Summary
Chitin, a major component of fungal cell walls, is a well‐known pathogen‐associated molecular pattern (PAMP) that triggers defense responses in several mammal and plant species. Here, we show that two chitooligosaccharides, chitin and chitosan, act as PAMPs in grapevine (Vitis vinifera) as they elicit immune signalling events, defense gene expression and resistance against fungal diseases. To identify their cognate receptors, the grapevine family of LysM receptor kinases (LysM‐RKs) was annotated and their gene expression profiles were characterized. Phylogenetic analysis clearly distinguished three V. vinifera LysM‐RKs (VvLYKs) located in the same clade as the Arabidopsis CHITIN ELICITOR RECEPTOR KINASE1 (AtCERK1), which mediates chitin‐induced immune responses. The Arabidopsis mutant Atcerk1, impaired in chitin perception, was transformed with these three putative orthologous genes encoding VvLYK1‐1, ‐2, or ‐3 to determine if they would complement the loss of AtCERK1 function. Our results provide evidence that VvLYK1‐1 and VvLYK1‐2, but not VvLYK1‐3, functionally complement the Atcerk1 mutant by restoring chitooligosaccharide‐induced MAPK activation and immune gene expression. Moreover, expression of VvLYK1‐1 in Atcerk1 restored penetration resistance to the non‐adapted grapevine powdery mildew (Erysiphe necator). On the whole, our results indicate that the grapevine VvLYK1‐1 and VvLYK1‐2 participate in chitin‐ and chitosan‐triggered immunity and that VvLYK1‐1 plays an important role in basal resistance against E. necator.
Keywords: pathogen‐associated molecular pattern, pattern recognition receptor, Vitis vinifera, immune responses, Erysiphe necator, resistance
Introduction
Plants are constantly exposed to potentially pathogenic microbes such as bacteria, fungi, oomycetes or viruses. However, plants have developed effective immune systems triggering various defense reactions against invading pathogens upon the perception of pathogen‐associated molecular patterns (PAMPs; Dodds and Rathjen, 2010). The recognition of these conserved microbial signatures is mediated by pattern recognition receptors (PRRs), which also detect plant endogenous molecules released by hydrolytic enzymes during interaction with the pathogen, and called damage‐associated molecular patterns (DAMPs; Boller and Felix, 2009; Boutrot and Zipfel, 2017). PRRs have a characteristic structure defined by the presence of a ligand‐binding ectodomain, a single transmembrane domain and, for some of them, an intracellular kinase domain. The structure of the ectodomain determines binding specificity: PRRs containing a leucine‐rich repeat ectodomain mostly bind peptides, such as flagellin or elongation factor Tu (EF‐Tu) from bacteria, whilst lysine motif (LysM)‐containing PRRs preferentially bind carbohydrates, such as chitin or peptidoglycans, from fungi and bacteria, respectively (Boutrot and Zipfel, 2017; Trdá et al., 2015). PAMP perception by PRRs leads to PAMP‐triggered immunity (PTI), which is characterized by a wide range of defense responses including the production of reactive oxygen species (ROS), calcium influx, mitogen‐ activated protein kinase (MAPK) phosphorylation and expression of defense‐related genes (Yu et al., 2017).
Several distinct microbial patterns are composed from N‐acetylglucosamine (GlcNAc) residues, including fungal chitin or bacterial peptidoglycan (PGN) present in microbial cell walls (Gust et al., 2012). Chitin, and its derivatives, are representative PAMPs from fungal cell walls known to induce immune responses in both monocots and dicots, indicating the presence of a conserved mechanism to perceive these chitooligosaccharides in a wide range of plant species (Shinya et al., 2015). In plants, chitin elicits a variety of defense responses including the activation of the phenylpropanoid pathway and production of pathogenesis‐related (PR) proteins such as peroxidases, chitinases, or thaumatin‐like proteins (Boller and Felix, 2009; Kaku et al., 2006; Miya et al., 2007). Chitosan, a deacetylated derivative of chitin, is also a potent elicitor of plant immunity (Aziz et al., 2006; Povero et al., 2011). In grapevine, chitosan elicits phytoalexin production, chitinase and glucanase activities leading to resistance against Botrytis cinerea and Plasmopara viticola, the causal agents of grey mould and downy mildew, respectively (Aziz et al., 2006).
The mechanism of chitin perception and signalling in plant cells was first characterized in rice with the identification of the chitin‐elicitor binding protein, CEBiP (Kaku et al., 2006), which contains three extracellular LysM motifs and is anchored to the plasma membrane via a glycosylphosphatidylinositol (GPI)‐anchor (Gong et al., 2017). Chitin perception in rice triggers the formation of a heterodimer complex between OsCEBiP and OsCERK1, a protein which contains an intracellular kinase domain required for signal transduction. Thus, two LysM proteins are required for chitin perception and signalling in rice (Hayafune et al., 2014; Shimizu et al., 2010). In Arabidopsis thaliana, AtCERK1/LYK1, a homolog of OsCERK1, has been shown to play a crucial role in both chitin signalling (Miya et al., 2007; Wan et al., 2008) and bacterial PGN perception (Gimenez‐Ibanez et al., 2009; Willmann et al., 2011). Homodimers of AtCERK1/LYK1 were shown to directly bind long chain chitin oligomers (Liu et al., 2012). However, more recent data suggest that other members of the LysM‐RK gene family in Arabidopsis may also be involved in chitin perception. For example, Cao et al. (2014) proposed that AtLYK5 (and/or AtLYK4), which have inactive kinase domains, may be the primary receptors for chitin, and that chitin perception may result in the formation of an AtLYK5‐AtLYK1 heterotetramer, triggering intracellular signal transduction.
The majority of commercially grown grapevine cultivars are derived from the species Vitis vinifera, which is highly susceptible to cryptogamic diseases, such as downy mildew (Plasmopara viticola), grey mould (Botrytis cinerea) and powdery mildew (Erysiphe necator). These two last pathogens are ascomycete fungi containing chitooligosaccharides in their cell walls. These diseases cause significant losses to viticultural production and control of these pathogens is heavily dependent on frequent fungicide application. The level of fungicide application has serious economic, environmental and potential health implications and has driven research efforts into alternative strategies (Trouvelot et al., 2014; Walters et al., 2013). Among them is the generation of new resistant varieties by introgression of downy and powdery mildew resistance (R) genes from wild North American grapevine species (Qiu et al., 2015). However, whilst R‐gene triggered resistance is very effective at controlling pathogens, widespread use of R‐genes may impose a selection pressure on parasites to evolve and evade R protein recognition, thereby compromising the durability of this control strategy (Jones and Dangl, 2006). Thus, characterization of new PRRs in a given plant species by identifying their cognate PAMPs and understanding their involvement in disease resistance may provide more durable and broad‐spectrum immunity (Piquerez et al., 2014), notably by promoting a PTI‐based crop protection (Boutrot and Zipfel, 2017; Wiesel et al., 2014).
In this study, we have investigated whether two chitooligosaccharides, chitin and chitosan, are active PAMPs in grapevine. We also report on the functional characterization of members of the VvLYK gene family, with particular focus on three orthologs of AtCERK1/LYK1 and OsCERK1, designated VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3. By functional complementation of the Arabidopsis Atcerk1 mutant, we demonstrate that VvLYK1‐1 and VvLYK1‐2 are involved in the chitooligosaccharide‐induced immune responses in V. vinifera. Moreover, VvLYK1‐1 was demonstrated to confer basal resistance against the grapevine powdery mildew E. necator when expressed in A. thaliana.
Results
Chitooligosaccharides trigger immune responses and induced resistance in grapevine
Chitooligosaccharides with a degree of polymerization (DP) ranging from 6 to 8 (hexamer to octamer) are the most effective at triggering ROS production and defense gene expression in rice and Arabidopsis, respectively (Miya et al., 2007; Petutschnig et al., 2010). In grapevine, chito‐oligosaccharides with a MW of 1500 (i.e. DP6) were shown to be the most effective at triggering phytoalexin production and expression of chitinase and glucanase, compared to chitooligosaccharides with a MW of 3000 and 10 000 (i.e. DP13–45) (Aziz et al., 2006). In this study, chitooligomers with a DP of 6 were used to test if their perception by grapevine triggers immune responses similar to that commonly observed in Arabidopsis or rice. To also investigate the importance of the degree of acetylation (DA), the early signalling events and defense gene expression induced by chitin hexamer (DA 99.9% and DP 6) or deacetylated chitosan hexamer (DA 0.1% and DP 6) were characterized in V. vinifera cell suspensions.
Contrary with what has been previously observed in Arabidopsis (Albert et al., 2006; Miya et al., 2007), chitin DP6 did not induce any oxidative burst in grapevine cells (Figure S1) whereas flg22 triggered the expected positive response (Trdá et al., 2014). Similarly, the fully deacetyled chitosan DP6 did not elicit any H2O2 production in grapevine cell suspension (Figure S1).
However, chitin DP6 induced a rapid and transient phosphorylation of two MAPKs with relative molecular masses of 45 and 49 kDa, which was not observed in water‐treated control cells (Figure 1a). Interestingly, chitosan DP6 also activated the phosphorylation of these two MAPKs but for a longer period (Figure 1a). In parallel, treatment of grapevine cells with unpurified crab shell chitin NA‐COS‐Y, previously used to elicit ROS production and defense gene expression in Brassica species (Lloyd et al., 2014), was also shown to activate these two MAPKs (Figure S2).
Figure 1.
