Abstract
MicroRNAs (miRNAs) are emerging as important in human embryo implantation, and we present here a review of the literature from a clinical perspective. Implantation involves complex interactions between the blastocyst and endometrium. miRNAs have been shown to be differentially expressed in implanted compared with non-implanted blastocysts and euploid compared with aneuploid blastocysts. Further, miRNAs are differentially expressed in proliferative compared with decidualized endometrium, and in receptive compared with pre-receptive endometrium. miRNAs are also differentially expressed in endometrium of women who failed implantation, and in endometrium of women with recurrent implantation failure. Due to the complexity of miRNA signaling, studies have suffered from inconsistency in reproducibility of results. However, miRNAs show potential as biomarkers in the pursuit of more reliable prediction of embryo implantation.
Electronic supplementary material
The online version of this article (10.1007/s10815-018-1326-y) contains supplementary material, which is available to authorized users.
Keywords: MicroRNA, Human embryo, Endometrium, Implantation, Pregnancy, Recurrent implantation failure
Introduction
Implantation, the process by which the rapidly dividing cells of the blastocyst successfully implant into the receptive maternal endometrium, is a crucial component of mammalian reproduction. The series of cellular changes, interactions, and signaling in implantation are carefully orchestrated by paracrine, autocrine, and juxtacrine communication. These events, primarily under the control of the hypothalamo-pituitary-gonadal axis, are temporally synchronized to create the “window of implantation” of the maternal endometrium. Simultaneously, the blastocyst requires activation in order to successfully attach to this receptive endometrium [1]. Signaling during implantation involves various genes and gene modifiers such as microRNA (miRNA) molecules, which modulate the intricate cross talk between embryo and endometrium.
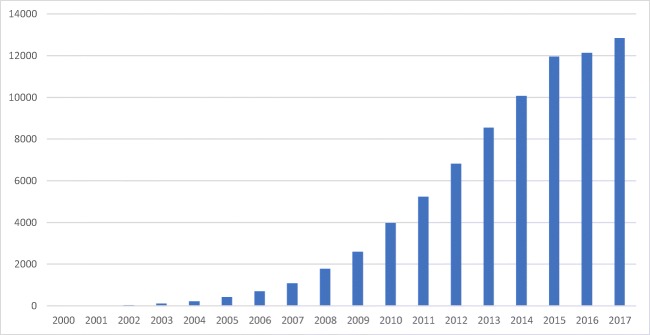
Interest in miRNAs has been steadily increasing over the past 17 years (Fig. 1). miRNAs have been shown to be involved in pathways of cellular signaling and have shown promise as novel prognostic markers and diagnostic tools. There are several reviews in the literature discussing miRNA and embryo implantation, as is warranted for such a quickly evolving subject [2–6]. Here, we discuss recent advances in the study of miRNAs in human embryo implantation.
Fig. 1.
The number of references found for each year of publication on the PubMed database using the keyword ‘microRNA’. In 2017, this number was 12,682. Image by author
Implantation
Implantation can be broadly divided into three steps: apposition, attachment, and invasion [7, 8].
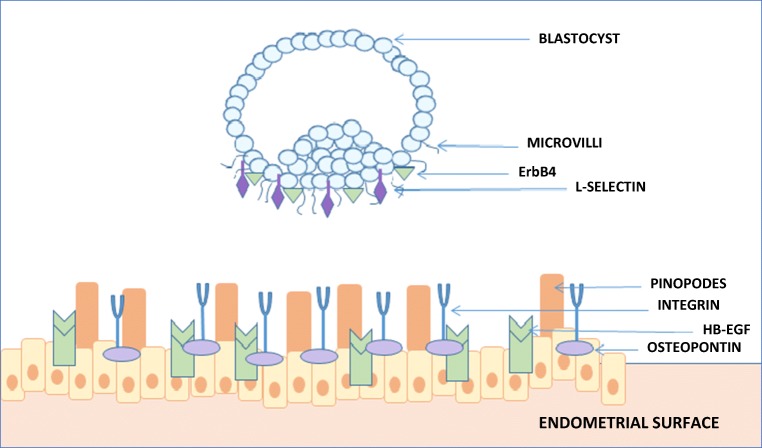
During apposition, numerous small pinopodes on the receptive endometrium developed by generalized stromal oedema on the inner uterine surface interlace with the microvilli on the outer surface of the cytotrophoblast [9] (Fig. 2). Further, Heparin-binding EGF-like growth factor (HB-EGF) is expressed by the receptive endometrium, and cells that express transmembrane HB-EGF adhere to blastocysts displaying ErbB4 on their cell surface [11]. Activated blastocysts upregulate their expression of HB-EGF, which via an auto-induction loop prompts its own gene expression in the endometrium at the site of blastocyst apposition [12]. HB-EGF expression is modulated by Lif and results in a reduction of COX-2, the deficiency of which results in implantation failure [13]. Gene expression in the Wnt/β-catenin pathway is also important at the site of implantation. Wnt/β-catenin signaling immediately before attachment requires activated blastocyst and preimplantation oestrogen secretion in transgenic mice models [14].
Fig. 2.
