Abstract
Deoxyhypusine synthase (DHS) is encoded by a nuclear gene and is the key enzyme involved in the post-translational activation of the eukaryotic translation initiation factor eIF5A. DHS plays important roles in plant growth and development. To gain a better understanding of DHS, the petunia (Petunia hybrida) PhDHS gene was isolated, and the role of PhDHS in plant growth was analyzed. PhDHS protein was localized to the nucleus and cytoplasm. Virus-mediated PhDHS silencing caused a sectored chlorotic leaf phenotype. Chlorophyll levels and photosystem II activity were reduced, and chloroplast development was abnormal in PhDHS-silenced leaves. In addition, PhDHS silencing resulted in extended leaf longevity and thick leaves. A proteome assay revealed that 308 proteins are upregulated and 266 proteins are downregulated in PhDHS-silenced plants compared with control, among the latter, 21 proteins of photosystem I and photosystem II and 12 thylakoid (thylakoid lumen and thylakoid membrane) proteins. In addition, the mRNA level of PheIF5A-1 significantly decreased in PhDHS-silenced plants, while that of another three PheIF5As were not significantly affected in PhDHS-silenced plants. Thus, silencing of PhDHS affects photosynthesis presumably as an indirect effect due to reduced expression of PheIF5A-1 in petunia.
Significance: PhDHS-silenced plants develop yellow leaves and exhibit a reduced level of photosynthetic pigment in mesophyll cells. In addition, arrested development of chloroplasts is observed in the yellow leaves.
Keywords: deoxyhypusine synthase, chloroplast development, sectored chlorotic leaf, photosynthesis, petunia
Introduction
Deoxyhypusine synthase (DHS) is an enzyme thought to be present in all eukaryotic cells (Jenkins et al., 2001). The eukaryotic translation initiation factor eIF5A is identified as the only hypusine-containing protein in eukaryotes (Gordon et al., 1987; Bartig et al., 1992; Kyrpides and Woese, 1998; Park et al., 2010). In animals and yeast, hypusine is formed via a posttranslational modification that involves two enzymes, DHS and deoxyhypusine hydroxylase (DOHH), which catalyzes the first and second steps in the formation of hypusine (Abbruzzese et al., 1986; Park et al., 2006). It has been proposed that plant DHS, similar to its mammalian and yeast counterparts, mediates the first of two reactions required for the post-translational activation of eIF-5A, specifically the formation of deoxyhypusine on the inactive eIF-5A protein (Wang et al., 2003). In plants, recombinant tomato (Solanum lycopersicum) and tobacco (Nicotiana tabacum) DHS are capable of catalyzing the formation of a deoxyhypusine residue in eIF-5A substrates, respectively (Ober and Hartmann, 1999; Wang et al., 2001).
In most eukaryotes, DHS is a single copy gene (Park et al., 2010). A haploid Saccharomyces cerevisiae strain with a disruption in the DHS gene was not viable (Sasaki et al., 1996; Park et al., 1998). Similarly, eIF5A depletion led to the growth arrest of enlarged cells in S. cerevisiae (Kang et al., 2007). In plants, suppression of DHS led to pleiotropic effects. Detached leaves from DHS-suppressed plants exhibited delayed post-harvest senescence in Arabidopsis thaliana (Wang et al., 2003). In A. thaliana, leaf-specific suppression of DHS dramatically increased growth and delayed leaf senescence without negative pleiotropic effects (Duguay et al., 2007). In tomato, DHS silencing delayed fruit softening and resulted in male sterility plants (Wang et al., 2005a). In canola (Brassica napus), transgenic plants with DHS suppression exhibited delayed natural leaf senescence, increased leaf size, and subsequent increases in seed yield and tolerance to chronic sublethal stress (Wang et al., 2005a). These studies provided evidence for senescence-induced DHS in tomato and A. thaliana tissues that may facilitate the translation of mRNA species required for programmed cell death (Wang et al., 2001, 2003, 2005a). Recent research shows that alterations in the biosynthesis of hypusine promoted by the DHS silencing in A. thaliana, result in a wide variety of phenotypes affecting many biological processes related with development such as control of flowering time, the shoot and root architecture and root hair phenotypes. Additionally this pathway is needed for the adaptation to challenging growth conditions (presence of salt, glucose and the plant hormone ABA in the growth medium) (Belda-Palazón et al., 2016). In addition, recombinant tomato and tobacco DHS are capable of catalyzing the formation of a deoxyhypusine residue in A. thaliana and tobacco eIF-5A substrates, respectively (Ober and Hartmann, 1999; Wang et al., 2001).
In yeast the key molecular function of eIF5A during translation has been elucidated and it is expected to be fully conserved in every eukaryotic cell (Zuk and Jacobson, 1998; Zanelli and Valentini, 2005; Gregio et al., 2009). eIF5A has been postulated as an RNA-binding protein involved in mRNA transport and metabolism (Xu and Chen, 2001; Xu et al., 2004; Li et al., 2010; Maier et al., 2010). Recent studies have uncovered the crucial function of eIF5A and EF-P, a prokaryotic structural homolog, within the ribosome as a sequence-specific translation factor required for translation of polyproline-rich proteins that may cause ribosome stalling (Doerfel et al., 2013; Gutierrez et al., 2013). In plant, the genetic approaches with transformation for either eIF5A overexpression or antisense have revealed some activities related to the control of cell death processes (Feng et al., 2007; Hopkins et al., 2008; Ren et al., 2013).
In higher plants, the normal development of chloroplasts is associated with leaf color variation (Lv et al., 2015). Chloroplast function is dependent upon proteins encoded both within the plastid and nuclear genomes (Næsted et al., 2004). Many nuclear genes affect overall chloroplast function and encode proteins involved in photosynthesis, plastid metabolism, chloroplast and host cell biogenesis or nuclear-chloroplast traffic and signaling (Barkan et al., 1995; Belcher et al., 2015). Disruptions in these genes lead to abnormal chloroplast development and the sectored chlorotic leaf phenotype. For example, IMMUTANS (IM) codes for the plastid terminal oxidase (PTOX) and is involved in regulation of the plastoquinone pool and carotenoid biosynthesis (Wetzel et al., 1994; Wu et al., 1999; Aluru and Rodermel, 2004). The filamentation temperature-sensitive H (FtsH) protease composed of type A (FtsH1 and FtsH5/VAR1) and B (FtsH2/VAR2 and FtsH8) subunits plays an important role in degradation of the photodamaged D1 protein in the PSII repair cycle and probably acts as a molecular chaperone in chloroplasts (Bailey et al., 2002; Sakamoto et al., 2003). Both ftsh2/var2 and im mutants display more severe leaf variegation in high light compared with low light (Rosso et al., 2009). Toc159 protein disruption results in a non-photosynthetic albino phenotype in A. thaliana mutant ppi2 (Agne et al., 2010). Defects in the vesicle-inducing protein in plastids 1 (VIPP1) gene lead to a pale green phenotype in A. thaliana hcf155 mutant at the early developmental stage (Vothknecht et al., 2012; Zhang et al., 2012). Defects in the thylakoid formation 1 (Thf1) gene cause impaired thylakoid formation and variegated leaves in A. thaliana (Wang et al., 2004). In cpSRP43 (Chloroplast Signal Recognition Particle 43) rice (Oryza sativa) mutants, chlorophyll and carotenoid levels are significantly reduced, and chloroplast development is impaired (Lv et al., 2015).
Here, full-length petunia PhDHS cDNA was cloned, and PhDHS protein was localized in the cytoplasm and nucleus. VIGS-mediated suppression of PhDHS reduced chlorophyll levels and resulted in abnormal chloroplast ultrastructure in leaves, suggesting the crucial function of PhDHS in chloroplast development. Proteome analysis revealed that the proteins involved in photosystem I (PSI) and photosystem II (PSII) were significantly reduced in PhDHS-silenced plants, indicating that PhDHS is associated with photosynthesis in petunia.
Materials and Methods
Plant Material
Petunia ‘Ultra’ plants purchased from Guangzhou Sanli Horticulture Co., Ltd. were grown under greenhouse conditions (22 ± 2°C, 14-h light/10-h dark). When the plants were ∼10 cm in height, roots, stems, and leaves were collected at the vegetative stage. At anthesis stages, 7–10 petunia flowers were harvested and placed in distilled water. Corollas were collected 1 day after flower opening. Petunia flowers were treated with ethylene according to previously described protocols (Tan et al., 2014). All tissues were frozen in liquid nitrogen and stored at −80°C until use in further experiments. All experiments were conducted at least thrice.
RNA Extraction, RT-PCR and Cloning of PhDHS and PhCH42 Genes
Total RNA was extracted and reverse transcription PCR (RT-PCR) was performed according to the previously described protocols (Liu et al., 2010). To clone the petunia PhDHS and PhCH42 cDNA by RT-PCR, degenerate primers were designed based on conserved sequences in DHS and CH42 from A. thaliana (AtDHS and AtCH42) and tomato (SlDHS and SlCH42) (Supplementary Table S1). The reaction produced 630- and 720-bp PCR products from petunia cDNA. The remaining 5′ and 3′ cDNA sequences were isolated by rapid-amplification of cDNA ends (RACE), and full-length cDNAs for these genes were isolated using the specific primers (Supplementary Table S1).
Sequence Analysis
The neighbour-joining trees at an amino acid level were generated using DNAMAN software (Lynnon Corporation, Quebec, Canada). The reliability of each branch of the tree was assessed using 1,000 bootstrap replications. An identity search for nucleotides and amino acids was performed using the National Center for Biotechnology Information (NCBI) BLAST network server1.
