Summary
The fundamental mechanisms of protein and lipid organization at the plasma membrane have continued to engage researchers for decades. Among proposed models, one idea has been particularly successful which assumes that sterol-dependent nanoscopic phases of different lipid chain order compartmentalize proteins, thereby modulating protein functionality. This model of membrane rafts has sustainably sparked the fields of membrane biophysics and biology, and shifted membrane lipids into the spotlight of research; by now, rafts have become an integral part of our terminology to describe a variety of cell biological processes. But is the evidence clear enough to continue supporting a theoretical concept which has resisted direct proof by observation for nearly twenty years? In this essay, we revisit findings that gave rise to and substantiated the raft hypothesis, discuss its impact on recent studies and present alternative mechanisms to account for plasma membrane heterogeneity.
Keywords: plasma membrane, membrane rafts, membrane heterogeneity, phase separation, lipids, membrane proteins, raft hypothesis
Introduction
The concept of membrane rafts is one of the most heatedly discussed topics in membrane biology. Part of this debate comes from a confusion about what a membrane raft actually is. Under the umbrella of the raft hypothesis, a vast variety of findings have amassed that are - to different degrees - compatible with its main postulation: that there is lipid-mediated phase separation in the plasma membrane serving to compartmentalize proteins and thus cellular functions [1] (see the Glossary in Box 1 for a more detailed explanation of key concepts described in this essay). On the one hand, these findings include protein and/or lipid assemblies on various length and time scales: nanometer [2–4] to micrometer [5,6], microseconds [7,8] to milliseconds [9] and seconds [10]. On the other hand, they span a broad range of cell biological, biochemical and biophysical observations: the cholesterol dependence of cellular processes [11], detergent insolubility of certain proteins and lipids [12], phase separation in model membranes composed of synthetic lipids [13], phase partitioning of proteins in plasma membrane vesicles [14], and differential sorting of proteins in polarized cells [15]. In general, the raft hypothesis seemed to offer an attractive explanation for a variety of functional cell biological data. So far, however, unequivocal proof for the postulated connection between these observations is missing: Numerous studies have shown that local and temporal heterogeneity of proteins and lipids exists in biological membranes, but while many of the observed phenomena could qualify as rafts, to day no study has yielded unambiguous evidence. In fact, in many cases segregation mechanisms other than lipid-mediated phase separation were identified [9,16–24].
Box 1. Glossary.
Raft hypothesis: The raft hypothesis distilled observations from biochemistry, cell biology and biophysics into a biological concept: sphingolipid and cholesterol clusters (rafts) are segregated from unsaturated lipids in the exoplasmic leaflet of the plasma membrane [1]. Proteins will segregate according to their affinity for raft and non-raft lipid phases. Rafts can act as sorting platforms for membrane traffic and relay stations for cell signaling.
Membrane raft: At a 2006 Keystone meeting, a new definition for rafts was adopted: “Membrane rafts are small (10–200 nm), heterogeneous, highly dynamic, sterol- and sphingolipid-enriched domains that compartmentalize cellular processes. Small rafts can sometimes be stabilized to form larger platforms through protein-protein and protein-lipid interactions”[25]. According to the raft hypothesis, the underlying mechanism of this phase separation is based on liquid-liquid immiscibility of certain lipid species. Lipid-protein assemblies arising from alternative phenomena, such as charge-mediated sequestering or specific binding of lipids to recognition sequences in proteins would thus not qualify as rafts.
Liquid ordered (Lo)/liquid disordered (Ld) phase separation: Lipid bilayers above the phase transition temperature exist in a fluid phase characterized by disordered lipid acyl chains and high lipid mobility. The presence of cholesterol has an ordering effect, leading to a more extended conformation and a tighter packing of the hydrocarbon chains, which results in a lower lateral mobility of lipids and an increase in membrane thickness [116]. Some synthetic lipid bilayers composed of a mixture of cholesterol, fully saturated lipids (e.g. sphingolipids), and unsaturated lipids show macroscopic phase separation into cholesterol-rich liquid ordered (Lo) and cholesterol-poor liquid disordered (Ld) domains [117]. This segregation is critically dependent on cholesterol, which, due to its rigid sterol backbone, disfavors interactions with the more bulky unsaturated lipid species and preferably associates with saturated lipids [118,119].
Glycosylphosphatidylinositol-anchored proteins (GPI-APs): In GPI-APs, the protein moiety is attached to the outer leaflet of the plasma membrane via a glycosylphosphatidylinositol lipid anchor. Often, the GPI-anchor contains two saturated acyl chains, but it can also feature unsaturated chains at the sn-2 position or even a third acyl chain [120]. The significance of the GPI-anchor is not yet fully understood; functions as an apical sorting signal and mediator of signal transduction have been proposed [32,121]. Based on the fact that GPI-APs are frequently isolated in detergent-resistant membrane (DRM) fractions and sorted to apical membranes, they have been considered to be archetypical “raft markers” [1].
Detergent-resistant membranes (DRMs): Upon extraction of the plasma membrane with cold non-ionic detergents such as Triton X-100, only a subset of its components gets solubilized. The remaining material (termed detergent-resistant) was found to be enriched in a characteristic set of proteins (e.g. GPI-APs), cholesterol and sphingolipids [15,32,122]. Likewise, Triton X-100 does not solubilize gel- or Lo-phase model membranes, but extracts lipids and GPI-APs from fluid membranes [12]. These similarities were taken as indication that – also in cell membranes – lipid phase separation occurs and that proteins segregate according to their Lo/Ld partition coefficients [12].
Giant plasma membrane vesicles (GPMVs): Upon treatment with formaldehyde and DTT, or N-ethylmaleimide, the plasma membrane is induced to produce blebs that can detach to form vesicles. At low temperature, phase separation into more ordered and more disordered phases can occur [14]. The difference in order and packing density between these membrane regions is not as high as in model membranes [86]. GPI-APs as well as most palmitoylated proteins have been shown to partition to the ordered phase in GPMVs [123]; it is assumed that this is mediated by their saturated lipid anchors.
Plasma membrane lipids: The most abundant lipid constituents of the mammalian plasma membrane are glycerophospholipids, sphingolipids, and cholesterol. The former two are categorized into distinct lipid classes defined by their headgroup, and further classified according to their acyl chain length and degree of saturation; this provides the potential of generating more than 100,000 different species [46]. Lipids and lipid-anchored proteins are distributed asymmetrically across the plasma membrane: the inner leaflet contains anionic, typically unsaturated phosphatidylserines and –inositols as well as phosphatidylethanolamines and palmitoylated proteins, whereas saturated sphingolipids and GPI-APs reside in the outer leaflet [124]. Among sphingolipids, gangliosides like GM1 take a special place as they have large carbohydrate headgroups and have a role in cell recognition, cell-to-cell contact formation, receptor binding and modulation and signal transduction [125]. The implications of this enormous compositional complexity for membrane-associated processes are unknown.
Moreover, we know today that important pillars that led to the proposition of the raft hypothesis are either outright artifacts [25], or at least ambiguous [26–30]. These new vistas, however, have had surprisingly little impact on the model itself. This is not necessarily cause for concern; as George E.P. Box famously wrote: “Essentially, all models are wrong, but some are useful” [31]. The question is: is the raft hypothesis right enough to be useful? In this essay we approach this question by separating it into two parts: 1) How right is the raft hypothesis? Here, we take a look at the past as we examine “milestones” that gave rise to or substantiated the raft hypothesis, and assess their validity in the light of evidence that we have today. While these milestones together represent the basis of the raft model today, we will attempt here a non-chronological dissection of its major tenets. 2) How useful is it? Over the years, new insights on plasma membrane organization and dynamics have been interpreted in view of the raft hypothesis. In the second part of this essay we will thus examine how the raft hypothesis itself has been shaping our view of the plasma membrane.
Part 1: The history of rafts revisited
Milestone 1: Detergent resistant membranes
Upon extraction of the plasma membrane with cold non-ionic detergents, sphingolipids, cholesterol, and glycosylphosphatidylinositol-anchored proteins (GPI-APs) remain insoluble [12,32]. Fueled by observations of lipid phase-separation in model membranes and apical sorting of GPI-APs, the idea seemed compelling that these detergent-resistant membranes (DRMs) reflect actual lipid-protein assemblies in living cells, which compartmentalize membrane proteins to functional domains [1]. It also sparked a huge amount of studies, resulting in the categorization of membrane proteins into raft and non-raft proteins based on their detergent-solubility (reviewed e.g. in [11]). However, problems with this classification system became apparent rather quickly: the group of proteins isolated in DRMs depends on the detergent and the conditions used [33]. Further, a generalization of the results was often not possible: for example, not all GPI-APs are actually found in DRMs [34]. Ultimately, a calorimetric study by Heerklotz suggested that the detergent Triton X-100 promotes growth of pre-existing liquid ordered (Lo) domains [35] and can even create ordered domains in an initially homogeneous, fluid model membrane. The latter finding, however, was challenged in a subsequent spectroscopic study [36]. In a 2006 Keystone Symposium on lipid rafts and cell function, DRM formation was considered as “an artificial and highly subjective approach that can induce the formation of membrane domains and hence does not provide physiologically relevant information” [25].
Conclusion: DRM experiments cannot be used to predict the protein or lipid composition of putative domains in live cell membranes.
