Abstract
Bemisia tabaci is one of the most threatening pests in agriculture, causing significant losses to many important crops on a global scale. The dramatic increase and availability of sequence data for B. tabaci species complex and its bacterial endosymbionts is critical for developing emerging sustainable pest management strategies which are based on pinpointing the global diversity of this important pest and its bacterial endosymbionts. To unravel the global genetic diversity of B. tabaci species complex focusing on its associated endosymbionts, along with Israeli whitefly populations collected in this study, we combined available sequences in databases, resulting in a total of 4,253 mitochondrial cytochrome oxidase I (mtCOI) sequences from 82 countries and 1,226 16S/23S rRNA endosymbiont sequences from 32 countries that were analyzed. Using Bayesian phylogenetic analysis, we identified two new B. tabaci groups within the species complex and described the global distribution of endosymbionts within this complex. Our analyses revealed complex divergence of the different endosymbiont sequences within the species complex, with overall one Hamiltonella, two Porteria (P1 and P2), two Arsenophonus (A1 and A2), two Wolbachia (super-groups O and B), four Cardinium (C1-C4) and three Rickettsia (R1-R3) groups were identified. Our comprehensive analysis provides an updated important resource for this globally important pest and its secondary symbionts, which have been a major subject for research in last three decades.
Introduction
The whitefly Bemisia tabaci (Gennadius) (Hemipetra: Aleyrodidae), is one of the most economically and agriculturally important insect pests worldwide. This pest causes serious economic damage by direct feeding and by transmitting plant viruses that belong to several virus families including Begomovirus, Crinivirus, Carlavirus, Torradovirus and Ipomovirus [1,2]. During the past two decades, begomoviruses and B. tabaci are considered the most economically important virus-vector complexes, threatening major vegetable, field and fiber crops on a worldwide scale especially in developing countries where stable food availability is scarce. B. tabaci was the first major global invasive pest in the Middle East during the 1980’s, and rapidly spread to other regions mainly via human activities and international trade.
The diversity of B. tabaci sibling species complex has been extensively studied and several biological and molecular characteristics such as plant host preference, fecundity, ability to transmit begomoviruses, dispersal, resistance to insecticide and mitochondrial cytochrome oxidase I (mtCOI) DNA sequences, have been used to compare the different genetic groups [3–10]. Recent molecular markers were used in phylogenetic analyses to identify the different worldwide populations of B. tabaci [7]. Sequencing a 657 bp portion of mtCOI helped identifying new species of B. tabaci and established the new term "cryptic species complex" [7,11]. On the basis of 3.5% pairwise divergence in mtCOI sequences within B. tabaci species, 42 distinct species have been reported: Africa, Asia I, Asia I-India, Asia II 1–12, Asia III, Asia IV, Asia V, Australia, Australia/Indonesia, China 1–5, Indian Ocean, Ru, Middle East Asia Minor I-II (MEAM), Mediterranean (MED), MEAM K, New World 1–2, Japan 1–2, Uganda, Italy 1, and Sub Saharan Africa 1–5 [7,10–14]. Recently, it was suggested that 4% genetic divergence is more realistic than the 3.5% in the identification of new species within this complex [15]. Although current studies provide a good understanding of this complex, the diversity of species within the complex is expected to be higher and more analyses are warranted for the discovery of such new species.
Similar to many other arthropods, B. tabaci species complex carries a primary endosymbiotic bacterium called Candidatus Portiera aleyrodidarum [16,17], which is fixed in populations and confined to bacteriocyte cells in all whitefly individuals. This bacterium is essential for host survival, development and has a long co-evolutionary history with all members of the subfamily Aleyrodidae [16,18,19]. In addition to the primary endosymbiont, seven different secondary endosymbionts namely Rickettsia [20], Wolbachia [21,22], Hamiltonella and Arsenophonus [16,17], Cardinium [23], Fritschea [24], and Hemipteriphilus [25] have been reported from B. tabaci populations around the world.
Except for Portiera, secondary symbionts differ in their 1) composition/infection frequencies in B. tabaci populations [16,26–32] 2) Spacial localization phenotypes/patterns in developmental and adult stages [26,30,31] and 3) Horizontal/vertical transfer [33–36] within and between populations. Several of these secondary endosymbionts interfere with host physiology, ecology and reproduction [37–41], rapid evolutionary shifts [42], thermotolerance [43], resistance to insecticides [44], host fitness [45], defense against pathogens [46] and influence virus transmission abilities by members of this species complex [33,47–49]. These studies suggest that identifying the infection status of endosymbionts in B. tabaci populations is highly relevant for understanding their association with B. tabaci genetic groups.
Several B. tabaci species populations around the world were surveyed for infection with endosymbionts and clear variations in the infection by the different endosymbionts within the Bemisia genetic groups were repeatedly observed [25–32,40]. For example, in populations tested from China Wolbachia, Rickettsia, Arsenophonus, Hamiltonella and Cardinium were detected in both MEAM1 and MED populations [50]. Another study from China detected Hamiltonella in both MEAM1 and MED, Rickettsia in Asia II 3, Asia II 7, China 1 and MEAM1. None of the MEAM1 and MED populations from China were infected with Wolbachia, Cardinium and Arsenophonus [25]. Arsenophonus, Cardinium, Rickettsia and Wolbachia were detected in native whiteflies of Africa [32], China [25] and India [51] but not Hamiltonella and Fritschea. Similarly, individual infection status of secondary symbionts from Tunisia identified Hamiltonella and Rickettsia from MEAM1 and only Hamiltonella from Q1 [29]. A recent study by Gorsane et al. [52] hypothesized that the presence of Cardinium in MEAM1 and Cardinium, Fritschea and Wolbachia in MED may explain the differences in infection status possibly due to plant hosts, site to site variations, year to year surveys, low titer of endosymbiont, influence of chemical insecticides, and technical detection problems like PCR, which could result in false negatives.
In this study, we sequenced part of mtCOI and 16S/23S rRNA genes from whitefly populations recently collected in Israel to identify and update the status of Bemisia genetic groups and their infection with bacterial endosymbionts, and to test whether the situation has changed since the last study conducted in this regard by Chiel et al. [40]. To put this study in a broader context, the sequences collected from Israel were combined with sequences available in databases, to identify B. tabaci worldwide species and their associated endosymbiont distribution. Among few molecular techniques, mtCOI and 16S/23S rRNA molecular markers are widely used to improve the robustness of the phylogenetic relationships among B. tabaci species identification, and genetically diverse distinct groups/strains of endosymbionts within the genetic groups of Bemisia. Our objectives were thus to determine 1) global diversity and geographic distribution of B. tabaci species complex and endosymbionts infection within Bemisia genetic groups worldwide and 2) provide baseline information on genetically diverse distinct groups/strains of endosymbionts within Bemisia genetic groups across the world, using Bayesian phylogenetic analysis.
Materials and methods
B. tabaci genotype identification
B. tabaci populations were collected as adults using a hand-held aspirator from several agricultural fields in Israel. After collection, the insects were transferred to glass bottle containing cotton leaves. About twenty whitefly adults were used to test the purity of the B/Q biotype population which was confirmed by PCR with Bem 23 Forward- CGGAGCTTGCGCCTTAGTC and Reverse- CGGCTTTATCATAGCTCTCGT specific microsattelite primers, that give expected PCR product of 200 bp for B and 400 bp for Q [53]. Whitefly species identity confirmation was performed using the primers C1-J-2195 (5’-TTGATTTTTTGGTCATCCAGAAGT-3’) and L2-N-3014 (5’-TCCAATGCACTAATCTGCCATATTA-3’) that amplify a fragment from the mitochondrial cytochrome oxidase I gene (mtCOI) [8]. Each species was maintained on cotton seedlings (Gossypium hirsutumL. cv. Acala) inside insect-proof cages and growth rooms under standard conditions of 25 ± 2°C, 60% relative humidity and a 14 h light/10 h dark photoperiod for further studies.
Detection of endosymbionts
The same DNA originated from each individual was used for screening of all primary (Portiera) and secondary symbionts (Hamiltonella, Rickettsia, Wolbachia, Arsenophonus, Cardinium and Fritschea) using genus-specific primers targeting the 16 S or 23 S rDNA genes was performed as previously described by Skaljac et al. [26–28]. PCR products were extracted from the gel and sequenced (Hylabs, Rehovot, Israel). Sequences were initially analyzed using BLASTn (www.ncbi.nlm.nih).
