138 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ram G, Chinen J (2011) Infections and immunodeficiency in Down syndrome. Clin Exp Immunol 164: 9–16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reich NC, Liu L (2006) Tracking STAT nuclear traffic. Nat Rev Immunol 6: 602–612 [DOI] [PubMed] [Google Scholar]
- Saito Y, Oka A, Mizuguchi M, Motonaga K, Mori Y, Becker LE, Arima K, Miyauchi J, Takashima S (2000) The developmental and aging changes of Down's syndrome cell adhesion molecule expression in normal and Down's syndrome brains. Acta Neuropathol 100: 654–664 [DOI] [PubMed] [Google Scholar]
- Savas JN, Ribeiro LF, Wierda KD, Wright R, DeNardo‐Wilke LA, Rice HC, Chamma I, Wang YZ, Zemla R, Lavallee‐Adam M, Vennekens KM, O'Sullivan ML, Antonios JK, Hall EA, Thoumine O, Attie AD, Yates JR III, Ghosh A, de Wit J (2015) The sorting receptor SorCS1 regulates trafficking of neurexin and AMPA receptors. Neuron 87: 764–780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schindelin J, Arganda‐Carreras I, Frise E, Kaynig V, Longair M, Pietzsch T, Preibisch S, Rueden C, Saalfeld S, Schmid B, Tinevez JY, White DJ, Hartenstein V, Eliceiri K, Tomancak P, Cardona A (2012) Fiji: an open‐source platform for biological‐image analysis. Nat Methods 9: 676–682 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmucker D, Clemens JC, Shu H, Worby CA, Xiao J, Muda M, Dixon JE, Zipursky SL (2000) Drosophila Dscam is an axon guidance receptor exhibiting extraordinary molecular diversity. Cell 101: 671–684 [DOI] [PubMed] [Google Scholar]
- Schramm RD, Li S, Harris BS, Rounds RP, Burgess RW, Ytreberg FM, Fuerst PG (2012) A novel mouse Dscam mutation inhibits localization and shedding of DSCAM. PLoS ONE 7: e52652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shao H, Xu X, Mastrangelo MA, Jing N, Cook RG, Legge GB, Tweardy DJ (2004) Structural requirements for signal transducer and activator of transcription 3 binding to phosphotyrosine ligands containing the YXXQ motif. J Biol Chem 279: 18967–18973 [DOI] [PubMed] [Google Scholar]
- Shindoh N, Kudoh J, Maeda H, Yamaki A, Minoshima S, Shimizu Y, Shimizu N (1996) Cloning of a human homolog of the Drosophila minibrain/rat Dyrk gene from “the Down syndrome critical region” of chromosome 21. Biochem Biophys Res Commun 225: 92–99 [DOI] [PubMed] [Google Scholar]
- Simicek M, Lievens S, Laga M, Guzenko D, Aushev VN, Kalev P, Baietti MF, Strelkov SV, Gevaert K, Tavernier J, Sablina AA (2013) The deubiquitylase USP33 discriminates between RALB functions in autophagy and innate immune response. Nat Cell Biol 15: 1220–1230 [DOI] [PubMed] [Google Scholar]
- Simmons AB, Bloomsburg SJ, Sukeena JM, Miller CJ, Ortega‐Burgos Y, Borghuis BG, Fuerst PG (2017) DSCAM‐mediated control of dendritic and axonal arbor outgrowth enforces tiling and inhibits synaptic plasticity. Proc Natl Acad Sci USA 114: E10224–E10233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soba P, Zhu S, Emoto K, Younger S, Yang SJ, Yu HH, Lee T, Jan LY, Jan YN (2007) Drosophila sensory neurons require Dscam for dendritic self‐avoidance and proper dendritic field organization. Neuron 54: 403–416 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Soundararajan M, Roos AK, Savitsky P, Filippakopoulos P, Kettenbach AN, Olsen JV, Gerber SA, Eswaran J, Knapp S, Elkins JM (2013) Structures of Down syndrome kinases, DYRKs, reveal mechanisms of kinase activation and substrate recognition. Structure 21: 986–996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spurkland A, Brinchmann JE, Markussen G, Pedeutour F, Munthe E, Lea T, Vartdal F, Aasheim HC (1998) Molecular cloning of a T cell‐specific adapter protein (TSAd) containing an Src homology (SH) 2 domain and putative SH3 and phosphotyrosine binding sites. J Biol Chem 273: 4539–4546 [DOI] [PubMed] [Google Scholar]
