Abstract
Progression of mitosis and cytokinesis depends on the reorganization of cytoskeleton, with microtubules driving the segregation of chromosomes and their partitioning to two daughter cells. In dividing plant cells, microtubules undergo global reorganization throughout mitosis and cytokinesis, and with the aid of various microtubule-associated proteins (MAPs), they form unique systems such as the preprophase band (PPB), the acentrosomal mitotic spindle, and the phragmoplast. Such proteins include nucleators of de novo microtubule formation, plus end binding proteins involved in the regulation of microtubule dynamics, crosslinking proteins underlying microtubule bundle formation and members of the kinesin superfamily with microtubule-dependent motor activities. The coordinated function of such proteins not only drives the continuous remodeling of microtubules during mitosis and cytokinesis but also assists the positioning of the PPB, the mitotic spindle, and the phragmoplast, affecting tissue patterning by controlling cell division plane (CDP) orientation. The affinity and the function of such proteins is variably regulated by reversible phosphorylation of serine and threonine residues within the microtubule binding domain through a number of protein kinases and phosphatases which are differentially involved throughout cell division. The purpose of the present review is to provide an overview of the function of protein kinases and protein phosphatases involved in cell division regulation and to identify cytoskeletal substrates relevant to the progression of mitosis and cytokinesis and the regulation of CDP orientation.
Keywords: microtubules, microtubule-associated proteins, mitotic spindle, phragmoplast, protein kinase, protein phosphatase
Introduction
Owing to their sedentary life style and their encasement within the barriers of rigid cell walls, plant cells adopted unique mechanisms for regulating fundamental eukaryotic processes such as cell division and cell division plane (CDP) orientation establishment (reviewed in Buschmann and Zachgo, 2016). In this respect, plant cells developed unique microtubule-based cytoskeletal structures which underlie the above processes. CDP orientation is marked by a plant-specific cortical microtubule ring, the preprophase band (PPB; Pickett-Heaps and Northcote, 1966) which determines spindle positioning (Schaefer et al., 2017) and coincides with the plane of cell plate deposition during cytokinesis (cell plate fusion site; CPFS; Marcus et al., 2005). The PPB exhibits a progressive narrowing and finally it disassembles shortly after the mitotic spindle is formed; however, the cortical site which was occupied by the PPB is marked by specific proteins throughout mitosis in a continuous or intermittent manner (e.g., Buschmann et al., 2006; Walker et al., 2007).
The plant mitotic spindle starts to assemble before nuclear envelope breakdown and in contrast to the mammalian or yeast spindle, and it forms in the absence of microtubule organizing center (reviewed in Buschmann and Zachgo, 2016).
Cytokinesis is hallmarked by the formation of another plant specific microtubule machinery, the phragmoplast. It is formed at the end of telophase between the reconstituting daughter nuclei. It comprises two sets of antiparallel microtubules, and it expands centrifugally toward the cell periphery. During its expansion, it guides the deposition of the cell plate until the latter merges with the parent cell wall, after which the phragmoplast disintegrates (Chen et al., 2018).
Throughout the cell cycle, precise temporal and spatial regulation of microtubule organization and dynamics is required for the formation, proper function, and structural transitions of these cytoskeletal structures (Dhonukshe and Gadella, 2003). Such regulation is achieved via microtubule-associated proteins (MAPs) involved in microtubule organization and dynamics. Among these proteins belong motor proteins from the kinesin (Müller et al., 2006; Lipka et al., 2014; Buschmann et al., 2015; de Keijzer et al., 2017) and the myosin superfamilies (Wu and Bezanilla, 2014), plus end-binding proteins and microtubule crosslinkers (Mao et al., 2005; Beck et al., 2010; Kohoutová et al., 2015; Lin et al., 2019). Many of such proteins exhibit a cell cycle dependent localization to mitotic and cytokinetic microtubule systems (Figure 1), and at large this is differentially regulated by protein kinases and phosphatases which become activated/deactivated in a similar cell cycle dependent manner.
Figure 1.
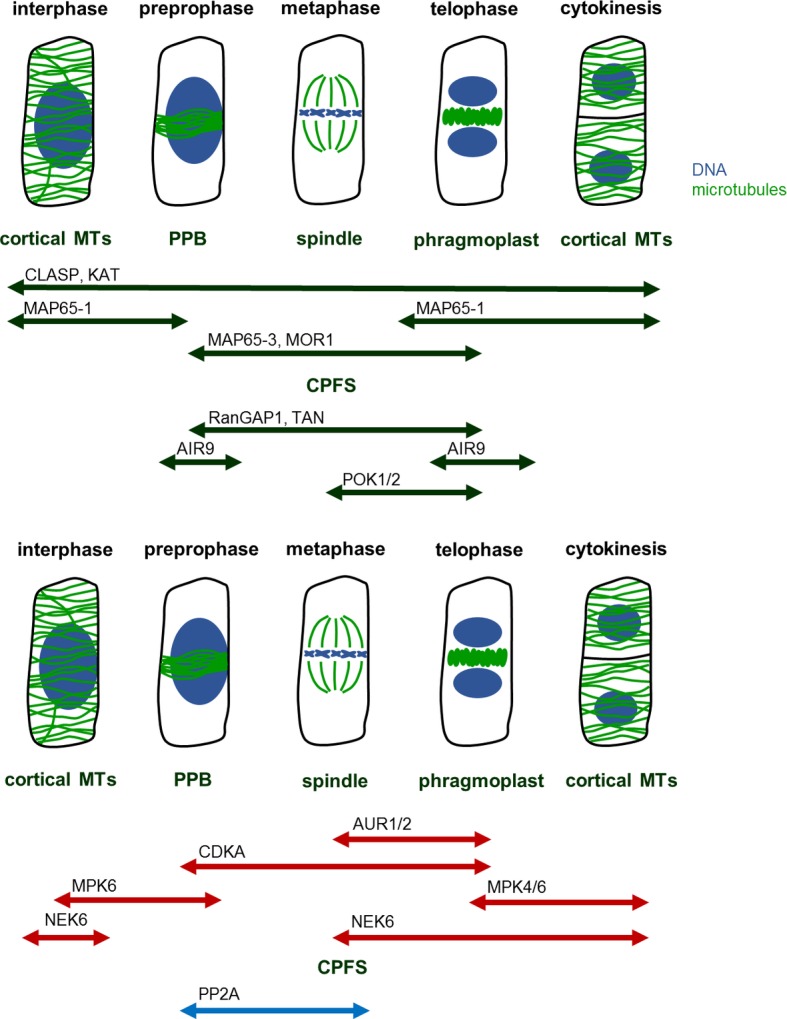
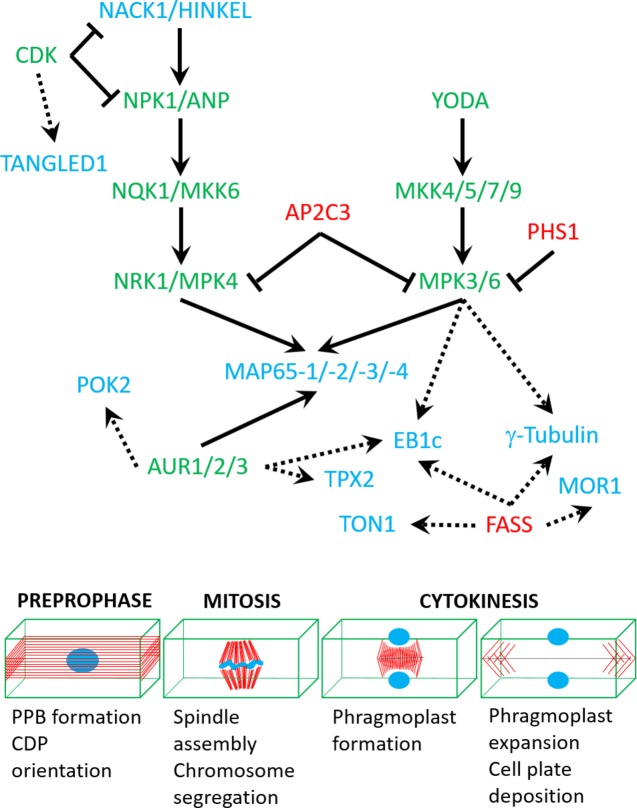
MAPs, kinases, and phosphatases regulating mitotic MT structures. Mitosis begins with preprophase, in which cortical MTs reorganize in preprophase band (PPB). PPB disassembles at the onset of metaphase, during which spindle forms. At this time, the former site of PPB remains marked as a future cell plate fusion site (CPFS) by various MAPs. After segregation of chromatids, at the late anaphase, phragmoplast begins to form at the center of cell. Phragmoplast serves as a scaffold for building cell plate and as the construction continues, phragmoplast expands until it reaches CPFS. At the end, in two daughter cells, MTs rearrange into cortical microarrays. Abbreviations: AIR9, auxin-induced root cultures; AUR, aurora kinase; CDKA, cell division kinase A; CLASP, cytoplasmic linker associated protein; CPFS, cell plate fusion site; KAT, katanin; MAP65, microtubule-associated protein 65; MOR1, microtubule organization 1; MPK, mitogen-activated protein kinase; MT, microtubule; NEK, never in a mitosis A-related kinase; PP2A, protein phosphatase type 2A; POK, phragmoplast orienting kinesin; PPB, preprophase band; RanGAP1, Ran GTPase activating protein; TAN, tangled.
