Abstract
Sirtuin-1 (SirT1) catalyzes NAD+-dependent protein lysine deacetylation and is a critical regulator of energy and lipid metabolism, mitochondrial biogenesis, apoptosis, and senescence. Activation of SirT1 mitigates metabolic perturbations associated with diabetes and obesity. Pharmacologic molecules, cellular redox, and nutritional states can regulate SirT1 activity.
Technical barriers against measuring endogenous SirT1 activity have limited characterization of SirT1 in disease and its activation by small molecules. Herein, we developed a relative quantitative mass spectrometry-based technique for measuring endogenous SirT1 activity (RAMSSAY/RelAtive Mass Spectrometry Sirt1 Activity assaY) in cell and tissue homogenates using a biotin-labeled, acetylated p53-derived peptide as a substrate.
We demonstrate that oxidative and metabolic stress diminish SirT1 activity in the hepatic cell line HepG2. Moreover, pharmacologic molecules including nicotinamide and EX-527 attenuate SirT1 activity; purported activators of SirT1, the polyphenol S17834, the polyphenol resveratrol, or the non-polyphenolic Sirtris compound SRT1720, failed to activate endogenous SirT1 significantly. Furthermore, we provide evidence that feeding a high fat high sucrose diet (HFHS) to mice inhibits endogenous SirT1 activity in mouse liver.
In summary, we introduce a robust, specific and sensitive mass spectrometry-based assay for detecting and quantifying endogenous SirT1 activity using a biotin-labeled peptide in cell and tissue lysates. With this assay, we determine how pharmacologic molecules and metabolic and oxidative stress regulate endogenous SirT1 activity. The assay may also be adapted for other sirtuin isoforms.
Abbreviations: BSA, bovine serum albumin; CysNO, S-nitrosocysteine; B6J, C57BL/6J mouse strain; DBC-1, deleted in breast cancer 1; GAPDH, glyceraldehyde-3-phosphate dehydrogenase; GSH, reduced glutathione; GSSG, oxidized glutathione; HEK-293, human embryonic kidney cell-293; HepG2, human hepatocellular carcinoma cell line; HFHS, high fat high sucrose diet; HPHG, high palmitate high glucose medium; HPLC, high performance liquid chromatography; IAM, iodoacetamide; IgG, immunoglobulin G; IP, immuno-precipitation; LacZ, beta-galactosidase; NAD+, nicotinamide adenine dinucleotide; ND, chow diet; MS, mass spectrometry; p53, tumor suppressor p53; RONS, reactive oxygen and nitrogen species; SDS-PAGE, sodium dodecyl sulfate polyacrylamide gel electrophoresis; SirBACO, Sirtuin-1 Bacterial Artificial Chromosome Overexpressor; SirT1, Sirtuin-1
Highlights
-
•
Fast, sensitive, and specific MALDI-TOF based sirtuin-1 activity assay applicable to cell and tissue lysates.
-
•
Oxidative and metabolic stress inhibit Sirtuin-1 deacetylase activity.
-
•
Purported activators of SirT1failed to significantly activate endogenous SirT1.
-
•
The activity assay is adaptable to other sirtuin isoforms using specific synthetic peptides and assay conditions.
1. Introduction
The mammalian sirtuin family consists of seven highly conserved class III NAD+-dependent deacetylases and ADP ribosyltransferases. Sirtuins are located in distinct cellular compartments, controlling important metabolic and signaling pathways [1,2]. The most prominent member sirtuin-1 (SirT1) activates processes relevant to caloric restriction and fasting [[3], [4], [5]]. Overexpression or activation of SirT1 [6,7] effectively alleviates metabolic and cardiovascular complications in animal models of metabolic diseases including type-2 diabetes and obesity [[8], [9], [10], [11], [12]]. Molecular targets of SirT1 include transcription factors that control essential cellular stress response pathways which improve metabolic functions such as mitochondrial biogenesis and fatty acid utilization [9,[13], [14], [15], [16], [17]].
The first non-histone SirT1 substrate identified was p53. SirT1 deacetylates the C-terminal Lys382 of p53, repressing its transcriptional activity, and consequently inhibits apoptosis and cell cycle arrest. Subsequently, numerous other targets have been characterized, including transcription factors, co-activators, histones and non-nuclear proteins [[18], [19], [20], [21]]. Assessment of the acetylation status of these proteins, including p53 and histone H3, using antibody-based analytical methods are standard measures of SirT1 activity in cells and tissues [11,12,22].
SirT1 activity is affected by NAD+ co-substrate availability [23,24], oligomerization status [25], association with regulatory proteins [26,27], and post-translational modifications [22,[28], [29], [30], [31], [32], [33]] including cysteine thiol oxidation. During the catalytic cycle, SirT1 deacetylates target proteins, converting NAD+ into nicotinamide and O-acetyl ADP-ribose [16,17,34]. Thus, primary assays use radioisotope labeled substrates including 14C-NAD+, 14C-acetylated p53 or 3H-acetylated histones [16,[35], [36], [37]]. More recent methods, however, employ a two-step fluorometric assays such as Fluor de Lys [[38], [39], [40], [41], [42], [43]] or high-performance liquid chromatography (HPLC) [[44], [45], [46], [47]] to detect deacetylated peptide levels. The fluorometric assays rely on substrate deacetylation because this gives trypsin access to cleave off the quenched fluorophore. The developed fluorescence is proportional to SirT1 activity. These assays are reliable for measuring the activity of purified SirT1, but generate variable results in cell and tissue samples, due to lack of sensitivity and specificity. To address these limitations, we used a biotin-labeled acetylated p53 peptide to develop a relative quantitative mass spectrometry-based method, which is specific and sensitive. This method allows differential quantification of the peak intensities of deacetylated-p53 vs. acetylated-p53 to measure in vivo SirT1 activity. Because custom-synthesized peptide substrates are commercially available, our strategy can also be applied for analysis of other sirtuin isoforms and peptide substrates. Employing this method, we investigated the impact of polyphenolic (S17834, resveratrol) or non-polyphenolic (SRT1720, EX-527) compounds, cellular redox potential (H2O2, CysNO, GSSG), and nutritional state (HPHG, high fat high sucrose diet) on SirT1 activity in cells and mice.
2. Materials and methods
2.1. Reagents, materials, and antibodies
S17834 (6,8-diallyl-5,7-dihydroxy-2-(2-allyl-3-hydroxyl-4-methoxyphenyl)1-H-benzo (b)pyran-4-one) and SRT1720 (N-{2-[3-(piperazine-1-ylmethyl)imidazo [2,1-b] [1,3]thiazol-6-yl]phenyl}-2-quinoxaline-carboxamide), EX-527 (6-chloro-2,3,4,9-tetrahydro-1-H-carbazole-1-carboxamide), were obtained from the Institut de Recherche Servier (Suresnes, France). The following antibodies were used: anti-Flag M2 (Sigma, St. Louis, MO; F1804), anti-Sirtuin-1 (Abcam, Cambridge, MA; ab110304), anti-GAPDH (Cell Signaling Technology, Danvers, MA; #2118). Anti-Flag M2 Affinity Gel was purchased from Sigma Aldrich, catalog number: A2220. Avidin agarose (cat # PI29200), streptavidin agarose (cat # 20347) and streptavidin magnetic beads (cat # 88816) were obtained from Thermo Fisher Scientific, Waltham, MA. Biotin-labeled Ac-Lys382-p53 peptide with a 6-carbon linker (cat # 65045) was synthesized by Anaspec, San Jose, CA. Zeba™ spin desalting columns (40K MWCO, 87767), Lipofectamine™ and cell culture media were bought from Life Technologies (Grand Island, NY).
2.2. Cell culture
HepG2 cells (ATCC, Manassas, VA) were maintained in Dulbecco's Modified Eagle Medium containing 10% fetal bovine serum and penicillin/streptomycin (Gibco, Grand Island, NY). Transfected cells were either incubated in control medium containing 5 mM glucose and 0.67% bovine serum albumin (BSA, fatty acid free, Sigma-Aldrich St. Louis, MO) or medium supplemented with high palmitate (0.4 mM palmitic acid and 0.67% BSA) and high glucose (25 mM glucose, referred to as HPHG) for 16 h.
2.3. Experimental animals
Male SirT1 Bacterial Artificial Chromosome Overexpressor (SirBACO) mice with C57BL6/NJ genetic background were obtained from Dr. Wei Gu, (Columbia University, NY). A cohort of 2-month-old male SirBACO mice and WT littermates were fed control or high fat and high sucrose diet (HFHS: 35.5% fat representing 60% calories, 16.4% sucrose) ad libitum for ten months (D09071702 and D09071703) to investigate the effects of metabolic stress. Mice were housed in rooms with 12-h light/dark cycle in groups of 3–4, whenever possible. The Institutional Animal Care and Use Committee at Boston University School of Medicine approved the animal protocol. Mice were euthanized after ten months on the diet and livers were perfused, excised, snap-frozen, and stored in liquid nitrogen or at −80 °C for later analysis.