Datasets and sequence analyses
B. tabaci mtCOI sequences available in GenBank (www.ncbi.nlm.nih.gov/) published until January 2017 with their geographical source, were retrieved. A total of 4,253 sequences from 82 countries were identified; 2,903 accessions were selected because they contained the 657 bp fragment of the 3’ end of mtCOI, required for further analysis. Clustal W algorithm was used to align the sequences and the identities were analyzed using BioEdit v7.0.9.0 (www.mbio.ncsu.edu/BioEdit), followed by visual inspection and manual adjustment. The sequences were translated into their corresponding amino acids to check for correct reading frame. To confirm the geographically distributed B. tabaci species, we compared the sequences with the 42 putative species sequences already published [7,10–14]. All 193 representative B. tabaci species from the different countries were recovered from GenBank sequences and are detailed in S1 Table.
All B. tabaci endosymbionts sequenced as part of this study, along with geographically available in the database (www.ncbi.nlm.nih.gov/). A total of 1,226 sequences were present in the database from 32 countries, of these 298 accessions were selected based on the sequences that containing alignment overlapping regions. For the purpose of detailed analysis, sequences from Bemisia genetic groups only considered for further analysis. Endosymbiont sequences were aligned using Clustal W algorithm to align the sequences and their identities using BioEdit v7.0.9.0 (www.mbio.ncsu.edu/BioEdit), followed by manual removing of sequences.
Phylogenetic analyses of B. tabaci and endosymbionts
To construct a comprehensive B. tabaci phylogenetic tree, we first collected mtCOI sequences from each country and conducted neighbor-joining tree with MEGA 6.0 [54]. Duplicated species were excluded; only representative sequences of each species from each country were included. Phylogenetic trees were constructed based on the 192-nucleotide mtCOI sequences of B. tabaci species and B. afer, B. atriplex, B. subdecipiens, Trialeurodes vaparariorum, T. abutilonea, T. ricini, T. lauri and Tetraleurodes acacia as outgroups. Genetic divergences were calculated from the 52 mtCOI sequences using P-distance and Kimura 2-parameter (K2P) distance models in MEGA 6.0 [54].
Multiple alignments of the mtCOI and 16S/23S rRNA sequences were generated using the program MAFFT 7 [55]. The alignments were then inspected and corrected manually. The appropriate model of evolution was estimated with Bayesian Information Criterion (BIC) with Jmodeltest 2.1.6 on CIPRES Science Gateway [56] the selected models were the same as for maximum likelihood inferences. The models selected were GTR+G for mtCOI, JC+G for Porteira (16S), HKY for Wolbachia (16S), GTR for Cardinium (16S), GTR+I for Rickettsia (16S) and Hamiltonella (16S), HKY+G for Arsenophonus (23S). Using these models phylogenetic reconstruction was carried out using Bayesian inference approach (BI). For BI analysis, two independent runs of Markov chain Monte Carlo (MCMC) were run for 10 million generations using eight chains and sampling frequency of 1000 generations by MrBayes ver. 3.2 [57]. The first 25 per cent samples were discarded by setting burn-in fraction to 0.25. This burn-in setting was shared across the SUMT command to discard 25 per cent of sampled trees. Bayesian posterior probabilities were then calculated from the sample points after the MCMC algorithm began to converge. To ensure that our analyses are not trapped in local optima, four independent MCMC runs were performed. Topologies and posterior clade probabilities from different runs were compared for congruence. The consensus tree generated was visualized using FigTree available at http://tre.bio.ed.ac.uk/software/figtree.
Results
B. tabaci species identification
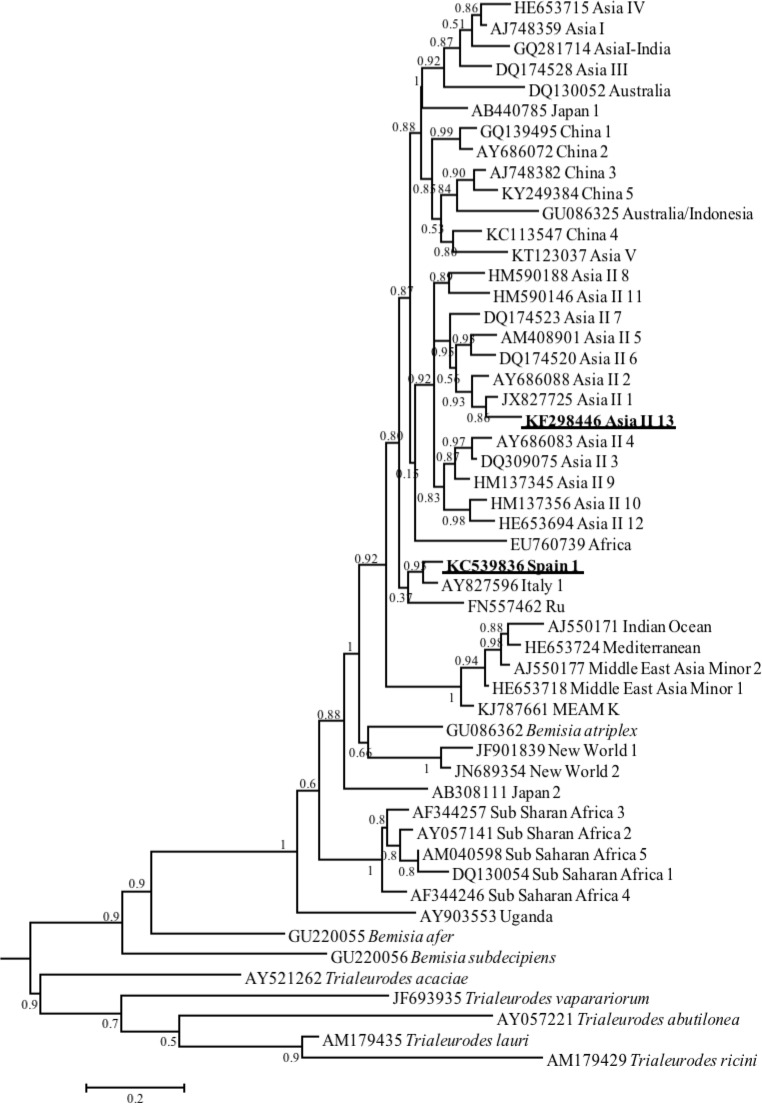
All together sixty whitefly populations collected in this study from Israel were identified initially based on Bem 23 F/R primers [53]. The same DNA was used to amplify the mtCOI sequences for determining the whitefly species. The phylogenetic analysis of mtCOI sequences revealed that 70% of the samples were B. tabaci MEAM1 while 30% were MED. Only representative sequences of MEAM1 and MED from Israel as well as representative endosymbiont sequences were included in the phylogenies generated afterwards for the worldwide analyses (Fig 1).
Fig 1. Bayesian phylogenetic tree inferred from mtCOI data using the GTR+G model.
Posterior probabilities for the branches are given. The proposed new species are indicated in bold and underlined.
mtCOI global B. tabaci sequence reanalysis and genetic diversity
To investigate the worldwide distribution of the putative B. tabaci species, 2,903 sequences were selected from 82 countries. Asia, Africa, North and South America, Europe and Australia were represented. Next, phylogenetic analyses were employed to infer the geographical distribution of B. tabaci putative species. Two sequences (accessions KC539836 from Spain and KF298446 from India) were separated from the other 42 species in the phylogenetic tree (Fig 1). The comparison conducted with the 42 B. tabaci putative species, showed that the above two sequences have pairwise divergences that exceed 4%, either with the consensus sequences or between themselves suggesting that they represent new species. KC539836 was clustered with Italy 1 (5%); and it was named Spain 1 in this study. KF298446 from India clustered with Asia II 1 (7%) species; and it was named Asia II 13 in this study. Therefore, two putative species, Asia II 13 and Spain 1, were identified and added to the previously reported 42 groups. The reclassification based on the 3.5 and 4% genetic divergence revealed that the 44 groups are clearly distinct (Fig 1, S2 Table, S1 Fig).
