Abstract
As complications associated with antibiotic resistance have intensified, copper (Cu) is attracting attention as an antimicrobial agent. Recent studies have shown that copper surfaces decrease microbial burden, and host macrophages use Cu to increase bacterial killing. Not surprisingly, microbes have evolved mechanisms to tightly control intracellular Cu pools and protect against Cu toxicity. Here, we identified two genes (copB and copL) encoded within the Staphylococcus aureus arginine-catabolic mobile element (ACME) that we hypothesized function in Cu homeostasis. Supporting this hypothesis, mutational inactivation of copB or copL increased copper sensitivity. We found that copBL are co-transcribed and that their transcription is increased during copper stress and in a strain in which csoR, encoding a Cu-responsive transcriptional repressor, was mutated. Moreover, copB displayed genetic synergy with copA, suggesting that CopB functions in Cu export. We further observed that CopL functions independently of CopB or CopA in Cu toxicity protection and that CopL from the S. aureus clone USA300 is a membrane-bound and surface-exposed lipoprotein that binds up to four Cu+ ions. Solution NMR structures of the homologous Bacillus subtilis CopL, together with phylogenetic analysis and chemical-shift perturbation experiments, identified conserved residues potentially involved in Cu+ coordination. The solution NMR structure also revealed a novel Cu-binding architecture. Of note, a CopL variant with defective Cu+ binding did not protect against Cu toxicity in vivo. Taken together, these findings indicate that the ACME-encoded CopB and CopL proteins are additional factors utilized by the highly successful S. aureus USA300 clone to suppress copper toxicity.
Keywords: copper, lipoprotein, metal homeostasis, methicillin-resistant Staphylococcus aureus (MRSA), structural biology, virulence factor, metal, arginine-catabolic mobile element (ACME), redox homeostasis
Introduction
Because of its ability to cycle between its reduced (Cu1+) and oxidized (Cu2+) states, copper (Cu)7 has catalytic roles in metalloenzymes such as dioxygen reductases (1), superoxide dismutases (2), laccases (3), and several proteins involved in denitrification (4). Cu can also have nonredox structural roles, such as in transcription factors (5). Despite its necessity, intracellular Cu accumulation is toxic due to, in part, its ability to compete with other transition metals. Cu can displace iron from iron–sulfur (FeS) clusters leading to cluster destruction and protein inactivation, as well as inhibit the assembly of FeS clusters by binding to assembly proteins that would typically bind FeS clusters (6–9). Cu poisoning may also occur as a result of Fenton-type chemistry, in which Cu1+ reacts with hydrogen peroxide leading to the formation of hydroxyl radicals (10), which in turn can damage proteins, membrane lipids, and DNA.
Copper has been used to sterilize wounds and drinking water and recently by hospitals to reduce microbial burden on touch surfaces (11, 12). The human innate immune system uses Cu to kill invading microorganisms. Upon challenge with bacteria, macrophages accumulate Cu within phagosomes, where it may synergize with reactive oxygen species produced by NADPH oxidase to increase killing (13, 14).
Staphylococcus aureus is a public health concern worldwide. S. aureus causes numerous infection types ranging from skin and soft tissue infections to more severe and life-threatening diseases, such as pneumonia, osteomyelitis, and bacteremia (15, 16). Methicillin-resistant S. aureus (MRSA) infections have become more prevalent in community settings, and this epidemic is widely attributed to the spread of the USA300 clone (17, 18).
The genome of the USA300 clone has various mobile genetic elements, including the arginine-catabolic mobile element (ACME) (19), which occupies a 31-kb region located adjacent to the SCCmecIV genetic element. Genes encoded within ACME provide an increased fitness advantage facilitating colonization and persistence of S. aureus on human skin. The ACME-encoded arginine-deiminase system (arc) improves survival in acidic conditions (20), and the speG gene product provides resistance to high levels of host-derived polyamines (20, 21).
Like other pathogens, S. aureus must employ strategies to tightly control intracellular Cu levels and avoid Cu toxicity. Membrane-spanning Cu exporters (22, 23), Cu chaperons (24), and intracellular metallothioneins (25) are the most common defense mechanisms employed by microorganisms. The low-molecular weight thiols bacillithiol and GSH can bind to Cu with relatively high affinity (26, 27), and mutant strains lacking these compounds display sensitivity to copper suggesting potential role(s) for low-molecular weight thiols in Cu buffering (28, 29).
In S. aureus, the Cu-sensitive operon repressor (CsoR) binds intracellular Cu leading to derepression of the copAZ operon (30). CopA is a transmembrane P1B-1-type ATPase Cu exporter (31), and CopZ is a cytoplasmic Atx1-like Cu-binding chaperon (32). Some S. aureus strains have an additional predicted P1B-3-subtype Cu transporter (CopB) and a Cu-dependent multicopper oxidase (Mco) (33). The described copB and mco genes are co-localized and located on mobile DNA in S. aureus (34). The copBmco genes provide increased Cu resistance and, like the copAZ operon, their transcription is regulated by CsoR (33, 34). The function of Mco is currently unknown (35).
In this work, we characterized the copBL operon located within the ACME region of the S. aureus USA300 clone, which encodes an additional Cu homeostatic mechanism. copL encodes for a membrane-bound, surface-exposed Cu-binding lipoprotein, and genetic evidence suggests that copB encodes a Cu exporter. The solution NMR structure of a B. subtilis CopL revealed that CopL has a unique Cu-binding architecture.
Results
Analyses of S. aureus genes involved in copper homeostasis
We analyzed the genome of the community-associated (CA)-MRSA strain USA300_FPR3757 (19) for genes involved in Cu homeostasis. We noted the presence of csoR, copA, and copZ. Further analysis identified the SAUSA300_0078 locus (CopB; Fig. 1A), which shows 36% identity with CopA. CopB contains most of the conserved structural elements of P1B-ATPases (Fig. S1), including a phosphatase domain (TGES), a conserved CPX metal-binding sequence, and an ATP-binding domain (MXGDGXNDXP) (36). Unlike CopA, CopB lacks the N-terminal metal–binding CXXC motifs, but instead contains a His-rich N terminus (Fig. S2). The CopB described herein shares 85 and 56% identity to the previously described S. aureus CopB (33) and Enterococcus hirae CopB (37), respectively. The N-terminal metal–binding domain of the CopB described herein is 52 and 42% identical to those of the N-terminal metal–binding domains of the previously described S. aureus CopB (33) and Enterococcus hirae CopB (37), respectively. A sequence alignment of the N-terminal metal–binding domains of these proteins found that they vary in the number of amino acids and in histidine content (Fig. S2).
Figure 1.
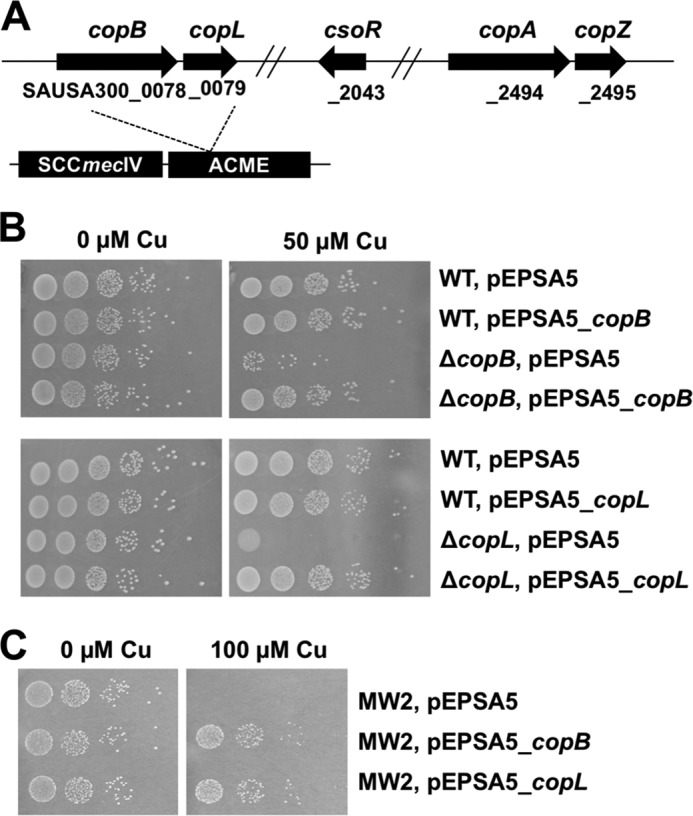
CopB and CopL protect against Cu toxicity in S. aureus. A, chromosomal location of genes involved in Cu homeostasis in S. aureus USA300_FPR3757. The copB and copL genes are located in the ACME, adjacent to the SCCmecIV genetic element. Locus tags are provided below the genes. B, copA, copB, and copL gene products are required to protect against Cu toxicity. Top, WT (JMB1100) and ΔcopB (JMB7900) strains containing pEPSA5 or pEPSA5_copB are shown. Bottom, WT and ΔcopL (JMB7711) strains containing pEPSA5 or pEPSA5_copL are shown. Strains were serially diluted and spot-plated on chemically defined media with or without 50 μm Cu. C, overexpression of copB or copL results in increased Cu resistance in the S. aureus USA400 strain MW2. The S. aureus USA400 clone MW2, which lacks genome-encoded copB and copL, containing pEPSA5, pEPSA5_copB, or pEPSA_copL is shown. Strains were serially diluted and spot-plated on chemically defined media without or with 100 μm Cu.
A second open reading frame (ORF) is located 17 bp downstream of copB. This ORF, which we and others named copL (copper-binding lipoprotein), encodes a putative lipoprotein containing two DUF1541 domains (38). The copBL genes are located within the ACME region.
We identified ∼200 CopL-like proteins in other microorganisms with the majority belonging to the Actinobacteria and Firmicutes phyla. The genomes containing copL also encoded for at least one additional Cu detoxification protein (CopA, CopZ, or CopB) (Table S1). The copL homologues are often co-localized near genes or within apparent operons encoding for genes involved in Cu homeostasis. copL is also located adjacent to copB in other staphylococci (Fig. S3).
S. aureus strains lacking CopB or CopL have increased sensitivity to Cu
We tested the hypothesis that CopB and CopL function in Cu homeostasis. We constructed ΔcopB and ΔcopL mutants in the S. aureus strain USA300_LAC (WT) (Fig. S4), which differs from the S. aureus strain USA300_FPR3757 by a few single-nucleotide polymorphisms (39). The WT, ΔcopB, and ΔcopL strains were spot-plated on chemically defined media containing varying concentrations of Cu. The ΔcopB and ΔcopL strains displayed decreased survival when cultured in the presence of Cu, but no growth abnormalities in the absence of Cu (Fig. 1B). Genetic complementation verified that mutational inactivation of copB or copL was resulting in the observed growth defects (Fig. 1B).
The S. aureus USA400 strain MW2 lacks copB and copL. We mobilized copB and copL to the MW2 strain via plasmid and examined Cu sensitivity. The MW2 strain containing copB or copL displayed increased resistance to Cu (Fig. 1C). Expression of copL in the S. aureus strains Newman, COL, and RN4220 also resulted in increased Cu resistance (Fig. S5).
Strains lacking CopB or CopL display exacerbated phenotypes in cells unable to export cytoplasmic Cu via CopA
We investigated whether CopB and CopL had a functional overlap with other genes involved in Cu homeostasis. Genes encoding proteins with functional overlap often display synergistic phenotypes when the gene products are absent or nonfunctional (40). An S. aureus copA mutant accumulates intracellular Cu and displays sensitivity to Cu (31). The copA::Tn and ΔcopB mutants had decreased growth on solid media containing >50 μm Cu, but no sensitivity was observed at lower (10 μm) Cu concentrations (Fig. 2A). The phenotypes associated with the copA::Tn ΔcopB mutations were synergistic for Cu sensitivity (Fig. 2A). Similarly, the copA::Tn ΔcopL double–mutant strain was more sensitive to Cu than the copA::Tn and ΔcopL single mutants (Fig. 2B). The copA::Tn ΔcopB ΔcopL triple mutant was more sensitive to Cu than the copA::Tn ΔcopB double–mutant strain (Fig. 2C).
Figure 2.
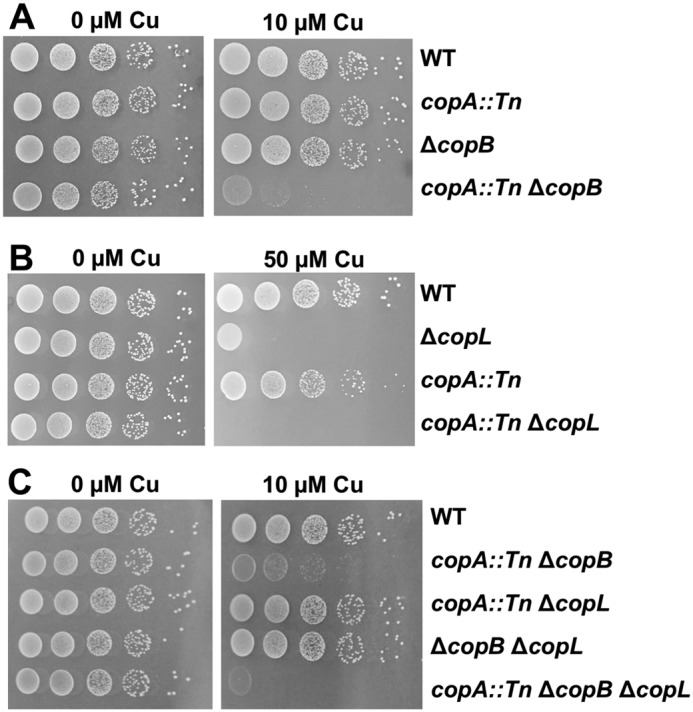
Intracellular Cu accumulation exacerbates the phenotypes of the copB and copL mutants and CopA, CopB, and CopL function independently. A, phenotypes associated with copA and copB mutations are synergistic. The WT (JMB1100), copA::Tn (JMB4084), ΔcopB (JMB7900), and copA::Tn ΔcopB (JMB8009) strains were serially diluted and spot-plated on chemically defined media without or with 50 μm Cu. B, phenotypes associated with copB and copL mutations are additive. The WT, ΔcopL (JMB7711), copA::Tn, and copA::Tn ΔcopL (JMB7803) strains were serially diluted and spot-plated on chemically defined media without or with 50 μm Cu. C, CopA, CopB, and CopL function independently to protect against Cu toxicity. The WT, copA::Tn ΔcopB, copA::Tn ΔcopL, ΔcopB ΔcopL (JMB7901), and copA::Tn ΔcopB ΔcopL (JMB7972) strains were serially diluted and spot-plated on chemically defined media without or with 10 μm Cu.
The S. aureus copZ::Tn mutant did not display a Cu sensitivity phenotype on solid media (data not shown). The copZ::Tn ΔcopB and copZ::Tn ΔcopL strains displayed Cu sensitivity phenotypes similar to the ΔcopB and ΔcopL single mutants (data not shown).