2.4. Homogenization and protein extraction of mouse liver
Homogenization and extraction of individual liver samples were carried out in NP-40 lysis buffer containing 50 mM Tris pH 7.4, 150 mM NaCl, 1 mM EDTA, 1% NP40, and a protease inhibitor cocktail (Roche Applied Science, Penzberg, Germany).
2.5. Preparation of S-nitrosocysteine
S-nitrosocysteine (Cys-NO) stock solutions were freshly prepared by mixing equimolar amounts of l-cysteine and NaNO2 under acidic conditions (0.25 M HCl) in the presence of 0.1 mM diethylenetriamine pentaacetate (DTPA). The concentration of Cys-NO in solution was measured spectrophotometrically at 334 nm, ε344 = 900 M−1 cm−1 and adjusted to 500 mM. Dilutions of Cys-NO were prepared in HEN buffer (250 mM HEPES, pH 7.7, 1 mM EDTA, 0.1 mM neocuproine) immediately before conducting experiments.
2.6. SirT1 activity measurement using p53-targeted mass spectrometry (RAMSSAY)
HepG2 cells (approx. 1.3 × 105 cells) were infected with 0.16 × 108 pfu WT SirT1 or dominant negative H355A mutant SirT1 (HA) adenovirus for 48 h. Cell or tissue lysates (800 μg of protein prepared in 500 μl NP-40 lysis buffer) were precleared with 5 μl streptavidin magnetic beads for 1 h at 4 °C on a rotator. Precleared lysates were incubated with biotin-labeled acetylated p53 peptide (Biotin-LC-KKGQSTSRHK-K(Ac)-LMFKTEG; 10 μM final concentration) and 100 μM NAD+ for 30 min at 37 °C. Streptavidin beads (10 μl of a 1:1 slurry) were added and incubated for 1 h at room temperature. Beads were washed three times with 300 μl PBS, three times with 300 μl Tris buffer (25 mM; pH 7.4), and three times with 300 μl of ddH2O. Biotin-tagged peptides were either eluted into 100 μl 25 mM Tris buffer with 5 mM biotin pH 8.0 at RT or into 100 μl ddH2O with 5 mM DTT heated at 90 °C for 5 min. The supernatant was dried down in a SpeedVac™ concentrator (Savant, Thermo Fisher Scientific), suspended in 15 μl ddH2O, and then desalted with a C18 ZipTip™ (EMD Millipore, Billerica, MA) before mass spectrometry analysis. The acetylated and deacetylated p53 peptides were analyzed with an UltrafleXtreme MALDI-TOF/TOF MS (Bruker Daltonics, Billerica, MA) using α-cyano-4-hydroxycinnamic acid as matrix. The MALDI-TOF mass spectra were summed from 2000 laser shots for good signal intensity with a resolution of 10,000 fwhm and mass accuracy <10 ppm at m/z 400. Concentration changes of the acetylated and deacetylated p53 were calculated by determining the difference in relative peak intensities observed for the [M + H]+ signal corresponding to each.
2.7. Statistical analysis
Statistical analysis was performed using Prism 5.0 (GraphPad Software). Means were compared between two groups by one-way ANOVA or multiple comparisons two-way ANOVA analysis with Bonferroni's post-test. A P value of <0.05 was considered statistically significant.
3. Results
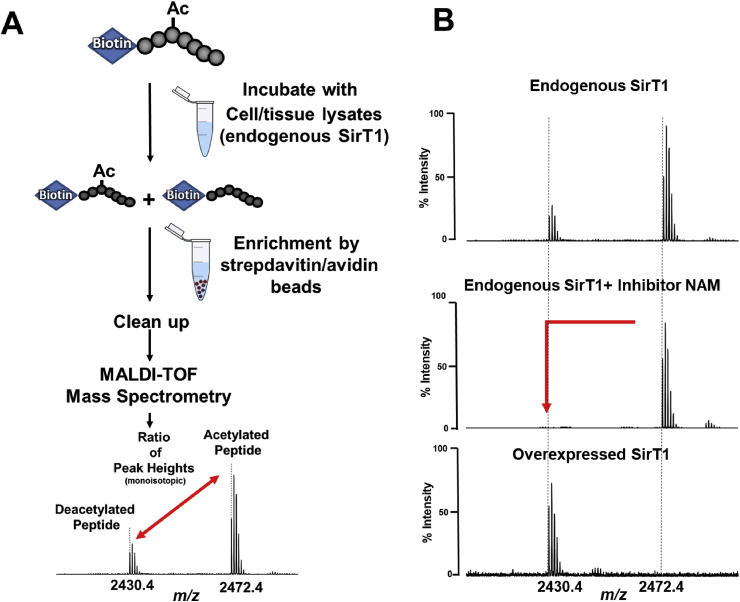
3.1. The principle of the relative quantitative mass spectrometry-based activity assay (RAMSSAY) using a biotin-tagged p53 peptide
We have selected matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS due to its wide availability, high sample throughput, relative ease of use, and tolerance to all classes of samples. Acetylated lysine 382 of the tumor suppressor p53 is a well-characterized SirT1 target. Therefore, we selected a readily acetylated peptide corresponding to amino acid residues 372–389 of p53 as a SirT1 substrate. Biotin, covalently attached to the N-terminus of the peptide, enables highly efficient enrichment and cleanup for MS analysis via streptavidin-avidin supports [48,49] (Fig. 1A). Because of the ease of custom peptide synthesis, the assay is adaptable to different peptide substrates for testing of newly discovered protein acetylation sites or other protein deacetylases. We selected streptavidin magnetic beads for fast sample preparation and tested various elution conditions. Mass spectrometry [50,51] is somewhat incompatible with detergents or chaotropic salts, which are required to denature and disrupt the highly stable biotin-streptavidin complex. Several reports showed that water heated above 70 °C efficiently disrupts the complexes [52]. With our assay, a combination of water and DTT released more biotinylated peptide than pure water (Supplemental Figs. 1A and B), but the recovery rate of biotinylated peptide remained below 10%. Competition with free biotin [[53], [54], [55]] in Tris buffer followed by desalting of the peptide with C18 Zip-tips resulted in a 50% higher recovery rate, so we used this protocol in all subsequent experiments (Supplemental Fig. 2 A and B). We detected acetylated and deacetylated peptide peaks with the expected mass shift of 42 Da and differentially quantified the abundances of these peaks in the same mass spectrum (Fig. 1B). The acetylated and non-acetylated peptides had comparable ionization and detection efficiencies. To determine SirT1 activity, we calculated the ratio of acetylated to deacetylated peptide peak intensities. Since the substrate concentration is known, the computation of specific activities is possible.
Fig. 1.
Principle of RAMSSAY. A) SirT1 activity was calculated by the peak intensity ratio of deacetylated (p53) versus acetylated p53 peptide substrate (Ac-p53). The [M + H]+ peaks of the deacetylated (m/z 2430.4) and acetylated (m/z 2472.4) p53 peptide are detected at isotopic resolution (cluster of peaks is derived from the natural abundance distribution of carbon 13 in the peptide). The height of the monoisotopic (first) peak in each cluster annotated by the dotted line was used for the calculation. B) The right panel shows typical examples of RAMSSAY with HepG2 cell lysates to measure endogenous (top and middle) or overexpressed SirT1 activity (bottom). Nicotinamide (10 mM; middle) inhibited SirT1 activity and prevented deacetylation of the peptide.
