Abstract
A large body of work has gone into understanding the effect of mutations on protein structure and function. Conventional treatments have involved quantifying the change in stability, activity and relaxation rates of the mutants with respect to the wild-type protein. However, it is now becoming increasingly apparent that mutational perturbations consistently modulate the packing and dynamics of a significant fraction of protein residues, even those that are located >10–15 Å from the mutated site. Such long-range modulation of protein features can distinctly tune protein stability and the native conformational ensemble contributing to allosteric modulation of function. In this review, I summarize a series of experimental and computational observations that highlight the incredibly pliable nature of proteins and their response to mutational perturbations manifested via the intra-protein interaction network. I highlight how an intimate understanding of mutational effects could pave the way for integrating stability, folding, cooperativity and even allostery within a single physical framework.
Introduction
Mutations in proteins occur via multiple well-understood molecular mechanisms primarily at the level of DNA contributing to variability in the population. Such variability is the cornerstone of evolution as functionally advantageous mutations get fixed in the presence of a selection pressure. Decades of work on mutations have revealed rich information on protein conformational behavior, binding site identities and thermodynamics, folding mechanisms and allostery. In parallel, understanding and modeling mutational effects has tremendous implications in not just designing proteins with enhanced solubility, stability, and catalytic efficiency, but also to understand evolutionary trajectories of proteins, enzyme evolvability and the contribution of mutant phenotypes to organismal fitness [1–5]. Numerous avenues are currently available to engineer proteins ranging from charge-charge interactions on the protein surface [6] to directed evolution [7] and saturation mutagenesis-based approaches [8,9].
A ‘neutral’ mutation is conventionally defined as a perturbation that has little effect on the organismal fitness (say, functioning of a protein or survival of the organism). However, the same mutation in conjunction with mutations at other sites can have a positive or a negative effect on the fitness landscape. How is this epistatic characteristic enabled? It is important to realize that the interior (surface) of a protein is a highly unique environment determined by the unique protein sequence with specific packing (electrostatic/polar) interactions. Therefore, any perturbation from a mutation is expected to be complex, as it would involve an abrupt reorganization of the evolutionarily tuned interaction network. In fact, it is well recognized in the field of protein NMR that mutations manifest as non-trivial effects on chemical shifts and order parameters of a majority of residues in the protein. However, there is an apparent disconnect between an NMR view of mutational effects (complex changes in multiple terms) and studies that merely quantify the change in stability or folding rate of the mutant compared to the wild-type. In this review, I highlight and summarize some of the recent developments towards resolving this apparent conflict and how a mere consideration of the intra-protein interaction network provides a convincing rationale with several testable predictions.
Mutational effects propagate beyond the firstshell of interactions
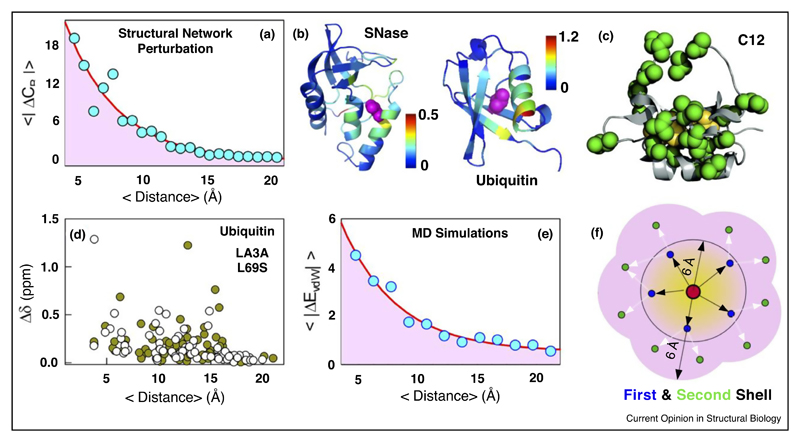
A reductionist approach in viewing proteins is to treat protein residues as nodes and the intra-protein interactions as edges [10]. When the edges are perturbed, either by deletion or by modulation of their strengths, the network properties are modulated not just in the immediate neighborhood but also at longer distances [11•] (Figure 1a). Such propagation and dissipation (used strictly in an equilibrium sense with no kinetic connotation) of perturbations with increasing distances from the source of perturbation is a robust feature of networks [12]. True to this expectation, point mutations in CI2 [13], Ubiquitin [14], T4 lysozyme [15], SSo7d [16], Staphylococcal nuclease [17•] and Protein L [18] all contribute to persistent modulation of chemical shifts, hydrogen-exchange protection factors or dynamics almost throughout the entire structure (for example, see Figure 1b, c). Such long-distance coupling of residues can affect catalysis in enzymes as shown through single-point and double-point mutations (>15 Å from the active site) in dihydrofolate reductase that modulate the rate of hydride transfer by up to three orders of magnitude [19,20]. By measuring chemical shift perturbations (CSPs) of β1-adrenergic receptor upon mutations and ligand binding, Grzesiek and coworkers identified that both the structural perturbations propagate to long distances determining the functionally relevant motions of the transmembrane helices [21•]. A study on Cyclophilin A, a peptidyl prolyl cis–trans isomerase, reported a 30% reduction in the rate of isomerization upon a conservative V29L mutation located nearly 15 Å away from the active site [22•]. While the activity modulation is minor in Cyclophilin, a 1000-fold increase in phosphotriesterase activity could be engineered in a bacterial lactonase by ‘tinkering’ with mutations in the second-shell of the active-site residues [23•]. Such extreme behaviors and context-dependence could be a manifestation of the robustness of the intra-protein interaction network to perturbations wherein the network readjusts to accommodate a residue by altering the dynamics and packing of distant residues. It also suggests that multiple mutations might be required in a protein-specific manner to irreversibly alter the interaction network or the correlated motions and hence the functional output.
Figure 1.