A schematic diagram of signaling between blastocyst and receptive maternal endometrium. Microvilli from the blastocyst aid in attachment to the endometrial pinopodes. Further, ErbB4 on the blastocyst surface adheres to transmembrane HB-EGF (modulated by Lif) on the receptive endometrium. Integrin avB3 is attached to the maternal endometrium via osteopontin. l-selectin molecules on the blastocyst attach to selectin oligosaccharide ligand on the maternal surface. Image by author, adapted from Davidson et al. 2016 [10]
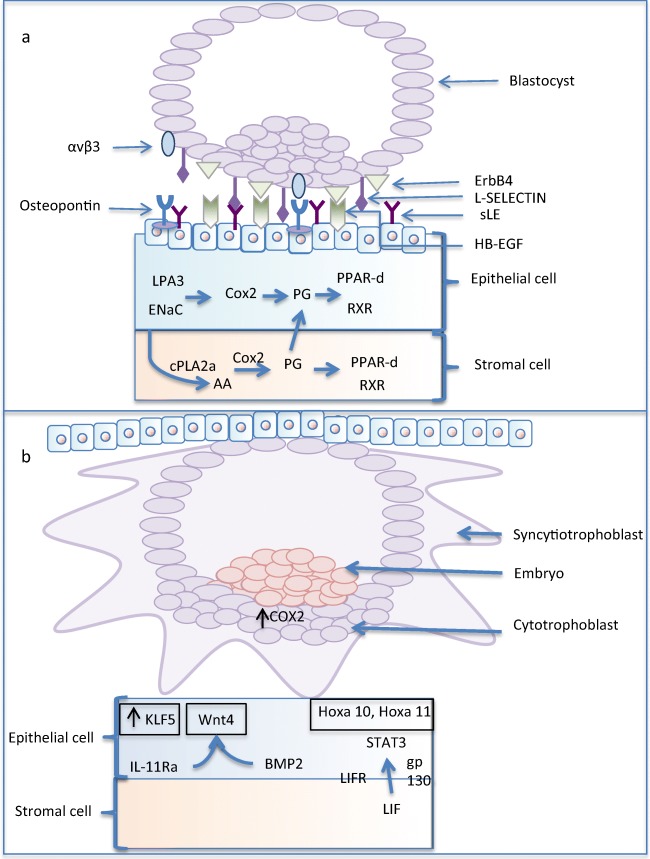
In the attachment phase, various glycoproteins, carbohydrate ligands, receptors, and integrins work together to adhere the embryo to the endometrial surface (Fig. 3). Several integrins are involved with implantation in varying capacities. During attachment α5β1, αvβ3, αvβ5, and αvβ6 are expressed by the embryo, and α1β1, α6β1, and α7β1 are subsequently involved in invasion [16]. The ligand osteopontin of epithelial origin acts to bind integrin αvβ3 on the maternal surface to support adhesion [17]. Further, during attachment, l-selectin molecules are presented on the blastocyst surface, and selectin oligosaccharide ligands are expressed in the primed endometrium [18]. Interestingly, the integrins are dynamic with α5β1 starting within the inner cell mass in early embryo development and then translocated to the trophoblast cells that invade the endometrium during implantation [15, 19] (Fig. 3). Invasion is dependent on endometrial vascular permeability, and decidualization is mediated by prostaglandin synthesis by COX1 and COX2. Various cellular molecules are essential for normal implantation and aberrant production or loss of them may be linked to unexplained infertility [20]. In fact, implantation has been considered so dependent on these mediators that integrins have been suggested as potential biomarkers of infertility in the future [21, 22].
Fig. 3.
A schematic diagram of a attachment and b implantation between the blastocyst and receptive maternal endometrium. ErbB4 on the blastocyst surface adheres to transmembrane HB-EGF (modulated by Lif) along with l-selectin ligands (sLE) expressed by the maternal endometrium epithelium to l-selectin receptors on the blastocyst. The other key signaling pathways for attachment and implantation are also shown. AA, arachidonic acid; BMP2, bone morphogenetic protein 2; Cox2, cyclooxygenase-2; ENaC, epithelium sodium channel; ErbB4; epidermal growth factor receptor 1/4; gp130, glycoprotein 130; HB-EGF, heparin-binding epidermal growth factor-like growth factor; Hoxa10/11, homeobox A10/11; KLF5, Kruppel-like factor 5; LIF, leukemia inhibitory factor; LIFR, LIF receptor; LPA3, lysophosphatidic acid receptor 3; PG, prostaglandin; PPAR-δ; peroxisome proliferators–activating receptor δ; Wnt4, wingless-type MMTV integration site family members 4. Image by author, adapted from Cha et al. [15]
Molecular signaling, hormone production, and gene expression are all related by genomic transcription. Gene expression can be altered in a variety of ways including by miRNAs. These small molecules are approximately 18–24 nucleotides of non-protein-coding RNA, and are secreted by cells via exosomes, apoptotic bodies, lipid-bound, or RNA-binding complex proteins such as Argonaute 2 in humans [23–25]. miRNAs act to regulate gene expression either negatively by mRNA cleavage, deadenylation, or inhibition of translational repression or positively through the targeting of gene promoters [26–29]. miRNA research in fertility has significantly increased in recent years to aid understanding of both normal and abnormal processes. In theory, they have the potential to be diagnostic tools as they are stable and present in the cells, saliva, urine, blood, uterine fluid, and cell culture media [30–32]. In reality, miRNAs are highly variable based on the needs of particular cells at specific times, and although interesting clinical correlations have been observed, the reproducibility of these findings and their application to clinical practice remain uncertain. In this review, we discuss miRNA as it is relevant to human embryo implantation from a clinical perspective.
MiRNA biogenesis
Small non-coding RNAs are broadly classified into three groups: microRNA (miRNA), short interfering RNA (siRNA), and PIWI-interacting RNA (piRNA). miRNAs are further divided into families based on identical sequences at nucleotides 2–8 of mature miRNA [33]. Small RNAs are distinct from long non-coding RNAs (lncRNAs), which are RNAs longer than 200 nucleotides that are not translated to protein but that may regulate epigenetic modifications or gene expression at the transcriptional or post-transcriptional levels [34].
The difference between groups of small non-coding RNAs begins with how they are synthesized. miRNAs are derived from single-stranded RNAs that form double-stranded hairpin structures that can be clipped and processed by Dicer, a double-stranded RNA exonuclease. siRNAs are produced by Dicer cleavage of an RNA duplex formed by two single-stranded RNAs. PIWI-interacting RNAs (piRNAs) are derived from single-stranded RNAs that are excised by an unknown nuclease [35]. miRNAs and siRNAs are loaded onto Argonaute family proteins (AGO). Typically, miRNA-AGO complexes inhibit translation of mRNA non-destructively, whereas siRNA-AGO complexes cause destruction of mRNA. piRNAs are loaded onto PIWI proteins and may inhibit transcription by histone methylation [35]. miRNAs have also been observed to be involved in epigenetic regulation [36].