Many kinases were directly shown to associate with cytoskeletal systems (Weingartner et al., 2001; Samaj et al., 2002, 2004; Lee et al., 2003; Oh et al., 2005, 2012) and indirect pharmacological (e.g., Binarova et al., 1996; Ayaydin et al., 2000) and subsequently more targeted studies (e.g., Mao et al., 2005; Brumbarova and Ivanov, 2016), establishing the functional reciprocity between protein kinases and cytoskeletal components. Plant microtubule systems can be targeted for phosphorylation-pendant regulation of their components after environmental stimulation (e.g., Ban et al., 2013; Bhaskara et al., 2017), or in a developmental context, which is the aim of this review.
Involvement of Maps in the Organization of Mitotic Structures
From numerous plant proteins related to the regulation of microtubule organization and dynamics, some have been inadvertently associated with the progress of mitotic and cytokinetic microtubule arrays and were shown to be regulated by reversible phosphorylation. These proteins are involved in all aspects of microtubule organization and dynamics.
Microtubule nucleation factors such as γ-tubulin and TPX2 (targeting protein for Xklp2) are essential for spindle formation and the establishment of spindle bipolarity (Petrovská et al., 2013), and it was suggested that they are regulated by mitogen-activated protein kinase (MAPK, MPK) and/or Aurora kinase (AUR) phosphorylation (Petrovská et al., 2012; https://string-db.org/cgi/network.pl?taskId=f13kHLYXYV1W). It is likely that γ-tubulin interacts with the FASS B″ subunit of protein phosphatase 2A (https://string-db.org/network/3702.AT5G18580.1; Figure 2). Notably, fass mutants exhibit altered geometry of microtubule nucleation at least in interphase microtubule arrays (Kirik et al., 2012).
Figure 2.
A speculative network of protein kinases (green), phosphatases (red), and targeted cytoskeletal proteins (blue) based on either published interaction studies (full arrows) or in silico predictions (dashed arrows; see text for more details). Lower panel shows mitotic stages which are regulated by the above network of interactions.
Microtubule dynamics are largely controlled by plus end binding proteins, including end-binding protein 1 isoforms (EB1a, b, and c; Komaki et al., 2010), SPIRAL1 (Sedbrook et al., 2004), CLASP (Ambrose et al., 2007), and GPT1 and 2 (growing plus-end tracking 1 and 2; Wong and Hashimoto, 2017). The plant-specific isoform EB1c was shown to be phosphorylated by MPK6 (Figure 2; Kohoutová et al., 2015), however, without apparent functional implications. Similarly, phosphorylation of CLASP was demonstrated only in the context of conditional signaling (Brumbarova and Ivanov, 2016).
Microtubule bundling via the MAP65 proteins is essential for the formation of the central spindle, its subsequent reorganization into phragmoplast and for support of its centrifugal expansion (Chang et al., 2005; Smertenko et al., 2008, 2018; Herrmann et al., 2018). From the nine members of the Arabidopsis MAP65 family, only MAP65-1, -2, -3, and -4 have been associated with the progression of mitosis and cytokinesis (Chang-Jie and Sonobe, 1993; Chan et al., 1999; Caillaud et al., 2008). MAP65-1 and MAP65-2 are nonessential as proven by the absence of cytokinetic phenotypes of single or double mutants (Lucas and Shaw, 2012). On the other hand, MAP65-3 and MAP65-4 appear to be essential for cytokinesis, in an additive manner (Müller et al., 2004; Li et al., 2017). MAP65-1 and MAP65-2 proteins differentially colocalize with microtubules and mediate their bundling during interphase and preprophase (Murata et al., 2013; Zhou et al., 2017). However, they are excluded from the mitotic spindle until telophase. This suggests that their colocalization with microtubule structures is subjected to temporal control during cell division (Gaillard et al., 2008). One possible mechanism controlling the differential localization of MAP65 proteins with mitotic microtubule systems is proteolytic degradation, since several map65 genes harbor a “destruction box” motif, which is a target for the ubiquitin degradation pathway. More importantly, at least MAP65-1, -2, and -3 are targeted for phosphorylation in their C-terminal microtubule binding domains by several kinases with cell cycle functions, such as cyclin-dependent kinases [CDKs (Smertenko et al., 2006), MAPKs (Kosetsu et al., 2010; Smékalová et al., 2014), and AURs (Boruc et al., 2017)]. Generally, phosphorylation downregulates microtubule binding of MAP65s; therefore, it may represent the means to abolish their localization from the mitotic spindle. This is supported by mutagenesis studies, showing that change of the consensus CDK-targeted site of MAP65-1 causes its localization at the mitotic spindle (Mao et al., 2005). As mentioned earlier, MAP65-1 and presumably MAP65-2 are nonessential for the mitotic and cytokinetic progress (Lucas and Shaw, 2012), and they may affect spindle and phragmoplast formation only when artificially overexpressed (Mao et al., 2005). MAP65-3, on the other hand, is essential for cell plate formation, since its genetic depletion results in the formation of giant, multinucleated cells with incomplete cell walls. Similar cytokinetic phenotypes have been observed in anp2anp3 and mpk4 mutants, which are related to MAPK signaling. The above mutants show reduced but not abolished MAP65-3 expression (Beck et al., 2010). In this case, it is speculated that the cytokinetic phenotype of anp2anp3 and mpk4 mutants maybe partially attributed to reduced phosphorylation of MAP65-3 (Beck et al., 2010). MAP65-4 alone has negligible cytokinetic phenotypes when depleted but contributes to the map65-3/pleiade phenotype in double mutants (Li et al., 2017). Its spatial localization coincides with that of MAP65-3 at the PPB and the phragmoplast midzone. However, MAP65-4 exhibits persistent localization at the cortical division zone throughout mitosis, unlike MAP65-3 (Li et al., 2017). Although the cytokinetic role of MAP65-4 was just recently described, it is also likely to be regulated by phosphorylation. Its carboxyl-terminal region harbors proline-directed serine or threonine residues, which are predicted targets of CDKs and MAPKs (based on prediction using GPS2.1.2; Xue et al., 2008). It is also predicted to interact with AUR (https://string-db.org/cgi/network.pl?taskId=qRfSrQ0dHJdK; Figure 2).
Several Arabidopsis microtubule motors of the kinesin superfamily, namely those related to the progress of mitosis and cytokinesis were shown to be regulated by phosphorylation. One example is the kinesin-like calmodulin-binding protein (KCBP), which is involved in the tethering of phragmoplast margins to the CPFS (Buschmann et al., 2015; Buschmann and Zachgo, 2016). KCBP was shown to be regulated by phosphorylation (Day et al., 2000; Humphrey et al., 2015). The mitotic kinesin NACK1 (NPK1-activating kinesin-like protein 1) is an activator of the NPK1-NQK1-NRK1 MAPK pathway in tobacco (similarly, its Arabidopsis homologue HINKEL also activates the ANP-MKK6-MPK4 MAPK pathway; Nishihama et al., 2002), which is involved in the regulation of cytokinesis. The interaction between NACK1 and NPK1 is negatively regulated by CDK phosphorylation in residues of both the motor domain and the stalk region of NACK1. CDKs also target the carboxyl-terminal regulatory domain of NPK1, which is engaged in NACK1-NPK1 interactions (Sasabe et al., 2011a). Interruption of NACK1-NPK1 interaction by CDK-mediated phosphorylation abolishes the recruitment of the NPK1-NQK1-NRK1 module to the mitotic spindle. When CDK activity declines during late anaphase, then the NPK1-NQK1-NRK1 module becomes localized to the phragmoplast (Sasabe et al., 2011a).
Interactions of kinesin motors with protein kinases may be reciprocal and may convey targeted transport or activity regulation to the kinase counterpart. For example, never in a mitosis (NIMA)-related kinase 6 (NEK6) is negatively regulated by the armadillo-repeat kinesin 1 (Eng et al., 2017). Also, NACK1 (and its Arabidopsis homologue HINKEL) directly activates the NPK1 MAPKKK (and its Arabidopsis homologues ANP1, 2, and 3; Nishihama et al., 2002; Takahashi et al., 2010).
