Abstract
We present the LiSEQ (Listeria SEQuencing) project, funded by the European Food Safety Agency (EFSA) to compare Listeria monocytogenes isolates collected in the European Union from ready-to-eat foods, compartments along the food chain (e.g. food-producing animals, food-processing environments) and humans. In this article, we report the molecular characterization of a selection of this data set employing whole-genome sequencing analysis. We present an overview of the strain diversity observed in different sampled sources, and characterize the isolates based on their virulence and resistance profile. We integrate into our analysis the global L. monocytogenes genome collection described by Moura and colleagues in 2016 to assess the representativeness of the LiSEQ collection in the context of known L. monocytogenes strain diversity.
Keywords: Listeria monocytogenes, whole-genome sequencing, genetic diversity, phylogeny, food, human
Data Summary
All fastq reads from this study have been deposited in the National Center for Biotechnology Information short-read archive (SRA) under the BioProject PRJNA475189.
Significance as a BioResource to the community.
The LiSEQ (Listeria SEQuencing) genome collection represents a valuable resource of Listeria monocytogenes for further study. It provides a framework to answer questions on genetic diversity amongst different sources assayed in this strain collection, as well as to explore possible epidemiological links between isolates from across Europe.
Introduction
Listeria monocytogenes is an opportunistic pathogen that causes a range of illnesses from mild febrile gastroenteritis to more severe invasive infections, including bacteraemia and meningitis (listeriosis) [1]. Whilst listeriosis is a relatively rare disease, it has a high fatality rate of 20–30 % and, therefore, the burden of the disease is high [2]. Some populations have an increased susceptibility, including the elderly, immunosuppressed patients, pregnant women, their foetuses and neonates [1]. The majority of cases appear to be sporadic, although outbreaks are not uncommon. In the European Union (EU) in 2016, a total of 2536 confirmed human cases were reported by 28 member states, corresponding to an EU notification rate of 0.47 cases per 100 000 population. The highest notification rates were observed in Finland, Belgium, Germany, Slovenia and Denmark, with 1.22, 0.92, 0.85, 0.73 and 0.70 cases per 100 000 population, respectively [3].
Cases of listeriosis are frequently associated with the consumption of contaminated ready-to-eat (RTE) food products, with meat and fish products and soft and semi-soft cheeses often identified as vehicles of infection [2]. L. monocytogenes can be found in both raw foods and in processed foods that are contaminated during and/or after processing. L. monocytogenes can survive and replicate at low temperatures (with a minimal growth temperature of −2 °C) and has the capacity to persist in food-processing environments, sometimes for years [4]. Contamination of food-processing environments is often the route by which RTE food becomes contaminated [5] and those foods with a relatively long shelf life are of particular public-health concern [6]. Identifying potential food vehicles and understanding how foods become contaminated is paramount to developing and implementing effective control and preventative measures, and the typing of L. monocytogenes isolates plays a crucial role in such investigations [7–9].
An EU-wide baseline survey (BLS) was conducted in 2010 and 2011 to estimate the prevalence of L. monocytogenes contamination in three RTE food categories at retail in accordance with EU decision 2010/678/EU: packaged (not frozen) smoked or gravad fish (3053 samples), packaged heat-treated meat products (3530 samples) and soft or semi-soft cheeses (3452 samples). The prevalence estimates were published in 2013 [10]. The percentage of fish in the EU contaminated with L. monocytogenes at the time of sampling was 10.4 % and at the end of shelf-life 10.3 %, whilst the level for contaminated meat and cheese samples at the end of shelf-life was 2.07 and 0.47 %, respectively. In the EU, the proportion of smoked or gravad fish samples with a L. monocytogenes count exceeding the level of 100 c.f.u. g−1 was 1.7 % at the end of shelf-life. For meat products, this proportion was 0.43 %, whilst for the cheese it was 0.06 %.
Several phenotypic and genotypic methods have been used worldwide for typing L. monocytogenes. Traditionally, serotyping, based on the agglutination of somatic (O) and flagellar (H) antigens, classifying L. monocytogenes into at least 13 serotypes, has been the first level of subtype discrimination [11]. However, as only three serotypes cause over 95 % of invasive infection, molecular typing methods are employed for greater strain discrimination with PFGE being, until recently, the gold-standard method for L. monocytogenes [12].
Multilocus sequence typing (MLST) has been used to study and describe the population structure and phylogeny of many bacterial pathogens, and has shown that L. monocytogenes forms a structured population consisting of four divergent lineages (I– IV) [11, 13]. Each lineage comprises specific serotypes: with lineage I containing serotypes 1/2b, 3b, 4b, 4e and 7; lineage II, serotypes 1/2a, 1/2 c, 3a and 3 c; lineage III, serotypes 4b, 1/2a, 4a and 4 c; and lineage IV, 4a and 4 c. The genetic lineages have different, although at times overlapping, genetic, phenotypic and epidemiological characteristics with the majority of human illness caused by strains in lineages I and II [11].
With the advent of whole-genome sequencing (WGS) technologies, entire bacterial genomes are now readily available for analysis affording the highest level of strain discrimination, the ability to infer phylogenetic relationships and access to a wealth of additional information such as virulence and resistance markers. A recently developed core genome MLST scheme has been described for L. monocytogenes by Moura and colleagues [7] encompassing 1748 loci, which has been used to describe the global population structure of the species. Furthermore, WGS has been used in several national studies for outbreak detection and investigations, e.g. in Austria [14], Australia [15], the USA [16], Denmark [17] and France [18]. The improvements in strain resolution obtained with WGS analyses provided robust genetic evidence for linking cases and more accurate case definitions than PFGE, enabling cases to be ruled in or out of outbreaks.
We present results derived from a study funded by the European Food Safety Agency (EFSA) to compare L. monocytogenes isolates collected in the EU from RTE foods, compartments along the food chain (e.g. food-producing animals, food-processing environments) and humans. In this article, we report the molecular characterization of a selection of L. monocytogenes isolates from the above sources, and human clinical cases employing WGS analysis.
