Abstract
Cluster of differentiation 38 (CD38) is the best-studied enzyme catalyzing the synthesis of the Ca2+ messenger cyclic ADP-ribose. It is a single-pass transmembrane protein, but possesses dual orientations. We have documented the natural existence of type III CD38 in cells and shown that it is regulated by a cytosolic activator, calcium- and integrin-binding 1 (CIB1). However, how type III CD38 can be folded correctly in the reductive cytosol has not been addressed. Using the yeast two-hybrid technique with CD38's catalytic domain (sCD38) as bait, here we identified a chaperone, Hsp70-interacting protein (Hip), that specifically interacts with both the type III CD38 and sCD38. Immunoprecipitation coupled with MS identified a chaperone complex associated specifically with sCD38. Pharmacological and siRNA-mediated knockdown of Hsp90 chaperones decreased the expression levels of both sCD38 and type III CD38, suggesting that these chaperones facilitate their folding. Moreover, knockdown of Hsc70 or DNAJA2 increased the levels of both CD38 types, consistent with the roles of these proteins in mediating CD38 degradation. Notably, Hip knockdown decreased type III CD38 substantially, but only marginally affected sCD38, indicating that Hip was selective for the former. More remarkably, DNAJA1 knockdown decreased sCD38 but increased type III CD38 levels. Mechanistically, we show that Hsc70 mediates lysosomal degradation of type III CD38, requiring the lysosomal receptor Lamp2A and the C19-motif in the C terminus of CD38. Our results indicate that folding and degradation of type III CD38 is effectively controlled in cells, providing further strong support of its physiological relevance.
Keywords: CD38, chaperone, heat shock protein (HSP), protein degradation, protein folding, cyclic ADP ribose (cADPR), heat shock protein 90 (Hsp90), cytosolic soluble CD38, hsp70, Hsp70-interacting protein (Hip), lysosomal degradation, type III CD38
Introduction
CD38 is usually considered as a cell surface marker and, in blood cells, is shown to be involved in HIV infection (1), malignancy in leukemias (2) and myelomas (3), as well as dysfunction of tumor-infiltrating CD8+ T cells (4). The discovery that it is also a signaling enzyme responsible for the metabolism of a calcium-mobilizing messenger, cyclic ADP-ribose (cADPR),4 raises a topological conundrum (5, 6). On the cell surface, CD38 is known to be a type II membrane protein with its N-terminal (N) tail inside the cytosol, whereas its carboxyl (C)-domain containing the catalytic site is on outside (Nin-Cout) (7). That the catalytic domain of CD38 is topological separated from its substrate and the endoplasmic target of its product, cADPR, has puzzled this field for two decades. We have provided a solution by documenting the natural existence of a type III CD38 (Cin-Nout) with its C-domain facing the cytosol and has ready access to its substrate NAD (8).
We further determine that the membrane orientation of CD38 is dictated by the positively charged amino acids flanking the transmembrane segment of the natural CD38 (8, 9). Their mutation to negatively charged residues directs the type II surface CD38 to express in the endoplasmic reticulum (ER) in an opposite type III orientation. Most intriguingly, the type III CD38, with its catalytic domain facing the reductive environment of the cytosol not only folds correctly but also is as active enzymatically as the WT CD38 (8, 9).
Type III CD38 is now established to be naturally present not only on cell surface but also internally. Immunostaining of live cells using externally applied antibody identify the ecto-facing N-terminal tail of CD38 on the cell surface (8). Internal type III CD38 is documented by a dual-epitope protein identification (DepID) technique using cytosolic nanobody-based probes transfected into cells (10). Type III CD38 not only resolves the topological conundrum but also points to cytosolic mechanisms regulating its activity. Indeed, yeast two-hybrid screening reveals a multifunctional regulator, CIB1, is associated with type III CD38 and modulates its cADPR-producing activity (10).
The dual membrane topology of CD38 necessitates the existence of elaborate mechanisms to ensure its correct folding and functioning in both the oxidative (outside) and reductive (cytosolic) environments. This is particularly critical for the type III orientation because the catalytic domain of CD38 has six disulfide bonds important for its enzymatic activity. To directly evaluate cytosolic disulfide formation of CD38, we constructed a soluble form of CD38 (sCD38) by fusing its catalytic domain with a soluble protein, EGFP, that directed the expression to the cytosol. The construct is shown to contain not only intact disulfides but also is enzymatically active and can effectively elevate cellular cADPR levels (11). The result is fully consistent with the finding that type III CD38 is more active in producing cADPR in cells than the dominant type II (10).
In this study, we probe the mechanism involved in the correct folding of type III CD38 by using the yeast two-hybrid screening and affinity-purification coupled MS (AP/MS) to identify a multi-chaperone complex, containing Hsp70s and their co-chaperones, Hsp90s and chaperonin TCP-1 (CCTs), that specifically associates not only with sCD38 but also with type III CD38. Loss-of-function studies show that the complex effectively modulates the correct folding. The degradation of the type III CD38 is in turn executed in the lysosomes and depends on the Lamp2A receptor and a motif identified in its C-terminal domain. Similar to the type III CD38, disulfide containing proteins present in the reductive cytosol are uncommon but do exist generally (12). Our results can shed light on how they can be folded and stabilized in the cytosol as well.
Results
Hip interacts with CD38 and positively regulates its levels
To identify the regulatory proteins that interact with the C-domain of the type III CD38, we employed the yeast two-hybrid approach, in which the 30-kDa C-domain of CD38 (sCD38) was used as a bait to screen the human spleen library (Clontech). Besides the protein CIB1, which we have shown playing important roles in regulating the enzymatic activity of type III CD38 (10), another positive clone encoding the cytosolic protein, suppression of tumorigenicity 13 (ST13), caught our attention. ST13, also named Hsp70 interacting protein, Hip, is reported to be down-regulated in several cancers (13). It functions as a co-chaperone of the 70 kDa heat shock cognate, Hsc70, and stimulates the assembly of Hsc70 with Hsp40 and the polypeptide substrate (14). It has been shown to play important roles in the maturation or trafficking of proteins like CXCR2 (15) and GR (16).
To verify the intracellular interaction between CD38 and Hip, we used the HEK-293T cell lines stably expressing the EGFP-tagged sCD38 in the cytosol (11) or the charge-mutated type III CD38 (mutCD38, described under “Experimental procedures”) (8). (Note: sCD38 in the following studies was all EGFP-tagged and labeled as sCD38 for simplicity.) The use of these constructs in the study resolved the complication that both type II and III CD38 naturally co-exist in cells. As both are sequence and immunologically identical, and type II being dominant, they cannot be independently analyzed. The use of mutCD38 (sCD38 also) in HEK-293T cells, which do not express CD38 normally, solved the problem and allowed focused investigations on type III CD38 without the interference from type II. MutCD38 not only has the same membrane topology as the endogenous type III CD38, but also is localized in the ER and is enzymatic active, with cADPR producing activity similar to WT CD38 (8, 9).
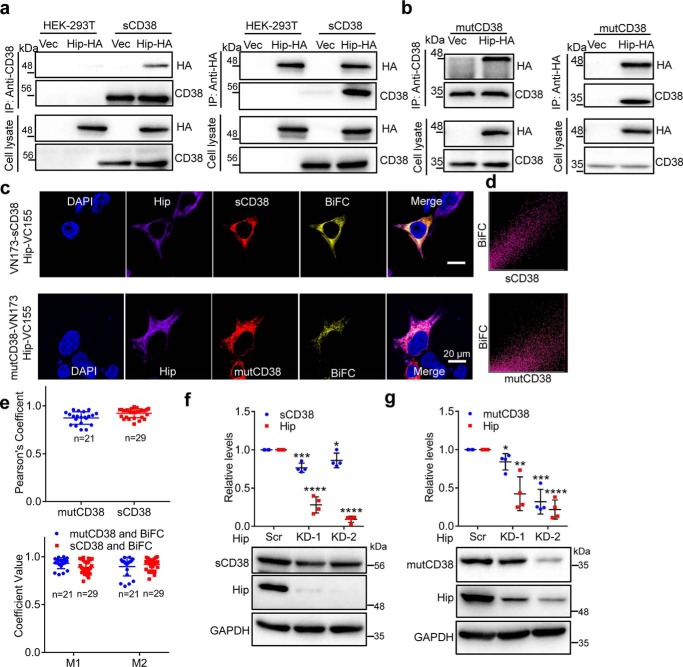
After transfection of the respective cell lines with HA-tagged Hip (Hip-HA), or vector as a control, Western blotting of the immunoprecipitants by anti-CD38 were probed with anti-HA. A prominent band with molecular mass of 50 kDa expected to be Hip was clearly present in both the cells expressing sCD38 (Fig. 1a, left panel, IP: anti-CD38; blot: anti-HA) or mutCD38 (Fig. 1b, left panel, IP: anti-CD38; blot: anti-HA), but not in the control (vector) HEK-293T cells. The precipitates and total cell lysate were also analyzed to confirm that the expression of both CD38 and Hip as well as the IP of CD38 were normal. Right panels of Fig. 1, a and b, show the results of the complementary experiments. The cell lysates were prepared from same cell lines transfected with either vector or Hip-HA, but the IP was done using anti-HA instead. Only cells expressing Hip-HA showed the presence of CD38 (Fig. 1, a and b, right panel, IP: anti-HA; blot: anti-CD38) in the precipitates. That the cytosolic protein, Hip, specifically associates with mutCD38 provides another support for its type III membrane topology.