Milestone 2: Apical sorting of GPI-APs
Intertwined with studies on their solubility in detergents, sphingolipids, cholesterol and GPI-APs were also found to follow specific secretory pathways [15,37]. It was argued that sphingolipids and cholesterol could form ordered domains in the Golgi apparatus, and recruit GPI-APs due to preferential lateral association with the long, saturated acyl chains of the GPI-moiety. Together, these components were thought to be sorted into specific vesicles for delivery to the apical membrane in polarized epithelial cells [12]. This view was not unchallenged: N-glycosylation of the protein, but not its GPI-anchor, was found to be responsible for apical targeting [38,39], and DRM-association did not necessarily correlate with apical sorting [39–41]. Today, the picture is still unclear, but it is known that the apical sorting of GPI-APs depends on their oligomerization [40,42]. This oligomerization, however, does not necessarily rely on the presence of cholesterol [42,43]; even when it does, GPI-APs with unsaturated lipid anchors are delivered to the plasma membrane just as efficiently [44]. Thus, depending on cell type and protein, GPI-APs can be sorted in an anchor-dependent or -independent manner to the apical or basolateral membrane.
Conclusion: Partitioning of GPI-APs into ordered domains in the Golgi apparatus is not a plausible mechanism for their sorting to the apical membrane in polarized cells.
Milestone 3: Phase separation in model membranes
In a bilayer, the presence of cholesterol can lead to phase separation of lipid components, producing a cholesterol-rich Lo phase and a cholesterol-poor liquid disordered (Ld) phase. The more pronounced the differences between the lipids’ physicochemical properties, the more pronounced is the phase separation [13]. Several observations indicated that a similar phase coexistence could also occur in cell membranes [12,37,45]. Indeed, the live cell plasma membrane seems predestined for phase separation as it contains all the necessary ingredients: saturated and unsaturated lipids, as well as cholesterol.
But how valid is the translation of findings in model membranes to the situation in a live cell? Due to chemical heterogeneities of the acyl chains, the headgroups, and the glycosylation patterns, a cell has the potential to produce up to 100,000 different lipid species [46]. Predicting the according phase diagram is virtually impossible. From Gibbs’ phase rule we know that – in equilibrium – a mixture of C components may produce up to C+1 coexisting phases, all of them containing a characteristic composition of lipids [47]; under non-equilibrium conditions there may be even more. In addition, the phase diagram contains regions which correspond to the coexistence of C phases, C-1 phases, etc. The enormous amount of different coexistence regions makes their size in the respective phase diagram fairly small. Essentially, the whole phase diagram consists of boundaries to the next coexistence region, and only marginal changes in the overall lipid composition lead to a transition to a different phase coexistence region. In general, this counteracts the formation of well-defined phases.
But the plasma membrane not only comprises a complex mixture of lipids, it is also subject to influences absent in model membranes: A substantial part of the plasma membrane area is occupied by proteins [48] that potentially act as direct binding partners for certain lipids, such as sphingomyelin, cholesterol or charged lipids [49–51]. In addition, hydrophobic or hydrophilic lipid-protein interactions result in slower lipid exchange from the protein-lipid interface; thereby, many proteins are surrounded by a shell of annular lipids that behave differently than bulk lipids [52]. Such lipids were found to be affected in their movement and their order parameter up to 3-4 nm away from the surface of a single Kv1.2 voltage gated ion channel [53]. In that study, the authors estimated that 50-100 lipid molecules diffuse in one dynamic complex with a single protein molecule, suggesting that virtually no unperturbed lipid molecule exists in the plasma membrane. Furthermore, lipids are continuously processed by enzymes and shuffled between the two leaflets of the plasma membrane. Anionic lipids in the inner leaflet can give rise to charge-mediated segregation, which has implications for protein clustering and many cellular processes [21,54,55].
In addition, proteins can interfere with phase separation simply by their abundance: steric pressure arising from collisions between proteins was found to destabilize Lo/Ld phase separation in model membranes, resulting in an optically homogeneous distribution of proteins and lipids over the membrane surface [56]. Also cortical actin has been shown to have a profound influence on overall membrane properties but also on protein and lipid dynamics [57,58]. Considering this complexity, it is not surprising that many observations, which were initially perceived as manifestations of raft-mediated phase separation in biological membranes, have today been identified to be of a different origin [3,4,17,19,33,43,59].
Conclusion: The presence of Lo/Ld phase separation in synthetic model systems does not provide conclusive evidence for its existence in cells.
Milestone 4: Cholesterol dependence
During the seventies, eighties and nineties of the past century, studies on DRMs, on lipid and protein sorting, and on phase-separated bilayers seemed to converge on one fundamental principle: the differential self-organization of lipids and membrane proteins based on their raft affinity. From the beginning, cholesterol was suggested to be a sine qua non component of rafts. In fact, the raft hypothesis has highlighted cholesterol in view of its influence on lipid phase separation, thereby creating an equivalency of the terms cholesterol-dependent and raft-dependent. A standard raft-experiment involves the alteration of cellular cholesterol levels, which is typically performed by addition of either methyl-β-cyclodextrin (MβCD), saponin or cholesterol oxidase. These treatments often resulted in dramatic changes of plasma membrane observables such as the organization of proteins in clusters [57,60,61], the dimerization of mobile components [9,10], and the immobilization of GPI-anchored proteins or lipids [4,62]. Functional consequences of cholesterol depletion were also frequently observed and attributed to dissolution of rafts [28].
Cholesterol depleting agents, however, are not very specific. MβCD extracts cholesterol, but also other lipids such as sphingolipids and phospholipids [63], removal of cholesterol using the detergent saponin can leave holes in the plasma membrane [64] and cholesterol oxidase produces cholestenone, which is membrane active by itself [65]. In addition to this, cholesterol has a profound influence on general lipid bilayer properties such as bilayer fluidity as well as permeability for ions and small non-ions [66–68]. The effects of cholesterol on proteins and their functions are manifold. The formation of supramolecular clusters of syntaxin, for instance, requires cholesterol, however, the association mechanism is mediated by homophilic SNARE-motif interactions and not by phase separation [69]. Cholesterol depletion has been found to affect the amount of key phospholipids regulating various steps of membrane transport pathways and PIP2 levels in the plasma membrane [70,71]. Cholesterol levels influence actin organization and dynamics [70,72], which in turn affects protein organization [73]. A direct and specific effect of cholesterol on protein function has been described for the β2-adrenergic receptor [74]. Apart from this, cholesterol-sensing domains have been identified in several transmembrane proteins, reviewed in [50], including caveolin-1 [75] and many scaffold proteins with key roles in protein networking [76,77]. For a more detailed discussion of this, we refer the reader to [27].
Conclusion: The importance of cholesterol for cellular processes cannot be reduced to its ability to induce Lo/Ld phase separation.
Milestone 5: Crosslinking and co-patching
Diffraction-limited fluorescence microscopy experiments show a rather homogeneous distribution of GPI-APs in the plasma membrane [78], yielding no direct support of raft-type segregation to membrane domains. This is why researchers used antibodies to crosslink GPI-APs to generate larger, optically resolvable platforms, and determined the recruitment of other membrane proteins [45]. The finding that crosslinking of DRM components induces cholesterol-dependent co-patching of other DRM-associated proteins and lipids – but not detergent-soluble proteins – was taken as indication for the consistency of the raft hypothesis [45]. Another intriguing effect of crosslinking of GPI-APs was the induction of T cell receptor signaling, which was thought to be mediated via the formation of Lo-like domains and concomitant recruitment of effector molecules [79].
One problem of these studies is a lack of coherence, as different cell types, proteins and/or experimental conditions yielded different results: While Mitchell, et al. [80] found co-patching of the GPI-AP CD59 with the sphingolipid GM1, Mrowczynska and Hagerstrand [81] did not; different viral glycoproteins showed co-patching, but only some of them were found in DRMs, and cholesterol-depletion did only slightly affect co-patching efficiency [82].
But why would proteins associate with other, crosslinked proteins? A clue for this comes from a study, where streptavidin attached to the plasma membrane via different lipid anchors was used to mimic GPI-APs [59]. As expected, streptavidin with fully saturated lipids as anchors was isolated in DRMs, whereas the unsaturated construct was not. Interestingly, the authors observed co-patching of their streptavidin constructs with DRM components irrespective of whether the lipid anchors were saturated or unsaturated. Also, crosslinking of either the saturated or unsaturated constructs led to activation of signaling events in Jurkat T cells, indicating that it is not phase partitioning of GPI-APs that is responsible for cell activation, as previously proposed [79]; in contrast, it seems to be a generic property of lipid-anchored proteins in the outer leaflet of the plasma membrane. Another study found that the ability to induce membrane curvature was critical for efficient co-patching of putative raft markers with the structure protein Gag [83]. Indeed, mere addition of short-chain phospholipids induced cellular extensions and filopodia that recruited the sphingolipid GM1 [84]. These observations suggest that the observed copatching may be based on phenomena related to the ectodomain rather than the membrane anchor, e.g. molecular crowding and induction of local membrane curvature. Such effects can easily be misinterpreted as local enrichment of membrane constituents: FRET measurements on Jurkat T cells revealed that the apparent recruitment of the raft markers GFP-GPI and Cholera toxin to sites of antibody-induced T cell receptor signaling was actually an imaging artifact caused by localized membrane ruffling [85].
Conclusion: The observed co-patching of proteins does not imply their co-partitioning to ordered membrane phases.
Milestone 6: Giant Plasma Membrane Vesicles (GPMVs)
GPMVs can be formed from the plasma membrane of live cells by treatment with formaldehyde and DTT, or N-ethylmaleimide. At low temperature, phase separation in optically resolvable more ordered and more disordered phases can be induced [14,86]. Protein and lipid partitioning into these phases often correlates with their detergent solubility. This was taken as an indication that the plasma membrane is actually poised towards large scale phase separation, which is actively prevented by cells resulting in nanoscopic phase separation [87].