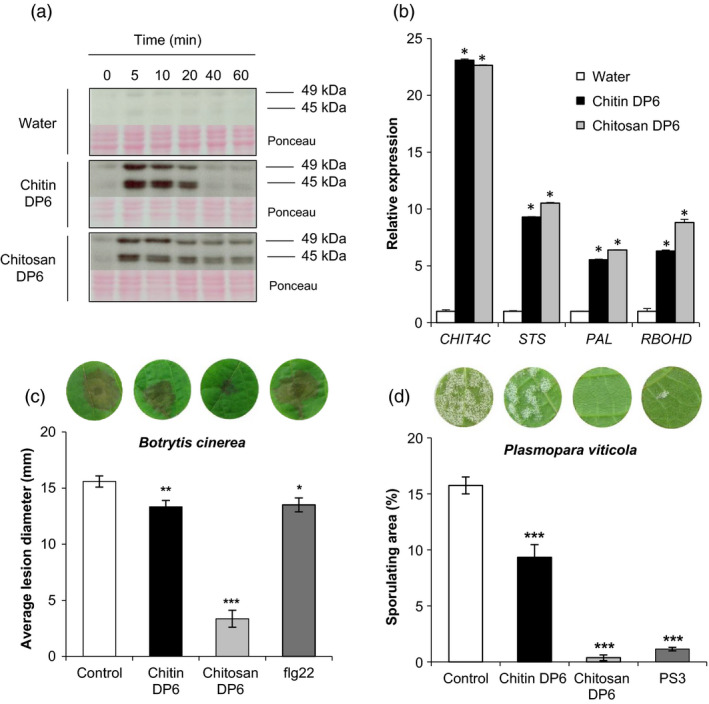
Chitin and chitosan induced defense responses and resistance to pathogens in grapevine. (a) Activation kinetics of two mitogen‐activated protein kinases (MAPKs) detected by immunoblotting with an antibody raised against the human phosphorylated extracellular regulated protein kinase 1/2 (α‐pERK1/2) in grapevine cells treated with chitin DP6 (100 μg/mL), chitosan DP6 (100 μg/mL) or water (negative control). Homogeneous loading was checked by Ponceau red staining. (b) Expression of defense genes encoding an acidic chitinase (Chit4C), a stilbene synthase ( STS ), a phenylalanine ammonia lyase ( PAL ) and a respiratory burst oxidase homolog D (RbohD) measured by quantitative polymerase chain reaction (qPCR) 1 h post‐treatment with chitin DP6 (100 μg/mL), chitosan DP6 (100 μg/mL) or water. Values represent the mean of triplicate data ±SE (n = 3) from one experiment out of three and data were normalized by the housekeeping gene EF1α and compared with water (negative control), set as 1. Asterisks (*) indicate statistically significant differences between water and chitooligosaccharide treatment, using an unpaired heteroscedastic Student's t test (P < 0.05). (c) Development of B. cinerea at 3 days post‐inoculation (dpi) on grapevine leaf discs treated 48 h before with chitin DP6 (1 mg/mL), chitosan DP6 (1 mg/mL) or flg22 (10 μm) previously solubilized in Dehscofix 0.1% and compared with control (adjuvant : Dehscofix 0.1%). Values represent the mean of lesion diameters ±SE (n ≥ 36 discs from three different plants) from one representative experiment out of three. (d) Sporulation caused by P. viticola at 8 dpi on grapevine leaf discs treated 48 h before inoculation with chitin DP6 (100 μg/mL), chitosan DP6 (100 μg/mL) or 2.5 mg/mL sulphated laminarin (PS3) previously solubilized in Dehscofix 0.1% and compared with control (adjuvant : Dehscofix 0.1%). Sporulating leaf area was evaluated by image analysis Visilog 6.9 software (Kim Khiook et al., 2013). Values represent the mean of percentage of sporulating area ±SE (n = 30 discs from three different plants) from one representative experiment out of three. Asterisks indicate a statistically significant difference between control and the elicitor treatment (Student's t‐test; *, P < 0.05, **, P < 0.01, ***, P < 0.001). A representative leaf disc for each treatment is shown. Similar results were obtained in at least three independent experiments.
In response to chitooligosaccharide treatment, the expression of defense genes known to be induced by different PAMPs in grapevine (Aziz et al., 2003; Dubreuil‐Maurizi et al., 2010; Poinssot et al., 2003; Trdá et al., 2014) was examined by qPCR. One hour post‐treatment (hpt), both chitin DP6 and chitosan DP6 markedly induced the expression of four selected grapevine defense genes (Figure 1b) encoding an acidic chitinase (CHIT4C), a stilbene synthase (STS), a phenylalanine ammonia lyase (PAL) and a respiratory burst oxidase homolog D (RBOHD).
To further characterize the immune responses triggered by chitooligosaccharides, we also investigated the efficacy of chitin‐ and chitosan‐induced resistance in grapevine. Leaf discs were treated with chitin DP6 and chitosan DP6 for 48 h prior to inoculation with either the necrotrophic fungus B. cinerea or with the biotrophic oomycete P. viticola. Chitin treatment induced a low but significant resistance against these pathogens (Figure 1c, d), whilst chitosan treatment significantly reduced B. cinerea lesion diameter and P. viticola sporulation (Figure 1c, d). Indeed, the reduced susceptibility to P. viticola infection, triggered by chitosan, was comparable to that obtained by pretreatment with the β‐1,3‐glucan sulphated laminarin (PS3), a potent resistance inducer in grapevine (Gauthier et al., 2014).
Phylogenetic analysis and characterization of grapevine LysM‐RKs (VvLYKs)
The results of Figure 1 demonstrate that grapevine cells are capable of detecting chitooligosaccharides, suggesting the presence of a perception system. To identify the CERK1/LYK1 ortholog(s) in grapevine, genes encoding LysM‐RKs were identified from the reference genome of Vitis vinifera cv. Pinot Noir PN40024 (Jaillon et al., 2007). A previous annotation of the VvLYK family based on the 8x grapevine genome predicted 12 gene family members (Zhang et al., 2009). However, our re‐annotation of the VvLYK gene family, based on the most recent version of the 12x genome, predicts the presence of 15 putative genes encoding VvLYK proteins in the V. vinifera genome (Table S1). A maximum‐likelihood phylogenetic tree indicated that of these 15 LysM‐RKs, three grapevine proteins are located in the same clade as the Arabidopsis AtCERK1/LYK1 and the rice ortholog OsCERK1 (Figure 2a), proteins that have been shown to be involved in chitin perception/signalling. These proteins, designated as VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3, share 60%, 57% and 56% amino acid identity with AtCERK1/LYK1, respectively (Table S2). VvLYK1‐1 and VvLYK1‐2 also show the highest percentage of amino acid identity with the rice chitin co‐receptor OsCERK1 (Table S2).
Figure 2.
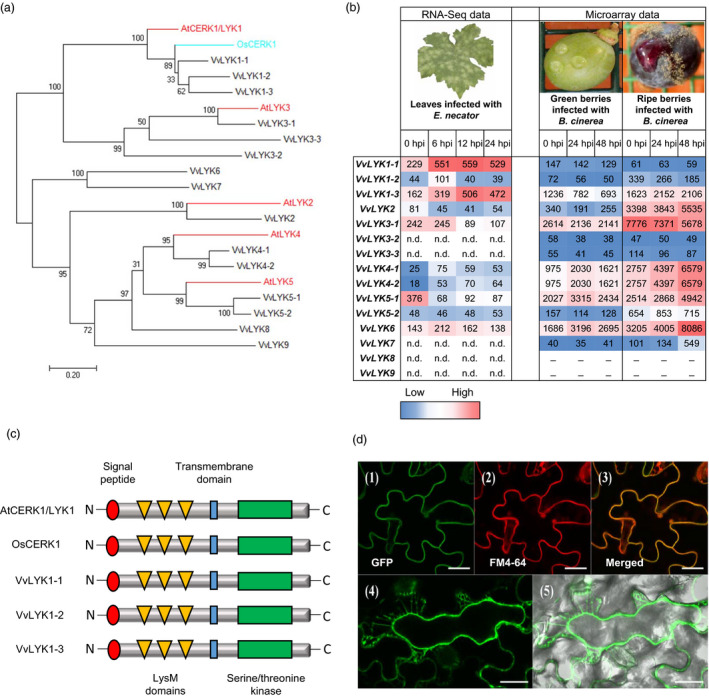
Phylogenetic analysis and characterization of grapevine LysM‐RKs (VvLYKs). (a) Maximum‐likelihood phylogenetic tree drawn with MEGA 7 (Kumar et al., 2016) showing the relationship between the Arabidopsis proteins AtCERK1/LYK1 and AtLYK2‐5 (red), the rice OsCERK1 (blue) and the most similar protein sequences of Vitis vinifera (black). Sequences used for the phylogenetic analysis were: AtCERK1/LYK1 (NP_566689), AtLYK2 (OAP05017), AtLYK3 (NP_175606), AtLYK4 (NP_179957), AtLYK5 (NP_180916), OsCERK1 (A0A0P0XII1), VvLYK1‐1 (XP_010657225), VvLYK1‐2 (XP_010655366), VvLYK1‐3 (XP_010655365), VvLYK2 (XP_019080819), VvLYK3‐1 (XP_002283628), VvLYK3‐2 (XP_019074828), VvLYK3‐3 (XP_002272814), VvLYK4‐1 (XP_002269408), VvLYK4‐2 (XP_010649202), VvLYK5‐1 (XP_002277331), VvLYK5‐2 (MF177034), VvLYK6 (XP_002280070), VvLYK7 (XP_002269472), VvLYK8 (XP_002281880) and VvLYK9 (XP_002276830). (b) VvLYK expression profiles during E. necator or B. cinerea infection. Results are expressed as Relative Expression Values. Colour range has been made independently from RNA‐Seq or microarray data. (n.d. = no full length transcript detected in RNA Seq; _ = no specific probe available in microarray). (c) Schematic structure of AtCERK1/LYK1, OsCERK1, VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3 based on the multiple alignment realized with T‐coffee (Figure S2). (d) Subcellular localization of VvLYK1‐1‐GFP in the line Atcerk1/p35S::VvLYK1‐1‐GFP . Leaves of Arabidopsis thaliana expressing VvLYK1‐1‐GFP were incubated with the plasma membrane dye FM4‐64. Confocal microscopy imaging revealed the green GFP‐tagged VvLYK1‐1 (1), the red FM4‐64 labelled plasma membrane (2) and the co‐localization of both probes in Arabidopsis leaves (3). (4) NaCl (1M) induced plasmolysis and confocal microscopy imaging revealed that VvLYK1‐1‐GFP fluorescence followed the plasma membrane shrinking (5). Bars, 20 μm.
The expression profile of each putative VvLYK gene was analysed using RNA‐Seq and microarray data obtained from time course infection experiments of leaves and berries with E. necator and B. cinerea (Kelloniemi et al., 2015), respectively. In response to inoculation with the fungal pathogen E. necator, only VvLYK1‐1 and VvLYK1‐3 were clearly up‐regulated across the entire 24 h period, while VvLYK1‐2 and VvLYK6 were transiently induced at 6 hpi (Figure 2b). During B. cinerea infection, in the clade of VvLYK1s, only VvLYK1‐3 was slightly induced in ripe susceptible berries (Figure 2b). Interestingly, VvLYK4‐1/2 (detected using the same Nimblegen probe), VvLYK5‐1 and VvLYK6 were strongly up‐regulated in berries during infection by B. cinerea (Figure 2b). VvLYK2 expression is also much higher in ripe berries than green berries, suggesting that it could have an as yet unknown function during grape berry ripening. VvLYK3‐1 appeared to be repressed during the infection by both pathogens. Of note, we found VvLYK3‐2, VvLYK3‐3, VvLYK7, VvLYK8 and VvLYK9 to only be expressed at very low levels or were undetectable in the tissues examined (Figure 2b). However, we cannot rule out the possibility that these genes are expressed at detectable levels in other tissues, such as roots or flowers, or in response to other biotic stresses.
As AtCERK1 and OsCERK1 are the key components that mediate chitin‐triggered signalling in Arabidopsis (Miya et al., 2007; Wan et al., 2008) and rice (Hayafune et al., 2014; Shimizu et al., 2010), we undertook further analysis of the three putative grapevine orthologs VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3. Sequencing of the cloned full‐length coding sequences (CDS) from V. vinifera cv Cabernet Sauvignon revealed that genes VvLYK1‐1, ‐2 or ‐3 consist of open‐reading frames of 1845, 1878 and 1866 bp, respectively (Table S1). All three VvLYK1 proteins contain a similar domain structure with a signal peptide, three extracellular LysM motifs, a single transmembrane domain and a RD‐type intracellular kinase domain (Figure 2c and Figure S3). Interestingly, the amino acids E110 and E114, shown to be involved in the binding of the N‐acetyl moieties of (GlcNAc)5 in AtCERK1/LYK1 (Liu et al., 2012) are mutated in the three VvLYK1 proteins (Figure S3). All three VvLYK1 protein sequences share a high degree of identity (Figure S3) and the kinase domains of VvLYK1‐1 and VvLYK1‐2 possess the highest identity with the kinase domains of AtCERK1/LYK1 and OsCERK1 (Figure S3, Table S2).
