ABSTRACT
Purpose: The recent success of anti-PD1 antibody in metastatic colorectal cancer (CRC) patients with microsatellite instability (MSI), known to be associated with an upregulated Th1/Tc1 gene signature, provides new promising therapeutic strategies. However, the partial objective response highlights a crucial need for relevant, easily evaluable, predictive biomarkers. Here we explore whether in situ assessment of Tbet+ tumor infiltrating lymphocytes (TILs) reflects a pre-existing functional antitumor Th1/Tc1/IFNγ response, in relation with clinicopathological features, microsatellite status and expression of immunoregulatory molecules (PD1, PDL1, IDO-1).
Methodology: In two independent cohorts of CRC (retrospective n = 80; prospective n = 27) we assessed TILs density (CD3, Tbet, PD1) and expression profile of PDL1 and IDO-1 by immunohistochemistry/image analysis. Furthermore, the prospective cohort allowed to perform ex vivo CRC explant cultures and measure by Elisa the IFNγ response, at baseline and upon anti-PD1 treatment.
Results: The density of Tbet+ TILs was significantly higher in MSI CRC, especially in the medullary subtype but also in a subgroup of MSS (microsatellite stable), and positively correlated with PD1 and PDL1 expression, but not with IDO-1. Finally, a high number of Tbet+ TILs was associated with a favorable overall survival. These Tbet+ TILs were functional as their density positively correlated with basal IFNγ levels. In addition, the combined score of Tbet+ PD1+ TILs coupled with IDO-1 expression predicted the magnitude of the IFNγ response upon anti-PD1.
Conclusion: Altogether, immunohistochemical quantification of Tbet+ TILs is a reliable and accurate tool to recapitulate a preexisting Th1/Tc1/IFNγ antitumor response that can be reinvigorated by anti-PD1 treatment.
KEYWORDS: Colorectal cancer, medullary carcinoma, Tbet, microsatellite instability, IFNγ response, explant culture, PD1/PDL1, IDO-1, PD1 blockade
Introduction
Cancer immunotherapy, consisting on the modulation of the immunosuppressive tumor microenvironment using antibodies targeting immune checkpoints such as the PD1/PDL1 axis, has changed the landscape of treatment strategy in diverse advanced tumors including colorectal cancer (CRC). Programmed Death Ligand 1 (PDL1) plays an important role in the inhibition of T-cell mediated immune response, mostly CD8+ cytotoxic T cells, via engagement of the PD1/PDL1 axis,1 leading to tumor immune escape and progression in several malignancies.2–6 The success of immune checkpoint blockade is dependent on the immunogenicity of the tumor.7–9 Thus, MicroSatellite Instable (MSI) or Mismatch Repair deficient (MMRd) CRC have been expected to be potential candidates for anti-PD1/PDL1 therapy due to a dense and vigorous immune microenvironment related to a higher mutation load compared with Microsatellite Stable (MSS) CRC.10,11 The results of the first proof-of-concept study,12 and of the current multicenter phase II clinical trial (Checkmate 142) testing the anti-PD1 monoclonal antibody, nivolumab, in a larger cohort of metastatic CRC patients, have validated the concept that targeting this host immune checkpoint leads to significant objective and durable response in MSI CRC patients, contrasting with a very low response in MSS CRC.13 Based on this promising clinical response, the FDA recently approved nivolumab for the treatment of patients with MSI metastatic CRC. However, responses to PD1 blockers appear to be uneven in these “immunogenic” MSI tumors, with 70% of nonresponders13 and only few MSS patients enrolled. Thus, these results remain perfectible, requiring a better understanding of the determinants of response to these therapies to identify new “fine tuned” predictive biomarkers, a critical challenge to manage MSI but also MSS CRC patients.
Multiple studies in murine models have clearly demonstrated the critical role of interferon gamma (IFNγ), the canonical Th1/Tc1 (T helper 1/type I cytotoxic T lymphocytes) cytokine, in coordinating the anti-tumor immune response by enhancing tumor cell immunogenicity and facilitating tumor recognition and elimination by lymphocytes. Indeed, the engagement of the TCR of tumor-infiltrating T lymphocytes (TILs) – CD8+ (Tc1) but also CD4+ (Th1) – through recognition of a specific tumor antigen/MHC complex induces the release of IFNγ that contributes to the recruitment of effector cells and coordinates the process of innate and adaptive antitumor immune responses.14–17 In humans, compelling evidence also suggests the major role of a Th1/IFNγ immune profile in CRC immunosurveillance. Indeed, based on gene expression profiling, a positive correlation has been found between the expression level of both IFNγ and Tbet – the hallmark Th1 transcription factor that controls the expression of IFNγ -, as well as other genes associated with Th1 adaptive immunity, and an improved prognosis,18,19 irrespective of the microsatellite status of the tumor.11 However, at the same time, IFNγ can compromise anti-tumor immune response by triggering a feedback inhibition loop. Indeed, IFNγ is known to induce the expression by tumor resident cells of immune suppressive molecules such as PDL1 which interacts with the inhibitory receptor PD1 on TILs,20,21 as well as IDO-1 (Indoleamine 2, 3 dioxygenase),22 both able to suppress local effector T-cell function. This dual effect of IFNγ can explain in part the positive correlation between the IFNγ/Th1 gene signature in pretreatment tumor biopsies and the objective responses to PDL1 or PD1 blockade across a wide variety of solid tumors including MSI CRC.23–26
Altogether, these clinical results suggest a major role of a preexisting IFNγ/Th1 tumor gene signature to predict both a favorable clinical outcome and the success of the anti-PD1/PDL1 immunotherapies. However, gene expression profile analyses in daily clinical practice are rarely performed due to some limitations, and the development of simpler and faster methods should be considered. Tbet is well known as a Th1-specific transcription factor required for the generation of CD4+ Th1 T cells, but also controls the generation of cytotoxic CD8+ effector T cells (Tc1) and directly activates IFNγ gene transcription.27,28 Therefore, the assessment of the density of Tbet+ TILs by immunohistochemistry in clinical practice could recapitulate this in situ Th1/Tc1/IFNγ antitumor immune response and predict both prognosis and response to immunotherapy.
To investigate this hypothesis, we performed a multiparametric analysis of the immune microenvironment of CRC, using a combination of quantitative immunohistochemistry and functional analysis on ex vivo explant cultures of CRC, based on two independent cohorts of CRC patients. We assessed the density of Tbet+ TILs in relation with i) the clinicopathological and molecular features of CRC, ii) the expression level of immunoregulatory molecules (PD1, PDL1, IDO-1), and iii) the IFNγ response measured in CRC explant culture supernatants, at baseline and upon PD1 blockade, in relation to the expression level of immunoregulatory molecules.
Results
Clinicopathological and molecular features of patients
Two independent cohorts of CRC patients were explored in this study: a retrospective cohort (n = 80, cohort 1) and a prospective cohort (n = 27, cohort 2) (Figure 1). For the purpose of this study, the retrospective cohort was enriched in medullary carcinomas due to their high density of TILs. Patient and tumor characteristics of these two cohorts are summarized in Table 1. In cohort 1, the majority of MSS CRC corresponded to well/moderately differentiated adenocarcinoma except one case classified as medullary carcinoma. MSI CRC, regardless of their histological subtypes, exhibited the features usually described in this group of CRC, including a majority of right side tumors, preferentially observed in women and elderly patients. Out of the 41 MSI CRC, 39 were sporadic cases, two patients had Lynch syndrome. The prospective cohort included 23 MSS and 4 MSI CRC. MSS CRC included 18 well/moderately differentiated adenocarcinoma, 1 poorly differentiated adenocarcinoma, 3 mucinous and 1 serrated adenocarcinoma. MSI CRC included two poorly differentiated and two mucinous adenocarcinomas.
Figure 1.
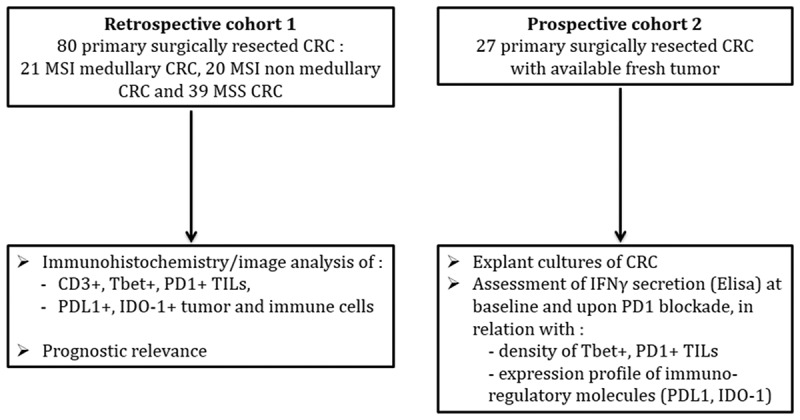
Study design.
Table 1.
Patients and tumor characteristics.
| |
Cohort 1 (n = 80) |
Cohort 2 (n = 27) |
||
|---|---|---|---|---|
| MSI MC(n = 21)N (%) | MSI non-MC(n = 20)N (%) | MSS(n = 39)N (%) | N (%) MSS n = 23MSI n = 4 | |
| Age | ||||
| Mean (range) | 76.2 (35–95) | 77.2 (61–89) | 66.4 (44–87) | 65.4 (34–88) |
| Gender | ||||
| Male | 8 (38.1) | 5 (25) | 22 (56.4) | 15 (56) |
| Female | 13 (61.9) | 15 (75) | 17 (43.6) | 12 (44) |
| Tumor site | ||||
| Right colon | 18 (85.7) | 16 (80) | 12 (30.8) | 8 (30) |
| Transverse colon | 2 (9.5) | 2 (10) | 5 (12.8) | 0 |
| Left colon | 0 | 2 (10) | 19 (48.7) | 16 (59) |
| Rectum | 0 | 0 | 3 (7.7) | 3 (11.1) |
| Small intestine | 1 (4.7) | 0 | 0 | 0 |
| Tumor size | ||||
| (cm) mean (range) | 7.52 (4–11) | 6.1 (3–10) | 5.03 (1.5–10) | 4.9 (1.6–10) |
| Histological subtype | ||||
| Adenocarcinoma NOS: | ||||
| well/mod differentiatedpoorly differentiated | 8 (40)0 | 30 (77) 2 (5) | 18 (66.7)3 (11.1) | |
| Mucinous | 10 (50) | 4 (10.2) | 5 (18.5) | |
| Medullary | 21(100) | 0 | 1 (2.5) | 0 |
| Polymorph | 2 (10) | 1 (2.5) | 0 | |
| Serrated | 0 | 1 (2.5) | 1 (3.7) | |
| pTNM stage | ||||
| I | 2 (9.5) | 0 | 6 (15.4) | 1 |
| II | 11 (52.5) | 7 (35) | 15 (38.6) | 3 |
| III | 6 (28.5) | 9 (45) | 10 (25.5) | 18 |
| IV | 2 (9.5) | 4 (20) | 8 (20.5) | 5 |
| Mutational status | ||||
| WT | 8/19 (42.1) | 6/18 (33.3) | 15/29 (51.7) | 10/27 (37) |
| RAS mutant | 1/19 (5.2) | 3/18 (16.6) | 11/29 (37.9) | 14/27 (51.9) |
| BRAF mutant | 10/19 (52.6) | 9/18 (50) | 2/29 (6.9) | 3/27 (10.3) |
| NI/ND | 2/21 (9.5) | 2/20 (10) | 11/39 (28) | 0 |
WT: wild-type; NI: not interpretable; ND: not done.
