Abstract
Mixed species biofilms are shaped and influenced by interactions between species. In the oral cavity, dysbiosis of the microbiome leads to diseases such as periodontitis. Porphyromonas gingivalis is a keystone pathogen of periodontitis. In this study, we showed that polymicrobial biofilm formation promoted the tolerance of Porphyromonas gingivalis to oxidative stress under micro-aerobic conditions. The presence of Streptococcus sanguinis, an oral commensal bacterium, inhibited the survival of P. gingivalis in dual-species biofilms via the secretion of hydrogen peroxide (H2O2). Interestingly, this repression could be attenuated by the presence of Aggregatibacter actinomycetemcomitans in tri-species biofilms. It was also shown that the katA gene, encoding a cytoplasmic catalase in A. actinomycetemcomitans, was responsible for the reduction of H2O2 produced by S. sanguinis, which consequently increased the biomass of P. gingivalis in tri-species biofilms. Collectively, these findings reveal that polymicrobial interactions play important roles in shaping bacterial community in biofilm. The existence of catalase producers may support the colonization of pathogens vulnerable to H2O2, in the oral cavity. The catalase may be a potential drug target to aid in the prevention of periodontitis.
Introduction
According to the 2016 global burden of disease study, periodontal disease is estimated to affect 750,847 million people worldwide, making it the 11th most prevalent human disease1. It is an inflammatory disorder, characterized by the destruction of tooth-supporting tissues such as gingiva, periodontal ligament and alveolar bone. Periodontitis is caused by the dysbiosis of the oral microbiome. In the pathogenesis of periodontitis, the subgingival microbiome switches from majority Gram-positive to majority Gram-negative bacterial species2.
Porphyromonas gingivalis is a Gram-negative bacterium which is regarded as one of the keystone pathogens in chronic periodontitis3,4. It produces virulence factors to disrupt host–microbial homeostasis, resulting in inflammation and bone loss5,6. Oral microbiome studies by 16s rRNA sequencing suggest that the abundance of periodontitis-associated species, such as P. gingivalis, Treponema denticola and Tannerella forsythia, is significantly increased in disease sites of periodontitis patients7,8. Many of these pathogens are anaerobic species that survive in deep dental pockets where oxygen is limited. Surprisingly, these anaerobic pathogens, including P. gingivalis, have also been reported in supragingival plaque, saliva and mucosa samples9,10, which are thought to be more micro-aerobic environments in the oral cavity. Several proteins have been reported to participate in the resistance to oxidative stress in P. gingivalis11–14 and they may promote the survival of P. gingivalis under micro-aerobic conditions.
The oxidative stress in the oral microbiome is not only related to oxygen concentration in the surroundings but reactive oxygen species (ROS) produced by eukaryotic cells and some oral commensal bacteria15–17. ROS leads to protein, DNA, and lipid damage, and results in an increased rate of mutagenesis and cell death18. Streptococcus sanguinis is a Gram-positive, facultative anaerobe bacterium that is able to inhibit the growth of P. gingivalis15 and produce a ROS, hydrogen peroxide (H2O2)19,20. It is a pioneering colonizer in the oral cavity and a key player in oral biofilm development21,22. Because the abundance was significantly decreased in the diseased subgingival microbiome, S. sanguinis was thought to be an oral health-associated species7,8. It is feasible that S. sanguinis maintains a healthy oral homeostasis by generating H2O2 in the oral cavity. Several genes have been reported to be responsible for H2O2 production in S. sanguinis SK3619,20.
P. gingivalis living in the oral cavity, particularly in sites under micro-aerobic conditions, must endure oxidative stress from the H2O2 produced by S. sanguinis. What are the mechanisms by which P. gingivalis tolerates oxidative stress under micro-aerobic conditions in oral microbiota?
Aggregatibacter actinomycetemcomitans is a Gram-negative, facultative anaerobe bacterium that is often found in chronic periodontitis23. The katA gene in A. actinomycetemcomitans encodes a cytoplasmic catalase that breaks down H2O224. When A. actinomycetemcomitans was co-cultured with Streptococcus gordonii, the expression of the katA gene was induced by the H2O2 released from S. gordonii. This regulation was mediated via the upstream regulator OxyR24. Both oxyR and katA were important for the survival of A. actinomycetemcomitans in the presence of S. gordonii24,25. Additionally, katA expression was higher in the biofilm state than in its planktonic state24. As A. actinomycetemcomitans can degrade H2O2, we proposed that A. actinomycetemcomitans might also confer protection to P. gingivalis from the damage of H2O2 produced by S. sanguinis in mixed species biofilm.
S. gordonii, another commensal bacterium and H2O2 producer in the oral cavity, is a well-studied species that interacts with P. gingivalis, though few papers discussed the interaction between S. sanguinis and P. gingivalis. FimA and Mfa1 fimbrial proteins mediate the attachment of P. gingivalis to S. gordonii26,27. Additionally, Mfa1 binds to streptococcal SspB protein28–30. The coadhesion between P. gingivalis and S. gordonii improves the biofilm formation of P. gingivalis on streptococcal substrates31. Moreover, S. gordonii generates streptococcal 4-aminobenzoate/para-amino benzoic acid (pABA), used for folate biosynthesis, that results in decreased stress and promotes expression of fimbrial adhesins in P. gingivalis32. These studies show that S. gordonii supports the biofilm formation of P. gingivalis. However, most of these studies were performed under anaerobic conditions. The generation of H2O2 may be limited under these conditions. Thus, the killing effect of P. gingivalis by S. gordonii may be attenuated under anaerobic conditions when compared with micro-aerobic conditions.
In this study, we showed that S. sanguinis inhibited the growth of P. gingivalis by producing H2O2 under micro-aerobic conditions. A. actinomycetemcomitans reduced the concentration of H2O2 and consequently aided the survival of P. gingivalis in S. sanguinis-P. gingivalis-A. actinomycetemcomitans tri-species biofilms.
