Abstract
Prior to the clinical phases of testing, safety, efficacy and pharmacokinetic profiles of lead compounds are evaluated in animal studies. These tests are primarily performed in rodents, such as mouse and rats. In order to reduce the number of animal experiments, computational models that predict the outcome of these studies and thus aid in prioritization of preclinical candidates are heavily needed. However, although computational models for human off-target interactions with decent quality are available, they cannot easily be transferred to rodents due to lack of respective data. In this study, we assess the transferability of human P-glycoprotein activity data for development of in silico models to predict in vivo effects in rats and mouse using a structure-based approach. P-glycoprotein (P-gp) is an ATP-dependent efflux transporter that transports xenobiotic compounds such as toxins and drugs out of cells and has a broad substrate and inhibitor specificity. Being mostly expressed at barriers, it influences the bioavailability of drugs and thus contributes also to toxicity. Comparison of the binding site interaction profiles of human, rat and mouse P-gp derived from docking studies with a set of common inhibitors suggests that the inhibitors share potentially similar binding modes. These findings encourage the use of in vitro human P-gp data for predicting in vivo effects in rodents and thus contributes to the 3Rs (Replace, Reduce and Refine) of animal experiments.
Keywords: Species differences, P-glycoprotein, Binding site comparison, Transmembrane domain, Protein-ligand interaction fingerprint
1. Introduction
The efflux transporter P-glycoprotein (P-gp) is a protein of high interest among other major anti-targets (Cramer et al., 2007). It is expressed in tissues such as intestine, liver, kidney, placenta, testis, and in the capillary endothelial cells of the brain (Seelig, 1998; Thiebaut et al., 1987), and plays an important role in the absorption, distribution and excretion of many drugs. Overexpression of P-gp has been implicated in resistance to multiple chemotherapeutic drugs and is a widely accepted mechanism underlying multidrug resistance (Aller et al., 2009; Fojo et al., 1987; Widmer et al., 2003). Co-administration of a P-gp inhibitor with a drug can lead to altered disposition of the latter, resulting in elevated plasma levels of the drug which could lead to adverse effects (Bussey, 1982; Tsuji, 2002; Verschraagen et al., 1999). Furthermore, the partial blockade of P-gp expressed in the blood–brain barrier or placenta could lead to an increased distribution of a co-administered drug in the corresponding organs. Thus, concomitant administration of substrates and P-gp inhibitors may lead to adverse drug reactions and organ toxicities (Balayssac et al., 2005). In this respect, the United States Food and Drug Administration (US FDA) guidance requires new drug candidates to be routinely screened against P-gp as part of the clinical drug interaction studies (“Clinical Drug Interaction Studies — Study Design, Data Analysis, and Clinical Implications Guidance for Industry,”, 2017; Klepsch et al., 2011). Therefore, computational methods that characterize P-gp interactions and thus guide the prioritization of compounds in the early phase of the drug discovery process are of considerable interest (Schneider, 2010).
In early stages of drug development, pharmacokinetic and toxicity profiles of a candidate drug are evaluated in animal models (typically rats or mouse) prior to the clinical phases of testing in humans. A substantial amount of experimental data against human P-gp is already available and has been utilized for the development of in silico models (see e.g. livertox.univie.ac.at). However, besides developing in silico models for the prediction of ligands for human P-gp, it would be beneficial to also establish models for rat and mouse P-gp in order to predict the outcomes of preclinical animal studies. Unfortunately, limited availability of experimental data for rat and mouse P-gp restricts the development of such models. In this context the question arises, whether predicted interaction profiles of ligands with human P-gp could be transferred to rodent P-gp. This would require a comprehensive comparison of the putative binding sites of the P-gp structures across species. Literature sheds little light on this, suggesting the need for exploration of species-related differences in P-gp mediated drug transport activity (Martignoni et al., 2006; Schwab et al., 2003; Suzuyama et al., 2007).
Inhibition of P-gp activity as a result of drug interactions has been reported in both animals and humans (Bussey, 1982; Choo et al., 2000; Pedersen, 1985), but only a few studies discussed species-related differences in the inhibitory effects on the P-gp function (Chu et al., 2013; Suzuyama et al., 2007; Zolnerciks et al., 2011). A few studies proposed moderate species differences, human vs. rat (Molden et al., 2000), human vs. mouse (Adachi et al., 2001; Lin and Yamazaki, 2003) and also among the three species (human vs. rat vs. mouse) (Katoh et al., 2006), while a few other studies reported no significant differences between human, rat and mouse P-gp (Chu et al., 2013; Feng et al., 2008; Hsiao and Unadkat, 2012). Nicklisch et al. (2016) compared the binding site residues of mouse and Thunnus albacares (Yellowfin tuna) P-gp. Despite the low similarity, of the 15 residues of mouse P-gp that interacted with the inhibitor BDE-100, 13 were conserved in the tuna P-gp sequence, and the remaining two did not play a major role in the binding site. However, it must be noted that only a small number of compounds were tested in these studies. It might thus well be that the inhibitory effects on P-gp-mediated drug transport are subjective to both the chemical structure of substrates/inhibitors and to the species. Moreover, it is not yet clear if the possible species differences in the inhibitory effects of P-gp activity are due to differences in binding site residues of P-gp, which is therefore worth investigating.
To the best of our knowledge, no computational study compared the binding site interaction profiles of P-gp across different species (human, rat and mouse) so far. In this study, we used a structure-based approach to compare their binding sites in order to derive information concerning potential species differences in P-gp-mediated drug transport. Since an X-ray crystal structure is available for mouse P-gp alone, homology modeling was performed to construct the models for human P-gp and for rat P-gp. Subsequently, docking of common inhibitors of rat, mouse and human P-gp was performed. Next, known inhibitors of human P-gp were docked into the models of the three species followed by an analysis of the interactions between the inhibitors and binding site residues. The interaction profiles of the P-gp binding sites of the three species were then compared to evaluate the transferability of in vitro human P-gp data for development of models to predict effects in rat and mouse.
2. Methodology
2.1. Dataset
A substantial amount of human P-gp data is made publicly available through previous literature reports (Broccatelli et al., 2011; Chen et al., 2011; Klepsch et al., 2014). However, due to the limited availability of rat P-gp data in public domain bioactivity databases such as ChEMBL (Gaulton et al., 2012; Willighagen et al., 2013) and BindingDB (Liu et al., 2007), an exhaustive literature search was performed. A total of 18 rat P-gp inhibitors could be identified that are known to also inhibit both human P-gp and mouse P-gp. Due to the inconsistencies in the assay conditions, these compounds unfortunately could not be utilized to compare inhibitory profiles across the species. Suzuyama et al. (2007) studied the species differences (human, monkey, canine, rat and mouse) in the inhibitory effects of the prototype P-gp inhibitors quinidine and verapamil. These two drugs served as the starting point for in silico comparison of binding site interaction profiles across the species. Further, we also extracted the human P-gp data from Broccatelli et al. (2011) in order to perform protein-ligand interaction fingerprint (PLIF) analysis and to identify the common functional group residue interactions among the three species. The dataset was standardized according to the procedure described in Pinto et al., 2012. (Pinto et al., 2012) The final dataset contained a total of 1161 compounds (612 inhibitors and 549 non-inhibitors).
2.2. Homology modeling
Based on sequence identity and resolution, the corrected mouse P-gp structure (mdr1a; PDB ID: 4M1M; UNIPROT ID: P21447) was selected as the most structurally related template protein for constructing the homology models for human P-gp (MDR1; UNIPROT ID: P08183), rat P-gp (MDR1a; UNIPROT ID: Q9JK64 and MDR1b; UNIPROT ID: P43245) and mouse P-gp (mdr1b; UNIPROT ID: P06795). Rat and mouse P-gp proteins are encoded by two paralogous genes namely MDR1a and MDR1b that show a sequence identity of 83% (Chu et al., 2013; Devault and Gros, 1990). Therefore, we constructed in total four homology models to consider the paralogs too. Homology models were constructed using MODELLER 9.13 (Eswar et al., 2007) and the Prime module in Maestro (Schrödinger, Inc. V-10.1.013) (Jacobson et al., 2004, 2002). The energy minimized models were further evaluated using DOPE score (Shen and Sali, 2006) and GA341 score (John and Sali, 2003; Melo et al., 2002). Quality of the stereochemical parameters and the normality of the structures were checked using the PROCHECK program, included in the PDBsum analysis (Laskowski et al., 1993). Ramachandran plot (Zhou et al., 2011) and G-factor (Engh and Huber, 1991), and finally the Q-score (Benkert et al., 2008, 2009) values were evaluated to identify the best homology models. The electrostatic potential surface (EPS) of each of the three best models for the three species was also calculated and compared using MOE 2013 (Molecular Operating Environment (MOE), 2013.08, 2013).