Methods
Strain selection
A total of 1143 L. monocytogenes isolates were selected to be part of the LiSEQ (Listeria SEQuencing) study. These encompassed and included those from the EU-wide RTE BLS [10] and were collected from: different compartments of the food-production chain (n=200); sporadic clinical cases (n=262) that were temporally and geographically matched to the RTE BLS (n=353); and isolates associated with outbreaks (n=105). The selected BLS isolates consisted of 353 strains originating from 22 member states and 1 non-member state with 297, 49 and 7 strains isolated, respectively, from RTE fishery products, meat products, and soft and semi-soft cheeses. To compensate for the excess of BLS isolates from RTE fish products, additional isolates (n=223) from RTE meat products and cheeses, collected during the years 2010–2011, were obtained from eight different EU member states to ensure a more equal distribution of isolates across each of the three RTE food categories. Clinical isolates from assumed sporadic human cases collected during the BLS period 2010–2011 were also included. Selection priority was according to availability of the isolate and the complementary typing data and country disease incidence. Additionally, isolates from raw food sampled at fish, meat and milk-product production sites, as well as environmental isolates from these sites, were also included. Isolates associated with nine retrospective outbreaks, including those from human cases and, where applicable, the confirmed food source, were selected, representing outbreaks with different sources and occurring in different geographical regions. A summary of the final set of strains included in the project is given in Table 1 and a complete table of meta data is provided in Table S1 (available with the online version of this article).
Table 1. Summary of the strains included in the LiSEQ study.
| Country | BLS | Other foods | Food-production chain | Clinical, sporadic | Outbreak | Total |
|---|---|---|---|---|---|---|
| A | 7 | 29 | 35 | 71 | ||
| B | 4 | 28 | 68 | 31 | 43 | 174 |
| C | 35 | 83 | 32 | 35 | 25 | 210 |
| D | 4 | 20 | 24 | |||
| E | 6 | 6 | ||||
| F | 15 | 8 | 23 | |||
| G | 4 | 4 | 8 | |||
| H | 5 | 5 | ||||
| J | 10 | 10 | ||||
| K | 14 | 14 | ||||
| L | 54 | 54 | ||||
| M | 2 | 2 | ||||
| N | 9 | 9 | ||||
| P | 3 | 4 | 7 | |||
| Q | 33 | 100 | 23 | 156 | ||
| R | 4 | 4 | ||||
| S | 4 | 4 | ||||
| T | 4 | 20 | 5 | 29 | ||
| U | 62 | 62 | ||||
| V | 6 | 28 | 34 | |||
| W | 7 | 15 | 22 | |||
| X | 38 | 34 | 35 | 32 | 139 | |
| Y | 8 | 20 | 28 | |||
| Z | 15 | 13 | 20 | 48 | ||
| Total | 353 | 223 | 200 | 262 | 105 | 1143 |
To assess the representativeness of the genetic diversity afforded by the isolates in this study, the LiSEQ results were placed into a global context via comparison with the L. monocytogenes collection of 1696 genomes from Moura and colleagues [7], which represent a larger geographically distributed data set.
Sequencing and bioinformatics analysis
Bacterial isolate growth was harvested into a 96-well processing plate and treated with lysozyme at 37 °C for 1 h, followed by proteinase K overnight at 56 °C with gentle shaking. Lysates were heated to 95–100 °C for 10 min to ensure inactivation of any non-lysed bacterial cells. Samples were then treated with ribonuclease A for 15 min at 37 °C, centrifuged and the supernatants transferred to an automated nucleic acid extraction platform, Qiagen’s QiaSymphony. The yield and purity of extracted DNA was assessed using the Life Technologies Quant-iT high sensitivity 96-well assay and the GloMax Multi+Detection and LabChip DX systems. DNA was diluted to 10–30 ng µl−1.
Paired-end libraries were generated using the Illumina Nextera XT sample preparation kit. Assessment of fragment sizes was performed on the Perkin Elmer LabChip GX after fragmentation and clean-up. After normalization, samples were pooled and library quantification was performed using the KAPA library quantification kit for Illumina sequencing, on an ABI ViiA7 system. Paired-end sequencing was performed on the Illumina HiSeq 2500 instrument using the TruSeq Rapid SBS kit (200 cycle) and TruSeq paired-end rapid cluster kit. The following cycle parameters were used for sequencing: read 1 : 101, index read 1 : 8, index read 2 : 8 and read 2 : 101. rta version 1.17.21.3 was used for generation of base call files. fastq creation and de-multiplexing via casava was performed on a dedicated high performance cluster (HPC). fastq reads were quality trimmed using Trimmomatic [19] with bases removed from the trailing end that fell below a phred score of 30. If the read length post-trimming was less than 50, the read and its pair were discarded. If the post-trimmed yield was less than 150 megabases, the sample was discarded. A kmer (a short string of DNA of length k) based approach was used (https://github.com/phe-bioinformatics/kmerid) to confirm the identity of the sample and to ensure the sequence was free from contamination. If any non-Listeria kmers were identified in the fastq reads, the sample was discarded. All fastq reads from this study can be found in the National Center for Biotechnology Information short-read archive under BioProject PRJNA475189.
The MLST sequence type (ST) as defined by the Pasteur scheme [13] was extracted from each sequence using most (https://github.com/phe-bioinformatics/MOST) [20] and assigned a clonal complex (CC) in accordance with the Institut Pasteur international MLST database for L. monocytogenes designation (http://bigsdb.pasteur.fr/listeria). One preselected isolate was found to be Listeria innocua; thus, it was excluded from further analysis.
Short reads were assembled using Spades assembly (version 3.5.0) run with Kmer 21, 33, 55, 77, 83, and the ‘only-assembler’ option [21]. Core genome SNP phylogenies were constructed on the obtained assemblies using Parsnp [22].
Resistance to tetracycline, penicillin, quaternary ammonium sanitizers and antiseptics (such as benzalkonium chloride) were assayed in silico from the genomes of the isolates in this study. Tetracycline resistance was inferred by the presence of tetM and tetS, and penicillin resistance by the presence of penA. Resistance to quaternary ammonium sanitizers and antiseptics was inferred by the presence of the bcrABC locus, the Tn6188_qac transposon, and/or the efflux pumps emrE [23] and qacA [24]. For detection of gene presence, ‘paired-end’ reads of each strain were mapped against the reference gene sequences using Bowtie2 v.2.2.5. [25]. The resulting alignment sam files were then converted into bam files and sorted by using SAMtools [26]. Genes were defined as detected if they covered greater than 80 % of the query sequence with greater than 80 % nucleotide identity. Genes with coverage less than 100 % were also classified as truncated.