Continental separation of B. tabaci populations
The phylogenetic analyses revealed that the Asian continent contains the highest number of B. tabaci species, with a total of 28 native and invasive species (Asia I, Asia I-India, Asia II 1–13, Asia III, Asia IV, MED, MEAM1, MEAM K, China 1–4, Australia/Indonesia, India, Japan 1–2) distributed across 13 countries (India, Pakistan, Bangladesh, Nepal, Cambodia, Indonesia, Malaysia, Myanmar, Singapore, China, Japan, South Korea and Taiwan). In addition to the MEAM1 (B biotype), the results demonstrated the geographic distribution of MED (Q biotype) across the Middle East (Egypt, Israel, Syria and Turkey), South East Asia (Cambodia and Malaysia), East Asia (China, Japan, South Korea and Taiwan), Africa (Algeria, Benin, Burkina Faso, Cameroon, Ghana, Ivory Coast, Morocco, Nigeria, Senegal, Sudan, South Africa, Tanzania, Togo, Tunisia, Uganda, Zimbabwe and Reunion Island), Europe (Bosnia Herzegovina, Croatia, Cyprus, Czech Republic, France, Greece, Italy, Netherlands, Portugal, Spain), North America (Guatemala, USA, Canada and Mexico) and South America (Argentina, Brazil and Uruguay). Furthermore, the putative Sub-Saharan African 1–5 species and the Indian Ocean were restricted to the Mediterranean area and Africa. The New World species which are native to the Americas were also detected in the Old World (Sudan). Altogether, MEAM1 was reported in 42 countries while MED from 44 countries, showing that these species are the most highly diverse and distributed on a worldwide scale. The species that are endemic to China were described in 4 countries, the Asian species in 14 countries, the Indian Ocean species in 6 countries, Sub-Saharan Africa in 19 countries and the New World species in 12 countries. The newly described species Asia I-India, Asia II 2, Asia II 3, Asia II 4, Asia II 8, Asia II 9, Asia II 10, Asia II 11, Asia II 12, Asia II 13, Asia IV, Asia V, Australia, Australia/Indonesia, China 4, China 5, MEAM K, Italy 1, Japan 1, Sub Saharan Africa 5 and Uganda, each was described in only one country (Table 1, Fig 2).
Table 1. Worldwide distribution of Bemisia tabaci species.
| No. | Country | B. tabaci species |
|---|---|---|
| Old World | ||
| South Asia | ||
| 1. | India | Asia I, Asia I-India, Asia II 1, Asia II 5, Asia II 7, Asia II 8, Asia II 11, Asia II 13, MEAM K, China 3, MEAM1 |
| 2. | Pakistan | Asia I, Asia II 1, Asia II 5, Asia II 7, MEAM1 |
| 3. | Bangladesh | Asia I, Asia II 1, Asia II 5, China 3 |
| 4. | Nepal | Asia II 1 |
| South East Asia | ||
| 5. | Cambodia | MED, Asia I |
| 6. | Indonesia | Australia/Indonesia, Asia II 7, Asia II 12 |
| 7. | Malaysia | Asia I, MED, China 2, Asia II 7 |
| 8. | Myanmar | Asia II 5 |
| 9. | Singapore | Asia I |
| East Asia | ||
| 10. | China | MED, MEAM1, Asia I, Asia II 1, Asia II 2, Asia II 3, Asia II 4, Asia II 6, Asia II 7, Asia II 9, Asia II 10, Asia IV, China 1, China 2, China 3, China 4 |
| 11. | Japan | MED, MEAM1, MEAM2, Asia I, Asia III, Japan 1, Japan 2, Asia II 1, Asia II 6 |
| 12. | South Korea | MED, MEAM1, Japan 2 |
| 13. | Taiwan | MED, MEAM1, Asia I, Asia III, Asia II 1, Asia II 6, Asia II 7 |
| Middle East | ||
| 14. | Egypt | MED, MEAM1 |
| 15. | Iran | MEAM1 |
| 16. | Iraq | MEAM1 |
| 17. | Israel | MED, MEAM1 |
| 18. | Jordan | MEAM1 |
| 19. | Kuwait | MEAM1 |
| 20. | Saudi Arabia | MEAM1 |
| 21. | Syria | Asia II 1, MED, MEAM1 |
| 22. | Turkey | MED, MEAM1, Asia I |
| 23. | United Arab Emirates (UAE) | MEAM1 |
| 24. | Yemen | MEAM1 |
| Africa | ||
| 25. | Algeria | MED |
| 26. | Benin | MED, Sub Saharan Africa 1 |
| 27. | Burkina Faso | MED |
| 28. | Burundi | Sub Saharan Africa 1 |
| 29. | Cameroon | Africa, MED, Sub Saharan Africa 2, Sub Saharan Africa 3, Sub Saharan Africa 4 |
| 30. | Democratic Republic of the Congo | Sub Saharan Africa 1, Sub Saharan Africa 3 |
| 31. | Ghana | MED, Sub Saharan Africa 3, Sub Saharan Africa 1 |
| 32. | Ivory Coast | MED |
| 33. | Kenya | Sub Saharan Africa 1, Sub Saharan Africa 2 |
| 34. | Madagascar | Indian Ocean |
| 35. | Malawi | Sub-Saharan Africa 1 |
| 36. | Mali | Sub Saharan Africa 2 |
| 37. | Mauritius | Indian Ocean |
| 38. | Morocco | MED, MEAM1, Spain 1 |
| 39. | Mozambique | Sub Saharan Africa 1 |
| 40. | Nigeria | MED, Sub Saharan Africa 2 |
| 41. | Senegal | MED, MEAM1 |
| 42. | Seychelles | Indian Ocean |
| 43. | Sudan | MED, New World |
| 44. | Swaziland | Sub-Saharan Africa 1 |
| 45. | South Africa | MED, MEAM1, Sub Saharan Africa 1 |
| 46. | Tanzania | MED, Sub Saharan Africa 1 |
| 47. | Togo | MED, Sub Saharan Africa 3 |
| 48. | Tunisia | MED, MEAM1, Sub Saharan Africa 2 |
| 49. | Uganda | Uganda, Indian Ocean, MED, Sub Saharan Africa 2, Sub Saharan Africa 5, Sub Saharan Africa 1 |
| 50. | Zambia | Sub Saharan Africa 1 |
| 51. | Zimbabwe | MED |
| 52. | Mayotte | MEAM1 |
| 53. | Reunion | MED, MEAM1, MEAM2, Indian Ocean |
| Europe | ||
| 54. | Bosnia and Herzegovina | MED |
| 55. | Croatia | MED, MEAM1 |
| 56. | Cyprus | MED, MEAM1 |
| 57. | Czech Republic | MED |
| 58. | France | MED, MEAM1, Indian Ocean, New World 1 |
| 59. | Greece | MED, MEAM1 |
| 60. | Italy | MED, MEAM1, Italy 1, Ru |
| 61. | Netherlands | MED |
| 62. | Netherlands Antilles | MEAM1 |
| 63. | Portugal | MED, Sub Saharan Africa 2 |
| 64. | Spain | MEAM1, MED, Sub Saharan Africa2, Sub Saharan Africa 3, Italy 2 |
| 65. | Australia | Australia, MEAM1 |
| New World | ||
| South America | ||
| 66. | Argentina | MEAM1, New World 2, MED |
| 67. | Bolivia | New World 2 |
| 68. | Brazil | MEAM1,MED, New World 1, New World 2 |
| 69. | Colombia | New World 1 |
| 70. | Uruguay | MED |
| 71. | Venezuela | New World 1, MEAM1 |
| North America | ||
| 72. | Cuba | MEAM1 |
| 73. | Belize | New World 1 |
| 74. | Trinidad and Tobago | MEAM1 |
| 75. | USA | MED, MEAM1, New World 1 |
| 76. | Canada | MED, MEAM1 |
| Central America | ||
| 77. | Honduras | New World 1 |
| 78. | Mexico | MED, MEAM1, New World 1 |
| 79. | Panama | New World 1 |
| 80. | Dominican Republic | MEAM1 |
| 81. | Guatemala | MED, MEAM1, New World 1 |
| Micronesia, Oceania | ||
| 82. | Nauru | Asia II 5 |
| Greater Antilles, Caribbean | ||
| 83. | Puerto Rico | MEAM1, New World 1 |
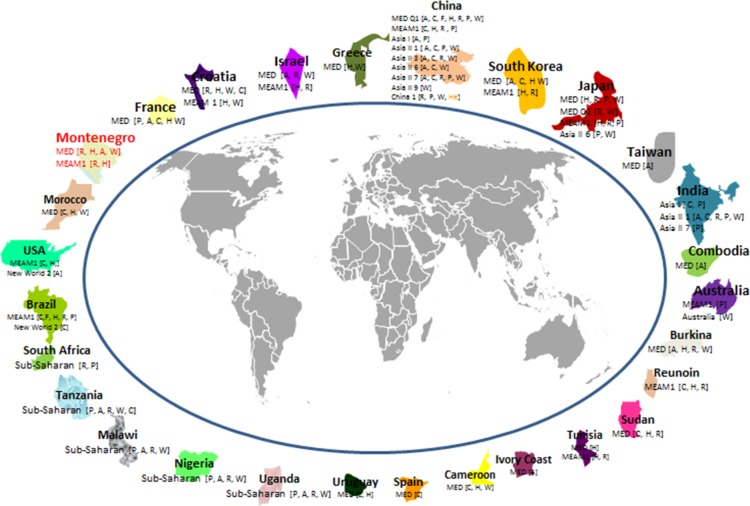
Fig 2. Schematic worldwide geographical distribution of Bemisia tabaci cryptic species and its bacterial endosymbionts.
