Abstract
The analysis of HIV-1 sequences has helped understand the viral molecular epidemiology, monitor the development of antiretroviral drug resistance, and design candidate vaccines. The introduction of single genome amplification (SGA) has been a major advancement in the field, allowing for the characterization of multiple sequences per patient while preserving linkage among polymorphisms in the same viral genome copy. Sequencing of SGA amplicons is performed by capillary Sanger sequencing, which presents low throughput, requires a high amount of template, and is highly sensitive to template/primer mismatching. In order to meet the increasing demand for HIV-1 SGA amplicon sequencing, we have developed a platform based on benchtop next-generation sequencing (NGS) (IonTorrent) accompanied by a bioinformatics pipeline capable of running on computer resources commonly available at research laboratories. During assay validation, the NGS-based sequencing of 10 HIV-1 env SGA amplicons was fully concordant with Sanger sequencing. The field test was conducted on plasma samples from 10 US Navy and Marine service members with recent HIV-1 infection (sampling interval: 2005–2010; plasma viral load: 5,884–194,984 copies/ml). The NGS analysis of 101 SGA amplicons (median: 10 amplicons/individual) showed within-individual viral sequence profiles expected in individuals at this disease stage, including individuals with highly homogeneous quasispecies, individuals with two highly homogeneous viral lineages, and individuals with heterogeneous viral populations. In a scalability assessment using the Ion Chef automated system, 41/43 tested env SGA amplicons (95%) multiplexed on a single Ion 318 chip showed consistent gene-wide coverage >50×. With lower sample requirements and higher throughput, this approach is suitable to support the increasing demand for high-quality and cost-effective HIV-1 sequences in fields such as molecular epidemiology, and development of preventive and therapeutic strategies.
Keywords: Next-generation sequencing, HIV-1, Single genome amplification, Bioinformatics, IonTorrent
1. Introduction
After more than three decades since the first identification of the Human Immunodeficiency Virus type 1 (HIV-1), the development of safe and effective preventive vaccines, antiretroviral treatments (ART), and cure strategies remain major public health priorities [1]. The study of viral sequences has been central to these efforts, spanning through different stages of product development. Molecular epidemiology analyses are widely employed to model viral evolution [2], inform immunogen selection and design [3], and monitor the circulation of ART resistance mutations [4]. In vaccine efficacy trials, the exploration of immune pressure signatures imprinted in viral genomes from breakthrough cases can help elucidate the mechanism of action [5].
Like other RNA viruses, HIV-1 populations are genetically diverse and behave as quasispecies (i.e., swarms of highly related but distinct viral sequences [6]), due to the high rates of viral replication, mutation, and recombination [7]. The plasticity of HIV-1 quasispecies allows them to infect different cell targets [8], escape host immunity [9], and resist inhibition by ART [10]. A major challenge for the study of this high level of genetic diversity is the limited capacity of bulk PCR/sequencing techniques to capture the complexity of the viral quasispecies [11].
The application of single genome amplification (SGA) of sub-genomic (i.e., 1.5–3 Kb) [12,13] and full-length genome HIV-1 (i.e., 9 Kb) [14,15] has been a major advancement in the field. This technique is based on serial dilution of a viral genome template (usually complementary DNA (cDNA) obtained from viral RNA (vRNA) by reverse transcription) followed by nested PCR of multiple (>10) replicates. Based on the Poisson distribution, the dilution that yields ≤30% positive reactions has an 80% probability of deriving from a single amplifiable template [12,13]. This approach allows for the study of multiple viral sequences per patient, preserving linkage among polymorphisms in the same viral genome copy, with limited impact from PCR-induced misincorporation/recombination or bacterial selection during cloning [14]. HIV-1 SGA amplicon derived sequences published to date have been obtained exclusively by capillary sequencing based on the Sanger method (the single exception is the PacBio-based sequencing of pooled single genome amplicons by Dilernia et al. [16] The Sanger sequencing technique provides reads of length ˜800 base pairs (bp) with low sequencing error, which allows for the straightforward generation of contigs de novo (i.e., without the need of a pre-existing reference sequence). This method is based on primer-directed sequencing [17], thus requiring prior knowledge of the target sequence. In the case of HIV-1, where inter-strain nucleotide sequence diversity can reach 20% [18], some sequencing reactions may fail due to mismatches between target and primer, and require the selection of a second set of sequencing primers to “fill in” the low-coverage areas in the contig. To achieve the desired level of bidirectional coverage (usually 4×), ∼6 μg of PCR amplicon is used as substrate for the multiple dye-termination sequencing reactions.
Recent years have seen an increase in demand for HIV-1 sequencing in large cohort studies [[19], [20], [21], [22], [23], [24], [25], [26], [27], [28], [29], [30], [31], [32]]. For instance, the sieve analysis of the RV144 vaccine efficacy trial generated >1000 HIV-1 env gene SGA amplicon sequences from 121 patients [33]. Consequently, interest is increasing in the field for reliable, cost-effective, and scalable alternatives to capillary Sanger sequencing. Here we describe the development, validation, and field-testing of an alternative HIV-1 SGA amplicon sequencing platform based on next-generation sequencing (NGS).
Unlike capillary sequencing, which allows for a maximum of 96 parallel reactions, NGS allows for millions of parallel reactions [34]. While the high cost of first-generation NGS instruments limited their availability to sequencing core facilities, by the early 2010′s Life Technologies and Illumina launched more affordable benchtop NGS sequencers (i.e., Ion Torrent PGM [35] and MiSeq [36], respectively) which has allowed for the wider spread of NGS technologies in research laboratories [37]. Sequence reads obtained by benchtop NGS instruments are of shorter length and lower quality [38] than capillary sequences, thus requiring a large reading redundancy to mitigate sequencing errors [39].
Here we propose a strategy that is based on benchtop NGS, including an accompanying bioinformatics pipeline that can run on conventional desktops/laptops. Overall, our results demonstrate that this NGS strategy performs with comparable accuracy to capillary sequencing. Properly incorporated, the NGS platform can accommodate the increasing needs of HIV-1 SGA amplicon sequencing with its advantages in cost, scalability and ease of data analysis.
2. Material and methods
2.1. Population under study
As a part of proactive public health management, we undertook a characterization of the contemporary HIV epidemic in the US Navy and Marine Corps [40]. Health system and occupational data as well as reposed sera from all Sailors and Marines identified as HIV-infected over a five-year period ending in 2010 were included (n = 496 service members). In addition to exploring holistic relationships which might inform public health engagement to reduce service member HIV infection risk, a cluster analysis was performed through molecular methods [41]. Also, a sub-group of the cohort volunteered and participated in a risk survey [42]. For the current work, samples from 10 random participants were used, meeting the following criteria: 1) plasma viral load >5000 copies/ml, and 2) available sample volume >2.0 ml (which would allow future work on leftover specimens) (Table 1).
Table 1.
Sample set used in the validation and field test of HIV-1 env SGA NGS.
| Patient | Plasma viral load |
SGA amplicons (n) |
||
|---|---|---|---|---|
| (copies/ml) | log10 | Validation | Field test | |
| A | 44,668 | 4.65 | 11 | |
| B | 169,824 | 5.23 | 9 | |
| C | 194,984 | 5.29 | 12 | |
| D | 5,884 | 3.77 | 9 | |
| E | 100,000 | 5.00 | 11 | |
| F | 85,114 | 4.93 | 13 | |
| G | 11,749 | 4.07 | 13 | |
| H | 47,863 | 4.68 | 9 | |
| I | 10,471 | 4.02 | 8 | |
| J | 154,882 | 5.19 | 10 | 6 |
2.2. Single genome amplification of HIV-1 env
vRNA was extracted from plasma samples using the QIAamp Viral RNA Mini Kit (QIAGEN, Valencia, CA). Single genome amplicons of full length HIV-1 env were retrieved from vRNA using reverse transcription (RT) followed by nested PCR as previously described [41]. Briefly, after RT, cDNA was titrated through nested-PCR of HIV-1 env of 10 replicates/dilution. The dilution that provided 3/10 positive reactions was used to generate HIV-1 env single genome amplicons [12,13].