Microtubule severing by the Arabidopsis KATANIN1 (Komis et al., 2017; Panteris et al., 2018) has not been shown to be regulated by phosphorylation yet. Moreover, the p60 catalytic subunit of the katanin holoenzyme is suspected to interact with FASS (https://string-db.org/cgi/network.pl?taskId=jBYrCeTF9nPv; Figure 2). In animals, phosphorylation is a major mechanism for the exclusion of katanin activity from the mitotic spindle and connected to spindle sizing (Loughlin et al., 2011; Whitehead et al., 2013).
Apart from several MAPs that have been proven or predicted to be regulated by phosphorylation, tubulin has also been identified as a target of the atypical kinase domain of the protein phosphatase PROPYZAMIDE HYPERSENSITIVE 1 (PHS1; Fujita et al., 2013). So far, tubulin phosphorylation is related to conditional microtubule depolymerization (Ban et al., 2013), but it will be of interest to identify tubulin phosphorylation as a regulator of mitotic microtubule transitions.
Kinases Regulating Map Activity
Since MAPs play a role in microtubule dynamics while being regulated by reversible phosphorylation, kinases and phosphatases are master regulators of microtubule reorganization throughout cell cycle. Several kinases were implicated in phosphorylating MAPs, namely CDKs, AURs, MAPKs, and NEKs.
Since CDKs are master regulators of cell cycle progression, they are also implicated cell cycle-dependent cytoskeletal reorganizations (Costa, 2017). The mode of action on CDKs on the microtubule cytoskeleton of dividing cells is not well understood, since the only known cytoskeletal CDK substrate is NACK1 kinesin (Sasabe et al., 2011a), while MAP65-1 was only shown in vitro to be phosphorylated by CDK (Smertenko et al., 2006). Localization observations and pharmacological and genetic evidence favor the implication of CDK in regulating microtubules. For example, Arabidopsis CDK was shown to colocalize with PPB, spindle, and phragmoplast (Stals et al., 1997). Second, plant CDKs were found to participate in regulating mitotic microtubule structures (Weingartner et al., 2001). Third, CDKs are known to regulate microtubule dynamics by phosphorylating animal homologues of plant MAPs (Ookata et al., 1997; Vasquez et al., 1999). Last, several plant MAPs contain a CDK phosphorylation site (Hussey et al., 2002; Smertenko et al., 2006). CDKs may be also involved in CDP orientation through the phosphorylation of cytoskeletal markers of the cell division zone such as the microtubule binding protein TANGLED1 (https://string-db.org/cgi/network.pl?taskId=4lbkQFdWZfbv; Figure 2).
Apart from CDKs, Aurora kinases are another component of cell cycle progression machinery. These Ser/Thr kinases are on lower hierarchical position than CDKs (Schecher et al., 2017). They themselves are regulated by phosphorylation and ubiquitin-dependent proteolysis (Castro et al., 2002a,b). Therefore, they are known to play a more direct role in cytoskeleton rearrangements than CDKs (Ritchey and Chakrabarti, 2014). In plants, not only do they associate with mitotic structures (Demidov et al., 2005) but also they interact with MAPs (Boruc et al., 2017). Since AUR does not possess microtubule-binding domains, its colocalization with mitotic structures is most likely related to its functional interactions with MAPs (Petrovská et al., 2012; Tomaštíková et al., 2015). Out of three members of Aurora kinase family in Arabidopsis (Kawabe et al., 2005), two of them, AUR1 and AUR2, are essential for regulating the orientation of formative cell divisions throughout plant ontogenesis (Van Damme et al., 2011). AUR phosphorylates MAP65-1 during metaphase (Boruc et al., 2017); however, the strength of AUR control over MAP65-1 is significantly weaker compared with the effect of another kinase, MAPK. Therefore, a hypothesis was presented, according to which the direct control of AUR over MAP65-1 is mild, yet the phosphorylation of MAP65-1 by AUR promotes phosphorylation by other kinases. This is in line with the observation that regulation of many eukaryotic proteins depends on multisite phosphorylation (Cohen, 2000; Repetto et al., 2018). Prediction studies show that other cytoskeletal regulators of mitosis and especially of CDP orientation like POK2 may interact and become regulated by Aurora kinases (https://string-db.org/cgi/network.pl?taskId=f13kHLYXYV1W; Figure 2).
MAPKs are well known to phosphorylate MAPs (Hoshi et al., 1992; Shiina et al., 1992). In Arabidopsis, MPK4 and MPK6 phosphorylate proteins from MAP65 family (Smertenko et al., 2006; Sasabe et al., 2011b; Smékalová et al., 2014; Zhou et al., 2017), and MPK6 also phosphorylates EB1c (Kohoutová et al., 2015). MAPKs are governed by MAPK kinases (MAPKKs), which, in turn, are regulated by MAPKK kinases (MAPKKKs). In plants, two MAPK signaling cascades were implicated in regulating microtubule dynamics during cell division and described in detail as follows. A third pathway which involves the MAPK MPK18 and the MAPK phosphatase PROPYZAMIDE HYPERSENSITIVE 1 (PHS1) is somehow elusive without knowledge on microtubule-associated substrates which may justify their role in microtubule regulation (Naoi and Hashimoto, 2004; Walia et al., 2009; Fujita et al., 2013). However, the role of PHS1 may be broader since it is presumably interacting and deactivating other MAPKs as well, including MPK3 and MPK6 (https://string-db.org/cgi/network.pl?taskId=j8pmY8S1UlbB; Figure 2).
The first MAPK cascade described to play a role in microtubule reorganization was the NACK-PQR pathway (Calderini et al., 1998, 2001; Bögre et al., 1999; Nishihama, 2001). In Arabidopsis, this pathway consists of ANP2/ANP3 (Arabidopsis nucleus and phragmoplast-localized kinase, MAP3K), MKK6, and MPK4/MPK6 (Krysan et al., 2002; Strompen et al., 2002), and it plays a crucial role during phragmoplast and cell plate formation (Takahashi et al., 2010). It affects the organization of mitotic structures via reversible phosphorylation of MAP65 proteins (Beck et al., 2010). Interestingly, activation of this MAP3K is negatively regulated by CDKs (Sasabe et al., 2011a). Moreover, CDKs also interfere with MAPK phosphorylating MAP65-1, since the single MAPK targeting motif in MAP65-1 overlaps with a CDK targeting site (Smertenko et al., 2006). In conclusion, this MAPK cascade controls microtubule organization and dynamics during phragmoplast and cell plate formation, and the temporal regulation of this module is facilitated by CDKs.
The other plant MAPK pathway, which is integral to cell division directionality, consists of YODA (YDA, MAP3K), MKK4/MKK5, and MPK3/MPK6. YODA is implicated in several types of asymmetrical divisions, e.g., first division of zygote and stomatal development (Lukowitz et al., 2004; Bergmann et al., 2004). However, the characterization of YODA mutants revealed its function in CDP orientation of regular cell divisions underlying tissue patterning of vegetative organs such as the root (Müller et al., 2010; López-Bucio et al., 2014). These observations are further supported by microscopic studies, which proved MPK6 colocalization with mitotic microtubule structures (Müller et al., 2010; Smékalová et al., 2014; Komis et al., 2018). Interaction studies showed that MAP65-1 is interacting with MPK6 and possibly phosphorylated by it (Smékalová et al., 2014). Interestingly, knockout mutants of YDA have deregulated transcript levels of CPFS markers (specifically TAN and phragmoplast orienting kinesin 1). Therefore, YODA may be involved at multiple levels of CPFS orientation (Smékalová et al., 2014).
The last family of kinases involved in regulation of mitotic microtubule structures is NEKs. This family of Ser/Thr protein kinases is highly conserved in eukaryotes, where it supervises crucial points in mitosis and cell division (O’Connell et al., 2003; Brieño-Enríquez et al., 2017). In plants, NEKs were shown to regulate cortical microtubules and, in turn, to affect cell expansion, organ growth, and stress responses (Vigneault et al., 2007; Agueci et al., 2012; Takatani et al., 2017). As for their role in rearrangement of microtubules during mitosis, NEK6 is known to associate with spindle and phragmoplast (Motose et al., 2011), but its function remains obscure.
Protein Phosphatases Regulating Map Activity
The reversibility of phosphorylation is ensured by cooperation between kinases and phosphatases. Numerous protein phosphatases were found in plants, with Ser/Thr specific phosphoprotein phosphatases (PPPs) being a prominent group among them. PPPs encompass large number of proteins, which can be grouped in several protein families. Three of these families were implicated to regulate microtubule dynamics during cytokinesis (Samofalova et al., 2017).