Figure 1.
Hip interacts with CD38 and regulates its protein levels. a and b, HEK-293T cells stably expressing EGFP-tagged sCD38 (a, sCD38-cells in short; note: sCD38 in the following studies were all EGFP-tagged and labeled as sCD38 for simplicity.) and mutCD38 (b, mutCD38-cells in short) were transfected with HA-Hip or vector (Vec) and the whole cell lysates were subjected to IP with anti-CD38 (left panel) or anti-HA (right panel), followed by blotting with anti-HA or anti-CD38. c, visualization of intracellular association between Hip (Hip-VC155) and soluble (VN173-sCD38, upper panel) or type III CD38 (mutCD38-VN173, lower panel) by BiFC technique. BiFC signal, yellow; immunostaining of CD38, red; Hip, purple; nuclear staining (DAPI), blue. The merged image showed superposition of BiFC, CD38, Hip, and DAPI signals. d, scatter-plot pixels of sCD38 and BiFC signals shown in the images in c. e, colocalization parameters including Pearson's correlation coefficient (upper chart) and Manders overlap coefficient (lower chart) were analyzed with JACoP. M1 is defined as the fraction of mutCD38 (blue dots) or sCD38 (red dots) overlapping BiFC signal; M2 is defined conversely. Mean ± S.D.; n = 21 (mutCD38); n = 29 (sCD38). f and g, sCD38- or mutCD38-cells were transfected with scramble or Hip-specific siRNAs. The whole lysates were blotted with anti-CD38 and anti-Hip, with anti-GAPDH as a loading control. Lower panel, one representative blot; upper panel, quantification of the protein levels in four independent experiments. The abundance of CD38 or Hip in one sample was firstly normalized with the corresponding GAPDH level to eliminate loading unevenness and further normalized with the expression from the scramble siRNA controls. *, p <0.05; **, p <0.01; ***, p <0.001; ****, p <0.0001 by Student's t test (n = 4).
To further substantiate and visualize the intracellular interaction of Hip and the type III CD38, we employed the BiFC technique (17), in which the candidate proteins are each fused with one of the two nonfluorescent fragments of the Venus, either the N-terminal (VN173) or the C-terminal (VC155) fragment, respectively. Fluorescence is produced if the complementary candidate proteins interact closely, such that the Venus fragments can recombine to reform the fluorescent probe. In our experiment, Hip was fused with VC155, whereas sCD38 or mutCD38 were each fused with VN173. HEK-293T cells were transfected with a pair of the constructs, VN173-sCD38/Hip-VC155 (Fig. 1c, upper panel) or mutCD38-VN173/Hip-VC155 (Fig. 1c, lower panel), respectively. Immunostaining with anti-CD38 and anti-HA confirmed the expression and localization of sCD38 (red, uniform in cytosol) or mutCD38 (red, punctate in ER) and Hip (purple, uniform in cytosol). Yellow BiFC signals were observed in the cells expressing either forms of CD38 and Hip, but the control cells transfected only one part of the BiFCs (Fig. S1), which validated the reliability of the BiFC signals. Merging the images of BiFC with either sCD38 (upper panel) or mutCD38 (lower panel) shows their colocalization. Pixel-by-pixel scattered plots of BiFC fluorescence versus that of sCD38, or mutCD38, show the signals fall mainly along the diagonal region, indicating colocalization (Fig. 1d). The colocalization of sCD38 or mutCD38 and the BiFC signal was further quantified by calculating the parameters, Pearson's coefficient (Fig. 1e, upper panel), and the M1, M2 values (Fig. 1e, lower panel) of the indicated number of cells using JACoP. All parameters were close to unity, indicating strong colocalization for both mutCD38 versus BiFC and sCD38 versus BiFC.
After confirming the intracellular interaction between CD38 and Hip, we studied the effects of knocking down Hip on the protein levels of the two forms of CD38. Two different siRNAs (Hip KD-1 and KD-2) were each transfected to HEK-293T cells stably expressing sCD38 or mutCD38, and the protein levels in the whole lysates were examined by Western blotting. As shown in the representative blots (lower panels) and quantitative charts (upper panels) of Fig. 1, f and g, both siRNA were effective, with Hip KD-2 being able to reduce Hip (red dots in upper panels) to 10–20% of control (Scr). Knockdown of Hip caused a slight decrease of sCD38 (Fig. 1f, blue dots in upper panel) as compared with the control (Scr), but much more substantial reduction to as low as about 30% was seen in mutCD38 (Fig. 1g, Hip KD-2, blue dots in upper panel). The results indicate that Hip is critical in maintaining the stability of the type III CD38 and that the effect is selective for mutCD38 over sCD38. Knocking down Hip thus increases the misfolding of mutCD38, leading to its degradation and reduction at protein level.
Identification of the chaperone complex associated with CD38 by the AP/MS
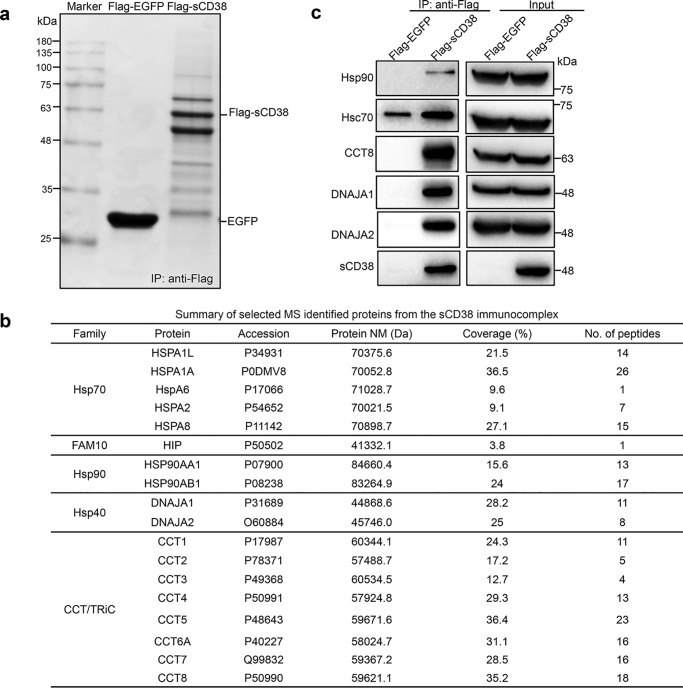
Because the knockdown of Hip only showed a slight influence on the protein levels of sCD38, we reasoned that other cytoplasmic chaperones or co-chaperones may be involved in regulating its stability. To evaluate this possibility, we employed the AP/MS strategy to identify the interacting proteins of sCD38. The HEK-293T cells stably expressing FLAG-sCD38 (10), or cells expressing FLAG-EGFP as a negative control, were permeabilized with 100 μm digitonin and the sCD38-associated protein complex was co-immunoprecipitated by anti-FLAG magnetic beads, and then eluted by 3×FLAG peptides for further analysis. SDS-PAGE of the eluents followed by Coomassie Blue staining shows several major bands in sCD38 immunoprecipitates (Fig. 2a), which were absent in the EGFP negative control, which showed only one major EGFP band. The gel bands of interest were subjected to in-gel protein digestion, and the profile of the interacting proteins of sCD38 was determined by LC-MS/MS as described in “Experimental procedures.” As listed in Fig. 2b, they turned out to be mainly chaperones, including five forms of Hsp70s, two Hsp90s, two Hsp40s, and eight CCTs, in addition to Hip described above. The identities of the proteins in the complex were further verified by Western blotting with specific antibodies. As shown in Fig. 2c, the chaperones in the complex were detected in the immunoprecipitates of sCD38, but not in the EGFP control (left panels, IP: anti-FLAG), although they were all expressed in both cells (right panels, Input).