The density of Tbet+ TILs is high in MSI and in a subgroup of MSS CRC
In cohort 1, the density of CD3+ and Tbet+ TILs was heterogeneous among CRC. MSI CRC contained a significantly higher number of Tbet+ TILs compared with MSS CRC (Figure 2(a-d)). Among MSI CRC, no significant difference was observed between MC and non-MC CRC (Figure 2(c-d)). Taking into account the Tbet/CD3 ratio, the density of Tbet+ TILs remained significantly higher in MSI than in MSS CRC (Figure 2(e)). The same results were observed in the prospective cohort 2 (Fig. S1). Interestingly, within MSS CRC of cohort 1, 12.8% (5/39) exhibited a high Tbet/CD3 ratio (mean 13.2% ± 1.4 vs 3.2% ± 0.4 in other MSS; Mann–Whitney test, p < 0.0001), exceeding the mean ratio of MSI CRC (8.8%) (Figure 2(e)). This subgroup of MSS CRC with a high Tbet/CD3 ratio corresponded to moderately differentiated adenocarcinomas NOS (n = 3), or mucinous adenocarcinomas (n = 2), mainly of early stage (2 stage I, 2 stage II and 1 stage IV), of the left colon (n = 4) and the majority of them were BRAF or RAS wild type except 1 harboring a RAS exon 2 mutation. In the entire cohort 1 and 2 of MSS CRC, the Tbet/CD3 ratio was quite similar between wild-type and BRAF or RAS mutated MSS CRC (10.5% vs 12%).
Figure 2.
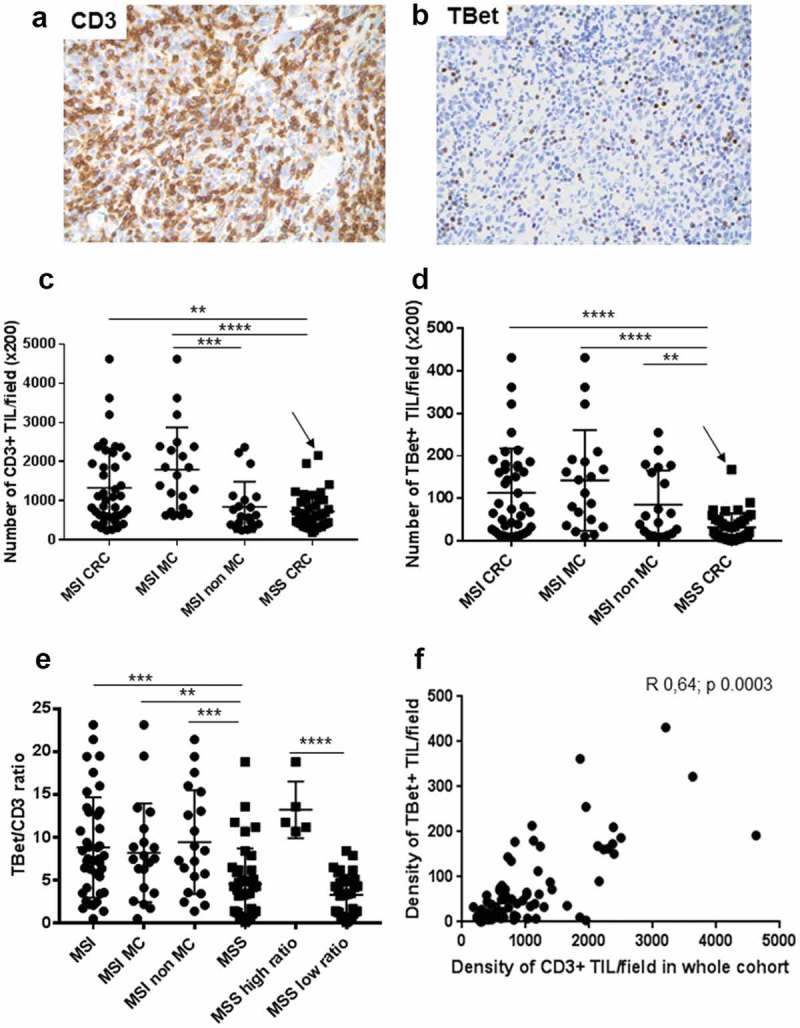
Density of CD3+ and Tbet+ TILs in CRC of cohort 1. (a) (b) Representative examples of an MSI medullary carcinoma infiltrated by numerous CD3+ or Tbet+ TILs (x 200 magnification). (c) (d) Density of CD3+ or Tbet+TILs depending on the microsatellite status and the histological subtypes of CRC. The arrow indicates the MSS medullary carcinoma in each graph. (e) Percentages of Tbet+ TILs among CD3+ TILs in CRC depending on the microsatellite status and histological subtypes. (f) Correlation between CD3+ and Tbet+ TILs (Spearman correlation test). Asterisks indicate the statistical significance between subgroups of CRC (Mann–Whitney test; **p < 0.01; ***p ≤ 0.001; ****p < 0.0001). Bars indicate mean ± SD.
Finally, we found a significant correlation between the densities of CD3+ and Tbet+ TILs in CRC (Figure 2(f)), with the strongest correlation, observed in MSI CRC (r 0.76, p < 0.0001 for all MSI CRC; 0.72, p < 0.0003 in MC and 0.76, p < 0.0001 in non-MC), but also observed in MSS CRC (r 0.46, p 0.002) especially those with a high Tbet/CD3 ratio (r 1, p 0.01). The correlation between CD3+ TILs and Tbet+ TILs in MSI and MSS CRC was also observed in the prospective cohort 2 (Fig. S2).
The density of Tbet+ TILs is an independent prognostic factor for overall survival
Due to the previously reported favorable prognostic impact of the Th1/Tc1 gene signature in CRC,18 we then analyzed the prognostic influence of the density of Tbet+ TILs in the retrospective cohort of CRC (cohort 1). After a 5-year follow-up, 29 patients had died (36.25%), and 20 exhibited a disease progression (25%). In univariate analysis, age (HR 1.05, p 0.008), pTNM stage IV (HR 3.41, p 0.001), and density of CD3+ (cut-off ≥ 591.6/field; HR 0.43, p 0.02) or Tbet+ TILs (median value, ≥ 40/field, HR 0.42, p 0.03) influenced the 5-year overall survival (OS). However, the microsatellite status (MSI, HR 1.86, p 0.1), the density of PD1+ TILs (irrespective of the cut-off value), or the expression of IDO-1 by tumor cells (HR 0.48, p 0.09) did not influence the OS. In multivariate analysis using the cox regression model and taking into account the confounding factors (age, stage, the density of Tbet+ TILs (median value) and microsatellite status), the density of Tbet+ TILs was an independent favorable prognostic factor (Table 2). The Kaplan Meier curves shown in Figure 3(a) confirmed the prognostic value of the density of Tbet+ TILs (threshold = 2nd quartile, median value) in terms of 5-year OS in the whole cohort but also in MSI and MSS subgroups (Figure 3(b, c)).
Table 2.
Multivariate analysis in the whole cohort of patients (Cox regression model).
| Variables | HR | CI | p-value |
|---|---|---|---|
| pTNM stage IV vs. others | 4.52 | 1.91;10.7 | 0.0006 |
| Age | 1.06 | 1.01;1.12 | 0.012 |
| MSI vs. MSS | 2.13 | 0.9;5.04 | 0.08 |
| Density of Tbet ≥40/field | 0.31 | 0.13;0.75 | 0.009 |
| Density of CD3 ≥ 591/field | 0.72 | 0.3;1.74 | 0.466 |
HR: Hazard Ratio; CI: Confidence Interval
Figure 3.
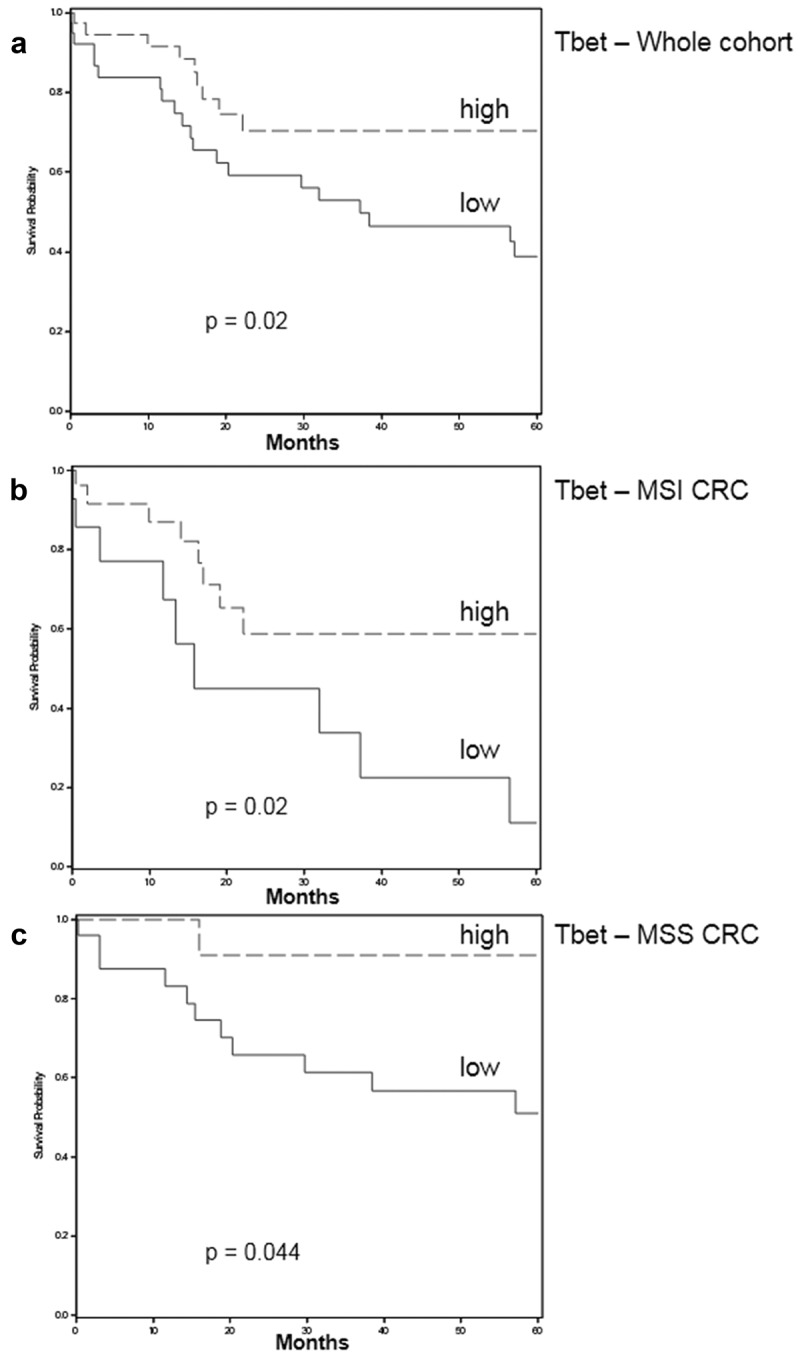
Five-year overall survival rates depending on the density of Tbet+ TILs (Kaplan Meier curves; cut-off: median value ≥ 40/field = high or < 40 = low) in the whole (a) cohort 1 of patients, in MSI (b), or in MSS (c) patients. The Logrank test was used to determine p-values.
Expression profiles of immunoregulatory molecules (IDO-1, PDL1, and PD1) among CRC
IDO-1 expression profile among CRC
In the normal colonic mucosa, IDO-1 was only expressed in some rare mononuclear cells and endothelial cells (Figure 4(a)). In CRC, IDO-1 was aberrantly expressed by tumor cells (Figure 4(b)), and also by some endothelial cells. In only 1 CRC, IDO-1 was also expressed by immune cells (Figure 4(c)). This aberrant expression profile of IDO-1 by tumor cells was observed in about 40% CRC, irrespective of the microsatellite status, and without any difference between MC and non-MC among MSI CRC (Figure 4(d)).
Figure 4.
