ABSTRACT
Zygotic genome activation (ZGA) is one of the most critical events at the beginning of mammalian preimplantation embryo development (PED). The mechanisms underlying mouse ZGA remain unclear although it has been widely studied. In the present study, we identified that tricho-rhino-phalangeal syndrome 1 (TRPS1), an atypical GATA family member, is an important factor for ZGA in mouse PED. We found that the Trps1 mRNA level peaked at the one-cell stage while TRPS1 protein did so at the two/four-cell stage. Knockdown of Trps1 by the microinjection of Trps1 siRNA reduced the developmental rate of mouse preimplantation embryos by approximately 30%, and increased the expression of ZGA marker genes MuERV-L and Zscan4d via suppressing the expression of major histone markers H3K4me3 and H3K27me3. Furthermore, Trps1 knockdown decreased the expression of Sox2 but increased Oct4 expression. We conclude that TRPS1 may be indispensable for zygotic genome activation during mouse PED.
KEYWORDS: Zygotic genome activation (ZGA), preimplantation embryo development (PED), tricho-rhino-phalangeal syndrome 1 (TRPS1)
Introduction
The terminally differentiated sperm and oocyte unite at fertilization to form a totipotent zygote that can differentiate into all kinds of cells and tissues in the mature individual. In mammals, the zygote undergoes several rounds of cleavage to form a multicellular blastocyst. This process is called preimplantation embryo development (PED), the very first step in mammalian development.
During mouse PED, a series of critical events occur in a strictly controlled fashion. Soon after fertilization, maternally stored materials promote the initiation of zygotic genome activation (ZGA), which appears in two waves: the minor one occurs at the mid-one-cell stage and the beginning of the two-cell stage, and the major one occurs at the mid-to-late two-cell stage [1–4]. Severe ZGA disorders result in two-cell arrest. ZGA emerges with the degradation of maternal deposits (including proteins and mRNAs) [5]. Upon reaching the four/eight-cell stage, products of ZGA have replaced most of the maternal materials and take control of further development. This replacement is an important milestone referred to as the maternal-to-zygote transition (MZT) [6].
Genes activated at the four/eight-cell stage subsequently promote blastocyst formation, which represents the first cell lineage differentiation. This is followed by the formation of an inner cell mass (ICM) from which embryonic stem cells (ESCs) are derived under in vitro manipulation, and trophectoderm (TE) which can invade the uterine wall under appropriate physiological conditions in vivo to take part in placenta formation [7]. At the late blastocyst stage, ICM further segregates into epiblast (EPI) giving rise to the embryo proper and primitive endoderm (PrE), which forms the yolk sac [8]. Thus, early critical events of PED such as ZGA are closely connected with the progression of later stages, as well as post-implantation embryonic development. Although some key factors in the regulation of ZGA and their associated signaling pathways have been extensively studied, the exact mechanism behind this regulation remains fully elucidated.
Mouse ZGA is probably controlled by epigenetic alterations (e.g., DNA demethylation, histone modification dynamics, and non-coding RNAs), cell cycle progression and transcription factors. High-throughput profiling and ChIP-seq studies have revealed highly dynamic expression patterns of numerous putative key factors [6,9]. Investigations of the functional mechanisms of these factors, by knockout or knockdown technologies, have also broadened our knowledge of ZGA regulation and cell fate determination. For example, by using ATAC-seq, Wu et al. investigated the chromatin accessibility landscape of mouse PED and identified several key transcription factors for early development, such as SOX2, POU5F1 (also known as OCT4), NANOG, and some GATA family members. Knocking down the expression of Gata4 at the one-cell stage by RNA interference (RNAi) resulted in the downregulation of PrE markers and Sox2, indicating the existence of intimate interplay between these factors [10]. Although maternal and zygotic knockout of Sox2 or Oct4 did not result in PED blockage [11–13], overexpression of either of them could lead to two-cell blockage and ZGA failure [14,15], implying the need for tight control of their expression during early development.
GATA proteins (including GATA1 to GATA6) are important transcription factors that contain two zinc fingers and can bind to consensus GATA motif on the cis-regulatory regions of target genes [16]. GATA3 is reported to take part in TE formation by regulating Cdx2 [17]. GATA4 and GATA6 are markers of PrE [7]. In mouse ESCs, GATA2 is reported to activate MuERV-L [18], which is an important retrotransposon highly expressed during mouse ZGA [19] and acts as a promoter for multiple neighboring genes including Zscan4d (one of the two-cell-specific ZGA markers) [20]. Both MuERV-L and Zscan4d are required for mouse preimplantation development [21,22]. However, knockout of Gata2 as well as other GATA members did not result in ZGA failure or early developmental blockage until peri-implantation stages, regardless of the active transcription of most GATA members during the two-/four-cell stages [16,23]. These results indicate the existence of other critical factors that control the expression of pluripotent factors such as Sox2 and Oct4, and restrict the functions of GATA proteins in early-stage preimplantation embryos. The nature of such factors has not previously been elucidated.
In 2000, Momeni et al. found that tricho-rhino-phalangeal syndrome 1 (TRPS1) is an atypical GATA protein containing two nuclear localization signals and zinc fingers including GATA-binding motifs and Ikaros-like domain (Figure 1(a)) [24]. TRPS1 can bind to GATA motifs and function as a potent repressor of other GATA members [25]. It has also been reported that TRPS1 works as an anti-metastasis factor by inhibiting epithelial-mesenchymal transition (EMT) in cancer cells [26–28], and takes part in the regulation of fetal kidney development [29,30], cartilage differentiation [31,32] and cell cycle progression [33,34]. Recently, TRPS1 was also reported to affect the histone H3 lysine 27 tri-methylation (H3K27me3) levels on the promoter region of target gene Cdh1 through the direct regulation of Suz12 expression [35]. Besides, TRPS1 is highly expressed in estrogen receptor-positive (ER+) breast cancer cells [36,37]. It was also found to recruit histone deacetylase (HDAC) complexes to enhancers and act as a key repressor of ERα [38], which was dynamically expressed during mouse PED [39–41] and affected the expression of MuERV-L at ZGA [42]. Furthermore, through regulating the expression of Gata3, ERα also functions in blastocyst formation [43]. Despite these findings, the possible roles of TRPS1 during PED have not been thoroughly explored.
Figure 1.
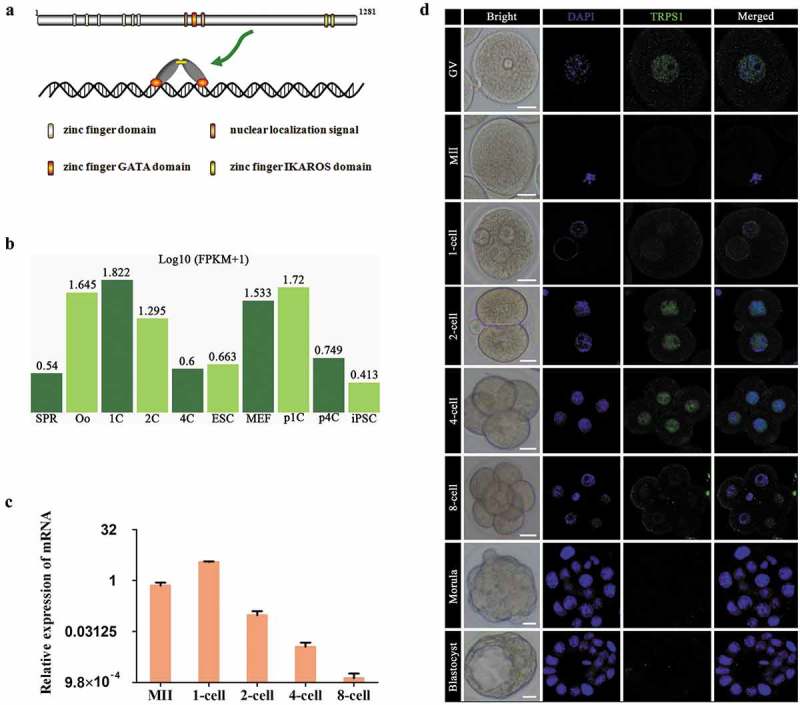
Stage-specific dynamic expression of TRPS1 during mouse PED. (a) Schematic diagram of TRPS1 protein. (b and c) Dynamic expression of Trps1 mRNA extracted from DBTMEE (b) and detected by real-time PCR analysis (c) using a log2 scale (-ΔΔCt) method. (d) Stage-specific localization of TRPS1 protein. At least 3 replicates were conducted for each developmental stage. Abbreviations: SPR, sperm; Oo, oocyte; 1C, 2C and 4C: one-cell, two-cell and four-cell embryos; ESC, embryonic stem cells; MEF, mouse embryonic fibroblast cells; p1C and p4C: parthenogenetic 1C and 4C; iPSC, induced pluripotent stem cells. Error bars indicate SEM. Scale bar, 20 μm.