2.3. Sequence alignment
Sequence alignment was performed using ClustalX (Larkin et al., 2007) and verified by including secondary structure predictions. Subsequently, the alignment was analyzed using Jalview (Supplementary Figs. S1–S4) (Clamp et al., 2004; Waterhouse et al., 2009).
2.4. Binding site identification and molecular docking
In order to avoid any bias, the binding site for all five structures (human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a, and mouse mdr1b) was defined as the complete transmembrane region, taking 20 Å around the coordinate of the center point to allow subsequent flexible docking of a series of P-gp inhibitors. The protein was prepared using the Protein Preparation Wizard of the Schrödinger Suite (2015) (Sastry et al., 2013; Schrödinger Release 2015-1: Maestro, version 10.1, Schrödinger, LLC, New York, NY, 2015). Hydrogen atoms were added, and optimal protonation states and ASN/GLN/HIS flips were determined. To assess their correct protonation states, ligands were prepared using the LigPrep module of the Schrödinger Suite, (Schrödinger Release 2015-1: LigPrep, version 3.3, Schrödinger, LLC, New York, NY,2015., 2015, Schrödinger Release 2015-1: Maestro, version 10.1, Schrödinger, LLC, New York, NY, 2015) which produces low-energy 3D structures that can be used for docking. The OPLS_2005 force field was used for minimization of the structures. Different ionization states were generated by adding or removing protons from the ligand at a target pH of 7.0 ± 2.0 using Epik version 3.1., (Greenwood et al., 2010; Shelley et al., 2007) and tautomers were generated for each ligand. To generate stereoisomers, the information on chirality from the input file for each ligand was retained as is for the entire calculation. All docking runs were performed in high-throughput mode with GlideXP (Friesner et al., 2006; Halgren et al., 2004) docking in Maestro. We also used the genetic algorithm-based GOLD suit (version 5.2.0) (Jones et al., 1997; Verdonk et al., 2003) for docking.
2.5. Protein ligand interaction fingerprint (PLIF)
A PLIF summarizes the interactions between a ligand and a protein using a molecular fingerprint scheme. We generated two types of PLIFs that differ in the information encoded. The first PLIF encodes residues involved in an interaction with the ligand at each bit position. The second type encodes the functional group of the ligand that interacts with the residue. For this, the substructure patterns of 100 functional groups (in SMARTS notation) were extracted from the Daylight website (http://www.daylight.com/dayhtml_tutorials/languages/smarts/smarts_examples.html#GROUP). All PLIF bits were calculated with the MOE 2013 (Molecular Operating Environment (MOE), 2013.08, 2013) built-in function CalculateRawInteractions using a 1% threshold for molecular interactions and a 20% threshold for surface contacts. The function was embedded in an in-house SVL script and was post-processed to enable calculation of functional group PLIFs.
3. Results and discussion
Predicting interactions of small molecules with membrane protein structures has always been challenging. Nevertheless, visualization of the 3D models contributes to the comprehension of the physical and chemical properties of these biomolecules, and of their intermolecular interactions with endogenous and exogenous compounds. Due to the lack of crystal structures for human and rat P-gp, homology modeling and computational ligand docking were used to generate structure-based hypotheses for protein-ligand-interactions.
3.1. Homology modeling
ABC transporters are transmembrane proteins that are in general difficult to be resolved via crystallization (Klepsch et al., 2010). In such cases, homology modeling is the method of choice for structure-based studies. The homology models generated in this study resemble the open-inward (or apo/ground) state of P-gp. This state was considered because it resembles the first step of the basic catalytic cycle for drug-binding in P-gp (Wilkens, 2015).
Since January 2014, a refined X-ray structure of a eukaryotic ABC efflux pump, ABCB1 (mouse) is available (Li et al., 2014) (PDB code: 4M1M, resolution: 3.8 Å). High sequence identities with human MDR1 (86%), rat MDR1a (94%), rat MDR1b (82%), mouse mdr1b (83%) and a moderate resolution of 3.8 Å renders 4M1M a reasonable template for homology modeling (Pajeva et al., 2009). Moreover, the secondary structure elements are also conserved among the species. When only the transmembrane domain (TMD) was analyzed, the sequence identity is greater than 85% for all structures (Supplementary Fig. S5). The best models had a normalized Dope score of less than −0.6, G-factors less than −0.12, and Qmean scores of greater than 0.60 (see Table 1). For all modelled structures, the Ramachandran plot (Supplementary Figs. S6–S9) showed excellent results with less than 1.9% of residues in generously allowed or disallowed regions. All of these residues are located in the nucleotide binding domains (NBD) or extracellular loops (ECL) and are therefore not involved in drug binding (Supplementary Figs. S10–S13). Table 1 summarizes the model assessment details for the best structure. The X-ray crystal structure and site directed mutagenesis studies on ABCB1 serve as validity tests for both helix orientation in the template (Ward et al., 2007), and the alignment used for ABC transporter modeling (Supplementary Figs. S1–S4). The homology models as well as the crystal structure displayed a V-shaped structure with analogos domain orientations.
Table 1. Results from the stereochemical validation of the homology models.
| Model | Dope score | G-factor | Qmean score | Residues in generously allowed or disallowed regions (%) |
|---|---|---|---|---|
| Human MDR1 | −0.633 | −0.13 | 0.68 | 1.7 |
| Rat MDR1a | −0.795 | −0.03 | 0.70 | 1.7 |
| Rat MDR1b | −0.703 | −0.16 | 0.65 | 1.8 |
| Mouse mdr1b | −0.808 | −0.06 | 0.69 | 2.0 |
3.2. Sequence alignment and binding site analysis
The amino acid sequence is highly conserved among the three species (Supplementary Fig. S5), suggesting a high structural similarity (see Table 2).
Table 2. Sequence identity/similarity [%] between human, rat and mouse P-gp.
| Human-MDR1 | Rat MDR1a | Rat MDR1b | Mouse mdr1a | Mouse mdr1b | |
|---|---|---|---|---|---|
| Human-MDR1 | 100 | 87/93 | 80/90 | 86/92 | 80/90 |
| Rat MDR1a | 87/93 | 100 | 84/91 | 94/97 | 84/92 |
| Rat MDR1b | 80/90 | 84/91 | 100 | 82/91 | 93/97 |
| Mouse mdr1a | 86/92 | 94/97 | 82/91 | 100 | 83/91 |
| Mouse mdr1b | 80/90 | 84/92 | 93/97 | 83/91 | 100 |
Experimental techniques such as cysteine and arginine scanning and photoaffinity labeling were previously employed to determine the drug binding sites of P-gp (Loo and Clarke, 2008; Pleban et al., 2005; Seeger and van Veen, 2009; Shilling et al., 2006). Multiple binding sites were identified and binding at different sites could lead to different inhibitory effects. Well characterized binding sites are the ones of Hoechst 33,342 and Rhodamine, the so called H-sites and R-site (Loo and Clarke, 2002; Qu and Sharom, 2002). Studies also suggest the presence of an allosteric regulatory site as well as a progesterone and prazosin binding region (Martin et al., 2000; Shapiro et al., 1999). The H-site and R-site residues, (characterized by Ferreira et al. (2013)) of the three species were compared and showed a high sequence identity. This would indicate the similar arrangement of the binding sites residues and thus further pointing to the presence of a similar binding/interaction profile of the inhibitors. Mostly identical or similar residues were present in the five structures. The H-site and R-site had 77% and 65% residues identical within the three species. Those residues which show a difference, have mostly similar properties. For example, Glu180 in mouse mdr1a is replaced with Aspartic Acid in mouse mdr1b and in ratMDR1b. Both residues are charged and have acidic properties. In a few instances, charged (basic) amino acids are replaced by polar (neutral) or hydrophobic (aliphatic) amino acids in the other species but most of these residues did not participate in interactions with docked ligands. In general, the H-site has a higher percentage of charged residues (lysine, histidine, and glutamic acid residues), while the R-site has a high number of glycine, glutamine, and proline residues. Interestingly, threonine and tyrosine were not found in the H-site and R-sites, respectively. A detailed comparison of the H-site and R-site residues of the five structures is shown in Supplementary Table S1. These observations signify the harmony of electrostatic properties and molecular features of the drug recognition site (central binding cavity) in the three species. Supplementary Figs. S14–S18 represents the electrostatic potential surface (EPS) of the substrate recognition area of each of the ABC-transporter models. The EPS of the substrate recognition area in the TMDs of the human model is neutral with negative and weakly positive areas, similar to the EPS of rat and mouse models.