Quantitative Real-Time PCR Assays
PCR analysis was performed using cDNA extracted from different samples as a template. Specific primers were designed using the sequences of PhDHS (Peaxi162Scf00178g01719.12), PhPDS (Accession no. AY593974), PhCH42, and four PheIF5A homologs, PheIF5A-1 (Peaxi162Scf00274g00004.1), PheIF5A-2 (Peaxi162Scf00062g00224.1), PheIF5A-3 (Peaxi162Scf00401g00830.1), and PheIF5A-4 (Peaxi162Scf00097g00108.1). Quantitative real-time PCR (qPCR) was performed on a LightCycler® 480 Real-Time PCR system (Roche). Samples were subjected to thermal-cycling conditions of DNA polymerase activation at 95°C for 4 min; 40 cycles of 45 s at 95°C, 45 s at 52°C or 55°C, 45 s at 72°C, and 45 s at 80°C; and a final elongation step of 7 min at 72°C. The amplicon was analyzed by electrophoresis and sequenced once for identity confirmation. Primer specificity was determined by melting curve analysis; a single, sharp peak in the melting curve ensured that a single, specific DNA species had been amplified. Quantification was based on analysis of the threshold cycle (Ct) value as described by Pfaffl (2001). Petunia Actin, which served as an internal reference gene (accession no. FN014209), was subject to quantitative PCR to quantify cDNA abundance (Supplementary Table S2).
Subcellular Localization
For protein subcellular localization, the ORF of PhDHS was inserted into the modified pSAT-1403TZ (Tzfira et al., 2005) plasmid (which contained GFP under the control of the CaMV 35S promoter) to form the pSAT-35S:GFP-DHS vector. The sequences of all the primers that were used for subcellular localization are described in Supplementary Table S3. The fusion vector and the control vector (35S:GFP) were separately transformed into petunia protoplasts. Rosette leaves of 5- to 6-week-old petunia plants were used for the isolation of protoplasts. Transformation was performed as described by Spitzer-Rimon et al. (2012). The relevant vectors were used in the polyethylene glycol-mediated transformation of the petunia protoplasts. The protoplasts were assayed for fluorescence 12–24 h after transformation. Digital images were captured using a confocal laser scanning system (Nikon ECLIPSE TE2000-E, Nikon Corporation, Tokyo, Japan).
Construction of VIGS Vectors
Specific forward and reverse primers of PhDHS, PhPDS, and PhCH42 were designed to clone approximately 250 bp of the 3′ and 5′ untranslated regions of these genes, and the PCR products were inserted into the pTRV2 vector to form pTRV2-PhDHS, pTRV2-PhDHS-5UTR, pTRV2-PhPDS and pTRV2-PhCH42 vectors, respectively (Supplementary Table S4). Agrobacterium tumefaciens (strain GV3101) was transformed with the pTRV2 and pTRV1 vectors, and the derivatives were prepared according to previously described protocols (Spitzer-Rimon et al., 2010). The Agrobacterium culture was grown overnight at 28°C in Luria-Bertani medium with 50 mg L−1 kanamycin and 200 mM acetosyringone. The cells were harvested and resuspended in inoculation buffer containing 10 mM MES, pH 5.5, 200 mM acetosyringone, and 10 mM MgCl2 to an OD600 of 10. Following an additional 3-h incubation at 28°C, bacteria containing pTRV1 were mixed with bacteria containing the pTRV2 derivatives in a 1:1 ratio. Next, 200 to 400 mL of this mixture was applied to the cut surface of 4-week-old petunia plantlets after the removal of the apical meristems.
Pigment Profiling
Four to five leaves were harvested and freeze-dried, and three replicate methanol extractions were prepared for each type of plant using the method of Wellburn (1994). The absorbance of the solution was red at 646.8, 663.2, and 470.0 nm against the solvent (acetone) blank. The individual concentrations of chlorophyll a, chlorophyll b, total chlorophyll, and total carotenoids (xanthophylls and carotenes) were measured by spectrophotometer and calculated using the equations given here (Lichtenthaler, 1987).
Chlorophyll Fluorescence Measurement
Chlorophyll fluorescence was measured using Dual-PAM-100/F (Walz, Effeltrich, Germany). The third fully expanded leaves from the top were dark-adapted for 30 min prior to measurement. Minimal fluorescence (F0) was measured under a weak pulse of modulating light, and maximal fluorescence (Fm) was induced by a saturating pulse of light (20,000 μmol m−2 s−1) applied. The values were used to calculate maximum quantum yield of photosystem II (PSII) photochemistry [Fv/Fm = (Fm − F0)/Fm].
Microscopic Examination
Confocal laser microscopic, transmission electron microscopic and optical microscopic examinations were performed according to previous studies (Waters et al., 2008; Yang et al., 2017).
Iodine and Trypan Blue Staining and Measurement of Ion Leakage
Starch was visualized in petunia rosettes by iodine staining as previously described (Bahaji et al., 2011; Ovecka et al., 2012), and trypan blue was used to reveal dead cells in petunia leaves using optical microscopy (Koch and Slusarenko, 1990).
Membrane ion leakage was determined by measuring electrolytes released from the leaves according to previous protocal (Song et al., 2016). Leaf samples were immersed into deionised water at 25°C with gentle shaking for 30 min. The conductivity was measured using a conductivity meter (CON 510, Oakton, VA, United States).
Paraffin Sections
Paraffin sections were prepared according to our previous studies (Yang et al., 2017). Briefly, the leaves from plants were cut into 5 mm × 5 mm × 5 mm pieces, fixed in FAA fixative solution (every 100 ml of FAA fixative solution contains 90 ml of 50% or 70% ethanol, 5 ml of acetic acid, and of 5 ml formalin) for 24 h at 25 ± 2°C, and then washed in running water for 24 h. Samples were stained with haematoxylin for 4 days, washed for another 24 h, and dehydrated in increasing grades of ethanol. A graded chloroform series was used for clearing, and the samples were embedded in paraffin. Paraffin sections were cut to a thickness of 8 μm on a Leica RM2235 followed by dewaxing with xylene. Finally, slides were sealed with neutral resin. Sections were observed and photographed with a Zeiss Scope.A1 microscope, and the thickness of the leaves was calculated according to the scale of the microscope.
Protein Extraction, Trypsin Digestion, TMT Labeling, HPLC Fractionation and LC-MS/MS Analysis
Protein extraction, trypsin digestion, TMT labeling and HPLC fractionation were performed according to our previous study (Guo et al., 2016). LC-MS/MS analysis was performed according to previously described protocols (Wu et al., 2015). Three biological replicates were performed.
Database Search
The resulting MS/MS data were processed with MaxQuant (v.1.4.1.2) (Cox and Mann, 2008). Tandem mass spectra were searched against a database (40,341 sequences) created based on the RNA sequencing of petunias in our previous study (Guo et al., 2016).
Bioinformatics Analysis
Bioinformatics analysis was performed according to previously described protocols (Xie et al., 2015). Gene Ontology (GO) term association and enrichment analysis were performed using DAVID (Database for Annotation, Visualization and Integrated Discovery3). The KEGG pathway database was used to identify enriched metabolic pathways4.
Protein Quantitative Ratio Analysis
Protein quantitative ratio was calculated as the median ratio of all unique peptides. Student’s t-test was performed to investigate the effects of differentially expressed proteins (DEPs). To meet the conditions for Student’s t-test, logarithmic transformation was performed to obtain the ratio of all peptides. Then, Student’s t-test was employed to calculate the p-value.
Western Blotting
Western blot analyses were performed according to the methods of Guo et al. (2017). The synthetic peptides (KRWLHFFMLFVPVT, PKIQGNLSSDEE-KYS and RIPVFCPGLTDGSLG) of three proteins, PsbD (Peaxi162Scf00060g00176.1), PsbQ1 (Peaxi162Scf00222g00618.1) and PhDHS (Peaxi162Scf00178g01719.1), were used as an antigen for antibody production in rabbits 5 (PTM BioLab, Inc.), and these antibodies were used for blotting analysis. Proteins were separated using SDS-polyacrylamide gel electrophoresis (PAGE; 10% acrylamide gels) and blotted onto nitrocellulose membranes. The membrane was blocked with 5% skim milk and 0.05% Tween 20 in Tris-buffered saline (50 mM Tris-HCl, pH 8.0, 150 mM NaCl). Purified antibodies were used at a concentration of 50 mg/ml. The membrane was washed with 0.05% Tween 20 in Tris-buffered saline and then reacted with horseradish peroxidase-conjugated goat anti-rabbit IgG (Pierce) at a dilution of 1:20,000. Detection was achieved using SuperSignal West FemtoTM (Pierce). Three biological replicates were performed.
Results
Isolation and Sequence Analyses of PhDHS cDNAs
Full-length DHS cDNA was isolated from petunia ‘Ultra’ and was designated as PhDHS, which was predicted to encode a 375-amino acid protein. Multiple sequence alignments of DHS of petunia, A. thaliana, S. cerevisiae, Drosophila melanogaster, and Homo sapiens are presented in Supplementary Figure S1A. The phylogenetic trees based on evolutionary distances were constructed from DHS amino acid sequences using the DNAMAN program (Supplementary Figure S1B).
The deduced amino acid sequence of PhDHS shares 74.2, 57.0, 54.7, and 59.7% identity with A. thaliana AtDHS, S. cerevisiae ScDHS, D. melanogaster DmDHS, and H. sapiens HsDHS, respectively (Supplementary Table S5). DHS protein alignment performed with the DNAMAN program is presented in Supplementary Figure S1A. These results revealed considerable conservation of sequence identity, especially in the C-terminal active site, which includes a conserved region of six amino acids from Glu 330 to Lys 335 (Supplementary Figure S1A) (Yan et al., 1996; Duguay et al., 2007). The NAD-binding site (positions 97–349) is strongly conserved in all species.