Mutational effects are consistently felt over long distances with evidence available from varied approaches. (a) Average changes in network connectivity, quantified by betweenness centrality (CB), plotted as a function of mean Cα-Cα distance from the mutated site from a ‘toy’ model of Ubiquitin that treats protein residues as nodes and their interactions as edges [11•]. (b) Experimental chemical shift perturbations on mutations mapped on to the structure of SNase (L125A; [17•]) and Ubiquitin (L43A; [14]), respectively. (c) Residues whose NMR order parameters are affected (green) on core mutations of residues shown in yellow in CI2 [13]. Reprinted with permission from Ref. [13], Copyright (2008), American Chemical Society. (d) Experimental chemical shift perturbations as a function of distance from the mutated site for specific ubiquitin mutations [11•,25•]. (e) Mean absolute changes in van der Waals packing interactions (ordinate in kJ mol–1) for seven core substitutions in ubiquitin plotted as a function of distance from the mutated site from all-atom MD simulations (circles) together with an exponential fit (red) [11•]. (f) A cartoon representation of how perturbation of a residue (red) affects not only its first-shell neighbors (blue and interactions as black arrows) but also the neighbors of neighbors (green and interactions as white arrows).
The fact that distal mutations can alter activity (to variable extents) raises questions on whether this is an intrinsic response of proteins to structural perturbations due to the fluid-like nature of the protein interior [24]. True to this expectation, a global analysis of 25 mutations from 12 different protein structures from the viewpoint of chemical shift perturbations revealed that the effect of mutations could be consistently felt even up till 10–20 Å from the mutation site, and is independent of the nature of the mutation, protein type or secondary structure content (for example, see Figure 1d) [25•]. Experimental double-mutant cycles that measure the degree of energetic coupling of one reside to another reveal a similar long-range coupling of residues [26,27]. In all of the cases above, an exponential-like dissipation of network parameters, coupling energy or chemical shift perturbations have been identified highlighting the possibility of a universal function form to describe mutational effects [25•]. The pervasive long-range coupling patterns explain the large conservation of even distal residues (as far as 20–27 Å) around the active site of enzymes [28•]. It is important to emphasize here that such conservation can have varied origins including functional requirements (either at the active site or binding of an effector at an allosteric site), stability (thermodynamic and kinetic), preventing aggregation and so on. As an aside, I would like to point out that long-range structural modulation is not only observed on mutational perturbations, but also on ligand-binding and phosphorylation [25•], similar to the domino-like propagative-cum-dissipative phenomenon observed in repeat proteins [29].
Truncation mutations primarily weaken native interactions
What are the molecular origins of destabilization induced by truncation mutations? An analysis of microsecond-long molecular dynamics trajectories of Ubiquitin WT and seven aliphatic truncation mutations revealed a distinct weakening of packing interactions across nearly the entire protein. The relative residue-level van der Waals interaction energy approaches zero (i.e. no perturbation) only at longer distances from the mutated site (Figure 1e) [11•]. Taking a cue from MD simulations, the effect of truncation mutations was recently modeled by considering two shells of interactions around the perturbed residue [11•]; the first shell accounts for the neighbors within a 6 Å distance from the mutated site, while the second shell accounts for the neighbors of neighbors and thus residues nearly 12 Å from the mutated site. By introducing different destabilization magnitudes in the first- and second-shells, accounting for the nature of mutation and parameterizing them into a statistical mechanical model [30,31], it was possible to reproduce the changes in stabilities of 375 truncation mutations in 19 different proteins with a correlation and slope comparable to that from the multi-parameter FOLDX energy function [11•]. In other words, on accounting for the nature of the mutation (merely from the ratio of atoms in the mutant compared to the WT), the first- and second-shell van der Waals interactions are weakened by 50% and 20%, respectively (Figure 1f). However, since there are many more interactions that define the second-shell, the energetic contributions are near equivalent but distributed throughout the structure. It is important to emphasize the implication of the above statement; for instance, L43, which is located in the hydrophobic core of ubiquitin is therefore connected to ~80% of the 76 residues in ubiquitin, underscoring the extent to which the intraprotein interaction network can be perturbed.
While the discussion above is primarily on truncation mutations, mutations that enhance the molecular volume also decrease protein stability though the mechanism is still unclear. Since the interior of a protein displays the packing density of a solid (despite displaying large dynamics [24,32]), any change in the protein interior environment (that is unique as the sequences themselves are unique) would disrupt interactions, as the protein molecule would struggle to fit in an amino acid with a larger volume. Not surprisingly, mutations that enhance molecular volume also contribute to an exponential-like dependence of the chemical shift perturbations as a function of distance from the mutated site [25•]. The same applies to mutations to glycine that dramatically enhance the basal backbone fluctuations (~4 kJ mol–1 destabilization at 310 K from merely single-site backbone entropy considerations [33]) and could manifest as nontrivial effects on the folding-functional landscape.
Population redistribution, partial unfolding and shifts in the native ensemble
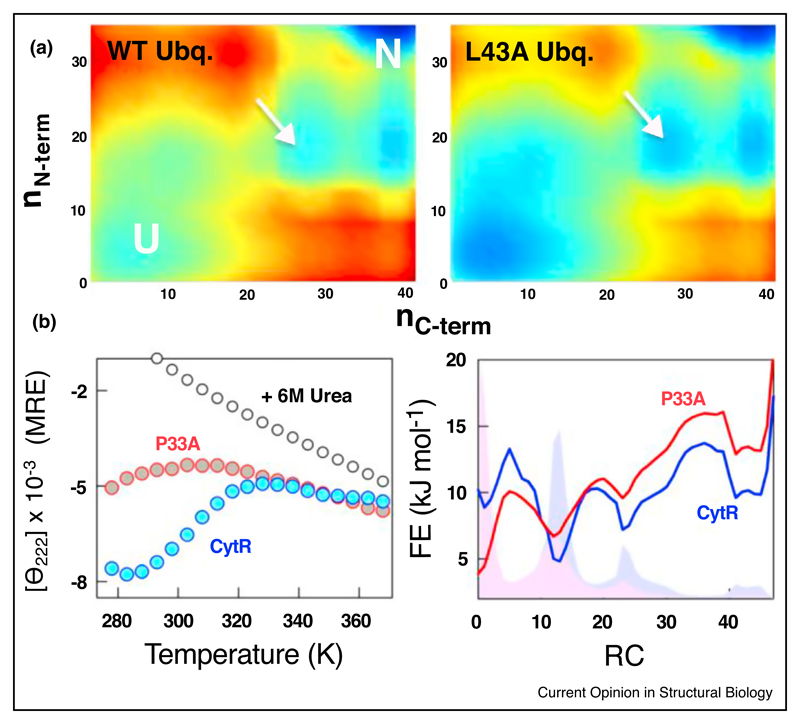
Protein native states are accurately defined as ensembles of multiple conformations or substates that are critical for function. Thermodynamic fluctuations are a feature intrinsic to polymeric protein chains arising from the finite size of protein molecules and the weak nature of the non-covalent interactions [34]. From a statistical mechanical perspective, it can therefore be immediately recognized that upon mutation the statistical weights and hence the probabilities of all the conformations in which the residue is structured would be modulated. This in turn would manifest as shifts in the distribution of conformations in the native ensemble and enhanced population of intermediate or excited states apart from higher unfolded state populations (Figure 2a). This immediately explains why mutational approaches to enhancing excited state populations or intermediates have been immensely successful as in the studies involving T4 lysozyme and Fyn-SH3 domains [15,35]. Similar mutation-induced population redistributions have also been reported in GPCRs [36,37], CAP [38], Ubiquitin [39•], U1A [40], Cyclophilin [22•] and Adenylate kinase [41], with distinct effects on function in each case.
