Abstract
This study reports the complete chloroplast sequences of three Spondias species. The genome sequences were obtained for Spondias tuberosa, Spondias bahienses, and Spondias mombin using the Illumina sequencing technology by a combination of de novo methods and a reference-guided assembly using Sapindus mukorossi as reference. The genomes of S. tuberosa, S. bahiensis, and S. mombin had 162,036, 162,218, and 162,302 bp, respectively. The coding regions exhibited 130 genes, including 34–35 tRNAs and 4 rRNAs. The results revealed synteny among the genomes, with high conservation in the gene order and content and CG content. The inverted repeat regions (IRA and IRB) and the large and small single copies were very similar among the three genomes. The phylogenomic analysis reported similar topologies as that of previous studies, which used partial chloroplast, wherein S. mombin was the first diverging lineage, while S. tuberosa and S. bahiensis were derived, indicating that the phylogenetic analysis using partial or complete genome produces similar results. In summary, (1) we presented the first complete chloroplast genome for the genus Spondias, (2) phylogenies analyzed using the complete chloroplast genomes revealed a robust phylogenetic topology for Spondias, and (3) gene order, content, and orientation in Spondias are highly conserved.
Keywords: plastome, evolution, Spondias
Introduction
The genus Spondias, belonging to the family Anacardiaceae (order: Sapindales), comprises about 20 species and is of economic, ecological, and social importance, as certain species are utilized in agriculture and industry for human and animal food. Some species are occur in Brazil, such as Spondias mombin Jacq, Spondias purpurea L., Spondias tuberosa Arruda Camara, Spondias venulosa Mart. ex Engl, Spondias bahiensis P. Carvalho, van den Berg & M. Machado, and Spondias dulcis Parkinson (Mitchell and Daly, 2015). These species have 2n = 32 chromosomes with similar chromosome morphology among the Spondias species (Almeida et al., 2007). Recent phylogenetic analysis has indicated that these species are closely related, wherein the species S. tuberosa, S. venulosa, and S. bahiensis are the most derived ones, and S. purpurea, S. mombin, and S. dulcis belong to a basal clade (Machado et al., 2015; Silva et al., 2015). However, these species are neglected in genetic studies, and only few genomic studies exist for the genus. In this context, sequencing the complete chloroplast genome is essential for studying the phylogeny and evolution of the genus, family Anacardiaceae, and order Sapindales. The complete chloroplast genome has been utilized for phylogenetic analysis with robust branch support in other taxa (Barret et al., 2016).
Chloroplasts are essential components of plants and are important for photosynthesis, biosynthesis, and carbon sequestration. These cytoplasmic organelles have a genome independent of the nuclear genome, which is inherited through the female plant only. Their genome is organized in a circular structure with a 100–200 kbp size range and a “quadripartite” structure comprising two large inverted repeats (IRs), which include the ribosomal genes and other plastid genes, separated by a large single copy (LSC) region and a small single copy (SSC) region. In general, the chloroplasts comprise 16S, 23S, 5S, and 27–31 tRNA genes, which are sufficient to translate all the amino acids, including three genes for the RNA polymerase subunit (similar to prokaryotes) and a majority of the genes for photosystem I, photosystem II, cytochrome, and ATP synthesis (revised by Green et al., 2011), totaling approximately 80 proteins (Huang et al., 2013). Plastid genomes are widely used for studies in taxonomy, phylogeny, phylogeography, and molecular identification of plants using rbcL and matK genes and the intergenic trnH–psbA spacer as DNA bar coding (Hollingsworth et al., 2009). As the chloroplast genomes are haploid and highly conserved regarding the genetic content and genomic structure, they have been widely used to study the evolutionary relationships of different taxonomic levels in plants. For Spondias, only two genes and one intergenic spacer have been used in the phylogenetic analyses, and the complete genome sequencing has a potential to resolve the hybrid origin for certain species.
With the advent of next-generation DNA sequencing technologies, there has been an increase in the number of chloroplast genomes sequenced; however, within the order Sapindales, which comprises 460 genera and 5670 species, only few plastid genomes have been sequenced. This study aimed to sequence the plastid genomes of S. tuberosa, S. bahiensis, S. mombin using high-throughput sequencing technology and to construct a phylogeny utilizing the complete chloroplast genomes of Spondias.