2.3. Library preparation and next-generation sequencing
For each single genome amplicon, 100 ng of second round PCR product was enzymatically sheared to 400 bp followed by barcoding using the Ion Xpress Plus Fragment Library & Ion Xpress Barcode Adapters kits (LifeTechnologies, ThermoFisher Scientific). Quantification was performed using a 2100 Bioanalyzer (DNA 1000 kit, Agilent Technologies, Sunnyvale, CA). DNA size-selection was performed using Blue Pippin 2% dye free cassette with internal standard marker V1 (Sage Science). The size-selected product was equalized using the Ion Equalizer kit (LifeTechnologies, ThermoFisher Scientific) following manufacturer’s instructions. All purifications used Agencourt AMPure XP Reagent (Beckman Coulter).
Emulsion PCR (ePCR) and enrichment for 400 bp sheared product used the Ion OneTouch 400 bp Template kit (LifeTechnologies, Carlsbad, CA) on the OneTouch and ES instruments. Sequencing was carried out using the Ion PGM 400 kit and Ion 316 chip v2 on the IonTorrent PGM platform (LifeTechnologies, Carlsbad, CA), following manufacturer’s instructions. For scalability experiments, the ePCR/enrichment of libraries derived from 43 different amplicons were carried out on the Ion Chef instrument with Ion PGM Hi-Q Chef kit (LifeTechnologies, Carlsbad, CA) and were sequenced on Ion 318 chips v2 BC.
2.4. Sanger sequencing
To validate the NGS-based platform, Sanger sequencing of HIV-1 env single genome amplicons was performed as previously described [41]. Contigs of Sanger reads were assembled with Sequencher 5.0 (Gene Codes Corporation, Ann Arbor, MI).
2.5. Data analysis
2.5.1. Quality control
FastQ files [43] were exported from the PGM using Torrent Suite 4.4 software (LifeTechnologies, ThermoFisher Scientific). Sequence quality control was performed using FastQC (courtesy of Dr. Simon Andrews, Babraham Institute, Cambridge, UK; URL: https://www.bioinformatics.babraham.ac.uk/projects/fastqc/). FastQ files [43] were then imported into CLC Genomics Workbench version 7.0.3 (Aarhus, Denmark) to remove sequencing adapters and trim sequences based on quality (limit = 0.05; maximal 2 ambiguous nucleotides) and length (minimum length = 200 nucleotides), followed by barcode-based demultiplexing.
We developed a pipeline, combining published and novel tools, to obtain the consensus sequence of single genome amplicons sequenced by NGS. The pipeline is composed of three modules. First, newly-developed tango was used to select a reference sequence to serve as seed for the initial reference-guided alignment. Then, previously-published Nautilus [44] and newly-developed SaGA were used for iterative determination of consensus sequence Finally, newly-developed ViKiNGS was employed for manual quality control and editing of the final consensus sequence. The description of these modules is presented in detail in the Results section, along with examples of their implementation. The software is accessible to interested users through the execution of individual software licenses with Henry M. Jackson Foundation for the Advancement of Military Medicine, Inc. Please contact the corresponding authors.
2.5.2. Reference-guided alignment
Filtered reads were aligned to reference using the Burrows-Wheeler Aligner (BWA) [45] implemented in tmap version 3.2.2 (by Nils Homer, distributed through https://github.com/iontorrent/TMAP) using the following parameters: command = map2; match score = 1; mismatch penalty = 3; gap open penalty = 5; gap extension penalty = 2; and soft-clip only the right portion of the read.
2.6. Statistical testing
Sequence alignments were generated using HIVAlign [46], and were manually edited using Geneious 3 (http://www.geneious.com) [47]. The HIV-1 subtype was determined using NCBI Genotyping tool (https://www.ncbi.nlm.nih.gov/projects/genotyping/formpage.cgi) and jumping profile Hidden Markov Model (jpHMM) tool [48] (http://jphmm.gobics.de).
The inter-variant sequence diversity within a sample was visualized using Highlighter tool [13]. Phylogenetic analyses were conducted using MEGA6.06 [49] (www.megasoftware.net). Prism version 6.0e (GraphPad Software) and JMP10 (SAS Institute, Cary, NC) were used for summary statistical analyses.
3. Results and discussion
3.1. Model selection
Since its first description, the SGA method has been applied to the study of the three structural HIV-1 genes (i.e., gag [1.5 kilobases (Kb)], pol [3 Kb], and env [2.6 Kb]) [12,13,50], as well as half-length (˜5 Kb) [51] and near full-length genomes (˜9 Kb) [14,51]. HIV-1 env has been a major focus of SGA amplicon sequencing efforts, due to the importance of the encoded glycoprotein (Gp160) in viral tropism and antibody-mediated immune responses [5,13]. Among HIV-1 structural genes, env presents the highest level of genetic diversity, with nucleotide sequences differing by up to 20% [http://www.hiv.lanl.gov/]. Moreover, env presents marked length polymorphism both intra- and inter-patient [52]. Thus, in the current paper we have focused on the NGS of env SGA amplicons, as a “worst-case scenario” for difficulty of sequence alignment, and the current platform can be applied to other subgenomic regions or full-length HIV-1.
3.2. Summary of SGA amplicon library preparation
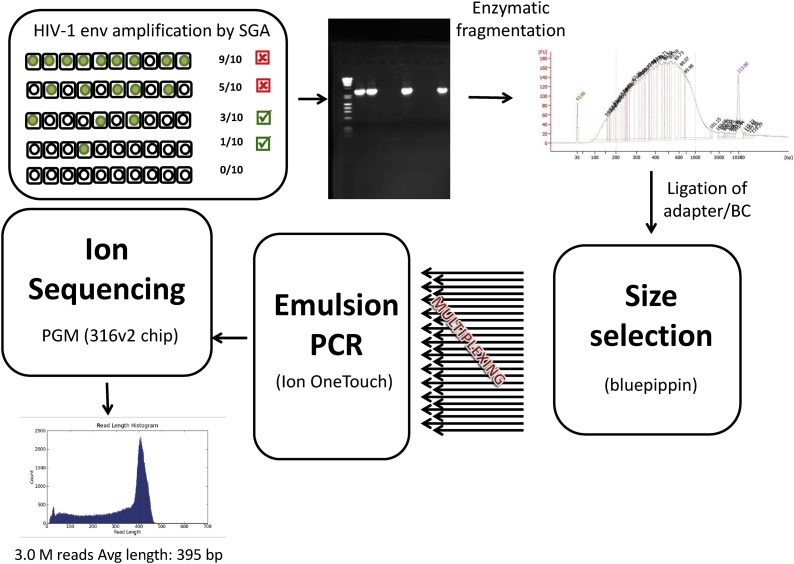
The preparation of HIV-1 env SGA amplicon NGS libraries is described in detail in the Materials and Methods section (Fig. 1). Briefly, HIV-1 env SGA amplicons were sheared to 400 bp and were subjected to adapter/barcode ligation. After emulsion PCR, the libraries were sequenced in IonTorrent PGM 316 chip v2. Reads were exported from the instrument using the Torrent Server. After filtering low-quality and short reads (<200 bp), sequences were exported in FastQ files for alignment.
Fig. 1.