Figure 2.
Cytoplasmic Hsp70s, Hsp90s, Hsp40s, chaperonins, and Hip were immunoprecipitated together with the soluble CD38. a, HEK-293T cells stably expressing FLAG-sCD38, or FLAG-EGFP as a negative control, were permeabilized by 100 μm digitonin and the cytosolic fractions were subjected to IP with anti-FLAG M2 Magnetic Beads, eluted by 3×FLAG peptide and analyzed by SDS-PAGE. b, the information of the chaperones identified by LC-MS/MS. Family: name of the protein family; Protein: reference name of protein documented in GenBank, NCBI; Accession: accession number in UniProt; Protein NM: molecular mass of the proteins; Coverage: percentage of the protein sequence that was covered by identified peptides; No. of peptides: number of peptide sequences unique to the identified proteins. c, Western blot analysis of both the immunoprecipitation (left panel) and input (right panel) by the antibodies labeled.
Chaperones regulate the folding and proteostasis of sCD38
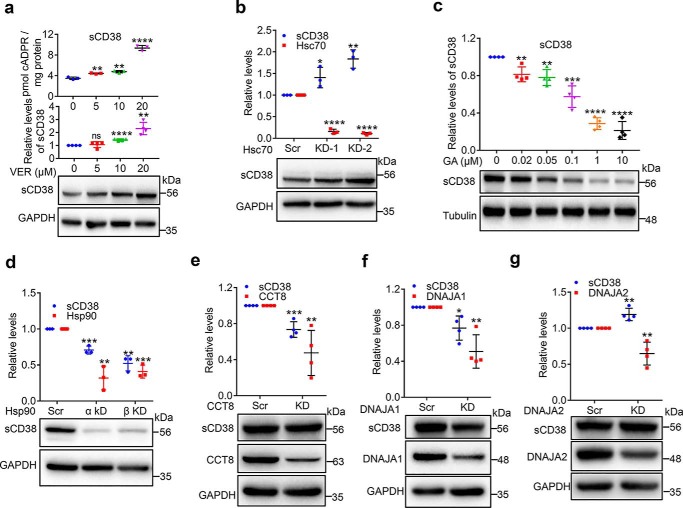
To investigate the roles of these chaperones in sCD38 proteostasis, we conducted loss-of-function experiments with both siRNA-knockdown and specific inhibitors. The first group is Hsp70 and its co-chaperones (Hsp40s and Hip), which are known to facilitate both folding and degradation (22). VER-155008 (VER) is an adenosine-derived inhibitor of Hsp70 that is selective over Hsp90 (18). As shown in Fig. 3a, inhibition of Hsp70s by VER for 24 h increased the protein levels of sCD38 in a dose-dependent manner (blots and middle chart). The increased amounts of sCD38 were folded correctly and enzymatically active, as the cellular cADPR contents showed corresponding elevation (upper chart). Consistently, increases in sCD38 level were also seen by knocking down Hsc70 (also named HSPA8, Fig. 2b), the major constitutive form of Hsp70, using two different siRNAs. This is shown both in the blots (Fig. 3b, lower panel) and the quantitative chart (Fig. 3b, upper panel). The knockdown efficiency of Hsc70 was confirmed by qRT-PCR (red dots in Fig. 3b) instead of Western blot analysis, because the commercial antibodies do not distinguish different members of Hsp70 (data not shown). The results indicate that the overall function of Hsp70s, particularly Hsc70, is to facilitate sCD38 degradation and thus its inhibition leads to increase in sCD38 level.
Figure 3.
Chaperones regulate the folding and proteostasis of cytosolic soluble CD38. a, sCD38-cells were treated with a series of concentrations of VER, a specific inhibitor of Hsp70s for 24 h. The protein levels of CD38, together with a housekeeping protein GAPDH were assayed by Western blotting and the cellular cADPR contents were measured by cycling assay. The representative blots (lower panel) are shown under the chart quantifying the relative expression of CD38 in at least three separate experiments (middle panel); the corresponding cADPR results are shown in the upper panel. b, sCD38-cells were transfected with Hsc70-specific or scramble siRNAs and the proteins or mRNA were analyzed by primers specific for Hsc70 or antibodies against CD38 or GAPDH. c, sCD38-cells were treated with different concentrations of geldanamycin (GA), an Hsp90 inhibitor for 3 h. The relative expression levels of CD38 were analyzed and presented as panel a. d–g, sCD38-cells were transfected with siRNAs specific for Hsp90α/β (d), CCT8 (e), DNAJA1 (f), or DNAJA2 (g) or scramble siRNA. The knockdown efficiency was evaluated by quantifying the target genes by qRT-PCR or Western blotting, and the relative expression levels of CD38 were analyzed and presented as panel a. Mean ± S.D.; n = 3 or 4; Student's t test, *, p <0.05; **, p <0.01; ***, p <0.001; ****, p <0.0001. All the protein or mRNA levels were normalized with the housekeeping genes such as GAPDH, tubulin, or β-actin and the relative levels were calculated by dividing the normalized levels by those from the control groups as described in Fig. 1.
Hsp90 acts as a proteostasis hub that promotes correct folding and controls a wide array of proteins of many important signaling pathways in eukaryotic cells (19). Geldanamycin (GA) is an antitumor antibiotic that binds to the ADP/ATP-binding pocket of Hsp90 and inhibits its function (20). Treating the cells for 3 h with increasing concentrations of GA dramatically decreased the levels of sCD38 in a dose-dependent manner (Fig. 3c). Hsp90 has two subunits, Hsp90α and Hsp90β, which form a functional clamp-shaped structure. As shown in Fig. 3d, knocking down either Hsp90α or Hsp90β (qRT-PCR, red dots in the upper chart) caused significant decrease of sCD38 (lower blot and blue dots in the upper chart), which is consistent with the pharmacological results (Fig. 3c). The data suggest that sCD38 is a client protein of Hsp90, which helps its folding and prevents degradation.
In the sCD38-associated complex, all eight subunits (CCT1–8) of CCT are present. They are believed to be double-ring complexes enclosing substrate proteins to facilitate their folding (21). Although all eight subunits are indispensable, CCT8 is particularly important in the assembly of the machinery (22). We used siRNA (CCT8-KD) to knock it down and assessed the importance of CCTs in the folding of sCD38. Results showed that the expression of CCT8 was effectively reduced (Fig. 3e, blots and red dots), leading to a corresponding reduction in sCD38 (Fig. 3e, blots and blue dots).
Hsp40s act as the co-chaperones of Hsp70s by binding the unfolded clients and transfer them to a particular Hsp70 for further folding or degradation. As such Hsp40s determine the client specificity of Hsp70 (23). To date, more than 45 different Hsp40s have been identified (23). In the sCD38-associated complex, we identified two Hsp40 proteins, DNAJA1 and DNAJA2. As shown in Fig. 3f, knocking down DNAJA1 decreased the sCD38 level, whereas the knockdown of DNAJA2 had an enhancing effect (Fig. 3g). The effects were significant but not extensive, most likely because of functional redundancy of other Hsp40s present in the cells.
Chaperones regulate the folding and proteostasis of type III CD38
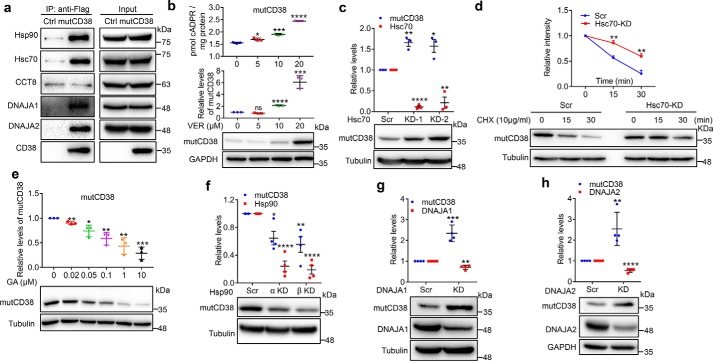
Results above establish a general understanding of the functions of the chaperones in the cytosolic sCD38 associating complex. We next turned to the membrane-bound mutCD38, a mimic of the natural signaling enzyme, the type III CD38 (8, 9). MutCD38 not only has the type III membrane topology but is also enzymatic active, with cADPR producing activity similar to WT CD38 (8, 9). As seen in Fig. 4a, all the chaperones described above, except CCT8, were also associated with mutCD38 (FLAG-tagged), as indicated by their co-immunoprecipitation with anti-FLAG (IP: anti-FLAG; blots: list specific antibodies used). The blots of the cell lysate with different antibodies serve as loading controls (Fig. 4a, Input).