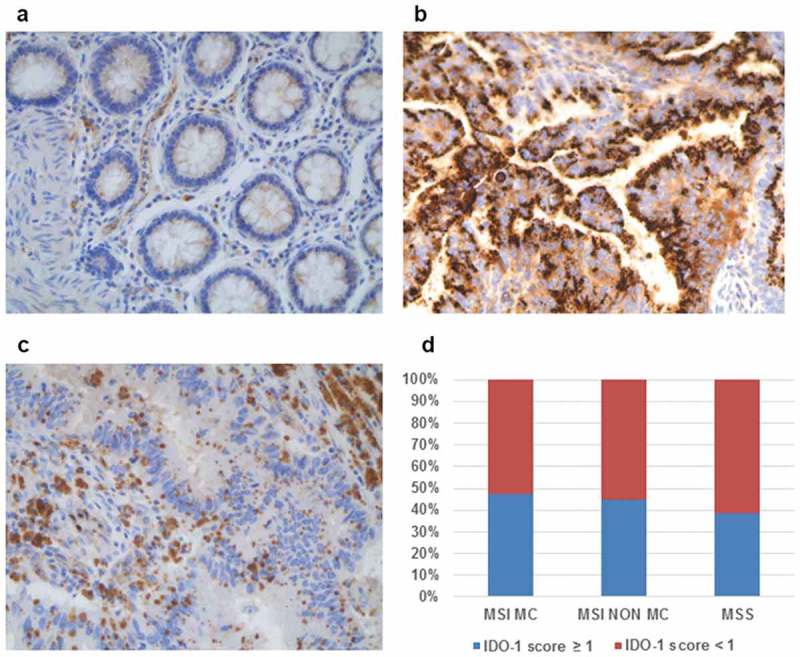
Expression profile of IDO-1 among CRC. (a) In the normal colonic mucosa, epithelial cells do not express IDO-1. (b) In some CRC, tumor cells aberrantly express IDO-1. (c) In rare CRC, IDO-1 was expressed by immune cells (x 200 magnification). (d) The percentage of IDO-1-positive CRC (score ≥ 1 in tumor cells) was similar in MSI (MC and non-MC) and in MSS CRC.
PDL1 expression profile among CRC
In the normal colonic mucosa, PDL1 expression was exclusively observed in some subepithelial macrophages of the lamina propria (Figure 5(a)). In CRC, in contrast to IDO-1, PDL1 expression was extremely heterogeneous among and within CRC. Representative PDL1 expression profiles are shown in Figure 5(b-g). Three expression patterns were observed: absence of PDL1 expression (17.5%), focal expression (51%) or diffuse expression (31%) (Table S1). PDL1 expression by tumor cells and immune cells (TC + IC) was preferentially observed in MSI CRC (78%), especially in MC (90%), compared with MSS CRC (46%) (Figure 6(a,b)). In order to precise the expression profile of PDL1, we investigated separately its expression on tumor cells and immune cells. PDL1-positive tumor cells were exclusively observed in MSI CRC, preferentially in MC frequently exhibiting a strong and diffuse expression pattern, often associated with PDL1+ immune cells (Figures 5(c) and 6(c-d)). In MSI non-MC, PDL1 expression always exhibited a focused pattern (scores 1 to 3) characterized by clusters of PDL1-positive tumor cells localized at the interface with the stroma, especially at the invasion front (Figure 5(e)). We then assessed the possible relation between the strong tumor cell pattern of PDL1 expression observed exclusively in MC and their mutational profile, since some in vitro experiments demonstrated a possible regulation of PDL1 expression by tumor cells through activation of the MEK/ERK pathway due to oncogenic driver mutations (PDL1-mediated innate immune resistance).29 Although the number of PDL1+ MC was higher among mutated (66%, 8/12) than wild-type tumors (33%), the difference was not statistically significant (p 0.17, Fisher exact test). PDL1 expression by immune cells was observed in almost the same proportions in MSI and MSS CRC, a finding contrasting with the tumor cell pattern (Figure 6(e)). PDL1+ immune cells most often surrounded tumor cell nests, in aggregates or in bands, at the invasive front but also in the center of the tumor (Figure 5(g)). The majority of these PDL1+ immune cells expressed CD163 (Figure 6(g)), an activation marker of M2-type Tumor-Associated Macrophages (TAM).30,31
Figure 5.
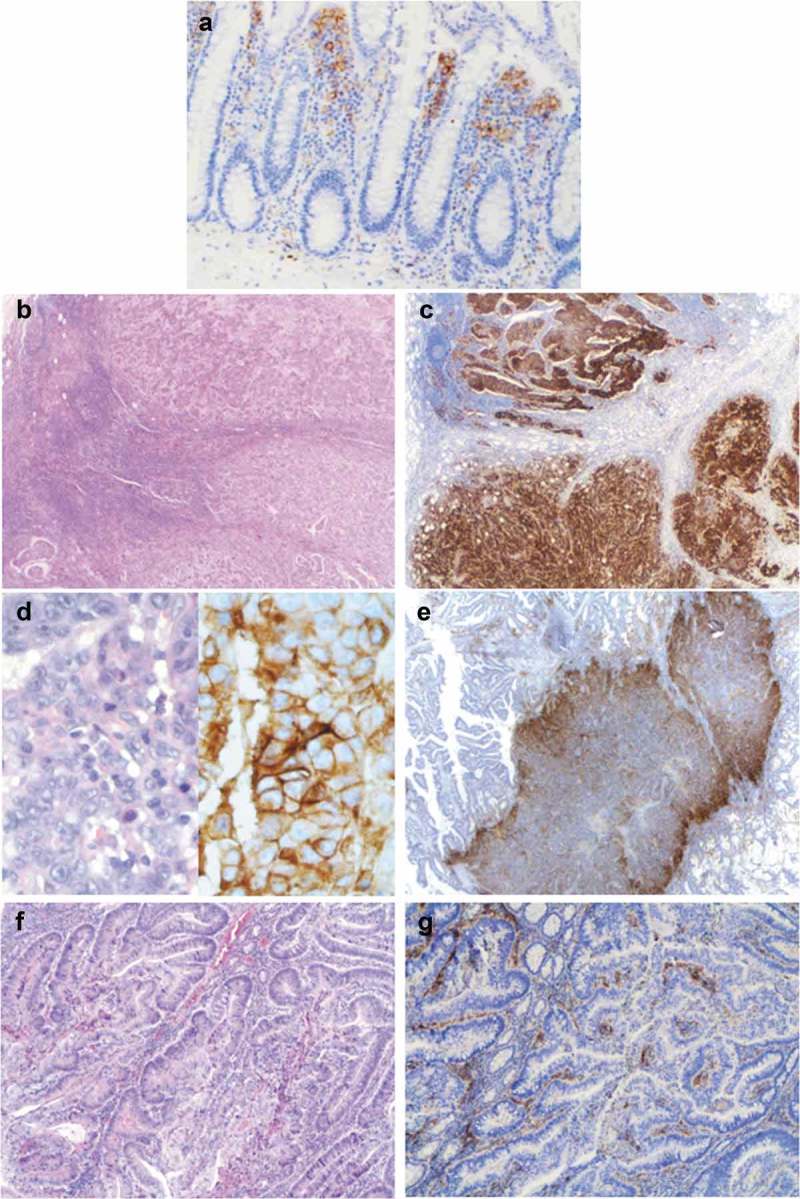
Representative examples of PDL1 expression patterns in CRC. (a) In the normal colonic mucosa, PDL1 was only expressed by subepithelial mononuclear cells, and not by epithelial cells. (b) (c) (d) Diffuse pattern of PDL1 expression by tumor cells (score 4) in an MSI medullary carcinoma (b, c: x250 magnification and d: x400 magnification). Heterogeneous expression of PDL1 by tumor cells (focal pattern) in an MSI, well- to moderately differentiated adenocarcinoma NOS (e: x250 magnification) and by immune cells in the stroma, around tumor glands in an MSS, well-differentiated adenocarcinoma NOS (f, g: x100 magnification).
Figure 6.
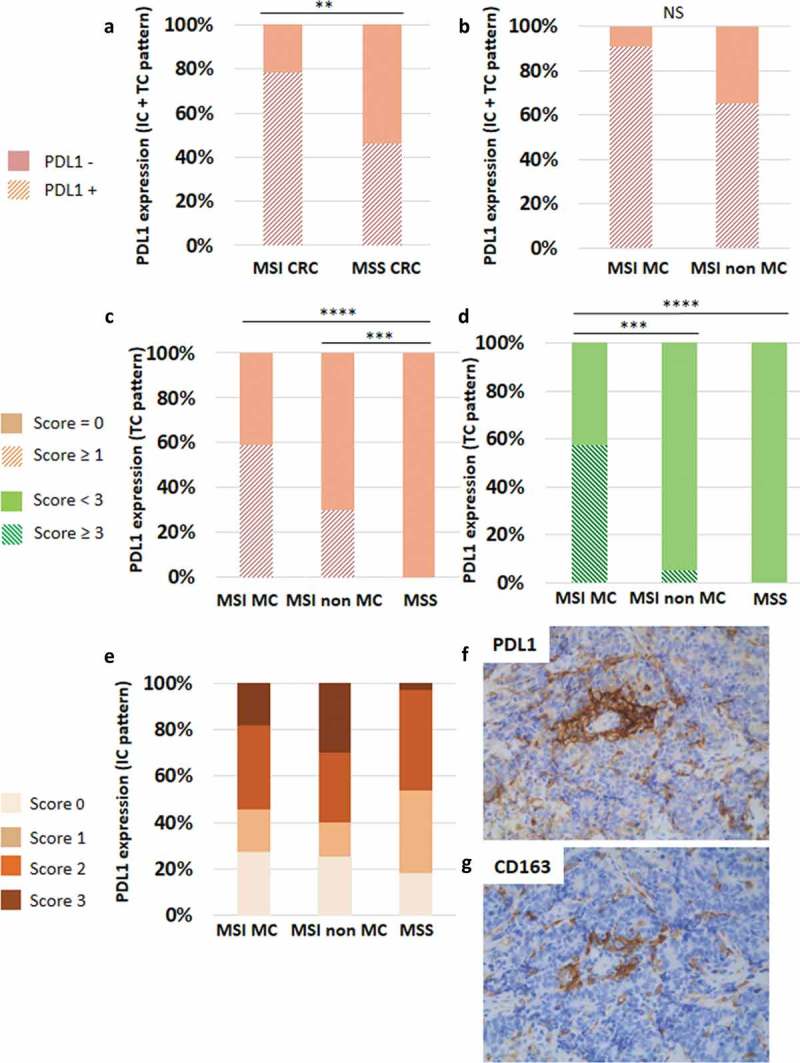
Heterogeneous expression profile of PDL1 in CRC. (a) PDL1 was preferentially expressed in MSI compared to MSS CRC. (b) Among MSI CRC, PDL1 expression was preferentially observed in the medullary subtype of CRC. Considering the expression of PDL1 by tumor cells (TC), only MSI CRC featured this expression pattern (c), especially the medullary subtype (d). PDL1 expression by immune cells (IC) was also heterogeneous. (e) Percentages of PDL1-positive IC among the three groups of CRC, depending on the scores (0 to 3). (f) A representative case of non-MC MSI CRC containing numerous PDL1-positive IC (score 3) surrounding PDL1-negative TC islets. (g) Most of PDL1-positive IC in close contact with TC are CD163+ M2 macrophages (x200 magnification). Asterisks indicate the statistical significance between subgroups of CRC (Mann–Whitney test; **p < 0.01; ***p ≤ 0.001; ****p < 0.0001).
By comparing the expression profiles of PDL1 and IDO-1 among CRC (Table 3), no significant association was found (p 0.5; Fisher exact test). We then assessed the relation between PDL1 or IDO-1 expression profiles and tumor characteristics. Overall, a strong expression of PDL1 (pattern TC + IC, score ≥3) was preferentially observed in large tumors, of early stage, of the right colon, in the medullary subtype, and of MSI/dMMR phenotype. IDO-1 was preferentially expressed in right-sided CRC. The main clinicopathological and molecular features of PDL1- or IDO1-positive tumors are summarized in Table S2.
Table 3.
