DNA-bound transcription factors (TFs) governing developmental gene regulation have been proposed to recruit polymerase II machinery at gene promoters through specific interactions with dedicated subunits of the evolutionarily conserved Mediator (MED) complex. However, whether such MED subunit-specific functions and partnerships have been conserved during evolution has been poorly investigated.
KEYWORDS: Drosophila development, GATA, Med1, Mediator complex, transcription factors, zinc fingers, gene regulation
ABSTRACT
DNA-bound transcription factors (TFs) governing developmental gene regulation have been proposed to recruit polymerase II machinery at gene promoters through specific interactions with dedicated subunits of the evolutionarily conserved Mediator (MED) complex. However, whether such MED subunit-specific functions and partnerships have been conserved during evolution has been poorly investigated. To address this issue, we generated the first Drosophila melanogaster loss-of-function mutants for Med1, known as a specific cofactor for GATA TFs and hormone nuclear receptors in mammals. We show that Med1 is required for cell proliferation and hematopoietic differentiation depending on the GATA TF Serpent (Srp). Med1 physically binds Srp in cultured cells and in vitro through its conserved GATA zinc finger DNA-binding domain and the divergent Med1 C terminus. Interestingly, GATA-Srp interaction occurs through the longest Med1 isoform, suggesting a functional diversity of MED complex populations. Furthermore, we show that Med1 acts as a coactivator for the GATA factor Pannier during thoracic development. In conclusion, the Med1 requirement for GATA-dependent regulatory processes is a common feature in insects and mammals, although binding interfaces have diverged. Further work in Drosophila should bring valuable insights to fully understand GATA-MED functional partnerships, which probably involve other MED subunits depending on the cellular context.
INTRODUCTION
Precise temporal and spatial regulation of gene transcription by RNA polymerase II (Pol II) is crucial to ensure the coordinated cell fate specification in multicellular organisms. To precisely control Pol II activity, metazoans have evolved an elaborate protein machinery, including the conserved multiprotein Mediator (MED) complex, which serves as a malleable interface between DNA-bound transcription factors (TFs) and the Pol II machinery (1). Dedicated MED subunits have been proposed to mediate specific TF activities. Whether these specific partnerships and binding interfaces have been conserved during evolution remains an open question.
The MED complex, conserved from yeast to human, contains 25 to 30 subunits organized into the head, middle, and tail modules as well as a dissociable cyclin-dependent kinase 8 (CDK8) module (2). The core MED, interacting directly with Pol II and its associated general transcription factors, contains essential head and middle module subunits. Conversely, more peripheral MED subunits belonging to the tail (e.g., Med15), CDK8 (e.g., Med12), and middle (e.g., Med1) modules are not required for cell viability and display more specific functions during cell differentiation. It is generally assumed that MED subunit specificity comes from their ability to interact directly with specific TFs, allowing Mediator recruitment to gene regulatory elements. For example, it has been shown that Med12 interacts directly with Sox9 and Sox10, whereas Med15 binds SMADs (3) and Med19 binds HOX (4) TFs. Another example is Med1, identified for its role as a major cofactor of hormone nuclear receptors (NRs) that directly bind its LXXL domain (5). Mammalian Med1 also mediates transcriptional activity of the GATA zinc finger (ZF) TF family. Physically interacting with at least five of the six mammalian GATAs (6, 7), Med1 is required for GATA1, GATA2, and GATA6 target gene expression in several developmental contexts, including erythropoiesis (8–11), and is recruited to specific GATA1 and GATA2 target genes (7, 9, 10, 12, 13). Whereas several MED subunit-TF partnerships have been characterized in mammals, it is not known to what extent these MED subunit-specific functions have been conserved in other species.
Drosophila melanogaster is an ideal model to analyze MED subunit-specific functions given that homologs of the 33 human subunits are encoded by single-copy genes (14, 15) and that overall MED complex structure has been conserved during evolution (16). Furthermore, several transcription factor families are strongly conserved both structurally and functionally in Drosophila. A good example is the GATA zinc finger factor family. In mice and humans, the GATA1/2/3 subfamily is required for blood cell lineage differentiation (17), and the GATA4/5/6 subfamily is involved in the meso-endoderm lineage, notably in cardiac development (18). In Drosophila, the GATA factor Serpent (Srp) is a central regulator of hematopoietic cell differentiation, controlling the formation of the two embryonic populations of blood cells (plasmatocytes and crystal cells), and the GATA factor Pannier (Pnr) is involved in embryonic heart development, dorsal thoracic closure, and sensory organ precursor development (19), revealing a functional conservation during bilaterian evolution.
Mammalian GATA factors generally contain two highly conserved Cys4-type ZFs (20). The C-terminal ZF (C-ZF) is both necessary and sufficient for sequence-specific DNA binding at [(A/T)GATA(G/A)] genomic sites (21, 22), while the N-terminal ZF (N-ZF) appears only to modulate DNA binding affinity, notably at palindromic double sites (22–24). Whereas Drosophila Pnr also displays two ZFs (25), srp encodes different isoforms containing either only a C-ZF (SrpC) or both a C- and an N-ZF (SrpNC), with the N-finger stabilizing the interaction of Srp with palindromic GATA sites (26).
The GATA N-ZF also mediates interactions with key coregulators, such as Friend-of-GATA (FOG) proteins (27), the LIM-only protein LMO2 (28, 29), and the basic helix-loop-helix (bHLH) factor SCL/TAL1 (30). GATA1 forms a pentameric transactivation complex with LMO2, the LIM-binding protein Ldb1, and the bHLH factors SCL and E1A, binding a composite E box/GATA enhancer sequence to transactivate erythroid gene expression. An equivalent pentameric complex has been characterized during Drosophila sensory organ precursor development, where the Achaete (Ac) bHLH protein and its obligatory cofactor, Daughterless (Da), associate with GATA/Pnr, dLMO, and the Lbd protein Chip for ac gene autoregulation (31, 32). Drosophila Srp also interacts with orthologues of mammalian GATA cofactors. Indeed, GATA/Srp associates with the RUNX cofactor Lozenge (Lz) or the FOG factor U-shaped (Ush) to induce or repress crystal cell differentiation, respectively (33, 34).
Thus, GATA factor functions, DNA binding interfaces, and transcriptional cofactors appear conserved in Drosophila, but less is known about how GATA factors contact the Mediator complex to activate their target genes. By a genome-wide RNA interference screen in cultured Drosophila blood cells, we previously identified several MED subunits (including Med1, Med12, and Med13) as modulators of GATA/Srp-induced transactivation (35). We further showed that Med12 and Med13 are indeed required in vivo for Srp-dependent crystal cell differentiation. Furthermore, a genome-wide expression profiling from Drosophila GATA/Srp- or Med12- or Med13-depleted cells revealed a significant overlap, notably concerning the innate immunity genes (36). Nevertheless, we were unable to detect a direct physical interaction in vitro between Srp and Med12 or Med13 (35), suggesting that GATA/Srp recruits the MED complex by contacting another subunit.
In this work, we address the issue of the conservation of Mediator subunit-specific functions across bilaterian evolution using as a model the Drosophila Med1 subunit whose mammalian orthologue is known as a GATA and NR cofactor. The generation of the first Med1 mutants in an insect reveals defects in GATA/Srp-dependent embryonic hematopoiesis. We further show that Srp forms a complex with Med1’s longest isoform in Drosophila cultured cells. Furthermore, the divergent, isoform-specific C terminus of Med1 interacts with the conserved zinc finger-containing domain of Srp in vitro. The generation of Med1 mutant clones indicates a Med1 requirement for cell proliferation control and for the expression of a GATA/Pnr target gene in larval imaginal tissues. Finally, we show a Med1 role in Pnr-dependent transactivation and a direct interaction between the GATA/Pnr ZF-containing domain and the Drosophila-specific Med1 C terminus. Taken together, our data reveal that the Med1 Mediator subunit has conserved GATA TF coactivator functions during bilaterian evolution through divergent binding interfaces.
RESULTS
Drosophila Med1 is an essential gene.
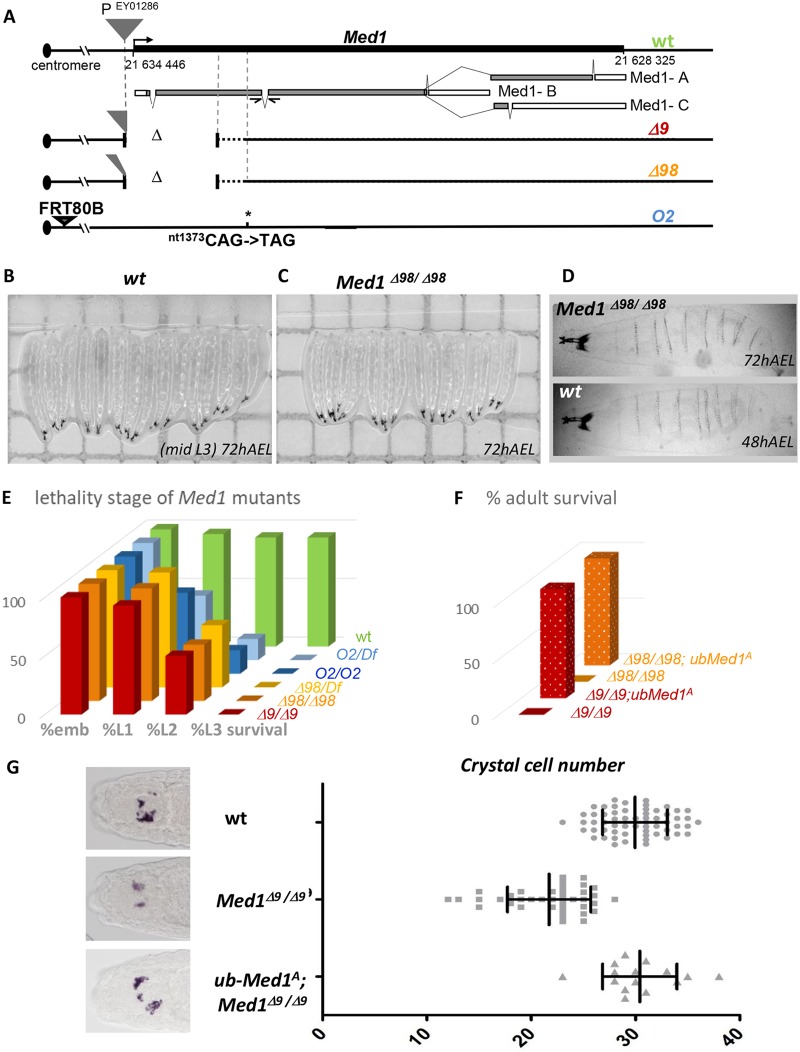
To analyze Med1 function in Drosophila, we took advantage of a viable P transposon insertion located 130 bp upstream of the Med1 transcription start site to generate Med1 loss-of-function mutations (Fig. 1A). Two homozygous lethal alleles, Med1Δ9 and Med1Δ98, were generated by imprecise excision of the P transposon and selected for their inability to complement a large chromosomal deficiency comprising Med1. Genetic analysis revealed that homozygous mutant embryos for both alleles hatch, but larvae display growth retardation (Fig. 1B and C), arrest their development at the L1 or L2 stage (Fig. 1E), survive several days, and do not show any external cuticular defects (Fig. 1D). These results indicate that Med1 is required for larval development.