Figure 4.
Chaperones regulate the folding and proteostasis of the artificial type III CD38, mutCD38. a, lysates from mutCD38-cells, with WT HEK-293T cells as a negative control, were subjected to IP with anti-FLAG M2 Magnetic Beads, followed by Western blotting with antibodies labeled. b and e, mutCD38-cells were treated with a series of concentrations of VER for 24 h (b) or GA for 3 h (e) and the expression levels of mutCD38 were assayed and presented as Fig. 3, a and c. c and f–h, the effects of knocking down different chaperones to the expression of mutCD38 were assayed and presented by the same methods as Fig. 3, b, d, f, and g. d, mutCD38-cells were transfected with siRNAs against Hsc70 (KD-2) for 24 h and treated with 10 μg/ml cycloheximide for 15 or 30 min. The cells were harvested and analyzed by Western blotting (for CD38 and tubulin) or qRT-PCR (for Hsc70) and analyzed as described in Fig. 1. All the above experiments were repeated at least three times and statistical analysis were done by GraphPad. Mean ± S.D.; n = 3; Student's t test, *, p <0.05; **, p <0.01; ***, p <0.001; ****, p <0.0001.
We then applied similar pharmacological and siRNA-knockdown interventions as described above on the associating chaperones in the mutCD38-expressing cells (Fig. 3). The results are summarized in Fig. 4, b–h. Inhibition of Hsp70s by VER led to the accumulation of the active mutCD38 enzyme, resulting in the corresponding elevation of cellular cADPR (Fig. 4b). Likewise, similar results were obtained by knocking down Hsc70 using siRNA (Fig. 4c). In the chase experiments, the degradation of mutCD38 in the presence of the translation inhibitor, cycloheximide, was significantly delayed when Hsc70 was knocked down (Fig. 4d), which confirmed the results of the steady expression (Fig. 4c). Inhibition of Hsp90 by GA (Fig. 4e) or gene knockdown (Fig. 4f) decreased the mutCD38 levels. Remarkably different from that observed with sCD38, knocking down either DNAJA1 (Fig. 4g and Fig. S2a for chase experiments) or DNAJA2 (Fig. 4h and Fig. S2b for chase experiments) induced dramatic accumulations of as high as 2- to 3-fold of mutCD38, consistent with the co-chaperones being responsible for transferring mutCD38 to Hsp70 for degradation.
Chaperone-mediated degradation of type III CD38
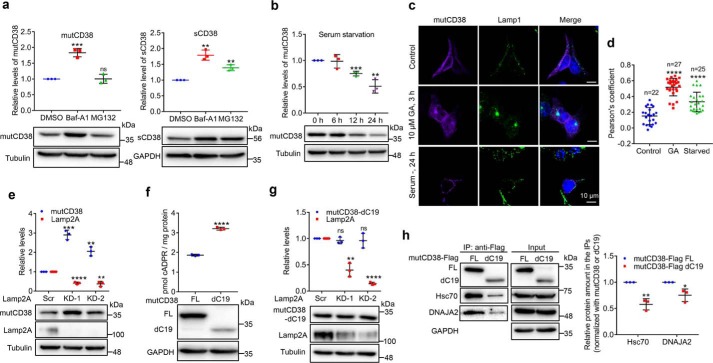
That the levels of mutCD38 and sCD38 show dramatically opposite responses to some of the chaperones is intriguing, especially considering the amino acid sequences of their cytosolic portions are identical. The distinction may be related to their degradation being mediated by different pathways. It was tested by treating the mutCD38-expressing HEK-293T cells with two specific inhibitors, bafilomycin A1 (Baf-A1) for the lysosomal (24) and MG-132 (25) for the proteasome pathways, respectively. As shown in Fig. 5a, Baf-A1 treatment doubled the level of mutCD38, whereas MG-132 had no effect (left panel), confirming the expectation that it is mainly degrades by the lysosomes. In contrast, both inhibitors increased the level of sCD38 (Fig. 5a), indicating that it is degraded through both the lysosomal and the proteosomal pathways.
Figure 5.
Chaperone-mediated lysosomal degradation of type III CD38. a, mutCD38-cells (left) or sCD38-cells (right) were treated with 1 μm Baf-A1 or 5 μm MG-132 for 6 h. The protein levels of CD38, together with a housekeeping protein tubulin or GAPDH were assayed by Western blotting. The relative expression levels of three independent experiments were analyzed and plotted by GraphPad as above. b, mutCD38-cells were cultured in zero-serum medium for 6, 12, or 24 h and the CD38 levels were analyzed as panel a. c, mutCD38-expressing cells were treated with 10 μm GA for 3 h, or zero-serum medium for 24 h and stained with anti-CD38 (purple), anti-Lamp1 (green), and DAPI (blue, nucleus). Scale bar: 10 μm. d, Pearson's correlation coefficient between CD38 and Lamp1 signals in the images from the same experiments shown in panel c by NIS-Elements AR analysis software. Signals from more than 20 cells were analyzed. e and g, HEK-293T cells stably expressing mutCD38 (e) or mutCD38-dC19 (g) were transfected with Lamp2A-specific siRNAs or scramble siRNA. The relative expression levels were analyzed as panel a. f, the cADPR levels and protein expression of mutCD38 (FL) and mutCD38-dC19–expressing cells were measured by cycling assay and Western blotting. h, mutCD38 (FL) and mutCD38-dC19–expressing cells were lysed and immunoprecipitated with anti-FLAG beads. The associated CD38, chaperones, and GAPDH were blotted with the specific antibodies. The relative amount of chaperones were calculated by normalization to the amount of mutCD38 or dC19 in the immunoprecipitates and the results were plotted in the right chart. All the above experiments were repeated at least three times and statistical analysis was done by GraphPad. Mean ± S.D.; n = 3; Student's t test, *, p <0.05; **, p <0.01; ***, p <0.001; ****, p <0.0001; ns, not significant.
It is known that Hsc70 facilitates protein degradation via the chaperone-mediated autophagy (CMA) in lysosomes (26). We hypothesized that mutCD38 may be degraded through this pathway, which is supported by our previous results showing that GA, which induces CMA (27), decreases mutCD38 (Fig. 4e). Another known way to activate CMA is by serum withdrawal (28). Indeed, the level of mutCD38 was significantly down-regulated after 12 h of starvation (Fig. 5b).
Confocal images in Fig. 5c show the colocalization between mutCD38 (purple) and the lysosomal marker, Lamp1 (green) and that the colocalization significantly increased after the activation of CMA by GA and serum starvation. This was further quantified by the statistical analyses using the Pearson's coefficient in Fig. 5d.
The best criterion for CMA degradation is the dependence on the lysosomal membrane receptor, Lamp2A (26). As shown in Fig. 5e, knocking down Lamp2A dramatically increased the levels of mutCD38 by 3-fold.
Analysis of the amino acid sequence of CD38 revealed the presence of 282DKFLQ sequence in the C-terminal α9-helix, which is a potential KFERQ-like motif (29) recognized by Hsc70 for lysosomal degradation. The sequence is known to be not critical for the enzymatic activity of CD38 (30). We constructed a truncation of mutCD38 by deleting the last 19 residues starting from the 285 residue, which disrupted its 282DKFLQ motif. The truncated mutCD38 (dC19) can be expressed and produce cADPR in HEK-293T cells as full-length mutCD38 (Fig. 5f). As shown in Fig. 5g, knocking down of Lamp2A now had essentially no effect on the protein levels of the truncated mutCD38 (mutCD38-dC19) in contrast to the 3-fold increase seen with mutCD38, indicating the motif sequence is indeed important for the degradation of type III CD38. Experiments showed that truncation of the C19-motif significantly decreased the immunoprecipitated contents of both Hsc70 and DNAJA2 in the mutCD38-associated complex (Fig. 5h), which further substantiated that this motif is important in mediating the binding with the chaperones. It is interesting to note that dC19 was more active enzymatically than full-length mutCD38 in making cADPR (Fig. 5f), suggesting that the binding of either Hsc70 or DNAJA2 or other proteins to the C-terminal α9-helix of the type III CD38 can negatively modulate its cADPR-producing activity.
