Abstract
Clinical application of siRNA-based therapeutics outside of the liver has been hindered by the inefficient delivery of siRNA effector molecules into extra-hepatic organs and cells of interest. To understand the parameters that enable RNAi activity in vivo, it is necessary to develop a systematic approach to identify which cells within a tissue are permissive to oligonucleotide internalization and activity. In the present study, we evaluate the distribution and activity within the lung of chemically stabilized siRNA to characterize cell-type tropism and structure-activity relationship. We demonstrate intratracheal delivery of fully modified siRNA for RNAi-mediated target knockdown in lung CD11c+ cells (dendritic cells, alveolar macrophages) and alveolar epithelial cells. Finally, we use an allergen-induced model of lung inflammation to demonstrate the capacity of inhaled siRNA to induce target knockdown in dendritic cells and ameliorate lung pathology.
Keywords: siRNA, delivery, lung, RNAi
Introduction
RNAi technology offers the ability to silence virtually any expressed mRNA, including molecular targets causal for human disease that are not amenable to conventional approaches.1 Clinical development of small interfering RNA (siRNA)-based therapeutics outside liver has been limited primarily by poor bioavailability to extra-hepatic tissues and cell types of interest. When delivered systemically, siRNA formulated within a lipid nanoparticle or carrying a hepatocyte-specific targeting ligand can trigger near-complete and highly specific mRNA silencing of numerous targets in the liver at well-tolerated doses.2, 3, 4 For certain indications, local delivery of siRNA (e.g., mucosal, intraocular, intrathecal) may be more desirable than the standard intravenous or subcutaneous administration, as it may enable higher oligonucleotide concentrations in relevant tissues and protection from systemic nucleases.5, 6 However, previous studies on local siRNA administration have yielded variable and unpredictable results, limiting its clinical relevance. These confounding results arise primarily by the need to achieve sufficient metabolic stability of siRNA while preserving its intrinsic RNAi activity and to avoid potential siRNA-induced innate immune responses, which have been historical challenges in the field. Overall, there is still a general paucity in understanding parameters that influence siRNA bioavailability and efficacy in non-liver tissues.
The lung is an accessible tissue with attractive therapeutic targets and unmet medical needs and has been the focus of several RNAi studies in recent years.7 In humans, ALN-RSV01, an unmodified siRNA targeting the RSV nucleocapsid (N) gene, advanced to phase IIb clinical trials, where it missed its primary endpoint but nevertheless showed a clinically meaningful treatment effect.8, 9 In preclinical studies, specificity of ALN-RSV01 was demonstrated in vivo by using mismatched controls, and a theoretical protective effect through innate immune activation was ruled out as immunogenic non-specific siRNAs lacked antiviral efficacy, while a chemically modified form of ALN-RSV01 with no detectable immunostimulatory capacity retained full activity in vivo.10 Furthermore, the RNAi mechanism of action of ALN-RSV01 was demonstrated in vivo by detecting site-specific cleavage product of the RSV mRNA. Additional studies report inhaled naked siRNA efficacy against parainfluenza virus (PIV), as well as lung xenograft tumor growth.11, 12 Other groups have described how intratracheal or intranasal delivery of siRNA targeting effectors of Th2-driven inflammation like SOCS3 and STAT6 improved disease outcome in rodents, although direct target engagement in pathology-relevant cells was not conclusively demonstrated.13, 14 In contrast with these reports, Moschos et al.15 concluded that intratracheally delivered siRNA could not be internalized by cells and therefore lacked efficacy in the lung. Importantly, it should be noted that the lung delivery reports so far have used unmodified or partially modified siRNAs that are expected to have poor metabolic stability.
To better translate the abundance of preclinical studies into the clinic, it will be necessary to fully understand the identity of cells in the lung permissive to oligonucleotide internalization and activity. The airway and lung are composed of a wide variety of cells with specific roles in pathology and homeostasis, including specialized epithelial cells, vascular endothelial cells, and different hematopoietic cell subpopulations with complex immunological functions. In particular, the lung immune compartment contains pharmacological targets that can be modulated for therapeutic benefit in asthma, COPD, and autoimmune disease. Flow cytometry and cell sorting technologies have allowed a better definition of lung parenchymal cells and resident leukocyte populations.16, 17 In the present study, we have taken advantage of these technologies, particularly advances in fully modified siRNAs to provide exceptional metabolic stability,18 to evaluate the distribution and activity of siRNA within the lung for a systematic characterization of cell-type tropism and structure-activity relationship of siRNA chemistry. Finally, we use the allergen-induced model of lung inflammation to demonstrate the capacity of inhaled siRNA to ameliorate lung pathology.
Results
Intratracheal Delivery of Chemically Modified siRNA Induces RNAi-Mediated Target mRNA Knockdown in the Lung
The siRNA sequences and chemical modification schemes used in this manuscript can be found in Figure S1. All constructs, except si-Ctnnb1 2′-OH, contain extensive 2′-fluoro/methoxy ribose modifications and position-specific phosphorothioate backbone linkages known to improve uptake, stability, and bioavailability of siRNA.18, 19, 20, 21 Additional chemistries utilized include inverted abasic ribose caps at both ends of the passenger (sense) strand and a stable phosphate mimic recently described.22, 23 Modification patterns used in the study, particularly in the guide strand, are largely conserved so that the specific pattern used does not have a major impact in their relative stability and RNAi activity.
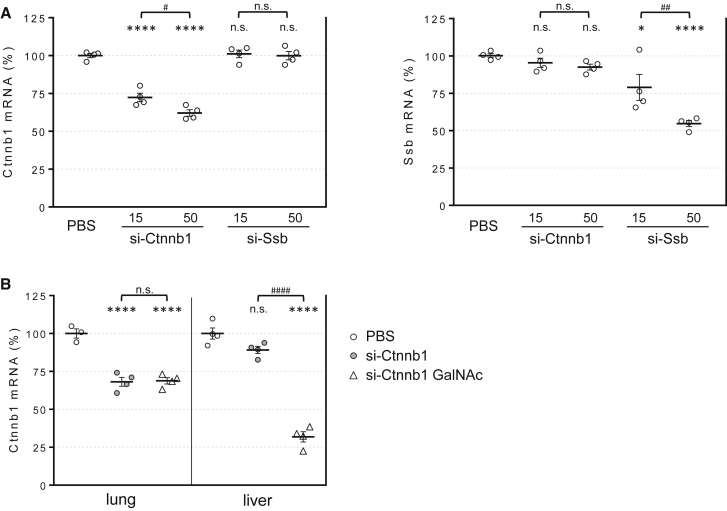
To evaluate if RNAi-mediated activity can be induced in the mouse lung after local siRNA administration, we generated siRNAs against two ubiquitously expressed gene targets, β-catenin (Ctnnb1), and Sjögren syndrome antigen B (Ssb). The sequences and chemical modification schemes used in this manuscript can be found in Figure S1. Synthesized siRNAs were dissolved in PBS and delivered without conjugation to cellular uptake agents (e.g., cholesterol) or association to any transfection-promoting reagents (e.g., liposomes) intratracheally using a microsprayer aerosolizer. Analysis of total lung RNA 3 days post-treatment revealed significant target knockdown in a dose-dependent fashion (up to 40% at 50 mg/kg) (Figure 1A). Importantly, crossover controls demonstrated that this effect was observed in each case only for the targeted gene, demonstrating mRNA knockdown occurs through an RNAi-mediated mechanism and not through non-specific inflammation-related events.
Figure 1.
siRNA-Induced Target Knockdown in the Lung Is Gene Specific, Dose Dependent, and Ligand Independent
(A) Total RNA was isolated from lungs of mice 3 days after intratracheal dosing with chemically modified siRNA targeting β-catenin (si-Ctnnb1) or Ssb (si-Ssb) at 15 or 50 mg/kg doses. Ctnnb1 and Ssb mRNA levels were determined by real-time PCR and expressed relative to Ppib loading control. (B) Ctnnb1 mRNA levels were determined in lungs and livers from mice dosed with untargeted (si-Ctnnb1) or GalNAc-conjugated (si-Ctnnb1 GalNAc) siRNA at 15 mg/kg. Lines indicate means ± SE. *p < 0.05, ****p < 0.0001 versus PBS-treated mice, #p < 0.05, ##p < 0.005, ####p < 0.0001; n.s., not significant.