There are several important differences between GPMVs and the live cell plasma membrane: First, lipid asymmetry between the two membrane leaflets is lost in GPMVs. Both head group charge and degree of acyl chain saturation are substantially different in the inner and outer leaflet of the plasma membrane [88]. Mixing of inner and outer leaflet lipids therefore may well affect the propensity of the new membrane for phase segregation. Second, in the plasma membrane a substantial fraction of proteins – and thus of the associated annular lipids – is immobilized to the cortical actin skeleton; those molecules do not participate in the equilibration of the live cell system, but they do equilibrate in the GPMVs. Third, membrane tension imposed by the cytoskeleton is lost, which can have consequences on lipid phase separation [89–91]. It should be noted that the specific conditions used to form GPMVs influence the phase partitioning of the studied proteins, partly due to different degrees of protein depalmitoylation during GPMV preparation [92,93].
The observation that large-scale phase separation can be induced in the plasma membrane by chemical treatment and temperature decrease per se is intriguing. It represents an amendment to DRM studies as it shows that phase separation can indeed occur and that proteins can be sorted in a context resembling the live cell plasma membrane in many ways. As DRMs, GPMV studies show what is biophysically possible, but the a priori assumption that observed phenomena also occur in a live cell does not seem justified.
Conclusion: The observation of microscale phase separation in GPMVs cannot give direct evidence for its existence on the nanoscale in living cells.
How right is the raft hypothesis?
For our first question “How right is the raft hypothesis?” we cannot provide a definite answer, as the existence of an undetectable phenomenon can neither be proven nor disproven. In view of the discrepancies and ambiguities associated with the milestones discussed above, however, it seems no longer empirically supported to suggest lipid-mediated phase separation in the plasma membrane as a fundamental basis for current cell biological models. The assumption that rafts exist does not seem more justified than the opposite.
Part 2: Alternative views on the plasma membrane
More than one mechanism for heterogeneity
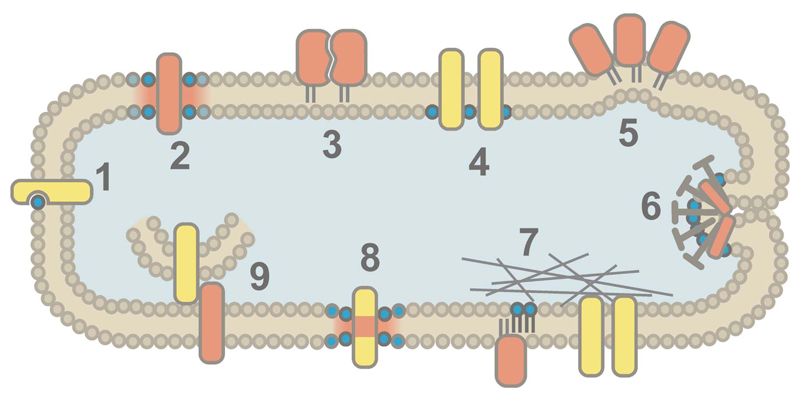
Among mechanisms accounting for heterogeneity of lipids and proteins in the plasma membrane, the raft-model is unrivaled in popularity. There are, however, several viable alternative mechanisms for phenomena currently attributed to rafts (Figure 1). One prominent example proposes that, conceptually analogous to hydration shells around proteins in aqueous solution, proteins are surrounded by a shell of annular lipids [52]. Favorable hydrophobic or hydrophilic lipid-protein, lipid-lipid, and/or lipid-glycan interactions can result in longer dwell times of certain lipids in the vicinity of proteins leading to the formation of dynamic lipid-protein complexes comprising up to 100 lipids [53,94]. A single protein molecule can thus affect the movement and order parameter of lipids up to 3-4 nm away from its surface, suggesting that virtually no unperturbed lipid molecule exists in the plasma membrane [53]. This motional restriction can be unspecific, but some proteins preferentially recruit lipids of a certain headgroup or acyl chain length [52]. The latter phenomenon is caused by a discrepancy in the length of a protein transmembrane domain and the thickness of the bilayer hydrophobic core. The energy penalty resulting from this ‘hydrophobic mismatch’ can thus lead to the recruitment of a suitable lipid environment but also cause membrane deformation, and/or promote protein clustering [95,96]. Some lipids engage in even stronger interactions with proteins by binding to specific recognition motifs in protein transmembrane regions and can be co-crystallized [49]. These lipids, often cholesterol, sphingolipids and phosphatidylinositols, can act as allosteric regulators of protein function, stabilizers or “molecular glue” between subunits of protein complexes [49–51].
Figure 1. Possible mechanisms for generating protein and lipid heterogeneity in the plasma membrane.
(1) Specific binding of lipids to recognition motifs in membrane proteins. (2) Transmembrane proteins surrounded by a shell of annular lipids. (3) Protein-protein interactions. (4) Charge-mediated sequestration of proteins and lipids. (5) Membrane bending induced by protein crowding. (6) Recruitment of specific lipids and proteins to endo- or exocytosis sites. (7) Protein and lipid diffusion confined by the cortical actin skeleton. (8) Hydrophobic mismatch between protein transmembrane domains and the hydrophobic core of the lipid bilayer. (9) Protein interactions and lipid transfer at contact sites between the ER and the plasma membrane.
Anionic lipids such as phosphatidylinositols or phosphatidylserines were found to profoundly influence protein organization via charge-mediated segregation. Divalent ions, membrane depolarization or interaction with positively charged proteins can result in the formation of nanodomains independently of cholesterol or lipid phases [20,21,55].
Anionic lipids are also key elements in endo- and exocytosis, such as clathrin-mediated endocytosis or synaptic vesicle fusion [97,98]. In all processes involving budding or fusion of vesicles, areas of high membrane curvature will be generated, which can be associated with the local enrichment of non-lamellar lipids such as phosphatidic acid or diacylglycerols that are formed by lipid-modifying enzymes [98,99]. Considering the abundance of proteins in the plasma membrane, it is quite conceivable that membrane bending can be caused merely due to lateral pressure generated by collisions between transmembrane as well as lipid-anchored membrane proteins [48,100]. This, in turn, will entail curvature-mediated sorting of lipids and proteins [101]. Of note, deviations from planar membrane topology can also lead to perceived clustering that is actually artefactual [85].
Also, other organelles can influence plasma membrane organization: the endoplasmic reticulum (ER) has been found to form tight contacts with the plasma membrane, facilitating non-vesicular exchange of lipids [102]. In addition, a phosphoinositide phosphatase located in the ER can, for instance, regulate phosphatidylinositol levels in the plasma membrane [103].
Last but not least, cortical actin can act to compartmentalize proteins and lipids in the plasma membrane, as proposed by the ‘fence and pickets’ model [73,104]: the diffusion of transmembrane proteins can be restricted directly by the actin cytoskeleton ‘fence’; proteins anchored to this fence can, in turn, impede the diffusion of other proteins and lipids due to steric hindrance at these obstacles. In addition to such actin corrals, dynamic actin assemblies were proposed to function as anchoring sites for protein and lipid clusters [18].
This is certainly not an exhaustive discussion of sources of plasma membrane heterogeneity, but offers a glimpse of the variety of influences proteins and lipids are subject to. These alternative models may have their own sets of limitations, but have to be integrated into the explanatory repertoire of cell biology. For more detailed information, we refer the reader to excellent reviews addressing one or more of the above-mentioned mechanisms [96,97,105–108].
What if there were no rafts?
We invite the reader to follow us on a thought experiment: If rafts had not been proposed twenty years ago, how would that change today’s picture of membrane biology? In a few recent high-impact studies high-resolution and ultrasensitive microscopy techniques were used to investigate plasma membrane organization; large parts of the scientific community understood those results under the premise of existing membrane rafts. It is, however, increasingly difficult for non-experts – as well as peer-reviewers – to judge a study [109]. We thus revisited four of these studies with respect to the experimental findings and their presentation, and tried to find consistent interpretations while avoiding the assumption of rafts (see BOX 2).
Box 2. Recent studies reinterpreted.
Finding 1
Kusumi and coworkers analyzed the diffusion behavior of single proteins with respect to the appearance of transient homo- or hetero-association events [9]. GPI-APs were shown to transiently co-diffuse for 50 to 300 ms. Colocalization lifetime decreased upon cholesterol depletion and increased with concentration, but was independent of protein N-glycosylation or the presence of actin. The replacement of the GPI-anchor with a transmembrane protein domain reduced the colocalization lifetime. When the GPI-APs CD59 and DAF were individually crosslinked with antibodies, these entities showed prolonged hetero-association.
Rafts: Protein ectodomain interactions are responsible for homodimer formation that serve as basic units for raft organization and can coalesce to larger hetero-associates via raft lipid interactions.
No rafts: Transient homo-association of GPI-APs is mediated by interactions of their ectodomains. Membrane anchoring of proteins per se has an effect on the binding constants by altering ectodomain orientation and accessibility. Indeed, GPI anchors were found to influence the conformation and structure of the protein to which they were attached. For example, antibodies to GPI-APs exhibit greatly reduced affinity toward the same proteins lacking the lipid tail [126,127]; for different GPI-APs the glycan moiety was found to be buried inside the protein, to assume an extended conformation on the bilayer surface or to pull the protein into close contact with the bilayer [128–130], all of which can have an impact on protein-protein interactions.
Comment: Quantifying brief molecular interactions via the analysis of co-diffusion times can be challenging [131]. Prolonged co-diffusion times may not only indicate the presence of an interaction between two entities, but could also reflect the increased duration of incidental co-diffusion of the two entities due to their reduced mobility. Since the diffusion of an antibody-crosslinked CD59 is almost three times slower than that of a single CD59 molecule labeled with a Fab fragment [132], a concomitant 3-fold increase in the co-diffusion times of antibody-crosslinked GPI-APs could in fact be expected. It is thus difficult to disentangle specific interactions and incidental co-diffusion of the fluorescent probe molecules.