All three VvLYK1 proteins have a predicted N‐terminal signal peptide (Figure S3). Confocal analysis of the Atcerk1 mutant expressing a VvLYK1‐1‐GFP fusion expression construct showed a GFP signal co‐localized with the red fluorescence of the plasma membrane‐specific probe FM4‐64 (Brandizzi et al., 2004) (Figure 2d). Furthermore, when plasmolysis was triggered by the addition of 1 m NaCl, VvLYK1‐1‐GFP fluorescence followed the movement of the plasma membrane away from the plant cell wall (Figure 2d). Both observations are consistent with VvLYK1‐1 being localized to the plasma membrane.
VvLYK1‐1 restores chitin‐induced MAPK activation and FRK1 expression in the Atcerk1 mutant
To investigate whether VvLYK1‐1, VvLYK1‐2 or VvLYK1‐3 are capable of activating chitooligosaccharide‐triggered defenses, expression constructs comprising each native VvLYK1 coding sequence (i.e. no C‐terminal tag) under the control of a constitutive 35S promoter were introduced into the Atcerk1 mutant. Semi‐quantitative PCR was performed on the leaves of T2 transgenic lines to test for the presence of the VvLYK1‐1, VvLYK1‐2 or VvLYK1‐3 transcripts. Six transgenic lines were positively identified as expressing the VvLYK1‐1 transgene and five transgenic lines were identified for VvLYK1‐3 (Figure 3a). However, analysis of five independent transgenic lines, confirmed to contain the VvLYK1‐2 construct by genomic PCR, indicated that transgene expression was either undetectable (lines #2, #12, #14) or at very low levels (lines #5 and #10) compared to VvLYK1‐1 and VvLYK1‐3 transgene expression (Figure 3a). The failure to positively identify lines highly expressing VvLYK1‐2 suggested that this gene is potentially lethal when expressed under a strong constitutive 35S promoter. This was confirmed by agro‐infiltration of the p35S::VvLYK1‐2 construct into N. benthamiana leaves resulting in patchy necrosis after 48 h compared to leaf segments infiltrated with Agrobacterium alone (Figure S4). As VvLYK1‐2 induces necrosis when over‐expressed, this suggests that it may have a crucial function in defense and its expression needs to be tightly regulated in planta. Based on these results, Arabidopsis lines transformed with the p35S::VvLYK1‐2 construct were excluded from complementation analysis using a constitutive expression system.
Figure 3.
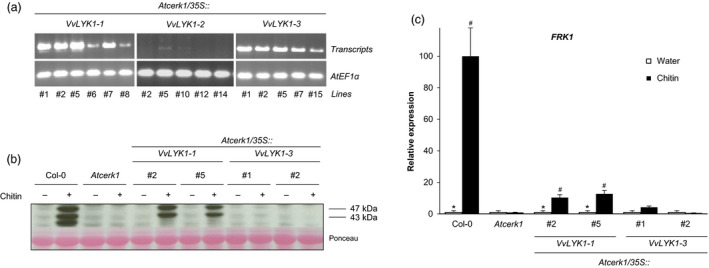
VvLYK1‐1 restores chitin‐induced immune responses in Atcerk1. (a) Semi‐quantitative RT‐PCR analysis of VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3 expression in leaf tissue of independently transformed Atcerk1 lines. AtEF1α (At5g60390) was used as an internal control. (b) Activation of two mitogen‐activated protein kinases (MAPKs) 10 min after chitin treatment (1 mg/mL) detected by immunoblotting with an antibody raised against the human phosphorylated extracellular regulated protein kinase 1/2 (α‐pERK1/2). Homogeneous loading was checked by Ponceau red staining. Similar results were obtained in three independent experiments. (c) Relative expression of a defense gene encoding flagellin‐induced receptor kinase1 (FRK1) measured by qPCR, 2 h after chitin treatment (1 mg/mL). Data show a representative experiment from three independent biological ones. Means of the triplicate data were normalized by the housekeeping gene At4g26410 and expressed as a percentage of the chitin‐treated WT Col‐0, set as 100%. Asterisks (*) indicate statistically significant differences between water and chitin treatment whereas hash marks (#) indicate statistically significant differences between WT or transgenic line and Atcerk1, using an unpaired heteroscedastic Student's t test (P < 0.05).
Transgenic Atcerk1/p35S::VvLYK1‐1 and Atcerk1/p35S::VvLYK1‐3 lines were first examined for restoration of early chitin‐induced events by analysing the phosphorylation of MAPKs in two independent lines following treatment with chitin (NA‐COS‐Y; Lloyd et al., 2014) for 10 min prior to protein extraction. Figure 3b shows that chitin treatment triggered the phosphorylation of two MAPKs, with relative molecular weights of 43 and 47 kDa, in WT Col‐0 seedlings but no MAPK phosphorylation was observed in Atcerk1, in agreement with the previous report of Miya et al. (2007). Chitin‐induced MAPK activation was restored in the two independent p35S::VvLYK1‐1 lines #2 and #5 (Figure 3b) but no MAPK phosphorylation was detected in protein samples extracted from the two independent p35S::VvLYK1‐3 lines #1 and #2 (Figure 3b).
The expression of the defense gene encoding flagellin‐induced receptor kinase 1 (FRK1) was also investigated 2 h after chitin treatment. Chitin induced a high level of expression of FRK1 in WT Col‐0 that was totally suppressed in the Atcerk1 mutant (Figure 3c). FRK1 expression was partly restored in the two p35S::VvLYK1‐1 lines #2 and #5, but remained close to the basal level in the two p35S::VvLYK1‐3 lines #1 and #2 (Figure 3c). Taken together, these results indicate that over‐expression of VvLYK1‐1 can restore chitin‐triggered immune responses in Atcerk1 but VvLYK1‐3 cannot.
VvLYK1‐1 expression restores penetration resistance in Atcerk1 against the non‐adapted powdery mildew Erysiphe necator
In addition to testing for complementation of MAPK activation and defense gene expression, the ability of VvLYK1‐1 and VvLYK1‐3 to restore resistance against a non‐adapted grapevine powdery mildew pathogen in the Atcerk1 mutant was also determined. Arabidopsis thaliana is a non‐host for the fungus E. necator. Although a proportion of E. necator spores placed onto a Col‐0 leaf will successfully penetrate the epidermal cell wall and form a haustorium under the first appressorium, the pathogen is unable to complete its life cycle on this host (Feechan et al., 2013).
Figure 4a shows that the Atcerk1 mutant is significantly more susceptible to penetration by E. necator than the WT Col‐0, showing the important role of AtCERK1 in non‐host resistance against non‐adapted powdery mildew species. More precisely, the penetration rate of WT Col‐0 leaves by E. necator spores ranged 35%–43% with a mean at 39% whereas in the Atcerk1 mutant, the penetration rates ranged 76%–88% with a mean of 82% which approaches the rate of penetration by the adapted powdery mildew species E. cichoracearum on Col‐0 (Feechan et al., 2013). Constitutive expression of VvLYK1‐1 in the Atcerk1 mutant significantly reduced the mean penetration rates of E. necator in the leaves of all Atcerk1/p35S::VvLYK1‐1 lines to levels comparable to the penetration rates on WT Col‐0 plants (31%–40%; Figure 4a). As additional negative controls, T2 lines #3 and #4 that had been generated through the same transformation procedure but had lost the introduced VvLYK1‐1 transgene through segregation, showed mean penetration rates of E. necator similar to Atcerk1 (77%–80%; Figure 4a). This demonstrates that the complementation of penetration resistance in the Atcerk1/p35S::VvLYK1‐1 lines is a result of VvLYK1‐1 expression and is not related to the transformation process. In contrast, expression of VvLYK1‐3 in the five independent Atcerk1/VvLYK1‐3 lines did not significantly reduce E. necator mean penetration rates (58%–72%; Figure 4b) in comparison to the Atcerk1 mutant or the negative T2 control lines (#9 and #10; Figure 4b).
Figure 4.
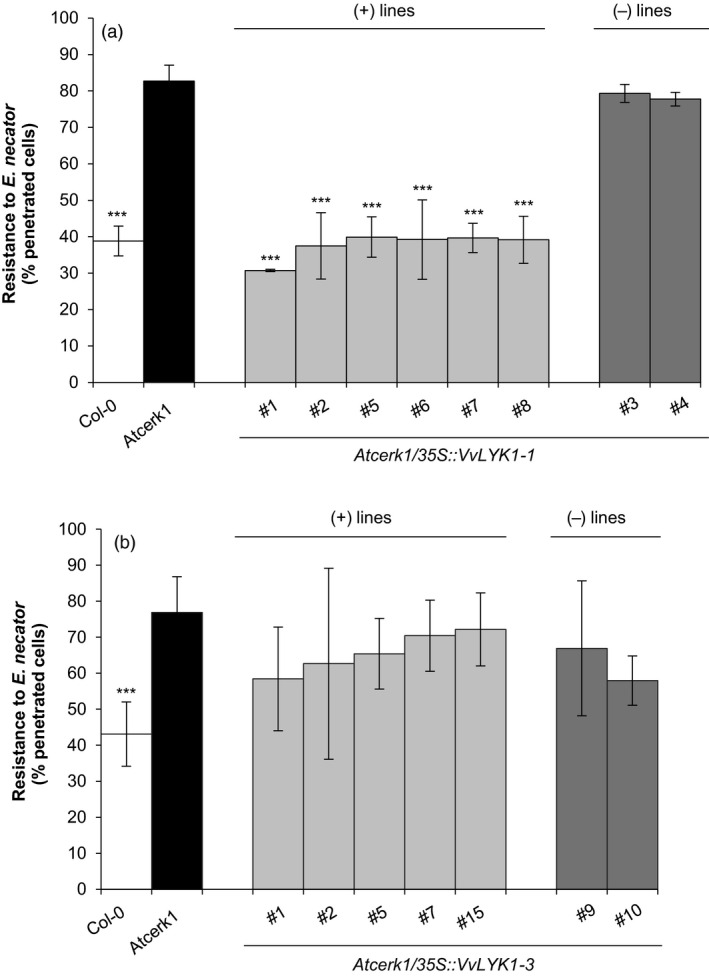
VvLYK1‐1 expression restores penetration resistance against the non‐adapted powdery mildew Erysiphe necator in Atcerk1. Penetration efficiency (i.e. haustorium formation) of the non‐adapted powdery mildew pathogen E. necator on Arabidopsis WT (Col‐0), Atcerk1 mutant and eight independent transgenic Atcerk1 lines transformed with the VvLYK1‐1 construct (a) or seven lines transformed with the VvLYK1‐3 construct (b). One hundred germinated conidia were scored per leaf, with three leaves inoculated per line. Each data point represents the mean of three independent experiments ±SE. WT Col‐0 and transgenic lines were compared to the mutant Atcerk1 with a Student t‐test (***, P < 0.001). (+) lines expressing the transgene. (−) lines with no detectable VvLYK1 transcripts.
Together, the ability of VvLYK1‐1 to restore MAPK activation, the expression of FRK1 and penetration resistance against E. necator in the Atcerk1 mutant background suggests that VvLYK1‐1 mediates chitin sensing and might be important for grapevine defense against E. necator.