Collectively, these results suggest the following: (a) CopA and CopB have a functional overlap and serve as Cu exporters; (b) CopL functions in Cu homeostasis independently of CopA and CopB; and (c) CopZ is not required to protect against Cu toxicity under the conditions examined.
copBL transcription increases in the presence of Cu
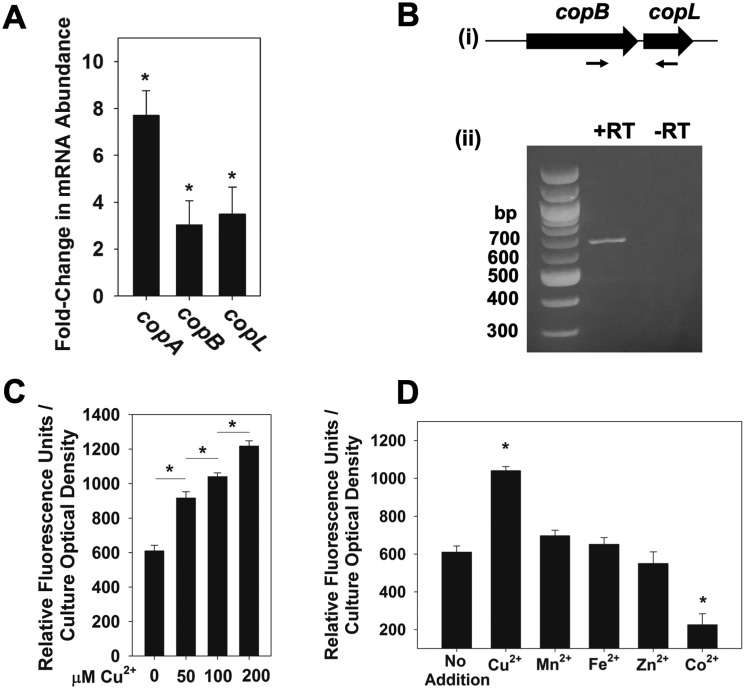
We tested the hypothesis that transcription of copB and copL increases in response to Cu. The abundances of the copB and copL transcripts were monitored in the WT strain grown with and without Cu. The S. aureus copA is induced during Cu stress (31, 33); therefore, we quantified abundance of the copA transcript as a control. The transcripts of copB and copL were increased ∼4-fold upon Cu stress, whereas the copA transcript was increased ∼8-fold (Fig. 3A).
Figure 3.
copBL operon is up-regulated under copper stress. A, copA, copB, and copL genes are induced by Cu. The S. aureus WT (JMB1100) was cultured in chemically defined media containing 0 or 100 μm Cu. RNA was isolated; cDNA was generated, and the abundance of the copA, copB, and copL transcripts was quantified using qPCR. Data show fold-induction of genes of interest upon addition of Cu with respect to no Cu addition. Data represent the average of biological triplicates with errors presented as standard deviations. B, copB and copL genes are co-transcribed. RNA was isolated from the WT grown in chemically defined medium with 100 μm Cu and cDNA libraries generated. Panel i, schematic showing the primer pair (ZRT21 and ZRT24) used to detect the copBL transcript; expected size: 643 bp. Panel ii, agarose gel electrophoresis was used to detect the copBL amplicon generated using cDNA libraries as template DNA. A reaction without reverse transcriptase (−RT) was included as a control for genomic DNA contamination. C, transcriptional activity of copB increases in synchrony with Cu concentration. Transcriptional activity of the copB reporter was monitored in the S. aureus USA300 WT grown in chemically defined media containing 0, 50, 100, or 200 μm Cu. D, transcriptional activity of copB is specific to copper stress. Transcriptional activity of copB was monitored in the WT grown in chemically defined media containing 100 μm Cu2+, 100 μm Mn2+, 100 μm Fe2+, 100 μm Zn2+, or 50 μm Co2+. C and D, fluorescence data were standardized to culture optical density (A590 nm), and data represent the average of biological triplicates with errors presented as standard deviations. Student's t test were performed on the data, and * indicates p ≤ 0.05.
We tested the hypothesis that copB and copL are co-transcribed. RNA was isolated from WT cultures grown in the presence of Cu, and cDNA libraries were generated. Using cDNA as a PCR template, and primers nested within copB and copL, we were able to obtain an amplicon that spanned copB and copL (Fig. 3B). No PCR products were obtained from control reactions lacking reverse transcriptase (−RT), confirming that the amplicon was not the result of genomic DNA contamination.
We created a transcriptional reporter to further analyze the regulation of copB. Transcriptional activity of copB increased in synchrony with the concentration of Cu added to the growth medium verifying the functionality of the reporter (Fig. 3C). Transcriptional activity of copB did not increase upon challenge with Mn2+, Fe2+, Zn2+, or Co2+ (Fig. 3D). In addition, the ΔcopB and ΔcopL mutant strains did not display increased sensitivity to these metals at concentrations that decreased the survival of the WT strain (data not shown).
CsoR regulates the copBL operon
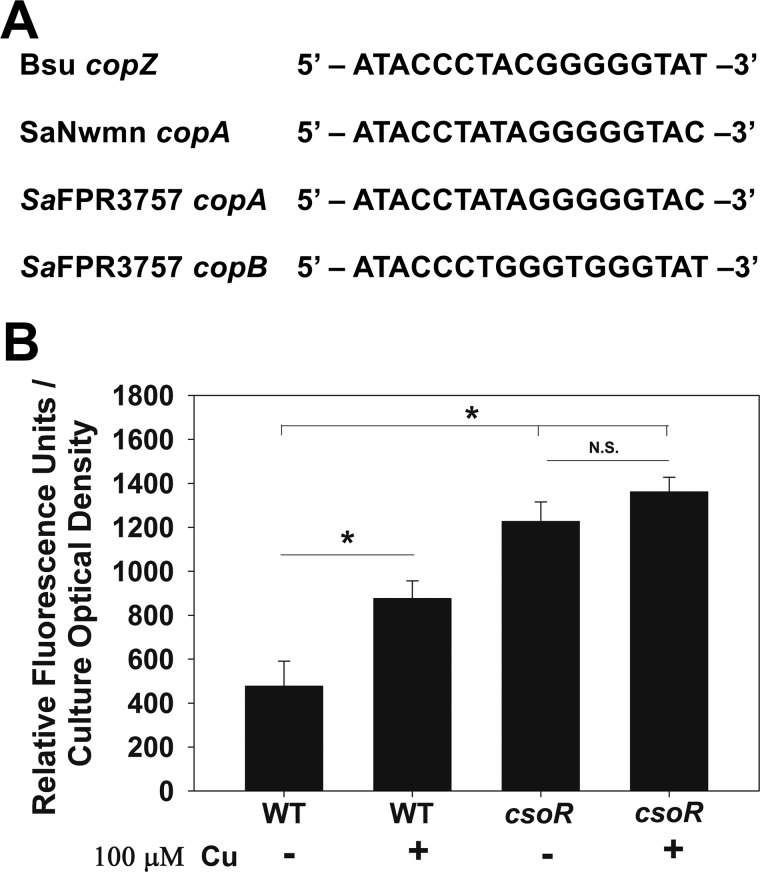
We tested the hypothesis that CsoR regulates the copBL operon. CsoR-binding sites have been identified in the promoters of copZA in Bacillus subtilis (41) and copAZ in S. aureus strain Newman (30), which are characterized by a pseudo-inverted repeat (TACCNNNNGGGGGTA) (Fig. 4A). We analyzed the promoter region of the copBL in strain FPR3757 and found that it contains a putative CsoR-binding site ∼100 bp upstream of the translational start site.
Figure 4.
Transcription of the copBL operon is regulated by CsoR. A, proposed CsoR-binding sites are shown for the S. aureus USA300_FPR3757 copA and copB promoter regions, as well as the B. subtilis copZ and S. aureus Newman copA promoter regions. B, transcriptional activity of copB is increased in a csoR mutant. Transcriptional activity of copB was monitored in the WT (JMB1100) and csoR::Tn (JMB6807) strains grown in chemically defined media containing 0 or 100 μm Cu. Fluorescence data were standardized to culture optical density (A590 nm). Data shown represent the average of biological triplicates with errors presented as standard deviations. Student's t test were performed on the data, and * indicates p ≤ 0.05, whereas N.S. denotes not significant.
We monitored the transcriptional activity of copB in the WT and csoR::Tn strains. The transcriptional activity of copB was increased in the csoR::Tn strain, suggesting that transcription of the copBL operon is repressed by CsoR (Fig. 4B). Addition of Cu led to increased copB transcriptional activity in the WT strain but not in the csoR::Tn strain (Fig. 4B). The S. aureus strain Newman lacks the copBL operon; however, the transcriptional activity of copB was also increased in an S. aureus Newman ΔcsoR mutant when compared with the parent strain (Fig. S6).
CopL is membrane-associated and surface-exposed
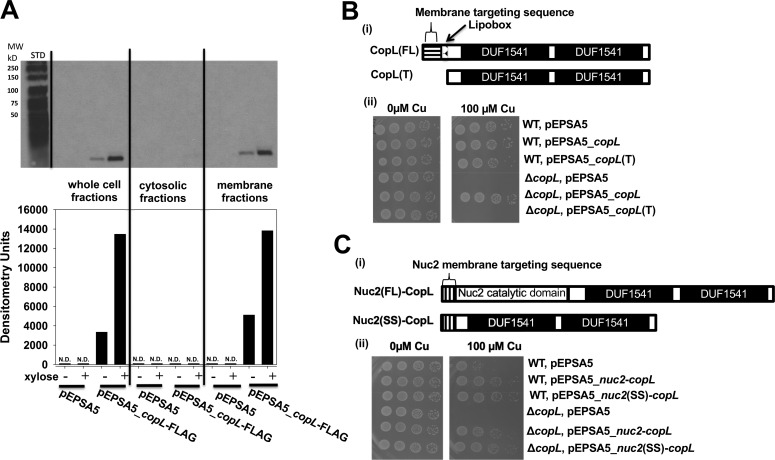
We tested the hypothesis that CopL is a surface-exposed lipoprotein. We conducted cell fractionation experiments to verify the cellular location of CopL. We utilized a C-terminal FLAG–tagged copL that was under the transcriptional control of a xylose-inducible promoter (xylRO) (pEPSA5_copL–FLAG). The copL–FLAG allele genetically complemented the Cu sensitivity phenotype of the ΔcopL mutant strain, verifying the functionality of the fusion (data not shown). Cultures of the ΔcopL strain harboring either pEPSA5 or pEPSA5_copL–FLAG were grown in the presence and absence of xylose. Cells were fractionated into cytosolic and membrane fractions. CopL–FLAG was detected in whole-cell extracts and membrane fractions, but not in cytosolic fractions (Fig. 5A). CopL–FLAG was also detected in noninduced samples, albeit at a much lower intensity, which is likely the result of leaky expression. CopL–FLAG was not detected in cells containing only empty vector.
Figure 5.
CopL is membrane-associated and surface-exposed. A, monitoring CopL abundance in S. aureus whole-cell extracts, cytosolic fractions, and membrane fractions. The ΔcopL (JMB7711) mutant containing the pEPSA or pEPSA5_copL–FLAG was cultured in the absence and presence of 0.2% xylose prior to fractionation and analysis. ND, not detected. B, membrane-anchor signal sequence is necessary for CopL function. Panel i, schematic showing the CopL variants utilized. The pEPSA_copL encoded saCopL and pEPSA_copL(T) encoded for the truncated saCopL(T), which lacked the proposed N terminus export signal-sequence and lipobox. Panel ii, WT and ΔcopL strains harboring the pEPSA5, pEPSA5_copL, or pEPSA5_copL(T) were serially diluted and spot-plated on chemically defined media without and with 100 μm Cu. C, cell-surface exposure is necessary for CopL function. Panel i, schematic showing the CopL-Nuc2 chimeric variants. The truncated copL gene, lacking the proposed N-terminal export–sequence and lipobox, was fused to either the full-length nuc2 or the nuc2 membrane-anchor signal sequence and cloned into the pEPSA5 plasmid to yield the pEPSA5_nuc2(FL)-copL and pEPSA5_nuc2(SS)-copL vectors, respectively. Panel ii, WT and ΔcopL strains harboring pEPSA5, pEPSA5_nuc2(FL)-copL, or pEPSA5_nuc2(SS)-copL were serially diluted and spot-plated on chemically defined media containing 0 or 100 μm Cu.
The TOPCONS algorithm (42) predicted that CopL contains an N-terminal membrane–targeting signal sequence (Fig. S1), which is a characteristic of proteins that are translocated across the cytoplasmic membrane (43). The LipoP server (44) identified a lipobox motif (sequence: LSAC) that is processed to provide a covalent lipid-linked membrane anchor. Lipoproteins in Gram-positive bacteria are anchored to the cytoplasmic membrane with the C termini facing the extracellular surface (45). We examined whether the functionality of the CopL protein depends on its cellular localization. We cloned a copL allele lacking the membrane translocation signal–sequence and lipobox, which we refer to as the truncated CopL (CopL(T)), into pEPSA5 (Fig. 5B). CopL(T) did not complement the Cu sensitivity phenotype of the ΔcopL strain, whereas the full-length CopL did (Fig. 5B). Cellular fractionation experiments using the ΔcopL strain containing pEPSA5_copL(T)-FLAG revealed that the CopL(T) accumulated in the soluble cytosolic fractions but not in the membrane fraction (data not shown). These results suggested that CopL does not function to protect against Cu toxicity unless localized to the membrane.
Nuc2 is a surface-exposed nuclease anchored to the membrane via its signal peptide (46). We created constructs encoding for chimeric proteins in which CopL(T) was fused to either Nuc2 (pEPSA5_nuc2-copL) or the described Nuc2 signal sequence (pEPSA5_nuc2(SS)-copL) (Fig. 5C). Both chimeric constructs genetically complemented the Cu sensitivity phenotype of the ΔcopL strain (Fig. 5C). Altogether, the data in Fig. 5 suggested that CopL is membrane-associated and surface-exposed and requires membrane association to protect against Cu toxicity.
S. aureus CopL (saCopL) binds copper in vitro
We tested the hypothesis that saCopL is a Cu-binding protein. Recombinant soluble saCopL(T) was purified from Escherichia coli (Fig. S7), and Cu1+ binding was examined using ultraviolet (UV)-visible absorption spectroscopy. Titrating Cu1+ into apo-saCopL(T) resulted in gradual increases of the absorbance in the UV region (A243 nm). The change of A243 was plotted as a function of the Cu1+/saCopL(T) ratio (Fig. 6A). The formation of Cu1+–saCopL(T) was linear and reached saturation after the addition of ∼4 molar eq of Cu1+.
Figure 6.