Type one protein phosphatases (TOPPs, also called PP1s) were predicted to be part of cell cycle regulation (Farkas et al., 2007; Boyer and Simon, 2015), which is supported by finding putative CDK recognition sites (Kwon et al., 1997) as well as noting crucial role of animal PP1s in cell cycle progression (Rodrigues et al., 2015). However, the functions of these proteins were not comprehensively studied to this date.
More progress was achieved in solving the function of protein phosphatase type 2A (PP2A). These PPs consist of three subunits—scaffolding (A), regulatory (B), and catalytic (C). They were characterized in both monocots (Wright et al., 2009) and dicots (Camilleri et al., 2002). Moreover, their animal homologs were found to be essential for regulating microtubule structures in both meiosis and mitosis (Tang et al., 2016; Enos et al., 2017; Varadkar et al., 2017). In plants, PP2A controls organization and dynamics of both cortical and mitotic microtubules (Figure 1; Camilleri et al., 2002; Yoon et al., 2018). This view is supported by observing knockout mutants displaying abnormal arrangement of cortical microtubules and severe problems with PPB formation and cell division plane orientation (Torres-Ruiz and Jurgens, 1994; Traas et al., 1995; McClinton and Sung, 1997). During mitosis, PP2A forms a complex with tonneau 1 (TON1) and TON1 recruiting motif proteins (TRMs) (Spinner et al., 2013). TON1 and TRMs recruit this complex to site where PPB forms (Drevensek et al., 2012), and there the complex governs PPB development. As it remains at this site even after PPB disassembly, it is possibly involved in CPFS maintenance (Wright et al., 2009). Targets of PP2A-driven dephosphorylation could be MAPs marking cytokinetic structures (specifically MOR1, TON1, and CLASP; Twell et al., 2002; Kawamura, 2005; Xu et al., 2008; Ambrose et al., 2011). PP2A could temporally and spatially restrict common MAP activities and this would allow microtubule stabilization and formation of PPB (Walker et al., 2007; Wright and Smith, 2007; Lipka et al., 2015).
Metallo-dependent protein phosphatases (PP2C) might be part of cell division machinery as well, since knockout mutants display improper cell division orientation (Song et al., 2006). These phosphatases are also implicated in regulating MAPKs and CDK (Meskiene et al., 1998; Umbrasaite et al., 2010). Currently, their role in cortical microtubule rearrangement in response to environmental stimuli has been explored (Bhaskara et al., 2017; Qu et al., 2018). However, details on how PP2C is integrated into the regulatory network of cytokinesis remain undisclosed.
Although plenty of research has been done in elucidating the role of kinases in microtubule reorganization during cell cycle, phosphatases involved in these events remain largely understudied. The cause of this lies mainly in the fact that these phosphatases form multiprotein complexes, which is a serious challenge for both analysis and evaluation. Nevertheless, the current advances in understanding the role of PP2A in regulating microtubule dynamics shows it is not an impossible task.
Conclusions/Future Directions
In summary, reversible phosphorylation of several different MAPs is essential for regulating microtubule dynamics and organization during the plant cell division. The affinity of MAPs for microtubules can be downregulated or restored pending on their phosphorylation status. To this extend, several protein kinases and phosphatases have been shown to target cytoskeletal proteins with various roles in the regulation of mitotic spindle and phragmoplast assembly and progression. However, many questions remain unanswered and are expected to be addressed in the near future:
How phosphorylation may affect microtubule nucleation during acentrosomal mitotic spindle formation?
Is phosphorylation related to the transition from PPB to mitotic spindle?
How phosphorylation really controls microtubule bundling during phragmoplast expansion?
Which phosphatases are reinstating microtubule binding of MAP65 proteins?
Which mechanisms allow the regulation of the same cytoskeletal proteins (e.g., MAP65-1, -2, and -3) by different protein kinases (e.g., MPK4 and MPK6 or auroras) with a different biological outcome (i.e., progression of cytokinesis and CDP orientation, respectively)?
How global phosphoproteomics analyses will help to decipher reversibly phosphorylatable cytoskeletal substrates in model cell suspension systems that can be synchronized?
How differential (phospho)proteomics comparing wild types with protein kinase/phosphatase mutants will advance our knowledge in the identification of cytoskeletal substrates?
Author Contributions
TV drafted the manuscript and drawn Figure 1. JŠ contributed critical evaluation and editing of the text. GK conceived the topic, supervised TV during drafting of the manuscript and edited its final form together with JŠ.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Acknowledgments
We thank all members of the JŠ lab for critical input on the manuscript.
Footnotes
Funding. This work was funded by a grant Nr. 16-24313S from the Czech Science Foundation GAČR and by ERDF project “Plants as a tool for sustainable global development” (CZ.02.1.01/0.0/0.0/16_019/0000827).
References
- Agueci F., Rutten T., Demidov D., Houben A. (2012). Arabidopsis AtNek2 kinase is essential and associates with microtubules. Plant Mol. Biol. Report. 30, 339–348. 10.1007/s11105-011-0342-1 [DOI] [Google Scholar]
- Ambrose C., Allard J. F., Cytrynbaum E. N., Wasteneys G. O. (2011). A CLASP-modulated cell edge barrier mechanism drives cell-wide cortical microtubule organization in Arabidopsis. Nat. Commun. 2430. 10.1038/ncomms1444, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambrose J. C., Shoji T., Kotzer A. M., Pighin J. A., Wasteneys G. O. (2007). The Arabidopsis CLASP gene encodes a microtubule-associated protein involved in cell expansion and division. Plant Cell 19, 2763–2775. 10.1105/tpc.107.053777, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ayaydin F., Vissi E., Mészáros T., Miskolczi P., Kovács I., Fehér A., et al. (2000). Inhibition of serine/threonine-specific protein phosphatases causes premature activation of cdc2MsF kinase at G2/M transition and early mitotic microtubule organisation in alfalfa. Plant J. 23, 85–96. 10.1046/j.1365-313x.2000.00798.x, PMID: [DOI] [PubMed] [Google Scholar]
- Ban Y., Kobayashi Y., Hara T., Hamada T., Hashimoto T., Takeda S., et al. (2013). α-tubulin is rapidly phosphorylated in response to hyperosmotic stress in rice and Arabidopsis. Plant Cell Physiol. 54, 848–858. 10.1093/pcp/pct065, PMID: [DOI] [PubMed] [Google Scholar]
- Beck M., Komis G., Müller J., Menzel D., Šamaj J. (2010). Arabidopsis homologs of nucleus- and phragmoplast-localized kinase 2 and 3 and mitogen-activated protein kinase 4 are essential for microtubule organization. Plant Cell 22, 755–771. 10.1105/tpc.109.071746, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bergmann D. C., Lukowitz W., Somerville C. R. (2004). Stomatal development and pattern controlled by a MAPKK kinase. Science 304, 1494–1497. 10.1126/science.1096014, PMID: [DOI] [PubMed] [Google Scholar]
- Bhaskara G. B., Wen T. -N., Nguyen T. T., Verslues P. E. (2017). Protein phosphatase 2Cs and microtubule-associated stress protein 1 control microtubule stability, plant growth, and drought response. Plant Cell 29, 169–191. 10.1105/tpc.16.00847, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Binarova P., Cihalikova C., Dolezel J., Gilmer S., Fowke L. C. (1996). Actin distribution in somatic embryos and embryogenic protoplasts of white spruce (Picea glauca). In Vitro-Plant 32, 59–65. 10.1007/BF02823132 [DOI] [Google Scholar]