3.3. Molecular docking
In order to analyze the putative binding pocket of the transport protein in the three species, we proceeded with docking of a set of inhibitors. Ligand docking is a commonly used approach to identify ligand-protein interactions. However, in case of P-gp, this appears to be challenging due to various reasons. Firstly, P-gp possesses a high degree of flexibility with a large binding cavity consisting of multiple binding sites. Secondly, it can harbor more than one ligand simultaneously (Loo et al., 2003a; Lugo and Sharom, 2005). And finally, lack of a high resolution crystal structure of human P-gp necessitates the use of homology models, which add additional layers of uncertainty. A large binding pocket could also be seen in a recent structure (PDB id: 4M1M) wherein large cyclopeptides bind at different sites with partially overlapping residues (Li et al., 2014). Some of these residues are identical to those involved in rhodamine or verapamil binding (Loo et al., 2006; Loo and Clarke, 1997). Other studies reported different prazosin binding sites in hamster (Isenberg et al., 2001) and human P-gp (Ambudkar et al., 2003). Overall, it is understood that P-gp possesses a huge binding pocket with at least four distinct binding sites, with TM 6 as the helix primarily involved in binding (Klepsch et al., 2010). Therefore, we considered the complete TMD as drug binding site (DBS) and generated a large number of docking poses to prevent any bias introduced by scoring functions.
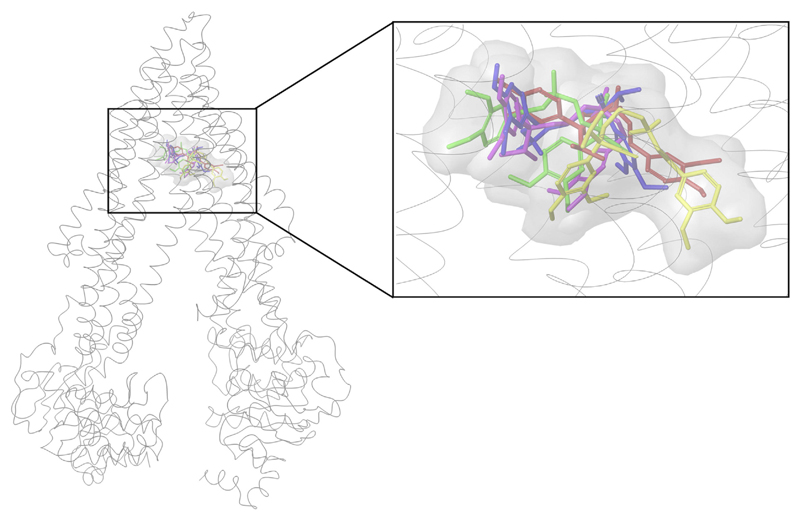
We started with docking of verapamil and quinidine into the binding pocket (complete TMD) of all models. These two compounds were chosen since IC50 values measured under the same assay conditions were available for all three species. Our study revealed that the top ranked docking poses of verapamil were found in the R-site of P-gp in all three species, which is in agreement with previous reports (Ferreira et al., 2013). The top scored docking pose for each of the five models was found in the same region of the binding pocket (R-site) are shown in Fig. 1.
Fig. 1.
Best scored docking pose of verapamil: green (human MDR1), yellow (rat MDR1a), pink (rat MDR1b), red (mouse mdr1a), blue (mouse mdr1b), secondary structure of human P-gp. (For interpretation of the references to color in this figure legend, the reader is referred to the web version of this article.)
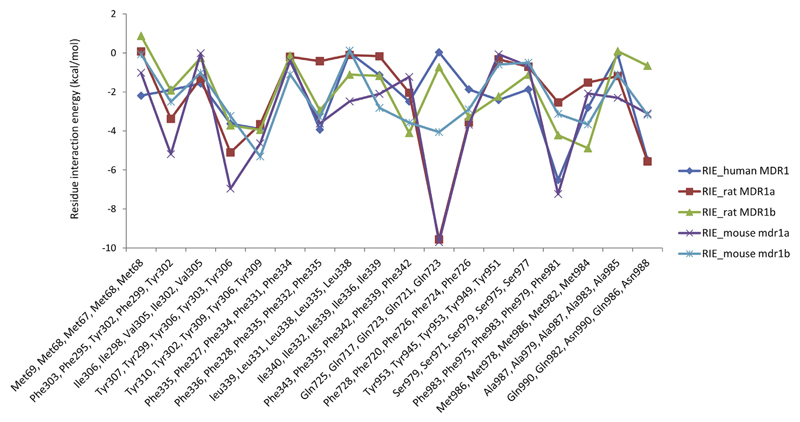
We used the GlideXP scoring function from Maestro (Friesner et al., 2006; Halgren et al., 2004) to evaluate the binding poses. GlideXP docking also provides the per residue interaction energies for a particular docking pose. For each model, the residue interaction energy (RIE) for the top scored docking poses was calculated. Phe303, Tyr307, Tyr310, Phe336, Phe343, Phe728, Phe983, Met986 and Gln990 (numbering according to human-MDR1) are residues that showed more negative interaction energy values in all three species, indicating their higher contribution to binding. Residue interaction energies for all residues which are involved in interactions with verapamil can be seen in Supplementary Table S2. For example, the residues corresponding to Tyr307 in human MDR1 are Tyr306 (RIE:-3.229 kcal/mol), Tyr306 (RIE:-3.714 kcal/mol), Try303 (RIE:-6.96 kcal/mol) and Tyr299 (RIE:-5.1 kcal/mol) in rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b, respectively. Each of these residues contributes to the binding with more negative interaction energy. We observed less negative RIE values with residues which are different within the species (e.g. human MDR1: Met68, rat MDR1a: Leu67, rat MDR1b: Leu66, mouse mdr1a and mdr1b: Met67), suggesting their small influence on - and involvement in - interactions with verapamil. Thus, the comparison of the per residue interaction energies of the best docking pose of the three species revealed that similar binding site residues (as per the alignment) are involved in strong interactions with the ligand (see Fig. 2).
Fig. 2.
Residue interaction energy for common interaction residues in human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b. x-axis denotes residue number in the order human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b, y-axis denotes the corresponding residue interaction energy (kcal/mol).
In case of quinidine, the human MDR1 residues Phe336 (RIE:-2.41 kcal/mol), Gln725 (RIE:-11.577 kcal/mol), Phe728 (RIE:-2.479 kcal/mol), Ser979 (RIE:-1.535 kcal/mol), Phe983 (RIE:-8.114 kcal/mol) and Met986 (RIE:-2.162 kcal/mol) interacted with greater negative interaction energies. The corresponding residues in rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b that demonstrated more negative interaction energies can be found in Supplementary Table S3. Supplementary Fig. S19 shows the RIE for common residues involved in interaction in human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b with the top scored docking pose of quinidine. Replacing a phenyl alanine in human (Phe303) and mouse (Phe299 in mdr1a and mdr1b) with another hydrophobic residue, for instance Tyr302 in MDR1a and MDR1b, still showed negative RIE values. Supplementary Fig. S20 shows the top scored docking poses of quinidine in the five models.