B. tabaci endosymbionts in square brackets: A-Arsenophonus, C-Cardinium, F-Fritschea, H-Hamiltonella, R-Rickettsia, P-Porteira, W-Wolbachia, He-Hemipteriphilus.
Infection and co-infection with secondary bacterial endosymbionts
Individual whiteflies were analyzed for primary and secondary endosymbionts by 16S and 23S rRNA primers. As expected, bacterial infections were detected in both MEAM1 and MED species. MEAM1 populations from Israel showed infection with P. aleyrodidarum, Rickettsia, and Hamiltonella, while MED populations showed the presence of P. aleyrodidarum, Rickettsia, Wolbachia and Arsenophonus. Cardinium and Fritschea were not detected in any population tested. BLAST analysis of these 16S and 23S rRNA sequences confirmed infection with the respective endosymbionts in GenBank. All the sequences generated in this work were deposited in NCBI GenBank with accession numbers from KY620201 to KY620209.
Worldwide distribution scenario of bacterial endosymbionts within the B. tabaci species
To investigate the worldwide endosymbiont diversity in B. tabaci species complex, we retrieved 16sRNA gene sequences of about 1,226 accessions of reported B. tabaci endosymbionts from GenBank. Of all these, a total of 298 sequences, including sequences from this study, were selected from 32 countries. All the six continents, Asia, Africa, North America, South America, Europe and Australia, were represented and provided an interesting geographical distribution and wide genetic diversity within the B. tabaci species complex. Based on the 16sRNA sequences available in the database, P. aleyrodidarum was reported from nine Bemisia cryptic species (MED, MEAM1, Asia I, Asia II 1, Asia II 6, Asia II 7, China 1, Japan, Sub-Saharan Africa), which were reported from fifteen countries, worldwide (China, Japan, France, Australia, Brazil, India, South Africa, Tanzania, Malawi, Uganda, Nigeria, USA, Mexico, Israel, Pakistan) (Fig 2). However, many B. tabaci populations around the world were studied without investigation of their P. aleyrodidarum sequences, thus the actual scenario of P. aleyrodidarum diversity might still be different from what is presented in this manuscript and we only relied our analysis on published sequences.
In case of the secondary symbionts, Rickettsia was associated with seven species (MED, MEAM1, Asia II 1, Asia II 3, Asia II 7, China 1 and Sub-Saharan Africa) in nineteen countries (China, Japan, Israel, Burkina, Montenegro, Croatia, South Korea, Sudan, Brazil, Tunisia, Reunion, Antilles, India, Bangladesh, Tanzania, Malawi, Uganda, Nigeria and Pakistan). Cardinium was associated with the species MED, MEAM1, Asia I, Asia II 1, Asia II 3, Asia II 6, Asia II 7, Japan, Sub-Sharan Africa and New World. Arsenophonus was associated with MED, Asia I, Asia II 1, Asia II 3, Asia II 6, Asia II 7, Sub-Sharan Africa and New World, while both Fritschea and Hamiltonella detected in individuals from MED, MEAM1 and New World 2. Hemipteriphilus was associated with China 1 species from China (Fig 2).
Overall, we found evidence for P. aleyrodidarum, Rickettsia, Wolbachia, Hamiltonella, Arsenophonus, Cardinium and Fritschea association with MED. In case of MEAM1, except Arsenophonus and Hemipteriphilus, the remaining endosymbionts were associated with this species. Interestingly, Fritschea and Hamiltonella were restricted to MED, MEAM1 and New World species, and were not associated with Bemisia new complex species. Accession numbers of endosymbiont sequences obtained from the Gen Bank database are detailed in the respective phylogenetic trees (Figs 3,4,5,6,7 and 8).
Fig 3. Molecular phylogenetic placements of Porteira (16S) from reported sequences of worldwide whitefly species.
The tree was constructed via Bayesian inference (BI) using an JC+G substitution model. Sequences from Israel are indicated in bold.
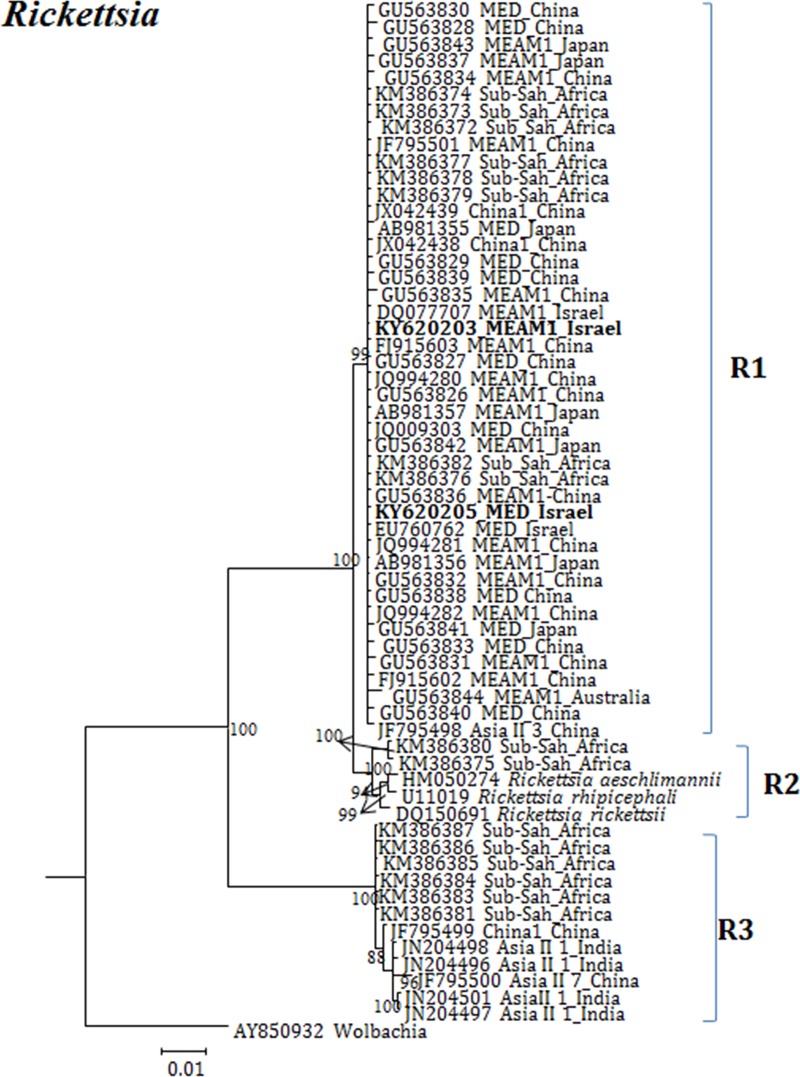
Fig 4. Molecular phylogenetic placements of Rickettsia (16S) from reported sequences of worldwide whitefly species.
The tree was constructed via Bayesian inference (BI) using an GTR+I substitution model. Sequences from Israel are indicated in bold.
Fig 5. Molecular phylogenetic placements of Hamiltonella (16S) from reported sequences of worldwide whitefly species.
The tree was constructed via Bayesian inference (BI) using an GTR+I substitution model. Sequences from Israel are indicated in bold.