A comprehensive set of 115 genes identified as putative or confirmed virulence factors were used as described in two previous studies [27; 28]. The gene sequences were extracted from L. monocytogenes EGD-e (accession no. NC_003210.1) except for the Listeria pathogenicity island (LIPI)-3/LIPI-4 cluster of genes, which was extracted from L. monocytogenes F2365. Genes were detected as above.
Results
Population structure
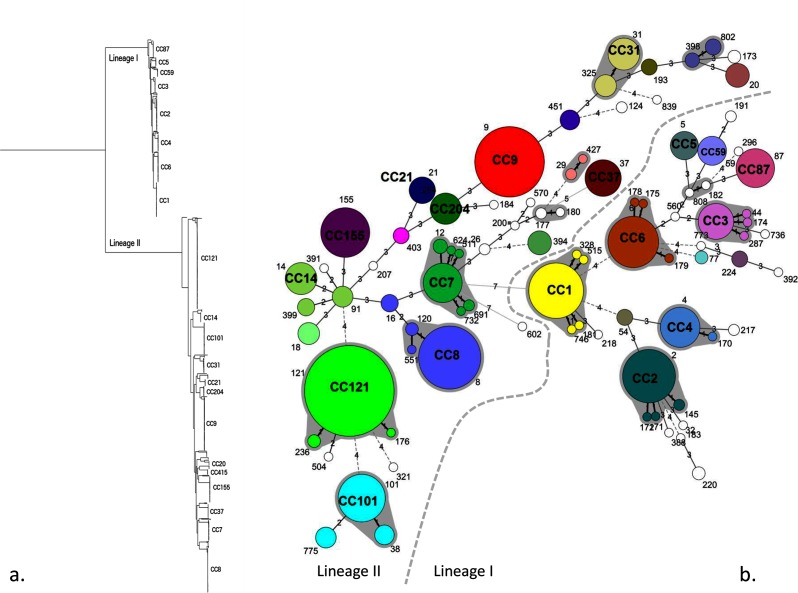
Of the 1143 isolates sequenced, 42 different CCs and 13 singleton STs (unassigned CCs) were identified. One isolate was L. innocua; thus, it was excluded from further analysis. One isolate could not be assigned to any ST or CC due to an incomplete MLST profile. Ten CCs accounted for 70 % of the isolates (Table 2). The MLST population structure of the isolates in this study is further described as a minimum spanning tree (Fig. 1b). All isolates in the study clustered in either lineage I or II and the population structure based on whole-genome SNPs is displayed in the phylogenetic tree in Fig. 1(a). From the phylogenetic analysis, it can be seen that there is a clear delineation between lineages and the MLST CCs within lineages.
Table 2. The CCs identified and the number of isolates by isolation context and listed in the strain selection information.
Minor CCs (i.e. CCs with less than 10 isolates) included CC398, CC11, CC193, CC224, CC403, CC54, CC177, CC19, CC220, CC29, CC77, CC217, CC26, CC379, CC207, CC218, CC388, CC475, CC88, CC89, ST184, ST200, ST32, ST382, ST392, ST560, ST570, ST602, ST736, ST773 and ST839 (ordered according to occurrence).
| CC | Lineage | RTE food | Food-chain processing | Clinical, sporadic | Outbreak related | Total |
|---|---|---|---|---|---|---|
| CC121 | II | 144 | 37 | 6 | 0 | 187 |
| CC9 | II | 81 | 15 | 14 | 0 | 110 |
| CC8 | II | 69 | 5 | 24 | 0 | 98 |
| CC1 | I | 10 | 4 | 50 | 8 | 72 |
| CC2 | I | 19 | 29 | 20 | 0 | 68 |
| CC101 | II | 10 | 41 | 16 | 0 | 67 |
| CC6 | I | 30 | 3 | 28 | 0 | 61 |
| CC155 | II | 32 | 1 | 8 | 13 | 54 |
| CC7 | II | 16 | 4 | 16 | 8 | 44 |
| CC14 | II | 13 | 2 | 9 | 13 | 37 |
| CC4 | I | 1 | 1 | 10 | 24 | 36 |
| CC87 | I | 10 | 0 | 4 | 19 | 33 |
| CC31 | II | 24 | 7 | 1 | 0 | 32 |
| CC3 | I | 18 | 7 | 6 | 0 | 31 |
| CC37 | II | 9 | 15 | 5 | 0 | 29 |
| CC204 | II | 17 | 3 | 1 | 0 | 21 |
| CC59 | I | 10 | 0 | 4 | 4 | 18 |
| CC5 | I | 7 | 6 | 4 | 0 | 17 |
| CC21 | II | 13 | 0 | 2 | 0 | 15 |
| CC20 | II | 8 | 2 | 2 | 0 | 12 |
| CC415 | II | 0 | 2 | 0 | 9 | 11 |
| CC18 | II | 0 | 6 | 4 | 0 | 10 |
| Minor CCs | LI=32 LII=47 |
35 | 9 | 28 | 7 | 79 |
| Total | 576 | 199 |
262 | 105 | 1142 |
Fig. 1.
(a) Core genome SNP maximum-likelihood phylogeny of L. monocytogenes genome sequences with the clades annotated by 7 loci MLST CC. (b) Minimum spanning tree of the isolates included in this study as described by 7 locus MLST. Each circle represents a single ST that is numbered on the tree. Major CCs defined by single locus variants are shaded in grey. The number of loci that differ between STs is labelled on the branches.
There was an uneven distribution in terms of origin of isolates (food, food-processing environment and clinical) between the two lineages (Chi squared test, P value 5×10−29). Across the CCs, within each lineage there was also an uneven distribution of food, food-processing environment and clinical isolates (lineage I, Chi squared test, P value 8.55×10−24; lineage II, Chi squared test, P value 1.95×10−30).