In the present study, we examined the stage-specific expression of TRPS1 in mouse preimplantation embryos and suppressed its expression, by using Trps1 siRNA microinjection to observe the developmental outcomes. The results demonstrated that TRPS1 plays indispensable roles during mouse ZGA through controlling the expression of ZGA markers MuERV-L and Zscan4d, transcription factors SOX2 and OCT4, and key histone markers H3K4me3 and H3K27me3.
Materials and methods
Experimental animals
Kunming (KM) mice (female, 4–6 w; male > 8 w) were purchased from SLRC Laboratory Animal Co., Ltd. (Shanghai, China), and housed for 3–5 days under conditions with controlled temperature (22 ± 1°C) and light cycle (07:00–19:00), to regulate the menstrual cycle of the female mice and acclimate them to the new environment prior to experimental treatment. All of the procedures concerning mouse handling were conducted under the guidance of the Institutional Animal Care and Use Committee (IACUC) of Fujian Medical University.
Main reagents
M2 medium, DEPC (Sigma-Aldrich, USA); KSOM medium (Merck Millipore, Germany); Pregnant mare serum gonadotropin (PMSG) (2nd Ningbo Hormone Production Company, China); Human chorionic gonadotropin (hCG) (ProSpec, Israel); Immunol Staining Fix Solution, 4ʹ, 6-diamidino-2-phenylindole (DAPI) (Beyotime, China); glycogen, Quick-RNA™ MicroPrepR1050 Kit and SYBR® Premix ExTaqTM (Roche, Switzerland); and Reverse Transcription Kit and dNTP Mix (Thermo Fisher Scientific, USA) were used in this study. The primary antibodies were anti-TRPS1 rabbit polyclonal antibody (Proteintech Group, USA); anti-RUNX1 rabbit polyclonal antibody (Genetex, USA); anti-SOX2 mouse monoclonal antibody (Santa Cruz Biotechnology, USA); anti-OCT4 rabbit polyclonal antibody (Abcam, UK); anti-H3K4me3 rabbit monoclonal antibody (Cell Signaling Technology, USA); anti-H3K27me3 rabbit monoclonal antibody (Cell Signaling Technology, USA). The secondary antibodies were Alexa Fluor® 488 labeled Donkey anti-Rabbit secondary antibody (Life Technology, USA); Alexa Fluor® 488 labeled donkey anti-mouse secondary antibody (Life Technology, USA); Trps1-siRNA (Santa Cruz, USA); and negative control siRNA (sense: 5ʹ-UUCUCCGAACGUGUCACGUTT-3ʹ; antisense: 5ʹ-ACGUGACACGUUCGGAGAATT-3ʹ) (GenePharma, China).
Mouse oocyte and preimplantation embryo collection
For the collection of germinal vesicle (GV) oocytes, female KM mice were intraperitoneally injected with 10 IU PMSG and sacrificed 48 h later. Their ovaries were separated and triturated in M2 medium (supplemented with IBMX) at room temperature (RT) to release the cumulus–oocyte complexes; by flushing with a mouth-controlled pipette, naked GV oocytes with intact germinal vesicles were obtained.
For the collection of MII oocytes, female KM mice were injected with PMSG followed by the injection of 6 IU hCG 46–48 h later, and sacrificed at 21 h post-hCG injection (p-hCG). The oviducts were then separated and cut with a syringe needle at the ampullar region to release cumulus–oocyte complexes, and granulosa cells were removed by incubating with hyaluronidase to obtain naked MII oocytes.
For the collection of embryos at the early and late one-cell, two-cell, four-cell, eight-cell, morula, and blastocyst stages, female KM mice were super-ovulated as mentioned above and mated with male KM mice at a 1:1 ratio. The female mice with vaginal plugs were sacrificed at 24, 27, 42, 54, 64, 72, and 90 h p-hCG; the preimplantation embryos were obtained by flushing oviducts (for early-stage embryos) or uteri (for morulae and blastocysts) with M2 medium.
Microinjection of one-cell embryos and in vitro cultivation
Mouse one-cell embryos were collected at 18 h p-hCG and pre-incubated in a 37°C, 5% CO2 incubator for 3 h before injection. Stock solution of Trps1-siRNA was diluted to a final concentration of 20 μM. The microinjection needles were pulled on a P-97 micropipette puller (Sutter Instrument, USA) from borosilicate glass BF100-78–10 (Sutter Instrument, USA).
The cells were injected using a microinjector (Nikon, Japan) and a micromanipulator (Nikon, Japan). Each one-cell embryo was placed in a 10 μL PVP-PBS microdrop. The holding pipette and the injection needle were descended to the same height in the same PVP-PBS microdrop; then, the injection needle was moved to an already prepared siRNA microdrop and about 5–7 pL of liquid was drawn into it. The one-cell embryo was held steady with the holding pipette, and microinjection was performed by penetrating the injection needle through the zona pellucida and cell membrane into the pronuclear region. The oil pressure controller was turned carefully; when the pronucleus started to swell, the injection needle was withdrawn.
After manipulation, the surviving cells were washed three times in KSOM medium and cultured in the incubator until the blastocyst stage (for continual observation of the developmental ratio) or until 43 h p-hCG (to recover samples for real-time PCR analysis).
Immunofluorescence staining
Mouse preimplantation embryos were washed three times with 0.1% PVA-PBS, treated with Tyrode’s solution to remove the zona pellucida, and fixed at RT for 1 h, followed by washing three times with 0.1% PVA-PBS. They were then permeabilized with 0.5% TritonX-100 for 30 min, washed with 0.2% BSA three times, and blocked in 0.2% BSA for 1 h at RT. After blocking, the cells were incubated with the primary antibodies at 4°C overnight. The cells were washed with PBST three times (10 min each time) and then incubated with secondary antibodies for 1 h at RT. Next, the cells were washed three times in PBST and incubated with DAPI (1:5000) for 1 h at RT. After DAPI counterstaining, the cells were washed three times with PBST and observed under a confocal microscope (Leica TCSSP5, Germany). SmartScape software (Shanghai Furi Science & Technology Co., Ltd., China) was used to analyze the gray values of immunostaining images. Briefly, images containing about 20 embryos each were normalized by the same parameter to subtract background staining. Then, gray values of the nucleus and cytoplasm of each cell were calculated separately. At least three independent experiments were conducted (and over 60 embryos were used) for each group.
Real-time PCR analysis
Total RNA from 50 mouse preimplantation embryos from each group was extracted in accordance with the manual of the Quick-RNA™ MicroPrepR1050 Kit. The concentration and purity of RNA were detected by using a NanoDrop ND-1000 (NanoDrop, USA). The cDNA was synthesized in accordance with the manual of the Reverse Transcription Kit using a PCR Amplifier (AB2720; Gene, USA). Sequence information of the PCR primers used in the present study is presented in Table 1. The internal housekeeping gene H2afz encoding H2A histone family member Z served as an internal control, as reported previously [42]. Amplification was performed on a real-time PCR Amplifier (PikoReal2.2.248; Thermo, USA) by employing SYBR® Premix Ex TaqTM with a cycling protocol consisting of an initial 10 min at 95°C, followed by 40 cycles of 15 s at 95°C and 1 min at 60°C. Samples were prepared in triplicate with at least three independent repeats.
Table 1.