FIG 1.
Drosophila Med1 is required for organism viability and hematopoietic crystal cell differentiation. (A) Med1 locus on chromosome 3 with the insertion site of the P element (EY01286; gray triangle) used for mutagenesis, the three Med1 alternatively spliced mRNAs (gray boxes) depicting the coding sequences, separated by intronic regions, and the molecular characterization of Med1 mutant alleles Δ9, Δ98, and O2. Deleted genomic sequences are represented by Δ. (B and C) Images of wild-type (wt) (B) versus Med1Δ98/Δ98 mutant (C) larvae at 72 h after egg laying (AEL). (D) A Med1Δ98 homozygous larva at 72 h AEL with a size equivalent to that of a 48-h AEL wt control and without external cuticular defects. (E) Complementation tests. Proportions of homozygous (Med1/Med1) or hemizygous (Med1/Df) Med1 mutant alleles dying at embryonic (emb) or larval stages L1, L2, and L3 are indicated. (F) Rescue tests. Proportion of ub-Med1A transgenic adults homozygous for Med1Δ9 or Med1Δ98 compared to the expected proportion of the F1 progeny for full rescue. (G) In situ hybridization of the crystal cell-specific PPO2 mRNA in wild-type, Med1Δ9/Δ9, or ub-Med1A; Med1Δ9Δ9 stage 14 embryos. Representative dorsal views of the embryo head region are shown on the left. The graph indicates the crystal cell number of each embryo of the given genotype.
Several lines of evidence indicate that both Δ9 and Δ98 are null Med1 alleles. First, these alleles lack the transcription start site, both being deleted of the first third of the Med1 coding sequence and retaining different portions of the P element 3′ extremity (Fig. 1A). Second, homozygotes for both alleles die approximately at the same developmental stage as hemizygotes (Med1−/Deficiency) (Fig. 1E), indicating that they behave genetically as null alleles. Furthermore, we established that the lethality is due solely to loss of Med1 function. Indeed, Med1Δ9 and Med1Δ98 homozygous mutant lethality is fully rescued by introducing one copy of a ubiquitin-Med1 transgene (ub-Med1A) that ubiquitously expresses the wild-type Med1A protein, giving rise to fertile adults (Fig. 1F).
In conclusion, genetic analysis of the first Med1 loss-of-function mutants in an insect indicates that Drosophila Med1 is an essential gene at least required for proper larval development.
Med1 is involved in Srp-dependent crystal cell development.
We have previously shown that Med1-depleted cultured Drosophila cells display Srp/Lz-dependent transactivation defects (35). We therefore asked whether Med1 is required in vivo for crystal cell differentiation, a developmental process that relies on GATA/Srp acting cooperatively with the RUNX factor Lozenge (Lz). Crystal cells, which can be easily visualized and counted (2 bilateral clusters of ∼15 cells) (Fig. 1G, left) (33, 37, 38), were analyzed by the expression of the specific marker PPO2 (also known as PO45/CG8193), a direct Srp-Lz target gene (38). In Med1Δ9 mutant embryos, differentiated crystal cells were present and localized correctly, but we observed a significant decrease in PPO2-positive cells compared to the level for the wild type (Fig. 1G, right graph). This decrease resulted from Med1 loss of function, since it was rescued by Med1 protein expressed from a ub-Med1A transgene (Fig. 1G). This partial loss of crystal cells could be attributed to a partial requirement of Med1 in this process or, more probably, to the maternal contribution of Med1 protein and mRNA deposited in the egg (39) that can partially rescue the zygotic loss of Med1 activity in mutant embryos.
Taken together, these data demonstrate that the Drosophila Med1 Mediator subunit is involved in vivo in hematopoietic crystal cell development, a process depending on GATA/Srp activity.
Med1 forms protein complexes with GATA/Srp in cultured cells.
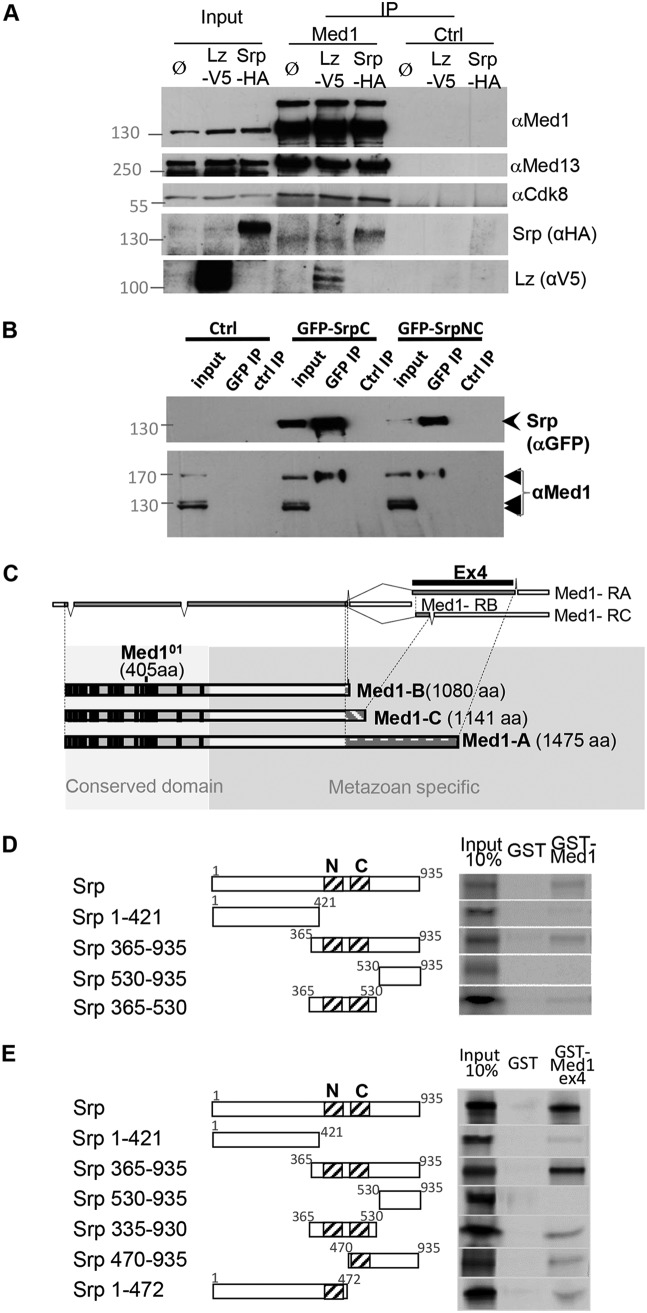
Given the GATA coactivator functions of Med1 shown in Drosophila cultured cells (35) and its mutant phenotypes in a GATA-dependent developmental process (described above), we asked whether Drosophila Med1 interacts physically with GATA factors. We first tested whether Med1-GATA protein complexes form within cells by performing coimmunoprecipitation (co-IP) experiments from cultured Drosophila Kc167 cells transfected with hemagglutinin (HA)-tagged Srp or its V5-tagged RUNX cofactor, Lz. Using an antibody recognizing Drosophila Med1, both Srp and Lz were efficiently coimmunoprecipitated with the endogenous Med1 protein (Fig. 2A). Two Mediator subunits, Med13 and Cdk8, were also coimmunoprecipitated, indicating that Med1 is indeed incorporated within the Mediator complex (Fig. 2A). Furthermore, none of these factors were immunoprecipitated in the control IP, and no specific signal was observed from cells that do not express the tagged protein of interest.
FIG 2.
Med1 interacts physically with GATA/Srp. (A) Coimmunoprecipitation experiments from Drosophila Kc167 cells transfected with pAc-Srp-HA or pAc-Lz-V5 using guinea pig anti-Med1 antibody or a nonrelevant (NR) guinea pig antibody (anti-Tyn) (ctrl IP) revealed by Western blotting against Med1, Med13, Cdk8, HA tag (Srp), or V5 tag (Lz). (B) Reverse coimmunoprecipitation experiments from Drosophila Kc167 cells transfected with pAc-GFP-SrpC or pAc-GFP-SrpNC using a rabbit anti-GFP antibody or a control rabbit preimmune serum (ctrl IP) revealed by Western blot analysis against the Med1 or GFP tag. (C) Map of the three alternatively spliced transcripts of Med1, with white boxes depicting untranslated sequences and gray boxes depicting the coding sequences, separated by intronic regions (lines). The exon 4 region (ex4) used to produce the Med1A isoform-specific protein fragment is indicated. In the lower part is a scheme of the three Med1 isoforms with amino acid numbers indicated. Black boxes represent highly conserved regions within the yeast Med1 orthologue, and white boxes represent the nonconserved metazoan-specific extension differing in their C termini, depending on the Med1 isoform considered. (D and E) GST pulldown binding assays of full-length 35S-SrpNC or protein fragments to immobilized GST-Med1 fusion (D) or GST-Med1ex4 fusion (E) corresponding to the C-terminal protein sequence specific to the longest Med1A isoform.
To provide further evidence for Med1-GATA/Srp interaction in Drosophila cultured cells, we also performed reverse co-IP experiments by immunoprecipitating green fluorescent protein (GFP)-tagged GATA/Srp. As shown in Fig. 2B, endogenous Med1 protein was effectively coprecipitated with both GFP-SrpC and GFP-SrpNC, corresponding to two Srp isoforms containing only C-ZF or both ZFs, respectively (26). The Med1 gene encodes three protein isoforms of 1,080, 1,141, and 1,475 amino acid (aa) residues, resulting from alternative splicing events (http://flybase.org/) (Fig. 2C). As shown in Fig. 2B (inputs), all three isoforms are expressed in Kc167 cultured cells, the shortest PB form being more strongly expressed. Strikingly, only the band corresponding to the longest Med1A isoform is present under coprecipitation with both Srp isoforms. Three conclusions can be drawn from these results. First, they provide complementary evidence for the presence of Med1-GATA protein complexes in Drosophila cells. Second, they indicate that the Srp N-ZF domain is not required for interaction with Med1 given that the latter interacted strongly with both SrpNC and SrpC forms. Third, they suggest that Srp binds only one of the three Med1 isoforms.