Discussion
The documentation of the natural existence of type III CD38 (8, 10) resolves the topological paradox that has puzzled the field for decades. With the catalytic domain facing the cytosol, type III CD38 not only has ready access to its substrate NAD and direct targeting of its product cADPR on the endoplasmic Ca2+-stores, but also allows regulation of its activity by cytosolic mechanisms. We have shown that type III CD38 is indeed enzymatically active and that the multifunctional regulator CIB1 is one such mechanism that can modulate its cADPR-producing activity (8, 11). The fact that the type III CD38 is active indicates its catalytic domain is folded correctly even in the reductive environment of the cytosol, despite having six critical disulfides (8–11). In this study, we document the cytosolic chaperones that affect the correct folding and degradation of the type III CD38.
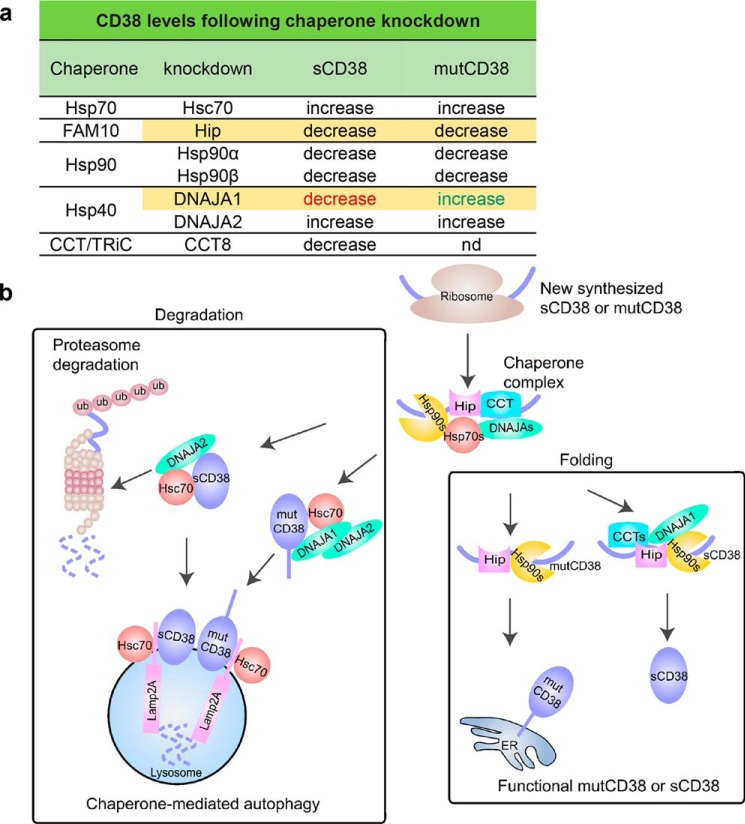
Chaperones ubiquitously control almost all aspects of cellular proteostasis and are central to maintaining the functional level of a wide range of proteins (31). We show in this study that it is also the case for the type III and the cytosolic CD38 (mutCD38 and sCD38). Using yeast two-hybrid and AP/MS, we identify a chaperone complex associated specifically with the two forms of CD38. As summarized in Fig. 6, functional studies using siRNA knockdown indicate Hip and Hsp90s facilitate correct folding of both sCD38 and the type III CD38. Their knockdown thus enhances degradation, leading to the decrease of the functional CD38 levels. On the other hand, knockdown of Hsc70 and DNAJA2s increases both CD38 levels, indicating they mainly act as house cleaners, targeting CD38 for degradation (cf. Fig. 4). A notable point of Hip is its differential effects on mutCD38 and sCD38. Knockdown of Hip decreases sCD38 level minimally, but induces more than 60% reduction in the level of mutCD38 (Fig. 1, f and g).
Figure 6.
a and b, summary (a) and working model (b).
Even more remarkable are the opposite effects of the DNAJA1 knockdown, decreasing sCD38 but increasing mutCD38 levels (Figs. 3f and 4g). The differential effects of DNAJA1 knockdown may well be related to the fact that mutCD38 is a transmembrane protein, whereas sCD38 is soluble. DNAJA1 is a co-chaperone of Hsc70 (32–34) and, together, has been shown to be key moderator of the degradation of hERG potassium channel (32), a transmembrane protein. Consistently, knockdown of either Hsc70 (Fig. 4, c and d) or DNAJA1 (Fig. 4g and Fig. S2a) increases mutCD38 quite dramatically, indicating both are involved in mediating the degradation of mutCD38, also a transmembrane protein. Their knockdown would thus reduce degradation and result in increased levels of mutCD38. Mechanistically, we show that the degradation of mutCD38 is through CMA and requires the lysosomal receptor Lamp2A (Fig. 5e). We further identified an Hsc70-recognition motif in the C-terminal portion of mutCD38 (Fig. 5, g and h). On the other hand, the degradation of sCD38, being a soluble protein, should be predominantly through the cytosolic proteasome pathway.
The degradation of the type III CD38 has many of the characteristics of CMA, in which substrates are mainly cytosolic proteins (35). Several membrane proteins such as RYR (36) and EGFR (37) have also been reported to be degraded by CMA, Exactly how membrane proteins are transported to lysosomes has not been elucidated. It could involve p97/Cdc48-mediated protein dislocation or intramembrane proteolysis extracting transmembrane segments and transporting to lysosomes (reviewed in Ref. 38). It is also possible that undefined mechanisms of lysosomal degradation for membrane proteins are involved that share common features with CMA such as Hsc70 and LAMP2A dependence.
For soluble proteins, the effect of Hsc70/DNAJA1 can be more diverse. Knockdown of DNAJA1 has been shown to facilitate the accumulation of tau, a microtubule-associated protein (39). However, using luciferase as a model for soluble protein, it has also been shown that the chaperones Hsc70/DNAJA1 mediate folding of the protein (33). Knockdown of DNAJA1, hampering folding, can thus lead to decreased levels. This is seen with a mutant form of p53, which shows increased degradation induced by the knockdown of DNAJA1 (40). Likewise, DNAJA1 depletion reduces the cellular levels of AID, an enzyme that deaminates deoxycytidine at the immunoglobulin genes (41). Consistently, we also observe that DNAJA1 knockdown reduces sCD38 levels (Fig. 3f). In any case, it is generally recognized that the action of chaperones is complex. Depending on the properties of the client proteins, as well as various co-chaperones present, the effects can be diverse, from promoting correct folding to inducing degradation.
This study is the first to identify the chaperone complex interacting with CD38 and documenting the diverse effects on its expression levels. That the cellular level of type III CD38 is effectively regulated by the cytosolic chaperone system provides a strong support for its physiological relevance. This study should pave the way for further and detailed analyses.
Experimental procedures
Reagents and cells
Geldanamycin, VER-155008, MG-132, and bafilomycin-A1 were purchased from Selleck Chemicals. Lipofectamine 2000, Lipofectamine RNAiMAX, FBS, DMEM, penicillin/streptomycin, trypsin, anti-HA Magnetic Beads, Alexa Fluor–conjugated anti-mouse, anti-rabbit and anti-goat were purchased from Thermo Fisher Scientific. NADase was prepared from Neurospora crassa. Diaphorase from Clostridium kluyveri, NAD, nicotinamide, alcohol dehydrogenase, resazurin, tri-n-octylamine, perchloric acid, digitonin, poly-l-lysine, DAPI, anti-FLAG M2 Magnetic Beads, 3×FLAG peptide, anti-FLAG and anti-HA antibodies and protease inhibitor mixture were obtained from Sigma-Aldrich. Cycloheximide was purchased from VWR Life Science AMRESCO. Anti-DNAJA1, anti-DNAJA2, and anti-CCT8 antibodies were purchased from Proteintech. Anti-Hip and Anti-Lamp2A were obtained from Abcam (catalog no. ab18528). Anti-Lamp1 was obtained from Cell Signaling Technology. Sheep polyclonal anti-CD38 antibody (for immunostaining in Fig. 5c) was purchased from R&D Systems and rabbit anti-CD38 were made by Absea. Anti-Hsc70 and anti-Hsp90α/β were purchased from Santa Cruz Biotechnology. Anti-β-tubulin, TRIzol, and qRT-PCR kit were obtained from TransGen Biotech. Anti-GAPDH was purchased from Sangon Biotech. siRNAs and primers were synthesized by GenePharma and Life Technologies, respectively.