Sequence information of the PCR primers.
| Gene Symbol | GenBank accession number | Primer sequences |
|---|---|---|
| H2afz | NM_016750 | F:5′- GTAAAGCGTATCACCCCTCGT -3′ |
| R:5′- TCAGCGATTTGTGGATGTGT -3′ | ||
| Trps1 | NM_001282903.2 | F:5′- GAGATCTCGAGACACTACAG -3′ |
| R:5′- CTCTTCGCCATTAGCAGTAG -3′ | ||
| ERα | NM_007956.4 | F:5′- TTCTGATGATTGGTCTCGTCTG -3′ |
| R:5′- ATGCCTTCCACACATTTACCTT -3′ | ||
| eIF1A | NM_010120 | F:5′- CCAAAGAATAAAGGCAAAGGAG -3′ |
| R:5′- CTCACACCGTCAAAGCACATT -3′ | ||
| Zscan4d | NM_001100186 | F:5′- CCATCTCATAGTTCTGGTGTGC -3′ |
| R:5′- GCTCCTTAGTCTGCTTTTCTGG -3′ | ||
| MuERV-L | Y12713 | F:5′- CGCACAGCAGCAGTCTATTAC -3′ |
| R:5′- TCTTCTCCTCTTCGGTCATTG -3′ | ||
| Hsp70.1 | NM_010478 | F:5′- AAGAGGAAGCACAAGAAGGACA -3′ |
| R:5′- GCGTGATGGATGTGTAGAAGTC -3′ | ||
| Sox2 | NM_011443.3 | F:5′- GAAAGAAAGGAGAGAAGTTTGGAG -3′ R:5′- ATCTGGCGGAGAATAGTTGGG -3′ |
| Oct4 | NM_013633.3 | F:5′- GAGAAGAGTATGAGGCTACAGGGAC- 3′ R:5′- CAGAGCAGTGACGGGAACAGAG -3′ |
Statistical analysis
The developmental rates of two-, four-, and eight-cell stage preimplantation embryos, morulae and blastocysts were calculated based on the total number of one-cell embryos. SPSS17.0 software was used to perform chi-square (χ2) test. Real-time PCR data were analyzed by the 2−ΔΔCt method to obtain relative mRNA expression levels. SPSS17.0 software was used to perform Student’s t-test. GraphPad Prism was used to draw histograms. Differences between experimental groups and the control groups with a P value < 0.05 were considered to be statistically significant.
Results
TRPS1 was highly expressed during mouse ZGA in a transcription–translation–uncoupled manner
TRPS1 is an important zinc finger transcription factor, whose function in mouse PED has not been disclosed. To reveal the expression of Trps1 mRNA in mouse preimplantation embryos at different stages, we first extracted public RNA sequencing data from DBTMEE [44] (a publicly accessible database based on large-scale whole-transcriptome profiling of mouse preimplantation embryos [45] with the accession number: DRA001066), which showed relatively high expression levels of Trps1 during early stages, especially in one-cell embryos (Figure 1(b)). The results from real-time PCR analysis confirmed that, compared with that in MII oocytes, the expression level of Trps1 mRNA was substantially elevated in one-cell mouse embryos (27 h p-hCG), declined sharply at the two-cell stage, and became almost undetectable at the eight-cell stage (Figure 1(c)).
Immunofluorescence staining showed that TRPS1 was detectable in the nucleus of GV oocytes, and early and late one-cell, two-cell, and four-cell embryos. In contrast to the expression pattern of Trps1 mRNA, the nuclear localization of TRPS1 was most apparent in two- and four-cell embryos, consistent with a previous observation that the transcription and translation were uncoupled during early mouse PED [46]. However, at the eight-cell, morula, and blastocyst stages, TRPS1 expression was almost undetectable (Figure 1(d)). These results indicate that, unlike typical GATA proteins that function at the peri-implantation stages, TRPS1 might have important roles in the early stages of mouse PED.
Knockdown of Trps1 impaired TRPS1 function
Viable early stage one-cell mouse embryos were collected and microinjected with different concentrations of Trps1-siRNA. Twenty-four hours later, two-cell embryos were used to examine the effect and function of Trps1 knockdown (Figure 2(a)). The results showed that the injection of Trps1-siRNA effectively decreased the Trps1 mRNA level (Figure 2(b)) and significantly reduced the TRPS1 protein level as well (Figure 2(c,d)). Furthermore, Trps1-siRNA significantly increased the expression level of RUNX1 (Figure 2(e,f)), which was reported to be negatively regulated by TRPS1 [47], confirming the efficiency of RNA interference (RNAi) in the present study.
Figure 2.
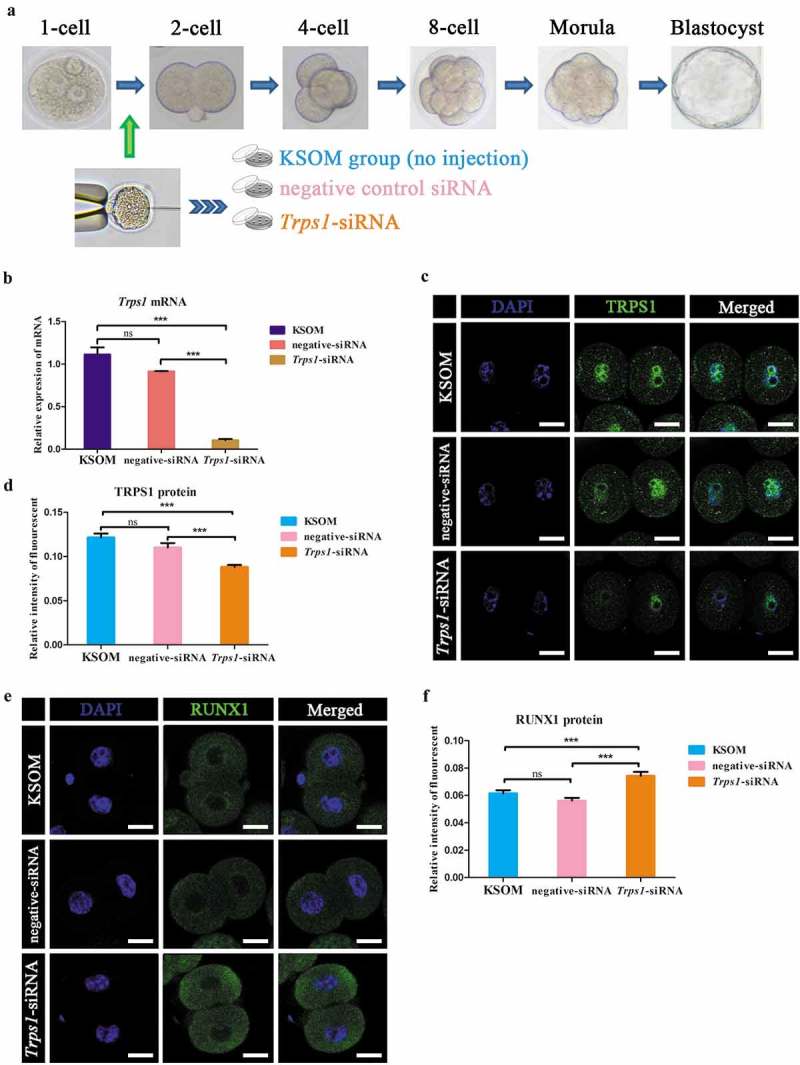
Trps1-siRNA injection efficiently disturbed the function of TRPS1. (a) Illustration of experimental groups. (b) Trps1-siRNA injection effectively knocked down the expression of Trps1 mRNA. (c and d) Immunostaining study show decreased expression level of TRPS1 protein under Trps1-siRNA injection. (e and f) Trps1-siRNA injection increased the expression level of RUNX1 in mouse two-cell embryos. N = 3, ***P < 0.001, Student’s t-test. Error bars indicate SEM. Scale bar, 20 μm.
Trps1 knockdown decreased the development potential of mouse preimplantation embryos
After Trps1-siRNA injection, the development rates of two-cell embryos (43 h p-hCG), four-cell embryos (66 h p-hCG), eight-cell embryos (90 h p-hCG) and blastocysts (120 h p-hCG) were determined. As expected, the blastocyst formation rate of the Trps1-siRNA group was significantly lower than that of the negative control siRNA group. Specifically, the development from one-cell embryos to two-cell embryos was relatively normal (91.60% versus 94.83% in the negative control siRNA group); however, the four-cell rate was significantly reduced in the RNAi group (72.27% versus 93.10% in the negative control siRNA group); while almost all of the embryos in the RNAi group that survived until the eight-cell stage could develop further to the blastocyst stage (74/76) (Figure 3 and Table 2). Furthermore, by injecting anti-TRPS1 antibody into mouse zygotes, the rate of two-cell to four-cell embryo transition was decreased to 28% or lower compared with that in the negative control group (non-specific normal rabbit IgG) (Figure S1). Thus, the most significant developmental blockage caused by Trps1-RNAi occurs at the two-cell stage, indicating the important roles of TRPS1 during mouse ZGA.