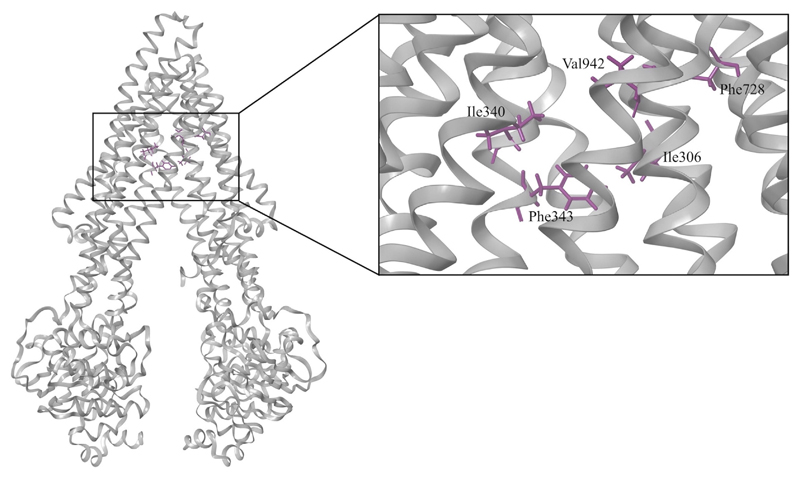
Thus, similar amino acids, as observed with verapamil and quinidine, also confirm the homogeneous nature of the binding site residues in different species. This would further support the hypothesis of similarity in their binding sites. Site directed mutagenesis studies on human ABCB1 also indicated that Ile306 (TMH5) (Loo et al., 2006; Loo and Clarke, 2005), Ile340 (TMH6) (Loo and Clarke, 2002), Phe343 (TMH6) (Loo et al., 2003b, 2006), Phe728 (TMH7) (Loo et al., 2006), and Val982 (TMH12) (Loo and Clarke, 2002, 2005) may participate in ligand binding. As shown in Fig. 3, these residues may form a substrate recognition site in the human ABCB1 model. The involvement of these residues in ligand binding was also confirmed by Li et al. (Aller et al., 2009; Li et al., 2014).
Fig. 3.
Key residues of the substrate recognition site in the human ABCB1 model from literature (Loo et al., 2003b, 2006; Loo and Clarke, 2002, 2005).
In our previous work, we demonstrated that the Chemscore scoring function from the GOLD docking suit facilitated docking-based classification of inhibitors and non-inhibitors for P-gp (Klepsch et al., 2014) and the bile salt export pump (BSEP) (Jain et al., 2017) with reasonable accuracies. Therefore, we used the Chemscore scoring function to perform docking of all human P-gp inhibitors into human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a, and mouse mdr1b structures in order to compare the interaction profiles of the binding site residues in the three species via PLIF analysis.
3.4. Protein ligand interaction fingerprint (PLIF)
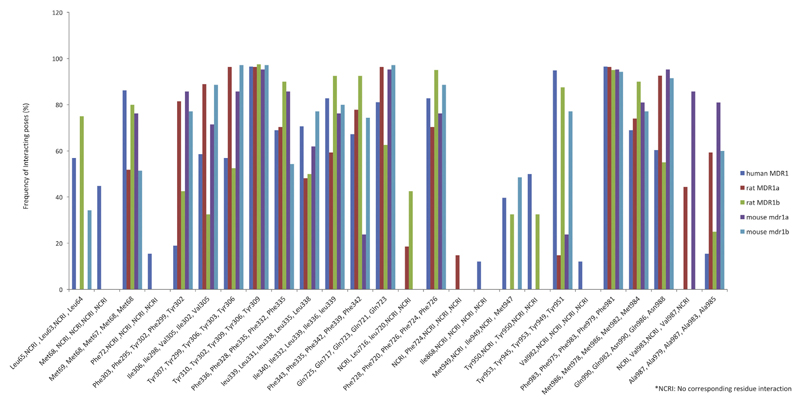
Maestro allows computation of different molecular interactions between binding site residues and a ligand in a specific pose. A PLIF summarizes the interactions between a ligand and a protein using a fingerprint scheme. It provides a detailed picture of the binding modes of different inhibitors. We retrospectively analyzed the PLIFs of complexes of verapamil and quinidine with structures of all three species (human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b) derived from docking, in order to compare their interaction profiles. In case of verapamil, in human MDR1, around 70% of the poses showed hydrophobic interactions with Phe336, Ile340, Phe343, Phe728 and Met986. Also, over 85% of the poses displayed interaction with Met69, Tyr310, Tyr 953, Phe983 (Fig. 4). In the rat structure, more than 73% of the residues showed interaction with Phe328, Phe335, Phe720, Met978 and over 85% residues showed interaction with Phe295, Ile298, Tyr299, Tyr302, Phe975 (Fig. 4). The percentage of binding poses in which specific residues are involved in interactions with verapamil in rat MDR1b, mouse mdr1a, and mouse mdr1b models can also be seen in Fig. 4. Supplementary Fig. S21 provides the same information for quinidine.
Fig. 4.
Hydrophobic interactions common in human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1bfor verapamil. X-axis denotes residue number in the order human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b, Y-axis denotes frequency of interacting residues (%).
For both verapamil and quinidine, the interacting residues that contributed to a significant number of binding poses were at similar positions in the 3D structures of the five transporters (Fig. 4, Supplementary Fig. S21). Specifically, for verapamil, Tyr310 (human MDR1), Tyr302 (rat MDR1a), Tyr310 (rat MDR1b), Tyr309 (mouse mdr1a and mdr1b) showed an interaction in more than 95% of the poses in all five docked structures. Interestingly, when an amino acid in one species was replaced with another amino acid in another species, a similar percentage of docking poses interacted with this residue. For instance, the exchange of Ile340 in human MDR1 with Leucine in rat MDR1b and mouse mdr1b showed hydrophobic interactions in almost 80% of the poses for verapamil, indicating that the interaction pattern did not change when two hydrophobic residues were interchanged. Similar PLIF-based observations could be inferenced after evaluation of the docking poses of quinidine for the three species. However, due to the lower degree of freedom (flexibility) of quinidine, relatively fewer docking poses could be obtained.
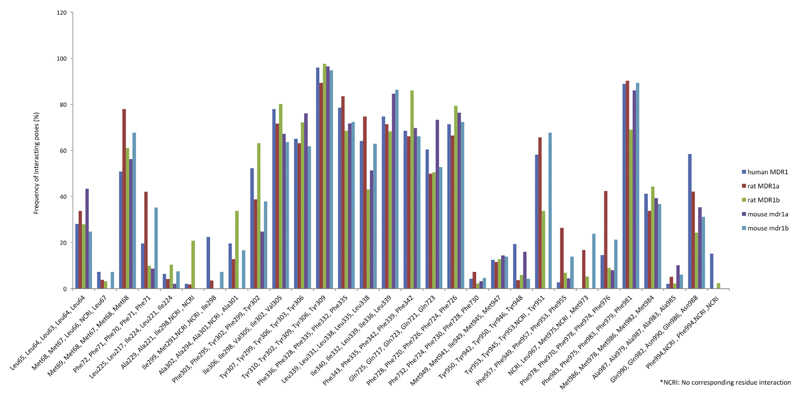
We also identified the interacting residues for a set of 612 human P-gp inhibitors that were docked into these five structures. For human P-gp, more than 70% of the inhibitors interacted with Ile306, Tyr310, Phe336, Phe343, Tyr953, Phe983 and Met986. Fig. 5 shows interacting residues common to human, rat and mouse structures. Supplementary Table S4 lists the occurrence of commonly interacting residues in the three species. PLIFs obtained from docking of a diverse set of human P-gp inhibitors into the five models revealed that similar residues were involved in the interactions, thereby further strengthening the existence of analogous binding site residues and interaction profiles in the three species.
Fig. 5.
Hydrophobic interactions common in human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b for human P-gp inhibitors. X-axis denotes residue number in the order human MDR1, rat MDR1a, rat MDR1b, mouse mdr1a and mouse mdr1b, y-axis denotes frequency of interacting residues (%).