Finding 2
In a series of studies on the clustering of GPI-APs, Mayor and coworkers determined the proximity of their fluorescent probes by measuring the decrease of fluorescence anisotropy due to homo-FRET [18,57,58]. They found that GPI-APs formed transient clusters of a few molecules that were immobile, concentration-independent, but actin- and cholesterol-dependent [2,57]. Theoretical work indicated that dynamic actin assemblies could induce clusters of transiently associated transmembrane proteins, but also GPI-anchored proteins by an unknown link [58]. Clustering was absent in cells expressing a GPI-AP with short or unsaturated acyl chains, and required the presence of long-chain phosphatidylserine on the inner leaflet of the plasma membrane [18].
Rafts: Transbilayer coupling of saturated long chain lipids within raft domains is the source of GPI-AP nanoclusters. These nanoclusters have Lo-like properties, and are maintained by the cortical actomyosin network in the cell.
No rafts: The observation of transbilayer coupling in the case of C16:0 and C18:0 lipids – but not in the case of C18:1 lipids – is not necessarily related to nanoscopic Lo phases. Unsaturated lipids are generally much more resistant to interdigitation and would thus be inefficient in inducing transbilayer coupling [133,134]. Indeed, in an earlier publication, the authors suggested that the observed effects upon cholesterol depletion may well arise from effects on the organization and dynamics of cortical actin [57].
Finding 3
Eggeling and co-workers used stimulated emission depletion fluorescence correlation spectroscopy (STED-FCS) to monitor the diffusion behavior of fluorescently labeled molecules on length scales down to 30 nm [4]. They found sphingomyelin, GM1, and GPI-APs to be transiently trapped within areas <20 nm in the plasma membrane in a cholesterol-dependent fashion. The trapping events lasted for 10-20 ms and were not observed for phosphatidylethanolamine lipids.
Rafts: The transient halts of GPI-APs and sphingolipids can be explained by dynamic partitioning into more ordered and slowly diffusing rafts with a size <20 nm.
No rafts: Sphingolipids are both donors and acceptors of hydrogen bonds [135], and can engage in interactions with each other, with cholesterol, and with proteins [136,137]; a recognition motif specifically for C18 sphingomyelin has been identified in several transmembrane proteins [49]. For GPI-APs, association to transient actin asters via lipid interdigitation has been proposed [18]. Other interpretations include trapping by transient binding to immobilized proteins, partitioning to lipid shells around proteins, or trapping at sites of crowding or of endocytosis.
Finding 4
Gaus and coworkers analyzed fluorescence lifetime decays of the environment-sensitive membrane dye Laurdan, which displays a decreased fluorescence lifetime in less hydrated environments [113]. Laurdan showed double-exponential fluorescence decays in the plasma membrane of live HeLa cells, indicative of two distinct membrane environments. Upon addition of 7-ketocholesterol, the environment featuring shorter Laurdan fluorescence lifetimes became more prevalent.
Rafts: Due to the tighter packing of lipids in ordered compared to disordered phases, the lipid-water interface region is less hydrated, which can be sensed by Laurdan. Quantitative analysis of Laurdan fluorescence lifetimes data suggests that the plasma membrane of HeLa cells consists of a mixture of nanoscopic Lo (76%) and Ld domains (26%).
No rafts: Laurdan lifetime does not directly report on lipid packing but on the hydration of the interface region [114]. Nanoscale domains of increased Laurdan fluorescence lifetimes could thus originate from e.g. annular lipids that exhibit slower rotational and lateral mobility [115]. Since ~20-30% of the plasma membrane area is occupied by proteins [48], the observed heterogeneity in hydration levels is not surprising. Even at far lower concentrations the presence of peptides has a pronounced effect on Laurdan fluorescence characteristics [115]. Further, Laurdan is expected to flip-flop rapidly in membranes [138] and could thus report on inhomogeneous distribution of charged lipids and proteins in the inner leaflet [54,55].
Comment: In this study, cholesterol extraction with MβCD had a very moderate effect and reduced the percentage of Lo phase from 76±6 to 70±3%, whereas addition of 7-ketocholesterol reduced it to 23±11%. There are indications, however, that the effect of 7-ketocholesterol is actually more related to lipid mobility than to the proposed conversion of an ordered state to a disordered: In a bilayer composed of 90% palmitoyl-oleoyl-phosphatidylcholine and 10% cholesterol, exchange of cholesterol for 7-ketocholesterol significantly decreased Laurdan fluorescence lifetimes while not decreasing the order parameter in the hydrophobic core [139,140].
Of two recent studies on the homo-association behavior of GPI-APs, one identified ectodomain interactions as the main underlying cause (BOX2 – Finding 1) [9], while the second proposed transient transbilayer interactions with actin assemblies (BOX2 – Finding 2) [18]. Both studies provide significant contributions to our understanding of membrane organization, but the connection to rafts is not obvious. Indeed, GPI-APs have frequently been reported to form homodimers or oligomers, but the findings are equivocal: For GPI-anchored monomeric GFP (mGFP-GPI) alone, lifetimes of the observed associates range from 70 ms [9] to seconds [10]; in some studies, the association has been found to require actin [2,57,110], others did not reveal any actin-dependence [9]; concentration dependence was observed [9,10] or not [57], and associates were found to be mobile [9,10] or immobile [57]. All studies agree on the cholesterol-dependence of the detected homo-association, but as discussed above, the disappearance of a phenomenon upon depletion of cellular cholesterol cannot give conclusive evidence. The observed discrepancies may reflect experimental limitations, idiosyncrasies of the cellular systems or GPI-anchor species used, as well as the multifaceted properties of the life-cycle of GPI-APs. It seems that a generalization of the association behavior of GPI-APs and the establishment of a relation to membrane rafts is currently not possible.
In a recent study, Eggeling, et al. [4] found that the diffusion behavior of sphingolipids and GPI-APs was characterized by transient cholesterol-dependent halts (BOX2 – Finding 3). Although the authors themselves were rather cautious about interpreting their findings, the community perceived the study as supportive of the raft hypothesis [87,107,111]. In more recent publications Eggeling and coworkers had extended their methodology and reported that the diffusion behavior of their probes was not determined by the degree of saturation of the lipid anchor [16,112]. Instead of partitioning into nanoscale more ordered domains, the authors thus proposed transient binding to immobile proteins and plasma membrane topology as possible explanations for the observed halts.
These studies highlight a general problem associated with the majority of raft studies: the experimentally determined parameters do not interrogate the presence of lipid-mediated phase separation, but merely report on the behavior of a putative raft- or non-raft molecule under specific conditions. Recalling that this dichotomous classification is problematic per se, it seems doubtful whether such studies can be employed to draw conclusions on raft properties.
A different approach was taken by Gaus and coworkers, who used the environment-sensitive dye Laurdan as a direct probe of lipid bilayer properties (BOX2 – Finding 4) [113]. Their study revealed a coexistence of nanoscopic ordered and disordered domains in the plasma membrane. In this case, however, the complexity of the plasma membrane compared to a simple lipid bilayer makes unequivocal conclusions very difficult: The detection of nanoscopic regions of different order could just as easily be explained by the mere presence of proteins, lipid shells around proteins, differences between inner and outer leaflet lipids and/or charge-mediated segregation of lipids and proteins [114,115].
Recent studies not in line with the raft model
Several recent studies have come up with results that are in outright discrepancy with major tenets of the raft hypothesis. Strong experimental evidence against the presence of cholesterol-enriched nano-domains in the plasma membrane came from studies using high-resolution secondary ion mass spectrometry. Kraft and coworkers indeed found that sphingolipids were clustered in 200 nm-sized domains in fixed fibroblasts [23], however, the distribution of cholesterol was homogeneous [22]. The authors concluded that favorable interactions between sphingolipids and cholesterol do not dictate their distribution in the plasma membrane. Using fluorescence cross-correlation spectroscopy, Groves and coworkers observed that the co-diffusion of lipid-anchored proteins at the inner leaflet of the plasma membrane followed complex and heterogeneous schemes incompatible with the simple phase separation model of the raft hypothesis [19].
Recently, our own group designed a study to specifically address the role of lipid-mediated phase separation in the organization of GPI-APs [24]. We immobilized GPI-anchored monomeric GFP (mGFP-GPI) in the plasma membrane of live cells within micrometer-sized patterns and used single-molecule tracking to determine the effect of mGFP-GPI enrichment on the local membrane properties. We did not find evidence, however, for the formation of a connective ordered membrane phase nor did we observe any enrichment of nanoscopic ordered domains within the mGFP-GPI micropatterned regions. Thus, our findings indicate that the association behavior of GPI-APs is governed by their ectodomains (as proposed in [9]) but does not involve phase partitioning.
How useful is the raft hypothesis?
This brings us to our second question: "How useful is the raft hypothesis?” Indeed, the postulate of lipid rafts has boosted research on biomembranes in general, and on the role of lipids in membrane protein interactions in particular. On the coat-tails of rafts, model membranes drew the attention of cell biologists, who started appreciating membranes as being more than just a matrix for protein interactions. In fact, current membrane biology would be incomplete without the concept of coexisting ordered and disordered phases, and without biophysicists who dive deeper into the mechanisms behind the formation of even nanoscale phases.
Moreover, the difficulty to verify the presence of membrane rafts in the live cell context drove the development of new high-resolution imaging techniques, which in turn have helped us to discover the nature of membranes at the nanoscale. For many years, it appeared to be just a matter of time until rafts would finally be proven.