Material and Methods
Plant material, DNA isolation, and high-throughput DNA sequencing
Spondias plant material was collected in the state of Alagoas, Brazil, and total DNA was extracted (including nuclear, chloroplast, and mitochondrial DNA) using approximately 2 cm2 of leaves following the cetyltrimethylammonium bromide (CTAB) extraction method (Doyle and Doyle, 1987). The quality and quantity of the extracted DNA were verified by visualization on 1% agarose gel and spectrophotometry, respectively. The DNA sample was fragmented into pieces of 400–500 bp to construct the sequencing library. The fragments were ligated with adapters using the Nextera DNA Sample Preparation kit (Illumina), and 100 nt single-end reads for S. bahiensis and 100 nt paired-end reads for S. tuberosa and S. mombin were obtained by Illumina HiSeq2500 sequencing, done at the Central Laboratory for High Performance Technologies in Life Sciences (LaCTAD-Laboratório Central de Tecnologias de Alto Desempenho em Ciências da Vida) at the State University of Campinas (UNICAMP, Campinas, SP, Brazil).
Chloroplast genome assembly and annotation
To generate the genomes, four million reads were mapped to the plastid genome of Sapindus mukorossi as reference (Table 1), using the software Geneious 9.1 (http://www.geneious.com) and the Map-to-Reference tool with a minimum 85% of identity, including 15% for gaps. The draft genomes were corrected using de novo contigs obtained from 165 million reads for S. tuberosa, 77 million reads for S. bahiensis, and 23 million reads for S. mombin, by means of the Ray software (Boisvert et al., 2012), with a minimum size of 500 nt and 8× coverage. The de novo contigs were then mapped using the draft genome as the reference, and the regions with gaps or errors were manually corrected.
Table 1. Chloroplast genomes sequenced in this study, and others utilized as reference or out-group in phylogenetic analysis.
| Taxon | GenBank | References |
|---|---|---|
| Spondias tuberosa | KU756562 | This study |
| Spodnias bahiensis | KU756561 | This study |
| Spondias mombin | KY828469 | This study |
| Pistacia vera | KY549635 | unpublished |
| Sapindus mukorossi | KM454982 | Yang et al., 2016 |
| Mangifera indica | KY635882 | Rabah et al., 2017 |
| Anacardium occidentale | KY635877 | Rabah et al., 2017 |
| Rhus chinensis | KX447140 | Lee et al., 2016 |
| Boswellia sacra | KU756561 | Khan et al., 2017 |
Validation was achieved by Sanger sequencing of the trnH–psbA, matK, trnD–trnT, accD–psaI, rblC, and trnK–rpd16 regions, using primers described by Scarcelli et al. (2011). PCR analyses were performed using a volume of 50 μL, containing 5 μL of a reaction buffer, 1.5 mM MgCl2, 0.2 mM dNTPs, 1 U Taq DNA polymerase, 0.5 μM of each primer, and 200 ng of DNA. Amplification was achieved with an initial denaturation at 94 °C for 3 min, followed by 40 cycles at 94 °C for 30 s, annealing at 55–60 °C for 30 s, and a final extension at 72 °C for 10 min. The PCR experiments were performed in a BioCycler thermocycler (Thermo Fisher Scientific), and the PCR products were subjected to electrophoresis on 1% agarose gels to confirm the amplification. The PCR products were then sequenced using BigDye® Terminator v3.1 Cycle Sequencing Kit (Applied Biosystems®) on a 3500 Genetic Analyzer (Applied Biosystems Inc., Foster City, CA, USA).
Genome annotation was achieved by the Geneious software, using S. mukorossi as the reference, and it was checked with the annotation achieved using Verdant (Mckain et al., 2016). For the annotation using Geneious, a minimum of 80% identity cutoff between the genomes was considered. The annotations were individually checked, and if necessary, were manually corrected for start and stop codons. A graphic representation of the plastomes was created using Organellar Genome DRAW (Lohse et al., 2013).
Genome comparison and phylogenetic analyses
The chloroplast genome sequences were aligned using the program MAFFT v7.017 (Katoh and Standley, 2013) implemented as the Multiple align tool in Geneious R9. The GTR model was determined using the Bayesian Information Criterion Evolutionary implemented in MEGA7 software ( Kumar et al., 2016). The evolutionary history was inferred by using the Maximum Likelihood method, and branch support was assessed with 1000 bootstrap replicates conducted in MEGA7 software. The three Spondias chloroplast genomes were compared using a BLAST Ring Image Generator assuming the default software settings (Alikhan et al., 2011), and the number of interspecific SNPs were identified using Geneious. Single sequence repeats (SSRs) or microsatellites were identified using Phobos software (Mayer, 2010).