Taken together, these results show that Med1 associates with Srp in cultured cells, suggesting a direct role for Med1 as an Srp cofactor.
GATA/Srp zinc finger domain interacts directly with a Med1 domain specific to the longest isoform.
We next asked whether Med1 is able to interact physically with Srp in vitro using glutathione S-transferase (GST) pulldown essays. We observed a faint but reproducible interaction between recombinant full-length GST-Med1 proteins and in vitro-translated GATA/Srp protein (Fig. 2D). To investigate which protein domain(s) is responsible for Med1 binding, we tested Srp truncated forms. As shown in Fig. 2D, binding to purified GST-Med1 was essentially retained by the Srp C-terminal half. After splitting this fragment into two parts, binding was still observed with the central Srp fragment (aa 365 to 530), containing the GATA N- and C-ZFs. These data suggest that physical interaction between Med1 and Srp involves the evolutionarily conserved GATA zinc finger domains.
We subsequently looked for the Srp-interacting domain(s) within its Med1 partner. The three Med1 isoforms contain the evolutionarily conserved N-terminal domain, shown to be sufficient for interaction with other Mediator subunits in mouse cells (40), and differ in their C-terminal part of 401, 6, or 66 aa residues for the A, B, and C isoforms, respectively (Fig. 2C). Since Srp only interacted with the longest A isoform within cultured cells (Fig. 2B), the Med1 interaction domain should lie within the A isoform-specific nonconserved C-terminal part. To test this hypothesis, we analyzed the ability of the Med1A-specific domain to physically bind GATA/Srp by in vitro GST pulldown experiments. As shown in Fig. 2E, a truncated GST-Med1 fusion protein containing the entire A isoform-specific domain (401 aa residues produced from the fourth exon of Med1A transcript, here called Med1ex4) bound in vitro-translated Srp. Analysis of truncated Srp forms showed that the Med1A-specific domain mainly interacted with the GATA ZF region (Fig. 2E), as previously shown for the full-length Med1. Splitting Srp in two halves containing N-ZF or C-ZF indicated that both ZF domains are able to bind Med1ex4 (Fig. 2E).
Taken together, coimmunoprecipitation and pulldown experiments indicate that the interaction between the Drosophila GATA factor Srp and Med1 occurs at least in part through the GATA zinc finger region binding to the Med1 nonconserved C-terminal domain specific to the longest isoform.
Med1 is required for proper cell proliferation and/or survival.
To further analyze Med1 requirements at different developmental stages, Drosophila offers a powerful genetic tool allowing the generation of mosaic animals bearing homozygous mutant clones within viable heterozygous individuals (Fig. 3A). This technique, based on FLP recombination target (FRT)/flippase-induced recombination (41), has three main advantages: (i) it reveals adult developmental functions hindered by earlier requirements, (ii) it provides access to total loss-of-function phenotypes without influence of early maternal contribution, and (iii) it allows the analysis of cell proliferation functions by comparing mutant to wild-type twin clone size (Fig. 3A and B). Owing to the close proximity of the Med1 locus to the third-chromosome centromere, we were unable to generate recombinant chromosomes combining the Med1Δ9 or Med1Δ98 allele with a centromeric FRT80B site. Thus, we performed an EMS (ethyl methane sulfonate) chemical mutagenesis of an isogenic FRT80B chromosome, followed by an F2 genetic screen to identify mutations that did not complement our initial Med1Δ9 allele (Fig. 1A). One such allele, called Med1O2, was obtained. It harbors a single point mutation within codon 406 (CAG to TAG), creating a stop codon which interrupts the three Med1 open reading frames (ORF) (Fig. 1A). This allele, which theoretically could produce a truncated Med1 protein of 405 amino acid residues lacking a portion of the evolutionarily conserved N-terminal part, does not display any dominant phenotype and behaves genetically as a null (Fig. 1B).
FIG 3.
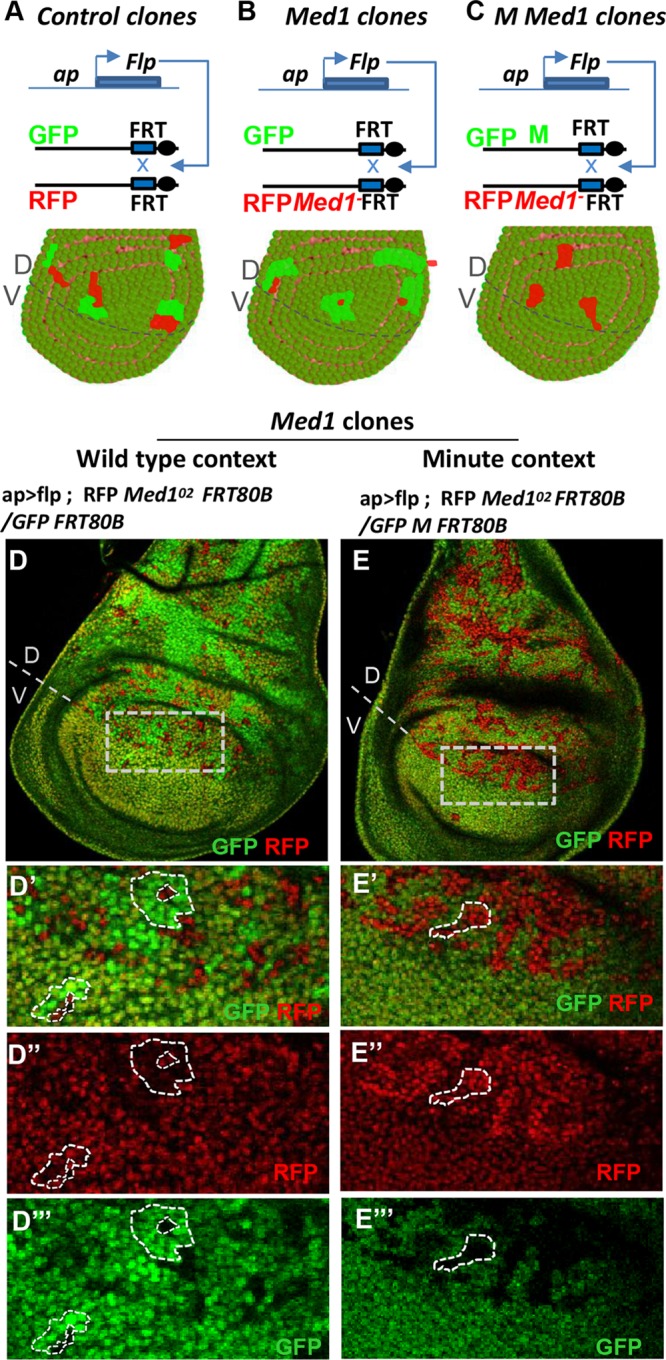
Med1 is partially required for cell proliferation/survival. (A) Twin spot clonal analysis. Principle of generation of somatic clones in heterozygous wing imaginal discs, using FRT/Flp-induced recombination and the dorsal compartment-specific ap-GAL4 driver. The lower panel shows a model of wing disc under control conditions, where homozygous RFP+/+ and GFP+/+ twin clones display equivalent areas. (B) Scheme of the recombination event leading to the generation of mutant clones for the Med1O2 allele marked with RFP associated with the wild-type twin clones marked with GFP. A representation of resulting clones is shown in the lower panel. (C) Principle of Med1O2 mutant clone induction in a Minute (M) context (upper) with schematic representation of a resulting wing disc (lower). Note that Minute homozygous clones are lethal and do not proliferate. (D to D′′′) Wing disc bearing Med1°2 mutant clones [RFP+ GFP−] generated as described for panel B. Magnifications of the zone of interest (D, dotted box) with overlay (D′) or single RFP (D″) or GFP (D′′′) channel are shown, and examples of mutant and control clones are surrounded by dashed lines. (E to E′′′) Wing disc bearing Med1°2 clones generated in a Minute context as described for panel C, with higher magnifications in panels E′ to E′′′. An example of a Med1− mutant clone is shown.
We subsequently generated Med1O2 homozygous mutant clones within wing imaginal discs of heterozygous third-instar larvae. Heterozygous cells expressed one copy of both GFP and red fluorescent protein (RFP) markers, whereas homozygous Med1 mutant or wild-type clones expressed two copies of RFP or GFP, respectively (Fig. 3A and B). As shown in Fig. 3B and D to D′′′, Med1−/− clones were often restricted to a single cell, while larger twin Med1+/+ territories were present. These data suggested a role for Med1 in cell viability. To obtain larger mutant clones, we introduced a Minute (M) mutation (42) which confers a growth disadvantage to surrounding heterozygous cells (Fig. 3C). In that case, larger Med1O2 clones were observed (Fig. 3C and E to E′′′). These results indicate that Med1 is not strictly necessary for cell viability but is required for proper cell proliferation and/or survival control.
Med1 is required for GATA/Pnr activity in vivo and in cultured cells.
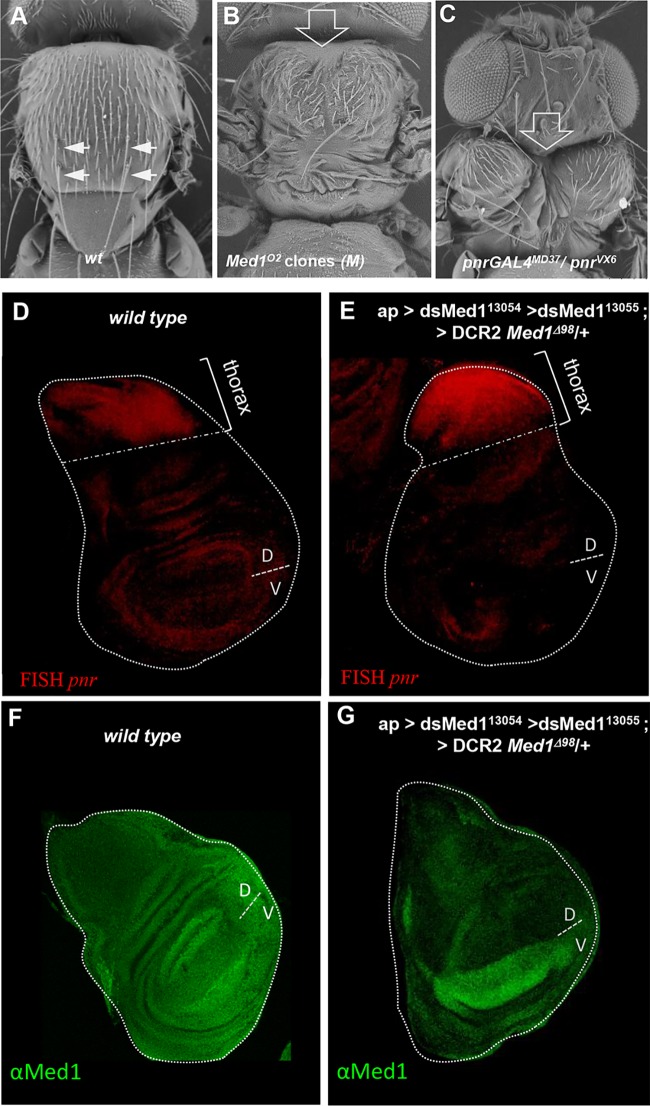
To identify adult developmental processes requiring Med1 function, we analyzed flies harboring mutant clones (Fig. 3C) in the thorax, notum, and wings, structures emerging from the dorsal part of the larval wing imaginal disc. Such clones gave rise to mosaic adult flies displaying a thoracic cleft and a loss of dorsocentral (DC) mechanosensory macrochaetae compared to that of wild-type thorax (Fig. 4A and B). These defects are reminiscent of loss-of-function phenotypes for pannier (Fig. 4C), whose function is required for central notum patterning (43, 44).