3.5. Analysis of the interactions of functional groups with protein residues
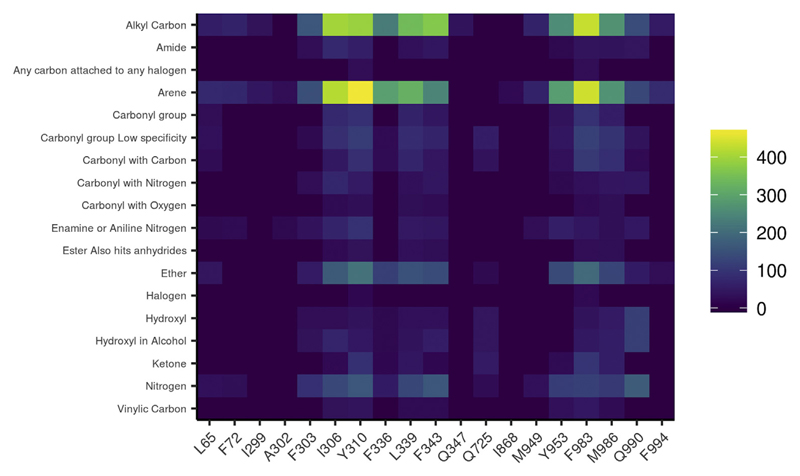
Investigation of the functional group-residue interactions for the set of 612 P-gp inhibitors docked revealed similar interaction patterns with all three species. Functional groups ether, carbonyl, alkyl carbon, nitrogen and arene showed more prominent interactions with Tyr310, Phe343, Phe983, and Met986 (numbering as per human MDR1). Corresponding residues in other species that participated in interactions are shown in Supplementary Table S5. To illustrate the outcomes, a heat map (Fig. 6, Supplementary Figs. S22–S25) plotting the interacting residues against the functional groups of ligands was generated. The color scale of the heat map represents the number of inhibitors involved in a particular interaction between a specific residue and a specific functional group. Thus, the most significant interactions could be visually identified.
Fig. 6.
Heat map illustrating the PLIF analysis of the human P-gp inhibitors for human MDR1. X-axis denotes contact residues. Y-axis denotes functional groups of the ligand which are showing an interaction with the residue. Color scale signifies the number of interacting ligands.
In the three species, the H-site and R-site showed high sequence identity, suggesting the presence of similar residues at specific positions in the 3D structure. Additionally, similar functional group-residue interaction patterns were observed for human (MDR1), rat (rat MDR1a, rat MDR1b) and mouse (mouse mdr1a, and mouse mdr1b) (Supplementary Figs. S22–S25). This further strengthens the idea of utilizing human P-gp activity data, collated from in vitro studies, for structure-based modeling of rodent ligand-target interactions.
Differences in the expression, inhibition and substrate specificity of P-gp across species have already been reported (Chu et al., 2013). As stated earlier, unlike the human P-gp, rat and mouse proteins are encoded by two paralogous genes. These genes are expressed in different organs in the respective species. In humans, MDR1 mRNA is predominantly expressed in the liver, adrenal gland and kidney. In mouse, mRNA analysis suggested that Mdr1a is predominantly expressed in intestine, brain, and testis while Mdr1b is prominently expressed in adrenal gland, ovaries, placenta, and kidney. However both these genes (Mdr1a and Mdr1b) are expressed in liver and heart (Chen et al., 2003; Schinkel et al., 1994). Expression of MDR1/Mdr1 genes is less pronounced in the liver and kidney as compared to other organs in the mouse. On the other hand, enriched expression in the gastro-intestinal tract was reported in rodents, when compared to humans (Bleasby et al., 2006). Ito et al. (2011) and Uchida et al. (2011) reported that the P-gp protein expression was approximately 3-fold higher in mouse as compared to the human brain microvessels. Further, Yamazaki et al. (2001) and Baltes et al. (2007) demonstrated that the antiepileptic drugs phenytoin and levetiracetam were directionally transported by mouse, but not by human P-gp. This would indicate a difference in the P-gp substrate susceptibility between the two species (Baltes et al., 2007). For instance, the lack of P-gp-mediated transport of levetiracetam in rats but not in mouse could be explained by the species differences (Baltes et al., 2007).
A study by Schwab et al. (2003) reported comparable IC50 values in a calcein-AM assay for human MDR1, mouse mdr1a and mouse mdr1b for 28 reference compounds. Zolnerciks et al. (2011) also observed comparable IC50 values for a set of compounds against human and rat P-gp transporter and also suggested that multiple P-gp substrates would be needed to accurately predict clinically significant P-gp drug interactions, in both in vitro and in vivo (including human) drug-drug interaction studies. As mentioned earlier, Suzuyama et al. (2007) evaluated the inhibitory effects of quinidine and verapamil on P-gp-mediated drug transport using MDR1 transfected cell lines of different species. As a common observation, although the IC50 values differ between the species, it was less than 10-fold. This along with our molecular docking and PLIF analysis results signify the possibility of similar interaction profiles in the three species (human, rat and mouse), suggesting the usability and transferability of in vitro human data for development of prediction models for rat and mouse.
4. Conclusion
P-glycoprotein is a transmembrane efflux transporter that plays an important role in drug absorption, disposition, metabolism, and toxicity. It is essential to investigate the interactions of P-gp with candidate drugs not only to understand the contribution of P-gp to the pharmacological properties of candidate drugs, but also to evaluate their drug-drug interaction (DDI) profiles and thereby their clinical implications. In this regard, it is important to understand the binding site interaction profiles of P-gp in rodents which is poorly addressed so far due to the limited availability of experimental data. In this communication, we compared the P-gp binding sites across human, rat and mouse using molecular docking and protein-ligand interaction fingerprint analysis. The suitability of heterogeneous in vitro and in vivo data to model different aspects of human toxicity (or activity), that remained quite challenging, was recently demonstrated (Taškova et al., 2018). However, transferability of human activity data for modeling in vitro and in vivo effects in rodents presents a contrasting perspective and has been so far unexplored. To the best of our knowledge, this is the first in silico study of its kind that compares the binding sites across three different species with emphasis on their inhibitory interaction profile. It is widely acknowledged that the wealth of preclinical toxicity data aggregated by pharmaceutical industry is not completely exploited. With significant increase in the drug development costs and the pressure to reduce the use of animals for experimental testing, development of in silico systems for predicting organ and in vivo toxicity was regarded as the primary goal of the eTOX consortium (www.etoxproject.eu) (Briggs et al., 2012; Hartmann and Pognan, 2017) that ultimately aimed at early identification of potential toxic effects and risk assessment during drug development. In the light of this, exploiting human in vitro data on transporters to model in in vivo effects in animals could significantly reduce the attrition rates as well as the number of animal experiments in preclinical studies.
Our results show a significant overlap between the binding site interacting residues across the three species. This strengthens the likelihood of similar binding mode of human, rat and mouse P-gp inhibitors, thus supporting the transferability of in vitro human P-gp data for development of computational models to predict effects in rat and mouse. As shown recently, the incorporation of predicted ligand transporter interaction profiles increases the performance of selected in vivo toxicity prediction models. The transferability of human P-gp data to rodent in silico models might thus increase the predictivity of rodent in vivo toxicological outcomes. This will subsequently improve the quality of drug candidates while lowering the attrition rate during subsequent phases of drug development, and, most remarkably, reduce the number of animal experiments in preclinical studies.
Supplementary Material
Supplementary data to this article can be found online at https://doi.org/10.1016/j.ejps.2018.06.022.
Acknowledgment
We gratefully acknowledge financial support provided by the Austrian Science Fund, grants #F03502 (SFB35), W1232 (MolTag) and by the Innovative Medicines Initiative Joint Undertaking under grant agreement n°115002 (eTOX).
Footnotes
Conflicts of interest
The authors have no conflicts of interest to declare.