The in vivo activity of siRNA in the absence of lipid or polymer transfection agents has only been conclusively demonstrated in liver so far24, 25 and requires conjugation to a hepatocyte-targeting ligand like multivalent N-acetylgalactosamine3 or a lipophilic moiety like cholesterol.26 Moschos et al.15 reported that intratracheally administered antisense oligonucleotides escaping the lung into blood circulation can accumulate and induce target knockdown in the liver. Correspondingly, we observed liver Ctnnb1 silencing after siRNA lung administration. Importantly, knockdown was observed only when siRNA molecules were conjugated to multimeric galactose N-acetyl (GalNAc) (Figure 1B). Overall, these results indicate the presence of cells in the lung that can passively take up siRNA (i.e., without a cell-targeting ligand) and are permissive to RNAi activity. Furthermore, the results demonstrate the potential of lung-administered siRNA to have an effect in distal tissues like liver.
Chemical siRNA Modification Is Required to Avoid Immune Activation in the Lung and Induce Target Gene Knockdown
The immunostimulatory activity of double-stranded RNA is caused primarily by activation of the type I interferon pathway by endosomal toll-like receptors.27 However, siRNA-induced immunostimulation can be extensively diminished by careful selection of the nucleotide sequence to avoid certain dinucleotide motifs28, 29 or chemical modifications.30, 31
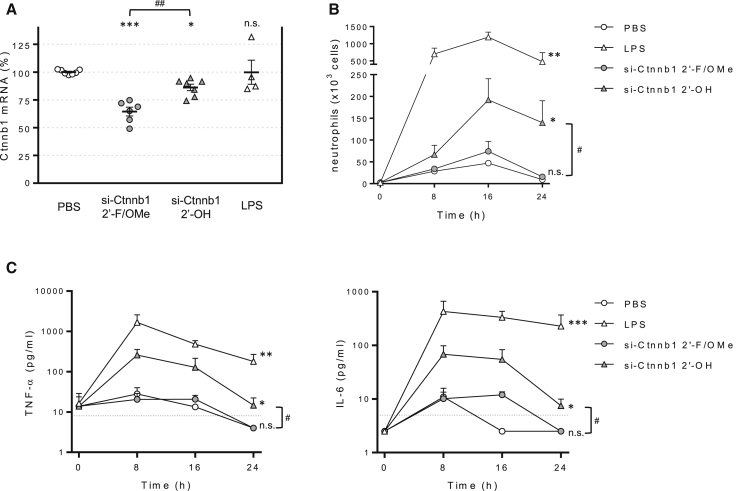
To confirm that our fully modified siRNA design strategy prevented activation of mucosal immunity, mice were dosed intratracheally with sequence-identical siRNAs carrying extensive chemical modifications (si-Ctnnb1 2′-F/OMe) or largely unmodified (siCtnnb1 2′-OH). As shown in Figure 2A, silencing activity triggered by chemically modified siRNA was significantly higher, suggesting a dependence on oligonucleotide stability for lung RNAi activity. We then compared the ability of si-Ctnnb1-2′-OH and si-Ctnnb1 to induce inflammation. A time course analysis, which included E. coli lipopolysaccharide (LPS) as a positive control, demonstrated that the neutrophilia (Figure 2B) and pro-inflammatory cytokine elevation in the BAL fluid (Figure 2C) induced by si-Ctnnb1 2′-OH were abrogated by the modifications included in the active si-Ctnnb1 siRNA. Intranasal LPS administration, which induces a strong inflammatory response, had no significant effect on Ctnnb1 levels. These data demonstrate that the modifications introduced in our siRNA design successfully prevent the activation of the innate immune system in the lung and rule out the possibility that non-specific inflammation contributes to the decrease in target mRNA levels.
Figure 2.
Extensive siRNA Chemical Modification Is Required to Induce RNAi Activity and Avoid an Immune Response
(A) Mice were dosed intratracheally with 50 mg/kg siRNA chemically modified (si-Ctnnb1 2′-F/OMe) or minimally modified (si-Ctnnb1 2′-OH) targeting the same nucleotide primary sequence. Plot represents Ctnnb1 mRNA in total lung relative to Ppib control 72 h after intratracheal dosing. Lines indicate means ± SE. (B) BAL neutrophil content after intratracheal dosing of PBS (empty circles), si-Ctnnb1 2′-F/OMe (filled circles), si-Ctnnb1 2′-OH (filled triangles), and LPS (empty triangles). (C) BAL cytokine levels were measured using the BD cytometric bead array (CBA) kit. Discontinued lines represent lower limit of detection (LOD). Data are means + SE. *p < 0.05, **p < 0.005, ***p < 0.001 versus PBS-treated mice, #p < 0.001, ##p < 0.005; n.s., not significant.
Visualization of Fluorescent siRNA Cellular Distribution in the Lung
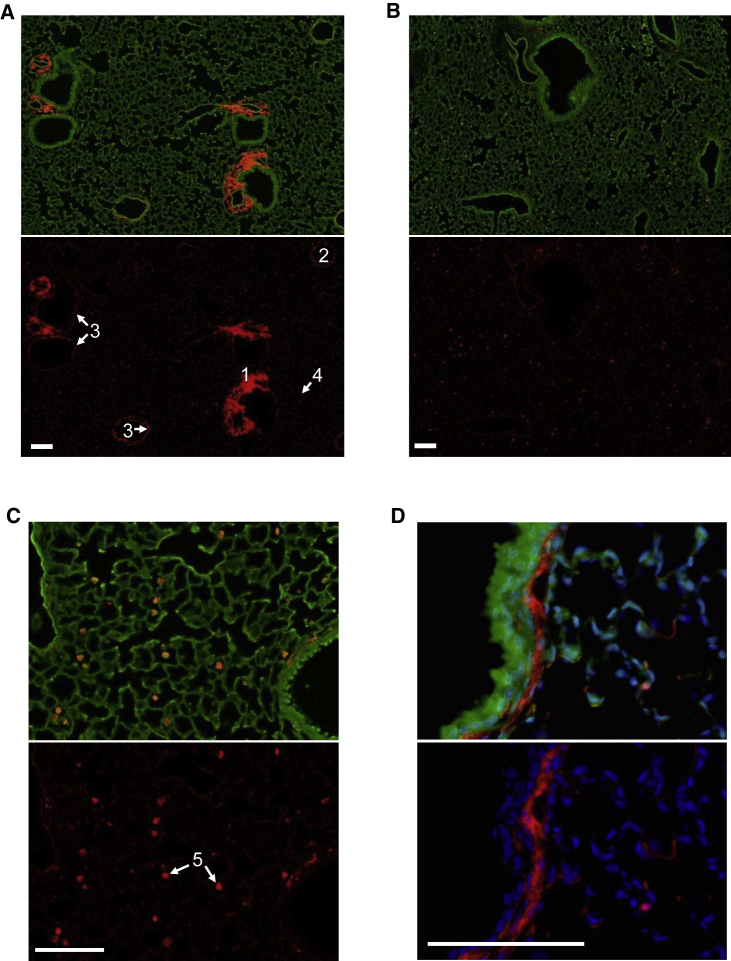
As a first step to characterize the suborgan distribution of siRNA, we examined lung histology sections from mice dosed with fluorescently labeled siRNA targeting Ctnnb1 (si-Ctnnb1 DyLight650). Analysis of mRNA levels showed that conjugation of the fluorescent label did not affect the RNAi activity of the siRNA (data not shown). The study revealed that early (2 h) after dosing by intratracheal delivery, siRNA localizes primarily to the muscular layer surrounding central arteries, with significant localization also to endothelial cells, the submucosal layer of upper airways, and alveolar epithelia (Figure 3A). This distribution pattern is subsequently lost, as the intense staining in the arterial region is significantly decreased 24 h post-dosing (Figure 3B). After 72 h, most of the fluorescent signal is observed in alveolar macrophages (Figure 3C). Importantly, we did not detect fluorescent siRNA in the bronchial epithelium at any of the time points analyzed. Higher magnification analysis at 2 h post-dose reveals siRNA localization to the lamina propria and smooth muscle layer area surrounding bronchi and bronchioles (Figure 3D). However, siRNAs designed against a smooth-muscle-specific gene marker (Pde4d) failed to induce gene knockdown activity at total lung RNA level, suggesting there is no productive siRNA delivery to smooth muscle cells (data not shown). Taken together, our data suggests that chemically modified unconjugated siRNA can distribute passively into lung cells, primarily alveolar macrophages and alveolar epithelia.