Expression profiles of PDL1 and IDO-1 in CRC (cohort 1).
| IDO-1 + PDL1+ | IDO-1 – PDL1 + | IDO-1 + PDL1 - | IDO-1- PDL1 - | |
|---|---|---|---|---|
| MSI MC | 43% (9/21) | 47% (10/21) | 4.7% (1/21) | 4.7% (1/21) |
| MSI non MC | 40% (8/20) | 35% (7/20) | 15% (3/20) | 10% (2/20) |
| MSS | 38% (15/39) | 43.5% (17/39) | 2.5% (1/39) | 15% (6/39) |
PD1 expression profile among CRC
The density of PD1+ TILs, as well as the ratio PD1/CD3, were higher in MSI than in MSS CRC (Figure 7(a-c)). Among MSI CRC, the density of PD1+ TILs was higher in MC than in non-MC (Figure 7(b)), whereas the PD1/CD3 ratio was quite similar between these two subgroups (Figure 7(c)). Interestingly, the subgroup of MSS CRC with a high Tbet/CD3 ratio was also infiltrated by a higher number of PD1+ TILs than MSS CRC with a low Tbet/CD3 ratio (5.74/field vs 3.4/field). As this co-inhibitory receptor PD1 interacts with PDL1 expressed by tumor cells or immune cells, we assessed the relation between these two markers. As expected, PDL1+ CRC were enriched in PD1+ TILs, surprisingly only when PDL1 was expressed by immune cells (Figure 7(d)). Moreover, we found a positive correlation between the extent of PDL1 expression (TC + IC) and the density of PD1+ TILs, significant only in MSS CRC (r 0.4, p 0.006 in MSS vs r 0.2, p 0.06 in MSI CRC).
Figure 7.
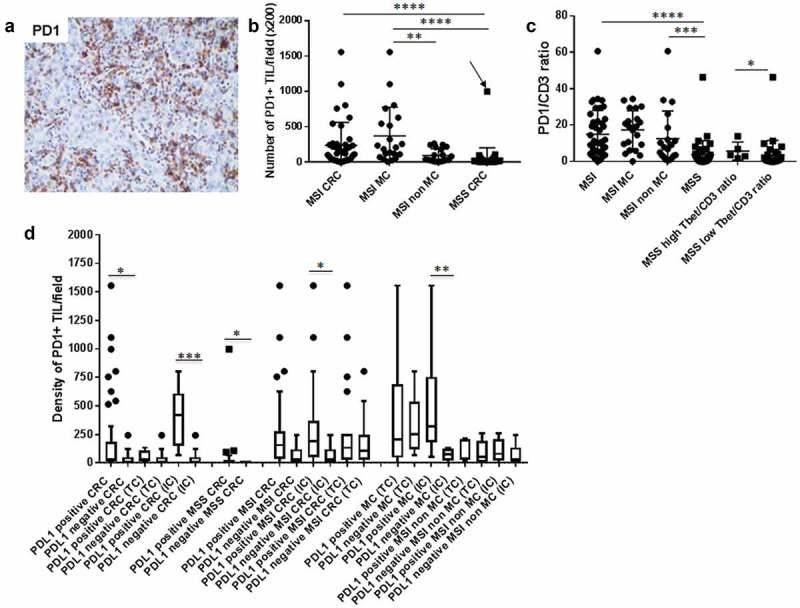
Density of PD1+ TILs among CD3 + T cells in CRC and correlation with PDL1 expression patterns. (a) A representative example of an MSI MC infiltrated by numerous PD1+ TILs. (b) (c) Density of PD1+ TILs and PD1+/CD3+ ratio depending on the microsatellite status and histological subtypes of CRC (arrow indicates the MSS MC). (d) Relationship between the density of PD1+ TILs and PDL1 expression patterns (TC + IC, TC or IC) in MSI and MSS CRC. Lines show medians; Tukey whisker plots. Asterisks indicate the statistical significance between subgroups of CRC (Mann–Whitney test; *p ≤ 0.05; **p < 0.01; ***p ≤ 0.001; ****p < 0.0001).
The density of Tbet+ TILs correlates with PD1/PDL1 expression, but not with IDO-1, irrespective of the microsatellite status of CRC
Since Tbet+ TILs could be the main source of IFNγ, able to induce both PDL1 and IDO-1 expression, we assessed the relation between these markers. As expected, the density of Tbet+ TILs was strongly associated with PDL1 expression, irrespective of the microsatellite status or the PDL1+ cell type (TC or IC) (Figure 8(a)). In addition, Spearman correlation test revealed a significant positive correlation between PDL1 expression and the density of Tbet+ TILs in MSI, but also and more noticeably in MSS CRC (Figure 8(b,c)). These data were confirmed in the prospective cohort 2 (Fig. S3). Moreover, we observed a strong correlation between the density of Tbet+ TILs and the density of PD1+ TILs, in each group of CRC (Figure 8(d-f)). This correlation was also observed in cohort 2 (Fig S2C).
Figure 8.
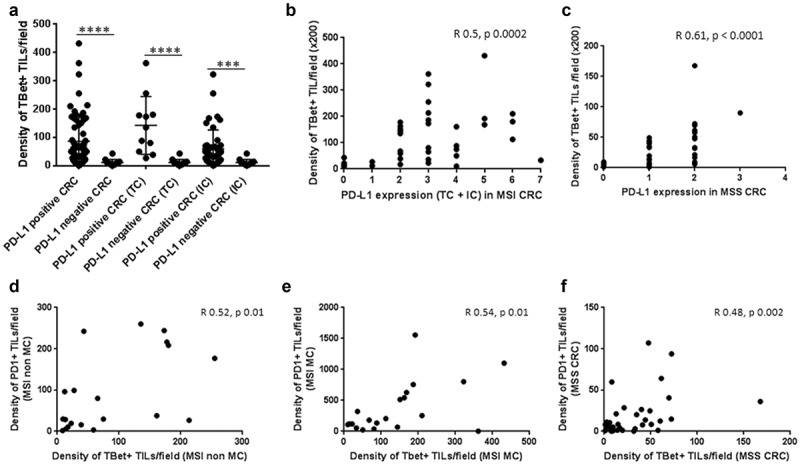
Relation between the density of Tbet+ TILs and the expression of the immunoregulatory markers PD1 and PDL1 among CRC. (a) Relation between Tbet+ TILs and PDL1 expression patterns (TC + IC, TC or IC; Mann–Whitney test; ***p ≤ 0.001; ****p < 0.0001) and correlation between these two markers in MSI (b) and MSS CRC (c). (d) (e) (f) Correlation between Tbet+ TILs and PD1+ TILs in the different subgroups of CRC was assessed by the Spearman correlation test.
However, no significant correlation was found between IDO-1 expression and the density of Tbet+ TILs or PD1+ TILs (data not shown).
The density of Tbet+ TILs correlates with the levels of IFNγ secreted at baseline and under PD1 blockade in CRC explant cultures
In order to confirm the link between the density of Tbet+ TILs and the magnitude of IFNγ response at baseline in CRC, we measured the levels of IFNγ secreted in the tumor microenvironment, in relation with Tbet+ TILs. To this end, we developed explant cultures of CRC, a 3-D model maintaining the interactions between tumor cells and the resident immune cells of the microenvironment. IFNγ was measured by Elisa in the supernatants of the 26 CRC explant cultures of the prospective cohort 2 (4 MSI and 22 MSS) after 24 h culture (baseline IFNγ levels), in relation with the density of Tbet+ TILs. We detected a basal IFNγ secretion in the supernatants of the majority of CRC (21/26) with a mean of 74pg/ml ± 23 (range 3–289). As expected, a positive correlation was observed between the density of Tbet+ TILs and the level of IFNγ secreted (Figure 9(a)). We next investigated the relation between IFNγ secretion and the expression profile of immunoregulatory molecules. Interestingly, we found a positive correlation between IFNγ secretion and PDL1 expression (TC + IC) (Figure 9(c)). In addition, the majority of CRC including three out of four MSI CRC, devoid of IFNγ secretion, exhibited a high density of PD1+ TILs (Figure 9(b)). In order to explore the involvement of the co-inhibitory receptor PD1 in TILs anergy/exhaustion, we blocked PD1 using neutralizing anti-PD1 antibody in eight CRC. As shown in Figure 10(a), PD1 blockade resulted either in an induction or an increase in basal IFNγ secretion, arguing for the presence of exhausted TILs. In addition, in most cases (6/8), the magnitude of IFNγ secretion under PD1 blockade was related both with high Tbet/CD3 and PD1/CD3 ratios. In all eight cases, PDL1 was expressed, mainly by immune cells (Figure 10(b)). However, in two cases, IFNγ was undetectable either at baseline or under PD1 blockade. In one case, the absence of IFNγ response was associated with the absence of Tbet+ TILs (CRC7). At the opposite, in the second case (CRC6), the absence of IFNγ response was associated with a high Tbet/CD3 ratio, no PD1+ TILs but a strong expression of IDO-1 (Figure 10(b)), suggesting the immunosuppressive effect of this enzyme on those TILs.
Figure 9.
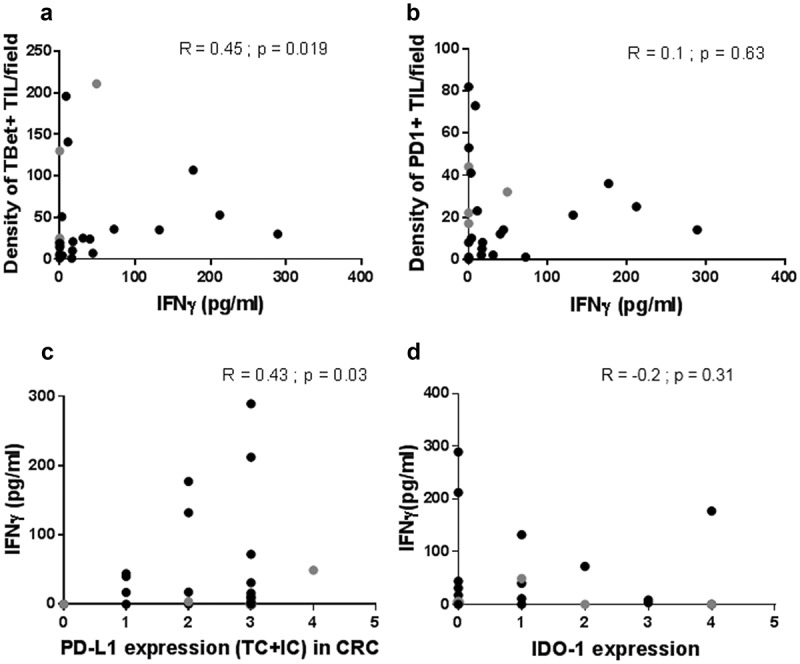
Relation between the IFNγ response of CRC explant cultures and the density of Tbet+ or PD1+ TILs, and the expression profile of PDL1 or IDO-1. The level of IFNγ secreted in 24 h explant cultures of CRC is positively correlated with the density of Tbet+ TILs (a) and the PDL1 expression profile (c). Most of CRC devoid of an IFNγ response displayed high numbers of PD1+ TILs (b) or a high IDO-1 score in tumor cells (d). Black dots: MSS CRC. Grey dots: MSI CRC. Spearman correlation test.
Figure 10.