3.2. RAMSSAY is specific for SirT1 activity
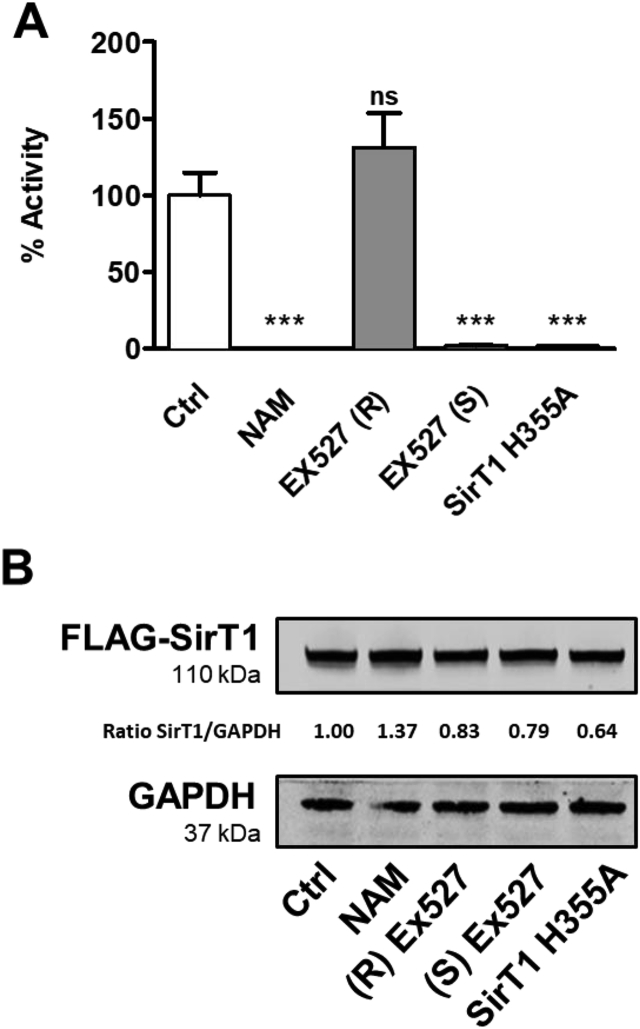
SirT1 activity requires the cofactor NAD+ (nicotinamide adenine dinucleotide) to deacetylate lysine residues of target proteins. The enzymatic reaction catalyzed by SirT1 produces the deacetylated substrate, nicotinamide (NAM) and O-acetyl-ADP-ribose (OAADPr) [56,57]. NAM acts as a competitive feedback inhibitor by binding to a conserved pocket adjacent to the NAD+ binding site, thereby blocking NAD+ hydrolysis [45]. EX-527 is a SirT1 specific inhibitor widely used in physiological studies [58,59]. The (S)-enantiomer ((S) EX-527) is a potent SirT1 inhibitor, whereas the (R)-enantiomer ((R) EX-527) is ineffective in inhibition, thus serves as control [59]. We used 1.6 × 107 pfu adenovirus to overexpress FLAG-tagged wild-type or dominant negative H355A mutant SirT1 (HA) [60,61], which lacks deacetylase activity, in HepG2 cells (1.3 × 105 cells). Cell lysates (800 μg) contained ∼1.5 μg SirT1 by comparison to a Coomassie-stained protein standard used in each assay (Supplemental Figs. 3A and B). We used 10 mM NAM and 10 μM (S) EX-527 to inhibit SirT1 activity in HepG2 cells (Fig. 2). Both inhibitors decreased SirT1 activity without affecting protein expression (Fig. 2 and Supplemental Fig. 4). Also, SirT1 H355A did not increase activity. The data indicate that the assay reliably measures SirT1 activity in cell lysates.
Fig. 2.
Effects of inhibitors on SirT1 activity in HepG2 cells. A) HepG2 cells overexpressing FLAG-SirT1 or dominant negative FLAG-SirT1 H355A were treated with 10 mM nicotinamide (NAM), or 10 μM active inhibitor EX-527 (S)-enantiomer, or 10 μM negative control EX-527 (R)-enantiomer. FLAG-tagged wild-type (Ctrl, NAM, EX527 (R), and EX527 (S)) or dominant negative SirT1 (SirT1 H355A), a mutant protein with inactivated deacetylase, were overexpressed at equal levels in HepG2 cells. p53/Ac-p53 peak intensity ratios were calculated from mass spectra shown in Supplemental Fig. 4 and expressed as percent activity relative to control. Data are presented as means ± SD of N = 3 and were analyzed with one-way ANOVA followed by Bonferroni's post-test (***p < 0.0001; ns = non-significant). B) Western blot analysis of FLAG-SirT1 expression levels in HepG2 cells. The ratio of SirT1 to GAPDH between the blots denotes the results of the densitometric analysis.
3.3. Effects of small molecule activators on SirT1 activity
Polyphenols interfere with common acetylated peptide substrates with covalently attached fluorophores such as Fluor-de-Lys or TAMRA-p53 in SirT1 deacetylation assays [47]. These peptide substrates suboptimally bind to the enzyme, and small molecule SirT1 activators [9,[62], [63], [64]] including resveratrol (RSV) improve this interaction, accelerating peptide turnover, which may result in SirT1 activity overestimation. However, SirT1 activity assays using native peptides failed to detect increased deacetylase activity in response to SirT1 activator treatment [47]. We treated HepG2 cells with S17834, RSV, or the non-polyphenolic compound SRT1720 to determine the effects of small molecule activators on SirT1 in our assay, but none significantly increased SirT1 activity (Supplemental Figs. 4A and B).
3.4. Oxidative and metabolic stress inhibit SirT1 activity in HepG2 cells
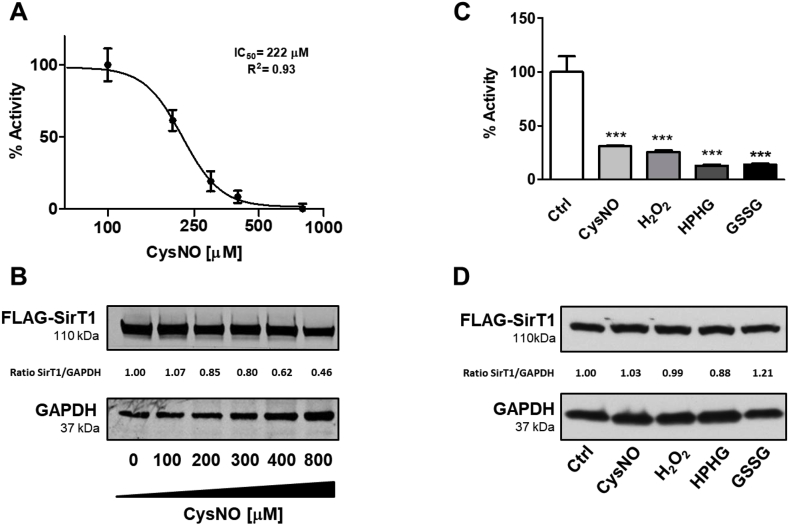
Through changes in intracellular NAD+ concentration, protein interactions, or post-translational modification, oxidative stress affects SirT1 activity [27,[65], [66], [67], [68]]. We previously showed that S-nitrosoglutathione (GSNO) inhibits SirT1 in vitro with an IC50 of 69 μM [68] by reversibly modifying several cysteines of SirT1. Removal of these modifications by chemical reduction with DTT fully restores SirT1 activity. Exposure of cells to S-nitrosocysteine (CysNO) or hydrogen peroxide also inhibit SirT1 through reversible cysteine oxidation [22]. Moreover, we demonstrated that mutant SirT1 with three cysteine-to-serine substitutions (C61S, C318S, and C613S) maintains full enzyme activity when exposed to oxidative or metabolic stress. Here, we incubated HepG2 cells overexpressing WT SirT1 with CysNO overnight. Increasing CysNO concentrations (100–800 μM) did not alter SirT1 protein expression (Fig. 3A and B, and Supplemental Fig. 6). However, our assay measured concentration-dependent SirT1 inhibition with an IC50 of ∼222 μM. Using endogenous acetylated p53 as a surrogate marker of SirT1 activity, we observed comparable results (Supplemental Fig. 7).
Fig. 3.
Effects of different stressors on SirT1 activity in HepG2 cells. A) HepG2 cells were treated with increasing concentrations of S-nitrosocysteine (CysNO) overnight. p53/Ac-p53 peak intensity ratios were calculated from mass spectra shown in Supplemental Fig. 6. Data are presented as means ± SD of N = 3. CysNO concentration was Log10 transformed, normalized, and curve fitted using the least squares method and variable slope. B) Western blot analysis of FLAG-SirT1 expression levels in HepG2 cells. The ratio of SirT1 to GAPDH between the blots denotes the results of the densitometric analysis. C) HepG2 cells were treated with 400 μM S-nitrosocysteine (CysNO), 500 μM oxidized glutathione (GSSG) ethyl ester, 100 μM hydrogen peroxide (H2O2), or 400 μM high palmitate high glucose (HPHG) overnight. p53/Ac-p53 peak intensity ratios were calculated from mass spectra shown in Supplemental Fig. 7 and normalized to Ctrl. Data are presented as means ± SD of N = 3 and were analyzed with one-way ANOVA followed by Bonferroni's post-test (***p < 0.0001). D) Western blot analysis of FLAG-SirT1 expression levels in HepG2 cells. The ratio of SirT1 to GAPDH between the blots denotes the results of the densitometric analysis.
Obesity, metabolic syndrome, and type-2 diabetes are associated with oxidative stress, leading to SirT1 inhibition [69,70]. We previously reported results obtained using an in vitro model which mimics metabolic stress and increases oxidants [22] in cultured cells by exposing them to high palmitate high glucose (HPHG). Similarly, loading cells with permeable GSSG diethyl ester mimics oxidative stress by disturbing the intracellular ratio of reduced-to-oxidized glutathione (GSH:GSSG) [71,72]. HepG2 cells exposed overnight to hydrogen peroxide, GSSG diethyl ester, or HPHG showed 70–80% diminished SirT1 activity without changing protein expression (Fig. 3C and D, and Supplemental Fig. 8).