Figure 3.
siRNA Accumulates in Alveolar Macrophages and Can Be Detected in Alveolar Epithelial Cells
(A) Sections from lungs excised 2 h after intratracheal dosing of fluorescently labeled siRNA (si-Ctnnb1 DyLight 650). (1) Arterial muscular fiber, (2) vein endothelial cells, (3) upper airway submucosa, (4) alveolar epithelia. (B) Lung sections 72 h after intratracheal dosing of labeled siRNA. (C) siRNA accumulation in alveolar macrophages (5) 72 h post-dosing. (D) siRNA localization to the submucosal layer 2 h post-dosing. Scale bars, 100 μm.
Lung Cell Fractionation Demonstrates siRNA-Mediated Target Gene Knockdown in CD11c+ Cells and Non-hematopoietic Cells
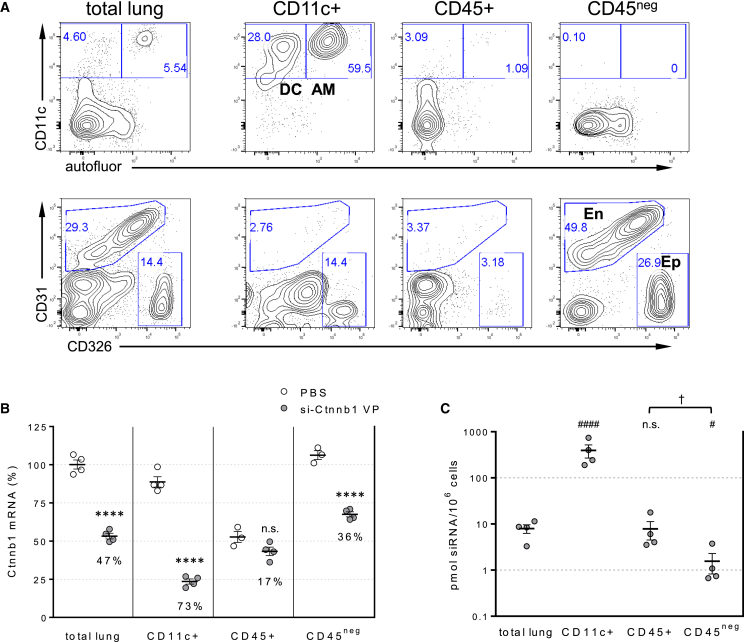
To directly assess siRNA distribution and activity in disease-relevant lung cell subpopulations, we developed a tissue disruption-cell-sorting protocol similar to the one described by Moschos et al.15 Mouse lungs were enzymatically dissociated, and the resulting single-cell lung mix suspension was sequentially fractionated using MACS microbead technology into CD11c+ cell, CD11c-depleted CD45+ cell, and CD45neg cell subsets (Figure 4A). As previously shown, the CD11c+ cell subset contains dendritic cells and alveolar macrophages that are easily identified by their different autofluorescence.32 Analysis of the CD45+ cell subset containing hematopoietic cells confirmed the lack of CD11c+ cells, demonstrating the success of the cell separation approach. Staining with pan-endothelial (CD31) and epithelial (CD326) cell markers segregates the CD45neg cell subset (non-hematopoietic cells) into CD31negCD326+ cells (containing most epithelial cells represented in lung), CD31+ cells (endothelial cells), and CD31negCD326neg cells (fibroblasts smooth muscle cells).
Figure 4.
siRNA Internalization and Activity in Discrete Lung Cell Subpopulations
(A) FACS analysis of dissociated lung tissues before (total lung) and after separation into alveolar macrophages and dendritic cells (CD11c+), CD11c-depleted hematopoietic cells (CD45+), and non-hematopoietic cells (CD45neg). AM, alveolar macrophages; D, dendritic cells; En, endothelial cells; Ep, epithelial cells. Numbers indicate percent of events gated in that area. (B) Plot represents Ctnnb1 mRNA in isolated cell subsets relative to Ppib mRNA control 72 h after 50 mg/kg siRNA intratracheal dosing. Numbers in the plot indicate Ctnnb1 mRNA knockdown relative to the respective PBS control. (C) siRNA content in isolated cell subsets was determined by stem-loop PCR. Lines indicate means ± SE. ****p < 0.0001 versus PBS-treated cell subset; ####p < 0.0001, #p < 0.05 versus total lung; †p < 0.05; n.s., not significant.
Analysis of Ctnnb1 mRNA levels in cell subsets isolated 3 days after 50 mg/kg siRNA dosing revealed significant differences in siRNA activity between different lung cell types. In contrast to the study from Moschos et al.,15 we observed strong target mRNA knockdown (>70%) in alveolar macrophages and dendritic cells (CD11c+), far higher than that observed in the starting total lung cell suspension. In addition, a lower but significant and reproducible knockdown (>35%) was detected in the non-hematopoietic (CD45neg) cell subset, while no significant activity was observed in CD11c-depleted hematopoietic cells (Figure 4B). A secondary observation from this analysis is that the CD45+ cells only contained about half of the Ctnnb1 target mRNA as the other populations, reflecting intrinsic differences in tissue-specific gene expression patterns.
Tissue siRNA levels, as measured by stem-loop qRT-PCR (Figure 4C), were significantly higher in in the CD11c+ subset compared to the total lung cell suspension (>20-fold), correlating well with the strong RNAi activity observed in this subpopulation. In contrast, the CD45neg (non-hematopoietic) population, which showed lower but reproducible RNAi activity, contained 5-fold lower siRNA than the total lung cell suspension. These tissue siRNA measurements also correlate well with the accumulation of fluorescent siRNA in alveolar macrophages (Figure 3). Taken together, these results demonstrate that different cell types within the lung are differentially permissive to siRNA internalization and RNAi activity.
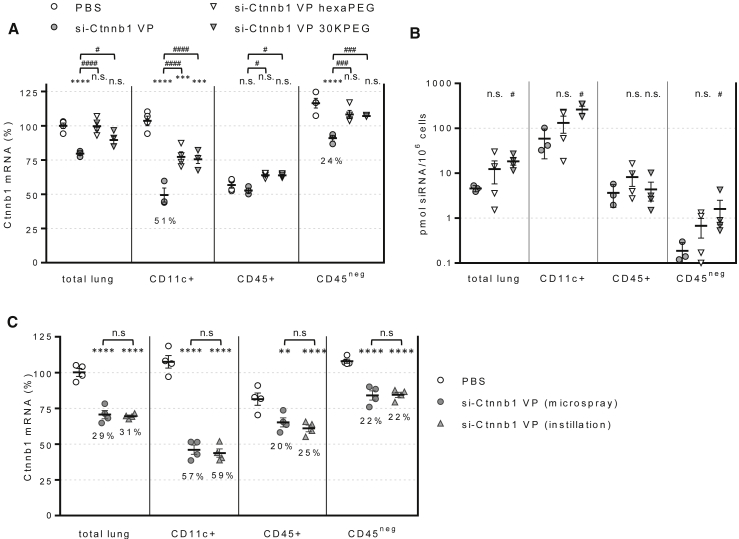
RNAi Activity in the Lung Is Inhibited by siRNA PEGylation
Attachment of polyethylene glycol (PEG) polymer is a common procedure in drug research aimed to improve pharmacokinetics of small molecules and biologics.33 For siRNA, PEGylation of the passenger strand has been shown to be irrelevant to intrinsic silencing activity while prolonging systemic circulation.34 We reasoned that the higher molecular weight of PEGylated siRNA could potentially increase its lung retention and activity by reducing escape through blood capillaries. To assess this possibility, we synthesized two siRNAs carrying PEG moieties of different molecular weight and configuration. Analysis of target mRNA levels 3 days post-dosing revealed that, despite carrying identical guide strands, passenger strand PEGylation rendered the siRNAs inactive (Figure 5A). Interestingly, the loss of activity did not correlate with lower siRNA content in the lung subsets analyzed. In fact, in support of our hypothesis, PEGylated siRNA content in the CD45neg cell population was up to 10-fold higher than control siRNA (Figure 5B). The results suggest that PEG modification results in changes to intracellular trafficking mechanisms that prevent RNAi activity.