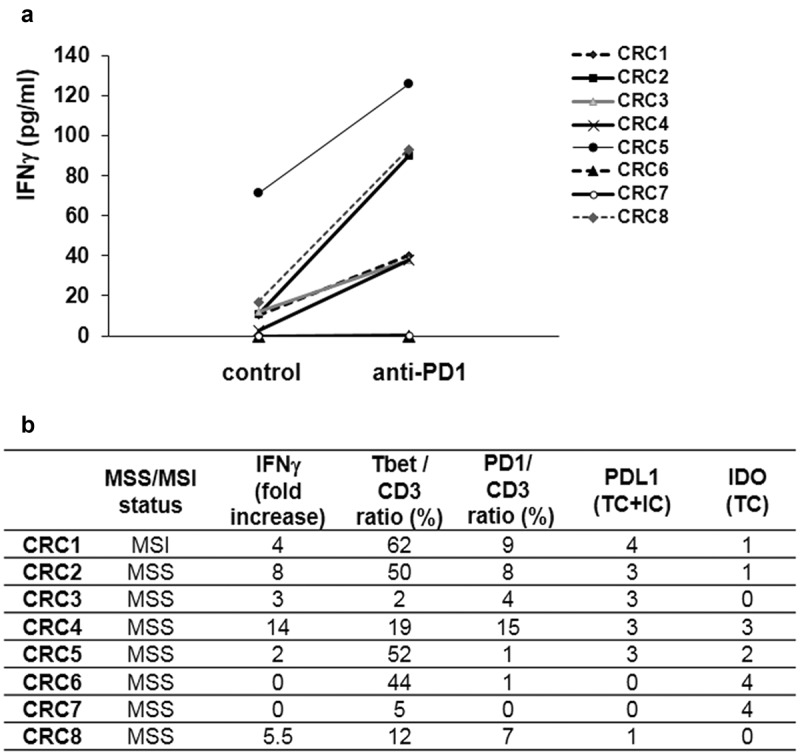
Effect of PD1 blockade on the IFNγ response of CRC explant cultures, in relation with the density of Tbet+ and PD1+ TILs, and with the expression profile of PDL1 and IDO-1. (a) In eight cases (seven MSS, one MSI), explant cultures of CRC were treated or not with an anti-PD1 neutralizing antibody (10 μg/ml for 24 h). (b) Fold increase IFNγ response upon anti-PD1 treatment versus control cultures, in relation with the density of Tbet+ or PD1+ among CD3+ TILs, and with the expression of PDL1 or IDO-1.
Finally, we did not observe any correlation between basal IFNγ secretion and IDO-1 expression (Figure 9(d)), a finding in line with the above-mentioned absence of correlation between the density of Tbet+ TILs and IDO-1 expression, arguing for the presence of other regulatory mechanisms controlling IDO-1 expression.
Discussion
The multiparametric in situ analysis of the immune microenvironment, in relation with the morphological and molecular features of the tumor, represents a major step to better understand the regulation of the antitumor immune response and its clinical relevance in CRC, in terms of tumor progression and therapeutic target in the era of immuno-oncology. More specifically, we are faced with a paradox showing a very low or lack of clinical benefit to PD1 blockade observed in MSS CRC patients and in almost 70% of MSI CRC12,13,32,33 while several compelling evidence suggest that the majority of CRC are immunogenic due to genetic and/or epigenetic alterations.34 However, these disappointing outcomes, especially in MSS CRC, probably resulted from the very limited number of MSS CRC patients enrolled in clinical studies. Importantly, despite this limited experience, disease control and even objective responses were recorded in MSS CRC patients treated with anti-PD1 antibodies, allowing a real promise of those immunotherapies in fine-tuned selected patients.12,13 This apparent paradox highlights the fact that microsatellite status is not perfectly predicting the response to anti-PD1 and thus, identification of new relevant predictive biomarkers is a critical issue to select patients that can likely benefit from this therapy. One of the main goals of immunotherapies is to re-invigorate the pre-existing effector T cells of the anti-tumor immune response, i.e. the activated Th1 and Tc1 in so-called “inflamed tumors”, that can be exhausted or anergized by the engagement of co-inhibitory receptors or other immunosuppressive molecules such as IDO-1. Indeed, as cytotoxic CD8 + T lymphocytes (Tc1) represent the main effectors of antitumor immunity, able to kill off tumor cells, tumor-specific CD4+ Th1 lymphocytes also play an important role for the maintenance, activation and expansion of Tc1 cells, in the draining lymph nodes during the priming phase, but also during the post priming phase at the tumor site. This phenomenon – called CD4 + T cell help –,35,36 involves direct cell–cell interactions between Th1 and Tc1. However, tumor-specific CD4+ Th1 per se may be direct effector cells of antitumor response in developing cytotoxic activity on tumor cells37 via IFNγ secretion. With this knowledge in mind, the simple and accurate in situ evaluation of these strategic immune effector cells – Tc1 and Th1 – could be a more direct indicator of antigenicity and ongoing antitumor immune response, and thus a more sensitive predictive biomarker for response to immune checkpoint inhibition and/or other immunotherapies. The present study provides key findings to support the clinical relevance of the assessment of the density of Tbet+ TILs – a marker that capture both Th1 and Tc1 TILs – by quantitative immunohistochemistry in a predictive and a prognostic setting.
The main findings of the current study, based on a multiparametric descriptive and functional analysis demonstrate that i) quantification of Tbet+ TILs among CD3+ TILs in the tumor microenvironment of primary tumors recapitulates, at the protein level, the prognostic and predictive roles of the Th1/Tc1/IFNγ immune response, irrespective of the microsatellite status, ii) preexisting Tbet+ TILs have a role in local IFNγ secretion that in turn can induce PDL1 expression and thus exhaustion of these effector TILs via engagement of the PD1 receptor and iii) a majority of CRC, irrespective of the microsatellite status, aberrantly express IDO-1 that could participate in the dysfunction of these effector TILs beyond the engagement of PD1.
The current study provides for the first time convincing arguments to support the role of preexisting Tbet+ TILs in local IFNγ secretion, in MSI as well as in MSS CRC. Indeed, we demonstrate a positive correlation between the density of Tbet+ among CD3+ TILs (both intra and peritumoral) and the magnitude of IFNγ secretion at baseline from ex vivo CRC explant cultures. We observed this functional profile in some MSI as expected, but also and most interestingly in a subgroup of MSS CRC (12%) featuring a high Tbet/CD3 ratio as high as in MSI CRC. Thus, our results are in line with and extend those of Galon et al. who clearly demonstrated that a subgroup of MSS CRC exhibits a high Th1/Tc1/IFNγ gene signature, as the majority of MSI CRC.11,18,38 Moreover, we recapture the favorable prognostic impact of this Th1/Tc1/IFNγ gene signature in MSI as well as in MSS CRC by quantifying the density of Tbet+ TILs. The identification of Tbet+ TILs by immunohistochemistry was previously reported in CRC in only few studies, with a higher density in MSI than in MSS CRC in line with our results.39,40 However, these studies did not precise the heterogeneous expression of this biomarker, especially among MSS CRC between inflammatory versus noninflammatory tumors, a major issue in this therapeutic and prognostic context. Furthermore, Ling et al. reported the favorable prognostic impact of Tbet+ TILs in MSI CRC, and observed the same trend (not statistically significant however) in MSS CRC.40 Thus, our results demonstrate that quantification of Tbet+ can recapitulate the favorable ongoing IFNγ response induced by Th1/Tc1 TILs, more frequent in MSI but also in some “inflamed” MSS CRC. To further characterize these “inflamed” MSS CRC, especially the potential immunogenicity of BRAF or RAS mutations in those tumors, we correlated the Tbet/CD3 ratio with the tumor BRAF or RAS mutational status but did not find any significant difference between mutated or wild-type MSS CRC. These findings suggest that BRAF or RAS mutations do not influence solely the number of in situ Th1/Tc1 TILs and that MSS inflamed CRC could harbor other more immunogenic mutations.
In addition, our ex vivo experiments clearly demonstrated for the first time that a high number of both preexistent Tbet+ and PD1+ TILs among CD3+ TILs is highly predictive of either an induction or an improvement of this IFNγ response under PD1 blockade, irrespective of the microsatellite status. Indeed, the majority of CRC devoid of IFNγ secretion exhibited either no preexisting Tbet+ TILs and could be low immunogenic CRC corresponding to “immune desert” tumors,41,42 or a high preexisting number of both Tbet+ and PD1+ TILs, reflecting an exhaustion of these Th1/Tc1 TILs by engagement of this co-inhibitory receptor. Thus, our ex vivo experiments argue that a combined assessment of Tbet (that recapitulates the adaptive Th1/Tc1/IFNγ response) and PD1 by a reproducible quantitative immunohistochemical analysis is more effective than the microsatellite status to predict the response to anti-PD1 therapy. Indeed, based on these ex vivo experiments, CRC patients exhibiting a high Tbet and PD1 immune score can be potentially eligible to anti-PD1 therapy. Our results deepen and complete, by ex vivo functional proofs, some previous descriptive and clinical data based on gene expression profiling. Indeed, the Th1/Tc1 gene signature is associated with the expression of multiple immune checkpoints including the PD1/PDL1 axis that counterbalance this antitumor immune response,10 in MSI but also in some MSS CRC.11 The relation between the clinical response to anti-PD1 and an upregulated Th1/Tc1 or IFNγ gene signature has been demonstrated in pretreatment biopsies in multiple tumor types including CRC.23–26,43
Thus, our findings are of major clinical importance as up to now, this adaptive Th1/Tc1/IFNγ immune response was only assessed by specific gene expression, a time-consuming and expensive approach, difficult to perform in daily clinical practice on FFPE tissue samples. Thus, this combined score – Tbet/CD3 and PD1/CD3 – measurable by an automatic computer-assisted method that minimizes observers’ bias, could be a valuable tool in clinical practice to define subsets of CRC that might be more amenable to PD1 blockade. Therefore, a prospective evaluation of this Tbet-PD1 score as a predictive biomarker could be investigated in ongoing or future clinical trials testing the efficacy of anti-PD1/PD-L1 immunotherapies, especially to define the most appropriate cut-off value of this combined score, in order to capture more likely responder patients.
PDL1 expression was highly heterogeneous among and within CRC, depending on the morphological subtypes of CRC, the microsatellite status and the PDL1+ cell type (tumor and/or immune cells). Indeed, our current results showing a high rate of PDL1+ CRC (78% of MSI CRC including 90% of medullary carcinomas, and 46% of MSS CRC) are not in agreement with previous studies, although the same monoclonal antibody (E1L3N) and the same 5% cut-off value have been used.44–47 Possible explanations for these conflicting results could be either technical issues (mainly Tissue Micro-Arrays vs whole tissue sections) and/or interpretation criteria (staining intensity, membranous vs cytoplasmic staining, tumor vs immune PDL1+ cells). Only the recent studies reported by Korehisa et al. and Valentini et al.48,49, based on whole tissue sections, are consistent with our results, showing that PDL1 expression should be evaluated on whole tissue sections to take into account its spatial heterogeneity. Furthermore, our results showing a significant association between PDL1 expression by immune cells, but not by tumor cells, and the density of PD1+ TILs, suggest the importance of assessing the PDL1 expression pattern by immune cells, more frequent than the tumor cell pattern in CRC (except in medullary carcinomas), to better appreciate its biological impact on the antitumor immune response. The lack of correlation that has been reported between the clinical response to anti-PD1 and PDL1 expression12,13,32 could be due both to the absence of a standardized score of PDL1 expression by immune cells and to its heterogeneity. In addition, assessing PDL1 expression in isolation probably offers only limited insight into the biology of the immune microenvironment.