HEK-293 and HEK-293T cells were purchased from the ATCC. The cells were cultured in DMEM containing 10% FBS supplemented with 1% penicillin/streptomycin and maintained at 37 °C in a humidified atmosphere of 5% CO2.
Yeast two-hybrid screening
The yeast two-hybrid screens were conducted with a Clontech Matchmarker Two-Hybrid system (cat. no. K1612-1) as described previously (10). The bait sCD38 (aa 46–300), fused to the GAL4 DNA-binding domain in the pGB vector was screened against a human spleen (Clontech, 638824) in pACT2-AD vector (Clontech, 638822) in the yeast strain Y190. Positive colonies were selected on the agar plate (S.D./-Leu/-Trp/-His/+3AT), tested by β-gal assay and sequenced.
Plasmids
To construct the lentiviral expression vectors for sCD38, FLAG-sCD38, and FLAG-sCD38(dC19), the corresponding DNAs were subcloned into pCHMWS-eGFP (11), gifted by HKU-Pasteur Research Centre. sCD38, short for EGFP-sCD38, contains a fragment (aa 46–300) of human CD38 and an EGFP tag in N terminus (11); FLAG-sCD38 is sCD38 tagged with a FLAG sequence in N terminus; FLAG-sCD38(dC19) was constructed based on FLAG-sCD38 by deleting the C-terminal 19-aa fragment (C19, DKFLQCVKNPEDSSCTSEI). The expression vectors for mutCD38-FLAG, mutCD38(dC19)-FLAG, and mutCD38(dC19) were constructed based on pLenti-puro (Addgene, no. 17452). mutCD38-FLAG is CD38 bearing the mutations of positively charged amino acids in the N-terminal fragment (8), with a FLAG in the C terminus; mutCD38(dC19)-FLAG is mutCD38-FLAG deleting the C19 fragment; mutCD38(dC19) is the truncated mutCD38 without FLAG-tag. To make the construct pKH3-Hip-HA for immunoprecipitation, Hip gene was subcloned into pKH3-HA (Addgene, no. 12555) with an HA-tag in C terminus. To make the construct pBiFC-Hip-VC155 for BiFC, Hip was subcloned into pBiFC-VC155 (Addgene, no. 22011). To make the construct pBiFC-mutCD38-VN173 and pBiFC-VN173-sCD38, mutCD38 and sCD38 were subcloned into pBiFC-VN173 (Addgene, no. 22010), respectively.
siRNAs
Sequences were as follows: Scr, 5′-UUCUCCGAACGUGUCACGUtt-3′; Hsc70–1, 5′-GGAGGUGUCUUCUAUGGUUtt-3′; Hsc70–2, 5′-CCAAAGAUGCUGGAACUAUtt-3′; Hip-1, 5′-GCUGCUAUUGAAGCCCUAAtt-3′; Hip-2, 5′-GCCAGUGUCUUCGUCAAAUtt-3′; Hsp90α, 5′-GGAAAGAGCUGCAUAUUAAtt-3′; Hsp90β, 5′-GGCUGAGGCCGACAAGAAUtt-3′; DNAJA1, 5′-GGAAGAUAGUUCGAGAGAAtt-3′; DNAJA2, 5′-CCUGAGAAGCGUGAGUUAUtt-3′; CCT-8, 5′-GGAAUUAGCUGAAGAACUUtt-3′; Lamp2A-1, 5′-GACUGCAGUGCAGAUGACGtt-3′; Lamp2A-2, 5′-GCACCAUCAUGCUGGAUAUtt-3′.
Transfection and lentivirus packaging
For siRNA transfection, cells were plated at 60–80% confluence in a 6-well plate and transfected with 3 μl of Lipofectamine RNAiMAX and 40 pmol siRNAs. After 24 h, cells were divided into two parts to detect the efficiency of gene knockdown and the expression of proteins. The knockdown efficiency was examined by qRT-PCR (24 h post transfection) or Western blotting (72 h post transfection).
Lentiviral particles were prepared as described previously (11). Briefly, HEK-293T cells were transfected with pLenti-puro or pCHMWS, pMD2.G, and psPAX2 with Lipofectamine 2000, and virus-containing media were harvested at 48–72 h post transfection.
Immunoprecipitation and Western blotting
Immunoprecipitation and Western blotting were performed as described previously (10). Briefly, cells were lysed in ice-cold lysis buffer (50 mm Tris, pH 7.4, 150 mm NaCl, 1 mm EDTA, 0.5% Triton X-100) containing protease inhibitor mixture and incubated with anti-FLAG M2 or anti-HA magnetic beads overnight at 4 °C. The immunoprecipitates were eluted with 3×FLAG peptide or solubilized in SDS-PAGE loading buffer. Ten percent of the total lysates were used as the input controls.
Protein profiling by affinity purification and LC-MS/MS
3 × 106 HEK-293T cells stably expressing FLAG-EGFP or FLAG-sCD38 were permeabilized with 100 μm digitonin in PBS containing protease inhibitors on ice for 10 min and the soluble fractions were collected. The target protein complexes were immunoprecipitated with anti-FLAG M2 Magnetic Beads overnight at 4 °C, eluted by 3×FLAG peptides, separated on SDS-PAGE followed by the MS-compatible Coomassie Brilliant Blue staining and in-gel digestion by trypsin.
MS data acquisition was performed by LC-MS/MS using a nanoLC.2D (Eksigent Technologies) coupled with a TripleTOF 5600+ System (AB SCIEX, Framingham, MA). First, samples were chromatographed using a 60-min gradient from 5 to 80% (mobile phase A: 0.1% (v/v) formic acid, 2% (v/v) acetonitrile; mobile phase B: 0.1% (v/v) formic acid, 98% (v/v) acetonitrile) after direct injection onto a nanoLC Column, 3C18-CL, 75 μm × 15 cm (Eksigent Technologies). The gradient was comprised of an increase from 2 to 22% mobile phase B over 40 min, 22 to 35% B in 12 min, and climbing to 80% B in 4 min then holding at 80% B for the last 4 min, all at a constant flow rate of 300 nanoliters/min on an Ekisgent NanoLC system. MS1 spectra were collected in the range 350–1500 m/z for 250 ms. The 50 most intense precursors with charge state 2–5 were selected for fragmentation, and MS2 spectra were collected in the range 100–2000 m/z for 100 ms; precursor ions were excluded from reselection for 15 s.
The original MS data were submitted to ProteinPilot Software (version 4.5, AB Sciex) for database searching against UniProt Homo Sapiens database (April 9, 2016, containing 160,566 sequences, http://www.uniprot.org/proteomes/UP000005640)5 concatenated with reverse decoy database. Trypsin/P was specified as cleavage enzyme allowing up to three missing cleavages, four modifications per peptide and two to five charges. Mass error was set to 20 ppm for first search, 5 ppm for main search, and 0.02 Da for fragmented ions. False discovery rate thresholds for protein, peptide, and modification site were specified at 1%. Minimum peptide length was set at seven. All the other parameters in ProteinPilot were set to default values. In addition, ProteinProspector (version 5.20.0) was used for search compare and protein quantification as described previously (42).
Quantitative RT-PCR
Total RNAs were extracted using TRIzol, reverse transcription was performed with random primers, and qPCR was done according to the manufacturer's instructions (TransGen Biotech). Primer pairs used in qPCR were 5′-CCTGGCACCCAGCACAAT-3′/5′-GGGCCGGACTCGTCATACT-3′ for β-actin; 5′-TGCTGCTGCTATTGCTTACG-3′/5′-TCAATAGTGAGGATTGACACATCA-3′ for Hsc70; 5′-GCTTGACCAATGACTGGGAAG-3′/5′-AGCTCCTCACAGTTATCCATGA-3′ for Hsp90α; 5′-CGAAGTTGGACAGTGGTAAAGAG-3′/5′-TGCCCAATCATGGAGATGTCT-3′ for Hsp90β.
Bimolecular fluorescence complementation assay and colocalization analysis
BiFC was performed as described previously (10, 17). Briefly, HEK-293T cells were transfected with the BiFC constructs as indicated and immunostained with anti-HA (VC155 fused with an HA tag) and anti-CD38, together with DAPI, staining nucleus. The resulting fluorescence signals, together with the BiFC signals (yellow) were visualized under a Nikon A1 confocal microscope. To estimate the colocalization (43), Pearson's coefficient and Manders' coefficients (M1 and M2) were calculated by JACoP and scatter plots generated by NIS-Elements AR analysis software (Nikon).