Results
Genome assembly and validation
The Illumina single-end and paired-end reads from Spondias species were mapped to the S. mukorossi chloroplast genome to obtain the draft genomes. The draft genomes revealed few errors and/or mapping failures of the reads (approximately 1–2% of the genomes), which were corrected using de novo contigs. After obtaining the final genomes, the reads were mapped with no errors and 100% identity, resulting in an average coverage of 150×. In addition, the trnH–psbA, matK, trnD–trnT, accD–psaI, rblC, and trnK–rpd16 regions were sequenced using the Sanger method and aligned with the genome, resulting in high identity as the regions indicated intraspecific variation.
Comparative chloroplast genome analysis
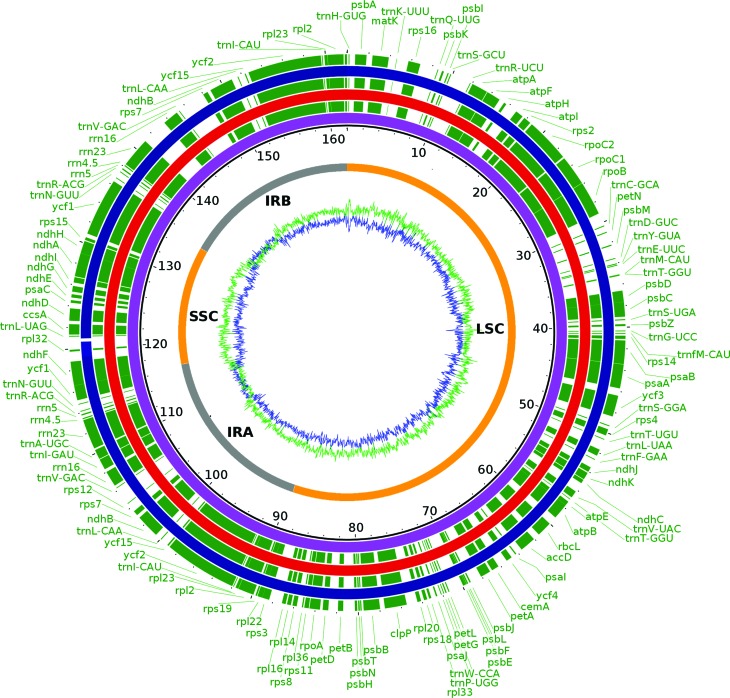
The plastid genome sizes of S. tuberosa, S. bahiensis, and S. mombin were 162,039, 162,218, and 162,302 bp, respectively (Table 2). All the genomes indicated a conserved structure with a pair of inverted repeats, IRA and IRB, separated by the LSC and SSC regions (Figures S1 (458.3KB, pdf) , S2 (445.7KB, pdf) , and S3 (461.6KB, pdf) ). For the Spondias species, the CG content was 37.6–37.7% and the length of the IR, LSC, and SSC regions were almost identical, revealing high similarity among the genomes (Table 2). The genomes comprised 130 genes, including 34–35 tRNAs and 4 rRNAs (Tables 2 and 3), and the arrangements of these regions were exclusively collinear (Figure 1). All protein-coding genes displayed AUG as the start codon; the coding sequences accounted for 63.8–64.2%, and the remaining regions included noncoding sequences (intergenic spacers).
Table 2. Summary of the chloroplast genome characteristics within the Anacardiaceae family. Genome size (bp), GC content (%), large single copy region - LSC (bp), small single copy region - SSC (bp), inverted repeat - IR (bp), N. of protein-coding genes, N. of tRNAs, and N. of rRNAs.
| Species | Characteristics | |||||||
|---|---|---|---|---|---|---|---|---|
| Size (bp) | GC (%) | LSC | SSC | IR | Genes | tRNAs | rRNAs | |
| Spondias tuberosa | 162,039 | 37.7 | 89,453 | 18,369 | 27,139 | 130 | 35 | 4 |
| Spondias bahiense | 162,218 | 37.7 | 89,606 | 18,381 | 27,156 | 130 | 34 | 4 |
| Spondias mombin | 162,302 | 37.6 | 89,938 | 18,094 | 27,135 | 130 | 35 | 4 |
| Pistacia vera | 160,674 | 37.9 | 88,236 | 19,086 | 26,676 | 126 | 37 | 4 |
Table 3. List of genes present in the Spondias tuberosa chloroplast genome, obtained by genome annotation using Sapindus mukorossi as reference.