Next-generation sequencing of HIV-1 single genome amplicons. cDNA of HIV-1 env is titrated using serial dilution followed by nested PCR. The dilution that yields ≤30% of positive reactions is used for downstream library preparation. Amplicons are subject to enzymatic fragmentation, which is visualized on a Bioanalyzer. Gel electrophoresis is used for size selection (˜400 bp), and ligation of barcodes and sequencing adapters allows for multiplexing dozens of samples in a single emulsion PCR (ePCR) run. Libraries are then loaded on a 316 chip v2, and Ion sequencing is performed on a PGM instrument. The histogram shows the NGS read length distribution of a typical run. See text for details.
3.3. Alignment
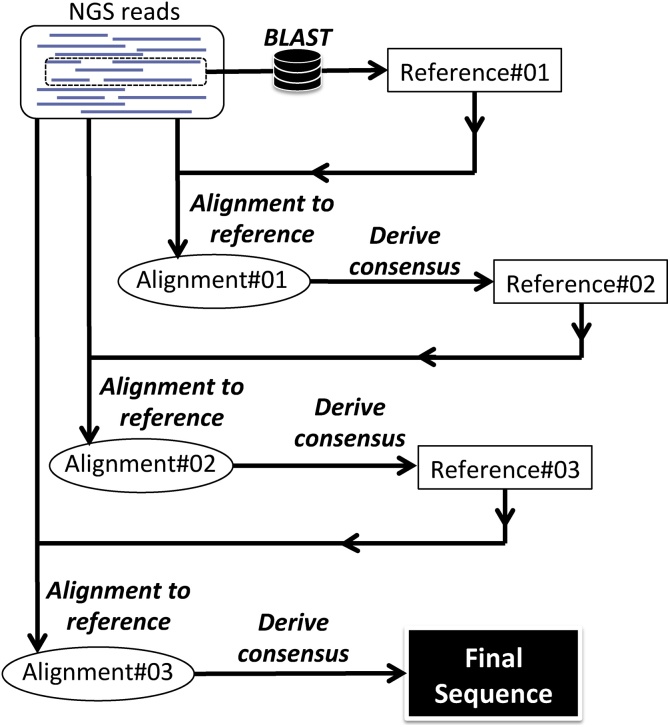
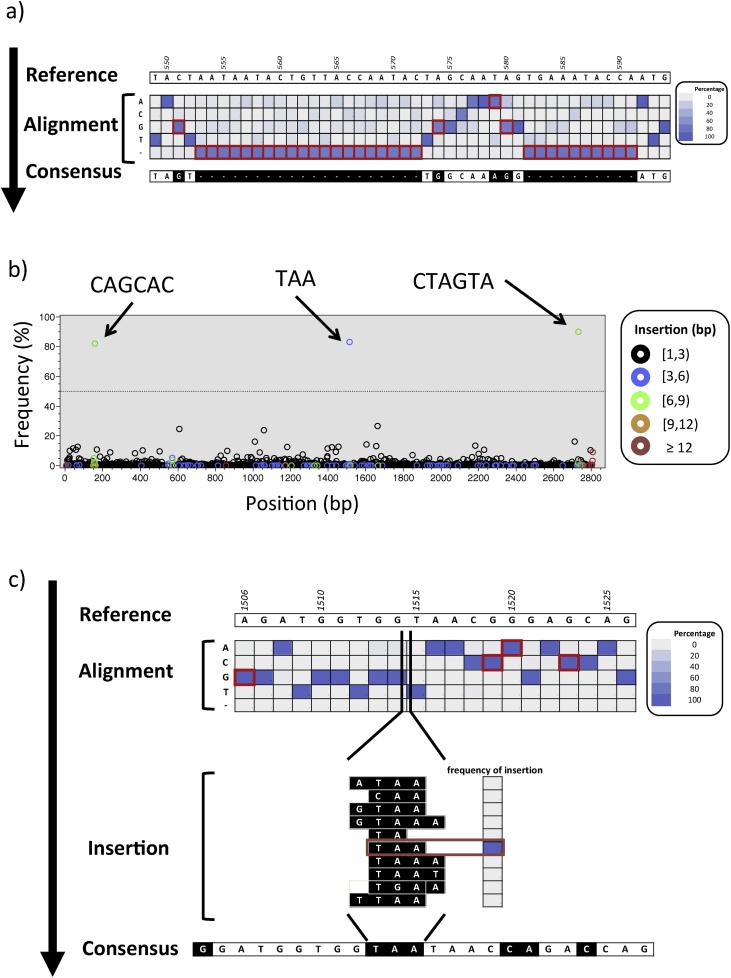
In order to allow for the current bioinformatics pipeline to run on a computer with capabilities usually found in research laboratories, we selected reference-guided alignment over de novo alignment. The challenge with the former method is that it requires a reference sequence to which align the reads. In the absence of an autologous (i.e., from the same patient) reference, we developed tango, an algorithm that uses a random sample of the NGS reads to select the closest sequence (“Reference#01” in Fig. 2), using BLAST [53], from a local database of published HIV-1 sequences, following the model proposed by Archer et al. [54]. All of the NGS reads were then aligned to Reference#01 using a BWA-based algorithm [45], resulting in a sam file: “Alignment#01”. Due to HIV-1 vast genetic diversity, Reference#01 will unavoidably represent an “imperfect” reference, which can differ from the query reads in substitutions, deletions and insertions. We used Nautilus [44] to explore Alignment#01, tallying the frequency of each nucleotide base at each position of the alignment (Fig. 3a), and we used SaGA to derive a consensus sequence depicting the base present at ≥ 50% in each position (in the case where no base was present at ≥ 50%, the position was represented by an “N”). In the cases where the query reads predominantly presented a deletion, the position was represented in the consensus by a gap (i.e., “-”). The Concise Idiosyncratic Gapped Alignment Report (CIGAR) field in the sam file [55] encodes for insertions in the query compared to the reference (the example in Fig. 3b shows the presence of three insertions in the query reads at 80%: one trinucleotide and two hexanucleotides). When insertions at a given position were present in ≥ 50% of the reads, the most frequent motif was inserted into the consensus (Fig. 3c). The consensus sequence thus generated, which reconciled differences between Reference#01 and the query reads, was made into a new reference sequence (Reference#02) that guided the alignment of NGS reads in a new iteration (Fig. 2). This process was then repeated, until it resulted in no further improvement; the obtained sequence was the “final sequence”.
Fig. 2.
Algorithm for deriving a consensus sequence from HIV-1 single genome amplicon Using the tango software, the closest published HIV-1 sequence that can serve as a reference is obtained by applying BLAST on a random sample of NGS reads (Reference#01). The complete set of NGS reads is then aligned to Reference#01 using an implementation of BWA [45]. Afterwards, the consensus of the alignment (Reference#02) is derived by analyzing the frequency of nucleotide bases, insertions, and deletions in the sam file using Nautilus and SaGA software. Reference#02 is used to guide the alignment of NGS reads and consensus derivation, in a new iteration, until the “final consensus sequence” is obtained. See text for details.
Fig. 3.
Example of derivation of single genome amplicon NGS consensus. A) At each alignment position (columns), the sequence of the reference (top) is compared with the frequency of nucleotide bases or gaps (rows) tallied based on the analysis of the sam file (to ease visualization, frequencies are here presented as a heat map). Whenever the most frequent base/gap differs from the reference (red border), the sequence of the consensus is modified accordingly (black boxes). B) By analyzing the CIGAR field in the sam alignment file it is possible to tally the sequences from the NGS reads encoded as “I”, which correspond to “insertion to the reference” (i.e., bases present in the NGS reads that do not have a corresponding position in the reference). The plot depicts, at position of the alignment (x-axis), the frequency of the insertions (y-axis). Data points are color-coded based on the length of the insertion. The arrows depict the sequences of three predominant insertions. Insertions above the operational threshold (dotted line at 50%) are followed up in downstream analysis, where C) the most common motif (in this case “TAA”) is inserted back into the new consensus sequence in the corresponding position.