Figure 2.
Stability changes and modulation of the folding-function landscape. (a) Projection of conformations onto a two-dimensional landscape generated from the WSME model for WT ubiquitin and its mutant L43A. nN-term and nC-term represent the number of residues structured in the N- and C-terminii, respectively. The arrow points to intermediate-like states in the landscape that are stabilized on mutations (N stands for native and U for unfolded macrostates). (b) (Left Panel) Changes in the secondary-structure upon mutating a proline to alanine (P33A) in a disordered protein CytR. Note that the proline is present in the loop region connecting two helices and not nucleating a helix. (Right Panel) One-dimensional free energy profiles as a function of the number of structured residues as the reaction coordinate (RC). The landscape is non-trivially modified with the population of a folded-like excited state decreasing on proline substitution [46].
Redistribution of populations in the native ensemble tune functions and downstream signaling responses revealing avenues by which molecular responses to environmental variables could be acquired, the molecular mechanisms of drug-resistance and onset of disease conditions. Work on p97 ATPase and NAD(P)H: quinone oxidoreductase 1 (NQO1) highlight that disease causing mutations shift the conformational substates in a graded manner thus compromising innate activity [42•,43]. Similarly, distal mutations on tryptophan synthase shift the conformational ensemble to the extent of modulating the rate-limiting catalytic step [44]. Recently, destabilizing distal and surface glycine mutations on Adenylate Kinase (AK) have been shown to influence both enzyme activity and substrate affinity [45•]; these mutations destabilize the native ensemble through partial unfolding of AK domains shedding light on how enzymes from psychrophilic organisms could tune their basal activity to compensate for lower thermal energy.
Note that such population redistributions upon mutations need not be restricted to folded proteins but even intrinsically disordered proteins. The outcome of such mutations in disordered proteins is expected to be non-trivial due to the heterogeneous nature of IDP ensembles manifesting as large signal changes in equilibrium [46,47] (Figure 2b), binding affinity [46–49], association and dissociation rate constants [48–51], altered binding transition-state ensembles [50] and even induce liquid-to-solid phase transition [52].
Allosteric mechanisms and paths from mutational perturbations?
It is well established that protein residues are coupled to each other through both the hydrogen-bond network and packing interactions leading to correlated motions or fluctuations in equilibrium that are critical for function. Such intra-molecular interaction-networks serve as channels for signal transmission playing a prominent role in dynamic allostery [53–55] (modulation of activity without changes in the overall structure upon distal perturbations). Since mutational effects are not localized it is natural to expect that they also modulate the communication network and hence influence function. In fact, a wide range of mutational tolerance and non-trivial functional outcomes has been observed in ubiquitin [56] and light-oxygen-voltage domain 2 [57]. Mutational studies on PDZ report that nearly the entire protein interaction network has evolved towards optimizing activity and potentially to minimize cross-reactivity while uncovering allosteric singling paths [58,59]. Similarly, extensive alanine-scanning mutagenesis has revealed that a significant fraction of residues (~30%) in the human liver pyruvate kinase can influence the binding to its substrate, PEP, in the presence of the activator Fru-1, 6-BP [60•].
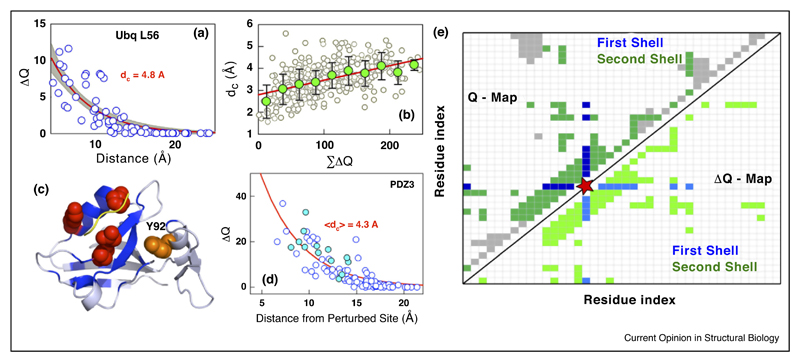
Since the precise mechanistic details of allostery are yet to be completely established with the possibility of large context dependence, a whole-protein mutagenesis (alanine-scanning, for example) might be a way forward to test mechanistic models without preconceived notions or biases in the analysis, as also argued for in a recent work [61]. An in silico version of this is the structural perturbation approach (SPA; [62•]) wherein every residue is mutated to alanine or glycine (perturbation) and the immediate environment is probed for the presence of strong packing (also see [63,64] for similar methods). Following this, two parameters are extracted for every residue — the coupling distance (dC) and the total perturbation (ΣΔQ) — that provide information on the extent to which the perturbed residue is coupled to its neighbors (Figure 3a, b). Interestingly, the SPA reproduces the results of statistical coupling analysis (SCA [65]) with just a single structure as input and from mere distance considerations (Figure 3c, d) while revealing additional allosteric sites that could be experimentally tested [62•].
Figure 3.