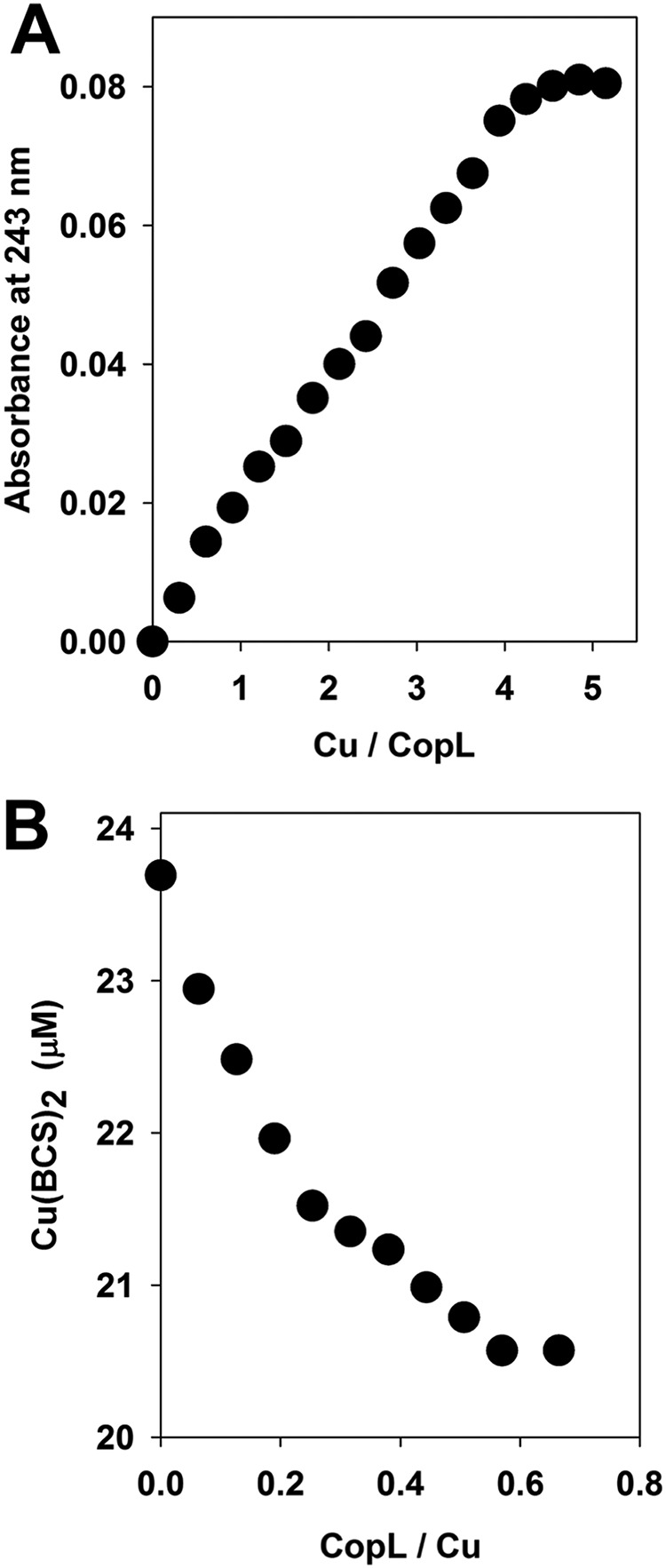
S. aureus CopL (saCopL) binds Cu1+in vitro. A, saCopL binds ∼4 molar eq of Cu1+. Apo-CopL (12.5 μm) was anaerobically titrated with Cu1+, and binding was monitored by measuring absorbance at 243 nm using UV absorption spectroscopy. B, affinity of saCopL for Cu1+ was monitored by BCS displacement. Solutions containing 0.5 mm BCS and 20 μm Cu1+ were titrated with apo-saCopL(T), and BCS displacement was monitored by measuring absorbance at 483 nm using visible absorption spectroscopy.
Competition experiments were conducted to determine the Cu1+-binding affinity of saCopL(T). Bathocuprione disulfonate (BCS) forms a complex with Cu1+ in a 2:1 ratio Cu1+/(BCS)2 that has an absorption maximum at A483 nm (47). To determine the effective Cu1+-binding affinity of saCopL(T), Cu1+ and BCS were combined anaerobically. A fixed concentration of saCopL(T) was then titrated into the sample, and absorption spectra were recorded. The formation of Cu1+-saCopL(T) was determined by monitoring the decrease in absorbance (A483 nm) after each addition (Fig. 6B).
At each of the saCopL(T) concentrations examined, Equation 6 of Xiao et al. (48) enabled calculation of the Kd for the protein–copper complex. The calculation yields the product of Kd and β2, where β2 is the association constant between copper and BCS. We used β2 = 1019.8 (47), and the resulting Kd value was 4.98 ± 0.20 × 10−18 m.
Structure of B. subtilis CopL (bsCopL) reveals a novel Cu-binding motif
B. subtilis ydhK (bscopL) encodes a homolog of S. aureus CopL, with sequence identity of ∼63% in the C-terminal region (Fig. S8). A B. subtilis ΔcopL mutant was more sensitive to growth in the presence of Cu than the parent strain (Fig. S9). As the unpublished 3D solution NMR structure of this homologous protein was already available from our National Institutes of Health Protein Structure Initiative (49) structural bioinformatics efforts, we used these NMR assignments and structure to validate that CopL does in fact bind Cu and predict Cu-binding ligands.
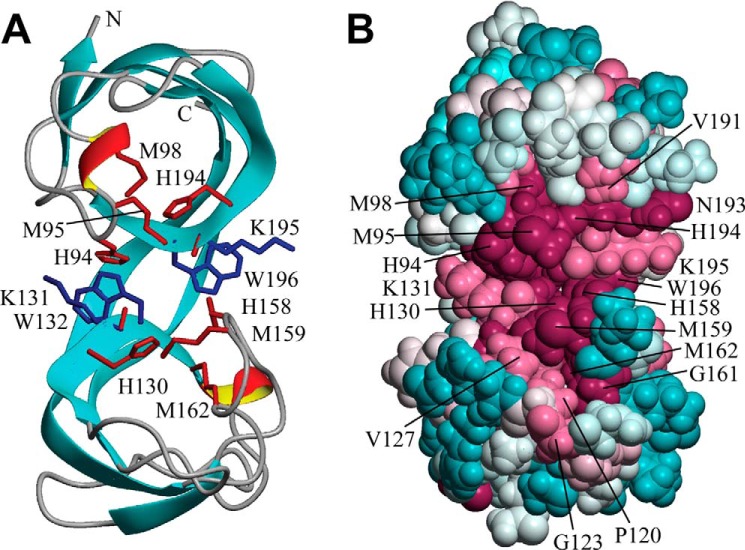
The solution NMR structure of the bsCopL fragment comprising residues 83–205, truncated to remove the signal peptide and lipobox motif, forms a well-ordered structure in solution (Fig. S10), consisting of a pair of distorted β-barrels joined by two shared β-strands, and includes two short turns with a 310-helix geometry (Figs. 7A and Fig. S10). The β-strands (A–H) consist of residues 87–90, 102–118, 127–134, 135–137, 141–153, 166–184, 189–198, and 202–204, whereas the 310-helical turns consist of residues 96–98 and 135–137. The two homologous DUF1541 domains are found to form a single structural unit, as each β-barrel is composed of polypeptide segments from both DUF1541 domains. An unusual structural feature of bsCopL is the cis conformation of the peptide bonds between two conserved residue pairs, Lys-131–Trp-132 and Lys-195–Trp-196 (Fig. 7A), which results in a stacked orientation of their side chains. The strong sequential Hα–Hα NOE peaks supporting the cis conformation of these two nonproline peptide bonds are presented in Fig. S11.
Figure 7.
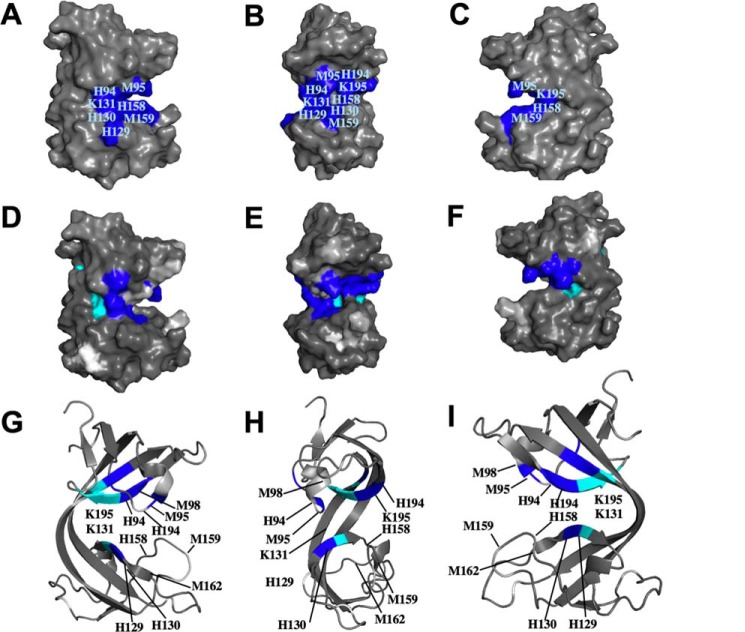
Solution NMR structure of B. subtilis CopL. A, ribbon diagram of the lowest-energy conformer of B. subtilis CopL: backbone of β-strands shown in cyan, short 310-helical segments in red/yellow, and other polypeptide segments in gray; the terminal Lys-83 and Lys-205 are labeled as “N” and “C.” Side chains of Lys-131–Trp-132 and Lys-195–Trp-196 residue pairs connected by unusual cis-peptide bonds are shown in blue. Moieties presumed to be involved in coordinating Cu, side chains of His-94, Met-95, Met-98, His-130, His-158, Met-159, Met-162, and His-194, as well as carbonyls of Lys-131 and Lys-195, are shown in red. B, space-filling representation of the same conformer showing the degree of residue conservation calculated with ConSurf. Conservation scores are arranged into nine groups with the corresponding colors ranging from cyan (most variable) to burgundy (most conserved). The numbering shown here is that of the UniProt entry BSU05790.
Residue conservation scores mapped onto the bsCopL structure indicate that the most conserved surface region, at the interface of the two β-barrels (Fig. 7B and Figs. S12 and S13), represents a likely binding site for copper atoms. Copper coordination in proteins is usually mediated by nitrogen atoms of histidine rings and sulfur atoms of cysteines and methionines; in less common cases it involves side-chain oxygen atoms of aspartic and glutamic acids, glutamine, or tyrosine, backbone carbonyl oxygens, and even indole rings of tryptophan in the form of π–cation interactions (50, 51). A search for conserved residues of this type identified residues His-94, Met-95, Met-98, His-129, His-130, His-158, Met-159, Met-162, and His-194 (Fig. 7A) as the most likely candidates. This is also consistent with the neutral ϵ1-protonated tautomer states of His-94, His-130, His-158, and His-194 side chains (Fig. S14). Additional potential contributions to Cu-binding may come from the backbone carbonyl oxygen atoms of Lys-131 and Lys-195, which are surface-exposed, and do not form intramolecular hydrogen bonds. These conserved residues do not appear to match any known type of protein copper center (50, 51).
A search of PDB90 (nonredundant subset of the PDB database with proteins sharing less than 90% sequence identity) for similar protein structures using DALI (52) produced 27 significant protein chain hits (Z-score ≥5). However, 25 of those the aligned segments were only between 45 and 69 residues long, with sequence identities to bsCopL between 10 and 23% and Cα atom r.m.s.d. between 1.3 and 3.8 Å. Here, the matching segments corresponded to only a single β-barrel of bsCopL. For the remaining two proteins, PDB codes 2QQR and 2MAM, the aligned segments were 82 and 91 residues long, respectively. Both are tandem tudor domain proteins of human origin, the former a histone demethylase and the latter a DNA-binding protein. However, the sequence identity to bsCopL was only 13 and 19%, respectively. Global Cα atom r.m.s.d. was quite high at 5.0 and 5.3 Å, due to a significantly different relative orientation of the β-barrels. None of the structurally similar proteins contained the conserved histidine and methionine residues of bsCopL. Accordingly, we conclude that bsCopL features a novel Cu-binding protein architecture.
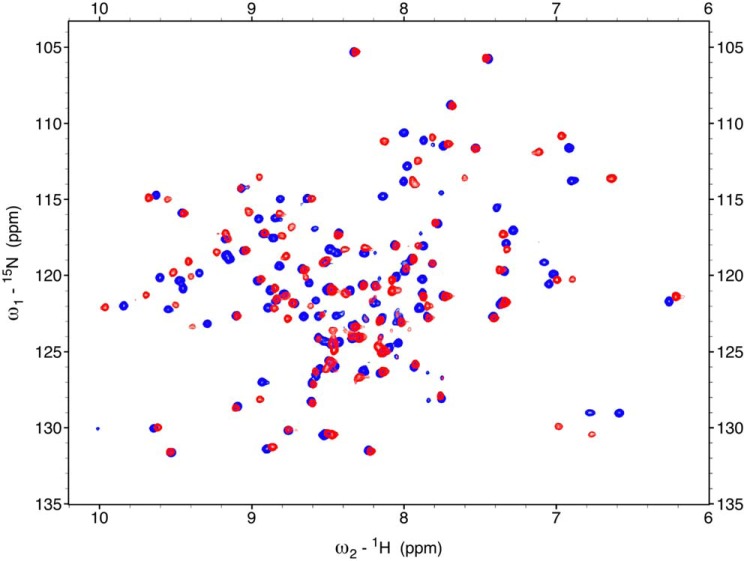
Backbone 1H, 15N chemical shift perturbation study of bsCopL interaction with Cu1+
For Cu1+-binding studies, the His6 purification tag was removed by TEV protease cleavage. The resulting protein includes three extra residues (SHM) at the N-terminal end of the bsCopL sequence. The 2D 15N,1H heteronuclear single-quantum correlation (HSQC) spectrum of bsCopL (residues 83–205) is shown in Fig. S15, with backbone and side-chain resonance assignments (deposited in the Biological Magnetic Resonance Bank (BMRB ID 16942)). Similar spectra were obtained upon addition of 1 mm EDTA (data not shown), demonstrating that under the conditions used for these NMR studies, no Cu or other related residual metal ions are associated with apo-bsCopL protein samples. The 15N,1H HSQC spectra of the apo-bsCopL with the C-terminal hexa-His tag and apo-bsCopL prepared with no hexa-His tag are essentially identical, aside from the small spectral differences due to the N- and C-terminal regions (Fig. S16).
To confirm the ability of bsCopL to bind copper in vitro, and to identify the Cu1+ binding sites, 8 mol/mol eq of Cu1+ were added to the apo-bsCopL protein, and the buffer was exchanged to remove adventitiously-associated Cu1+ ions. Sequence-specific backbone NMR resonance assignments were then determined using standard triple-resonance NMR experiments (Table S3), and deposited in the Biological Magnetic Resonance Bank (BMRB ID 27741).
Comparison of the overlay of the assigned 2D 15N,1H HSQC spectra of apo-bsCopL and holo-bsCopL (Fig. 8) revealed that Cu1+ addition results in specific chemical shift perturbations. The numerous spectral changes induced upon copper addition unambiguously demonstrate Cu1+ association with bsCopL. A plot of chemical shift perturbations along the protein sequence is shown in Fig. S17. When mapped onto the 3D structure of B. subtilis apo-bsCopL (Fig. 9), these data demonstrate that many residues with significant chemical shift perturbations are located in the conserved surface region at the interface of the two β-barrels, which, as outlined above, we predict based on amino acid composition and conservation to include copper-binding sites.
Figure 8.
Chemical shift perturbations of B. subtilis CopL due to Cu1+ binding. Overlay of 600 MHz 15N,1H HSQC spectra of 0.3 mm apo-bsCopL-N (blue) and 0.3 mm holo-bsCopL-N with 6 mol/mol eq of Cu1+ (red) is shown. The sequence-specific 15N,1H HSQC resonance assignments for apo-bsCopL-N are shown in Fig. S15.
Figure 9.
Cu1+ binding to B. subtilis CopL. A–C, surface molecular representation of B. subtilis CopL, in three orientations, showing the locations of conserved residues His-94, Met-95, Met-98 (buried), His-129, His-130, Lys-131, His-158, Met-159, Met-162 (buried), His-194, and Lys-195, presumed to be involved in coordinating Cu1+. Only residues that can be seen on the surface of the 3D structure are labeled. D–F, surface representation of B. subtilis CopL, in same three orientations, showing chemical shift perturbations due to Cu1+ binding. Color code: dark blue (ΔδNH >0.25 ppm, viz. Thr-92, Met-95, His-130, His-131, Met-179, Val-180, Asn-193, His-194, Lys-195); cyan, residues with HN resonances that are broadened due to Cu1+ binding (viz. Trp-132, Tyr-178, Trp-196, Val-197, and Thr-198); light gray, residues with HN resonances that are absent in the 15N,1H HSQC spectrum of apo-CopL and not assigned (viz. Lys-83, His-94, Lys-96, Gly-97, Lys-160, Gly-161, and Ser-187); and white for proline residues that lack backbone amide protons (viz. Pro-120 and Pro-148). G–I, ribbon representation of B. subtilis CopL, in same three orientations, showing conformational perturbations due to Cu1+ binding using the same color code as in D–F. G–I, only the residues with HN resonances that are affected by Cu1+ binding are labeled. The numbering shown here is that of the UniProt entry BSU05790.