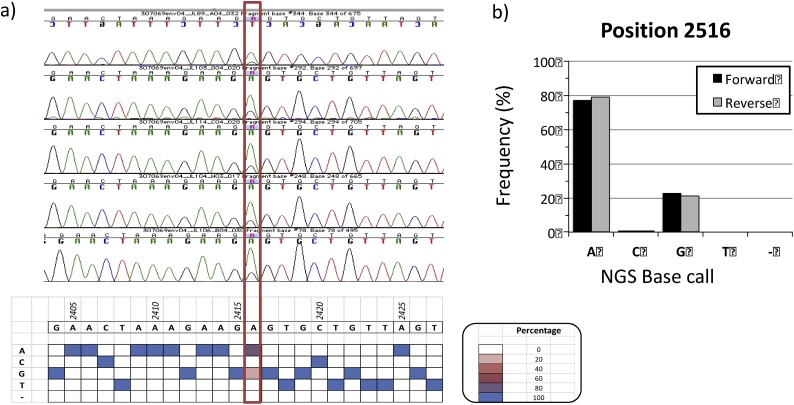
In some instances, SGA amplicon sequences can present ≥1 mixed bases, due to the presence of >1 template in the original PCR reaction [14]. Also, Salazar-Gonzalez et al. have reported on the presence of mixed bases due to misincorporation by Taq polymerase during initial PCR cycles [14], emphasizing the importance of using high-fidelity polymerases. In capillary sequencing, mixed bases are evidence as overlapping peaks in the chromatogram, but it is not possible to reliably estimate the frequency of the different variants. Using the current NGS platform, it is possible to quantify the number of forward and reverse reads supporting each variant (Fig. 4). This information is particularly useful when ruling out miscalls due to strand bias. We have designed a GUI, ViKiNGS, to support the quality control and manual editing of the “final sequences” (Suppl Fig. 1).
Fig. 4.
NGS-based analysis of mixed bases. A) In amplicon #04 from participant “J” 5/5 Sanger capillary sequencing chromatograms covering position 2516 (red border) show overlapping peaks for A and G, which would result in an “R” base calling based on the IUPAC nomenclature. The presence of “A” and “G” are also supported in the NGS reads (heat map on the bottom). B) NGS data shows ˜80% of reads supporting the A and 20% supporting the G, with no directional bias. Quantitation of NGS reads allows to discriminate between events that should be base called as single vs. mixed bases, in a more consistent manner.
3.4. Validation
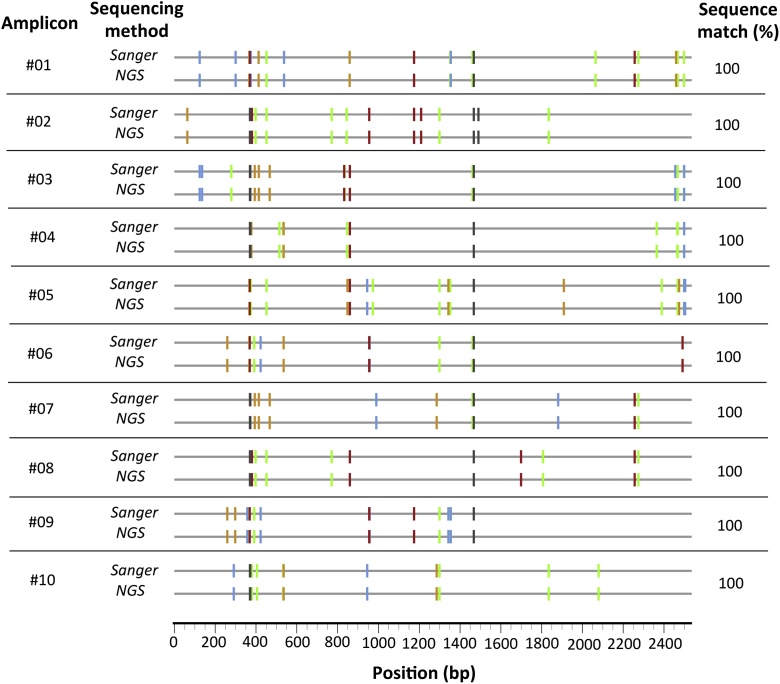
In order to assess the accuracy of the NGS platform, we used SGA amplicons that had been sequenced with capillary Sanger sequencing. The validation set represented 10 HIV-1 env SGA amplicons from the same individual (participant "J”) with median inter-variant genetic distance of 0.86% (range: 0.35–1.13%), distinguishable by substitutions and insertions/deletions. In 10/10 cases, the NGS results fully match the capillary Sanger sequences (Fig. 5).
Fig. 5.
Validation of NGS of HIV-1 single genome amplicons. Ten env amplicons from participant “J” were subject to Sanger capillary sequencing and NGS. To ease visualization, the highlighter plot is shown, where each sequence was compared to the consensus of the participant, and differences from the consensus are denoted with color-coded tic marks using the LANL Highlighter convention (i.e., green = A, blue = C, orange = G, red = T, and gray = deletion) [13]. Each cognate pair shows identical patterns indicating 100% concordance between the two techniques.
3.5. Field test
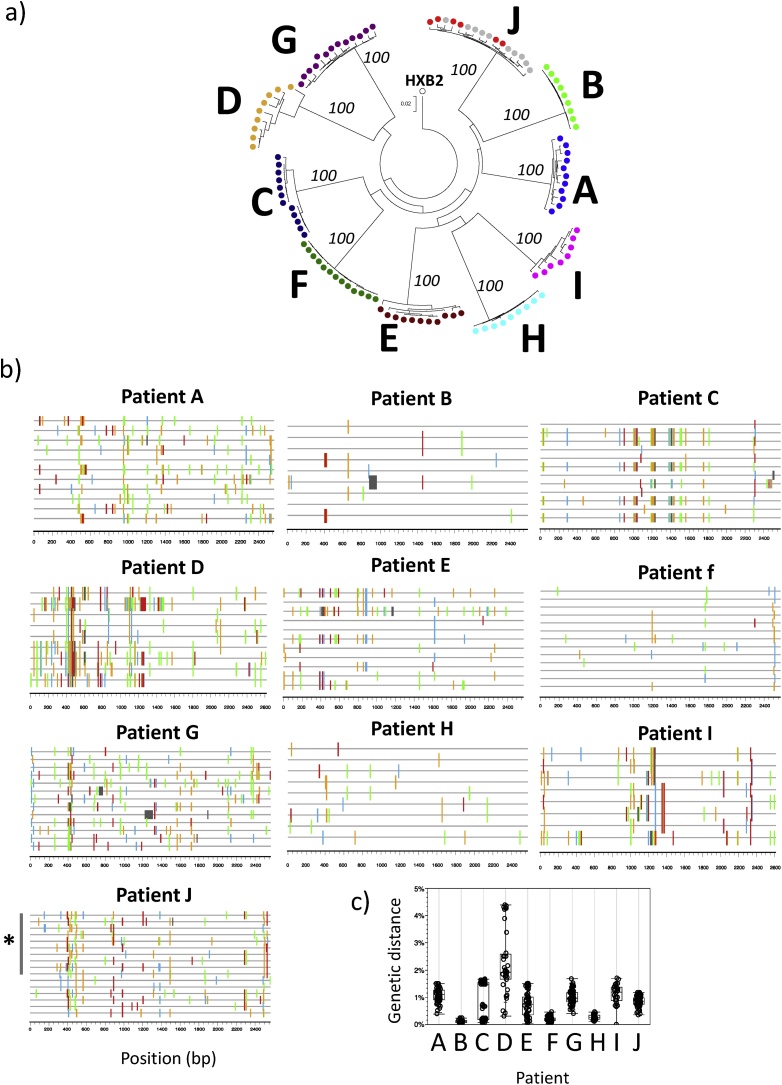
The applicability of the NGS platform was assessed on a field test of additional 101 HIV-1 env SGA amplicons from 10 individuals with plasma viral loads ranging 5,884-194,984 copies/ml (Table 1). This sample set included 6 additional SGA amplicons from the same individual used in the assay validation (participant “J”). All of the sequences were subtype B (Suppl Fig. 2) and the phylogenetic analysis supported the grouping of sequences from each individual in separate and distinct clusters (Fig. 6a). The observed profiles included individuals with highly homogeneous viral quasispecies (e.g., participant “B”), individuals with two highly homogeneous viral lineages (e.g., participant “C”), and individuals with heterogeneous viral populations (e.g., participant “G”) (Fig. 6b and c). Of note, 9/9 SGA amplicons from participant “H” presented the same premature stop codon in gp160 (codon 24). The capillary Sanger sequencing of the SGA amplicon showed an open reading frame, while the frame shift observed in NGS was due to a single-base deletion at codon 9. A detailed inspection of the NGS data showed that only a minority of the reads (13%) presented the open reading frame sequence (Suppl Fig. 3). The reason for the aberrant NGS sequence is unknown, but the presence of an unusually high GC-content in the vicinity of the affected region (GC = 75%, compared to genome-wide average GC = 43%) might have affected NGS reading.