The regulatory nature of miRNAs in embryo development was first described in Caenorhabditis elegans and lin-4, but has been described in numerous species since [37]. It has been estimated that miRNAs may modulate up to three fifths of protein-coding genes at the translation phase in the human genome [33].
The formation of miRNAs is complex involving both nuclear and cytoplasmic phases. The traditional canonical pathway is responsible for approximately ~ 99% of miRNA synthesis whereas the non-canonical pathway is hypothesized to be responsible for the remaining miRNA [38]. The miRNA sequences have been located in the entirety of the genome including within introns of non-coding and coding regions, and in exonic regions [38].
By the canonical pathway, miRNAs are transcribed by RNA polymerase II into primary miRNAs that include a hairpin structure containing the miRNA sequences. Nuclear processing is completed by the microprocessor complex, composed of RNase III Drosha and DGCR8 (Digeorge Syndrome Critical Region 8). The microprocessor complex isolates the stem-loop of the primary miRNAs to produce the hairpin-shaped pre-cursor miRNA (pre-miRNA). The pre-miRNA is then exported out of the nucleus into the cytoplasm by Exportin-5. In the cytoplasm, an RNase III endonuclease, Dicer, crops the pre-miRNAs into specific RNA duplexes. These RNA sequences are loaded onto AGO [39]. Once loaded onto AGOs, the miRNAs induce their cleavage and/or inhibit their translation, and they are unwound to become mature miRNAs [29] (Fig. 4).
Fig. 4.
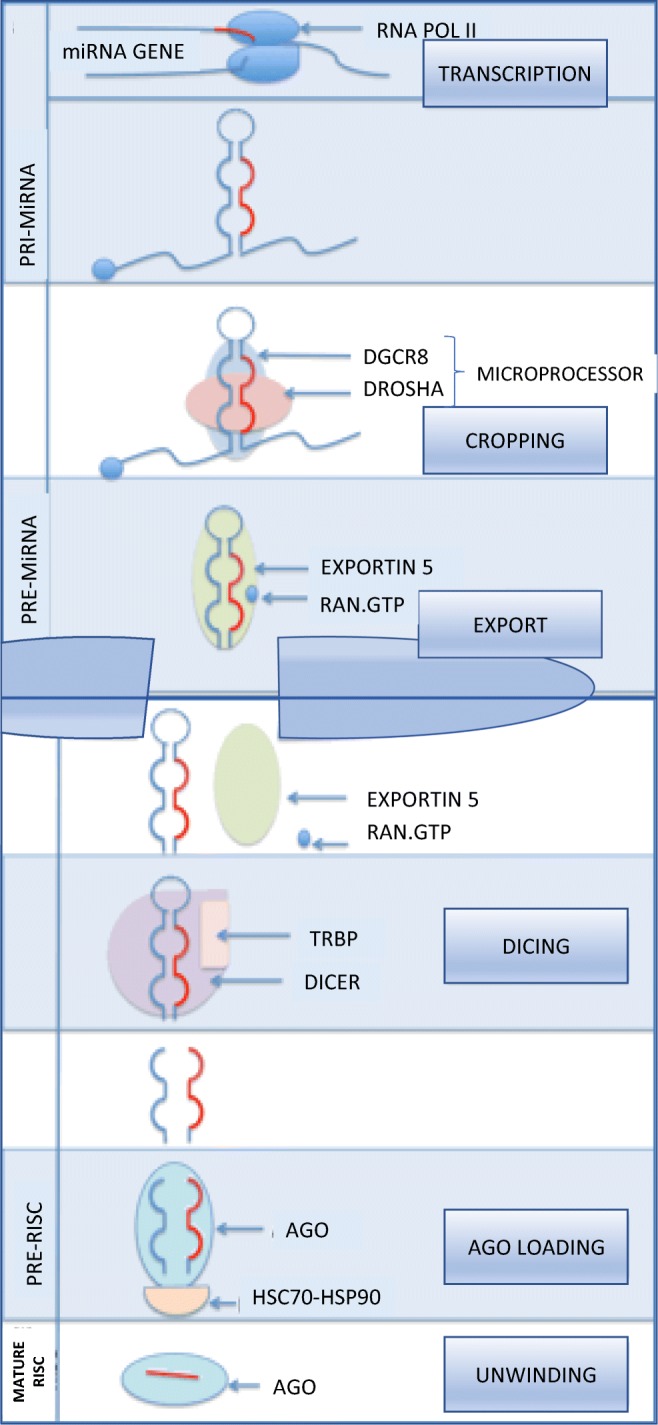
Nuclear events in miRNA canonical biogenesis. Schematic diagram of miRNA gene transcription by RNA polymerase II to form pre-miRNA. This is processed by the microprocessor complex of Drosha and DGCR8 (DiGeorge Syndrome Critical Region Gene 8) and cropped to form pre-miRNA. The pre-miRNA is exported from the nucleus via exportin 5 in complex with RAN.GTP. In the cytoplasm, after Dicer-processing RNA duplex is loaded onto AGO (Argonaute proteins) stabilized by a heatshock protein (HSC70-HSP90). The remaining “passenger” strand is discarded, and the mature miRNA strand is loaded onto the AGO protein. Image by author, adapted from Ha and Kim [38]
The non-canonical pathway refers to pathways that do not follow this sequence of complexes for synthesis. This includes those independent of DGCR8, Drosha, and Dicer and those dependent on the enzyme TUTase. The distinction between canonical pathway miRNA and non-canonical pathway miRNA is not always clear. Kim et al. recently revealed miRNAs known to be synthesized via canonical pathway were synthesized in Drosher and Dicer knockout mice revealing a potential gap in regulation that is not fully understood [40].
MiRNA and human embryo implantation
Blastocyst
Implantation involves elaborate communication between the endometrium and blastocyst. Throughout this process, miRNAs are secreted from both the blastocyst and endometrium and act in signaling to alter gene expression.