Expression Profile of PhDHS Gene
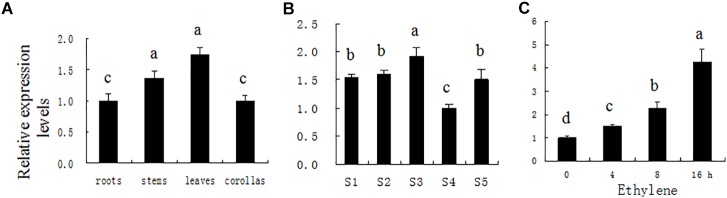
PhDHS expression was examined in different plant organs at different leaf developmental stages and in response to ethylene, a flower-senescence hormone by quantitative real-time PCR (qPCR), given that a previous study suggested that DHS is involved in fruit softening and senescence (Wang et al., 2005a). PhDHS transcription was strong in the leaves, and no significant difference was noted between roots and corollas (Figure 1A).
FIGURE 1.
PhDHS expression patterns determined by quantitative real-time PCR in different organs (A) during leaf development (B) and in leaf in response to 2 μl l−1 of exogenous ethylene (C). Petunia Actin (accession no. FN014209) served as an internal reference gene. S1, young leaves at 0.5 cm; S2, young leaves at 1.0 cm; S3, young leaves at 2.5 cm; S4, mature leaves; S5, aging leaves. The relative expression levels are presented as fold-change values. The data are presented as the mean ± SD (n = 3). Different letters indicate significant differences at the P = 0.05 level.
To assess PhDHS expression during leaf development, leaf development is divided into five stages: S1 (young leaf, 0.5 cm in length), S2 (young leaf, 1.0 cm in length), S3 (young leaf, 2.5 cm in length), S4 (mature leaf, approximately 4.5 cm), and S5 (aging leaf). High PhDHS expression is noted in young and senescence stages, whereas low expression is observed in the mature stage (Figure 1B). In addition, 2 μL L−1 ethylene treatment significantly increased PhDHS expression in leaves (Figure 1C).
PhDHS Protein Is Localized to the Cytoplasm and Nucleus
To examine the cellular localization of PhDHS in plant cells, the full-length form of the protein fused to GFP at the C-terminal end was constructed and transiently expressed in petunia leaf protoplasts under the control of the CaMV 35S promoter. After 16–24 h, accumulation of the PhDHS-GFP fusion protein was detected in the cytoplasm and nucleus of petunia protoplasts (Figure 2A,B). To further verify the fluorescence signal of PhDHS-GFP fusion protein in the nucleus, we use EOBII-RFP (red fluorescent protein) as a nuclear marker given that it was previously demonstrated that EOBI are nuclear-localized MYB factors (Spitzer-Rimon et al., 2010, 2012; Liu et al., 2017). Localization of the PhDHS-GFP fusion protein was determined by visualization with a fluorescence microscope. The results showed that the fluorescence signal of the PhDHS-GFP fusion protein appears in the cytoplasm and nucleus (Figure 2C).
FIGURE 2.
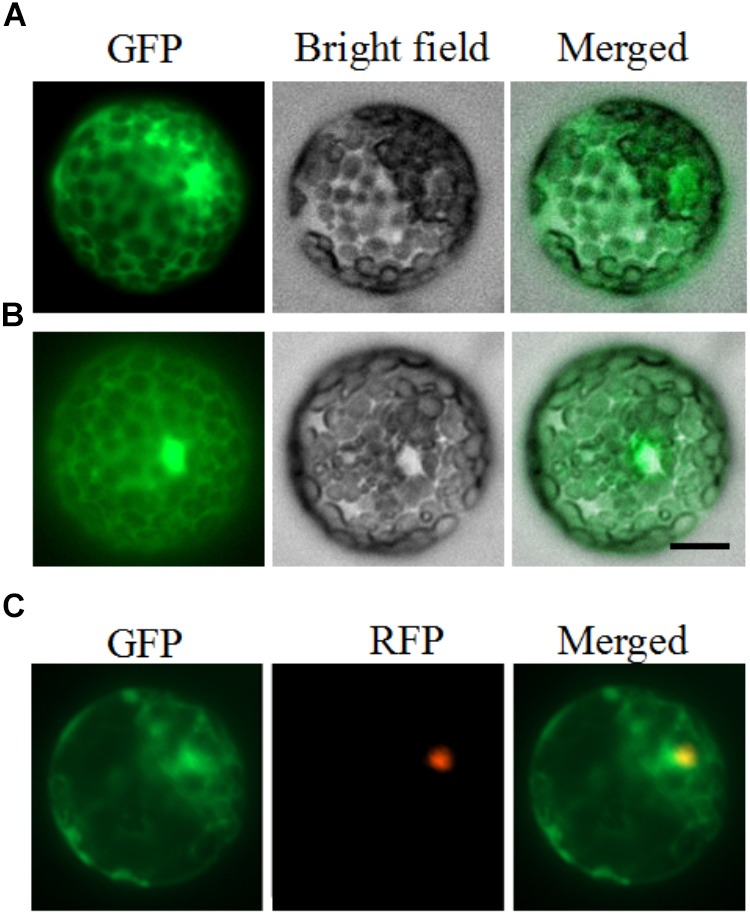
Subcellular localization of PhDHS. Petunia protoplasts were transfected with a construct carrying GFP or PhDHS-GFP under the control of CaMV 35S promoter to assess subcellular localization. (A) Expression of PhDHS-GFP fusion protein. (B) Expression of GFP protein. (C) Subcellular localization of PhDHS-GFP with EOBI-RFP as makers. All images were captured with a confocal laser scanning system. Scale bar: 5 μm.
Phenotypes of PhDHS-Silenced Plants
To suppress PhDHS expression, a VIGS system based on the pTRV2 vector was used in petunia ‘Ultra’ (Tan et al., 2014). An approximately 250-bp fragment of the 3′ untranslated sequence of PhDHS cDNA was inserted into the pTRV2 vector to form the pTRV2-PhDHS vector. Thirty to thirty-five petunia plants were used for infection. PhDHS expression in both mRNA and protein level in leaves of pTRV2-PhDHS-infected plants decreased significantly compared with pTRV2-infected plants (Supplementary Figure S2).
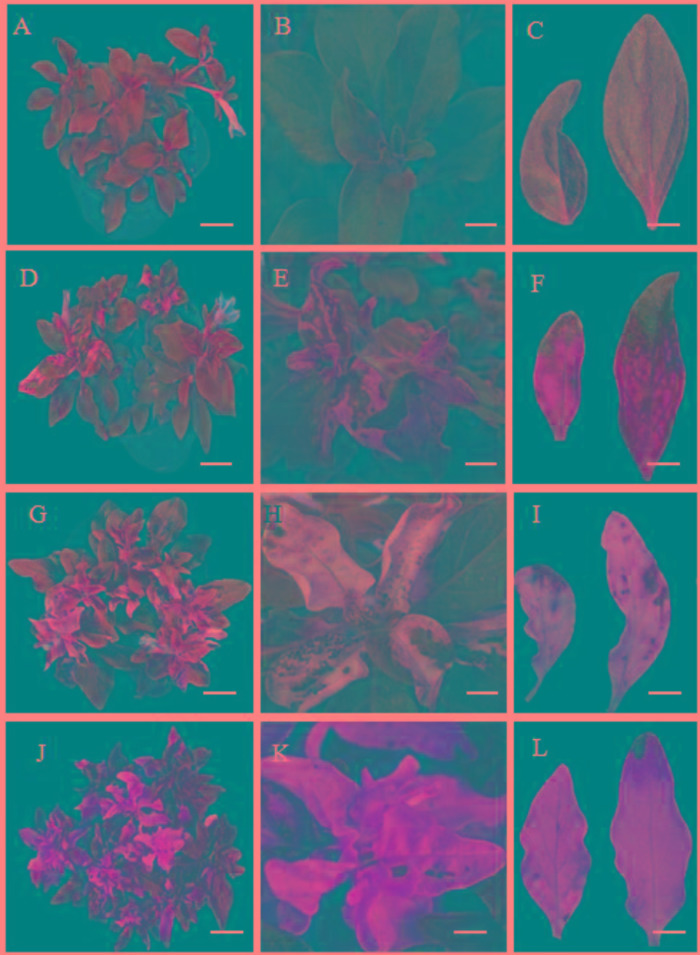
Four to five weeks after petunia seedlings were infected with a mixture of A. tumefaciens transformed with the pTRV2-PhDHS vector and A. tumefaciens was transformed with pTRV1 according to the protocol of Liu et al. (2002), the newly emerged leaves of pTRV2 empty vector-infected plants (control) remained green (Figure 3A–C), whereas those of pTRV2-PhDHS-infected plants exhibited a chlorotic leaf phenotype (Figure 3D–F). Some leaves were completely yellow. Some leaves exhibited reduced chlorophyll in the vicinity of vascular bundles of the systemic leaves. Some plants had leaves with yellow sectors (Figure 3D–F). Additionally, sepals also appeared yellow in PhDHS-silenced plants (Supplementary Figure S3).
FIGURE 3.
Phenotype of pTRV2 empty vector-infected plants (control) and PhDHS-, PhPDS-, and PhCH42-silenced plants. Seven-week-old plants exhibiting different leaf color in pTRV2 control (A–C), PhDHS- (D–F), PhPDS- (G–I), and PhCH42 (J–L)-silenced plants. (A,D,G,J) Scale bar, 6.0 cm. (B,E,H,K) Scale bar, 1.3 cm. (C,F,I,L) Scale bar, 1.0 cm.
As a control, we silenced the petunia Phytoene desaturase (PhPDS) gene (Accession no. AY593974), which results in a photobleaching phenotype due to inhibition of carotenoid biosynthesis (Ratcliff et al., 2001; Liu et al., 2002; Turnage et al., 2002), and the homolog of the A. tumefaciens Chlorata42 gene Petunia Chlorata42 (PhCH42, Peaxi162Scf00269g00514.1), which produces a yellow-leaf phenotype (Ratcliff et al., 2001; Liu et al., 2002; Turnage et al., 2002). PDS and CH42 have been used as marker genes for the effectiveness of VIGS in several studies (Burch-Smith et al., 2006).