A structural perturbation approach (SPA), an in silico version of alanine-scanning mutagenesis, towards understanding allosteric coupling [62•]. (a) Changes in packing density (∆Q) as a function of distance upon L56A substitution in ubiquitin extracted through the SPA. The red curve is a single-exponential fit highlighting a coupling distance (dC) of 4.8 Å. (b) The coupling distance is approximately linearly related (red line) to the overall perturbation magnitude (abscissa). This indicates that larger residues that are located in the protein interior are extensively coupled, the perturbation of which can contribute to significant changes in packing density and hence dynamics-folding-function behaviors. The gray circles are from a perturbation analysis of all residues in six different proteins while the green circles are block averages. (c) Perturbation of the residues in red in PDZ3 reveals strong coupling to several residues (blue) that can potentially modulate the binding of the peptide (yellow). Note that Y92, a PTM site, is located in vicinity of the perturbed residues indicating how information on PTM could be transmitted to the binding site. (d) An SPA of the three residues shown in red again results in an exponential dependence of ∆Q (blue circles). The coupled residues identified using the statistical coupling analysis (SCA) is shown as filled circles are in good agreement with that predicted from the SPA. (e) A schematic of how longrange coupling can be extracted from the SPA. The perturbed residue is shown in red, unperturbed residues in gray, the contact map as the upper left triangular matrix and the ∆Q-map as the lower right triangular matrix. On perturbing a residue in red, apart from the first-shell neighbors (dark blue), the second shell is also affected (dark green) that constitutes a significant fraction in a small single-domain protein. The effective number of interactions that are lost are shown in the ∆Q-map (a uniform coloring code is employed for the sake of clarity).
Deducing allosteric communication paths is almost entirely the purview of computational methods [66–71]. Since signal transmission should occur through the network of non-covalent interactions in the protein interior (any surface propagation would be quickly damped by solvent collisions) it is necessary that such methods also reproduce mutational destabilization thermodynamics. This would provide an independent test for the relative strengths of non-covalent interactions at different regions of the protein structure required to model signal propagation as put forth by Ernesto Freire [72]: “ … the propagation of binding signals should obey precise thermodynamic rules, and the location of allosteric sites should be dictated by thermodynamic stability criteria within the protein.” The overall features of the signaling paths should not only be consistent with the available chemical shift perturbations upon mutations (at least at the level of relative trends) but also the mutation-induced destabilization providing a sound equilibrium-thermodynamic framework for modeling allosteric communication networks.
On folding mechanisms and cooperativity
Putative transition state structures and hence protein folding mechanisms are generally inferred from Φ-value analysis that involves measuring the changes in stability and (un)folding rates upon point mutations [73]. One of the primary assumptions of this approach is that the recommended truncation mutations in the protein interior [73] influence only the nearest neighbors (or the first-shell) providing an intimate view of the degree of structure in the transition state. Given that mutational effects consistently propagate beyond the first shell of interactions and modulate both packing and dynamics of distant residues, it is likely that Φ-values represent an effective average of multiple energetic and entropic terms and not just the extent of local structure. The frequently observed folding Φ-value of ~0.3 (independent of protein type, structure or mutation [74]) could therefore represent the fraction of stabilization free energy gained during folding [75,76]. It remains to be seen if it is possible to disentangle the energetic and structural contributions to Φ-values from computational studies. Moreover, a two-state-like treatment does not account for altered dynamics or population redistributions within the native well, necessitating a shift towards the use of more detailed thermodynamic models that could potentially provide exciting insights.
Cooperativity is feature intrinsic to systems held together by weak non-covalent interactions, and in proteins it is quantified in terms of the slope of the unfolding curve, folding barrier heights or other extent of similarity of atomic-level unfolding curves [77–79]. Cooperativity features are thus intrinsically related to the contact environment of residues in the protein. Therefore, it should be possible to perform a series of alanine-scanning experiments and iteratively identify the extent to which the interaction shell radius (or the coupling distance [11•,25•,62•]; Figure 3a) needs to be modified to account for the destabilization thermodynamics. In fact, the structural perturbation approach (SPA) can be extended to generate a ΔQ-map (similar to the contact map, or Q-map; Figure 3e) that highlights the degree of coupling of every residue with its neighbors [62•]; such maps emphasize that second-shell interactions around a residue should be formed to consider the residue to be ‘folded’ (Figure 3e). It remains to be seen if such perturbation-based approaches alone are sufficient to generate cooperativity indices or regions of structure that are more coupled (or more locally stable and hence more cooperative) than others.
Concluding remarks
Recent experiments and computational works highlight that mutations in the protein interior manifest as altered chemical-shifts, order parameters (dynamics), HX protection factors, and modulation of packing interactions involving a significant fraction of protein residues. Accordingly, mutational perturbations are better understood in terms of their impact on the underlying interaction network or correlated motions and likely serve as the evolutionary first-step towards altered protein activity, diseased states, functional promiscuity and fold-switching. Given the robustness of the interaction network to mutations, successful engineering of enzymatic activity might require multiple perturbations [80–82] while in some cases single or double-mutations alone have been successful highlighting a certain degree of context-dependence. Deciphering this context dependence could be the way forward to engineer protein function at will.
Surface mutations also contribute to complex alteration of folded and unfolded ensembles apart from folding mechanisms [83–86]. It is therefore possible that even apparently neutral mutations modulate specific features of the native conformational ensemble which is however invisible or challenging to identify in the absence of a functional output. These observations underline the need to expand the outlook on mutational outcomes to include perturbation of native conformational ensembles, populated intermediate- and excited-states and redistribution of dynamics. Since mutations alter the evolutionarily constrained intra-molecular network of interactions, they are also expected to reshape the folding funnel and hence tune folding mechanisms, aspects that could be explored with advanced computational protocols and experiments. However, there is still a distance to travel in understanding the impact of mutations at a distal site in more functional terms — will a specific mutation at a distant site enhance or diminish binding affinity to the substrate? Detailed and intimate characterization of mutational effects supplemented with quantitative modeling could thus open up new vistas with implications in protein design, function and allostery.
Acknowledgements
This work was supported by the Wellcome Trust/DBT India Alliance Intermediate Fellowship to A. N. N. (IA/I/15/1/501837).
Footnotes
Conflict of interest
The author declares no conflict of interest.