The inducible expression of VvLYK1‐2 also restores chitin‐triggered responses in the Atcerk1 mutant
Due to the toxicity of constitutively expressed VvLYK1‐2, new constructs were generated in which VvLYK1‐2 expression was driven by an inducible promoter. The pABindGFP vector (Bleckmann et al., 2010) permitted the inducible expression of a C‐terminally tagged VvLYK1‐2‐GFP fusion protein regulated by the β‐estradiol LexA promoter in the Atcerk1 mutant background. Two independent hygromycin‐resistant T3 lines Atcerk1/LexA::VvLYK1‐2‐GFP #27 and #28 were selected to be homozygous and containing only one copy of the transgene.
Following β‐estradiol treatment, confocal microscopy confirmed the presence of the VvLYK1‐2‐GFP protein at the cell periphery (Figure 5a) suggesting a localization at the plasma membrane similar to VvLYK1‐1 (Figure 2d).
Figure 5.
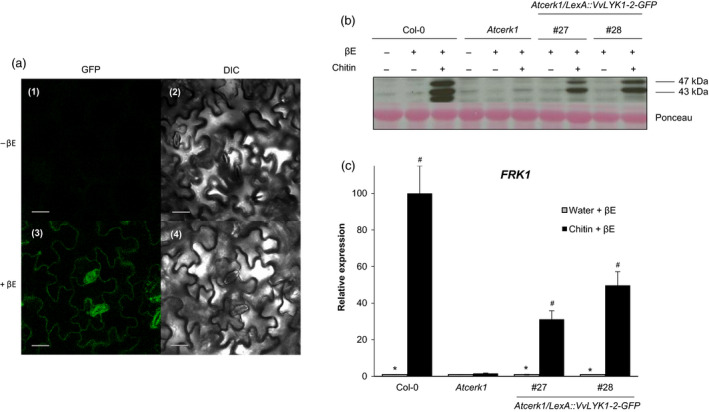
The inducible expression of VvLYK1‐2 also restores chitin‐triggered responses in Atcerk1. (a) Subcellular localization of VvLYK1‐2‐GFP visualized by confocal microscopy 4 h post‐treatment with β‐estradiol (βE). DIC, differential interference contrast. Bars, 20 μm. (b) Activation of two mitogen‐activated protein kinases (MAPKs) detected 10 min after chitin treatment (1 mg/mL) by immunoblotting with an antibody raised against the human phosphorylated extracellular regulated protein kinase 1/2 (α‐pERK1/2). Homogeneous loading was checked by Ponceau red staining. Similar results were obtained in three independent experiments. (c) Relative expression of the defense gene encoding flagellin‐induced receptor kinase1 (FRK1) measured by qPCR, 2 h post‐chitin treatment (1 mg/mL). Data show a representative experiment from three independent biological ones. Means of the triplicate data were normalized by the housekeeping gene At4g26410 and expressed as a percentage of the transcript level in WT Col‐0 plants treated by chitin + β‐estradiol, set as 100%. Asterisks (*) indicate statistically significant differences between water and chitin treatment whereas hash marks (#) indicate statistically significant differences between WT or transgenic line and Atcerk1, using an unpaired heteroscedastic Student's t test (P < 0.05). For (b) and (c), inducible transgenic lines Atcerk1/LexA::VvLYK1‐2‐GFP were treated 1 h before chitin treatment with 10 μm β‐estradiol (βE).
To investigate whether VvLYK1‐2 can also restore chitin‐induced signalling and immune responses in the Atcerk1 mutant, MAPK activation and defense gene expression were analysed. Figure 5b shows that β‐estradiol pre‐treatment alone did not induce MAPK phosphorylation in the WT Col‐0 or in the Atcerk1 mutant. However, β‐estradiol pre‐treatment followed by a chitin treatment lead to the restoration of MAPK phosphorylation in the two independent Atcerk1/LexA::VvLYK1‐2‐GFP lines #27 and #28 (Figure 5b). Similarly, the chitin‐induced expression of the defense gene FRK1 was also restored in both lines Atcerk1/pLexA::VvLYK1‐2‐GFP #27 and #28 (Figure 5c). These data indicate that VvLYK1‐2, like VvLYK1‐1, also restores MAPK activation and immune gene expression in the Atcerk1 mutant. Unfortunately, the use of this transient β‐estradiol‐inducible expression system did not permit us to obtain reproducible results concerning the putative role of VvLYK1‐2 in the resistance against E. necator.
VvLYK1‐1 and VvLYK1‐2 expression restore chitosan‐triggered responses in the Atcerk1 mutant
To further characterize these new grapevine PRRs, we also tested the responses triggered by chitosan in Atcerk1/VvLYK1 transgenic lines (Figure 6). Like chitin (Figure 3), chitosan was able to strongly induce the phosphorylation of MAPKs in WT Col‐0 and this signalling pathway was highly compromised in the Atcerk1 mutant (Figure 6a). Expression of VvLYK1‐1 in the Atcerk1 mutant also restored chitosan‐induced MAPK activation but VvLYK1‐3 did not (Figure 6a). Similarly, the chitosan‐induced expression of the defense gene FRK1 was also restored at the WT or higher level in both lines Atcerk1/p35S::VvLYK1‐1 #2 and #5 whereas the FRK1 transcript level in lines Atcerk1/p35S::VvLYK1‐3 #1 and #2 was comparable to the one in Atcerk1 (Figure 6b). Of note, the FRK1 expression level in Atcerk1 treated by chitosan is significantly higher than in the water control (Figure 6b). Chitosan‐induced phosphorylation of MAPKs (Figure 6c) and FRK1 defense gene expression (Figure 6d) were also complemented in the two independent lines Atcerk1/pLexA::VvLYK1‐2‐GFP #27 and #28. Thus, VvLYK1‐1 and VvLYK1‐2 also restore chitosan‐triggered responses in Atcerk1.
Figure 6.
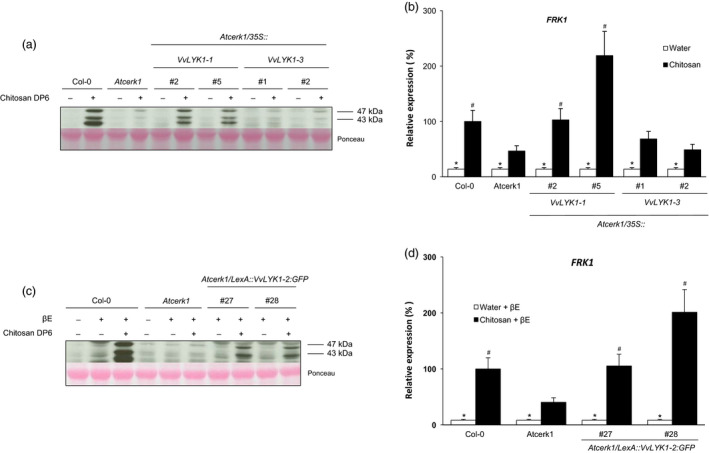
VvLYK1‐1 and VvLYK1‐2 expression restores chitosan‐triggered responses in the Atcerk1 mutant. (a, c) Activation of two mitogen‐activated protein kinases (MAPKs) detected 10 min after treatment with chitosan DP6 (1 mg/mL) by immunoblotting with an antibody raised against the human phosphorylated extracellular regulated protein kinase 1/2 (α‐pERK1/2). Homogeneous loading was checked by Ponceau red staining. (b, d) Expression of a defense gene encoding the flagellin‐induced receptor‐like protein kinase 1 ( FRK1) measured by qPCR 2 h after chitosan treatment. Data show an average of three biological experiments that were normalized by housekeeping gene At4g26410 and compared with Col‐0 treated with chitosan, set as 100%. Asterisks (*) indicate statistically significant differences between water and chitosan treatment whereas hash marks (#) indicate statistically significant differences between WT or transgenic line and Atcerk1, using an unpaired heteroscedastic Student's t test (P < 0.05). (c, d) All lines were pretreated 1 h before elicitor treatment with β‐estradiol (βE; 10 μm), when indicated.
Discussion
Chitin is a well‐known PAMP which elicits typical immune responses in Arabidopsis (Cao et al., 2014; Miya et al., 2007; Petutschnig et al., 2010; Wan et al., 2008) and a number of other plant species (Akamatsu et al., 2013; Ao et al., 2014; Felix et al., 1998; Hayafune et al., 2014; Kaku et al., 2006; Liu et al., 2016; Shimizu et al., 2010; Zeng et al., 2012). However, little is known about chitin perception in grapevine. Here, we clearly demonstrate that chitin and chitosan, its deacetylated derivative, trigger grapevine immune responses such as phosphorylation of MAPKs and the expression of defense genes including CHIT4C, STS, PAL and RBOHD. Up‐regulation of genes encoding chitinases and PAL was also observed in Arabidopsis and rice upon chitin treatment (Kaku et al., 2006; Miya et al., 2007). Surprisingly, these chitooligosaccharides did not induce any detectable H2O2 production in grapevine, in contrast with Arabidopsis (Miya et al., 2007) and rice (Hayafune et al., 2014). However, this lack of H2O2 production clearly does not prevent the phosphorylation of MAPKs showing independence between these two pathways, in accordance with results previously obtained in N. benthamiana and Arabidopsis (Segonzac et al., 2011; Xu et al., 2014).
We also show that chitin enhances the resistance of grapevine leaves to the necrotrophic fungus B. cinerea and the obligate biotrophic oomycete P. viticola (Figure 1), as previously demonstrated following treatment with chitosan, flg22 or sulphated β‐1,3‐glucan (Aziz et al., 2006; Gauthier et al., 2014; Trdá et al., 2014). Similarly, treatment of rice plants with chitin reduced the susceptibility to the fungal pathogen Magnaporthe oryzae (Tanabe et al., 2006). More recently, chitin treatment was also shown to reduce the susceptibility of Arabidopsis to the bacterial pathogen Pseudomonas syringae pv tomato (Pto) DC3000 and the fungus Alternaria brassicicola (Cao et al., 2014). All of these results confirm that stimulation of plant immune responses with PAMPs can trigger enhanced resistance against different plant pathogens.
The degree of acetylation (DA) of chitooligosaccharides appeared to have no effect on the amplitude of the immune responses in grapevine although the duration of the chitosan‐triggered MAPK activation was longer (Figure 1). Similarly, in Arabidopsis, the DA of chitooligosaccharides had no effect on the activation of PAL (Cabrera et al., 2006). In contrast, in wheat, chitosan oligomers with a DA of 50% were better able to induce PAL activity than those possessing a DA of 0% (Vander et al., 1998). Thus, the structure/activity of chitooligomers might differ depending on the plant species used (Yin et al., 2016).
The activation of MAPKs and defense gene expression in grapevine cells treated with chitin demonstrates that grapevine possesses the cognate PRRs. Zhang et al. (2009) previously proposed the grapevine LYK family to be comprised of 12 members. We undertook a re‐examination of the predicted LYK gene family in grapevine in combination with published EST data and our own RNA‐Seq data. This revealed a number of errors in the original Zhang et al. (2009) predictions both in terms of the predicted ORFs and gene number. For example, the previously annotated single VvLYK10 gene (Zhang et al., 2009) was found to contain a tandemly arrayed LYK gene pair. The two ORFs encode proteins with a 74% amino acid similarity to each other and homology to AtLYK5 (49% and 51% amino acid similarity). Tandem LYK gene pairs have also been identified in legume and poplar plants (Zhang et al., 2009). We are therefore proposing a new annotation scheme for the grapevine VvLYK gene family which uses a naming convention based on sequence similarity to Arabidopsis LYK gene family (Figure 2a, Table S1).