FIG 4.
Thoracic Med1 mutant clone phenotypes resemble GATA pnr loss of function. (A to C) Scanning electron microscopy images of Drosophila notum of wild-type (A), ap>flp; H2A-RFP Med1°2 FRT 80B/M ub-GFP FRT 80B (B), or pnr-GAL4MD37/pnrVX6 (C) flies. Filled arrows point to dorsocentral (DC) mechanosensory bristles, and open arrows point to thoracic clefts. (D and E) In situ hybridization against pnr mRNA in wild-type (D) versus Med1-deficient wing discs expressing dsRNA against Med1 (dsMed1) together with DCR2 in the dorsal wing disc of Med1Δ98/+ heterozygous flies (E). Disc areas giving rise to the presumptive thorax and corresponding to the Pnr expression domain are shown. (F and G) αMed1 immunolocalization in wild-type (F) versus Med1-deficient (G) wing discs, showing ubiquitous Med1 protein expression in the whole wing disc (F) and strong protein depletion in the dsRNA-expressing dorsal part (G).
Med1-induced dorsal thoracic phenotypes could be due to defective GATA/Pnr expression or to the loss of Pnr target gene expression. To test the first hypothesis, we examined pnr gene expression in Med1-deficient wing discs by fluorescent in situ hybridization (FISH) (Fig. 4D and E). We generated RNA interference (RNAi)-mediated knockdown of Med1 using the ap-GAL4 driver depleting Med1 in the whole dorsal compartment of the wing disc (Fig. 4F and G), including the presumptive thorax territory, giving rise to adults with thoracic closure phenotypes (described above). The use of anti-Med1 antibody showed first a ubiquitous and globally homogeneous nuclear expression of Med1 in imaginal discs (as previously observed for other Drosophila MED subunits) (45) (Fig. 4D) and, second, that the RNAi treatment efficiently depletes the protein in the dorsal wing compartment (Fig. 4E). Nevertheless, we observed that pnr is still transcribed in Med1-depleted wing discs (Fig. 4D and E), indicating that Med1 mutant phenotypes cannot be explained by a loss of pnr expression.
We then addressed the second hypothesis, proposing that Med1 functions as a GATA/Pnr cofactor by analyzing target gene expression in vivo. The GATA factor Pnr is known to activate the achaete (ac) and scute (sc) gene complex via direct binding to the DC enhancer (46). We first used a DC-ac-lacZ reporter (DC enhancer of the achaete-scute complex and the ac promoter), shown to respond to pnr depletion in vivo and to be directly activated upon Pnr binding to the DC enhancer (46). Compared with the wild type (Fig. 5A to A″), DC-ac-lacZ expression was strongly reduced (Fig. 5B to B″, RFP staining) in Med1O2 mutant clones visualized by the absence of GFP. This indicates that Med1 is required for the expression of the GATA/Pnr target gene ac, which could explain the loss of DC bristles observed in Med1 loss-of-function mosaic flies. To further test the Med1 requirement for Pnr-driven transcriptional activation, we examined the expression of another Pnr target gene: wingless (wg) (46). As shown in Fig. 5D to D″, Wg protein was still expressed in Med1 mutant clones. The detection of Wg in mutant cells cannot be explained by the diffusion of the Wingless morphogen coming from adjacent wild-type cells, because this extracellular fraction of Wingless is hardly detected by conventional staining protocols (47). Thus, these results indicate that Med1 is not required for the expression of all Pnr target genes.
FIG 5.
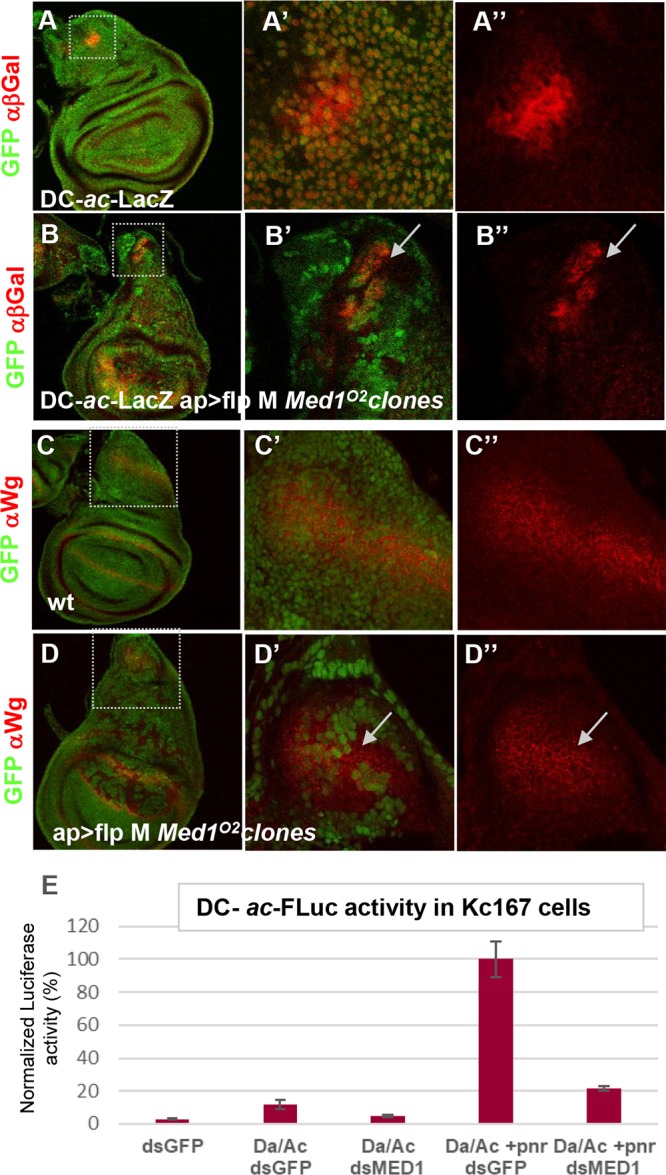
Med1 is differentially required for GATA/Pnr target gene activation. (A to A″) Expression analysis of Pnr target gene ac-sc in control wing discs. The DC-ac-LacZ transgene expression visualized with anti-β-galactosidase (red channel) recapitulates ac-sc expression in the DC cluster. (B to B″) DC-ac-lacZ expression is cell autonomously lost in Med1O2 mitotic clones (GFP−) generated in the wing disc dorsal compartment. (C to C″) Immunofluorescence analysis of the Pnr target gene wg in a control wing disc revealed with anti-Wg antibody (red). (D to D″) Wg protein is still present in Med1O2 mitotic clones (GFP−). Magnifications of the DC and Wg regions are shown in the middle column, with the red channel shown separately in the right column. (E) Functional transcription assay in Drosophila Kc167 cells left untransfected or transfected with plasmids expressing Pnr and/or its cofactors Ac and Da and treated with Med1 dsRNA or control GFP dsRNA. The expression of firefly luciferase was standardized to a transfection control, pAc-Rluc. Three independent transfections were performed. Standard deviations are indicated by error bars.
To further investigate Med1 function as a Pnr coactivator, we asked whether Med1 is required for Pnr-dependent transactivation in Drosophila cultured cells. We performed a transcriptional assay using cultured cells transfected with a luciferase reporter driven by the DC enhancer of the achaete-scute complex and the ac promoter (Fig. 5E). As previously shown (48, 49), strong stimulation of the luciferase reporter gene was observed in Drosophila Kc167 cells when Pnr and the Da-Ac bHLH heterodimer were expressed together, while the Da-Ac complex alone induced lower levels of luciferase activity (Fig. 5E). Importantly, Pnr-induced reporter activation was drastically reduced upon double-stranded RNA (dsRNA)-mediated depletion of Med1 (Fig. 5E).
Taken together, these results show that Med1 is required for two developmental processes depending on GATA/Pannier (thoracic closure and DC bristle formation) and for the activation of at least one direct Pnr target gene in vivo and in cultured cells. That Med1 regulates Pnr activity but not pnr expression suggests that Med1 acts as a GATA/Pnr cofactor, which implies direct binding.
Med1 isoform A-specific domain interacts with GATA/Pnr zinc finger domain.
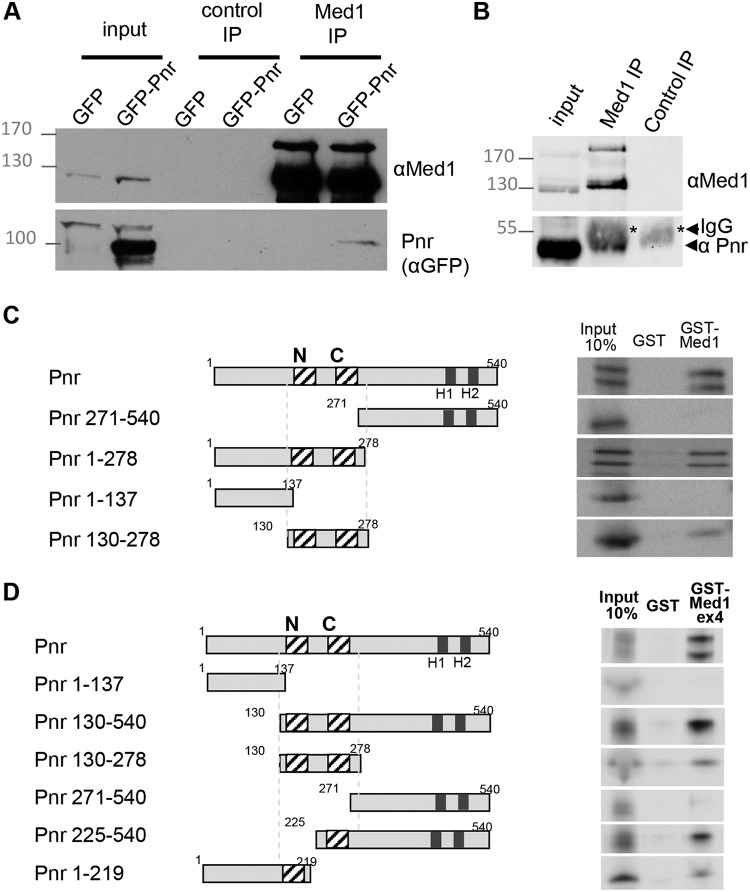
As previously shown for Srp, we first tested whether Med1-Pnr protein complexes exist within cells by performing co-IP experiments from Drosophila Kc167 cells transfected with GFP-tagged Pnr. Using our antibody recognizing Drosophila Med1, we observed that the transgenic protein GFP-Pnr was efficiently coimmunoprecipitated with endogenous Med1 protein (Fig. 6A). Using cultured S2 Drosophila cells naturally expressing pannier, we further observed that the endogenous Pnr protein could also be coprecipitated with our antibody directed against Med1 (Fig. 6B).