Results
The impact of S. sanguinis and A. actinomycetemcomitans on the biomass of P. gingivalis in multi-species biofilms
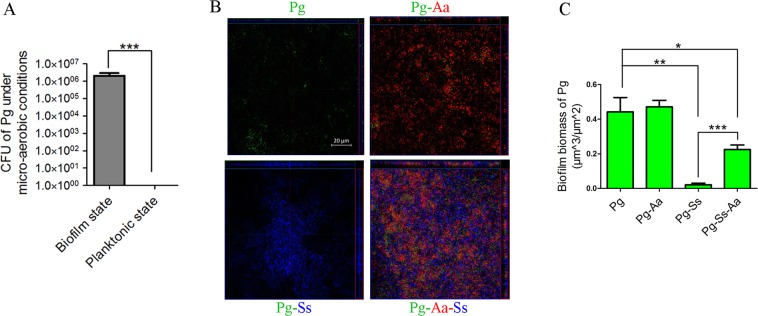
It has been reported that many anaerobic species, including P. gingivalis, are widely distributed in the oral cavity9,10. We performed experiments to evaluate the survival of P. gingivalis in 3 environments: 14 mL test tubes with shaking at 100 rpm under micro-aerobic conditions (6% oxygen, gas mixture), and 4-well chambers without shaking (static) under either anaerobic (0% oxygen, gas mixture) conditions or micro-aerobic conditions. The incubator shaking was used to inhibit biofilm growth and thus encourage planktonic growth while the lack of shaking was used to facilitate biofilm formation. All three environments had an initial inoculation of 1 × 108 P. gingivalis ATCC 33277 (Pg) cells into CDM and were incubated for four days at 37 °C. When Pg was cultured in the 14 mL tubes environment, it was not able to survive (Fig. 1A). Pg survived in both 4-well chamber environments; the biomass under micro-aerobic conditions was significantly lower than that under anaerobic conditions (P ≤ 0.01) (Fig. S1). Pg cells from the micro-aerobic 4-well chamber were then inoculated onto agar plates and grown in order to verify that the detected fluorescent signal was from live cells. The growth of 2.03 × 106 ± 9.71 × 105 colony-forming units (CFUs) confirmed the existence of live biofilm cells in static micro-aerobic conditions (Fig. 1A). Although the CFU of Pg grown under micro-aerobic conditions in 4-well chambers was lower than that of the initial inoculation, it was still greater than the CFU of Pg grown in the 14 mL test tube, suggesting that the biofilm formation increased the tolerance of Pg to oxidative stress from the presence of environmental oxygen (Fig. 1A). Because there have already been a number of reports illustrating that biofilm formation increase the tolerance of bacteria to oxidative stress33, the next experiment was designed to focus on the effect of mixed-species biofilm bacterial interactions on the oxidative stress tolerance of P. gingivalis.
Figure 1.
Confocal microscopical analysis of Pg biomass in 4-day old mixed-species biofilms under micro-aerobic conditions (6% O2). (A) Pg was incubated under static conditions for biofilm growth (left bar) and under shaking conditions (100 rpm) to prevent biofilm formation and to maintain a planktonic state (right bar is absent due to lack of growth). After 4 days, the CFUs from both samples were tested. (B) Single and multiplex staining of Pg (green), Aa (red) and Ss (blue) with FISH probes specific to the conserved 16s ribosomal (rRNA) genes of these bacteria (scale bar shown in top left panel = 20 µm). (C) Pg biomass in (B) was calculated for single and multiplex biofilms. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t-test. Means and standard deviations from triplicate experiments are shown.
To examine Pg survival in multi-species biofilms, four groups of bacterial mixes were tested: Pg only, Pg and A. actinomycetemcomitans 652 (Aa), Pg and S. sanguinis SK36 (Ss) and a mixture of all three species. Each bacterial mix group was incubated in CDM medium under micro-aerobic conditions in 4-well chambers. Biofilms were first grown for four days, after which they were stained by fluorescence in situ hybridization (FISH) and were visualized using confocal laser scanning microscopy (CLSM). Biofilm biomass was quantified by COMSTAT script in Matlab software33.
All four groups of biofilms were successfully detected using FISH probes. Under these micro-aerobic conditions, the biomass of Pg in biofilm was not significantly changed by the presence of Aa when compared to the Pg single species biofilm control (Fig. 1B,C). In contrast, the presence of Ss significantly lowered the biomass of Pg in Ss-Pg dual-species biofilms, suggesting that Ss was dominant and somehow inhibited survival of Pg (0.442 ± 0.083 µm3/µm2 in Pg only biofilm and 0.021 ± 0.009 µm3/µm2 in Pg-Ss dual species biofilm (P ≤ 0.001)) (Fig. 1B,C). Interestingly, the biomass of Pg in Pg-Aa-Ss tri-species biofilms was significantly increased compared to that in Pg-Ss dual-species biofilms, implying that Pg survival inhibition by Ss could be partially attenuated by the presence of Aa (Fig. 1B,C). As there was no significant difference between the biomass of Pg from Pg single species biofilms and Pg-Aa dual-species biofilms, it is feasible to suggest that Aa could interact with Ss, counteracting the influence of Ss on Pg, indirectly promoting the survival of Pg.
Ss-produced H2O2 reduced the biomass of Pg
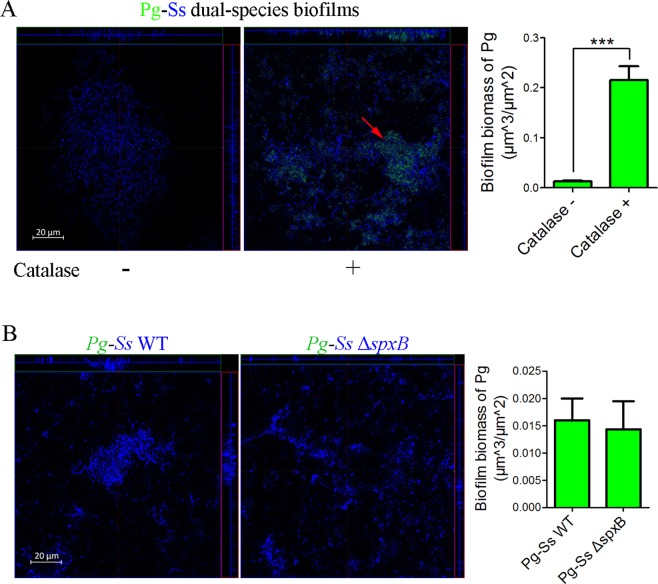
H2O2 is a well-studied inhibitory mechanism that S. sanguinis uses to compete with Streptococcus mutans19,34. It can be generated by a pyruvate oxidase (SpxB) in Ss via a reaction converting pyruvate to acetyl phosphate. During this catalytic process, oxygen is consumed35,36. Pg-Ss dual-species biofilms were grown under micro-aerobic conditions. We tested the inhibitory ability of Ss-produced H2O2 by decomposing H2O2 with 10,000 U/mL of catalase (Catalase from bovine liver, Sigma). More Pg was present in the dual-species biofilms when catalase was supplemented in the medium (P ≤ 0.001) (Fig. 2A). This result suggested that H2O2 was essential for Ss to inhibit the survival of Pg under micro-aerobic conditions. Pg appeared to preferentially colocalize with Ss in Pg-Ss dual-species biofilms when catalase was supplemented. This phenomenon implied that Pg might coaggregate with Ss in multi-species biofilms, which was similar to the interaction between P. gingivalis and S. gordonii29.