References
- Adachi Y, Suzuki H, Sugiyama Y. Comparative studies on in vitro methods for evaluating in vivo function of MDR1 P-glycoprotein. Pharm Res. 2001;18:1660–1668. doi: 10.1023/a:1013358126640. [DOI] [PubMed] [Google Scholar]
- Aller SG, Yu J, Ward A, Weng Y, Chittaboina S, Zhuo R, Harrell PM, Trinh YT, Zhang Q, Urbatsch IL, Chang G. Structure of P-glycoprotein reveals a molecular basis for poly-specific drug binding. Science. 2009;323:1718–1722. doi: 10.1126/science.1168750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambudkar SV, Kimchi-Sarfaty C, Sauna ZE, Gottesman MM. P-glycoprotein: from genomics to mechanism. Oncogene. 2003;22:7468–7485. doi: 10.1038/sj.onc.1206948. [DOI] [PubMed] [Google Scholar]
- Balayssac D, Authier N, Cayre A, Coudore F. Does inhibition of P-glycoprotein lead to drug–drug interactions? Toxicol Lett. 2005;156:319–329. doi: 10.1016/j.toxlet.2004.12.008. [DOI] [PubMed] [Google Scholar]
- Baltes S, Gastens AM, Fedrowitz M, Potschka H, Kaever V, Löscher W. Differences in the transport of the antiepileptic drugs phenytoin, levetiracetam and carbamazepine by human and mouse P-glycoprotein. Neuropharmacology. 2007;52:333–346. doi: 10.1016/j.neuropharm.2006.07.038. [DOI] [PubMed] [Google Scholar]
- Benkert P, Tosatto SCE, Schomburg D. QMEAN: a comprehensive scoring function for model quality assessment. Proteins. 2008;71:261–277. doi: 10.1002/prot.21715. [DOI] [PubMed] [Google Scholar]
- Benkert P, Künzli M, Schwede T. QMEAN server for protein model quality estimation. Nucleic Acids Res. 2009;37:W510–W514. doi: 10.1093/nar/gkp322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bleasby K, Castle JC, Roberts CJ, Cheng C, Bailey WJ, Sina JF, Kulkarni AV, Hafey MJ, Evers R, Johnson JM, Ulrich RG, et al. Expression profiles of 50 xenobiotic transporter genes in humans and pre-clinical species: a resource for investigations into drug disposition. Xenobiotica Fate Foreign Compd Biol Syst. 2006;36:963–988. doi: 10.1080/00498250600861751. [DOI] [PubMed] [Google Scholar]
- Briggs K, Cases M, Heard DJ, Pastor M, Pognan F, Sanz F, Schwab CH, Steger-Hartmann T, Sutter A, Watson DK, Wichard JD. Inroads to predict in vivo toxicology—an introduction to the eTOX project. Int J Mol Sci. 2012;13:3820–3846. doi: 10.3390/ijms13033820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Broccatelli F, Carosati E, Neri A, Frosini M, Goracci L, Oprea TI, Cruciani G. A novel approach for predicting P-glycoprotein (ABCB1) inhibition using molecular interaction fields. J Med Chem. 2011;54:1740–1751. doi: 10.1021/jm101421d. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bussey HI. The influence of quinidine and other agents on digitalis glycosides. Am Heart J. 1982;104:289–302. doi: 10.1016/0002-8703(82)90205-8. [DOI] [PubMed] [Google Scholar]
- Chen C, Liu X, Smith BJ. Utility of Mdr1-gene deficient mice in assessing the impact of P-glycoprotein on pharmacokinetics and pharmacodynamics in drug discovery and development. Curr Drug Metab. 2003;4:272–291. doi: 10.2174/1389200033489415. [DOI] [PubMed] [Google Scholar]
- Chen L, Li Y, Zhao Q, Peng H, Hou T. ADME evaluation in drug discovery. 10. Predictions of P-glycoprotein inhibitors using recursive partitioning and naive Bayesian classification techniques. Mol Pharm. 2011;8:889–900. doi: 10.1021/mp100465q. [DOI] [PubMed] [Google Scholar]
- Choo EF, Leake B, Wandel C, Imamura H, Wood AJ, Wilkinson GR, Kim RB. Pharmacological inhibition of P-glycoprotein transport enhances the distribution of HIV-1 protease inhibitors into brain and testes. Drug Metab Dispos Biol Fate Chem. 2000;28:655–660. [PubMed] [Google Scholar]
- Chu X, Bleasby K, Evers R. Species differences in drug transporters and implications for translating preclinical findings to humans. Expert Opin Drug Metab Toxicol. 2013;9:237–252. doi: 10.1517/17425255.2013.741589. [DOI] [PubMed] [Google Scholar]
- Clamp M, Cuff J, Searle SM, Barton GJ. The Jalview Java alignment editor. Bioinforma Oxf Engl. 2004;20:426–427. doi: 10.1093/bioinformatics/btg430. [DOI] [PubMed] [Google Scholar]
- Clinical Drug Interaction Studies — Study Design. Data Analysis, and Clinical Implications Guidance for Industry. 2017. [DOI] [PubMed]
- Cramer J, Kopp S, Bates SE, Chiba P, Ecker GF. Multispecificity of drug transporters: probing inhibitor selectivity for the human drug efflux transporters ABCB1 and ABCG2. ChemMedChem. 2007;2:1783–1788. doi: 10.1002/cmdc.200700160. [DOI] [PubMed] [Google Scholar]
- Devault A, Gros P. Two members of the mouse mdr gene family confer multidrug resistance with overlapping but distinct drug specificities. Mol Cell Biol. 1990;10:1652–1663. doi: 10.1128/mcb.10.4.1652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Engh R, Huber R. Accurate bond and angle parameters for {X}-ray protein structure refinement. Acta Crystallogr. 1991;A47:392–400. [Google Scholar]
- Eswar N, Webb B, Marti-Renom MA, Madhusudhan MS, Eramian D, Shen M-Y, Pieper U, Sali A. Comparative protein structure modeling using MODELLER. Curr Protoc Protein Sci. 2007 doi: 10.1002/0471140864.ps0209s50. 2, 9. Editor. Board John E Coligan Al Chapter 2, Unit. [DOI] [PubMed] [Google Scholar]
- Feng B, Mills JB, Davidson RE, Mireles RJ, Janiszewski JS, Troutman MD, de Morais SM. In vitro P-glycoprotein assays to predict the in vivo interactions of P-glycoprotein with drugs in the central nervous system. Drug Metab Dispos Biol Fate Chem. 2008;36:268–275. doi: 10.1124/dmd.107.017434. [DOI] [PubMed] [Google Scholar]
- Ferreira RJ, Ferreira M-JU, dos Santos DJVA. Molecular docking characterizes substrate-binding sites and efflux modulation mechanisms within P-glycoprotein. J Chem Inf Model. 2013;53:1747–1760. doi: 10.1021/ci400195v. [DOI] [PubMed] [Google Scholar]
- Fojo AT, Ueda K, Slamon DJ, Poplack DG, Gottesman MM, Pastan I. Expression of a multidrug-resistance gene in human tumors and tissues. Proc Natl Acad Sci U S A. 1987;84:265–269. doi: 10.1073/pnas.84.1.265. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friesner RA, Murphy RB, Repasky MP, Frye LL, Greenwood JR, Halgren TA, Sanschagrin PC, Mainz DT. Extra precision glide: docking and scoring incorporating a model of hydrophobic enclosure for protein–ligand complexes. J Med Chem. 2006;49:6177–6196. doi: 10.1021/jm051256o. [DOI] [PubMed] [Google Scholar]
- Gaulton A, Bellis LJ, Bento AP, Chambers J, Davies M, Hersey A, Light Y, McGlinchey S, Michalovich D, Al-Lazikani B, Overington JP. ChEMBL: a large-scale bioactivity database for drug discovery. Nucleic Acids Res. 2012;40:D1100–D1107. doi: 10.1093/nar/gkr777. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwood JR, Calkins D, Sullivan AP, Shelley JC. Towards the comprehensive, rapid, and accurate prediction of the favorable tautomeric states of drug-like molecules in aqueous solution. J Comput Aided Mol Des. 2010;24:591–604. doi: 10.1007/s10822-010-9349-1. [DOI] [PubMed] [Google Scholar]
- Halgren TA, Murphy RB, Friesner RA, Beard HS, Frye LL, Pollard WT, Banks JL. Glide: a new approach for rapid, accurate docking and scoring. 2. Enrichment factors in database screening. J Med Chem. 2004;47:1750–1759. doi: 10.1021/jm030644s. [DOI] [PubMed] [Google Scholar]
- Hartmann TS, Pognan F. The eTOX consortium: to improve the safety assessment of new drug candidates. Pharm Med. 2017;2017 [Google Scholar]