| Group of genes | Name of genes | |
|---|---|---|
| Protein synthesis and DNA replication | Transfer RNAs | trnA-UGC (2x), trnC-GCA, trnD-GUC, trnE-UUC, trnF-GAA, trnfM-CAU, trnG-UCC, trnH-GUG, trnI-CAU (2x), trnI-GAU (3x) trnK-UUU, trnL-CAA(2x), trnL-UAA, trnL-UAG, trnM-CAU, trnN-GUU (2x), trnP-UGG, trnQ-UUG, trnR-ACG (2x), trnR-UCU, trnS-GCU, trnS-GGA, trnS-UGA, trnT-GGU (2x), trnT-UGU, trnV-GAC (2x), trnV-UAC, trnW-CCA, trnY-GUA |
| Ribossomal RNAs (16S, 23S, 4.5S, 5S) | rrn16 (2x), rrn23 (2x), rrn4.5 (2x), rrn5 (2x) | |
| Ribossomal Protein small subunit | rps2, rps3, rps4, rps7 (2x), rps8, rps11, rps12, rps14, rps15, rps16, rps18, rps19 (2x) | |
| Ribossomal Protein large subunit | rpl2 (2x), rpl14, rpl16, rpl20, rpl22, rpl23 (2x), rpl32, rpl33, rpl36 | |
| Subunits (α, β, β‘, β“) of the DNA-dependent RNA polymerase | rpoA, rpoB, rpoC1, rpoC2 | |
| Photosynthesis | Photosystem I | psaA, psaB, psaC, psaI, psaJ |
| Photosystem II | psbA, psbB, psbC, psbD, psbE, psbF, psbH, psbI, psbJ, psbK, psbL, psbM, psbN, psbT, psbZ | |
| Cythochrome b/f complex | petA, petB, petD, petG, petL, petN | |
| ATP synthase | atpA, atpB, atpE, atpF, atpH, atpI | |
| NADH-dehydrogenase | ndhA, ndhB (2×), ndhC, ndhD, ndhE, ndhF, ndhG, ndhH, ndhI, ndhJ, ndhK | |
| Large subunit RUBISCO | rbcL | |
| Miscellaneous | Acetyl-CoA carboxylase | accD |
| Cythochrome c biogenesis | ccsA | |
| Maturase | matK | |
| ATP-dependent protease | clpP | |
| Inner membrane protein | cemA | |
| Pseudogene unknown function | Conserved hypothetical chloroplast ORFs | ycf1 (2×), ycf2 (2×), ycf3, ycf4, ycf15 (2x) |
Figure 1. Complete gene map of Spondias chloroplast genomes. Gene annotations are represented in green. The chloroplast genomes are represented in purple (S. tuberosa), red (S. bahiensis), and blue (S. mombin). LSC: large single copy region; SSC: small single copy region; IR: inverted repeat. The green ring represents the A+T contents and the blue ring indicates C+G contents. The numbers near to S. tuberosa (purple circle) represent the nucleotide positions (in kbp).