Figure 2.
Confocal microscopical images of 4-day old Pg-Ss dual-species biofilms to analyze the effect of H2O2 on Pg biomass under micro-aerobic conditions. (A) Pg-Ss dual-species biofilms (Pg = green, Ss = blue) with/without the treatment of 10,000 U/mL of catalase. (B) Pg-Ss wild type (WT) and Pg-Ss ΔspxB dual-species biofilms. Samples were stained by FISH probes. Orthogonal CLSM images were shown in the left panel. Scale bars were indicated on the corresponding images. In the right panel, the biomass of Pg from images in the left panel was quantified by COMSTAT analysis. ***P ≤ 0.001, Student’s t-test. Means and standard deviations from triplicate experiments are shown.
When Pg was co-cultured with Ss ΔspxB, the biomass of Pg was still greatly inhibited by Ss ΔspxB (Fig. 2B). One probability was that, in contrast with Ss wild type (WT), the ΔspxB mutant could still produce about 25% the concentration of H2O219, which might be enough for the inhibition of Pg growth.
Since more Pg survived in the Pg-Aa-Ss tri-species biofilm, the supplementation of Aa might have a better effect than the deletion of spxB on reducing H2O2 concentration. To test this hypothesis, Ss WT and Ss ΔspxB biofilms with/without the addition of Aa were cultured. The amount of H2O2 in the supernatant of the 4-day old biofilms was measured using Hydrogen Peroxide Assay. Indeed, the H2O2 concentration in the Aa-Ss WT dual-species biofilm was much lower than that in the Ss ΔspxB single species biofilm (P ≤ 0.0001), which supported the hypothesis (Fig. 3A). Additionally, in comparison to the Ss WT single species biofilm, the Ss ΔspxB single species biofilm contained less H2O2 in the supernatant (P ≤ 0.0001), which was consistent with the result in the previous study showing that the spxB gene deletion decreased H2O2 production in Ss (Fig. 3A)19.
Figure 3.
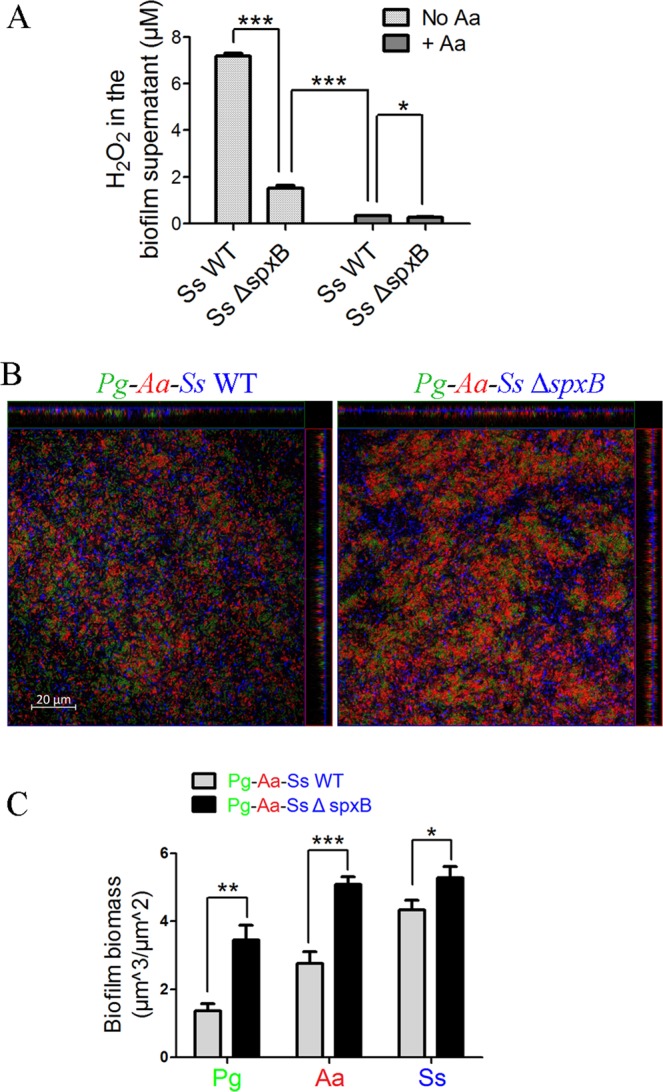
The effect of the spxB gene deletion on Pg biomass in Pg-Aa-Ss tri-species biofilms under micro-aerobic conditions. (A) Ss WT and Ss ΔspxB biofilms with/without the supplement of Aa were cultured. The H2O2 concentration in the supernatant of these biofilms was measured by the Hydrogen Peroxide Assay as described in Materials and methods. (B) Pg-Aa-Ss WT (left) and Pg-Aa-Ss ΔspxB tri-species (right) (Pg = green, Ss = blue, Aa = red) biofilms were shown. (C) The biomass of Pg, Aa and Ss in B was quantified by COMSTAT and shown as a bar chart. Scale bars were indicated on the corresponding images. *P ≤ 0.05, **P ≤ 0.01, ***P ≤ 0.001, Student’s t-test. Means and standard deviations from triplicate experiments are shown.
The Aa-Ss ΔspxB dual-species biofilm contained less H2O2 than the Aa-Ss WT biofilm, which indicated that the spxB gene deletion might promote Pg survival in Pg-Aa-Ss tri-species biofilms (Fig. 3A). 4-day old Pg-Aa-Ss WT and Pg-Aa-Ss ΔspxB tri-species biofilms were treated with FISH and observed by CLSM. The biomass of Pg in the Pg-Aa-Ss ΔspxB tri-species biofilm was more than that in the Pg-Aa-Ss WT biofilm (P ≤ 0.01), which suggested that the H2O2 produced by Ss played an important role in inhibiting Pg growth (Fig. 3B,C).