Fig. 6.
Field test of NGS of HIV-1 single genome amplicons. 101 env amplicons from 10 HIV-1 infected individuals were subject to NGS sequencing. A) the phylogenetic tree shows separate clustering of sequences from each individual, supported by 100% bootstrap values. The 10 sequences from participant “J” that had been used in the assay validation are shown in grey. HIV-1 reference sequence HXB2 is depicted. B) Highlighter plot of sequences from each participant show different within-individual diversity profiles, ranging from highly homogeneous (e.g., participant “B”) to more diverse (e.g., participant “D”). Color-coding is as in Fig. 5. The 10 sequences from participant “J” that had been used in the assay validation are depicted by the grey bar. C) Pair-wise sequence diversity within each participant.
3.6. Scalability
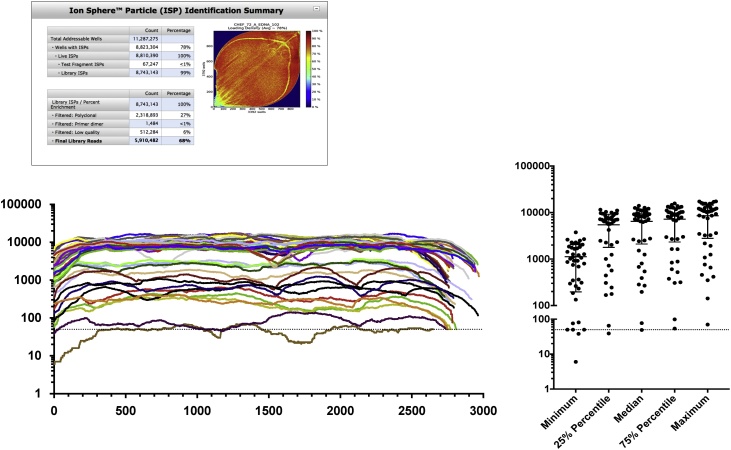
Next, we assessed the capacity of the NGS platform to scale up, in order to accommodate the increasing demand for SGA amplicon sequencing. The assay validation and field test had been run on IonTorrent 316 chip v2, and in the following experiment we tested the 318 chip v2 BC, which provides ˜1.8× sequencing capacity compared to the 316v2 chip. Also, to assist with ePCR/enrichment and chip loading, we employed the Ion Chef automated system. We ran 43 different HIV-1 env SGA amplicons, which had already been sequenced during the assay validation and field test. The run resulted in 5.9 million reads, of which 3.9 million (66.1%) had a length >300 bp. The number of reads per barcode ranged 704-187,350 (median: 108,548 reads per barcode; inter-quartile range: 26,132-127,354.5). When the NGS reads were ran through the genotyping pipeline, 41/43 (95%) of the SGA amplicons presented consistent gene-wide coverage >50× (the operational cut-off depth following guidelines by Pelak et al. [56]), with 37/43 (86%) of the SGA amplicons having a median coverage >500× (Fig. 7). Consensus sequences matched those obtained in the previous experiment using 316 chip v2.
Fig. 7.
Scale up of NGS of HIV-1 single genome amplicons. 43 different env amplicons used in the assay validation were multiplexed using distinct barcodes were run together on a 318 chip v2 BC. The top panel shows the loading efficiency and run metrics. The bottom-left graph shows the per-base coverage for each sequence, represented by a different line color and the bottom-right graph summarizes the statistics of the 43 sequences. The dotted line indicates 50x coverage.
3.7. Overall comparison of NGS- and Sanger-based SGA amplicon sequencing platforms
Motivated by the increase in the demand for HIV-1 SGA amplicon sequencing, we have developed an NGS-based SGA amplicon sequencing platform. Among the advantages of the NGS-based system are:
-
1)
Lower DNA input: the gold standard Sanger sequencing platform requires ∼6 μg of 2nd round PCR product to support bidirectional sequencing of HIV-1 env (at 4x coverage), which is usually achieved by running multiple PCRs in parallel. In contrast, the NGS-based platform has a much lower template requirement (˜100 ng), which represents a substantial reduction in the cost of library preparation.
-
2)
No need of HIV-1 sequencing primers: HIV-1 sequence diversity is a major challenge for Sanger-based sequencing. Mismatching between primers and target sequences usually results in gaps in the contigs, which require the selection of new sets of primers by a trained operator and the running of additional sequencing reactions to provide complete coverage. In contrast, the NGS-based platform does not depend on HIV-1 primers for sequencing; however, it does necessitate an HIV-1 sequence for reference-guided alignment. In the current bioinformatics pipeline, the query for a suitable reference capitalizes from the vast public sequence database (to date 53,666 HIV-1 env sequences have been deposited in the Los Alamos National Laboratory HIV database, URL:http://www.hiv.lanl.gov/, accessed 03JAN18).
-
3)
Higher throughput: The large sequencing landscape within a chip combined with the capacity to multiplex samples using distinguishable barcodes lends the NGS platform the capacity to process dozens of different samples per run [56]. The throughput of this platform can be further increased by leveraging automation systems for library preparation and more efficient bioinformatics pipelines for sequence handling.
-
4)
Platform independence: While the data presented in the current study were obtained using the IonTorrent platform, similar analyses could be perform on other benchtop NGS platforms (i.e., Illumina’s MiSeq). Moreover, the bioinformatics pipeline is implemented in Java, which allows for implementation on Windows, Linux, and Mac OS.
The per-base cost of NGS has been rapidly decreasing (Data from the NHGRI Genome Sequencing Program. URL: https://www.genome.gov/27541954/dna-sequencing-costs-data/ accessed on 11JAN19), making the current platform an affordable alternative to Sanger capillary sequencing.
Despite the abovementioned advantages of the NGS-based platform, it is important to consider that Sanger capillary sequencing is a mature technology, widely employed for more than 2 decades, which can substantially ease assay development and troubleshooting. As shown in the current field test, NGS-based systems can also be affected by artifacts. Moreover, the use of short-read NGS limits the capacity to assess the linkage/phasing of polymorphisms, which could be needed to distinguish if the presence of multiple instances of mixed bases in the same sequence are due to multiple templates in the PCR or misincorporation by Taq polymerase. As the new technology expands, it will be important to remain vigilant to systematic errors and the search for technical ways to mitigate them.