After twenty years without clear-cut evidence, however, it seems that - instead of facilitating new insights - the dominance of the raft hypothesis rather obstructs an unbiased scientific approach. Often, straightforward interpretation and presentation of new data within the raft paradigm is accepted more easily by the community than alternative models, making it difficult for more detailed concepts to receive the appropriate attention. As a matter of fact, it was the broad scope of the raft model which set a foundation for its success and generated a plethora of functional studies that in turn could consistently be interpreted within the framework of rafts. But many of these studies were specifically tailored to the raft model, and actually did not attempt to put it to the test. To us it therefore does not seem reasonable, if one considers the sheer amount of consistent studies as additional justification of the model.
A remarkable feature of rafts is the flexibility towards new data: whenever fundamental conflicts between new observations and the raft hypothesis arose, amendments to the model allowed to incorporate the new findings. Expressed positively, continuous refinements of the model will indeed ultimately yield a fairly reasonable approximation of the truth. For scientific communication, however, we need to keep track of the version of the raft model: a 3.0 raft from 2016 will likely be incommensurable with the 2.0 Keynote raft from 2006, or the original 1.0 launch from 1997.
Concluding remarks
As we have tried to convey in this essay, a thorough re-examination of previous milestone experiments does not yield sufficient evidence for the a priori assumption that membrane rafts exist and function to compartmentalize cellular processes. Certainly, there are DRM-associated proteins, Lo-phase-partitioning proteins, or cholesterol-dependent protein clusters, but these observations cannot be condensed into the one underlying principle of membrane organization the raft hypothesis suggests. Beyond that, we think that the unparalleled popularity of the raft model has generated an unwanted side-effect: there is a tendency to interpret one’s results within the raft context, while alternative explanations are neglected. There are, however, no obvious grounds for treating the raft model differently to other proposed membrane models, until advanced methods can provide conclusive evidence. Until then, as we continue to unravel more and more aspects of plasma membrane organization and dynamics, it seems conducive to welcome them in an unbiased manner, instead of reshaping the raft hypothesis to accommodate new findings.
Acknowledgements
The authors gratefully acknowledge support from the Austrian Science Fund (FWF projects P26337-B21 and P 25730-B21), and the Austrian Research Promotion Agency FFG (842379).
Abbreviations
- GPI-AP
glycosylphosphatidylinositol-anchored protein
- DRM
detergent-resistant membrane
- GPMV
giant plasma membrane vesicle
- MβCD,
methyl-β-cyclodextrin
- FRET
Förster resonance energy transfer
- ER
endoplasmic reticulum
Footnotes
Conflict of interest
The authors declare that there are no conflicts of interest.
References
- 1.Simons K, Ikonen E. Functional rafts in cell membranes. Nature. 1997;387:569–72. doi: 10.1038/42408. [DOI] [PubMed] [Google Scholar]
- 2.Sharma P, Varma R, Sarasij R, Ira, et al. Nanoscale organization of multiple GPI-anchored proteins in living cell membranes. Cell. 2004;116:577–89. doi: 10.1016/s0092-8674(04)00167-9. [DOI] [PubMed] [Google Scholar]
- 3.Douglass AD, Vale RD. Single-Molecule Microscopy Reveals Plasma Membrane Microdomains Created by Protein-Protein Networks that Exclude or Trap Signaling Molecules in T Cells. Cell. 2005;121:937–50. doi: 10.1016/j.cell.2005.04.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Eggeling C, Ringemann C, Medda R, Schwarzmann G, et al. Direct observation of the nanoscale dynamics of membrane lipids in a living cell. Nature. 2009;457:1159–62. doi: 10.1038/nature07596. [DOI] [PubMed] [Google Scholar]
- 5.Schütz GJ, Kada G, Pastushenko VP, Schindler H. Properties of lipid microdomains in a muscle cell membrane visualized by single molecule microscopy. Embo J. 2000;19:892–901. doi: 10.1093/emboj/19.5.892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Gaus K, Gratton E, Kable EP, Jones AS, et al. Visualizing lipid structure and raft domains in living cells with two-photon microscopy. Proc Natl Acad Sci U S A. 2003;100:15554–9. doi: 10.1073/pnas.2534386100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dietrich C, Yang B, Fujiwara T, Kusumi A, et al. Relationship of lipid rafts to transient confinement zones detected by single particle tracking. Biophys J. 2002;82:274–84. doi: 10.1016/S0006-3495(02)75393-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Dahan M, Levi S, Luccardini C, Rostaing P, et al. Diffusion dynamics of glycine receptors revealed by single-quantum dot tracking. Science. 2003;302:442–5. doi: 10.1126/science.1088525. [DOI] [PubMed] [Google Scholar]
- 9.Suzuki KG, Kasai RS, Hirosawa KM, Nemoto YL, et al. Transient GPI-anchored protein homodimers are units for raft organization and function. Nat Chem Biol. 2012;8:774–83. doi: 10.1038/nchembio.1028. [DOI] [PubMed] [Google Scholar]
- 10.Brameshuber M, Weghuber J, Ruprecht V, Gombos I, et al. Imaging of Mobile Long-lived Nanoplatforms in the Live Cell Plasma Membrane. J Biol Chem. 2010;285:41765–71. doi: 10.1074/jbc.M110.182121. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simons K, Ehehalt R. Cholesterol, lipid rafts, and disease. The Journal of clinical investigation. 2002;110:597–603. doi: 10.1172/JCI16390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schroeder R, London E, Brown D. Interactions between saturated acyl chains confer detergent resistance on lipids and glycosylphosphatidylinositol (GPI)-anchored proteins: GPI-anchored proteins in liposomes and cells show similar behavior. Proc Natl Acad Sci U S A. 1994;91:12130–4. doi: 10.1073/pnas.91.25.12130. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Veatch SL, Keller SL. A closer look at the canonical 'Raft Mixture' in model membrane studies. Biophys J. 2003;84:725–6. doi: 10.1016/S0006-3495(03)74891-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baumgart T, Hammond AT, Sengupta P, Hess ST, et al. Large-scale fluid/fluid phase separation of proteins and lipids in giant plasma membrane vesicles. Proc Natl Acad Sci U S A. 2007;104:3165–70. doi: 10.1073/pnas.0611357104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Brown DA, Crise B, Rose JK. Mechanism of membrane anchoring affects polarized expression of two proteins in MDCK cells. Science. 1989;245:1499–501. doi: 10.1126/science.2571189. [DOI] [PubMed] [Google Scholar]
- 16.Honigmann A, Mueller V, Ta H, Schoenle A, et al. Scanning STED-FCS reveals spatiotemporal heterogeneity of lipid interaction in the plasma membrane of living cells. Nature Communications. 2014;5 doi: 10.1038/ncomms6412. [DOI] [PubMed] [Google Scholar]
- 17.Lin WC, Iversen L, Tu HL, Rhodes C, et al. H-Ras forms dimers on membrane surfaces via a protein-protein interface. Proc Natl Acad Sci U S A. 2014;111:2996–3001. doi: 10.1073/pnas.1321155111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Raghupathy R, Anilkumar AA, Polley A, Singh PP, et al. Transbilayer lipid interactions mediate nanoclustering of lipid-anchored proteins. Cell. 2015;161:581–94. doi: 10.1016/j.cell.2015.03.048. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Triffo S, Huang H, Smith A, Chou E, et al. Monitoring lipid anchor organization in cell membranes by PIE-FCCS. Journal of the American Chemical Society. 2012;134:10833–42. doi: 10.1021/ja300374c. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.van den Bogaart G, Meyenberg K, Risselada HJ, Amin H, et al. Membrane protein sequestering by ionic protein-lipid interactions. Nature. 2011;479:552–5. doi: 10.1038/nature10545. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zhou Y, Wong CO, Cho KJ, van der Hoeven D, et al. SIGNAL TRANSDUCTION. Membrane potential modulates plasma membrane phospholipid dynamics and K-Ras signaling. Science. 2015;349:873–6. doi: 10.1126/science.aaa5619. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Frisz JF, Klitzing HA, Lou K, Hutcheon ID, et al. Sphingolipid domains in the plasma membranes of fibroblasts are not enriched with cholesterol. J Biol Chem. 2013;288:16855–61. doi: 10.1074/jbc.M113.473207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Frisz JF, Lou K, Klitzing HA, Hanafin WP, et al. Direct chemical evidence for sphingolipid domains in the plasma membranes of fibroblasts. Proc Natl Acad Sci U S A. 2013;110:E613–E22. doi: 10.1073/pnas.1216585110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sevcsik E, Brameshuber M, Folser M, Weghuber J, et al. GPI-anchored proteins do not reside in ordered domains in the live cell plasma membrane. Nat Commun. 2015;6 doi: 10.1038/ncomms7969. 6969. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Pike LJ. Rafts Defined. J Lipid Res. 2006;47:1597–8. doi: 10.1194/jlr.E600002-JLR200. [DOI] [PubMed] [Google Scholar]