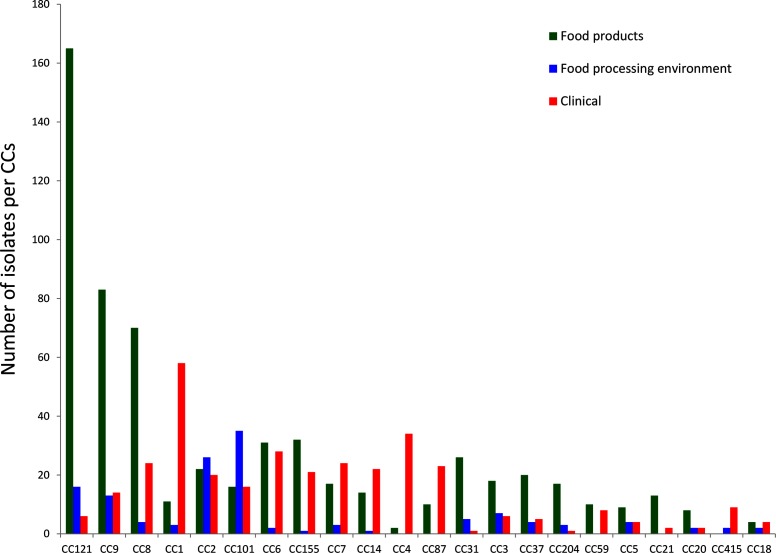
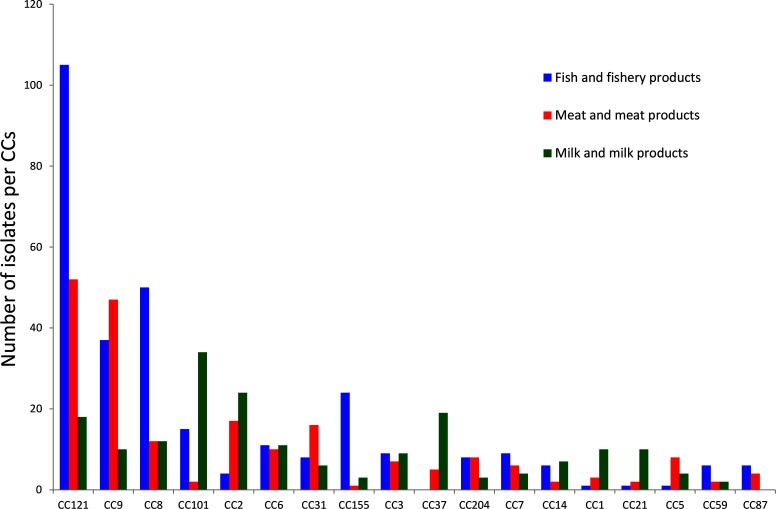
A total of 51 % of the isolates in lineage I were from humans, compared to only 19 % of the isolates from lineage II. The proportion of food isolates in lineage I was 37% and in lineage II it was 69 %. Across the sampled population, some CCs were significantly enriched with food isolates (e.g. CC121) and others were more associated with human cases (CC1, CC4) (Fig. 2). Some CCs were more representative of a food category, for example CC121 for fish/fishery products and CC101 for milk/milk products (Fig. 3).
Fig. 2.
Distribution of CCs in RTE food and from human clinical infections.
Fig. 3.
Distribution of CCs from the three major food-product categories, including isolates from food-processing environments.
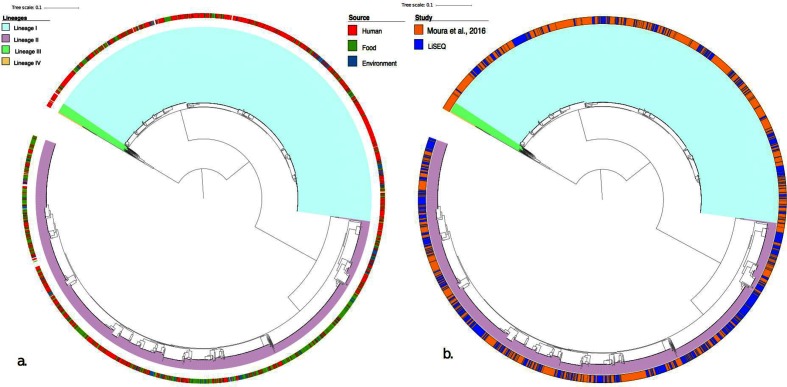
To assess how the sampled genetic diversity in this study corresponded to that described in a global context, the 1696 genomes from the study by Moura et al. [7] were integrated into the analysis. Fig. 4 shows a core SNP phylogeny of the combined LiSEQ and global data set. The LiSEQ isolates cluster within the global diversity and all major CCs are represented in the study data. A similar distribution of food and clinical isolates are also observed in the data set, with a predominance of clinical isolates found within lineage I and an excess of food isolates found within lineage II (Figs S1 and S2).
Fig. 4.
Core SNP tree built with Parsnp showing lineages for L. monocytogenes. (a) The external ring shows the source of isolates. (b) The external ring describes the study origin of the isolates: orange, from the study by Moura et al. [7]; and blue, from LiSEQ.
Resistance and virulence
Table 3 shows the percentage of strains in the study collection harbouring the assayed resistance genes. The resistance profile for each strain is included in Table S2. Less than 1 % of isolates harboured tetracycline-resistance genes (tetM) with no detection of tetS. Benzalkonium chloride-resistance genes were found with 18.5 % of isolates carrying the Tn6188_qac transposon and approximately 5 % of isolates carrying bcrABC loci. Less than 1 % of isolates harboured the efflux proteins emrE and qacA, whilst the efflux protein qacE was found in 18.3 % of isolates and generally found in conjunction with Tn6188_qac. No isolates showed the presence of penA, which is involved in resistance to penicillin.
Table 3. Percentage of isolates in the study harbouring the assayed resistance genes.
| Gene | Moura et al. [718]
(% detection) |
LiSEQ
(% detection) |
|---|---|---|
| tetM | 0.3 | 0.6 |
| tetS | 0 | 0 |
| bcrA | 4.6 | 4.5 |
| bcrB | 4.5 | 4.5 |
| bcrC | 4.5 | 4.4 |
| emrE | 0.8 | 0.3 |
| qacA | 0.2 | 0.5 |
| qacE | 14.9 | 18.7 |
| Tn6188_qac | 15.0 | 18.9 |
| penA | 0 | 0 |
The proportion of resistance determinants observed in the LiSEQ data displays a high correlation with the Moura et al. [7] data set. The highest percentage difference was observed with the gene qacE and the transposon Tn6188_qac efflux mediated benzalkonium chloride resistance, which is approximately 3 % higher in the LiSEQ data set.