Finally, a third generation of sequencing technologies has been developed, characterized by longer reads (>10 Kb) of single molecules (e.g., SMRT by Pacific Biosciences and MinION by Oxford Nanopore Technologies). Reports published thus far show promising results regarding the sequencing of complex HIV-1 quasi-species [16,[57], [58], [59]].
4. Conclusion
In the current study, we have demonstrated the applicability of benchtop NGS platforms for the sequencing of HIV-1 single genome amplicons. With lower sample requirements and higher throughput, this approach is suitable to support the increasing demand for high-quality HIV-1 sequences in fields such as molecular epidemiology, and development of preventive and therapeutic strategies.
Conflict of interests
The authors declare no conflict of interests.
Gustavo Kijak is currently an employee of GSK Vaccines, Rockville, MD, USA.
Financial support
This project was funded in part by the U.S. Navy Bureau of Medicine and Surgery and the Military Infectious Diseases Research Program, project MIDRP-H014010OTPPOC.These studies were supported by a cooperative agreement (W81XWH-07-2-0067) between the Henry M. Jackson Foundation for the Advancement of Military Medicine and the Department of Defense.
Acknowledgements
This public health activity was reviewed by the Walter Reed Army Institute of Research (WRAIR) Institutional Review Board and termed RV314. The authors would like to thank Lydia Bonar, Bahar Ahani, Shana Howell and Emma De Neef for their technical support. The views expressed in this article are those of the authors and do not reflect the official policy or position of the Departments of the Army, Navy, or Defense, the Uniformed Services University, or any agency of the U.S. Government. Mention of trade names, commercial products, or organizations does not imply endorsement by the U.S. government. An author is an employee of the U.S. Government and as such under the provisions of 17 U.S.C. 105, copyright protection is not available for this work.
Handled by Justin O’Grady
Footnotes
Supplementary data associated with this article can be found, in the online version, at https://doi.org/10.1016/j.bdq.2019.01.002.
Appendix A. Supplementary data
The following are Supplementary data to this article:
References
- 1.Fauci A.S., Marston H.D. Focusing to achieve a world without AIDS. JAMA. 2015;313(4):357–358. doi: 10.1001/jama.2014.17454. PubMed PMID: 25626032. [DOI] [PubMed] [Google Scholar]
- 2.Kijak G.H., McCutchan F.E. HIV diversity, molecular epidemiology, and the role of recombination. Curr. Infect. Dis. Rep. 2005;7(6):480–488. doi: 10.1007/s11908-005-0051-8. PubMed PMID: 16225787. [DOI] [PubMed] [Google Scholar]
- 3.Korber B., Hraber P., Wagh K., Hahn B.H. Polyvalent vaccine approaches to combat HIV-1 diversity. Immunol. Rev. 2017;275(1):230–244. doi: 10.1111/imr.12516. PubMed PMID: 28133800; PubMed Central PMCID: PMCPMC5362114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Rhee S.Y., Blanco J.L., Jordan M.R., Taylor J., Lemey P., Varghese V. Geographic and temporal trends in the molecular epidemiology and genetic mechanisms of transmitted HIV-1 drug resistance: an individual-patient- and sequence-level meta-analysis. PLoS Med. 2015;12(4) doi: 10.1371/journal.pmed.1001810. PubMed PMID: 25849352; PubMed Central PMCID: PMCPMC4388826. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Rolland M., Edlefsen P.T., Larsen B.B., Tovanabutra S., Sanders-Buell E., Hertz T. Increased HIV-1 vaccine efficacy against viruses with genetic signatures in Env V2. Nature. 2012;490(7420):417–420. doi: 10.1038/nature11519. PubMed PMID: 22960785; PubMed Central PMCID: PMCPMC3551291. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Domingo E., Sheldon J., Perales C. Viral quasispecies evolution. Microbiol. Mol. Biol. Rev. 2012;76(2):159–216. doi: 10.1128/MMBR.05023-11. PubMed PMID: 22688811; PubMed Central PMCID: PMCPMC3372249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Coffin J.M. HIV population dynamics in vivo: implications for genetic variation, pathogenesis, and therapy. Science. 1995;267(5197):483–489. doi: 10.1126/science.7824947. PubMed PMID: 7824947. [DOI] [PubMed] [Google Scholar]
- 8.Council O.D., Joseph S.B. Evolution of host target cell specificity during HIV-1 infection. Curr. HIV Res. 2018;16(1):13–20. doi: 10.2174/1570162X16666171222105721. PubMed PMID: 29268687. [DOI] [PubMed] [Google Scholar]
- 9.Kijak G.H., Sanders-Buell E., Chenine A.L., Eller M.A., Goonetilleke N., Thomas R. Rare HIV-1 transmitted/founder lineages identified by deep viral sequencing contribute to rapid shifts in dominant quasispecies during acute and early infection. PLoS Pathog. 2017;13(7) doi: 10.1371/journal.ppat.1006510. PubMed PMID: 28759651; PubMed Central PMCID: PMCPMC5552316. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Sluis-Cremer N., Wainberg M.A., Schinazi R.F. Resistance to reverse transcriptase inhibitors used in the treatment and prevention of HIV-1 infection. Future Microbiol. 2015;10(11):1773–1782. doi: 10.2217/fmb.15.106. PubMed PMID: 26517190; PubMed Central PMCID: PMCPMC4813512. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Gunthard H.F., Wong J.K., Ignacio C.C., Havlir D.V., Richman D.D. Comparative performance of high-density oligonucleotide sequencing and dideoxynucleotide sequencing of HIV type 1 pol from clinical samples. AIDS Res. Hum. Retroviruses. 1998;14(10):869–876. doi: 10.1089/aid.1998.14.869. PubMed PMID: 9671215. [DOI] [PubMed] [Google Scholar]
- 12.Palmer S., Kearney M., Maldarelli F., Halvas E.K., Bixby C.J., Bazmi H. Multiple, linked human immunodeficiency virus type 1 drug resistance mutations in treatment-experienced patients are missed by standard genotype analysis. J. Clin. Microbiol. 2005;43(1):406–413. doi: 10.1128/JCM.43.1.406-413.2005. PubMed PMID: 15635002; PubMed Central PMCID: PMCPMC540111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Keele B.F., Giorgi E.E., Salazar-Gonzalez J.F., Decker J.M., Pham K.T., Salazar M.G. Identification and characterization of transmitted and early founder virus envelopes in primary HIV-1 infection. Proc. Natl. Acad. Sci. U. S. A. 2008;105(21):7552–7557. doi: 10.1073/pnas.0802203105. PubMed PMID: 18490657; PubMed Central PMCID: PMCPMC2387184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Salazar-Gonzalez J.F., Salazar M.G., Keele B.F., Learn G.H., Giorgi E.E., Li H. Genetic identity, biological phenotype, and evolutionary pathways of transmitted/founder viruses in acute and early HIV-1 infection. J. Exp. Med. 2009;206(6):1273–1289. doi: 10.1084/jem.20090378. PubMed PMID: 19487424; PubMed Central PMCID: PMCPMC2715054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herbeck J.T., Rolland M., Liu Y., McLaughlin S., McNevin J., Zhao H. Demographic processes affect HIV-1 evolution in primary infection before the onset of selective processes. J. Virol. 2011;85(15):7523–7534. doi: 10.1128/JVI.02697-10. PubMed PMID: 21593162; PubMed Central PMCID: PMCPMC3147913. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Dilernia D.A., Chien J.T., Monaco D.C., Brown M.P., Ende Z., Deymier M.J. Multiplexed highly-accurate DNA sequencing of closely-related HIV-1 variants using continuous long reads from single molecule, real-time sequencing. Nucleic Acids Res. 2015;43(20) doi: 10.1093/nar/gkv630. PubMed PMID: 26101252; PubMed Central PMCID: PMCPMC4787755. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Sanger F., Nicklen S., Coulson A.R. DNA sequencing with chain-terminating inhibitors. Proc. Natl. Acad. Sci. U. S. A. 1977;74(12):5463–5467. doi: 10.1073/pnas.74.12.5463. PubMed PMID: 271968; PubMed Central PMCID: PMCPMC431765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Taylor B.S., Sobieszczyk M.E., McCutchan F.E., Hammer S.M. The challenge of HIV-1 subtype diversity. N. Engl. J. Med. 2008;358(15):1590–1602. doi: 10.1056/NEJMra0706737. PubMed PMID: 18403767; PubMed Central PMCID: PMCPMC2614444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Moyo S., Vandormael A., Wilkinson E., Engelbrecht S., Gaseitsiwe S., Kotokwe K.P. Analysis of viral diversity in relation to the recency of HIV-1C infection in Botswana. PLoS One. 2016;11(8) doi: 10.1371/journal.pone.0160649. PubMed PMID: 27552218; PubMed Central PMCID: PMCPMC4994946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Salazar-Gonzalez J.F., Salazar M.G., Tully D.C., Ogilvie C.B., Learn G.H., Allen T.M. Use of dried blood spots to elucidate full-length transmitted/founder HIV-1 genomes. Pathog. Immun. 2016;1(1):129–153. doi: 10.20411/pai.v1i1.116. PubMed PMID: 27819061; PubMed Central PMCID: PMCPMC5096837. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Boltz V.F., Rausch J., Shao W., Hattori J., Luke B., Maldarelli F. Ultrasensitive single-genome sequencing: accurate, targeted, next generation sequencing of HIV-1 RNA. Retrovirology. 2016;13(1):87. doi: 10.1186/s12977-016-0321-6. PubMed PMID: 27998286; PubMed Central PMCID: PMCPMC5175307. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Barton K., Hiener B., Winckelmann A., Rasmussen T.A., Shao W., Byth K. Broad activation of latent HIV-1 in vivo. Nat. Commun. 2016;7:12731. doi: 10.1038/ncomms12731. PubMed PMID: 27605062; PubMed Central PMCID: PMCPMC5025526 clinical trial of vorinostat described in this manuscript. Payment was made to their institution. The remaining authors declare no competing financial interests. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Lange C.M., Hue S., Violari A., Cotton M., Gibb D., Babiker A. Single genome analysis for the detection of linked multiclass drug resistance mutations in HIV-1-Infected children after failure of protease inhibitor-based first-line therapy. J. Acquir. Immune Defic. Syndr. 2015;69(2):138–144. doi: 10.1097/QAI.0000000000000568. PubMed PMID: 25923117; PubMed Central PMCID: PMCPMC4679142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.de Azevedo S.S.D., Caetano D.G., Cortes F.H., Teixeira S.L.M., Dos Santos Silva K., Hoagland B. Highly divergent patterns of genetic diversity and evolution in proviral quasispecies from HIV controllers. Retrovirology. 2017;14(1):29. doi: 10.1186/s12977-017-0354-5. PubMed PMID: 28464889; PubMed Central PMCID: PMCPMC5414336. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Laird Smith M., Murrell B., Eren K., Ignacio C., Landais E., Weaver S. Rapid sequencing of complete env genes from primary HIV-1 samples. Virus Evol. 2016;2(2) doi: 10.1093/ve/vew018. PubMed PMID: 29492273; PubMed Central PMCID: PMCPMC5822884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sanborn K.B., Somasundaran M., Luzuriaga K., Leitner T. Recombination elevates the effective evolutionary rate and facilitates the establishment of HIV-1 infection in infants after mother-to-child transmission. Retrovirology. 2015;12(96) doi: 10.1186/s12977-015-0222-0. PubMed PMID: 26573574; PubMed Central PMCID: PMCPMC4647327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chen M., Ma Y., Yang C., Yang L., Chen H., Dong L. The combination of phylogenetic analysis with epidemiological and serological data to track HIV-1 transmission in a sexual transmission case. PLoS One. 2015;10(3) doi: 10.1371/journal.pone.0119989. PubMed PMID: 25807147; PubMed Central PMCID: PMCPMC4373787. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Evering T.H., Kamau E., St Bernard L., Farmer C.B., Kong X.P., Markowitz M. Single genome analysis reveals genetic characteristics of Neuroadaptation across HIV-1 envelope. Retrovirology. 2014;11:65. doi: 10.1186/s12977-014-0065-0. 10.1186/PREACCEPT-1509273248119831. PubMed PMID: 25125210; PubMed Central PMCID: PMCPMC4145222. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Todesco E., Wirden M., Calin R., Simon A., Sayon S., Barin F. Caution is needed in interpreting HIV transmission chains by ultra-deep sequencing. AIDS. 2018 doi: 10.1097/QAD.0000000000002105. PubMed PMID: 30585843. [DOI] [PubMed] [Google Scholar]
- 30.Yu F., Wen Y., Wang J., Gong Y., Feng K., Ye R. The transmission and evolution of HIV-1 quasispecies within one couple: a follow-up study based on next-generation sequencing. Sci. Rep. 2018;8(1):1404. doi: 10.1038/s41598-018-19783-3. PubMed PMID: 29362487; PubMed Central PMCID: PMCPMC5780463. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ratmann O., Wymant C., Colijn C., Danaviah S., Essex M., Frost S.D.W. HIV-1 full-genome phylogenetics of generalized epidemics in sub-Saharan Africa: impact of missing nucleotide characters in next-generation sequences. AIDS Res. Hum. Retroviruses. 2017 doi: 10.1089/aid.2017.0061. PubMed PMID: 28540766; PubMed Central PMCID: PMCPMC5597042. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Zhao Q., Zhang C., Jiang Y., Wen Y., Pan P., Li Y. Short communication: investigating a chain of HIV transmission events due to homosexual exposure and blood transfusion based on a next generation sequencing method. AIDS Res. Hum. Retroviruses. 2015;31(12):1225–1229. doi: 10.1089/aid.2015.0178. PubMed PMID: 26355677. [DOI] [PubMed] [Google Scholar]
- 33.Edlefsen P.T., Rolland M., Hertz T., Tovanabutra S., Gartland A.J., deCamp A.C. Comprehensive sieve analysis of breakthrough HIV-1 sequences in the RV144 vaccine efficacy trial. PLoS Comput. Biol. 2015;11(2) doi: 10.1371/journal.pcbi.1003973. PubMed PMID: 25646817; PubMed Central PMCID: PMCPMC4315437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Goodwin S., McPherson J.D., McCombie W.R. Coming of age: ten years of next-generation sequencing technologies. Nat. Rev. Genet. 2016;17(6):333–351. doi: 10.1038/nrg.2016.49. PubMed PMID: 27184599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Rothberg J.M., Hinz W., Rearick T.M., Schultz J., Mileski W., Davey M. An integrated semiconductor device enabling non-optical genome sequencing. Nature. 2011;475(7356):348–352. doi: 10.1038/nature10242. PubMed PMID: 21776081. [DOI] [PubMed] [Google Scholar]
- 36.Eyre D.W., Golubchik T., Gordon N.C., Bowden R., Piazza P., Batty E.M. A pilot study of rapid benchtop sequencing of Staphylococcus aureus and Clostridium difficile for outbreak detection and surveillance. BMJ Open. 2012;2(3) doi: 10.1136/bmjopen-2012-001124. PubMed PMID: 22674929; PubMed Central PMCID: PMCPMC3378946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Loman N.J., Misra R.V., Dallman T.J., Constantinidou C., Gharbia S.E., Wain J. Performance comparison of benchtop high-throughput sequencing platforms. Nat. Biotechnol. 2012;30(5):434–439. doi: 10.1038/nbt.2198. PubMed PMID: 22522955. [DOI] [PubMed] [Google Scholar]