The current embryo grading system is subjective with significant inter-observer and intra-observer variabilities [41]. By studying miRNAs associated with blastocysts that ultimately implanted and comparing them to blastocysts that did not implant, investigators have sought to identify patterns that may be able to non-invasively predict the outcome of future embryos (Table 1). Cuman et al. measured miRNA expression in blastocyst media and demonstrated miR-661 was significantly overexpressed in blastocysts that failed to implant [32]. From their data, they further hypothesized a possible role for Argonaute 1 in the transport of miR-661, as abnormal expression of either was associated with failed implantation [32]. Rosenbluth et al. found significantly increased expression of miR-372 and miR-191 in the blastocyst media of blastocysts that failed implanted vs those that successfully implanted [44]. Cuman et al. also identified miR-372 in blastocyst media of blastocysts that failed to implant, but they did not include miR-191 in their array panel [32]. A pilot study by Borges et al. studied miR-142-3p as a potential biomarker of blastocyst implantation failure in blastocyst culture media. This study confirmed that higher expression of miR-142-3p in blastocysts that implanted compared with those that did not [43]. Differential expression of certain miRNAs has been associated with implantation failure in various studies. However, miRNA profiles have not been reproducible between studies and this warrants further study.
Table 1.
Studies on the involvement of miRNAs on blastocyst and implantation in humans
| Sample | Groups compared | miRNA measurement* | miRNA | Regulation | Reference |
|---|---|---|---|---|---|
| BCM | Implanted vs non-implanted euploid blastocysts | TaqMan | miR-20a, miR-30c | Upregulated in implanted | [42] |
| BCM | Implanted vs non-implanted blastocysts | TaqMan | miR-661, miR-372 | Upregulated in non-implanted | [32] |
| BCM | Implanted vs non-implanted blastocysts | TaqMan | miR-142-3p | Upregulated in implanted | [43] |
| BCM | Implanted vs non-implanted blastocysts Euploid vs aneuploid blastocysts |
TaqMan | miR-372, miR-191 miR-191 |
Upregulated in non-implanted Upregulated in aneuploid |
[44] |
| BB | Euploid vs aneuploid blastocysts | TaqMan | miR-141, miR-27b, miR-339-3p, miR-345 | Upregulated in euploid | [45] |
| BB | Blastocysts from MF patients vs fertile donors | TaqMan | let-7a, miR-24 | Downregulated in MF | [46] |
| BB | Blastocysts from PCOS patients vs fertile donors | TaqMan | let-7a, miR-24, miR-92, miR-93, miR-19a, miR-19b | Downregulated in PCOS | [46] |
BB, blastocyst biopsy; BCM, blastocyst culture media; PCOS, polycystic ovarian syndrome; MF, male factor infertility; ICSI, intra-cytoplasmic sperm injection
*See supplemental Table S1 for description of miRNA measurement methods
miRNA profiles may differ in euploid compared with aneuploid embryos. Rosenbluth et al. found in an analysis of miRNA in blastocyst biopsies that miR-141, miR-27b, miR-339-3p, and miR-345 were more highly expressed in euploid compared with aneuploid embryos [45]. Of note, this study did not include blastocyst grade in the analysis and had small sample sizes. As miRNA populations may influence development, differences in embryo quality may reflect different miRNA populations. Rosenbluth et al. also found in another study that miRNA-191 expression was upregulated in blastocyst culture media of aneuploid embryos [44].
Certain pathologic parental states may also influence blastocyst miRNA expression. McCallie et al. compared the expression of blastocyst miRNAs in blastocyst biopsies from fertile women with morphologically similar blastocysts from couples with either male factor infertility or polycystic ovarian syndrome (PCOS) [46]. They found that expression of let-7a and miR-24 were decreased in both groups, and that expression of four other miRNAs (miR-92, miR-93, miR-19a, and miR-19b) were decreased in the PCOS group [46]. Of note, circulating miR-93 has been described as a biomarker for PCOS [47]. The demonstration of this miRNA in blastocysts of PCOS women is thus interesting but of unclear significance [47].
It is important to note that very few similarities in blastocyst miRNA expression are noted in the above studies. This could be due to methodological differences in RNA extraction, differences in miRNA array panels, differences in blastocyst culture media, or differences between fresh and frozen embryos. Furthermore, miRNA expression is dynamic and likely varies tremendously with the gene expression required at particular junctures of development. The inherent difficulty in accounting for the myriad differences between unique embryos means this will continue to be a challenge in understanding the roles of miRNA in the blastocyst.
Endometrium
Implantation of the embryo requires an optimal environment in the endometrium, which may include signaling by miRNA (Table 2). Profiling of differential miRNA expression in the menstrual cycle has revealed significant differences between the secretory and proliferative phases linking hormonal and cyclical variation to miRNA expression [48, 49]. Kuokkanen et al. observed upregulated miR-30b and miR-30d in mid-secretory endometrial epithelial cells when compared with epithelial cells from the late proliferative phase in a small sample [48]. miR-30b and miR-30d were also both found to be upregulated in mid-secretory endometrium by Kresowik et al. [49], as was miR-31. miR-31 has a significant inverse association with FOXP3, a transcription factor for T regulatory cells and CXCL12, a chemoattractant for uterine natural killer cells, which has potential implications on creating an immune-tolerant environment in the secretory phase.
Table 2.