Some experimental studies in murine models or tumor cell lines have previously demonstrated the key role of IFNγ in the induction of immunoregulatory molecules, especially IDO-1 or PDL1. This mechanism, first described in melanoma,50 corresponds to an adaptive immune resistance that inhibits the adaptive antitumor immune response via PD1 engagement and favors tumor progression.20,21 In addition, it was also demonstrated in Esophageal Squamous Cell lines that the EGFR-Ras-Raf-Erk signaling pathway participates in the regulation of PDL1 expression,51 and that BRAF inhibitors in melanoma patients induce a down-regulation of PDL1 expression.52 Some of our results support the first hypothesis since PDL1 expression was positively correlated with both basal levels of IFNγ and the density of Tbet+ TILs, and since no correlation was observed between PDL1 expression and BRAF/RAS mutational status, a finding in line with other studies.49 However, the strong and specific tumor cell pattern of PDL1 expression in almost all MSI medullary carcinoma, previously observed by others but at a lower and variable frequency (18% to 62% cases)46,49,53, raises the issue of its oncogenic regulation, taking into account the high rate of BRAF mutations we found in these CRC. Indeed, this association has been previously reported by Valentini et al.49 However, we failed to demonstrate such an association between PD-L1 expression and BRAF mutations in our series of MC CRC. Therefore, we cannot exclude that BRAF mutations modulate PDL1 expression in tumor cells in MC through activation of the MAPK signaling pathway. Beyond this potential oncogenic regulation, the hypothesis of an immune-regulated mechanism is sustained by our demonstration of a high density of effector TILs especially Tbet+ TILs and PD1+ TILs, in MC CRC, also associated with a high number of PDL1+ immune cells. In any case, with this “ideal” adaptive immune profile – Tbet high, PD1 high and PDL1 high – associated with a high BRAF mutation rate, medullary carcinoma could represent a target of choice for anti-PD1 therapy, as BRAF-mutated CRC exhibit a high clinical response rate upon nivolumab.13
Finally, regarding IDO-1, an immunosuppressive enzyme that catabolizes tryptophan, an essential amino acid for vital cellular function, and induces consequently the suppression of T cell function, its expression in cancer might be a potential new candidate for immunotherapy. Here we show that a large proportion of CRC among MSI as well as MSS subgroups aberrantly overexpress IDO-1, especially in tumor cells, a result in line with previous studies using the same monoclonal antibody.54 Surprisingly, as IFNγ is widely considered to be the major inducer of IDO-1 in most cells,55 especially in tumor cell lines,22,56 we did not find any correlation between IDO-1 expression and IFNγ levels (either basal or induced) or Tbet+ TILs density, suggesting that IDO-1 expression is not a surrogate marker of an ongoing antitumor Th1/Tc1 immune response, and that the adaptive Th1/Tc1/IFNγ immune microenvironment is not the only modulator of IDO-1 expression by tumor cells in CRC. Most interestingly, from a therapeutic point of view, our ex vivo experiments suggest the role of IDO-1 in promoting Th1/Tc1 immune dysfunction since, among CRC explants, one case with no detectable IFNγ contained a high number of Tbet+ TILs, no PD1+ TILs, but a strong expression of IDO-1. Yet, experimental data have demonstrated that IDO-1+ tumor cells can inhibit cytotoxic T cells.57 Thus, this rich Tbet+ immune profile could be first switched on by the new IDO-1 inhibitors that are currently being tested in other solid tumors.
In conclusion, the current study provides evidence that: i) a preexisting Th1/Tc1 immune response is present in some MSI but also in a subgroup of MSS CRC and positively impacts prognosis; ii) this Th1/Tc1 immune response can be recapitulated by the quantification of Tbet+ TILs among CD3+ TILs by immunohistochemistry/image analysis, and iii) a combined assessment of Tbet+ and PD1+ TILs along with IDO-1 expression can predict the exhaustion of this preexisting Th1/Tc1 immune response and thus effectiveness of PD1 blockade and/or IDO-1 inhibition. Altogether, our results highlight the need for an in situ multiparametric approach to identify the most appropriate immune profile able to predict the response to immunotherapy. The clinical relevance of this combined immune profile associating Tbet, PD1, and IDO-1 should be investigated in the future clinical trials testing anti-PD1 or other immunotherapies in order to reinvigorate the preexisting Th1/Tc1 antitumor immune response.
Patients and methods
Patients and samples
One hundred and seven patients were included in the current study in two independent cohorts (Figure 1). For the purpose of the study, the retrospective cohort (cohort 1) was enriched in medullary carcinomas, a morphological subtype of CRC, due to their known high number of TILs. To this end, the database of the Department of Pathology at the CHU of Nantes was searched to identify all cases of surgically resected CRCs of the medullary subtype between 2002 and 2015 using the keywords « medullary carcinoma» (MC) or « poorly differentiated adenocarcinoma ». The cases were reviewed by two specialized gastrointestinal pathologists (JFM, CB) to confirm the diagnosis of MC based on a strict interpretation of the WHO criteria which required that at least 80% of the invasive tumor featured a solid and syncytial growth pattern containing medium-sized cells with vesicular nuclei and prominent nucleoli.58 Finally, we collected a total of 21 MC. In addition, 20 non-MC MSI CRC and 39 MSS CRC resected in our institution served as control groups. The clinicopathological data collected included gender, age at surgery, tumor localization and size, pTNM, histological subtype, and overall survival (OS) data.
For functional experiments, an independent prospective cohort (cohort 2) of patients was enrolled in this study, including 27 successive patients with surgically removed CRC in our institution between 2017 and 2018.
This study was approved by the institutional board of the University Hospital of Nantes. This tissue biocollection has been registered by the French Ministry for Higher Education and Research (DC-2014–2206) with approval from EC (CPP ouest IV – Nantes). Our study was conducted in accordance with Helsinki declaration. Each patient included in this study signed an informed consent.
Immunohistochemistry
Five-micrometer-thick full sections of formalin-fixed, paraffin-embedded tissue were cut from a representative block of tumor for each case included in this study. Immunohistochemistry was performed on an automated platform (DAKO autostainer) according to a standard protocol with the following antibodies: Tbet (clone 4B10) to identify Th1/Tc1 TILs, PD-L1 (clone E1L3N), a clone already validated,59 PD1 (clone nat 105), CD3 (polyclonal rabbit antibody, DAKO), IDO-1 (mouse monoclonal, clone 10.1) and CD163 (clone 10D6) a specific marker of monocyte/macrophage lineage.60 Some paired normal colonic mucosae taken at distance from the tumor were used as controls.
Evaluation of immunostaining scores
PDL1 immunostainings with high or moderate intensity were scored using a semi-quantitative evaluation of the percentage of PDL1+ cells. Membranous PDL1 staining, the most biologically relevant pattern of expression of this transmembrane molecule, was scored separately for tumor cells (score 0 to 4) and stromal cells (score 0 to 3) in a five- and four-tiered scale, respectively (Table 4). IDO-1 immunostainings with high or moderate intensity were also scored using a semi-quantitative evaluation of percentage of IDO-1+ tumor cells (Table 5). Indeed, IDO-1 positive tumor cells were only considered since scarce IDO-1 positive immune cells were noticed within the tumor except in one case. The number of CD3+, Tbet+ and PD1+ TILs were counted with the Digital Pathology (DP) software (Leica), after scanning the 3 most representative microscope fields at a x200 magnification. The average count per field was calculated and recorded, without analyzing separately the center and the invasive front of the tumor since the clinical impact of Tbet+ TILs seems to be independent of the intratumoral subsite.40 Two pathologists (EO and CB) independently scored all cases and discrepant cases were reviewed on a multiheaded microscope.
Table 4.
Semi-quantitative assessment of PDL1 expression by tumor cells or immune cells in CRC.
| Score | Tumor cells | Immune cells |
|---|---|---|
| 0 | <5% | <4 clusters of at least 50 stained immune cells/cm2 or isolated cells |
| 1 | 5–25% | 4 to 5 clusters of at least 50 stained immune cells/cm2 |
| 2 | 26–50% | >5 clusters of at least 50 stained immune cells/cm2 with heterogeneous staining |
| 3 | 51–70% | Diffuse staining |
| 4 | >70% |
Table 5.
Semi-quantitative assessment of IDO-1 expression by tumor cells in CRC.
| Score | Tumor cells |
|---|---|
| 0 | <5% |
| 1 | 6%–25% |
| 2 | 26%–50% |
| 3 | 51%–70% |
| 4 | >70% |
Microsatellite instability and mismatch repair status
Microsatellite instability (MSI) or mismatch repair (MMR) status was performed as part of the diagnosis or for the purpose of the study by PCR or immunohistochemistry, respectively. The microstatellite instability status was determined using a pentaplex PCR with five markers: BAT-25, BAT-26, NR-21, NR-22, NR-24.61 Briefly, genomic DNA was extracted from 10 µm thick tissue sections of formalin-fixed, paraffin-embedded colorectal tumor tissue after manual macrodissection using iPrepTM ChargeSwitch® Forensic kit (Invitrogen), and according to the manufacturer’s instructions. A CRC was considered as MSI (MSI-high) if at least two of these five markers showed MSI.62 The mismatch repair status was assessed by immunohistochemistry using the following antibodies: MLH1 (clone E505), MSH2 (clone FE11), MSH6 (clone EP49) and PMS2 (clone EP51).
Mutational analysis of RAS and BRAF
Mutational analysis of RAS and BRAF genes was completed for CRC that was not previously tested in clinical practice. Ten µm tissue sections were prepared from Formalin Fixed Paraffin Embedded (FFPE) tumors. DNA was extracted using the Maxwell DNA FFPE kit (Promega, France) according to the manufacturer’s recommendations. Molecular testing was performed by Sanger sequencing of KRAS and NRAS exons 2, 3 and 4. For the latest samples tested, a prescreening was performed using the RAS mutations screening panel (RAS-RT50, Entrogen, Woodland Hills, CA). This PCR-based assay uses allele-specific probes to identify the presence of mutations in KRAS (19 mutations) and NRAS (13 mutations). This assay is approved for in vitro diagnosis (CE-IVD) and is used in routine practice in our laboratory, which is accredited in accordance with the International Standard ISO15189.
Ex vivo CRC explant cultures
Fresh fragments of tumor were collected from the 27 CRC patients included prospectively in the study. These fragments (40 mg, n = 3 to 4 per patient) were maintained in culture for 24 h in RPMI/HAMF12 + Bovine Serum Albumin (BSA) and antibiotics (penicillin/streptomycin) in 95% O2 – 5% CO2 humid atmosphere on a rocking platform at low speed, according to a protocol adapted from that of human normal colonic mucosa we previously validated.63,64 In eight cases of these CRC with a sufficient amount of tissue, the effects of anti-PD1 blocking antibody (Biolegend; 10μg/ml) were assessed in 24 h explant cultures. The supernatants of all explant cultures were collected, centrifuged, stored at −80°C and further analyzed for IFNγ secretion.
IFNγ ELISA
IFNγ was measured in the explant culture supernatants using an ELISA (Diaclone). Results are expressed as pg/ml as the mean of 3 to 4 explants per condition and per patient. IFNγ levels were correlated with the number of CD3+, Tbet+ or PD1+ TILs and the expression profile of PDL1 and IDO-1, assessed by immunohistochemistry and scored as described above.
Statistics
Clinical and pathological information, as well as the results from the quantification of CD3+, Tbet+, PD1+ TILs and PDL1, IDO-1 expression, were entered into a database. The relationship between the number of CD3+, Tbet+, PD1+ TILs, PDL1 and IDO-1 status, clinicopathological features, microsatellite status, and mutational status were assessed using Chi-square test or Fisher’s exact test for qualitative variables and a nonparametric Mann–Whitney test for continuous variables. Correlations between PDL1 or IDO-1 expression and the density of the different TILs were tested by the Spearman test. All statistical analyses were performed using GraphPad Prism 7 software. Five-year OS was measured from date of surgery to date of death related to CRC or latest follow-up. The prognostic influence of the different parameters (stage, age, PDL1 or IDO-1 expression and CD3+, Tbet+ or PD1+ TILs) in terms of OS was evaluated using the Kaplan–Meier method and compared by the log-rank test according to two cut-offs (median and first quartile) and the most significant was chosen. For CD3+ TILs, as the median value was not the best cut-off value, we searched for a new cut-point for patient stratification. Strategy has consisted in determining the appropriateness of a cut-point model (graphical diagnostic plot) and, if relevant, estimate this cut point based on the method proposed by Contal and O’Quigley (macro SAS % findcut).65
Cox regression model was used to perform univariate analysis, and multivariate analysis was performed using a Cox proportional hazards regression model including all factors with p < 0.1. A p value of less than 0.05 was considered as statistically significant. *p ≤ 0.05; **p < 0.01; ***p ≤ 0.001; ****p < 0.0001.
Funding Statement
This work was supported by the Cancéropole Grand Ouest [Réseau Immunothérapie - Amgen RC16-0212-1];Ligue contre le cancer [Grand Ouest (44, 37, 29)], and DHU Oncogreffe CHU Nantes [RC14-0416-1]. LB is the recipient of a CIFRE PhD grant from Roche and ANRT [Cifre PhD thesis].
Acknowledgments
The authors thank the Department of Pathology of CHU de Nantes for expert technical assistance in immunohistochemistry and the MicroPicell facility (SFR Bonamy, Nantes) for assistance in image analysis.