- Hsiao P, Unadkat JD. P-glycoprotein-based loperamide-cyclosporine drug interaction at the rat blood-brain barrier: prediction from in vitro studies and extrapolation to humans. Mol Pharm. 2012;9:629–633. doi: 10.1021/mp200563a. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Isenberg B, Thole H, Tümmler B, Demmer A. Identification and localization of three photobinding sites of iodoarylazidoprazosin in hamster P-glycoprotein. Eur J Biochem. 2001;268:2629–2634. doi: 10.1046/j.1432-1327.2001.02155.x. [DOI] [PubMed] [Google Scholar]
- Ito K, Uchida Y, Ohtsuki S, Aizawa S, Kawakami H, Katsukura Y, Kamiie J, Terasaki T. Quantitative membrane protein expression at the blood-brain barrier of adult and younger cynomolgus monkeys. J Pharm Sci. 2011;100:3939–3950. doi: 10.1002/jps.22487. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Friesner RA, Xiang Z, Honig B. On the role of the crystal environment in determining protein side-chain conformations. J Mol Biol. 2002;320:597–608. doi: 10.1016/s0022-2836(02)00470-9. [DOI] [PubMed] [Google Scholar]
- Jacobson MP, Pincus DL, Rapp CS, Day TJF, Honig B, Shaw DE, Friesner RA. A hierarchical approach to all-atom protein loop prediction. Proteins. 2004;55:351–367. doi: 10.1002/prot.10613. [DOI] [PubMed] [Google Scholar]
- Jain S, Grandits M, Richter L, Ecker GF. Structure based classification for bile salt export pump (BSEP) inhibitors using comparative structural modeling of human BSEP. J Comput Aided Mol Des. 2017;31:507–521. doi: 10.1007/s10822-017-0021-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- John B, Sali A. Comparative protein structure modeling by iterative alignment, model building and model assessment. Nucleic Acids Res. 2003;31:3982–3992. doi: 10.1093/nar/gkg460. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones G, Willett P, Glen RC, Leach AR, Taylor R. Development and validation of a genetic algorithm for flexible docking. J Mol Biol. 1997;267:727–748. doi: 10.1006/jmbi.1996.0897. [DOI] [PubMed] [Google Scholar]
- Katoh M, Suzuyama N, Takeuchi T, Yoshitomi S, Asahi S, Yokoi T. Kinetic analyses for species differences in P-glycoprotein-mediated drug transport. J Pharm Sci. 2006;95:2673–2683. doi: 10.1002/jps.20686. [DOI] [PubMed] [Google Scholar]
- Klepsch F, Jabeen I, Chiba P, Ecker GF. Pharmacoinformatic approaches to design natural product type ligands of ABC-transporters. Curr Pharm Des. 2010;16:1742–1752. doi: 10.2174/138161210791163992. [DOI] [PubMed] [Google Scholar]
- Klepsch F, Chiba P, Ecker GF. Exhaustive sampling of docking poses reveals binding hypotheses for Propafenone type inhibitors of P-glycoprotein. PLoS Comput Biol. 2011;7:e1002036. doi: 10.1371/journal.pcbi.1002036. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Klepsch F, Vasanthanathan P, Ecker GF. Ligand and structure-based classification models for prediction of P-glycoprotein inhibitors. J Chem Inf Model. 2014;54:218–229. doi: 10.1021/ci400289j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larkin MA, Blackshields G, Brown NP, Chenna R, McGettigan PA, McWilliam H, Valentin F, Wallace IM, Wilm A, Lopez R, Thompson JD, et al. Clustal W and Clustal X version 2.0. Bioinformatics. 2007;23:2947–2948. doi: 10.1093/bioinformatics/btm404. [DOI] [PubMed] [Google Scholar]
- Laskowski R, Macarthur M, Moss D, Thornton J. {PROCHECK}: a program to check the stereochemical quality of protein structures. J Appl Crystallogr. 1993;26:283–291. [Google Scholar]
- Li J, Jaimes KF, Aller SG. Refined structures of mouse P-glycoprotein. Protein Sci Publ Protein Soc. 2014;23:34–46. doi: 10.1002/pro.2387. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin JH, Yamazaki M. Role of P-glycoprotein in pharmacokinetics: clinical implications. Clin Pharmacokinet. 2003;42:59–98. doi: 10.2165/00003088-200342010-00003. [DOI] [PubMed] [Google Scholar]
- Liu T, Lin Y, Wen X, Jorissen RN, Gilson MK. BindingDB: a web-accessible database of experimentally determined protein–ligand binding affinities. Nucleic Acids Res. 2007;35:D198–D201. doi: 10.1093/nar/gkl999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Identification of residues in the drug-binding site of human P-glycoprotein using a thiol-reactive substrate. J Biol Chem. 1997;272:31945–31948. doi: 10.1074/jbc.272.51.31945. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Location of the rhodamine-binding site in the human multidrug resistance P-glycoprotein. J Biol Chem. 2002;277:44332–44338. doi: 10.1074/jbc.M208433200. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Recent progress in understanding the mechanism of P-glycoprotein-mediated drug efflux. J Membr Biol. 2005;206:173–185. doi: 10.1007/s00232-005-0792-1. [DOI] [PubMed] [Google Scholar]
- Loo TW, Clarke DM. Mutational analysis of ABC proteins. Arch Biochem Biophys. 2008;476:51–64. doi: 10.1016/j.abb.2008.02.025. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Simultaneous binding of two different drugs in the binding pocket of the human multidrug resistance P-glycoprotein. J Biol Chem. 2003a;278:39706–39710. doi: 10.1074/jbc.M308559200. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Methanethiosulfonate derivatives of rhodamine and verapamil activate human P-glycoprotein at different sites. J Biol Chem. 2003b;278:50136–50141. doi: 10.1074/jbc.M310448200. [DOI] [PubMed] [Google Scholar]
- Loo TW, Bartlett MC, Clarke DM. Transmembrane segment 7 of human P-glycoprotein forms part of the drug-binding pocket. Biochem J. 2006;399:351–359. doi: 10.1042/BJ20060715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lugo MR, Sharom FJ. Interaction of LDS-751 and rhodamine 123 with P-glycoprotein: evidence for simultaneous binding of both drugs. Biochemistry (Mosc) 2005;44:14020–14029. doi: 10.1021/bi0511179. [DOI] [PubMed] [Google Scholar]
- Martignoni M, Groothuis GMM, de Kanter R. Species differences between mouse, rat, dog, monkey and human CYP-mediated drug metabolism, inhibition and induction. Expert Opin Drug Metab Toxicol. 2006;2:875–894. doi: 10.1517/17425255.2.6.875. [DOI] [PubMed] [Google Scholar]
- Martin C, Berridge G, Higgins CF, Mistry P, Charlton P, Callaghan R. Communication between multiple drug binding sites on P-glycoprotein. Mol Pharmacol. 2000;58:624–632. doi: 10.1124/mol.58.3.624. [DOI] [PubMed] [Google Scholar]
- Melo F, Sánchez R, Sali A. Statistical potentials for fold assessment. Protein Sci Publ Protein Soc. 2002;11:430–448. doi: 10.1002/pro.110430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Molden E, Christensen H, Sund RB. Extensive metabolism of diltiazem and P-glycoprotein-mediated efflux of desacetyl-diltiazem (M1) by rat jejunum in vitro. Drug Metab Dispos Biol Fate Chem. 2000;28:107–109. [PubMed] [Google Scholar]
- Molecular Operating Environment (MOE): 2013.08. Chemical Computing Group Inc., 1010 Sherbooke St. West, Suite #910; Montreal, QC, Canada: 2013. [Google Scholar]
- Nicklisch SCT, Rees SD, McGrath AP, Gökirmak T, Bonito LT, Vermeer LM, Cregger C, Loewen G, Sandin S, Chang G, Hamdoun A. Global marine pollutants inhibit P-glycoprotein: environmental levels, inhibitory effects, and co-crystal structure. Sci Adv. 2016;2:e1600001. doi: 10.1126/sciadv.1600001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pajeva IK, Globisch C, Wiese M. Combined pharmacophore modeling, docking, and 3D QSAR studies of ABCB1 and ABCC1 transporter inhibitors. ChemMedChem. 2009;4:1883–1896. doi: 10.1002/cmdc.200900282. [DOI] [PubMed] [Google Scholar]