Similar to Pg, the biomass of Aa was also increased in the Pg-Aa-Ss ΔspxB biofilm (P ≤ 0.001) (Fig. 3B,C). Surprisingly, the biofilm biomass of Ss was increased in the Pg-Aa-Ss ΔspxB biofilms than that in the Pg-Aa-Ss WT tri-species biofilm (P ≤ 0.05) (Fig. 3B,C), despite Ss ΔspxB had a reduced biofilm formation in Ss-Pg dual-species biofilms (Figs 2B and S2). When we treated tri-species biofilms using FISH protocol, we observed that the Pg-Aa-Ss WT biofilm was more fragile than the Pg-Aa-Ss ΔspxB biofilm, indicating that H2O2 might affect inter-species attachment. It has been shown that P. gingivalis utilizes fimbrillin to bind to glyceraldehyde-3-phosphate dehydrogenase, a cell surface protein of S. sanguinis37, implying that S. sanguinis may not only inhibit the growth of P. gingivalis but also coaggregate with P. gingivalis. Due to the deletion of spxB, a reduced antagonism in Pg-Aa-Ss tri-species biofilm might be beneficial for the co-aggregation between Pg and Ss, and as a result, might increase the biomass of Ss. Though a similar relationship may exist between S. sanguinis and A. actinomycetemcomitans, the current knowledge on their interactions is limited, and the mechanism of such phenomenon needs further exploration.
Aa degraded H2O2 and protected Pg from H2O2 attack
The data in Fig. 3A implied that Aa might impact Ss and indirectly promoted the survival of Pg through degrading H2O2. To further elucidate whether Aa protected Pg from H2O2, the concentration of H2O2 was measured using Hydrogen Peroxide Assay. Briefly, cells were resuspended in fresh CDM and mixed with Hydrogen Peroxide Assay solution. The final reaction solutions were incubated under room atmospheric conditions at 37 °C. The optical density for cell growth and fluorescent signal for H2O2 concentration were monitored by the plate reader.
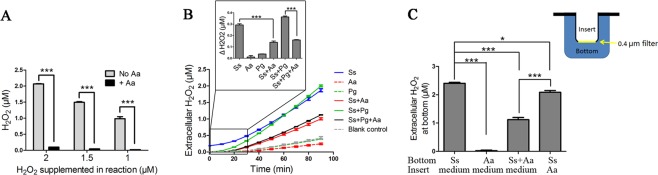
Firstly, the Hydrogen Peroxide Assay solution was supplemented with H2O2 (2, 1.5 or 1 μM). It was mixed or not mixed with Aa suspension. The H2O2 concentration was recorded after 30 minutes of reaction. The presence of Aa greatly reduced H2O2 concentrations for all variants of the Hydrogen Peroxide Assay solution, which demonstrated that Aa has the ability to degrade H2O2 (Fig. 4A).
Figure 4.
The effect of Aa on the concentration of H2O2. (A) Aa suspensions were mixed with H2O2 solutions. H2O2 concentrations were then tested after 30 minutes of reaction. (B) H2O2 concentrations of different bacterial mixtures were measured at 10-minute intervals. The enlarged section of the graph, illustrated as a bar chart, showed the H2O2 concentration after 30 minutes of reaction. CDM media without bacteria was used as a blank control. (C) Wells were divided into two parts by a transwell system. Ss, Aa or CDM medium were located at different places as shown in the figure. The H2O2 concentration in the bottom section was measured. *P ≤ 0.05, ***P ≤ 0.001, Student’s t-test. Means and standard deviations from triplicate experiments are shown.
Subsequently, the H2O2 produced by Ss, Pg and Aa was tested. Compared with the blank control (CDM without bacteria), neither Pg nor Aa produced H2O2 (Fig. 4B). The H2O2 concentration in Aa was even lower than the concentration in the blank control (Fig. 4B). Ss produced almost 0.3 μM of H2O2 after 30 minutes of reaction, which could be attenuated by the addition of Aa (P ≤ 0.001) but not Pg (Fig. 4B). The presence of Aa decreased the H2O2 concentration by nearly half in both the dual-species (Ss-Aa) and tri-species (SS-Pg-Aa) suspensions (Fig. 4B). There was no significant change in cell density during the 30 minutes of experimentation, indicating that cell growth did not influence the results of H2O2 concentrations (Fig. S3). Similar results were reported in previous works, which utilized scanning electrochemical microscopy to do real-time mapping of H2O2 concentrations on bacteria biofilms38. They reported that the H2O2 generated by S. gordonii, another oral commensal bacterium and H2O2 producer, could be reduced by A. actinomycetemcomitans38. These data suggested that Aa degraded H2O2 produced by Ss and implied that Aa might be able to promote the survival of Pg in Pg-Aa-Ss tri-species biofilm by reducing H2O2 concentration.
Bacteria living in biofilms are surrounded by matrix composed of polysaccharide, eDNA and proteins39. As materials may slow penetrate and transverse a biofilm39, cell to cell distance may impact the interaction between Aa and Ss. To test the contribution of cell-cell distance to the Aa–Ss interaction, Aa and Ss were cultured in a transwell system, where they were separated by a 0.4 μm filter. Ss was cultured at the bottom and Aa was either incubated in the insert or mixed with Ss at the bottom. After 30 minutes of reaction, the H2O2 concentration at the bottom of the well was measured. The H2O2 concentration at the bottom was 2.404 ± 0.035 μM when the insert was filled with CDM medium and the bottom was Ss. When Aa was put in the insert and Ss was set at the bottom of the well, Aa slightly but significantly decreased the H2O2 concentration at the bottom to 2.087 ± 0.061 μM (P ≤ 0.05) (Fig. 4C). However, the reduction was much lower than that in the well where Ss and Aa mixed directly at the bottom (P ≤ 0.001) (Fig. 4C). This result showed that a closer distance between Ss and Aa was beneficial for Aa to reduce H2O2 produced by Ss. Aa might have limited function to degrade H2O2 when it was far away from Ss. In an in vitro study, Aggregatibacter has been shown to close contact with Streptococcus40, indicating that A. actinomycetemcomitans might exist near to S. sanguinis in vivo to detoxify H2O2.