Studies on the involvement of miRNAs on the endometrium and implantation in humans
| Sample | Groups compared | miRNA measurement* | miRNA | Regulation | Reference |
|---|---|---|---|---|---|
| EB | Proliferative vs mid-secretory phase endometrium | TaqMan | miR-29b, miR-29c, miR-30b, miR-30d, miR-31, miR193a-3p, miR-203, miR-204, miR200c, miR210, miR-582-5p, miR-345 | Upregulated in mid-secretory endometrium | [48] |
| EB | Proliferative vs mid-secretory phase endometrium | TaqMan | miR-30b, miR-30d, miR-31, miR-203 miR-503, miR-145 |
Upregulated in mid-secretory endometrium Downregulated in mid-secretory endometrium |
[49] |
| EB | hESC decidualized in vitro vs control hESC | mRNA microarray and RT-qPCR | miR-483-3p miR-503, miR-542-3p, miR-155, miR-145, miR-424 |
Upregulated in decidualized hESC Downregulated in decidualized hESC |
[50] |
| EB | hESC decidualized in vitro vs control hESC | miScript SYBR Green | miR-181, miR-183, and miR-200 | Downregulated in decidualized hESC | [51] |
| EB | Receptive vs pre-receptive endometrium in fertile patients | TaqMan | miR-30b, miR-30d miR-494, miR-923 |
Upregulated in receptive endometrium Downregulated in receptive endometrium |
[52] |
| ECA | Receptive endometrial cavity aspirate of successful vs failed implantation | TaqMan | miR-891a, miR-522, miR-198 | Downregulated in failed implantation | [53] |
| EB | Receptive endometrium in RIF patients vs infertile patients who conceived | miRNA Complete labeling and Hyb |
miR-30b, miR-374a-5p, miR-145-5p, miR-196b-5p, miR-199a-5p, miR-449a, miR-424-5p, miR-125b-5p, miR-21-5p miR-1207-5p, miR-4306, miR-572, miR-5739, miR-6088 |
Upregulated in RIF group Downregulated in RIF group |
[54] |
| EB | Receptive endometrium in RIF patients vs fertile patients | TaqMan | miR-145, miR-23b, miR-99a | Upregulated in RIF patients | [55] |
EB, endometrial biopsy; ECA, endometrial cavity aspirate; PCOS, polycystic ovarian syndrome; MF, male factor infertility; ICSI, intra-cytoplasmic sperm injection; RIF, recurrent implantation failure
*See supplemental Table S1 for description of miRNA measurement methods
In other studies, human endometrial stem cells (hESC) have been decidualized in vitro to explore the effect of decidualization on miRNA expression. Studies by Tochigi et al. and Estella et al. found miRNA expression profiles of decidualized hESC vs control hESC were different [50, 51]. Tochigi et al. further demonstrated that hESC transfection of miR-542-3p suppressed the gene expression of IGFBP-1 which led to suppression of PRL and WNT4 and inhibition of decidualization in human endometrial stroma cells [50]. This suggests an important role of miR-542-3p in regulation of endometrial decidualization. Estrella et al. found a total of 26 upregulated and 17 downregulated miRNAs following in vitro decidualization of human endometrial stromal cells [51]. Interestingly, only miR-155 was commonly downregulated in both the Tochigi and Estrella studies, and no change was found in the expression of miR-542-3p in the Estrella study [51]. This may be in part due to the difference in methodology of induced decidualization, as the Estella et al. methodology used 17-estradiol and progesterone for decidualization of the endometrial stromal cells, whereas the Tochigi et al. study used 9-bromo-cyclic adenosine monophosphate and medroxyprogesterone acetate. There may be differences induced by in vitro decidualization that may not be generalizable to physiologic decidualization. These studies show that the miRNA profiles of proliferative vs secretory phase endometrium do likely differ, but there is a significant variability between studies.
This cyclical variation of miRNA expression offers an opportunity to identify miRNA changes during the window of implantation, when the endometrium is receptive to blastocyst implantation during the progesterone-dependent phase of the menstrual cycle. This window typically occurs 5–7 days after ovulation, or on day 19–21 of a 28-day menstrual cycle. Altmäe et al. studied the receptive and pre-receptive endometrial miRNA expression in fertile women and demonstrated miR-30b miR-30d were significantly upregulated and miR-494 and miR-923 were downregulated during the receptive phase [52]. Of note, miR-30b and miR-30d were consistently found in other studies to be elevated in the mid-secretory as compared with the proliferative phase.
Some studies sought to compare miRNA expression in endometrium of women who failed embryo implantation with that of women who experienced successful implantation. Comparing secretory endometrium of fertile women and women with recurrent implantation failure undergoing in vitro fertilization, Revel et al. found that miR-145, miR-23b, and miR-99a were upregulated in women with recurrent implantation failure [55]. Shi et al. compared endometrium collected during the window of implantation (WOI) of recurrent implantation failure (RIF) patients with that of infertile patients who conceived after one embryo transfer. Shi et al. also found miR-145 was upregulated in RIF patients, though there was much variability of other miRNAs analyzed. MiR-145 targets ERa, mucin 1, and RTKN; all of which have been implicated in implantation failure [54]. Park et al. analyzed miRNA expression profile in uterine aspirate taken 24 h ahead of frozen embryo transfer. They described 29 miRNAs upregulated with successful implantation and found that miR-891a, miR-522, miR-198 were downregulated in failed implantation [53]. Though there is significant variability in studies at present, miRNA expression profile may be associated with a successful rate of implantation.
Conclusions and future implications
As the world of reproductive technology continues into the era of genetics, physicians and patients are turning to genetic technologies such as embryo preimplantation genetic screening or endometrial receptivity assay in an effort to achieve the highest possible pregnancy and live birth rates.
miRNA analysis is an emerging frontier of diagnostics. Measured by minimally invasive body fluid or tissue analysis, miRNAs have opened a new and exciting pathway through which pathology, cellular communication, and control of gene expression may be understood. miRNAs have proven invaluable in the diagnosis of certain diseases previously based on a constellation of clinical suspicion and suggestive but not conclusive diagnostic tests. Though miRNAs were only discovered in 2001, research into them has exponentially increased and diagnostic miRNA testing is currently undergoing phase 2 clinical trials [56].