In Arabidopsis, AtCERK1/LYK1 has been demonstrated to play a key role in chitin‐induced signalling. Grapevine encodes three putative orthologs of AtCERK1/LYK1, designated VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3. Our data demonstrate that the constitutive expression of VvLYK1‐1 or the inducible expression of VvLYK1‐2 in the Atcerk1 mutant restores chitooligosaccharide‐induced immune responses such as MAPK activation and expression of the defense gene FRK1. Thus our results demonstrate that these two independent grapevine proteins are functional orthologs of AtCERK1/LYK1, suggesting duplication events during the evolution of the ancestral genome of V. vinifera (Jaillon et al., 2007). VvLYK1‐1 and/or VvLYK1‐2 also restore chitosan perception by the Atcerk1 mutant, as indicated by MAPK activation, suggesting that in grapevine the same PRRs can mediate both chitin and chitosan signalling. Similarly, an AtCERK1 protein band shift was detected in Arabidopsis after treatment with chitin or chitosan and the ectodomain of AtCERK1 has been shown to bind chitosan DP6 (Petutschnig et al., 2010). However, the fact that ROS production in Arabidopsis is induced by chitin DP6 but not by the fully deacetylated chitosan DP6 (Figure S1) confirmed previous results of Gubaeva et al. (2018) indicating that some downstream signalling events may be divergent. When the fact that a weak signal for MAPKs activation and a significant FRK1 transcript accumulation are observed in the Atcerk1 mutant following chitosan treatment (Figure 6) is considered together with previous results demonstrating AtCERK1/LYK1‐independent defense gene expression (Povero et al., 2011), it suggests that different AtLYK proteins may be involved in detecting different chitooligosaccharides. A preliminary investigation of the response of five different Atlyk mutants to chitosan DP6 treatment shows that MAPK activation is weaker in the Atlyk5 and Atlyk3 mutants compared to WT Col‐0 (Figure S5) suggesting that the AtLYK3 and AtLYK5 proteins might also participate in the perception of chitosan oligomers in combination with AtCERK1/LYK1.
Interestingly, we were unable to obtain Arabidopsis lines with high levels of constitutively expressed VvLYK1‐2. Furthermore, we observed an induction of cell death following transient expression of VvLYK1‐2 in tobacco (Figure S4), confirming gene toxicity. Cell death in response to heterologous LysM‐RK expression in N. benthamiana has previously been observed when AtCERK1 was fused with the yellow fluorescent protein variant, sYFP2 and transiently expressed under the control of a 35S promoter (Pietraszewska‐Bogiel et al., 2013), demonstrating the importance of regulating LYK expression levels.
In order to confirm the results obtained from complementation studies in Arabidopsis, we also attempted to confirm the function of VvLYK1‐1 and VvLYK1‐2 in chitin and chitosan perception in grapevine by generating grapevine transgenics in which these genes had been silenced. However, no transformed calli were recovered in three independent agrobacterium‐mediated transformations of somatic grapevine embryos with p35S::antisense‐VvLYK1‐1 and p35S::antisense‐VvLYK1‐2 constructs whereas parallel control transformations with a p35S::GFP construct were successful (data not shown).
VvLYK1‐1 expression in the Atcerk1 mutant background was demonstrated to restore non‐host resistance against grapevine powdery mildew suggesting that VvLYK1‐1 may participate in anti‐fungal basal resistance also in grapevine. As the Atcerk1 mutant is more susceptible to the non‐adapted pathogen E. necator, it also indicates that AtCERK1/LYK1 plays a role in the non‐host resistance against this grapevine pathogen. Paparella et al. (2014) previously showed that an Atlyk3‐1 mutant was more resistant to B. cinerea suggesting that AtLYK3 negatively regulates certain immune responses such as the production of phytoalexins, suggesting that different members of the AtLYK gene family may play a role in the basal resistance of Arabidopsis against different fungal pathogens. It is interesting to note that VvLYK4‐1/2, VvLYK5‐1 and VvLYK6 genes are highly up‐regulated during B. cinerea infection of grapevine berries (Figure 2). Thus, it is plausible that other members of the large VvLYK family may exhibit specificity to the different ligands released during the interactions of grapevine with this kind of pathogen.
Cao et al. (2014) recently showed that AtLYK5 is able to bind chitin at a greater affinity than AtCERK1 and that chitin perception leads to the formation of an AtCERK1‐AtLYK5 dimer which is required for AtCERK1 phosphorylation. These observations led them to propose that AtLYK5, and not AtCERK1, is the primary receptor for chitin perception and which has been proposed to be responsible for the activation of defense responses (Cao et al., 2014). Thus, one explanation of our complementation data is that VvLYK1‐1 or VvLYK1‐2 could dimerize with AtLYK5 in the presence of chitin, but that the interaction is not sufficiently effective to obtain a full restoration of MAPK activation and defense gene expression back to wild‐type levels. This suggests the existence of molecular complexes for chitooligosaccharides perception in grapevine, as previously shown for rice (Hayafune et al., 2014) and Arabidopsis (Cao et al., 2014).
In summary, we present a re‐annotation of the VvLYK gene family and demonstrate that two AtCERK1/LYK1 orthologs, VvLYK1‐1 and VvLYK1‐2, are involved in chitooligosaccharide signalling. Elucidating components of PAMP‐triggered immunity in grapevine opens the possibility of developing grapevine varieties with durable resistance against fungal pathogens. E. necator has adapted to successfully infect grapevine by evolving host‐specific effector proteins that target and re‐programme the signalling pathways that lead to PAMP‐triggered immunity. The introduction of PRRs from a closely related species that can function in V. vinifera but are not modulated by E. necator's specific effector suite has the potential to restore PAMP‐triggered immunity against this adapted pathogen (Heath, 2000; Lee et al., 2016). The proof of concept for this approach was demonstrated by the expression of the Arabidopsis PRR EFR in N. benthamiana, tomato, rice and wheat plants which conferred greater resistance against a range of phytopathogenic bacteria (Lacombe et al., 2010; Lu et al., 2015; Schoonbeek et al., 2015; Zipfel et al., 2006). Several components of the chitin‐signalling network are known targets of numerous pathogen effector proteins (van den Burg et al., 2006; van Esse et al., 2007, 2008; Gimenez‐Ibanez et al., 2009; Mentlak et al., 2012; Yamaguchi et al., 2013; Zeng et al., 2012), supporting the hypothesis that the chitin‐signalling network is an excellent candidate for enhancing the grapevine immune response.
Thus, further experiments will be necessary to gain a better understanding of how grapevine cells specifically perceive different chitooligosaccharides via these complex receptors and to determine the role of each member of the VvLYK multigene family, particularly during its interactions with both beneficial and pathogenic microbes.
Experimental procedures
Plant, cell culture and fungal materials
Arabidopsis thaliana wild‐type (WT) Columbia (Col‐0), mutant Atcerk1 (GABI‐Kat_096F09, allele Atcerk1‐2; (Gimenez‐Ibanez et al., 2009) or transgenic lines Atcerk1/35S::VvLYK1‐1/3 and Atcerk1/LexA::VvLYK1‐2‐GFP were grown under a 10/14‐h day/night cycle at 20/18 °C (Trdá et al., 2014). For in vitro culture, Arabidopsis plants were grown on solid or in liquid half Murashige and Skoog (MS) medium including Nitsch vitamins (M0256; Duchefa, Haarlem, the Netherlands) supplied with 10 g/L sucrose. Seedlings were grown at 20 °C (day) or 18 °C (night) with a 14‐h photoperiod.
Grapevine (V. vinifera cvs Cabernet Sauvignon and Marselan) cuttings were grown in a greenhouse until they had developed 6–8 leaves. The second and third youngest adult leaves from each plant were used for experiments, as previously indicated (Steimetz et al., 2012). Grapevine cells (V. vinifera cv. Gamay) were cultivated as described in Vandelle et al. (2006). For all experiments, 7‐day‐old cultures were diluted twice with new medium 24 h prior to use.
Grapevine powdery mildew (E. necator – isolate APC) was maintained detached leaves of V. vinifera cv. Cabernet Sauvignon as previously described (Donald et al., 2002). Grapevine downy mildew (P. viticola – isolate collected from a Burgundy vineyard) was routinely maintained on V. vinifera cv. Marselan plants as previously described (Steimetz et al., 2012).
Elicitors
Chitin and chitosan hexamer, with a degree of acetylation (DA) of 99.9% and 0.1%, respectively, were provided by Elicityl (Crolles, France). They were extracted from exoskeletons of crustaceans, hydrolysed, purified by chromatography and finally their degree of polymerization (DP) and DA were verified by 1H NMR analysis. The crab shell chitin NA‐COS‐Y (Lloyd et al., 2014), was obtained from Yaizu Suisankagaku Industry Co. (Yaizu, Japan). All the above mentioned chitooligosaccharides were dissolved in sterile ultrapure water (pH 8.5) at a concentration of 1 or 10 mg/mL. Sulphated laminarin (PS3), used as a potent inducer of grapevine resistance (Gauthier et al., 2014), was provided by Goëmar Laboratories and dissolved in sterile ultrapure water.
The flagellin‐derived flg22 peptide from Xanthomonas campestris pv campestris strain 305 (QRLSSGLRINSAKDDAAGLAIS) was purchased from Proteogenix and dissolved in sterile ultra‐pure water at 1 mm, as previously described (Trdá et al., 2014).
MAPK activation
Grapevine cells were equilibrated as described in Dubreuil‐Maurizi et al. (2010), then treated with chitooligosaccharides (100 μg/mL) or water (as control) and harvested at 0, 5, 10, 20, 40 and 60 min post‐treatment. MAPK activation was detected after immunoblotting of the extracted proteins using anti‐p42/44‐phospho‐ERK antibody (Cell Signaling, Danvers, MA). Transfer quality and homogeneous loading were checked by Ponceau red staining.
For Arabidopsis plantlets, 10‐ to 15‐day‐old liquid‐grown seedlings were equilibrated for 24 h in fresh half MS medium. β‐estradiol (10 μm) was added 1 h before elicitor treatment (1 mg/mL) for inducible transgenic lines Atcerk1/LexA::VvLYK1‐2‐GFP. Seedling samples were harvested 10 min after chitin or chitosan treatment.
Analysis of defense gene expression by quantitative polymerase chain reaction (qPCR)
For defense gene expression kinetics using grapevine cell suspensions, the cell culture density was adjusted to 0.1 g FWC/mL with NN medium, 16 h prior to experiment. Cells were then treated with 100 μg/mL chitooligosaccharides or water (as control) and harvested at 1 h post‐treatment by filtration on GF/A filters.
For Arabidopsis, 10‐ to 15‐day‐old seedlings grown on solid half MS medium were transferred in liquid medium 2 days before treatment in a 24‐well microtitre plate. β‐estradiol (10 μm) was added 1 h before treatment with 1 mg/mL of chitooligosaccharides for 2 h.
For both cells and seedlings, tissues were briefly ground before the addition of TRIzol® (Invitrogen, Life Technologies, Saint‐Aubin, France). RNA extraction was then carried out following the manufacturer's instructions (Invitrogen). Reverse transcription was performed using Superscript III (Invitrogen) for cells or M‐MLV reverse transcriptase (Invitrogen) for seedlings, following the manufacturer's protocol. Real‐time qPCR was carried out as described previously (Trdá et al., 2014), except that a 1:100 dilution of cDNA was used. The relative transcript level was calculated using the comparative ΔΔCt method (Livak and Schmittgen, 2001) with the previously validated grapevine VvEF1α (Dubreuil‐Maurizi et al., 2010; Reid et al., 2006) or the Arabidopsis At4g26410 (Czechowski et al., 2005) housekeeping gene as internal control for normalization (AtOLI in Table S3).