KatA has been reported to produce catalase in A. actinomycetemcomitans strain VT1169 (Aa VT1169) to detoxify H2O2 and is essential for the survival of A. actinomycetemcomitans during co-infection with S. gordonii24,25. It was hypothesized that KatA was essential for Aa to improve the survival of Pg.
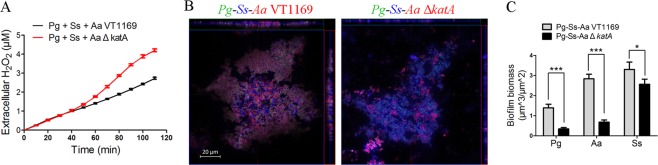
Using the Hydrogen Peroxide Assay Kit, H2O2 concentration and cell density of Pg + Ss + Aa VT1169 and Pg + Ss + A. actinomycetemcomitans VT1169 ΔkatA (Aa ΔkatA) suspensions were monitored. Compared to the katA deletion mutant, Aa VT1169 had the greater ability to repress H2O2 production (P ≤ 0.001 at the time point of 110 minutes), implying that KatA was important for Aa to reduce the H2O2 generated by Ss (Fig. 5A). There was no significant difference in cell density between Pg + Ss + Aa VT1169 and Pg + Ss + Aa ΔkatA, suggesting that the difference in H2O2 concentration was not caused by a difference in cell growth (Fig. S4).
Figure 5.
The influence of the katA gene of Aa VT1169 on Pg-Ss -Aa VT1169 tri-species biofilms. (A) The H2O2 concentrations of Pg-Ss-Aa VT1169 and Pg-Ss-Aa ΔkatA were recorded at 10-minute intervals using the Hydrogen Peroxide Assay. (B) The biofilms of Pg-Ss-Aa VT1169 and Pg-Ss-Aa ΔkatA were stained by FISH and observed by CLSM. Pg, Aa VT1169 and Ss were marked as green, red and blue, respectively. (C) The biomass of Pg, Aa VT1169 and Ss in (B) were quantified by COMSTAT. Scale bars were indicated on the corresponding images. *P ≤ 0.05, ***P ≤ 0.001, Student’s t-test. Means and standard deviations from triplicate experiments are shown.
Pg-Ss-Aa VT1169 and Pg-Ss-Aa ΔkatA Tri-species biofilms were stained by FISH and observed by CLSM as described above. Compared with the biofilm of Pg-Ss-Aa VT1169, the Pg-Ss-Aa ΔkatA biofilm contained less Pg and Aa (P ≤ 0.001 for both comparisons) (Fig. 5B,C), which suggested that the catalase of A. actinomycetemcomitans was essential for the survival of both Pg and A. actinomycetemcomitans in the tri-species biofilms and further confirmed the hypothesis that A. actinomycetemcomitans protected P. gingivalis from H2O2 damage. The biofilm biomass of Ss in Pg-Ss-Aa ΔkatA was slightly decreased (P ≤ 0.05) (Fig. 5B,C). This phenomenon where Ss biofilm biomass decreased in conditions that also led to decreased biofilm biomass of Aa and Pg, was similar to the observed phenomenon in Fig. 3B,C allowing for the possible hypothesis that the biomass of Ss might have been impacted by Pg and/or A. actinomycetemcomitans in tri-species biofilms.
The VT1169 and ΔkatA single species biofilms were stained by SYTO9 and observed by CLSM. The morphology of ΔkatA biofilm was different from that of the wild type strain. The biofilm of ΔkatA was much thicker (Fig. S5A). It contained larger aggregations and bigger gaps between aggregations (Fig. S5A). However, the biofilm biomass of these two strains were similar, which confirmed that the reduction of Aa biomass in Pg-Ss-Aa ΔkatA tri-species was not caused by an attenuated biofilm formation ability of Aa ΔkatA (Fig. S5).
Discussion
In this study, it was shown that Aa degraded H2O2 produced by Ss, which consequently aided the survival of Pg in Pg-Aa-Ss tri-species biofilms under micro-aerobic conditions. KatA, which produces catalase in Aa, was also shown to participate in this interaction. There have been many epidemiological studies showing that anaerobic bacteria such as P. gingivalis exist in supragingival, salivary and mucosal samples9,10. One possibility is that micro-environment may exist in oral cavity, which allows these anaerobic bacteria to survive in micro-aerobic conditions. The results of this study presented the possibility that catalase producers in oral microbiota attenuate the oxidative stress and help the survival of anaerobic species under micro-aerobic conditions.
The pathogenesis of periodontitis has been thought to possess polymicrobial synergistic interactions41,42. A temporal dynamics study showed that facultative anaerobic bacteria, especially Streptococcus, were dominant in the early stage of oral biofilm formation43. Subsequently, the ‘healthy’ biofilm composition was replaced with a population of gram-negative anaerobic bacteria43. In our study, we observed essentially no Pg presence in Pg-Ss dual species biofilms but Pg presence was obvious in Pg-Aa-Ss tri-species biofilms, suggesting that the existence of Aa was important for Pg survival. This result indicated that the earlier colonization of bacteria species with catalase activity than anaerobic species in oral biofilms might be necessary to generate suitable surroundings for the survival of anaerobic microorganisms. Further studies need to be performed to test this hypothesis. Additionally, our study illustrated that KatA of Aa VT1169 was important for the growth of both Pg and Aa VT1169, implying that catalase might be a promising drug target to prevent periodontitis.
Welch et al. utilized FISH technology to stain supragingival dental plaque40. They hypothesized that the Porphyromonas growing at the periphery of biofilm samples might not be P. gingivalis because the outer shell of the biofilm was in a presumably aerobic environment40. Here, we demonstrated that P. gingivalis was able to survive in a micro-aerobic environment and had better survival in the presence of A. actinomycetemcomitans, which implied that it was possible that the bacteria at the periphery of supragingival biofilm samples, seen in the Welch’s study, was P. gingivalis. In their study, they showed that both Porphyromonas and Haemophilus/Aggregatibacter were in close contact with Streptococcus cells40. Furthermore, Aggregatibacter was not found adjacent to cells of Porphyromonas in the absence of Streptococcus40. Their results indicated that P. gingivalis, A. actinomycetemcomitans and S. sanguinis might be close to each other in vivo and a similar interaction between these three species might also exist in vivo.