- 26.Munro S. Lipid rafts: elusive or illusive? Cell. 2003;115:377–88. doi: 10.1016/s0092-8674(03)00882-1. [DOI] [PubMed] [Google Scholar]
- 27.Kraft M. Plasma membrane organization and function: moving past lipid rafts. Molecular biology of the cell. 2013;24:2765–8. doi: 10.1091/mbc.E13-03-0165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Kenworthy AK. Have we become overly reliant on lipid rafts? Talking Point on the involvement of lipid rafts in T-cell activation. EMBO Rep. 2008;9:531–5. doi: 10.1038/embor.2008.92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Shaw AS. Lipid rafts: now you see them, now you don't. Nat Immunol. 2006;7:1139–42. doi: 10.1038/ni1405. [DOI] [PubMed] [Google Scholar]
- 30.Klotzsch E, Schütz GJ. A critical survey of methods to detect plasma membrane rafts. Phil Trans R Soc B. 2013;368 doi: 10.1098/rstb.2012.0033. 20120033. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Box GEP, Draper NR. Empirical Model Building and Response Surfaces. New York, NY: John Wiley & Son; 1987. [Google Scholar]
- 32.Brown DA, Rose JK. Sorting of GPI-anchored proteins to glycolipid-enriched membrane subdomains during transport to the apical cell surface. Cell. 1992;68:533–44. doi: 10.1016/0092-8674(92)90189-j. [DOI] [PubMed] [Google Scholar]
- 33.Shogomori H, Brown DA. Use of detergents to study membrane rafts: the good, the bad, and the ugly. Biol Chem. 2003;384:1259–63. doi: 10.1515/BC.2003.139. [DOI] [PubMed] [Google Scholar]
- 34.Legler DF, Doucey MA, Schneider P, Chapatte L, et al. Differential insertion of GPI-anchored GFPs into lipid rafts of live cells. Faseb J. 2005;19:73–5. doi: 10.1096/fj.03-1338fje. [DOI] [PubMed] [Google Scholar]
- 35.Heerklotz H. Triton promotes domain formation in lipid raft mixtures. Biophys J. 2002;83:2693–701. doi: 10.1016/S0006-3495(02)75278-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Pathak P, London E. Measurement of lipid nanodomain (raft) formation and size in sphingomyelin/POPC/cholesterol vesicles shows TX-100 and transmembrane helices increase domain size by coalescing preexisting nanodomains but do not induce domain formation. Biophys J. 2011;101:2417–25. doi: 10.1016/j.bpj.2011.08.059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.van Meer G, Simons K. Viruses budding from either the apical or the basolateral plasma membrane domain of MDCK cells have unique phospholipid compositions. EMBO J. 1982;1:847–52. doi: 10.1002/j.1460-2075.1982.tb01258.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Pang S, Urquhart P, Hooper NM. N-glycans, not the GPI anchor, mediate the apical targeting of a naturally glycosylated, GPI-anchored protein in polarised epithelial cells. J Cell Sci. 2004;117:5079–86. doi: 10.1242/jcs.01386. [DOI] [PubMed] [Google Scholar]
- 39.Benting JH, Rietveld AG, Simons K. N-Glycans mediate the apical sorting of a GPI-anchored, raft-associated protein in Madin-Darby canine kidney cells. J Cell Biol. 1999;146:313–20. doi: 10.1083/jcb.146.2.313. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Paladino S, Sarnataro D, Pillich R, Tivodar S, et al. Protein oligomerization modulates raft partitioning and apical sorting of GPI-anchored proteins. J Cell Biol. 2004;167:699–709. doi: 10.1083/jcb.200407094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lipardi C, Nitsch L, Zurzolo C. Detergent-insoluble GPI-anchored proteins are apically sorted in fischer rat thyroid cells, but interference with cholesterol or sphingolipids differentially affects detergent insolubility and apical sorting. Mol Biol Cell. 2000;11:531–42. doi: 10.1091/mbc.11.2.531. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Paladino S, Lebreton S, Tivodar S, Formiggini F, et al. Golgi sorting regulates organization and activity of GPI proteins at apical membranes. Nat Chem Biol. 2014;10:350–7. doi: 10.1038/nchembio.1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Imjeti NS, Lebreton S, Paladino S, de la Fuente E, et al. N-Glycosylation instead of cholesterol mediates oligomerization and apical sorting of GPI-APs in FRT cells. Mol Biol Cell. 2011;22:4621–34. doi: 10.1091/mbc.E11-04-0320. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Jaensch N, Correa IR, Jr, Watanabe R. Stable cell surface expression of GPI-anchored proteins, but not intracellular transport, depends on their fatty acid structure. Traffic. 2014;15:1305–29. doi: 10.1111/tra.12224. [DOI] [PubMed] [Google Scholar]
- 45.Harder T, Scheiffele P, Verkade P, Simons K. Lipid domain structure of the plasma membrane revealed by patching of membrane components. J Cell Biol. 1998;141:929–42. doi: 10.1083/jcb.141.4.929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Yetukuri L, Ekroos K, Vidal-Puig A, Oresic M. Informatics and computational strategies for the study of lipids. Mol Biosyst. 2008;4:121–7. doi: 10.1039/b715468b. [DOI] [PubMed] [Google Scholar]
- 47.Veatch SL, Keller SL. Seeing spots: complex phase behavior in simple membranes. Biochim Biophys Acta. 2005;1746:172–85. doi: 10.1016/j.bbamcr.2005.06.010. [DOI] [PubMed] [Google Scholar]
- 48.Dupuy AD, Engelman DM. Protein area occupancy at the center of the red blood cell membrane. Proc Natl Acad Sci U S A. 2008;105:2848–52. doi: 10.1073/pnas.0712379105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Contreras FX, Ernst AM, Haberkant P, Bjorkholm P, et al. Molecular recognition of a single sphingolipid species by a protein's transmembrane domain. Nature. 2012;481:525–9. doi: 10.1038/nature10742. [DOI] [PubMed] [Google Scholar]
- 50.Fantini J, Barrantes FJ. How cholesterol interacts with membrane proteins: an exploration of cholesterol-binding sites including CRAC, CARC, and tilted domains. Front Physiol. 2013;4:31. doi: 10.3389/fphys.2013.00031. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hansen SB, Tao X, MacKinnon R. Structural basis of PIP2 activation of the classical inward rectifier K+ channel Kir2.2. Nature. 2011;477:495–8. doi: 10.1038/nature10370. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Anderson RG, Jacobson K. A role for lipid shells in targeting proteins to caveolae, rafts, and other lipid domains. Science. 2002;296:1821–5. doi: 10.1126/science.1068886. [DOI] [PubMed] [Google Scholar]
- 53.Niemela PS, Miettinen MS, Monticelli L, Hammaren H, et al. Membrane proteins diffuse as dynamic complexes with lipids. J Am Chem Soc. 2010;132:7574–5. doi: 10.1021/ja101481b. [DOI] [PubMed] [Google Scholar]
- 54.Hammond GR, Fischer MJ, Anderson KE, Holdich J, et al. PI4P and PI(4,5)P2 are essential but independent lipid determinants of membrane identity. Science. 2012;337:727–30. doi: 10.1126/science.1222483. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Yeung T, Gilbert GE, Shi J, Silvius J, et al. Membrane phosphatidylserine regulates surface charge and protein localization. Science. 2008;319:210–3. doi: 10.1126/science.1152066. [DOI] [PubMed] [Google Scholar]
- 56.Scheve C, Gonzales P, Momin N, Stachowiak J. Steric pressure between membrane-bound proteins opposes lipid phase separation. Journal of the American Chemical Society. 2013;135:1185–8. doi: 10.1021/ja3099867. [DOI] [PubMed] [Google Scholar]
- 57.Goswami D, Gowrishankar K, Bilgrami S, Ghosh S, et al. Nanoclusters of GPI-anchored proteins are formed by cortical actin-driven activity. Cell. 2008;135:1085–97. doi: 10.1016/j.cell.2008.11.032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gowrishankar K, Ghosh S, Saha S, C R, et al. Active Remodeling of Cortical Actin Regulates Spatiotemporal Organization of Cell Surface Molecules. Cell. 2012;149:1353–67. doi: 10.1016/j.cell.2012.05.008. [DOI] [PubMed] [Google Scholar]
- 59.Wang TY, Leventis R, Silvius JR. Artificially lipid-anchored proteins can elicit clustering-induced intracellular signaling events in Jurkat T-lymphocytes independent of lipid raft association. J Biol Chem. 2005;280:22839–46. doi: 10.1074/jbc.M502920200. [DOI] [PubMed] [Google Scholar]
- 60.Saka SK, Honigmann A, Eggeling C, Hell SW, et al. Multi-protein assemblies underlie the mesoscale organization of the plasma membrane. Nat Commun. 2014;5:4509. doi: 10.1038/ncomms5509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.van Zanten TS, Gomez J, Manzo C, Cambi A, et al. Direct mapping of nanoscale compositional connectivity on intact cell membranes. Proc Natl Acad Sci U S A. 2010;107:15437–42. doi: 10.1073/pnas.1003876107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Lenne PF, Wawrezinieck L, Conchonaud F, Wurtz O, et al. Dynamic molecular confinement in the plasma membrane by microdomains and the cytoskeleton meshwork. Embo J. 2006;25:3245–56. doi: 10.1038/sj.emboj.7601214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zidovetzki R, Levitan I. Use of cyclodextrins to manipulate plasma membrane cholesterol content: evidence, misconceptions and control strategies. Biochim Biophys Acta. 2007;1768:1311–24. doi: 10.1016/j.bbamem.2007.03.026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Jamur MC, Oliver C. Permeabilization of cell membranes. Methods Mol Biol. 2010;588:63–6. doi: 10.1007/978-1-59745-324-0_9. [DOI] [PubMed] [Google Scholar]