In this review, we reviewed work analyzing the potential of miRNA isolated from blastocyst and endometrium in predicting the window of implantation or likelihood of successful implantation. Freis et al. explored microRNA in serum during the window of implantation and found four miRNAs that could be possible biomarkers for implantation success [57]. However, studies profiling miRNA expression show inconsistent results due to the complexity of miRNA signaling, its rapid variation with time, and variation between individuals. Due to this, panel approaches will likely be most successful in predicting implantation. miRNA panels have also been studied for miscarriage prediction with one panel of seven miRNAs achieving a sensitivity of 87.5% and a specificity of 89.5% for miscarriage in IVF [28]. Panels of miRNA may increase the sensitivity and specificity of outcome prediction as compared with single miRNA testing; however, further study with larger power is required to ensure accuracy and reproducibility allowing for biological variation between individuals. Expanded miRNA panels and bioinformatics approaches may be on the horizon to make more reliable predictions around implantation.
miRNA functions are generally speculative based of expression patterns, and real-time functional research is lacking in mammalian development. Even if these studies are feasible, most miRNA functional research is based on cell culture models, and thus, the results may not be representative of complex physiological processes in vivo. The majority of miRNA extraction and quantification research is based on known miRNA assay and further qPCR confirmation. New methodology utilizing nanotechnology, such as DNA-carbon quantum dots, has been proposed for miRNA extraction but the probes used are specific to a miRNA [58]. Further research is required to discover new investigative technologies to extract very small amounts of miRNA and accurately identify unknown molecules. Though technical challenges abound, the world of miRNAs promises exciting vistas to the future of assisted reproduction.
Electronic supplementary material
(DOCX 15 kb)
Compliance with ethical standards
Conflict of interest
The authors declare that they have no conflict of interest
References
- 1.Paria BC, Huet-Hudson YM, Dey SK. Blastocyst’s state of activity determines the “window” of implantation in the receptive mouse uterus. Proc Natl Acad Sci U S A. 1993;90(21):10159–10162. doi: 10.1073/pnas.90.21.10159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu W, Niu Z, Li Q, Pang RTK, Chiu PCN, Yeung WSB. MicroRNA and embryo implantation. American Journal of Reproductive Immunology. 2016. [DOI] [PubMed]
- 3.Evans J, Salamonsen LA, Winship A, Menkhorst E, Nie G, Gargett CE, Dimitriadis E. Fertile ground: human endometrial programming and lessons in health and disease. Nat Rev Endocrinol. 2016;12:654–667. doi: 10.1038/nrendo.2016.116. [DOI] [PubMed] [Google Scholar]
- 4.Salamonsen LA, Evans J, Nguyen HPT, Edgell TA. The microenvironment of human implantation: Determinant of reproductive success. American Journal of Reproductive Immunology. 2016. [DOI] [PubMed]
- 5.Liang J, Wang S, Wang Z. Role of microRNAs in embryo implantation. Reprod Biol Endocrinol. 2017, 15; [DOI] [PMC free article] [PubMed]
- 6.Galliano D, Pellicer A. Fertility and sterility. 2014. MicroRNA and implantation; pp. 1531–1544. [DOI] [PubMed] [Google Scholar]
- 7.Enders AC, Schlafke S, Hendrickx AG. Differentiation of the embryonic disc, amnion, and yolk sac in the rhesus monkey. Am J Anat. 1986;177:161–185. doi: 10.1002/aja.1001770205. [DOI] [PubMed] [Google Scholar]
- 8.Diedrich K, Fauser BCJM, Devroey P, Griesinger G. The role of the endometrium and embryo in human implantation. Hum Reprod Update. 2007, 13, 365, 377; [DOI] [PubMed]
- 9.Bentin-Ley U, Sjogren A, Nilsson L, Hamberger L, Larsen JF, Horn T. Presence of uterine pinopodes at the embryo-endometrial interface during human implantation in vitro. Hum Reprod. 1999;14(2):515–520. doi: 10.1093/humrep/14.2.515. [DOI] [PubMed] [Google Scholar]
- 10.Davidson LM, Coward K. Molecular mechanisms of membrane interaction at implantation. Birth Defects Res C Embryo Today. 2016;108(1):19–32. doi: 10.1002/bdrc.21122. [DOI] [PubMed] [Google Scholar]
- 11.Chobotova K, Spyropoulou I, Carver J, Manek S, Heath JK, Gullick WJ, Barlow DH, Sargent IL, Mardon HJ. Heparin-binding epidermal growth factor and its receptor ErbB4 mediate implantation of the human blastocyst. Mech Dev. 2002;119(2):137–144. doi: 10.1016/S0925-4773(02)00342-8. [DOI] [PubMed] [Google Scholar]
- 12.Hamatani T, Carter MG, Sharov AA, Ko MSH. Dynamics of global gene expression changes during mouse preimplantation development. Dev Cell. 2004;6(1):117–131. doi: 10.1016/S1534-5807(03)00373-3. [DOI] [PubMed] [Google Scholar]
- 13.Song H, Lim H, Das SK, Paria BC, Dey SK. Dysregulation of EGF family of growth factors and COX-2 in the uterus during the preattachment and attachment reactions of the blastocyst with the luminal epithelium correlates with implantation failure in LIF-deficient mice. Mol Endocrinol. 2000;14(8):1147–1161. doi: 10.1210/mend.14.8.0498. [DOI] [PubMed] [Google Scholar]