Confocal microscopy
Confocal microscopy was performed using a Leica TCS SP2‐AOBS confocal laser scanning microscope with a 40X oil‐immersion objective (numerical aperture 1.25; Leica, Nanterre, France). Inducible transgenic lines were sprayed with 200 μm β‐estradiol, 4 h before visualization. Leaf segments were mounted in ultra‐pure water or in 1 m NaCl solution for plasmolysis experiments. For FM4‐64 staining, samples were incubated in 8 μm FM4‐64 solution in water for 10 min prior to observation. Fluorescent markers were visualized at 488 nm. GFP and FM4‐64 emissions were bandpass filtered at 500–525 nm and 616–694 nm, respectively.
Botrytis and downy mildew assays
Leaves from the second and third adult top leaves of at least three grapevine plants were first sprayed on both sides with elicitor solution in 0.1% surfactant (Deshcofix) or surfactant alone (control) for 48 h.
For B. cinerea infection assays, 36 leaf discs (1.9 cm diameter) were incubated on moist Whatmann paper and inoculated on the upper surface with 1000 conidia in a 20 μL‐droplet of potato dextrose broth (PDB), ¼ diluted. Inoculated discs were placed in a plastic box maintained in 100% humidity under a 10/14 h day/night cycle at 20/18 °C. Infection intensity was assessed 3 days post‐inoculation (dpi) by measuring the macerated lesion diameter.
For P. viticola infection, the lower leaf surface was sprayed with a freshly prepared suspension (2.104 sporangia/mL) and plants were maintained in 100% humidity for 2 h. Leaf discs (1 cm diameter) were cut, transferred onto moist Whatmann paper in a plastic box and maintained in 100% humidity under a 10/14 h day/night cycle at 20/18 °C. Infection intensity was assessed at 8 dpi by measuring the sporulating area using image analysis Visilog 6.9 software (Kim Khiook et al., 2013).
Powdery mildew penetration assay on transgenic Arabidopsis
Four‐week‐old Arabidopsis plants were used to assess powdery mildew penetration efficiency. Two leaves per plant were infected with E. necator using a fine paintbrush. Detached leaf material was sampled 48 hpi and stained with trypan blue according to Koch and Slusarenko (1990). Fungal structures were visualized using a Zeiss (Göttingen, Germany) Axioscop 2 light microscope. A minimum of 100 germinated spores were scored on each leaf. Successful penetration of epidermal cells (% penetrated cells) was indicated by the presence of a haustorium or a secondary hyphae.
Phylogenetic analysis of the VvLYK family
Proteins were aligned with the CLUSTAL W program (Tables S1 and S2). The Maximum Likelihood phylogenetic tree was generated with the MEGA7 software (Kumar et al., 2016), using a bootstrapping of 1000 replications.
Expression analysis of VvLYK genes in pathogen‐infected grape tissues
Young glossy V. vinifera cv. Cabernet Sauvignon leaves of similar developmental stage (~6 cm in diameter) were inoculated with E. necator conidia as described previously (Donald et al., 2002). The leaves were incubated at 23 °C under a 16 h light/8 h dark cycle and sampled at 0, 6, 12 and 24 hpi into liquid nitrogen. Total RNA was extracted from two independent leaves at each time point using the Spectrum Plant Total RNA Kit (Sigma‐Aldrich, St. Louis, MO) and DNase‐treated according to the manufacturer's instructions. RNA quantity and quality were assessed using a 2100 Bioanalyzer (Agilent Technologies, Waldbronn, Germany). Library construction and Illumina RNA sequencing (single end, 100 bp reads) were carried out at the Australian Genome Research Facility (Melbourne, Australia). Reads were mapped to the coding sequences of each predicted VvLYK cDNA sequence using CLC Genomics Workbench v6.0.1. Reads were normalized according to (i) length of the VvLYK reference sequence and (ii) mean relative expression of V. vinifera cv Cabernet sauvignon housekeeping genes: elongation factor 1‐alpha (XM_002284888); glyceraldehyde‐3‐phosphate dehydrogenase (XM_002263109), phosphoenolpyruvate carboxylase (XM_010658735), to produce a Relative Expression Value (REV) at each time point of infection.
For microarray data, RNA was extracted from grape berries infected with B. cinerea then microarray hybridization and data analysis were performed as described in Kelloniemi et al. (2015). All microarray expression data are available at GEO under the entry GSE65969.
Generation of the Atcerk1/VvLYK1 transgenic lines
The coding sequences of VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3 from V. vinifera cv. Cabernet Sauvignon were amplified from grapevine leaf cDNA prepared as previously described (Feechan et al., 2013). Gene‐specific primers were designed with 5′‐Xho I or Xba I restriction sites to facilitate subcloning (Table S3). Amplified products of the expected size were cloned into pCR‐BLUNT vector and verified by sequencing. The coding sequences were subcloned into pART7 vector (Gleave, 1992) between the 35S promoter and OCS terminator sequences. The 35S‐VvLYK1‐OCS expression cassettes were subcloned as Not I fragments into the binary vector pART27 and then transferred into Agrobacterium tumefaciens strain EHA‐105 for Arabidopsis transformation or A. tumefaciens strain GV3101 for agroinfiltration experiments (Williams et al., 2016).
The GFP‐tagged constructs were amplified using primers designed to replace the stop codon with an Ala codon (GCC nucleotides, Table S3). PCR products of the expected size were first directionally subcloned into pENTR™/D‐TOPO® vector (Invitrogen), then inserted into Gateway expression vectors (Karimi et al., 2002) by using Gateway LR Clonase™ II enzyme mix (Invitrogen). The three full‐length coding sequences of VvLYK1‐1, VvLYK1‐2 and VvLYK1‐3 were cloned into pK7FWG2 (kanamycin resistance) to obtain a constitutive overexpression construct (p35S::VvLYK1‐1/‐2/‐3‐GFP) or in pABindGFP (Bleckmann et al., 2010; hygromycin resistance) for a β‐estradiol inducible gene expression (pLexA::VvLYK1‐2‐GFP).
The Arabidopsis Atcerk1 mutant (Gimenez‐Ibanez et al., 2009) was transformed using the floral dip method (Clough and Bent, 1998). Antibiotic resistant transgenic plants were screened in the T1 generation as described previously (Zipfel et al., 2006).
For analysis of VvLYK1‐1/‐2/‐3 transgene expression in the T2 generation, seed collected from selfed T1 lines was sown into soil and plants grown in a controlled growth chamber under a 10/14 h day/night cycle at 24 °C. Leaf material (~50 mg) was sampled from individual T2 segregating lines and the presence of the VvLYK1‐1/‐2/‐3 transgene confirmed by genomic PCR. Positive lines were resampled for total RNA extraction, cDNA synthesis and semi‐quantitative PCR analysis of VvLYK1‐1, VvLYK1‐2 or VvLYK1‐3 transcript expression using primers listed in Table S3.
Accession numbers
Vitis vinifera cv. Cabernet Sauvignon sequences: VvLYK1‐1 (MF177032), VvLYK1‐2 (MF177033), VvLYK1‐3 (MF177034). VvLYK sequences fused with Cter‐GFP tag: VvLYK1‐1‐GFP (MF537036), VvLYK1‐2‐GFP (MF537037) and VvLYK1‐3‐GFP (MF537038).
Funding
This work has been financially supported by ANR (PATRIC project, grant ANR‐13‐KBBE‐0001) (to BP), the Regional Council of Bourgogne Franche‐Comté (PARI grant 2016‐9201AAO050S01636 and FEDER grant BG0005888) and INRA for the funding of Justine Claverie's PhD (grants 2015‐9201AAO048502578 and 29000907), by the Gatsby Charitable Foundation (to CZ), and by the Biotechnology and Biological Sciences Research Council (BBSRC) grants BB/G024936/1 (ERA‐PG PRR CROP (to CZ).
Author contributions
DB, CV, LJD performed most of the experiments; LT, FB, CZ and BP conceived the original screening and research plans; JC, AC, BD and LS provided technical assistance; MCH, FB, CZ and BP supervised the experiments, LT, DB, LJD and PT designed the experiments and analysed the data; DB, LJD, IBD and BP conceived the project and wrote the article with contributions of all the authors; FB, MA and CZ supervised and complemented the writing.
Conflict of interest
The authors declare no conflict of interest.
Supporting information
Figure S1 Chitin and chitosan DP6 did not induce similarly ROS production in Arabidopsis and grapevine cells.
Figure S2 The crab shell chitin also induced defense responses in grapevine cells.
Figure S3 Alignment of AtCERK1/LYK1, the rice OsCERK1 and its putative orthologs in grapevine (VvLYK1‐1/‐2/‐3).
Figure S4 Necrosis observed in response to the over‐expression of VvLYK1‐2 in Nicotiana benthamiana.
Figure S5 Immunodetection of MAPKs in Arabidopsis mutants Atlyk1‐5 in response to chitosan.
Table S1 The Vitis vinifera VvLYK family contains 15 putative genes in the grapevine genome.
Table S2 Percentage of amino acid identity or similarity between VvLYK1‐1/‐2/‐3 and AtCERK1/LYK1 or OsCERK1.
Table S3 Primers used in this study.
Acknowledgements
We thank Angelica Jermakow, Nayana Arunasiri, Adam Wells, Soufiane Nassiri, Agnès Klinguer and Lucile Jacquens for excellent technical assistance. This work has benefited of the expertise of Christine Arnould and Elodie Noirot from the regional Centre of Microscopy/DImaCell platform (Dijon, France). We thank Gary Stacey for the gift of homozygous seeds of GABI‐Kat_096F09 Atcerk1 mutant line. We also thank Andrea Gust and Frederic Brunner for the gift of homozygous seeds of the Atlyk2‐5 mutant lines.
Contributor Information
Ian B. Dry, Email: ian.dry@csiro.au.
Benoit Poinssot, Email: benoit.poinssot@inra.fr.