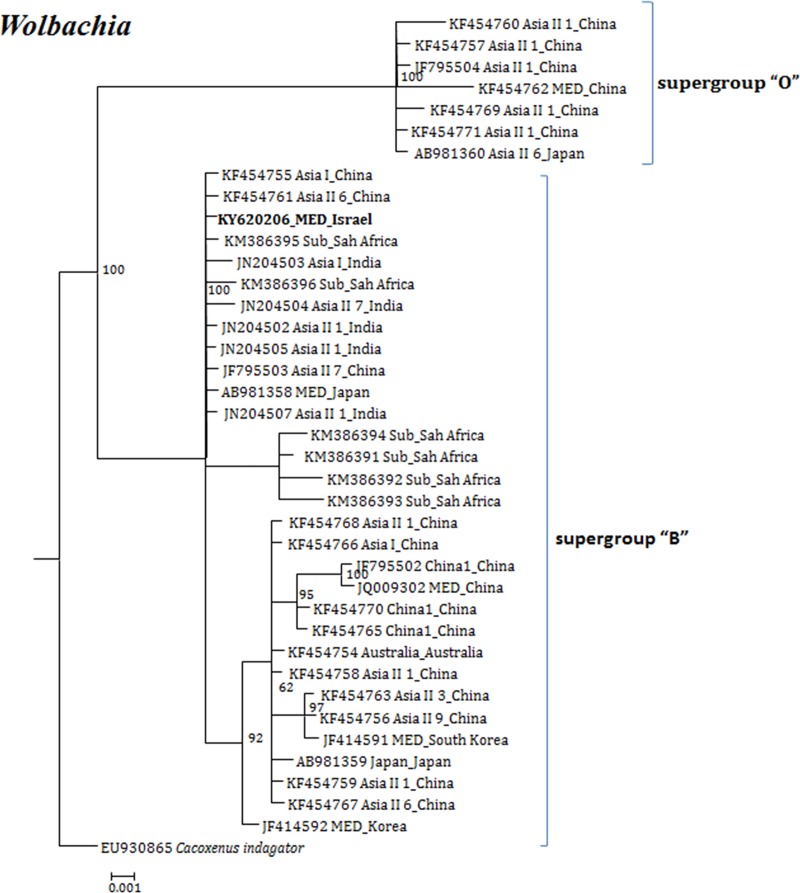
Fig 6. Molecular phylogenetic placements of Wolbachia (16S) from reported sequences of worldwide whitefly species.
The tree was constructed via Bayesian inference (BI) using an HKY substitution model. Sequences from Israel are indicated in bold.
Fig 7. Molecular phylogenetic placements of Cardinium (16S) from reported sequences of worldwide whitefly species.
The tree was constructed via Bayesian inference (BI) using an GTR substitution model. Sequences from Israel are indicated in bold.
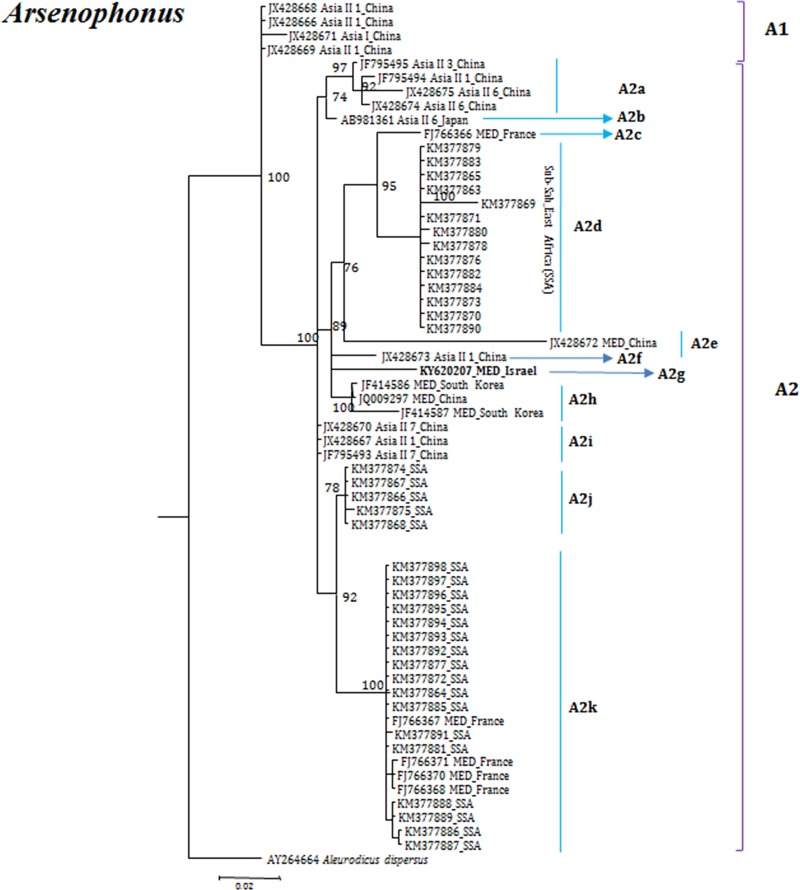
Fig 8. Molecular phylogenetic placements of Aresenophonus (23S) from reported sequences of worldwide whitefly species.
The tree was constructed via Bayesian inference (BI) using an HKY+G substitution model. Sequences from Israel are indicated in bold.
Phylogenetic analysis of the primary endosymbiont of B. tabaci cryptic species
The phylogenetic relationships among the endosymbionts were first estimated using the 16s rDNA sequences obtained from GenBank. The P. aleyrodidarum grouped into two major clades P1 and P2 with bootstrap scores of >70% (Fig 3). Both P1 and P2 includes both invasive and indigenous species from six countries. Interestingly, the percentage nucleotide identities of selected Porteira from each species from different countries showed a peculiar scenario; in which the bacterium from different species in India share >95% identity with species from Japan, Brazil and Sub-Saharan Africa, Australia, and vice versa. Portiera from China also shares <95% identity with Portiera reported from B. tabaci species of the above countries. Porteira in all species (includes both Indigenous and invasive species) that originated from China share the highest percentage similarities compared with other countries (S3 Table).
Phylogenetic analysis of secondary endosymbionts of B. tabaci cryptic species
Rickettsia was grouped in to three major clusters, R1, R2 and R3. The top clade R1 (Fig 4) consists of Rickettsia from both invasive and indigenous members from Australia, China, Japan, Israel and Sub-Saharan African countries. Clade R2 consists of Rickettsia only from Sub-Saharan Africa species. Clade R3 consists of Rickettsia only from indigenous species that originated from Sub-Saharan Africa, Asia II 7 Asia II 1 from China and India.
Interestingly, published sequences for Hamiltonella from the invasive species (MED and MEAM1) all belonged to only one major cluster (H1) from Brazil, Japan, USA, China and South Korea (Fig 5).
In the case of Wolbachia, 16 super-groups are so far successively named A-Q and are infecting a wide range of arthropods and filarial nematodes [58]. In our analysis, two super-groups B and O were observed in the overall analysis of B. tabaci Wolbachia sequences. The O super-group was observed in MED and Asian species from China and Japan. The B super-group was found in both MED and MEAM1 invasive species from Asia, Sub-Sharan Africa, China 1, Japan and Australia, and in indigenous species. Interestingly, both super-groups were observed in MED from China (Fig 6).
The phylogenetic tree of Cardinium resulted in four major clusters (C1-C4). The C1 consists, invasive MED species from China; C2 consists, Asian, MEAM1, Sub-Sharan African species from 4 countries (India, china, Sub-Saharan Africa and USA). In case of C3, invasive MEAM1 species from USA and C4 consists MED, MEAM1, Japan, Sub-Saharan Africa, Asian and New World species from 10 countries (China, Japan, Sudan, Spain, South Korea, Morocco, France, Cameroon, Uruguay and USA) (Fig 7). Interestingly, isolates that belong to the three clades C1, C2 and C4 were present in China. The isolates that appeared in C4 included MED, MEAM1 and Asia II 1 and Asia II 6. Clade C2 consists only indigenous species (Asia II 1, Asia II 7 and Sub-Saharan Africa).
Arsenophonus phylogenetic analyses supported its grouping into two major groups (A1 and A2), the top clade A1 consisted only indigenous Asian species from China. Interestingly, the second clade A2 consists eleven subclades named as A2 (a-k). In these clades, Chinese Arsenophonus strains from invasive species observed in the two subclades A2e and A2h and indigenous species in three subclades A2a, A2f and A2i. Similarly, strains from Sub-Sahran Africa were more distinct and diverse within the Sub-Saharan Africa species. In this analysis, all isolates from Sub-Saharan Africa were observed only in the major clade A2, in which they sub-grouped into three clades A2d, A2j and A2k (Fig 8).
The overall analysis for all endosymbionts showed that, except Hamiltonella (Fig 5), the rest of the secondary endosymbionts were more diverse and appeared in different genetic groups in invasive and indigenous B. tabaci species than previously assumed. Interestingly, Hamiltonella was observed in both invasive species (MEAM1 and MED species worldwide) and in the New World 2 species from Brazil. In contrast, Fritschea was only observed in MED, MEAM1 and New World 2 species while Hemipteriphilus was detected in China1 species from China.