- 65.Neuvonen M, Manna M, Mokkila S, Javanainen M, et al. Enzymatic oxidation of cholesterol: properties and functional effects of cholestenone in cell membranes. PLoS One. 2014;9:e103743. doi: 10.1371/journal.pone.0103743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Owicki JC, McConnell HM. Lateral diffusion in inhomogeneous membranes. Model membranes containing cholesterol. Biophys J. 1980;30:383–97. doi: 10.1016/S0006-3495(80)85103-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Grunze M, Deuticke B. Changes of membrane permeability due to extensive cholesterol depletion in mammalian erythrocytes. Biochim Biophys Acta. 1974;356:125–30. doi: 10.1016/0005-2736(74)90300-9. [DOI] [PubMed] [Google Scholar]
- 68.Finkelstein A. Water and nonelectrolyte permeability of lipid bilayer membranes. J Gen Physiol. 1976;68:127–35. doi: 10.1085/jgp.68.2.127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Sieber JJ, Willig KI, Kutzner C, Gerding-Reimers C, et al. Anatomy and dynamics of a supramolecular membrane protein cluster. Science. 2007;317:1072–6. doi: 10.1126/science.1141727. [DOI] [PubMed] [Google Scholar]
- 70.Kwik J, Boyle S, Fooksman D, Margolis L, et al. Membrane cholesterol, lateral mobility, and the phosphatidylinositol 4,5-bisphosphate-dependent organization of cell actin. Proceedings of the National Academy of Sciences of the United States of America. 2003;100:13964–9. doi: 10.1073/pnas.2336102100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Mayinger P. Phosphoinositides and vesicular membrane traffic. Biochim Biophys Acta. 2012;1821:1104–13. doi: 10.1016/j.bbalip.2012.01.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Chadda R, Howes MT, Plowman SJ, Hancock JF, et al. Cholesterol-sensitive Cdc42 activation regulates actin polymerization for endocytosis via the GEEC pathway. Traffic. 2007;8:702–17. doi: 10.1111/j.1600-0854.2007.00565.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Kusumi A, Suzuki KG, Kasai RS, Ritchie K, et al. Hierarchical mesoscale domain organization of the plasma membrane. Trends Biochem Sci. 2011;36:604–15. doi: 10.1016/j.tibs.2011.08.001. [DOI] [PubMed] [Google Scholar]
- 74.Zocher M, Zhang C, Rasmussen SG, Kobilka BK, et al. Cholesterol increases kinetic, energetic, and mechanical stability of the human beta2-adrenergic receptor. Proc Natl Acad Sci U S A. 2012;109:E3463–72. doi: 10.1073/pnas.1210373109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Hoop CL, Sivanandam VN, Kodali R, Srnec MN, et al. Structural characterization of the caveolin scaffolding domain in association with cholesterol-rich membranes. Biochemistry. 2012;51:90–9. doi: 10.1021/bi201356v. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Sheng R, Chen Y, Yung Gee H, Stec E, et al. Cholesterol modulates cell signaling and protein networking by specifically interacting with PDZ domain-containing scaffold proteins. Nat Commun. 2012;3 doi: 10.1038/ncomms2221. 1249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Wang PY, Weng J, Anderson RG. OSBP is a cholesterol-regulated scaffolding protein in control of ERK 1/2 activation. Science. 2005;307:1472–6. doi: 10.1126/science.1107710. [DOI] [PubMed] [Google Scholar]
- 78.Maxfield FR, Mayor S. Cell surface dynamics of GPI-anchored proteins. Adv Exp Med Biol. 1997;419:355–64. doi: 10.1007/978-1-4419-8632-0_47. [DOI] [PubMed] [Google Scholar]
- 79.Stefanova I, Horejsi V, Ansotegui IJ, Knapp W, et al. GPI-anchored cell-surface molecules complexed to protein tyrosine kinases. Science. 1991;254:1016–9. doi: 10.1126/science.1719635. [DOI] [PubMed] [Google Scholar]
- 80.Mitchell JS, Kanca O, McIntyre BW. Lipid microdomain clustering induces a redistribution of antigen recognition and adhesion molecules on human T lymphocytes. J Immunol. 2002;168:2737–44. doi: 10.4049/jimmunol.168.6.2737. [DOI] [PubMed] [Google Scholar]
- 81.Mrowczynska L, Hagerstrand H. Patching of ganglioside(M1) in human erythrocytes - distribution of CD47 and CD59 in patched and curved membrane. Mol Membr Biol. 2008;25:258–65. doi: 10.1080/09687680802043638. [DOI] [PubMed] [Google Scholar]
- 82.Favoreel HW, Mettenleiter TC, Nauwynck HJ. Copatching and lipid raft association of different viral glycoproteins expressed on the surfaces of pseudorabies virus-infected cells. J Virol. 2004;78:5279–87. doi: 10.1128/JVI.78.10.5279-5287.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hogue IB, Grover JR, Soheilian F, Nagashima K, et al. Gag induces the coalescence of clustered lipid rafts and tetraspanin-enriched microdomains at HIV-1 assembly sites on the plasma membrane. J Virol. 2011;85:9749–66. doi: 10.1128/JVI.00743-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Larive RM, Baisamy L, Urbach S, Coopman P, et al. Cell membrane extensions, generated by mechanical constraint, are associated with a sustained lipid raft patching and an increased cell signaling. Biochim Biophys Acta. 2010;1798:389–400. doi: 10.1016/j.bbamem.2009.11.016. [DOI] [PubMed] [Google Scholar]
- 85.Glebov O, Nichols B. Lipid raft proteins have a random distribution during localized activation of the T-cell receptor. Nature cell biology. 2004;6:238–43. doi: 10.1038/ncb1103. [DOI] [PubMed] [Google Scholar]
- 86.Sezgin E, Kaiser HJ, Baumgart T, Schwille P, et al. Elucidating membrane structure and protein behavior using giant plasma membrane vesicles. Nat Protoc. 2012;7:1042–51. doi: 10.1038/nprot.2012.059. [DOI] [PubMed] [Google Scholar]
- 87.Simons K, Gerl MJ. Revitalizing membrane rafts: new tools and insights. Nat Rev Mol Cell Biol. 2010;11:688–99. doi: 10.1038/nrm2977. [DOI] [PubMed] [Google Scholar]
- 88.Llorente A, Skotland T, Sylvanne T, Kauhanen D, et al. Molecular lipidomics of exosomes released by PC-3 prostate cancer cells. Biochimica Et Biophysica Acta-Molecular and Cell Biology of Lipids. 2013;1831:1302–9. doi: 10.1016/j.bbalip.2013.04.011. [DOI] [PubMed] [Google Scholar]
- 89.Liu AP, Fletcher DA. Actin polymerization serves as a membrane domain switch in model lipid bilayers. Biophys J. 2006;91:4064–70. doi: 10.1529/biophysj.106.090852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Veksler A, Gov NS. Calcium-actin waves and oscillations of cellular membranes. Biophys J. 2009;97:1558–68. doi: 10.1016/j.bpj.2009.07.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Chen D, Santore MM. Large effect of membrane tension on the fluid-solid phase transitions of two-component phosphatidylcholine vesicles. Proc Natl Acad Sci U S A. 2014;111:179–84. doi: 10.1073/pnas.1314993111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Levental I, Grzybek M, Simons K. Raft domains of variable properties and compositions in plasma membrane vesicles. Proc Natl Acad Sci U S A. 2011;108:11411–6. doi: 10.1073/pnas.1105996108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Johnson SA, Stinson BM, Go MS, Carmona LM, et al. Temperature-dependent phase behavior and protein partitioning in giant plasma membrane vesicles. Biochim Biophys Acta. 2010;1798:1427–35. doi: 10.1016/j.bbamem.2010.03.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Marsh D. Protein modulation of lipids, and vice-versa, in membranes. Biochimica et biophysica acta. 2008;1778:1545–75. doi: 10.1016/j.bbamem.2008.01.015. [DOI] [PubMed] [Google Scholar]
- 95.Kaiser HJ, Orlowski A, Rog T, Nyholm TK, et al. Lateral sorting in model membranes by cholesterol-mediated hydrophobic matching. Proc Natl Acad Sci U S A. 2011;108:16628–33. doi: 10.1073/pnas.1103742108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Killian JA. Hydrophobic mismatch between proteins and lipids in membranes. Biochim Biophys Acta. 1998;1376:401–15. doi: 10.1016/s0304-4157(98)00017-3. [DOI] [PubMed] [Google Scholar]
- 97.Ewers H, Helenius A. Lipid-mediated endocytosis. Cold Spring Harb Perspect Biol. 2011;3:a004721. doi: 10.1101/cshperspect.a004721. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Suetsugu S, Kurisu S, Takenawa T. Dynamic shaping of cellular membranes by phospholipids and membrane-deforming proteins. Physiol Rev. 2014;94:1219–48. doi: 10.1152/physrev.00040.2013. [DOI] [PubMed] [Google Scholar]
- 99.Tu-Sekine B, Goldschmidt H, Raben DM. Diacylglycerol, phosphatidic acid, and their metabolic enzymes in synaptic vesicle recycling. Adv Biol Regul. 2015;57:147–52. doi: 10.1016/j.jbior.2014.09.010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Stachowiak JC, Schmid EM, Ryan CJ, Ann HS, et al. Membrane bending by protein-protein crowding. Nat Cell Biol. 2012;14:944–9. doi: 10.1038/ncb2561. [DOI] [PubMed] [Google Scholar]
- 101.Heinrich M, Tian A, Esposito C, Baumgart T. Dynamic sorting of lipids and proteins in membrane tubes with a moving phase boundary. Proc Natl Acad Sci U S A. 2010;107:7208–13. doi: 10.1073/pnas.0913997107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Elbaz Y, Schuldiner M. Staying in touch: the molecular era of organelle contact sites. Trends Biochem Sci. 2011;36:616–23. doi: 10.1016/j.tibs.2011.08.004. [DOI] [PubMed] [Google Scholar]