References
- Akamatsu, A. , Wong, H.L. , Fujiwara, M. , Okuda, J. , Nishide, K. , Uno, K. , Imai, K. et al. (2013) An OsCEBiP/OsCERK1‐OsRacGEF1‐OsRac1 module is an essential early component of chitin‐induced rice immunity. Cell Host Microbe, 13, 465–476. [DOI] [PubMed] [Google Scholar]
- Albert, P. , Miya, A. , Hiratsuka, K. , Kawakami, N. and Shibuya, N. (2006) A high‐throughput evaluation system for Arabidopsis mutants for defense signaling. Plant Biotechnol. 23, 459–466. [Google Scholar]
- Ao, Y. , Li, Z. , Feng, D. , Xiong, F. , Liu, J. , Li, J.F. , Wang, M. et al. (2014) OsCERK1 and OsRLCK176 play important roles in peptidoglycan and chitin signaling in rice innate immunity. Plant J. 80, 1072–1084. [DOI] [PubMed] [Google Scholar]
- Aziz, A. , Poinssot, B. , Daire, X. , Adrian, M. , Bezier, A. , Lambert, B. , Joubert, J.M. et al. (2003) Laminarin elicits defense responses in grapevine and induces protection against Botrytis cinerea and Plasmopara viticola . Mol. Plant Microbe Interact. 16, 1118–1128. [DOI] [PubMed] [Google Scholar]
- Aziz, A. , Trotel‐Aziz, P. , Dhuicq, L. , Jeandet, P. , Couderchet, M. and Vernet, G. (2006) Chitosan oligomers and copper sulfate induce grapevine defense reactions and resistance to gray mold and downy mildew. Phytopathology, 96, 1188–1194. [DOI] [PubMed] [Google Scholar]
- Bleckmann, A. , Weidtkamp‐Peters, S. , Seidel, C.A. and Simon, R. (2010) Stem cell signaling in Arabidopsis requires CRN to localize CLV2 to the plasma membrane. Plant Physiol. 152, 166–176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boller, T. and Felix, G. (2009) A renaissance of elicitors: perception of microbe‐associated molecular patterns and danger signals by pattern‐recognition receptors. Annu. Rev. Plant Biol. 60, 379–406. [DOI] [PubMed] [Google Scholar]
- Boutrot, F. and Zipfel, C. (2017) Function, discovery, and exploitation of plant pattern recognition receptors for broad‐spectrum disease resistance. Annu. Rev. Phytopathol. 55, 257–286. [DOI] [PubMed] [Google Scholar]
- Brandizzi, F. , Irons, S.L. , Johansen, J. , Kotzer, A. and Neumann, U. (2004) GFP is the way to glow: bioimaging of the plant endomembrane system. J. Microsc. 214, 138–158. [DOI] [PubMed] [Google Scholar]
- van den Burg, H.A. , Harrison, S.J. , Joosten, M.H. , Vervoort, J. and de Wit, P.J. (2006) Cladosporium fulvum Avr4 protects fungal cell walls against hydrolysis by plant chitinases accumulating during infection. Mol. Plant Microbe Interact. 19, 1420–1430. [DOI] [PubMed] [Google Scholar]
- Cabrera, J.C. , Messiaen, J. , Cambier, P. and Van Cutsem, P. (2006) Size, acetylation and concentration of chitooligosaccharide elicitors determine the switch from defence involving PAL activation to cell death and water peroxide production in Arabidopsis cell suspensions. Physiol. Plant. 127, 44–56. [Google Scholar]
- Cao, Y. , Liang, Y. , Tanaka, K. , Nguyen, C.T. , Jedrzejczak, R.P. , Joachimiak, A. and Stacey, G. (2014) The kinase LYK5 is a major chitin receptor in Arabidopsis and forms a chitin‐induced complex with related kinase CERK1. eLife, 3, e03766. 10.7554/eLife.03766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clough, S. and Bent, A. (1998) Floral dip: a simplified method for Agrobacterium‐mediated transformation of Arabidopsis thaliana . Plant J. 16, 735–743. [DOI] [PubMed] [Google Scholar]
- Czechowski, T. , Stitt, M. , Altmann, T. , Udvardi, M.K. and Scheible, W.R. (2005) Genome‐wide identification and testing of superior reference genes for transcript normalization in Arabidopsis. Plant Physiol. 139, 5–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodds, P. and Rathjen, J. (2010) Plant immunity: towards an integrated view of plant‐pathogen interactions. Nat. Rev. Genet. 11, 539–548. [DOI] [PubMed] [Google Scholar]
- Donald, T.M. , Pellerone, F. , Adam‐Blondon, A.F. , Bouquet, A. , Thomas, M.R. and Dry, I.B. (2002) Identification of resistance gene analogs linked to a powdery mildew resistance locus in grapevine. Theor. Appl. Genet. 104, 610–618. [DOI] [PubMed] [Google Scholar]
- Dubreuil‐Maurizi, C. , Trouvelot, S. , Frettinger, P. , Pugin, A. , Wendehenne, D. and Poinssot, B. (2010) beta‐Aminobutyric acid primes an NADPH oxidase‐dependent reactive oxygen species production during grapevine‐triggered immunity. Mol. Plant Microbe Interact. 23, 1012–1021. [DOI] [PubMed] [Google Scholar]
- van Esse, H.P. , Bolton, M.D. , Stergiopoulos, I. , de Wit, P.J. and Thomma, B.P. (2007) The chitin‐binding Cladosporium fulvum effector protein Avr4 is a virulence factor. Mol. Plant Microbe Interact. 20, 1092–1101. [DOI] [PubMed] [Google Scholar]
- van Esse, H.P. , Van't Klooster, J.W. , Bolton, M.D. , Yadeta, K.A. , van Baarlen, P. , Boeren, S. , Vervoort, J. et al. (2008) The Cladosporium fulvum virulence protein Avr2 inhibits host proteases required for basal defense. Plant Cell, 20, 1948–1963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Feechan, A. , Jermakow, A.M. , Ivancevic, A. , Godfrey, D. , Pak, H. , Panstruga, R. and Dry, I.B. (2013) Host cell entry of powdery mildew is correlated with endosomal transport of antagonistically acting VvPEN1 and VvMLO to the papilla. Mol. Plant Microbe Interact. 26, 1138–1150. [DOI] [PubMed] [Google Scholar]
- Felix, G. , Baureithel, K. and Boller, T. (1998) Desensitization of the perception system for chitin fragments in tomato cells. Plant Physiol. 117, 643–650. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gauthier, A. , Trouvelot, S. , Kelloniemi, J. , Frettinger, P. , Wendehenne, D. , Daire, X. , Joubert, J.M. et al. (2014) The sulfated laminarin triggers a stress transcriptome before priming the SA‐ and ROS‐dependent defenses during grapevine's induced resistance against Plasmopara viticola . PLoS ONE, 9, e88145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gimenez‐Ibanez, S. , Ntoukakis, V. and Rathjen, J.P. (2009) The LysM receptor kinase CERK1 mediates bacterial perception in Arabidopsis. Plant Signal. Behav. 4, 539–541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gleave, A.P. (1992) A versatile binary vector system with a T‐DNA organisational structure conducive to efficient integration of cloned DNA into the plant genome. Plant Mol. Biol. 20, 1203–1207. [DOI] [PubMed] [Google Scholar]
- Gong, B.Q. , Xue, J. , Zhang, N. , Xu, L. , Yao, X. , Yang, Q.J. , Yu, Y. et al. (2017) Rice chitin receptor OsCEBiP is not a transmembrane protein but targets the plasma membrane via a GPI anchor. Mol. Plant, 10, 767–770. [DOI] [PubMed] [Google Scholar]
- Gubaeva, E. , Gubaev, A. , Melcher, R. , Cord‐Landwehr, S. , Singh, R. , Gueddari, N.E.E. and Moerschbacher, B.M. (2018) ‘Slipped sandwich’ model for chitin and chitosan perception in Arabidopsis. Mol. Plant Microbe Interact. 10.1094/MPMI-04-18-0098-R. [DOI] [PubMed] [Google Scholar]
- Gust, A.A. , Willmann, R. , Desaki, Y. , Grabherr, H.M. and Nürnberger, T. (2012) Plant LysM proteins: modules mediating symbiosis and immunity. Trends Plant Sci. 17, 495–502. [DOI] [PubMed] [Google Scholar]
- Hayafune, M. , Berisio, R. , Marchetti, R. , Silipo, A. , Kayama, M. , Desaki, Y. , Arima, S. et al. (2014) Chitin‐induced activation of immune signaling by the rice receptor CEBiP relies on a unique sandwich‐type dimerization. Proc. Natl Acad. Sci. USA, 111, E404–E413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath, M.C. (2000) Nonhost resistance and nonspecific plant defenses. Curr. Opin. Plant Biol. 3, 315–319. [DOI] [PubMed] [Google Scholar]
- Jaillon, O. , Aury, J.M. , Noel, B. , Policriti, A. , Clepet, C. , Casagrande, A. , Choisne, N. et al. (2007) The grapevine genome sequence suggests ancestral hexaploidization in major angiosperm phyla. Nature, 449, 463–U465. [DOI] [PubMed] [Google Scholar]
- Jones, J.D.G. and Dangl, J.L. (2006) The plant immune system. Nature, 444, 323–329. [DOI] [PubMed] [Google Scholar]
- Kaku, H. , Nishizawa, Y. , Ishii‐Minami, N. , Akimoto‐Tomiyama, C. , Dohmae, N. , Takio, K. , Minami, E. et al. (2006) Plant cells recognize chitin fragments for defense signaling through a plasma membrane receptor. Proc. Natl Acad. Sci. USA, 103, 11086–11091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Karimi, M. , Inzé, D. and Depicker, A. (2002) GATEWAY vectors for Agrobacterium‐mediated plant transformation. Trends Plant Sci. 7, 193–195. [DOI] [PubMed] [Google Scholar]
- Kelloniemi, J. , Trouvelot, S. , Héloir, M.C. , Simon, A. , Dalmais, B. , Frettinger, P. , Cimerman, A. et al. (2015) Analysis of the molecular dialogue between gray mold (Botrytis cinerea) and grapevine (Vitis vinifera) reveals a clear shift in defense mechanisms during berry ripening. Mol. Plant Microbe Interact. 28, 1167–1180. [DOI] [PubMed] [Google Scholar]
- Kim Khiook, I.L. , Schneider, C. , Heloir, M.C. , Bois, B. , Daire, X. , Adrian, M. and Trouvelot, S. (2013) Image analysis methods for assessment of H2O2 production and Plasmopara viticola development in grapevine leaves: application to the evaluation of resistance to downy mildew. J. Microbiol. Methods, 95, 235–244. [DOI] [PubMed] [Google Scholar]
- Koch, E. and Slusarenko, A. (1990) Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell, 2, 437–445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kumar, S. , Stecher, G. and Tamura, K. (2016) MEGA7: molecular evolutionary genetics analysis version 7.0 for bigger datasets. Mol. Biol. Evol. 33, 1870–1874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lacombe, S. , Rougon‐Cardoso, A. , Sherwood, E. , Peeters, N. , Dahlbeck, D. , van Esse, H.P. , Smoker, M. et al. (2010) Interfamily transfer of a plant pattern‐recognition receptor confers broad‐spectrum bacterial resistance. Nat. Biotechnol. 28, 365–369. [DOI] [PubMed] [Google Scholar]
- Lee, S. , Whitaker, V.M. and Hutton, S.F. (2016) Mini review: potential applications of non‐host resistance for crop improvement. Front. Plant Sci. 7, 997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu, T. , Liu, Z. , Song, C. , Hu, Y. , Han, Z. , She, J. , Fan, F. et al. (2012) Chitin‐induced dimerization activates a plant immune receptor. Science, 336, 1160–1164. [DOI] [PubMed] [Google Scholar]
- Liu, S. , Wang, J. , Han, Z. , Gong, X. , Zhang, H. and Chai, J. (2016) Molecular mechanism for fungal cell wall recognition by rice chitin receptor OsCEBiP. Structure, 24, 1192–1200. [DOI] [PubMed] [Google Scholar]
- Livak, K.J. and Schmittgen, T.D. (2001) Analysis of relative gene expression data using real‐time quantitative PCR and the 2(T)(‐Delta Delta C) method. Methods, 25, 402–408. [DOI] [PubMed] [Google Scholar]