References and recommended reading
Papers of particular interest, published within the period of review, have been highlighted as:
• of special interest
- 1.Tokuriki N, Tawfik DS. Stability effects of mutations and protein evolvability. Curr Opin Struct Biol. 2009;19:596–604. doi: 10.1016/j.sbi.2009.08.003. [DOI] [PubMed] [Google Scholar]
- 2.Serohijos AWR, Shakhnovich EI. Merging molecular mechanism and evolution: theory and computation at the interface of biophysics and evolutionary population genetics. Curr Opin Struct Biol. 2014;26:84–91. doi: 10.1016/j.sbi.2014.05.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bershtein S, Serohijos AW, Shakhnovich EI. Bridging the physical scales in evolutionary biology: from protein sequence space to fitness of organisms and populations. Curr Opin Struct Biol. 2017;42:31–40. doi: 10.1016/j.sbi.2016.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Sikosek T, Chan HS. Biophysics of protein evolution and evolutionary protein biophysics. J R Soc Interface. 2014;11 doi: 10.1098/rsif.2014.0419. 20140419. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Bastolla U, Dehouck Y, Echave J. What evolution tells us about protein physics, and protein physics tells us about evolution. Curr Opin Struct Biol. 2017;42:59–66. doi: 10.1016/j.sbi.2016.10.020. [DOI] [PubMed] [Google Scholar]
- 6.Sanchez-Ruiz JM, Makhatadze GI. To charge or not to charge? Trends Biotechnol. 2001;19:132–135. doi: 10.1016/s0167-7799(00)01548-1. [DOI] [PubMed] [Google Scholar]
- 7.Goldsmith M, Tawfik DS. Directed enzyme evolution: beyond the low-hanging fruit. Curr Opin Struct Biol. 2012;22:406–412. doi: 10.1016/j.sbi.2012.03.010. [DOI] [PubMed] [Google Scholar]
- 8.Fowler DM, Araya CL, Fleishman SJ, Kellogg EH, Stephany JJ, Baker D, Fields S. High-resolution mapping of protein sequence-function relationships. Nat Methods. 2010;7:741–746. doi: 10.1038/nmeth.1492. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gupta K, Varadarajan R. Insights into protein structure, stability and function from saturation mutagenesis. Curr Opin Struct Biol. 2018;50:117–125. doi: 10.1016/j.sbi.2018.02.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Vijayabaskar MS, Vishveshwara S. Interaction energy based protein structure networks. Biophys J. 2010;99:3704–3715. doi: 10.1016/j.bpj.2010.08.079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Rajasekaran N, Suresh S, Gopi S, Raman K, Naganathan AN. A general mechanism for the propagation of mutational effects in proteins. Biochemistry. 2017;56:294–305. doi: 10.1021/acs.biochem.6b00798. [• This work describes a multi-model treatment of mutational effects including graph theory, MD simulations and a statistical model, apart from re-analysis of experimental NMR data. The analysis consistently reveals that mutational effects are not localized and influence distant regions of a protein.] [DOI] [PubMed] [Google Scholar]
- 12.Maslov S, Ispolatov I. Propagation of large concentration changes in reversible protein-binding networks. Proc Natl Acad Sci U S A. 2007;104:13655–13660. doi: 10.1073/pnas.0702905104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Whitley MJ, Zhang J, Lee AL. Hydrophobic core mutations in CI2 globally perturb fast side-chain dynamics similarly without regard to position. Biochemistry. 2008;47:8566–8576. doi: 10.1021/bi8007966. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Haririnia A, Verma R, Purohit N, Twarog MZ, Deshaies RJ, Bolon D, Fushman D. Mutations in the hydrophobic core of ubiquitin differentially affect its recognition by receptor proteins. J Mol Biol. 2008;375:979–996. doi: 10.1016/j.jmb.2007.11.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Bouvignies G, Vallurupalli P, Hansen DF, Correia BE, Lange O, Bah A, Vernon RM, Dahlquist FW, Baker D, Kay LE. Solution structure of a minor and transiently formed state of a T4 lysozyme mutant. Nature. 2011;477:111–114. doi: 10.1038/nature10349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Consonni R, Santomo L, Fusi P, Tortora P, Zetta L. A single-point mutation in the extreme heat- and pressure-resistant sso7d protein from sulfolobus solfataricus leads to a major rearrangement of the hydrophobic core. Biochemistry. 1999;38:12709–12717. doi: 10.1021/bi9911280. [DOI] [PubMed] [Google Scholar]
- 17.Roche J, Caro JA, Dellarole M, Guca E, Royer CA, Garcia-Moreno BE, Garcia AE, Roumestand C. Structural, energetic, and dynamic responses of the native state ensemble of staphylococcal nuclease to cavity-creating mutations. Proteins. 2013;81:1069–1080. doi: 10.1002/prot.24231. [• A detailed experimental dissection of mutational effects in SNase highlights the pervasive modulation of multiple energetic and dynamic terms far from the mutated site.] [DOI] [PubMed] [Google Scholar]
- 18.Millet O, Mittermaier A, Baker D, Kay LE. The effects of mutations on motions of side-chains in protein L studied by 2H NMR dynamics and scalar couplings. J Mol Biol. 2003;329:551–563. doi: 10.1016/s0022-2836(03)00471-6. [DOI] [PubMed] [Google Scholar]
- 19.Wang L, Tharp S, Selzer T, Benkovic SJ, Kohen A. Effects of a distal mutation on active site chemistry. Biochemistry. 2006;45:1383–1392. doi: 10.1021/bi0518242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Wang L, Goodey NM, Benkovic SJ, Kohen A. Coordinated effects of distal mutations on environmentally coupled tunneling in dihydrofolate reductase. Proc Natl Acad Sci U S A. 2006;103:15753–15758. doi: 10.1073/pnas.0606976103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Isogai S, Deupi X, Opitz C, Heydenreich FM, Tsai CJ, Brueckner F, Schertler GF, Veprintsev DB, Grzesiek S. Backbone NMR reveals allosteric signal transduction networks in the beta1-adrenergic receptor. Nature. 2016;530:237–241. doi: 10.1038/nature16577. [• The authors employ NMR experiments, ligand-mutation and point-mutation induced changes in chemical shifts to map the allosteric network of a transmembrane signaling protein, β1-adrenergic receptor.] [DOI] [PubMed] [Google Scholar]