Figure 3.
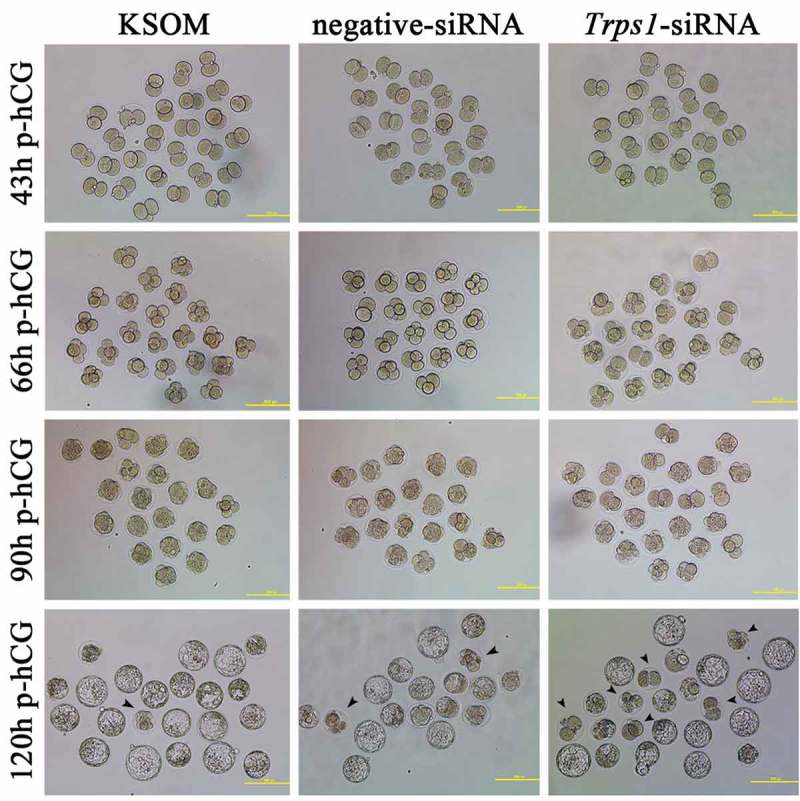
Representative images of stage-specific development rate in each group. Arrowheads indicate blocked embryos observed at blastocyst stage. Scale bar, 150 μm.
Table 2.
Effects of Trps1-siRNA micro-injection on mouse preimplantation embryo development.
| Group | 1-cell (%) | 2-cell (%) | 4-cell (%) | 8-cell (%) | Bl (%) |
|---|---|---|---|---|---|
| KSOM | 131 | 129(98.47) | 127(96.95) | 127(96.95) | 124(94.90) |
| negative siRNA | 116 | 110(94.83) | 108(93.10) | 107(92.24) | 105(90.52) |
| Trps1 siRNA | 119 | 109(91.60) | 86(72.27) * | 76(63.87) * | 74(62.18) * |
Chi-square (χ2)-test, *P < 0.001. Bl: Blastocysts.
Knockdown of Trps1 elevated the expression levels of Zscan4d and MuERV-L
To investigate the connections between TRPS1 and ERα during mouse PED, we evaluated the changes in ERα mRNA expression after Trps1-siRNA injection. As shown in Figure 4(a), ERα mRNA expression level in the experimental group did not change significantly relative to control groups, whereas coinjection of ERα-siRNA with Trps1-siRNA into zygotes significantly elevated the rate of developmental blockage (Figure S2), suggesting that ERα may not work downstream of TRPS1, but cooperatively with it.
Figure 4.
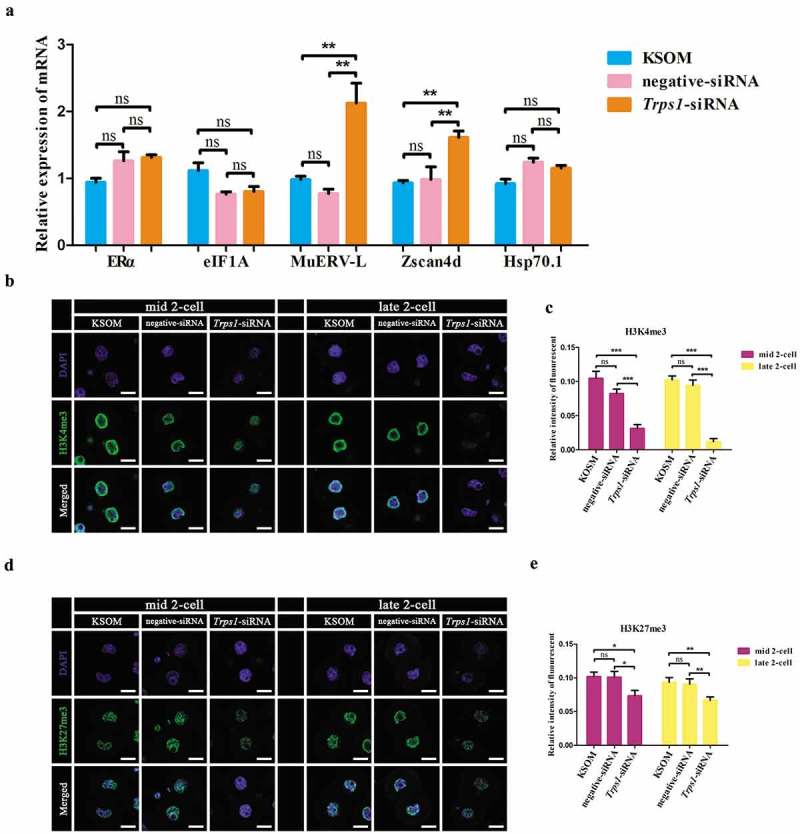
TRPS1 regulated mouse ZGA and histone H3 trimethylation in two-cell embryos. (a) Effects of Trps1-siRNA on the mRNA expression of ERα, eIF1A, MuERV-L, Zscan4d and Hsp70.1. About 140 embryos were used in each experimental group (N = 6). (b and d) Trps1 knockdown resulted in down-regulation of H3K4me3 (b) and H3K27me3 (d) in both mid and late two-cell embryos. (c and e) Gray value analysis results for (b) and (d), respectively. Mid two-cell, 20 h post injection; late two-cell, 29 h post injection. About 80 embryos were used in each experimental group (N = 3). * P < 0.05, ** P < 0.01, ***P < 0.001, Student’s t-test. Error bars indicate SEM. Scale bar, 20 μm.
Several genes such as MuERV-L, Zscan4d, Hsp70.1 and eIF1A are regarded as “marker” genes in studies assessing ZGA status [42,48–53]. To investigate whether the observed two-cell blockage correlated with ZGA, alterations in the expression of these ZGA markers in two-cell embryos after Trps1-siRNA were examined. Trps1 knockdown significantly increased the expression levels of MuERV-L and Zscan4d mRNA while the levels of eIF1A and Hsp70.1 did not change significantly (Figure 4(a)). In addition, JASPAR [54] motif analysis identified both ESR1 (i.e., ERα) motifs and GATA motifs (which can be potently bound by TRPS1) on the promoter region of Zscan4d, but not on that of eIF1A and Hsp70.1 (Table S1-3 and Figure S3). TRPS1 was recently reported to be able to affect the expression of the H3K27me3 regulator Suz12 [35] and recruit the HDAC complex [38] (both of which are regulators of local chromatin conditions), indicating that in mouse two-cell embryos, TRPS1 restrains the expression levels of Zscan4d and MuERV-L possibly by regulating histone modifications.