To further confirm reversible copper association, the Cu1+ chelator BCS was added to the holo-bsCopL protein sample followed by recording a 15N,1H HSQC spectrum. As expected, the spectrum of the BCS-treated sample almost fully returned to match that of the apo-bsCopL (Fig. S18).
A CopL variant with decreased Cu binding does not protect against Cu toxicity in vivo
We tested the hypothesis that Cu binding by CopL is necessary to protect from Cu toxicity. We created a construct that encoded for the S. aureus copL mutant allele (called copL*) that encoded for the following directed changes to CopL: H70A, M71A, H134A, and M135A (His-94, Met-95, His-158, and Met-159 in bsCopL) (Fig. 10A). These are strictly conserved residues that NMR studies suggested may function in Cu1+ ligation. The copL* allele was unable to correct the Cu-dependent growth defects of the S. aureus ΔcopL mutant (Fig. 10A).
Figure 10.
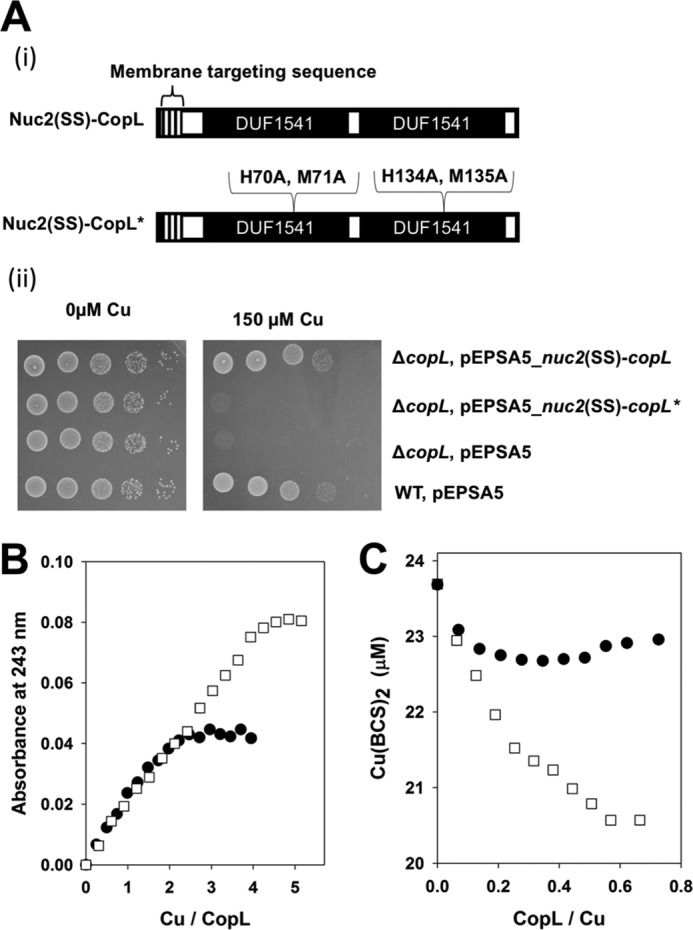
Effective Cu binding is necessary for CopL to protect against Cu toxicity. A, copL* allele does not correct the Cu sensitivity phenotype of the S. aureus ΔcopL mutant. Panel i, schematic of the copL and copL* constructs utilized. Panel ii, WT and ΔcopL strains harboring pEPSA5, pEPSA5_nuc2(SS)-copL, or pEPSA5_nuc2(SS)-copL* were serially diluted and spot-plated on chemically defined media without and with 100 μm Cu. B, saCopL(T)* binds less Cu1+ than saCopL(T). 12.5 μm apo-saCopL(T) or apo-saCopL(T)* were anaerobically titrated with Cu1+. Binding was monitored by measuring absorbance at 243 nm using UV absorption spectroscopy. C, saCopL(T)* has a lower affinity for Cu1+ than saCopL(T). Solutions containing 0.5 mm BCS and 20 μm Cu1+ were titrated with either apo-saCopL(T) or apo-saCopL(T)*. Copper binding to CopL was examined using BCS displacement by monitoring sample absorbance at 483 nm, which is the absorption maxima for the Cu1+–BCS complex. The data for the saCopL(T) are also presented in Fig. 6 and included here for comparison.
We purified saCopL(T)* (viz. H70A, M71A, H134A, M135A-CopL(T)) and examined the near UV CD spectra of the apo-saCopL(T) and apo-saCopL(T)* proteins. The spectra were nearly identical indicating little difference in secondary structure between the variants (Fig. S19). We next titrated with Cu1+ into saCopL(T)* and monitored spectral changes using UV absorption spectroscopy. Cu1+ titration resulted in a linear increase in absorbance at 243 nm, and the absorbance plateaued after approximately 2 eq of Cu1+ per saCopL(T)* (Fig. 10B). These data indicate that the saCopL(T)* variant binds less Cu1+ than saCopL(T). We next titrated saCopL(T)* into a solution of BCS and Cu1+ and monitored absorbance at 483 nm. The addition of saCopL(T)* decreased absorbance at 483 nm but not to the extent of that of saCopL(T) (Fig. 10C). saCopL(T)* binds Cu1+ with a Kd of 1.35 ± 0.17 × 10−17 m, which is ∼3-fold lower than the affinity of saCopL(T) for Cu1+.
Discussion
This study was initiated to further investigate the mechanisms of copper (Cu) homeostasis in the epidemic CA-MRSA S. aureus USA300 clone. The work presented has re-affirmed the roles of CopA and CsoR in Cu efflux and intracellular Cu sensing, respectively. We have also assigned roles for copB and copL in Cu homeostasis. These data, as well as published work on CopA (31), CopZ (32), and CsoR (53), resulted in a working model for Cu homeostasis in the S. aureus USA300_LAC, which is illustrated in Fig. 11. Upon sensing Cu in the cytosol, CsoR derepresses transcription of the copAZ and copBL operons. The binding of apo-CsoR to the copB promoter was recently confirmed by Purves et al. (54). CopZ binds Cu and acts as an intracellular Cu buffer delivering it to CopA. The CopA and CopB proteins function to efflux Cu from the cytosol. CopL is a membrane-associated, surface–exposed protein that binds Cu on the outside of the cell preventing it from entering the cytosol and binds Cu after efflux by CopA or CopB.
Figure 11.
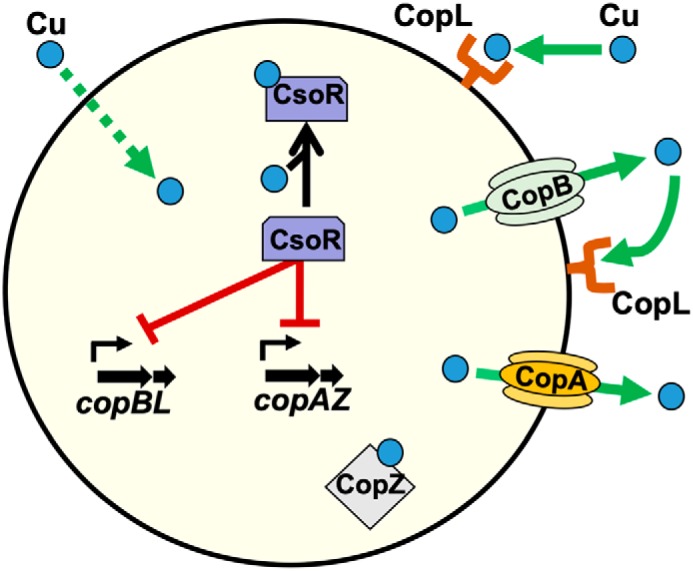
Working model for copper homeostasis in S. aureus. The CsoR transcriptional regulator binds intracellular copper, leading to derepression of the copAZ and copBL operons. CopA and CopB are transmembrane copper exporters. CopZ is a chaperon protein that binds copper and acts as a cytosolic buffer. CopL is a membrane-bound, surface-exposed, and copper-binding lipoprotein. CopL likely functions in preventing copper uptake by binding extracellular Cu to prevent it from entering or re-entering the cell after export by CopB or CopA.
CopL tightly binds Cu1+ with a dissociation constant of 4.98 ± 0.20 × 10−18 m. For comparison, the reported Cu1+-binding affinities of the S. aureus (30), B. subtilis (55), and Mycobacterium tuberculosis (53) CsoR proteins, HAH1 from Homo sapiens (56), and N-terminal metal–binding domain of CopA (57) and CopZ from B. subtilis (58) are of the same order of magnitude as CopL. The CusCFBA system protects the E. coli cytosol and periplasm from Cu (59). The metallochaperone of this system (CusF) has a Cu1+ Kd of 5.0 ± 1.2 × 10−15 m (60).
It should be emphasized that small differences in binding affinity among the individual CopL-binding sites could exist, but this could not be confirmed by the present data. The Kd values reported here should therefore be considered weighted averages of the sites. It is clear nevertheless that the binding is tight and comparable with that of other copper-binding proteins. It is also clear (Fig. 10C) that the CopL* variant binds copper with less affinity than the WT CopL.
One aim of the work presented was to validate that CopL, from both S. aureus or B. subtilis, binds Cu1+. This was achieved using Cu1+-binding data on S. aureus CopL and NMR studies on the B. subtilis CopL. Although there may in fact be structural differences in the Cu1+-binding networks of S. aureus and B. subtilis CopL, the basic residues in the Cu1+-binding site of B. subtilis CopL form a highly conserved network that is common to other homologs of this two-domain protein family, including S. aureus CopL.
The solution NMR structure of bsCopL revealed a novel copper-binding architecture. The overall structure of the Cu1+-bound form is similar to that of the apo-form because most distant residues do not exhibit significant perturbations of chemical shifts upon Cu1+ binding. To preserve these NMR data for more comprehensive future studies, we have deposited these chemical shift data in the BioMagRes Database (BMRB ID 27741).
The 3D structure of bsCopL(T) contains unusual cis peptide bonds between two conserved residue pairs, Lys-131–Trp-132 and Lys-195–Trp-196. Nonproline cis peptide bonds are rare and occur only in about 0.03% of all peptide bonds in known protein structures (61). Their occurrence is significantly more frequent in X-ray crystal structures determined at high resolution than in structures determined at medium and low resolution, suggesting that these bonds may be more abundant than generally recognized (61). The cis-trans isomerization of peptide bonds before proline residues can play an important kinetic role in controlling protein folding (62, 63). This isomerization can be catalyzed by enzymes known as prolyl-cis/trans isomerases that have been shown to act on Xaa–Pro peptide bonds, but not on nonproline peptide bonds (64). Nonproline cis peptide bonds tend to be buried and not greatly accessible compared with Xaa–Pro cis peptide bonds, which suggests that they may form early during the folding process (65). They tend to be observed near active sites indicating that they may sometimes be important for protein activity (61). The majority of the cis peptide bonds occur in regular secondary structures, more specifically in β-strands (65). In the case of bsCopL, the two non-Pro cis peptide bonds are located in β-strands β3 and β6, adjacent to the proposed Cu-binding residues His-130 and His-194, respectively. These cis peptide bond conformations result in unique local side-chain structure, with stacked orientation of their side chains. Although there may be functional consequences of such non-Pro cis peptide bonds, such structure–function studies are difficult to carry out because mutations that prevent cis peptide bonds also tend to have structurally-disruptive effects.
The CA-MRSA epidemic is widely attributed to the spread of the USA300 clone (17, 18). The majority of the genetic differences between USA300 and other staphylococcal strains of clinical importance is the presence of mobile genetic elements, including ACME (19). Expression of the ACME-encoded arginine-deiminase system (arc) improves survival in acidic conditions (20), and the ACME-encoded speG gene provides resistance to high levels of host-derived polyamines and survival within murine abscesses (20, 21, 66).
While this manuscript was being revised, Purves et al. (54) reported that S. aureus strains lacking copB (called copX in their study) or copL are sensitive to growth in the presence of Cu. Interestingly, they were not able to genetically complement the copL mutant and were only able to complement in the copB mutant in one of the two media utilized. They found that the copB or copL mutants, but not the copA mutant, had slightly increased cell-associated Cu. The authors used these data to suggest a role for CopBL in Cu efflux. Similar to the findings reported herein, they found that CsoR controls transcription of the copAZ and copBL operons. They also reported that apo-CsoR binds to the copB promoter suggesting that it directly controls transcription of copBL. Importantly, Purves et al. (54) showed that strains lacking CopB or CopL, but not a strain lacking CopA, had decreased survival in murine macrophages. These data highlight a potentially important roles of CopB and CopL in pathogenesis.
The human skin commensal Staphylococcus epidermidis contains the copB, mco, and copL genes in an apparent operon, as well as two additional Cu efflux proteins (67). Phylogenetic analyses suggest that the genes comprising ACME were assembled into a single genetic locus in S. epidermidis before they were transferred to S. aureus, which occurred prior to the epidemic expansion of the USA300 clone (66). Our bioinformatics analysis (Table S1) revealed that Staphylococcus xylosus, Staphylococcus capitis, and Staphylococcus haemolyticus, which are skin commensals, also have copB and copL. The acquisition of genetic elements from species that share the same niche is a strategy employed by S. aureus to adapt to new environments (68, 69). It is tempting to speculate that additional Cu detoxification mechanisms provided by the ACME-encoded copBL operon further promote survival on skin; however, this speculation awaits further examination.
A USA300 Latin American variant (USA300-LV) has become one of the most prevalent clones associated with MRSA infections in community settings in South America. Most of the genomic differences between USA300-LV and USA300 can be attributed to mobile genetic elements and, specifically, to the absence of ACME in USA300-LV (70). Despite this difference, the USA300-LV genome contains copBL. Similar to the location of copBL in USA300, copBL in USA300-LV are located adjacent to SCCmec on a mobile genetic element, which has been designated as the copper and mercury resistance mobile element (COMER) (70).
Mobile genetic elements containing copper detoxification islands have been discovered in other organisms (71). Copper-resistance mechanisms acquired via transposable elements include copper transporters, multicopper oxidases, or as reported in this study, membrane-bound lipoproteins. These studies provide additional evidence that copper exposure (immune system, healthcare settings, and/or diet) may not only promote the dissemination of genetic elements that result in the development of metal-resistant microorganisms but also strains that are hypervirulent and resistant to antimicrobials.
In summary, the work presented here describes an additional strategy by which S. aureus protects against copper toxicity. The ACME-encoded copBL may contribute to the high success of USA300, by providing protection from Cu-dependent killing. Moreover, the fact that copBL is encoded on mobile DNA alongside virulence factors suggests that the maintenance of Cu homeostasis could be a selective pressure for mobilization of alternative virulence factors or antibiotic resistance genes.
Experimental procedures
Reagents
Restriction enzymes, quick DNA ligase kit, deoxynucleoside triphosphates, and Phusion DNA polymerase were purchased from New England Biolabs. Primers were obtained from Integrated DNA Technologies and listed in Table S2. Plasmid mini-prep and gel extraction kits were purchased from Qiagen. Lysostaphin was purchased from Ambi Products. Tryptic Soy Broth (TSB) was purchased from MP Biomedicals. The ELC chemiluminescent detection kit was purchased from Pierce. Pierce protease and phosphatase inhibitor mini tablets were purchased from ThermoFisher Scientific. GSTrap 4B columns and PreScission Protease were purchased from GE Healthcare. Unless specified, all other chemicals were purchased from Sigma and were of the highest purity available.