In Fig. 2A, Pg appeared to preferentially colocalize with Ss in Pg-Ss dual-species biofilms when catalase was supplemented. Additionally, the biomass of Ss in both Figs 3B and 5B were positively related with the biomass of Pg and Aa. All the phenomena above indicated that Ss might also cooperate with Pg and/or Aa in multi-species biofilms. The antagonism and the cooperation between commensal bacteria and pathogens may exist in equilibrium in oral microbiota. Whenever the antagonism was weakened, or the cooperation was strengthened either by other microorganisms or environmental conditions, dysbiosis may happen and lead to diseases such as periodontitis.
Materials and Methods
Bacterial strains, growth and antibiotics
Strains used in this study are listed in Table S1. Unless otherwise stated, Pg, A. actinomycetemcomitans strains and Ss cells from −80 °C frozen glycerol stocks were 0.5% inoculated into TSB medium (tryptic soy broth supplemented with yeast extract (5 mg/ml), hemin (5 µg/ml) and menadione (1 µg/ml)) and incubated statically under anaerobic conditions (10% CO2, 10% H2 and 80% N2) at 37 °C using an Anoxomat® system (Spiral Biotech, Norwood, MA). Spectinomycin was used at 50 μg/mL for the culture of Aa ΔkatA. No antibiotic was added to multi-species biofilms. The CFUs of Pg were tested by growing Pg on sheep blood agar plates (Trypticase™ Soy Agar (TSA II™) with Sheep Blood, BD BBL™) under anaerobic conditions. All media was incubated in anaerobic jars for at least 2 days before experiments to equilibrate.
Biofilm assay
Pg, A. actinomycetemcomitans strains and Ss were initially incubated separately for 48 hours, 24 hours and overnight respectively, in TSB medium under anaerobic conditions to early stationary phase. The resultant growth was then resuspended in fresh CDM, followed by 10% inoculation into CDM medium and incubation under micro-aerobic conditions for biofilm formation. CDM was prepared as previously described (10.0 mM of NaH2PO4, 10.0 mM of KCl, 2.0 mM of citric acid, 1.25 mM of MgCl2, 100 µM of FeCl3, 20.0 µM of CaCl2, 0.1 µM of Na2MoO4, 25.0 µM of ZnC12, 50.0 µM of MnC12, 5.0 µM of CuCl2, 10.0 µM of CoCl2, 5.0 µM of H2BO3, 1% (w/v) Tryptone, 7.67 µM of Hemin and 2.91 µM of Menadione)44. Biofilms were incubated in 4-chambered glass coverslip wells (Chambered Coverglass, Nunc™ Lab-Tek™) for 4 days at 37 °C. Cultures were grown anaerobically (0% O2, 10% CO2, 10% H2 and 80% N2) or micro-aerobically (6% O2, 7.2% CO2, 7.2% H2 and 79.6% N2) in jars using the Anoxomat® system (Spiral Biotech, Norwood, MA). Single-species biofilms were stained using SYTO 9 (SYTO™ 9 Green Fluorescent Nucleic Acid Stain, Invitrogen™ Molecular Probes™) and FISH was used to analyze and characterize the composition in multi-species biofilms.
FISH assay
FISH was performed as previously described40. FISH probes used in the study were ordered from Integrated DNA Technologies, Inc. and the sequences were listed in Table S2. Biofilms were grown in 4-well chambers for 4 days in 1 mL of CDM medium. 800 µL of supernatant was discarded by pipetting and then 4-well chambers were slowly turned over on paper towels to discard remaining supernatant. Biofilms were gently washed by 200 µL of 1× PBS buffer and fixed by 2% (wt/vol) paraformaldehyde on ice for at least 1.5 hours. After fixation, samples were gently washed again in 1× PBS for 15 min. Next, PBS was discarded and 10 µL of hybridization solution (900 mM of NaCl, 20 mM of Tris, pH 7.5, 0.01% of SDS, 20% (vol/vol) of formamide, each probe at a final concentration of 0.1 µM) was dropped on biofilm samples and stained at 46 °C for 4 hours in a chamber humidified with 20% (vol/vol) formamide. Samples were then gently washed in wash buffer (215 mM of NaCl, 20 mM of Tris, pH 7.5, 5 mM of EDTA) at 48 °C for 15 minutes. Finally, biofilms were gently washed by cold water, and mounted in ProLong Gold Antifade Solution (ThermoFisher) for CLSM observation.
CLSM and biomass quantification
FISH-treated biofilms were observed by a Zeiss LSM710 confocal laser scanning microscope (Zeiss, Germany) (VCU Core Facilities) and quantified by COMSTAT in Matlab software33. The fluorescent dyes were listed in Table S2. Three images of each sample were quantified to calculate the means and standard deviations.
Hydrogen Peroxide Assay
H2O2 concentration was measured by Hydrogen Peroxide Assay Kit (Red Hydrogen Peroxide/Peroxidase Assay Kit, Amplex™). Operations followed a standard protocol of the kit. For testing H2O2 concentration in biofilm supernatant, 80 ul of biofilm supernatant was centrifuged. Subsequently, 50 ul of supernatant was mixed with Hydrogen Peroxide Assay solution. After 30 minutes of reaction, the fluorescent signal (excitation 560 nm/emission 590 nm) was recorded by a Synergy H1 Hybrid Reader. The preparation of a standard curve for quantifying H2O2 concentration followed the standard protocol of the kit. To get data presented in Fig. 4B, Pg, A. actinomycetemcomitans strains and Ss were grown for 48 hours, 24 hours and overnight respectively in TSB to early stationary phase under anaerobic conditions. Cells were resuspended in fresh CDM and 10% inoculated into fresh CDM to get bacterial suspensions. 50 ul of bacteria suspensions were mixed with 50 ul of Hydrogen Peroxide Assay solution and incubated under laboratory atmospheric conditions at 37 °C using the Synergy H1 Hybrid Reader. The optical density (OD600) for cell growth and fluorescent signal for H2O2 concentration were monitored continuously by the reader. For testing H2O2 concentration in the transwell system (96 Well Permeable Support System transwell, Corning™ HTS Transwell™), Aa and Ss cells were grown to early stationary phase in TSB medium, followed by resuspension in fresh CDM. 50 ul of bacteria suspension and 50 ul of Hydrogen Peroxide Assay solution were mixed at the bottom of the well. The insert was filled with 50 ul of CDM or 50 ul of Aa suspension. After 30 minutes of reaction, the insert was discarded and the H2O2 concentration at the bottom of the well was measured. Three replicates were performed to calculate the means and standard deviations.