Figure 5.
siRNA Activity Is Affected by siRNA PEGylation and Does Not Require Aerosolization
(A) Plot represents Ctnnb1 mRNA in isolated cell subsets relative to Ppib control 72 h after 15 mg/kg siRNA intratracheal dosing. (B) siRNA content in isolated cell subsets determined by stem-loop PCR. (C) Ctnnb1 mRNA levels were determined 72 h after dosing 50 mg/kg siRNA intratracheally with a microsprayer syringe or by direct instillation. Lines indicate means ± SE. **p < 0.005, ***p < 0.001, ****p < 0.0001 versus PBS-treated cell subset; #p < 0.05, ###p < 0.001, ####p < 0.0001 versus si-Ctnnb1 VP-dosed subsets; n.s., not significant.
siRNA Activity and Distribution in the Lung Is Independent of Dose Aerosolization
Intratracheal delivery via microsprayer aerosolizer allows homogeneous drug distribution into the lower airways.35 In an effort to achieve siRNA delivery into the upper airways epithelium, mice were dosed by liquid bolus intratracheal instillation with a pipette. However, the results revealed no differences in target gene knockdown in any of the isolated cell subsets, indicating that microspraying and instillation delivery results in similar siRNA distribution and activity in the lung. Importantly, the unchanged gene target knockdown in the CD45neg population indicates that bronchial epithelial cells are not permissive to RNAi, at least not with this siRNA structural class (Figure 5C).
siRNA Activity Observed in Non-hematopoietic Lung Cells Occurs Primarily in CD326+ Epithelial Cells
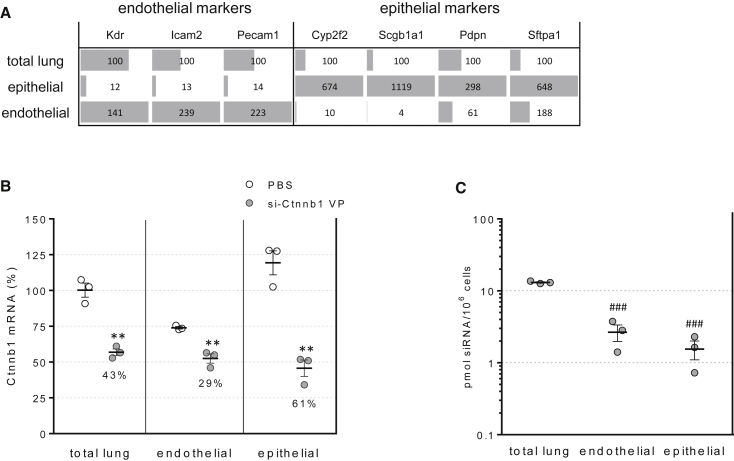
To establish the identity of cells in the lung non-hematopoietic (CD45neg) subset that are permissive to siRNA activity, we isolated endothelial (defined as CD45neg CD31+) and epithelial (CD45neg CD31neg CD326+) cell subsets with over 90% purity using a fluorescence-activated cell sorter (FACS) from mouse lungs 3 days after siRNA dosing. Analysis of mRNA expression of pan-endothelial markers Vegfr2/Kdr, CD102/Icam2, and CD31/Pecam1 demonstrated the effective separation of endothelial cells from the mixed lung population (Figure 6A). Likewise, analysis of Cyp2f2 and Scgb1a1 (club cells), Pdpn (alveolar epithelium I), and Sftpa1 (alveolar epithelium II) demonstrated the successful separation of a subset of epithelial cells. Lung-cell-specific markers Tubb4 (ciliated cells), Acta2 (smooth muscle cells), and Muc5ac (goblet cells) did not segregate into the epithelial or endothelial subsets, indicating cells expressing these markers are present in the triple-negative (CD45neg CD31neg CD326neg) subset (data not shown).
Figure 6.
siRNA Internalization and Activity in FACS-Isolated Lung Epithelial and Endothelial Cells
(A) Expression of cell-specific markers for endothelial cells (Kdr, Icam2, Pecam1), club cells (Cyp2f2, Scg1a1), type I (Pdpn), and type II (Sftpa1) epithelial cells in dissociated lung tissue before (total lung) and after FACS-purification of CD45negCD326+CD31neg (epithelial) and CD45negCD31+ (endothelial) cell subsets. (B) Ctnnb1 mRNA levels were determined in dissociated lung tissue and FACS-isolated subsets 72 h post-dosing of 15 mg/kg siRNA. (C) siRNA content was determined by stem-loop PCR. Lines indicate means ± SE. **p < 0.005 versus PBS-treated cell subset, ###p < 0.0005 versus total lung cells.
Analysis of Ctnnb1 mRNA levels revealed target mRNA knockdown in both the epithelial (61%) and the endothelial cell subsets (29%) (Figure 6B), in agreement with the localization of fluorescently labeled siRNA to alveolar epithelial cells previously discussed (Figure 3). Despite the 3-fold higher knockdown observed in epithelial cells, siRNA levels were not higher in the epithelial cell subset (Figure 6C), suggesting that the difference in RNAi activity between the endothelial and epithelial cells were likely caused by differential subcellular trafficking and cytosolic partitioning, which ultimately determines the siRNA concentration accessible to the RNA-induced silencing complex (RISC).4
Administration of siRNA against OX40L Inhibits Th2-Mediated Memory Response in the OVA-Induced Lung Inflammation Model
Previous studies have established the role of dendritic-cell-expressed OX40L as a downstream amplifier of the allergen-triggered Th2 lung inflammation cascade.36 For example, administration of a blocking anti-OX40L monoclonal antibody inhibited Th2 responses in the ovalbumin (OVA)-induced allergic lung inflammation mouse model. Importantly, this inhibitory effect occurs in the recall and not the primary effector response, suggesting that OX40L likely plays a major role in vivo during reactivation of memory Th2 responses.37, 38 To evaluate the effectiveness of RNAi-based technology in modulating lung pathology, we tested an OX40L-targeting siRNA in this phenotypic model.
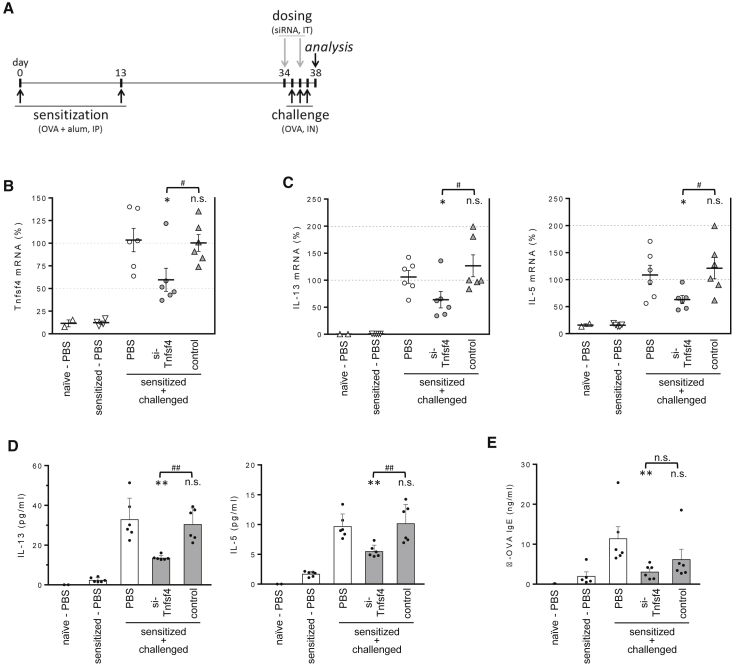
Mice were sensitized by intraperitoneal injection of OVA protein plus alum on days 0 and 13 and challenged intranasally with soluble OVA on days 35–37. Two siRNA (50 mg/kg) doses were given on days 34 and 36, and lungs were collected on day 37 to assess OX40L knockdown and Th2 recall responses (Figure 7A). As treatment controls, mice were dosed with a non-specific siRNA or PBS only.