The sectored chlorotic leaves and sepals were obvious in PhPDS- and PhCH42-silenced plants. The silencing of PhPDS and PhCH42 produces typical white and yellow colors, respectively, in leaves, which occurs in the absence of the genes products (Figure 3G–L). Of note, PhCH42-silenced plant exhibit more yellow leaves compared with PhDHS-silenced plants (Figure 3J–L). No visible symptoms of pTRV2 infection were observed in these plants, and these plants were indistinguishable from pTRV2-infected plants. The levels of gene silencing were monitored by qPCR, and an 83 and 85% reduction in PhPDS and PhCH42 transcriptional levels, respectively, was noted in sectored chlorotic leaves (Supplementary Figures S4A, B).
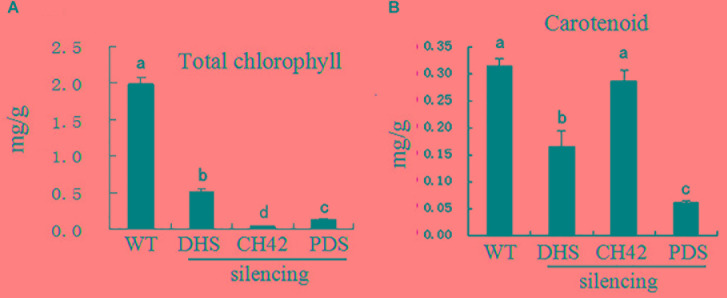
Leaf color is typically associated with chlorophyll and carotenoid levels (Kim and An, 2013). We thus investigated chlorophyll and carotenoid levels in leaves from PhDHS-, PhPDS-, and PhCH42-silenced plants. As shown in Figure 4, the silencing of three genes significantly reduced chlorophyll content in leaves compared with the control. Leaves from PhPDS- and PhCH42-silenced plants exhibited reduced chlorophyll content compared with PhDHS-silenced plants. Both PhDHS and PhPDS silencing reduced carotenoid content in leaves compared with the control, whereas PhCH42 silencing produced the same carotenoid level as the control. In addition, leaves from PhPDS-silenced plants exhibited less carotenoid levels compared with PhDHS-silenced plants.
FIGURE 4.
Effects of PhDHS, PhPDS and PhCH42 silencing on the photosynthetic pigment content of leaf mesophyll cells from 5-week-old plants. (A) Total chlorophyll; (B) Carotenoid. Data are presented as the mean ± SD (n = 3). Different letters indicate significant differences at the P = 0.05 level.
Effects of PhDHS silencing on maximum quantum yield of PSII photochemistry (Fv/Fm) in petunia leaves were examined. Compared to the control, there was a significant decrease in Fv/Fm in leaves of PhDHS silencing and the values of Fv/Fm were decreased by 18.55% (Supplementary Figure S5), which showed PSII activity in the leaves of PhDHS silencing were reduced.
To visualize starch accumulation, we used iodine staining of young leaves at the S3 stage and mature leaves from 5-week-old PhDHS-silenced and pTRV2-infected plants. The staining revealed that the control leaves retained more starch compared with PhDHS-silenced plants (Supplementary Figure S6).
In addition, PhDHS silencing resulted in extended leaf longevity. The detached leaves exhibited a normal configuration in PhDHS-silenced plants, whereas leaf senescence was noted 10 days after harvest in pTRV2-infected plants (Supplementary Figure S7A). Trypan blue staining of leaves revealed an increased number of dead cells in pTRV2-infected plants compared with PhDHS-silenced leaves 10 days after harvest (Supplementary Figure S7B). Moreover, measurement of ion leakage, which indicates the plasma membrane integrity, revealed that the plasma membrane of leaves of pTRV2-PhDHS-infected plants was more intact than leaves of pTRV2-infected plants 5 weeks after infection, which further confirmed that PhDHS silencing delayed senescence (Supplementary Figure S8).
The cross-sections of leaves pTRV2- and pTRV2-PhDHS-infected plants were prepared, revealing that mature leaves of pTRV2-PhDHS-infected plant were thicker compared with control (Supplementary Figure S9). The thickness of leaves in PhDHS-silenced and pTRV2-infected plants was 83 ± 6 μm and 63 ± 7 μm, respectively. In addition, the height of pTRV2-PhDHS-infected plants (22.76 ± 0.90 cm) did not show significantly difference compared with that of the pTRV2-PhDHS-infected plants (22.13 ± 1.84 cm).
Although the proper gene silencing for PhDHS was showed, the phenotypes uncovered and assigned to the expression level of PhDHS cannot be allocated exclusively to down-regulation of PhDHS gene expression as off-targets can be generated by the approach used. So, a 253-bp fragment of the 5′ untranslated sequence of PhDHS cDNA was inserted into the pTRV2 vector to form the pTRV2-PhDHS-5UTR vector. Four to five weeks after infection, a phenotype similar to the plants treated with pTRV2-PhDHS was exhibited in the petunia plants treated with pTRV2-PhDHS-5UTR (Supplementary Figure S2). The newly emerged leaves of pTRV2-PhDHS-5UTR-infected plants also exhibited a chlorotic leaf phenotype (Supplementary Figure S10). PhDHS expression in leaves of pTRV2-PhDHS-5UTR-infected plants decreased significantly compared with pTRV2-infected plants (Supplementary Figure S11).
PhDHS Suppression Affects Chloroplast Development
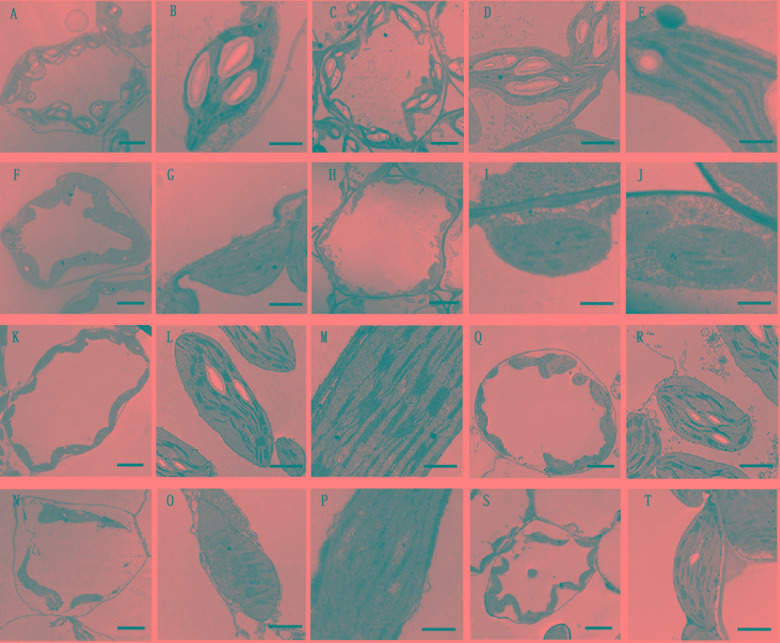
Given that the chlorophyll content deceased in PhDHS-silenced plants, we hypothesized that the PhDHS silencing phenotype resulted from the abnormal development of chloroplasts in mesophyll cells. To test this hypothesis, we observed the ultrastructure of chloroplasts of PhDHS-silenced and pTRV2-infected plants at four developmental stages, S1, S2, S3 and S4, by transmission electron microscopy (Figure 5).
FIGURE 5.
Effects of PhDHS silencing on chloroplast development. (A–J) Chloroplast ultrastructure of mesophyll chloroplasts in the S1 (A,B,F,G) and S2 (C–E,H–J) stages of leaves in 5-week-old pTRV2 empty vector-infected (A–E) and PhDHS-silenced (F–J) plants. (A,C,F,H) Overviews depicting the chloroplast morphology and arrangement within the cells (scale bar, 5 μm). (B,D,G,I) Inside chloroplast views depicting thylakoid arrangement (scale bar, 1 μm). (E,I) Higher magnification depicting the grana stacking (scale bar, 0.5 μm). (K–P) Chloroplast ultrastructure of mesophyll chloroplasts in the S3 stage of leaves in 5-week-old pTRV2 empty vector-infected (K–M) and PhDHS-silenced (N–P) plants. (K,N) Overviews depicting the chloroplast morphology and arrangement within the cells (scale bar, 5 μm). (L,O) Inside chloroplast views depicting thylakoid arrangement (scale bar, 1 μm). (M,P) Higher magnification depicting the grana stacking (scale bar, 0.5 μm). (Q–T) Chloroplast ultrastructure in the leaves of S4 stage in pTRV2 empty vector-infected (Q,R) and PhDHS-silenced (S,T) plants. (Q,S) Overviews depicting the chloroplast morphology and arrangement within the cells (scale bar, 5 μm). (R,T) Inside chloroplast views depicting thylakoid arrangement (scale bar, 1 μm).
In the S1 and S2 stages of leaves, the thylakoid arrangement in chloroplasts of middle parts of leaves in PhDHS-silenced and pTRV2-infected plants was regular (Figure 5A–J). However, the size of grana in chloroplasts and starch granules in chloroplasts in PhDHS-silenced plants were reduced compared with the control.
In S3 stage leaves from pTRV2-infected plants, individual ellipsoidal chloroplasts of the middle leaves were aligned along the cytoplasmic membrane. These chloroplasts contained starch granules, and their internal structures were completely developed (Figure 5K–M). The lamellae in chloroplasts were oriented along the convex side facing the interior of the cell (Figure 5K). In contrast, the chloroplasts of yellow areas of PhDHS-silenced leaves were detached from the plasma membrane (Figure 5N). Grana thylakoids were significantly decreased (or even completely lost) with discontinuous distribution in PhDHS-silenced plants. A loose arrangement of grana was noted, and some grana were even arranged in the vertical axis of the chloroplast in PhDHS-silenced plants (Figure 5O,P). Fewer starch granules in chloroplasts were observed in PhDHS-silenced plants (Figure 5N,P).