- Lloyd, S.R. , Schoonbeek, H.J. , Trick, M. , Zipfel, C. and Ridout, C.J. (2014) Methods to study PAMP‐triggered immunity in Brassica species. Mol. Plant Microbe Interact. 27, 286–295. [DOI] [PubMed] [Google Scholar]
- Lu, F. , Wang, H. , Wang, S. , Jiang, W. , Shan, C. , Li, B. , Yang, J. et al. (2015) Enhancement of innate immune system in monocot rice by transferring the dicotyledonous elongation factor Tu receptor EFR. J. Integr. Plant Biol. 57, 641–652. [DOI] [PubMed] [Google Scholar]
- Mentlak, T.A. , Kombrink, A. , Shinya, T. , Ryder, L.S. , Otomo, I. , Saitoh, H. , Terauchi, R. et al. (2012) Effector‐mediated suppression of chitin‐triggered immunity by magnaporthe oryzae is necessary for rice blast disease. Plant Cell, 24, 322–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miya, A. , Albert, P. , Shinya, T. , Desaki, Y. , Ichimura, K. , Shirasu, K. , Narusaka, Y. et al. (2007) CERK1, a LysM receptor kinase, is essential for chitin elicitor signaling in Arabidopsis. Proc. Natl Acad. Sci. USA, 104, 19613–19618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Paparella, C. , Savatin, D.V. , Marti, L. , De Lorenzo, G. and Ferrari, S. (2014) The Arabidopsis LYSIN MOTIF‐CONTAINING RECEPTOR‐LIKE KINASE3 regulates the cross talk between immunity and abscisic acid responses. Plant Physiol. 165, 262–276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petutschnig, E. , Jones, A. , Serazetdinova, L. , Lipka, U. and Lipka, V. (2010) The lysin motif receptor‐like kinase (LysM‐RLK) CERK1 is a major chitin‐binding protein in Arabidopsis thaliana and subject to chitin‐induced phosphorylation. J. Biol. Chem. 285, 28902–28911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pietraszewska‐Bogiel, A. , Lefebvre, B. , Koini, M.A. , Klaus‐Heisen, D. , Takken, F.L. , Geurts, R. , Cullimore, J.V. et al. (2013) Interaction of Medicago truncatula lysin motif receptor‐like kinases, NFP and LYK3, produced in Nicotiana benthamiana induces defence‐like responses. PLoS ONE, 8, e65055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Piquerez, S.J. , Harvey, S.E. , Beynon, J.L. and Ntoukakis, V. (2014) Improving crop disease resistance: lessons from research on Arabidopsis and tomato. Front. Plant Sci. 5, 671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Poinssot, B. , Vandelle, E. , Bentéjac, M. , Adrian, M. , Levis, C. , Brygoo, Y. , Garin, J. et al. (2003) The endopolygalacturonase 1 from Botrytis cinerea activates grapevine defense reactions unrelated to its enzymatic activity. Mol. Plant Microbe Interact. 16, 553–564. [DOI] [PubMed] [Google Scholar]
- Povero, G. , Loreti, E. , Pucciariello, C. , Santaniello, A. , Di Tommaso, D. , Di Tommaso, G. , Kapetis, D. et al. (2011) Transcript profiling of chitosan‐treated Arabidopsis seedlings. J. Plant. Res. 124, 619–629. [DOI] [PubMed] [Google Scholar]
- Qiu, W. , Feechan, A. and Dry, I. (2015) Current understanding of grapevine defense mechanisms against the biotrophic fungus (Erysiphe necator), the causal agent of powdery mildew disease. Hortic. Res. 2, 15020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reid, K.E. , Olsson, N. , Schlosser, J. , Peng, F. and Lund, S.T. (2006) An optimized grapevine RNA isolation procedure and statistical determination of reference genes for real‐time RT‐PCR during berry development. BMC Plant Biol. 6, 27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schoonbeek, H.J. , Wang, H.H. , Stefanato, F.L. , Craze, M. , Bowden, S. , Wallington, E. , Zipfel, C. et al. (2015) Arabidopsis EF‐Tu receptor enhances bacterial disease resistance in transgenic wheat. New Phytol. 206, 606–613. [DOI] [PubMed] [Google Scholar]
- Segonzac, C. , Feike, D. , Gimenez‐Ibanez, S. , Hann, D.R. , Zipfel, C. and Rathjen, J.P. (2011) Hierarchy and roles of pathogen‐associated molecular pattern‐induced responses in Nicotiana benthamiana . Plant Physiol. 156, 687–699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shimizu, T. , Nakano, T. , Takamizawa, D. , Desaki, Y. , Ishii‐Minami, N. , Nishizawa, Y. , Minami, E. et al. (2010) Two LysM receptor molecules, CEBiP and OsCERK1, cooperatively regulate chitin elicitor signaling in rice. Plant J. 64, 204–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shinya, T. , Nakagawa, T. , Kaku, H. and Shibuya, N. (2015) Chitin‐mediated plant‐fungal interactions: catching, hiding and handshaking. Curr. Opin. Plant Biol. 26, 64–71. [DOI] [PubMed] [Google Scholar]
- Steimetz, E. , Trouvelot, S. , Gindro, K. , Bordier, A. , Poinssot, B. , Adrian, M. and Daire, X. (2012) Influence of leaf age on induced resistance in grapevine against Plasmopara viticola . Physiol. Mol. Plant Pathol. 79, 89–96. [Google Scholar]
- Tanabe, S. , Okada, M. , Jikumaru, Y. , Yamane, H. , Kaku, H. , Shibuya, N. and Minami, E. (2006) Induction of resistance against rice blast fungus in rice plants treated with a potent elicitor, N‐acetylchitooligosaccharide. Biosci. Biotechnol. Biochem. 70, 1599–1605. [DOI] [PubMed] [Google Scholar]
- Trdá, L. , Fernandez, O. , Boutrot, F. , Héloir, M.C. , Kelloniemi, J. , Daire, X. , Adrian, M. et al. (2014) The grapevine flagellin receptor VvFLS2 differentially recognizes flagellin‐derived epitopes from the endophytic growth‐promoting bacterium Burkholderia phytofirmans and plant pathogenic bacteria. New Phytol. 201, 1371–1384. [DOI] [PubMed] [Google Scholar]
- Trdá, L. , Boutrot, F. , Claverie, J. , Brulé, D. , Dorey, S. and Poinssot, B. (2015) Perception of pathogenic or beneficial bacteria and their evasion of host immunity: pattern recognition receptors in the frontline. Front. Plant Sci. 6, 219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Trouvelot, S. , Héloir, M.C. , Poinssot, B. , Gauthier, A. , Paris, F. , Guillier, C. , Combier, M. et al. (2014) Carbohydrates in plant immunity and plant protection: roles and potential application as foliar sprays. Front. Plant Sci. 5, 592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vandelle, E. , Poinssot, B. , Wendehenne, D. , Bentejac, M. and Pugin, A. (2006) Integrated signaling network involving calcium, nitric oxide, and active oxygen species but not mitogen‐activated protein kin in BcPG1‐elicited grapevine defenses. Mol. Plant Microbe Interact. 19, 429–440. [DOI] [PubMed] [Google Scholar]
- Vander, P. , Vårum, K.M. , Domard, A. , Eddine El Gueddari, N. and Moerschbacher, B.M. (1998) Comparison of the ability of partially N‐acetylated chitosans and chitooligosaccharides to elicit resistance reactions in wheat leaves. Plant Physiol. 118, 1353–1359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walters, D.R. , Ratsep, J. and Havis, N.D. (2013) Controlling crop diseases using induced resistance: challenges for the future. J. Exp. Bot. 64, 1263–1280. [DOI] [PubMed] [Google Scholar]
- Wan, J. , Zhang, X. , Neece, D. , Ramonell, K. , Clough, S. , Kim, S. , Stacey, M. et al. (2008) A LysM receptor‐like kinase plays a critical role in chitin signaling and fungal resistance in Arabidopsis. Plant Cell, 20, 471–481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wiesel, L. , Newton, A.C. , Elliott, I. , Booty, D. , Gilroy, E.M. , Birch, P.R. and Hein, I. (2014) Molecular effects of resistance elicitors from biological origin and their potential for crop protection. Front. Plant Sci. 5, 655. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Williams, S.J. , Yin, L. , Foley, G. , Casey, L.W. , Outram, M.A. , Ericsson, D.J. , Lu, J. et al. (2016) Structure and function of the TIR domain from the grape NLR protein RPV1. Front. Plant Sci. 7, 1850. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willmann, R. , Lajunen, H. , Erbs, G. , Newman, M. , Kolb, D. , Tsuda, K. , Katagiri, F. et al. (2011) Arabidopsis lysin‐motif proteins LYM1 LYM3 CERK1 mediate bacterial peptidoglycan sensing and immunity to bacterial infection. Proc. Natl Acad. Sci. USA, 108, 19824–19829. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu, J. , Xie, J. , Yan, C. , Zou, X. , Ren, D. and Zhang, S. (2014) A chemical genetic approach demonstrates that MPK3/MPK6 activation and NADPH oxidase‐mediated oxidative burst are two independent signaling events in plant immunity. Plant J. 77, 222–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamaguchi, K. , Yamada, K. , Ishikawa, K. , Yoshimura, S. , Hayashi, N. , Uchihashi, K. , Ishihama, N. et al. (2013) A receptor‐like cytoplasmic kinase targeted by a plant pathogen effector is directly phosphorylated by the chitin receptor and mediates rice immunity. Cell Host Microbe, 13, 347–357. [DOI] [PubMed] [Google Scholar]
- Yin, H. , Du, Y. and Dong, Z. (2016) Chitin oligosaccharide and chitosan oligosaccharide: two similar but different plant elicitors. Front. Plant Sci. 7, 522. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yu, X. , Feng, B. , He, P. and Shan, L. (2017) From chaos to harmony: responses and signaling upon microbial pattern recognition. Annu. Rev. Phytopathol. 55, 109–137. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zeng, L. , Velásquez, A.C. , Munkvold, K.R. , Zhang, J. and Martin, G.B. (2012) A tomato LysM receptor‐like kinase promotes immunity and its kinase activity is inhibited by AvrPtoB. Plant J. 69, 92–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang, X.C. , Cannon, S.B. and Stacey, G. (2009) Evolutionary genomics of LysM genes in land plants. BMC Evol. Biol. 9, 183. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zipfel, C. , Kunze, G. , Chinchilla, D. , Caniard, A. , Jones, J.D.G. , Boller, T. and Felix, G. (2006) Perception of the bacterial PAMP EF‐Tu by the receptor EFR restricts Agrobacterium‐mediated transformation. Cell, 125, 749–760. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1 Chitin and chitosan DP6 did not induce similarly ROS production in Arabidopsis and grapevine cells.
Figure S2 The crab shell chitin also induced defense responses in grapevine cells.
Figure S3 Alignment of AtCERK1/LYK1, the rice OsCERK1 and its putative orthologs in grapevine (VvLYK1‐1/‐2/‐3).
Figure S4 Necrosis observed in response to the over‐expression of VvLYK1‐2 in Nicotiana benthamiana.
Figure S5 Immunodetection of MAPKs in Arabidopsis mutants Atlyk1‐5 in response to chitosan.
Table S1 The Vitis vinifera VvLYK family contains 15 putative genes in the grapevine genome.
Table S2 Percentage of amino acid identity or similarity between VvLYK1‐1/‐2/‐3 and AtCERK1/LYK1 or OsCERK1.
Table S3 Primers used in this study.