- 14.Mohamed OA, Jonnaert M, Labelle-Dumais C, Kuroda K, Clarke HJ, Dufort D. Uterine Wnt/beta-catenin signaling is required for implantation. Proc Natl Acad Sci U S A. 2005;102(24):8579–8584. doi: 10.1073/pnas.0500612102. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cha J, Sun X, Dey SK. Mechanisms of implantation: strategies for successful pregnancy. Nat Med. 2012;18:1754–1767. doi: 10.1038/nm.3012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bloor DJ, Metcalfe AD, Rutherford A, Brison DR, Kimber SJ. Expression of cell adhesion molecules during human preimplantation embryo development. Mol Hum Reprod. 2002;8(3):237–245. doi: 10.1093/molehr/8.3.237. [DOI] [PubMed] [Google Scholar]
- 17.Kang Y-J, Forbes K, Carver J, Aplin JD. The role of the osteopontin-integrin alphavbeta3 interaction at implantation: functional analysis using three different in vitro models. Hum Reprod. 2014;29(4):739–749. doi: 10.1093/humrep/det433. [DOI] [PubMed] [Google Scholar]
- 18.Genbacev OD, Prakobphol A, Foulk RA, Krtolica AR, Ilic D, Singer MS, et al. Trophoblast L-selectin-mediated adhesion at the maternal-fetal interface. Science. 2003;299(5605):405–408. doi: 10.1126/science.1079546. [DOI] [PubMed] [Google Scholar]
- 19.Clark K, Pankov R, Travis MA, Askari JA, Mould AP, Craig SE, et al. A specific alpha5beta1-integrin conformation promotes directional integrin translocation and fibronectin matrix formation. J Cell Sci. 2005;118(Pt 2):291–300. doi: 10.1242/jcs.01623. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lessey BA, Castelbaum AJ, Sawin SW, Sun J. Integrins as markers of uterine receptivity in women with primary unexplained infertility. Fertil Steril. 1995;63(3):535–542. doi: 10.1016/S0015-0282(16)57422-6. [DOI] [PubMed] [Google Scholar]
- 21.Chen G, Xin A, Liu Y, Shi C, Chen J, Tang X, Chen Y, Yu M, Peng X, Li L, Sun X. Integrins beta1 and beta3 are biomarkers of uterine condition for embryo transfer. J Transl Med. 2016;14(1):303. doi: 10.1186/s12967-016-1052-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Dorostghoal M, Ghaffari H-O-A, Shahbazian N, Mirani M. endometrial expression of beta3 integrin, calcitonin and plexin-B1 in the window of implantation in women with unexplained infertility. Int J Reprod Biomed (Yazd, Iran). 2017;15(1):33–40. doi: 10.29252/ijrm.15.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Valadi H, Ekstrom K, Bossios A, Sjostrand M, Lee JJ, Lotvall JO. Exosome-mediated transfer of mRNAs and microRNAs is a novel mechanism of genetic exchange between cells. Nat Cell Biol. 2007;9(6):654–659. doi: 10.1038/ncb1596. [DOI] [PubMed] [Google Scholar]
- 24.Arroyo JD, Chevillet JR, Kroh EM, Ruf IK, Pritchard CC, Gibson DF, Mitchell PS, Bennett CF, Pogosova-Agadjanyan EL, Stirewalt DL, Tait JF, Tewari M. Argonaute2 complexes carry a population of circulating microRNAs independent of vesicles in human plasma. Proc Natl Acad Sci U S A. 2011;108(12):5003–5008. doi: 10.1073/pnas.1019055108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Vickers KC, Palmisano BT, Shoucri BM, Shamburek RD, Remaley AT. MicroRNAs are transported in plasma and delivered to recipient cells by high-density lipoproteins. Nat Cell Biol. 2011;13(4):423–433. doi: 10.1038/ncb2210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ambros V. MicroRNAs: tiny regulators with great potential. Cell. 2001;107(7):823–826. doi: 10.1016/S0092-8674(01)00616-X. [DOI] [PubMed] [Google Scholar]
- 27.Bagga S, Bracht J, Hunter S, Massirer K, Holtz J, Eachus R, Pasquinelli AE. Regulation by let-7 and lin-4 miRNAs results in target mRNA degradation. Cell. 2005;122(4):553–563. doi: 10.1016/j.cell.2005.07.031. [DOI] [PubMed] [Google Scholar]
- 28.Wu L, Fan J, Belasco JG. MicroRNAs direct rapid deadenylation of mRNA. Proc Natl Acad Sci U S A. 2006;103(11):4034–4039. doi: 10.1073/pnas.0510928103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. doi: 10.1016/S0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 30.Hanke M, Hoefig K, Merz H, Feller AC, Kausch I, Jocham D, Warnecke JM, Sczakiel G. A robust methodology to study urine microRNA as tumor marker: microRNA-126 and microRNA-182 are related to urinary bladder cancer. Urol Oncol. 2010;28(6):655–661. doi: 10.1016/j.urolonc.2009.01.027. [DOI] [PubMed] [Google Scholar]
- 31.Ng YH, Rome S, Jalabert A, Forterre A, Singh H, Hincks CL, Salamonsen LA. Endometrial exosomes/microvesicles in the uterine microenvironment: a new paradigm for embryo-endometrial cross talk at implantation. PLoS One. 2013;8(3):e58502. doi: 10.1371/journal.pone.0058502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Cuman C, Van Sinderen M, Gantier MP, Rainczuk K, Sorby K, Rombauts L, et al. Human blastocyst secreted microRNA regulate endometrial epithelial cell adhesion. EBioMedicine. 2015;2(10):1528–1535. doi: 10.1016/j.ebiom.2015.09.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Bartel DP. MicroRNAs: target recognition and regulatory functions. Cell. 2009;136(2):215–233. doi: 10.1016/j.cell.2009.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Kung JTY, Colognori D, Lee JT. Long noncoding RNAs: past, present, and future. Genetics. 2013;193(3):651–669. doi: 10.1534/genetics.112.146704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Burgess DJ. Gene regulation: multiple mechanisms of small RNAs. Nat Rev Genet. 2015;16(2):70. doi: 10.1038/nrg3902. [DOI] [PubMed] [Google Scholar]
- 36.Osella M, Riba A, Testori A, Corà D, Caselle M. Interplay of microRNA and epigenetic regulation in the human regulatory network. Vol. 5, Frontiers in Genetics. 2014. [DOI] [PMC free article] [PubMed]
- 37.Olsen PH, Ambros V. The LIN-4 regulatory RNA controls developmental timing in Caenorhabditis elegans by blocking LIN-14 protein synthesis after the initiation of translation. Dev Biol. 1999;216(2):671–680. doi: 10.1006/dbio.1999.9523. [DOI] [PubMed] [Google Scholar]