In S4 stage leaves, grana thylakoids and starch granules in some chloroplasts were slightly increased compared with S3 stage leaves in PhDHS-silenced plants; however, the levels were slightly reduced compared with the control (Figure 5Q–T).
Effects of PhDHS Silencing on the mRNA Levels of PheIF5As
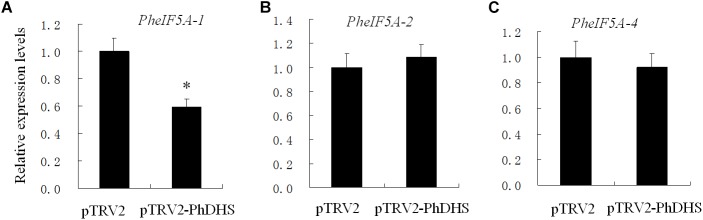
Given that previous studies suggested that eIF5A is the only substrate of DHS, the effect of PhDHS silencing on PheIF5A expression was examined. Four (PheIF5A-1, PheIF5A-2, PheIF5A-3, and PheIF5A-4) and three (AteIF5A-1, At1g13950; AteIF5A-2, At1g26630; AteIF5A-3, and At1g69410) eIF5A homologs are present in the petunia and A. thaliana genome, respectively (Hopkins et al., 2008). The phylogenetic tree demonstrated that the predicted four PheIF5A proteins were classified as one group, whereas three AteIF5A proteins belong to another group (Supplementary Figure S12). The qPCR assays showed that in the leaves of PhDHS–silenced plants, PheIF5A-2 and PheIF5A-4 mRNA levels did not significantly change compared with levels in pTRV2-infected plants, whereas PheIF5A-1 expression was significantly reduced (Figure 6). In addition, PheIF5A-3 mRNA was not detected in leaves of both pTRV2-infected and pTRV2-PhDHS-infected plants. We further examined the expression patterns of PheIF5A-1, PheIF5A-2, and PheIF5A-4 in different organs. The results revealed that PheIF5A-1 and PheIF5A-2 have higher expression level in corollas and leaves than that in stems and roots, while PheIF5A-4 show highest expression level in roots (Supplementary Figure S13).
FIGURE 6.
Effects of PhDHS silencing on the expression of PheIF5A-1 (A), PheIF5A-2 (B), and PheIF5A-4 (C) by quantitative real-time PCR. Petunia Actin (accession no. FN014209) served as an internal reference gene. Relative expression levels are presented as fold-change values (1 = time 0). Data are presented as the means ± SD (n = 3).
PhDHS Suppression Resulted in the Reduction of Proteins Associated With Photosynthesis
To characterize the sectored chlorotic leaves in PhDHS-silenced plants, we quantitatively investigated the petunia proteome of S3 stage leaves in pTRV2-infected plants and PhDHS-silenced plants by iTRAQ. Tandem mass spectra were searched against the transcriptome sequences (SRA accession: SRP077541) we previously constructed to analyze the proteome (Guo et al., 2016). In the proteome, a total of 3763 proteins were identified, among which 3101 proteins were quantified. The fold-change cut-off established a quantitative ratio greater than 1.5 or less than 0.66 as significant. Among the quantified proteins, 308 proteins were upregulated and 266 proteins were downregulated in PhDHS-silenced plants compared with control with high repeatability (Supplementary Figure S14 and Supplementary Data 1).
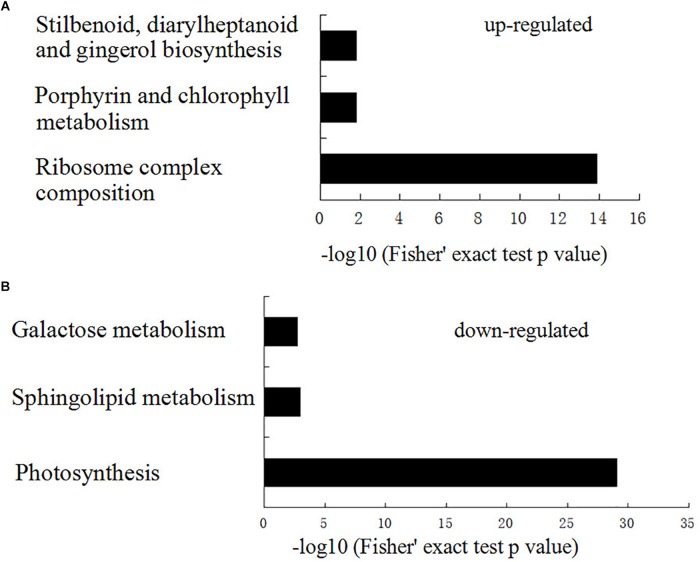
To elucidate the functional differences of the upregulated and downregulated proteins, the quantified proteins were analyzed for KEGG pathway enrichment based on clustering analysis (Supplementary Data 2). KEGG pathway enrichment-based clustering analysis revealed that ribosome complex composition, porphyrin and chlorophyll metabolism, and stilbenoid, diarylheptanoid, and gingerol biosynthesis were the most prominent pathways enriched in quantiles with increased protein levels in PhDHS-silenced leaves (Figure 7). In contrast, photosynthesis, sphingolipid metabolism, and galactose metabolism pathways were reduced in PhDHS-silenced leaves (Figure 7 and Supplementary Figures S15–S20).
FIGURE 7.
KEGG pathway-based enrichment analysis of up- (A) and down- (B) regulated proteins in PhDHS-silenced plants compared with control.
Given that DHS catalyzes eIF5A function in protein translation, we focused on our attention on the downregulated proteins. Ten proteins among 16 subunit proteins in the PSI complex (PsaC, PsaD1, PsaD2, PsaE1, PsaE2, PsaF, PsaK, PsaL1, PsaL2, and PsaN) and 11 proteins among 27 subunit proteins in the PSII complex (Psb27, PsbB, PsbC, PsbD, PsbO2A, PsbO2B, PsbP2, PsbQ1, PsbQ2A, PsbQ2B, and PsbR) were significantly reduced (Supplementary Table S6). Among them, PsbO2A and PsaE protein levels in PhDHS-silenced plants were reduced to one-fifth of those in control. In addition, twelve thylakoid membrane or lumenal proteins were also significantly reduced in PhDHS-silenced leaves compared with control (Supplementary Table S7).
Confirmation of the Petunia Proteome by Western Blotting
To confirm the petunia proteome results of S3 stage leaves in pTRV2-infected plants and PhDHS-silenced plants by iTRAQ, we random selected 2 protein, photosystem II D2 protein (PsbD, Peaxi162Scf00060g00176.1) and PsbQ1 (Peaxi162Scf00222g00618.1), to perform western blotting using the antibodies raised against these proteins. The results showed that protein abundance was reduced in PhDHS-silenced plants compared with plants treated with pTRV2 vector (Supplementary Figure S21), which is consistent with that in the petunia proteome as demonstrated by iTRAQ.
Discussion
In the present study, the PhDHS-silencing phenotype in petunia was obtained by using the VIGS system, and we demonstrated that DHS plays important roles in chloroplast development.
DHS and eIF5A full-length cDNAs were isolated from A. thaliana, canola, tomato, tobacco and rice (Chamot and Kuhlemeier, 1992; Mehta et al., 1994; Wang et al., 2001; Wang et al., 2005b). Studies to date have indicated that DHS is a single gene, whereas eIF5A is encoded by a multi-gene family (Wang et al., 2005a; Duguay et al., 2007). In this study, full-length petunia PhDHS cDNA was isolated. The amino acid identity between the petunia PhDHS and the H. sapiens enzyme is 59.7%. The identity between the petunia PhDHS and the D. melanogaster enzyme is 54.7%, the lowest identity among the petunia, H. sapiens, the A. thaliana, D. melanogaster, and S. cerevisiae enzyme, suggesting that DHS is highly conserved in eukaryotes.
Similar to the expression of AtDHS and SlDHS observed in A. thaliana and tomato (Wang et al., 2005a), PhDHS expression changes both spatially and temporally as development progresses in petunia. PhDHS was expressed at high levels in young and senescent leaves (Figure 1B). This finding is consistent with the finding that suppression of DHS resulted in abnormal development of chloroplasts in leaves and delayed leaf senescence (Figure 5 and Supplementary Figure S7). Treatment with the senescence hormone ethylene increased PhDHS expression, which further implicated the probable involvement of PhDHS in leaf senescence. This finding is consistent with previous studies demonstrating that DHS protein levels in leaves were upregulated in response to both natural senescence and stress-induced premature senescence in tomato and A. thaliana (Wang et al., 2001, 2003, 2005a).
Although VIGS-mediated silencing of PhDHS, PhPDS, and PhCH42 resulted in sectored chlorotic leaf phenotype, the leaf color in the plants with three genes silenced was not completely identical. Moreover, the carotenoid and chlorophyll content in PhDHS-silenced plants differed from that in PhPDS- and PhCH42-silenced plants. These results indicate that the molecular mechanism of the sectored chlorotic leaves differed among plants with these genes silenced. PhPDS and PhCH42 are the key enzymes of carotenoid and chlorophyll biosynthesis, respectively (Ratcliff et al., 2001; Liu et al., 2002; Turnage et al., 2002).
Measurements of photosynthetic pigments revealed that carotenoid levels in PhDHS-silenced plants were 50% of that in pTRV2-infected plants, whereas chlorophyll a and b levels in PhDHS-silenced plants were approximately 27 and 20% of those in pTRV2-infected plants, respectively. This finding indicated that loss of DHS function has a greater effect on chlorophyll levels than carotenoid levels. Iodine staining of leaves revealed that the chloroplasts of pTRV2-infected leaves retained more starch granules compared with PhDHS-silenced plants (Supplementary Figure S6). The reduced starch granules in the chloroplasts of leaves of PhDHS-silenced plants might be attributed to the reduction in total chlorophyll levels and abnormal chloroplast development.