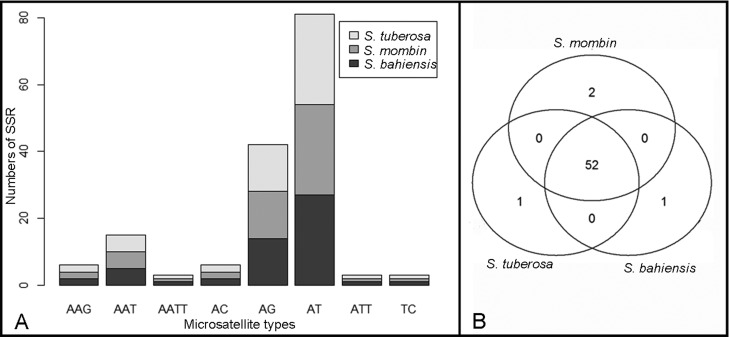
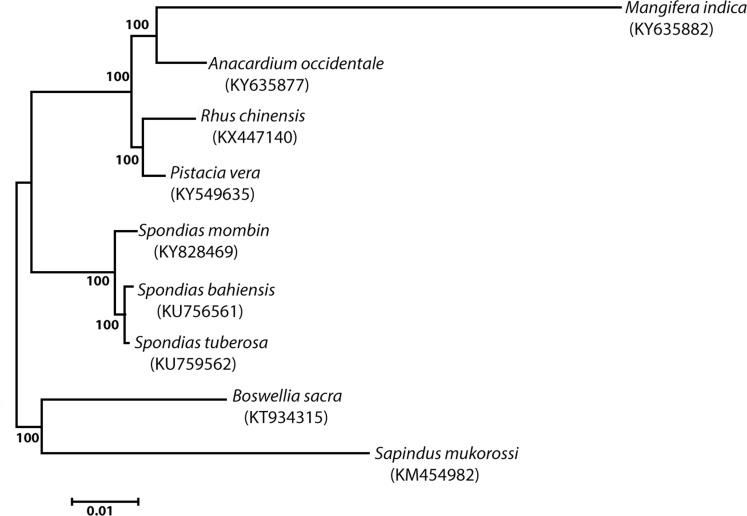
In the three genomes of the genus Spondias we found 159 SSRs, which were evenly distributed (Figure 2A), and the motifs AT and AG were more abundant. The Spondias species indicated similar SSR distribution, wherein S. tuberosa and S. bahiensis had one species-specific SSR each, and S. mombin had two species-specific SSRs (Figure 2B). The comparison among Spondias species indicated 856 SNPs between S. tuberosa and S. bahiensis, 3044 SNPs between S. tuberosa and S. mombin, and 3289 SNPs between S. bahiensis and S. mombin. The phylogenetic analysis revealed three clades, of which one clade was formed by S. tuberosa and S. bahiensis, the second clade by S. mombin, and the outgroup clade was formed by other species (Figure 3).
Figure 2. Comparative analysis of microsatellites in the chloroplast genomes of Spondias. (A) Microsatellite type distribution in three Spondias species. (B) Venn diagram showing the number of SSR that are shared among S. bahiensis, S. tuberosa, and S. mombin.
Figure 3. Molecular phylogenetic analysis by maximum likelihood method, with supported values estimated by bootstrap.
Discussion
The assembly of the chloroplast genomes using single- or paired-end reads resulted in genomes with few errors due to the differences between the reads of the species and the reference genome utilized; however, these errors were easily identified and corrected by the alignment of the de novo contigs. Genetic analysis of the genus Spondias was performed using molecular (Machado et al., 2015; Silva et al., 2015) and cytogenetic data (Almeida et al., 2007). These studies have utilized the partial chloroplast regions or expressed sequences for the phylogenetic analysis; however, the advent of next-generation sequencing technologies has increased the number of complete chloroplast genomes sequencing, allowing robust phylogenetic analysis. In this context, the complete chloroplast genome sequences are important to assess the phylogenetic relationships. The chloroplast genomes have been utilized for phylogenetic analyses in Arecaceae and they provide robust branch support for deep phylogenetic relationships among tribes of the subfamily (Barrett et al., 2016).
The chloroplast genomes of the genus Spondias are highly similar, with only one deletion of tRNA in S. bahiensis. The order and structure of the IR, LSC, and SSC regions were exclusively collinear. The genome size, CG content, and the length of the IR, SLC, and SSC regions were almost identical, revealing high similarity among the genomes and suggesting low diversity within the genus Spondias. Remarkably, the chloroplast genomes of S. tuberosa and S. bahiensis are highly similar when compared with S. mombin, suggesting a close genetic relationship and indicating a possible hybrid origin for S. bahiensis. S. bahiensis has been described as a hybrid between S. tuberosa and S. mombin (Almeida et al., 2007). Studies using chromosome banding and genomic in situ hybridization could not determine if S. bahiensis is a hybrid (Almeida et al., 2007); however, molecular studies using phylogeny with chloroplast regions (Machado et al., 2015; Silva et al., 2015) and ESTs (Machado et al., 2015) suggest that S. bahiensis is a new species and a hybrid between S. tuberosa and S. venulosa. In the present study, the comparison between S. tuberosa and S. bahiensis suggests the hybrid origin of S. bahiensis, with S. tuberosa as the female genitor; the SNP comparison displayed few SNPs between S. bahiensis and S. tuberosa, whereas S. bahiensis and S. mombin revealed several SNPs. The SSRs revealed high conservation in the three species, indicating low evolution of SSRs in the genus.