- Bögre L., Calderini O., Binarova P., Mattauch M., Till S., Kiegerl S., et al. (1999). A MAP kinase is activated late in plant mitosis and becomes localized to the plane of cell division. Plant Cell 11, 101–114. 10.1105/tpc.11.1.101, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boruc J., Weimer A. K., Stoppin-Mellet V., Mylle E., Kosetsu K., Cedeño C., et al. (2017). Phosphorylation of MAP65-1 by Arabidopsis Aurora kinases is required for efficient cell cycle progression. Plant Physiol. 173, 582–599. 10.1104/pp.16.01602, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boyer F., Simon R. (2015). Asymmetric cell divisions constructing Arabidopsis stem cell niches: the emerging role of protein phosphatases. Plant Biol. 17, 935–945. 10.1111/plb.12352, PMID: [DOI] [PubMed] [Google Scholar]
- Brieño-Enríquez M. A., Moak S. L., Holloway J. K., Cohen P. E. (2017). NIMA-related kinase 1 (NEK1) regulates meiosis I spindle assembly by altering the balance between α-Adducin and Myosin X. PLoS One 12e0185780. 10.1371/journal.pone.0185780, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brumbarova T., Ivanov R. (2016). Differential gene expression and protein phosphorylation as factors regulating the state of the Arabidopsis SNX1 protein complexes in response to environmental stimuli. Front. Plant Sci. 71456. 10.3389/fpls.2016.01456, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Buschmann H., Zachgo S. (2016). The evolution of cell division: from streptophyte algae to land plants. Trends Plant Sci. 21, 872–883. 10.1016/j.tplants.2016.07.004, PMID: [DOI] [PubMed] [Google Scholar]
- Buschmann H., Chan J., Sanchez-Pulido L., Andrade-Navarro M. A., Doonan J. H., Lloyd C. W. (2006). Microtubule-associated AIR9 recognizes the cortical division site at preprophase and cell-plate insertion. Curr. Biol. 16, 1938–1943. 10.1016/j.cub.2006.08.028, PMID: [DOI] [PubMed] [Google Scholar]
- Buschmann H., Dols J., Kopischke S., Peña E. J., Andrade-Navarro M. A., Heinlein M., et al. (2015). Arabidopsis KCBP interacts with AIR9 but stays in the cortical division zone throughout mitosis via its MyTH4-FERM domain. J. Cell Sci. 128, 2033–2046. 10.1242/jcs.156570 [DOI] [PubMed] [Google Scholar]
- Caillaud M. -C., Lecomte P., Jammes F., Quentin M., Pagnotta S., Andrio E., et al. (2008). MAP65-3 microtubule-associated protein is essential for nematode-induced giant cell ontogenesis in Arabidopsis. Plant Cell 20, 423–437. 10.1105/tpc.107.057422, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calderini O., Bogre L., Vicente O., Binarova P., Heberle-Bors E., Wilson C. (1998). A cell cycle regulated MAP kinase with a possible role in cytokinesis in tobacco cells. J. Cell Sci. 111, 3091–3100. [DOI] [PubMed] [Google Scholar]
- Calderini O., Glab N., Bergounioux C., Heberle-Bors E., Wilson C. (2001). A novel tobacco mitogen-activated protein (MAP) kinase kinase, NtMEK1, activates the cell cycle-regulated p43Ntf6 MAP kinase. J. Biol. Chem. 276, 18139–18145. 10.1074/jbc.M010621200, PMID: [DOI] [PubMed] [Google Scholar]
- Camilleri C., Azimzadeh J., Pastuglia M., Bellini C., Grandjean O., Bouchez D. (2002). The Arabidopsis TONNEAU2 gene encodes a putative novel protein phosphatase 2A regulatory subunit essential for the control of the cortical cytoskeleton. Plant Cell 14, 833–845. 10.1105/tpc.010402, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A., Arlot-Bonnemains Y., Vigneron S., Labbé J. -C., Prigent C., Lorca T. (2002a). APC/fizzy-related targets Aurora-A kinase for proteolysis. EMBO Rep. 3, 457–462. 10.1093/embo-reports/kvf095 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Castro A., Vigneron S., Bernis C., Labbé J. -C., Prigent C., Lorca T. (2002b). The D-Box-activating domain (DAD) is a new proteolysis signal that stimulates the silent D-Box sequence of Aurora-A. EMBO Rep. 3, 1209–1214. 10.1093/embo-reports/kvf241 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan J., Jensen C. G., Jensen L. C. W., Bush M., Lloyd C. W. (1999). The 65-kDa carrot microtubule-associated protein forms regularly arranged filamentous cross-bridges between microtubules. Proc. Natl. Acad. Sci. 96, 14931–14936. 10.1073/pnas.96.26.14931 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chang H. -Y., Smertenko A. P., Igarashi H., Dixon D. P., Hussey P. J. (2005). Dynamic interaction of NtMAP65-1a with microtubules in vivo. J. Cell Sci. 118, 3195–3201. 10.1242/jcs.02433, PMID: [DOI] [PubMed] [Google Scholar]
- Chang-Jie J., Sonobe S. (1993). Identification and preliminary characterization of a 65 kDa higher-plant microtubule-associated protein. J. Cell Sci. 105, 891–901. [DOI] [PubMed] [Google Scholar]
- Chen H. -W., Persson S., Grebe M., McFarlane H. E. (2018). Cellulose synthesis during cell plate assembly. Physiol. Plant. 164, 17–26. 10.1111/ppl.12703, PMID: [DOI] [PubMed] [Google Scholar]
- Cohen P. (2000). The regulation of protein function by multisite phosphorylation – a 25 year update. Trends Biochem. Sci. 25, 596–601. 10.1016/S0968-0004(00)01712-6, PMID: [DOI] [PubMed] [Google Scholar]
- Costa S. (2017). Are division plane determination and cell-cycle progression coordinated? New Phytol. 213, 16–21. 10.1111/nph.14261, PMID: [DOI] [PubMed] [Google Scholar]
- Day I. S., Miller C., Golovkin M., Reddy A. S. (2000). Interaction of a kinesin-like calmodulin-binding protein with a protein kinase. J. Biol. Chem. 275, 13737–13745. 10.1074/jbc.275.18.13737, PMID: [DOI] [PubMed] [Google Scholar]
- de Keijzer J., Kieft H., Ketelaar T., Goshima G., Janson M. E. (2017). Shortening of microtubule overlap regions defines membrane delivery sites during plant cytokinesis. Curr. Biol. 27, 514–520. 10.1016/j.cub.2016.12.043 [DOI] [PubMed] [Google Scholar]
- Demidov D., Damme D. V., Geelen D., Blattner F. R., Houben A. (2005). Identification and dynamics of two classes of Aurora-like kinases in Arabidopsis and other plants. Plant Cell 17, 836–848. 10.1105/tpc.104.029710, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dhonukshe P., Gadella T. W. J. (2003). Alteration of microtubule dynamic instability during preprophase band formation revealed by yellow fluorescent protein–CLIP170 microtubule plus-end labeling. Plant Cell 15, 597–611. 10.1105/tpc.008961, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drevensek S., Goussot M., Duroc Y., Christodoulidou A., Steyaert S., Schaefer E., et al. (2012). The Arabidopsis TRM1–TON1 interaction reveals a recruitment network common to plant cortical microtubule arrays and eukaryotic centrosomes. Plant Cell. 24, 178–191. 10.1105/tpc.111.089748, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Eng R. C., Halat L. S., Livingston S. J., Sakai T., Motose H., Wasteneys G. O. (2017). The ARM domain of ARMADILLO-REPEAT KINESIN 1 is not required for microtubule catastrophe but can negatively regulate NIMA-RELATED KINASE 6 in Arabidopsis thaliana. Plant Cell Physiol. 58, 1350–1363. 10.1093/pcp/pcx070, PMID: [DOI] [PubMed] [Google Scholar]
- Enos S. J., Dressler M., Ferreira Gomes B., Hyman A. A., Woodruff J. B. (2017). Phosphatase PP2A and microtubule pulling forces disassemble centrosomes during mitotic exit. Biol. Open. 10.1242/bio.029777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Farkas I., Dombrádi V., Miskei M., Szabados L., Koncz C. (2007). Arabidopsis PPP family of serine/threonine phosphatases. Trends Plant Sci. 12, 169–176. 10.1016/j.tplants.2007.03.003 [DOI] [PubMed] [Google Scholar]
- Fujita S., Pytela J., Hotta T., Kato T., Hamada T., Akamatsu R., et al. (2013). An atypical tubulin kinase mediates stress-induced microtubule depolymerization in Arabidopsis. Curr. Biol. 23, 1969–1978. 10.1016/j.cub.2013.08.006 [DOI] [PubMed] [Google Scholar]
- Gaillard J., Neumann E., Van Damme D., Stoppin-Mellet V., Ebel C., Barbier E., et al. (2008). Two microtubule-associated proteins of Arabidopsis MAP65s promote antiparallel microtubule bundling. Mol. Biol. Cell 19, 4534–4544. 10.1091/mbc.e08-04-0341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Herrmann A., Livanos P., Lipka E., Gadeyne A., Hauser M. T., Van Damme D., et al. (2018). Dual localized kinesin-12 POK2 plays multiple roles during cell division and interacts with MAP65-3. EMBO Rep. 19e46085. 10.15252/embr.201846085, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoshi M., Ohta K., Gotoh Y., Mori A., Murofushi H., Sakai H., et al. (1992). Mitogen-activated-protein-kinase-catalyzed phosphorylation of microtubule-associated proteins, microtubule-associated protein 2 and microtubule-associated protein 4, induces an alteration in their function. Eur. J. Biochem. 203, 43–52. [DOI] [PubMed] [Google Scholar]