FIG 6.
GATA/Pnr physically interacts with Med1. (A) Coimmunoprecipitation experiments from stable Drosophila lines expressing GFP-Pnr or GFP alone using anti-Med1 antibody or preimmune serum (control IP) revealed by Western blotting against Med1 or GFP tag. (B) Coimmunoprecipitation experiments from Drosophila S2 cells endogenously expressing pnr, using anti-Med1 antibody or preimmune serum. (C and D) GST pulldown binding assays of full-length 35S-Pnr or protein fragments to GST-Med1 fusion (C) or to GST-Med1ex4 fusion (D) corresponding to the Med1A-specific extension. The doublets observed with full-length Pnr or some truncated forms are due to alternative initiations at the regular or internal initiator AUG during the in vitro transcription/translation reaction.
We then asked whether Med1 was also able to bind GATA/Pnr in vitro. As shown in Fig. 6C, in vitro-translated full-length Pnr bound purified GST-Med1. Pnr was then split into two halves (Fig. 6C), with the N-terminal half specifically binding GST-Med1. Smaller truncated versions were tested, revealing that as for GATA/Srp, GST-Med1 retained the Pnr ZF-containing domain. Taken together, our data indicate that Drosophila Med1 interacts in vitro with both GATA/Srp and GATA/Pnr and do so through their ZF-containing domain.
Thus, we wondered whether the Med1-Pnr interaction occurs through the isoform A-specific domain of the longest Med1 isoform, as previously shown for Srp. As shown in Fig. 6D, purified GST-Med1Ex4 (Fig. 2C) interacted strongly with in vitro-translated full-length Pnr and retained the Pnr subfragments containing the ZFs. Both truncated Pnr versions harboring either C- or N-ZF bound GST-Med1Ex4, suggesting that both ZFs are sufficient but not necessary for Med1 binding.
Taken together, these data indicate that interaction between Med1 and the zinc fingers of Drosophila GATA factor Pnr occurs, at least in part, through the C-terminal domain of the longest Med1 isoform, as is the case for GATA/Srp.
DISCUSSION
Med1 displays essential functions during development.
In this work, we report the first Med1 loss-of-function mutants in an insect. We showed that Med1 is an essential gene which is not strictly required for cell viability but is clearly involved in context-dependent proliferation or cell survival processes. As opposed to Drosophila, mammalian Med1 is not essential for cell viability given that primary embryonic fibroblasts can be derived from null Med1 mutants in mice (50–52). Nevertheless, mouse Med1 mutant cells display impaired cell cycle regulation (53), suggesting a conserved Med1 function in cell proliferation control that deserves to be analyzed in the future using Drosophila as a model.
The take-home message of our work is the functional partnership between Drosophila Med1 and GATA transcription factors (Fig. 7A). We show that Med1 is involved in at least two developmental processes, embryonic crystal cell differentiation and larval thoracic development, depending on two GATA transcription factors, Serpent and Pannier, respectively. Med1 is required for the expression of at least one Srp and one Pnr target gene in vivo. The partial loss of crystal cells observed in Med1 zygotic mutant embryos could reflect a partial requirement for Med1 in this differentiation process or could be due to partial rescue by the Med1 maternal contributions (39). Furthermore, we show that Med1 interacts physically with Srp and Pnr both in vitro and in cultured cells. Taken together, these results reveal evolutionarily conserved functions of Drosophila Med1 as a GATA cofactor involving a divergent Med1 region binding the conserved GATA zinc fingers. This Med1-GATA partnership appears restricted to one Med1 isoform, suggesting a new layer of regulation by the Mediator complex through a diversity of MED populations. Finally, Med1 is not required for all GATA/Pannier activity, suggesting the use of alternative MED subunits depending on the cellular context.
FIG 7.
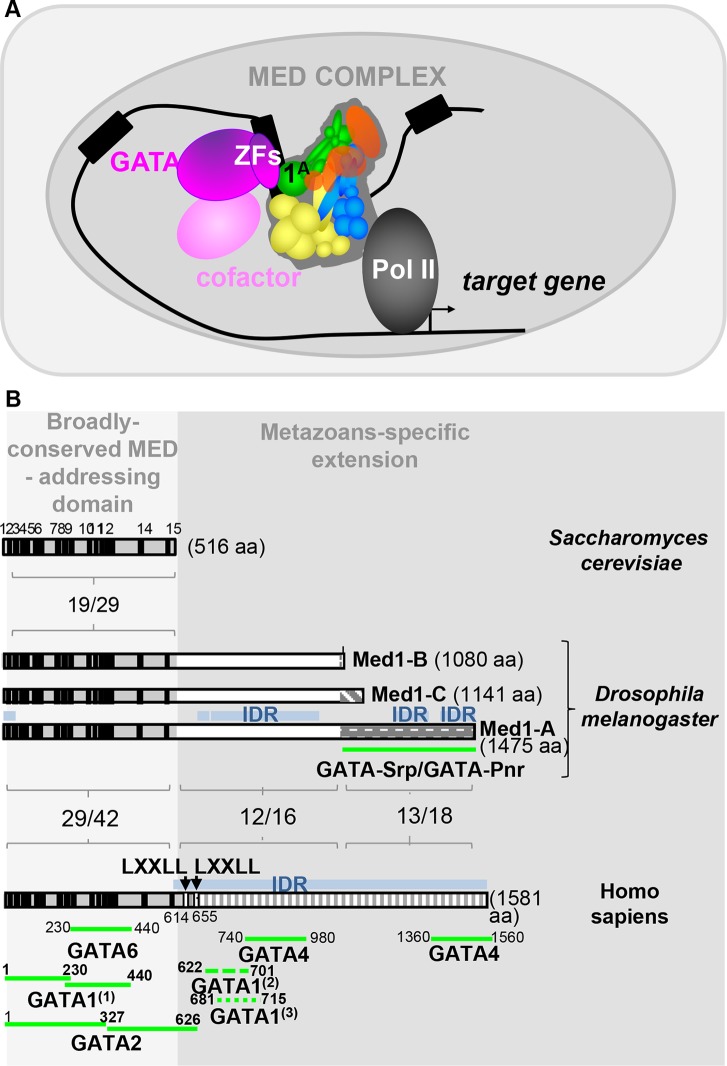
MED1-GATA molecular partnership. (A) Model of Med1-GATA partnership. GATA transcription factors are represented by fuchsia, with GATA binding domains indicated as ZF, and corresponding cofactors are in light pink. Mediator complex is composed of four modules, tail (yellow), head (blue), CDK8 (red), and middle (green), with the latter including the Med1 subunit isoforms. (B) Med1-GATA interacting domains in Saccharomyces cerevisiae, Drosophila melanogaster, and Homo sapiens. The light gray part emphasizes their N-terminal region, comprising 15 short evolutionarily conserved motifs (black boxes), which have been shown to be sufficient for Med1 integration into the mammalian MED complex. Conversely, the darker gray part emphasizes the divergent long metazoan-specific extensions. Percentage of identity/similarity between homologous protein domains is indicated. Blue boxes represent intrinsically disordered regions (IDR) for which no conserved motifs could be identified. Their richness in serine residues (21% to 22%) accounts for most of the amino acid identity observed in the metazoan-specific extension. Position of GATA-interacting domains identified to date for human GATA6 (6), GATA2 (9), GATA4 (7), and GATA1 (55) and Drosophila Srp and Pnr (this work) are indicated below. In the case of mammalian GATA1, note that two independent analyses from Stumpf et al. (7) and Mizuta et al. (55) identified two overlapping domains at the beginning of the divergent metazoan-specific extension, whereas Crawford and collaborators (6) identified two other regions within the conserved and structured N-terminal part that revealed some discrepancies.
Diversity of the GATA-Med1 interfaces throughout evolution.
We show here that Med1 activity as a GATA cofactor is not restricted to vertebrates, since it also acts in Drosophila for at least two different GATA factors, suggesting an ancient GATA-Med1 partnership among bilaterians (Fig. 7B). The MED-TF partnership conservation along evolution is not the rule, since the LXXL motif of mammalian Med1 interacting with hormone nuclear receptors is not present in Drosophila Med1, whereas it is in two other Drosophila Mediator subunits, Cdk8 and Med14, which have been shown to be necessary for ecdysone nuclear receptor activity (54).
What about the conservation of binding interfaces between Med1 and GATA factors? Our results indicate that two Drosophila GATAs, like mammalian GATA1, -2, -3, and -6 and chicken GATA1, -2, and -3 (6, 7, 9, 55), bind Med1 at least through their ZF-containing domains. Nevertheless, some discrepancies exist concerning N- or C-ZF involvement in mammals (7, 55). In Drosophila, we show that either N-ZF or C-ZF interacts with Med1ex4 in vitro and that Srp N-ZF is dispensable for binding in cultured cells. More diversity is observed for GATA-interacting domains within the Med1 protein. Med1 primary sequence conservation lies essentially within the N-terminal part, corresponding to the entire yeast orthologue, whereas the long additional C-terminal part diverges in each metazoan species (Fig. 7B). In Drosophila, we show here that both Srp and Pnr interact with the Med1 isoform A-specific domain lacking sequence homology with mammalian Med1 (Fig. 7B). In mammals, GATA-interacting domains differ depending on the GATA paralogue considered and are distributed throughout the Med1 sequence (Fig. 7B). In conclusion, despite functional conservation of the Med1-GATA partnership, the GATA-binding interface within Med1 has not been fixed during bilaterian evolution, even among paralogs. Such versatility is clearly favored by the enrichment of intrinsically disordered regions (IDRs) within many metazoan MED subunits (56). Indeed, IDRs, which do not fold into stable three-dimensional globular structures, would be a natural way for the Mediator complex to adapt to the increasing diversity of transcriptional regulators during evolution (56, 57). Shown to interact with many TFs, Med1 has the largest IDR among MED subunits (57). Interestingly, partially conserved IDRs lie within a specific fragment of the longest Drosophila isoform, which we identified as the Srp and Pnr interacting domain (Fig. 7B) (57). It has been proposed recently that mammalian Med1 IDRs can form phase-separated droplets that compartmentalize and concentrate the transcription apparatus at superenhancers to drive robust gene expression (58). Thus, we propose that GATA interaction with Med1 IDRs concentrates MED-Pol II clusters at GATA-bound enhancers to activate transcription (59).