Trps1-siRNA downregulated the levels of both H3K4me3 and H3K27me3 at the two-cell stage
MuERV-L repeats have been reported to closely associate with histone modifications such as H3K4 methylation [55,56], which leads to gene activation, and H3K27me3, which is associated with gene silencing [57]. However, their dynamic enrichment on Zscan4d during mouse PED has not been addressed in detail. According to public ChIP-seq data (GSE73952), at the two-cell stage the promoter region of Zscan4d is modified by a slightly higher level of broad H3K4me3, which also appears at the MII stage and almost completely disappears at later stages. Notably, increased enrichment of H3K4me3 was observed on the intron of Zscan4d at the four-cell stage (shaded area of Figure S4A). In combination with finding in a previous report [22], Zscan4d is highly specifically expressed at the late two-cell stage (Figure S4B, extracted from GSE71434). Thus, promoter enrichment (but not the overall expression level) of H3K4me3 partially explains the expression pattern of Zscan4d. While almost no H3K27me3 peak was observed on Zscan4d promoter, the region upstream of Zscan4d contains a long (over 5 kb) regulatory MuERV-L element [57] that is modified by stage-specific H3K27me3 peaks and broad H3K4me3 (Figure S4A); this region also contains multiple GATA binding sites (Table S1 and Figure S3). This indicates that the binding of TRPS1 on GATA motifs might affect the local histone modification feature of target genes. Consistent with this, knockdown of Trps1 led to global downregulation of both H3K4me3 (Figure 4(b,c)) and H3K27me3 (Figure 4(d,e)) in two-cell embryos.
In breast cancer cells, TRPS1 is reported to affect the expression of histone lysine 27 acetylation (H3K27ac) [38], which is known for activating gene expression. According to the public ChIP-seq data (GSE72784), the stage-specific expression of Zscan4d is highly correlated with H3K27ac modification (Figure S5A). However, Trps1-siRNA did not alter the expression levels of H3K27ac in both mid- and late two-cell stages (Figure S5B and S5C). Besides, Trps1-siRNA did not change the expression levels of H3K36me3, which is also associated with active gene transcription (Figure S5D and S5E). Noncanonical broad H3K4me3 has also been reported to silence gene expression before the two-cell stage [58,59]. Thus, TRPS1 might work through maintaining the enrichment of broad H3K4me3 and H3K27me3 to prevent the overexpression of Zscan4d and MuERV-L at the two-cell stage.
Trps1-siRNA decreased Sox2 expression but increased Oct4 expression in two-cell embryos
Sox2 and Oct4 are important regulators of pluripotency in ESCs; their overexpression in mouse zygotes has been shown to result in ZGA failure [14,15]. To this end, we further examined the changes of Sox2 and Oct4 expression after Trps1 knockdown to address whether TRPS1 plays a part in controlling these two critical factors. As shown in Figure 5(a,b), Trps1 siRNA halved the expression of Sox2 mRNA, whereas Oct4 mRNA levels were significantly increased by over threefold at the mid-two-cell stage. However, at this time point, the expression of SOX2 protein (mainly cytoplasm-localized) did not change markedly (Figure 5(c,d)). Interestingly, at the late two-cell stage, SOX2 translocated into the nucleus, and a significant decrease of nuclear SOX2 expression level was observed after Trps1 knockdown (Figure 5(c,e)). Consistent with the elevated expression of Oct4 mRNA, Trps1-siRNA injection increased cytoplasmic OCT4 protein levels at both mid- and late two-cell stages, while the nuclear expression levels of OCT4 protein did not change significantly (Figure 5(f,h)). Moreover, the expression of another key pluripotent factor, NANOG, was not changed by Trps1 RNAi (Figure S6A and S6B). These results indicate that TRPS1 can restrain the expression of Oct4 in two-cell embryos while sustaining the level of Sox2.
Figure 5.
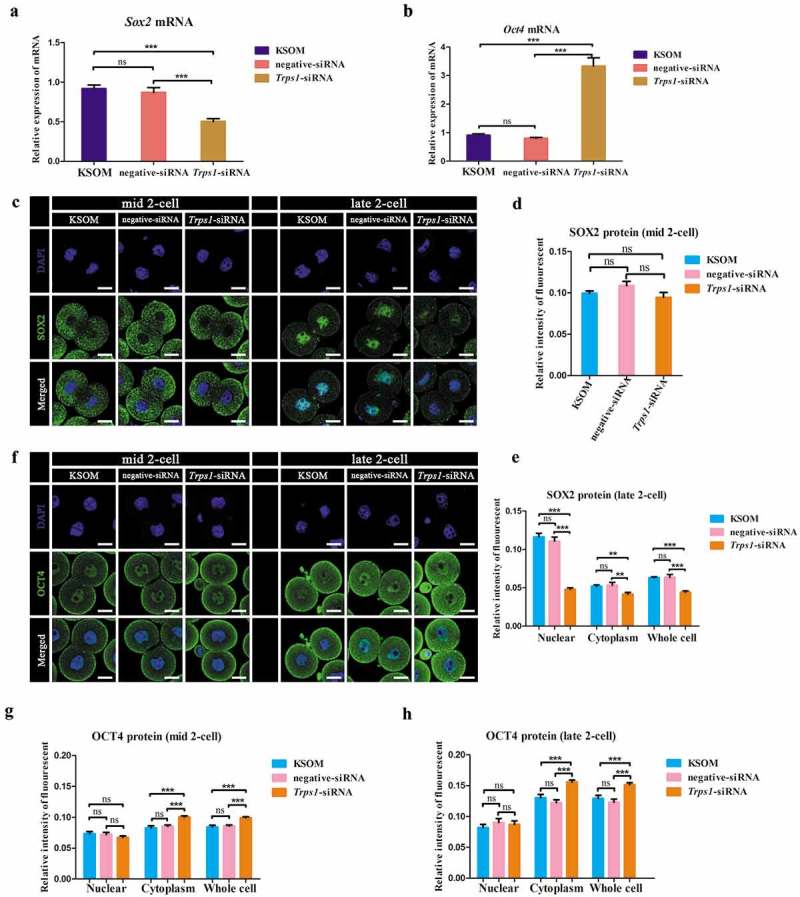
TRPS1 differentially regulated the expression of Sox2 and Oct4. (a and b) Trps1-siRNA significantly changed the mRNA expression level of Sox2 (a) and Oct4 (b). About 60 embryos were used in each experimental group (N = 3). (c, d and e) Trps1 knockdown resulted in less nuclear localization of SOX2 protein at late two-cell stage (C and E), but no change was observed in early two-cell embryos (c and d). (f, g and h) Trps1 knockdown resulted in elevated cytoplasm expression level of OCT4 protein at mid (f and g) and late (f and h) two-cell stage. Mid two-cell, 20 h post injection; late two-cell, 29 h post injection. About 80 embryos were used in each experimental group (N = 3). * P < 0.05, ** P < 0.01, ***P < 0.001, Student’s t-test. Error bars indicate SEM. Scale bar, 20 μm.
Discussion
The preimplantation stage is one of the most critical periods during mammalian development. However, the underlying mechanisms remain unclear. TRPS1 is an important atypical GATA transcription factor. Its possible functions in mouse preimplantation embryos have not been reported previously. In the present study, we found the unique high expression of Trps1 mRNA in one-cell embryos, an increase of TRPS1 nuclear localization in two/four-cell embryos, and that Trps1 knockdown induced effects that include partial blockage of mouse PED at the two-cell stage, increased expression of ZGA markers (Zscan4d and MuERV-L), decreased expression of H3K4me3 and H3K27me3, downregulation of Sox2 and up-regulation of Oct4 mRNA levels. These results clearly suggest that TRPS1 plays important roles in the early stages of mouse preimplantation development.
Mouse ZGA occurs in two waves. Minor ZGA that occurs at the one-cell stage is promiscuous and over 90% of genes (including extensive intergenic regions) are transcribed, mainly with poor splicing and 3ʹ processing. Major ZGA takes place at the two-cell stage, where less than 80% of genes are selectively transcribed, probably due to a condensed chromatin state [60]. Recently, it was confirmed that minor ZGA is critical for mouse PED [61]. In the present study, we showed that the expression level of Trps1 mRNA is relatively high in MII oocytes and highest in one-cell embryos. After this stage, the Trps1 mRNA expression level decreased markedly and it became almost undetectable at the eight-cell stage (Figure 1(b)). These results indicate that, while Trps1 mRNA is maternally stored, it is also transcribed during minor ZGA and is cleared at later stages of PED. However, TRPS1 protein was intensely stained in two- and four-cell embryos (Figure 1(d)), suggesting its importance during early PED, especially around major ZGA.