3.5. Detection of SirT1 activity in hepatic lysates
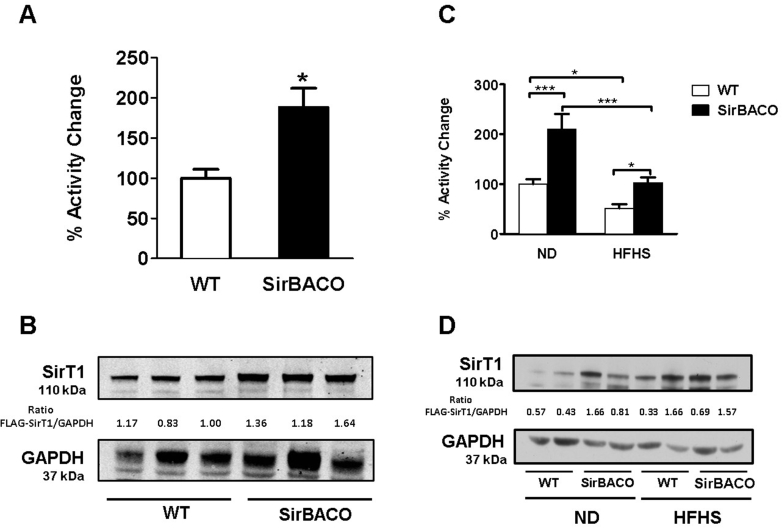
SirT1 bacterial artificial chromosome overexpressor (SirBACO) mice [73] overexpress SirT1 up to 2-fold in the liver, compared to WT mice (Fig. 4A and B). Because of high non-specific deacetylase activity in hepatic lysates, the Fluor-de-Lys assay failed to detect changes in SirT1 activity in SirBACO mice (data not shown), leading investigators to use Western blot-based methods to measure SirT1 activation in these mice [74]. Our MS-based assay, however, measured an ∼2-fold increase in SirT1 activity, consistent with SirT1 protein expression levels in SirBACO mice (Fig. 4A and Supplemental Fig. 9).
Fig. 4.
Endogenous SirT1 activity in liver of WT and SirBACO mice fed normal and high fat high sucrose diet. A) SirT1 activity was measured in liver homogenates from WT (n = 4) and SirBACO mice (n = 4). p53/Ac-p53 peak intensity ratios were calculated from mass spectra shown in Supplemental Fig. 8. Data are presented as means ± SD of N = 4 and were analyzed with unpaired t-test (*p < 0.05). B) Western blot analysis of SirT1 expression levels in liver from WT and SirBACO mice. The ratio of SirT1 to GAPDH between the blots denotes the results of the densitometric analysis. C) SirT1 activity was measured in WT (n = 3) and SirBACO mice (n = 3) fed normal chow (ND) or HFHS diet. p53/Ac-p53 peak intensity ratios were calculated from mass spectra shown in Supplemental Fig. 9. Data are presented as means ± SD of N = 3 and were analyzed with one-way ANOVA followed by Bonferroni's post-test (*p < 0.05, ***p < 0.001). D) Western blot analysis of SirT1 expression levels in mouse liver. The ratio of SirT1 to GAPDH between the blots denotes the results of the densitometric analysis.
We next investigated the nutritional effects on liver SirT1 activity in mice fed regular chow (ND) or HFHS for ten months. HFHS increased SirT1 protein expression in both WT and SirBACO mice (Fig. 4C and D). Although SirT1 expression in HFHS-fed WT mice increased to levels comparable to SirBACO mice, the activity dropped to ∼50% of the level observed in ND WT mice. As expected, SirT1 activity was markedly higher in ND-fed SirBACO mice compared to ND-fed WT controls, and HFHS significantly decreased SirT1 activity in both (Fig. 4C and D, and Supplemental Fig. 10). The data are in accordance with our previous findings [11,12] that HFHS inhibits SirT1 by oxidative inactivation.
4. Discussion
We developed a specific and sensitive mass spectrometry-based assay to measure endogenous SirT1 activity. The assay offers significant improvement over current methods, because it avoids potential artifacts associated with methods that indirectly detect SirT1 activity, including Western blots and fluorophore-containing peptide-based measurements [36,47,75]. Because of the higher sensitivity and specificity of the RAMSSAY method, we can now reliably determine SirT1 activity in cell and tissue lysates [[44], [45], [46]].
To measure SirT1 deacetylase activity in this assay, we used a synthetic p53-derived peptide that was acetylated and biotin-tagged for efficient recovery from lysates. To overcome recognized technical limitations to the efficient recovery of biotinylated peptides, we developed this assay by investigating different elution conditions. Streptavidin/avidin coated supports are highly selective to retrieve biotinylated molecules from lysates. However, the non-covalent bond between biotin and streptavidin/avidin with a Kd of ∼10−15 M hampers complex dissociation [52,76]. Bioengineered streptavidin/avidin such as nitrated CaptAvidin [77], deglycosylated neutravidin, or monomeric avidin [78] have lower binding affinities for biotin and require less stringent conditions for elution. Furthermore, most detergents in lysis buffers interfere with mass spectrometry [54], so their use should be minimized if not avoided. Elution with free biotin followed by ZipTip desalting yielded the best results.
We used known SirT1 inhibitors to verify the specificity of our assay. EX-527 (IC50 = 38 nM) specifically inhibits SirT1 deacetylase activity and is more potent than nicotinamide (NAM; IC50 = 40–50 μM) [40,42,58]. Both EX-527 and nicotinamide inhibited deacetylation of the peptide substrate in HepG2 cells. Overexpression of dominant-negative inactive mutant SirT1 (SirT1 H355A) had a similar effect and almost erased SirT1 activity (98% vs. control; Fig. 2A and B). The results of these experiments confirm that our assay measures specifically SirT1 deacetylase activity in HepG2 cells.
Polyphenols [13] and structurally unrelated Sirtris compounds [9] are believed to stimulate SirT1 activity directly. Small molecule SirT1 activators such as SRT1720 improve insulin sensitivity, inhibit atherogenesis, increase mitochondrial content, and prolong survival of obese mice [[62], [63], [64],[79], [80], [81]]. Recent reports demonstrated that pharmacological activation of SirT1 by these small molecules might falsely indicate indirect SirT1 activation in vitro due to an assay artifact [47]. Several studies have shown that resveratrol and SRT1720 increase SirT1 activity only when assayed with peptide substrates containing a covalently attached fluorophore [36,47,75]. On the contrary, and consistent with other reports [36,75,82], both SirT1 activators failed to increase deacetylase activity in HepG2 cells when measured with our assay (Supplemental Figs. 4A and B). The polyphenol S17834 developed by Servier showed a trend of increased endogenous SirT1 activity. Regulation of SirT1 activity by polyphenols largely depends on the cell type [83,84]. In many cases, indirect factors including changes in SirT1 expression, protein interactions, post-translational modifications or antioxidant effects of polyphenols influence deacetylase activity. Thus, measurements of SirT1 activity, expression level, and downstream targets are pivotal to thoroughly characterizing the pharmacodynamics of small molecule SirT1 activators.
Nutritional status, inflammation, stress response, and tissue damage are closely associated with changes in cellular redox homeostasis and SirT1 activity. Conflicting effects on SirT1 expression and activity have been reported in some cell and mouse models [85]. Cells challenged by oxidative or genotoxic stress have significantly decreased SirT1 expression and activity [66,[86], [87], [88]]. Other reports, including our studies, showed unchanged SirT1 expression levels while observed SirT1 activity decreased due to oxidative post-translational modifications or protein-protein interaction [11,12,22,27,69,89]. Here we similarly reported unaltered SirT1 expression in HepG2 cells exposed to oxidative or metabolic stress in association with decreased activity (Fig. 3A–D). In contrast, increased SirT1 expression has been reported for fasting and caloric restriction [3,89,90], although oxidants increased in muscle, liver, and heart of fasted mice [3,19,89] and aged monkey hearts [91]. Consistent with the fact that SirT1 activity does not always correlate with its expression levels, we found that HFHS diet upregulated SirT1 expression in mouse livers while decreasing its activity (Fig. 4C and D). Also, SirBACO mice, an in vivo model of SirT1 overexpression, showed decreased SirT1 activity after feeding despite even greater increased hepatic SirT1 expression (Fig. 4A–D).
The assay described in this manuscript only measures SirT1 enzyme deacetylase activity and its modulation by post-translational modifications. However, the biology of SirT1 is far more complex and requires additional control experiments including measuring the intracellular NAD+:NADH ratio [43,56], a co-substrate that may become limiting in pathophysiology including aging, ischemia, and metabolic disease [57,92]. SirT1 is the largest member of the sirtuin family and its N- and C-terminal unstructured regions interact with various proteins that function as scaffolds [93], enzyme modulators (aros), or transcription factors [94]. Thus, surrogate markers of SirT1 activity including acetylated-histone H3 or acetylated p53 are excellent endogenous indicators of SirT1 activity. In cardiac myocytes and other cells, SirT1 may translocate from the cytoplasm to the nucleus [95,96]. Thus, intracellular localization could be another factor affecting SirT1 biology.