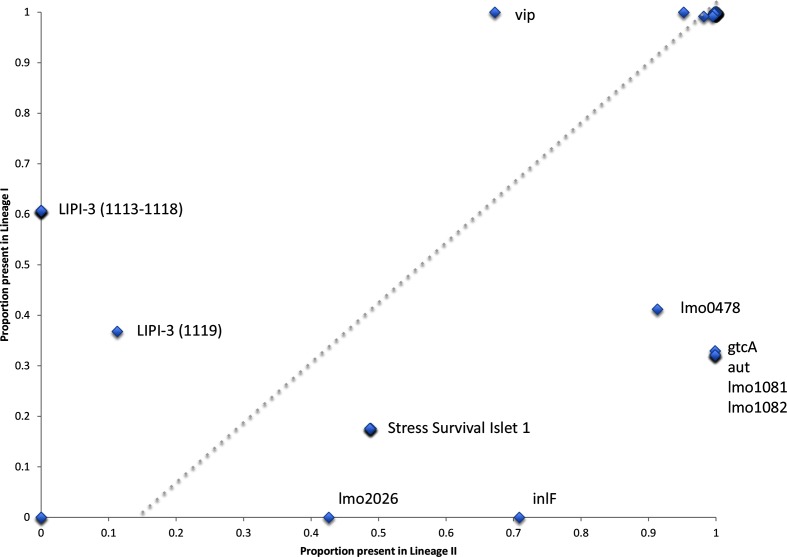
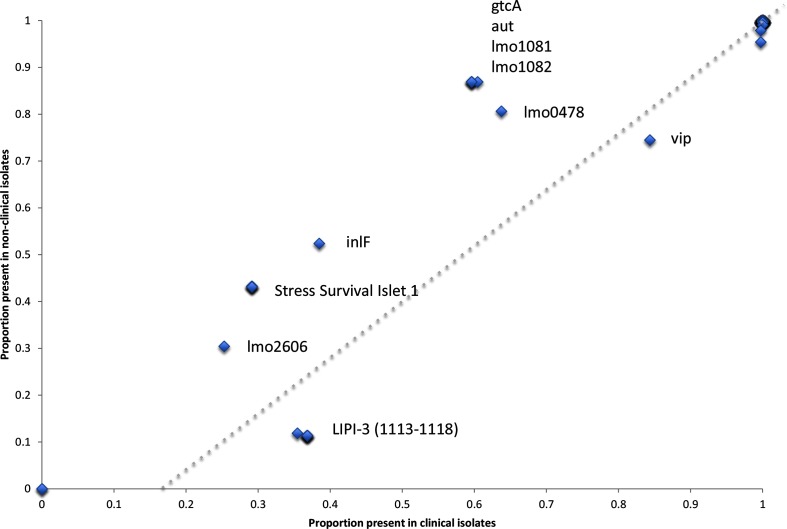
Table S2 shows the presence and absence of 115 putative virulence markers across the strain collection. Of the 115 markers, 2 were absent across all isolates, conversely 92 markers were present in greater than 95 % of isolates. Fig. 5 shows for each virulence marker the proportion present in linage I and lineage II isolates, and Fig. 6 shows for each virulence marker the proportion present in clinical and non-clinical isolates.
Fig. 5.
Scatter plot showing the proportion of each of the 115 putative virulence markers found in lineage I or lineage II (only significant results have been labelled).
Fig. 6.
Scatter plot showing the proportion of each of the 115 putative virulence markers found in clinical or non-clinical isolates (only significant results have been labelled).
In total, 21 putative virulence markers had significant variability in their detection across the strain collection. As described by Maury et al. [28], the Listeria pathogenicity island 3 (LIPI-3) was found in 60 % of isolates from lineage I (ubiquitous in CC1, CC3, CC4 and CC6), but completely absent in lineage II isolates. LIPI-3 loci 1119 (IIsP gene LMOf2365_1119) showed a different presence and absence profile to the other LIPI-3 alleles, being found in a minority of lineage II isolates (12/187 CC121, 11/54 CC155, 14/98 CC8 and 11/110 CC9) in the absence of the other LIPI-3 loci. Conversely, in lineage I some isolates do not possess LIPI-3 loci 1119 (IIsP gene) and have an otherwise intact LIPI-3. The Listeria pathogenicity island 4 (LIPI-4) was found in only 81 strains (0.07 % of the data set). It was detected in all the CC4 isolates that were tested, in agreement with the findings of Maury et al [28], and also in all the CC87 isolates that were investigated.
The known virulence surface protein Vip [29] was found across all isolates in lineage I, but only in 70 % of lineage II isolates (absent in CC204, CC21, CC31 and CC37, and 1/43 of CC7 isolates and 3/98 CC8 isolates). Several putative virulence factors were found in a greater proportion in lineage II isolates compared to lineage I isolates. These included the internalins lmo2026 (absent in lineage I and ubiquitous in CC155, CC18, CC20, CC204, CC21, CC37, CC415, CC7 and CC9 lineage II isolates) and inlF (absent in lineage I and only absent in CC121 and CC14 of lineage II), which have previously been shown to be detected variably in different serotypes [30]. The five gene locus termed the stress survival islet (SSI-1) [31, 32], which has previously been associated with growth of L. monocytogenes under sub-optimal conditions, contributing to survival of certain strains in food environments, was over-represented in lineage II isolates. However, when we consider the number of CCs, this association is less clear. SSI-1 is present in CC3 and CC5 of lineage I, and conversely absent in lineage II CCs 101, 121, 14, 20, 21, 415 and 7.