We demonstrate that the gene order, content, and orientation in Spondias are highly conserved, and that this observation is similar to other taxa, such as in Aconitum (Ranunculaceae) ( Park et al., 2017), Salix (Salicaceae) (Huang et al., 2017), and in the gymnosperm genus Pinus (Asaf et al., 2018), indicating that the chloroplast genomes reveal high synteny in close species; however, when analyzed across distant species of the Malpighiales order, three inversions were found in the LSC region (Cauz-Santos et al., 2017). In summary, (1) we presented the first complete chloroplast genome sequences for the genus Spondias; (2) phylogenies analyzed using the complete chloroplast genomes revealed a robust phylogenetic topology for Spondias; and (3) gene order, content, and orientation in Spondias are highly conserved.
Acknowledgments
We thank the Federal University of Alagoas for the laboratory and scientific support and the Fundação de Apoio à Pesquisa de Alagoas (FAPEAL) for funding this Project.
Supplementary material
The following online material is available for this article:
Footnotes
Associate Editor: Guilherme Corrêa de Oliveira
Conflict of interest
The authors declare that they have no conflict of interest.
Author contributions
VS conducted the experiments and CA analyzed the data and wrote the manuscript.
References
- Alikhan NF, Petty NK, Ben Zakour NL, Beatson S. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402. [DOI] [PMC free article] [PubMed] [Google Scholar]; Alikhan NF, Petty NK, Ben Zakour NL and Beatson S (2011) BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics 12:402. [DOI] [PMC free article] [PubMed]
- Almeida CC, De Lemos Carvalho PC, Guerra M. Karyotype differentiation among Spondias species and the putative hybrid Umbu-cajá (Anacardiaceae) Bot J Linn Soc. 2007;155:541–547. [Google Scholar]; Almeida CC, De Lemos Carvalho PC and Guerra M (2007) Karyotype differentiation among Spondias species and the putative hybrid Umbu-cajá (Anacardiaceae). Bot J Linn Soc 155:541–547.
- Asaf S, Khan AL, Khan MA, Shahzad R, Kang SM, Al-harrasi A, Al-rawahi A, Lee I. Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species. PLoS One. 2018;13:e192966. doi: 10.1371/journal.pone.0192966. [DOI] [PMC free article] [PubMed] [Google Scholar]; Asaf S, Khan AL, Khan MA, Shahzad R, Kang SM, Al-harrasi A, Al-rawahi A and Lee I (2018) Complete chloroplast genome sequence and comparative analysis of loblolly pine (Pinus taeda L.) with related species. PLoS One 13:e192966. [DOI] [PMC free article] [PubMed]
- Barrett CF, Baker WJ, Comer JR, Conran JG, Lahmeyer SC, Leebens-Mack JH, Li J, Lim GS, Mayfield-Jones DR, Perez L, et al. Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol. 2016;209:855–870. doi: 10.1111/nph.13617. [DOI] [PubMed] [Google Scholar]; Barrett CF, Baker WJ, Comer JR, Conran JG, Lahmeyer SC, Leebens-Mack JH, Li J, Lim GS, Mayfield-Jones DR, Perez L et al. (2016) Plastid genomes reveal support for deep phylogenetic relationships and extensive rate variation among palms and other commelinid monocots. New Phytol 209:855–870. [DOI] [PubMed]
- Boisvert S, Raymond F, Godzaridis E, Laviolette F, Corbeil J. Ray Meta: Scalable de novo metagenome assembly and profiling. Genome Biol. 2012;13:R122. doi: 10.1186/gb-2012-13-12-r122. [DOI] [PMC free article] [PubMed] [Google Scholar]; Boisvert S, Raymond F, Godzaridis E, Laviolette F and Corbeil J (2012) Ray Meta: Scalable de novo metagenome assembly and profiling. Genome Biol 13:R122. [DOI] [PMC free article] [PubMed]
- Cauz-Santos LA, Munhoz CF, Rodde N, Cauet S, Santos AA, Penha HA, Dornelas MC, Varani AM, Oliveira GCX, Bergès H, et al. The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: Structural organization and phylogenomic studies in Malpighiales. Front Plant Sci. 2017;8:1–17. doi: 10.3389/fpls.2017.00334. [DOI] [PMC free article] [PubMed] [Google Scholar]; Cauz-Santos LA, Munhoz CF, Rodde N, Cauet S, Santos AA, Penha HA, Dornelas MC, Varani AM, Oliveira GCX, Bergès H et al. (2017) The chloroplast genome of Passiflora edulis (Passifloraceae) assembled from long sequence reads: Structural organization and phylogenomic studies in Malpighiales. Front Plant Sci 8:1–17. [DOI] [PMC free article] [PubMed]
- Doyle JJ, Doyle JL. A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull. 1987;19:11–15. [Google Scholar]; Doyle JJ and Doyle JL (1987) A rapid DNA isolation procedure for small quantities of fresh leaf tissue. Phytochem Bull 19:11-15.