Bacterial strains and growth conditions
Bacterial strains used in this work are listed in Table 1. Unless otherwise noted, the S. aureus strains used in this study are derived from the community-associated MRSA USA300 LAC strain that was cured of the pUSA03 plasmid, which confers erythromycin resistance (72). S. aureus strains were cultured in TSB or a defined medium, and E. coli strains were grown in Luria Broth (LB). Unless otherwise specified, all bacterial strains were cultured at 37 °C. The chemically defined medium was previously described (73) and contained the following: 1 g of (NH4)2SO4, 4.5 g of KH2PO4, 10.5 g of K2HPO4, 110 mm NaCl, 30 mm KCl, 50 μg of nicotinic acid, 50 μg of pantothenic acid, 50 μg of thiamine, 0.3 μg of biotin, and 2.5 mg of each of the 20 amino acids per 100 ml. When supplemented to the media, chemicals were added at the following concentrations: 10–300 μm CuSO4; 50–200 μm Fe2(NH4)2(SO4)2; 5–200 μm CoCl; 10–300 μm MnSO4; 10–300 μm ZnSO4. When appropriate, antibiotics were added at the following concentrations: 150 μg ml−1 ampicillin (Amp), 6 or 30 μg ml−1 chloramphenicol (Cm) (defined or complex media, respectively), 10 μg ml−1 erythromycin, 3 μg ml−1 tetracycline (Tet), and 150 ng ml−1 anhydrotetracycline. Overnight cultures were grown in 7-ml culture tubes containing 2 ml of TSB shaken at 200 rpm. When growing overnight cultures of strains containing pEPSA-derived plasmids, 2% xylose (w/v) was added to the media.
Table 1.
Construction of mutant strains and plasmids
Chromosomal DNA from JMB1100 was used as the template for PCRs used in the construction of plasmids. All plasmids were isolated from E. coli DH5α and transformed into electrocompetent S. aureus RN4220 using a standard protocol (74). Phage α80 was used for plasmid and chromosomal transductions (75). All bacterial strains and DNA constructs were verified by PCR, genetic complementation, or DNA sequencing prior to use. DNA sequencing was conducted by Genewiz (South Plainfield, NJ).
Mutational inactivation of the S. aureus copB and copL genes was achieved by chromosomal deletion to yield the ΔcopB, ΔcopL, and ΔcopBL mutant strains as described previously (76). For the ΔcopB mutant, upstream and downstream regions of the copB gene (SAUSA300_0078) were PCR-amplified using the following primers: ZRC199 and ZRC200; ZRC164 and ZRC201. PCR products were gel-purified and fused by PCR using the ZRC199 and ZRC201 primers. For the ΔcopL mutant, upstream and downstream regions of copL (SAUSA300_0079) were PCR-amplified using the following primers: ZRC166 and ZRC167; ZRC168 and ZRC169. PCR products were gel-purified and fused by PCR using the ZRC166 and ZRC169 primers. For the ΔcopBL double mutant, upstream and downstream regions of the copBL operon were PCR-amplified using the following primers: ZRC185 and ZRC186; ZRC168 and ZRC169. PCR products were gel-purified and fused by PCR using the ZRC185 and ZRC169 primers. The ΔcopB, ΔcopL, and ΔcopBL PCR products were digested with EcoRI and SalI and ligated into similarly digested pJB38 (77). The recombinant vectors were transformed into chemically competent E. coli DH5α. PCR was used to screen for E. coli colonies harboring the recombinant plasmids using ZRC196 and ZRC201 primers for pJB38_ΔcopB, ZRC196 and ZRC169 for pJB38_ΔcopL, and ZRC196 and ZRC169 for pJB38_ΔcopBcopL. The plasmids were isolated and mobilized into RN4220 and subsequently JMB1100. Colonies that were Cm-sensitive were screened using PCR for the double-recombination event. The ΔcopB mutant strain was verified using the ZRC139 and ZRC201 primers. The ΔcopL and ΔcopBL mutants were verified using the ZRC139 and ZRC169 primers.
The pTET plasmid was used to construct the copA::Tn(tet) and csoR::Tn(tet) in the USA300 LAC background by allelic exchange as described previously (78). Mutational inactivation was confirmed using PCR with the following primers: copA::Tn(ermB), copA::Tn(tet), and copZ::Tn(ermB) with ZRC133 and ZRC134; csoR::Tn(ermB) and csoR::Tn(tet) with ZRC153 and ZRC155.
For complementation and expression studies, genes were cloned into pEPSA5 (79). The ZRC146 and ZRC141 primers were used to PCR amplify the copB gene. The ZRC149 and ZRC150 primers were used to PCR amplify the full-length copL. The ZRC184 and ZRC150 primers were used to PCR amplify the truncated copL. PCR products were digested with BamHI and SalI and ligated into similarly digested pEPSA5 to yield pEPSA5_copB, pEPSA5_copL, and pEPSA_copL(T) vectors. Plasmid-containing strains were PCR-verified, using the pEPSA5upveri and ZRC141 (pEPSA_copB) or the pEPSA5upveri and ZRC150 ((pEPSA5_copL or pEPSA_copL(T)).
The pEPSA5_copL–FLAG and pEPSA5_copL(T)-FLAG vectors were constructed by using the ZRC149–ZRC181 and ZRC ZRC184–ZRC181, respectively. The inserts were digested with BamHI and NheI and then ligated into similarly digested pEPSA5_CitB-FLAG (80). Strains containing the pEPSA5_copL–FLAG plasmid were PCR-verified using the pEPSA5upveri and ZRC181 primers. The pGEX-6P-1_copL was constructed using the ZRC198 and ZRC178 primers. The PCR product was digested with BamHI and XhoI and then ligated into similarly digested pGEX-6P-1(GE Healthcare). The pGEX-6P-1_copL* was created using the pEPSA5_nuc2(SS)-copL* as a PCR template for copL*. The copB transcriptional reporter was created by amplifying the copB promoter using the ZRC139 and ZRC140 primers. The PCR product was digested with HindIII and KpnI and ligated into similarly digested pCM11 (81).
The pEPSA5_nuc2-copL and pEPSA5_nuc2(SS)-copL vectors were created by using yeast homologous recombination cloning in Saccharomyces cerevisiae FY2 as described previously (82, 83). The pEPSA5_CitB-FLAG vector was linearized with NheI. The amplicon for the pEPSA5_nuc2(FL)-copL was created using the following primer pairs: ZRC188 and ZRC189; ZRC191 and ZRC193. The amplicon for the pEPSA5_nuc2(SS)-copL was created using the following primer pairs: ZRC188 and ZRC190; ZRC192 and ZRC193. Strains containing the pEPSA5_nuc2-copL and pEPSA5_nuc2(SS)-copL plasmids were PCR-verified using the ZRC188 and ZRC193 primers. The pEPSA5_nuc2(SS)-copL* was created using YCC cloning as outlined above using the additional following primers: CblSDM1for, CblSDM1rev, CblSDM2for, and CblSDM2rev.
qRT-PCR
RNA isolation and quantitative real-time PCR were performed as described previously with a few modifications (84). The WT (JMB1100) strain was cultured overnight in TSB in biological triplicates. Cells were pelleted by centrifugation and resuspended in PBS before diluting 1:100 into chemically defined media without and with 100 μm Cu. Cells were harvested 6 h post-inoculation (OD ∼1, A600 nm) by centrifugation, treated with RNAProtect (Qiagen) for 10 min at room temperature, and stored at −80 °C. Pellets were thawed and washed twice with 0.5 ml of lysis buffer (50 mm RNase-free Tris, pH 8). Cells were lysed with 20 μg of DNase and 20 μg of lysostaphin for 30 min at 37 °C. RNAs were isolated using TRIzol reagent (Ambion) as per the manufacturer's protocol. DNA was digested with the TURBO DNA-free kit (Ambion, Life Technologies, Inc.) and RNA-quantified using a Nanodrop (ND-1000) spectrophotometer. cDNA libraries were constructed using isolated RNA as a template with a high capacity RNA-to-cDNA kit (Applied Biosystems). Power SYBR Green PCR Master Mix (Applied Biosystems) was used to perform qRT-PCR in an Applied Biosystems StepOnePlus thermocycler. Data were analyzed using the ΔΔCt method. CopARTfwd and CopARTrev primers were used to detect copA transcripts; CopBRTfwd and CopBRTrev primers were used to detect copB transcripts; CblRTfwd and CblRTrev primers were used to detect copL transcripts. Transcripts corresponding to the 16S rRNA gene were detected with 16sfwdRT and 16srevRT primers. RT primers were designed using the Primer Express 3.0 software from Applied Biosystems.
Transcriptional reporter assays
Strains containing the pCM11-derived reporters were grown overnight in TSB-Erm. The overnight cultures (>16 h) were pelleted and resuspended in PBS. Washed cells were subcultured into 5 ml of fresh chemically defined media (1:100) in 30-ml culture tubes with and without copper. Culture aliquots were periodically removed (200 μl), and culture optical density (A590 nm), and fluorescence was monitored using a PerkinElmer Life Sciences HTS 7000 Plus Bio Assay Reader. GFP was excited at 485 nm, and emission was read at 535 nm. Relative fluorescence units were normalized with respect to the culture optical density at each time point.
Cell fractionation
Overnight cultures were diluted to 0.1 OD (A600 nm) in fresh TSB with Cm. Cultures were induced with 0 or 0.2% xylose at 1 OD (A600 nm), incubated for an additional 2 h, and harvested by centrifugation. Cultures were resuspended and washed with PBS. Cells were lysed (PBS with 10 μg of DNase, 10 μg of lysostaphin, protease, and phosphatase inhibitor mini tablets) at 37 °C for ∼45 min. The cell fractionation protocol was followed as described previously with modifications (85). Cell lysates were spun for 10 min at (12,000 × g, 4 °C) to remove unbroken cells and debris. Supernatants (whole cell, crude lysates) were spun at 100,000 × g for 2 h at 4 °C in Beckman polyallomer centrifuge tubes using a Beckman Optima TLX ultracentrifuge and TLA 120.2 rotor. The resulting supernatant was saved as the cytoplasmic fraction. The pellet (crude membrane fraction) was resuspended in membrane buffer (100 mm Tris-HCl, pH 7.5, 100 mm NaCl, 10 mm MgCl2, 10% glycerol, 0.1% SDS) to solubilize membrane proteins and spun at 100,000 × g for 2 h at 4 °C to remove the detergent-insoluble material. Supernatants were saved as the membrane-soluble fractions.
Western blot analysis
A total of 40 μg of total protein per sample were separated using SDS-polyacrylamide gel. Proteins were transferred to a polyvinylidene difluoride membrane and incubated with mouse monoclonal anti-FLAG primary antibody (Sigma) (1:4000 dilution) and subsequently horseradish peroxidase–conjugated secondary antibody (Bio-Rad) (1:12,000 dilution). The blots were developed using chemiluminescent detection (Pierce) and scanned as TIFF images.
Protein concentration determination
Protein concentration was determined using a bicinchoninic acid assay modified for a 96-well plate (86) with BSA as a protein standard. Apo-saCopL(T) concentrations were also estimated spectrophotometrically using ϵ280 = 19,940 m−1 cm−1. Apo-bsCopL(T) concentrations were estimated spectrophotometrically using ϵ280 = 18,450 m−1 cm−1.
Recombinant saCopL(T) expression and purification
E. coli BL21 DE3 containing pGEX-6P-1_copL or pGEX-6P-1_copL* was cultured overnight in LB-Amp and used to inoculate 1 liter of 2× YT media (16 g of tryptone, 10 g of yeast extract, and 5 g of NaCl, pH 7.0) to 0.1 OD (A600 nm). Cultures were grown shaking at 30 °C, induced with 1 mm isopropyl 1-thio-β-d-galactopyranoside at 0.8 OD (A600 nm), and incubated for additional 4 h. Cultures were harvested by centrifugation, resuspended in cold PBS, and stored at −80 °C. For lysis, thawed cell pastes were passed through a French press three times, and cell lysates were clarified by centrifugation (15,000 × g for 30 min at 4 °C). Cell extracts were loaded onto GSTrap 4B columns pre-equilibrated with binding buffer (PBS, pH 7.4) and then washed with 30 column volumes of binding buffer. The column was washed with PreScission cleavage buffer (50 mm Tris-HCl, 150 mm NaCl, 1 mm EDTA, 1 mm DTT, pH 7.5) and then incubated overnight at 4 °C with the PreScission Protease before eluting the recombinant protein with PreScission cleavage buffer. Fractions were analyzed for purity by SDS-PAGE. Protein was concentrated using YM-3 Centriplus Centrifugal Concentrators (Millipore). CD spectra were recorded using an AVIV model 62A (Aviv Associates, Lakewood, NJ) spectropolarimeter.
Copper binding and BCS competition assays
All biochemical assays were conducted under strict anaerobic conditions, either in a Coy anaerobic chamber (Grass Lake, MI) or using sealed cuvettes and Hamilton gas-tight syringes. Cu1+ binding was conducted as described previously (55). After purification, apo-saCopL(T) was transferred to the anaerobic chamber and buffer-exchanged (buffer R: 50 mm MOPS, 50 mm NaCl, pH 7.4) using a PD-10 column (GE Healthcare), concentrated, and allowed to incubate overnight to establish anaerobiosis. A 0.5 mm CuCl stock was prepared anaerobically in 10 mm HCl, 1 m NaCl, and 1 mm ascorbic acid. For Cu1+-binding assays, 12.5 μm saCopL(T) and 100 μm ascorbic acid in a total volume of 600 μl buffer R was placed in an anaerobic cuvette. The absorbance of the sample at 243 nm was determined using a Beckman-Coulter DU800 spectrophotometer after each 4-μl addition of 0.5 mm Cu1+.
Competition assays with the Cu1+-specific chelator BCS were conducted as described previously (55). Anaerobic solutions containing 0.5 mm BCS, 20 μm Cu1+, 0.2 mm ascorbic acid, 50 mm NaCl, 50 mm MOPS, pH 7.4, were combined in a 700-μl sealed gas-tight cuvette. A series of additions of 80 μm saCopL(T) in buffer R were added to the cuvette. After each addition, and a subsequent 3-min incubation, the absorbance at 483 nm was recorded using a Beckman-Coulter DU800 spectrophotometer. For dilution correction, experiments were also performed in an identical manner using only buffer R. BCS forms a complex with Cu1+ in a 2:1 ratio Cu1+/(BCS)2, which can be monitored by changes in absorbance at 483 nm with an overall association constant of β2 = 1019.8 (47).