Statistical analysis
All data were obtained from at least three biological replicates. Student’s t-test was applied to analyze data on biofilm biomass, H2O2 concentration and CFU.
Supplementary information
Acknowledgements
We are extremely grateful to Dr. Marvin Whiteley (Georgia Institute of Technology) for providing the strains A. actinomycetemcomitans VT1169 and A. actinomycetemcomitans VT1169 ΔkatA, and to Dr. Donald Demuth (University of Louisville School of Dentistry) for providing the strain A. actinomycetemcomitans 652. CLSM was performed at Virginia Commonwealth University, Department of Anatomy & Neurobiology Microscopy Facility, supported, in part, by funding from NIH-NINDS Center Core Grant 5 P30 NS047463 and, in part, by funding from the NIH-NCI Cancer Center Support Grant P30 CA016059. This work was supported by National Institutes of Health grants R01DE023078 and R01DE018138 (PX). The funders had no role in study design, data collections and interpretation, or the decision to submit the work for publication.
Author Contributions
B.Z. and P.X. conceived and designed this study. B.Z. carried out all of the experiments with the assistance of L.M. and E.N., B.Z. and P.X. analyzed the data and wrote this manuscript. All authors reviewed and discussed the manuscript.
Competing Interests
The authors declare no competing interests.
Footnotes
Publisher’s note: Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1038/s41598-019-41467-9.
References
- 1. Global, regional, and national incidence, prevalence, and years lived with disability for 328 diseases and injuries for 195 countries, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. Lancet (London, England)390, 1211–1259, 10.1016/s0140-6736(17)32154-2 (2017). [DOI] [PMC free article] [PubMed]
- 2.Marsh PD. Microbial ecology of dental plaque and its significance in health and disease. Advances in dental research. 1994;8:263–271. doi: 10.1177/08959374940080022001. [DOI] [PubMed] [Google Scholar]
- 3.Darveau RP. Periodontitis: a polymicrobial disruption of host homeostasis. Nature reviews. Microbiology. 2010;8:481–490. doi: 10.1038/nrmicro2337. [DOI] [PubMed] [Google Scholar]
- 4.Olsen I, Yilmaz O. Modulation of inflammasome activity by Porphyromonas gingivalis in periodontitis and associated systemic diseases. Journal of oral microbiology. 2016;8:30385. doi: 10.3402/jom.v8.30385. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.How KY, Song KP, Chan KG. Porphyromonas gingivalis: An Overview of Periodontopathic Pathogen below the Gum Line. Frontiers in microbiology. 2016;7:53. doi: 10.3389/fmicb.2016.00053. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Olsen I, Lambris JD, Hajishengallis G. Porphyromonas gingivalis disturbs host-commensal homeostasis by changing complement function. Journal of oral microbiology. 2017;9:1340085. doi: 10.1080/20002297.2017.1340085. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Griffen AL, et al. Distinct and complex bacterial profiles in human periodontitis and health revealed by 16S pyrosequencing. The ISME journal. 2012;6:1176–1185. doi: 10.1038/ismej.2011.191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Abusleme L, et al. The subgingival microbiome in health and periodontitis and its relationship with community biomass and inflammation. The ISME journal. 2013;7:1016–1025. doi: 10.1038/ismej.2012.174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hintao J, Teanpaisan R, Chongsuvivatwong V, Ratarasan C, Dahlen G. The microbiological profiles of saliva, supragingival and subgingival plaque and dental caries in adults with and without type 2 diabetes mellitus. Oral microbiology and immunology. 2007;22:175–181. doi: 10.1111/j.1399-302X.2007.00341.x. [DOI] [PubMed] [Google Scholar]
- 10.Diaz PI, et al. Using high throughput sequencing to explore the biodiversity in oral bacterial communities. Molecular oral microbiology. 2012;27:182–201. doi: 10.1111/j.2041-1014.2012.00642.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.McKenzie RM, Henry LG, Boutrin MC, Ximinies A, Fletcher HM. Role of the Porphyromonas gingivalis iron-binding protein PG1777 in oxidative stress resistance. Microbiology (Reading, England) 2016;162:256–267. doi: 10.1099/mic.0.000213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Dou, Y. et al. Role of extracytoplasmic function sigma factor PG1660 (RpoE) in the oxidative stress resistance regulatory network of Porphyromonas gingivalis. 33, 89–104, 10.1111/omi.12204 (2018). [DOI] [PMC free article] [PubMed]
- 13.Dou Y, et al. Involvement of PG2212 zinc finger protein in the regulation of oxidative stress resistance in Porphyromonas gingivalis W83. Journal of bacteriology. 2014;196:4057–4070. doi: 10.1128/jb.01907-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Leclerc J, et al. The Cytochrome bd Oxidase of Porphyromonas gingivalis Contributes to Oxidative Stress Resistance and Dioxygen Tolerance. PloS one. 2015;10:e0143808. doi: 10.1371/journal.pone.0143808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Herrero ER, et al. Dysbiosis by neutralizing commensal mediated inhibition of pathobionts. Scientific reports. 2016;6:38179. doi: 10.1038/srep38179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Leke N, Grenier D, Goldner M, Mayrand D. Effects of hydrogen peroxide on growth and selected properties of Porphyromonas gingivalis. FEMS microbiology letters. 1999;174:347–353. doi: 10.1111/j.1574-6968.1999.tb13589.x. [DOI] [PubMed] [Google Scholar]
- 17.Storz G, Tartaglia LA, Farr SB, Ames BN. Bacterial defenses against oxidative stress. Trends in genetics: TIG. 1990;6:363–368. doi: 10.1016/0168-9525(90)90278-E. [DOI] [PubMed] [Google Scholar]