We established a new specific assay to measure endogenous SirT1 activity in cell and tissue lysates (Fig. 1). Our assay protocol can be used as a template for the testing of a variety of custom synthesized substrate peptides, in addition to the p53 peptide employed by us. In this example reported here, putative small molecule SirT1 activators RSV, SRT1720, or S17834 failed to increase SirT1 deacetylase activity. We confirmed that metabolic and oxidative stress in cells and mice markedly inhibited SirT1 activity despite increased protein expression. Our study underlines the importance of sensitive and specific methods to measure SirT1 activity, in addition to SirT1 protein expression, in order to reliably elucidate the biological function of SirT1, particularly in the settings of metabolic and oxidative stresses, and, most importantly, for drug development of SirT1 activators.
Author disclosure statement
No competing financial interests exist.
Acknowledgments
This work was supported by NIH grants P01 HL068758, R37 HL104017, R01 DK076942, R01 HL136311. R01 DK103750, R01 HL133013, R01 HL115955, S10 OD010724, NIH CTSI award 1UL1TR001430, NIH-NHLBI contract no. HHSN268201000031C (N01-HV-00239), American Heart Association ‘Grant in Aid’ 16GRNT27660006 and European Cooperation in Science and Technology (COST Action BM1203/EU-ROS). The manuscript contents are solely the responsibility of the authors and do not necessarily represent the official views of the awarding offices. D. Shao was supported by an American Heart Association Scientist Postdoctoral Fellowship Award 15POST21790006. The work was supported by a Strategic Alliance with Institut de Recherche Servier and the Metabolic Clinical Research Collaborative at Boston University. We thank Drs. J. Han, M. Zang, M. Kirber, T. Balon, L. Deng, and the Boston University School of Medicine Analytical Instrumentation, Immunohistochemistry, Cellular Imaging, and Metabolic Phenotyping Cores for their technical support.
Footnotes
Supplementary data to this article can be found online at https://doi.org/10.1016/j.redox.2019.101150.
Appendix A. Supplementary data
The following is the supplementary data to this article:
References
- 1.Haigis M.C., Sinclair D.A. Mammalian sirtuins: biological insights and disease relevance. Annu. Rev. Pathol. 2010;5:253–295. doi: 10.1146/annurev.pathol.4.110807.092250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Michan S., Sinclair D. Sirtuins in mammals: insights into their biological function. Biochem. J. 2007;404:1–13. doi: 10.1042/BJ20070140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Rodgers J.T., Puigserver P. Fasting-dependent glucose and lipid metabolic response through hepatic sirtuin 1. Proc. Natl. Acad. Sci. U. S. A. 2007;104:12861–12866. doi: 10.1073/pnas.0702509104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cohen H.Y., Miller C., Bitterman K.J., Wall N.R., Hekking B., Kessler B., Howitz K.T., Gorospe M., de Cabo R., Sinclair D.A. Calorie restriction promotes mammalian cell survival by inducing the SIRT1 deacetylase. Science. 2004;305:390–392. doi: 10.1126/science.1099196. [DOI] [PubMed] [Google Scholar]
- 5.Wolf G. Calorie restriction increases life span: a molecular mechanism. Nutr. Rev. 2006;64:89–92. doi: 10.1301/nr.2006.feb.89-92. http://www.ncbi.nlm.nih.gov/pubmed/16536186 [DOI] [PubMed] [Google Scholar]
- 6.Bordone L., Cohen D., Robinson A., Motta M.C., van Veen E., Czopik A., Steele A.D., Crowe H., Marmor S., Luo J., Gu W., Guarente L. SIRT1 transgenic mice show phenotypes resembling calorie restriction. Aging Cell. 2007;6:759–767. doi: 10.1111/j.1474-9726.2007.00335.x. [DOI] [PubMed] [Google Scholar]
- 7.Pfluger P.T., Herranz D., Velasco-Miguel S., Serrano M., Tschöp M.H. Sirt1 protects against high-fat diet-induced metabolic damage. Proc. Natl. Acad. Sci. U. S. A. 2008;105:9793–9798. doi: 10.1073/pnas.0802917105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Elliott P.J., Jirousek M. Sirtuins: novel targets for metabolic disease. Curr. Opin. Investig. Drugs. 2008;9:371–378. http://www.ncbi.nlm.nih.gov/pubmed/18393104 [PubMed] [Google Scholar]
- 9.Milne J.C., Lambert P.D., Schenk S., Carney D.P., Smith J.J., Gagne D.J., Jin L., Boss O., Perni R.B., Vu C.B., Bemis J.E., Xie R., Disch J.S., Ng P.Y., Nunes J.J., Lynch A.V., Yang H., Galonek H., Israelian K., Choy W., Iffland A., Lavu S., Medvedik O., Sinclair D.A., Olefsky J.M., Jirousek M.R., Elliott P.J., Westphal C.H. Small molecule activators of SIRT1 as therapeutics for the treatment of type 2 diabetes. Nature. 2007;450:712–716. doi: 10.1038/nature06261. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Potente M., Dimmeler S. Emerging roles of SIRT1 in vascular endothelial homeostasis. Cell Cycle. 2008;7:2117–2122. doi: 10.4161/cc.7.14.6267. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18641460 [DOI] [PubMed] [Google Scholar]
- 11.Fry J.L., Al Sayah L., Weisbrod R.M., Van Roy I., Weng X., Cohen R.A., Bachschmid M.M., Seta F. Vascular smooth muscle sirtuin-1 protects against diet-induced aortic stiffness novelty and significance. Hypertension. 2016:68. doi: 10.1161/HYPERTENSIONAHA.116.07622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Weisbrod R.M., Shiang T., Al Sayah L., Fry J.L., Bajpai S., Reinhart-King C.A., Lob H.E., Santhanam L., Mitchell G., Cohen R.A., Seta F. Arterial stiffening precedes systolic hypertension in diet-induced obesity. Hypertension. 2013;62:1105–1110. doi: 10.1161/HYPERTENSIONAHA.113.01744. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Baur J.A., Pearson K.J., Price N.L., Jamieson H.A., Lerin C., Kalra A., Prabhu V.V., Allard J.S., Lopez-Lluch G., Lewis K., Pistell P.J., Poosala S., Becker K.G., Boss O., Gwinn D., Wang M., Ramaswamy S., Fishbein K.W., Spencer R.G., Lakatta E.G., Le Couteur D., Shaw R.J., Navas P., Puigserver P., Ingram D.K., de Cabo R., Sinclair D.A. Resveratrol improves health and survival of mice on a high-calorie diet. Nature. 2006;444:337–342. doi: 10.1038/nature05354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Lagouge M., Argmann C., Gerhart-Hines Z., Meziane H., Lerin C., Daussin F., Messadeq N., Milne J., Lambert P., Elliott P., Geny B., Laakso M., Puigserver P., Auwerx J. Resveratrol improves mitochondrial function and protects against metabolic disease by activating SIRT1 and PGC-1alpha. Cell. 2006;127:1109–1122. doi: 10.1016/j.cell.2006.11.013. [DOI] [PubMed] [Google Scholar]
- 15.Rothgiesser K.M., Erener S., Waibel S., Luscher B., Hottiger M.O. SIRT2 regulates NF-kappaB dependent gene expression through deacetylation of p65 Lys310. J. Cell Sci. 2010;123:4251–4258. doi: 10.1242/jcs.073783. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=21081649 [DOI] [PubMed] [Google Scholar]
- 16.Luo J., Nikolaev A.Y., Imai S., Chen D., Su F., Shiloh A., Guarente L., Gu W. Negative control of p53 by Sir2alpha promotes cell survival under stress. Cell. 2001;107:137–148. doi: 10.1016/s0092-8674(01)00524-4. http://www.ncbi.nlm.nih.gov/pubmed/11672522 [DOI] [PubMed] [Google Scholar]
- 17.Nemoto S., Fergusson M.M., Finkel T. SIRT1 functionally interacts with the metabolic regulator and transcriptional coactivator PGC-1{alpha} J. Biol. Chem. 2005;280:16456–16460. doi: 10.1074/jbc.M501485200. [DOI] [PubMed] [Google Scholar]
- 18.Brunet A., Sweeney L.B., Sturgill J.F., Chua K.F., Greer P.L., Lin Y., Tran H., Ross S.E., Mostoslavsky R., Cohen H.Y., Hu L.S., Cheng H.-L., Jedrychowski M.P., Gygi S.P., Sinclair D.A., Alt F.W., Greenberg M.E. Stress-dependent regulation of FOXO transcription factors by the SIRT1 deacetylase. Science. 2004;303 doi: 10.1126/science.1094637. 2011–5. [DOI] [PubMed] [Google Scholar]