Differential roles for Med1 isoforms?
To our knowledge, our work reveals for the first time a TF partnership apparently restricted to one isoform of a Mediator subunit. Indeed, we have shown that (i) GATA/Srp only coprecipitates with the longest Med1A isoform in cultured cells, (ii) a domain specific to isoform A is sufficient to bind both Srp and Pnr in vitro, and (iii) a transgene ubiquitously expressing the Med1A isoform fully rescues the crystal cell differentiation defects of Med1-depleted embryos. More work is required to determine whether all of the GATA transactivation function of Med1 resides within the longest isoform or whether the shorter ones can partially fulfill this role or display antagonizing activity. With our antibody directed against a common protein portion, we showed that all three Drosophila Med1 isoforms are incorporated into the MED complex (4) and that Med1 is ubiquitously expressed in imaginal tissues, but we could not evaluate whether or not Med1 isoforms are differentially expressed. Nevertheless, this hypothesis is supported by the fact that the ratio between Med1 transcripts, as well as their relative levels, has been shown to change during development and in different cell lines (60). These data strongly suggest a physiological relevance of alternative transcript production for Med1 and pave the way for future research. If MED complexes containing different Med1 isoforms are functionally distinct, the view of MED as a unique ubiquitous entity would be challenged. Mediator would then be the name of a heterogeneous population of complexes with different regulatory specificities.
Med1 is not an obligatory partner of GATA factors.
We showed that Drosophila Med1 acts as a cofactor for the GATA factors Pnr and Srp, physically binding both TFs and GATA-type ZFs and mediating their target gene’s transactivation. This suggests that Med1 is an obligatory partner of GATA factors necessary to recruit the Pol II transcription machinery to GATA-activated promoters, as was proposed in mammals in earlier studies (6, 7). Nevertheless, we found here that Med1 is critical for achaete- but not for wingless-induced transactivation by Pnr. Similarly, it was shown that mammalian Med1 regulates only a limited subset of GATA1-dependent genes in erythroid cells (8) and that GATA1 recruits Med1 at activated genes but not at repressed loci (13). In addition, the analysis of different blood cell types produced from conditional Med1 knockout mice showed that Med1 participates in GATA1-dependent erythropoiesis but is dispensable for other GATA-dependent processes (12). These authors suggested that GATA factors, despite binding Med1 in vitro, contact other MED subunits to regulate their target genes in vivo. Along these lines, Med14, Med17, and Med25 have also been proposed as GATA1 interactors (7, 8).
It is now clear that the view of MED action as a binary partnership, i.e., one subunit to one TF, is too simplistic. It has been postulated that MED subunits act in a concerted manner, in a positive or negative way, by interacting simultaneously with one or several transcription factors and cofactors bound at gene enhancers, as well as promoters, to finely regulate gene expression in response to TFs. This attractive view of MED action as an integrative molecular hub device, transforming complex combinatorial inputs (TFs, cofactors, chromatin modifiers, etc.) into a simple transcriptional output, has rarely been tackled experimentally, particularly in metazoans. The use of Drosophila and the GATA-MED paradigm should allow us to explore this view in vivo.
MATERIALS AND METHODS
Drosophila stocks, genetic mosaics, and phenotypic analyses.
Stocks and crosses were maintained at 25°C on standard yeast-agar-cornmeal medium. The following stocks were used: P(EPgy2)EY01286, Df(3L)BSC418, FRT80B, UAS-Med1dsRNA number 13054 and number 13055 from VDRC, UAS-Med19VC, UAS-Med1VC (4), and DC-ac-lacZ (gift from P. Heitzler).
Mitotic clones were generated using the Flp-FRT system with the Med1O2 FRT80B chromosome. The Flp recombinase was expressed in the dorsal part of the wing using the ap-GAL4 driver (61) recombined with UAS-Flp. Clones were visualized using the Ub-GFP FRT80B chromosome or M(3)RpS17 ub-GFP FRT80B (BDSC) with the RFP-marked Med1O2 mutant chromosome.
Adult phenotypes were analyzed by light microscopy (Zeiss Axiophot) of dissected samples mounted in Hoyer's medium or by scanning electron microscopy (TM-1000 tabletop model; Hitachi) of frozen adults.
Genetic screens for generation of Med1 mutants and rescue experiments.
Mutant chromosomes were generated by mobilizing the P element (EY01286) inserted 118 bp upstream from the Med1 transcription start site, with the Δ2-3 P transposase source (FlyBase) and white-eyed flies selected. Recessive lethal chromosomes were subjected to complementation tests with a large deficiency removing Med1 [Df(3L)BSC418]. Two selected mutant chromosomes, Med1Δ9 and Med1Δ98, were further characterized genetically and molecularly by Southern blotting, PCR, and reverse transcription-PCR.
For the second-round mutagenesis screen, 1,000 isogenized FRT80B males were starved for 4 h, placed for 16 h on a piece of tissue soaked with 2 ml of 2% sucrose–100 mM EMS, and then individually crossed with TM3/TM6B females. F1 progeny was then tested for complementation in front of the previous Δ9 or Δ98 Med1 allele. One recessive-lethal Med1 mutant (Med1O2) was obtained. It was cleaned by recombination with a wild-type chromosome (bearing an RFP transgenic marker) to eliminate other potential EMS-induced mutations on the same chromosomal arm and was further tested for rescue by a Ub-Med1 transgene.
For rescue experiments, full-length Med1A cDNA (from the Berkeley Drosophila Genome Project) was inserted downstream of a general (ubiquitin63E) or an inducible (UAS) promoter into pUbHB1 and pUAST-attB vectors, respectively, and the corresponding constructs were used to generate transgenic lines by P-element transformation. Med1Δ9, Med1Δ98, and Med1O2 homozygotes were rescued by a single copy of a Ub-Med1A insert (number 1.2 on the 2nd chromosome).
Immunostaining and in situ hybridization.
Histology and antibody staining on imaginal discs were done as described previously (62), using rabbit anti-β-galactosidase (1/2,500; Cappel), mouse anti-Wg (1/100; from DSHB), and guinea pig anti-Med1 (1:1,000) (4). For in situ hybridization experiments, digoxigenin-UTP-labeled antisense RNA probes against pannier, spanning the whole ORF, were used. Fast-RED revelation was carried out at 4°C overnight. PPO2 in situ detection in embryos was done as described previously (38).
Pulldown experiments.
Bacterial cultures and preparation of GST fusion proteins, preparation of [35S]methionine-labeled protein probes, and pulldowns were carried out essentially as described previously (63). Chimeric GST-Med1 and GST-Med1ex4 fuse the GST moiety to the Med1A longest isoform or to its specific C-terminal part, respectively. Radiolabeled Pnr, Srp, or subfragments have been produced from Pnr cDNA corresponding to the PnrA isoform (25) and Srp cDNA used previously (26), producing a functional SrpB isoform lacking the first 300 amino acid residues and containing both N- and C-ZFs using in vitro transcription/translation-coupled reactions using rabbit reticulocyte extracts (TnT; Promega).
Cell culture, dsRNA treatment, and transfection.
Drosophila Kc167 cells were grown at 25°C in Schneider’s medium (Invitrogen) supplemented with 10% fetal bovine serum (PAA Cell Culture Company) and 50 μg of penicillin-streptomycin (Invitrogen). dsRNA treatment was performed as follows. Cells were incubated with dsRNA (1 μg/well for 96-well plate assays and 16 μg/well for 6-well plate assays) in serum-free medium for 40 min; serum-containing medium was subsequently added, and after 24 h of incubation cells were transfected using Effectene (Qiagen) according to the supplier’s instructions.
For immunoprecipitation experiments, Kc167 cells were transiently transfected with pAc-SrpNC-HA, pAc-Lz-V5, or pAc-GFP-Pnr-RA. Inducible cell lines expressing GFP-SrpC or GFP-SrpNC were established by cotransfection of pCoBlast and pRM-GFP-SrpC or pRM-GFP-SrpNC and by selection on 25 μg/ml Blasticidin medium according to the Drosophila expression system protocol (Invitrogen). Srp expression were induced with copper sulfate treatment (500 μM).
Luciferase reporter assay.
A mix of 20 ng of pAc-Rluc normalization plasmid and 50 ng of pGL3b-DC-ac-Fluc reporter (49) was cotransfected in each well (96-well plate) with various amounts of expression plasmids. DC-ac-Fluc reporter was induced with either 50 ng of pAc-V5 alone, 10 ng pAc-myc-Da/10 ng pAc-Flag-Ac/30 ng control pAc-V5, or 10 ng pAc-myc-Da/10 ng pAc-Flag-Ac/30 ng pAc-pnr per well (plasmids are kind gifts from P. Heitzler), with the exception of the pAc-V5 control (50 ng of empty pAcV5 per well). Cells were lysed 72 h after transfection, and activities of Firefly and Renilla luciferases were measured using the Dual-Luciferase reporter assay system (Promega). Each transfection was performed at least in triplicates.
Immunoprecipitations.
Kc167 cells were collected, washed in phosphate-buffered saline (PBS), and incubated for 30 min on 200 μl of IP buffer (150 mM NaCl, 0.5% NP-40, 50 mM Tris-HCl, pH 8.0, 1 mM EGTA supplemented with protease inhibitor cocktail; Roche). The extracts were cleared by centrifugation at 13,000 × g for 15 min at 4°C. Amounts of 1.5 (Med1 IP) (Fig. 2A), 2.5 (GFP-Srp IP) (Fig. 2B), or 3 mg (Med1 IP) (Fig. 6A and B) of proteins were preadsorbed with 30 μl of Sepharose bead slurry (Sigma) for 1 h at 4°C. Supernatants were recovered before being incubated with 1 μl of antibody at 4°C. After 1 h of incubation, 10 μl of protein A- or G-Sepharose beads (Sigma) was added for an overnight incubation. The beads were spun down and washed in IP buffer. Immunoprecipitated proteins were processed for SDS-PAGE and Western blot analyses (40 μg of proteins for input proteins and the totality of immunoprecipitated proteins) using standard techniques. The blots were developed by a photoluminescence procedure using Lumi-LightPLUS Western blotting substrate (64) and chemiluminescence film (GE Healthcare). The following antibodies were used for Western blotting: guinea pig anti-Med1 (1:1,000), rabbit anti-Med13 (1:5,000; from J. Treisman), guinea pig anti-Cdk8 (1:2,500) (65), mouse anti-Pnr (1:1,000) (42), guinea pig anti-Trinity (control IP; from F. Payre), mouse anti-HA (1:1,000; Covance), mouse anti-V5 (1:2,000; Invitrogen), rabbit anti-GFP (1:5,000; Torrey), rabbit IgG (Santa Cruz), donkey anti-rabbit antibody coupled to horseradish peroxidase (HRP) (1:100,000), and goat anti-guinea pig antibody (1:10,000) coupled to HRP (Jackson ImmunoResearch).