Discussion
The whitefly B. tabaci species complex constitutes the most economically and agriculturally important insect pest worldwide. This pest is considered a super-vector and possibly is the most important insect vector transmitting viruses in agricultural crops worldwide [59,60]. It transmits more than 100 plant viruses and those are considered the most important in terms of damage they cause on a worldwide scale. This complex of cryptic species or biotypes differ in their behavior, plant host adaptations, ability to develop resistance to pesticides and induce plant disorders, ability to transmit plant viruses, the bacterial endosymbionts they harbor [26–28], and their genetic make-up, as has recently been described in the genomes of the two most widespread members of this complex: MEAM1 and MED species [61,62]. Most importantly, B. tabaci is rated among the 100 most invasive species worldwide. One of the most studied aspects of this complex, is the bacterial endosymbionts that infect members of this complex. Those bacteria were shown to greatly impact many aspects of the biology of B. tabaci including the ability to vector plant viruses, which in turn have significant impact on the damage caused by members of this species complex, especially the MEAM1 and MED species [63,64]. We thus attempted to collect all published sequences of bacterial symbionts from populations reported around the world and analyze their relationships. These analyses might shed light of the origins and relationships between different B. tabaci species and populations within and between countries and continents.
Several recent studies have suggested that biological and molecular differences between members of B. tabaci complex of cryptic species or biotypes warrants further analyses to clarify whether those members can be considered different species. Global phylogenetic reconstruction of the B. tabaci complex species was conducted with 366 sequences and around 12 major genetic groups were generated [12]. Later, De Barro et al. [11] and Dinsdale et al. [7] suggested that the term "cryptic species" be used to distinguish the whitefly population based on 3.5% mitochondrial cytochrome oxidase gene sequence divergence. Based on this criterion, 24 B. tabaci cryptic species were determined and nominated. Recently, Lee et al. [15] observed that a 4.0% genetic boundary was more realistic than 3.5% in distinguishing the B. tabaci species complex members. Though few controversies did occur [65,66], reclassification based on the 3.5% and 4% genetic divergence revealed that 42 groups are clearly distinct (Fig 1).
The newly described morphologically indistinguishable species (which include Africa, Asia I, Asia I-India, Asia II 1–12, Asia III, Asia IV, Asia V, Australia, Australia/Indonesia, China 1–5, Indian Ocean, Ru, Middle East Asia Minor I-II (MEAM), Mediterranean (MED), MEAM K, New World 1–2, Japan 1–2, Uganda, Italy 1, and Sub Saharan Africa 1–5) have been currently delimited at the global level [7,10–14]. The worldwide B. tabaci mtCOI sequence analysis that we conducted in this study supports the existence of two new B. tabaci species: Asia II 13 and Spain 1. This is the first report that shows the existence of 44 B. tabaci species worldwide.
Our analyses focused on comprehensively studying and analyzing the bacterial endosymbiont communities that infect wide geographic range of the B. tabaci species complex members, undertaken to gain more thorough understanding of endosymbiont diversity and complexity. Our results revealed complex divergence of the different endosymbiont sequences analyzed, except for Hamiltonella, which showed only one genetic group across the different worldwide populations analyzed. Two Porteria groups were identified in this analysis: P1 and P2. The sequences belonging to the P1 group were present in Australia, Brazil, China and Japan, whereas P2 group was identified in China, India, Japan and Sub-Sharan African populations. Similarly, total of four groups of Cardinium (C1-C4) were identified, in which C1 from China, C2- China, India and Sub-Saharan Africa, C3- USA, Cameroon, China, France, Japan, South Korea, Spain, Sudan and Uruguay, while the C4 was also widespread in a worldwide scale. Wolbachia super-groups O and B were identified. The B group members were identified in Australia, China, Japan, India, Sub-Saharan Africa and South Korea, while the O members in China and Japan. Other groups of Wolbachia that were reported from other arthropods were not identified in our study, suggesting that these two groups were acquired in earlier events of B. tabaci speciation and coevolved with the different members of the insect species complex, as they appear in many members of the group.
An important observation that could be drawn from our analysis that although more than 40 cryptic species of B. tabaci are identified, endosymbionts which infect phylogenetically remote members of this complex are sometimes grouped in the same clades. This applies to almost all symbionts except for Hamiltonella which showed only one group infecting all reported species. This observation, combined with the fact that evidence for direct horizontal transmission of the majority of these symbionts does not exist, although some evidence has been shown [67], suggests that the majority of the symbionts were acquired before the start of B. tabaci complex speciation from other whitefly species. Thus, their spread in whitefly populations, including B. tabaci species, occurred after this speciation. Their co-evolution with B. tabaci species and the intimate symbiotic associations they have developed with the whitefly also limited the evolved variations in their genomes. This observation further supports the fact that those symbionts are unevenly distributed between the different species complex members as shown in previous studies [26–29,40]. This is evident from the variation observed in the infection of the different symbionts in the different species complex members, and that not all reported symbionts infect all B. tabaci species complex members. The fact that although many of these members share many of the host plants and yet are significantly different in their symbiotic complements suggest that horizontal transmission via plants in indeed unlikely, as has been shown for Rickettsia [36,40].
It is known among B. tabaci endosymbionts, but also among other arthropods, that mixed infections with more than one symbiont exist, and B. tabaci harbors the highest number of mixed infection in one insect [28,29]. The different endosymbionts presented in this paper were reported to coexist in B. tabaci populations, sometimes inside the same organ [29,33]. This coexistence however, is also sometimes the cause for the uneven distribution of these symbionts in B. tabaci populations, as it is the cause for competition for space and resources in the insect by these symbionts, and the cause for immune system responses in the insect for maintaining levels of the symbionts inside the insect that do not interfere with its proper physiology and development. Although there is some chance that multiple horizontal events may be the cause for mixed infection, as mentioned above, the ability of horizontal transfer was only reported for Rickettsia. It thus remains more likely that early acquisition events of these symbionts is the cause for the diversity observed in the B. tabaci species complex, combined with the low probability that some of these symbionts could be distributed via horizontal transfers. Horizontal transfer could also occur between different species of whiteflies for example B. tabaci and T. vaporariurom, which also share the same host plants and have been shown to be infected with the same endosymbiont species [26–28].
In summary, we have performed combined analyses for the largest available datasets of both B. tabaci species and its endosymbiont sequences which resulted in a global diversity and geographic distribution of this important insect pest and its associated bacterial endosymbionts. These results will be helpful in understanding the successful invasions of B. tabaci species complex members across the globe, and the possible contribution of their associated symbionts in their invasions and abilities to cause damage in agricultural ecosystems.
Supporting information
(XLSX)
(XLSX)
(XLSX)
The bootstrap values are indicated. More Asia II 1, Spain 1 and Italy 1 sequences were included.
(PDF)
Data Availability
All relevant data are within the manuscript and its Supporting Information files.
Funding Statement
SK was supported with a Postdoctoral Fellowship # 2014–2016 from ARO. Research in the Ghanim lab is supported by grants from the Israel Science Foundation (ISF), Binational Agricultural Research and Development Fund (BARD) and the Chief Scientist of the Ministry of Agriculture in Israel. The funders had no role in study design, data collection and analysis, decision to publish, or preparation of the manuscript.