Recombinant expression and purification of bsCopL(T)
B. subtilis bsCopL(T) (NESG Target ID SR518) was cloned, expressed, and purified following the standard NESG protocols (87). Briefly, the construct SR518-83-205-21.8, containing the 83–205-residue fragment with a C-terminal affinity purification tag LEHHHHHH and an N-terminal methionine, was cloned into a pET21 (Novagen) derivative vector. E. coli BL21 (DE3) pMGK cells, a rare codon-enhanced strain, were transformed with SR518-83-205-21.8 and cultured in MJ9 minimal media (92) with [15N]ammonium sulfate and/or [13C]glucose as sole nitrogen and carbon sources. Uniformly 13C,15N-labeled CopL (CopL-NC) was purified using an AKTAxpress (GE Healthcare)-based two-step protocol consisting of IMAC (HisTrap HP) and gel-filtration (HiLoad 26/60 Superdex 75) chromatography. The final yield of purified CopL-NC (>95% homogeneous by SDS-PAGE; 15.4 kDa by MALDI-TOF MS) was about 73 mg/liter. The final sample of CopL-NC for structure determination with NMR spectroscopy was prepared at a concentration of 0.4 mm in 10 mm Tris-HCl buffer solution, pH 7.5, containing 5% D2O, 100 mm NaCl, 5 mm DTT, 0.02% NaN3, and proteinase inhibitors (Roche Applied Science). A uniformly 15N-labeled and 5% biosynthetically-directed fractionally 13C-labeled sample CopL-NC5 was prepared in the same manner using a mixture of 95% natural abundance and 5% [U-13C]glucose as a carbon source yielding the final concentration of 0.7 mm. NMR samples were prepared by exchange via spin columns into 10 mm Tris-HCl buffer at pH 7.5, containing 100 mm NaCl and 5% v/v D2O, and concentrating to about 0.12 mm.
This material was used to produce anisotropic NMR samples, CopL-NC5-PEG and CopL-NC5-Pf1, by adding 4% (w/v) C12E5–PEG/hexanol at 1:1 molar ratio or 12.5 mg/ml Pf1 phage, respectively. For measurement of 15N-1H RDCs, additional anisotropic NMR samples, CopL-NC5-PEG and CopL-NC5-Pf1, were prepared from CopL-NC5 material by adding 4% (w/v) C12E5–PEG/hexanol at 1:1 molar ratio or 12.5 mg/ml Pf1 phage (ASLA Biotech), respectively.
For Cu-binding studies the same gene fragment (residues 83–205) was cloned into a pET15 (Novagen) derivative vector, pET15TEV_NESG (88), yielding the construct SR518-83-205-TEV. The corresponding N-terminal His6–fusion protein was expressed and purified as described above, with uniform 15N-enrichment. The His6 purification tag was then removed by TEV protease cleavage, followed by IMAC (HisTrap HP) purification and gel-filtration chromatography, as described elsewhere (89). The resulting protein includes three extra residues (SHM) at the N-terminal end of the bsCopL sequence.
To prepare the holo-bsCopL-N protein sample, CuCl was prepared anaerobically in 10 mm HCl, 1 m NaCl, and 1 mm ascorbic acid. 2 mol/mol eq of CuCl was added to 1.4 ml of uniformly 15N-enriched apo-bsCopL (0.11 mm), to prevent protein precipitation. The sample was then concentrated to ∼0.1 ml and then diluted to 1.4 ml with 10 mm Tris-HCl buffer at pH 7.5, containing 100 mm NaCl and 1 mm ascorbic acid. This was repeated three times (8 eq of Cu1+). Subsequently, the buffer of the sample was exchanged to remove any excess or adventitiously associated Cu1+. The final sample was adjusted to contain 5% D2O in 500 μl (∼0.25 mm). The BCS-treated sample holo-bsCopL-N-BCS was prepared by adding 1 mm anaerobically prepared BCS to the holo-bsCopL-N protein sample. All the above procedures were carried out at room temperature and under aerobic conditions using sealed NMR tubes (Wilmad Glass).
NMR spectroscopy and structure determination of bsCopL
NMR data acquisition was carried out on Bruker AVANCE 600, 700, 800, and 900 MHz as well as Varian INOVA 600 and 750 MHz spectrometers, all equipped with cryogenic inverse-detected triple-resonance probes. All NMR experiments were carried out at 25 °C and are summarized in Table S3. 1D 15N relaxation HSQC spectra were processed with VNMRJ 4.2A (Agilent Technologies) and integrated over the 1H range 10.0–8.5 ppm. 15N relaxation times t1 = 740 ± 30 ms and t2 = 85 ± 9 ms were extracted by fitting exponential decay curves. Rotational correlation time (τc) was calculated using the “slow molecular tumbling” approximation of the complete relaxation Equation 1 (90),
| (Eq. 1) |
where νN represents the 15N resonance frequency. The resulting τc value of 7 ns indicates that CopL-NC is monomeric in solution. This finding was confirmed by analytical gel filtration (GE Healthcare) followed by static light scattering (Wyatt Technology).
2D and 3D NMR spectra were processed with TopSpin (Bruker BioSpin), NMRPipe (91), or PROSA (92). Visualization and analysis of NMR spectra were performed with the programs CARA (93), XEASY (94), and Sparky (95). Chemical shifts of 1H spins were referenced to internal 4,4-dimethyl-4-silapentane-1-sulfonic acid (DSS), whereas13C and 13N chemical shift were referenced indirectly via their gyromagnetic ratios according to IUPAC recommendations (96). Sequence-specific assignments of backbone (1HN, 15N, 13CO, and 13Cα) and 13Cβ resonances were obtained in a semi-automated manner by analyzing HNCO, HN(CA)CO, CBCA(CO)NH, and HNCACB peak lists with the program PINE (97). Side-chain resonance assignment was performed interactively in CARA using HBHA(CO)NH, (H)CCH-COSY, and (H)CCH-TOCSY, and 15N,13C-resolved 1H,1H-NOESY (mixing time 70 ms) spectra 2D 13C,1H constant-time HSQC of CopL-NC5 was used to obtain stereospecific resonance assignments of valine and leucine methyl groups (98). Protonation and tautomeric states of His side chains were determined from long-range 2D 15N,1H HSQC (99) revealing that His-129 is positively charged, whereas His-94, His-130, His-158, and His-194 are Nϵ2-protonated. Chemical shifts, NOE peak lists, 15N-1H RDCs, and time-domain NOESY data were deposited in the BioMagResBank (100) under accession number 16942.
1H-1H upper distance constraints for structure determination were obtained from 3D 15N,13C-resolved 1H,1H-NOESY. Constraints for ϕ and ψ dihedral angle constraints were derived from backbone chemical shifts using TALOS+ (101). Automated iterative NOE peak assignment and structure calculation was initially performed with AutoStructure 2.2.1 (102) and CYANA 3.0 (103) using dihedral angle constraints and stereospecific assignments of Val and Leu methyl groups. The resulting consensus NOE assignments were verified and corrected by interactive spectral analysis. Subsequently, calculations were performed iteratively with CYANA, with iterations used to verify and complete resonance assignments, refine NOESY peak lists, and optimize the distance calibration constants. Backbone 15N-1H RDCs for two alignment media were determined from J-modulated spectra (104) and used as orientational constraints for the folded core at the later stages of refinement. The final 20 conformers out of 100 were further refined by restrained molecular dynamics in explicit water (105) using the program CNS 1.2 (106) with the PARAM19 force field.
The bsCopL structure was determined using 1725 NOE-based conformation-restricting distance restraints, together with 130 restraints on backbone dihedral angle φ and ψ based on chemical shift data, and an additional 164 15N-1H residual dipolar coupling measurements. These data include 15.4 distance restraints per residue, and 7.1 long-range distance restraints per residue. Structural statistics and global quality factors were computed with PSVS 1.4 (107) and are summarized in Table S4. The goodness-of-fit between the final ensemble of conformers and the NOESY peak lists (108) was calculated with RPF module of AutoStructure 2.2.1 (102). The NMR-derived structures are well-converged and have structure quality scores, including model versus data metrics for back-calculated NOESY spectra and knowledge-based packing scores, typical of very high quality, accurate NMR structures. The resulting coordinates of bsCopL were deposited in the Protein Data Bank with PDB code 2KY9.
The structural presentations of the protein were produced with the programs Molmol (109) and PyMOL(110). Residue conservation analysis was performed with the ConSurf server (111). A total of 399 nonredundant protein sequences with sequence identity between 35 and 95% were selected from the UniRef90 database after three iterations of CSI-BLAST search with E-value cutoff of 0.0001 by using B. subtilis fragment 83–205 as a query and aligned with MAFFT algorithm.
For Cu1+ studies, the construct used did not include a C-terminal hexaHis tag and incorporated two additional N-terminal residues, compared with the construct used for structure determination. Sequence-specific assignments of backbone (1HN, 15N, 13CO, and 13Cα) and 13Cβ resonances were obtained for Cu1+-loaded holo-CopL by manual interactive analysis of HNCO, HNCA, HN(CO)CA, CBCA(CO)NH, and HNCACB spectra. Chemical shift perturbations (absolute value) of 1H and 15N backbone amides, ΔδNH, were then calculated relative to the same construct of apo-CopL lacking the C-terminal hexaHis tag using Equation 2 (112),
| (Eq. 2) |
where ΔδH and ΔδN are the chemical shift changes in the 1HN and 15N dimensions, respectively.
Author contributions
Z. R.-C. and J. M. B. conceptualization; Z. R.-C., A. E., N. S. D., and G. S. data curation; Z. R.-C., A. E., N. S. D., P. C. K., G. T. M., and J. M. B. formal analysis; Z. R.-C., A. E., N. S. D., H. A.-T., P. C. K., G. T. M., and J. M. B. investigation; Z. R.-C., A. E., N. S. D., T. S., G. T. M., and J. M. B. writing-original draft; Z. R.-C., A. E., H. A.-T., P. C. K., G. T. M., and J. M. B. writing-review and editing; N. S. D., H. A.-T., G. S., P. C. K., and J. M. B. methodology; T. S., G. T. M., and J. M. B. supervision; T. S., G. T. M., and J. M. B. funding acquisition; T. S., G. T. M., and J. M. B. project administration.
Supplementary Material
Acknowledgments
NMR instrumentation used in this work was supported in part by National Institutes of Health shared instrumentation Grant 1S10-OD018207. When the NMR data acquisition took place, Prof. T. Szyperski was a member of the New York Structural Biology Center; the Center is a STAR Center supported by the New York Office of Science, Technology and Academic Research. 900 MHz spectrometer was purchased with the funds from the National Institutes of Health and the Keck Foundation. We thank R. Xiao, D. Lee, C. Ciccosanti, S. Sahdev, and T. B. Acton for producing B. subtilis CopL constructs and protein samples, G. Liu for assistance in NMR studies, and H.-W. Lee and Prof J. H. Prestegard for providing RDC data. We also thank Prof. W. Belden and Prof. G. Zylstra for the use of their real-time thermocycler and spectrophotometer, respectively.
This work was supported in part by Rutgers University, the Charles and Johanna Busch Foundation, the United States Department of Agriculture MRF Project NE-1028 (to J. M. B.), National Institute of Allergy and Infectious Diseases (NIAID) grant 1R01AI139100-01 (to J. M. B.) and as a Community Outreach Project of the National Institutes of Health NIGMS Protein Structure Initiative Grant U54 GM094597 (to G. T. M. and T. S.). Support was also provide by NIGMS Grants R01 GM120574 (to G. T. M.) and S10 OD018207 (to G. T. M.). G. T. M. is a founder of Nexomics Biosciences, Inc. The content is solely the responsibility of the authors and does not necessarily represent the official views of the National Institutes of Health.
This article contains Figs. S1–S19, Tables S1–S4, and supporting Refs. 1–18.
The atomic coordinates and structure factors (code 2KY9) have been deposited in the Protein Data Bank (http://wwpdb.org/).
The NMR chemical shift data of this paper are available from the Biological Magnetic Resonance Data Bank under BMRB accession numbers 16942 and 27741.
- Cu
- copper
- MRSA
- methicillin-resistant S. aureus
- CA-MRSA
- community-associated–MRSA
- PDB
- Protein Data Bank
- TEV
- tobacco etch virus
- RDC
- residual dipolar coupling
- BCS
- bathocuprione disulfonate
- HSQC
- 2-dimensiional heteronuclear single-quantum correlation spectroscopy
- Amp
- ampicillin
- Cm
- chloramphenicol
- Tet
- tetracycline
- IMAC
- immobilized metal affinity chromatography.