Figure 7.
Administration of siRNA Targeting OX40L Impairs Th2 Response in the OVA-Induced Lung Inflammation Model
(A) Experimental timeline. (B) Tnfsf4 (OX40L) mRNA levels were determined in total lung RNA from mice dosed with siRNA targeting Tnfsf4 or siRNA control. (C) Th2 cytokine mRNA levels in BAL fluid were determined by RT-PCR. Lines indicate means ± SE. (D) Th2 cytokine protein levels in BAL fluid. (E) Serum OVA-specific IgE were determined by ELISA. Data was analyzed using one-way ANOVA with post-hoc Dunnett’s test. Data are means + SE. *p < 0.05, **p < 0.005 versus challenged, PBS-dosed control group; #p < 0.05, ##p < 0.005, n.s., not significant.
As expected, OX40L mRNA levels in total lung were almost undetectable in sensitized-only (no challenge) mice and were upregulated (>100-fold) upon intranasal challenge. Importantly, mice dosed with OX40L-specific siRNA showed significant downregulation of the target gene, while control siRNA had no effect (Figure 7B). As expected due to the effect of OX40L modulating T cell memory response, siRNA-treated mice with reduced OX40L expression had significant lower levels of Th2-associated cytokines interleukin-13 (IL-13) and IL-5 mRNA in total lung (Figure 7C) and protein in bronchoalveolar lavage (BAL) fluid (Figure 7D). Serum antigen-specific immunoglobulin E (IgE) was also lower in OX40L-siRNA-dosed mice, further demonstrating inhibition of the Th2 recall response (Figure 7E). While we did not detect a statistical significant decrease in lung IL-4 and eosinophil recruitment, all together these results recapitulate published studies analyzing the effect of blocking antibody38 and demonstrate the capacity of local siRNA delivery to modulate biological outcomes in the lung.
Discussion
Induction of RNAi activity is dependent on endogenous cellular machinery. Once internalized and engaged by cytosolic RNA induced silencing complex (RISC), the RNA duplex is unraveled and the guide (antisense) strand is bound to Ago2 protein, the endonuclease responsible for site-specific degradation of the target mRNA. Because siRNAs are large, polar, non-membrane-permeable molecules, they typically require a lipid or polymer-based transfection agent or a targeting ligand to achieve efficient cellular uptake. Internalization and cytosolic release of single-stranded oligonucleotides heavily modified with phosphorothioate backbone can occur without the use of special delivery vehicles but remains an inefficient process.39 It is well established, however, that some cell types are far more permissive than others to oligonucleotide delivery,40 making necessary the characterization of siRNA biodistribution at a cellular resolution rather than solely at the organ level.
Early studies analyzing intratracheal and intranasal siRNA delivery reported downregulation of mRNA targets that induced biological effects in animal models of lung pathology, suggesting the presence of cells in the lung permissive to siRNA internalization and activity.11, 41 In particular, it was demonstrated that ALN-RSV101 showed antiviral activity via RNAi mechanism, but it failed to advance in clinic.8, 10 These findings contrast with later studies concluding that naked siRNA is not delivered productively to lung cells after topical administration and cannot mediate RNA interference.15, 42, 43 Furthermore, many of the initial reports on siRNA lung activity have been attributed to non-specific effects, particularly activation of innate immune response.44, 45 The conflicting reports underscore the need for a systematic approach to characterize the intrinsic siRNA permissibility of cells in the lung.
Addressing this problem, Moschos et al.15 used tissue disruption and cell sorting to conclude that intratracheally delivered unmodified siRNA could not internalize into cells and therefore could not mediate RNAi activity in the lung. In the same study, chemically modified antisense oligonucleotides were shown to accumulate in alveolar macrophages and lung epithelia but did not induce target knockdown. We now conclusively demonstrate that chemically modified siRNA can indeed mediate RNAi activity in multiple cells in the lung while avoiding immune system activation. The conflicting results can be explained by our optimization of chemical modifications that render the siRNA more resistant to lung nucleases and/or lysosomal degradation, increasing its in vivo half-life and facilitating cytosolic release, respectively. In correspondence with this idea, we observed that lung mRNA silencing is highly dependent on such chemical modifications, which confer resistance to circulating and intracellular nucleases. This finding agrees with the reports of extensive chemical modifications needed for intracellular stability of siRNA conjugates in liver.3
To our knowledge, the work presented here represents the most comprehensive characterization of lung siRNA distribution and activity and demonstrates that intratracheally delivered siRNA results in target knockdown 3 days post-dosing in lung cells, particularly CD11c+ cells (dendritic cells, alveolar macrophages) and alveolar epithelia. Importantly, no targeting or other large functional group was required on the molecule to achieve pharmacological activity. Similar results were observed 7 days post-dosing or using an siRNA against a different target (Ssb), thus demonstrating the sequence-specific effect and by using a bolus instillation instead of intratracheal delivery. Importantly, unlike unmodified siRNAs, delivery of chemically modified siRNAs did not induce inflammation in the lung or induce marked histological changes in the lung, nor did we observe any significant change in relative cell numbers or staining pattern in the lungs of dosed animals, except for increased autofluorescence of alveolar macrophages.
Stem-loop PCR analysis revealed that most of the siRNA in the lung 3 days post-dosing was contained in CD11c+ cells (400 pmol/106 cells), so it is not surprising that this is the cell subset with the highest target knockdown observed (70%). However, a direct correlation between cellular siRNA content and mRNA knockdown cannot be established, since the epithelial (CD45negCD326+CD31neg) cell subset showed similar mRNA knockdown (60%) with 200-fold lower siRNA levels. Likewise, PEG conjugation abolished the activity of siRNA molecules while actually inducing a significant increase in cellular siRNA content. The results clearly demonstrate how cell-intrinsic differences in siRNA internalization and trafficking mechanisms that can be regulated by the oligonucleotide design define RNAi enablement at the cellular level.
Multiple studies disrupting the OX40L-OX40 signaling axis and employing dendritic cell (DC) depletion approaches have demonstrated a crucial role for DC-expressed OX40L in recall responses in the lung.36, 37 Using a protocol similar to those that established the effectiveness of anti-OX40L-blocking antibodies,38 we have demonstrated that intratracheal delivery of siRNA induces gene silencing within the DC-containing lung CD11c+ cell population and modulates lung pathology. Furthermore, we have shown these effects were dependent on the siRNA modification pattern employed, demonstrating that siRNA stabilization by chemical modifications is absolutely required for in vivo activity. In correspondence with the role of DC-expressed OX40L in memory Th2 responses, we observed a downregulation of IL-13, IL-5, and IgE in siRNA-dosed mice. However, we did not see statistically significant decrease in IL-4 or lung eosinophilia that would be expected by a complete downregulation of the Th2 recall response. Seshasayee et al. have proposed that study protocols using two sensitization doses instead of one result in longer-lasting effector responses that are not fully suppressed by OX40L-blocking reagents.38 Considering this possibility, we started challenging mice 21 days after our second sensitization dose. For comparison, a Hoshino et al.46 study challenging mice starting at 10 days after the second dose reported no significant suppression of the Th2 recall response by OX40L-blocking reagents. Therefore, it is possible that study protocols with longer prechallenge periods would result in even stronger downregulation of Th2-associated parameters. Alternatively, a combination approach with another agent targeting a different stage in the Th2 pathway could have a synergistic protective effect.47 The systematic characterization of siRNA tropism and in vivo activity as described here, together with recent advances in optimal modifications for improved in vivo potency, can pave the way for the development of newer siRNAs as inhaled reagents for human therapy.48
Materials and Methods
Animals
BALB/c mice (6–8 weeks old) were purchased from Taconic and housed in the animal facilities of Merck & Co., Inc., West Point, PA, USA. All animal experiments conform to the NIH guidelines for the care and use of animals in biomedical research and were approved by the Institutional Animal Care and Use Committee of Merck & Co., Inc., Kenilworth, NJ, USA.