- 22.Doshi U, Holliday MJ, Eisenmesser EZ, Hamelberg D. Dynamical network of residue-residue contacts reveals coupled allosteric effects in recognition, catalysis, and mutation. Proc Natl Acad Sci U S A. 2016;113:4735–4740. doi: 10.1073/pnas.1523573113. [• This combined experimental-simulation study highlights how distal regions in Cyclophilin A are coupled to the active site through the intra-molecular contact network. The allosteric coupling is further validated employing experimental mutational studies and graph theoretic modeling.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Yang G, Hong N, Baier F, Jackson CJ, Tokuriki N. Conformational tinkering drives evolution of a promiscuous activity through indirect mutational effects. Biochemistry. 2016;55:4583–4593. doi: 10.1021/acs.biochem.6b00561. [• The authors demonstrate how second-shell mutations can have a dramatic impact on the activity of an enzyme mediated through the intramolecular interaction network.] [DOI] [PubMed] [Google Scholar]
- 24.DuBay KH, Bowman GR, Geissler PL. Fluctuations within folded proteins: implications for thermodynamic and allosteric regulation. Acc Chem Res. 2015;48:1098–1105. doi: 10.1021/ar500351b. [DOI] [PubMed] [Google Scholar]
- 25.Rajasekaran N, Sekhar A, Naganathan AN. A universal pattern in the percolation and dissipation of protein structural perturbations. J Phys Chem Lett. 2017;8:4779–4784. doi: 10.1021/acs.jpclett.7b02021. [• A large-scale analysis of chemical shift perturbation data upon mutations, ligand binding and post-translational modifications reveals an exponential dissipation pattern uncovering avenues to understand and model allostery.] [DOI] [PubMed] [Google Scholar]
- 26.Fodor AA, Aldrich RW. On evolutionary conservation of thermodynamic coupling in proteins. J Biol Chem. 2004;279:19046–19050. doi: 10.1074/jbc.M402560200. [DOI] [PubMed] [Google Scholar]
- 27.Chi CN, Elfstrom L, Shi Y, Snall T, Engstrom A, Jemth P. Reassessing a sparse energetic network within a single protein domain. Proc Natl Acad Sci U S A. 2008;105:4679–4684. doi: 10.1073/pnas.0711732105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Jack BR, Meyer AG, Echave J, Wilke CO. Functional sites induce long-range evolutionary constraints in enzymes. PLoS Biol. 2016;14 doi: 10.1371/journal.pbio.1002452. e1002452. [• A sequence-structure study reveals that active sites in enzymes induce conservation of residues even far from it. The reported distance (~27Å) potentially identifies the maximal extent to which active-site residues can be coupled to its surrounding neighbors through the intra-molecular interaction network.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sivanandan S, Naganathan AN. A disorder-induced domino-like destabilization mechanism governs the folding and functional dynamics of the repeat protein IκBα. PLoS Comput Biol. 2013;9 doi: 10.1371/journal.pcbi.1003403. e1003403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wako H, Saito N. Statistical mechanical theory of protein conformation. 2. Folding pathway for protein. J Phys Soc Jpn. 1978;44:1939–1945. [Google Scholar]
- 31.Muñoz V, Eaton WA. A simple model for calculating the kinetics of protein folding from three-dimensional structures. Proc Natl Acad Sci U S A. 1999;96:11311–11316. doi: 10.1073/pnas.96.20.11311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Bowman GR, Geissler PL. Extensive conformational heterogeneity within protein cores. J Phys Chem B. 2014;118:6417–6423. doi: 10.1021/jp4105823. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Daquino JA, Gomez J, Hilser VJ, Lee KH, Amzel LM, Freire E. The magnitude of the backbone conformational entropy change in protein folding. Proteins. 1996;25:143–156. doi: 10.1002/(SICI)1097-0134(199606)25:2<143::AID-PROT1>3.0.CO;2-J. [DOI] [PubMed] [Google Scholar]
- 34.Cooper A. Thermodynamic fluctuations in protein molecules. Proc Natl Acad Sci USA. 1976;73:2740–2741. doi: 10.1073/pnas.73.8.2740. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Korzhnev DM, Salvatella X, Vendruscolo M, Di Nardo AA, Davidson AA, Dobson CM, Kay LE. Low-populated folding intermediates of Fyn SH3 characterized by relaxation dispersion NMR. Nature. 2005;430:586–590. doi: 10.1038/nature02655. [DOI] [PubMed] [Google Scholar]
- 36.Lu ZL, Coetsee M, White CD, Millar RP. Structural determinants for ligand-receptor conformational selection in a peptide G protein-coupled receptor. J Biol Chem. 2007;282:17921–17929. doi: 10.1074/jbc.M610413200. [DOI] [PubMed] [Google Scholar]
- 37.Abrol R, Trzaskowski B, Goddard WA, 3rd, Nesterov A, Olave I, Irons C. Ligand- and mutation-induced conformational selection in the CCR5 chemokine G protein-coupled receptor. Proc Natl Acad Sci U S A. 2014;111:13040–13045. doi: 10.1073/pnas.1413216111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Tzeng SR, Kalodimos CG. Protein activity regulation by conformational entropy. Nature. 2012;488:236–240. doi: 10.1038/nature11271. [DOI] [PubMed] [Google Scholar]
- 39.Michielssens S, Peters JH, Ban D, Pratihar S, Seeliger D, Sharma M, Giller K, Sabo TM, Becker S, Lee D, et al. A designed conformational shift to control protein binding specificity. Angew Chem Int Ed Engl. 2014;53:10367–10371. doi: 10.1002/anie.201403102. [• An integrated experimental-simulation approach is presented where distal core mutations induce a conformational shift in ubiquitin thus altering binding specificity.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Guzman I, Ghaemi Z, Baranger A, Luthey-Schulten Z, Gruebele M. Native conformational dynamics of the spliceosomal U1A protein. J Phys Chem B. 2015;119:3651–3661. doi: 10.1021/jp511760m. [DOI] [PubMed] [Google Scholar]