Measurement of the intracellular cADPR
5 × 105 HEK-293T cells stably expressing mutCD38 or sCD38 were seeded in a 6-well plate and treated with different concentrations of VER-155008 for 24 h, and the cells were lysed with 0.6 m perchloric acid. After centrifugation, cADPR in the supernatant was extracted and measured by a cycling assay (44). The cell pellets were solubilized in 1 m NaOH and the protein amount was quantified by Bradford assay. Results were presented as picomoles of cADPR per milligram of total proteins.
Quantification and statistical analysis
All experiments were performed at least three times independently. The band intensity in Western blotting was quantified with ImageJ software. Statistical analysis was done with the GraphPad Prism software and the significance of differences was analyzed by the unpaired Student's t test. Data shown in the figures are mean ± S.D. from at least three repeats.
Author contributions
Y. W., J. Z., and Y. J. Z. data curation; Y. W., J. Z., L. F., H.-C. L., and Y. J. Z. formal analysis; Y. W., J. Z., H.-C. L., and Y. J. Z. investigation; Y. W., J. Z., H.-C. L., and Y. J. Z. methodology; Y. W., J. Z., L. F., H.-C. L., and Y. J. Z. writing-review and editing; L. F. and Y. J. Z. resources; L. F., H.-C. L., and Y. J. Z. supervision; L. F., H.-C. L., and Y. J. Z. funding acquisition; H.-C. L. and Y. J. Z. conceptualization; Y. J. Z. validation; Y. J. Z. writing-original draft; Y. J. Z. project administration.
Supplementary Material
This study was supported by Shenzhen Governments Grants JCYJ20160608091848749 (to Y. J. Z.) and JCYJ20170412150609690 (to H. C. L.); National Science Foundation of China Grants 31571438 (to Y. J. Z.), 31671463 (to H. C. L.), and 31500664 and 31770838 (to L. F.); Natural Science Foundation of Jiangsu Province Grant BK20171338 (to L. F.); and the Fundamental Research Funds for the Central Universities Grant 021414380334 (to L. F.). The authors declare that they have no conflicts of interest with the contents of this article.
Please note that the JBC is not responsible for the long-term archiving and maintenance of this site or any other third party hosted site.
- cADPR
- cyclic ADP-ribose
- ER
- endoplasmic reticulum
- EGFP
- enhanced GFP
- AP/MS
- affinity-purification coupled mass spectrometry
- CCT
- chaperonin containing TCP-1
- Hsc
- heat shock cognate
- IP
- immunoprecipitation
- BiFC
- bimolecular fluorescence complementation
- JACoP
- a plugin of ImageJ software
- VER
- VER-155008
- qRT-PCR
- quantitative RT-PCR
- GA
- geldanamycin
- Baf-A1
- bafilomycin A1
- CMA
- chaperone-mediated autophagy
- DAPI
- 4′,6-diamidino-2-phenylindole
- aa
- amino acid.
References
- 1. Giorgi J. V., Ho H. N., Hirji K., Chou C. C., Hultin L. E., O'Rourke S., Park L., Margolick J. B., Ferbas J., and Phair J. P. (1994) CD8+ lymphocyte activation at human immunodeficiency virus type 1 seroconversion: Development of HLA-DR+ CD38− CD8+ cells is associated with subsequent stable CD4+ cell levels. The Multicenter AIDS Cohort Study Group. J. Infect. Dis. 170, 775–781 10.1093/infdis/170.4.775 [DOI] [PubMed] [Google Scholar]
- 2. Dürig J., Naschar M., Schmücker U., Renzing-Köhler K., Hölter T., Hüttmann A., and Dührsen U. (2002) CD38 expression is an important prognostic marker in chronic lymphocytic leukaemia. Leukemia 16, 30–35 10.1038/sj.leu.2402339 [DOI] [PubMed] [Google Scholar]
- 3. Ruiz-Argüelles G. J., and San Miguel J. F. (1994) Cell surface markers in multiple myeloma. Mayo Clin. Proc. 69, 684–690 10.1016/S0025-6196(12)61350-0 [DOI] [PubMed] [Google Scholar]
- 4. Philip M., Fairchild L., Sun L., Horste E. L., Camara S., Shakiba M., Scott A. C., Viale A., Lauer P., Merghoub T., Hellmann M. D., Wolchok J. D., Leslie C. S., and Schietinger A. (2017) Chromatin states define tumour-specific T cell dysfunction and reprogramming. Nature 545, 452–456 10.1038/nature22367 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Howard M., Grimaldi J. C., Bazan J. F., Lund F. E., Santos-Argumedo L., Parkhouse R. M., Walseth T. F., and Lee H. C. (1993) Formation and hydrolysis of cyclic ADP-ribose catalyzed by lymphocyte antigen CD38. Science 262, 1056–1059 10.1126/science.8235624 [DOI] [PubMed] [Google Scholar]
- 6. Takasawa S., Tohgo A., Noguchi N., Koguma T., Nata K., Sugimoto T., Yonekura H., and Okamoto H. (1993) Synthesis and hydrolysis of cyclic ADP-ribose by human leukocyte antigen CD38 and inhibition of the hydrolysis by ATP. J. Biol. Chem. 268, 26052–26054 [PubMed] [Google Scholar]
- 7. Jackson D. G., and Bell J. I. (1990) Isolation of a cDNA encoding the human CD38 (T10) molecule, a cell surface glycoprotein with an unusual discontinuous pattern of expression during lymphocyte differentiation. J. Immunol. 144, 2811–2815 [PubMed] [Google Scholar]
- 8. Zhao Y. J., Lam C. M., and Lee H. C. (2012) The membrane-bound enzyme CD38 exists in two opposing orientations. Sci. Signal. 5, ra67 10.1126/scisignal.2002700 [DOI] [PubMed] [Google Scholar]
- 9. Zhao Y. J., Zhu W. J., Wang X. W., Zhang L. H., and Lee H. C. (2015) Determinants of the membrane orientation of a calcium signaling enzyme CD38. Biochim. Biophys. Acta 1853, 2095–2103 10.1016/j.bbamcr.2014.10.028 [DOI] [PubMed] [Google Scholar]
- 10. Liu J., Zhao Y. J., Li W. H., Hou Y. N., Li T., Zhao Z. Y., Fang C., Li S. L., and Lee H. C. (2017) Cytosolic interaction of type III human CD38 with CIB1 modulates cellular cyclic ADP-ribose levels. Proc. Natl. Acad. Sci. U.S.A. 114, 8283–8288 10.1073/pnas.1703718114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Zhao Y. J., Zhang H. M., Lam C. M., Hao Q., and Lee H. C. (2011) Cytosolic CD38 protein forms intact disulfides and is active in elevating intracellular cyclic ADP-ribose. J. Biol. Chem. 286, 22170–22177 10.1074/jbc.M111.228379 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12. Cumming R. C., Andon N. L., Haynes P. A., Park M., Fischer W. H., and Schubert D. (2004) Protein disulfide bond formation in the cytoplasm during oxidative stress. J. Biol. Chem. 279, 21749–21758 10.1074/jbc.M312267200 [DOI] [PubMed] [Google Scholar]
- 13. Mo Y., Zheng S., and Shen D. (1996) Differential expression of HSU17714 gene in colorectal cancer and normal colonic mucosa [In Chinese]. Zhonghua zhong liu za zhi [Chin. J. Oncol.] 18, 241–243 [PubMed] [Google Scholar]
- 14. Höhfeld J., Minami Y., and Hartl F. U. (1995) Hip, a novel cochaperone involved in the eukaryotic Hsc70/Hsp40 reaction cycle. Cell 83, 589–598 10.1016/0092-8674(95)90099-3 [DOI] [PubMed] [Google Scholar]
- 15. Fan G. H., Yang W., Sai J., and Richmond A. (2002) Hsc/Hsp70 interacting protein (hip) associates with CXCR2 and regulates the receptor signaling and trafficking. J. Biol. Chem. 277, 6590–6597 10.1074/jbc.M110588200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Nelson G. M., Prapapanich V., Carrigan P. E., Roberts P. J., Riggs D. L., and Smith D. F. (2004) The heat shock protein 70 cochaperone hip enhances functional maturation of glucocorticoid receptor. Mol. Endocrinol. 18, 1620–1630 10.1210/me.2004-0054 [DOI] [PubMed] [Google Scholar]
- 17. Shyu Y. J., Liu H., Deng X., and Hu C. D. (2006) Identification of new fluorescent protein fragments for bimolecular fluorescence complementation analysis under physiological conditions. BioTechniques 40, 61–66 10.2144/000112036 [DOI] [PubMed] [Google Scholar]