Design and In Vivo Delivery of siRNA
Primary sequences and chemical modification schemes of siRNA targeting mouse Ctnnb1, Ssb, Tnsf4, and siRNA controls employed in our studies are summarized in Figure S1. Oligonucleotides were dissolved and dosed in 50 μL PBS. Animals sedated by inhaled isofluorane were dosed intratracheally using a microsprayer aerosolizer high pressure syringe (Penn-Century).
Lung Imaging
Animals were dosed with si-Ctnnb1 siRNA labeled with DyLight650 fluorophore (Thermo Scientific) and sacrificed 2 or 72 h post-dosing. Tracheas were cannulated with a 20G teflon angiocath catheter (BD Biosciences) to inflate the lungs with 1 mL of 10% neutral-buffered formalin solution (Sigma-Aldrich) and tied with surgical suture. Lungs were excised and immersed in additional 10% neutral-buffered formalin solution for 24 h, paraffin-processed, and sectioned at 5 μm. Lung section slides were stained with DAPI immediately before imaging. Images were acquired in a sequential mode for the blue (DAPI), green (autofluorescence), and far-red (fluorescently labeled si-Ctnnb1) channels using the appropriate excitation and emission settings in an Ariol system (Leica Biosystems) using a Leica DM6000 microscope.
Lung Dissociation
Immediately after euthanasia, mouse tracheas were cannulated and lungs were inflated with 2 mL collagenase A (Roche, 0.8 mg/mL) and 10% fetal bovine serum (FBS) in RPMI solution. Lungs were excised into a 50-mL conical tube with additional collagenase A solution (5 mL) and placed in a 37°C water bath without shaking. After 25 min incubation, cold PBS was added to 25 mL and vigorously shaken until complete dissociation of lung tissue. The suspension was filtered through a 100-μm strainer into a new 50-mL conical tube and mixed with 25 mL cold red blood cell (RBC) lysis solution (BioLegend). Cells were immediately pelleted and resuspended in 2% FBS/0.2 mM EDTA/PBS (10 mL), filtered through a 40 μm strainer, and counted in a Vi-CELL cell viability analyzer.
Antibodies and Flow Cytometry
Dissociated lung cells were treated with Fc block (BD Biosciences) before staining with the following fluorophore-conjugated antibodies obtained from eBioscience: CD326-PE, CD45.2-PerCP-Cy5.5, CD11c-PE-Cy7, and CD31-APC. Viability was assessed with LIVE/DEAD fixable near-IR cell stain (Life Technologies). No fluorescein isothiocyanate (FITC)-conjugated antibody was employed to help identify autofluorescent alveolar macrophages. Flow cytometry analysis was performed on a BD FACSCanto system.
Lung Cell Subset Isolation
Cell suspensions from each mouse lung were resuspended in 1 mL of cold sort buffer (2% FBS/1.5 mM EDTA/PBS) containing Fc block (BD Biosciences). After 5 min incubation on ice, 100 μL of CD11c microbeads (Miltenyi Biotec) were added and incubated on ice for another 15 min. Cells were washed, resuspended in cold sort buffer (500 μL), and fractionated with an AutoMACS Pro (Miltenyi Biotec) using the Possel_s setting. Cells in the positive-selected fraction (CD11c+ cells) were counted and lysed with 250 μL TRIzol (Life Technologies). The negative-selected fraction was mixed with 100 μL of CD45 microbeads (Miltenyi Biotec), incubated for 15 min on ice, and fractionated with the AutoMACS Pro using the Possel_s setting. Cells in the positive-selected fraction (CD11c-depleted CD45+ cells) and the negative-selected fraction (CD45-negative cells) were counted and lysed with 250 μL and 1 mL of TRIzol, respectively. Cell subset purity was assessed to be over 95% by flow cytometry analysis. Alternatively, for isolation of endothelial and epithelial cell subsets, lung cell suspensions were stained with viability dye and fluorophore-conjugated antibodies and sorted into TRIzol LS reagent (Life Technologies) using a BD FACSJazz system. An epithelial cells subset was identified as CD45negCD326+CD31neg cells, and endothelial cells were identified as CD45negCD326+ cells.
Quantification of siRNA Uptake and Activity
siRNA guide-strand quantitation was performed by stem-loop RT-qPCR from total tissue homogenate as described.49 For mRNA quantitation, total cellular RNA was extracted from TRIzol following manufacturer’s instructions, and cDNA was synthesized with the high capacity cDNA reverse transcription kit (Applied Biosystems). TaqMan gene expression assays were purchased from Applied Biosystems for analysis on a 7900HT FAST real-time PCR system. All samples were run in duplicate, and relative mRNA expression levels were determined after normalizing each value to Ppib mRNA, as described previously.50
BAL Fluid Analysis
BAL fluid was isolated by four consecutive washes (0.5 mL, 0.7 mL, 1 mL, 1 mL) with 0.5 mM EDTA/cOmplete protease inhibitor cocktail (Roche) in Ca2+/Mg2+-free Dulbecco’s PBS (DPBS). Fluid recovered from the first two washes were pooled and centrifuged. Supernatant was kept for cytokine analysis using BD cytometric bead array (BD Biosciences), and pelleted cells were pooled with cells from the additional two washes. Differential counts were performed as described by van Rijt et al.51
OVA-Induced Allergic Lung Inflammation
BALB/c mice were sensitized on days 0 and 13 by intraperitoneal injection of 50 μg OVA protein (chicken egg albumin, Sigma-Aldrich) emulsified in 2 mg Imject alum adjuvant (aluminum hydroxide, Thermo Scientific) in PBS. On days 35, 36, and 37, mice were anesthetized with inhaled isofluorane (Forane, Abbot) and challenged by intranasal instillation of 50 μg of OVA protein in 50 μL of PBS alone. On days 34 and 36, mice were dosed with siRNA in sterile PBS or PBS alone by intratracheal microspraying. Finally, all animals were sacrificed 24 h after the last OVA administration for analysis.
Statistical Analysis
All statistical analyses were performed using Prism 5.0 software (GraphPad Software). Each dataset is presented as the mean ± SEM unless indicated otherwise. The significance of the difference between two mean group values was analyzed using unpaired, two-tailed Student t tests or one-way ANOVA with Dunnett test analyses for more than two groups, to determine if there was a statistical significance of at least α = 0.05. Asterisks indicate a statistically significant difference between the group and its respective control group, unless otherwise indicated by a line.
Author Contributions
B.N., T.C.-M., Y.W., V.D., and J.D. performed experiments, contributed to experiment design, and assisted in data interpretation; J.B. helped design experiments and assisted in data interpretation and statistical analysis; J.S. and D.B. designed and optimized siRNAs; V.J. and L.S.-L. supervised planning of experiments and data interpretation and contributed to the preparation of the manuscript; P.J.C. wrote the manuscript, designed and performed experiments, and analyzed the data.
Conflicts of Interest
The authors declare no competing financial interests.
Footnotes
Supplemental Information can be found with this article online at https://doi.org/10.1016/j.omtn.2019.02.013.