- 103.Stefan CJ, Manford AG, Baird D, Yamada-Hanff J, et al. Osh proteins regulate phosphoinositide metabolism at ER-plasma membrane contact sites. Cell. 2011;144:389–401. doi: 10.1016/j.cell.2010.12.034. [DOI] [PubMed] [Google Scholar]
- 104.Kusumi A, Sako Y. Cell surface organization by the membrane skeleton. Curr Opin Cell Biol. 1996;8:566–74. doi: 10.1016/s0955-0674(96)80036-6. [DOI] [PubMed] [Google Scholar]
- 105.Trimble WS, Grinstein S. Barriers to the free diffusion of proteins and lipids in the plasma membrane. J Cell Biol. 2015;208:259–71. doi: 10.1083/jcb.201410071. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Contreras FX, Ernst AM, Wieland F, Brugger B. Specificity of intramembrane protein-lipid interactions. Cold Spring Harb Perspect Biol. 2011;3 doi: 10.1101/cshperspect.a004705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Sonnino S, Prinetti A. Lipids and membrane lateral organization. Front Physiol. 2010;1:153. doi: 10.3389/fphys.2010.00153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Goni FM. The basic structure and dynamics of cell membranes: an update of the Singer-Nicolson model. Biochim Biophys Acta. 2014;1838:1467–76. doi: 10.1016/j.bbamem.2014.01.006. [DOI] [PubMed] [Google Scholar]
- 109.Huang S. When peers are not peers and don't know it The Dunning-Kruger effect and self-fulfilling prophecy in peer-review. Bioessays. 2013;35:414–6. doi: 10.1002/bies.201200182. [DOI] [PubMed] [Google Scholar]
- 110.Sengupta P, Jovanovic-Talisman T, Skoko D, Renz M, et al. Probing protein heterogeneity in the plasma membrane using PALM and pair correlation analysis. Nat Methods. 2011;8:969–75. doi: 10.1038/nmeth.1704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Mouritsen OG. The liquid-ordered state comes of age. Biochim Biophys Acta. 2010;1798:1286–8. doi: 10.1016/j.bbamem.2010.02.017. [DOI] [PubMed] [Google Scholar]
- 112.Mueller V, Ringemann C, Honigmann A, Schwarzmann G, et al. STED nanoscopy reveals molecular details of cholesterol- and cytoskeleton-modulated lipid interactions in living cells. Biophysical journal. 2011;101:1651–60. doi: 10.1016/j.bpj.2011.09.006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Owen DM, Williamson DJ, Magenau A, Gaus K. Sub-resolution lipid domains exist in the plasma membrane and regulate protein diffusion and distribution. Nat Commun. 2012;3 doi: 10.1038/ncomms2273. 1256. [DOI] [PubMed] [Google Scholar]
- 114.Amaro M, Sachl R, Jurkiewicz P, Coutinho A, et al. Time-resolved fluorescence in lipid bilayers: selected applications and advantages over steady state. Biophys J. 2014;107:2751–60. doi: 10.1016/j.bpj.2014.10.058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Machan R, Jurkiewicz P, Olzynska A, Olsinova M, et al. Peripheral and integral membrane binding of peptides characterized by time-dependent fluorescence shifts: focus on antimicrobial peptide LAH(4) Langmuir. 2014;30:6171–9. doi: 10.1021/la5006314. [DOI] [PubMed] [Google Scholar]
- 116.Yeagle PL. Cholesterol and the cell membrane. Biochim Biophys Acta. 1985;822:267–87. doi: 10.1016/0304-4157(85)90011-5. [DOI] [PubMed] [Google Scholar]
- 117.Brown DA, London E. Structure and origin of ordered lipid domains in biological membranes. J Membr Biol. 1998;164:103–14. doi: 10.1007/s002329900397. [DOI] [PubMed] [Google Scholar]
- 118.Subczynski WK, Kusumi A. Dynamics of raft molecules in the cell and artificial membranes: approaches by pulse EPR spin labeling and single molecule optical microscopy. Biochim Biophys Acta. 2003;1610:231–43. doi: 10.1016/s0005-2736(03)00021-x. [DOI] [PubMed] [Google Scholar]
- 119.Ipsen JH, Karlstrom G, Mouritsen OG, Wennerstrom H, et al. Phase equilibria in the phosphatidylcholine-cholesterol system. Biochim Biophys Acta. 1987;905:162–72. doi: 10.1016/0005-2736(87)90020-4. [DOI] [PubMed] [Google Scholar]
- 120.Paulick MG, Bertozzi CR. The glycosylphosphatidylinositol anchor: a complex membrane-anchoring structure for proteins. Biochemistry. 2008;47:6991–7000. doi: 10.1021/bi8006324. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 121.Robinson PJ. Signal transduction via GPI-anchored membrane proteins. Adv Exp Med Biol. 1997;419:365–70. doi: 10.1007/978-1-4419-8632-0_48. [DOI] [PubMed] [Google Scholar]
- 122.Yu J, Fischman DA, Steck TL. Selective solubilization of proteins and phospholipids from red blood cell membranes by nonionic detergents. J Supramol Struct. 1973;1:233–48. doi: 10.1002/jss.400010308. [DOI] [PubMed] [Google Scholar]
- 123.Levental I, Lingwood D, Grzybek M, Coskun U, et al. Palmitoylation regulates raft affinity for the majority of integral raft proteins. Proc Natl Acad Sci U S A. 2010;107:22050–4. doi: 10.1073/pnas.1016184107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.van Meer G, Voelker DR, Feigenson GW. Membrane lipids: where they are and how they behave. Nat Rev Mol Cell Biol. 2008;9:112–24. doi: 10.1038/nrm2330. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Yu RK, Tsai YT, Ariga T, Yanagisawa M. Structures, biosynthesis, and functions of gangliosides--an overview. J Oleo Sci. 2011;60:537–44. doi: 10.5650/jos.60.537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Barboni E, Rivero BP, George AJ, Martin SR, et al. The glycophosphatidylinositol anchor affects the conformation of Thy-1 protein. J Cell Sci. 1995;108(Pt 2):487–97. doi: 10.1242/jcs.108.2.487. [DOI] [PubMed] [Google Scholar]
- 127.Butikofer P, Malherbe T, Boschung M, Roditi I. GPI-anchored proteins: now you see 'em, now you don't. FASEB J. 2001;15:545–8. doi: 10.1096/fj.00-0415hyp. [DOI] [PubMed] [Google Scholar]
- 128.Qu Q, Sharom FJ. Proximity of bound Hoechst 33342 to the ATPase catalytic sites places the drug binding site of P-glycoprotein within the cytoplasmic membrane leaflet. Biochemistry. 2002;41:4744–52. doi: 10.1021/bi0120897. [DOI] [PubMed] [Google Scholar]
- 129.Rademacher TW, Edge CJ, Dwek RA. Dropping anchor with the lipophosphoglycans. Curr Biol. 1991;1:41–2. doi: 10.1016/0960-9822(91)90123-e. [DOI] [PubMed] [Google Scholar]
- 130.Homans SW, Edge CJ, Ferguson MA, Dwek RA, et al. Solution structure of the glycosylphosphatidylinositol membrane anchor glycan of Trypanosoma brucei variant surface glycoprotein. Biochemistry. 1989;28:2881–7. doi: 10.1021/bi00433a020. [DOI] [PubMed] [Google Scholar]
- 131.Ruprecht V, Brameshuber M, Schütz GJ. Two-color single molecule tracking combined with photobleaching for the detection of rare molecular interactions in fluid biomembranes. Soft Matter. 2010;6:568–81. [Google Scholar]
- 132.Wieser S, Moertelmaier M, Fuertbauer E, Stockinger H, et al. (Un)Confined Diffusion of CD59 in the Plasma Membrane Determined by High-Resolution Single Molecule Microscopy. Biophys J. 2007;92:3719–28. doi: 10.1529/biophysj.106.095398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 133.Ichimori H, Hata T, Matsuki H, Kaneshina S. Effect of unsaturated acyl chains on the thermotropic and barotropic phase transitions of phospholipid bilayer membranes. Chemistry and Physics of Lipids. 1999;100:151–64. [Google Scholar]
- 134.Tada K, Goto M, Tamai N, Matsuki H, et al. Pressure effect on the bilayer phase transition of asymmetric lipids with an unsaturated acyl chain. Ann N Y Acad Sci. 2010;1189:77–85. doi: 10.1111/j.1749-6632.2009.05203.x. [DOI] [PubMed] [Google Scholar]
- 135.Pascher I. Molecular arrangements in sphingolipids. Conformation and hydrogen bonding of ceramide and their implication on membrane stability and permeability. Biochim Biophys Acta. 1976;455:433–51. doi: 10.1016/0005-2736(76)90316-3. [DOI] [PubMed] [Google Scholar]
- 136.Niemela PS, Hyvonen MT, Vattulainen I. Atom-scale molecular interactions in lipid raft mixtures. Biochim Biophys Acta. 2009;1788:122–35. doi: 10.1016/j.bbamem.2008.08.018. [DOI] [PubMed] [Google Scholar]
- 137.Mombelli E, Morris R, Taylor W, Fraternali F. Hydrogen-bonding propensities of sphingomyelin in solution and in a bilayer assembly: a molecular dynamics study. Biophys J. 2003;84:1507–17. doi: 10.1016/S0006-3495(03)74963-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 138.Dinic J, Biverstahl H, Maler L, Parmryd I. Laurdan and di-4-ANEPPDHQ do not respond to membrane-inserted peptides and are good probes for lipid packing. Biochim Biophys Acta. 2011;1808:298–306. doi: 10.1016/j.bbamem.2010.10.002. [DOI] [PubMed] [Google Scholar]
- 139.Massey JB, Pownall HJ. The polar nature of 7-ketocholesterol determines its location within membrane domains and the kinetics of membrane microsolubilization by apolipoprotein A-I. Biochemistry. 2005;44:10423–33. doi: 10.1021/bi0506425. [DOI] [PubMed] [Google Scholar]
- 140.Kulig W, Olzynska A, Jurkiewicz P, Kantola AM, et al. Cholesterol under oxidative stress-How lipid membranes sense oxidation as cholesterol is being replaced by oxysterols. Free Radic Biol Med. 2015;84:30–41. doi: 10.1016/j.freeradbiomed.2015.03.006. [DOI] [PubMed] [Google Scholar]