- 19.Rodgers J.T., Lerin C., Haas W., Gygi S.P., Spiegelman B.M., Puigserver P. Nutrient control of glucose homeostasis through a complex of PGC-1alpha and SIRT1. Nature. 2005;434:113–118. doi: 10.1038/nature03354. [DOI] [PubMed] [Google Scholar]
- 20.Mattagajasingh I., Kim C.-S., Naqvi A., Yamamori T., Hoffman T.A., Jung S.-B., DeRicco J., Kasuno K., Irani K. SIRT1 promotes endothelium-dependent vascular relaxation by activating endothelial nitric oxide synthase. Proc. Natl. Acad. Sci. U. S. A. 2007;104:14855–14860. doi: 10.1073/pnas.0704329104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jung S.-B., Kim C.-S., Kim Y.-R., Naqvi A., Yamamori T., Kumar S., Kumar A., Irani K. Redox factor-1 activates endothelial SIRTUIN1 through reduction of conserved cysteine sulfhydryls in its deacetylase domain. PLoS One. 2013;8 doi: 10.1371/journal.pone.0065415. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shao D., Fry J.L., Han J., Hou X., Pimentel D.R., Matsui R., Cohen R.A., Bachschmid M.M. A redox-resistant sirtuin-1 mutant protects against hepatic metabolic and oxidant stress. J. Biol. Chem. 2014;289:7293–7306. doi: 10.1074/jbc.M113.520403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lin S.-J., Ford E., Haigis M., Liszt G., Guarente L. Calorie restriction extends yeast life span by lowering the level of NADH. Genes Dev. 2004;18:12–16. doi: 10.1101/gad.1164804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chen D., Bruno J., Easlon E., Lin S.-J., Cheng H.-L., Alt F.W., Guarente L. Tissue-specific regulation of SIRT1 by calorie restriction. Genes Dev. 2008;22:1753–1757. doi: 10.1101/gad.1650608. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Guo X., Kesimer M., Tolun G., Zheng X., Xu Q., Lu J., Sheehan J.K., Griffith J.D., Li X. The NAD(+)-dependent protein deacetylase activity of SIRT1 is regulated by its oligomeric status. Sci. Rep. 2012;2:640. doi: 10.1038/srep00640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kim J.E., Chen J., Lou Z. DBC1 is a negative regulator of SIRT1. Nature. 2008;451:583–586. doi: 10.1038/nature06500. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=18235501 [DOI] [PubMed] [Google Scholar]
- 27.Escande C., Chini C.C.S., Nin V., Dykhouse K.M., Novak C.M., Levine J., Van Deursen J., Gores G.J., Chen J., Lou Z., Chini E.N. Deleted in breast cancer – 1 regulates SIRT1 activity and contributes to high-fat diet – induced liver steatosis in mice. J. Clin. Investig. 2010;120:545–558. doi: 10.1172/JCI39319. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Yang S.-R., Wright J., Bauter M., Seweryniak K., Kode A., Rahman I. Sirtuin regulates cigarette smoke-induced proinflammatory mediator release via RelA/p65 NF- B in macrophages in vitro and in rat lungs in vivo: implications for chronic inflammation and aging, AJP lung cell. Mol. Physiol. 2006;292:L567–L576. doi: 10.1152/ajplung.00308.2006. [DOI] [PubMed] [Google Scholar]
- 29.Guo X., Williams J.G., Schug T.T., Li X. DYRK1A and DYRK3 promote cell survival through phosphorylation and activation of SIRT1. J. Biol. Chem. 2010;285:13223–13232. doi: 10.1074/jbc.M110.102574. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nasrin N., Kaushik V.K., Fortier E., Wall D., Pearson K.J., de Cabo R., Bordone L. JNK1 phosphorylates SIRT1 and promotes its enzymatic activity. PLoS One. 2009;4 doi: 10.1371/journal.pone.0008414. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sasaki T., Maier B., Koclega K.D., Chruszcz M., Gluba W., Stukenberg P.T., Minor W., Scrable H. Phosphorylation regulates SIRT1 function. PLoS One. 2008;3 doi: 10.1371/journal.pone.0004020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zschoernig B., Mahlknecht U. Carboxy-terminal phosphorylation of SIRT1 by protein kinase CK2. Biochem. Biophys. Res. Commun. 2009;381:372–377. doi: 10.1016/j.bbrc.2009.02.085. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=19236849 [DOI] [PubMed] [Google Scholar]
- 33.Bachschmid M.M.M., Schildknecht S., Matsui R., Zee R., Haeussler D., Cohen R.A., Pimental D., van der Loo B.V.D.B., Cohen R.A., Pimental D., van der Loo B.V.D.B. Vascular aging: chronic oxidative stress and impairment of redox signaling-consequences for vascular homeostasis and disease. Ann. Med. 2013;45:17–36. doi: 10.3109/07853890.2011.645498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peterson C.L., Laniel M.-A. Histones and histone modifications. Curr. Biol. 2004;14:R546–R551. doi: 10.1016/j.cub.2004.07.007. [DOI] [PubMed] [Google Scholar]
- 35.Bedalov A., Gatbonton T., Irvine W.P., Gottschling D.E., Simon J.A. Identification of a small molecule inhibitor of Sir2p. Proc. Natl. Acad. Sci. U. S. A. 2001;98:15113–15118. doi: 10.1073/pnas.261574398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Borra M.T., Smith B.C., Denu J.M. Mechanism of human SIRT1 activation by resveratrol. J. Biol. Chem. 2005;280:17187–17195. doi: 10.1074/jbc.M501250200. [DOI] [PubMed] [Google Scholar]
- 37.McDonagh T., Hixon J., DiStefano P.S., Curtis R., Napper A.D. Microplate filtration assay for nicotinamide release from NAD using a boronic acid resin. Methods. 2005;36:346–350. doi: 10.1016/j.ymeth.2005.03.005. [DOI] [PubMed] [Google Scholar]
- 38.Heltweg B., Dequiedt F., Verdin E., Jung M. Nonisotopic substrate for assaying both human zinc and NAD+-dependent histone deacetylases. Anal. Biochem. 2003;319:42–48. doi: 10.1016/s0003-2697(03)00276-8. http://www.ncbi.nlm.nih.gov/pubmed/12842105 [DOI] [PubMed] [Google Scholar]
- 39.Heltweg B., Jung M. A homogeneous nonisotopic histone deacetylase activity assay. J. Biomol. Screen. 2003;8:89–95. doi: 10.1177/1087057102239644. [DOI] [PubMed] [Google Scholar]
- 40.Marcotte P.A., Richardson P.L., Richardson P.R., Guo J., Barrett L.W., Xu N., Gunasekera A., Glaser K.B. Fluorescence assay of SIRT protein deacetylases using an acetylated peptide substrate and a secondary trypsin reaction. Anal. Biochem. 2004;332:90–99. doi: 10.1016/j.ab.2004.05.039. [DOI] [PubMed] [Google Scholar]
- 41.Wegener D., Hildmann C., Schwienhorst A. Recent progress in the development of assays suited for histone deacetylase inhibitor screening. Mol. Genet. Metabol. 2003;80 doi: 10.1016/j.ymgme.2003.08.008. http://www.ncbi.nlm.nih.gov/pubmed/14567963 138–47. [DOI] [PubMed] [Google Scholar]
- 42.Bitterman K.J., Anderson R.M., Cohen H.Y., Latorre-Esteves M., Sinclair D.A. Inhibition of silencing and accelerated aging by nicotinamide, a putative negative regulator of yeast sir2 and human SIRT1. J. Biol. Chem. 2002;277:45099–45107. doi: 10.1074/jbc.M205670200. [DOI] [PubMed] [Google Scholar]
- 43.Smith J.S., Brachmann C.B., Celic I., Kenna M.A., Muhammad S., Starai V.J., Avalos J.L., Escalante-Semerena J.C., Grubmeyer C., Wolberger C., Boeke J.D. A phylogenetically conserved NAD+-dependent protein deacetylase activity in the Sir2 protein family. Proc. Natl. Acad. Sci. U. S. A. 2000;97:6658–6663. doi: 10.1073/pnas.97.12.6658. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=10841563 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Hoffmann K., Brosch G., Loidl P., Jung M. A non-isotopic assay for histone deacetylase activity. Nucleic Acids Res. 1999;27 doi: 10.1093/nar/27.9.2057. http://www.ncbi.nlm.nih.gov/pubmed/10198441 2057–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Jackson M.D., Denu J.M. Structural identification of 2’- and 3’-O-acetyl-ADP-ribose as novel metabolites derived from the Sir2 family of beta -NAD+-dependent histone/protein deacetylases. J. Biol. Chem. 2002;277:18535–18544. doi: 10.1074/jbc.M200671200. [DOI] [PubMed] [Google Scholar]