Disclosure of Potential Conflicts of Interest
No potential conflicts of interest were disclosed.
Supplementary material
Supplemental data for this article can be accessed on the publisher’s website.
References
- 1.Topalian SL, Taube JM, Anders RA, Pardoll DM.. Mechanism-driven biomarkers to guide immune checkpoint blockade in cancer therapy. Nat Rev Cancer. 2016;16(5):275–287. PMID:27079802. doi: 10.1038/nrc.2016.36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Ghebeh H, Mohammed S, Al-Omair A, Qattan A, Lehe C, Al-Qudaihi G, Elkum N, Alshabanah M, Bin Amer S, Tulbah A, et al. The B7-H1 (PD-L1) T lymphocyte-inhibitory molecule is expressed in breast cancer patients with infiltrating ductal carcinoma: correlation with important high-risk prognostic factors. Neoplasia. 2006;8(3):190–198. PMID:16611412. doi: 10.1593/neo.05733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Wu C, Zhu Y, Jiang J, Zhao J, Zhang XG, Xu N. Immunohistochemical localization of programmed death-1 ligand-1 (PD-L1) in gastric carcinoma and its clinical significance. Acta Histochem. 2006;108(1):19–24. PMID:16530813. doi: 10.1016/j.acthis.2006.01.003. [DOI] [PubMed] [Google Scholar]
- 4.Hamanishi J, Mandai M, Iwasaki M, Okazaki T, Tanaka Y, Yamaguchi K, Higuchi T, Yagi H, Takakura K, Minato N, et al. Programmed cell death 1 ligand 1 and tumor-infiltrating CD8+ T lymphocytes are prognostic factors of human ovarian cancer. Proc Natl Acad Sci USA. 2007;104(9):3360–3365. PMID:17360651. doi: 10.1073/pnas.0611533104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Nomi T, Sho M, Akahori T, Hamada K, Kubo A, Kanehiro H, Nakamura S, Enomoto K, Yagita H, Azuma M, et al. Clinical significance and therapeutic potential of the programmed death-1 ligand/programmed death-1 pathway in human pancreatic cancer. Clin Cancer Res. 2007;13(7):2151–2157. PMID:17404099. doi: 10.1158/1078-0432.CCR-06-2746. [DOI] [PubMed] [Google Scholar]
- 6.Velcheti V, Schalper KA, Carvajal DE, Anagnostou VK, Syrigos KN, Sznol M, Herbst RS, Gettinger SN, Chen L, Rimm DL. Programmed death ligand-1 expression in non-small cell lung cancer. Lab Invest. 2014;94(1):107–116. PMID: 24217091. doi: 10.1038/labinvest.2013.130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Gubin MM, Zhang X, Schuster H, Caron E, Ward JP, Noguchi T, Ivanova Y, Hundal J, Arthur CD, Krebber WJ, et al. Checkpoint blockade cancer immunotherapy targets tumour-specific mutant antigens. Nature. 2014;515(7528):577–581. PMID:25428507. doi: 10.1038/nature13988. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Linnemann C, van Buuren MM, Bies L, Verdegaal EM, Schotte R, Calis JJ, Behjati S, Velds A, Hilkmann H, Atmioui DE, et al. High-throughput epitope discovery reveals frequent recognition of neo-antigens by CD4+ T cells in human melanoma. Nat Med. 2015;21(1):81–85. PMID:25531942. doi: 10.1038/nm.3773. [DOI] [PubMed] [Google Scholar]
- 9.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348(6230):124–128. PMID: 25765070. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Llosa NJ, Cruise M, Tam A, Wicks EC, Hechenbleikner EM, Taube JM, Blosser RL, Fan H, Wang H, Luber BS, et al. vigorous immune microenvironment of microsatellite instable colon cancer is balanced by multiple counter-inhibitory checkpoints. Cancer Discov. 2015;5(1):43–51. PMID: 25358689. doi: 10.1158/2159-8290.CD-14-0863. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Mlecnik B, Bindea G, Angell HK, Maby P, Angelova M, Tougeron D, Church SE, Lafontaine L, Fischer M, Fredriksen T, et al. Integrative analyses of colorectal cancer show immunoscore is a stronger predictor of patient survival than microsatellite instability. Immunity. 2016;44(3):698–711. PMID: 26982367. doi: 10.1016/j.immuni.2016.02.025. [DOI] [PubMed] [Google Scholar]
- 12.Le DT, Uram JN, Wang H, Bartlett BR, Kemberling H, Eyring AD, Skora AD, Luber BS, Azad NS, Laheru D, et al. PD-1 blockade in tumors with mismatch-repair deficiency. N Engl J Med. 2015;372(26):2509–2520. PMID:26028255. doi: 10.1056/NEJMoa1500596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Overman MJ, McDermott R, Leach JL, Lonardi S, Lenz HJ, Morse MA, Desai J, Hill A, Axelson M, Moss RA, et al. Nivolumab in patients with metastatic DNA mismatch repair-deficient or microsatellite instability-high colorectal cancer (CheckMate 142): an open-label, multicentre, phase 2 study. Lancet Oncol. 2017;18(9):1182–1191. PMID:28734759. doi: 10.1016/S1470-2045(17)30422-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Shankaran V, Ikeda H, Bruce AT, White JM, Swanson PE, Old LJ, Schreiber RD. IFNgamma and lymphocytes prevent primary tumour development and shape tumour immunogenicity. Nature. 2001;410(6832):1107–1111. PMID:11323675. doi: 10.1038/35074122. [DOI] [PubMed] [Google Scholar]
- 15.Ikeda H, Old LJ, Schreiber RD. The roles of IFN gamma in protection against tumor development and cancer immunoediting. Cytokine Growth Factor Rev. 2002;13(2):95–109. PMID: 11900986. [DOI] [PubMed] [Google Scholar]
- 16.Dunn GP, Ikeda H, Bruce AT, Koebel C, Uppaluri R, Bui J, Chan R, Diamond M, White JM, Sheehan KC, et al. Interferon-gamma and cancer immunoediting. Immunol Res. 2005;32(1–3):231–245. PMID: 16106075. [DOI] [PubMed] [Google Scholar]
- 17.Ostroumov D, Fekete-Drimusz N, Saborowski M, Kühnel F, Woller N. CD4 and CD8 T lymphocyte interplay in controlling tumor growth. Cell Mol Life Sci. 2018;75(4):689–713. PMID:29032503. doi: 10.1007/s00018-017-2686-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Galon J, Costes A, Sanchez-Cabo F, Kirilovsky A, Mlecnik B, Lagorce-Pagès C, Tosolini M, Camus M, Berger A, Wind P, et al. Type, density, and location of immune cells within human colorectal tumors predict clinical outcome. Science. 2006;313(5795):1960–1964. PMID:17008531. doi: 10.1126/science.1129139. [DOI] [PubMed] [Google Scholar]
- 19.Tosolini M, Kirilovsky A, Mlecnik B, Fredriksen T, Mauger S, Bindea G, Berger A, Bruneval P, Fridman WH, Pagès F, et al. Clinical impact of different classes of infiltrating T cytotoxic and helper cells (Th1, th2, treg, th17) in patients with colorectal cancer. Cancer Res. 2011;71(4):1263–1271. PMID: 21303976. doi: 10.1158/0008-5472.CAN-10-2907. [DOI] [PubMed] [Google Scholar]
- 20.Abiko K, Matsumura N, Hamanishi J, Horikawa N, Murakami R, Yamaguchi K, Yoshioka Y, Baba T, Konishi I, Mandai M. IFN-γ from lymphocytes induces PD-L1 expression and promotes progression of ovarian cancer. Br J Cancer. 2015;112(9):1501–1509. PMID:25867264. doi: 10.1038/bjc.2015.101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bellucci R, Martin A, Bommarito D, Wang K, Hansen SH, Freeman GJ, Ritz J. Interferon-γ-induced activation of JAK1 and JAK2 suppresses tumor cell susceptibility to NK cells through upregulation of PD-L1 expression. Oncoimmunology. 2015;4(6):e1008824 PMID:26155422. doi: 10.1080/2162402X.2015.1008371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Spranger S, Spaapen RM, Zha Y, Williams J, Meng Y, Ha TT, Gajewski TF. Up-regulation of PD-L1, IDO, and T(regs) in the melanoma tumor microenvironment is driven by CD8(+) T cells. Sci Transl Med. 2013;5(200):200ra116 PMID:23986400. doi: 10.1126/scitranslmed.3006504. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Herbst RS, Soria JC, Kowanetz M, Fine GD, Hamid O, Gordon MS, Sosman JA, McDermott DF, Powderly JD, Gettinger SN, et al. Predictive correlates of response to the anti-PD-L1 antibody MPDL3280A in cancer patients. Nature. 2014;515(7528):563–567. PMID:25428504. doi: 10.1038/nature14011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fehrenbacher L, Spira A, Ballinger M, Kowanetz M, Vansteenkiste J, Mazieres J, Park K, Smith D, Artal-Cortes A, Lewanski C, et al. Atezolizumab versus docetaxel for patients with previously treated non-small-cell lung cancer (POPLAR): a multicentre, open-label, phase 2 randomised controlled trial. Lancet. 2016;387(10030):1837–1846. PMID:26970723. doi: 10.1016/S0140-6736(16)00587-0. [DOI] [PubMed] [Google Scholar]
- 25.Ayers M, Lunceford J, Nebozhyn M, Murphy E, Loboda A, Kaufman DR, Albright A, Cheng JD, Kang SP, Shankaran V, et al. IFN-γ-related mRNA profile predicts clinical response to PD-1 blockade. J Clin Invest. 2017;127(8):2930–2940. PMID:28650338. doi: 10.1172/JCI91190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Karachaliou N, Gonzalez-Cao M, Crespo G, Drozdowskyj A, Aldeguer E, Gimenez-Capitan A, Teixido C, Molina-Vila MA, Viteri S, Karachaliou N, et al. Interferon gamma, an important marker of response to immune checkpoint blockade in non-small cell lung cancer and melanoma patients. Ther Adv Med Oncol. 2018;10:1758834017749748 PMID:29383037. doi: 10.1177/1758834017749748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Szabo SJ, Kim ST, Costa GL, Zhang X, Fathman CG, Glimcher LH. A novel transcription factor, T-bet, directs Th1 lineage commitment. Cell. 2000;100(6):655–669. PMID: 10761931. [DOI] [PubMed] [Google Scholar]
- 28.Sullivan BM, Juedes A, Szabo SJ, von Herrath M, Glimcher LH. Antigen-driven effector CD8 T cell function regulated by T-bet. Proc Natl Acad Sci U S A. 2003;100(26):15818–15823. PMID:14673093. doi: 10.1073/pnas.2636938100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Liu J, Hamrouni A, Wolowiec D, Coiteux V, Kuliczkowski K, Hetuin D, Saudemont A, Quesnel B. Plasma cells from multiple myeloma patients express B7-H1 (PD-L1)and increase expression after stimulation with IFNgamma and TLR ligands via a MyD88, TRAF6-, and MEK-dependent pathway. Blood. 2007;110(1):296–304. PMID:17363736. doi: 10.1182/blood-2006-10-051482. [DOI] [PubMed] [Google Scholar]
- 30.Gordon S. Alternative activation of macrophages. Nat Rev Immunol. 2003;3(1):23–35. PMID:12511873. doi: 10.1038/nri978. [DOI] [PubMed] [Google Scholar]
- 31.Gordon S, Martinez FO. Alternative activation of macrophages: mechanism and functions. Immunity. 2010;32(5):593–604. PMID: 20510870. doi: 10.1016/j.immuni.2010.05.007. [DOI] [PubMed] [Google Scholar]
- 32.Overman MJ, Lonardi S, Wong KYM, Lenz HJ, Gelsomino F, Aglietta M, Morse MA, Van Cutsem E, McDermott R, Hill A, et al. Durable clinical benefit with nivolumab plus ipilimumab in DNA mismatch repair-deficient/microsatellite instability-high metastatic colorectal cancer. J Clin Oncol. 2018;36(8):773–779. PMID:29355075. doi: 10.1200/JCO.2017.76.9901. [DOI] [PubMed] [Google Scholar]