References
- 1. Sousa F. L., Alves R. J., Ribeiro M. A., Pereira-Leal J. B., Teixeira M., and Pereira M. M. (2012) The superfamily of heme-copper oxygen reductases: types and evolutionary considerations. Biochim. Biophys. Acta 1817, 629–637 10.1016/j.bbabio.2011.09.020 [DOI] [PubMed] [Google Scholar]
- 2. Osman D., Patterson C. J., Bailey K., Fisher K., Robinson N. J., Rigby S. E., and Cavet J. S. (2013) The copper supply pathway to a Salmonella Cu,Zn-superoxide dismutase (SodCII) involves P(1B)-type ATPase copper efflux and periplasmic CueP. Mol. Microbiol. 87, 466–477 10.1111/mmi.12107 [DOI] [PubMed] [Google Scholar]
- 3. Claus H. (2003) Laccases and their occurrence in prokaryotes. Arch. Microbiol. 179, 145–150 10.1007/s00203-002-0510-7 [DOI] [PubMed] [Google Scholar]
- 4. Tavares P., Pereira A. S., Moura J. J., and Moura I. (2006) Metalloenzymes of the denitrification pathway. J. Inorg. Biochem. 100, 2087–2100 10.1016/j.jinorgbio.2006.09.003 [DOI] [PubMed] [Google Scholar]
- 5. Rutherford J. C., and Bird A. J. (2004) Metal-responsive transcription factors that regulate iron, zinc, and copper homeostasis in eukaryotic cells. Eukaryot. Cell 3, 1–13 10.1128/EC.3.1.1-13.2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Macomber L., and Imlay J. A. (2009) The iron–sulfur clusters of dehydratases are primary intracellular targets of copper toxicity. Proc. Natl. Acad. Sci. U.S.A. 106, 8344–8349 10.1073/pnas.0812808106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Chillappagari S., Seubert A., Trip H., Kuipers O. P., Marahiel M. A., and Miethke M. (2010) Copper stress affects iron homeostasis by destabilizing iron–sulfur cluster formation in Bacillus subtilis. J. Bacteriol. 192, 2512–2524 10.1128/JB.00058-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Fung D. K., Lau W. Y., Chan W. T., and Yan A. (2013) Copper efflux is induced during anaerobic amino acid limitation in Escherichia coli to protect iron–sulfur cluster enzymes and biogenesis. J. Bacteriol. 195, 4556–4568 10.1128/JB.00543-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Tan G., Cheng Z., Pang Y., Landry A. P., Li J., Lu J., and Ding H. (2014) Copper binding in IscA inhibits iron-sulphur cluster assembly in Escherichia coli. Mol. Microbiol. 93, 629–644 10.1111/mmi.12676 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Gunther M. R., Hanna P. M., Mason R. P., and Cohen M. S. (1995) Hydroxyl radical formation from cuprous ion and hydrogen peroxide: a spin-trapping study. Arch. Biochem. Biophys. 316, 515–522 10.1006/abbi.1995.1068 [DOI] [PubMed] [Google Scholar]
- 11. Mikolay A., Huggett S., Tikana L., Grass G., Braun J., and Nies D. H. (2010) Survival of bacteria on metallic copper surfaces in a hospital trial. Appl. Microbiol. Biotechnol. 87, 1875–1879 10.1007/s00253-010-2640-1 [DOI] [PubMed] [Google Scholar]
- 12. Schmidt M. G., Attaway H. H., Sharpe P. A., John J. Jr., Sepkowitz K. A., Morgan A., Fairey S. E., Singh S., Steed L. L., Cantey J. R., Freeman K. D., Michels H. T., and Salgado C. D. (2012) Sustained reduction of microbial burden on common hospital surfaces through introduction of copper. J. Clin. Microbiol. 50, 2217–2223 10.1128/JCM.01032-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. White C., Lee J., Kambe T., Fritsche K., and Petris M. J. (2009) A role for the ATP7A copper-transporting ATPase in macrophage bactericidal activity. J. Biol. Chem. 284, 33949–33956 10.1074/jbc.M109.070201 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Achard M. E., Stafford S. L., Bokil N. J., Chartres J., Bernhardt P. V., Schembri M. A., Sweet M. J., and McEwan A. G. (2012) Copper redistribution in murine macrophages in response to Salmonella infection. Biochem. J. 444, 51–57 10.1042/BJ20112180 [DOI] [PubMed] [Google Scholar]
- 15. Klevens R. M., Morrison M. A., Nadle J., Petit S., Gershman K., Ray S., Harrison L. H., Lynfield R., Dumyati G., Townes J. M., Craig A. S., Zell E. R., Fosheim G. E., McDougal L. K., Carey R. B., et al. (2007) Invasive methicillin-resistant Staphylococcus aureus infections in the United States. JAMA 298, 1763–1771 10.1001/jama.298.15.1763 [DOI] [PubMed] [Google Scholar]
- 16. Otto M. (2010) Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Annu. Rev. Microbiol. 64, 143–162 10.1146/annurev.micro.112408.134309 [DOI] [PubMed] [Google Scholar]
- 17. Tenover F. C., McDougal L. K., Goering R. V., Killgore G., Projan S. J., Patel J. B., and Dunman P. M. (2006) Characterization of a strain of community-associated methicillin-resistant Staphylococcus aureus widely disseminated in the United States. J. Clin. Microbiol. 44, 108–118 10.1128/JCM.44.1.108-118.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Talan D. A., Krishnadasan A., Gorwitz R. J., Fosheim G. E., Limbago B., Albrecht V., Moran G. J., and EMERGEncy ID Net Study Group. (2011) Comparison of Staphylococcus aureus from skin and soft-tissue infections in US emergency department patients, 2004 and 2008. Clin. Infect. Dis. 53, 144–149 10.1093/cid/cir308 [DOI] [PubMed] [Google Scholar]
- 19. Diep B. A., Gill S. R., Chang R. F., Phan T. H., Chen J. H., Davidson M. G., Lin F., Lin J., Carleton H. A., Mongodin E. F., Sensabaugh G. F., and Perdreau-Remington F. (2006) Complete genome sequence of USA300, an epidemic clone of community-acquired methicillin-resistant Staphylococcus aureus. Lancet 367, 731–739 10.1016/S0140-6736(06)68231-7 [DOI] [PubMed] [Google Scholar]
- 20. Thurlow L. R., Joshi G. S., Clark J. R., Spontak J. S., Neely C. J., Maile R., and Richardson A. R. (2013) Functional modularity of the arginine-catabolic mobile element contributes to the success of USA300 methicillin-resistant Staphylococcus aureus. Cell Host Microbe 13, 100–107 10.1016/j.chom.2012.11.012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Joshi G. S., Spontak J. S., Klapper D. G., and Richardson A. R. (2011) Arginine-catabolic mobile element encoded speG abrogates the unique hypersensitivity of Staphylococcus aureus to exogenous polyamines. Mol. Microbiol. 82, 9–20 10.1111/j.1365-2958.2011.07809.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Argüello J. M., González-Guerrero M., and Raimunda D. (2011) Bacterial transition metal P(1B)-ATPases: transport mechanism and roles in virulence. Biochemistry 50, 9940–9949 10.1021/bi201418k [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Argüello J. M., Patel S. J., and Quintana J. (2016) Bacterial Cu+-ATPases: models for molecular structure–function studies. Metallomics 8, 906–914 10.1039/C6MT00089D [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Singleton C., and Le Brun N. E. (2007) Atx1-like chaperones and their cognate P-type ATPases: copper-binding and transfer. Biometals 20, 275–289 10.1007/s10534-006-9068-1 [DOI] [PubMed] [Google Scholar]
- 25. Ding C., Festa R. A., Chen Y. L., Espart A., Palacios Ò., Espín J., Capdevila M., Atrian S., Heitman J., and Thiele D. J. (2013) Cryptococcus neoformans copper detoxification machinery is critical for fungal virulence. Cell Host Microbe 13, 265–276 10.1016/j.chom.2013.02.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Kay K. L., Hamilton C. J., and Le Brun N. E. (2016) Mass spectrometry of B. subtilis CopZ: Cu(i)-binding and interactions with bacillithiol. Metallomics 8, 709–719 10.1039/C6MT00036C [DOI] [PubMed] [Google Scholar]
- 27. Osterberg R., Ligaarden R., and Persson D. (1979) Copper(I) complexes of penicillamine and glutathione. J. Inorg. Biochem. 10, 341–355 10.1016/S0162-0134(00)80200-7 [DOI] [PubMed] [Google Scholar]
- 28. Rajkarnikar A., Strankman A., Duran S., Vargas D., Roberts A. A., Barretto K., Upton H., Hamilton C. J., and Rawat M. (2013) Analysis of mutants disrupted in bacillithiol metabolism in Staphylococcus aureus. Biochem. Biophys. Res. Commun. 436, 128–133 10.1016/j.bbrc.2013.04.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Helbig K., Bleuel C., Krauss G. J., and Nies D. H. (2008) Glutathione and transition-metal homeostasis in Escherichia coli. J. Bacteriol. 190, 5431–5438 10.1128/JB.00271-08 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Grossoehme N., Kehl-Fie T. E., Ma Z., Adams K. W., Cowart D. M., Scott R. A., Skaar E. P., and Giedroc D. P. (2011) Control of copper resistance and inorganic sulfur metabolism by paralogous regulators in Staphylococcus aureus. J. Biol. Chem. 286, 13522–13531 10.1074/jbc.M111.220012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Sitthisak S., Knutsson L., Webb J. W., and Jayaswal R. K. (2007) Molecular characterization of the copper transport system in Staphylococcus aureus. Microbiology 153, 4274–4283 10.1099/mic.0.2007/009860-0 [DOI] [PubMed] [Google Scholar]
- 32. Radford D. S., Kihlken M. A., Borrelly G. P., Harwood C. R., Le Brun N. E., and Cavet J. S. (2003) CopZ from Bacillus subtilis interacts in vivo with a copper exporting CPx-type ATPase CopA. FEMS Microbiol. Lett. 220, 105–112 10.1016/S0378-1097(03)00095-8 [DOI] [PubMed] [Google Scholar]
- 33. Baker J., Sengupta M., Jayaswal R. K., and Morrissey J. A. (2011) The Staphylococcus aureus CsoR regulates both chromosomal and plasmid-encoded copper resistance mechanisms. Environ. Microbiol. 13, 2495–2507 10.1111/j.1462-2920.2011.02522.x [DOI] [PubMed] [Google Scholar]
- 34. Zapotoczna M., Riboldi G. P., Moustafa A. M., Dickson E., Narechania A., Morrissey J. A., Planet P. J., Holden M. T. G., Waldron K. J., and Geoghegan J. A. (2018) Mobile-genetic-element-encoded hypertolerance to copper protects Staphylococcus aureus from killing by host phagocytes. MBio 9, e00550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Sitthisak S., Howieson K., Amezola C., and Jayaswal R. K. (2005) Characterization of a multicopper oxidase gene from Staphylococcus aureus. Appl. Environ. Microbiol. 71, 5650–5653 10.1128/AEM.71.9.5650-5653.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Argüello J. M., Eren E., and González-Guerrero M. (2007) The structure and function of heavy metal transport P1B-ATPases. Biometals 20, 233–248 10.1007/s10534-006-9055-6 [DOI] [PubMed] [Google Scholar]
- 37. Odermatt A., Suter H., Krapf R., and Solioz M. (1993) Primary structure of two P-type ATPases involved in copper homeostasis in Enterococcus hirae. J. Biol. Chem. 268, 12775–12779 [PubMed] [Google Scholar]
- 38. Marchler-Bauer A., Derbyshire M. K., Gonzales N. R., Lu S., Chitsaz F., Geer L. Y., Geer R. C., He J., Gwadz M., Hurwitz D. I., Lanczycki C. J., Lu F., Marchler G. H., Song J. S., Thanki N., et al. (2015) CDD: NCBI's conserved domain database. Nucleic Acids Res. 43, D222–D226 10.1093/nar/gku1221 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Li M., Diep B. A., Villaruz A. E., Braughton K. R., Jiang X., DeLeo F. R., Chambers H. F., Lu Y., and Otto M. (2009) Evolution of virulence in epidemic community-associated methicillin-resistant Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 5883–5888 10.1073/pnas.0900743106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Pérez-Pérez J. M., Candela H., and Micol J. L. (2009) Understanding synergy in genetic interactions. Trends Genet. 25, 368–376 10.1016/j.tig.2009.06.004 [DOI] [PubMed] [Google Scholar]
- 41. Smaldone G. T., and Helmann J. D. (2007) CsoR regulates the copper efflux operon copZA in Bacillus subtilis. Microbiology 153, 4123–4128 10.1099/mic.0.2007/011742-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Bernsel A., Viklund H., Hennerdal A., and Elofsson A. (2009) TOPCONS: consensus prediction of membrane protein topology. Nucleic Acids Res. 37, W465–W468 10.1093/nar/gkp363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Hutchings M. I., Palmer T., Harrington D. J., and Sutcliffe I. C. (2009) Lipoprotein biogenesis in Gram-positive bacteria: knowing when to hold 'em, knowing when to fold 'em. Trends Microbiol. 17, 13–21 10.1016/j.tim.2008.10.001 [DOI] [PubMed] [Google Scholar]
- 44. Rahman O., Cummings S. P., Harrington D. J., and Sutcliffe I. C. (2008) Methods for the bioinformatic identification of bacterial lipoproteins encoded in the genomes of Gram-positive bacteria. World J. Microbiol. Biotechnol. 24, 2377–2382 10.1007/s11274-008-9795-2 [DOI] [Google Scholar]
- 45. Navarre W. W., Daefler S., and Schneewind O. (1996) Cell wall sorting of lipoproteins in Staphylococcus aureus. J. Bacteriol. 178, 441–446 10.1128/jb.178.2.441-446.1996 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Kiedrowski M. R., Crosby H. A., Hernandez F. J., Malone C. L., McNamara J. O. 2nd, and Horswill A. R. (2014) Staphylococcus aureus Nuc2 is a functional, surface-attached extracellular nuclease. PLoS ONE 9, e95574 10.1371/journal.pone.0095574 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Xiao Z., Loughlin F., George G. N., Howlett G. J., and Wedd A. G. (2004) C-terminal domain of the membrane copper transporter Ctr1 from Saccharomyces cerevisiae binds four Cu(I) ions as a cuprous-thiolate polynuclear cluster: sub-femtomolar Cu(I) affinity of three proteins involved in copper trafficking. J. Am. Chem. Soc. 126, 3081–3090 10.1021/ja0390350 [DOI] [PubMed] [Google Scholar]
- 48. Xiao Z., Gottschlich L., van der Meulen R., Udagedara S. R., and Wedd A. G. (2013) Evaluation of quantitative probes for weaker Cu(i) binding sites completes a set of four capable of detecting Cu(i) affinities from nanomolar to attomolar. Metallomics 5, 501–513 10.1039/c3mt00032j [DOI] [PubMed] [Google Scholar]
- 49. Montelione G. T. (2012) The protein structure initiative: achievements and visions for the future. F1000 Biol. Rep. 4, 7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Bertini I., Cavallaro G., and McGreevy K. S. (2010) Cellular copper management–a draft user's guide. Coord. Chem. Rev. 254, 506–524 10.1016/j.ccr.2009.07.024 [DOI] [Google Scholar]
- 51. Rubino J. T., and Franz K. J. (2012) Coordination chemistry of copper proteins: how nature handles a toxic cargo for essential function. J. Inorg. Biochem. 107, 129–143 10.1016/j.jinorgbio.2011.11.024 [DOI] [PubMed] [Google Scholar]
- 52. Holm L., and Laakso L. M. (2016) Dali server update. Nucleic Acids Res. 44, W351–W355 10.1093/nar/gkw357 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Ma Z., Cowart D. M., Ward B. P., Arnold R. J., DiMarchi R. D., Zhang L., George G. N., Scott R. A., and Giedroc D. P. (2009) Unnatural amino acid substitution as a probe of the allosteric coupling pathway in a mycobacterial Cu(I) sensor. J. Am. Chem. Soc. 131, 18044–18045 10.1021/ja908372b [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Purves J., Thomas J., Riboldi G. P., Zapotoczna M., Tarrant E., Andrew P. W., Londoño A., Planet P. J., Geoghegan J. A., Waldron K. J., and Morrissey J. A. (2018) A horizontally gene transferred copper resistance locus confers hyper-resistance to antibacterial copper toxicity and enables survival of community acquired methicillin-resistant Staphylococcus aureus USA300 in macrophages. Environ. Microbiol. 20, 1576–1589 10.1111/1462-2920.14088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55. Ma Z., Cowart D. M., Scott R. A., and Giedroc D. P. (2009) Molecular insights into the metal selectivity of the copper(I)-sensing repressor CsoR from Bacillus subtilis. Biochemistry 48, 3325–3334 10.1021/bi900115w [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Badarau A., and Dennison C. (2011) Copper trafficking mechanism of CXXC-containing domains: insight from the pH-dependence of their Cu(I) affinities. J. Am. Chem. Soc. 133, 2983–2988 10.1021/ja1091547 [DOI] [PubMed] [Google Scholar]
- 57. Singleton C., and Le Brun N. E. (2009) The N-terminal soluble domains of Bacillus subtilis CopA exhibit a high affinity and capacity for Cu(I) ions. Dalton Trans. 2009, 688–696 [DOI] [PubMed] [Google Scholar]