- 38.Quail M.A., Smith M., Coupland P., Otto T.D., Harris S.R., Connor T.R. A tale of three next generation sequencing platforms: comparison of Ion Torrent, Pacific Biosciences and Illumina MiSeq sequencers. BMC Genomics. 2012;13(341) doi: 10.1186/1471-2164-13-341. PubMed PMID: 22827831; PubMed Central PMCID: PMCPMC3431227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Sims D., Sudbery I., Ilott N.E., Heger A., Ponting C.P. Sequencing depth and coverage: key considerations in genomic analyses. Nat. Rev. Genet. 2014;15(2):121–132. doi: 10.1038/nrg3642. PubMed PMID: 24434847. [DOI] [PubMed] [Google Scholar]
- 40.Brett-Major D.M., Hakre S., Naito N.A., Armstrong A., Bower E.A., Michael N.L. Epidemiology of contemporary seroincident HIV infection in the Navy and Marine corps. Mil. Med. 2012;177(11):1328–1334. doi: 10.7205/milmed-d-12-00299. PubMed PMID: 23198509. [DOI] [PubMed] [Google Scholar]
- 41.Heipertz R.A., Jr., Sanders-Buell E., Kijak G., Howell S., Lazzaro M., Jagodzinski L.L. Molecular epidemiology of early and acute HIV type 1 infections in the United States Navy and Marine Corps, 2005–2010. AIDS Res. Hum. Retroviruses. 2013;29(10):1310–1320. doi: 10.1089/AID.2013.0087. PubMed PMID: 23972100. [DOI] [PubMed] [Google Scholar]
- 42.Hakre S., Armstrong A.W., O’Connell R.J., Michael N.L., Scott P.T., Brett-Major D.M. A pilot online survey assessing risk factors for HIV acquisition in the Navy and Marine Corps, 2005-2010. J. Acquir. Immune Defic. Syndr. 2012;61(2):125–130. doi: 10.1097/QAI.0b013e31826a15db.. PubMed PMID: 23007117. [DOI] [PubMed] [Google Scholar]
- 43.Cock P.J., Fields C.J., Goto N., Heuer M.L., Rice P.M. The Sanger FASTQ file format for sequences with quality scores, and the Solexa/Illumina FASTQ variants. Nucleic Acids Res. 2010;38(6):1767–1771. doi: 10.1093/nar/gkp1137. PubMed PMID: 20015970; PubMed Central PMCID: PMCPMC2847217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kijak G.H., Pham P., Sanders-Buell E., Harbolick E.A., Eller L.A., Robb M.L. Nautilus: a bioinformatics package for the analysis of HIV type 1 targeted deep sequencing data. AIDS Res. Hum. Retroviruses. 2013;29(10):1361–1364. doi: 10.1089/aid.2013.0175. PubMed PMID: 23809062; PubMed Central PMCID: PMCPMC3785804. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Li H., Durbin R. Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics. 2009;25(14):1754–1760. doi: 10.1093/bioinformatics/btp324. PubMed PMID: 19451168; PubMed Central PMCID: PMCPMC2705234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Gaschen B., Kuiken C., Korber B., Foley B. Retrieval and on-the-fly alignment of sequence fragments from the HIV database. Bioinformatics. 2001;17(5):415–418. doi: 10.1093/bioinformatics/17.5.415. PubMed PMID: 11331235. [DOI] [PubMed] [Google Scholar]
- 47.Kearse M., Moir R., Wilson A., Stones-Havas S., Cheung M., Sturrock S. Geneious Basic: an integrated and extendable desktop software platform for the organization and analysis of sequence data. Bioinformatics. 2012;28(12):1647–1649. doi: 10.1093/bioinformatics/bts199. PubMed PMID: 22543367; PubMed Central PMCID: PMCPMC3371832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Schultz A.K., Zhang M., Leitner T., Kuiken C., Korber B., Morgenstern B. A jumping profile Hidden Markov Model and applications to recombination sites in HIV and HCV genomes. BMC Bioinformatics. 2006;7(265) doi: 10.1186/1471-2105-7-265. PubMed PMID: 16716226; PubMed Central PMCID: PMCPMC1525204. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Tamura K., Stecher G., Peterson D., Filipski A., Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol. Biol. Evol. 2013;30(12):2725–2729. doi: 10.1093/molbev/mst197. PubMed PMID: 24132122; PubMed Central PMCID: PMCPMC3840312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rolland M., Tovanabutra S., deCamp A.C., Frahm N., Gilbert P.B., Sanders-Buell E. Genetic impact of vaccination on breakthrough HIV-1 sequences from the STEP trial. Nat. Med. 2011;17(3):366–371. doi: 10.1038/nm.2316. PubMed PMID: 21358627; PubMed Central PMCID: PMCPMC3053571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Tovanabutra S., Sanders E.J., Graham S.M., Mwangome M., Peshu N., McClelland R.S. Evaluation of HIV type 1 strains in men having sex with men and in female sex workers in Mombasa, Kenya. AIDS Res. Hum. Retroviruses. 2010;26(2):123–131. doi: 10.1089/aid.2009.0115. PubMed PMID: 20156095. [DOI] [PubMed] [Google Scholar]
- 52.Curlin M.E., Zioni R., Hawes S.E., Liu Y., Deng W., Gottlieb G.S. HIV-1 envelope subregion length variation during disease progression. PLoS Pathog. 2010;6(12) doi: 10.1371/journal.ppat.1001228. PubMed PMID: 21187897; PubMed Central PMCID: PMCPMC3002983. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Altschul S.F., Gish W., Miller W., Myers E.W., Lipman D.J. Basic local alignment search tool. J. Mol. Biol. 1990;215(3):403–410. doi: 10.1016/S0022-2836(05)80360-2. PubMed PMID: 2231712. [DOI] [PubMed] [Google Scholar]
- 54.Archer J., Weber J., Henry K., Winner D., Gibson R., Lee L. Use of four next-generation sequencing platforms to determine HIV-1 coreceptor tropism. PLoS One. 2012;7(11) doi: 10.1371/journal.pone.0049602. PubMed PMID: 23166726; PubMed Central PMCID: PMCPMC3498215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N. The sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25(16):2078–2079. doi: 10.1093/bioinformatics/btp352. PubMed PMID: 19505943; PubMed Central PMCID: PMCPMC2723002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Pelak K., Shianna K.V., Ge D., Maia J.M., Zhu M., Smith J.P. The characterization of twenty sequenced human genomes. PLoS Genet. 2010;6(9) doi: 10.1371/journal.pgen.1001111. PubMed PMID: 20838461; PubMed Central PMCID: PMCPMC2936541. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caskey M., Schoofs T., Gruell H., Settler A., Karagounis T., Kreider E.F. Antibody 10-1074 suppresses viremia in HIV-1-infected individuals. Nat. Med. 2017;23(2):185–191. doi: 10.1038/nm.4268. PubMed PMID: 28092665; PubMed Central PMCID: PMCPMC5467219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Brese R.L., Gonzalez-Perez M.P., Koch M., O’Connell O., Luzuriaga K., Somasundaran M. Ultradeep single-molecule real-time sequencing of HIV envelope reveals complete compartmentalization of highly macrophage-tropic R5 proviral variants in brain and CXCR4-using variants in immune and peripheral tissues. J. Neurovirol. 2018;24(4):439–453. doi: 10.1007/s13365-018-0633-5. PubMed PMID: 29687407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Gonzalez C., Gondola J., Ortiz A.Y., Castillo J.A., Pascale J.M., Martinez A.A. Barcoding analysis of HIV drug resistance mutations using Oxford Nanopore MinION (ONT) sequencing. bioRxiv. 2017:240077. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.