- 46.Tanner K.G., Landry J., Sternglanz R., Denu J.M. Silent information regulator 2 family of NAD- dependent histone/protein deacetylases generates a unique product, 1-O-acetyl-ADP-ribose. Proc. Natl. Acad. Sci. U. S. A. 2000;97:14178–14182. doi: 10.1073/pnas.250422697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pacholec M., Bleasdale J.E., Chrunyk B., Cunningham D., Flynn D., Garofalo R.S., Griffith D., Griffor M., Loulakis P., Pabst B., Qiu X., Stockman B., Thanabal V., Varghese A., Ward J., Withka J., Ahn K. SRT1720, SRT2183, SRT1460, and resveratrol are not direct activators of SIRT1. J. Biol. Chem. 2010;285:8340–8351. doi: 10.1074/jbc.M109.088682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Sethuraman M., McComb M.E., Huang H., Huang S., Heibeck T., Costello C.E., Cohen R.A. Isotope-coded affinity tag (ICAT) approach to redox proteomics: identification and quantitation of oxidant-sensitive cysteine thiols in complex protein mixtures. J. Proteome Res. 2004;3:1228–1233. doi: 10.1021/pr049887e. [DOI] [PubMed] [Google Scholar]
- 49.Hou S., Shi L., Lei H. Biotin-streptavidin affinity purification of RNA-protein complexes assembled in vitro. Methods Mol. Biol. 2016;1421:23–34. doi: 10.1007/978-1-4939-3591-8_3. [DOI] [PubMed] [Google Scholar]
- 50.De La Fuente E.K., Dawson C.A., Nelin L.D., Bongard R.D., McAuliffe T.L., Merker M.P. Biotinylation of membrane proteins accessible via the pulmonary circulation in normal and hyperoxic rats. Am. J. Physiol. 1997;272:L461–L470. doi: 10.1152/ajplung.1997.272.3.L461. http://www.ncbi.nlm.nih.gov/pubmed/9124603 [DOI] [PubMed] [Google Scholar]
- 51.Brändli A.W., Parton R.G., Simons K. Transcytosis in MDCK cells: identification of glycoproteins transported bidirectionally between both plasma membrane domains. J. Cell Biol. 1990;111:2909–2921. doi: 10.1083/jcb.111.6.2909. http://www.ncbi.nlm.nih.gov/pubmed/2269660 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Holmberg A., Blomstergren A., Nord O., Lukacs M., Lundeberg J., Uhlén M. The biotin-streptavidin interaction can be reversibly broken using water at elevated temperatures. Electrophoresis. 2005;26:501–510. doi: 10.1002/elps.200410070. [DOI] [PubMed] [Google Scholar]
- 53.Rösli C., Rybak J.-N., Neri D., Elia G. Quantitative recovery of biotinylated proteins from streptavidin-based affinity chromatography resins. Methods Mol. Biol. 2008;418:89–100. doi: 10.1007/978-1-59745-579-4_8. http://www.ncbi.nlm.nih.gov/pubmed/18287652 [DOI] [PubMed] [Google Scholar]
- 54.Rybak J.-N., Scheurer S.B., Neri D., Elia G. Purification of biotinylated proteins on streptavidin resin: a protocol for quantitative elution. Proteomics. 2004;4:2296–2299. doi: 10.1002/pmic.200300780. [DOI] [PubMed] [Google Scholar]
- 55.Austin R.J., Smidansky H.M., Holstein C.A., Chang D.K., Epp A., Josephson N.C., Martin D.B. Proteomic analysis of the androgen receptor via MS-compatible purification of biotinylated protein on streptavidin resin. Proteomics. 2012;12:43–53. doi: 10.1002/pmic.201100348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Imai S., Armstrong C.M., Kaeberlein M., Guarente L. Transcriptional silencing and longevity protein Sir2 is an NAD-dependent histone deacetylase. Nature. 2000;403:795–800. doi: 10.1038/35001622. [DOI] [PubMed] [Google Scholar]
- 57.Landry J., Slama J.T., Sternglanz R. Role of NAD(+) in the deacetylase activity of the SIR2-like proteins. Biochem. Biophys. Res. Commun. 2000;278:685–690. doi: 10.1006/bbrc.2000.3854. [DOI] [PubMed] [Google Scholar]
- 58.Solomon J.M., Pasupuleti R., Xu L., McDonagh T., Curtis R., DiStefano P.S., Huber L.J. Inhibition of SIRT1 catalytic activity increases p53 acetylation but does not alter cell survival following DNA damage. Mol. Cell Biol. 2006;26:28–38. doi: 10.1128/MCB.26.1.28-38.2006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Napper A.D., Hixon J., McDonagh T., Keavey K., Pons J.-F., Barker J., Yau W.T., Amouzegh P., Flegg A., Hamelin E., Thomas R.J., Kates M., Jones S., Navia M.A., Saunders J.O., DiStefano P.S., Curtis R. Discovery of indoles as potent and selective inhibitors of the deacetylase SIRT1. J. Med. Chem. 2005;48:8045–8054. doi: 10.1021/jm050522v. [DOI] [PubMed] [Google Scholar]
- 60.Vaziri H., Dessain S.K., Ng Eaton E., Imai S.I., Frye R.A., Pandita T.K., Guarente L., Weinberg R.A. hSIR2(SIRT1) functions as an NAD-dependent p53 deacetylase. Cell. 2001;107:149–159. doi: 10.1016/s0092-8674(01)00527-x. http://www.ncbi.nlm.nih.gov/pubmed/11672523 [DOI] [PubMed] [Google Scholar]
- 61.Rajamohan S.B., Pillai V.B., Gupta M., Sundaresan N.R., Birukov K.G., Samant S., Hottiger M.O., Gupta M.P. SIRT1 promotes cell survival under stress by deacetylation-dependent deactivation of poly(ADP-ribose) polymerase 1. Mol. Cell Biol. 2009;29:4116–4129. doi: 10.1128/MCB.00121-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Wang H.D., Xu S., Johns D.G., Du Y., Quinn M.T., Cayatte A.J., Cohen R.A. Role of NADPH oxidase in the vascular hypertrophic and oxidative stress response to angiotensin II in mice. Circ. Res. 2001;88:947–953. doi: 10.1161/hh0901.089987. http://www.ncbi.nlm.nih.gov/pubmed/11349005 [DOI] [PubMed] [Google Scholar]
- 63.Xu S., Jiang B., Hou X., Shi C., Bachschmid M.M.M., Zang M., Verbeuren T.J.T.J., Cohen R.A.R.A. High-fat diet increases and the polyphenol, S17834, decreases acetylation of the sirtuin-1-dependent lysine-382 on p53 and apoptotic signaling in atherosclerotic lesion-prone aortic endothelium of normal mice. J. Cardiovasc. Pharmacol. 2011;58:263–271. doi: 10.1097/FJC.0b013e3182239eb7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Zang M., Xu S., Maitland-Toolan K.A., Zuccollo A., Hou X., Jiang B., Wierzbicki M., Verbeuren T.J., Cohen R.A. Polyphenols stimulate AMP-activated protein kinase, lower lipids, and inhibit accelerated atherosclerosis in diabetic LDL receptor-deficient mice. Diabetes. 2006;55:2180–2191. doi: 10.2337/db05-1188. [DOI] [PubMed] [Google Scholar]
- 65.Jones D.P. Redefining oxidative stress. Antioxidants Redox Signal. 2006;8 doi: 10.1089/ars.2006.8.1865. 1865–79. [DOI] [PubMed] [Google Scholar]
- 66.de Kreutzenberg S.V., Ceolotto G., Papparella I., Bortoluzzi A., Semplicini A., Dalla Man C., Cobelli C., Fadini G.P., Avogaro A. Downregulation of the longevity-associated protein sirtuin 1 in insulin resistance and metabolic syndrome: potential biochemical mechanisms. Diabetes. 2010;59:1006–1015. doi: 10.2337/db09-1187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Chini E.N. CD38 as a regulator of cellular NAD: a novel potential pharmacological target for metabolic conditions. Curr. Pharmaceut. Des. 2009;15:57–63. doi: 10.2174/138161209787185788. http://www.ncbi.nlm.nih.gov/pubmed/19149603 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Zee R.S., Yoo C.B., Pimentel D.R., Perlman D.H., Burgoyne J.R., Hou X., McComb M.E., Costello C.E., Cohen R.A., Bachschmid M.M. Redox regulation of sirtuin-1 by S-glutathiolation. Antioxidants Redox Signal. 2010;13:1023–1032. doi: 10.1089/ars.2010.3251. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sebastián C., Satterstrom F.K., Haigis M.C., Mostoslavsky R. From sirtuin biology to human diseases: an update. J. Biol. Chem. 2012;287:42444–42452. doi: 10.1074/jbc.R112.402768. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Yuan J., Luo K., Liu T., Lou Z. Regulation of SIRT1 activity by genotoxic stress. Genes Dev. 2012;26:791–796. doi: 10.1101/gad.188482.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Zitka O., Skalickova S., Gumulec J., Masarik M., Adam V., Hubalek J., Trnkova L., Kruseova J., Eckschlager T., Kizek R. Redox status expressed as GSH:GSSG ratio as a marker for oxidative stress in paediatric tumour patients. Oncol. Lett. 2012;4:1247–1253. doi: 10.3892/ol.2012.931. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Yamada H., Arai T., Endo N., Yamashita K., Fukuda K., Sasada M., Uchiyama T. LPS-induced ROS generation and changes in glutathione level and their relation to the maturation of human monocyte-derived dendritic cells. Life Sci. 2006;78:926–933. doi: 10.1016/j.lfs.2005.05.106. [DOI] [PubMed] [Google Scholar]