Sequence alignments were generated with SIAS (http://imed.med.ucm.es/Tools/sias.html).
ACKNOWLEDGMENTS
We are grateful to Cindy Cavelier, Benjamin Klapholtz, and Raphael Aguillon for participation in Med1 mutant generation and analysis, Marie Anais Thibiergen, Elodie Prince, and Julien Favier for aid in generating transgenic lines, and Alain Vincent, Francois Payre, Michèle Crozatier, Emmanuelle Guillou, and Caroline Monod for critical comments and valuable help. We thank the Toulouse RIO imaging platform for assistance with confocal microscopy. We deeply thank P. Heitzler, J. Treisman, the Bloomington Stock Center, and the Developmental Studies Hybridoma Bank for providing us with antibodies, plasmids, or fly stocks.
This work was supported by grants from the Ligue Nationale contre le Cancer (C.I. and A.P.), the Association pour la Recherche sur le Cancer (ARC PJA 20141201932), and the Agence Nationale de Recherche (ANR-16 CE12-0021-01), and we benefitted from the support of the Centre National de Recherche Scientifique (CNRS) and Toulouse III University.
REFERENCES
- 1.Jeronimo C, Robert F. 2017. The Mediator complex: at the nexus of RNA polymerase II transcription. Trends Cell Biol 27:765–783. doi: 10.1016/j.tcb.2017.07.001. [DOI] [PubMed] [Google Scholar]
- 2.Soutourina J. 2018. Transcription regulation by the Mediator complex. Nat Rev Mol Cell Biol 19:262–274. doi: 10.1038/nrm.2017.115. [DOI] [PubMed] [Google Scholar]
- 3.Yin JW, Wang G. 2014. The Mediator complex: a master coordinator of transcription and cell lineage development. Development 141:977–987. doi: 10.1242/dev.098392. [DOI] [PubMed] [Google Scholar]
- 4.Boube M, Hudry B, Immarigeon C, Carrier Y, Bernat-Fabre S, Merabet S, Graba Y, Bourbon HM, Cribbs DL. 2014. Drosophila melanogaster Hox transcription factors access the RNA polymerase II machinery through direct homeodomain binding to a conserved motif of Mediator subunit Med19. PLoS Genet 10:e1004303. doi: 10.1371/journal.pgen.1004303. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Fondell JD. 2013. The Mediator complex in thyroid hormone receptor action. Biochim Biophys Acta 1830:3867–3875. doi: 10.1016/j.bbagen.2012.02.012. [DOI] [PubMed] [Google Scholar]
- 6.Crawford SE, Qi C, Misra P, Stellmach V, Rao MS, Engel JD, Zhu Y, Reddy JK. 2002. Defects of the heart, eye, and megakaryocytes in peroxisome proliferator activator receptor-binding protein (PBP) null embryos implicate GATA family of transcription factors. J Biol Chem 277:3585–3592. doi: 10.1074/jbc.M107995200. [DOI] [PubMed] [Google Scholar]
- 7.Stumpf M, Waskow C, Krötschel M, van Essen D, Rodriguez P, Zhang X, Guyot B, Roeder RG, Borggrefe T. 2006. The mediator complex functions as a coactivator for GATA-1 in erythropoiesis via subunit Med1/TRAP220. Proc Natl Acad Sci U S A 103:18504–18509. doi: 10.1073/pnas.0604494103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Pope NJ, Bresnick EH. 2013. Establishment of a cell-type-specific genetic network by the mediator complex component Med1. Mol Cell Biol 33:1938–1955. doi: 10.1128/MCB.00141-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gordon DF, Tucker EA, Tundwal K, Hall H, Wood WM, Ridgway EC. 2006. MED220/thyroid receptor-associated protein 220 functions as a transcriptional coactivator with Pit-1 and GATA-2 on the thyrotropin-beta promoter in thyrotropes. Mol Endocrinol 20:1073–1089. doi: 10.1210/me.2005-0115. [DOI] [PubMed] [Google Scholar]
- 10.Wu D, Sunkel B, Chen Z, Liu X, Ye Z, Li Q, Grenade C, Ke J, Zhang C, Chen H, Nephew KP, Huang TH, Liu Z, Jin VX, Wang Q. 2014. Three-tiered role of the pioneer factor GATA2 in promoting androgen-dependent gene expression in prostate cancer. Nucleic Acids Res 42:3607–3622. doi: 10.1093/nar/gkt1382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Nakamura Y, Xing Y, Sasano H, Rainey WE. 2009. The mediator complex subunit 1 enhances transcription of genes needed for adrenal androgen production. Endocrinology 150:4145–4153. doi: 10.1210/en.2009-0006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Stumpf M, Yue X, Schmitz S, Luche H, Reddy JK, Borggrefe T. 2010. Specific erythroid-lineage defect in mice conditionally deficient for Mediator subunit Med1. Proc Natl Acad Sci U S A 107:21541–21546. doi: 10.1073/pnas.1005794107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Pope NJ, Bresnick EH. 2010. Differential coregulator requirements for function of the hematopoietic transcription factor GATA-1 at endogenous loci. Nucleic Acids Res 38:2190–2200. doi: 10.1093/nar/gkp1159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Boube M, Joulia L, Cribbs DL, Bourbon HM. 2002. Evidence for a mediator of RNA polymerase II transcriptional regulation conserved from yeast to man. Cell 110:143–151. [DOI] [PubMed] [Google Scholar]
- 15.Bourbon HM. 2008. Comparative genomics supports a deep evolutionary origin for the large, four-module transcriptional mediator complex. Nucleic Acids Res 36:3993–4008. doi: 10.1093/nar/gkn349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tsai KL, Yu X, Gopalan S, Chao TC, Zhang Y, Florens L, Washburn MP, Murakami K, Conaway RC, Conaway JW, Asturias FJ. 2017. Mediator structure and rearrangements required for holoenzyme formation. Nature 544:196–201. doi: 10.1038/nature21393. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Crispino JD, Horwitz MS. 2017. GATA factor mutations in hematologic disease. Blood 129:2103–2110. doi: 10.1182/blood-2016-09-687889. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stefanovic S, Christoffels VM. 2015. GATA-dependent transcriptional and epigenetic control of cardiac lineage specification and differentiation. Cell Mol Life Sci 72:3871–3881. doi: 10.1007/s00018-015-1974-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Sorrentino RP, Gajewski KM, Schulz RA. 2005. GATA factors in Drosophila heart and blood cell development. Semin Cell Dev Biol 16:107–116. doi: 10.1016/j.semcdb.2004.10.005. [DOI] [PubMed] [Google Scholar]
- 20.Zhang SJ, Ma LY, Huang QH, Li G, Gu BW, Gao XD, Shi JY, Wang YY, Gao L, Cai X, Ren RB, Zhu J, Chen Z, Chen SJ. 2008. Gain-of-function mutation of GATA-2 in acute myeloid transformation of chronic myeloid leukemia. Proc Natl Acad Sci U S A 105:2076–2081. doi: 10.1073/pnas.0711824105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Evans T, Reitman M, Felsenfeld G. 1988. An erythrocyte-specific DNA-binding factor recognizes a regulatory sequence common to all chicken globin genes. Proc Natl Acad Sci U S A 85:5976–5980. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Martin DI, Orkin SH. 1990. Transcriptional activation and DNA binding by the erythroid factor GF-1/NF-E1/Eryf 1. Genes Dev 4:1886–1898. [DOI] [PubMed] [Google Scholar]
- 23.Whyatt DJ, deBoer E, Grosveld F. 1993. The two zinc finger-like domains of GATA-1 have different DNA binding specificities. EMBO J 12:4993–5005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Trainor CD, Omichinski JG, Vandergon TL, Gronenborn AM, Clore GM, Felsenfeld G. 1996. A palindromic regulatory site within vertebrate GATA-1 promoters requires both zinc fingers of the GATA-1 DNA-binding domain for high-affinity interaction. Mol Cell Biol 16:2238–2247. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Ramain P, Heitzler P, Haenlin M, Simpson P. 1993. Pannier, a negative regulator of achaete and scute in Drosophila, encodes a zinc finger protein with homology to the vertebrate transcription factor GATA-1. Development 119:1277–1291. [DOI] [PubMed] [Google Scholar]
- 26.Waltzer L, Bataillé L, Peyrefitte S, Haenlin M. 2002. Two isoforms of Serpent containing either one or two GATA zinc fingers have different roles in Drosophila haematopoiesis. EMBO J 21:5477–5486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Tsang AP, Visvader JE, Turner CA, Fujiwara Y, Yu C, Weiss MJ, Crossley M, Orkin SH. 1997. FOG, a multitype zinc finger protein, acts as a cofactor for transcription factor GATA-1 in erythroid and megakaryocytic differentiation. Cell 90:109–119. [DOI] [PubMed] [Google Scholar]
- 28.Rodriguez P, Bonte E, Krijgsveld J, Kolodziej KE, Guyot B, Heck AJ, Vyas P, de Boer E, Grosveld F, Strouboulis J. 2005. GATA-1 forms distinct activating and repressive complexes in erythroid cells. EMBO J 24:2354–2366. doi: 10.1038/sj.emboj.7600702. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Tripic T, Deng W, Cheng Y, Zhang Y, Vakoc CR, Gregory GD, Hardison RC, Blobel GA. 2009. SCL and associated proteins distinguish active from repressive GATA transcription factor complexes. Blood 113:2191–2201. doi: 10.1182/blood-2008-07-169417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Wilkinson-White L, Gamsjaeger R, Dastmalchi S, Wienert B, Stokes PH, Crossley M, Mackay JP, Matthews JM. 2011. Structural basis of simultaneous recruitment of the transcriptional regulators LMO2 and FOG1/ZFPM1 by the transcription factor GATA1. Proc Natl Acad Sci U S A 108:14443–14448. doi: 10.1073/pnas.1105898108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Heitzler P, Vanolst L, Biryukova I, Ramain P. 2003. Enhancer-promoter communication mediated by Chip during Pannier-driven proneural patterning is regulated by Osa. Genes Dev 17:591–596. doi: 10.1101/gad.255703. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Asmar J, Biryukova I, Heitzler P. 2008. Drosophila dLMO-PA isoform acts as an early activator of achaete/scute proneural expression. Dev Biol 316:487–497. doi: 10.1016/j.ydbio.2008.01.040. [DOI] [PubMed] [Google Scholar]