Recent research shows that AtDHS silencing resulted in early flowering by the increasing of expression of AtFT gene, one of key genes involved in the transition process, and alterations of shoot and root architecture (Belda-Palazón et al., 2016). PhDHS silencing did not significantly change the height of plants and flowering time and it is probably because the seedlings used to infect by using VIGS method have already bloomed and are tall enough in this study.
Transmission electron microscopy revealed abnormal chloroplast development in PhDHS-silenced plants (Figure 5). These results suggested that PhDHS was required for chloroplast development. Moreover, PhDHS mRNA levels were high in young leaves when chloroplasts were developing. In addition, the mesophyll chloroplasts of leaves in PhDHS-silenced plants exhibited frequent changes in their thylakoid orientation and fewer thylakoids per granal stack (Figure 5).
The proteome assay demonstrated that PhDHS silencing resulted in the reduction of proteins involved in photosynthesis pathways, suggesting that PhDHS is associated with photosynthesis. PSI and PSII complexes are located in the thylakoid membrane, and the assembly of PSI and PSII complexes is associated with thylakoid formation and chloroplast development (Pogson and Albrecht, 2011). In addition, lipid and protein biosynthetic pathways are involved in the integration and formation of thylakoids (Benning, 2009). In this study, 21 proteins in PSI and PSII complexes and 12 thylakoid membrane or lumenal proteins were significantly reduced in PhDHS-silenced leaves compared with control, suggesting that the reduction of photosynthesis proteins could result in sectored chlorotic leaves and abnormal chloroplast development in PhDHS-silenced plants. These results further supported the involvement of PhDHS in chloroplast development. PhDHS silencing significantly decreased the expression of PheIF5A-1 and did not significantly change the transcription levels of PheIF5A-2 and PheIF5A-4 (Figure 6), while in the proteome of this study, the protein levels of PheIF5A1, PheIF5A2, and PheIF5A4 in the leaves of PhDHS-silenced plants increase by 1.224, 1.362, and 1.537 times compared with those in control, while the peptides of PheIF5A3 were not detected, respectively (Supplementary Data 1). These results show that PhDHS silencing promotes the translation of PheIF5A1, PheIF5A2, and PheIF5A4 mRNA since PhDHS silencing did not increase their mRNA levels. The up-regulation of the protein levels of PheIF5A1, PheIF5A2, and PheIF5A4 might be attributed to the decrease of hypusinated PheIF5As resulting from PhDHS silencing in petunia and this may indicate that feedback control is involved in the regulation of PheIF5As translation. PheIF5A-1, PheIF5A-2, PheIF5A-4, and PhDHS exhibit high mRNA levels in petunia leaves (Figure 1 and Supplementary Figure S12). Similar to PhDHS, AteIF5A-2 is localized to the cytoplasm and nucleus (Hopkins et al., 2008). It is not ruled out that PheIF5A-1, PheIF5A-2, and PheIF5A-4, as possible substrates of PhDHS, might play a role in abnormal chloroplast development resulting from PhDHS silencing in petunia.
In addition, it is shown here proteins of PSI and PSII are downregulated in PhDHS-silenced plants and most PSI and PSII proteins are encoded in the chloroplast and translated there, while PhDHS is localized to the cytoplasm and nucleus. Presumably the defects in photosynthesis in this study are a secondary effect. It is possible that the down-regulation of PheIF5A-1 mRNA in PhDHS-silenced plants affects the elongation or termination of the protein encoding by the nuclear genes involved in thylakoid formation, which resulted in the reduction of chloroplast encoded proteins.
PhDHS silencing resulted in extended leaf longevity and increased thickness in leaves. These effects are consistent with DHS silencing in tomato, A. thaliana and canola (Duguay et al., 2007). Similarly, suppression of DHS also delays postharvest senescence of cut carnation flowers (Hopkins et al., 2007). In addition, A. thaliana eIF5A, specifically AteIF5A-2, is involved in plant growth and development by regulating cell division, cell growth, and cell death and pathogen-induced cell death and development of disease symptoms (Feng et al., 2007; Hopkins et al., 2008; Ren et al., 2013). Thus, hypusination of eIF5A appears to be an important element of the regulation of senescence or cell death.
The sectored chlorotic leaves, which appeared in PhDHS-silenced petunias in this study, was not reported in constitutive suppression of DHS in A. thaliana, tomato and canola (Wang et al., 2003, 2005a,b; Duguay et al., 2007). In A. thaliana and tomato, both chlorophyll content and PSII activity proved to be higher for DHS-suppressed leaves than for corresponding wild-type leaves (Wang et al., 2003, 2005a). In contrast, both chlorophyll content and PSII activity in the leaves of PhDHS silencing were reduced compared with the control leaves. These results indicated functional differences of DHSs among different species.
Based on previous studies (Wang et al., 2003; Park et al., 2010), eIF5As are the only substrate of DHS. Although there are four and three eIF5A homologs in petunia and A. thaliana genome, respectively, there is only one DHS isoform, implying that the single isoform of DHS is capable of modifying all four isoforms of eIF-5A. Suppression of DHS and reduction of eIF-5A levels in A. thaliana and/or tomato has dramatic effects on growth and development (Wang et al., 2003, 2005b). These pleiotropic effects arising from the suppression of DHS in A. thaliana and/or tomato indicate that eIF-5A plays a central role in plant growth and development. The functional differences of DHSs between different species might result from different eIF5A substrates or different downstream events of DHS enzyme and possible novel specific functions for eIF5A could explain the phenotypes of PhDHS silencing, and further studies are required.
It seems to be contradictory that PhDHS removal results in chlorophyll loss and chloroplast disruption, which reduces photosynthesis and sugar and starch accumulation and should be associated with accelerated senescence of leaves (Oda-Yamamizo et al., 2016) and that PhDHS silencing delayed senescence. It is possible that sugar and starch output from leaves is reduced, whereas the retained portion in the leaves is not significantly reduced in PhDHS-silenced plants (Wang et al., 2005a). Alternatively, PhDHS silencing results in increased thickness in leaves (Supplementary Figure S8) or likely inhibits the expression of senescence-associated genes at transcriptional and translational levels, which adequately compensates for the effects of the reduction in energy.
The proteome assay showed that besides photosynthesis pathways, the metabolism pathways of sphingolipid, which plays an important role in modulating plant programmed cell death associated with defense (Wang et al., 2008), and the metabolism pathways of galactose, which is involved in xyloglucan synthesis in Arabidopsis (Kong et al., 2015), were reduced in PhDHS-silenced leaves (Supplementary Figures S15–S20). In addition, ribosome complex composition, porphyrin and chlorophyll metabolism, and stilbenoid, diarylheptanoid and gingerol biosynthesis were the most prominent pathways enriched in quantiles with increased protein levels in PhDHS-silenced leaves.
Accession Numbers
The mass spectrometry proteomics data have been deposited to the ProteomeXchange Consortium (Vizcaíno et al., 2010) via the Proteomics Identification Database partner repository with the dataset identifier PXD012227.
Author Contributions
YY and JL planned and designed the research. JL, XC, SZ, BD, LP, JM, and QW performed the experiments, conducted fieldwork, analyzed the data etc. YY and JL wrote the manuscript.
Conflict of Interest Statement
The authors declare that the research was conducted in the absence of any commercial or financial relationships that could be construed as a potential conflict of interest.
Funding. This study was supported by National Natural Science Foundation of China (31770737, 31701953, 31870692, and 31661143047) and National Key Research and Development Plan (SQ2018YFD100015).