- 38.Ha M, Kim VN. Regulation of microRNA biogenesis. Nat Rev Mol Cell Biol. 2014;15(8):509–524. doi: 10.1038/nrm3838. [DOI] [PubMed] [Google Scholar]
- 39.Yi R, Qin Y, Macara IG, Cullen BR. Exportin-5 mediates the nuclear export of pre-microRNAs and short hairpin RNAs. Genes Dev. 2003;17(24):3011–3016. doi: 10.1101/gad.1158803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Kim Y-K, Kim B, Kim VN. Re-evaluation of the roles of DROSHA, export in 5, and Dicer in microRNA biogenesis. Proc Natl Acad Sci U S A. 2016;113(13):E1881–E1889. doi: 10.1073/pnas.1602532113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Baxter Bendus AE, Mayer JF, Shipley SK, Catherino WH. Interobserver and intraobserver variation in day 3 embryo grading. Fertil Steril. 2006;86(6):1608–1615. doi: 10.1016/j.fertnstert.2006.05.037. [DOI] [PubMed] [Google Scholar]
- 42.Capalbo A, Ubaldi FM, Cimadomo D, Noli L, Khalaf Y, Farcomeni A, Ilic D, Rienzi L. MicroRNAs in spent blastocyst culture medium are derived from trophectoderm cells and can be explored for human embryo reproductive competence assessment. Fertil Steril. 2016;105(1):225–235e3. doi: 10.1016/j.fertnstert.2015.09.014. [DOI] [PubMed] [Google Scholar]
- 43.Borges E, Setti AS, Braga DPAF, Geraldo M V., Figueira R de CS, Iaconelli A. miR-142-3p as a biomarker of blastocyst implantation failure - a pilot study. J Bras Reprod Assist 2016;20(4):200–205. [DOI] [PMC free article] [PubMed]
- 44.Rosenbluth EM, Shelton DN, Wells LM, Sparks AET, Van Voorhis BJ. Human embryos secrete microRNAs into culture media - a potential biomarker for implantation. Fertil Steril 2014;101(5):1493–1500. [DOI] [PubMed]
- 45.Rosenbluth EM, Shelton DN, Sparks AET, Devor E, Christenson L, Van Voorhis BJ. MicroRNA expression in the human blastocyst. Fertil Steril. 2013;99(3):855–861.e3. doi: 10.1016/j.fertnstert.2012.11.001. [DOI] [PubMed] [Google Scholar]
- 46.McCallie B, Schoolcraft WB, Katz-Jaffe MG. Aberration of blastocyst microRNA expression is associated with human infertility. Fertil Steril. 2010;93(7):2374–2382. doi: 10.1016/j.fertnstert.2009.01.069. [DOI] [PubMed] [Google Scholar]
- 47.Sathyapalan T, David R, Gooderham NJ, Atkin SL. Increased expression of circulating miRNA-93 in women with polycystic ovary syndrome may represent a novel, non-invasive biomarker for diagnosis. Sci Rep. 2015;5:16890. doi: 10.1038/srep16890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kuokkanen S, Chen B, Ojalvo L, Benard L, Santoro N, Pollard JW. Genomic profiling of microRNAs and messenger RNAs reveals hormonal regulation in microRNA expression in human endometrium. Biol Reprod. 2010;82(4):791–801. doi: 10.1095/biolreprod.109.081059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Kresowik JDK, Devor EJ, Van Voorhis BJ, Leslie KK. MicroRNA-31 is significantly elevated in both human endometrium and serum during the window of implantation: a potential biomarker for optimum receptivity1. Biol Reprod 2014;91(1):1–6. [DOI] [PMC free article] [PubMed]
- 50.Tochigi H, Kajihara T, Mizuno Y, Mizuno Y, Tamaru S, Kamei Y, Okazaki Y, Brosens JJ, Ishihara O. Loss of miR-542-3p enhances IGFBP-1 expression in decidualizing human endometrial stromal cells. Sci Rep. 2017;7:40001. doi: 10.1038/srep40001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Estella C, Herrer I, Moreno-Moya JM, Quiñonero A, Martínez S, Pellicer A, et al. MiRNA signature and Dicer requirement during human endometrial stromal decidualization in vitro. PLoS One. 2012;7(7). [DOI] [PMC free article] [PubMed]
- 52.Altmäe S, Martinez-Conejero JA, Esteban FJ, Ruiz-Alonso M, Stavreus-Evers A, Horcajadas JA, Salumets A. MicroRNAs miR-30b, miR-30d, and miR-494 regulate human endometrial receptivity. Reprod Sci. 2013;20(3):308–317. doi: 10.1177/1933719112453507. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Parks JC, McCallie BR, Strieby A, McReynolds S, Schoolcraft WB, Katz-Jaffe MG. Non-invasive omics analysis of endometrial secretions 24 hours prior to frozen embryo transfer is predictive of implantation outcome. Fertil Steril. 2014;102(3):e134–e135. doi: 10.1016/j.fertnstert.2014.07.460. [DOI] [Google Scholar]
- 54.Shi C, Shen H, Fan LJ, Guan J, Zheng XB, Chen X, et al. Endometrial microRNA signature during the window of implantation changed in patients with repeated implantation failure. Chin Med J. 2017;130(5):566–573. doi: 10.4103/0366-6999.200550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Revel A, Achache H, Stevens J, Smith Y, Reich R. MicroRNAs are associated with human embryo implantation defects. Hum Reprod. 2011;26(10):2830–2840. doi: 10.1093/humrep/der255. [DOI] [PubMed] [Google Scholar]
- 56.Haouzi D, Drissennek L, Antoine Y, Entezami F, Gala A, Mullet T, Vincens C, Hamamah S. Identification of human endometrial microRNAs associated with repeated implantation failures. Fertil Steril. 2016;106(3):e218. doi: 10.1016/j.fertnstert.2016.07.627. [DOI] [Google Scholar]
- 57.Freis A, Keller A, Ludwig N, Meese E, Jauckus J, Rehnitz J, Capp E, Strowitzki T, Germeyer A. Altered miRNA-profile dependent on ART outcome in early pregnancy targets Wnt-pathway. Reproduction. 2017;154(6):799–805. doi: 10.1530/REP-17-0396. [DOI] [PubMed] [Google Scholar]
- 58.Khakbaz F, Mahani M. Micro-RNA detection based on fluorescence resonance energy transfer of DNA-carbon quantum dots probes. Anal Biochem. 2017;523:32–38. doi: 10.1016/j.ab.2017.01.025. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(DOCX 15 kb)