- Pedersen KE. Digoxin interactions. The influence of quinidine and verapamil on the pharmacokinetics and receptor binding of digitalis glycosides. Acta Medica Scand Suppl. 1985;697:1–40. [PubMed] [Google Scholar]
- Pinto M, Trauner M, Ecker GF. An in silico classification model for putative ABCC2 substrates. Mol Inf. 2012;31:547–553. doi: 10.1002/minf.201200049. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pleban K, Kopp S, Csaszar E, Peer M, Hrebicek T, Rizzi A, Ecker GF, Chiba P. P-glycoprotein substrate binding domains are located at the transmembrane domain/transmembrane domain interfaces: a combined photoaffinity labeling-protein homology modeling approach. Mol Pharmacol. 2005;67:365–374. doi: 10.1124/mol.104.006973. [DOI] [PubMed] [Google Scholar]
- Qu Q, Sharom FJ. Proximity of bound Hoechst 33342 to the ATPase catalytic sites places the drug binding site of P-glycoprotein within the cytoplasmic membrane leaflet. Biochemistry (Mosc) 2002;41:4744–4752. doi: 10.1021/bi0120897. [DOI] [PubMed] [Google Scholar]
- Sastry GM, Adzhigirey M, Day T, Annabhimoju R, Sherman W. Protein and ligand preparation: parameters, protocols, and influence on virtual screening enrichments. J Comput Aided Mol Des. 2013;27:221–234. doi: 10.1007/s10822-013-9644-8. [DOI] [PubMed] [Google Scholar]
- Schinkel AH, Smit JJ, van Tellingen O, Beijnen JH, Wagenaar E, van Deemter L, Mol CA, van der Valk MA, Robanus-Maandag EC, te Riele HP. Disruption of the mouse mdr1a P-glycoprotein gene leads to a deficiency in the blood-brain barrier and to increased sensitivity to drugs. Cell. 1994;77:491–502. doi: 10.1016/0092-8674(94)90212-7. [DOI] [PubMed] [Google Scholar]
- Schneider G. Virtual screening: an endless staircase? Nat Rev Drug Discov. 2010;9:273–276. doi: 10.1038/nrd3139. [DOI] [PubMed] [Google Scholar]
- Schrödinger Release 2015-1: LigPrep. version 3.3. Schrödinger, LLC; New York, NY: 2015. [Google Scholar]
- Schrödinger Release 2015-1: Maestro. version 10.1. Schrödinger, LLC; New York, NY: 2015. [Google Scholar]
- Schwab D, Fischer H, Tabatabaei A, Poli S, Huwyler J. Comparison of in vitro P-glycoprotein screening assays: recommendations for their use in drug discovery. J Med Chem. 2003;46:1716–1725. doi: 10.1021/jm021012t. [DOI] [PubMed] [Google Scholar]
- Seeger MA, van Veen HW. Molecular basis of multidrug transport by ABC transporters. Biochim Biophys Acta. 2009;1794:725–737. doi: 10.1016/j.bbapap.2008.12.004. [DOI] [PubMed] [Google Scholar]
- Seelig A. A general pattern for substrate recognition by P-glycoprotein. Eur J Biochem. 1998;251:252–261. doi: 10.1046/j.1432-1327.1998.2510252.x. [DOI] [PubMed] [Google Scholar]
- Shapiro AB, Fox K, Lam P, Ling V. Stimulation of P-glycoprotein-mediated drug transport by prazosin and progesterone. Evidence for a third drug-binding site. Eur J Biochem. 1999;259:841–850. doi: 10.1046/j.1432-1327.1999.00098.x. [DOI] [PubMed] [Google Scholar]
- Shelley JC, Cholleti A, Frye LL, Greenwood JR, Timlin MR, Uchimaya M. Epik: a software program for pK(a) prediction and protonation state generation for drug-like molecules. J Comput Aided Mol Des. 2007;21:681–691. doi: 10.1007/s10822-007-9133-z. [DOI] [PubMed] [Google Scholar]
- Shen M, Sali A. Statistical potential for assessment and prediction of protein structures. Protein Sci Publ Protein Soc. 2006;15:2507–2524. doi: 10.1110/ps.062416606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shilling RA, Venter H, Velamakanni S, Bapna A, Woebking B, Shahi S, van Veen HW. New light on multidrug binding by an ATP-binding-cassette transporter. Trends Pharmacol Sci. 2006;27:195–203. doi: 10.1016/j.tips.2006.02.008. [DOI] [PubMed] [Google Scholar]
- Suzuyama N, Katoh M, Takeuchi T, Yoshitomi S, Higuchi T, Asashi S, Yokoi T. Species differences of inhibitory effects on P-glycoprotein-mediated drug transport. J Pharm Sci. 2007;96:1609–1618. doi: 10.1002/jps.20787. [DOI] [PubMed] [Google Scholar]
- Taškova K, Fontaine J-F, Mrowka R, Andrade-Navarro MA. Evaluation of in vivo and in vitro models of toxicity by comparison of toxicogenomics data with the literature. Methods San Diego Calif. 2018;132:57–65. doi: 10.1016/j.ymeth.2017.07.010. [DOI] [PubMed] [Google Scholar]
- Thiebaut F, Tsuruo T, Hamada H, Gottesman MM, Pastan I, Willingham MC. Cellular localization of the multidrug-resistance gene product P-glycoprotein in normal human tissues. Proc Natl Acad Sci U S A. 1987;84:7735–7738. doi: 10.1073/pnas.84.21.7735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsuji A. Transporter-mediated drug interactions. Drug Metab Pharmacokinet. 2002;17:253–274. doi: 10.2133/dmpk.17.253. [DOI] [PubMed] [Google Scholar]
- Uchida Y, Ohtsuki S, Katsukura Y, Ikeda C, Suzuki T, Kamiie J, Terasaki T. Quantitative targeted absolute proteomics of human blood-brain barrier transporters and receptors. J Neurochem. 2011;117:333–345. doi: 10.1111/j.1471-4159.2011.07208.x. [DOI] [PubMed] [Google Scholar]
- Verdonk ML, Cole JC, Hartshorn MJ, Murray CW, Taylor RD. Improved protein–ligand docking using GOLD. Proteins Struct Funct Bioinforma. 2003;52:609–623. doi: 10.1002/prot.10465. [DOI] [PubMed] [Google Scholar]
- Verschraagen M, Koks CH, Schellens JH, Beijnen JH. P-glycoprotein system as a determinant of drug interactions: the case of digoxin-verapamil. Pharmacol Res. 1999;40:301–306. doi: 10.1006/phrs.1999.0535. [DOI] [PubMed] [Google Scholar]
- Ward A, Reyes CL, Yu J, Roth CB, Chang G. Flexibility in the ABC transporter MsbA: alternating access with a twist. Proc Natl Acad Sci U S A. 2007;104:19005–19010. doi: 10.1073/pnas.0709388104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waterhouse AM, Procter JB, Martin DMA, Clamp M, Barton GJ. Jalview version 2–a multiple sequence alignment editor and analysis workbench. Bioinforma Oxf Engl. 2009;25:1189–1191. doi: 10.1093/bioinformatics/btp033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Widmer N, Colombo S, Buclin T, Decosterd LA. Functional consequence of MDR1 expression on imatinib intracellular concentrations. Blood. 2003;102:1142. doi: 10.1182/blood-2003-03-0993. [DOI] [PubMed] [Google Scholar]
- Wilkens S. Structure and mechanism of ABC transporters. F1000Prime Rep. 2015;7:14. doi: 10.12703/P7-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Willighagen EL, Waagmeester A, Spjuth O, Ansell P, Williams AJ, Tkachenko V, Hastings J, Chen B, Wild DJ. The ChEMBL database as linked open data. Aust J Chem. 2013;5:23. doi: 10.1186/1758-2946-5-23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yamazaki M, Neway WE, Ohe T, Chen I, Rowe JF, Hochman JH, Chiba M, Lin JH. In vitro substrate identification studies for p-glycoprotein-mediated transport: species difference and predictability of in vivo results. J Pharmacol Exp Ther. 2001;296:723–735. [PubMed] [Google Scholar]
- Zhou AQ, O'Hern C, Regan L. Revisiting the Ramachandran plot from a new angle. Protein Sci Publ Protein Soc. 2011;20:1166–1171. doi: 10.1002/pro.644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zolnerciks JK, Booth-Genthe CL, Gupta A, Harris J, Unadkat JD. Substrate- and species-dependent inhibition of P-glycoprotein-mediated transport: implications for predicting in vivo drug interactions. J Pharm Sci. 2011;100:3055–3061. doi: 10.1002/jps.22566. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.