- 18. Williamson D. S., Borgognoni J., Clay A., Daniels Z., Dokurno P., Drysdale M. J., Foloppe N., Francis G. L., Graham C. J., Howes R., Macias A. T., Murray J. B., Parsons R., Shaw T., Surgenor A. E., Terry L., Wang Y., Wood M., and Massey A. J. (2009) Novel adenosine-derived inhibitors of 70 kDa heat shock protein, discovered through structure-based design. J. Med. Chem. 52, 1510–1513 10.1021/jm801627a [DOI] [PubMed] [Google Scholar]
- 19. Taipale M., Jarosz D. F., and Lindquist S. (2010) HSP90 at the hub of protein homeostasis: emerging mechanistic insights. Nat. Rev. Mol. Cell Biol. 11, 515–528 10.1038/nrm2918 [DOI] [PubMed] [Google Scholar]
- 20. Whitesell L., Mimnaugh E. G., De Costa B., Myers C. E., and Neckers L. M. (1994) Inhibition of heat shock protein HSP90-pp60v-src heteroprotein complex formation by benzoquinone ansamycins: Essential role for stress proteins in oncogenic transformation. Proc. Natl. Acad. Sci. U.S.A. 91, 8324–8328 10.1073/pnas.91.18.8324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Lopez T., Dalton K., and Frydman J. (2015) The mechanism and function of group II chaperonins. J. Mol. Biol. 427, 2919–2930 10.1016/j.jmb.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Noormohammadi A., Khodakarami A., Gutierrez-Garcia R., Lee H. J., Koyuncu S., König T., Schindler C., Saez I., Fatima A., Dieterich C., and Vilchez D. (2016) Somatic increase of CCT8 mimics proteostasis of human pluripotent stem cells and extends C. elegans lifespan. Nat. Commun. 7, 13649 10.1038/ncomms13649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Craig E. A., and Marszalek J. (2017) How do J-proteins get Hsp70 to do so many different things? Trends Biochem. Sci. 42, 355–368 10.1016/j.tibs.2017.02.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Yamamoto A., Tagawa Y., Yoshimori T., Moriyama Y., Masaki R., and Tashiro Y. (1998) Bafilomycin A1 prevents maturation of autophagic vacuoles by inhibiting fusion between autophagosomes and lysosomes in rat hepatoma cell line, H-4-II-E cells. Cell Struct. Funct. 23, 33–42 10.1247/csf.23.33 [DOI] [PubMed] [Google Scholar]
- 25. Lee D. H., and Goldberg A. L. (1998) Proteasome inhibitors: Valuable new tools for cell biologists. Trends Cell Biol. 8, 397–403 10.1016/S0962-8924(98)01346-4 [DOI] [PubMed] [Google Scholar]
- 26. Kaushik S., and Cuervo A. M. (2018) The coming of age of chaperone-mediated autophagy. Nat. Rev. Mol. Cell Biol. 19, 365–381 10.1038/s41580-018-0001-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27. Finn P. F., Mesires N. T., Vine M., and Dice J. F. (2005) Effects of small molecules on chaperone-mediated autophagy. Autophagy 1, 141–145 10.4161/auto.1.3.2000 [DOI] [PubMed] [Google Scholar]
- 28. Cuervo A. M., Knecht E., Terlecky S. R., and Dice J. F. (1995) Activation of a selective pathway of lysosomal proteolysis in rat liver by prolonged starvation. Am. J. Physiol. 269, C1200–C1208 10.1152/ajpcell.1995.269.5.C1200 [DOI] [PubMed] [Google Scholar]
- 29. Dice J. F. (1982) Altered degradation of proteins microinjected into senescent human fibroblasts. J. Biol. Chem. 257, 14624–14627 [PubMed] [Google Scholar]
- 30. Hoshino S., Kukimoto I., Kontani K., Inoue S., Kanda Y., Malavasi F., and Katada T. (1997) Mapping of the catalytic and epitopic sites of human CD38/NAD+ glycohydrolase to a functional domain in the carboxyl terminus. J. Immunol. 158, 741–747 [PubMed] [Google Scholar]
- 31. Radons J. (2016) The human HSP70 family of chaperones: Where do we stand? Cell Stress Chaperones 21, 379–404 10.1007/s12192-016-0676-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Walker V. E., Wong M. J., Atanasiu R., Hantouche C., Young J. C., and Shrier A. (2010) Hsp40 chaperones promote degradation of the HERG potassium channel. J. Biol. Chem. 285, 3319–3329 10.1074/jbc.M109.024000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Terada K., and Oike Y. (2010) Multiple molecules of Hsc70 and a dimer of DjA1 independently bind to an unfolded protein. J. Biol. Chem. 285, 16789–16797 10.1074/jbc.M110.101501 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Terada K., and Mori M. (2000) Human DnaJ homologs dj2 and dj3, and bag-1 are positive cochaperones of hsc70. J. Biol. Chem. 275, 24728–24734 10.1074/jbc.M002021200 [DOI] [PubMed] [Google Scholar]
- 35. Galluzzi L., Baehrecke E. H., Ballabio A., Boya P., Bravo-San Pedro J. M., Cecconi F., Choi A. M., Chu C. T., Codogno P., Colombo M. I., Cuervo A. M., Debnath J., Deretic V., Dikic I., Eskelinen E. L., et al. (2017) Molecular definitions of autophagy and related processes. EMBO J. 36, 1811–1836 10.15252/embj.201796697 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Pedrozo Z., Torrealba N., Fernández C., Gatica D., Toro B., Quiroga C., Rodriguez A. E., Sanchez G., Gillette T. G., Hill J. A., Donoso P., and Lavandero S. (2013) Cardiomyocyte ryanodine receptor degradation by chaperone-mediated autophagy. Cardiovasc. Res. 98, 277–285 10.1093/cvr/cvt029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Shen S., Zhang P., Lovchik M. A., Li Y., Tang L., Chen Z., Zeng R., Ma D., Yuan J., and Yu Q. (2009) Cyclodepsipeptide toxin promotes the degradation of Hsp90 client proteins through chaperone-mediated autophagy. J. Cell Biol. 185, 629–639 10.1083/jcb.200810183 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Avci D., and Lemberg M. K. (2015) Clipping or extracting: Two ways to membrane protein degradation. Trends Cell Biol. 25, 611–622 10.1016/j.tcb.2015.07.003 [DOI] [PubMed] [Google Scholar]
- 39. Abisambra J. F., Jinwal U. K., Suntharalingam A., Arulselvam K., Brady S., Cockman M., Jin Y., Zhang B., and Dickey C. A. (2012) DnaJA1 antagonizes constitutive Hsp70-mediated stabilization of tau. J. Mol. Biol. 421, 653–661 10.1016/j.jmb.2012.02.003 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40. Parrales A., Ranjan A., Iyer S. V., Padhye S., Weir S. J., Roy A., and Iwakuma T. (2016) DNAJA1 controls the fate of misfolded mutant p53 through the mevalonate pathway. Nat. Cell Biol. 18, 1233–1243 10.1038/ncb3427 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Orthwein A., Zahn A., Methot S. P., Godin D., Conticello S. G., Terada K., and Di Noia J. M. (2012) Optimal functional levels of activation-induced deaminase specifically require the Hsp40 DnaJa1. EMBO J. 31, 679–691 10.1038/emboj.2011.417 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Fang L., Kaake R. M., Patel V. R., Yang Y., Baldi P., and Huang L. (2012) Mapping the protein interaction network of the human COP9 signalosome complex using a label-free QTAX strategy. Mol. Cell. Proteomics 11, 138–147 10.1074/mcp.M111.016352 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Dunn K. W., Kamocka M. M., and McDonald J. H. (2011) A practical guide to evaluating colocalization in biological microscopy. Am. J. Physiol. Cell Physiol. 300, C723–C742 10.1152/ajpcell.00462.2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Graeff R., and Lee H. C. (2002) A novel cycling assay for cellular cADP-ribose with nanomolar sensitivity. Biochem. J. 361, 379–384 10.1042/bj3610379 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.