- 58. Singleton C., Hearnshaw S., Zhou L., Le Brun N. E., and Hemmings A. M. (2009) Mechanistic insights into Cu(I) cluster transfer between the chaperone CopZ and its cognate Cu(I)-transporting P-type ATPase, CopA. Biochem. J. 424, 347–356 10.1042/BJ20091079 [DOI] [PubMed] [Google Scholar]
- 59. Delmar J. A., Su C. C., and Yu E. W. (2013) Structural mechanisms of heavy-metal extrusion by the Cus efflux system. Biometals 26, 593–607 10.1007/s10534-013-9628-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Bagchi P., Morgan M. T., Bacsa J., and Fahrni C. J. (2013) Robust affinity standards for Cu(I) biochemistry. J. Am. Chem. Soc. 135, 18549–18559 10.1021/ja408827d [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Jabs A., Weiss M. S., and Hilgenfeld R. (1999) Non-proline cis peptide bonds in proteins. J. Mol. Biol. 286, 291–304 10.1006/jmbi.1998.2459 [DOI] [PubMed] [Google Scholar]
- 62. Brandts J. F., Halvorson H. R., and Brennan M. (1975) Consideration of the Possibility that the slow step in protein denaturation reactions is due to cis-trans isomerism of proline residues. Biochemistry 14, 4953–4963 10.1021/bi00693a026 [DOI] [PubMed] [Google Scholar]
- 63. Schmidpeter P. A., and Schmid F. X. (2015) Prolyl isomerization and its catalysis in protein folding and protein function. J. Mol. Biol. 427, 1609–1631 10.1016/j.jmb.2015.01.023 [DOI] [PubMed] [Google Scholar]
- 64. Scholz C., Scherer G., Mayr L. M., Schindler T., Fischer G., and Schmid F. X. (1998) Prolyl isomerases do not catalyze isomerization of non-prolyl peptide bonds. Biol. Chem. 379, 361–365 [DOI] [PubMed] [Google Scholar]
- 65. Pal D., and Chakrabarti P. (1999) Cis peptide bonds in proteins: residues involved, their conformations, interactions and locations. J. Mol. Biol. 294, 271–288 10.1006/jmbi.1999.3217 [DOI] [PubMed] [Google Scholar]
- 66. Planet P. J., LaRussa S. J., Dana A., Smith H., Xu A., Ryan C., Uhlemann A. C., Boundy S., Goldberg J., Narechania A., Kulkarni R., Ratner A. J., Geoghegan J. A., Kolokotronis S. O., and Prince A. (2013) Emergence of the epidemic methicillin-resistant Staphylococcus aureus strain USA300 coincides with horizontal transfer of the arginine-catabolic mobile element and speG-mediated adaptations for survival on skin. MBio 4, e00889–13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Zhang Y. Q., Ren S. X., Li H. L., Wang Y. X., Fu G., Yang J., Qin Z. Q., Miao Y. G., Wang W. Y., Chen R. S., Shen Y., Chen Z., Yuan Z. H., Zhao G. P., Qu D., et al. (2003) Genome-based analysis of virulence genes in a non-biofilm-forming Staphylococcus epidermidis strain (ATCC 12228). Mol. Microbiol. 49, 1577–1593 10.1046/j.1365-2958.2003.03671.x [DOI] [PubMed] [Google Scholar]
- 68. Lowder B. V., Guinane C. M., Ben Zakour N. L., Weinert L. A., Conway-Morris A., Cartwright R. A., Simpson A. J., Rambaut A., Nübel U., and Fitzgerald J. R. (2009) Recent human-to-poultry host jump, adaptation, and pandemic spread of Staphylococcus aureus. Proc. Natl. Acad. Sci. U.S.A. 106, 19545–19550 10.1073/pnas.0909285106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Resch G., François P., Morisset D., Stojanov M., Bonetti E. J., Schrenzel J., Sakwinska O., and Moreillon P. (2013) Human-to-bovine jump of Staphylococcus aureus CC8 is associated with the loss of a β-hemolysin converting prophage and the acquisition of a new staphylococcal cassette chromosome. PLoS ONE 8, e58187 10.1371/journal.pone.0058187 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. Planet P. J., Diaz L., Kolokotronis S. O., Narechania A., Reyes J., Xing G., Rincon S., Smith H., Panesso D., Ryan C., Smith D. P., Guzman M., Zurita J., Sebra R., Deikus G., et al. (2015) Parallel epidemics of community-associated methicillin-resistant Staphylococcus aureus USA300 infection in North and South America. J. Infect. Dis. 212, 1874–1882 10.1093/infdis/jiv320 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Hao X., Lüthje F. L., Qin Y., McDevitt S. F., Lutay N., Hobman J. L., Asiani K., Soncini F. C., German N., Zhang S., Zhu Y. G., and Rensing C. (2015) Survival in amoeba–a major selection pressure on the presence of bacterial copper and zinc resistance determinants? Identification of a “copper pathogenicity island”. Appl. Microbiol. Biotechnol. 99, 5817–5824 10.1007/s00253-015-6749-0 [DOI] [PubMed] [Google Scholar]
- 72. Pang Y. Y., Schwartz J., Bloomberg S., Boyd J. M., Horswill A. R., and Nauseef W. M. (2014) Methionine sulfoxide reductases protect against oxidative stress in Staphylococcus aureus encountering exogenous oxidants and human neutrophils. J. Innate Immun. 6, 353–364 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Mashruwala A. A., Bhatt S., Poudel S., Boyd E. S., and Boyd J. M. (2016) The DUF59 containing protein SufT is involved in the maturation of iron–sulfur (FeS) proteins during conditions of high FeS cofactor demand in Staphylococcus aureus. PLoS Genet. 12, e1006233 10.1371/journal.pgen.1006233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Kreiswirth B. N., Löfdahl S., Betley M. J., O'Reilly M., Schlievert P. M., Bergdoll M. S., and Novick R. P. (1983) The toxic shock syndrome exotoxin structural gene is not detectably transmitted by a prophage. Nature 305, 709–712 10.1038/305709a0 [DOI] [PubMed] [Google Scholar]
- 75. Novick R. P. (1991) Genetic systems in Staphylococci. Methods Enzymol. 204, 587–636 10.1016/0076-6879(91)04029-N [DOI] [PubMed] [Google Scholar]
- 76. Rosario-Cruz Z., Chahal H. K., Mike L. A., Skaar E. P., and Boyd J. M. (2015) Bacillithiol has a role in Fe-S cluster biogenesis in Staphylococcus aureus. Mol. Microbiol. 98, 218–242 10.1111/mmi.13115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Bose J. L., Fey P. D., and Bayles K. W. (2013) Genetic tools to enhance the study of gene function and regulation in Staphylococcus aureus. Appl. Environ. Microbiol. 79, 2218–2224 10.1128/AEM.00136-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Mashruwala A. A., Guchte A. V., and Boyd J. M. (2017) Impaired respiration elicits SrrAB-dependent programmed cell lysis and biofilm formation in Staphylococcus aureus. Elife 6, e23845 10.7554/eLife.23845 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Forsyth R. A., Haselbeck R. J., Ohlsen K. L., Yamamoto R. T., Xu H., Trawick J. D., Wall D., Wang L., Brown-Driver V., Froelich J. M., C K. G., King P., McCarthy M., Malone C., Misiner B., Robbins D., et al. (2002) A genome-wide strategy for the identification of essential genes in Staphylococcus aureus. Mol. Microbiol. 43, 1387–1400 10.1046/j.1365-2958.2002.02832.x [DOI] [PubMed] [Google Scholar]
- 80. Mashruwala A. A., Pang Y. Y., Rosario-Cruz Z., Chahal H. K., Benson M. A., Mike L. A., Skaar E. P., Torres V. J., Nauseef W. M., and Boyd J. M. (2015) Nfu facilitates the maturation of iron–sulfur proteins and participates in virulence in Staphylococcus aureus. Mol. Microbiol. 95, 383–409 10.1111/mmi.12860 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Malone C. L., Boles B. R., Lauderdale K. J., Thoendel M., Kavanaugh J. S., and Horswill A. R. (2009) Fluorescent reporters for Staphylococcus aureus. J. Microbiol. Methods 77, 251–260 10.1016/j.mimet.2009.02.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Joska T. M., Mashruwala A., Boyd J. M., and Belden W. J. (2014) A universal cloning method based on yeast homologous recombination that is simple, efficient, and versatile. J. Microbiol. Methods 100, 46–51 10.1016/j.mimet.2013.11.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83. Mashruwala A. A., and Boyd J. M. (2016) De novo assembly of plasmids Using yeast recombinational cloning. Methods Mol. Biol. 1373, 33–41 10.1007/7651_2015_275 [DOI] [PubMed] [Google Scholar]
- 84. Roberts C. A., Al-Tameemi H. M., Mashruwala A. A., Rosario-Cruz Z., Chauhan U., Sause W. E., Torres V. J., Belden W. J., and Boyd J. M. (2017) The Suf iron–sulfur cluster biosynthetic system is essential in Staphylococcus aureus and decreased Suf function results in global metabolic defects and reduced survival in human neutrophils. Infect. Immun. 85, e00100–17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Ranjit D. K., Endres J. L., and Bayles K. W. (2011) Staphylococcus aureus CidA and LrgA proteins exhibit holin-like properties. J. Bacteriol. 193, 2468–2476 10.1128/JB.01545-10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Olson B. J., and Markwell J. (2007) Assays for determination of protein concentration. Curr. Protoc. Protein Sci. Chapter 3, Unit 3.4 10.1002/0471140864.ps0304s48 [DOI] [PubMed] [Google Scholar]
- 87. Xiao R., Anderson S., Aramini J., Belote R., Buchwald W. A., Ciccosanti C., Conover K., Everett J. K., Hamilton K., Huang Y. J., Janjua H., Jiang M., Kornhaber G. J., Lee D. Y., Locke J. Y., et al. (2010) The high-throughput protein sample production platform of the Northeast Structural Genomics Consortium. J. Struct. Biol. 172, 21–33 10.1016/j.jsb.2010.07.011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Jansson M., Li Y. C., Jendeberg L., Anderson S., Montelione G. T., and Nilsson B. (1996) High-level production of uniformly 15N- and 13C-enriched fusion proteins in Escherichia coli. J. Biomol. NMR 7, 131–141 [DOI] [PubMed] [Google Scholar]
- 89. Acton T. B., Xiao R., Anderson S., Aramini J., Buchwald W. A., Ciccosanti C., Conover K., Everett J., Hamilton K., Huang Y. J., Janjua H., Kornhaber G., Lau J., Lee D. Y., Liu G., et al. (2011) Preparation of protein samples for NMR structure, function, and small-molecule screening studies. Methods Enzymol. 493, 21–60 10.1016/B978-0-12-381274-2.00002-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Kay L. E., Torchia D. A., and Bax A. (1989) Backbone dynamics of proteins as studied by 15N inverse detected heteronuclear NMR spectroscopy: application to staphylococcal nuclease. Biochemistry 28, 8972–8979 10.1021/bi00449a003 [DOI] [PubMed] [Google Scholar]
- 91. Delaglio F., Grzesiek S., Vuister G. W., Zhu G., Pfeifer J., and Bax A. (1995) NMRPipe: a multidimensional spectral processing system based on UNIX pipes. J. Biomol. NMR 6, 277–293 [DOI] [PubMed] [Google Scholar]
- 92. Güntert P., Dötsch V., Wider G., and Wüthrich K. (1992) Processing of multi-dimensional NMR data with the new software PROSA. J. Biomol. NMR. 2, 619–629 10.1007/BF02192850 [DOI] [Google Scholar]
- 93. Keller R. L. J. (2004) The Computer Aided Resonance Assignment Tutorial, pp. 1–73. CANTINA Verlag, Switzerland [Google Scholar]
- 94. Bartels C., Xia T. H., Billeter M., Güntert P., and Wüthrich K. (1995) The program XEASY for computer-supported NMR spectral analysis of biological macromolecules. J. Biomol. NMR 6, 1–10 10.1007/BF00417486 [DOI] [PubMed] [Google Scholar]
- 95. Goddard T. D., and Knelle D. G. (2008) SPARKY 3. University of California, San Francisco, CA [Google Scholar]
- 96. Harris R. K., Becker E. D., Cabral de Menezes S. M., Granger P., Hoffman R. E., and Zilm K. W. (2008) Further conventions for NMR shielding and chemical shifts (IUPAC Recommendations 2008). Pure Appl. Chem. 80, 59–84 10.1351/pac200880010059 [DOI] [PubMed] [Google Scholar]
- 97. Bahrami A., Assadi A. H., Markley J. L., and Eghbalnia H. R. (2009) Probabilistic interaction network of evidence algorithm and its application to complete labeling of peak lists from protein NMR spectroscopy. PLoS Comput. Biol. 5, e1000307 10.1371/journal.pcbi.1000307 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Neri D., Szyperski T., Otting G., Senn H., and Wüthrich K. (1989) Stereospecific nuclear magnetic resonance assignments of the methyl groups of valine and leucine in the DNA-binding domain of the 434 repressor by biosynthetically directed fractional 13C labeling. Biochemistry 28, 7510–7516 10.1021/bi00445a003 [DOI] [PubMed] [Google Scholar]
- 99. Pelton J. G., Torchia D. A., Meadow N. D., and Roseman S. (1993) Tautomeric states of the active-site histidines of phosphorylated and unphosphorylated IIIGlc, a signal-transducing protein from Escherichia coli, using two-dimensional heteronuclear NMR techniques. Protein Sci. 2, 543–558 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100. Ulrich E. L., Akutsu H., Doreleijers J. F., Harano Y., Ioannidis Y. E., Lin J., Livny M., Mading S., Maziuk D., Miller Z., Nakatani E., Schulte C. F., Tolmie D. E., Kent Wenger R., Yao H., and Markley J. L. (2008) BioMagResBank. Nucleic Acids Res. 36, D402–D408 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101. Shen Y., Delaglio F., Cornilescu G., and Bax A. (2009) TALOS+: a hybrid method for predicting protein backbone torsion angles from NMR chemical shifts. J. Biomol. NMR 44, 213–223 10.1007/s10858-009-9333-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Huang Y. J., Tejero R., Powers R., and Montelione G. T. (2006) A topology-constrained distance network algorithm for protein structure determination from NOESY data. Proteins 62, 587–603 [DOI] [PubMed] [Google Scholar]
- 103. Güntert P., Mumenthaler C., and Wüthrich K. (1997) Torsion angle dynamics for NMR structure calculation with the new program DYANA. J. Mol. Biol. 273, 283–298 10.1006/jmbi.1997.1284 [DOI] [PubMed] [Google Scholar]
- 104. Tjandra N., Grzesiek S., and Bax A. (1996) Magnetic field dependence of nitrogen-proton J splittings in 15N-enriched human ubiquitin resulting from relaxation interference and residual dipolar coupling. J. Am. Chem. Soc. 118, 6264–6272 10.1021/ja960106n [DOI] [Google Scholar]
- 105. Linge J. P., Williams M. A., Spronk C. A., Bonvin A. M., and Nilges M. (2003) Refinement of protein structures in explicit solvent. Proteins 50, 496–506 10.1002/prot.10299 [DOI] [PubMed] [Google Scholar]
- 106. Brünger A. T., Adams P. D., Clore G. M., DeLano W. L., Gros P., Grosse-Kunstleve R. W., Jiang J. S., Kuszewski J., Nilges M., Pannu N. S., Read R. J., Rice L. M., Simonson T., and Warren G. L. (1998) Crystallography and NMR system: a new software suite for macromolecular structure determination. Acta Crystallogr. D Biol. Crystallogr. 54, 905–921 10.1107/S0907444998003254 [DOI] [PubMed] [Google Scholar]
- 107. Bhattacharya A., Tejero R., and Montelione G. T. (2007) Evaluating protein structures determined by structural genomics consortia. Proteins 66, 778–795 [DOI] [PubMed] [Google Scholar]
- 108. Huang Y. J., Powers R., and Montelione G. T. (2005) Protein NMR recall, precision, and F-measure scores (RPF scores): structure quality assessment measures based on information retrieval statistics. J. Am. Chem. Soc. 127, 1665–1674 10.1021/ja047109h [DOI] [PubMed] [Google Scholar]
- 109. Koradi R., Billeter M., and Wüthrich K. (1996) MOLMOL: a program for display and analysis of macromolecular structures. J. Mol. Graph. 14, 51–55, 29–32 10.1016/0263-7855(96)00009-4 [DOI] [PubMed] [Google Scholar]
- 110. Schrödinger L. (2015) The PyMOL Molecular Graphics System, Version 1.8, Schrödinger, LLC, New York [Google Scholar]
- 111. Ashkenazy H., Erez E., Martz E., Pupko T., and Ben-Tal N. (2010) ConSurf 2010: calculating evolutionary conservation in sequence and structure of proteins and nucleic acids. Nucleic Acids Res. 38, W529–W533 10.1093/nar/gkq399 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112. Farmer B. T. 2nd., Constantine K. L., Goldfarb V., Friedrichs M. S., Wittekind M., Yanchunas J. Jr., Robertson J. G., and Mueller L. (1996) Localizing the NADP+ binding site on the MurB enzyme by NMR. Nat. Struct. Biol. 3, 995–997 10.1038/nsb1296-995 [DOI] [PubMed] [Google Scholar]
- 113. Fey P. D., Endres J. L., Yajjala V. K., Widhelm T. J., Boissy R. J., Bose J. L., and Bayles K. W. (2013) A genetic resource for rapid and comprehensive phenotype screening of nonessential Staphylococcus aureus genes. MBio 4, e00537–12 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.