Trps1-siRNA injection was applied in the present study to knock down both maternal and zygotic Trps1 mRNA, leading to blockage of the development of about 30% of preimplantation embryos (Figure 3 and Table 1). Maternally inherited TRPS1 protein in early PED may mask its roles in preimplantation stage when applying traditional knockout technologies that caused neonatal death of homogeneous Trps1-KO mice from respiratory failure [62]. (Thus Trps1-KO preimplantation embryos cannot be obtained through mating of homogeneous mice). In line with this assumption, the injection of anti-TRPS1 antibody significantly increased the arrest rate of mouse PED (Figure S1). Haploinsufficiency of the Trps1 gene is responsible for tricho-rhino-phalangeal syndrome, which is extremely rare [63]. In addition, Trps1 gene amplification occurs in 28% of human breast cancer cases [38]. Thus, the level of Trps1 should also be tightly controlled. Naturally, about half of the mammalian preimplantation embryos fail to implant [64,65], which suggests the existence of a protective surveillance mechanism during PED that prevents possible developmental defects, possibly by setting the responsible developmental genes as gears of the “zygotic clock.”
In the present study, we found that Trps1 knockdown significantly elevated the expression levels of MuERV-L and Zscan4d (Figure 4(a)). Both MuERV-L and Zscan4d are highly expressed in two-cell embryos, and decline sharply at later stages [19,22]. Active MuERV-L in two-cell embryos is reported to be closely connected with open chromatin [10], and promotes the expression of hundreds of neighboring genes including Zscan4d [20]. In addition, Zscan4 cluster also activates MuERV-L expression [53]. Furthermore, expression of Zscan4 and MuERV-L in ESCs can promote the activation of many two-cell-specific genes and turn mouse ESCs into a “two-cell like” state that is transiently totipotent and important for cell potency [55,66]. Thus, the increased expression of these two markers implies the elevated expression of multiple ZGA genes induced by Trps1 knockdown. TRPS1 is an important repressor of other GAGA family members [25] as well as ERα [38]. Inhibition of ERα was previously shown to decrease the expression of MuERV-L in two-cell embryos [42]. Hence, the functional mechanisms of TRPS1 during early development might be associated with negative gene regulation, which restricts the expression of stage-specific genes to an appropriate level and within the correct time window.
Chromatin state is important for the control of selective gene expression. Recent studies have revealed that preimplantation stage-specific broad H3K4me3 domains are enriched in genes highly expressed at the specific stage, and correlate with ZGA [67]. Other studies revealed that ~22% of the oocyte genome is modified with “noncanonical” broad H3K4me3, which is associated with gene silencing in late-stage oocytes, zygotes, and early two-cell embryos. Broad H3K4me3 domains are erased at the late two-cell stage when H3K4me3 is confined to promoter regions and distal peaks of these genes [58,59]. These results revealed that the regulatory effects of H3K4me3 enrichment during maternal-to-zygotic transition are specific to the cell context and rely on the transcription of critical genes. H3K4me3 and other kinds of histone modifications such as H3K27me3 (whose occupation is resisted by broad H3K4me3 [67]) and H3K27ac (which correlates with open chromatin [10]) cooperate in a tightly regulated manner to determine specific gene expression during major ZGA. Key factors that regulate their synchronous actions have not been addressed. In the present study, we showed that Trps1 knockdown resulted in decreased expression levels of both H3K4me3 and H3K27me3 (Figure 4), while H3K27ac (and H3K36me3) did not change significantly (Figure S5), suggesting that TRPS1 might work through maintaining H3K27me3 and noncanonical H3K4me3 on target genes (e.g., Zscan4d and MuERV-L) and prevent their overexpression.
ZGA failure was observed when mouse two-cell embryos were overexpressed with Sox2 or Oct4, both of which are key pluripotency regulators that can interact with each other to form a SOX2–OCT4 complex and are absent in Zscan4+/MERVL+ ESCs [55,66]. It has been reported that the long-lived SOX2 predicts the cell fate of mouse four-cell embryos because blastomeres with more stable SOX2-DNA binding led to inner cell allocation, during the first lineage specification [68]. Another study revealed that heterogeneity in OCT4 and SOX2 targets (like Sox21) also predicts four-cell embryo fate [69]. Thus, the transcription, translation, and nuclear translocation of Sox2 and Oct4 during two-cell to four-cell transition are critical. Our results showed that the expression level of Sox2 mRNA was decreased by Trps1 knockdown, while Oct4 expression increased significantly (Figure 5). Notably, SOX2 translocated from cytoplasm to nucleus at the late two-cell stage, which could be blocked by Trps1 knockdown. These results suggest the differential regulation of these two genes by TRPS1. The precise levels of Sox2 and Oct4 are critical for the maintenance of pluripotency during embryogenesis after blastocyst formation. Complicated crosstalk may occur between Sox2 and Oct4, although negative feedback loops have been shown to delicately balance their expression [70]. It is reasonable to assert that the differential regulation of Sox2 and Oct4 by TRPS1 might block the function of the SOX2–OCT4-centered signaling network of pluripotency in two-cell embryos, maintain the totipotent state during this stage, and also determine the long-lived DNA binding of SOX2 (but not OCT4) for cell fate at the four-cell stage [68], when TRPS1 expression is maintained at a high level.
Taking these findings together, elevated nuclear localization of TRPS1 exclusively during two-cell to four-cell embryo transition plays an indispensable role in controlling the expression of three highly correlated critical pairs: (1) repeat element MuERV-L with its associated two-cell-specific gene Zscan4d; (2) bivalent histone modifiers, H3K4me3 and H3K27me3; and (3) core regulatory partners of pluripotency, Sox2 and Oct4. We propose that through maintaining the occupation of H3K27me3 and H3K4me3 on cis-regulatory regions of Zscan4d and MuERV-L, TRPS1 restrains their expression to a narrow time window important for further development and blocks the function of the SOX2–OCT4-centered signaling network of pluripotency critical for totipotency establishment during the mouse two-cell stage, thus regulating the progression of ZGA (Figure 6).
Figure 6.
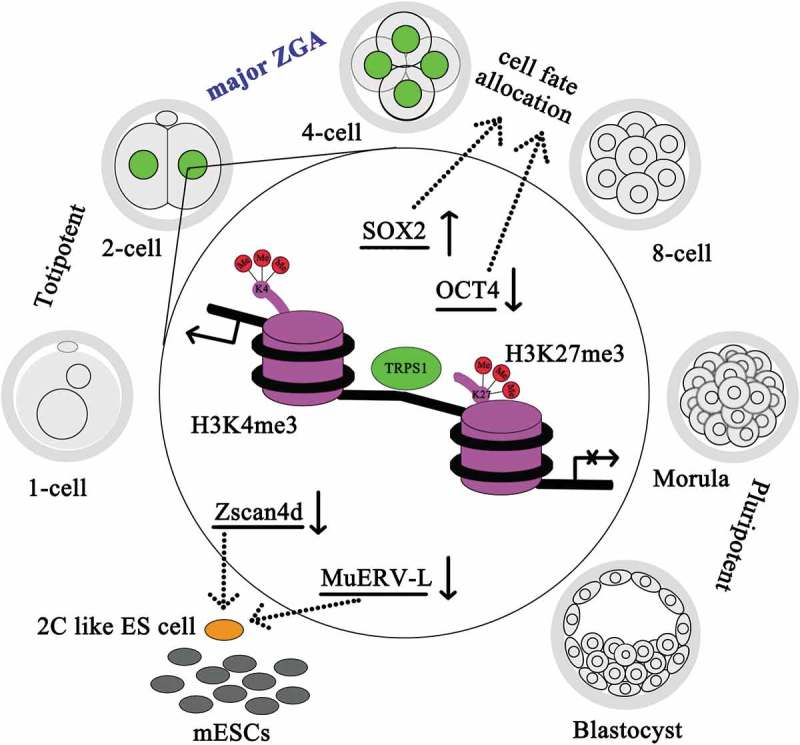
Regulatory model of TRPS1 during mouse early embryogenesis. During mouse preimplantation embryonic development, nuclear localization of TRPS1 (green) was most apparent in two-cell and four-cell embryos and regulated the expression of Zscan4d, MuERV-L, H3K4me3, H3K27me3, Sox2 and Oct4. The effects of TRPS1 on each of them are indicated by solid arrows (results of the present study). Dotted arrows indicate the roles of SOX2 and OCT4 that determine four-cell fate, and the roles of Zscan4d and MuERV-L in establishing two-cell like (“2C”) state in mouse embryonic stem (ES) cells.