Ubiquitous amongst lineage II isolates was the rmlACBD l-rhamnose biosynthesis loci (lmo1081 and lmo182) [33] and gtcA [34], both of which are involved in L. monocytogenes cell wall teichoic acid production. The former involved in providing protection against the activity of antimicrobial peptides and the later in teichoic acid glycosylation. The autolysin aut (lmo1076), which has a proposed role in entry of L. monocytogenes into non-phagocytic mammalian cells [35], was found across all lineage II isolates, but only in CC3, CC5, CC59 and CC87 of lineage I; however, the shorter variant LMOF2365_RS00075 was found across all lineage I isolates. Finally, the surface adhesion lapB required for entry into mammalian cells is present across all lineages, but absent in all isolates of CC31.
Loss of function through partial gene deletion or miss-sense mutations is also known to be important in virulence attenuation. To explore this, genes with less than 100 % coverage of the query sequence were designated as truncated (see Table S2). Several genes had a loss of function truncation in lineage II but were found intact in lineage I, these included the already described inlA deletions [28], as well as lmo0257, the terminal SSI loci lmo0478, the autolysin gene ami and the actin-assembly inducing protein precursor gene actA. Conversely several genes were disrupted in lineage I, but intact in lineage II isolates. These included the internalins inlH, inlJ, lmo1290, the stress protein clpB and the flagellar motor switch protein lmo0698.
Discussion
The main objective of this study was to compare L. monocytogenes isolates collected in the EU from RTE foods, compartments along the food chain and from human cases, and highlights the value of revisiting well-structured surveys. A total of 1142 L. monocytogenes isolates were analysed, including 333 human clinical isolates and 809 isolates from the food chain.
Phylogenetic analysis showed a clear delineation between L. monocytogenes lineages and between CCs within lineages. The association of isolate type was unevenly distributed across the genetic diversity, with CCs within lineage I strongly associated with clinical cases and lineage II strongly associated with isolates from food. The diversity and distribution observed in this study were consistent with those previously described in a globally representative data set [28, 36–38].
As well as affording high-resolution typing and phylogenetic context, WGS provides immediate access to a wealth of additional data. Antimicrobial resistance in Listeria sp. has been studied in various food, environmental and clinical settings [39, 40, 41, 42]. L. monocytogenes has generally been shown to be more susceptible to antimicrobial agents than other species in the genus, such as L. innocua [31]. In this study, we found remarkable low-prevalence genes encoding resistances to tetracycline (<0.1 %) and penicillin (1 %). Genes conferring resistance to detergents and antiseptics via efflux activity were detected at a prevalence approaching 20 %. Whilst it is encouraging that the isolates in this study show potentially low levels of antimicrobial resistance, it is important to remain vigilant for emerging resistance. WGS allows antimicrobial-resistance monitoring to be done at no additional cost if WGS is part of routine microbial surveillance and, therefore, allows this potential threat to be monitored going forward.
WGS data were also assessed for the presence of 115 putative markers of virulence. More than 80 % of markers were present in more than 95 % of the isolates suggesting that most putative markers described in the literature are ubiquitous across L. monocytogenes lineages I and II. The majority of markers not present in all isolates were over-represented in food and/or lineage II isolates, with markers associated with stress survival or cell wall modification being particularly enriched. Conversely, the recently discovered Listeria pathogenicity island 3 and the surface protein VIP were more likely to be found in clinical and/or lineage I isolates. Although most virulence markers were present in all strains, it is not known whether the genes are expressed. Further work is needed, including the determination of truncation and non-sense mutations that have been shown to be associated with changes in virulence in particular that associated with the internalin genes [28]. Several truncations were identified in virulence genes across the data set, with some having an increased propensity for truncation dependent on lineage.
The WGS data generated represents a valuable resource for further studies. The LiSEQ isolates have all been typed using current molecular methods and, thus, can be used to demonstrate the back compatibility of WGS with historical data and also to assess bioinformatic programmes that are able to predict such typing results from WGS data. WGS has allowed us to define the population of L. monocytogenes from this study to an unprecedented resolution. It has provided the framework to answer questions on genetic diversity amongst different sources assayed in this strain collection, as well as to explore possible epidemiological links between isolates.
Another application of WGS data is related to the improvement of quantitative microbial risk assessment [43, 44]. It has been recently proposed that more targeted risk assessments focused on subpopulations that pose the greatest risk should be performed, e.g. those that have an enhanced ability to survive or grow in the food chain or those considered to be more pathogenic [44–46]. The characterization of CCs, and virulence, stress and antibiotic markers of strains circulating in the EU in RTE foods, as described in this study, provides the opportunity for improved risk assessments for L. monocytogenes exposure [2].
Funding information
This work was funded by EFSA, contract number OC/EFSA/BIOCONTAM/2014/01-CT 1 on “Closing gaps for performing a risk assessment on Listeria monocytogenes in ready‐to‐eat (RTE) foods: activity 3, the comparison of isolates from different compartments along the food chain, and from humans using whole genome sequencing (WGS) analysis’, EFSA‐Q‐2014‐00 026.
Acknowledgements
A. P., T. D. and K. G. are affiliated to the National Institute for Health Research – Health Protection Research Unit (NIHR HPRU) in Gastrointestinal Infections at University of Liverpool in partnership with Public Health England, in collaboration with the University of East Anglia, the University of Oxford and the Quadram Institute. A. P., T. D. and K. G. are based at Public Health England. The views expressed are those of the author(s) and not necessarily those of the NHS, the NIHR, the Department of Health or Public Health England.
Conflicts of interest
The authors declare that there are no conflicts of interest.
Footnotes
Abbreviations: BLS, baseline survey; CC, clonal complex; EFSA, European Food Safety Agency; EU, European Union; MLST, multilocus sequence typing; RTE, ready-to-eat; ST, sequence type; WGS, whole-genome sequencing.
All supporting data, code and protocols have been provided within the article or through supplementary data files. Two supplementary figures and two supplementary tables are available with the online version of this article.