- 41.Aviram HY, Pirchi M, Mazal H, Barak Y, Riven I, Haran G. Direct observation of ultrafast large-scale dynamics of an enzyme under turnover conditions. Proc Natl Acad Sci U S A. 2018;115:3243–3248. doi: 10.1073/pnas.1720448115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Schuetz AK, Kay LE. A dynamic molecular basis for malfunction in disease mutants of p97/VCP. eLife. 2016;5 doi: 10.7554/eLife.20143. [• Evidence is presented for how mutations far from the active site can shift conformational ensembles through propagation of structural changes thus contributing to disease.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Muñoz IG, Morel B, Medina-Carmona E, Pey AL. A mechanism for cancer-associated inactivation of NQO1 due to P187S and its reactivation by the consensus mutation H80R. FEBS Lett. 2017;591:2826–2835. doi: 10.1002/1873-3468.12772. [DOI] [PubMed] [Google Scholar]
- 44.Buller AR, van Roye P, Cahn JKB, Scheele RA, Herger M, Arnold FH. Directed evolution mimics allosteric activation by stepwise tuning of the conformational ensemble. J Am Chem Soc. 2018;140:7256–7266. doi: 10.1021/jacs.8b03490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Saavedra HG, Wrabl JO, Anderson JA, Li J, Hilser VJ. Dynamic allostery can drive cold adaptation in enzymes. Nature. 2018;558:324–328. doi: 10.1038/s41586-018-0183-2. [• The authors show that distal mutations promote partial unfolding of specific domains in Adenylate Kinase. This in turn modulates the dynamics and hence function revealing insights into how specific sequence changes drive cold adaptation.] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Munshi S, Rajendran D, Naganathan AN. Entropic control of an excited folded-like conformation in a disordered protein ensemble. J Mol Biol. 2018;430:2688–2694. doi: 10.1016/j.jmb.2018.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Borcherds W, Theillet FX, Katzer A, Finzel A, Mishall KM, Powell AT, Wu H, Manieri W, Dieterich C, Selenko P, et al. Disorder and residual helicity alter p53-Mdm2 binding affinity and signaling in cells. Nat Chem Biol. 2014;10:1000–1002. doi: 10.1038/nchembio.1668. [DOI] [PubMed] [Google Scholar]
- 48.Crabtree MD, Borcherds W, Poosapati A, Shammas SL, Daughdrill GW, Clarke J. Conserved helix-flanking prolines modulate intrinsically disordered protein: target affinity by altering the lifetime of the bound complex. Biochemistry. 2017;56:2379–2384. doi: 10.1021/acs.biochem.7b00179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Dahal L, Kwan TOC, Hollins JJ, Clarke J. Promiscuous and selective: how intrinsically disordered BH3 proteins interact with their pro-survival partner MCL-1. J Mol Biol. 2018;430:2468–2477. doi: 10.1016/j.jmb.2018.04.004. [DOI] [PubMed] [Google Scholar]
- 50.Karlsson OA, Chi CN, Engstrom A, Jemth P. The transition state of coupled folding and binding for a flexible beta-finger. J Mol Biol. 2012;417:253–261. doi: 10.1016/j.jmb.2012.01.042. [DOI] [PubMed] [Google Scholar]
- 51.Iesmantavicius V, Dogan J, Jemth P, Teilum K, Kjaergaard M. Helical propensity in an intrinsically disordered protein accelerates ligand binding. Angew Chem Int Ed. 2014;53:1548–1551. doi: 10.1002/anie.201307712. [DOI] [PubMed] [Google Scholar]
- 52.Patel A, Lee HO, Jawerth L, Maharana S, Jahnel M, Hein MY, Stoynov S, Mahamid J, Saha S, Franzmann TM, et al. A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell. 2015;162:1066–1077. doi: 10.1016/j.cell.2015.07.047. [DOI] [PubMed] [Google Scholar]
- 53.Cooper A, Dryden DT. Allostery without conformational change. A plausible model. Eur Biophys J. 1984;11:103–109. doi: 10.1007/BF00276625. [DOI] [PubMed] [Google Scholar]
- 54.Popovych N, Sun S, Ebright RH, Kalodimos CG. Dynamically driven protein allostery. Nat Struct Mol Biol. 2006;13:831–838. doi: 10.1038/nsmb1132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Reynolds KA, McLaughlin RN, Ranganathan R. Hotspots for allosteric regulation on protein surfaces. Cell. 2011;147:1564–1575. doi: 10.1016/j.cell.2011.10.049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Lee SY, Pullen L, Virgil DJ, Castaneda CA, Abeykoon D, Bolon DN, Fushman D. Alanine scan of core positions in ubiquitin reveals links between dynamics, stability, and function. J Mol Biol. 2014;426:1377–1389. doi: 10.1016/j.jmb.2013.10.042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Zayner JP, Antoniou C, French AR, Hause RJ, Jr, Sosnick TR. Investigating models of protein function and allostery with a widespread mutational analysis of a light-activated protein. Biophys J. 2013;105:1027–1036. doi: 10.1016/j.bpj.2013.07.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gianni S, Haq SR, Montemiglio LC, Jurgens MC, Engstrom A, Chi CN, Brunori M, Jemth P. Sequence-specific long range networks in PSD-95/discs large/ZO-1 (PDZ) domains tune their binding selectivity. J Biol Chem. 2011;286:27167–27175. doi: 10.1074/jbc.M111.239541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Hultqvist G, Haq SR, Punekar AS, Chi CN, Engstrom A, Bach A, Stromgaard K, Selmer M, Gianni S, Jemth P. Energetic pathway sampling in a protein interaction domain. Structure. 2013;21:1193–1202. doi: 10.1016/j.str.2013.05.010. [DOI] [PubMed] [Google Scholar]
- 60.Tang Q, Fenton AW. Whole-protein alanine-scanning mutagenesis of allostery: a large percentage of a protein can contribute to mechanism. Hum Mutat. 2017;38:1132–1143. doi: 10.1002/humu.23231. [• This work highlights through mutational studies that nearly 30% of protein residues can influence the functioning of human liver Pyruvate Kinase] [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Carlson GM, Fenton AW. What mutagenesis can and cannot reveal about allostery. Biophys J. 2016;110:1912–1923. doi: 10.1016/j.bpj.2016.03.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rajasekaran N, Naganathan AN. A self-consistent structural perturbation approach for determining the magnitude and extent of allosteric coupling in proteins. Biochem J. 2017;474:2379–2388. doi: 10.1042/BCJ20170304. [• A rapid alanine-scanning protocol is presented to explore the strength and extent of coupling of a particular residue with its immediate neighborhood. The perturbation approach is shown to reproduce the thermodynamics of protein destabilization, identify allosterically-coupled residues and generate cooperativity indices consistent with experiments] [DOI] [PubMed] [Google Scholar]