Funding Statement
This work was supported by the National Natural Science Foundation of China [81170624]; National Natural Science Foundation of China [81671526]; Natural Science Foundation of Fujian Province [2018J01728]; Startup Foundation of Fujian Medical University [2017XQ2001]; Foundation for High-level Talents of Fujian Medical University [XRCZX2017014].
Acknowledgments
We are grateful for the kind assistance offered by Mrs. L. Lin on confocal imaging. This work was supported by grants from National Natural Science Foundation of China (81671526, 81170624), Natural Science Foundation of Fujian Province (2018J01728), Startup Foundation of Fujian Medical University (2017XQ2001) and Foundation for High-level Talents of Fujian Medical University (XRCZX2017014).
Author Contributions
S. W. designed the study, Y. L. and L. X. conducted most of the microinjection experiments; S. X. and S. W. wrote the manuscript; Y. L., X. L., Y. S. and Y. Z. conducted real-time PCR analysis; S. X. and H. Z. analyzed GEO datasets; R. L., K. M., X. W. and S. X. conducted immunostaining; Y. L. and S. X. analyzed the data; S. W. supervised the study.
Disclosure statement
No potential conflict of interest was reported by the authors.
Supplementary material
Supplemental data for this article can be accessed here.
References
- [1].Flach G, Johnson MH, Braude PR, et al. The transition from maternal to embryonic control in the two-cell mouse embryo. Embo J. 1982;1:681–686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Svoboda P. Mammalian zygotic genome activation. Semin Cell Dev Biol. 2017;84:118–126. [DOI] [PubMed] [Google Scholar]
- [3].Matsumoto K, Anzai M, Nakagata N, et al. Onset of paternal gene activation in early mouse embryos fertilized with transgenic mouse sperm. Mol Reprod Dev. 1994;39:136–140. [DOI] [PubMed] [Google Scholar]
- [4].Aoki F, Worrad DM, Schultz RM.. Regulation of transcriptional activity during the first and second cell cycles in the preimplantation mouse embryo. Dev Biol. 1997;181:296–307. [DOI] [PubMed] [Google Scholar]
- [5].DeRenzo C, Seydoux G.. A clean start: degradation of maternal proteins at the oocyte-to-embryo transition. Trends Cell Biol. 2004;14:420–426. [DOI] [PubMed] [Google Scholar]
- [6].Schultz RM, Stein P, Svoboda P.. The oocyte-to-embryo transition in mouse: past, present, and future. Biol Reprod. 2018;99:160–174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Frum T, Ralston A. Cell signaling and transcription factors regulating cell fate during formation of the mouse blastocyst. Trends Genet. 2015;31:402–410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Plusa B, Piliszek A, Frankenberg S, et al. Distinct sequential cell behaviours direct primitive endoderm formation in the mouse blastocyst. Development. 2008;135:3081–3091. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [9].Eckersley-Maslin MA, Alda-Catalinas C, Reik W. Dynamics of the epigenetic landscape during the maternal-to-zygotic transition. Nat Rev Mol Cell Biol. 2018;19:436–450. [DOI] [PubMed] [Google Scholar]
- [10].Wu J, Huang B, Chen H, et al. The landscape of accessible chromatin in mammalian preimplantation embryos. Nature. 2016;534:652–657. [DOI] [PubMed] [Google Scholar]
- [11].Wicklow E, Blij S, Frum T, et al. HIPPO pathway members restrict SOX2 to the inner cell mass where it promotes ICM fates in the mouse blastocyst. PLoS Genet. 2014;10:e1004618. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Wu G, Han D, Gong Y, et al. Establishment of totipotency does not depend on Oct4A. Nat Cell Biol. 2013;15:1089–1097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Frum T, Halbisen MA, Wang C, et al. Oct4 cell-autonomously promotes primitive endoderm development in the mouse blastocyst. Dev Cell. 2013;25:610–622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pan H, Schultz RM. Sox2 modulates reprogramming of gene expression in two-cell mouse embryos. Biol Reprod. 2011;85:409–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].Fukuda A, Mitani A, Miyashita T, et al. Spatiotemporal dynamics of OCT4 protein localization during preimplantation development in mice. Reproduction. 2016;152:417–430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Chen Y, Wang K, Leach R. GATA transcription factors in pregnancy. Med J Obstet Gynecol. 2013;1:1013. [PMC free article] [PubMed] [Google Scholar]
- [17].Home P, Ray S, Dutta D, et al. GATA3 is selectively expressed in the trophectoderm of peri-implantation embryo and directly regulates Cdx2 gene expression. J Biol Chem. 2009;284:28729–28737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Choi YJ, Lin CP, Risso D, et al. Deficiency of microRNA miR-34a expands cell fate potential in pluripotent stem cells. Science. 2017;355:eaag1927. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].Svoboda P, Stein P, Anger M, et al. RNAi and expression of retrotransposons MuERV-L and IAP in preimplantation mouse embryos. Dev Biol. 2004;269:276–285. [DOI] [PubMed] [Google Scholar]
- [20].Schoorlemmer J, Perez-Palacios R, Climent M, et al. Regulation of mouse retroelement MuERV-L/MERVL expression by REX1 and epigenetic control of stem cell potency. Front Oncol. 2014;4:14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Kigami D, Minami N, Takayama H, et al. MuERV-L is one of the earliest transcribed genes in mouse one-cell embryos. Biol Reprod. 2003;68:651–654. [DOI] [PubMed] [Google Scholar]
- [22].Falco G, Lee SL, Stanghellini I, et al. Zscan4: a novel gene expressed exclusively in late two-cell embryos and embryonic stem cells. Dev Biol. 2007;307:539–550. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Bai H, Sakurai T, Godkin JD, et al. Expression and potential role of GATA factors in trophoblast development. J Reprod Dev. 2013;59:1–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Momeni P, Glockner G, Schmidt O, et al. Mutations in a new gene, encoding a zinc-finger protein, cause tricho-rhino-phalangeal syndrome type I. Nat Genet. 2000;24:71–74. [DOI] [PubMed] [Google Scholar]
- [25].Malik TH, Shoichet SA, Latham P, et al. Transcriptional repression and developmental functions of the atypical vertebrate GATA protein TRPS1. Embo J. 2001;20:1715–1725. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Hu J, Su P, Jia M, et al. TRPS1 expression promotes angiogenesis and affects VEGFA expression in breast cancer. Exp Biol Med (Maywood). 2014;239:423–429. [DOI] [PubMed] [Google Scholar]
- [27].Hong J, Sun J, Huang T. Increased expression of TRPS1 affects tumor progression and correlates with patients’ prognosis of colon cancer. Biol Med Res Int. 2013;2013:454085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Liang H, Cheung LW, Li J, et al. Whole-exome sequencing combined with functional genomics reveals novel candidate driver cancer genes in endometrial cancer. Genome Res. 2012;22:2120–2129. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Gai Z, Zhou G, Gui T, et al. Trps1 haploinsufficiency promotes renal fibrosis by increasing Arkadia expression. J Am Soc Nephrol. 2010;21:1468–1476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Gai Z, Zhou G, Itoh S, et al. Trps1 functions downstream of Bmp7 in kidney development. J Am Soc Nephrol. 2009;20:2403–2411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].Piscopo DM, Johansen EB, Derynck R. Identification of the GATA factor TRPS1 as a repressor of the osteocalcin promoter. J Biol Chem. 2009;284:31690–31703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Wuelling M, Kaiser FJ, Buelens LA, et al. Trps1, a regulator of chondrocyte proliferation and differentiation, interacts with the activator form of Gli3. Dev Biol. 2009;328:40–53. [DOI] [PubMed] [Google Scholar]