Supplementary Data
References
- 1.Allerberger F, Wagner M. Listeriosis: a resurgent foodborne infection. Clin Microbiol Infect. 2010;16:16–23. doi: 10.1111/j.1469-0691.2009.03109.x. [DOI] [PubMed] [Google Scholar]
- 2.Ricci A, Allende A, Bolton D, Chemaly M., EFSA Panel on Biological Hazards (BIOHAZ) et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018;16:5134. doi: 10.2903/j.efsa.2018.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.EFSA, ECDC The European Union summary report on trends and sources of zoonoses, zoonotic agents and food-borne outbreaks in 2016. EFSA J. 2017;15:5077. doi: 10.2903/j.efsa.2017.5077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Holch A, Webb K, Lukjancenko O, Ussery D, Rosenthal BM, et al. Genome sequencing identifies two nearly unchanged strains of persistent Listeria monocytogenes isolated at two different fish processing plants sampled 6 years apart. Appl Environ Microbiol. 2013;79:2944–2951. doi: 10.1128/AEM.03715-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Carpentier B, Cerf O. Review-Persistence of Listeria monocytogenes in food industry equipment and premises. Int J Food Microbiol. 2011;145:1–8. doi: 10.1016/j.ijfoodmicro.2011.01.005. [DOI] [PubMed] [Google Scholar]
- 6.Allerberger F, Bagó Z, Huhulescu S, Pietzka A. Listeriosis: the dark side of refrigeration and ensiling. In: Sing A, editor. Zoonoses – Infections Affecting Humans and Animals. Dordrecht: Springer Netherlands; 2015. pp. 249–286. (editor) [Google Scholar]
- 7.Moura A, Criscuolo A, Pouseele H, Maury MM, Leclercq A, et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat Microbiol. 2016;2:16185. doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Camargo AC, Woodward JJ, Nero LA. The continuous challenge of characterizing the foodborne pathogen Listeria monocytogenes. Foodborne Pathog Dis. 2016;13:405–416. doi: 10.1089/fpd.2015.2115. [DOI] [PubMed] [Google Scholar]
- 9.Chen Y, Luo Y, Carleton H, Timme R, Melka D, et al. Whole genome and core genome multilocus sequence typing and single nucleotide polymorphism analyses of Listeria monocytogenes associated with an outbreak linked to cheese, United States, 2013. Appl Environ Microbiol. 2017;83:e00633-17. doi: 10.1128/AEM.00633-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.European Food Safety Authority Analysis of the baseline survey on the prevalence of Listeria monocytogenes in certain ready-to-eat foods in the EU, 2010–2011. Part A: Listeria monocytogenes prevalence estimates. EFSA J. 2013;11:3241. [Google Scholar]
- 11.Orsi RH, den Bakker HC, Wiedmann M. Listeria monocytogenes lineages: genomics, evolution, ecology, and phenotypic characteristics. Int J Med Microbiol. 2011;301:79–96. doi: 10.1016/j.ijmm.2010.05.002. [DOI] [PubMed] [Google Scholar]
- 12.Burall LS, Grim CJ, Mammel MK, Datta AR. A comprehensive evaluation of the genetic relatedness of Listeria monocytogenes serotype 4b variant strains. Front Public Health. 2017;5:241. doi: 10.3389/fpubh.2017.00241. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ragon M, Wirth T, Hollandt F, Lavenir R, Lecuit M, et al. A new perspective on Listeria monocytogenes evolution. PLoS Pathog. 2008;4:e1000146. doi: 10.1371/journal.ppat.1000146. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Rychli K, Müller A, Zaiser A, Schoder D, Allerberger F, et al. Genome sequencing of Listeria monocytogenes "Quargel" listeriosis outbreak strains reveals two different strains with distinct in vitro virulence potential. PLoS One. 2014;9:e89964. doi: 10.1371/journal.pone.0089964. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kwong JC, Mercoulia K, Tomita T, Easton M, Li HY, et al. Prospective whole-genome sequencing enhances national surveillance of Listeria monocytogenes. J Clin Microbiol. 2016;54:333–342. doi: 10.1128/JCM.02344-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Jackson BR, Tarr C, Strain E, Jackson KA, Conrad A, et al. Implementation of nationwide real-time whole-genome sequencing to enhance listeriosis outbreak detection and investigation. Clin Infect Dis. 2016;63:380–386. doi: 10.1093/cid/ciw242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Kvistholm Jensen A, Nielsen EM, Björkman JT, Jensen T, Müller L, et al. Whole-genome sequencing used to investigate a nationwide outbreak of listeriosis caused by ready-to-eat delicatessen meat, Denmark, 2014. Clin Infect Dis. 2016;63:64–70. doi: 10.1093/cid/ciw192. [DOI] [PubMed] [Google Scholar]
- 18.Moura A, Tourdjman M, Leclercq A, Hamelin E, Laurent E, et al. Real-time whole-genome sequencing for surveillance of Listeria monocytogenes, France. Emerg Infect Dis. 2017;23:1462–1470. doi: 10.3201/eid2309.170336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bolger AM, Lohse M, Usadel B. Trimmomatic: a flexible trimmer for Illumina sequence data. Bioinformatics. 2014;30:2114–2120. doi: 10.1093/bioinformatics/btu170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tewolde R, Dallman T, Schaefer U, Sheppard CL, Ashton P, et al. MOST: a modified MLST typing tool based on short read sequencing. PeerJ. 2016;4:e2308. doi: 10.7717/peerj.2308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bankevich A, Nurk S, Antipov D, Gurevich AA, Dvorkin M, et al. SPAdes: a new genome assembly algorithm and its applications to single-cell sequencing. J Comput Biol. 2012;19:455–477. doi: 10.1089/cmb.2012.0021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Treangen TJ, Ondov BD, Koren S, Phillippy AM. The Harvest suite for rapid core-genome alignment and visualization of thousands of intraspecific microbial genomes. Genome Biol. 2014;15:524. doi: 10.1186/s13059-014-0524-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Charpentier E, Courvalin P. Antibiotic resistance in Listeria spp. Antimicrob Agents Chemother. 1999;43:2103–2108. doi: 10.1128/AAC.43.9.2103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Lungu B, O'Bryan CA, Muthaiyan A, Milillo SR, Johnson MG, et al. Listeria monocytogenes: antibiotic resistance in food production. Foodborne Pathog Dis. 2011;8:569–578. doi: 10.1089/fpd.2010.0718. [DOI] [PubMed] [Google Scholar]