- Humphrey T. V., Haasen K. E., Aldea-Brydges M. G., Sun H., Zayed Y., Indriolo E., et al. (2015). PERK-KIPK-KCBP signalling negatively regulates root growth in Arabidopsis thaliana. J. Exp. Bot. 66, 71–83. 10.1093/jxb/eru390, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hussey P. J., Hawkins T. J., Igarashi H., Kaloriti D., Smertenko A. (2002). The plant cytoskeleton: recent advances in the study of the plant microtubule-associated proteins MAP-65, MAP-190 and the Xenopus MAP215-like protein, MOR1. Plant Mol. Biol. 50, 915–924. 10.1023/A:1021236307508, PMID: [DOI] [PubMed] [Google Scholar]
- Kawabe A., Matsunaga S., Nakagawa K., Kurihara D., Yoneda A., Hasezawa S., et al. (2005). Characterization of plant Aurora kinases during mitosis. Plant Mol. Biol. 58, 1–13. 10.1007/s11103-005-3454-x, PMID: [DOI] [PubMed] [Google Scholar]
- Kawamura E. (2005). MICROTUBULE ORGANIZATION 1 regulates structure and function of microtubule arrays during mitosis and cytokinesis in the Arabidopsis root. Plant Physiol. 140, 102–114. 10.1104/pp.105.069989, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kirik A., Ehrhardt D. W., Kirik V. (2012). TONNEAU2/FASS regulates the geometry of microtubule nucleation and cortical array organization in interphase Arabidopsis cells. Plant Cell 24, 1158–1170. 10.1105/tpc.111.094367, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kohoutová L., Kourová H., Nagy S. K., Volc J., Halada P., Mészáros T., et al. (2015). The Arabidopsis mitogen-activated protein kinase 6 is associated with γ-tubulin on microtubules, phosphorylates EB1c and maintains spindle orientation under nitrosative stress. New Phytol. 207, 1061–1074. 10.1111/nph.13501, PMID: [DOI] [PubMed] [Google Scholar]
- Komaki S., Abe T., Coutuer S., Inzé D., Russinova E., Hashimoto T. (2010). Nuclear-localized subtype of end-binding 1 protein regulates spindle organization in Arabidopsis. J. Cell Sci. 123, 451–459. 10.1242/jcs.062703, PMID: [DOI] [PubMed] [Google Scholar]
- Komis G., Luptovčiak I., Ovečka M., Samakovli D., Šamajová O., Šamaj J. (2017). Katanin effects on dynamics of cortical microtubules and mitotic arrays in Arabidopsis thaliana revealed by advanced live-cell imaging. Front. Plant Sci. 8866. 10.3389/fpls.2017.00866 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Komis G., Novák D., Ovečka M., Šamajová O., Šamaj J. (2018). Advances in imaging plant cell dynamics. Plant Physiol. 176, 80–93. 10.1104/pp.17.00962 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kosetsu K., Matsunaga S., Nakagami H., Colcombet J., Sasabe M., Soyano T., et al. (2010). The MAP kinase MPK4 is required for cytokinesis in Arabidopsis thaliana. Plant Cell 22, 3778–3790. 10.1105/tpc.110.077164, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krysan P. J., Jester P. J., Gottwald J. R., Sussman M. R. (2002). An Arabidopsis mitogen-activated protein kinase kinase kinase gene family encodes essential positive regulators of cytokinesis. Plant Cell 14, 1109–1120. 10.1105/tpc.001164, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwon Y. -G., Lee S. Y., Choi Y., Greengard P., Nairn A. C. (1997). Cell cycle-dependent phosphorylation of mammalian protein phosphatase 1 by cdc2 kinase. Proc. Natl. Acad. Sci. 94, 2168–2173. 10.1073/pnas.94.6.2168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Das A., Yamaguchi M., Hashimoto J., Tsutsumi N., Uchimiya H., et al. (2003). Cell cycle function of a rice B2-type cyclin interacting with a B-type cyclin-dependent kinase. Plant J. 34, 417–425. 10.1046/j.1365-313X.2003.01736.x, PMID: [DOI] [PubMed] [Google Scholar]
- Li H., Sun B., Sasabe M., Deng X., Machida Y., Lin H., et al. (2017). Arabidopsis MAP65-4 plays a role in phragmoplast microtubule organization and marks the cortical cell division site. New Phytol. 215, 187–201. 10.1111/nph.14532, PMID: [DOI] [PubMed] [Google Scholar]
- Lin F., Krishnamoorthy P., Schubert V., Hause G., Heilmann M., Heilmann I. (2019). A dual role for cell plate-associated PI4Kβ in endocytosis and phragmoplast dynamics during plant somatic cytokinesis. EMBO J. (In Press). 10.15252/embj.2018100303, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E., Gadeyne A., Stöckle D., Zimmermann S., Jaeger G. D., Ehrhardt D. W., et al. (2014). The phragmoplast-orienting kinesin-12 class proteins translate the positional information of the preprophase band to establish the cortical division zone in Arabidopsis thaliana. Plant Cell. 26, 2617–2632. 10.1105/tpc.114.124933, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lipka E., Herrmann A., Mueller S. (2015). Mechanisms of plant cell division. Wiley Interdiscip. Rev. Dev. Biol. 4, 391–405. 10.1002/wdev.186, PMID: [DOI] [PubMed] [Google Scholar]
- López-Bucio J. S., Dubrovsky J. G., Raya-González J., Ugartechea-Chirino Y., López-Bucio J., Luna-Valdez D., et al. (2014). Arabidopsis thaliana mitogen-activated protein kinase 6 is involved in seed formation and modulation of primary and lateral root development. J. Exp. Bot. 65, 169–183. 10.1093/jxb/ert368, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loughlin R., Wilbur J. D., McNally F. J., Nédélec F. J., Heald R. (2011). Katanin contributes to interspecies spindle length scaling in Xenopus. Cell 147, 1397–1407. 10.1016/j.cell.2011.11.014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lucas J. R., Shaw S. L. (2012). MAP65-1 and MAP65-2 promote cell proliferation and axial growth in Arabidopsis roots. Plant J. 71, 454–463. 10.1111/j.1365-313X.2012.05002.x, PMID: [DOI] [PubMed] [Google Scholar]
- Lukowitz W., Roeder A., Parmenter D., Somerville C. (2004). A MAPKK kinase gene regulates extra-embryonic cell fate in Arabidopsis. Cell 116, 109–119. 10.1016/S0092-8674(03)01067-5, PMID: [DOI] [PubMed] [Google Scholar]
- Mao T., Jin L., Li H., Liu B., Yuan M. (2005). Two microtubule-associated proteins of the Arabidopsis MAP65 family function differently on microtubules. Plant Physiol. 138, 654–662. 10.1104/pp.104.052456 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marcus A. I., Dixit R., Cyr R. J. (2005). Narrowing of the preprophase microtubule band is not required for cell division plane determination in cultured plant cells. Protoplasma 226, 169–174. 10.1007/s00709-005-0119-1, PMID: [DOI] [PubMed] [Google Scholar]
- McClinton R. S., Sung Z. R. (1997). Organization of cortical microtubules at the plasma membrane in Arabidopsis. Planta 201, 252–260. 10.1007/s004250050064, PMID: [DOI] [PubMed] [Google Scholar]
- Meskiene I., Bogre L., Glaser W., Balog J., Brandstotter M., Zwerger K., et al. (1998). MP2C, a plant protein phosphatase 2C, functions as a negative regulator of mitogen-activated protein kinase pathways in yeast and plants. Proc. Natl. Acad. Sci. 95, 1938–1943. 10.1073/pnas.95.4.1938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Motose H., Hamada T., Yoshimoto K., Murata T., Hasebe M., Watanabe Y., et al. (2011). NIMA-related kinases 6, 4, and 5 interact with each other to regulate microtubule organization during epidermal cell expansion in Arabidopsis thaliana. Plant J. 67, 993–1005. 10.1111/j.1365-313X.2011.04652.x [DOI] [PubMed] [Google Scholar]
- Müller J., Beck M., Mettbach U., Komis G., Hause G., Menzel D., et al. (2010). Arabidopsis MPK6 is involved in cell division plane control during early root development, and localizes to the pre-prophase band, phragmoplast, trans-Golgi network and plasma membrane. Plant J. 61, 234–248. 10.1111/j.1365-313X.2009.04046.x, PMID: [DOI] [PubMed] [Google Scholar]