Supplemental Information
References
- 1.de Fougerolles A., Vornlocher H.P., Maraganore J., Lieberman J. Interfering with disease: a progress report on siRNA-based therapeutics. Nat. Rev. Drug Discov. 2007;6:443–453. doi: 10.1038/nrd2310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Leung A.K., Tam Y.Y., Cullis P.R. Lipid nanoparticles for short interfering RNA delivery. Adv. Genet. 2014;88:71–110. doi: 10.1016/B978-0-12-800148-6.00004-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Nair J.K., Willoughby J.L., Chan A., Charisse K., Alam M.R., Wang Q., Hoekstra M., Kandasamy P., Kel’in A.V., Milstein S. Multivalent N-acetylgalactosamine-conjugated siRNA localizes in hepatocytes and elicits robust RNAi-mediated gene silencing. J. Am. Chem. Soc. 2014;136:16958–16961. doi: 10.1021/ja505986a. [DOI] [PubMed] [Google Scholar]
- 4.Shi B., Abrams M. Technologies for investigating the physiological barriers to efficient lipid nanoparticle-siRNA delivery. J. Histochem. Cytochem. 2013;61:407–420. doi: 10.1369/0022155413484152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Krebs M.D., Alsberg E. Localized, targeted, and sustained siRNA delivery. Chemistry. 2011;17:3054–3062. doi: 10.1002/chem.201003144. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vicentini F.T., Borgheti-Cardoso L.N., Depieri L.V., de Macedo Mano D., Abelha T.F., Petrilli R., Bentley M.V. Delivery systems and local administration routes for therapeutic siRNA. Pharm. Res. 2013;30:915–931. doi: 10.1007/s11095-013-0971-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Koli U., Krishnan R.A., Pofali P., Jain R., Dandekar P. SiRNA-based therapies for pulmonary diseases. J. Biomed. Nanotechnol. 2014;10:1953–1997. doi: 10.1166/jbn.2014.1928. [DOI] [PubMed] [Google Scholar]
- 8.Gottlieb J., Zamora M.R., Hodges T., Musk A.W., Sommerwerk U., Dilling D., Arcasoy S., DeVincenzo J., Karsten V., Shah S. ALN-RSV01 for prevention of bronchiolitis obliterans syndrome after respiratory syncytial virus infection in lung transplant recipients. J. Heart Lung Transplant. 2016;35:213–221. doi: 10.1016/j.healun.2015.08.012. [DOI] [PubMed] [Google Scholar]
- 9.Zamora M.R., Budev M., Rolfe M., Gottlieb J., Humar A., Devincenzo J., Vaishnaw A., Cehelsky J., Albert G., Nochur S. RNA interference therapy in lung transplant patients infected with respiratory syncytial virus. Am. J. Respir. Crit. Care Med. 2011;183:531–538. doi: 10.1164/rccm.201003-0422OC. [DOI] [PubMed] [Google Scholar]
- 10.Alvarez R., Elbashir S., Borland T., Toudjarska I., Hadwiger P., John M., Roehl I., Morskaya S.S., Martinello R., Kahn J. RNA interference-mediated silencing of the respiratory syncytial virus nucleocapsid defines a potent antiviral strategy. Antimicrob. Agents Chemother. 2009;53:3952–3962. doi: 10.1128/AAC.00014-09. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bitko V., Musiyenko A., Shulyayeva O., Barik S. Inhibition of respiratory viruses by nasally administered siRNA. Nat. Med. 2005;11:50–55. doi: 10.1038/nm1164. [DOI] [PubMed] [Google Scholar]
- 12.Fujita Y., Kuwano K., Ochiya T. Development of small RNA delivery systems for lung cancer therapy. Int. J. Mol. Sci. 2015;16:5254–5270. doi: 10.3390/ijms16035254. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Healey G.D., Lockridge J.A., Zinnen S., Hopkin J.M., Richards I., Walker W. Development of pre-clinical models for evaluating the therapeutic potential of candidate siRNA targeting STAT6. PLoS ONE. 2014;9:e90338. doi: 10.1371/journal.pone.0090338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Zafra M.P., Mazzeo C., Gámez C., Rodriguez Marco A., de Zulueta A., Sanz V., Bilbao I., Ruiz-Cabello J., Zubeldia J.M., del Pozo V. Gene silencing of SOCS3 by siRNA intranasal delivery inhibits asthma phenotype in mice. PLoS ONE. 2014;9:e91996. doi: 10.1371/journal.pone.0091996. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Moschos S.A., Frick M., Taylor B., Turnpenny P., Graves H., Spink K.G., Brady K., Lamb D., Collins D., Rockel T.D. Uptake, efficacy, and systemic distribution of naked, inhaled short interfering RNA (siRNA) and locked nucleic acid (LNA) antisense. Mol. Ther. 2011;19:2163–2168. doi: 10.1038/mt.2011.206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Bantikassegn A., Song X., Politi K. Isolation of epithelial, endothelial, and immune cells from lungs of transgenic mice with oncogene-induced lung adenocarcinomas. Am. J. Respir. Cell Mol. Biol. 2015;52:409–417. doi: 10.1165/rcmb.2014-0312MA. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Messier E.M., Mason R.J., Kosmider B. Efficient and rapid isolation and purification of mouse alveolar type II epithelial cells. Exp. Lung Res. 2012;38:363–373. doi: 10.3109/01902148.2012.713077. [DOI] [PubMed] [Google Scholar]
- 18.Carr, B.A., Jadhav, V.R. Kenski, D.M., Tellers, D.M., and Willingham, A.T. (2012). Short Interfering Nucleic Acid (siNA) Compositions. International patent WO2013165816A3, filed April 26, 2013, and published November 7, 2013.
- 19.Allerson C.R., Sioufi N., Jarres R., Prakash T.P., Naik N., Berdeja A., Wanders L., Griffey R.H., Swayze E.E., Bhat B. Fully 2′-modified oligonucleotide duplexes with improved in vitro potency and stability compared to unmodified small interfering RNA. J. Med. Chem. 2005;48:901–904. doi: 10.1021/jm049167j. [DOI] [PubMed] [Google Scholar]
- 20.Detzer A., Sczakiel G. Phosphorothioate-stimulated uptake of siRNA by mammalian cells: a novel route for delivery. Curr. Top. Med. Chem. 2009;9:1109–1116. doi: 10.2174/156802609789630884. [DOI] [PubMed] [Google Scholar]
- 21.Manoharan M. RNA interference and chemically modified small interfering RNAs. Curr. Opin. Chem. Biol. 2004;8:570–579. doi: 10.1016/j.cbpa.2004.10.007. [DOI] [PubMed] [Google Scholar]
- 22.Parmar R., Willoughby J.L., Liu J., Foster D.J., Brigham B., Theile C.S., Charisse K., Akinc A., Guidry E., Pei Y. 5′-(E)-Vinylphosphonate: A Stable Phosphate Mimic Can Improve the RNAi Activity of siRNA-GalNAc Conjugates. ChemBioChem. 2016;17:985–989. doi: 10.1002/cbic.201600130. [DOI] [PubMed] [Google Scholar]
- 23.Prakash T.P., Kinberger G.A., Murray H.M., Chappell A., Riney S., Graham M.J., Lima W.F., Swayze E.E., Seth P.P. Synergistic effect of phosphorothioate, 5′-vinylphosphonate and GalNAc modifications for enhancing activity of synthetic siRNA. Bioorg. Med. Chem. Lett. 2016;26:2817–2820. doi: 10.1016/j.bmcl.2016.04.063. [DOI] [PubMed] [Google Scholar]
- 24.Fitzgerald K., White S., Borodovsky A., Bettencourt B.R., Strahs A., Clausen V., Wijngaard P., Horton J.D., Taubel J., Brooks A. A Highly Durable RNAi Therapeutic Inhibitor of PCSK9. N. Engl. J. Med. 2017;376:41–51. doi: 10.1056/NEJMoa1609243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Morrissey D.V., Blanchard K., Shaw L., Jensen K., Lockridge J.A., Dickinson B., McSwiggen J.A., Vargeese C., Bowman K., Shaffer C.S. Activity of stabilized short interfering RNA in a mouse model of hepatitis B virus replication. Hepatology. 2005;41:1349–1356. doi: 10.1002/hep.20702. [DOI] [PubMed] [Google Scholar]
- 26.Wolfrum C., Shi S., Jayaprakash K.N., Jayaraman M., Wang G., Pandey R.K., Rajeev K.G., Nakayama T., Charrise K., Ndungo E.M. Mechanisms and optimization of in vivo delivery of lipophilic siRNAs. Nat. Biotechnol. 2007;25:1149–1157. doi: 10.1038/nbt1339. [DOI] [PubMed] [Google Scholar]