References
- 1.Jones DR. Plant viruses transmitted by whiteflies. Eur J Plant Pathol. 2003; 109: 195–219. [Google Scholar]
- 2.Navas-Castillo J, Fiallo-Olivé E, Sánchez-Campos S. Emerging virus diseases transmitted by whiteflies. Ann Rev Phytopathol. 2011; 49: 219–248. [DOI] [PubMed] [Google Scholar]
- 3.Simón B, Cenis JL, Beitia F, Khalid S, Moreno IM, Fraile A. et al. Genetic structure of field populations of begomoviruses and of their vector Bemisia tabaci in Pakistan. Phytopathology. 2003; 93: 1422–1429. 10.1094/PHYTO.2003.93.11.1422 [DOI] [PubMed] [Google Scholar]
- 4.Zang LS, Chen WQ, Liu SS. Comparison of performance on different host plants between the B biotype and a non‐B biotype of Bemisia tabaci from Zhejiang, China. Entomol Exp Appl. 2006; 121, 221–227. [Google Scholar]
- 5.Tay WT, Evans GA, Boykin LM, De Barro PJ. Will the real Bemisia tabaci please stand up?. PLoS ONE. 2012; 7: e50550 10.1371/journal.pone.0050550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Perring TM. The Bemisia tabaci species complex. Crop Prot. 2001; 20: 725–737. [Google Scholar]
- 7.Dinsdale A, Cook L, Riginos C, Buckley YM, De Barro P. Refined global analysis of Bemisia tabaci (Hemiptera: Sternorrhyncha: Aleyrodoidea: Aleyrodidae) mitochondrial cytochrome oxidase 1 to identify species level genetic boundaries. Ann Entomol Soc Am. 2010; 103: 196–208. [Google Scholar]
- 8.Frohlich DR, Torres‐Jerez I, Bedford ID, Markham PG, Brown JK. A phylogeographical analysis of the Bemisia tabaci species complex based on mitochondrial DNA markers. Mol Ecol. 1999; 8: 1683–1691. [DOI] [PubMed] [Google Scholar]
- 9.Liu SS, Colvin J, De Barro PJ. Species concepts as applied to the whitefly Bemisia tabaci systematics: how many species are there?. J Integr Agr. 2012; 11: 176–186. [Google Scholar]
- 10.Firdaus S, Vosman B, Hidayati N, Supena EDJ, Visser RG, van Heusden AW. The Bemisia tabaci species complex: additions from different parts of the world. Insect Sci. 2013; 20, 723–733. 10.1111/1744-7917.12001 [DOI] [PubMed] [Google Scholar]
- 11.De Barro PJ, Liu SS, Boykin LM, Dinsdale AB. Bemisia tabaci: a statement of species status. Ann Rev Entomol. 2011; 56: 1–19. [DOI] [PubMed] [Google Scholar]
- 12.Boykin LM, Shatters RG Jr, Rosell RC, McKenzie CL, Bagnall RA, De Barro P, et al. Global relationships of Bemisia tabaci (Hemiptera: Aleyrodidae) revealed using Bayesian analysis of mitochondrial COI DNA sequences. Mol Phylogenet Evol. 2007; 44: 1306–1319. 10.1016/j.ympev.2007.04.020 [DOI] [PubMed] [Google Scholar]
- 13.Hu J, Zhang X, Jiang Z, Zhang F, Liu Y, Li Z, et al. New putative cryptic species detection and genetic network analysis of Bemisia tabaci (Hempitera: Aleyrodidae) in China based on mitochondrial COI sequences. Mitochondrial DNA Part A. 2018; 29: 474–484. [DOI] [PubMed] [Google Scholar]
- 14.Roopa HK, Asokan R, Rebijith KB, Hande RH, Mahmood R, Kumar NK, et al. Prevalence of a new genetic group, MEAM-K, of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae) in Karnataka, India, as evident from mtCOI sequences. Florida Entomol. 2015; 98, 1062–1071. [Google Scholar]
- 15.Lee W, Park J, Lee GS, Lee S, Akimoto SI. Taxonomic status of the Bemisia tabaci complex (Hemiptera: Aleyrodidae) and reassessment of the number of its constituent species. PLoS ONE. 2013; 8: e63817 10.1371/journal.pone.0063817 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thao ML, Baumann P. Evidence for multiple acquisition of Arsenophonus by whitefly species (Sternorrhyncha: Aleyrodidae) Curr Microbiol. 2004; 48: 140–144. 10.1007/s00284-003-4157-7 [DOI] [PubMed] [Google Scholar]
- 17.Sloan DB, Moran NA. Endosymbiotic bacteria as a source of carotenoids in whiteflies. Biol Lett. 2012; 8: 986–989. 10.1098/rsbl.2012.0664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Moran NA, Telang A. Bacteriocyte-associated symbionts of insects. Bioscience. 1998; 48: 295–304. [Google Scholar]
- 19.Baumann P. Biology of bacteriocyte-associated endosymbionts of plant sap-sucking insects. Ann Rev Microbiol. 2005; 59: 155–189. [DOI] [PubMed] [Google Scholar]
- 20.Gottlieb Y, Ghanim M, Chiel E, Gerling D, Portnoy V, Steinberg S, et al. Identification and localization of a Rickettsia sp. in Bemisia tabaci (Homoptera: Aleyrodidae) Appl Environ Microbiol. 2006; 72: 3646–3652. 10.1128/AEM.72.5.3646-3652.2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Zchori-Fein E, Brown JK. Diversity of prokaryotes associated with Bemisia tabaci (Gennadius)(Hemiptera: Aleyrodidae) Ann Entomol Soc Am. 2002; 95: 711–718. [Google Scholar]
- 22.Nirgianaki A, Banks GK, Frohlich DR, Veneti Z, Braig HR, Miller TA, et al. Wolbachia infections of the whitefly Bemisia tabaci. Curr Microbiol. 2003; 47: 0093–0101. [DOI] [PubMed] [Google Scholar]
- 23.Weeks AR, Velten R, Stouthamer R. Incidence of a new sex–ratio–distorting endosymbiotic bacterium among arthropods. P Roy Soc Lond B Bio. 2003; 270: 1857–1865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Everett KD, Thao M, Horn M, Dyszynski GE, Baumann P. Novel chlamydiae in whiteflies and scale insects: endosymbionts ‘Candidatus Fritschea bemisiae’ strain Falk and ‘Candidatus Fritschea eriococci’strain Elm. Int J Syst Evol Micr. 2005; 55: 1581–1587. [DOI] [PubMed] [Google Scholar]
- 25.Bing XL, Yang J, Zchori-Fein E, Wang XW, Liu SS. Characterization of a newly discovered symbiont of the whitefly Bemisia tabaci (Hemiptera: Aleyrodidae). Appl Env Microbiol. 2013; 79: 569–575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Škaljac M, Žanić K, Ban SG, Konstedalov S, Ghanim M. Co-infection and localization of secondary symbionts in two whitefly species. BMC Microbiol. 2010; 10: 142–1. 10.1186/1471-2180-10-142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Škaljac M, Žanić K, Hrnčić S, Radonjić S, Perović T, Ghanim M. Diversity and localization of bacterial symbionts in three whitefly species (Hemiptera: Aleyrodidae) from the east coast of the Adriatic Sea. B Entomol Res. 2013; 103: 48–59. [DOI] [PubMed] [Google Scholar]
- 28.Skaljac M, Kanakala S, Zanic K, Puizina J, Pleic IL, Ghanim M. Diversity and phylogenetic analyses of bacterial symbionts in three whitefly species from Southeast Europe. Insects. 2017; 8: 113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Gueguen G, Vavre F, Gnankine O, Peterschmitt M, Charif D, Chiel E, et al. Endosymbiont metacommunities, mtDNA diversity and the evolution of the Bemisia tabaci (Hemiptera: Aleyrodidae) species complex. Mol Ecol. 2010; 19: 4365–4376. 10.1111/j.1365-294X.2010.04775.x [DOI] [PubMed] [Google Scholar]
- 30.Marubayashi JM, Kliot A, Yuki VA, Rezende JAM, Krause-Sakate R, Pavan MA, et al. Diversity and localization of bacterial endosymbionts from whitefly species collected in Brazil. PLoS ONE. 2014; 9: e108363 10.1371/journal.pone.0108363 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Zchori-Fein E, Lahav T, Freilich S. Variations in the identity and complexity of endosymbiont combinations in whitefly hosts. Front Microbiol. 2014; 5: 310 10.3389/fmicb.2014.00310 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ghosh S, Bouvaine S, Maruthi MN. Prevalence and genetic diversity of endosymbiotic bacteria infecting cassava whiteflies in Africa. BMC Microbiol. 2015; 15: 93 10.1186/s12866-015-0425-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Gottlieb Y, Ghanim M, Gueguen G, Kontsedalov S, Vavre F, Fleury F, et al. Inherited intracellular ecosystem: symbiotic bacteria share bacteriocytes in whiteflies. FASEB J. 2008; 22: 2591–2599. 10.1096/fj.07-101162 [DOI] [PubMed] [Google Scholar]
- 34.Vautrin E, Genieys S, Charles S, Vavre F. Do vertically transmitted symbionts co‐existing in a single host compete or cooperate? A modelling approach. J Evol Biol. 2008; 21: 145–161. 10.1111/j.1420-9101.2007.01460.x [DOI] [PubMed] [Google Scholar]
- 35.Vautrin E, Vavre F. Interactions between vertically transmitted symbionts: cooperation or conflict?. Trends Microbiol. 2009; 17: 95–99. 10.1016/j.tim.2008.12.002 [DOI] [PubMed] [Google Scholar]