- Müller S., Han S., Smith L. G. (2006). Two kinesins are involved in the spatial control of cytokinesis in Arabidopsis thaliana. Curr. Biol. 16, 888–894. 10.1016/j.cub.2006.03.034, PMID: [DOI] [PubMed] [Google Scholar]
- Müller S., Smertenko A., Wagner V., Heinrich M., Hussey P. J., Hauser M. -T. (2004). The plant microtubule-associated protein AtMAP65-3/PLE is essential for cytokinetic phragmoplast function. Curr. Biol. 14, 412–417. 10.1016/j.cub.2004.02.032, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murata T., Sano T., Sasabe M., Nonaka S., Higashiyama T., Hasezawa S., et al. (2013). Mechanism of microtubule array expansion in the cytokinetic phragmoplast. Nat. Commun. 41967. 10.1038/ncomms2967 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Naoi K., Hashimoto T. (2004). A semidominant mutation in an Arabidopsis mitogen-activated protein kinase phosphatase-like gene compromises cortical microtubule organization. Plant Cell 16, 1841–1853. 10.1105/tpc.021865, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R. (2001). The NPK1 mitogen-activated protein kinase kinase kinase is a regulator of cell-plate formation in plant cytokinesis. Genes Dev. 15, 352–363. 10.1101/gad.863701, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nishihama R., Soyano T., Ishikawa M., Araki S., Tanaka H., Asada T., et al. (2002). Expansion of the cell plate in plant cytokinesis requires a kinesin-like protein/MAPKKK complex. Cell 109, 87–99. 10.1016/S0092-8674(02)00691-8, PMID: [DOI] [PubMed] [Google Scholar]
- O’Connell M. J., Krien M. J. E., Hunter T. (2003). Never say never. The NIMA-related protein kinases in mitotic control. Trends Cell Biol. 13, 221–228. 10.1016/S0962-8924(03)00056-4, PMID: [DOI] [PubMed] [Google Scholar]
- Oh S. A., Allen T., Kim G. J., Sidorova A., Borg M., Park S. K., et al. (2012). Arabidopsis fused kinase and the kinesin-12 subfamily constitute a signalling module required for phragmoplast expansion. Plant J. 72, 308–319. 10.1111/j.1365-313X.2012.05077.x, PMID: [DOI] [PubMed] [Google Scholar]
- Oh S. A., Johnson A., Smertenko A., Rahman D., Park S. K., Hussey P. J., et al. (2005). A divergent cellular role for the FUSED kinase family in the plant-specific cytokinetic phragmoplast. Curr. Biol. 15, 2107–2111. 10.1016/j.cub.2005.10.044 [DOI] [PubMed] [Google Scholar]
- Ookata K., Hisanaga S., Sugita M., Okuyama A., Murofushi H., Kitazawa H., et al. (1997). MAP4 is the in vivo substrate for CDC2 kinase in HeLa cells: identification of an M-phase specific and a cell cycle-independent phosphorylation site in MAP4. Biochemist 36, 15873–15883. 10.1021/bi971251w, PMID: [DOI] [PubMed] [Google Scholar]
- Panteris E., Diannelidis B. -E., Adamakis I. -D. S. (2018). Cortical microtubule orientation in Arabidopsis thaliana root meristematic zone depends on cell division and requires severing by katanin. J. Biol. Res.Thessalon. 2512. 10.1186/s40709-018-0082-6, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Petrovská B., Cenklová V., Pochylová Ž., Kourová H., Doskočilová A., Plíhal O., et al. (2012). Plant Aurora kinases play a role in maintenance of primary meristems and control of endoreduplication. New Phytol. 193, 590–604. 10.1111/j.1469-8137.2011.03989.x, PMID: [DOI] [PubMed] [Google Scholar]
- Petrovská B., Jerábková H., Kohoutová L., Cenklová V., Pochylová Ž., Gelová Z., et al. (2013). Overexpressed TPX2 causes ectopic formation of microtubular arrays in the nuclei of acentrosomal plant cells. J. Exp. Bot. 64, 4575–4587. 10.1093/jxb/ert271, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pickett-Heaps J. D., Northcote D. H. (1966). Organization of microtubules and endoplasmic reticulum during mitosis and cytokinesis in wheat meristems. J. Cell Sci. 1, 109–120. PMID: [DOI] [PubMed] [Google Scholar]
- Qu Y., Song P., Hu Y., Jin X., Jia Q., Zhang X., et al. (2018). Regulation of stomatal movement by cortical microtubule organization in response to darkness and ABA signaling in Arabidopsis. Plant Growth Regul. 84, 467–479. 10.1007/s10725-017-0353-5 [DOI] [Google Scholar]
- Repetto M. V., Winters M. J., Bush A., Reiter W., Hollenstein D. M., Ammerer G., et al. (2018). CDK and MAPK synergistically regulate signaling dynamics via a shared multi-site phosphorylation region on the scaffold protein Ste5. Mol. Cell 69, 938–952.e6. 10.1016/j.molcel.2018.02.018, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritchey L., Chakrabarti R. (2014). Aurora A kinase modulates actin cytoskeleton through phosphorylation of Cofilin: implication in the mitotic process. Biochim. Biophys. Acta BBA Mol. Cell Res. 1843, 2719–2729. 10.1016/j.bbamcr.2014.07.014, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rodrigues N. T. L., Lekomtsev S., Jananji S., Kriston-Vizi J., Hickson G. R. X., Baum B. (2015). Kinetochore-localized PP1–Sds22 couples chromosome segregation to polar relaxation. Nature 524, 489–492. 10.1038/nature14496, PMID: [DOI] [PubMed] [Google Scholar]
- Samaj J., Baluska F., Hirt H. (2004). From signal to cell polarity: mitogen-activated protein kinases as sensors and effectors of cytoskeleton dynamicity. J. Exp. Bot. 55, 189–198. 10.1093/jxb/erh012, PMID: [DOI] [PubMed] [Google Scholar]
- Samaj J., Ovecka M., Hlavacka A., Lecourieux F., Meskiene I., Lichtscheidl I., et al. (2002). Involvement of the mitogen-activated protein kinase SIMK in regulation of root hair tip growth. EMBO J. 21, 3296–3306. 10.1093/emboj/cdf349, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Samofalova D. O., Karpov P. A., Raevsky A. V., Blume Y. B. (2017). Protein phosphatases potentially associated with regulation of microtubules, their spatial structure reconstruction and analysis. Cell Biol. Int. 10.1002/cbin.10810, PMID: [DOI] [PubMed] [Google Scholar]
- Sasabe M., Boudolf V., Veylder L. D., Inzé D., Genschik P., Machida Y. (2011a). Phosphorylation of a mitotic kinesin-like protein and a MAPKKK by cyclin-dependent kinases (CDKs) is involved in the transition to cytokinesis in plants. Proc. Natl. Acad. Sci. 108, 17844–17849. 10.1073/pnas.1110174108 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasabe M., Kosetsu K., Hidaka M., Murase A., Machida Y. (2011b). Arabidopsis thaliana MAP65-1 and MAP65-2 function redundantly with MAP65-3/PLEIADE in cytokinesis downstream of MPK4. Plant Signal. Behav. 6, 743–747. 10.4161/psb.6.5.15146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schaefer E., Belcram K., Uyttewaal M., Duroc Y., Goussot M., Legland D., et al. (2017). The preprophase band of microtubules controls the robustness of division orientation in plants. Science 356, 186–189. 10.1126/science.aal3016 [DOI] [PubMed] [Google Scholar]
- Schecher S., Walter B., Falkenstein M., Macher-Goeppinger S., Stenzel P., Krümpelmann K., et al. (2017). Cyclin K dependent regulation of Aurora B affects apoptosis and proliferation by induction of mitotic catastrophe in prostate cancer: cyclin K in prostate cancer. Int. J. Cancer 141, 1643–1653. 10.1002/ijc.30864, PMID: [DOI] [PubMed] [Google Scholar]
- Sedbrook J. C., Ehrhardt D. W., Fisher S. E., Scheible W. R., Somerville C. R. (2004). The Arabidopsis sku6/spiral1 gene encodes a plus end-localized microtubule-interacting protein involved in directional cell expansion. Plant Cell 16, 1506–1520. 10.1105/tpc.020644, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shiina N., Moriguchi T., Ohta K., Gotoh Y., Nishida E. (1992). Regulation of a major microtubule-associated protein by MPF and MAP kinase. EMBO J. 11, 3977–3984. 10.1002/j.1460-2075.1992.tb05491.x, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smékalová V., Luptovčiak I., Komis G., Šamajová O., Ovečka M., Doskočilová A., et al. (2014). Involvement of YODA and mitogen activated protein kinase 6 in Arabidopsis post-embryogenic root development through auxin up-regulation and cell division plane orientation. New Phytol. 203, 1175–1193. 10.1111/nph.12880, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A. P., Chang H. -Y., Sonobe S., Fenyk S., Weingartner M., Bögre L., et al. (2006). Control of the AtMAP65-1 interaction with microtubules through the cell cycle. J. Cell Sci. 119, 3227–3237. 10.1242/jcs.03051, PMID: [DOI] [PubMed] [Google Scholar]