- 33.Le DT, Kavan P, Kim TW, Burge ME, Cutsem EV, Hara H, McKay Boland P, Van Laethem JL, Geva R, Taniguchi H, et al. Pembrolizumab for patients with advanced microsatellite instability high (MSI-H) colorectal cancer. J Clin Oncol. 2018;36(suppl; abstr 3514):3514. doi: 10.1200/JCO.2018.36.15_suppl.3514. [DOI] [Google Scholar]
- 34.de Vries NL, Swets M, Vahrmeijer AL, Hokland M, Kuppen PJ. The Immunogenicity of colorectal cancer in relation to tumor development and treatment. Int J Mol Sci. 2016;17(7):1030 PMID:27367680. doi: 10.3390/ijms17071030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Keene JA, Forman K. Helper activity is required for the in vivo generation of cytotoxic T cells. J Exp Med. 1982;155:768–782. PMID:6801178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yo-Ping L, Chung-Jiuan J, Shu-Ching C. The roles of CD4+ T cells in tumor immunity. ISRN Immunology. 2011;2011:Article ID 497397, 6. doi: 10.5402/2011/497397. [DOI] [Google Scholar]
- 37.Quezada SA, Simpson TR, Peggs KS, Merghoub T, Vider J, Fan X, Blasberg R, Yagita H, Muranski P, Antony PA, et al. Tumor-reactive CD4(+) T cells develop cytotoxic activity and eradicate large established melanoma after transfer into lymphopenic hosts. J Exp Med. 2010;207(3):637–650. PMID:20156971. doi: 10.1084/jem.20091918. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marisa L, Svrcek M, Collura A, Becht E, Cervera P, Wanherdrick K, Buhard O, Goloudina A, Jonchère V, Selves J, et al. The balance between cytotoxic T-cell lymphocytes and immune checkpoint expression in the prognosis of colon tumors. J Natl Cancer Inst. 2018;110(1). PMID:28922790. doi: 10.1093/jnci/djx136. [DOI] [PubMed] [Google Scholar]
- 39.Boissière-Michot F, Lazennec G, Frugier H, Jarlier M, Roca L, Duffour J, Du Paty E, Laune D, Blanchard F, Le Pessot F, et al. Characterization of an adaptive immune response in microsatellite-instable colorectal cancer. Oncoimmunology. 2014;3:e29256 PMID: 25101223. doi: 10.4161/onci.29256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ling A, Lundberg IV, Eklöf V, Wikberg ML, Öberg Å, Edin S, Palmqvist R. The infiltration, and prognostic importance, of Th1 lymphocytes vary in molecular subgroups of colorectal cancer. J Pathol Clin Res. 2015;2(1):21–31. PMID:27499912. doi: 10.1002/cjp2.31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Teng MW, Ngiow SF, Ribas A, Smyth MJ. Classifying cancers based on T-cell infiltration and PD-L1. Cancer Res. 2015;75(11):2139–2145. PMID: 25977340. doi: 10.1158/0008-5472.CAN-15-0255. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chen DS, Mellman I. Elements of cancer immunity and the cancer-immune set point. Nature. 2017;541(7637):321–330. PMID:28102259. doi: 10.1038/nature21349. [DOI] [PubMed] [Google Scholar]
- 43.Ribas A, Robert C, Hodi FS, Wolchok JD, Joshua AM, Hwu WJ, Weber JS, Zarour HM, Kefford R, Loboda A, et al. Association of response to programmed death receptor 1 (PD-1) blockade with pembrolizumab (MK-3475) with an interferon-inflammatory immune gene signature. J Clin Oncol. 2015;33:15_suppl, 3001. doi: 10.1200/jco.2015.33.15_suppl.3001. [DOI] [Google Scholar]
- 44.Kim JH, Park HE, Cho NY, Lee HS, Kang GH. Characterisation of PD-L1-positive subsets of microsatellite-unstable colorectal cancers. Br J Cancer. 2016. August 9;115(4):490–496. MID: 27404452. doi: 10.1038/bjc.2016.211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Lee LH, Cavalcanti MS, Segal NH, Hechtman JF, Weiser MR, Smith JJ, Garcia-Aguilar J, Sadot E, Ntiamoah P, Markowitz AJ, et al. Patterns and prognostic relevance of PD-1 and PD-L1 expression in colorectal carcinoma. Mod Pathol. 2016;29(11):1433–1442. PMID: 27443512. doi: 10.1038/modpathol.2016.139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Rosenbaum MW, Bledsoe JR, Morales-Oyarvide V, Huynh TG, Mino-Kenudson M. PDL1 expression in colorectal cancer is associated with microsatellite instability, BRAF mutation, medullary morphology and cytotoxic tumor-infiltrating lymphocytes. Mod Pathol. 2016;29(9):1104–1112. PMID: 27198569. doi: 10.1038/modpathol.2016.95. [DOI] [PubMed] [Google Scholar]
- 47.Inaguma S, Lasota J, Wang Z, Felisiak-Golabek A, Ikeda H, Miettinen M. Clinicopathologic profile, immunophenotype, and genotype of CD274(PD-L1)-positive colorectal carcinomas. Mod Pathol. 2017;30(2):278–285. PMID: 27813511. doi: 10.1038/modpathol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Korehisa S, Oki E, Iimori M, Nakaji Y, Shimokawa M, Saeki H, Okano S, Oda Y, Maehara Y. Clinical significance of programmed cell death-ligand 1 expression and the immune microenvironment at the invasive front of colorectal cancers with high microsatellite instability. Int J Cancer. 2018;142(4):822–832. PMID: 29044503. doi: 10.1002/ijc.31107. [DOI] [PubMed] [Google Scholar]
- 49.Valentini AM, Di Pinto F, Cariola F, Guerra V, Giannelli G, Caruso ML, Pirrelli M. PD-L1 expression in colorectal cancer defines three subsets of tumor immune microenvironments. Oncotarget. 2018;9(9):8584–8596. PMID: 29492219. doi: 10.18632/oncotarget.24196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Taube JM, Anders RA, Young GD, Xu H, Sharma R, McMiller TL, Chen S, Klein AP, Pardoll DM, Topalian SL, et al. Colocalization of inflammatory response with B7-h1 expression in human melanocytic lesions supports an adaptive resistance mechanism of immune escape. Sci Transl Med. 2012;4(127):127ra37 PMID: 22461641. doi: 10.1126/scitranslmed.3003689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Zhang W, Pang Q, Yan C, Wang Q, Yang J, Yu S, Liu X, Yuan Z, Wang P, Xiao Z. Induction of PD-L1 expression by epidermal growth factor receptor-mediated signaling in esophageal squamous cell carcinoma. Onco Targets Ther. 2017;10:763–771. PMID: 28243112. doi: 10.2147/OTT.S118982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jiang X, Zhou J, Giobbie-Hurder A, Wargo J, Hodi FS. The activation of MAPK in melanoma cells resistant to BRAF inhibition promotes PD-L1 expression that is reversible by MEK and PI3K inhibition. Clin Cancer Res. 2013;19(3):598–609. PMID:23095323. doi: 10.1158/1078-0432.CCR-12-2731. [DOI] [PubMed] [Google Scholar]
- 53.Friedman K, Brodsky AS, Lu S, Wood S, Gill AJ, Lombardo K, Yang D, Resnick MB. Medullary carcinoma of the colon: a distinct morphology reveals a distinctive immunoregulatory microenvironment. Mod Pathol. 2016;29(5):528–541. PMID:26965581. doi: 10.1038/modpathol.2016.54. [DOI] [PubMed] [Google Scholar]
- 54.Engin A, Gonul II, Engin AB, Karamercan A, Sepici Dincel A, Dursun A. Relationship between indoleamine 2,3-dioxygenase activity and lymphatic invasion propensity of colorectal carcinoma. World J Gastroenterol. 2016;22(13):3592–3601. PMID:27053851. doi: 10.3748/wjg.v22.i13.3592. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Chon SY, Hassanain HH, Gupta SL. Cooperative role of interferon regulatory factor 1 and p91 (STAT1) response elements in interferon-gamma-inducible expression of human indoleamine 2,3-dioxygenase gene. J Biol Chem. 1996;271(29):17247–17252. PMID:8663541. [DOI] [PubMed] [Google Scholar]
- 56.Takikawa O, Habara-Ohkubo A, Yoshida R. Induction of indoleamine 2,3-dioxygenase in tumor cells transplanted into allogeneic mouse: interferon-gamma is the inducer. Adv Exp Med Biol. 1991;294:437–444. PMID:1772076. [DOI] [PubMed] [Google Scholar]
- 57.Brochez L, Chevolet I, Kruse V. The rationale of indoleamine 2,3 dioxygenase inhibition for cancer therapy. Eur J Cancer. 2017;76:167–182. PMID:28324751. doi: 10.1016/j.ejca.2017.01.011. [DOI] [PubMed] [Google Scholar]
- 58.Knox RD, Luey N, Sioson L, Kedziora A, Clarkson A, Watson N, Toon CW, Cussigh C, Pincott S, Pillinger S, et al. Medullary colorectal carcinoma revisited: a clinical and pathological study of 102 cases. Ann Surg Oncol. 2015;22(9):2988–2996. PMID:25572685. doi: 10.1245/s10434-014-4355-5. [DOI] [PubMed] [Google Scholar]
- 59.McLaughlin J, Schalper K, Carvajal-Hausdorf D, Velcheti V, Haack H, Silver M, Goldberg SB, Herbst RS, Rimm DL. Domain-specific PD-L1 protein measurement in non-small cell lung cancer (NSCLC). J Clin Oncol. 2014;5(suppl, abst 8064):8064. [Google Scholar]
- 60.Lau SK, Chu PG, Weiss LM. CD163: a specific marker of macrophages in paraffin-embedded tissue samples. Am J Clin Pathol. 2004;122(5):794–801. PMID:15491976. doi: 10.1309/QHD6-YFN8-1KQX-UUH6. [DOI] [PubMed] [Google Scholar]
- 61.Suraweera N, Duval A, Reperant M, Vaury C, Furlan D, Leroy K, Seruca R, Iacopetta B, Hamelin R. Evaluation of tumor microsatellite instability using five quasimonomorphic mononucleotide repeats and pentaplex PCR. Gastroenterology. 2002;123(6):1804–1811. PMID:12454837. doi: 10.1053/gast.2002.37070. [DOI] [PubMed] [Google Scholar]
- 62.Umar A, Boland CR, Terdiman JP, Syngal S, de la Chapelle A, Rüschoff J, Fishel R, Lindor NM, Burgart LJ, Hamelin R, et al. Revised Bethesda guidelines for hereditary nonpolyposis colorectal cancer (Lynch syndrome) and microsatellite instability. J Natl Cancer Inst. 2004;96(4):261–268. PMID:14970275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Jarry A, Bossard C, Bou-Hanna C, Masson D, Espaze E, Denis MG, Laboisse CL. Mucosal IL-10 and TGF-beta play crucial roles in preventing LPS-driven,IFN-gamma-mediated epithelial damage in human colon explants. J Clin Invest. 2008;118(3):1132–1142. PMID:18259614. doi: 10.1172/JCI32140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jarry A, Bossard C, Sarrabayrouse G, Mosnier JF, Laboisse CL. Loss ofinterleukin-10 or transforming growth factor β signaling in the human colon initiates a T-helper 1 response via distinct pathways. Gastroenterology. 2011;141(5):1887–96.e1–2 PMID:21839042. doi: 10.1053/j.gastro.2011.08.002. [DOI] [PubMed] [Google Scholar]
- 65.Mandrekar JN, Mandrekar SJ, Cha SS. Cutpoint determination methods in survival analysis using SAS®. Proceedings of the 28th SAS Users Group International Conference (SUGI), Philadelphia, PA, 2003; Paper 261–28. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