- 63.Zheng W, Brooks BR, Thirumalai D. Low-frequency normal modes that describe allosteric transitions in biological nanomachines are robust to sequence variations. Proc Natl Acad Sci U S A. 2006;103:7664–7669. doi: 10.1073/pnas.0510426103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Gerek ZN, Ozkan SB. Change in allosteric network affects binding affinities of PDZ domains: analysis through perturbation response scanning. PLoS Comput Biol. 2011;7 doi: 10.1371/journal.pcbi.1002154. e1002154. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Lockless SW, Ranganathan R. Evolutionarily conserved pathways of energetic connectivity in protein families. Science. 1999;286:295–299. doi: 10.1126/science.286.5438.295. [DOI] [PubMed] [Google Scholar]
- 66.Liu T, Whitten ST, Hilser VJ. Ensemble-based signatures of energy propagation in proteins: a new view of an old phenomenon. Proteins: Struct Funct Bioinf. 2006;62:728–738. doi: 10.1002/prot.20749. [DOI] [PubMed] [Google Scholar]
- 67.Chennubhotla C, Bahar I. Signal propagation in proteins and relation to equilibrium fluctuations. PLoS Comput Biol. 2007;3:1716–1726. doi: 10.1371/journal.pcbi.0030172. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zheng W, Brooks BR, Thirumalai D. Allosteric transitions in biological nanomachines are described by robust normal modes of elastic networks. Curr Protein Pept Sci. 2009;10:128–132. doi: 10.2174/138920309787847608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dokholyan NV. Controlling allosteric networks in proteins. Chem Rev. 2016;116:6463–6487. doi: 10.1021/acs.chemrev.5b00544. [DOI] [PubMed] [Google Scholar]
- 70.Schueler-Furman O, Wodak SJ. Computational approaches to investigating allostery. Curr Opin Struct Biol. 2016;41:159–171. doi: 10.1016/j.sbi.2016.06.017. [DOI] [PubMed] [Google Scholar]
- 71.Amor BR, Schaub MT, Yaliraki SN, Barahona M. Prediction of allosteric sites and mediating interactions through bond-to-bond propensities. Nat Commun. 2016;7 doi: 10.1038/ncomms12477. 12477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Luque I, Leavitt SA, Freire E. The linkage between protein folding and functional cooperativity: two sides of the same coin? Annu Rev Biophys Biomol Struct. 2002;31:235–256. doi: 10.1146/annurev.biophys.31.082901.134215. [DOI] [PubMed] [Google Scholar]
- 73.Fersht AR, Matouschek A, Serrano L. the folding of an enzyme. 1. Theory of protein engineering analysis of stability and pathway of protein folding. J Mol Biol. 1992;224:771–782. doi: 10.1016/0022-2836(92)90561-w. [DOI] [PubMed] [Google Scholar]
- 74.Naganathan AN, Muñoz V. Insights into protein folding mechanisms from large scale analysis of mutational effects. Proc Natl Acad Sci U S A. 2010;107:8611–8616. doi: 10.1073/pnas.1000988107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Akmal A, Muñoz V. The nature of the free energy barriers to two-state folding. Proteins. 2004;57:142–152. doi: 10.1002/prot.20172. [DOI] [PubMed] [Google Scholar]
- 76.Muñoz V, Sadqi M, Naganathan AN, de Sancho D. Exploiting the downhill folding regime via experiment. HFSP J. 2008;2:342–353. doi: 10.2976/1.2988030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Kaya H, Chan HS. Polymer principles of protein calorimetric two-state cooperativity. Proteins. 2000;40:637–661. doi: 10.1002/1097-0134(20000901)40:4<637::aid-prot80>3.0.co;2-4. [DOI] [PubMed] [Google Scholar]
- 78.Li MS, Klimov DK, Thirumalai D. Thermal denaturation and folding rates of single domain proteins: size matters. Polymer. 2004;45:573–579. [Google Scholar]
- 79.Muñoz V, Campos LA, Sadqi M. Limited cooperativity in protein folding. Curr Opin Struct Biol. 2016;36:58–66. doi: 10.1016/j.sbi.2015.12.001. [DOI] [PubMed] [Google Scholar]
- 80.Risso VA, Gavira JA, Mejia-Carmona DF, Gaucher EA, Sanchez-Ruiz JM. Hyperstability and substrate promiscuity in laboratory resurrections of Precambrian beta-lactamases. J Am Chem Soc. 2013;135:2899–2902. doi: 10.1021/ja311630a. [DOI] [PubMed] [Google Scholar]
- 81.Blomberg R, Kries H, Pinkas DM, Mittl PR, Grutter MG, Privett HK, Mayo SL, Hilvert D. Precision is essential for efficient catalysis in an evolved Kemp eliminase. Nature. 2013;503:418–421. doi: 10.1038/nature12623. [DOI] [PubMed] [Google Scholar]
- 82.Kaltenbach M, Burke JR, Dindo M, Pabis A, Munsberg FS, Rabin A, Kamerlin SCL, Noel JP, Tawfik DS. Evolution of chalcone isomerase from a noncatalytic ancestor. Nat Chem Biol. 2018;14:548–555. doi: 10.1038/s41589-018-0042-3. [DOI] [PubMed] [Google Scholar]
- 83.Zarrine-Afsar A, Zhang ZQ, Schweiker KL, Makhatadze GI, Davidson AR, Chan HS. Kinetic consequences of native state optimization of surface-exposed electrostatic interactions in the Fyn SH3 domain. Proteins: Struct Func Bioinf. 2012;80:858–870. doi: 10.1002/prot.23243. [DOI] [PubMed] [Google Scholar]
- 84.Naganathan AN. Predictions from an Ising-like statistical mechanical model on the dynamic and thermodynamic effects of protein surface electrostatics. J Chem Theor Comput. 2012;8:4646–4656. doi: 10.1021/ct300676w. [DOI] [PubMed] [Google Scholar]
- 85.Xiao S, Patsalo V, Shan B, Bi Y, Green DF, Raleigh DP. Rational modification of protein stability by targeting surface sites leads to complicated results. Proc Natl Acad Sci U S A. 2013;110:11337–11342. doi: 10.1073/pnas.1222245110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Naganathan AN, Sanchez-Ruiz JM, Munshi S, Suresh S. Are protein folding intermediates the evolutionary consequence of functional constraints? J Phys Chem B. 2015;119:1323–1333. doi: 10.1021/jp510342m. [DOI] [PubMed] [Google Scholar]