- Green BR. Chloroplast genomes of photosynthetic eukaryotes. Plant J. 2011;66:34–44. doi: 10.1111/j.1365-313X.2011.04541.x. [DOI] [PubMed] [Google Scholar]; Green BR (2011) Chloroplast genomes of photosynthetic eukaryotes. Plant J 66:34–44. [DOI] [PubMed]
- Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ, et al. A DNA barcode for land plants. Proc Natl Acad Sci U S A. 2009;106:12794–12797. doi: 10.1073/pnas.0905845106. [DOI] [PMC free article] [PubMed] [Google Scholar]; Hollingsworth PM, Forrest LL, Spouge JL, Hajibabaei M, Ratnasingham S, Bank M, Chase MW, Cowan RS, Erickson DL, Fazekas AJ et al. (2009) A DNA barcode for land plants. Proc Natl Acad Sci U S A 106:12794-12797. [DOI] [PMC free article] [PubMed]
- Huang YY, Matzke AJM, Matzke M. Complete sequence and comparative analysis of the chloroplast genome of coconut palm (Cocos nucifera) PLoS One. 2013;8:e74736. doi: 10.1371/journal.pone.0074736. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang YY, Matzke AJM and Matzke M (2013) Complete sequence and comparative analysis of the chloroplast genome of coconut palm (Cocos nucifera). PLoS One 8:e74736. [DOI] [PMC free article] [PubMed]
- Huang Y, Wang J, Yang Y, Fan C, Chen J. Phylogenomic analysis and dynamic evolution of chloroplast genomes in Salicaceae. Front Plant Sci. 2017;8:1050. doi: 10.3389/fpls.2017.01050. [DOI] [PMC free article] [PubMed] [Google Scholar]; Huang Y, Wang J, Yang Y, Fan C and Chen J (2017) Phylogenomic analysis and dynamic evolution of chloroplast genomes in Salicaceae. Front Plant Sci 8: 1050. [DOI] [PMC free article] [PubMed]
- Katoh K, Standley DM. MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol. 2013;30:772–780. doi: 10.1093/molbev/mst010. [DOI] [PMC free article] [PubMed] [Google Scholar]; Katoh K and Standley DM (2013) MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol Biol Evol 30:772–780. [DOI] [PMC free article] [PubMed]
- Kumar S, Stecher G, Tamura K. MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol. 2016;33:1870–1874. doi: 10.1093/molbev/msw054. [DOI] [PMC free article] [PubMed] [Google Scholar]; Kumar S, Stecher G and Tamura K (2016) MEGA7: Molecular Evolutionary Genetics Analysis version 7.0 for bigger datasets. Mol Biol Evol 33:1870-1874 [DOI] [PMC free article] [PubMed]
- Lee YS, Kim I, Kim JK, Park JY, Joh HJ, Park H, Lee HO, Lee SC, Hur YJ, Yan TJ. The complete chloroplast genome sequence of Rhus chinensis Mill (Anacardiaceae) Mitochondrial DNA Part B Resour. 2016;1:696–697. doi: 10.1080/23802359.2016.1222250. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lee YS, Kim I, Kim JK, Park JY, Joh HJ, Park H, Lee HO, Lee SC, Hur YJ and Yan TJ (2016) The complete chloroplast genome sequence of Rhus chinensis Mill (Anacardiaceae). Mitochondrial DNA Part B Resour 1:696-697. [DOI] [PMC free article] [PubMed]
- Lohse M, Drechsel O, Kahlau S, Bock R. Organellar Genome DRAW— a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res. 2013;41:575–581. doi: 10.1093/nar/gkt289. [DOI] [PMC free article] [PubMed] [Google Scholar]; Lohse M, Drechsel O, Kahlau S and Bock R (2013) Organellar Genome DRAW— a suite of tools for generating physical maps of plastid and mitochondrial genomes and visualizing expression data sets. Nucleic Acids Res 41:575-581. [DOI] [PMC free article] [PubMed]
- Machado M, Carvalho P, van den Berg C. Domestication, hybridization, speciation, and the origins of an economically important tree crop of Spondias (Anacardiaceae) from the Brazilian Caatinga dry forest. Neodiversity. 2015;8:8–49. [Google Scholar]; Machado M, Carvalho P and van den Berg C (2015) Domestication, hybridization, speciation, and the origins of an economically important tree crop of Spondias (Anacardiaceae) from the Brazilian Caatinga dry forest. Neodiversity 8:8–49.