- 33.Waltzer L, Ferjoux G, Bataillé L, Haenlin M. 2003. Cooperation between the GATA and RUNX factors Serpent and Lozenge during Drosophila hematopoiesis. EMBO J 22:6516–6525. doi: 10.1093/emboj/cdg622. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fossett N, Hyman K, Gajewski K, Orkin SH, Schulz RA. 2003. Combinatorial interactions of serpent, lozenge, and U-shaped regulate crystal cell lineage commitment during Drosophila hematopoiesis. Proc Natl Acad Sci U S A 100:11451–11456. doi: 10.1073/pnas.1635050100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Gobert V, Osman D, Bras S, Augé B, Boube M, Bourbon HM, Horn T, Boutros M, Haenlin M, Waltzer L. 2010. A genome-wide RNA interference screen identifies a differential role of the mediator CDK8 module subunits for GATA/RUNX-activated transcription in Drosophila. Mol Cell Biol 30:2837–2848. doi: 10.1128/MCB.01625-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kuuluvainen E, Hakala H, Havula E, Sahal Estimé M, Rämet M, Hietakangas V, Mäkelä TP. 2014. Cyclin-dependent kinase 8 module expression profiling reveals requirement of Mediator subunits 12 and 13 for transcription of Serpent-dependent innate immunity genes in Drosophila. J Biol Chem 289:16252–16261. doi: 10.1074/jbc.M113.541904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Bataillé L, Augé B, Ferjoux G, Haenlin M, Waltzer L. 2005. Resolving embryonic blood cell fate choice in Drosophila: interplay of GCM and RUNX factors. Development 132:4635–4644. doi: 10.1242/dev.02034. [DOI] [PubMed] [Google Scholar]
- 38.Ferjoux G, Augé B, Boyer K, Haenlin M, Waltzer L. 2007. A GATA/RUNX cis-regulatory module couples Drosophila blood cell commitment and differentiation into crystal cells. Dev Biol 305:726–734. doi: 10.1016/j.ydbio.2007.03.010. [DOI] [PubMed] [Google Scholar]
- 39.Graveley BR, Brooks AN, Carlson JW, Duff MO, Landolin JM, Yang L, Artieri CG, van Baren MJ, Boley N, Booth BW, Brown JB, Cherbas L, Davis CA, Dobin A, Li R, Lin W, Malone JH, Mattiuzzo NR, Miller D, Sturgill D, Tuch BB, Zaleski C, Zhang D, Blanchette M, Dudoit S, Eads B, Green RE, Hammonds A, Jiang L, Kapranov P, Langton L, Perrimon N, Sandler JE, Wan KH, Willingham A, Zhang Y, Zou Y, Andrews J, Bickel PJ, Brenner SE, Brent MR, Cherbas P, Gingeras TR, Hoskins RA, Kaufman TC, Oliver B, Celniker SE. 2011. The developmental transcriptome of Drosophila melanogaster. Nature 471:473–479. doi: 10.1038/nature09715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Ge K, Cho YW, Guo H, Hong TB, Guermah M, Ito M, Yu H, Kalkum M, Roeder RG. 2008. Alternative mechanisms by which mediator subunit MED1/TRAP220 regulates peroxisome proliferator-activated receptor gamma-stimulated adipogenesis and target gene expression. Mol Cell Biol 28:1081–1091. doi: 10.1128/MCB.00967-07. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Golic KG. 1991. Site-specific recombination between homologous chromosomes in Drosophila. Science 252:958–961. [DOI] [PubMed] [Google Scholar]
- 42.Morata G, Ripoll P. 1975. Minutes: mutants of drosophila autonomously affecting cell division rate. Dev Biol 42:211–221. [DOI] [PubMed] [Google Scholar]
- 43.Heitzler P, Haenlin M, Ramain P, Calleja M, Simpson P. 1996. A genetic analysis of pannier, a gene necessary for viability of dorsal tissues and bristle positioning in Drosophila. Genetics 143:1271–1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Calleja M, Herranz H, Estella C, Casal J, Lawrence P, Simpson P, Morata G. 2000. Generation of medial and lateral dorsal body domains by the pannier gene of Drosophila. Development 127:3971–3980. [DOI] [PubMed] [Google Scholar]
- 45.Boube M, Faucher C, Joulia L, Cribbs DL, Bourbon HM. 2000. Drosophila homologs of transcriptional mediator complex subunits are required for adult cell and segment identity specification. Genes Dev 14:2906–2917. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.García-García MJ, Ramain P, Simpson P, Modolell J. 1999. Different contributions of pannier and wingless to the patterning of the dorsal mesothorax of Drosophila. Development 126:3523–3532. [DOI] [PubMed] [Google Scholar]
- 47.Strigini M, Cohen SM. 2000. Wingless gradient formation in the Drosophila wing. Curr Biol 10:293–300. [DOI] [PubMed] [Google Scholar]
- 48.Ramain P, Khechumian R, Khechumian K, Arbogast N, Ackermann C, Heitzler P. 2000. Interactions between chip and the achaete/scute-daughterless heterodimers are required for pannier-driven proneural patterning. Mol Cell 6:781–790. [DOI] [PubMed] [Google Scholar]
- 49.Biryukova I, Heitzler P. 2008. Drosophila C-terminal binding protein, dCtBP is required for sensory organ prepattern and sharpens proneural transcriptional activity of the GATA factor Pnr. Dev Biol 323:64–75. doi: 10.1016/j.ydbio.2008.08.014. [DOI] [PubMed] [Google Scholar]
- 50.Ito M, Yuan CX, Okano HJ, Darnell RB, Roeder RG. 2000. Involvement of the TRAP220 component of the TRAP/SMCC coactivator complex in embryonic development and thyroid hormone action. Mol Cell 5:683–693. [DOI] [PubMed] [Google Scholar]
- 51.Malik S, Guermah M, Yuan CX, Wu W, Yamamura S, Roeder RG. 2004. Structural and functional organization of TRAP220, the TRAP/mediator subunit that is targeted by nuclear receptors. Mol Cell Biol 24:8244–8254. doi: 10.1128/MCB.24.18.8244-8254.2004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Taatjes DJ, Tjian R. 2004. Structure and function of CRSP/Med2; a promoter-selective transcriptional coactivator complex. Mol Cell 14:675–683. doi: 10.1016/j.molcel.2004.05.014. [DOI] [PubMed] [Google Scholar]
- 53.Ito M, Yuan CX, Malik S, Gu W, Fondell JD, Yamamura S, Fu ZY, Zhang X, Qin J, Roeder RG. 1999. Identity between TRAP and SMCC complexes indicates novel pathways for the function of nuclear receptors and diverse mammalian activators. Mol Cell 3:361–370. [DOI] [PubMed] [Google Scholar]
- 54.Xie XJ, Hsu FN, Gao X, Xu W, Ni JQ, Xing Y, Huang L, Hsiao HC, Zheng H, Wang C, Zheng Y, Xiaoli AM, Yang F, Bondos SE, Ji JY. 2015. CDK8-cyclin C mediates nutritional regulation of developmental transitions through the ecdysone receptor in Drosophila. PLoS Biol 13:e1002207. doi: 10.1371/journal.pbio.1002207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Mizuta S, Minami T, Fujita H, Kaminaga C, Matsui K, Ishino R, Fujita A, Oda K, Kawai A, Hasegawa N, Urahama N, Roeder RG, Ito M. 2014. CCAR1/CoCoA pair-mediated recruitment of the Mediator defines a novel pathway for GATA1 function. Genes Cells 19:28–51. doi: 10.1111/gtc.12104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Tóth-Petróczy A, Oldfield CJ, Simon I, Takagi Y, Dunker AK, Uversky VN, Fuxreiter M. 2008. Malleable machines in transcription regulation: the mediator complex. PLoS Comput Biol 4:e1000243. doi: 10.1371/journal.pcbi.1000243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Nagulapalli M, Maji S, Dwivedi N, Dahiya P, Thakur JK. 2016. Evolution of disorder in Mediator complex and its functional relevance. Nucleic Acids Res 44:1591–1612. doi: 10.1093/nar/gkv1135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Sabari BR, Dall'Agnese A, Boija A, Klein IA, Coffey EL, Shrinivas K, Abraham BJ, Hannett NM, Zamudio AV, Manteiga JC, Li CH, Guo YE, Day DS, Schuijers J, Vasile E, Malik S, Hnisz D, Lee TI, Cisse II, Roeder RG, Sharp PA, Chakraborty AK, Young RA. 2018. Coactivator condensation at super-enhancers links phase separation and gene control. Science 361:eaar3958. doi: 10.1126/science.aar3958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Cho WK, Spille JH, Hecht M, Lee C, Li C, Grube V, Cisse II. 2018. Mediator and RNA polymerase II clusters associate in transcription-dependent condensates. Science 361:412–415. doi: 10.1126/science.aar4199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Tikhonov M, Georgiev P, Maksimenko O. 2013. Competition within introns: splicing wins over polyadenylation via a general mechanism. Acta Naturae 5:52–61. [PMC free article] [PubMed] [Google Scholar]
- 61.Calleja M, Moreno E, Pelaz S, Morata G. 1996. Visualization of gene expression in living adult Drosophila. Science 274:252–255. [DOI] [PubMed] [Google Scholar]
- 62.Baanannou A, Mojica-Vazquez LH, Darras G, Couderc JL, Cribbs DL, Boube M, Bourbon HM. 2013. Drosophila distal-less and Rotund bind a single enhancer ensuring reliable and robust bric-a-brac2 expression in distinct limb morphogenetic fields. PLoS Genet 9:e1003581. doi: 10.1371/journal.pgen.1003581. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Mojica-Vázquez LH, Benetah MH, Baanannou A, Bernat-Fabre S, Deplancke B, Cribbs DL, Bourbon HM, Boube M. 2017. Tissue-specific enhancer repression through molecular integration of cell signaling inputs. PLoS Genet 13:e1006718. doi: 10.1371/journal.pgen.1006718. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Lin AC, Roche AE, Wilk J, Svensson EC. 2004. The N termini of Friend of GATA (FOG) proteins define a novel transcriptional repression motif and a superfamily of transcriptional repressors. J Biol Chem 279:55017–55023. doi: 10.1074/jbc.M411240200. [DOI] [PubMed] [Google Scholar]
- 65.Loncle N, Boube M, Joulia L, Boschiero C, Werner M, Cribbs DL, Bourbon HM. 2007. Distinct roles for Mediator Cdk8 module subunits in Drosophila development. EMBO J 26:1045–1054. doi: 10.1038/sj.emboj.7601566. [DOI] [PMC free article] [PubMed] [Google Scholar]