- 36.Caspi-Fluger A, Inbar M, Mozes-Daube N, Katzir N, Portnoy V, Belausov E. et al. Horizontal transmission of the insect symbiont Rickettsia is plant-mediated. P Roy Soc Lond B Bio. 2011; 279: 1791–1796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zchori-Fein E, Gottlieb Y, Kelly SE, Brown JK, Wilson JM, Karr TL. et al. A newly discovered bacterium associated with parthenogenesis and a change in host selection behavior in parasitoid wasps. P Natl Acad Sci USA. 2001; 98: 12555–12560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Hunter MS, Perlman SJ, Kelly SE. A bacterial symbiont in the Bacteroidetes induces cytoplasmic incompatibility in the parasitoid wasp Encarsia pergandiella. P Roy Soc Lon B Bio. 2003; 270: 2185–2190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Zchori‐Fein EINAT, Perlman SJ. Distribution of the bacterial symbiont Cardinium in arthropods. Mol Ecol. 2004; 13: 2009–2016. 10.1111/j.1365-294X.2004.02203.x [DOI] [PubMed] [Google Scholar]
- 40.Chiel E, Gottlieb Y, Zchori-Fein E, Mozes-Daube N, Katzir N, Inbar M, et al. Biotype-dependent secondary symbiont communities in sympatric populations of Bemisia tabaci. B Entomol Res. 2007; 97: 407–413. [DOI] [PubMed] [Google Scholar]
- 41.Su Q, Xie W, Wang S, Wu Q, Liu B, Fang Y, et al. The endosymbiont Hamiltonella increases the growth rate of its host Bemisia tabaci during periods of nutritional stress. PLoS ONE. 2014; 9: e89002 10.1371/journal.pone.0089002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Himler AG, Adachi-Hagimori T, Bergen JE, Kozuch A, Kelly SE, Tabashnik BE, et al. Rapid spread of a bacterial symbiont in an invasive whitefly is driven by fitness benefits and female bias. Science. 2011; 332, 254–256. 10.1126/science.1199410 [DOI] [PubMed] [Google Scholar]
- 43.Brumin M, Kontsedalov S, Ghanim M. Rickettsia influences thermotolerance in the whitefly Bemisia tabaci B biotype. Insect Sci. 2011; 18: 57–66. [Google Scholar]
- 44.Kontsedalov S, Zchori‐Fein E, Chiel E, Gottlieb Y, Inbar M, Ghanim M. The presence of Rickettsia is associated with increased susceptibility of Bemisia tabaci (Homoptera: Aleyrodidae) to insecticides. Pest Manag Sci. 2008; 64: 789–792. 10.1002/ps.1595 [DOI] [PubMed] [Google Scholar]
- 45.Shan HW, Zhang CR, Yan TT, Tang HQ, Wang XW, Liu SS, et al. Temporal changes of symbiont density and host fitness after rifampicin treatment in a whitefly of the Bemisia tabaci species complex. Insect Sci. 2016; 23: 200–214. 10.1111/1744-7917.12276 [DOI] [PubMed] [Google Scholar]
- 46.Hendry TA, Hunter MS, Baltrus DA. The facultative symbiont Rickettsia protects an invasive whitefly against entomopathogenic Pseudomonas syringae strains. Appl Environ Microb. 2014; AEM-02447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Morin S, Ghanim M, Zeidan M, Czosnek H, Verbeek M, van den Heuvel JF. A GroEL homologue from endosymbiotic bacteria of the whitefly Bemisia tabaci is implicated in the circulative transmission of tomato yellow leaf curl virus. Virology. 1999; 256: 75–84. 10.1006/viro.1999.9631 [DOI] [PubMed] [Google Scholar]
- 48.Morin S, Ghanim M, Sobol I, Czosnek H. The GroEL protein of the whitefly Bemisia tabaci interacts with the coat protein of transmissible and nontransmissible begomoviruses in the yeast two-hybrid system. Virology. 2000; 276: 404–416. 10.1006/viro.2000.0549 [DOI] [PubMed] [Google Scholar]
- 49.Rana VS, Singh ST, Priya NG, Kumar J, Rajagopal R. Arsenophonus GroEL interacts with CLCuV and is localized in midgut and salivary gland of whitefly B. tabaci. PLoS ONE. 2012; 7: e42168 10.1371/journal.pone.0042168 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chu D, Gao CS, De Barro P, Zhang YJ, Wan FH, Khan IA. Further insights into the strange role of bacterial endosymbionts in whitefly, Bemisia tabaci: Comparison of secondary symbionts from biotypes B and Q in China. B Entomol Res. 2011; 101: 477–486. [DOI] [PubMed] [Google Scholar]
- 51.Singh ST, Priya NG, Kumar J, Rana VS, Ellango R, Joshi A, et al. Diversity and phylogenetic analysis of endosymbiotic bacteria from field caught Bemisia tabaci from different locations of North India based on 16S rDNA library screening. Infect Genet Evol. 2012; 12: 411–419. 10.1016/j.meegid.2012.01.015 [DOI] [PubMed] [Google Scholar]
- 52.Gorsane F, Ben Halima A, Ben Khalifa M, Bel-Kadhi MS, Fakhfakh H. Molecular characterization of Bemisia tabaci populations in Tunisia: genetic structure and evidence for multiple acquisition of secondary symbionts. Environ Entomol. 2011; 40: 809–817. 10.1603/EN10162 [DOI] [PubMed] [Google Scholar]
- 53.De Barro PJ, Scott KD, Graham GC, Lange CL, Schutze MK. Isolation and characterization of microsatellite loci in Bemisia tabaci. Mol Ecol Notes. 2003; 3: 40–43. [Google Scholar]
- 54.Tamura K, Stecher G, Peterson D, Filipski A, Kumar S. MEGA6: molecular evolutionary genetics analysis version 6.0. Mol Biol Evol. 2013; 30: 2725–2729. 10.1093/molbev/mst197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: improvements in performance and usability. Mol Biol Evol. 2013; 30: 772–780. 10.1093/molbev/mst010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Miller MA, Pfeiffer W, Schwartz T. Creating the CIPRES Science Gateway for inference of large phylogenetic trees. In gateway computing environments workshop (GCE). 2010; 14: 1–8. [Google Scholar]
- 57.Ronquist F, Teslenko M, Van Der Mark P, Ayres DL, Darling A, Höhna S, et al. MrBayes 3.2: efficient Bayesian phylogenetic inference and model choice across a large model space. Syst Biol. 2012; 61: 539–542. 10.1093/sysbio/sys029 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Glowska E, Dragun-Damian A, Dabert M, Gerth M. New Wolbachia supergroups detected in quill mites (Acari: Syringophilidae) Infect Genet Evol. 2015; 30: 140–146. 10.1016/j.meegid.2014.12.019 [DOI] [PubMed] [Google Scholar]
- 59.Czosnek H, Hariton-Shalev A, Sobol I, Gorovits R, Ghanim M. The incredible journey of begomoviruses in their whitefly vector. Viruses. 2017; 273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kanakala S, Ghanim M. Whitefly-transmitted begomoviruses and advances in the control of their vectors In: Pail B, editor. Genes, genetics and transgenics for virus resistance in plants. United Kingdom: Caister Academic Press; 2018. pp. 201–220. [Google Scholar]
- 61.Chen W, Hasegawa DK, Kaur N, Kliot A, Pinheiro PV, Luan J, et al. The draft genome of whitefly Bemisia tabaci MEAM1, a global crop pest, provides novel insights into virus transmission, host adaptation, and insecticide resistance. BMC Biol. 2016; 14: 110 10.1186/s12915-016-0321-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Xie W, Chen C, Yang Z, Guo L, Yang X, Wang D, et al. Genome sequencing of the sweetpotato whitefly Bemisia tabaci MED/Q. Gigascience. 2017; 6: 1–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ghanim M. A review of the mechanisms and components that determine the transmission efficiency of Tomato yellow leaf curl virus (Geminiviridae; Begomovirus) by its whitefly vector. Virus Res. 2014; 186: 47–54. 10.1016/j.virusres.2014.01.022 [DOI] [PubMed] [Google Scholar]
- 64.Pinherio PV, Kliot A, Ghanim M, Cilia M. Is there a role for symbiotic bacteria in plant virus transmission by insects?. Curr Opin Insect Sci. 2015; 8: 69–78. [DOI] [PubMed] [Google Scholar]
- 65.Mouton L, Gnankiné O, Henri H, Terraz G, Ketoh G, Martin T, et al. Detection of genetically isolated entities within the Mediterranean species of Bemisia tabaci: new insights into the systematics of this worldwide pest. Pest Manag Sci. 2015; 71: 452–458. 10.1002/ps.3834 [DOI] [PubMed] [Google Scholar]
- 66.Qin L, Pan LL, Liu SS. Further insight into reproductive incompatibility between putative cryptic species of the Bemisia tabaci whitefly complex. Insect Sci. 2016; 23, 215–224. 10.1111/1744-7917.12296 [DOI] [PubMed] [Google Scholar]
- 67.Mouton L, Thierry M, Henri H, Baudin R, Gnankine O, Reynaud B, et al. Evidence of diversity and recombination in arsenophonus symbionts of the Bemisia tabaci species complex. BMC Microbiol. 2012; 12. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
(XLSX)
(XLSX)
(XLSX)
The bootstrap values are indicated. More Asia II 1, Spain 1 and Italy 1 sequences were included.
(PDF)
Data Availability Statement
All relevant data are within the manuscript and its Supporting Information files.