- 18.Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nature reviews. Microbiology. 2013;11:443–454. doi: 10.1038/nrmicro3032. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Chen L, et al. Identification of hydrogen peroxide production-related genes in Streptococcus sanguinis and their functional relationship with pyruvate oxidase. Microbiology (Reading, England) 2011;157:13–20. doi: 10.1099/mic.0.039669-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Zhu B, Macleod LC, Kitten T, Xu P. Streptococcus sanguinis biofilm formation & interaction with oral pathogens. Future microbiology. 2018;13:915–932. doi: 10.2217/fmb-2018-0043. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Caufield PW, et al. Natural history of Streptococcus sanguinis in the oral cavity of infants: evidence for a discrete window of infectivity. Infection and immunity. 2000;68:4018–4023. doi: 10.1128/IAI.68.7.4018-4023.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Aas JA, Paster BJ, Stokes LN, Olsen I, Dewhirst FE. Defining the normal bacterial flora of the oral cavity. Journal of clinical microbiology. 2005;43:5721–5732. doi: 10.1128/jcm.43.11.5721-5732.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Henderson B, Ward JM, Ready D. Aggregatibacter (Actinobacillus) actinomycetemcomitans: a triple A* periodontopathogen? Periodontology 2000. 2010;54:78–105. doi: 10.1111/j.1600-0757.2009.00331.x. [DOI] [PubMed] [Google Scholar]
- 24.Ramsey MM, Whiteley M. Polymicrobial interactions stimulate resistance to host innate immunity through metabolite perception. Proceedings of the National Academy of Sciences of the United States of America. 2009;106:1578–1583. doi: 10.1073/pnas.0809533106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Stacy A, et al. Bacterial fight-and-flight responses enhance virulence in a polymicrobial infection. Proceedings of the National Academy of Sciences of the United States of America. 2014;111:7819–7824. doi: 10.1073/pnas.1400586111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yoshimura F, Takahashi K, Nodasaka Y, Suzuki T. Purification and characterization of a novel type of fimbriae from the oral anaerobe Bacteroides gingivalis. Journal of bacteriology. 1984;160:949–957. doi: 10.1128/jb.160.3.949-957.1984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Demuth DR, Irvine DC, Costerton JW, Cook GS, Lamont RJ. Discrete protein determinant directs the species-specific adherence of Porphyromonas gingivalis to oral streptococci. Infection and immunity. 2001;69:5736–5741. doi: 10.1128/IAI.69.9.5736-5741.2001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Brooks W, Demuth DR, Gil S, Lamont RJ. Identification of a Streptococcus gordonii SspB domain that mediates adhesion to Porphyromonas gingivalis. Infection and immunity. 1997;65:3753–3758. doi: 10.1128/iai.65.9.3753-3758.1997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Park Y, et al. Short fimbriae of Porphyromonas gingivalis and their role in coadhesion with Streptococcus gordonii. Infection and immunity. 2005;73:3983–3989. doi: 10.1128/iai.73.7.3983-3989.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Forsgren N, Lamont RJ, Persson K. Two intramolecular isopeptide bonds are identified in the crystal structure of the Streptococcus gordonii SspB C-terminal domain. Journal of molecular biology. 2010;397:740–751. doi: 10.1016/j.jmb.2010.01.065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lamont RJ, et al. Role of the Streptococcus gordonii SspB protein in the development of Porphyromonas gingivalis biofilms on streptococcal substrates. Microbiology (Reading, England) 2002;148:1627–1636. doi: 10.1099/00221287-148-6-1627. [DOI] [PubMed] [Google Scholar]
- 32.Kuboniwa, M. et al. Metabolic crosstalk regulates Porphyromonas gingivalis colonization and virulence during oral polymicrobial infection. 2, 1493–1499, 10.1038/s41564-017-0021-6 (2017). [DOI] [PMC free article] [PubMed]
- 33.Heydorn A, et al. Quantification of biofilm structures by the novel computer program COMSTAT. Microbiology (Reading, England) 2000;146(Pt 10):2395–2407. doi: 10.1099/00221287-146-10-2395. [DOI] [PubMed] [Google Scholar]
- 34.Kreth J, Merritt J, Shi W, Qi F. Competition and coexistence between Streptococcus mutans and Streptococcus sanguinis in the dental biofilm. Journal of bacteriology. 2005;187:7193–7203. doi: 10.1128/jb.187.21.7193-7203.2005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Zheng LY, Itzek A, Chen ZY, Kreth J. Oxygen dependent pyruvate oxidase expression and production in Streptococcus sanguinis. International journal of oral science. 2011;3:82–89. doi: 10.4248/ijos11030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Carlsson J, Edlund MB, Lundmark SK. Characteristics of a hydrogen peroxide-forming pyruvate oxidase from Streptococcus sanguis. Oral microbiology and immunology. 1987;2:15–20. doi: 10.1111/j.1399-302X.1987.tb00264.x. [DOI] [PubMed] [Google Scholar]
- 37.Maeda K, et al. Oral streptococcal glyceraldehyde-3-phosphate dehydrogenase mediates interaction with Porphyromonas gingivalis fimbriae. Microbes and infection. 2004;6:1163–1170. doi: 10.1016/j.micinf.2004.06.005. [DOI] [PubMed] [Google Scholar]
- 38.Liu X, et al. Real-time mapping of a hydrogen peroxide concentration profile across a polymicrobial bacterial biofilm using scanning electrochemical microscopy. Proceedings of the National Academy of Sciences of the United States of America. 2011;108:2668–2673. doi: 10.1073/pnas.1018391108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Stewart PS, Costerton JW. Antibiotic resistance of bacteria in biofilms. Lancet (London, England) 2001;358:135–138. doi: 10.1016/S0140-6736(01)05321-1. [DOI] [PubMed] [Google Scholar]
- 40.Mark Welch JL, Rossetti BJ, Rieken CW, Dewhirst FE, Borisy GG. Biogeography of a human oral microbiome at the micron scale. Proceedings of the National Academy of Sciences of the United States of America. 2016;113:E791–800. doi: 10.1073/pnas.1522149113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hajishengallis G. Periodontitis: from microbial immune subversion to systemic inflammation. Nature reviews. Immunology. 2015;15:30–44. doi: 10.1038/nri3785. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Hajishengallis G. The inflammophilic character of the periodontitis-associated microbiota. Molecular oral microbiology. 2014;29:248–257. doi: 10.1111/omi.12065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wake N, et al. Temporal dynamics of bacterial microbiota in the human oral cavity determined using an in situ model of dental biofilms. NPJ biofilms and microbiomes. 2016;2:16018. doi: 10.1038/npjbiofilms.2016.18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Davey ME. Techniques for the growth of Porphyromonas gingivalis biofilms. Periodontology 2000. 2006;42:27–35. doi: 10.1111/j.1600-0757.2006.00183.x. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.