Supplementary Material
The Supplementary Material for this article can be found online at: https://www.frontiersin.org/articles/10.3389/fpls.2019.00284/full#supplementary-material
References
- Abbruzzese A., Park M. H., Folk J. E. (1986). Deoxyhypusine hydroxylase from rat testis. Partial purification and characterization. J. Biol. Chem. 261 3085–3089. [PubMed] [Google Scholar]
- Agne B., Andrès C., Montandon C., Christ B., Ertan A., Jung F., et al. (2010). The acidic A-domain of Arabidopsis TOC159 occurs as a hyperphosphorylated protein. Plant Physiol. 153 1016–1030. 10.1104/pp.110.158048 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Aluru M. R., Rodermel S. R. (2004). Control of chloroplast redox by the IMMUTANS terminal oxidase. Physiol. Plant. 120 4–11. 10.1111/j.0031-9317.2004.0217.x [DOI] [PubMed] [Google Scholar]
- Bahaji A., Ovecka M., Bárány I., Risueño M. C., Muñoz F. J., Baroja-Fernández E., et al. (2011). Dual targeting to mitochondria and plastids of AtBT1 and ZmBT1, two members of the mitochondrial carrier family. Plant Cell Physiol. 52 597–609. 10.1093/pcp/pcr019 [DOI] [PubMed] [Google Scholar]
- Bailey S., Thompson E., Nixon P. J., Horton P., Mullineaux C. W., Robinson C., et al. (2002). A critical role for the Var2 FtsH homologue of Arabidopsis thaliana in the photosystem II repair cycle in vivo. J. Biol. Chem. 277 2006–2011. 10.1074/jbc.M105878200 [DOI] [PubMed] [Google Scholar]
- Barkan A., Voelker R., Mendel Hartvig J., Johnson D., Walker M. (1995). Genetic analysis of chloroplast biogenesis in higher plants. Physiol. Plant. 93 163–170. 10.1034/j.1399-3054.1995.930123.x [DOI] [Google Scholar]
- Bartig D., Lemkemeier K., Frank J., Lottspeich F., Klink F. (1992). The archaebacterial hypusine-containing protein. Eur. J. Biochem. 204 751–758. 10.1111/j.1432-1033.1992.tb16690.x [DOI] [PubMed] [Google Scholar]
- Belcher S., Williams-Carrier R., Stiffler N., Barkan A. (2015). Large-scale genetic analysis of chloroplast biogenesis in maize. Biochim. Biophys. Acta 1847 1004–1016. 10.1016/j.bbabio.2015.02.014 [DOI] [PubMed] [Google Scholar]
- Belda-Palazón B., Almendáriz C., Martí E. (2016). Relevance of the axis spermidine/eIF5A for plant growth and development. Front. Plant Sci. 7:245. 10.3389/fpls.2016.00245 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Benning C. (2009). Mechanisms of lipid transport involved in organelle biogenesis in plant cells. Annu. Rev. Cell Dev. 25 71–91. 10.1146/annurev.cellbio.042308.113414 [DOI] [PubMed] [Google Scholar]
- Burch-Smith T. M., Schiff M., Liu Y., Dinesh-Kumar S. P. (2006). Efficient virus-induced gene silencing in Arabidopsis. Plant Physiol. 142 21–27. 10.1104/pp.106.084624 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chamot D., Kuhlemeier C. (1992). Differential expression of genes encoding the hypusine-containing translation initiation factor, elF-5A, in tobacco. Nucleic Acids Res. 20 665–669. 10.1093/nar/20.4.665 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox J., Mann M. (2008). MaxQuant enables high peptide identification rates, individualized p.p.b.-range mass accuracies and proteome-wide protein quantification. Nat. Biotechnol. 26 1367–1372. 10.1038/nbt.1511 [DOI] [PubMed] [Google Scholar]
- Doerfel L. K., Wohlgemuth I., Kothe C., Peske F., Urlaub H., Rodnina M. V. (2013). EF-P is essential for rapid synthesis of proteins containing consecutive proline residues. Science 339 85–88. 10.1126/science.1229017 [DOI] [PubMed] [Google Scholar]
- Duguay J., Jamal S., Liu Z., Wang T., Thompson J. E. (2007). Leaf-specific suppression of deoxyhypusine synthase in Arabidopsis thaliana enhances growth without negative pleiotropic effects. J. Plant Physiol. 164 408–420. 10.1016/j.jplph.2006.02.001 [DOI] [PubMed] [Google Scholar]
- Feng H., Chen Q., Feng J., Zhang J., Yang X., Zuo J. (2007). Functional characterization of the Arabidopsis eukaryotic translation initiation factor 5A-2 that plays a crucial role in plant growth and development by regulating cell division, cell growth, and cell death. Plant Physiol. 144 1531–1545. 10.1104/pp.107.098079 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gordon E. D., Mora R., Meredith S. C., Lee C., Lindquist S. L. (1987). Eukaryotic initiation factor 4D, the hypusine-containing protein, is conserved among eukaryotes. J. Biol. Chem. 262 16585–16589. [PubMed] [Google Scholar]
- Gregio A. P. B., Cano V. P. S., Avaca J. S., Valentini S. R., Zanelli C. F. (2009). eIF5A has a function in the elongation step of translation in yeast. Biochem. Biophys. Res. Commun. 380 785–790. 10.1016/j.bbrc.2009.01.148 [DOI] [PubMed] [Google Scholar]
- Guo J., Liu J., Wei Q., Wang R., Yang W., Ma Y., et al. (2016). Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation. Plant Physiol. 173 668–687. 10.1104/pp.16.00795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Guo J., Liu J., Wei Q., Wang R., Yang W., Ma Y., et al. (2017). Proteomes and ubiquitylomes analysis reveals the involvement of ubiquitination in protein degradation in petunias. Plant Physiol. 173 668–687. 10.1104/pp.16.00795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gutierrez E., Shin B. S., Woolstenhulme C. J., Kim J. R., Saini P., Buskirk A. R., et al. (2013). eIF5A promotes translation of polyproline motifs. Mol. Cell 51 35–45. 10.1016/j.molcel.2013.04.021 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M., Lampi Y., Wang T. W., Liu Z., Thompson J. E. (2008). Eukaryotic translation initiation factor 5A is involved in pathogen-induced cell death and development of disease symptoms in Arabidopsis. Plant Physiol. 148 479–489. 10.1104/pp.108.118869 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hopkins M., Taylor C., Liu Z., Ma F., McNamara L., Wang T. W., et al. (2007). Regulation and execution of molecular disassembley and catabolism during senescence. New Phytol. 175 201–214. 10.1111/j.1469-8137.2007.02118.x [DOI] [PubMed] [Google Scholar]
- Jenkins Z. A., Haag P. G., Johansson H. E. (2001). Human eIF5A2 on chromosome 3q25-q27 is a phylogenetically conserved vertebrate variant of eukaryotic translation initiation factor 5A with tissue-specific expression. Genomics 71 101–109. 10.1006/geno.2000.6418 [DOI] [PubMed] [Google Scholar]
- Kang K. R., Kim Y. S., Wolff E. C., Park M. H. (2007). Specificity of the deoxyhypusine hydroxylase-eukaryotic translation initiation factor (eIF5A) interaction identification of amino acid residues of the enzyme required for binding of its substrate, deoxyhypusine-containing eIF5A. J. Biol. Chem. 282 8300–8308. 10.1074/jbc.M607495200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim S., An G. (2013). Rice chloroplast-localized heat shock protein 70, OsHsp70CP1, is essential for chloroplast development under high-temperature conditions. J. Plant Physiol. 170 854–863. 10.1016/j.jplph.2013.01.006 [DOI] [PubMed] [Google Scholar]
- Koch E., Slusarenko A. (1990). Arabidopsis is susceptible to infection by a downy mildew fungus. Plant Cell 2 437–445. 10.1105/tpc.2.5.437 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kong Y., Pena M. J., Renna L., Avci U., Pattathil S., Tuomivaara S. T., et al. (2015). Galactose-depleted xyloglucan is dysfunctional and leads to dwarfism in Arabidopsis. Plant Physiol. 167 1296–1306. 10.1104/pp.114.255943 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kyrpides N. C., Woese C. R. (1998). Universally conserved translation initiation factors. Proc. Natl. Acad. Sci. U.S.A. 95 224–228. 10.1073/pnas.95.1.224 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li C. H., Ohn T., Ivanov P., Tisdale S., Anderson P. (2010). eIF5A promotes translation elongation, polysome disassembly and stress granule assembly. PLoS One 5:e9942. 10.1371/journal.pone.0009942 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lichtenthaler H. K. (1987). Chlorophylls and carotenoids: pigments of photosynthetic biomembranes. Methods Enzymol. 148 350–382. 10.1016/0076-6879(87)48036-1 [DOI] [Google Scholar]
- Liu F., Xiao Z., Yang L., Chen Q., Shao L., Liu J., et al. (2017). PhERF6, interacting with EOBI, negatively regulates fragrance biosynthesis in petunia flowers. New Phytol. 215 1490–1502. 10.1111/nph.14675 [DOI] [PubMed] [Google Scholar]
- Liu J., Li J., Wang H., Fu Z., Liu J., Yu Y. (2010). Identification and expression analysis of ERF transcription factor genes in petunia during flower senescence and in response to hormone treatments. J. Exp. Bot. 62 825–840. 10.1093/jxb/erq324 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu Y., Schiff M., Dinesh Kumar S. P. (2002). Virus-induced gene silencing in tomato. Plant J. 31 777–786. 10.1046/j.1365-313X.2002.01394.x [DOI] [PubMed] [Google Scholar]
- Lv X., Shi Y., Xu X., Wei Y., Wang H., Zhang X., et al. (2015). Oryza sativa chloroplast signal recognition particle 43 (OscpSRP43) is required for chloroplast development and photosynthesis. PLoS One 10:e0143249. 10.1371/journal.pone.0143249 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maier B., Ogihara T., Trace A. P., Tersey S. A., Robbins R. D., Chakrabarti S. K., et al. (2010). The unique hypusine modification of eIF5A promotes islet β cell inflammation and dysfunction in mice. J. Clin. Invest. 120 2156–2170. 10.1172/JCI38924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mehta A. M., Saftner R. A., Mehta R. A., Davies P. J. (1994). Identification of post translationally modified 18-kilodalton protein from rice as eukaryotic translation initiation factor 5A. Plant Physiol. 106 1413–1419. 10.1104/pp.106.4.1413 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Næsted H., Holm A., Jenkins T., Nielsen H. B., Harris C. A., Beale M. H., et al. (2004). Arabidopsis VARIEGATED 3 encodes a chloroplast-targeted, zinc-finger protein required for chloroplast and palisade cell development. J. Cell Sci. 117 4807–4818. 10.1242/jcs.01360 [DOI] [PubMed] [Google Scholar]
- Ober D., Hartmann T. (1999). Deoxyhypusine synthase from tobacco. cDNA isolation, characterization, and bacterial expression of an enzyme with extended substrate specificity. J. Biol. Chem. 274 32040–32047. 10.1074/jbc.274.45.32040 [DOI] [PubMed] [Google Scholar]