- 25.Langmead B, Salzberg SL. Fast gapped-read alignment with Bowtie 2. Nat Methods. 2012;9:357–359. doi: 10.1038/nmeth.1923. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Li H, Handsaker B, Wysoker A, Fennell T, Ruan J, 1000 Genome Project Data Processing Subgroup et al. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Camejo A, Carvalho F, Reis O, Leitão E, Sousa S, et al. The arsenal of virulence factors deployed by Listeria monocytogenes to promote its cell infection cycle. Virulence. 2011;2:379–394. doi: 10.4161/viru.2.5.17703. [DOI] [PubMed] [Google Scholar]
- 28.Maury MM, Tsai YH, Charlier C, Touchon M, Chenal-Francisque V, et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat Genet. 2016;48:308–313. doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cabanes D, Sousa S, Cebriá A, Lecuit M, García-del Portillo F, et al. Gp96 is a receptor for a novel Listeria monocytogenes virulence factor, Vip, a surface protein. Embo J. 2005;24:2827–2838. doi: 10.1038/sj.emboj.7600750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Chen J, Luo X, Jiang L, Jin P, Wei W, et al. Molecular characteristics and virulence potential of Listeria monocytogenes isolates from Chinese food systems. Food Microbiol. 2009;26:103–111. doi: 10.1016/j.fm.2008.08.003. [DOI] [PubMed] [Google Scholar]
- 31.Gómez D, Azón E, Marco N, Carramiñana JJ, Rota C, et al. Antimicrobial resistance of Listeria monocytogenes and Listeria innocua from meat products and meat-processing environment. Food Microbiol. 2014;42:61–65. doi: 10.1016/j.fm.2014.02.017. [DOI] [PubMed] [Google Scholar]
- 32.Ryan S, Begley M, Hill C, Gahan CG. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J Appl Microbiol. 2010;109:984–995. doi: 10.1111/j.1365-2672.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 33.Carvalho F, Atilano ML, Pombinho R, Covas G, Gallo RL, et al. L-Rhamnosylation of Listeria monocytogenes Wall Teichoic Acids Promotes Resistance to Antimicrobial Peptides by Delaying Interaction with the Membrane. PLoS Pathog. 2015;11:e1004919. doi: 10.1371/journal.ppat.1004919. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Promadej N, Fiedler F, Cossart P, Dramsi S, Kathariou S. Cell wall teichoic acid glycosylation in Listeria monocytogenes serotype 4b requires gtcA, a novel, serogroup-specific gene. J Bacteriol. 1999;181:418–425. doi: 10.1128/jb.181.2.418-425.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Cabanes D, Dussurget O, Dehoux P, Cossart P. Auto, a surface associated autolysin of Listeria monocytogenes required for entry into eukaryotic cells and virulence. Mol Microbiol. 2004;51:1601–1614. doi: 10.1111/j.1365-2958.2003.03945.x. [DOI] [PubMed] [Google Scholar]
- 36.Ebner R, Stephan R, Althaus D, Brisse S, Maury M, et al. Phenotypic and genotypic characteristics of Listeria monocytogenes strains isolated during 2011–2014 from different food matrices in Switzerland. Food Control. 2015;57:321–326. doi: 10.1016/j.foodcont.2015.04.030. [DOI] [Google Scholar]
- 37.Félix B, Feurer C, Maillet A, Guillier L, Boscher E, et al. Population genetic structure of Listeria monocytogenes strains isolated from the pig and pork production chain in France. Front Microbiol. 2018;9:684. doi: 10.3389/fmicb.2018.00684. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Henri C, Leekitcharoenphon P, Carleton HA, Radomski N, Kaas RS, et al. An assessment of different genomic approaches for inferring phylogeny of Listeria monocytogenes. Front Microbiol. 2017;8:2351. doi: 10.3389/fmicb.2017.02351. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Bertrand S, Huys G, Yde M, D'Haene K, Tardy F, et al. Detection and characterization of tet(M) in tetracycline-resistant Listeria strains from human and food-processing origins in Belgium and France. J Med Microbiol. 2005;54:1151–1156. doi: 10.1099/jmm.0.46142-0. [DOI] [PubMed] [Google Scholar]
- 40.Morvan A, Moubareck C, Leclercq A, Hervé-Bazin M, Bremont S, et al. Antimicrobial resistance of Listeria monocytogenes strains isolated from humans in France. Antimicrob Agents Chemother. 2010;54:2728–2731. doi: 10.1128/AAC.01557-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Granier SA, Moubareck C, Colaneri C, Lemire A, Roussel S, et al. Antimicrobial resistance of Listeria monocytogenes isolates from food and the environment in France over a 10-year period. Appl Environ Microbiol. 2011;77:2788–2790. doi: 10.1128/AEM.01381-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Jamali H, Paydar M, Ismail S, Looi CY, Wong WF, et al. Prevalence, antimicrobial susceptibility and virulotyping of Listeria species and Listeria monocytogenes isolated from open-air fish markets. BMC Microbiol. 2015;15:144. doi: 10.1186/s12866-015-0476-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.den Besten HMW, Amézquita A, Bover-Cid S, Dagnas S, Ellouze M, et al. Next generation of microbiological risk assessment: potential of omics data for exposure assessment. Int J Food Microbiol. 2018;287:18–27. doi: 10.1016/j.ijfoodmicro.2017.10.006. [DOI] [PubMed] [Google Scholar]
- 44.Rantsiou K, Kathariou S, Winkler A, Skandamis P, Saint-Cyr MJ, et al. Next generation microbiological risk assessment: opportunities of whole genome sequencing (WGS) for foodborne pathogen surveillance, source tracking and risk assessment. Int J Food Microbiol. 2018;287:3–9. doi: 10.1016/j.ijfoodmicro.2017.11.007. [DOI] [PubMed] [Google Scholar]
- 45.Franz E, Gras LM, Dallman T. Significance of whole genome sequencing for surveillance, source attribution and microbial risk assessment of foodborne pathogens. Curr Opin Food Sci. 2016;8:74–79. doi: 10.1016/j.cofs.2016.04.004. [DOI] [Google Scholar]
- 46.Pielaat A, Boer MP, Wijnands LM, van Hoek AH, Bouw E, et al. First step in using molecular data for microbial food safety risk assessment; hazard identification of Escherichia coli O157:H7 by coupling genomic data with in vitro adherence to human epithelial cells. Int J Food Microbiol. 2015;213:130–138. doi: 10.1016/j.ijfoodmicro.2015.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.