- Smertenko A. P., Kaloriti D., Chang H. -Y., Fiserova J., Opatrny Z., Hussey P. J. (2008). The C-terminal variable region specifies the dynamic properties of Arabidopsis microtubule-associated protein MAP65 isotypes. Plant Cell 20, 3346–3358. 10.1105/tpc.108.063362, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smertenko A., Hewitt S. L., Jacques C. N., Kacprzyk R., Liu Y., Marcec M. J., et al. (2018). Phragmoplast microtubule dynamics - a game of zones. J. Cell Sci. 131, 1–11. 10.1242/jcs.203331, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Song S. -K., Lee M. M., Clark S. E. (2006). POL and PLL1 phosphatases are CLAVATA1 signaling intermediates required for Arabidopsis shoot and floral stem cells. Development 133, 4691–4698. 10.1242/dev.02652, PMID: [DOI] [PubMed] [Google Scholar]
- Spinner L., Gadeyne A., Belcram K., Goussot M., Moison M., Duroc Y., et al. (2013). A protein phosphatase 2A complex spatially controls plant cell division. Nat. Commun. 41863. 10.1038/ncomms2831 [DOI] [PubMed] [Google Scholar]
- Stals H., Bauwens S., Traas J., Montagu M. V., Engler G., Inzé D. (1997). Plant CDC2 is not only targeted to the pre-prophase band, but also co-localizes with the spindle, phragmoplast, and chromosomes. FEBS Lett. 418, 229–234. 10.1016/S0014-5793(97)01368-9, PMID: [DOI] [PubMed] [Google Scholar]
- Strompen G., El Kasmi F., Richter S., Lukowitz W., Assaad F. F., Jürgens G., et al. (2002). The Arabidopsis HINKEL gene encodes a kinesin-related protein involved in cytokinesis and is expressed in a cell cycle-dependent manner. Curr. Biol. 12, 153–158. 10.1016/S0960-9822(01)00655-8 [DOI] [PubMed] [Google Scholar]
- Takahashi Y., Soyano T., Kosetsu K., Sasabe M., Machida Y. (2010). HINKEL kinesin, ANP MAPKKKs and MKK6/ANQ MAPKK, which phosphorylates and activates MPK4 MAPK, constitute a pathway that is required for cytokinesis in Arabidopsis thaliana. Plant Cell Physiol. 51, 1766–1776. 10.1093/pcp/pcq135, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Takatani S., Ozawa S., Yagi N., Hotta T., Hashimoto T., Takahashi Y., et al. (2017). Directional cell expansion requires NIMA-related kinase 6 (NEK6)-mediated cortical microtubule destabilization. Sci. Rep. 77826. 10.1038/s41598-017-08453-5, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tang A., Shi P., Song A., Zou D., Zhou Y., Gu P., et al. (2016). PP2A regulates kinetochore-microtubule attachment during meiosis I in oocyte. Cell Cycle 15, 1450–1461. 10.1080/15384101.2016.1175256, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tomaštíková E., Demidov D., Jeřábková H., Binarová P., Houben A., Doležel J., et al. (2015). TPX2 protein of Arabidopsis activates Aurora kinase 1, but not Aurora kinase 3 in vitro. Plant Mol. Biol. Report. 33, 1988–1995. 10.1007/s11105-015-0890-x [DOI] [Google Scholar]
- Torres-Ruiz R. A., Jurgens G. (1994). Mutations in the FASS gene uncouple pattern formation and morphogenesis in Arabidopsis development. Development 120, 2967–2978. [DOI] [PubMed] [Google Scholar]
- Traas J., Bellini C., Nacry P., Kronenberger J., Bouchez D., Caboche M. (1995). Normal differentiation patterns in plants lacking microtubular pre-prophase bands. Nature 375, 676–677. 10.1038/375676a0 [DOI] [Google Scholar]
- Twell D., Park S. K., Hawkins T. J., Schubert D., Schmidt R., Smertenko A., et al. (2002). MOR1/GEM1 has an essential role in the plant-specific cytokinetic phragmoplast. Nat. Cell Biol. 4, 711–714. 10.1038/ncb844, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Umbrasaite J., Schweighofer A., Kazanaviciute V., Magyar Z., Ayatollahi Z., Unterwurzacher V., et al. (2010). MAPK phosphatase AP2C3 induces ectopic proliferation of epidermal cells leading to stomata development in Arabidopsis. PLoS One 5e15357. 10.1371/journal.pone.0015357, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Damme D. V., De Rybel B., Gudesblat G., Demidov D., Grunewald W., De Smet I., et al. (2011). Arabidopsis α Aurora kinases function in formative cell division plane orientation. Plant Cell. 23, 4013–4024. 10.1105/tpc.111.089565, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Varadkar P., Abbasi F., Takeda K., Dyson J. J., McCright B. (2017). PP2A-B56γ is required for an efficient spindle assembly checkpoint. Cell Cycle 16, 1210–1219. 10.1080/15384101.2017.1325042, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vasquez R. J., Gard D. L., Cassimeris L. (1999). Phosphorylation by CDK1 regulates XMAP215 function in vitro. Cell Motil. 43, 310–321. [DOI] [PubMed] [Google Scholar]
- Vigneault F., Lachance D., Cloutier M., Pelletier G., Levasseur C., Séguin A. (2007). Members of the plant NIMA-related kinases are involved in organ development and vascularization in poplar, Arabidopsis and rice. Plant J. 51, 575–588. 10.1111/j.1365-313X.2007.03161.x, PMID: [DOI] [PubMed] [Google Scholar]
- Walia A., Lee J. S., Wasteneys G., Ellis B. (2009). Arabidopsis mitogen-activated protein kinase MPK18 mediates cortical microtubule functions in plant cells. Plant J. 59, 565–575. 10.1111/j.1365-313X.2009.03895.x, PMID: [DOI] [PubMed] [Google Scholar]
- Walker K. L., Müller S., Moss D., Ehrhardt D. W., Smith L. G. (2007). Arabidopsis TANGLED identifies the division plane throughout mitosis and cytokinesis. Curr. Biol. 17, 1827–1836. 10.1016/j.cub.2007.09.063, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weingartner M., Binarova P., Drykova D., Schweighofer A., David J. -P., Heberle-Bors E., et al. (2001). Dynamic recruitment of Cdc2 to specific microtubule structures during mitosis. Plant Cell 13, 1929–1943. 10.1105/tpc.13.8.1929, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whitehead E., Heald R., Wilbur J. D. (2013). N-terminal phosphorylation of p60 katanin directly regulates microtubule severing. J. Mol. Biol. 425, 214–221. 10.1016/j.jmb.2012.11.022, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wong J. H., Hashimoto T. (2017). Novel Arabidopsis microtubule-associated proteins track growing microtubule plus ends. BMC Plant Biol. 1733. 10.1186/s12870-017-0987-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wright A. J., Smith L. G. (2007). “Division plane orientation in plant cells” in Cell division control in plants plant cell monographs. (Berlin, Heidelberg: Springer; ), 33–57. [Google Scholar]
- Wright A. J., Gallagher K., Smith L. G. (2009). Discordia1 and alternative discordia1 function redundantly at the cortical division site to promote preprophase band formation and orient division planes in maize. Plant Cell 21, 234–247. 10.1105/tpc.108.062810 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu S. -Z., Bezanilla M. (2014). Myosin VIII associates with microtubule ends and together with actin plays a role in guiding plant cell division. elife 3e03498. 10.7554/eLife.03498, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xu X. M., Zhao Q., Rodrigo-Peiris T., Brkljacic J., He C. S., Muller S., et al. (2008). RanGAP1 is a continuous marker of the Arabidopsis cell division plane. Proc. Natl. Acad. Sci. 105, 18637–18642. 10.1073/pnas.0806157105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xue Y., Ren J., Gao X., Jin C., Wen L., Yao X. (2008). GPS 2.0, a tool to predict kinase-specific phosphorylation sites in hierarchy. Mol. Cell. Proteomics 7, 1598–1608. 10.1074/mcp.M700574-MCP200, PMID: [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yoon J. -T., Ahn H. -K., Pai H. -S. (2018). The subfamily II catalytic subunits of protein phosphatase 2A (PP2A) are involved in cortical microtubule organization. Planta 248, 1–17. 10.1007/s00425-018-3000-0, PMID: [DOI] [PubMed] [Google Scholar]
- Zhou S., Chen Q., Li X., Li Y. (2017). MAP65-1 is required for the depolymerization and reorganization of cortical microtubules in the response to salt stress in Arabidopsis. Plant Sci. 264, 112–121. 10.1016/j.plantsci.2017.09.004, PMID: [DOI] [PubMed] [Google Scholar]