- Oda-Yamamizo C., Mitsuda N., Sakamoto S., Ogawa D., Ohme-Takagi M., Ohmiya A. (2016). The NAC transcription factor ANAC046 is a positive regulator of chlorophyll degradation and senescence in Arabidopsis leaves. Sci. Rep. 6:23609. 10.1038/srep23609 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ovecka M., Bahaji A., Muñoz F. J., Almagro G., Ezquer I., Baroja-Fernández E., et al. (2012). A sensitive method for confocal fluorescence microscopic visualization of starch granules in iodine stained samples. Plant Signal. Behav. 7 1146–1150. 10.4161/psb.21370 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park J., Aravind L., Wolff E. C., Kaevel J., Kim Y. S., Park M. H. (2006). Molecular cloning, expression, and structural prediction of deoxyhypusine hydroxylase: a HEAT-repeat-containing metalloenzyme. Proc. Natl. Acad. Sci. U.S.A. 103 51–56. 10.1073/pnas.0509348102 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Park M. H., Joe Y. A., Kang K. R. (1998). Deoxyhypusine synthase activity is essential for cell viability in the yeast Saccharomyces cerevisiae. J. Biol. Chem. 273 1677–1683. 10.1074/jbc.273.3.1677 [DOI] [PubMed] [Google Scholar]
- Park M. H., Nishimura K., Zanelli C. F., Valentini S. R. (2010). Functional significance of eIF5A and its hypusine modification in eukaryotes. Amino Acids 38 491–500. 10.1007/s00726-009-0408-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pfaffl M. W. (2001). A new mathematical model for relative quantification in real-time RT-PCR. Nucleic Acids Res. 29:e45 10.1093/nar/29.9.e45 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pogson B. J., Albrecht V. (2011). Genetic dissection of chloroplast biogenesis and development: an overview. Plant Physiol. 155 1545–1551. 10.1104/pp.110.170365 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ratcliff F., Martin Hernandez A. M., Baulcombe D. C. (2001). Technical advance: tobacco rattle virus as a vector for analysis of gene function by silencing. Plant J. 25 237–245. 10.1046/j.0960-7412.2000.00942.x [DOI] [PubMed] [Google Scholar]
- Ren B., Chen Q., Hong S., Zhao W., Feng J., Feng H., et al. (2013). The Arabidopsis eukaryotic translation initiation factor eIF5A-2 regulates root protoxylem development by modulating cytokinin signaling. Plant Cell 25 3841–3857. 10.1105/tpc.113.116236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosso D., Bode R., Li W., Krol M., Saccon D., Wang S., et al. (2009). Photosynthetic redox imbalance governs leaf sectoring in the Arabidopsis thaliana variegation mutants immutans, spotty, var1, and var2. Plant Cell 21 3473–3492. 10.1105/tpc.108.062752 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sakamoto W., Zaltsman A., Adam Z., Takahashi Y. (2003). coordinated regulation and complex formation of yellow variegated1 and yellow variegated2, chloroplastic ftsh metalloproteases involved in the repair cycle of photosystem II in Arabidopsis thylakoid membranes. Plant Cell 15 2843–2855. 10.1105/tpc.017319 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sasaki K., Abid R., Miyazaki M. (1996). Deoxyhypusine synthase gene is essential for cell viability in the yeast Saccharomyces cerevisiae. FEBS Lett. 384 151–154. 10.1016/0014-5793(96)00310-9 [DOI] [PubMed] [Google Scholar]
- Song Y., Xiang F., Zhang G., Miao Y., Miao C., Song C.-P. (2016). Abscisic acid as an internal integrator of multiple physiological processes modulates leaf senescence onset in Arabidopsis thaliana. Front. Plant Sci. 7:181. 10.3389/fpls.2016.00181 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B., Marhevka E., Barkai O., Marton I., Edelbaum O., Masci T., et al. (2010). EOBII, a gene encoding a flower-specific regulator of phenylpropanoid volatiles’ biosynthesis in petunia. Plant Cell 22 1961–1976. 10.1105/tpc.109.067280 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Spitzer-Rimon B., Farhi M., Albo B., Cna’ani A., Ben Zvi M. M., Masci T., et al. (2012). The R2R3-MYB-like regulatory factor EOBI, acting downstream of EOBII, regulates scent production by activating ODO1 and structural scent-related genes in petunia. Plant Cell 24 5089–5105. 10.1105/tpc.112.105247 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tan Y., Liu J., Huang F., Zhong S., Yu Y. (2014). PhGRL2 protein, interacting with PhACO1, is involved in flower senescence in the petunia. Mol. Plant 7 1384–1387. 10.1093/mp/ssu024 [DOI] [PubMed] [Google Scholar]
- Turnage M. A., Muangsan N., Peele C. G., Robertson D. (2002). Geminivirus-based vectors for gene silencing in Arabidopsis. Plant J. 30 107–114. 10.1046/j.1365-313X.2002.01261.x [DOI] [PubMed] [Google Scholar]
- Tzfira T., Tian G.-W., Lacroix B., Vyas S., Li J., Leitner-Dagan Y., et al. (2005). pSAT vectors: a modular series of plasmids for autofluorescent protein tagging and expression of multiple genes in plants. Plant Mol. Biol. 57 503–516. 10.1007/s11103-005-0340-5 [DOI] [PubMed] [Google Scholar]
- Vizcaíno J. A., Côté R., Reisinger F., Barsnes H., Foster J. M., Rameseder J., et al. (2010). The proteomics identifications database: 2010 update. Nucleic Acids Res. 38 D736–D742. 10.1093/nar/gkp964 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vothknecht U. C., Otters S., Hennig R., Schneider D. (2012). Vipp1: a very important protein in plastids? J. Exp. Bot. 63 1699–1712. 10.1093/jxb/err357 [DOI] [PubMed] [Google Scholar]
- Wang Q., Sullivan R. W., Kight A., Henry R. L., Huang J., Jones A. M., et al. (2004). Deletion of the chloroplast-localized Thylakoid formation 1 gene product in Arabidopsis leads to deficient thylakoid formation and variegated leaves. Plant Physiol. 136 3594–3604. 10.1104/pp.104.049841 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang T., Lu L., Wang D., Thompson J. E. (2001). Isolation and characterization of senescence-induced cDNAs encoding deoxyhypusine synthase and eucaryotic translation initiation factor 5A from tomato. J. Biol. Chem. 276 17541–17549. 10.1074/jbc.M008544200 [DOI] [PubMed] [Google Scholar]
- Wang T., Lu L., Zhang C., Taylor C., Thompson J. E. (2003). Pleiotropic effects of suppressing deoxyhypusine synthase expression in Arabidopsis thaliana. Plant Mol. Biol. 52 1223–1235. 10.1023/B:PLAN.0000004332.80792.4d [DOI] [PubMed] [Google Scholar]
- Wang T. W., Wu W., Zhang C. G., Nowack L. M., Liu Z., Thompson J. E. (2005a). Antisense suppression of deoxyhypusine synthase by vacuum-infiltration of Agrobacterium enhances growth and seed yield of canola. Physiol. Plant. 124 493–503. 10.1111/j.1399-3054.2005.00524.x [DOI] [Google Scholar]
- Wang T. W., Zhang C., Wu W., Nowack L. M., Madey E., Thompson J. E. (2005b). Antisense suppression of deoxyhypusine synthase in tomato delays fruit softening and alters growth and development. Plant Physiol. 138 1372–1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang W., Yang X., Tangchaiburana S., Ndeh R., Markham J. E., Tsegaye Y., et al. (2008). An inositolphosphorylceramide synthase is involved in regulation of plant programmed cell death associated with defense in Arabidopsis. Plant Cell 20 3163–3179. 10.1105/tpc.108.060053 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Waters M. T., Moylan E. C., Langdale J. A. (2008). GLK transcription factors regulate chloroplast development in a cell-autonomous manner. Plant J. 56 432–444. 10.1111/j.1365-313X.2008.03616.x [DOI] [PubMed] [Google Scholar]
- Wellburn A. R. (1994). The spectral determination of chlorophylls a and b, as well as total carotenoids, using various solvents of spectrophotometers of different resolution. J. Plant Physiol. 144 307–313. 10.1016/S0176-1617(11)81192-2 [DOI] [Google Scholar]
- Wetzel C. M., Jiang C. Z., Meehan L. J., Voytas D. F., Rodermel S. R. (1994). Nuclear-organelle interactions: the immutans variegation mutant of Arabidopsis is plastid autonomous and impaired in carotenoid biosynthesis. Plant J. 6 161–175. 10.1046/j.1365-313X.1994.6020161.x [DOI] [PubMed] [Google Scholar]
- Wu D., Wright D. A., Wetzel C., Voytas D. F., Rodermel S. (1999). The IMMUTANS variegation locus of Arabidopsis defines a mitochondrial alternative oxidase homolog that functions during early chloroplast biogenesis. Plant Cell 11 43–55. 10.1105/tpc.11.1.43 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu Q., Cheng Z., Zhu J. (2015). Suberoylanilide hydroxamic acid treatment reveals crosstalks among proteome, ubiquitylome and acetylome in non-small cell lung cancer A549 cell line. Sci. Rep. 5:9520. 10.1038/srep09520 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Xie X., Kang H., Liu W., Wang G. (2015). Comprehensive profiling of the rice ubiquitome reveals the significance of lysine ubiquitination in young leaves. J. Proteome Res. 14 2017–2025. 10.1021/pr5009724 [DOI] [PubMed] [Google Scholar]
- Xu A., Chen K. Y. (2001). Hypusine is required for a sequence-specific interaction of eukaryotic initiation factor 5A with postsystematic evolution of ligands by exponential enrichment RNA. J. Biol. Chem. 276 2555–2561. 10.1074/jbc.M008982200 [DOI] [PubMed] [Google Scholar]
- Xu A., Jao D. L.-E., Chen K. Y. (2004). Identification of mRNA that binds to eukaryotic initiation factor 5A by affinity co-purification and differential display. Biochem. J. 384 585–590. 10.1042/BJ20041232 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yan Y. P., Yong T., Chen K. Y. (1996). Molecular cloning and functional expression of human deoxyhypusine synthase cDNA based on expressed sequence tag information. Biochem. J. 315 429–434. 10.1042/bj3150429 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang W., Cai Y., Hu L., Wei Q., Chen G., Bai M., et al. (2017). PhCESA3 silencing inhibits elongation and stimulates radial expansion in petunia. Sci. Rep. 7:41471. 10.1038/srep41471 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zanelli C. F., Valentini S. R. (2005). Pkc1 acts through Zds1 and Gic1 to suppress growth and cell polarity defects of a yeast eIF5A mutant. Genetics 171 1571–1581. 10.1534/genetics.105.048082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang L., Kato Y., Otters S., Vothknecht U. C., Sakamoto W. (2012). Essential role of VIPP1 in chloroplast envelope maintenance in Arabidopsis. Plant Cell 24 3695–3707. 10.1105/tpc.112.103606 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zuk D., Jacobson A. (1998). A single amino acid substitution in yeast eIF-5A results in mRNA stabilization. EMBO J. 17 2914–2925. 10.1093/emboj/17.10.2914 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.