- Stahl N, Farruggella TJ, Boulton TG, Zhong Z, Darnell JE Jr, Yancopoulos GD (1995) Choice of STATs and other substrates specified by modular tyrosine‐based motifs in cytokine receptors. Science 267: 1349–1353 [DOI] [PubMed] [Google Scholar]
- Sterne GR, Kim JH, Ye B (2015) Dysregulated Dscam levels act through Abelson tyrosine kinase to enlarge presynaptic arbors. eLife 4: e05196 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Struhl G, Adachi A (2000) Requirements for presenilin‐dependent cleavage of notch and other transmembrane proteins. Mol Cell 6: 625–636 [DOI] [PubMed] [Google Scholar]
- Sun W, You X, Gogol‐Doring A, He H, Kise Y, Sohn M, Chen T, Klebes A, Schmucker D, Chen W (2013) Ultra‐deep profiling of alternatively spliced Drosophila Dscam isoforms by circularization‐assisted multi‐segment sequencing. EMBO J 32: 2029–2038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taniguchi Y, Kim SH, Sisodia SS (2003) Presenilin‐dependent “gamma‐secretase” processing of deleted in colorectal cancer (DCC). J Biol Chem 278: 30425–30428 [DOI] [PubMed] [Google Scholar]
- Thompson KR, Otis KO, Chen DY, Zhao Y, O'Dell TJ, Martin KC (2004) Synapse to nucleus signaling during long‐term synaptic plasticity; a role for the classical active nuclear import pathway. Neuron 44: 997–1009 [DOI] [PubMed] [Google Scholar]
- Tinti M, Kiemer L, Costa S, Miller ML, Sacco F, Olsen JV, Carducci M, Paoluzi S, Langone F, Workman CT, Blom N, Machida K, Thompson CM, Schutkowski M, Brunak S, Mann M, Mayer BJ, Castagnoli L, Cesareni G (2013) The SH2 domain interaction landscape. Cell Rep 3: 1293–1305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Watson FL, Puttmann‐Holgado R, Thomas F, Lamar DL, Hughes M, Kondo M, Rebel VI, Schmucker D (2005) Extensive diversity of Ig‐superfamily proteins in the immune system of insects. Science 309: 1874–1878 [DOI] [PubMed] [Google Scholar]
- Xu D, Farmer A, Chook YM (2010) Recognition of nuclear targeting signals by Karyopherin‐beta proteins. Curr Opin Struct Biol 20: 782–790 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR (2008) Dscam and Sidekick proteins direct lamina‐specific synaptic connections in vertebrate retina. Nature 451: 465–469 [DOI] [PubMed] [Google Scholar]
- Yamagata M, Sanes JR (2010) Synaptic localization and function of Sidekick recognition molecules require MAGI scaffolding proteins. J Neurosci 30: 3579–3588 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamakawa K, Huot YK, Haendelt MA, Hubert R, Chen XN, Lyons GE, Korenberg JR (1998) DSCAM: a novel member of the immunoglobulin superfamily maps in a Down syndrome region and is involved in the development of the nervous system. Hum Mol Genet 7: 227–237 [DOI] [PubMed] [Google Scholar]
- Yang L, Garbe DS, Bashaw GJ (2009) A frazzled/DCC‐dependent transcriptional switch regulates midline axon guidance. Science 324: 944–947 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaseen NR, Blobel G (1997) Cloning and characterization of human karyopherin beta3. Proc Natl Acad Sci USA 94: 4451–4456 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Appendix
Expanded View Figures PDF
Source Data for Expanded View
Review Process File
Source Data for Figure 1J
Source Data for Figure 2F
Data Availability Statement
RNA‐seq raw data have been deposited in NCBI's Gene Expression Omnibus (Edgar et al, 2002) and are accessible through GEO Series accession number GSE122568 (https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE122568).