- [33].Wu L, Wang Y, Liu Y, et al. A central role for TRPS1 in the control of cell cycle and cancer development. Oncotarget. 2014;5:7677–7690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Wuelling M, Pasdziernik M, Moll CN, et al. The multi zinc-finger protein Trps1 acts as a regulator of histone deacetylation during mitosis. Cell Cycle. 2013;12:2219–2232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Hu J, Su P, Jiao M, et al. TRPS1 suppresses breast cancer epithelial-mesenchymal transition program as a negative regulator of SUZ12. Transl Oncol. 2018;11:416–425. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Radvanyi L, Singh-Sandhu D, Gallichan S, et al. The gene associated with trichorhinophalangeal syndrome in humans is overexpressed in breast cancer. Proc Natl Acad Sci U S A. 2005;102:11005–11010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Chen JQ, Litton J, Xiao L, et al. Quantitative immunohistochemical analysis and prognostic significance of TRPS-1, a new GATA transcription factor family member, in breast cancer. Horm Cancer. 2010;1:21–33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Serandour AA, Mohammed H, Miremadi A, et al. TRPS1 regulates oestrogen receptor binding and histone acetylation at enhancers. Oncogene. 2018;37(39):2581–2591. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hiroi H, Momoeda M, Inoue S, et al. Stage-specific expression of estrogen receptor subtypes and estrogen responsive finger protein in preimplantational mouse embryos. Endocr J. 1999;46:153–158. [DOI] [PubMed] [Google Scholar]
- [40].Wu TC, Wang L, Wan YJ. Expression of estrogen receptor gene in mouse oocyte and during embryogenesis. Mol Reprod Dev. 1992;33:407–412. [DOI] [PubMed] [Google Scholar]
- [41].Xu S, Lian X, Cheng X, et al. Dynamic subcellular localization of estrogen receptor alpha during the first two cleavages of mouse preimplantation embryos. Acta Histochem. 2016;118:317–321. [DOI] [PubMed] [Google Scholar]
- [42].Zhang Y, Jiang Y, Lian X, et al. Effects of ERalpha-specific antagonist on mouse preimplantation embryo development and zygotic genome activation. J Steroid Biochem Mol Biol. 2015;145:13–20. [DOI] [PubMed] [Google Scholar]
- [43].Cheng X, Xu S, Song C, et al. Roles of ERalpha during mouse trophectoderm lineage differentiation: revealed by antagonist and agonist of ERalpha. Dev Growth Differ. 2016;58:327–338. [DOI] [PubMed] [Google Scholar]
- [44].Park SJ, Shirahige K, Ohsugi M, et al. DBTMEE: a database of transcriptome in mouse early embryos. Nucleic Acids Res. 2015;43:D771–D776. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Park SJ, Komata M, Inoue F, et al. Inferring the choreography of parental genomes during fertilization from ultralarge-scale whole-transcriptome analysis. Genes Dev. 2013;27:2736–2748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Nothias JY, Miranda M, DePamphilis ML. Uncoupling of transcription and translation during zygotic gene activation in the mouse. Embo J. 1996;15:5715–5725. [PMC free article] [PubMed] [Google Scholar]
- [47].Kanno S, Gui T, Itoh S, et al. Aberrant expression of the P2 promoter-specific transcript Runx1 in epiphyseal cartilage of Trps1-null mice. Exp Mol Pathol. 2011;90:143–148. [DOI] [PubMed] [Google Scholar]
- [48].Sonehara H, Nagata M, Aoki F. Roles of the first and second round of DNA replication in the regulation of zygotic gene activation in mice. J Reprod Dev. 2008;54:381–384. [DOI] [PubMed] [Google Scholar]
- [49].Shin SW, Tokoro M, Nishikawa S, et al. Inhibition of the ubiquitin-proteasome system leads to delay of the onset of ZGA gene expression. J Reprod Dev. 2010;56:655–663. [DOI] [PubMed] [Google Scholar]
- [50].Zhu Y, Jiang YH, He YP, et al. Knockdown of regulator of G-protein signalling 2 (Rgs2) leads to abnormal early mouse embryo development in vitro. Reprod Fert Dev. 2015;27:557–566. [DOI] [PubMed] [Google Scholar]
- [51].Chen J, Lian X, Du J, et al. Inhibition of phosphorylated Ser473-Akt from translocating into the nucleus contributes to two-cell arrest and defective zygotic genome activation in mouse preimplantation embryogenesis. Dev Growth Differ. 2016;58:280–292. [DOI] [PubMed] [Google Scholar]
- [52].Ye R, Xu S, Liu Y, et al. Protective effect of Icariin on the development of preimplantation mouse embryos against hydrogen peroxide-induced oxidative injury. Oxid Med Cell Longev. 2017;2017:2704532. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Ko MS. Zygotic genome activation revisited: looking through the expression and function of Zscan4. Curr Top Dev Biol. 2016;120:103–124. [DOI] [PubMed] [Google Scholar]
- [54].Sandelin A, Alkema W, Engstrom P, et al. JASPAR: an open-access database for eukaryotic transcription factor binding profiles. Nucleic Acids Res. 2004;32:D91–D94. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Macfarlan TS, Gifford WD, Driscoll S, et al. Embryonic stem cell potency fluctuates with endogenous retrovirus activity. Nature. 2012;487:57–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [56].Macfarlan TS, Gifford WD, Agarwal S, et al. Endogenous retroviruses and neighboring genes are coordinately repressed by LSD1/KDM1A. Gene Dev. 2011;25:594–607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Hayashi M, Maehara K, Harada A, et al. Chd5 regulates MuERV-L/MERVL expression in mouse embryonic stem cells via H3K27me3 modification and histone H3.1/H3.2. J Cell Biochem. 2016;117:780–792. [DOI] [PubMed] [Google Scholar]
- [58].Zhang B, Zheng H, Huang B, et al. Allelic reprogramming of the histone modification H3K4me3 in early mammalian development. Nature. 2016;537:553–557. [DOI] [PubMed] [Google Scholar]
- [59].Dahl JA, Jung I, Aanes H, et al. Broad histone H3K4me3 domains in mouse oocytes modulate maternal-to-zygotic transition. Nature. 2016;537:548–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Funaya S, Aoki F. Regulation of zygotic gene activation by chromatin structure and epigenetic factors. J Reprod Dev. 2017;63:359–363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Abe KI, Funaya S, Tsukioka D, et al. Minor zygotic gene activation is essential for mouse preimplantation development. Proc Natl Acad Sci U S A. 2018;115:E6780–E8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Malik TH, Von Stechow D, Bronson RT, et al. Deletion of the GATA domain of TRPS1 causes an absence of facial hair and provides new insights into the bone disorder in inherited tricho-rhino-phalangeal syndromes. Mol Cell Biol. 2002;22:8592–8600. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Solc R, Klugerova M, Vcelak J, et al. Mutation analysis of TRPS1 gene including core promoter, 5ʹUTR, and 3ʹUTR regulatory sequences with insight into their organization. Clin Chim Acta. 2017;464:30–36. [DOI] [PubMed] [Google Scholar]
- [64].Hertz-Picciotto I, Samuels SJ. Incidence of early loss of pregnancy. New Engl J Med. 1988;319:1483–1484. [DOI] [PubMed] [Google Scholar]
- [65].Wilcox AJ, Weinberg CR, O’Connor JF, et al. Incidence of early loss of pregnancy. New Engl J Med. 1988;319:189–194. [DOI] [PubMed] [Google Scholar]
- [66].Eckersley-Maslin MA, Svensson V, Krueger C, et al. MERVL/Zscan4 network activation results in transient genome-wide DNA demethylation of mESCs. Cell Rep. 2016;17:179–192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Liu X, Wang C, Liu W, et al. Distinct features of H3K4me3 and H3K27me3 chromatin domains in pre-implantation embryos. Nature. 2016;537:558–562. [DOI] [PubMed] [Google Scholar]
- [68].White MD, Angiolini JF, Alvarez YD, et al. Long-lived binding of Sox2 to DNA predicts cell fate in the four-cell mouse embryo. Cell. 2016;165:75–87. [DOI] [PubMed] [Google Scholar]
- [69].Goolam M, Scialdone A, Graham SJL, et al. Heterogeneity in Oct4 and Sox2 targets biases cell fate in four-cell mouse embryos. Cell. 2016;165:61–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [70].Rizzino A, Wuebben EL. Sox2/Oct4: A delicately balanced partnership in pluripotent stem cells and embryogenesis. BBA-Gene Regul Mech. 2016;1859:780–791. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