- 27.Dalpke A., Helm M. RNA mediated Toll-like receptor stimulation in health and disease. RNA Biol. 2012;9:828–842. doi: 10.4161/rna.20206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Goodchild A., Nopper N., King A., Doan T., Tanudji M., Arndt G.M., Poidinger M., Rivory L.P., Passioura T. Sequence determinants of innate immune activation by short interfering RNAs. BMC Immunol. 2009;10:40. doi: 10.1186/1471-2172-10-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Sioud M. Induction of inflammatory cytokines and interferon responses by double-stranded and single-stranded siRNAs is sequence-dependent and requires endosomal localization. J. Mol. Biol. 2005;348:1079–1090. doi: 10.1016/j.jmb.2005.03.013. [DOI] [PubMed] [Google Scholar]
- 30.Judge A.D., Bola G., Lee A.C., MacLachlan I. Design of noninflammatory synthetic siRNA mediating potent gene silencing in vivo. Mol. Ther. 2006;13:494–505. doi: 10.1016/j.ymthe.2005.11.002. [DOI] [PubMed] [Google Scholar]
- 31.Morrissey D.V., Lockridge J.A., Shaw L., Blanchard K., Jensen K., Breen W., Hartsough K., Machemer L., Radka S., Jadhav V. Potent and persistent in vivo anti-HBV activity of chemically modified siRNAs. Nat. Biotechnol. 2005;23:1002–1007. doi: 10.1038/nbt1122. [DOI] [PubMed] [Google Scholar]
- 32.Vermaelen K., Pauwels R. Accurate and simple discrimination of mouse pulmonary dendritic cell and macrophage populations by flow cytometry: methodology and new insights. Cytometry A. 2004;61:170–177. doi: 10.1002/cyto.a.20064. [DOI] [PubMed] [Google Scholar]
- 33.Ishihara H. Current status and prospects of polyethyleneglycol-modified medicines. Biol. Pharm. Bull. 2013;36:883–888. doi: 10.1248/bpb.b13-00087. [DOI] [PubMed] [Google Scholar]
- 34.Iversen F., Yang C., Dagnæs-Hansen F., Schaffert D.H., Kjems J., Gao S. Optimized siRNA-PEG conjugates for extended blood circulation and reduced urine excretion in mice. Theranostics. 2013;3:201–209. doi: 10.7150/thno.5743. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Fujita Y., Takeshita F., Kuwano K., Ochiya T. RNAi Therapeutic Platforms for Lung Diseases. Pharmaceuticals (Basel) 2013;6:223–250. doi: 10.3390/ph6020223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Liu Y.J. Thymic stromal lymphopoietin and OX40 ligand pathway in the initiation of dendritic cell-mediated allergic inflammation. J. Allergy Clin. Immunol. 2007;120:238–244. doi: 10.1016/j.jaci.2007.06.004. quiz 245–246. [DOI] [PubMed] [Google Scholar]
- 37.Salek-Ardakani S., Song J., Halteman B.S., Jember A.G., Akiba H., Yagita H., Croft M. OX40 (CD134) controls memory T helper 2 cells that drive lung inflammation. J. Exp. Med. 2003;198:315–324. doi: 10.1084/jem.20021937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Seshasayee D., Lee W.P., Zhou M., Shu J., Suto E., Zhang J., Diehl L., Austin C.D., Meng Y.G., Tan M. In vivo blockade of OX40 ligand inhibits thymic stromal lymphopoietin driven atopic inflammation. J. Clin. Invest. 2007;117:3868–3878. doi: 10.1172/JCI33559. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Habibian M., Yahyaee-Anzahaee M., Lucic M., Moroz E., Martín-Pintado N., Di Giovanni L.D., Leroux J.C., Hall J., González C., Damha M.J. Structural properties and gene-silencing activity of chemically modified DNA-RNA hybrids with parallel orientation. Nucleic Acids Res. 2018;46:1614–1623. doi: 10.1093/nar/gky024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Geary R.S., Norris D., Yu R., Bennett C.F. Pharmacokinetics, biodistribution and cell uptake of antisense oligonucleotides. Adv. Drug Deliv. Rev. 2015;87:46–51. doi: 10.1016/j.addr.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 41.Li B.J., Tang Q., Cheng D., Qin C., Xie F.Y., Wei Q., Xu J., Liu Y., Zheng B.J., Woodle M.C. Using siRNA in prophylactic and therapeutic regimens against SARS coronavirus in Rhesus macaque. Nat. Med. 2005;11:944–951. doi: 10.1038/nm1280. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Glud S.Z., Bramsen J.B., Dagnaes-Hansen F., Wengel J., Howard K.A., Nyengaard J.R., Kjems J. Naked siLNA-mediated gene silencing of lung bronchoepithelium EGFP expression after intravenous administration. Oligonucleotides. 2009;19:163–168. doi: 10.1089/oli.2008.0175. [DOI] [PubMed] [Google Scholar]
- 43.Griesenbach U., Kitson C., Escudero Garcia S., Farley R., Singh C., Somerton L., Painter H., Smith R.L., Gill D.R., Hyde S.C. Inefficient cationic lipid-mediated siRNA and antisense oligonucleotide transfer to airway epithelial cells in vivo. Respir. Res. 2006;7:26. doi: 10.1186/1465-9921-7-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Kleinman M.E., Yamada K., Takeda A., Chandrasekaran V., Nozaki M., Baffi J.Z., Albuquerque R.J., Yamasaki S., Itaya M., Pan Y. Sequence- and target-independent angiogenesis suppression by siRNA via TLR3. Nature. 2008;452:591–597. doi: 10.1038/nature06765. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Robbins M., Judge A., Ambegia E., Choi C., Yaworski E., Palmer L., McClintock K., MacLachlan I. Misinterpreting the therapeutic effects of small interfering RNA caused by immune stimulation. Hum. Gene Ther. 2008;19:991–999. doi: 10.1089/hum.2008.131. [DOI] [PubMed] [Google Scholar]
- 46.Hoshino A., Tanaka Y., Akiba H., Asakura Y., Mita Y., Sakurai T., Takaoka A., Nakaike S., Ishii N., Sugamura K. Critical role for OX40 ligand in the development of pathogenic Th2 cells in a murine model of asthma. Eur. J. Immunol. 2003;33:861–869. doi: 10.1002/eji.200323455. [DOI] [PubMed] [Google Scholar]
- 47.Choi M., Gu J., Lee M., Rhim T. A new combination therapy for asthma using dual-function dexamethasone-conjugated polyethylenimine and vitamin D binding protein siRNA. Gene Ther. 2017;24:727–734. doi: 10.1038/gt.2017.83. [DOI] [PubMed] [Google Scholar]
- 48.Foster D.J., Brown C.R., Shaikh S., Trapp C., Schlegel M.K., Qian K., Sehgal A., Rajeev K.G., Jadhav V., Manoharan M. Advanced siRNA Designs Further Improve In Vivo Performance of GalNAc-siRNA Conjugates. Mol. Ther. 2018;26:708–717. doi: 10.1016/j.ymthe.2017.12.021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Seitzer J., Zhang H., Koser M., Pei Y., Abrams M. Effect of biological matrix and sample preparation on qPCR quantitation of siRNA drugs in animal tissues. J. Pharmacol. Toxicol. Methods. 2011;63:168–173. doi: 10.1016/j.vascn.2010.09.005. [DOI] [PubMed] [Google Scholar]
- 50.Tadin-Strapps M., Peterson L.B., Cumiskey A.M., Rosa R.L., Mendoza V.H., Castro-Perez J., Puig O., Zhang L., Strapps W.R., Yendluri S. siRNA-induced liver ApoB knockdown lowers serum LDL-cholesterol in a mouse model with human-like serum lipids. J. Lipid Res. 2011;52:1084–1097. doi: 10.1194/jlr.M012872. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.van Rijt L.S., Kuipers H., Vos N., Hijdra D., Hoogsteden H.C., Lambrecht B.N. A rapid flow cytometric method for determining the cellular composition of bronchoalveolar lavage fluid cells in mouse models of asthma. J. Immunol. Methods. 2004;288:111–121. doi: 10.1016/j.jim.2004.03.004. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.