- 73.Banks A.S., Kon N., Knight C., Matsumoto M., Gutiérrez-Juárez R., Rossetti L., Gu W., Accili D. SirT1 gain of function increases energy efficiency and prevents diabetes in mice. Cell Metabol. 2008;8:333–341. doi: 10.1016/j.cmet.2008.08.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Fry J.L., Al Sayah L., Weisbrod R.M., Van Roy I., Weng X., Cohen R.A., Bachschmid M.M., Seta F. Vascular smooth muscle sirtuin-1 protects against diet-induced aortic stiffness. Hypertension. 2016;68:775–784. doi: 10.1161/HYPERTENSIONAHA.116.07622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Kaeberlein M., McDonagh T., Heltweg B., Hixon J., Westman E.A., Caldwell S.D., Napper A., Curtis R., DiStefano P.S., Fields S., Bedalov A., Kennedy B.K. Substrate-specific activation of sirtuins by resveratrol. J. Biol. Chem. 2005;280:17038–17045. doi: 10.1074/jbc.M500655200. [DOI] [PubMed] [Google Scholar]
- 76.Green N.M. Avidin. Adv. Protein Chem. 1975;29:85–133. doi: 10.1016/s0065-3233(08)60411-8. http://www.ncbi.nlm.nih.gov/pubmed/237414 [DOI] [PubMed] [Google Scholar]
- 77.Morag E., Bayer E.A., Wilchek M. Reversibility of biotin-binding by selective modification of tyrosine in avidin. Biochem. J. 1996;15(316):193–199. doi: 10.1042/bj3160193. http://www.ncbi.nlm.nih.gov/pubmed/8645205 (Pt 1), PMID: 8645205; PMCID: PMC1217322. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Ellerbroek S.M., Wu Y.I., Overall C.M., Stack M.S. Functional interplay between type I collagen and cell surface matrix metalloproteinase activity. J. Biol. Chem. 2001;276:24833–24842. doi: 10.1074/jbc.M005631200. [DOI] [PubMed] [Google Scholar]
- 79.Facchini F.S., Hua N., Abbasi F., Reaven G.M. Insulin resistance as a predictor of age-related diseases. J. Clin. Endocrinol. Metab. 2001;86:3574–3578. doi: 10.1210/jcem.86.8.7763. [DOI] [PubMed] [Google Scholar]
- 80.Civitarese A.E., Carling S., Heilbronn L.K., Hulver M.H., Ukropcova B., Deutsch W.A., Smith S.R., Ravussin E., CALERIE Pennington Team Calorie restriction increases muscle mitochondrial biogenesis in healthy humans. PLoS Med. 2007;4 doi: 10.1371/journal.pmed.0040076. e76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Barzilai N., Banerjee S., Hawkins M., Chen W., Rossetti L. Caloric restriction reverses hepatic insulin resistance in aging rats by decreasing visceral fat. J. Clin. Investig. 1998;101:1353–1361. doi: 10.1172/JCI485. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Beher D., Wu J., Cumine S., Kim K.W., Lu S.-C., Atangan L., Wang M. Resveratrol is not a direct activator of SIRT1 enzyme activity. Chem. Biol. Drug Des. 2009;74:619–624. doi: 10.1111/j.1747-0285.2009.00901.x. [DOI] [PubMed] [Google Scholar]
- 83.Howitz K.T., Bitterman K.J., Cohen H.Y., Lamming D.W., Lavu S., Wood J.G., Zipkin R.E., Chung P., Kisielewski A., Zhang L.-L., Scherer B., Sinclair D.A. Small molecule activators of sirtuins extend Saccharomyces cerevisiae lifespan. Nature. 2003;425:191–196. doi: 10.1038/nature01960. [DOI] [PubMed] [Google Scholar]
- 84.de Boer V.C.J., de Goffau M.C., Arts I.C.W., Hollman P.C.H., Keijer J. SIRT1 stimulation by polyphenols is affected by their stability and metabolism. Mech. Ageing Dev. 2006;127:618–627. doi: 10.1016/j.mad.2006.02.007. [DOI] [PubMed] [Google Scholar]
- 85.Kwon H.-S., Ott M. The ups and downs of SIRT1. Trends Biochem. Sci. 2008;33:517–525. doi: 10.1016/j.tibs.2008.08.001. [DOI] [PubMed] [Google Scholar]
- 86.Caito S., Rajendrasozhan S., Cook S., Chung S., Yao H., Friedman A.E., Brookes P.S., Rahman I. SIRT1 is a redox-sensitive deacetylase that is post-translationally modified by oxidants and carbonyl stress. FASEB J. 2010;24:3145–3159. doi: 10.1096/fj.09-151308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yang Y., Fu W., Chen J., Olashaw N., Zhang X., V Nicosia S., Bhalla K., Bai W. SIRT1 sumoylation regulates its deacetylase activity and cellular response to genotoxic stress. Nat. Cell Biol. 2007;9:1253–1262. doi: 10.1038/ncb1645. http://www.ncbi.nlm.nih.gov/entrez/query.fcgi?cmd=Retrieve&db=PubMed&dopt=Citation&list_uids=17934453 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Gao Z., Zhang J., Kheterpal I., Kennedy N., Davis R.J., Ye J. Sirtuin 1 (SIRT1) protein degradation in response to persistent c-Jun N-terminal kinase 1 (JNK1) activation contributes to hepatic steatosis in obesity. J. Biol. Chem. 2011;286:22227–22234. doi: 10.1074/jbc.M111.228874. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Nemoto S., Fergusson M.M., Finkel T. Nutrient availability regulates SIRT1 through a forkhead-dependent pathway. Science. 2004;306:2105–2108. doi: 10.1126/science.1101731. [DOI] [PubMed] [Google Scholar]
- 90.Prozorovski T., Schulze-Topphoff U., Glumm R., Baumgart J., Schröter F., Ninnemann O., Siegert E., Bendix I., Brüstle O., Nitsch R., Zipp F., Aktas O. Sirt1 contributes critically to the redox-dependent fate of neural progenitors. Nat. Cell Biol. 2008;10:385–394. doi: 10.1038/ncb1700. [DOI] [PubMed] [Google Scholar]
- 91.Alcendor R.R., Gao S., Zhai P., Zablocki D., Holle E., Yu X., Tian B., Wagner T., Vatner S.F., Sadoshima J. Sirt1 regulates aging and resistance to oxidative stress in the heart. Circ. Res. 2007;100:1512–1521. doi: 10.1161/01.RES.0000267723.65696.4a. [DOI] [PubMed] [Google Scholar]
- 92.Lin S.J., Defossez P.A., Guarente L. Requirement of NAD and SIR2 for life-span extension by calorie restriction in Saccharomyces cerevisiae. Science. 2000;289:2126–2128. doi: 10.1126/science.289.5487.2126. http://www.ncbi.nlm.nih.gov/pubmed/11000115 [DOI] [PubMed] [Google Scholar]
- 93.North B.J., Marshall B.L., Borra M.T., Denu J.M., Verdin E. The human Sir2 ortholog, SIRT2, is an NAD+-dependent tubulin deacetylase. Mol. Cell. 2003;11:437–444. doi: 10.1016/s1097-2765(03)00038-8. http://www.ncbi.nlm.nih.gov/pubmed/12620231 [DOI] [PubMed] [Google Scholar]
- 94.Cheng H.-L., Mostoslavsky R., Saito S., Manis J.P., Gu Y., Patel P., Bronson R., Appella E., Alt F.W., Chua K.F. Developmental defects and p53 hyperacetylation in Sir2 homolog (SIRT1)-deficient mice. Proc. Natl. Acad. Sci. U. S. A. 2003;100 doi: 10.1073/pnas.1934713100. 10794–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95.Tanno M., Sakamoto J., Miura T., Shimamoto K., Horio Y. Nucleocytoplasmic shuttling of the NAD+-dependent histone deacetylase SIRT1. J. Biol. Chem. 2007;282:6823–6832. doi: 10.1074/jbc.M609554200. [DOI] [PubMed] [Google Scholar]
- 96.Jin Q., Yan T., Ge X., Sun C., Shi X., Zhai Q. Cytoplasm-localized SIRT1 enhances apoptosis. J. Cell. Physiol. 2007;213:88–97. doi: 10.1002/jcp.21091. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.