- Mitchell JD, Daly DC. A revision of Spondias (Anacardiaceae) in the Neotropics. Phytotaxa. 2015;92:1–92. doi: 10.3897/phytokeys.55.8489. [DOI] [PMC free article] [PubMed] [Google Scholar]; Mitchell JD and Daly DC (2015) A revision of Spondias (Anacardiaceae) in the Neotropics. Phytotaxa 92:1–92. [DOI] [PMC free article] [PubMed]
- Park I, Kim WJ, Yang S, Yeo SM, Li H, Moon BC. The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS One. 2017;12:e184257. doi: 10.1371/journal.pone.0184257. [DOI] [PMC free article] [PubMed] [Google Scholar]; Park I, Kim WJ, Yang S, Yeo SM, Li H and Moon BC (2017) The complete chloroplast genome sequence of Aconitum coreanum and Aconitum carmichaelii and comparative analysis with other Aconitum species. PLoS One 12:e184257. [DOI] [PMC free article] [PubMed]
- Rabah SO, Lee C, Hajrah NH, Makki RM, Alharby HF, Alhebshi AM, Sabir JSM, Jansen RK, Ruhlman TA. Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome. 2017;10:1–14. doi: 10.3835/plantgenome2017.03.0020. [DOI] [PubMed] [Google Scholar]; Rabah SO, Lee C, Hajrah NH, Makki RM, Alharby HF, Alhebshi AM, Sabir JSM, Jansen RK and Ruhlman TA (2017) Plastome sequencing of ten nonmodel crop species uncovers a large insertion of mitochondrial DNA in cashew. Plant Genome 10:1-14. [DOI] [PubMed]
- Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, d’Anfray A, Vigouroux Y, Pintaud JC. A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. PLoS One. 2011;6:e19954. doi: 10.1371/journal.pone.0019954. [DOI] [PMC free article] [PubMed] [Google Scholar]; Scarcelli N, Barnaud A, Eiserhardt W, Treier UA, Seveno M, d’Anfray A, Vigouroux Y and Pintaud JC (2011) A set of 100 chloroplast DNA primer pairs to study population genetics and phylogeny in monocotyledons. PLoS One 6:e19954. [DOI] [PMC free article] [PubMed]
- Silva JN, Costa A, Silva JV, Almeida C. DNA barcoding and phylogeny in Neotropical species of the genus Spondias. Biochem Syst Ecol. 2015;61:240–243. [Google Scholar]; Silva JN, Costa A, Silva JV and Almeida C (2015) DNA barcoding and phylogeny in Neotropical species of the genus Spondias. Biochem Syst Ecol 61:240–243.
- Yang B, Li M, Ma J, Fu Z, Xu X, Chen Q. The complete chloroplast genome sequence of Sapindus mukorossi. Mitochondrial DNA Part A. 2016;27:1825–1826. doi: 10.3109/19401736.2014.971243. [DOI] [PubMed] [Google Scholar]; Yang B, Li M, Ma J, Fu Z, Xu X and Chen Q (2016) The complete chloroplast genome sequence of Sapindus mukorossi. Mitochondrial DNA Part A 27:1825-1826. [DOI] [PubMed]
Internet Resources
- Mayer C. Phobos 3.3.11, 2006-2010. 2010. http://www.rub.de/spezzoo/cm/cm_phobos.htm.; Mayer C, (2010) Phobos 3.3.11, 2006-2010, http://www.rub.de/spezzoo/cm/cm_phobos.htm
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.